Abstract
G proteins, which bind and hydrolyze GTP, are involved in regulating a variety of critical cellular processes, including the process of protein synthesis. Many members of the subfamily of elongation factor class G proteins interact with the ribosome and function to regulate discrete steps during the process of protein synthesis. Despite sequence similarity to factors involved in translation, a role for the yeast Hbs1 protein has not been defined. In this work we have identified a genetic relationship between genes encoding components of the translational apparatus and HBS1. HBS1, while not essential for viability, is important for efficient growth and protein synthesis under conditions of limiting translation initiation. The identification of an Hbs1p-interacting factor, Dom34p, which shares a similar genetic relationship with components of the translational apparatus, suggests that Hbs1p and Dom34p may function as part of a complex that facilitates gene expression. Dom34p contains an RNA binding motif present in several ribosomal proteins and factors that regulate translation of specific mRNAs. Thus, Hbs1p and Dom34p may function together to help directly or indirectly facilitate the expression either of specific mRNAs or under certain cellular conditions.
G proteins are involved in regulating a variety of critical cellular processes, including protein synthesis, cytoskeleton assembly, vesicular transport, and signal transduction (reviewed in reference 9). The superfamily of G proteins includes three main families: Ras and its homologs, the heterotrimeric G proteins, and the translation elongation factors. The G proteins' property of cycling between active (GTP-bound) and inactive (GDP-bound) conformations is key in their ability to function as molecular switches and to make G proteins uniquely suitable for the regulation of diverse cellular processes. Crystallographic analysis of G proteins, first achieved with the GDP-bound form of translation elongation factor 1A (EF1A) from Escherichia coli (37) and later with higher-resolution EF1A (33) and the p21Ras structures (39, 40), identified a common structural core and five loops designated G-1 through G-5 (10) involved in guanine nucleotide binding. The G-1 (GXXXXGK[S/T]), G-3 (DXXG), and G-4 (NKXD) loops contain the best-defined consensus residues (19) and serve as sequence indicators of putative G-protein function.
The HBS1 gene encodes a nonessential protein in the yeast Saccharomyces cerevisiae. Hbs1p contains the G-protein consensus elements in addition to regions with sequence similarity to translation elongation factor eEF1A and the translation release factor eRF3 (25, 53). eEF1A delivers aminoacyl-tRNA to the A site of the ribosome during the process of translation elongation (13), while eRF3 functions during the translation termination process (30). Hbs1p also shares sequence similarity with Ski7p, a protein involved in exosome-mediated mRNA degradation (52). The identification of putative HBS1 orthologs from mouse and human (53), and more recently from Drosophila melanogaster, Schizosaccharomyces pombe, and Candida albicans (25), suggests that HBS1 is evolutionarily conserved.
HBS1 was originally identified as a gene dosage suppressor of the cold-sensitive (Cs−) growth defect of strains deficient in the Hsp70 proteins, Ssb1p and Ssb2p (38). In the yeast S. cerevisiae, the Ssb proteins are a major, but distinct, class of cytosolic Hsp70 chaperones (28). The Ssbs are not induced upon heat shock but instead are constitutively active chaperones, the majority of which are found localized with actively translating ribosomes (38, 43). During translation, the Ssbs likely function as core components of the ribosome, facilitating the folding of nascent polypeptide chains as they are being synthesized on the translating ribosome (38, 43).
Since Hbs1p was a putative GTPase with similarity to both eEF1A and eRF3 (25, 53), initial models for Hbs1p function suggested that, in the absence of Ssbps, movement of the nascent peptide through the channel of the large subunit was slowed and the polypeptide “backed up” (38). Consequently, this reduction of the rate at which the nascent chain could feed through the channel could, in turn, perturb the translocation step of protein synthesis and reduce the accessibility of the A site for eEF1A delivery of charged tRNA to the ribosome. Utilizing our understanding of the translation elongation factor class of G proteins, we have sought to examine the function of Hbs1p with regard to the process of protein synthesis. The homology of Hbs1p to known translation factors and a genetic relationship to components of the translation apparatus suggested that Hbs1p may provide an evolutionarily conserved function that facilitates the process of protein synthesis. In this work, we demonstrate that Hbs1p is a bona fide G protein important for efficient growth and protein synthesis under conditions of limiting translation initiation. The eEF1A-like domain of Hbs1p mediates its function and is sufficient for its interaction with Dom34p. Dom34p is also important for efficient growth and protein synthesis under conditions of limiting translation initiation, suggesting that these factors may function together as a complex to directly or indirectly facilitate protein synthesis.
MATERIALS AND METHODS
Strains, media, and growth assays.
E. coli strain DH5α was used for plasmid preparation, and BL21(DE3) was used for protein expression. S. cerevisiae strains used in these studies are listed in Table 1. Standard yeast genetic methods were employed (49). Yeast cells were grown in either yeast extract-peptone-dextrose (1% Bacto yeast extract, 2% peptone, and 2% dextrose) or defined synthetic complete media (C) supplemented with 2% dextrose as a carbon source unless otherwise noted. Yeast was transformed by the lithium acetate method (26). Growth was examined by streaking or spotting 10-fold serial dilutions of yeast cultures to an A600 of 0.5 on appropriate media, followed by incubation at 13, 24, 30, or 37°C. To assess the status of the general control response, strains were grown in C-His until saturated, diluted to an A600 of 0.5, spotted onto C-His supplemented with 40 mM leucine and either 0, 10, 20, or 50 mM 3-aminotriazole (3-AT) (17), and grown for 3 days at 30°C.
TABLE 1.
