Abstract
p73 is a p53-related tumor suppressor but is also induced by oncogene products such as E2F-1, raising a question as to whether p73 is a tumor suppressor gene or oncogene. Unlike p53, p73 has several variants, including ΔNp73, which lacks the NH2-terminal transactivation domain. Although, in developing neurons, ΔNp73 is expressed abundantly and seems to inhibit the proapoptotic function of p53, the role of p73 and ΔNp73 and their regulatory mechanism in cell growth and differentiation are poorly understood. Here we report that p73, but not p53, directly activates the transcription of endogenous ΔNp73 by binding to the p73-specific target element located at positions −76 to −57 within the ΔNp73 promoter region. The activation of ΔNp73 promoter by p63 was marginal. ΔNp73 was associated with p73α, p73β, and p53, as demonstrated by immunoprecipitation assays, and inhibited their transactivation activities when we used reporters of Mdm2, Bax, or ΔNp73 itself in SAOS-2 cells. Furthermore, induction or overexpression of ΔNp73 promoted cell survival by competing with p53 and p73 itself. Thus, our results suggest that the negative feedback regulation of p73 by its target ΔNp73 is a novel autoregulatory system for modulating cell survival and death.
The p73 gene encodes a protein with a significant sequence homology and a functional similarity with the tumor suppressor p53 (24). However, unlike p53, p73 is expressed as multiple alternatively spliced forms (5, 6, 24, 51). The overexpression of p73 in cultured cells promotes a growth arrest and/or apoptosis similarly to p53. These biological activities are linked to its sequence-specific transactivation function, and both p53 and p73 are believed to recognize and bind to the p53-responsive sequence in the promoter region of each target gene (23, 24). However, the precise mechanism is poorly understood.
The p73 gene has been mapped to chromosome 1p36.2-3, a region which shows frequent loss of heterozygosity in a variety of human cancers, and the protein product induces cell cycle arrest and/or apoptosis, showing that p73 acts as a tumor suppressor (24, 45, 54). However, unlike p53, p73 is infrequently mutated in many human cancers (21). Furthermore, p73-deficient mice do not show a tumorigenic phenotype (57). These findings suggest that p73 does not function as a classic Knudson-type tumor suppressor. On the other hand, some human neoplasms, including breast and ovarian cancers, express high levels of p73 compared to the corresponding normal tissues (3, 58). In addition, adenovirus-induced cellular transformation causes an increased level of p73 expression (43). Recently, it has also been reported that the deregulated overexpression of cellular and viral oncogene products such as E2F-1, c-myc, and E1A induces expression of the endogenous p73 (22, 31, 48, 59). Thus, these observations have suggested that the growth-regulatory function of p73 is disturbed by complex mechanisms and have raised the question whether p73 is a tumor suppressor or an oncoprotein.
Accumulating evidence has suggested that p53 family members do not always function similarly. The viral oncoproteins, such as the simian immunodeficiency virus large T antigen, adenovirus E1B, and human papillomavirus E6, which efficiently inhibit p53 function, are unable to inactivate p73 (19, 32, 43). On the other hand, the adenovirus early protein E4orf6 represses the activity of p73 but has no effect on p53 (47). MDM2, an important regulator determining the half-life of p53, stimulates the ubiquitin-proteasome-dependent degradation of p53 to block the p53-mediated transcription (17, 20, 26), whereas MDM2 inhibits p73 by disrupting the physical and functional interaction with p300/CBP without promoting p73 degradation (9, 60). These findings imply that the function of p73 is regulated through a mechanism distinct from that used for p53.
Recently, Yang et al. have reported that p73 is enriched in the nervous system and that the p73-deficient mice, which do not exhibit an increased susceptibility to spontaneous tumorigenesis, have neurological and immunological defects (57). Other studies have shown that p73 contributes to the induction of neuronal differentiation in vitro and that the T-cell receptor-mediated apoptosis is dependent on both E2F-1 and p73 (7, 31). More recently, Pozniak et al. have found that ΔNp73 is expressed predominantly in mouse developing brain and sympathetic neurons and inhibits the neuronal apoptosis by blocking the proapoptotic function of p53 (41). These observations strongly suggest that p73 as well as ΔNp73 is one of the key regulators to determine cellular differentiation and apoptosis.
Here, we show that the cisplatin-induced neuronal apoptosis is associated with the upregulation of both p73 and ΔNp73. The overexpression of p73, but not p53 or p63, induces expression of ΔNp73 mRNA and we have identified the functional p73-specific responsive element within the ΔNp73 promoter region. ΔNp73 has hetero-oligomerized with p73 or p53 to repress the transactivation activity. ΔNp73 has also inhibited its own promoter activity by directly binding to p73. Thus, we have found a novel autoregulatory mechanism to modulate cell survival and death, in which p73 function is negatively regulated by its target ΔNp73.
MATERIALS AND METHODS
Cell culture and transfections.
Human neuroblastoma cell lines SH-SY5Y, SK-N-AS, and SK-N-BE and human lung carcinoma H1299 cells were grown in RPMI 1640 medium supplemented with 50 μg of kanamycin/ml and 10% (vol/vol) heat-inactivated fetal bovine serum. Human osteosarcoma SAOS-2 cells, embryonic kidney 293 cells, and COS7 cells were cultured in Dulbecco modified Eagle medium containing 10% (vol/vol) heat-inactivated fetal bovine serum and antibiotics. Cultures were maintained in a humidified atmosphere containing 5% CO2 at 37°C. SAOS-2 cells were transfected with Lipofectamine (Gibco-BRL) according to the manufacturer's protocol. 293, COS7, and H1299 cells were transfected with FuGENE 6 (Roche Molecular Biochemicals).
RNA isolation and reverse transcription-PCR (RT-PCR) analysis.
Total RNA was prepared by using Trizol reagent (Gibco-BRL) or the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. cDNA was created by using it as a template in the 20-μl cDNA synthesis reaction mixture containing random primers and Moloney murine leukemia virus reverse transcriptase (SuperScript II; Gibco-BRL) at 42°C for 1 h, followed by 15 min of denaturation at 70°C and then quick cooling. The cDNA was amplified in the 20-μl PCR mixture containing 100 μM concentrations of each deoxynucleoside triphosphate, 1× PCR buffer, 1 μM concentrations of each primer, and 0.2 U of rTaq or LA-Taq DNA polymerase (Takara). PCR primers were as follows: HA-p73, 5′-TGGCTTACCCATACGATGTTC-3′ (sense) and 5′-GTGCTGGACTGCTGGAAAGT-3′ (antisense); p73, 5′-TCTGGAACCAGACAGCACCT-3′ (sense) and 5′-GTGCTGGACTGCTGGAAAGT-3′ (antisense); ΔNp73, 5′-CGCCTACCATGCTGTACGTC-3′ (sense) and 5′-GTGCTGGACTGCTGGAAAGT-3′ (antisense); and GAPDH, 5′-ACCTGACCTGCCGTCTAGAA-3′ (sense) and 5′-TCCACCACCCTGTTGCTGTA-3′ (antisense).
Plasmids and adenovirus-mediated gene transfer.
The mammalian expression plasmid encoding hemagglutinin (HA)-tagged p73α or p73β was a gift from M. Kaghad, and the expression plasmid for HA-p63γ was obtained from S. Ikawa. A 494-bp cDNA encoding the NH2-terminal region of ΔNp73 was amplified by RT-PCR with total RNA derived from SH-SY5Y cells which were infected with Ad-p73α and the primers 5′-GGATTCAGCCAGTTGACAGAACTA-3′ (sense) and 5′-GTGCTGGACTGCTGGAAAGT-3′ (antisense). The sense oligonucleotide primer was designed based on the similar sequence found in both human ΔNp73 genomic sequence and mouse ΔNp73 cDNA (accession number Y19235). The amplified product was subcloned into the pGEM-T Easy vector (Promega) to give pGEM-ΔNp73, and the identity of the construct was verified by DNA sequencing. pGEM-ΔNp73 was then partially digested with NarI, blunt ended, and completely digested with NarI, and the restriction fragment for the extreme NH2-terminal region was subcloned into the enzymatically modified KpnI and NarI sites of pcDNA3-HA-p73α or pcDNA3-HA-p73β to give pcDNA3-ΔNp73α or pcDNA3-ΔNp73β, respectively. For construction of the adenovirus expression vectors, the HindIII-XbaI restriction fragment derived from pcDNA3-p53, pcDNA3-HA-p73α, pcDNA3-HA-p73β, or pcDNA3-ΔNp73α was filled in with Klenow fragment and inserted into the enzymatically modified NotI site of the shuttle vector pHMCMV6 (34, 35). Each shuttle vector was digested with I-CeuI and PI-SceI and ligated into the identical restriction sites of the adenovirus expression vector pAdHM4. Each of these recombinant adenovirus constructs was digested with PacI and transfected into 293 cells to generate recombinant adenovirus.
