Abstract
Vav2, like all Dbl family proteins, possesses tandem Dbl homology (DH) and pleckstrin homology (PH) domains and functions as a guanine nucleotide exchange factor for Rho family GTPases. Whereas the PH domain is a critical positive regulator of DH domain function for a majority of Dbl family proteins, the PH domains of the related Vav and Vav3 proteins are dispensable for DH domain activity. Instead, Vav proteins contain a cysteine-rich domain (CRD) critical for DH domain function. We evaluated the contribution of the PH domain and the CRD to Vav2 guanine nucleotide exchange, signaling, and transforming activity. Unexpectedly, we found that mutations of the PH domain impaired Vav2 signaling, transforming activity, and membrane association. However, these mutations do not influence exchange activity on Rac and only slightly affect exchange on RhoA and Cdc42. We also found that the CRD was critical for the exchange activity in vitro and contributed to Vav2 membrane localization. Finally, we found that phosphoinositol 3-kinase activation synergistically enhanced Vav2 transforming and signaling activity by stimulating exchange activity but not membrane association. In conclusion, the PH domain and CRD are mechanistically distinct, positive modulators of Vav2 DH domain function in vivo.
Rho family proteins are members of the Ras superfamily of small GTPases. Currently, 18 mammalian Rho family proteins have been identified, with Rac1, Cdc42, and RhoA being the best characterized (37, 45). Rho family GTPases are guanine nucleotide binding proteins that function as molecular switches that cycle between active GTP-bound and inactive GDP-bound states. Dbl family proteins serve as guanine nucleotide exchange factors (GEFs), which accelerate the intrinsic GDP/GTP exchange activity of Rho GTPases to cause formation of the active GTP-bound protein (8, 39). The activated Rho GTPases then interact with a wide spectrum of downstream effector proteins to mediate cellular activities that include regulation of actin cytoskeletal organization, gene expression, and cellular proliferation (3). A second class of regulatory proteins, Rho family-specific GTPase-activating proteins, stimulate intrinsic GTPase activity to return these small GTPases to their inactive GDP-bound state and to terminate downstream signaling (37).
Vav proteins (Vav, Vav2, and Vav3) are mammalian members of the Dbl family of proteins (7). All Vav proteins have similar structural organizations. Like all Dbl family proteins, Vav proteins possess a Dbl homology (DH) domain followed by a COOH-terminal pleckstrin homology (PH) domain (8, 39). Previous studies indicate that the DH domain interacts directly with Rho family GTPases to catalyze GDP release (8, 39). The Vav DH domains exhibit broad GTPase specificity and serve as GEFs for multiple Rho GTPases (RhoA, RhoG, Rac1, and Cdc42), although different studies have reached contrasting conclusions regarding the specific GTPases targeted by Vav (1, 12, 17, 23, 28, 35, 43).
The invariant topography of DH and PH domains (DH/PH domain) found in all Dbl family proteins suggests a critical role for the PH domain in regulation of DH domain function. Extensive structure-function analyses of the DH/PH domains of various Dbl family proteins suggest that the PH domain may serve two distinct functions to modulate DH domain activity (8, 39). First, the PH domain may act as a positive modulator of the intrinsic catalytic activity of the DH domain. For example, a comparison of the catalytic activities of the DH and DH/PH domains derived from several Dbl family proteins (e.g., Dbl, Trio, and Dbs) showed that the GEF activity exhibited by a PH-containing protein was up to 100-fold greater than that measured for the DH domain alone in vitro (24, 34, 44). Second, it may serve a membrane-targeting function and regulate DH domain interaction with its membrane-bound GTPase substrates. For example, the loss of function caused by mutation of the PH domains of Lfc and Dbs could be reversed by addition of a plasma membrane targeting sequence (40, 41).
In contrast to what has been observed for many Dbl family proteins, the PH domains of Vav proteins appear to serve as negative regulators of DH domain function and no role in membrane targeting has been described. Han and colleagues determined that the PH domain of Vav serves as a negative regulator of DH domain GEF activity in vitro (18). This negative regulatory function is promoted by phosphatidylinositol 4,5-phosphate (PIP2), a substrate of phosphatidylinositol 3-kinase (PI3K), and is antagonized by the PI3K product, phosphatidylinositol 3,4,5-phosphate (PIP3). Hence, PI3K activation is proposed to facilitate the activation of Vav. Consistent with a negative regulatory function for the PH domain, Ma et al. determined that a variant of Vav with its PH domain deleted was activated constitutively in vivo (26). In evaluations of PH domain function in NH2-terminally truncated and constitutively activated versions of Vav proteins, mutation of the PH domains of Vav and Vav3 did not cause significant alteration in GEF activity in vitro or growth and/or morphological transforming activity in vivo, suggesting that the PH domain is not a critical regulator of Vav DH domain function (18, 28). Since the role of the PH domain in Vav2 function has not been addressed, it has not been previously established whether the PH domains of Vav family proteins exhibit similar or distinct functions.
Vav proteins also contain additional protein-protein or protein-lipid interaction domains that flank the DH/PH domains. A calponin homology domain and an acidic amino acid-rich domain positioned NH2 terminal to the DH and PH domains serve negative regulatory roles in Vav function (7). Tyrosine phosphorylation of the NH2-terminal region relieves this negative regulatory function. Hence, mutant versions of all three Vav proteins with the NH2 termini deleted are activated constitutively in a phosphorylation-independent fashion and exhibit potent transforming activity (2, 20). COOH terminal to the PH domain are a cysteine-rich domain (CRD) and an Src homology 2 (SH2) domain that is flanked by two SH3 domains. The role of the SH domains is to facilitate the phosphorylation of Vav (7). Consistent with this role, the SH domains are dispensable for the transforming activities of the NH2-terminally truncated and transforming versions of Vav (35). In contrast, mutational analyses determined that the CRD is critical for the growth and/or morphological transforming activity of Vav and Vav3 (11, 28). While the precise role of the CRD in Vav function is not known, the impaired GEF activity in vitro of a Vav3 CRD mutant, coupled with the capacity of the isolated Vav3 CRD to bind to RhoA in vitro, argues that the CRD may facilitate DH domain interaction with its GTPase substrates (28). Thus, the CRD may substitute functionally for the PH domain and hence render the PH domain dispensable for Vav function. Whether the CRD can also regulate Vav association with membranes has not been determined.
In light of the incomplete or conflicting observations regarding the role of the PH domain and CRD in Vav function, we have evaluated the biological and biochemical contributions of the PH domain and the CRD to Vav2 catalytic activity in vitro and signaling and transforming activity in vivo. In contrast to previous observations with Vav and Vav3, we found that an intact PH domain is not required for the catalytic activity of the DH domain in vitro but is required for complete biological and catalytic activity of Vav2 in vivo and facilitates Vav2 association with membranes. Similarly, mutation of the CRD also impaired Vav2 signaling and transformation, in part, by impairing the intrinsic catalytic activity of the DH domain. Finally, we found that PI3K activation enhanced the signaling and transforming activity of Vav2; this enhancement involves the PH domain but does not promote Vav2 association with membranes. Taken together, our observations suggest the PH domain and the CRD are both essential for complete Vav2 activity but that they serve distinct functions in regulating DH domain activity.
MATERIALS AND METHODS
Molecular constructs.
