Abstract
Cell morphogenesis requires dynamic reorganization of the actin cytoskeleton, a process that is tightly regulated by the Rho family of small GTPases. These GTPases act as molecular switches by shuttling between their inactive GDP-bound and active GTP-bound forms. Here we show that Nir2, a novel protein related to Drosophila retinal degeneration B (RdgB), markedly affects cell morphology through a novel Rho-inhibitory domain (Rid) which resides in its N-terminal region. Rid exhibits sequence homology with the Rho-binding site of formin-homology (FH) proteins and leads to an apparent loss of F-actin staining when ectopically expressed in mammalian cells. We also show that Rid inhibits Rho-mediated stress fiber formation and lysophosphatidic acid-induced RhoA activation. Biochemical studies demonstrated that Nir2, via Rid, preferentially binds to the inactive GDP-bound form of the small GTPase Rho. Microinjection of antibodies against Nir2 into neuronal cells markedly attenuates neurite extension, whereas overexpression of Nir2 in these cells attenuates Rho-mediated neurite retraction. These results implicate Nir2 as a novel regulator of the small GTPase Rho in actin cytoskeleton reorganization and cell morphogenesis.
Cell morphogenesis is highly dependent on fine coordination among membrane biosynthesis, intracellular transport, and cytoskeletal organization (4, 50). Small GTPases of the Rho family are essential mediators of a variety of morphogenetic events required for normal development of multicellular organisms (39). Their role in cell morphogenesis is strongly related to their ability to reorganize the actin cytoskeleton (16). The members of this family of proteins, which includes Rho, Rac, and Cdc42, elicit their cellular function by shuttling between their inactive GDP-bound and their active GTP-bound forms (42, 45). The activation state of Rho, like that of the other family members, is tightly regulated by numerous intracellular proteins that can be functionally divided into three groups: the GDIs (GDP-dissociation inhibitors), the GAPs (GTPase-activating proteins), and the GEFs (GDP/GTP exchange factors). These upstream regulators negatively or positively regulate Rho activity, while Rho downstream effectors mediate Rho signaling to elicit Rho-regulated cellular functions (6, 7). Over the last few years, several Rho effector proteins have been identified (5). Among them, PKN, rhophilin, and rhotekin possess a conserved Rho-binding motif (35), while a second Rho-binding motif has been recently identified in ROCK-I and ROKα (15). A Rho-binding site has also been mapped to the N-terminal portion of three formin-homology (FH) proteins. Although no consensus Rho-binding site has been identified among these family members, their N-terminal portion contains a conserved motif that falls within the Rho-interaction domain defined elsewhere for Bnr1p and mDia1 (47).
We recently isolated a novel family of human genes designated Nirs (Nir1, Nir2, and Nir3), which are the mammalian homologues of the Drosophila retinal degeneration B (rdgB) gene (26). Nirs belong to a growing family of highly conserved proteins that share structural features. Genes encoding Nir-related proteins have been recently identified in worms, mammals, flies, and fish (1, 10, 12, 26, 29, 46). They possess several putative functional domains, including an N-terminal phosphatidylinositol (PI) transfer domain that specifically transfers PI and phosphatidylcholine (32), an acidic region that binds calcium, six hydrophobic stretches, and a C-terminal PYK2-binding domain (26). Although the Nir family members are highly expressed in the retina and may be involved in inherited human retinal degeneration diseases (1, 10, 26), their cellular functions, as well as those of their Drosophila melanogaster homologue RdgB, remain largely unknown.
Nir2 is expressed in various tissues and cell types, particularly neuronal, epithelial, and hematopoietic cells (26). In the present study we investigated the role of Nir2 in cell morphogenesis. We show that Nir2 markedly affects cell morphology through a novel Rho-inhibitory domain (Rid) which resides in its N-terminal region. Rid exhibits sequence homology with the Rho-binding site of FH proteins and leads to an apparent loss of F-actin staining when ectopically expressed in mammalian cells. We also show that Rid inhibits Rho-mediated stress fiber formation and lysophosphatidic acid (LPA)-induced Rho activation. Biochemical studies demonstrate that Rid exclusively binds to the dominant negative mutants of Rho and Rac, suggesting that its effect on the actin cytoskeleton is mediated by binding to the inactive form of Rho. An in vitro binding assay using full-length Nir2 confirmed these results and demonstrated that Nir2 preferentially binds to the inactive Rho-GDP. Microinjection of antibodies against Nir2 into neuronal cells markedly attenuates neurite extension, whereas overexpression of Nir2 in these cells attenuates Rho-mediated neurite retraction. Furthermore, we show that the entire N-terminal domain retains its binding specificity for the dominant negative mutants of Rho and Rac but has no obvious effect on cell or F-actin morphology. Thus, the PI transfer domain somehow suppresses the negative effect of Rid on the actin cytoskeleton. These results delineate the molecular basis underlying the role of Nir2 in cell morphogenesis and introduce Nir2 as a novel regulator of the small GTPase Rho.
MATERIALS AND METHODS
Plasmids, expression vectors, and antibodies.
The ΔPI Nir2 construct was made by replacing an EcoRI-BglII DNA fragment of the full-length Nir2 with a PCR product amplified by the sense and antisense oligonucleotide primers 5′-GGCGAATTCAAGGAGGTACGACCATGGCTGACATCCGGGCAC-3′and 5′-TCGGCCGCGTGGAAGAGGTTGTAGATCT-3′, respectively. The same strategy was applied to construct the ΔN1 and ΔN2 truncated mutants. The sense oligonucleotide primer used in each PCR was 5′-GCGAATTCAAGGAGGTACGACCATGTCTGAGAACAGCTCCGAG-3′ for the ΔN1 mutant and 5′-GCGAATTCAAGGAGGTACGACCATGGACTCAGGCCCTGGAGAC-3′ for the ΔN2 mutant. The antisense oligonucleotide primer was identical to that described for the ΔPI construct. The ΔPB truncated mutant was constructed by deleting a PvuII DNA fragment from the C-terminal region of Nir2. A hemagglutinin (HA) epitope tag (YPYDVPDYAS) was fused in frame to the C-terminal ends of these truncated mutants. PCR was used to amplify the N-terminal region (amino acids [aa] 2 to 424) of Nir2, Rid (aa 205 to 424), RidL (aa 241 to 424), and RidS (aa 205 to 244). The amplified DNA fragments were subcloned in frame into pCMV-Neo expression vector containing either an N-terminal Myc or an HA epitope tag. The same PCR products were subcloned in frame into pGEX-2T to generate a fusion protein with glutathione S-transferase (GST). The Nir2 constructs were verified by restriction enzyme analysis and sequencing. Complete cloning details are available upon request. Additional constructs used in this study were pGEX-RhoA, as well as pXJ40-HA-V14-RhoA, pXJ40-HA-V12-Cdc42, and pXJ40-HA-V12-Rac, kindly provided by E. Manser (Glaxo-IMCB Laboratory, Institute of Molecular and Cell Biology, Singapore). All other Cdc42, Rac, RhoA, and C3 exoenzyme constructs were generous gifts from A. Hall (MRC Laboratory for Molecular Cell Biology, University College, London, United Kingdom). Anti-Nir2 antibody was raised in rabbits against peptide derived from the Nir2 N-terminal region (aa 287 to 300, TPDGPEAPPGPDAS). The antibody was affinity purified by standard procedures. The specificity of these antibodies for Nir2 was confirmed by immunoprecipitation and immunoblotting analysis. Anti-Myc monoclonal antibody, anti-HA polyclonal antibody, and anti-RhoA monoclonal antibody were obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.), and anti-HA monoclonal antibody was purchased from Boehringer Mannheim Biotechnology.
Cell culture, transfection, and fluorescence microscopy.
NIH 3T3, COS-7, HEK 293, TE671, and HeLa cells from the American Type Culture Collection were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 mg/ml). Transfection of HeLa cells was carried out using Lipofectamine according to the manufacturer's standard protocol (GIBCO/BRL); HEK 293 cells were transfected using the calcium phosphate method as previously described (28). HeLa cells grown on glass coverslips were transfected as indicated and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature. The fixed cells were permeabilized with 0.1% (vol/vol) Triton X-100 in PBS for 5 min and then incubated for 30 min in blocking buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 2% bovine serum albumin, 1% glycine, 10% goat serum, and 0.1% Triton X-100). Various primary antibodies were added for 1 h at room temperature, and after three washes with PBS the cells were incubated with Oregon Green donkey anti-rabbit antibody (Molecular Probes), Cy3-conjugated goat anti-mouse immunoglobulin G (IgG), or both, as indicated. In some experiments, Alexa 488-conjugated donkey anti-mouse IgG (Molecular Probes) or Cy5-conjugated goat anti-rabbit IgG (Jackson Laboratories) was used. Tetramethyl-rhodamine isocyanate (TRITC)-phalloidin (Sigma) was used to visualize filamentous actin. Coverslips were mounted in Moviol 4-88 (Calbiochem) and analyzed by a Zeiss Axioplan fluorescence microscope equipped with a plan-Neofluor 100×, 1.3-numerical-aperture oil-immersion lens. Fluorescence images were obtained with a DVC-1310 digital camera (DVC Company, Inc.) and XCAP imaging software (EPIX, Inc.). Confocal images were collected by a Zeiss 510 confocal microscope equipped with filters for fluorescein and Cy3 epifluorescence. Both 488-nm and 543-nm laser lines were used for excitation, and 505- to 530-nm band-pass and 560-nm long-pass filters were used for emission. In some experiments (triplet staining; see Fig. 3) the specimens were analyzed by three laser channels for excitation, 633 nm (Cy5), 543 nm (TRITC), and 488 nm (Alexa 488), and 650-nm long-pass, 560- to 615-nm band-pass, and 505- to 530-nm band-pass filters were used for emission. Image analysis was performed using the standard system operating software provided with the Zeiss 510 microscope (version 2.01).