Production of a polyclonal anti-ΔNp73 antibody.
The polyclonal anti-ΔNp73 antibody was raised against a peptide “Cys” plus containing the amino acid sequences between positions 2 and 14 of ΔNp73. The peptide and the polyclonal antibody were generated by Biologica Co.
Immunoblot analysis.
Cells were placed on ice, washed twice with phosphate-buffered saline, and lysed in EBC buffer (50 mM Tris [pH 8.0], 120 mM NaCl, 0.5% [vol/vol] Nonidet P-40) (23) supplemented with 1 mM phenylmethylsulfonyl fluoride. Lysates were clarified by centrifugation at 15,000 × g for 15 min at 4°C. Protein concentrations were determined by using a Bio-Rad protein assay. For immunoblot analysis, proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred onto a nitrocellulose membrane. The membrane filter was blocked with 5% powdered milk in TBST (Tris-buffered saline containing 0.1% [vol/vol] Tween 20) for 1 h at room temperature and then incubated with a primary antibody including a monoclonal anti-HA (12CA5; Roche Molecular Biochemicals), monoclonal anti-p73α (Ab-1; Oncogene Research Products), monoclonal anti-FLAG (M2; Sigma), polyclonal anti-ΔNp73, or monoclonal anti-p53 (DO-1; Oncogene Research Products) antibody for 1 h at room temperature. The membrane filter was then incubated with a goat anti-mouse or anti-rabbit secondary antibody conjugated to horseradish peroxidase (Pierce) for 1 h at room temperature, and bound secondary antibody was detected by enhanced chemiluminescence (Amersham Pharmacia Biotech) according to the manufacturer's protocol. For normalization of protein loading, blots were stripped and reprobed with a polyclonal anti-actin antibody (20-33; Sigma).
Immunoprecipitation.
293, COS7, or H1299 cells were transfected with the indicated expression plasmid. At 48 h posttransfection, cells were lyzed in EBC buffer, sonicated briefly, and centrifuged at 15,000 × g for 15 min at 4°C to remove cell debris. After preclearing, immunoprecipitation was carried out by incubating the whole-cell lysate with anti-HA polyclonal antibody (Medical and Biological Laboratories), with anti-ΔNp73 antibody, or with a mixture of anti-p53 monoclonal antibodies (DO-1 and PAb1801; Oncogene Research Products). Immunocomplexes were precipitated with protein A or with protein G-Sepharose beads (Sigma). The immunoprecipitates were then washed three times with EBC buffer, resuspended in 30 μl of 2× SDS sample buffer (28), and boiled for 5 min. The immunoprecipitated proteins were separated on an SDS-10% polyacrylamide gel under reducing conditions and analyzed by immunoblotting with anti-FLAG M2 antibody, anti-ΔNp73 antibody, or anti-p53 antibody (DO-1).
Cloning the ΔNp73 promoter region.
The ΔNp73 promoter region including ΔNp73 exon 3′ was amplified by PCR with the PAC clone (dJ363H11) and the primers 5′-CCAGGGAGGATCTGTAGCTG-3′ (sense) and 5′-TGAACCCTACACTGCAGCAA-3′ (antisense). The amplified PCR product (2.9 kb) was cloned directly into the pGEM-T Easy vector (Promega) to give pGEM-ΔNp73P. The sequence of this construct was confirmed by DNA sequencing. To generate a series of 5′-deletion constructs, the 2.9-kb fragment was subcloned into the enzymatically modified XhoI site of pGL2-Basic luciferase reporter (Promega) to give pGL2-ΔNp73PF. pGL2-ΔNp73PF was then digested with SmaI, NcoI, or StuI, blunt ended, and self-ligated to generate pGL2-ΔNp73P(−1082), pGL2-ΔNp73P(−911), or pGL2-ΔNp73P(−63), respectively. In addition, pGL2-ΔNp73PF was digested partially with ApaI, and each of the restriction fragments was recovered, blunt ended, and self-ligated to give pGL2-ΔNp73P(−1245), pGL2-ΔNp73P(−786), pGL2-ΔNp73P(−619), and pGL2-ΔNp73P(−203), respectively. To generate ΔNp73P(−184), ΔNp73P(−100), ΔNp73P(−80), ΔNp73P(−76), ΔNp73P(−71), ΔNp73P(−66), or ΔNp73P(+1), the corresponding regions were amplified by PCR with pGEM-ΔNp73P as a template. The resulting PCR products were then subcloned into the SmaI site of pGL2-Basic luciferase reporter, and the nucleotide sequences of these constructs were confirmed by sequencing.
EMSAs.
p73α, p73β, or p53 was generated in vitro by using the TNT Quick Coupled Transcription/Translation System (Promega) according to the manufacturer's instructions. Electrophoretic mobility shift assays (EMSAs) were performed as described previously (39, 61). Briefly, double-stranded oligonucleotide was 32P labeled by using T4 polynucleotide kinase. DNA-protein binding was carried out at room temperature in a reaction mixture containing the reticulocyte lysate (4 μl), 12.5 mM Tris (pH 7.5), 50 mM KCl, 3.125 mM MgCl2, 0.25 mM EDTA, 0.5 mM dithiothreitol, 5% glycerol, and 100 μg of poly(dI-dC) (Amersham Pharmacia Biotech)/ml. The reaction mixtures were resolved on a 5% native polyacrylamide gel (acrylamide/bisacrylamide ratio of 29:1) in 1× Tris-borate-EDTA buffer at room temperature. After the electrophoresis, the gel was dried and exposed to an X-ray film at −70°C with an intensifying screen.
Luciferase reporter assay.
For luciferase assays, SAOS-2 cells were seeded in triplicates into 12-well plates (5 × 104 cells/well) 24 h prior to transfection. Cells were cotransfected with 200 ng of the indicated reporter plasmid, 20 ng of pRL-TK encoding Renilla luciferase cDNA, and 200 ng of the indicated expression plasmid (p53, p73α, or p73β) in the presence or absence of ΔNp73α or ΔNp73β. The total amount of DNA transfected was kept constant (0.6 μg) with parental pcDNA3 (Invitrogen). At 48 h after transfection, luciferase activity was measured by a dual luciferase reporter assay system (Promega), and the transfection efficiency was standardized against Renilla luciferase activity.
Flow cytometry.
SH-SY5Y cells were seeded in 6-cm-diameter cell culture dishes and treated with increasing amounts of cisplatin. At 24 h after the treatment, cells were collected and fixed with ice-cold 70% ethanol for 4 h. The cells were then collected by centrifugation and resuspended in phosphate-buffered saline containing 25 μg of propidium iodide/ml and 0.1% RNase A (1). Thirty minutes later, the DNA content was measured by using a FACScan (Becton Dickinson). The data were plotted by using CellQuest software (Becton Dickinson).
Cell survival assays.
SH-SY5Y and SK-N-BE cells were seeded into 96-well plates (5 × 103 and 2 × 104 cells/well, respectively) 24 h prior to viral infection. Cells were infected with Ad-lacZ, Ad-ΔNp73α or Ad-p73α at the indicated multiplicity of infection (MOI) for the indicated time periods. Survival was determined by using cell counting kit 8 (Wako) with WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2 H-tetrazolium, monosodium salt] as a substrate.
RESULTS
Cisplatin induces expression of both p73 and ΔNp73 in neuroblastoma cells.