The pCGN-hygro mammalian expression vector regulates the expression of introduced cDNA sequences from a cytomegalovirus promoter, encodes hygromycin resistance, and introduces an NH2-terminal hemagglutinin (HA) epitope tag into the encoded protein (14). Human Vav2 truncation mutants were generated by PCR-mediated approaches using wild-type full-length humanVav2 cDNA (pCMV5-Vav2; a generous gift from Dan Broek) as the template. A 5′ primer containing a BamHI site and sequences encoding eight Vav2 NH2-terminal residues corresponding to residues 192 to 200 was used in conjunction with a 3′ primer containing a BamHI restriction site and sequences encoding eight Vav2 COOH-terminal residues corresponding to residues 845 to 851 to generate a 2-kb fragment encoding the sequence from the DH domain to the end of the protein. Three additional 3′ primers containing eight Vav2 COOH-terminal residues corresponding to residues 565 to 573, 510 to 518, or 368 to 377 were used to generate a 1.7-kb fragment encoding the DH and PH domains and the CRD (residues 192 to 573 [Vav2-DPC]), a 1.2-kb fragment encoding the DH and PH domains (Vav2-DP), and a 0.7-kb fragment encoding the DH domain (Vav2-D), respectively. All constructs were digested with BamHI and ligated into the BamHI site of pCGN-hygro in frame with the HA epitope tag at the NH2 terminus. cDNA sequences encoding missense mutations in the PH domain (K407A and W503L) or the CRD (K533A, K538A, K563A, and V568E) of Vav2-DPC were introduced with the Quickchange site-directed mutagenesis kit (Stratagene) in accordance with the manufacturer's protocol. All constructs were sequenced in their entirety to eliminate the possibility of extra mutations.
Cell culture and transformation assay.
NIH 3T3 mouse fibroblasts were cultured in Dulbecco's modified Eagle's medium supplemented with 10% calf serum. DNA transfections for transformation assays were performed by using the calcium phosphate precipitation method as described previously (9). For each assay, the cognate empty vector was used as a control. The transfected cultures were maintained in culture media for 14 days, fixed, and stained with crystal violet (0.5%), and the number of foci of transformed cells was then quantitated. NIH 3T3 cells stably expressing the Vav2 mutants were established by transfection of pCGN-vav2 constructs and isolation of drug-resistant colonies after growth in medium supplemented with hygromycin (400 μg/μl). Multiple drug-resistant colonies (>100) were pooled together to establish stable cell lines for use in the various assays described.
Transient expression reporter gene assays.
NIH 3T3 cells were transfected by Lipofectamine Plus according to the manufacturer's protocol. Forty-eight hours posttransfection, cells were starved for 12 to 14 h with Dulbecco's modified Eagle's medium supplemented with 0.5% bovine calf serum. Analyses of the cell lysates of the transiently transfected NIH 3T3 cells were performed using enhanced chemiluminescence reagents and a Monolight 2010 luminometer (Analytical Luminescence). The (SREm)2 reporter plasmid contains a luciferase gene whose expression is under the control of a serum response factor (SRF)-responsive promoter as has been described previously (38). All assays were performed at least in triplicate.
Guanine nucleotide exchange assays.
cDNA fragments encoding either human Vav2-DPC (residues 192 to 573) or the Vav2 PH domain (K407A) and CRD (K533A, K538A, K563A, and V568E) mutants were generated by PCR and inserted into the NcoI/XhoI sites of bacterial expression vector pET-28a (Novagen). The bacterial expression constructs were transformed into Escherichia coli strain BL21 (DE3), and protein expression was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 22°C. The recombinant proteins were His6 tagged at their COOH termini and were purified from bacterial lysate on a nickel-nitrilotriacetic acid agarose column (Qiagen). Bacterially expressed glutathione S-transferase (GST)-RhoA(F25N), GST-Rac1(WT), and GST-Cdc42(WT) proteins were kindly provided by J. Sondek.
Fluorescence spectroscopy analysis of N-methylanthraniloyl (mant)-GDP incorporation into GDP-preloaded Rac1, Cdc42, and RhoA was carried out with a Perkin-Elmer LS 50 B spectrometer at 20°C as described previously (1). Exchange reaction mixtures containing 20 mM Tris, pH 7.5, 50 mM NaCl, 10% glycerol, 400 nM mant-GDP (Biomol), and 2 μM GTPase were prepared and allowed to equilibrate with continuous stirring. After equilibration, each Vav2 polypeptide was added to 100 nM, and the relative mant fluorescence (excitation λ [λex] = 360 nm; emission λ [λem] = 440 nm) was monitored. All experiments were performed in duplicate.
RESULTS
Vav2 transforming and signaling activity requires an intact PH domain.
An intact PH domain is critical for the transforming activity of some Dbl family proteins by promoting DH domain catalytic function and/or by facilitating its association with the plasma membrane (8). Surprisingly, mutational analyses suggest that the PH domains of Vav family proteins serve as negative regulators of DH domain function and hence are dispensable for transforming activity (18, 26, 28). Instead, Vav and Vav3 biological activity has been found to require an intact CRD adjacent to the PH domain (11, 28). Therefore, in the present study, we assessed whether the PH domain is dispensable and whether the CRD may serve analogous functions as an important regulator of DH domain function and as a regulator of membrane association.
We determined previously that NH2-terminal deletion of all sequences upstream of the DH domain of Vav2 (amino acids 1 to 191) results in a constitutively activated, highly transforming Vav2 mutant (1). In the present study, we determined that the COOH-terminal SH3-SH2-SH3 domain sequence is also dispensable for Vav2 transforming and signaling activity (Fig. 1 and 2). Thus, as has been reported for Vav (35), the fragment of Vav2 that contains the DH, PH, and CRD regions alone (designated Vav2-DPC) defines a minimal functional, transforming unit of Vav2. Therefore, we introduced mutations into an HA epitope-tagged version of Vav2-DPC to evaluate the role of the PH domain and CRD in Vav2 transforming activity.
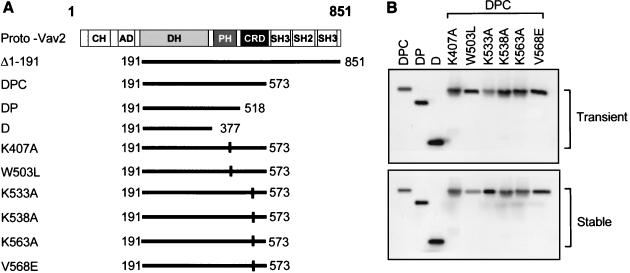
FIG. 1.
Primary structure and protein expression levels of the human Vav2 truncation and point mutants. (A) Schematic structure of full-length human Vav2, NH2-terminal and COOH-terminal truncation mutants, and PH domain and CRD point mutants. Horizontal lines below full-length Vav2 show the predicted translational product of each truncation or point mutant initiated at the indicated amino acid. NH2-terminally truncated Vav2 (Δ1-191) is a strongly transforming variant of Vav2, and further COOH-terminal truncation to generate the Vav2-DPC fragment (DPC) did not reduce this activity. Therefore, to avoid potential complications in interpretation, all PH and CRD point mutants were generated in the background of the highly transforming Vav2-DPC fragment. Vertical lines, mutations in the PH domain (K407A and W503L) and the CRD (K533A, K538A, K563A, and V568E). Truncation mutants containing the DH and PH domains (DP) or DH domain (D) were also generated. The Δ1-191 and wild-type and mutant Vav2-DPC, -DP, and -D sequences were fused in frame to an HA epitope tag at the NH2 terminus. (B) Expression of the Vav2-DPC, truncation, and point mutants in stably and transiently transfected NIH 3T3 cells.
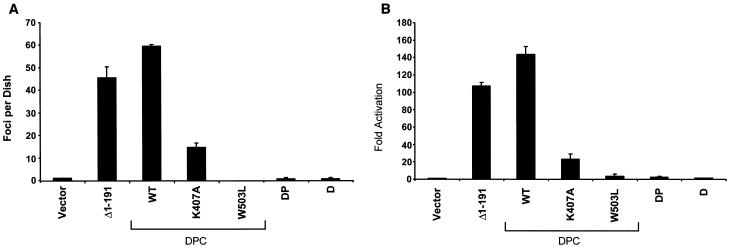
FIG. 2.