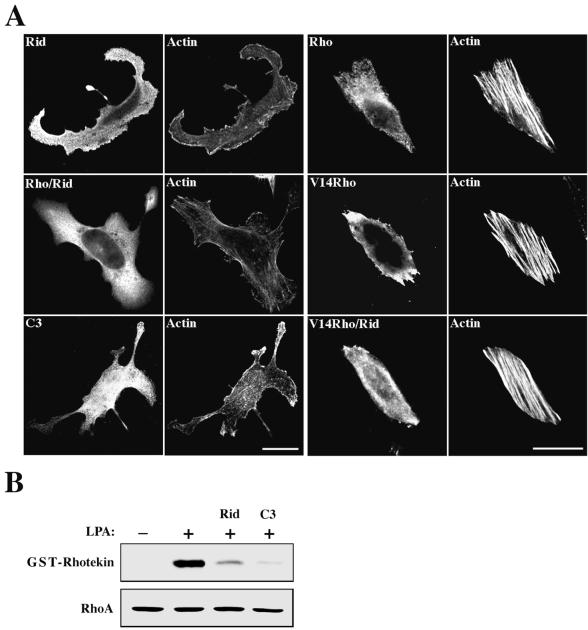
FIG. 3.
Rid inhibits Rho. (A) Rid inhibits Rho-mediated stress fiber formation. HeLa cells grown on coverslips were transiently transfected with an expression vector encoding either HA-Rid (aa 205 to 424), HA-C3 exoenzyme, Myc-tagged wild-type RhoA, or Myc-tagged Val14-Rho or were cotransfected with HA-Rid and Myc-RhoA or with HA-Rid and Myc-Val14-Rho as indicated. Cells were fixed 18 h later and immunostained with either anti-HA (Rid and C3 exoenzyme) or anti-Myc (RhoA and Val14-Rho) or costained with anti-Myc and anti-HA (Rid/Rho and Rid/Val14-Rho) antibodies. Actin morphology was visualized by TRITC-phalloidin staining. Cells coexpressing Rid and Rho were detected by triplet staining with Cy5-conjugated goat anti-rabbit IgG, Alexa 488-conjugated donkey anti-mouse IgG, and TRITC-phalloidin. Bars, 10 μm. (B) Rid inhibits LPA-induced RhoA activation. HeLa cells were transiently transfected with expression vectors encoding either Rid or C3 exoenzyme or were mock transfected. The cells were serum starved for 12 h and then stimulated with 10 μM LPA for 5 min. Cell lysates were incubated with GST-rhotekin immobilized on glutathione-agarose beads, and the bound Rho-GTP was analyzed by Western blotting with anti-RhoA antibodies. The level of RhoA expression in each sample was determined by Western analysis using anti-RhoA antibodies (lower panel).
Microinjection.
Microinjection of TE671 cells grown on coverslips was performed on a Zeiss Axiovert 135 microscope equipped with an Eppendorf microinjection system. Antibodies were dialyzed against PBS and concentrated to 1 mg/ml. Immediately after microinjection, coverslips were washed with PBS and incubated in growth medium for at least 1 h before treatment with dibutyryl cyclic AMP (dbcAMP) (1 mM) for the indicated periods of time.
GST fusion proteins and coimmunoprecipitation assay.
GST fusion proteins were expressed in bacteria and purified by standard procedures (Pharmacia Biotech). The purified GST fusion proteins immobilized on glutathione-agarose beads were incubated with cell lysate of HEK 293 cells as indicated. HEK 293 cells were typically transfected with the appropriate expression vector and 36 h later lysed in lysis buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM dithiothreitol (DTT), 5 mM MgCl2, 50 mM NaF, 1 mM EDTA, 1 mM Na3VO4, 10% glycerol, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, and 10 μg of aprotinin/ml. Cell lysates were incubated with the glutathione-agarose beads containing the appropriate fusion protein (1 to 3 μg) for 2 h at 4°C. The beads were washed three times with ice-cold PBS containing 0.1% Triton X-100 and 5 mM MgCl2. The beads were boiled in sodium dodecyl sulfate (SDS) sample buffer and separated by SDS-polyacrylamide gel electrophoresis (PAGE). Western blotting was performed essentially as described previously (27). For coimmunoprecipitation assays, cells were transfected and lysed as described above. Cell lysates were incubated with either polyclonal antibodies bound to Sepharose-protein A (Pharmacia) or monoclonal antibodies bound to anti-mouse IgG conjugated to agarose beads (Sigma), as indicated. The beads were washed, boiled in SDS sample buffer, and separated by SDS-PAGE as described above.
Affinity precipitation of Nir2 with GST-Rho.
Recombinant GST-RhoA fusion protein was loaded with GDP or GTPγS essentially as previously described (18) with the following modifications. Purified recombinant GST-RhoA fusion protein (∼1 μg/sample) immobilized on glutathione-agarose beads was incubated in buffer A (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1% Triton X-100, 1 mM DTT, 10 mM EDTA) for 1 h at room temperature to establish a guanine nucleotide-depleted state. The beads were then washed with loading buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1% Triton X-100, 1 mM DTT, 10 mM MgCl2) and resuspended in loading buffer containing either 100 μM GDP or 100 μM GTPγS. After 30 min of incubation at 30°C, the beads were incubated with cell lysate of HEK 293 cells expressing Nir2. After 2 h of incubation at 4°C, the beads were washed with PBS containing 0.1% Triton X-100 and 5 mM MgCl2, boiled in SDS sample buffer, and separated by SDS-PAGE as described above.
Overlay assay.
Affinity-purified recombinant GST, GST-rhotekin, and GST-Rid proteins (0.25 to 8 μg) were loaded on neutral nylon transfer membranes (Nytran, 0.45-μm pore size; Schleicher & Schuell) by applying the SRC 96 Minifold I dot blotter (Schleicher & Schuell) according to the manufacturer's instructions. The membranes were air dried for 45 min at room temperature and then incubated in blocking buffer (PBS containing 5% dry milk, 0.1% Triton X-100, and 10 mM MgCl2) for an additional 1 h. Purified recombinant GST-RhoA (10 μM) was preloaded with either 240 μM GTPγS or 240 μM GDPβS for 30 min at 30°C in PBS containing 2 mM DTT, 5 mM EDTA, and 30 μM bovine serum albumin. Nucleotide exchange was stopped by the addition of 10 mM MgCl2, and incubation was done for 10 min on ice (14). The nylon membranes were incubated for 1 h at 4°C with binding buffer containing PBS, 10 mM MgCl2, and 1 mg of nucleotide preloaded recombinant GST-Rho/ml. The membranes were washed with PBS containing 10 mM MgCl2 for 15 min at room temperature and incubated with anti-RhoA antibody, followed by anti-mouse IgG conjugated to horseradish peroxidase. Rho binding was detected by enhanced chemiluminescence (ECL) and autoradiography.
RESULTS
Nir2 truncated mutants differentially affect cell morphology.