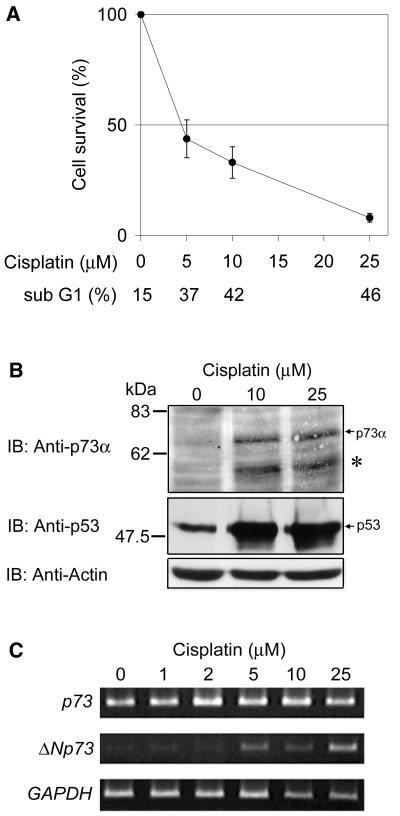
Since one of the main target organs of p73 knockout mice is the central and peripheral nervous system (57), we have chosen human neuroblastoma cells to investigate the functional difference and/or significance of p73 and p53 in induction of neuronal cell death. When SH-SY5Y neuroblastoma cells were treated with cisplatin, they underwent apoptosis in a dose-dependent manner as measured by cell survival assays and changes in the number of cells with sub-G1 DNA content (Fig. 1A). We then examined the changes in protein levels of p53 and p73 with anti-p53 antibody (DO-1) and anti-p73α antibody (Ab-1) which did not recognize p73β, respectively. The equal protein loading was confirmed by reprobing the p73α blot with an antibody against actin. As shown in Fig. 1B, SH-SY5Y cells accumulated p53 and p73α at protein levels after the treatment with cisplatin, whereas expression of p73 mRNA was not induced (Fig. 1C), as reported previously (15, 16). On the Western blot, however, we repeatedly noticed the presence of the faster-migrating band than p73α that appeared to correspond to the predicted molecular mass of ΔNp73α (62 kDa [41]) (Fig. 1B). To confirm this possibility, we performed semiquantitative RT-PCR with the ΔNp73-specific primers, which revealed that ΔNp73 mRNA expression was induced by the cisplatin treatment in a dose-dependent manner (Fig. 1C). This raised the possibility that the treatment of the cells with cisplatin induced the expression of ΔNp73α at both mRNA and protein levels in parallel with accumulation of p73α protein. Therefore, we hypothesized that the cisplatin-induced p53 or p73 or other proteins triggered the expression of ΔNp73 mRNA.
FIG. 1.
Induction of p73α and ΔNp73 in cisplatin-treated neuroblastoma cells. (A) Cell survival assays of SH-SY5Y cells treated with increasing amounts of cisplatin for 24 h. Data are presented as the mean values ± the standard deviation (SD) of six independent experiments. At the same time, numbers of cells with sub-G1 DNA content were counted. (B) Induction of p73α and p53 by cisplatin treatment in SH-SY5Y cells. SH-SY5Y cells were cultured in the absence or presence of cisplatin (10 or 25 μM) for 36 h. Total cellular proteins (40 μg) were extracted from each culture, and the expression level of p73α or p53 was determined by immunoblot analysis with the p73α-specific (Ab-1) or anti-p53 (DO-1) antibody, respectively. An asterisk marks the faster-migrating band recognized by anti-p73α antibody. The p73α blot was reprobed for actin to ensure equal loading. Size markers are indicated on the left. (C) Cisplatin treatment induces the expression of ΔNp73. SH-SY5Y cells were treated with increasing amounts of cisplatin as indicated. Using the RNA prepared 24 h after the treatment, semiquantitative RT-PCR analysis for expression of p73 and ΔNp73 was performed under linear amplification conditions. Expression of GAPDH is shown as a control.
ΔNp73 is a target of p73.
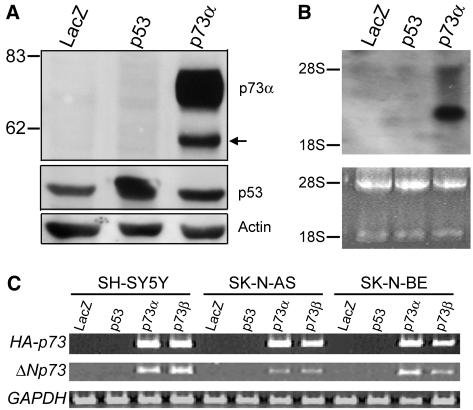
To determine whether p53 or p73α stimulated the expression of ΔNp73α, SH-SY5Y cells were infected with recombinant adenovirus expressing p53 (Ad-p53) or HA-tagged p73α (Ad-p73α). As shown in Fig. 2A, overproduction of HA-p73α markedly enhanced the expression of ΔNp73α, whereas p53 had no effect on it. Upregulation of ΔNp73 mRNA expression was also confirmed by Northern blot analysis with a ΔNp73-specific probe (Fig. 2B). Furthermore, similar results were also obtained in two other human neuroblastoma cell lines, SK-N-AS and SK-N-BE, by semiquantitative RT-PCR analysis (Fig. 2C).
FIG. 2.
Induction of ΔNp73 by p73α. (A) Western blot analysis. SH-SY5Y cells were infected with the recombinant adenovirus for LacZ (Ad-LacZ), p53 (Ad-p53), or HA-p73α (Ad-p73α) at an MOI of 10. At 36 h after the infection, whole-cell lysates (40 μg) were prepared and then subjected to Western blotting with anti-p73α (top) or anti-p53 (middle) antibody. The p73α blot was reprobed for actin to ensure equal protein loading (bottom). (B) Northern blot analysis of ΔNp73 expression. SH-SY5Y cells were infected with an MOI of 10 of adenovirus expressing LacZ, p53, or HA-p73α. Total RNA (20 μg) was prepared 24 h postinfection and subjected to Northern blotting with the ΔNp73-specific cDNA probe. Ethidium bromide staining of 28S and 18S rRNAs is shown to allow comparison of RNA loaded. (C) RT-PCR analysis of HA-p73 or ΔNp73 expression in adenovirus-infected SH-SY5Y, SK-N-AS, or SK-N-BE cells (MOI = 10). GAPDH expression is shown as a control.
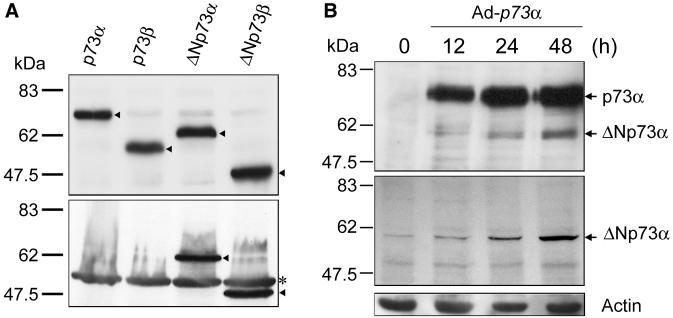
To confirm these results, we raised a ΔNp73-specific antibody against the sequence in the amino-terminal region of ΔNp73 and then examined its specificity. The in vitro translation products of FLAG-tagged p73α, p73β, ΔNp73α, and ΔNp73β were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with the monoclonal anti-FLAG antibody. A notable difference in the amount of each in vitro translation product was not observed (Fig. 3A, top panel). Then, these in vitro translation products were analyzed by immunoblotting with the ΔNp73-specific antibody. This antibody did not bind to p73α and p73β but recognized ΔNp73α and ΔNp73β (Fig. 3A, bottom panel). The results demonstrated that our antibody was highly specific for ΔNp73 and potentially useful for detecting endogenous ΔNp73. We then examined the endogenous ΔNp73α in SH-SY5Y cells upon the infection of the recombinant adenovirus possessing HA-p73α. At the indicated time periods after the infection, whole-cell lysates were prepared, separated by SDS-PAGE, and immunoblotted with the anti-p73α or with the anti-ΔNp73 antibody (Fig. 3B). As expected, the band migrating faster than p73α, which was detected by the anti-p73α antibody, comigrated with ΔNp73α, and overexpression of p73α resulted in the significant accumulation of ΔNp73α.
FIG. 3.
Specificity of the anti-ΔNp73 antibody and identification of ΔNp73α in SH-SY5Y cells infected with recombinant adenovirus for HA-p73α. (A) FLAG-tagged p73α, p73β, ΔNp73α, and ΔNp73β were generated in vitro by using the rabbit reticulocyte lysate, subjected to SDS-PAGE (10% polyacrylamide), and transferred to a nitrocellulose membrane, and the membrane was probed with the monoclonal anti-FLAG antibody at a dilution of 1:3,000 (top). Arrowheads indicate the position of each product. Similarly, the in vitro-translated products were immunoblotted with the polyclonal anti-ΔNp73 antibody at a dilution of 1:10,000 (bottom). Arrowheads indicate the positions of ΔNp73α and ΔNp73β. The asterisk indicates a nonspecific protein. The positions of molecular mass markers are marked at the left of each panel in kilodaltons. (B) At the indicated times after infection with recombinant adenovirus for HA-p73α, SH-SY5Y cell lysates were prepared, subjected to SDS-8% PAGE, and immunoblotted with the monoclonal anti-p73α antibody (Ab-1; Oncogene Research Products) (top) or with the polyclonal anti-ΔNp73 antibody (middle). The p73α blot was stripped and reprobed with the anti-actin antibody to ensure equal protein loading (bottom). The positions of the molecular size standards are indicated on the left in kilodaltons.