Mutation of the Vav2 PH domain decreases Vav2 transforming and signaling activity. (A) NIH 3T3 cells were transfected with the empty pCGN-hygro plasmid (Vector) or pCGN-hygro plasmids encoding the various PH domain mutants (Fig. 1; 1 μg per 60-mm-diameter dish). The appearance of transformed foci was quantitated on day 14. The values represent the averages ± standard errors of three dishes and are representative of at least three independent assays. WT, wild type. (B) NIH 3T3 cells were transiently transfected with the empty pCGN-hygro plasmid or pCGN-hygro plasmids encoding the various PH domain mutants (100 ng per 60-mm-diameter dish), together with a mutant serum response element luciferase reporter plasmid to determine stimulation of SRF. Fold activation was determined by the number of relative luciferase units relative to the number of units seen with the empty vector control. Data are representative of at least three independent assays performed on duplicate plates.
We introduced two missense mutations in the PH domain of Vav2-DPC (Fig. 1A). First, we substituted a leucine for the invariant tryptophan residue that occurs in all PH domains and that is essential for the transforming ability of several Dbl family oncoproteins (30, 40) [designated Vav2-DPC(W503L)]. This mutation is expected to disrupt the global PH fold and completely abolish PH domain function (22). Second, we introduced mutation K407A, analogous to one introduced into Vav (K404A), which impairs PIP2 binding and decreases exchange activity toward Rac1 in vitro (18).
We evaluated the expression of each mutant protein to determine if these point mutants were altered in protein stability. First, we assessed the levels of protein expression when Vav2-DPC and each mutant were expressed in transiently transfected NIH 3T3 cells. Second, we transfected NIH 3T3 cells with the empty pCGN-hygro mammalian expression vector and pCGN-hygro encoding Vav2-DPC and the W503L and K407A mutant proteins to establish mass populations of stably transfected cells. We detected comparable levels of protein expression for all mutants in the transiently or stably transfected NIH 3T3 cells, suggesting that the different amino acid substitutions did not cause a significant alteration in protein stability (Fig. 1B).
We next performed focus formation transformation assays with NIH 3T3 cells to evaluate the effect of the PH domain mutations on Vav2 transforming activity. Surprisingly, we found that, compared to that of Vav2-DPC, the transforming activity of the K407A mutant was decreased greater than threefold whereas the W503L mutant completely lost transforming activity (Fig. 2A). These results contrast with those made with Vav and Vav3 (26, 28) and demonstrate that a functional PH domain, within the context of the intact Vav2-DPC cassette, is required for full Vav2 transforming activity.
We next determined whether mutations in the PH domain correlated with decreased activation of the SRF signaling pathway mediated by Rac1, Cdc42, and RhoA activation. NIH 3T3 cells were transiently transfected with expression vectors encoding Vav2-DPC and the PH domain mutant proteins. We found that the decreased transforming potential of the PH domain mutants correlated directly with a reduced ability to stimulate SRF activation (Fig. 2B). Vav2-DPC expression greatly enhanced SRF activity (70-fold), whereas the K407A mutant exhibited an approximately 10-fold-reduced ability to stimulate SRF activity and the W503L mutant was completely inactive.
Vav2 does not require an intact PH domain for the activation of GTPases in vitro, but a PH domain is required for facilitating membrane association
Based on observations from structure-function analyses of other Dbl family proteins, the PH domain may be critical for the intrinsic catalytic activity of the DH domain (24, 34, 44) or, alternatively, may regulate the subcellular location of Vav2 to facilitate DH domain interaction with its membrane-bound GTPase substrates (40, 41). To distinguish between these two possibilities, we next investigated the guanine nucleotide exchange activity of wild-type and mutant Vav2-DPC in vitro. For these analyses, we purified bacterially expressed hexahistidine-tagged versions of wild-type Vav2-DPC and Vav2-DPC PH domain mutants and analyzed the activity of these proteins by quantitation of mant-GDP incorporation into bacterially expressed Rac1, Cdc42, and RhoA proteins. Although both mutant proteins were expressed at equivalent levels in E. coli, the W503L mutant protein was completely insoluble when isolated, thereby preventing any detailed biochemical analysis. Consequently, we were only able to evaluate the consequences of the K407A mutant for exchange activity in vitro.
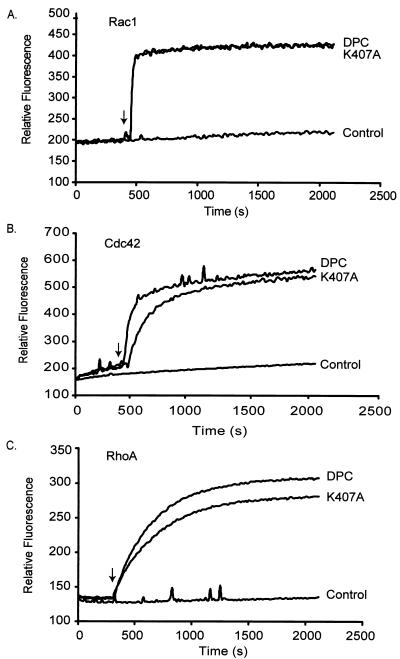
RhoA, Rac1, and Cdc42 alone did not exhibit significant intrinsic GDP dissociation activity in vitro (Fig. 3). As we found for Vav2-D and Vav2-DP (1), Vav2-DPC stimulated rapid GDP dissociation from Rac1, Cdc42, and RhoA, which was complete within approximately 20 min (Fig. 3). Interestingly, the wild-type and K407A mutant Vav2-DPCs exhibited identical exchange activities on Rac1 (Fig. 3A). We also found that the K407A mutant displayed slightly decreased but comparable exchange activities toward Cdc42 and RhoA (Fig. 3). Therefore, mutation of residue K407 which was designed to eliminate phosphoinositide binding and is necessary for activity in vivo did not alter the GTPase specificity or catalytic activity of the Vav2-DPC fragment in vitro.
FIG. 3.
Stimulation of mant-GDP incorporation into Rac1, Cdc42, and RhoA by the Vav2 PH domain mutants in vitro. The abilities of bacterially expressed wild-type Vav2-DPC and Vav2-DPC PH domain mutants (50 nM) to stimulate the incorporation of mant-GDP into bacterially expressed (2 μM) Rac1 (A), Cdc42 (B), and RhoA (C) were measured by fluorescence spectroscopy (λex = 360 nm; λem = 440 nm). Arrows, time point (300 s) at which the Vav2 PH or CRD polypeptides were added to the exchange reaction mixture. Results are representative of two independent assays.
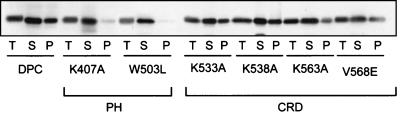
Since the PH domain has been shown to contribute to the membrane association of at least some Dbl family proteins (40, 41), we assessed the possibility that mutations in the PH domain impaired membrane association. For these analyses, we performed crude subcellular fractionation (100,000 × g) with cell lysates from NIH 3T3 cells stably expressing the wild-type or mutant Vav2-DPC proteins (Fig. 4). Control Western blot analyses were done to verify that the fractionations of all cell lysates were equivalent, and we found that equal amounts of endogenous RhoGDI were detected only in the cytosolic fraction and that equal amounts of endogenous Ras were detected only in the membrane-containing fraction (data not shown). Whereas a significant fraction (approximately 40%) of Vav2-DPC was found in the P100 membrane-containing fraction, the two Vav2-DPC PH domain mutants (K407A and W503L mutants) exhibited a significant loss of association with the P100 fraction. In addition, these results support a role for the PH domain in contributing to Vav2 membrane association. Thus the loss of transforming activity due to mutation of the PH domain may be a consequence of impaired DH domain interaction with its membrane-bound GTPase substrates in vivo.
FIG. 4.