S. cerevisiae strains used
| Strain | Genotype | Source or reference |
|---|---|---|
| PJ43-2B | MATα ade2-1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 | P. James (unpublished) |
| CP1 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 | This study |
| CP5 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 hbs1::LEU2 | This study |
| CP60 | MATa/MATα ade2-1/ade2-1 ade3-Δ1/ade3-Δ1 ura3-1/ura3-1 leu2-3,112/leu2-3,112 lys2-Δ2/LYS2 trp1-1/trp1-1 met2-Δ1/MET2 his3-11,15/his3-11,15 can1-100/can1-100 hbs1::LEU2/HBS1 | This study |
| CP61 | MATα ade2-1 ade3-Δ1 ura3-1 LYS2 leu2-3,112 trp1-1 met2-Δ1 his3-11,15 can1-100 hbs1::LEU2 | This study |
| CP62 | MATα ade2-1 ade3-Δ1 ura3-1 LYS2 leu2-3,112 trp1-1 met2-Δ1 his3-11,15 can1-100 HBS1 | This study |
| CP50 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 hbs1::LEU2 rps30A [pRS316-ADE3-HBS1] | This study |
| CP57 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 hbs1::LEU2 rps17B [pRS316-ADE3-HBS1] | This study |
| CP76 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 hbs1::LEU2 rps14A [pRS316-ADE3-HBS1] | This study |
| CP182 | MATα ade2-1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 rps14A::URA3 | This study |
| CP180 | MATα ade2-1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 his3-11,15 hbs1::LEU2 rps14A::URA3 | This study |
| CP177 | MATα ade2-1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 met2-Δ1 his3-11,15 rps14B::LEU2 | This study |
| CP194 | MATα ade2-1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 met2-Δ1 his3-11,15 hbs1::LEU2 rps14B::LEU2 | This study |
| TKY422 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 rps17b::HIS3 | This study |
| TKY423 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 hbs1::LEU2 rps17b::HIS3 | This study |
| TKY514 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 hbs1::LEU2 dom34::LEU2 | This study |
| TKY522 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 dom34::LEU2 rps30A::HIS3 | This study |
| TKY525 | MATα ade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 dom34::LEU2 | This study |
| TKY535 | MATα ade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 hbs1::LEU2 dom34::LEU2 rps30A::HIS3 | This study |
| TKY608 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 rps30A::HIS3 | This study |
| TKY609 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2his3-11,15 can1-100 hbs1::LEU2 rps30A::HIS3 | This study |
| TKY610 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 rps30b::HIS3 | This study |
| TKY611 | MATaade2-1 ade3-Δ1 ura3-1 lys2-Δ2 leu2-3,112 trp1-1 MET2 his3-11,15 can1-100 hbs1::LEU2 rps30b::HIS3 | This study |
| pJ69-4A | MATatrp1-901 leu2-3,112 ura3-52, his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | 27 |
Yeast strain construction.
HBS1 was disrupted in CP1 by one-step disruption using a HindIII fragment (38) carrying HBS1 disrupted by insertion of LEU2 into its unique EcoRI site producing CP5. In other strain backgrounds, HBS1 does not suppress the Cs− defect of an ssb1Δssb2Δ strain, indicating that the original strain may have an additional mutation (38). The rps14A::URA3 and rps14B::LEU2 disruptions in CP1 and CP5 were generated by one-step disruption as previously described (41). A dom34::LEU2 strain (TKY525) was created as previously described by using pTK3 (16). Conversion of the his3-to-11,15-null allele was carried out by transformation of linearized BamHI-XhoI fragments of HIS3 digested from pJJ217 (29) to produce HIS3 strains TKY600, -601, and -602 for all GCN2p experiments.
PCR-based gene disruptions of RPS30A, RPS30B, and RPS17B were generated in CP1 and CP5 strains. The HIS3 gene was amplified from vector pJJ217 utilizing the primers specified below and was selected on C-His. PCR-based gene disruptions utilized the following primers: rps30a5′ (5′-GCC ACT GTA ATA ATC TTC CAT ATC CCC ATA CAA AAA CTA CGC AAA TAT GGG GAA GTC ATA ACA CAG TCC-3′) and rps30a3′ (5′-GCT CCT TTT CCA CAA CTG TTA ATT TTC TTA TTG GAC GGA TGG ACC CGT ATA GAA TGA TGC ATT ACC-3′) for rps30A::HIS3, rps30b5′ (5′-GCT CGA TTT TCC TCC AAT TAG TTT TGT CCC GTA TTC ACC ATG GGA AGT CAT AAC ACA GTC C-3′) and rps30b3′ (5′-GCA GTT CGA ATC AAA AAC CCA TGT AAA AAG GGT AAA TAT TTC CCG TAT AGA ATG ATG CAT TAC C-3′) for rps30B::HIS3, and 5′rp51BHIS (5′-GCA GAT AAA AAT GGT ACG TAC CAC GAG ATG TTG ATG AAG CCG GAT ATG ATG GGG AAG TCA TAA CAC ATG CC-3′) and 3′rp51BHIS (5′-CGT CTG TCT CTT TGA GCG GAA ACG TTG ATG ACA GAT AAT GGC AAC TTC AAA CCC GTA TAG AAT GAT GC-3′) for rps17B::HIS3.
Disruptions were confirmed by Southern blot analysis utilizing the following DNA fragments as probes: the 1.4-kb BamHI-XhoI fragment of HIS3 digested from pJJ217; the 1.7-kb SalI-ScaI fragment of DOM34 from B462 (16); the 1.4-kb SspI-SspI LEU2 fragment digested from pRS425; the 1.0-kb PstI-ClaI RPS30A fragment from pTKB356; the 2.3-kb XhoI-EcoRI RPS17B fragment digested from pTKB349; and the 2.1-kb ScaI fragment of HBS1 from pTKB330. Probes were prepared as per the Gene Images Random Prime Labeling Module (Amersham) and were detected with the CDP Star Gene Images Southern Kit (Amersham).
Additional strains were constructed by mating and sporulating confirmed deletion strains as listed in Table 1. When required, mating-type switching was performed by introduction of the HO gene under a GAL1-inducible promoter (pTKB321). Strains were grown on C-Ura plus galactose overnight, streaked to yeast-peptone-dextrose, checked for loss of the plasmid on 5-fluoroorotic acid (5-FOA) (7), and confirmed by mating assay.
Identification of mutations synthetic with hbs1Δ.