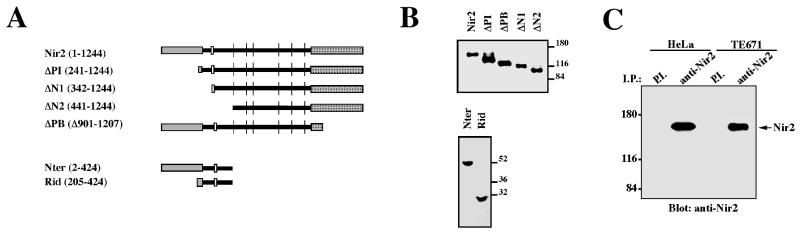
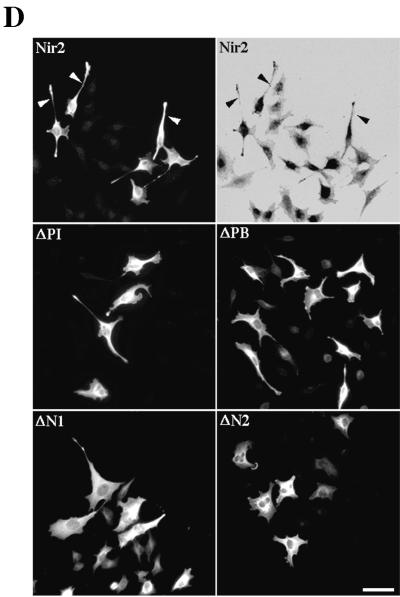
We observed that overexpression of Nir2 in a variety of mammalian cell lines (NIH 3T3, COS-7, and HeLa) leads to a profound change in cell morphology. To further characterize this observation, we constructed a set of Nir2 truncated mutants (Fig. 1A), in which different structural domains of Nir2 had been deleted. The truncated mutants were transiently expressed in HeLa cells, and Western blot analysis revealed that each mutant was expressed as a protein of the expected size (Fig. 1B). Indirect immunofluorescence analysis was carried out to assess the effect of these mutants on HeLa cell shape, as HeLa cells express relatively high levels of endogenous Nir2 (Fig. 1C). As shown in Fig. 1D, overexpression of wild-type Nir2, as well as of the ΔPI and ΔPB truncated mutants, markedly affected cell morphology. The cells exhibited an unusual extended phenotype that was distinct from the surrounding nontransfected cells. The long extensions or protrusions sometimes extended more than 100 μm from the cell body and were also observed in other cell types, such as NIH 3T3 fibroblasts (data not shown). Cellular extensions were also evident in ΔN1-expressing cells, but the effect was not as dramatic as that obtained for the wild type or for the ΔPI and ΔPB truncated Nir2 mutants. In contrast, expression of the ΔN2 truncated mutant had no pronounced effect on HeLa cell shape. The ratio between the long and short axes of ΔN2-expressing cells (1.7 ± 0.01, n = 50, P < 0.0001) was almost identical to that of the mock-transfected cells (1.61 ± 0.02, n = 50, P < 0.0001), whereas this ratio was significantly greater for wild-type Nir2 and ΔPI and ΔPB truncated mutants (Table 1). The extended phenotype of the Nir2 transfectants was reminiscent of that obtained upon overexpression of the p190 RhoGAP protein in NIH 3T3 fibroblasts (43). Moreover, it has been previously shown that inactivation of the small GTPase Rho is usually associated with the formation of protrusions at the cell periphery (19, 43) and that a constitutively active Rho induces a retracted cell shape (21). The small GTPase Rho is an important regulator of actin cytoskeleton organization and is required for stress fiber formation (33). Therefore, we examined the effect of wild-type Nir2 and its truncated mutants on actin stress fiber morphology by using TRITC-phalloidin staining. Overexpression of wild-type Nir2 or its truncated mutants had no obvious effect on stress fiber formation. The extended phenotype obtained coincided with long actin-based extensions, and in many cases, actin-based filopodium-like extensions were observed. Other cytoskeletal elements, such as microtubules, were not affected by overexpression of Nir2 or its truncated mutants (data not shown).
FIG. 1.
Nir2 truncated mutants differentially affect cell morphology. (A) Structure and expression of the wild-type Nir2 and its truncated mutants. Shown are the different Nir2 structural domains: the N-terminal PI transfer domain (gray), the acidic domain (white), the C-terminal PYK2-binding domain (patterned box), and the six hydrophobic stretches (vertical lines). Numbers denote amino acid positions. (B) An HA epitope tag was fused in frame to the C-terminal ends of the wild-type Nir2 and its truncated mutants. The different Nir2 truncated mutants were expressed as proteins of the expected sizes as determined by Western analysis using anti-HA antibodies. Numbers at right of the panels show molecular mass in kilodaltons. (C) Expression of endogenous Nir2 in HeLa and TE671 cells. Shown is a Western analysis of Nir2 immunoprecipitated (I.P.) from HeLa or TE671 cells. Preimmune serum (P.I.) was used as the control. Numbers at the left show molecular mass in kilodaltons. (D) Overexpression of Nir2 and its truncated mutants affects cell morphology. HeLa cells were transiently transfected with expression vectors encoding the different Nir2 truncated mutants as indicated. Cell morphology was determined by indirect immunofluorescence analysis using anti-HA antibodies. Shown are the different phenotypes of the transfected cells. Nir2 transfectants (arrowheads) exhibited an extended phenotype that was distinct from that of the surrounding nontransfected cells, as shown in the phase-contrast picture (top right panel). Bar, 50 μm.
TABLE 1.
Quantitative analysis of the ratio between the long and short axes of HeLa cells expressing Nir2 and its truncated mutantsa
| Cell type | Long axis (μm) | Short axis (μm) | Ratio (L/S) |
|---|---|---|---|
| Control | 46.75 ± 0.40 | 29.33 ± 0.31 | 1.61 ± 0.02 |
| ΔPI | 82.81 ± 0.49 | 30.30 ± 0.14 | 2.75 ± 0.02 |
| ΔPB | 83.49 ± 0.51 | 30.39 ± 0.14 | 2.88 ± 0.02 |
| ΔN1 | 61.43 ± 0.27 | 30.22 ± 0.07 | 2.02 ± 0.01 |
| ΔN2 | 48.35 ± 0.16 | 29.49 ± 0.14 | 1.70 ± 0.01 |
| Nir2 | 84.96 ± 0.51 | 29.07 ± 0.16 | 2.96 ± 0.02 |
Quantitative analysis of the ratio between the long and short axes of HeLa cells expressing Nir2 and its truncated mutants. HeLa cells were either mock transfected (control) or transfected with expression vectors encoding the different Nir2 truncated mutants as indicated. After 24 h, the cells were immunostained with anti-HA antibodies and analyzed by fluorescence microscopy. The lengths of the long and short axes of 30 to 50 cells expressing the indicated protein were measured by Zeiss 510 confocal microscope operating software (version 2.01). Shown are the means ± standard errors of the means of the long and short axes along with the calculated ratio between them (L/S).
The marked differences in cell morphology obtained by overexpression of the Nir2 truncated mutants suggested that the N-terminal domain of Nir2 is responsible for the induction of these changes. However, since the ΔPI transfectants exhibited an extended phenotype, we assume that most of the PI transfer domain is dispensable for this phenotype.
Nir2 exhibits homology to FH proteins.
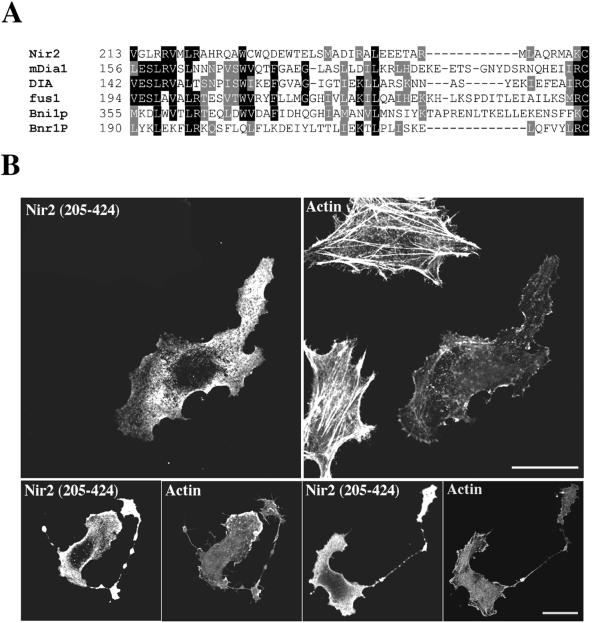
Inspection of the primary sequence of Nir2 revealed that the N-terminal region contains, in addition to the PI transfer domain, a region that shows homology to the N-terminal portion of FH proteins. FH proteins participate in a wide range of actin-mediated processes affecting cell polarity and shape and are implicated in the regulation of cytokinesis (47). Several proteins belonging to this family have been shown elsewhere to directly bind the small GTPase Rho (13, 24, 49). Multiple alignment analysis of Nir2 with several FH proteins revealed homology within the N-terminal domain of Nir2 at a region that partially overlaps with the end of the PI transfer domain (Fig. 2A). This region was found to align with a conserved motif that falls within the Rho-interaction domain of FH family members.
FIG. 2.
The N-terminal region of Nir2 contains a Rid. (A) Sequence alignment of Nir2 and the N-terminal region of several FH proteins. The different protein sequences used in this alignment are for Nir2 (Homo sapiens, GenBank accession no. AF334584), mDia1 (Mus musculus; GenBank accession no. U96963), DIA (D. melanogaster, GenBank accession no. U11288), fus1 (Schizosaccharomyces pombe, GenBank accession no. Z71547), Bni1p (Saccharomyces cerevisiae, GenBank accession no. Z71547), and Bnr1P (S. cerevisiae, GenBank accession no. Z47047). Identical residues among at least three of the aligned sequences are boxed in black, and residues representing conserved substitutions are boxed in gray. Numbers indicate the position of the first residue relative to the amino acid sequence of the indicated proteins. Sequences were aligned using the ClustalW algorithm. (B) Effect of Nir2 variant on the organization of the actin cytoskeleton. A short variant of Nir2 (aa 205 to 424) was expressed in HeLa cells as an HA-tagged fusion protein, and its effect on F-actin and cell morphology was analyzed by fluorescence microscopy. Shown are representative phenotypes of the transfected cells stained with anti-HA antibodies and the filamentous actin in the same field stained with TRITC-phalloidin. The unusual bead-like extensions are shown (lower panels). Bars, 10 μm.