The Vav2 PH domain, but not the CRD, influences subcellular distribution. NIH 3T3 cells were transiently transfected with the empty pCGN mammalian expression vector and pCGN plasmids encoding the various PH domain and CRD mutants (Fig. 1; 1 μg per 60-mm-diameter dish). Subcellular fractions were prepared 48 h after transfection by lysis of cells in hypotonic buffer (0.1 M Tris [pH 7.4], 0.5 M MgCl2, 1 mM Pefabloc, 1 μM leupeptin, 2 μM pepstatin, 0.1% aprotinin) and addition of NaCl to adjust the ionic strength to 0.15 M, followed by ultracentrifugation for 30 min at 100,000 × g as described previously (5). Proteins in the crude S100 (S) and P100 (P) fractions were precipitated with 5 ml of acetone for 1 h at 4°C, collected by centrifugation at 2,000 × g for 30 min, and resuspended in 100 μl of electrophoresis sample buffer. Equal volumes of each fraction were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and protein expression was determined by Western blot analysis using the anti-HA epitope antibody.
Mutation of the CRD disrupts Vav2 signaling and transformation.
Previous studies determined that missense mutations that completely disrupt the structural integrity of the CRD result in a complete loss of Vav and Vav3 biological activity (11, 28). Similarly, we determined that deletion of the CRD from Vav2-DPC also resulted in a complete loss of transforming and signaling activity (Fig. 2). However, the functional consequences of these approaches to disrupt CRD function may be indirect, due to nonspecific disruption of the function of flanking domains. Furthermore, as we have seen with the CRD of Raf-1, multiple ligands can interact with the CRD and have either positive (e.g., Ras) or negative (14-3-3) regulatory roles 10; J. K. Drugan, G.-S. Yi, J. G. Williams, G. J. Clark, H. R. Mott, C. J. Der, and S. L. Campbell, submitted for publication). Thus, a complete disruption of CRD function may not reveal such a complexity of regulation.
To better characterize the role of the CRD in Vav2-mediated transformation and signaling, we introduced missense mutations into the CRD of Vav2-DPC at conserved, charged, surface-exposed regions that are not predicted to affect the structural integrity of the CRD. These point mutations were generated based on homology sequence alignments of CRD sequences for the various Vav family members, Raf-1, and protein kinase C (PKC) (19); known ligand-binding sequences of other CRDs; and our previously determined nuclear magnetic resonance solution structure of the Raf-1 CRD (Fig. 5) (27). Structural studies on the Raf-1 CRD revealed binding determinants in the CRD that selectively interfere with ligand binding interactions. Based on these analyses, we changed lysines 533 and 538 to alanines [mutants Vav2-DPC(K533A) and Vav2-DPC(K538), respectively] (Fig. 1A and 5). These residues in Vav2 correspond to residues L149 and F151 in Raf-1, which appear to directly contact Ras but not 14-3-3 or phosphatidylserine (42). We also mutated lysine 563 to alanine and valine 568 to glutamic acid [mutants Vav2-DPC(K563A) and Vav2-DPC(V568E), respectively] (Fig. 1A). These residues correspond to a region in Raf-1 that, for the most part, has not been characterized.
FIG. 5.
Alignment of representative CRDs. The conserved cysteine and histidine residues in the different Raf serine/threonine kinases and Vav protein CRDs were aligned. Black shading, identical residues; gray shading, highly conserved residues, dashes, gaps in the alignment with PKCδ C1A. Residues in PKCδ C1A involved in diacylglycerol (DAG) and membrane binding (membrane) are indicated (19). Residues in human c-Raf-1 involved in Ras and 14-3-3 binding are also indicated (19). Arrows, residues that were mutated in human Vav2.
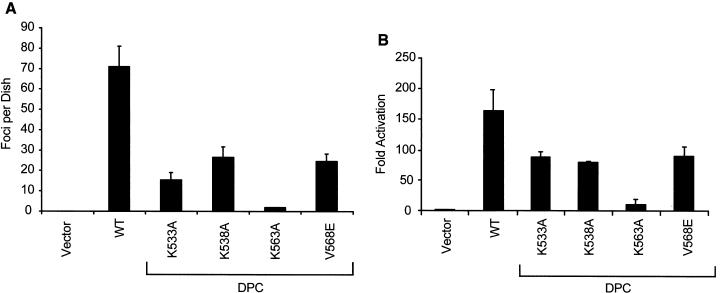
Western blot analysis of transiently or stably transfected NIH 3T3 cells confirmed that the CRD mutations did not perturb protein stability (Fig. 1B). We then performed focus formation assays to evaluate the effect of each of the CRD mutations on Vav2 transforming activity (Fig 6A). We found that, compared to that of the parental Vav2-DPC protein, the transforming activities of the K533A, K538A, and V568E mutants were impaired significantly (62 to 79% reduction), whereas the K563A mutant completely lost focus-forming activity (Fig. 6A).
FIG. 6.
Mutation of the Vav2 CRD decreases Vav2 transforming and signaling activity. (A) NIH 3T3 cells were transfected with the empty pCGN-hygro plasmid or pCGN-hygro plasmids encoding the various CRD mutants (Fig. 1; 1 μg per 60-mm-diameter dish). The appearance of transformed foci was quantitated on day 14. The values represent the averages ± standard errors of three dishes and are representative of at least three independent assays. WT, wild type. (B) NIH 3T3 cells were transiently transfected with empty pCGN plasmid or pCGN plasmids encoding the various CRD mutants (100 ng per 60-mm-diameter dish), together with the SRF luciferase reporter plasmid. Fold activation was determined by the number of relative luciferase units relative to the number of units seen with the empty vector control. Data are representative of at least three independent assays performed on duplicate plates.
We also compared the abilities of the CRD mutants to stimulate transcriptional activation of SRF. We found that the degree of stimulation seen with SRF correlated directly with transforming activity (Fig. 6B). The weakly transforming K533A, K538A, and V568E mutants showed a weak ability to stimulate transcription from SRF: levels were two- to threefold less than the activity seen with the nonmutated Vav2-DPC protein. The nontransforming K563A mutant showed minimal ability to activate SRF.
Vav2 requires an intact CRD for the activation of GTPases in vitro.
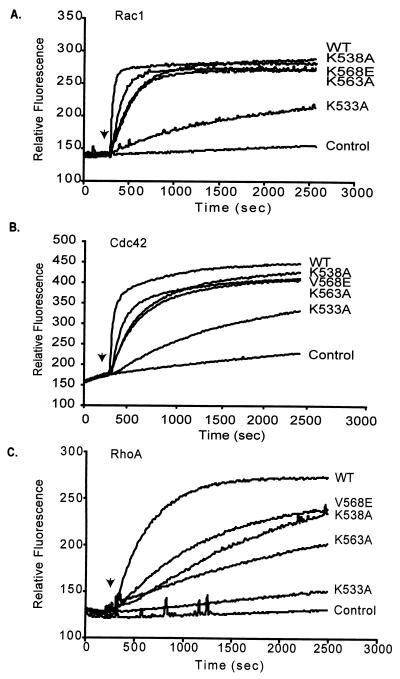
An intact CRD may be critical for the intrinsic catalytic activity of the DH domain (28) or, alternatively, by analogy to the CRD of PKC, may regulate subcellular location to facilitate DH domain interaction with its membrane-bound GTPase substrates (19). To address these two possibilities, we evaluated the GEF activity and specificity of bacterially expressed Vav2-DPC proteins containing the CRD mutations. We observed two unexpected results from these analyses. First, although all four CRD mutations compromised GEF activity in vitro, the degree of impairment was not uniform for all three Rho GTPases and was most severe for RhoA. For example, whereas K538A, K563A, and K568E mutants showed a slight but reproducible decrease in exchange on Rac and Cdc42 (Fig. 7A and B), all three were severely compromised in their ability to mediate exchange on RhoA (Fig. 7C). Substitution of K533, a residue in the region that corresponds to the phorbol ester binding site of PKC, caused the most-severe reductions in exchange activity toward all three GTPases. Vav2-DPC harboring this substitution retained a small amount of activity toward Rac1 and Cdc42 but was almost completely inactive on RhoA (Fig. 7). Second, the degree of impairment in GEF activity in vitro did not correlate directly with the impairment in signaling and transforming activity. Although K563E caused the most drastic reduction in transforming activity, it was the K533A mutant that was the most impaired in GEF activity. These results indicate that the CRD is critical for regulating the intrinsic catalytic activity of the DH domain, most critically for RhoA, and the consequences of the CRD mutations for transforming activity cannot be ascribed solely to perturbation of the intrinsic guanine nucleotide exchange activity.