Mutations synthetic with the loss of Hbs1p were identified using an ade2 ade3 color-sectoring assay (6). An hbs1Δ strain carrying pRS316-ADE3-HBS1 was grown overnight at 30°C in C-Ura to an optical density at 600 nm of 0.7. Cells were washed twice with water, diluted, and plated at approximately 650 cells/plate. Approximately 65,000 cells were UV irradiated to 15 or 35% survival and were incubated at 25°C for 8 to 10 days. Cells from colonies that lacked obvious sectoring (Sect−) were restreaked onto yeast-peptone-dextrose. False positives were eliminated using 5-FOA counterscreening to identify cells that had lost the URA3-ADE3-HBS1 plasmid (7). Specificity for the HBS1 gene was demonstrated by introduction of a pRS314-HBS1 plasmid, resulting in Sect+ and resistance to FOA, while cells transformed with the pRS314-hbs1::LEU2 plasmid were of the Sect− phenotype. Three candidate strains passed all tests, and the genes that complemented the Sect− and Cs− defects were identified using a yeast genomic library (45).
Assays of translational status of yeast strains.
Yeast polyribosome profile analysis was performed as previously described (4) with the following modifications. Yeast cultures were grown at 30°C, divided, and extracted or shifted to 13°C for 2 h and extracted. Cultures were extracted at an A600 of 0.8 to 0.9. Thirty-five A260 units of extract were layered on a 35-ml, 7 to 47% sucrose gradient and were centrifuged for 4 h at 27,000 rpm in an SW28 rotor. The A254 was monitored and was recorded using a Model 185 density gradient fractionator (ISCO, Inc., Lincoln, Nebr.). The location of ribosomal subunits and polyribosomes was determined by Western blot analysis of fractions across a gradient with an antibody to S. cerevisiae rpl5p (Jonathan Warner, Albert Einstein College of Medicine). In vivo [35S]methionine incorporation was performed as previously described (11). rRNA processing was monitored essentially as described earlier (32), with total RNA extracted as described earlier (47).
Preparation of GST-HBS1 alleles.
Recombinant DNA techniques were performed as described earlier (46). Restriction endonucleases and DNA- modifying enzymes were obtained from Roche Biochemicals or Gibco BRL. A series of CUP1 promoter yeast expression constructs encoding N-terminal, glutathione transferase (GST)-tagged, full-length and truncated forms of Hbs1p were prepared utilizing vector pCBGST1 (5). Plasmid pTKB370 consists of the full-length HBS1 coding sequence amplified from plasmid pTKB330 using primers 5′HBSNCO (5′-GGG AAT TCC ATG GCT TAC AGT GAC TAC AGC GAT GGA GCA GAC G-3′) and 3′HBSNCO (5′-GGG AAT TCC ATG GCC CTT CTA CTG AGT TAT TTC GGA TAT TTT ACC-3′) and cloned into pCBGST1. The HBS1 sequence corresponding to amino acids (aa) 1 to 160 was PCR amplified from pTKB330 using primers 5′HBSNCO and HBS1Nterm (5′-GCA GCA GCT GCT AAA ATG CAG AA TAT CAT GAG G-3′) and was cloned into pCBGST1 to create pTKB385. The HBS1 sequence corresponding to aa 161 to 611 was PCR amplified from pTKB330 using 5′primer HBS1CTERM (5′-GGA ACC ATG GTT AAA TCT GCC TTA CC) and 3′ primer 3′HBSNCO (see above) and was cloned into pCBGST1 to create pTKB389. Expression of the tagged proteins in yeast was confirmed by Western blot analysis using polyclonal anti-GST antibodies (Nancy Walworth, Robert Wood Johnson Medical School) and relying on the enhanced chemiluminescence immunoblot analysis kit (Amersham). The addition of exogenous copper sulfate to C-Trp media was not necessary to see expression of the GST-Hbs1 proteins.
Mutations in GST-HBS1 were created in pTKB370 utilizing the PCR-based Quik Change Kit (Stratagene). pTKB383 encoding the V176G mutation was created using primers 5′V176G (5′-CCT CAC TTA AGT TTT GTT GTT CTT GGC CAT GGT GAT GCG GGA AAA TC-3′) and 3′V176G (5′-GAT TTT CCC GCA TCA CCA TGG CCA AGA ACA ACA AAA CTT AAG TGA GG-3′). pTKB384 encoding the H255E mutation was created using primers 5′H255E (5′-GCA AAT TTT ACT ATT GTG GAT GCC CCG GGC GAG AGA GAT TTT GTT CC-3′) and 3′H255E (5′-GGA ACA AAA TCT CTC TCG CCC GGG GCA TCC ACA ATA GTA AAA TTT GC-3′). pTKB382 encoding the N313D mutation was created using primers 5′N313D (5′-CTG ATT ATT GCC ATG GAT AAG ATG GAT AAT GTT GAC TGG TCC C-3′) and 3′N313D (5′-GGG ACC AGT CAA CAT TAT CCA TCT TAT CCA TGG CAA TAA TCA G-3′). All mutations were confirmed by restriction digest and sequence analysis. Expression of the tagged proteins in yeast was confirmed by Western blot analysis as above.
Preparation of two-hybrid HBS1 construct.
To express a Gal4 DNA binding domain-Hbs1 fusion protein (Gal4DBD-Hbs1p), the HBS1 coding sequence was amplified from pTKB330 utilizing primers 5′HBSNCO (5′-GGG AAT TCC ATG GCT TAC AGT GAC TAC AGC GAT GGA GCA GAC G-3′) and 3′HBSNCO (5′-GGG AAT TCC ATG GCC CTT CTA CTG AGT TAT TTC GGA TAT TTT ACC-3′) and was subcloned into pAS1CYH2 to create pTKB420. Expression of Gal4DBD-Hbs1p in pJ69-4A was confirmed by Western blot analysis with an affinity-purified polyclonal antibody to Hbs1p.
Preparation of full-length and truncated DOM34 two-hybrid constructs.