Based on this homology and on the morphological changes induced by the different Nir2 truncated mutants, we constructed an epitope-tagged variant of Nir2 encompassing aa 205 to 424. This region partially overlaps with the end of the PI transfer domain and continues to the end of the N-terminal region (Fig. 1A). The effect of this variant on cell and F-actin morphology was assessed by immunofluorescence analysis. As shown in Fig. 2B, its overexpression in HeLa cells resulted in complete loss of F-actin staining. The cells exhibited an unusual morphology, and many of them sent out long, bead-like extensions. Likewise, HeLa cells expressing the C3 exoenzyme, which causes the inactivation of the small GTPase Rho (38), showed a loss of F-actin staining (Fig. 3A). Similar effects on F-actin morphology have previously been obtained by overexpression of the mDia1 N-terminal region, which contains the Rho-binding domain (RBD). It was proposed previously that this variant sequesters activated Rho, thereby inhibiting stress fiber formation (48).
Nir2 inhibits Rho via Rid.
To investigate whether the Nir2 variant (aa 205 to 424) can functionally inhibit Rho, we coexpressed it with wild-type RhoA in HeLa cells and examined the morphology of the actin cytoskeleton by phalloidin staining (Fig. 3A). This variant did in fact inhibit the effect of wild-type RhoA on stress fiber formation and was therefore designated Rid (Rho-inhibitory domain). Coexpression of Rid and wild-type RhoA resulted in the loss of stress fibers, whereas Rid had no effect on stress fiber formation induced by the activated RhoA, Val14-Rho (Fig. 3A).
To further demonstrate the inhibitory effect of Rid on Rho, we examined the effect of Rid on LPA-induced RhoA activation utilizing a biochemical approach. To affinity precipitate Rho-GTP, we used the RBD of rhotekin immobilized on agarose beads as previously described (36). HeLa cells were transiently transfected with expression vector encoding either Rid or C3 exoenzyme or were mock transfected. The cells were serum starved for 12 h and then stimulated with LPA for 5 min. Cell lysates were incubated with recombinant GST-rhotekin-RBD immobilized on glutathione-agarose beads, and the bound Rho-GTP was analyzed by Western blotting with anti-RhoA antibodies (Fig. 3B). As shown, Rid overexpression significantly decreased the level of activated RhoA, consistent with the morphological results shown in Fig. 3A.
Nir2 inhibits Rho-mediated neurite retraction.
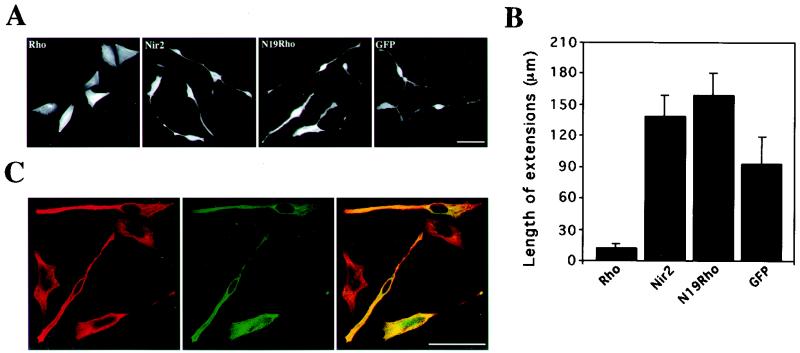
The inhibitory effect of Nir2 on Rho could also be demonstrated in neuronal cells. Activated Rho is well known to induce rapid growth cone collapse and neurite retraction (8, 25). Cells of the human cerebellar medulloblastoma line TE671 rapidly undergo a distinctive morphological transformation, characterized by neurite extension, in response to dbcAMP (40). A constitutively active mutant of Rho, Val14-Rho, inhibits dbcAMP-induced neurite extensions in TE671 cells, whereas C3 exoenzyme exerts the opposite effect (data not shown). Similar to Val14-Rho, overexpression of wild-type RhoA in TE671 cells inhibited neurite extensions in response to dbcAMP, whereas the dominant negative mutant Asn19-Rho slightly accelerated neurite extensions upon dbcAMP treatment (Fig. 4A and B). To assess the effect of Nir2 on Rho-mediated neurite retraction, TE671 cells were cotransfected with different ratios of vectors encoding wild-type RhoA and Nir2. The cells were treated with dbcAMP for different time periods, costained with anti-Nir2 and anti-Rho antibodies, and analyzed by confocal microscopy. Overexpression of Nir2 with Rho attenuated the inhibitory effect of Rho on neurite extensions (Fig. 4C). Almost all the cells that expressed only Rho exhibited a rounded morphology similar to that of undifferentiated TE671 cells (Fig. 4A). This retracted phenotype was still observed 12 h after dbcAMP treatment (data not shown). However, coexpression of Nir2 with Rho markedly changed the cell morphology: the Rho-expressing cells were elongated and developed neurites in response to dbcAMP. These morphological changes were more pronounced in cells that strongly expressed Nir2 than in cells that did not (Fig. 4C) and are consistent with the inhibitory effect of Rid on Rho-mediated stress fiber formation (Fig. 3A).
FIG. 4.
Nir2 attenuates Rho-mediated neurite retraction in TE671 cells. TE671 cells grown on coverslips were transiently transfected with expression vectors encoding either wild-type RhoA (Myc tagged), dominant negative Rho (N19Rho; Myc tagged), Nir2 (HA tagged), or green fluorescent protein (GFP) or cotransfected with Nir2 and RhoA. Fifteen hours later, the cells were treated with dbcAMP for 8 h, fixed, and either immunostained with anti-HA or anti-Myc antibodies or costained with anti-Myc and anti-Rho antibodies. (A and B) Representative phenotypes of cells expressing RhoA, Nir2, N19Rho, or GFP (A) (bar, 50 μm), along with a quantitative analysis of neurite extensions in cells expressing the indicated proteins (B). Neurite extensions were measured with Zeiss 510 confocal microscope operating software (version 2.01). Shown are the means ± standard deviations obtained from more than 30 cells expressing the indicated proteins. (C) Representative confocal images of cells that coexpress Nir2 (green) and RhoA (red) and the merged images. Coexpression appears in yellow. Note that neurite extensions are more pronounced in Rho-expressing cells that strongly express Nir2. Bar, 50 μm.
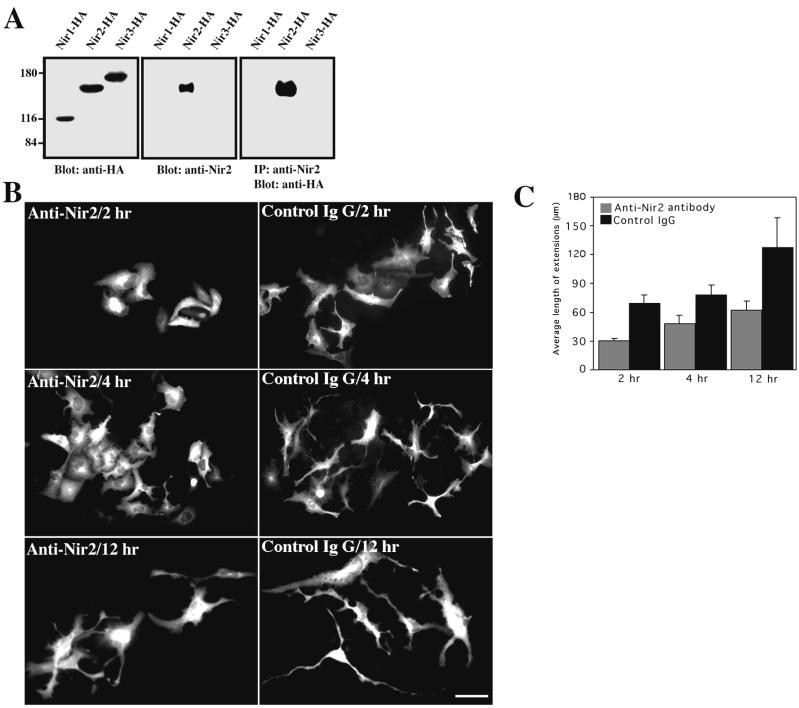
Since endogenous Nir2 is relatively highly expressed in TE671 cells (Fig. 1C) and upregulated upon their differentiation (data not shown), we further investigated its role in regulating neuronal morphogenesis by applying microinjection of anti-Nir2 antibody. A polyclonal anti-Nir2 antibody was raised in rabbits as described in Materials and Methods. This antibody is specific for Nir2 and does not cross-react with the other family members (Nir1 and Nir3) (Fig. 5A). TE671 cells were microinjected with an affinity-purified anti-Nir2 antibody or control rabbit IgG. The injected cells were treated with dbcAMP for different time periods and analyzed by fluorescence microscopy. As shown in Fig. 5B and C, anti-Nir2 antibodies significantly attenuated neurite extension in response to dbcAMP (∼30 to 55%) compared to the control rabbit IgG (Fig. 5C). These results suggest that Nir2 plays a regulatory role in neuronal morphogenesis. This role might be related to its ability to inhibit Rho; nevertheless, we cannot exclude the possibility that other cellular components are also involved.
FIG. 5.