FIG. 7.
Stimulation of mant-GDP incorporation into Rac1, Cdc42, and RhoA by the Vav2 CRD mutants in vitro. The ability of bacterially expressed wild-type (WT) Vav2-DPC (50 nM) and Vav2-DPC with the listed substitutions within the CRD (50 nM) to stimulate the incorporation of mant-GDP into bacterially expressed (2 μM) Rac1 (A), Cdc42 (B), and RhoA (C) was measured by fluorescence spectroscopy (λex = 360 nm; λem = 440 nm). Arrows, time point (300 s) at which the Vav2 CRD polypeptides were added to the exchange reaction mixture. Results are representative of two independent assays.
CRDs have been shown to promote the membrane association of PKC and other signaling proteins (19). Therefore, we assessed the possibility that mutations in the CRD may also impair Vav2-DPC activity as a consequence of impaired membrane association. In contrast to our observations with the PH domain mutants, which were largely cytosolic (∼95%), the Vav2 CRD K533A, K538A, and V568E mutants exhibited only slight reductions in the amount of protein in the membrane-containing fraction (30 to 40% in P100) compared to Vav2-DPC (Fig. 4). However, Vav2-DPC(K563A) had a significantly decreased amount of protein in the P100 fraction (20% of total protein). Thus, these results support a role for the CRD in contributing to Vav2 membrane association and suggest that the impaired transforming activity of the K563A mutant is due, in part, to impaired intrinsic catalytic activity as well as impaired interaction with membranes in vivo
PI3K activation enhances Vav2 activity via the PH domain.
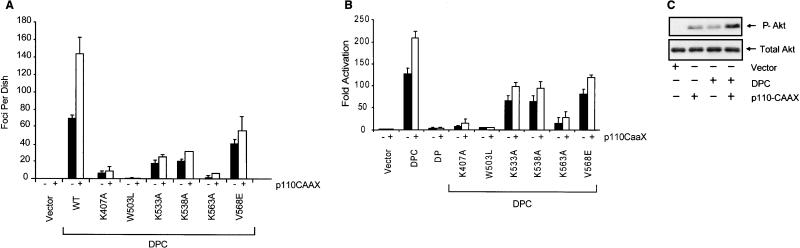
Our mutational analyses showed that the PH domain regulates DH domain activity in vivo but not in vitro. Based on the observations made with the PH domain of Vav and other Dbl family proteins, one possible role of the Vav2 PH domain may be to bind products of PI3K that in turn facilitate Vav2 activation (18). To determine if PI3K products are regulators of Vav2 activity, we tested whether coexpression of activated PI3K would cooperate with Vav2-DPC to cause enhanced signaling and focus-forming activity. NIH 3T3 cells were transfected with the expression vector encoding Vav2-DPC, either alone or with an expression vector encoding a plasma membrane-targeted and constitutively activated variant of the p110 catalytic subunit of PI3K (p110-CAAX) (33). Expression of p110-CAAX alone did not result in any focus formation, whereas Vav2-DPC alone caused the appearance of 67 foci per dish; coexpression of p110-CAAX caused a twofold synergistic enhancement of this activity (Fig. 8A). Activated PI3K alone did not activate SRF and also caused a similar twofold enhancement of Vav2-DPC stimulation of SRF (Fig. 8B). These results suggest that products of PI3K appear to stimulate Vav2-DPC activity in vivo. Finally, to verify that p110-CAAX was active, we monitored the phosphorylation state of a PI3K target, Akt/protein kinase B, using phosphospecific Akt antibodies (Fig. 8C). Expression of p110-CAAX caused an increase in Akt phosphorylation. Interestingly, Vav2-DPC also caused Akt phosphorylation, which was enhanced by coexpression of p110-CAAX.
FIG. 8.
Coexpression of activated PI3K causes enhancement of the transforming and signaling activities of Vav2 DPC but has minimal effect on the Vav2 PH or CRD mutants. Assays were performed as described in the legends for Fig. 2 and 5 except that 500 ng of the pZIP-NeoSV(x)1 empty vector (black bars) or pZIP-NeoSV(x)1 encoding activated PI3K (p110-CAAX) (open bars) was cotransfected with 100 ng of pCGN-hygro plasmid DNA encoding the indicated Vav2 protein. (A) NIH 3T3 cells were cotransfected with control plasmid pCGN-hygro (vector) encoding the indicated Vav2 PH domain mutant proteins (Fig. 1; 100 ng per 60-mm-diameter dish), together with pZIP-NeoSV(x)1 (vector) or pZIP-p110-CAAX (500 ng per 60-mm-diameter dish). Values are the averages ± standard errors for two dishes and are representative of three independent assays. WT, wild type. (B) NIH 3T3 cells were transiently transfected with the above plasmids along with a luciferase reporter construct for SRF transcriptional activity. Data are representative of at least three independent assays performed on duplicate plates. (C) The expression of p110-CAAX in the SRF reporter assay (B) was analyzed by Western blotting 20 μg of total cell lysates with an anti-Akt and an anti-phospho-Akt (P-Akt) antibody.
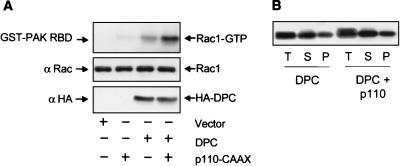
The ability of PI3K activation to cooperate with Vav2-DPC may result from the cooperation of two independent signaling pathways or, alternatively, may be due to direct enhancement of Vav2-DPC catalytic function. We found that coexpression of p110-CAAX with Vav2 caused an enhanced formation of GTP-bound Rac1, suggesting that PI3K activation may result in enhanced Vav2 DH domain catalytic activity in vivo (Fig. 9A). We then determined whether the enhanced exchange activity was facilitated by the increased association of Vav2-DPC with membranes. However, fractionation analyses showed that coexpression of p110-CAAX did not increase the fraction of Vav2-DPC associated with the membrane fraction (Fig. 9B). Thus, PI3K stimulation of Vav2 DH domain activity may be due to modulation of the intrinsic catalytic function of the DH domain.
FIG. 9.
Coexpression of activated PI3K and Vav2-DPC caused a synergistic increase in the level of Rac1-GTP but did not increase Vav2-DPC membrane association. (A) NIH 3T3 cells stably expressing Vav2-DPC, p110CAAX, or both were cultured for 24 h in low serum (0.1%) and lysed, and the lysates were used in GST pull-down assays using GST-PAK Rac binding domain (RBD) immobilized on glutathione agarose beads. Bound proteins and total cell lysates were analyzed by Western blotting with anti-Rac1 antibodies. The expression of Vav2-DPC was analyzed by Western blotting total cell lysates with an anti-HA antibody. The expression of p110-CAAX was analyzed by Western blotting total cell lysates with an anti-phospho-Akt antibody (data not shown). (B) PI3K activation does not enhance Vav2-DPC association with membranes. NIH 3T3 cells were cotransfected with the control plasmid pCGN (vector) encoding Vav2-DPC (1 μg per 60-mm-diameter dish), together with pZIP-NeoSV(x)1 (vector) or pZIP-p110CAAX (500 ng per 60-mm-diameter dish). Subcellular fractionation was performed as described for Fig. 4.