Truncations of Dom34p were prepared as N-terminal hemagglutinin (HA) epitope-tagged Gal4 activation domain fusion proteins for use in the two-hybrid system. The DOM34 sequence corresponding to aa 1 to 268 was PCR amplified from pTKB465 using primers DOM5′BAM (5′-CGG GAT CCG AAT GAA GGT TAT TAG TCT GAA AAA GG-3′) and DOMXHO268 (5′-GGC GTG GAG CTA ATT TTT CAA AAC TTC ATT AAT GCC-3′) and was cloned into pACTII, creating pTKB570. The DOM34 sequence corresponding to aa 1 to 348 was PCR amplified from pTKB465 using primers DOM5′BAM (see above) and DOMXHO348 (5′-GGC CTC GAG CTA ATT GCT CTC TAC GGA GTC C-3′) and was cloned into pACTII to create pTKB571. Expression of the Gal4ACT-Dom34p truncations in pJ69-4A was confirmed by Western blot analysis utilizing monoclonal anti-HA and anti-Gal4ACT antibodies (Clontech).
Two-hybrid analysis.
Two-hybrid analysis was performed as previously described (23, 27). Expression of Gal4DBD-Hbs1p in two-hybrid screen strain pJ69-4A (27) did not activate transcription of either the GAL1-HIS3 or GAL2-ADE2 reporter genes either alone or in the presence of Gal4ACT-Snf4p fusion protein. pJ69-4A expressing Gal4DBD-Hbs1p was transformed with a Gal4ACT fusion cDNA library, and 10,000 colonies were screened. Cells were directly plated onto C-Trp-Leu-Ade, and 91 candidate colonies were isolated, 83 of which also grew on C-Trp-Leu-His + 1 mM 3-AT. cDNA library plasmids were extracted, shuttled through E. coli, and reintroduced into pJ69-4A expressing either Gal4DBD-Hbs1p (pTKB420) or pJ69-4A expressing the nonspecific control Gal4DBD-Snf1p. One library plasmid (pTKB460) that passed all controls and that activated transcription of the GAL2-ADE2 and GAL1-HIS3 reporter in a Gal4DBD-Hbs1p-dependent manner was sequenced and identified as DOM34. An internal primer, DOM34INT (5′GGG ATA CAC GTA GTC AAG GG-3′), was used to sequence over the GAL4ACT-DOM34 junction, and Western blot analysis utilizing monoclonal antibodies to the GAL4 activation domain (Clontech) confirmed the production of the Gal4ACT-Dom34 fusion protein.
Purification of Hbs1p from E. coli.
The HBS1 coding sequence was PCR amplified from pTKB330 using primers GEXHBS5′ (5′-GAG CGG ATC CAT GGC TTA CAG TGA CTA CAG C-3′) and GEXHBS3′ (5′-GAG CCT CGA GCT ATG AGT TAT TCG GAT ATT TTA CC3′) and was cloned into pGEX-6P-1 (Pharmacia) to produce pTKB519. Strain BL21(DE3) was transformed with pTKB519, grown at 37°C in 2× YT plus 100 μg of ampicillin/ml to an optical density at 600 nm of 0.6, and induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (0.1 mM) for 3 h. Cells were collected and washed once in 1× phosphate-buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 [pH 7.3]), and pellets were frozen in liquid nitrogen. Cells were thawed and resuspended in 50 μl of PBS with 0.5 mM phenylmethylsulfonyl fluoride (PMSF) per ml of culture and were lysed by sonication. Triton X-100 was added to a final concentration of 1%, and lysates were mixed for 30 min at 4°C and centrifuged at 12,000 × g for 20 min at 4°C. Cleared sonicate was passed over a column of 1 ml per 100 ml of sonicate glutathione Sepharose 4B and washed three times with 10 bed volumes of 1× PBS, and the protein was eluted in 50 mM Tris, pH 8.0, and 10 mM glutathione. Eluates were collected and dialyzed overnight against buffer A-50 (20 mM Tris, pH 7.5, 0.1 mM EDTA, 1 mM dithiothreitol (DTT), 50 mM KCl, 15% glycerol, and 0.2 mM PMSF). Purified Hbs1 protein was monitored by Western blot analysis utilizing affinity-purified anti-Hbs1p antibodies.
UV cross-linking of Hbs1p to GTP.
UV cross-linking of [α-32P]GTP to purified protein was performed as previously described (42). Briefly, 20-μl reactions in buffer B (20 mM HEPES, pH 7.5, 100 mM potassium acetate, 2 mM DTT, and 2.5 mM magnesium acetate) containing 3.2 μg of purified GST-Hbs1p from E. coli and 100 μCi of [α-32P]GTP and 33 pmol of cold GTP were incubated for 7 min at 30°C and irradiated on ice for 30 min. Competition assays were performed by introduction of increasing concentrations of unlabeled GTP (330 or 3,300 pmol) or unlabeled ATP (3,300 pmol) prior to incubation and irradiation. Samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, and detected by a Typhoon phosphorimager.
GST pulldown rxperiments.
Yeast extracts were prepared by glass bead lysis in TEDG buffer (10 mM Tris HCl, pH 7.4, 2 mM EDTA, 5 mM DTT, 50 mM KCl, and 1 mM PMSF) from strains expressing triple-HA-tagged Dom34p (16) and either a GST-tagged, full-length or truncated version of Hbs1p (pTKB370, pTKB385, and pTKB389) or GST alone (pCBGST1). Two-hundred-microliter reactions containing 20 μg of total protein and 20 μl of a 50% glutathione Sepharose 4B slurry (Sigma) in KETN150 buffer (150 mM KCl, 1 mM EDTA, 20 mM Tris, pH 8.0, 0.5% Nonidet, and 1 mM PMSF) were mixed at 4°C for 1 h. Beads were washed three times with KETN buffer with either 150 or 300 mM KCl. Samples were resolved by SDS-PAGE, and Western analysis was carried out with anti-HA or anti-GST antibodies as described above.
RESULTS
Hbs1p is not redundant with other G proteins involved in protein synthesis.