Microinjection of anti-Nir2 antibody attenuates neurite extension. (A) Specificity of anti-Nir2 antibody. Polyclonal anti-Nir2 antibody was raised in rabbits as described in Materials and Methods. Anti-Nir2 antibody bound to Sepharose-protein A was incubated with cell lysate of HEK 293 cells expressing an HA-tagged Nir1, Nir2, or Nir3 protein. The samples were incubated for 2 h at 4°C, washed, separated by SDS-PAGE, and immunoblotted with anti-HA antibodies. Western blot analysis of total cell lysate of HEK 293 cells expressing each of the Nir proteins was carried out using either anti-HA or anti-Nir2 antibodies as indicated. IP, immunoprecipitation. Numbers at left show molecular mass in kilodaltons. (B) TE671 cells grown on coverslips were microinjected with either antibodies against Nir2 or control rabbit IgG. Cells were treated with dbcAMP for the indicated time periods, fixed, and immunostained with Alexa 488-conjugated goat anti-rabbit IgG. Shown are representative images of microinjected cells obtained in three independent experiments. Bar, 50 μm. (C) Quantitative analysis of neurite extensions of more than 50 microinjected cells was performed as described for Fig. 4B.
Nir2 binds Rho-GDP via Rid.
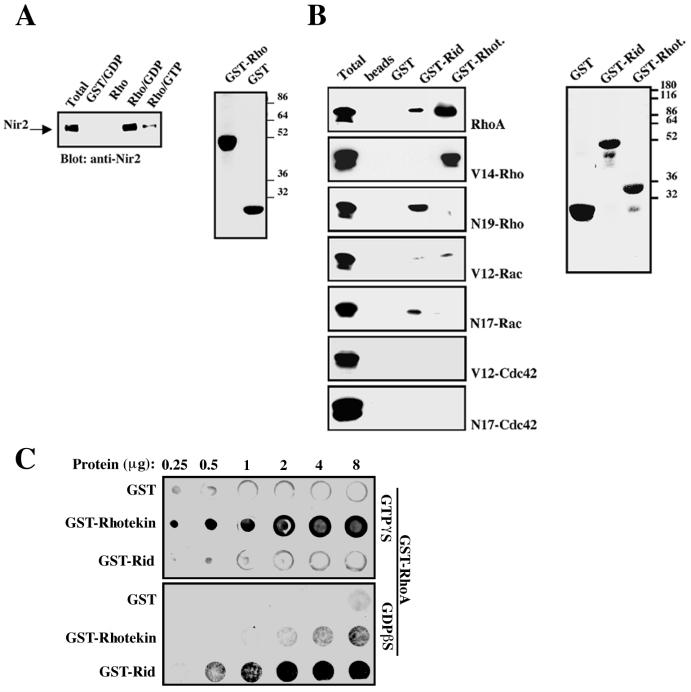
Since Rid possesses homology to the Rho-binding site of FH proteins, we hypothesized that it may bind to the small GTPase Rho. To explore this possibility, we first tested whether Nir2 could bind Rho. An in vitro binding assay was carried out by incubating GST-RhoA fusion protein, bound to glutathione-agarose beads, with lysate of HEK 293 cells expressing Nir2. Nir2 binding to Rho was detected by Western blotting (Fig. 6A), and their association was shown to be nucleotide dependent: no binding of Nir2 to either nucleotide-free GST-Rho or GST was detected. In contrast, Nir2 was found to interact with the GDP-bound GST-Rho and to a lesser extent with the GTP-bound Rho. These results suggested that Nir2 preferentially interacts with the inactive form of the small GTPase Rho under the experimental conditions described.
FIG. 6.
Nir2 preferentially binds the inactive form of RhoA via its Rid. (A) In vitro binding of Nir2 to GDP-bound GST-RhoA. Bacterially expressed GST or GST-RhoA fusion proteins were purified on glutathione-agarose beads. The GST-RhoA fusion protein was either preloaded with GDP or GTPγS or left free of nucleotides. The purified recombinant proteins were then incubated with cell lysate of HEK 293 cells expressing the wild-type Nir2, and binding of Nir2 was detected by Western blotting. (B) In vitro binding of Rho-family small GTPases to recombinant Rid. Rid and the RBD of rhotekin (GST-Rhot.) (32) were expressed as GST fusion proteins in bacteria. The recombinant proteins were purified on glutathione-agarose beads and incubated with cell lysate of HEK 293 cells expressing the indicated small GTPase. The same binding assays were carried out using glutathione beads or recombinant GST as a control. Binding of the small GTPases was determined by Western blotting with the appropriate antibodies. The purified GST fusion proteins used in these binding experiments were resolved on an SDS-15% polyacrylamide gel and stained with Coomassie brilliant blue (right panels in panels A and B). Numbers at the extreme right of panels A and B show molecular mass in kilodaltons. (C) Direct binding of Rho to Rid with an overlay assay. Equal amounts of purified GST, GST-rhotekin, and GST-Rid recombinant proteins (0.25 to 8 μg) were loaded on a nitrocellulose membrane and probed with GST-Rho bound to either GDP or GTP as described in Materials and Methods. Following washing, the membranes were probed with anti-Rho antibody and then with horseradish peroxidase-conjugated anti-mouse IgG. The bound Rho was detected by ECL.
To further characterize the interaction between Nir2 and Rho, we expressed Rid as a GST fusion protein in bacteria. GST-Rid was purified on glutathione-agarose beads and incubated with cell lysates of HEK 293 cells expressing Rho-family small GTPases (Fig. 6B). The same binding assays were performed with GST or GST-rhotekin as control. As shown in Fig. 6B, GST-rhotekin bound exclusively to the wild-type RhoA and to the constitutively active mutant Val14-Rho. These results are consistent with previous reports demonstrating binding of rhotekin to activated Rho (32). In contrast to rhotekin, Rid exclusively interacted with the wild-type RhoA and the dominant negative mutants of Rho and Rac. Neither the Val14-Rho, Val12-Rac, or Val12-Cdc42 mutant (Fig. 6B) nor the L63-RhoA, L61-Rac, or L61-Cdc42 mutant (data not shown) had a detectable interaction with GST-Rid. The interaction of Rid with wild-type RhoA probably occurs with GDP-bound RhoA, as Rho cycles between inactive GDP-bound and active GTP-bound forms. Therefore, binding of Rho-GDP to Rid probably maintains it in an inactive state, thereby inhibiting its conversion to active Rho-GTP. Furthermore, this interaction appears to be direct as determined by an overlay assay using recombinant GST-Rho loaded with GDP or GTP as a probe (Fig. 6C). Equal amounts of purified GST, GST-rhotekin, or GST-Rid recombinant proteins were loaded onto nitrocellulose membranes and incubated with recombinant Rho-GDP or Rho-GTP as described in Materials and Methods. The bound Rho protein was detected by anti-Rho antibodies and ECL. As shown, GST failed to interact with either Rho-GDP or Rho-GTP, whereas GST-rhotekin strongly interacted with Rho-GTP, and Rid preferentially interacted with Rho-GDP. These results are consistent with those of the other binding experiments shown in Fig. 6A and B.
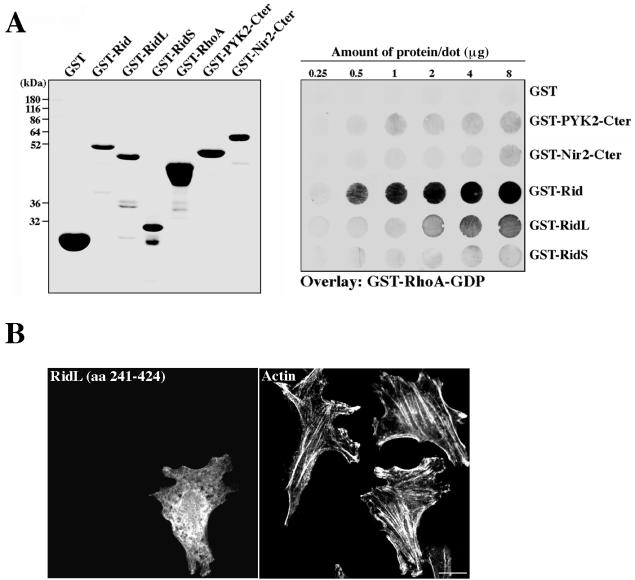
The binding of Rid to Rho was further characterized using two additional truncated Rid constructs: a long one, RidL (aa 241 to 424), and a short one, RidS (aa 205 to 244). These truncated mutants were expressed in bacteria as GST fusion proteins, and their binding to Rho-GDP was assessed by an overlay assay. As shown in Fig. 7A, Rid (aa 205 to 424) exhibits specific binding to Rho-GDP, consistent with the results shown in Fig. 6C. On the other hand, the long truncated mutant RidL (aa 241 to 424) exhibits very weak binding, and no detectable binding was observed for the short construct (aa 205 to 244). These results suggest that the first 36 aa of Rid are important for Rho binding, as their deletion affects its binding affinity. However, the short construct containing this region failed to interact with Rho, suggesting that it may not contain the full binding site or that it is inappropriately folded. To test whether these truncated mutants have any effect on the actin cytoskeleton and cell morphology, they were subcloned into a mammalian expression vector and transiently expressed in HeLa cells. The short mutant, RidS, was not stable and rapidly degraded, whereas the long one, RidL, was appropriately expressed as Myc-tagged fusion protein. Confocal microscopy analysis of RidL-expressing cells indicated that this mutant had no obvious effect on the actin cytoskeleton or cell morphology (Fig. 7B). Since Rid markedly affects F-actin staining and cell shape (Fig. 2B), these results suggest that aa 205 to 240 are critical for Rid's phenotype. In conclusion, both the binding properties and the phenotypes of Rid and its truncated mutants suggest that Rid binds Rho and that this binding is critical for Rid's phenotype.