Finally, we determined if the ability of PI3K activity to modulate Vav2-DPC activity is dependent on the PH domain or the CRD. We found that coexpression of p110-CAAX did not enhance the signaling or transforming activity of either PH domain mutant, indicating that an intact PH domain was important for PI3K to modulate Vav2 function (Fig. 8A and B). The inability of PI3K to stimulate the activity of the K407A mutant is also consistent with previous studies demonstrating the importance of this residue in the PH domain of Vav for binding to PIP3 in vitro (18). In contrast, we found that the transforming and signaling activities of the K533A, K538A, and K563A mutants retained responsiveness to activated PI3K to a limited degree (Fig. 7A and B). Thus, PI3K stimulation of Vav2-DPC appears to be dependent on an intact PH domain but not CRD.
DISCUSSION
The tandem DH/PH domain structure of all Dbl family proteins suggests that the PH domain is a critical regulator of DH domain catalytic activity (8, 39). Consistent with such a role, the PH domain has been found to be a critical positive regulator of DH domain function for the majority of Dbl family proteins. In contrast, the PH domains of Vav and Vav3 were found to be dispensable for DH domain activity. Instead, a CRD adjacent to the PH domain was demonstrated to be essential for Vav DH domain function in vitro and in vivo (11, 28), suggesting that the CRD may substitute for PH domain function. In the present study, we evaluated the role of the PH domain and CRD in regulating the function of the DH domain of Vav2. Unexpectedly, we found that the PH domain was critical for Vav2 signaling and transformation. An intact PH domain was dispensable for the intrinsic catalytic activity of the DH domain in vitro and, instead, facilitated Vav2 membrane association. We found that missense mutations in the CRD abolished or impaired Vav2 transforming and signaling activity. We also found that all mutations of the CRD impaired catalytic activity in vitro. However, the lack of a direct correlation between impaired transforming activity and impaired catalytic activity in vitro argues that the CRD must facilitate DH domain function by other means, including regulation of membrane association. Finally, we found that PI3K activation synergistically enhanced Vav2 transforming and signaling activity by stimulating a PH domain-dependent increase in the intrinsic catalytic activity of the DH domain that did not involve promotion of membrane association. We conclude that the PH domain and CRD serve distinct but positive regulatory roles in Vav2 DH domain function.
Previous mutagenesis studies evaluated the importance of the PH domain in Vav and Vav3 function and concluded that the PH domain served either a negative regulatory, or dispensable, role in Vav DH domain function. For example, Ma et al. found that deletion of the PH domain in full-length Vav resulted in a constitutively activated variant (26). Movilla and Bustelo observed that mutation of the invariant tryptophan residue in the PH domain of Vav3, in the context of the isolated DH/PH/CRD fragment, did not alter exchange activity in vitro or biological activity in NIH 3T3 cells (28). Thus, we were surprised to find that introduction of missense mutations in the PH domain impaired or abolished Vav2-DPC signaling and transforming activity. Why do our observations differ from those of other studies? One possibility is that the PH domain may have functions in Vav2 distinct from those in Vav and Vav3. The PH domain is one of the less conserved domains among Vav family proteins (6). However, we have also found that mutation of the PH domain of Vav, in the context of the DH/PH/CRD fragment, also abolishes signaling and transforming activity (T. Palmby, K. Abe, and C. J. Der, submitted for publication). Therefore, in our analyses, the PH domain has equivalent and necessary roles in the functions of two Vav family proteins. Thus, at present, we do not have a straightforward explanation for the different observations. Nevertheless, our conclusion that the PH domain serves a positive regulatory role in Vav2 DH domain function is similar to observations made for other Dbl family proteins. Finally, we should also stress that our studies evaluated PH domain function in a truncated, constitutively activated Vav2 protein variant. Whether the PH domain also serves a positive regulatory role in the full-length protein remains unresolved.
Structure-function analyses of other Dbl family proteins established two possible roles for the PH domain in regulating DH domain function. First, a comparison of the catalytic activities of the isolated DH and DH/PH domains of several Dbl family proteins (Dbl, Trio, and Dbs) found that the presence of the PH domain caused up to a 100-fold-enhanced guanine nucleotide exchange activity in vitro (24, 34, 44). However, we found that the K407A mutation in the PH domain did not significantly perturb catalytic activity in vitro. While we were unable to evaluate this issue with the more drastic W503L mutation, which is expected to completely perturb PH domain structure, Movilla and Bustelo found that the equivalent mutation in the PH domain of Vav3 also did not impair catalytic activity in vitro (28). Thus, in contrast to results of observations for other Dbl family proteins (24, 34, 44), the PH domain does not greatly influence the intrinsic catalytic activity of the DH domain. Finally, the PH domains of some Dbl family proteins were observed to be critical for protein function yet could be replaced by a plasma membrane-targeting sequence (40, 41). Our observation that the two PH domain mutations greatly impaired Vav2-DPC association with the membrane fraction is consistent with a role for the PH domain in promoting DH domain interaction and activation of its membrane-associated GTPase substrates. In support for such a role, we found that addition of a membrane-targeting sequence partially restored the transforming activity of the K407A mutant version of Vav (Palmby et al., submitted). However, addition of a membrane-targeting sequence to the more drastic W503L version of Vav did not restore the ability of this mutant Vav to transform NIH 3T3 cells or activate SRF. Thus, the PH domains of Vav proteins must serve a function independent of membrane targeting.
Previous studies showed that deletion of the CRD or introduction of missense mutations that disrupt the structural integrity of this domain eliminated the transforming activity of Vav and Vav3 in vivo and in vitro (11, 28). Consistent with these studies, we have shown that truncation of the CRD also completely eliminates the biological activity of the Vav2-DPC cassette. While these analyses clearly demonstrate the critical role of the CRD for the function of all Vav family proteins, they do not provide much insight into what that role may be.
Similar to the CRD of the Raf-1 serine/threonine kinase, the CRDs of Vav family proteins are atypical in that they do not bind diacylglycerol or phorbol esters and instead may bind small GTPases and other membrane lipids. However, we and others have shown that the Raf-1 CRD interacts with multiple, distinct ligands that regulate Raf-1 kinase activity (4, 13, 15, 16, 25, 42). Therefore, we further investigated the functional contributions of the Vav2 CRD by introducing specific missense mutations at surface-exposed residues not predicted to disrupt the global fold of the CRD. These mutations were based on our previously determined nuclear magnetic resonance solution structure of the CRD of Raf-1 and on mutagenesis analyses of the CRD of PKC and Raf-1 (27, 32, 36). These mutants all decreased Vav2 transforming and signaling activity to various degrees. Interestingly, the impairment in catalytic exchange activity for these mutants did not correlate directly with the impairment in biological activity. In particular, the K533A mutant was significantly reduced in its catalytic activity toward Rac, Cdc42, and RhoA in vitro, while the K538A and V568E mutants showed decreased activity toward RhoA alone. Despite this difference, all three showed comparable, modest reductions in transforming activity. Thus, while our results indicate that the Vav2 CRD is involved in mediating DH domain catalytic activity, by analogy to the CRD of Raf-1, we also suggest that the Vav2 CRD may also facilitate interaction with other molecules that can influence DH domain function in vivo.
Movilla and Bustelo observed that the CRD of Vav3 could bind to its GTPase substrates in vitro and suggested that the CRD may facilitate DH domain interaction with its substrates (28). Our observation that mutation of the CRD impaired Vav2 catalytic activities in vitro is also consistent with this observation. Therefore, experiments to determine if the Vav2 CRD mutants are deficient in substrate binding would help elucidate the exact residues that may be important for specific GTPase recognition or other protein ligands that are required for complete catalytic activation.