To address whether Hbs1p might share an overlapping function with eEF1A, we examined whether overexpression of HBS1 from a 2μm plasmid could suppress the conditional growth defects associated with mutant alleles of either eEF1A (TEF2-4) (20) or its guanine nucleotide exchange factor eEF1Bα (tef5-7), which slows global translation (12). Overexpression of HBS1 did not result in suppression, nor did it synthetically enhance the growth defects in either mutant strain (data not shown).
In addition to similarity to eEF1A, Hbs1p also shares similar homology to eRF3, which functions in translation termination. Previously, it was demonstrated that overexpression of HBS1 failed to complement the conditional growth defects or influence the efficiency of nonsense suppression of a strain expressing a mutant form of eRF3 (sup35-57) (53). In addition, Hbs1p failed to interact with the translation termination factor eRF1 or eRF3 (53). In yeast, the translation termination factors interact with additional factors involved in the process of nonsense-mediated mRNA decay (reviewed in reference 15). Hbs1p failed to interact with Upf1p, Upf2p, Upf3p, Mof2p, Xrn1p, or Hrp1p by two-hybrid analysis (data not shown; constructs provided by Stuart Peltz, Robert Wood Johnson Medical School). These results suggest that Hbs1p likely functions within the cell in a distinct manner from eEF1A and eRF3.
The loss of HBS1 produces synthetic slow growth with null alleles of genes encoding 40S ribosomal subunit proteins.
Unlike other elongation factor G proteins, including eEF1A and eRF3, Hbs1p is not essential for viability in S. cerevisiae and deletion of HBS1 confers no obvious phenotypic effect (38). In order to gain a better insight into the function of Hbs1p, a synthetic lethal screen was performed with an HBS1 null. By using a combination of the red/white (ade2/ade3) sectoring assay (6) and counterscreening on 5-FOA (7), three Sect− (red) candidates were isolated after UV mutagenesis (CP50, CP57, and CP76). All three candidates depended upon an HBS1-containing plasmid for wild-type growth and passed all screens for false positives as described in Materials and Methods. Each candidate, upon 5-FOA counterscreening, was viable but demonstrated dramatic slow-growth and Cs− phenotypes. Thus, all three mutants require HBS1 for efficient growth but not for viability. The three mutations comprised three complementation groups and were all recessive (data not shown).
In order to identify the gene responsible for the synthetic slow-growth and Cs−phenotypes, each of the mutant strains was transformed with a genomic S. cerevisiae library. All clones were selected based on their ability to rescue the Cs−, slow-growth, and Sect− phenotypes. The ribosomal protein genes RPS30A, RPS17B, and RPS14A were determined to be sufficient to restore the growth rate, Cs+, and Sect+ phenotypes of the CP50, CP57, and CP76 mutant strains, respectively.
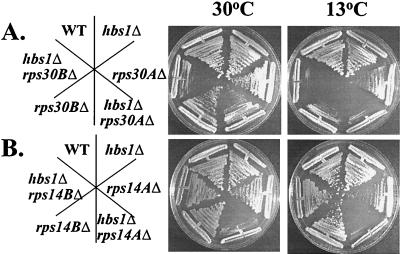
RPS14A, RPS17B, and RPS30A are each one of a pair of duplicated genes that encode a specific, small-ribosomal-subunit protein. Expression of at least one of the duplicated genes is required for viability (1, 41, 54). To determine if the mutants harbor null alleles of the rp genes RPS14A, RPS17B, and RPS30A, individual deletion strains were constructed in isogenic wild-type and hbs1Δ strains (Table 1). As shown in Fig. 1, a strain harboring an HBS1-null allele does not demonstrate a growth defect and has a doubling time of 1.8 h at 30°C, essentially the same as that for a wild-type strain (1.7 h). Strains harboring a deletion of either the RPS30A or RPS14A showed no change in doubling time from a wild-type strain, while a strain lacking RPS30B conferred slightly slower growth at 30°C (1.9 h). A strain lacking RPS30A or RPS14A showed a slight Cs− phenotype, while loss of RPS30B was clearly Cs− (Fig. 1). A reduction in ribosomal protein gene dosage has been previously demonstrated to confer slight slow-growth and Cs− phenotypes in some cases (1, 41). Deletion of HBS1 served to exaggerate both the slow growth at 30°C of strain harboring null alleles of RPS30A (2.6 h), RPS30B (2.8 h), or RPS14A (2.7 h) and Cs− defects of strain harboring null alleles of RPS30A, RPS30B, or RPS14A (Fig. 1), and comparable results were obtained with an rps17B hbs1Δ strain (data not shown).
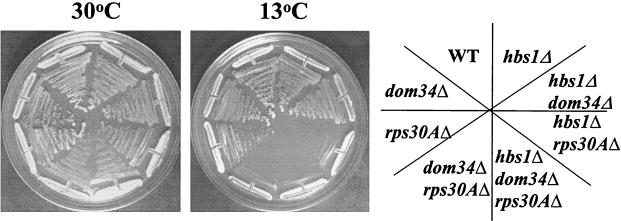
FIG. 1.
Deletion of HBS1 synthetically enhances the growth defects of strains harboring ribosomal protein gene deletions. Wild-type (WT) (CP1) and hbs1Δ (CP5) were streaked with rps30AΔ (TKY516), hbs1Δ rps30AΔ (TKY517), rps30BΔ (TKY610), and hbs1Δ rps30BΔ (TKY611) (A) or rps14AΔ (CP182), hbs1Δ rps14AΔ (CP180), rps14BΔ (CP177), and hbs1Δ rps14BΔ (CP194) (B). Shown are streaks of yeast strains on rich media grown at either 30°C (2 days) or 13°C (7 days).