Taken together, these findings suggested that p73, but not p53, selectively regulated the expression of ΔNp73.
Identification of the p73-target element within the ΔNp73 promoter region.
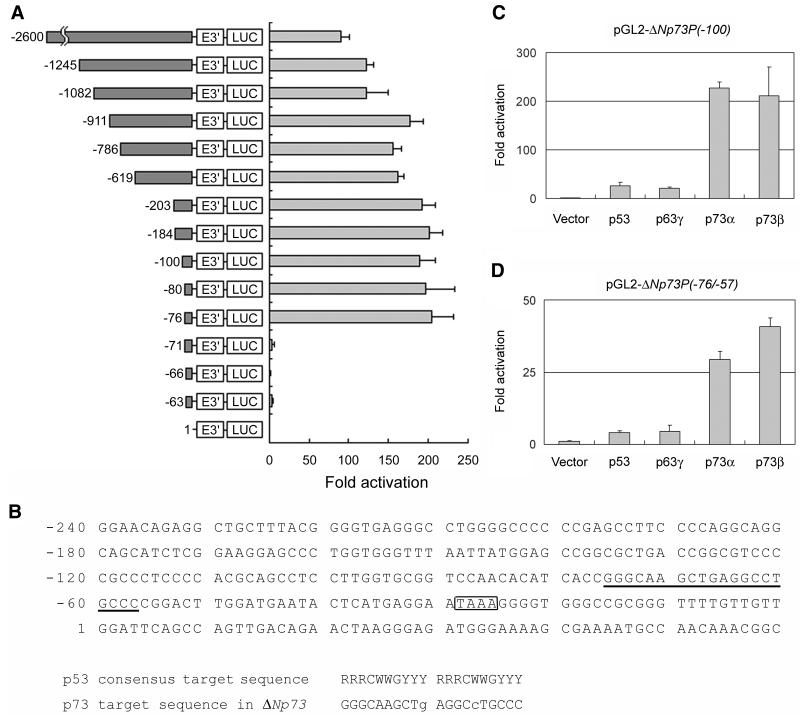
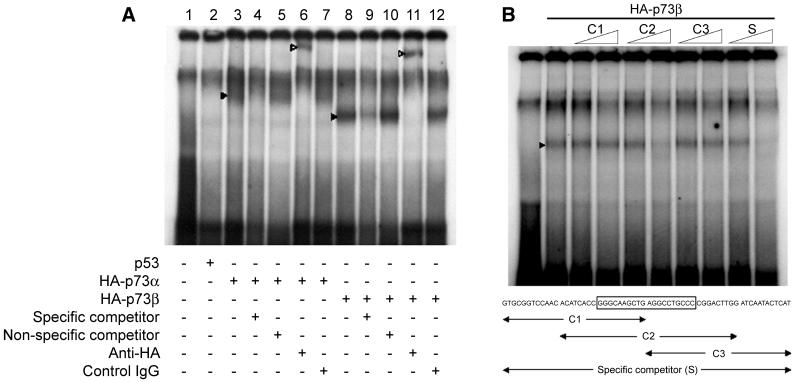
We then examined whether or not p73 directly activates the ΔNp73 promoter. As described previously, ΔNp73 mRNA was derived from an alternative promoter located in intron 3 of the p73 gene (41, 57). To obtain the promoter region of ΔNp73, we screened the human PAC library and finally cloned a 2.9-kb genomic fragment including exon 3′ of ΔNp73, which was ligated with the luciferase reporter to make a pGL2-ΔNp73PF construct. In SAOS-2 cells carrying a homozygous deletion of p53, coexpression of p73α and pGL2-ΔNp73PF containing a 2.6-kb sequence upstream from the exon 3′ exhibited an ∼90-fold increase in luciferase expression above that of the promoterless vector (Fig. 4A). Sequence analysis revealed that a putative p73-responsive element similar to the p53 consensus sequence was present in the region immediately upstream of the exon 3′ (Fig. 4B). Progressive deletion analysis showed that the deletion up to position −71 which disrupted the putative p73-responsive element (at position −76 to −57) dramatically reduced the luciferase activity (Fig. 4A), indicating that the putative p73-responsive element we found was necessary for the p73-mediated stimulation of the ΔNp73 promoter. Intriguingly, both p73α and p73β transactivated the ΔNp73P(−100) promoter-luciferase reporter, whereas p53 was far less effective on the activation of this promoter (Fig. 4C). In addition, the effect of p63γ, the other p53 family member (38, 56), on the ΔNp73 promoter was also minimal and comparable to that of p53. Under these experimental conditions, a notable difference in the amounts of ectopically expressed proteins was not detected (data not shown). To further determine whether p73 recognizes and activates the p73-binding sequence of the ΔNp73 promoter, we inserted a double-stranded synthetic segment comprising nucleotides −76 to −57 into the luciferase reporter vector pGL2-promoter to give pGL2-ΔNp73P(−76/−57). As seen in Fig. 4D, p73α as well as p73β transactivated the ΔNp73P(−76/−57) promoter-luciferase reporter. In contrast, p53 and p63γ activated transcription of the same promoter to a far smaller degree. Thus, the putative p73-binding element is responsible for the p73-specific activation of the ΔNp73 promoter. This notion was further supported by the results obtained in the EMSAs. As shown in Fig. 5A, both p73α and p73β specifically bound to the radiolabeled oligonucleotide spanning sequences between −96 and −36, whereas the p53-DNA complex could not be detected under these experimental conditions. Similarly, p53 did not bind to this radiolabeled probe DNA even in the presence of the anti-p53 antibody PAb421 (data not shown). To delineate the p73-binding region, we performed the competition assays (Fig. 5B). The p73β-DNA complex was efficiently competed for by C2 competitor DNA containing the entire putative p73-binding site. In contrast, both C1 and C3 competitor DNAs (which carried the 5′ and 3′ parts of the putative p73-binding site, respectively) resulted in loss of competition. p73α also showed similar results (data not shown). These data suggested that p73 directly activated transcription of ΔNp73 by binding to the p73-specific binding site in the ΔNp73 promoter.
FIG. 4.
Identification of the p73-specific binding sequence in the ΔNp73 promoter region. (A) Luciferase reporter assay. The left panel shows a schematic drawing of various fragments of the 5′-flanking sequence of p73 exon 3′ subcloned upstream of the luciferase reporter plasmid pGL2-Basic. The positions relative to the first nucleotide of the p73 exon 3′ (+1) are indicated. Each of the reporter constructs (180 ng) was cotransfected in triplicate in SAOS-2 cells, together with pRL-TK for Renilla luciferase cDNA (20 ng) and 300 ng of the expression plasmid encoding HA-p73α or else an empty vector. At 48 h after transfection, cells were harvested and assayed for luciferase activity. The data shown represent the averages of at least three independent experiments with the SDs (right panel). (B) Nucleotide sequence of the ΔNp73 promoter region. A canonical p73-binding site and the putative TATA element are indicated by underlining and boxed, respectively. The sequence comparison between the consensus p53-target sequence and the putative p73-binding sequence in the ΔNp73 promoter is show below. R, purine; Y, pyrimidine; W, adenine or thymidine. Two mismatches are indicated by lowercase letters. (C and D) The putative p73-responsive element was required for the activation of ΔNp73 promoter. The potential 20-bp p73-binding site (5′-GGGCAAGCTGAGGCCTGCCC-3′) was subcloned upstream of the luciferase reporter plasmid pGL2-promoter to generate pGL2-ΔNp73P(−76/−57). SAOS-2 cells were cotransfected with 380 ng of pGL2-ΔNp73P(−100) (C) or pGL2-ΔNp73P(−76/−57) (D), along with pRL-TK (20 ng) and 100 ng of the expression plasmid for p53, HA-p63γ, HA-p73α, or HA-p73β. Luciferase activity was measured as described above. The data shown are the mean values ± SD.
FIG. 5.