FIG. 7.
Binding of Rho to Rid is critical for Rid's phenotype. (A) Binding of Rho-GDP to Rid and to its truncated mutants. Rid (aa 205 to 424) and its two truncated mutants, RidS (aa 205 to 244) and RidL (aa 241 to 424), were expressed as GST fusion proteins in bacteria and purified on glutathione-agarose beads. Their binding to Rho-GDP was assessed by an overlay assay, essentially as described in the legend to Fig. 6C. The following purified recombinant proteins were used as negative controls: GST, GST fused to the C-terminal region of the tyrosine kinase PYK2 (GST-PYK2-Cter; aa 800 to 1009), and GST fused to the C-terminal region of Nir2 (GST-Nir2-Cter; aa 912 to 1244). Coomassie brilliant blue staining of the purified GST fusion proteins is shown in the left panel. The results of the binding experiment (right panel) indicate that Rid directly and specifically binds Rho-GDP, whereas RidL exhibits weak binding to Rho-GDP, and no detectable binding is observed for RidS. (B) RidL has no effect on actin or cell morphology. Myc-tagged RidL was transiently expressed in HeLa cells, and its effect on the cell and actin morphology was assessed by confocal laser microscopy. As shown, overexpression of this mutant has no obvious effect on either F-actin staining or cell morphology, suggesting that the first 36 aa of Rid are critical for the Rid-induced phenotype. Bar, 10 μm.
The PI transfer domain negatively regulates the effect of Rid on the actin cytoskeleton.
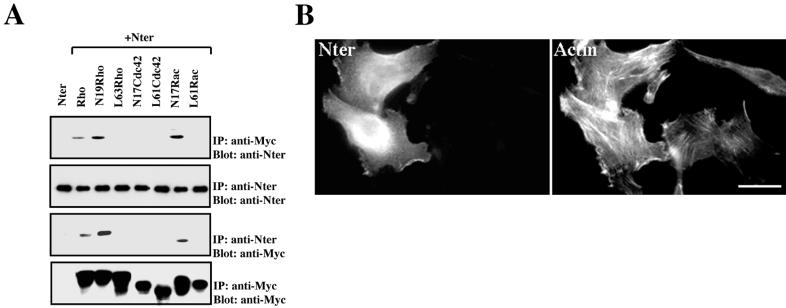
Next, we assessed whether the entire N-terminal region, containing the PI transfer and Rids, retains the binding specificity of Rid to the small GTPase Rho. An epitope-tagged construct encoding the entire N-terminal region of Nir2 (aa 2 to 424 [Fig. 1A]) was expressed in HEK 293 cells, either alone or with Rho-family small GTPases, and their interaction was determined by coimmunoprecipitation assay. The N-terminal region interacted exclusively with the wild-type RhoA and the dominant negative mutants, Asn19-Rho and Asn17-Rac (Fig. 8A). These results are consistent with the binding properties of Rid observed in vitro, suggesting that the N-terminal region retains the binding specificities of Rid. Therefore, we expected that overexpression of the N-terminal region would cause a loss of F-actin, similar to the effect obtained by overexpressing Rid. However, expression of the entire N-terminal region of Nir2 had no obvious effect on stress fiber formation (Fig. 8B), suggesting that the PI transfer domain of Nir2 somehow suppresses this particular effect of Rid, without affecting its binding properties to the Rho small GTPases. Furthermore, overexpression of the N-terminal region also had no profound effect on cell morphology. The ratio between the long and short axes of N-terminal-region-expressing cells (1.6 ± 0.02, n = 50, P < 0.0001) was very similar to that of untransfected HeLa cells.
FIG. 8.
The N-terminal region of Nir2 binds the inactive Rho but has no effect on F-actin morphology. (A) Binding of the Rho-family small GTPases to the N-terminal region of Nir2. The entire N-terminal region of Nir2 (aa 2 to 424; Nter) was coexpressed with the indicated Myc-tagged small GTPases in HEK 293 cells. Cell lysates were subjected to immunoprecipitation (IP) with either anti-Myc or anti-Nir2 antibodies. After extensive washing the samples were boiled, separated by SDS-PAGE, and transferred to nitrocellulose. The membranes were probed with the indicated antibodies. (B) The N-terminal region of Nir2 has no effect on F-actin morphology. The Nir2 N-terminal region (Nter) was expressed in HeLa cells as an HA-tagged fusion protein, and its effect on F-actin morphology was analyzed by fluorescence microscopy. Shown are the transfected cells stained with anti-HA antibodies and the filamentous actin in the same field stained with TRITC-phalloidin. Bar, 10 μm.
DISCUSSION
The Nir/RdgB family members have been highly conserved throughout evolution. Although little is known of their cellular functions, it is well established that mutations within the Drosophila rdgB gene dramatically affect photoreceptor cell morphology. Typically, rdgB mutant flies begin to exhibit the morphological hallmarks of photoreceptor cell degeneration several days after eclosion and a profound loss in electroretinogram amplitude shortly after initial light exposure (17, 41). While it is not yet clear how RdgB affects photoreceptor cell morphology, several lines of evidence suggest that its PI transfer domain plays a critical role in this process (32). In the last few years, four different mammalian genes encoding RdgB homologues have been cloned (1, 10, 26, 29). Nir2 (also known as mRdgB) shares high sequence homology with the other family members, particularly within the N-terminal PI transfer domain and the C-terminal region (26). Although mRdgB can rescue the phenotype of rdgB mutant flies (10), its cellular function in mammalian cells remains unknown.
In the present study, we provide evidence that Nir2 is an important regulator of cell morphogenesis. We showed that ectopic expression of Nir2 markedly affects cell morphology (Fig. 1D). By using different Nir2 truncated mutants and sequence alignment analysis, we defined a specific domain, confined to the Nir2 N-terminal region, that shares sequence homology with the Rho-binding site of FH proteins (Fig. 2A). This domain causes an apparent loss of F-actin when expressed in mammalian cells (Fig. 2B). Since this domain also inhibits Rho-mediated stress fiber formation (Fig. 3A) and LPA-induced Rho activation (Fig. 3B), it was designated Rid (Rho-inhibitory domain). In vitro binding experiments using Rid as a fusion protein demonstrated its exclusive binding to the wild-type Rho and the dominant negative mutants of Rho and Rac (Fig. 6B). Nir2 was also shown to preferentially bind the inactive GDP-bound form of RhoA (Fig. 6A). Thus, Nir2 binds to Rho via its Rid.
The inhibitory effect of Nir2 on Rho was also demonstrated in neuronal cells (Fig. 4). In those cells, Rho appears to regulate growth cone collapse and neurite retraction (21, 22, 44). These morphological changes are driven by Rho-mediated contraction of the actomyosin-based cytoskeleton (19, 21). Overexpression of V14-Rho or wild-type RhoA in TE671 cells dramatically inhibited neurite extension in response to dbcAMP (Fig. 4), whereas C3 exoenzyme significantly accelerated neurite extension upon dbcAMP treatment (data not shown). Coexpression of RhoA and Nir2 inhibited Rho-mediated neurite retraction (Fig. 4C), and microinjection of anti-Nir2 antibodies markedly attenuated neurite extension in response to dbcAMP (Fig. 5B). These results suggest that Nir2 is essential for neurite extension, probably due to its inhibitory effect on Rho. Nevertheless, at present, we cannot exclude the possibility that additional signaling components upstream or downstream of Nir2 are also involved in this process.
Other members of the Rho family of small GTPases have been previously shown to regulate neuronal cell morphology (30). Drosophila Rac1 was recently shown to be essential for photoreceptor morphogenesis, and overexpression of a dominant negative mutant of Drac1 resulted in retinal degeneration. It was proposed previously that Drac1 links rhodopsin to photoreceptor morphogenesis by providing a structural support through organization of the actin cytoskeleton (9). Likewise, Drosophila RdgB, via a Rid-related region, may regulate Rho or Rac function and thereby affect photoreceptor morphology. This possibility needs to be further investigated. It is worth nothing that Rid shares sequence homology with the other Nir/RdgB family members (26). Multiple alignment analysis of Rid's sequence with the corresponding regions of the mammalian proteins Nir3 and mRdgB2 (29) and their invertebrate counterparts the Drosophila RdgB and the Caenorhabditis elegans homologues revealed 11% identity and 33% similarity. Among the mammalian proteins, the Rid sequence is highly related to Nir3, exhibiting 46% identity and 73% similarity. The high homology of Rid with the corresponding region of Nir3 suggests that Nir3 may also affect cell morphogenesis via a Rid-related region. However, this possibility needs to be examined further, since Rid contains a unique repeated motif (aa 260 to 272, NTGSEGSEAQPPG, and aa 284 to 296, NTGTPDGPEAPPG) that is not found in the corresponding regions of the other family members (including Nir3), and its homology to the RBD of FH proteins appears to be stronger. In this regard, it is worth noting that the homology between Nir2 and FH proteins is not restricted to the RBD but could also be found in three additional regions by Block Maker analysis.