Another possible role for the CRD of Vav2 may be to serve as a lipid binding, membrane-targeting sequence. The tandem CRDs from various PKC isoforms have been shown to promote translocation to the plasma membrane upon interaction with diacylglycerol and phorbol ester (19). Although the CRD of Vav2 does not bind phorbol esters, it may function to target Vav2 to the plasma membrane by interacting with membrane-associated phospholipids or other protein ligands. However, specific mutations in the CRD (K533A, K538A, and V568E) that impaired Vav2 function did not cause a significant alteration in membrane association. Furthermore, we found that the loss of function due to mutation of the CRD of Vav could not be corrected by addition of a plasma membrane-targeting sequence (Palmby et al., submitted). Thus, it seems unlikely that the CRD facilitates the membrane association of Vav or Vav2. Studies to determine if the CRD mutants can interact with the 14-3-3 adapter protein or with other cellular proteins would help determine whether the CRD serves as a protein interaction domain, analogous to the Raf-1 kinase.
Finally, previous studies suggest that the products of PI3K, such as PIP3, can associate with the PH domains of Vav and Sos to promote the activation of these Dbl family proteins in vitro and in vivo (18, 29). Our in vitro exchange analyses showed that the K407A PH domain mutant, which is expected to be impaired in phosphoinositide binding, retained strong catalytic activity. Thus, we anticipated that products of PI3K would not enhance Vav2 activity in vivo. However, we found that PI3K activation synergistically stimulated Vav2 transforming and signaling activity. Since expression of PI3K with Vav2 resulted in enhanced formation of GTP-bound Rac1, this indicates that PI3K products may enhance the intrinsic catalytic activity of the DH domain in vivo. Thus, our results are consistent with the observations of Han and colleagues, who propose that PIP3 association with the PH domain enhances the intrinsic catalytic activity of the DH domain of Vav (18).
Interestingly, the PI3K modulation of biological activity was independent of membrane association, as our fractionation analyses showed no difference in subcellular localization of the Vav2-DPC fragment in the presence of a constitutively activated PI3K. According to previous studies (21), in order to promote a PI3K-mediated membrane targeting event, PI3K products must exhibit high-affinity binding and have at least a 25-fold-higher affinity for PIP3 than for PIP2 for a PH domain to function as a PI3K-stimulated membrane-targeting domain, such as the PH domain of Akt. Therefore, our observation that PI3K activation enhanced Vav2 activity in vivo independent of promoting plasma membrane association was not surprising. Our observations argue that the PH domain will only facilitate membrane association by cooperating with other membrane-targeting domains. This mechanism is illustrated by findings that efficient recruitment of the β-adrenergic receptor kinase (βARK) to membranes requires the cooperative binding of the COOH terminus of βARK to Gβγ subunits and of the PH domain of βARK to PIP2 (31). Additionally, we found that, unlike the PH domain of the Akt serine/threonine kinase, the isolated PH domain of Vav did not show PI3K activation-mediated association with the plasma membrane (T. Palmby and C. J. Der, unpublished data). Thus, Vav2 catalytic activity and function, but not subcellular location, appear to be positively modulated by activation of PI3K, and this enhanced activity requires intact CRD and PH domains.
In summary, our observations show that, unlike other Dbl family members, Vav2 requires both a PH domain and a CRD in addition to its catalytic DH domain to transform NIH 3T3 cells. Our studies further demonstrate that PI3K lipid second messengers synergistically enhance Vav2 transforming and signaling activity in the context of Vav2-DPC but have no effect on polypeptides harboring missense mutations in either the PH domain or the CRD. Finally, these analyses suggest the both the PH domain and the CRD contribute a second essential function independent of membrane association in order to facilitate guanine exchange activity of the DH domain and that this function may encompass binding to other protein or lipid ligands.
Acknowledgments
We thank Kent Rossman, John Sondek, and Ernesto Fuentes for reagents and help in purifying recombinant Vav2 proteins; Heena Mehta and Staeci Morita for technical support; Misha Rand for preparation of figures; Brenda Temple and the Structural BioInformatics Core Facility for sequence alignments; and David Siderovski for critical appraisal of the manuscript.
Our research was supported by grants from the National Institutes of Health (NIH) to C.J.D. (CA55008 and CA63071) and S.L.C. (CA84480-01A1), and M.A.B. was supported by an NIH training grant fellowship and by a Leukemia & Lymphoma Society postdoctoral fellowship.
REFERENCES
- 1.Abe, K., K. L. Rossman, B. Liu, K. D. Ritola, D. Chiang, S. L. Campbell, K. Burridge, and C. J. Der. 2000. Vav2 is an activator of Cdc42, Rac1, and RhoA. J. Biol. Chem. 275:10141-10149. [DOI] [PubMed] [Google Scholar]
- 2.Abe, K., I. P. Whitehead, J. P. O'Bryan, and C. J. Der. 1999. Involvement of N-terminal sequences in the negative regulation of vav signaling and transforming activity. J. Biol. Chem. 274:30410-30418. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, A. L., and A. Hall. 2000. Rho GTPases and their effector proteins. Biochem. J. 348:241-255. [PMC free article] [PubMed] [Google Scholar]
- 4.Brtva, T. R., J. K. Drugan, S. Ghosh, R. S. Terrell, S. Campbell-Burk, R. M. Bell, and C. J. Der. 1995. Two distinct Raf domains mediate interaction with Ras. J. Biol. Chem. 270:9809-9812. [DOI] [PubMed] [Google Scholar]
- 5.Buss, J. E., P. A. Solski, J. P. Schaeffer, M. J. MacDonald, and C. J. Der. 1989. Activation of the cellular proto-oncogene product p21Ras by addition of a myristylation signal. Science 243:1600-1603. [DOI] [PubMed] [Google Scholar]
- 6.Bustelo, X. R. 1996. The VAV family of signal transduction molecules. Crit. Rev. Oncog. 7:65-88. [DOI] [PubMed] [Google Scholar]
- 7.Bustelo, X. R. 2000. Regulatory and signaling properties of the Vav family. Mol. Cell. Biol. 20:1461-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerione, R. A., and Y. Zheng. 1996. The Dbl family of oncogenes. Curr. Opin. Cell Biol. 8:216-222. [DOI] [PubMed] [Google Scholar]
- 9.Clark, G. J., A. D. Cox, S. M. Graham, and C. J. Der. 1995. Biological assays for Ras transformation. Methods Enzymol. 255:395-412. [DOI] [PubMed] [Google Scholar]
- 10.Clark, G. J., J. K. Drugan, K. L. Rossman, J. W. Carpenter, K. Rogers-Graham, H. Fu, C. J. Der, and S. L. Campbell. 1997. 14-3-3 zeta negatively regulates Raf-1 activity by interactions with the Raf-1 cysteine-rich domain. J. Biol. Chem. 272:20990-20993. [DOI] [PubMed] [Google Scholar]
- 11.Coppola, J., S. Bryant, T. Koda, D. Conway, and M. Barbacid. 1991. Mechanism of activation of the vav protooncogene. Cell Growth Differ. 2:95-105. [PubMed] [Google Scholar]
- 12.Crespo, P., K. E. Schuebel, A. A. Ostrom, J. S. Gutkind, and X. R. Bustelo. 1997. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature 385:169-172. [DOI] [PubMed] [Google Scholar]