EMSAs. (A) A 61-bp oligonucleotide fragment containing the potential p73-binding site (5′-GTGCGGTCCAACACATCACCGGGCAAGCTGAGGCCTGCCCCGGACTTGGATGAATACTCAT-3′) was labeled with [γ-32P]ATP and incubated with in vitro-translated p53 (lane 2), HA-p73α (lanes 3 to 7), HA-p73β (lanes 8 to 12), or unprogrammed reticulocyte lysate (lane 1). Binding reaction mixtures in lanes 4 and 9 contained a 10-fold molar excess of the specific competitor, and binding reaction mixtures in lanes 5 and 10 included a 10-fold molar excess of the nonspecific competitor. For supershifting the protein-DNA complex, anti-HA antibody was added to the reaction mixtures (lanes 6 and 11). Preimmune serum was used as a negative control (lanes 7 and 12). The protein-DNA complexes and the supershift complexes are indicated by filled and open arrowheads, respectively. (B) Gel retardation assays were done as described for panel A. Reaction mixtures contained unprogrammed reticulocyte lysate (lane 1) or in vitro translated HA-p73β (lanes 2 to 10). Unlabeled competitor oligonucleotides used are indicated (the potential p73-binding site is boxed) and present in 1- and 10-fold molar excess (indicated by ramps). The position of the protein-DNA complex is indicated by the arrowhead.
Physical and functional interaction between p73 and ΔNp73.
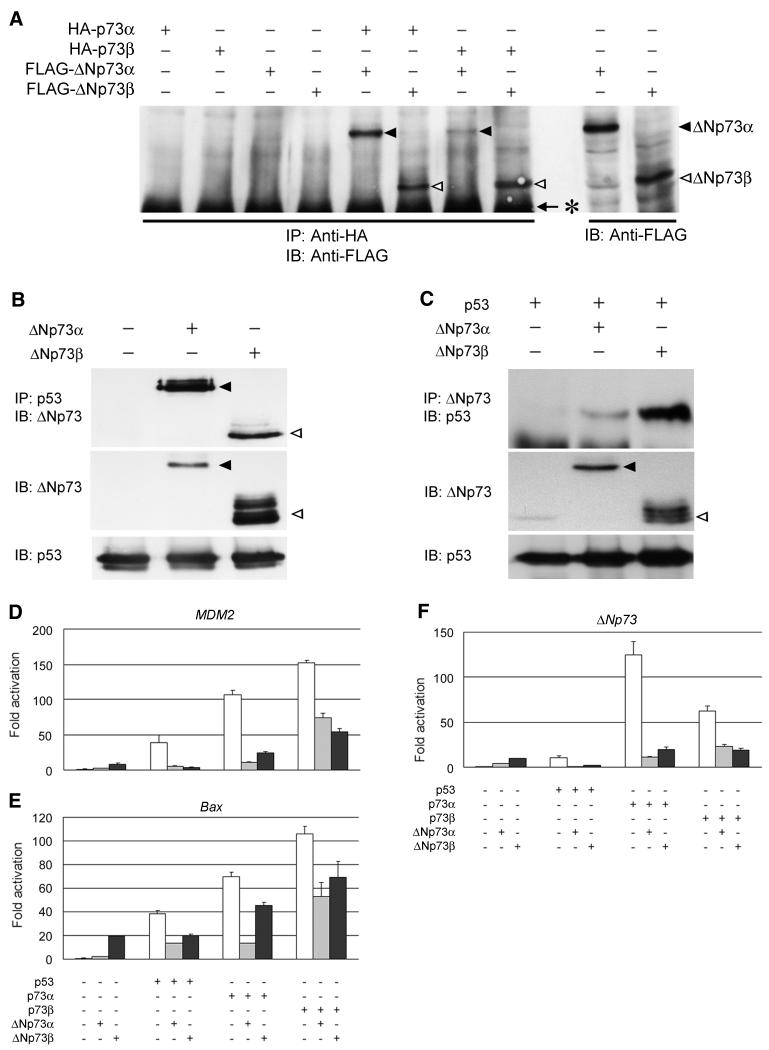
To understand the physiological role of ΔNp73 induction by p73, we next sought to determine whether there is interaction between ΔNp73 and p73 or p53. So far, it has already been shown that the mutant form of p53, but not the wild-type, makes a hetero-oligomer with p73 (4, 8, 33, 50) and that ΔNp73 directly binds to the wild-type p53 and inhibits its apoptosis-promoting activity (41). Our immunoprecipitation and Western blot analyses revealed that both of the ΔNp73 isoforms interacted with p73α or p73β (Fig. 6A), whereas p53 was bound, though not strongly, to ΔNp73α or ΔNp73β under our experimental conditions (data not shown). We then examined the physical interaction between p53 and ΔNp73 in COS7 cells with a high transfection efficiency. As shown in Fig. 6B, both of the ΔNp73 isoforms coprecipitated with the endogenous p53. Similar results were also obtained in p53-deficient H1299 cells ectopically expressing either p53 and ΔNp73α or p53 and ΔNp73β (Fig. 6C). These suggested that ΔNp73 directly induced by p73 interacted with p73 itself as well as the wild-type p53.
FIG. 6.
Functional interactions between p73 and ΔNp73. (A) Immunoprecipitation and Western blot analysis. 293 cells were transiently transfected with the indicated expression plasmids. Whole-cell lysates (400 μg of protein) were subjected to immunoprecipitation (IP) with anti-HA antibody, and the precipitated proteins were analyzed by immunoblotting (IB) with anti-FLAG M2 antibody. ΔNp73α and ΔNp73β are indicated by closed and open arrowheads, respectively. The asterisk indicates the position of heavy-chain immunoglobulin G. (B) p53 interacts with ΔNp73α or ΔNp73β in the COS7 cells. The cells were transfected with 8 μg each of the indicated expression plasmids. At 48 h after transfection, whole-cell lysates (1.5 mg of protein) were prepared, followed by immunoprecipitation with anti-p53 (DO-1/PAb1801) antibodies and immunoblotting with the anti-ΔNp73 antibody (top). ΔNp73α and ΔNp73β are indicated by closed and open arrowheads, respectively. The expression of ΔNp73 and endogenous p53 was examined by immunoblotting with the anti-ΔNp73 and anti-p53 antibodies, respectively (middle and bottom, respectively). (C) p53 interacts with ΔNp73α or ΔNp73β in H1299 cells. The cells were transiently transfected with 4 μg each of the indicated expression plasmids. At 48 h after transfection, whole-cell lysates (1.5 mg of protein) were prepared, followed by immunoprecipitation with the anti-ΔNp73 antibody and immunoblotting with the anti-p53 antibody (top). The expression of ΔNp73 and p53 was examined by immunoblotting with the anti-ΔNp73 and anti-p53 antibody, respectively (middle and bottom, respectively). ΔNp73α and ΔNp73β are indicated by closed and open arrowheads, respectively. For luciferase assays, SAOS-2 cells were cotransfected with the indicated expression plasmids, together with a reporter plasmid containing the MDM2 (D), Bax (E), or ΔNp73 (F) promoter driving luciferase expression. At 48 h posttransfection, cells were lysed and subjected to the luciferase assays. The data shown are mean values ± SD.
We then examined the functional significance of the interaction between p73 and ΔNp73 by using the reporter assay systems. Under our experimental conditions, the amount of ectopically expressed p53, p73α, or p73β was unaffected in the presence of ΔNp73 (data not shown). Cotransfection of HA-p73α with ΔNp73α or ΔNp73β resulted in a reduction of the p73α-induced transcriptional activation of the MDM2 or Bax promoter in SAOS-2 cells (Fig. 6D and E). Similarly, both of these ΔNp73 isoforms suppressed p73β- or p53-dependent transcriptional activation (Fig. 6D and E). These findings were similar to the result reported by Fillippovich et al. that p53 transactivation of the effector gene p21Waf1 was inhibited in cells transfected with p73Δexon2 (14). Intriguingly, overexpression of p73α along with ΔNp73α or ΔNp73β led to the marked decrease in the ΔNp73 promoter activity (Fig. 6F), a finding consistent with our observations that the increasing p73α protein levels resulted in a dose-dependent decrease in the expression level of ΔNp73 (data not shown). Thus, it has become possible that p73 and its target ΔNp73 form a negative autoregulatory loop in regulating transcriptional activity.
Overexpression of ΔNp73 inhibits apoptosis induced by p73.