Although Nir2 shares sequence homology with the Rho-binding site of FH proteins (Fig. 2A), their binding properties are distinct. Several proteins belonging to the FH family, including mDia1, mDia2, Bni1p, and Bnr1p, have been previously shown to preferentially bind the GTP-bound form of Rho small GTPases (47). By using pull-down experiments and/or the two-hybrid assay, it was shown elsewhere that mDia1 preferentially binds the activated form of Rho (48, 49), whereas mDia2 binds to the activated form of Rho and Cdc42 in vitro (2). Bnr1p binds the activated form of Rho4p (20), and Bni1p preferentially interacts with the activated form of Rho1p and Cdc42 (9, 24). Nevertheless, recent studies have shown that the FH protein FRL interacts with both the active and the inactive forms of the small GTPase Rac (52). This interaction is mediated by its FH3 domain, which partially overlaps with the RBD of mDia1 and mDia2 (34). Thus, it could be that differences within the GTPase-binding site of FH proteins affect their binding specificity toward different Rho GTPases and their affinity toward the active or inactive forms. Although the alignment shown in Fig. 2A falls within the GTPase-binding domain of several FH proteins, no consensus Rho-binding motif has been identified among these family members. Nevertheless, the results shown in Fig. 6C clearly demonstrate direct binding of Rho to Rid by an overlay-binding assay. These results indicate that this region (aa 205 to 424) contains a Rho-binding site.
Unfortunately, there is currently little information regarding the specific structural requirements of the FH protein GTPase-binding domains. It was suggested elsewhere that this interaction is complex and may not be restricted to a limited peptide domain but may rather involve multiple binding sites (3). This may also be the case for Nir2. Deletion of the first 36 aa of Rid markedly affected Rho binding but completely abolished Rid's phenotype (Fig. 7), suggesting that these residues are critical for Rho binding and Rid's phenotype. On the other hand, RidL (aa 241 to 424) exhibits a weak binding affinity to Rho (Fig. 7A). Similarly, the ΔPI mutant, which lacks the first 240 aa of Nir2, exhibits an extended phenotype (Fig. 1A). These results suggest that the interaction of Nir2 with Rho is complex and may not be restricted to a limited peptide within Rid but may rather require the entire Rid sequence.
In contrast to Rid, overexpression of full-length Nir2 had no obvious effect on the actin cytoskeleton. Similar results have been previously obtained for mDia1: while overexpression of the RBD resulted in a loss of F-actin staining, the full-length protein had no apparent effect on the actin cytoskeleton (48). This negligible effect of mDia1 appears to be due to a carboxy-terminal autoregulatory domain that physically interacts with the N-terminal RBD (3, 48). This intramolecular autoregulatory mechanism is not restricted to mDia1 and has been identified elsewhere in other signaling proteins including WASP (23, 37), the protein kinase PAK1 (53), and the Src family of protein tyrosine kinases (51), among others. We are currently investigating whether a similar autoregulatory mechanism exists in Nir2. This kind of mechanism might be considered as one explanation for the different effects of the full-length Nir2 (Fig. 1D) and the N-terminal region (Fig. 8B) on cell morphology. If Nir2 contains a C-terminal autoregulatory domain that interacts with its N-terminal region, it may affect the function of the PI transfer domain and/or Rid, thereby causing Nir2 to differently affect cell morphology compared to the N-terminal region. This mechanism might also explain the minor effect of the ΔPI mutant on the actin cytoskeleton. If the PI transfer domain suppresses the phenotype of Rid, as shown in Fig. 8B, then it is expected to produce a Rid-like phenotype. However, the phenotypes of ΔPI and Rid are distinct, suggesting that an additional inhibitory domain may exist. At present, we cannot exclude the possibility that other as yet uncharacterized domains of Nir2 may affect the phenotypes of the different truncated mutants. However, we show here that the N-terminal region contains two defined domains: the PI transfer domain, which was previously identified (26), and Rid. Although this region retained the binding properties of Rid (Fig. 8A), it had no obvious effects on actin or cell morphology (Fig. 8B). These results suggest that the PI transfer domain somehow inhibits the effect of Rid on the actin cytoskeleton. We have previously shown that Nir2 contains a functional PI transfer domain (26), and other studies have demonstrated the role of PI transfer proteins in PI 4,5-bisphosphate (PIP2) synthesis (11). PI transfer proteins have been proposed elsewhere to present PIs to lipid kinases, thereby regulating PIP2 synthesis (11). The ability of PIP2 to bind and regulate several actin-binding proteins, including those involved in filament capping, cross-linking, and severing, suggests that PIP2 synthesis is a critical regulatory step in actin polymerization (31). Therefore, the PI transfer domain of Nir2 may be required for the local release of PIP2, which is essential for actin polymerization, whereas Rid inhibits Rho and thereby suppresses actin polymerization. This might explain the negligible effect of the entire N-terminal region on the actin cytoskeleton (Fig. 8B). These two opposite functions, mediated by the PI transfer domain and its adjacent Rid, may work sequentially in cellular processes, such as cell motility or morphogenesis, which require dynamic assembly and disassembly of filamentous actin.
In conclusion, in the present study we provide evidence that Nir2 is an important regulator of cell morphogenesis. We defined a novel domain designated Rid that plays a role in this process through its inhibitory effect on the small GTPase Rho. Our findings describe, for the first time, a function of Nir/RdgB family members in mammalian cells. Further studies are expected to shed light on the molecular mechanisms underlying the role of Nir/RdgB proteins in cell morphogenesis, including their potential involvement in regulating photoreceptor cell morphology and retinal degeneration.
Acknowledgments
We thank B. Geiger, S. Bershadsky, and Y. Koch for critical reading of the manuscript.
Sima Lev is an incumbent of the Helena Rubinstein Career Development Chair. This work was supported by the Binational Science Foundation (grant no. 98-00352), the Israel Science Foundation (grant no. 649/00-1), the Heineman Foundation, the Minerva Foundation, and the Israel Cancer Research Foundation.
REFERENCES
- 1.Aikawa, Y., H. Hara, and T. Watanabe. 1997. Molecular cloning and characterization of mammalian homologues of the Drosophila retinal degeneration B gene. Biochem. Biophys. Res. Commun. 236:559-564. [DOI] [PubMed] [Google Scholar]
- 2.Alberts, A. S., N. Bouquin, L. H. Johnston, and R. Treisman. 1998. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 273:8616-8622. [DOI] [PubMed] [Google Scholar]
- 3.Alberts, A. S. 2001. Identification of a carboxyl-terminal Diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 276:2824-2830. [DOI] [PubMed] [Google Scholar]
- 4.Arellano, M., P. M. Coll, and P. Perez. 1999. RHO GTPases in the control of cell morphology, cell polarity, and actin localization in fission yeast. Microsc. Res. Tech. 47:51-60. [DOI] [PubMed] [Google Scholar]
- 5.Aspenstrom, P. 1999. Effectors for the Rho GTPases. Curr. Opin. Cell Biol. 11:95-102. [DOI] [PubMed] [Google Scholar]
- 6.Bar-Sagi, D., and A. Hall. 2000. Ras and Rho GTPases: a family reunion. Cell 103:227-238. [DOI] [PubMed] [Google Scholar]
- 7.Bishop, A. L., and A. Hall. 2000. Rho GTPases and their effector proteins. Biochem. J. 348:241-255. [PMC free article] [PubMed] [Google Scholar]
- 8.Bito, H., T. Furuyashiki, H. Ishihara, Y. Shibasaki, K. Ohashi, K. Mizuno, M. Maekawa, T. Ishizaki, and S. Narumiya. 2000. A critical role for a Rho-associated kinase, p160ROCK, in determining axon outgrowth in mammalian CNS neurons. Neuron 26:431-441. [DOI] [PubMed] [Google Scholar]
- 9.Chang, H. Y., and D. F. Ready. 2000. Rescue of photoreceptor degeneration in rhodopsin-null Drosophila mutants by activated Rac1. Science 290:1978-1980. [DOI] [PubMed] [Google Scholar]
- 10.Chang, J. T., S. Milligan, Y. Li, C. E. Chew, J. Wiggs, N. G. Copeland, N. A. Jenkins, P. A. Campochiaro, D. R. Hyde, and D. J. Zack. 1997. Mammalian homolog of drosophila retinal degeneration B rescues the mutant fly phenotype. J. Neurosci. 17:5881-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham, E., G. M. Thomas, A. Ball, I. Hiles, and S. Cockcroft. 1995. Phosphatidylinositol transfer protein dictates the rate of inositol trisphosphate production by promoting the synthesis of PIP2. Curr. Biol. 5:775-783. [DOI] [PubMed] [Google Scholar]