Generation of an rps14BΔ hbs1Δ strain revealed that HBS1 does not share a synthetic relationship with RPS14B (Fig. 1). RPS14A and RPS14B genes are expressed at approximately a 10:1 ratio within the cell (41). Deletion of RPS14A leads to increased expression of RPS14B; however, a deficiency of 40S subunits remains. The inactivation of RPS14B, however, does not create a 40S subunit deficiency or a slow-growth defect (41). The different results with RPS14A- and RPS14B-null alleles suggested that the genetic relationship between HBS1 and the small-ribosomal-protein genes may not reflect specificity for the ribosomal protein gene product; instead, it may reflect the relative expression of the ribosomal protein gene or the degree to which inactivation of the gene produces a 40S subunit deficiency. To analyze the genetic relationship between HBS1 and the small ribosomal proteins, subsequent analysis is limited to the RPS30A gene, unless otherwise stated.
Absence of HBS1 does not alter ribosomal subunit levels or rRNA processing.
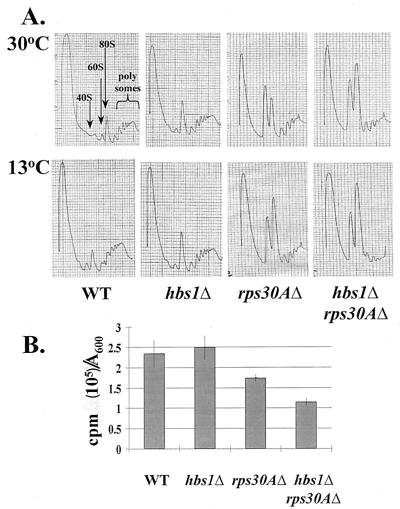
Reductions in gene dosage of ribosomal proteins have the potential to alter the efficiency of rRNA processing events, thus leading to growth defects (54). Strains deficient in HBS1 failed to demonstrate alterations in 35S pre-rRNA processing or the presence of aberrant processing intermediates as monitored by pulse-chase labeling of rRNA (Materials and Methods, data not shown). rRNA processing was slightly delayed in hbs1Δ rps30AΔ relative to that for rps30AΔ; however, the effect was slight and was observed after 2-h shifts to the nonpermissive 13°C temperature prior to RNA extraction (data not shown). Polyribosome profile analysis demonstrated that HBS1-null strains do not show alterations in the profile or levels of free 40S or 60S ribosomal subunits, 80S monoribosomes, and polyribosomes relative to the wild-type strain (Fig. 2A). These results suggest that Hbs1p does not play a critical role in the process of ribosome biogenesis and that the defects associated with the hbs1Δ rps30AΔ strain may instead arise from a defect in protein synthesis.
FIG. 2.
Deletion of HBS1 synthetically enhances the translation defects of the rps30AΔ and rpl6BΔ strains. (A) Wild-type (WT) yeast (CP1) or cells lacking hbs1Δ (CP5), rps30AΔ (TKY516), and hbs1Δ rps30AΔ (TKY517) were grown at 30°C to mid-log phase. Cells were extracted at 30°C or shifted to 13°C for 2 h prior to extraction. Polyribosome content (polysomes) was analyzed on a 7 to 47% sucrose gradient, and subunits and ribosomes are located as indicated in the first panel. (B) Incorporation of [35S]methionine is reduced in rps30AΔ and hbs1Δ rps30AΔ strains. Wild-type yeast (CP1) or cells lacking hbs1Δ (CP5), rps30AΔ (TKY516), and hbs1Δ rps30AΔ (TKY517) were grown at 30°C in media lacking methionine to an A600 of 0.5 to 0.7. Fifty micromolar cold methionine and 1μCi of [35S]methionine/ml were added to each culture. A600 and cold trichloroacetic acid precipitable radioactivity were monitored at the zero-time point and at 15-min intervals for a total of 45 min. The data presented represent the incorporation after 30 min.
Hbs1p is important for efficient translation under conditions of limiting ribosomal subunits.
To determine whether the loss of the Hbs1 protein altered the rate of protein synthesis in vivo, the efficiency of protein synthesis in the hbs1Δ strain was examined relative to that for the wild-type strain. Incorporation of [35S]methionine into newly synthesized proteins was monitored at 15-min intervals over a 45-min period at the permissive temperature of 30°C. After 30 min of incubation, the rates of [35S]methionine incorporation for the wild-type and hbs1Δ mutant were similar (Fig. 2B); however, incorporation by the rps30AΔ strain was reduced approximately 35% relative to that by the wild type. Furthermore, the rate of [35S]methionine incorporation during global protein synthesis was reduced to approximately 50% in the hbs1Δ rps30AΔ mutant (Fig. 2B). These results suggest that the global protein synthesis defect observed for the rps30A-null strain is synthetically enhanced by the loss of Hbs1p function. The translation defects appear to correlate with the synthetic growth defects observed for these strains.
To address at which phase during translation the loss of Hbs1p function impairs the rps30AΔ strain, the polyribosome content of extracts prepared from the wild-type, single-null strains, or double-null strains was examined by sucrose gradient centrifugation. Samples were analyzed before and after the cells were shifted from the permissive (30°C) to the restrictive temperature (13°C) for 2 h. No significant alterations in profiles between the wild-type and hbs1Δ strain were noted following a shift to 13°C, as mentioned above (Fig. 2A). The rps30AΔ strain demonstrated a reduced level of 40S ribosomal subunits, a concomitant increase in free 60S subunits, and a slight reduction in heavier polyribosomes (Fig. 2A and data not shown). These defects are typical of a small- ribosomal-protein deletion strain profile (2) and are enhanced upon a shift to the nonpermissive temperature (Fig. 2A). The hbs1Δ rps30AΔ strain demonstrated a pronounced increase in the size of the 80S peak, at the expense of heavier polyribosomes, relative to the rps30AΔ profile. This effect was more apparent for strains grown at 13°C prior to extraction (Fig. 2A). The decrease in height of the heavier polyribosome peaks indicates that it is most likely translation initiation which is reduced to a greater extent in the double-null strain, rather than translation elongation.
The C terminus of Hbs1p is sufficient for Hbs1p complementation in vivo.