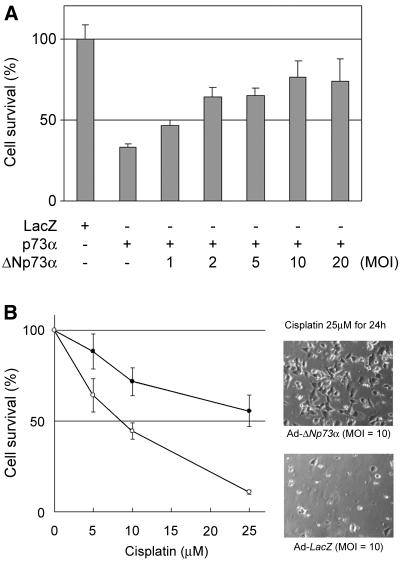
The effect of a dose-dependent expression of ΔNp73α on the p73α-induced neuronal cell death was tested. SK-N-BE cells were infected with Ad-p73α (MOI = 10) along with various MOIs of Ad-ΔNp73α, and the numbers of cells undergoing cell death were monitored by cell survival assay. As shown in Fig. 7A, increasing ΔNp73α levels led to a substantial reduction in the extent of p73α-induced cell death. Furthermore, cisplatin-induced apoptosis of SH-SY5Y cells was inhibited by the infection of Ad-ΔNp73α (Fig. 7B). These data suggested that the proapoptotic ability of p73α interfered with the ΔNp73α isoform at the cellular level. These results also appeared to be related with the recent report that Ad-p53 infection-induced apoptosis is suppressed by coinfection with Ad-ΔNp73 in mouse sympathetic neurons (41).
FIG. 7.
ΔNp73 inhibits p73- or cisplatin-induced apoptosis. (A) ΔNp73α suppresses apoptosis induced by p73α. SK-N-BE cells (2 × 104/well) were coinfected with Ad-p73α (MOI = 10), along with increasing amounts of Ad-ΔNp73α, as indicated. At 48 h after infection, the cell viability was determined by cell survival assays. The graph (mean ± SD of six experiments) indicates relative viability based on the percent viable cells compared to the control infection (Ad-lacZ). (B) Cisplatin-induced apoptosis is inhibited in the presence of ΔNp73α overexpressed. SH-SY5Y cells (5 × 103/well) were infected with Ad-LacZ (○) or Ad-ΔNp73α (•) (MOI = 10). Six hours after the infection, cells were treated with the indicated concentrations of cisplatin for 24 h, and then the cell viability was determined by cell survival assays. The graph presents mean values (±SD) from six experiments. Cells infected with Ad-LacZ or Ad-ΔNp73α were photographed 24 h after the treatment of cisplatin (25 μM).
DISCUSSION
In the present study, we have identified p73-specific target element spanning from position −76 to position −57 within the ΔNp73 promoter region. p73 but not p53 directly recognizes this cis-regulatory element and induces the expression of ΔNp73. The induction of p73α protein with the proapoptotic function by treating the neuronal cells with cisplatin has in turn induced expression of ΔNp73α with antiapoptotic function. We have also determined the physical and functional interaction between ΔNp73 and p73 or p53, which may inhibit the proapoptotic pathway. Thus, we have shown new evidence that there is a negative feedback regulation, in which the activation of p73 directly induces its own negative autoregulator, ΔNp73, in cells.
It has been known that both p53 and p73, as well as p63, activate the transcription of a common set of p53-target genes such as Bax, p21Waf1, and Mdm2. However, there is a promoter preference between them (29, 62). For example, p73 activates the transcription of 14-3-3σ much more efficiently than p53, even though it was identified as one of the p53-target genes (18). On the other hand, the transcription of the p53-regulated gene, BTG2, was only weakly induced by p73 (44, 62). The promoters of Bax, p21Waf1, and Mdm2 were commonly used as targets of the reporter analyses, but their activities were different among the p53 family members (6, 8, 13, 56). This raised a possibility that there exists a preferable target or target sequence or even one that is specific to each family member. Our initial finding that the induction of p73 protein, but not of p53, increased the expression of ΔNp73 at both mRNA and protein levels led us to identify the p73-specific target element within the promoter region of ΔNp73.
According to the human genomic sequence information, the nucleotide sequences in the promoter region of p73 and ΔNp73 are absolutely different (accession number AL136528). Based on the finding that ΔNp73 mRNA is transcribed under the control of an alternative promoter located within the intron 3 of the p73 gene (41, 57), we identified the predicted p73-responsive element (at positions −76 to −57), which is similar to the consensus p53-binding site (11, 25), immediately upstream of the exon 3′. Like the p53-binding site, this predicted p73-responsive element contained two copies of a 10-bp motif, and each copy was not separated by random sequence. Indeed, p73-mediated activation of the ΔNp73 transcription has been confirmed to be dependent on this element by using the transient luciferase reporter analyses and the EMSAs. It should be noted that overexpression of p73α activated the promoter activity of pGL2-ΔNp73P(−100) ∼8-fold more than that of pGL2-ΔNp73P(−76/−57). Similarly, the introduction of p73β expression vector resulted in a ∼4-fold-enhanced luciferase activity from pGL2-ΔNp73P(−100) compared to that from pGL2-ΔNp73P(−76/−57), suggesting that the degree of transactivation of the ΔNp73 promoter by p73 might be dependent on the sequences upstream and/or downstream of the putative p73-binding site. Consistent with the observation that the overexpression of p53 failed to induce the expression of endogenous ΔNp73, p53 could not bind to and activate this potential p73-target site. Of interest, activation of the ΔNp73 promoter by p63γ, the other p53 family member, was also very weak, although p63γ has been shown to exhibit strong transactivation of promoters containing p53-responsive elements (56). Thus, our results have suggested that the transcriptional activation of ΔNp73 by p73 was specific. Comparing with the consensus p53-binding site, the putative p73-responsive element that we have found, carries only two mismatches in noncritical positions. It is unclear at this time whether the structural alteration caused by these mismatches or another factor(s) could be involved in the p73-specific activation of the ΔNp73 promoter.
In accordance with the previous results (14, 57), our data showed that ΔNp73 hetero-oligomerized with p73 or p53. The ΔNp73-dependent inhibition of the transactivation and apoptosis-inducing activities of p73 or p53 might be partly due to the formation of inactive hetero-oligomers as described previously (41). Intriguingly, Yang et al. found that ΔNp73 bound specifically to the p53-binding site (57). In addition, it has been shown that ΔNp63, the other member of the p53 family lacking the amino-terminal transactivation domain, bound specifically to the p53-binding site (56). Therefore, it is possible that there exists an alternative inhibitory mechanism for ΔNp73 in competing with p53 for the p53-binding site.
The most important point of our observation was that ΔNp73, which was induced by p73, in turn inhibited p73 by a direct interaction. Ours may be the first report showing that, in the autoregulatory system of the p53 family members, proapoptotic p73 function is negatively regulated by its own target ΔNp73, whose function is antiapoptotic. A similar negative feedback regulation is known in the case of p53 and Mdm2. Mdm2, which is encoded by a p53-responsive oncogene, interacts with p53 and abolish the transactivation activity of p53 through stimulating its ubiquitination by its E3 ligase activity and promoting its nuclear export and degradation in cytoplasmic proteasomes (17, 20, 26, 30, 42). Thus, the amount of p53 is regulated by an autoregulatory feedback loop. Alternatively, E2F-1 transcription factor, a major effector of the retinoblastoma (pRB) tumor suppressor pathway, elevates the expression level of the tumor suppressor protein p14ARF (mouse p19ARF), which promotes sequestration and degradation of Mdm2 (2, 40, 46, 49, 53). p14ARF interacts directly with Mdm2 in a region distinct from the p53-binding domain without disrupting the interaction between them, inhibits Mdm2-mediated degradation of p53, and thereby induces the accumulation of p53. O'Connor et al. found that there exists a physical and functional interaction between E2F-1 and p53 and that the E2F-1-dependent transcriptional activity is blocked by the complex formation with p53 (37). Of interest, the physical interaction between E2F-1 and p53 contributes to block the p53-mediated transactivation (36, 37, 52). Recently, it has been shown that p14ARF directly binds to E2F-1 and inhibits its transactivation function (12). However, in the p53-Mdm2, E2F-1-p53, or E2F-1-p14ARF pathway, the target is the different gene and the system is not an autoregulation. By luciferase reporter and cell survival analyses, we have shown that overexpression of ΔNp73 resulted in the significant reduction of its own promoter activity through abolishing the transactivation function of p73 and that cisplatin- or p73-induced apoptosis was inhibited in the presence of ΔNp73. Thus, our present results might give a clue to solve the recently raised question of whether p73 is an oncogene or a tumor suppressor gene.