- 13.Drugan, J. K., R. Khosravi-Far, M. A. White, C. J. Der, Y.-J. Sung, Y.-W. Huang, and S. L. Campbell. 1996. Ras interaction with two distinct binding domains in Raf-1 may be required for Ras transformation. J. Biol. Chem. 271:233-237. [DOI] [PubMed] [Google Scholar]
- 14.Fiordalisi, J. J., A. Ulku, R. L. Johnson, C. J. Der, and A. D. Cox. 2000. Mammalian expression vectors for ras family proteins: generation and use of expression constructs to analyze Ras family function. Methods Enzymol. 332:3-36. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh, S., and R. M. Bell. 1994. Identification of discrete segments of human Raf-1 kinase critical for high affinity binding to Ha-Ras. J. Biol. Chem. 269:30785-30788. [PubMed] [Google Scholar]
- 16.Ghosh, S., W. Q. Xie, A. F. G. Quest, G. M. Mabrouk, J. C. Strum, and R. M. Bell. 1994. The cysteine-rich region of Raf-1 kinase contains zinc, translocates to liposomes, and is adjacent to a segment that binds GTP-Ras. J. Biol. Chem. 269:10000-10007. [PubMed] [Google Scholar]
- 17.Han, J., B. Das, W. Wei, L. Van Aelst, R. D. Mosteller, R. Khosravi-Far, J. K. Westwick, C. J. Der, and D. Broek. 1997. Lck regulates Vav activation of members of the Rho family of GTPases. Mol. Cell. Biol. 17:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han, J., K. Luby-Phelps, B. Das, X. Shu, Y. Xia, R. D. Mosteller, U. M. Krishna, J. R. Falck, M. A. White, and D. Broek. 1998. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science 279:558-560. [DOI] [PubMed] [Google Scholar]
- 19.Hurley, J. H., and S. Misra. 2000. Signaling and subcellular targeting by membrane-binding domains. Annu. Rev. Biophys. Biomol. Struct. 29:49-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katzav, S., J. L. Cleveland, H. E. Heslop, and D. Pulido. 1991. Loss of the amino-terminal helix-loop-helix domain of the vav proto-oncogene activates its transforming potential. Mol. Cell. Biol. 11:1912-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemmon, M. A., and K. M. Ferguson. 2000. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 350:1-18. [PMC free article] [PubMed] [Google Scholar]
- 22.Lemmon, M. A., K. M. Ferguson, and J. Schlessinger. 1996. PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell 85:621-624. [DOI] [PubMed] [Google Scholar]
- 23.Liu, B. P., and K. Burridge. 2000. Vav2 activates Rac1, Cdc42, and RhoA downstream from growth factor receptors but not β1 integrins. Mol. Cell. Biol. 20:7160-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, X., H. Wang, M. Eberstadt, A. Schnuchel, E. T. Olejniczak, R. P. Meadows, J. M. Schkeryantz, D. A. Janowick, J. E. Harlan, E. A. Harris, D. E. Staunton, and S. W. Fesik. 1998. NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell 95:269-277. [DOI] [PubMed] [Google Scholar]
- 25.Luo, Z., B. Diaz, M. S. Marshall, and J. Avruch. 1997. An intact Raf zinc finger is required for optimal binding to processed Ras and for Ras-dependent Raf activation in situ. Mol. Cell. Biol. 17:46-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma, A. D., A. Metjian, S. Bagrodia, S. Taylor, and C. S. Abrams. 1998. Cytoskeletal reorganization by G protein-coupled receptors is dependent on phosphoinositide 3-kinase γ, a Rac guanosine exchange factor, and Rac. Mol. Cell Biol. 18:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mott, H. R., J. W. Carpenter, S. Zhong, S. Ghosh, R. M. Bell, and S. L. Campbell. 1996. The solution structure of the Raf-1 cysteine-rich domain: a novel Ras and phospholipid binding site. Proc. Natl. Acad. Sci. USA 93:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Movilla, N., and X. R. Bustelo. 1999. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol. Cell. Biol. 19:7870-7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nimnual, A. S., B. A. Yatsula, and D. Bar-Sagi. 1998. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science 279:560-563. [DOI] [PubMed] [Google Scholar]
- 30.Olson, M. F., P. Sterpetti, K.-I. Nagata, D. Toksoz, and A. Hall. 1997. Distinct roles for DH and PH domains in the Lbc oncogene. Oncogene 15:2827-2831. [DOI] [PubMed] [Google Scholar]
- 31.Pitcher, J. A., K. Touhara, E. S. Payne, and R. J. Lefkowitz. 1995. Pleckstrin homology domain-mediated membrane association and activation of the β-adrenergic receptor tyrosine kinase requires coordinate interaction with Gβy subunits and lipid. J. Biol. Chem. 270:11707-11710. [DOI] [PubMed] [Google Scholar]
- 32.Quest, A. F. G., E. S. G. Bardes, and R. M. Bell. 1994. A phorbol ester binding domain of protein kinase Cζ. J. Biol. Chem. 269:2961-2970. [PubMed] [Google Scholar]
- 33.Rodriguez-Viciana, P., P. H. Warne, A. Khwaja, B. M. Marte, D. Pappin, P. Das, M. D. Waterfield, A. Ridley, and J. Downward. 1997. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89:457-467. [DOI] [PubMed] [Google Scholar]
- 34.Rossman, K. L., and S. L. Campbell. 2000. Bacterial expressed DH and DH/PH domains. Methods Enzymol. 325:25-38. [DOI] [PubMed] [Google Scholar]
- 35.Schuebel, K. E., N. Movilla, J. L. Rosa, and X. R. Bustelo. 1998. Phosphorylation-dependent and constitutive activation of Rho proteins by wild-type and oncogenic Vav-2. EMBO J. 17:6608-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharkey, N. A., K. L. Leach, and P. M. Blumberg. 1984. Competitive inhibition by diacylglycerol of specific phorbol ester binding. Proc. Natl. Acad. Sci. USA 81:607-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Aelst, L., and C. D'Souza-Schorey. 1997. Rho GTPases and signaling networks. Genes Dev. 11:2295-2322. [DOI] [PubMed] [Google Scholar]
- 38.Westwick, J. K., Q. T. Lambert, G. J. Clark, M. Symons, L. Van Aelst, R. G. Pestell, and C. J. Der. 1997. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol. Cell. Biol. 17:1324-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitehead, I. P., S. Campbell, K. L. Rossman, and C. J. Der. 1997. Dbl family proteins. Biochim. Biophys. Acta 1332:F1-F23. [DOI] [PubMed] [Google Scholar]
- 40.Whitehead, I. P., H. Kirk, and R. Kay. 1995. Expression cloning of oncogenes by retroviral transfer of cDNA libraries. Mol. Cell. Biol. 15:704-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitehead, I. P., Q. T. Lambert, J. A. Glaven, K. Abe, K. L. Rossman, G. M. Mahon, J. M. Trzaskos, R. Kay, S. L. Campbell, and C. J. Der. 1999. Dependence of Dbl and Dbs transformation on MEK and NF-KB activation. Mol. Cell. Biol. 19:7759-7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams, J. G., J. K. Drugan, G. S. Yi, G. J. Clark, C. J. Der, and S. L. Campbell. 2000. Elucidation of binding determinants and functional consequences of Ras/Raf-cysteine-rich domain interactions. J. Biol. Chem. 275:22172-22179. [DOI] [PubMed] [Google Scholar]
- 43.Zeng, L., P. Sachdev, L. Yan, J. L. Chan, T. Trenkle, M. McClelland, J. Welsh, and L. H. Wang. 2000. Vav3 mediates receptor protein tyrosine kinase signaling, regulates GTPase activity, modulates cell morphology, and induces cell transformation. Mol. Cell. Biol. 20:9212-9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng, Y., M. J. Hart, and R. A. Cerione. 1995. Guanine nucleotide exchange catalyzed by dbl oncogene product. Methods Enzymol. 256:77-84. [DOI] [PubMed] [Google Scholar]
- 45.Zohn, I. M., S. L. Campbell, R. Khosravi-Far, K. L. Rossman, and C. J. Der. 1998. Rho family proteins and Ras transformation: the RHOad less traveled gets congested. Oncogene 17:1415-1438. [DOI] [PubMed] [Google Scholar]