- 12.Elagin, V. A., R. B. Elagina, C. J. Doro, T. S. Vihtelic, and D. R. Hyde. 2000. Cloning and tissue localization of a novel zebrafish RdgB homolog that lacks a phospholipid transfer domain. Vis. Neurosci. 17:303-311. [DOI] [PubMed] [Google Scholar]
- 13.Evangelista, M., K. Blundell, M. S. Longtine, C. J. Chow, N. Adames, J. R. Pringle, M. Peter, and C. Boone. 1997. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276:118-122. [DOI] [PubMed] [Google Scholar]
- 14.Flynn, P., H. Mellor, R. Palmer, G. Panayotou, and P. J. Parker. 1998. Multiple interactions of PRK1 with RhoA. Functional assignment of the Hr1 repeat motif. J. Biol. Chem. 273:2698-2705. [DOI] [PubMed] [Google Scholar]
- 15.Fujusawa, K., A. Fujita, T. Ishizaki, Y. Saito, and S. Narumiya. 1996. Identification of the Rho-binding domain of p160ROCK, a Rho-associated coiled-coil containing protein kinase. J. Biol. Chem. 271:23022-23028. [DOI] [PubMed] [Google Scholar]
- 16.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 17.Harris, W. A., and W. S. Stark. 1977. Hereditary retinal degeneration in Drosophila melanogaster. A mutant defect associated with the phototransduction process. J. Gen. Physiol. 69:261-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart, M. J., A. Eva, D. Zangrilli, S. A. Aaronson, T. Evans, R. A. Cerione, and Y. Zheng. 1994. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J. Biol. Chem. 269:62-65. [PubMed] [Google Scholar]
- 19.Hirose, M., T. Ishizaki, N. Watanabe, M. Uehata, O. Kranenburg, W. H. Moolenaar, F. Matsumura, M. Maekawa, H. Bito, and S. Narumiya. 1998. Molecular dissection of the Rho-associated protein kinase (p160ROCK)-regulated neurite remodeling in neuroblastoma N1E-115 cells. J. Cell Biol. 141:1625-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamura, H., K. Tanaka, T. Hihara, M. Umikawa, T. Kamei, K. Takahashi, T. Sasaki, and Y. Takai. 1997. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16:2745-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jalink, K., E. J. van Corven, T. Hengeveld, N. Morii, S. Narumiya, and W. H. Moolenaar. 1994. Inhibition of lysophosphatidate- and thrombin-induced neurite retraction and neuronal cell rounding by ADP ribosylation of the small GTP-binding protein Rho. J. Cell Biol. 126:801-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh, H., J. Aoki, A. Ichikawa, and M. Negishi. 1998. p160 RhoA-binding kinase ROKα induces neurite retraction. J. Biol. Chem. 273:2489-2492. [DOI] [PubMed] [Google Scholar]
- 23.Kim, A. S., L. T. Kakalis, N. Abdul-Manan, G. A. Liu, and M. K. Rosen. 2000. Autoinhibition and activation mechanisms of Wiskott-Aldrich syndrome protein. Nature 404:151-158. [DOI] [PubMed] [Google Scholar]
- 24.Kohno, H., K. Tanaka, A. Mino, M. Umikawa, H. Imamura, T. Fujiwara, Y. Fujita, K. Hotta, H. Qadota, T. Watanabe, Y. Ohya, and Y. Takai. 1996. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15:6060-6068. [PMC free article] [PubMed] [Google Scholar]
- 25.Kranenburg, O., M. Poland, F. P. van Horck, D. Drechsel, A. Hall, and W. H. Moolenaar. 1999. Activation of RhoA by lysophosphatidic acid and Gα12/13 subunits in neuronal cells: induction of neurite retraction. Mol. Biol. Cell 10:1851-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lev, S., J. Hernandez, R. Martinez, A. Chen, G. Plowman, and J. Schlessinger. 1999. Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mol. Cell. Biol. 19:2278-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lev, S., H. Moreno, R. Martinez, P. Canoll, E. Peles, J. M. Musacchio, G. D. Plowman, B. Rudy, and J. Schlessinger. 1995. Protein tyrosine kinase PYK2 involved in Ca(2+)-induced regulation of ion channel and MAP kinase functions. Nature 376:737-745. [DOI] [PubMed] [Google Scholar]
- 28.Litvak, V., D. Tian, Y. D. Shaul, and S. Lev. 2000. Targeting of PYK2 to focal adhesions as a cellular mechanism for convergence between integrins and G protein-coupled receptor signaling cascades. J. Biol. Chem. 275:32736-32746. [DOI] [PubMed] [Google Scholar]
- 29.Lu, C., T. S. Vihtelic, D. R. Hyde, and T. Li. 1999. A neuronal-specific mammalian homolog of the Drosophila retinal degeneration B gene with expression restricted to the retina and dentate gyrus. J. Neurosci. 19:7317-7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo, L., L. Y. Jan, and Y. N. Jan. 1997. Rho family GTP-binding proteins in growth cone signalling. Curr. Opin. Neurobiol. 7:81-86. [DOI] [PubMed] [Google Scholar]
- 31.Martin, T. F. 1998. Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu. Rev. Cell Dev. Biol. 14:231-264. [DOI] [PubMed] [Google Scholar]
- 32.Milligan, S. C., J. G. J. Alb, R. B. Elagina, V. A. Bankaitis, and D. R. Hyde. 1997. The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J. Cell Biol. 139:351-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nobes, C. D., and A. Hall. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81:53-62. [DOI] [PubMed] [Google Scholar]
- 34.Petersen, J., O. Nielsen, R. Egel, and I. M. Hagan. 1998. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 141:1217-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reid, T., T. Furuyashiki, T. Ishizaki, G. Watanabe, N. Watanabe, K. Fujisawa, N. Morii, P. Madaule, and S. Narumiya. 1996. Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, and rhophilin in the rho-binding domain. J. Biol. Chem. 271:13556-13560. [DOI] [PubMed] [Google Scholar]
- 36.Ren, X.-D., W. B. Kiosses, and M. A. Schwartz. 1999. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohatgi, R., H. Y. Ho, and M. W. Kirschner. 2000. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 150:1299-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekine, A., M. Fujiwara, and S. Narumiya. 1989. Asparagine residue in the rho gene product is the modification site for botulinum ADP-ribosyltransferase. J. Biol. Chem. 264:8602-8605. [PubMed] [Google Scholar]
- 39.Settleman, J. 1999. Rho GTPases in development. Prog. Mol. Subcell. Biol. 22:201-229. [DOI] [PubMed] [Google Scholar]
- 40.Siegel, H. N., and R. J. Lukas. 1988. Morphological and biochemical differentiation of the human medulloblastoma cell line TE671. Brain Res. Dev. Brain Res. 44:269-280. [DOI] [PubMed] [Google Scholar]
- 41.Stark, W. S., and R. Sapp. 1987. Ultrastructure of the retina of Drosophila melanogaster: the mutant ora (outer rhabdomeres absent) and its inhibition of degeneration in rdgB (retinal degeneration-B). J. Neurogenet. 4:227-240. [PubMed] [Google Scholar]
- 42.Symons, M., and J. Settleman. 2000. Rho family GTPases: more than simple switches. Trends Cell Biol. 10:415-419. [DOI] [PubMed] [Google Scholar]
- 43.Tatsis, N., D. A. Lannigan, and I. G. Macara. 1998. The function of the p190 Rho GTPase-activating protein is controlled by its N-terminal GTP binding domain. J. Biol. Chem. 273:34631-34638. [DOI] [PubMed] [Google Scholar]
- 44.Tigyi, G., D. J. Fischer, A. Sebok, C. Yang, D. L. Dyer, and R. Miledi. 1996. Lysophosphatidic acid-induced neurite retraction in PC12 cells: control by phosphoinositide-Ca2+ signaling and Rho. J. Neurochem. 66:537-548. [DOI] [PubMed] [Google Scholar]
- 45.Van Aelst, L., and C. D'Souza-Schorey. 1997. Rho GTPases and signaling networks. Genes Dev. 11:2295-2322. [DOI] [PubMed] [Google Scholar]
- 46.Vihtelic, T. S., D. R. Hyde, and J. E. O'Tousa. 1991. Isolation and characterization of the Drosophila retinal degeneration B (rdgB) gene. Genetics 127:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wasserman, S. 1998. FH proteins as cytoskeletal organizers. Trends Cell Biol. 8:111-115. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe, N., T. Kato, A. Fujita, T. Ishizaki, and S. Narumiya. 1999. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1:136-143. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe, N., P. Madaule, T. Reid, T. Ishizaki, G. Watanabe, A. Kakizuka, Y. Saito, K. Nakao, B. M. Jockusch, and S. Narumiya. 1997. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 16:3044-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welch, M. D., A. Mallavarapu, J. Rosenblatt, and T. J. Mitchison. 1997. Actin dynamics in vivo. Curr. Opin. Cell Biol. 9:54-61. [DOI] [PubMed] [Google Scholar]
- 51.Williams, J. C., R. K. Wierenga, and M. Saraste. 1998. Insights into Src kinase functions: structural comparisons. Trends Biochem. Sci. 23:179-184. [DOI] [PubMed] [Google Scholar]
- 52.Yayoshi-Yamamoto, S., I. Taniuchi, and T. Watanabe. 2000. FRL, a novel formin-related protein, binds to Rac and regulates cell motility and survival of macrophages. Mol. Cell. Biol. 20:6872-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao, Z. S., E. Manser, X. Q. Chen, C. Chong, T. Leung, and L. Lim. 1998. A conserved negative regulatory region in αPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol. Cell. Biol. 18:2153-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]