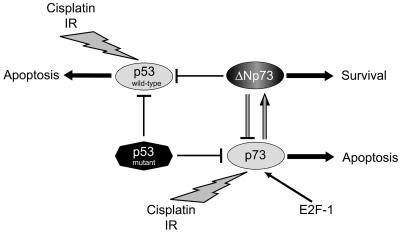
It may be possible that the balance between intracellular levels of p73 and ΔNp73 determines the cell's fate to survive or to die. Figure 8 shows the schematic interaction between p73, ΔNp73, wild-type p53, and mutant p53. p73, which is infrequently mutated in human cancers and has a proapoptotic function, inhibits the apoptosis-inducing activity of both wild-type p53 and p73 itself through the induction of ΔNp73, while p53 eliminates the function of both wild-type p53 and p73, through its loss-of-function mutations that frequently occur in many cancers. Thus, death and survival of many cell types in various organs could be regulated by a subtle balance between p53 family members and their isoforms, including the antagonizing variants such as ΔNp73 and ΔNp63, as suggested previously (41).
FIG. 8.
Schematic representation of interactions between p73, ΔNp73, wild-type p53, or mutant type p53.
It has been shown that some human neoplasms, including breast and ovarian cancers, express relatively high levels of p73 compared to the corresponding normal tissues (3, 58). In these malignant tissues, the growth-limiting and/or apoptosis-inducing activity of p73 may be blocked by mutant forms of p53 and/or its dominant-negative form ΔNp73. In this connection, the specific disruption of ΔNp73 by ribozyme (27) or RNAi (10, 55) may provide a novel strategy for anticancer treatment.
Acknowledgments
We thank M. Kaghad (Sanofi Recherche) and S. Ikawa (Tohoku University) for providing reagents used in this study, S. Sakiyama (Chiba Cancer Center Research Institute) for critical reading of the manuscript, and M. Ohira and A. Morohashi (Chiba Cancer Center Research Institute) for PAC screening and DNA sequencing.
This work was supported in part by a Grant-in-Aid from the Ministry of Health and Welfare for a New 10-Year Strategy for Cancer Control and Grant-in-Aids for Scientific Research on Priority Areas and for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science, and Technology (Japan).
T.N., M.T., and T.O. contributed equally to this work.
REFERENCES
- 1.Bai, L., and J. L. Merchant. 2001. ZBP-89 promotes growth arrest through stabilization of p53. Mol. Cell. Biol. 21:4670-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, S., A. C. Phillips, P. A. Clark, F. Scott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124-125. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C. L., S. M. Ip, D. Cheng, L. C. Wong, and H. Y. Ngan. 2000. p73 gene expression in ovarian cancer tissues and cell lines. Clin. Cancer Res. 6:3910-3915. [PubMed] [Google Scholar]
- 4.Davison, T. S., C. Vagner, M. Kaghad, A. Ayed, D. Caput, and C. H. Arrowsmith. 1999. p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J. Biol. Chem. 274:18709-18714. [DOI] [PubMed] [Google Scholar]
- 5.De Laurenzi, V., A. Costanzo, D. Barcaroli, A. Terrinoni, M. Falco, M. Annicchiarico-Petruzzelli, M. Levrero, and G. Melino. 1998. The new p73 splice variants, γ and δ, with different transcriptional activity. J. Exp. Med. 188:1763-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Laurenzi, V., M. Catani, A. Costanzo, A. Terrinoni, M. Corazzari, M. Levrero, R. Knight, and G. Melino. 1999. Additional complexity in p73: induction by mitogens in lymphoid cells and identification of two new splicing variants ε and ζ. Cell Death Differ. 6:389-390. [DOI] [PubMed] [Google Scholar]
- 7.De Laurenzi, V., G. Raschella, D. Barcaroli, M. Annicchiarico-Petruzzelli, M. Ranalli, M. V. Catani, B. Tanno, A. Costanzo, M. Levrero, and G. Melino. 2000. Induction of neuronal differentiation by p73 in a neuroblastoma cell line. J. Biol. Chem. 275:15226-15231. [DOI] [PubMed] [Google Scholar]
- 8.Di Como, C. J., C. Gaiddon, and C. Prives. 1999. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol. Cell. Biol. 19:1438-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobbelstein, M., S. Wienzek, C. Konig, and J. Roth. 1999. Inactivation of the p53-homologue p73 by the mdm2-oncoprotein. Oncogene 18:2101-2106. [DOI] [PubMed] [Google Scholar]
- 10.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry, W. S., S. E. Kern, J. A. Pietenpol, K. W. Kinzler, and B. Vogelstein. 1992. Definition of a consensus binding site for p53. Nat. Genet. 1:45-49. [DOI] [PubMed] [Google Scholar]
- 12.Eymin, B., L. Karayan, P. Seite, C. Brambilla, E. Brambilla, C.-J. Larsen, and S. Gazzeri. 2001. Human ARF binds E2F1 and inhibits its transcriptional activity. Oncogene 20:1033-1041. [DOI] [PubMed] [Google Scholar]
- 13.Fang, L., S. W. Lee, and S. A. Aaronson. 1999. Comparative analysis of p73 and p53 regulation and effector functions. J. Cell Biol. 147:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fillippovich, I., N. Sorokina, M. Gatei, Y. Haupt, K. Hobson, E. Moallem, K. Spring, M. Mould, M. A. McGuckin, M. F. Lavin, and K. K. Khanna. 2001. Transactivation-deficient p73α (p73Δexon2) inhibits apoptosis and competes with p53. Oncogene 20:514-522. [DOI] [PubMed] [Google Scholar]
- 15.Fritsche, M., C. Haessler, and G. Brandner. 1993. Induction of nuclear accumulation of the tumor-suppressor protein p53 by DNA-damaging agents. Oncogene 8:307-318. [PubMed] [Google Scholar]
- 16.Gong, J., A. Costanzo, H.-Q. Yang, G. Melino, W. G. Kaelin, M. Levrero, and J. Y. J. Wang. 1999. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399:806-809. [DOI] [PubMed] [Google Scholar]
- 17.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. MDM2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 18.Hermeking, H., C. Lengauer, K. Polyak, T.-C. He, L. Zhang, S. Thiagalingam, K. W. Kinzler, and B. Vogelstein. 1997. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol. Cell 1:3-11. [DOI] [PubMed] [Google Scholar]
- 19.Higashino, F., J. M. Pipas, and T. Shenk. 1998. Adenovirus E4orf6 oncoprotein modulates the function of the p53-related protein, p73. Proc. Natl. Acad. Sci. USA 95:15683-15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 21.Ikawa, S., A. Nakagawara, and Y. Ikawa. 1999. p53 family genes: structural comparison, expression and mutation. Cell Death Differ. 6:1154-1161. [DOI] [PubMed] [Google Scholar]
- 22.Irwin, M., M. C. Marin, A. C. Phillips, R. S. Seelan, D. I. Smith, W. Liu, E. R. Flores, K. Y. Tsai, T. Jacks, K. H. Vousden, and W. G. Kaelin. 2000. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407:645-648. [DOI] [PubMed] [Google Scholar]
- 23.Jost, C., M. Marin, and W. G. Kaelin. 1997. p73 is a human p53-related protein that can induce apoptosis. Nature 389:191-194. [DOI] [PubMed] [Google Scholar]
- 24.Kaghad, M., H. Bonnet, A. Yang, L. Creancier, J. C. Biscan, A. Valent, A. Minty, P. Chalon, J. M. Lelias, X. Dumont, P. Ferrara, F. McKeon, and D. Caput. 1997. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90:809-819. [DOI] [PubMed] [Google Scholar]
- 25.Kern, S. E., K. W. Kinzler, A. Bruskin, D. Jarosz, P. Friedman, C. Prives, and B. Vogelstein. 1991. Identification of p53 as a sequence-specific DNA-binding protein. Science 252:1708-1711. [DOI] [PubMed] [Google Scholar]
- 26.Kubbutat, M. H. G., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 27.Kuwabara, T., M. Warashina, T. Tanabe, K. Tani, S. Asano, and K. Taira. 1998. A novel allosterically trans-activated ribozyme, the maxizyme, with exceptional specificity in vitro and in vivo. Mol. Cell 2:617-627. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lee, C.-W., and N. B. L. Thangue. 1999. Promoter specificity and stability control of the p53-related protein p73. Oncogene 18:4171-4181. [DOI] [PubMed] [Google Scholar]