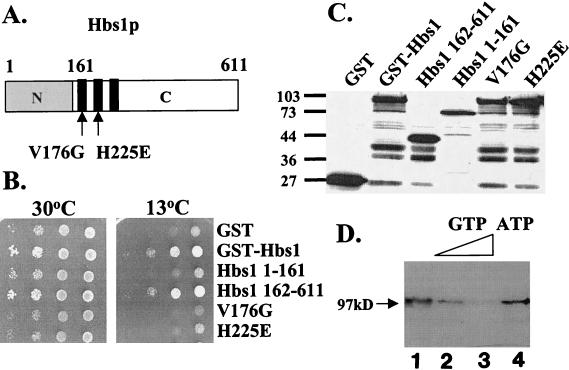
To address the domain of Hbs1p required for its activity in vivo, a series of GST-tagged, full-length and truncated forms of Hbs1p were expressed in the hbs1Δ rps30AΔ strain and their ability to complement the growth defects of the strain was determined. Hbs1p contains two domains: a 161-aa N-terminal domain, which has no obvious motifs or significant homology to known proteins, and a 450-aa C-terminal eEF1A-like domain, which contains the GTP binding consensus elements (Fig. 3A). As shown in Fig. 3B, the full-length protein and C-terminal domain (aa 162 to 611) of Hbs1p complemented the hbs1Δ rps30AΔ growth defect. Complementation was not observed with the N-terminal domain of Hbs1p (aa 1 to 161), even though all proteins are stably produced (Fig. 3C). Thus, expression of the eEF1A-like portion of Hbs1p is sufficient to restore Hbs1p function in strains with reduced RPS30A gene dosage.
FIG. 3.
Hbs1p is a GTP binding protein and requires intact GTP binding motifs for function in vivo. (A) Schematic diagram of the Hbs1p protein. Hbs1p contains a divergent N terminus (grey) and an eEF1A-like C terminus (white). The black blocks depict the three main GTP binding consensus elements, and point mutations are indicated with arrows. (B) A strain lacking hbs1Δ rps30AΔ (TKY517) was transformed with plasmids expressing wild-type, truncated, and mutant GST-tagged forms of Hbs1p from the CUP1 promoter. Transformants were grown on C-Trp to mid-log phase, and equal numbers of cells were spotted as 10-fold serial dilutions on C-Trp and grown at 30°C (2 days) or 13°C (7 days). (C) Western blot analysis of the strains from panel B. Equal amounts of total cell extract were separated by SDS-PAGE and probed with an antibody to GST. Migration of molecular weight standards is indicated in kilodaltons. (D) GST-Hbs1p purified from E. coli was UV cross-linked to [α-32P]GTP (lane 1). Nucleotide specificity of the cross-link signals was determined by competition with nonlabeled nucleotide using 10- and 100-fold excesses of cold GTP (lanes 2 and 3) and a 100-fold excess of ATP (lane 4). The 97-kDa protein corresponds to GST-Hbs1p.
Hbs1p is a GTP binding protein.
To understand the biochemical function of Hbs1p, the prediction that it would be capable of binding GTP was investigated by UV cross-linking of radiolabeled GTP to E. coli-purified, GST-tagged-Hbs1p. As shown in Fig. 3D, lane 1, cross-linking of the recombinant Hbs1p was detected. In order to investigate the specificity of the UV cross-link, competition experiments with nonlabeled nucleoside triphosphates up to 100-fold-molar excess over the labeled GTP were performed. The recombinant Hbs1p was determined to bind GTP specifically, as increasing concentrations of cold GTP reduced the cross-link signals significantly, while a 100-fold excess of ATP did not (Fig. 3D).
Single amino acid substitutions in the conserved GTP binding motifs of Hbs1p are sufficient to abolish Hbs1p function in vivo.
To test whether a wild-type GTP binding site was critical for Hbs1p function in vivo, point mutations based on prior work in other G proteins were introduced within the highly conserved guanine nucleotide binding motifs GXXXXGKS/T and DXXG of GST-tagged, full-length Hbs1p. The V176G mutation in the G174HVDAGKST182 motif or the H255E mutation of the D250APGH255 motif, both of which are critical for the interaction of the G protein with the α and β phosphate residues of GTP and GDP, resulted in proteins that failed to complement the hbs1Δ rps30AΔ growth defect (Fig. 3B). The inability of mutant alleles of Hbs1p to functionally complement does not arise from reduced expression levels (Fig. 3C). The inability of Hbs1p-V176G or Hbs1p-H255E to complement HBS1 function in vivo is consistent with the effects of the corresponding mutations resulting in reducing the activity of various G proteins, including EF1A, Ras2, and E. coli and human IF2/eIF5B (10, 35). Thus, Hbs1p is a bona fide G protein, and the presence of a wild-type GTP binding motif in Hbs1p is required for its function.
Two-hybrid analysis identifies a potential Hbs1p-interacting protein.
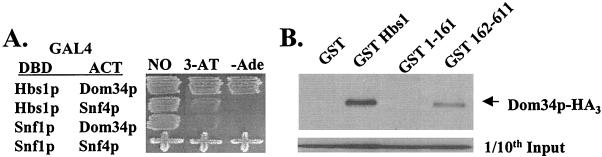
In order to identify factors that interact with Hbs1p in vivo, the GAL4-based two-hybrid screen was employed (23). The Gal4DBD-Hbs1p fusion protein was constructed, determined to be stably expressed (data not shown), and found unable to nonspecifically stimulate expression of the integrated reporter genes (Fig. 4A). Screening of 104 transformants of a yeast Gal activation domain cDNA library led to the isolation of a single plasmid that stimulated expression from the GAL2-ADE2 and GAL1-HIS3 integrated reporters in a Gal4DBD-Hbs1p fusion protein-specific and -dependent manner (Fig. 4A). Sequence analysis identified the cDNA library clone as encoding a truncated form of Dom34p (aa 20 to 386) expressed in frame as a fusion with Gal4ACT. A full-length Gal4ACT-Dom34p fusion was prepared, and confirmed full-length Gal4ACT-Dom34p stimulated expression from the integrated reporters in a Gal4DBD-Hbs1p-specific and -dependent manner (data not shown).