- 30.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 31.Lissy, N. A., P. K. Davis, M. Irwin, W. G. Kaelin, and S. F. Dowdy. 2000. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature 407:642-645. [DOI] [PubMed] [Google Scholar]
- 32.Marin, M. C., C. A. Jost, M. S. Irwin, J. A. DeCaprio, D. Caput, and W. G. Kaelin. 1998. Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol. Cell. Biol. 18:6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marin, M. C., C. A. Jost, L. A. Brooks, M. S. Irwin, J. O'Nions, J. A. Tidy, N. James, J. M. McGregor, C. A. Harwood, I. G. Yulug, K. H. Vousden, M. J. Allday, B. Gusterson, S. Ikawa, P. W. Hinds, T. Crook, and W. G. Kaelin. 2000. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat. Genet. 25:47-54. [DOI] [PubMed] [Google Scholar]
- 34.Mizuguchi, H., and M. A. Kay. 1998. Efficient construction of a recombinant adenovirus vector by an improved in vitro ligation method. Hum. Gene Ther. 9:2577-2583. [DOI] [PubMed] [Google Scholar]
- 35.Mizuguchi, H., and M. A. Kay. 1999. A simple method for constructing E1- and E1/E4-deleted recombinant adenoviral vectors. Hum. Gene Ther. 10:2013-2017. [DOI] [PubMed] [Google Scholar]
- 36.Nip, J., D. K. Strom, C. M. Eischen, J. L. Cleveland, G. P. Zambetti, and S. W. Hiebert. 2001. E2F-1 induces the stabilization of p53 but blocks p53-mediated transactivation. Oncogene 20:910-920. [DOI] [PubMed] [Google Scholar]
- 37.O'Connor, D. J., E. W.-F. Lam, S. Griffin, S. Zhong, L. C. Leighton, S. A. Burbidge, and X. Lu. 1995. Physical and functional interactions between p53 and cell cycle co-operating transcription factors, E2F- and DP1. EMBO J. 14:6184-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osada, M., M. Ohba, C. Kawahara, C. Ishioka, R. Kanamaru, I. Katoh, Y. Ikawa, Y. Nimura, A. Nakagawara, M. Obinata, and S. Ikawa. 1998. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat. Med. 4:839-843. [DOI] [PubMed] [Google Scholar]
- 39.Ozaki, T., M. Naka, N. Takada, M. Tada, S. Sakiyama, and A. Nakagawara. 1999. Deletion of the COOH-terminal region of p73α enhances both its transactivation function and DNA-binding activity but inhibits induction of apoptosis in mammalian cells. Cancer Res. 59:5902-5907. [PubMed] [Google Scholar]
- 40.Pomerantz, J., N. Schreiber-Agus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, H.-W. Lee, C. Cordon-Cardo, and R. A. DePinho. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92:713-723. [DOI] [PubMed] [Google Scholar]
- 41.Pozniak, C. D., S. Radinovic, A. Yang, F. McKeon, D. R. Kaplan, and F. D. Miller. 2000. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science 289:304-306. [DOI] [PubMed] [Google Scholar]
- 42.Prives, C. 1998. Signaling to p53: breaking the MDM2-p53 circuit. Cell 95:5-8. [DOI] [PubMed] [Google Scholar]
- 43.Roth, J., C. Konig, S. Wienzek, S. Weigel, S. Ristea, and M. Dobbelstein. 1998. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J. Virol. 72:8510-8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rouault, J. P., N. Falette, F. Guehenneux, C. Guillot, R. Rimokh, O. Wang, C. Berthet, C. Moyret-Lalle, P. Savatier, B. Pain, P. Shaw, R. Berger, J. Samarut, J. P. Magaud, M. Ozturk, C. Ssmarut, and A. Puisieux. 1996. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat. Genet. 14:482-486. [DOI] [PubMed] [Google Scholar]
- 45.Schwab, M., C. Praml, and L. C. Amler. 1996. Genomic instability in 1p and human malignancies. Genes Chromosom. Cancer 16:211-229. [DOI] [PubMed] [Google Scholar]
- 46.Sherr, C. J. 1998. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12:2984-2991. [DOI] [PubMed] [Google Scholar]
- 47.Steegenga, W. T., A. Shvarts, N. Riteco, J. L. Bos, and A. G. Jochemsen. 1999. Distinct regulation of p53 and p73 activity by adenovirus E1A, E1B, and E4orf6 proteins. Mol. Cell. Biol. 19:3885-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stiewe, T., and B. M. Putzer. 2000. Role of the p53-homologue p73 in E2F-1-induced apoptosis. Nat. Genet. 26:464-469. [DOI] [PubMed] [Google Scholar]
- 49.Stott, F. J., S. Bates, M. C. James, B. B. McConnell, M. Starborg, S. Brookes, I. Palmero, K. Ryan, E. Hara, K. H. Vousden, and G. Peters. 1998. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17:5001-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strano, S., E. Munarriz, M. Rossi, B. Cristofanelli, Y. Shaul, L. Castagnoli, A. J. Levine, A. Sacchi, G. Cesareni, M. Oren, and G. Blandino. 2000. Physical and functional interaction between p53 mutants and different isoforms of p73. J. Biol. Chem. 275:29503-29512. [DOI] [PubMed] [Google Scholar]
- 51.Ueda, Y., M. Hijikata, S. Takagi, T. Chiba, and K. Shimotohno. 1999. New p73 variants with altered C-terminal structures have varied transcriptional activities. Oncogene 18:4993-4998. [DOI] [PubMed] [Google Scholar]
- 52.Vaishnav, Y. N., and V. Pant. 1999. Differential regulation of E2F transcription factors by p53 tumor suppressor protein. DNA Cell Biol. 18:911-922. [DOI] [PubMed] [Google Scholar]
- 53.Vousden, K. H., and G. F. Woude. 2000. The ins and outs of p53. Nat. Cell Biol. 2:E178-E180. [DOI] [PubMed] [Google Scholar]
- 54.White, P. S., J. M. Maris, C. Beltinger, C. Sulman, H. N. Marshall, M. Fujimori, B. A. Kaufman, J. A. Biegel, C. Allen, C. Hilliard, M. B. Valentine, A. T. Look, H. Enomoto, S. Sakiyama, and G. M. Brodeur. 1995. A region of consistent deletion in neuroblastoma maps within 1p36.2-3. Proc. Natl. Acad. Sci. USA 92:5520-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wianny, F., and M. Zernicka-Goetz. 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol. 2:70-75. [DOI] [PubMed] [Google Scholar]
- 56.Yang, A., M. Kaghad, Y. Wang, E. Gillett, M. D. Fleming, V. Dotsch, N. C. Andrews, D. Caput, and F. McKeon. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2:305-316. [DOI] [PubMed] [Google Scholar]
- 57.Yang, A., N. Walker, R. Bronson, M. Kaghad, M. Oosterwegel, J. Bonnin, C. Vagner, H. Bonnet, P. Dikkes, A. Scharpe, F. McKeon, and D. Caput. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99-103. [DOI] [PubMed] [Google Scholar]
- 58.Zaika, A. I., S. Kovalev, N. Marchenko, and U. M. Moll. 1999. Overexpression of the wild type p73 gene in breast cancer tissues and cell lines. Cancer Res. 59:3257-3263. [PubMed] [Google Scholar]
- 59.Zaika, A., M. Irwin, C. Sansome, and U. M. Moll. 2001. Oncogenes induce and activate endogenous p73 protein. J. Biol. Chem. 276:11310-11316. [DOI] [PubMed] [Google Scholar]
- 60.Zeng, X., L. Chen, C. A. Jost, R. Maya, D. Keller, X. Wang, W. G. Kaelin, M. Oren, J. Chen, and H. Lu. 1999. MDM2 suppresses p73 function without promoting p73 degradation. Mol. Cell. Biol. 19:3257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou, X., X. W. Wang, L. Xu, K. Hagiwara, M. Nagashima, R. Wolkowicz, I. Zurer, V. Rotter, and C. C. Harris. 1999. COOH-terminal domain of p53 modulates p53-mediated transcriptional transactivation, cell growth, and apoptosis. Cancer Res. 59:843-848. [PubMed] [Google Scholar]
- 62.Zhu, J., J. Jiang, W. Zhou, and X. Chen. 1998. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 58:5061-5065. [PubMed] [Google Scholar]