FIG. 4.
Hbs1p interacts with Dom34p. (A) Gal4DBD-Hbs1p and Gal4ACT-Dom34p fusion proteins were assayed for interaction in two-hybrid reporter strain pJ69-4A on C-Leu-Trp-His + 1 mM 3-AT and on C-Leu-Trp-Ade plates. Gal4DBD-Snf1p and Gal4ACT-Snf4p serve as positive control for an interaction and as negative controls for the interaction with the Hbs1p and Dom34p fusions. (B) Twenty micrograms of protein extracts was prepared from yeast strain TKY514 expressing Dom34p-triple-HA-tag (HA3) and either the full-length, aa-1-to-161, or aa-162-to-611 form of GST-tagged Hbs1p, incubated with 20 μl of 50% glutathione Sepharose 4B slurry, washed three times with 150 mM KCl buffer, and analyzed by Western blotting.
Dom34p interacts with the functional domain of Hbs1p in vivo.
To further investigate the potential interaction between Hbs1p and Dom34p, GST pulldown experiments were performed utilizing yeast extracts of strains expressing full-length GST-Hbs1p fusion protein and triple-HA-tagged, full-length Dom34p. As shown in Fig. 4B, immobilized GST-Hbs1p but not GST alone (Fig. 4B, lane GST) bound full-length Dom34p in the presence of 150 mM KCl. This result supports the two-hybrid data indicating that Hbs1p and Dom34p interact in vivo. To further define the interaction between the two proteins, truncated forms of GST-tagged Hbs1p were tested for their ability to bind Dom34p (Fig. 4B). Expression of the C domain of Hbs1p (aa 162 to 611) of Hbs1p was sufficient for interaction with Dom34p, while expression of the N domain (aa 1 to 161) was not (Fig. 4B).
Loss of DOM34 shows synthetic slow growth with a RPS30A-null allele.
In order to determine if a genetic relationship between Dom34p and Hbs1p exists, a DOM34-null allele was first analyzed in the wild-type (CP1) strain background. Consistent with previous reports, the dom34Δ strain was viable (16). However, the dom34Δ strain did not demonstrate the dramatic growth and Cs− defects previously reported for a dom34Δ strain (16). These effects likely reflect a strain-specific phenomenon, as a dom34Δ strain obtained from the Saccharomyces Genome Deletion Project (Research Genetics, Inc.) also did not display any phenotypic growth defects (data not shown). Construction of a dom34Δ hbs1Δ strain also failed to yield a strain with an obvious phenotypic growth effect (Fig. 5). Since the interaction of Dom34p and Hbs1p is mediated through the functional domain of Hbs1p sufficient for complementation of the hbs1Δ rps30AΔ growth defects, we sought to address the requirement for Dom34p under conditions of limiting 40S ribosomal subunits. The rps30AΔ dom34Δ strain exhibited slower growth than and increased Cs− relative to the rps30AΔ strain (Fig. 5). The phenotypic effects were consistent with the growth defects associated with the hbs1Δ rps30AΔ strain. DOM34 or RPS30A was able to complement the growth and Cs− defects of the dom34Δ rps30AΔ strain (data not shown).
FIG. 5.
Loss of DOM34, like HBS1, results in synthetic slow growth in combination with an RPS30A-null allele. Wild-type (WT) yeast (CP1) or cells lacking hbs1Δ (CP5), rps30AΔ (TKY516), hbs1Δ rps30AΔ (TKY517), dom34Δ (TKY525), hbs1Δ dom34Δ (TKY514), dom34Δ rps30AΔ (TKY522), and dom34Δ hbs1Δ rps30AΔ (TKY535) were streaked on yeast extract-peptone-dextrose and grown at either 30°C (2 days) or 13°C (7 days).
A triple-knockout strain was constructed to determine if deletion of HBS1 in the dom34Δ rps30AΔ strain serves to synthetically enhance the growth defects of the strain, with a doubling time of 2.9 h at 30°C similar to that of the hbs1Δ rps30AΔ strain. Lending genetic support to the biochemical evidence that Hbs1p and Dom34p function together as a complex, deletion of hbs1Δ failed to synthetically enhance the defects of the dom34Δ rps30AΔ strain (Fig. 5). Overexpression of HBS1 from either a low-copy-number (pTKB330) or high-copy-number (pTKB332) plasmid failed to suppress the growth defects of the dom34Δ rps30AΔ strain (data not shown). In addition, overexpression of DOM34 from low-copy-number (pTKB465 or -466) or high-copy-number (pTKB467) plasmids did not suppress or enhance the growth defects of the hbs1Δ rps30AΔ strain (data not shown).
Absence of Dom34p enhances the translation initiation defect of strains deficient in 40S subunits.
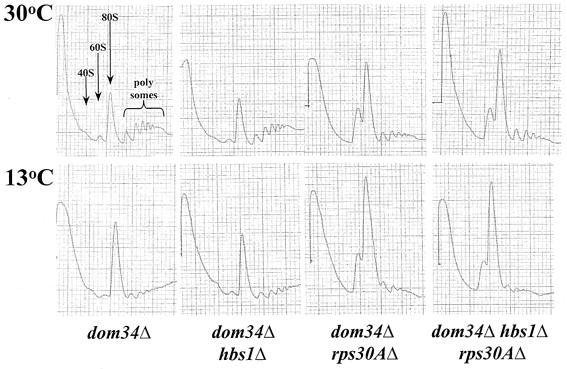
To ascertain whether the synthetic growth defects of the dom34Δ rps30AΔ strain arise from a similar translation initiation defect associated with the hbs1Δ rps30AΔ strain, the polyribosome content of extracts from the dom34Δ and dom34Δ rps30AΔ strains were examined by sucrose gradient centrifugation. Extracts were analyzed from strains grown at the permissive temperature (30°C) and were shifted to 13°C for 2 h. Wild-type and dom34Δ strains demonstrated similar profiles; however, upon a shift to the nonpermissive temperature, there was a slight increase in the 80S peak in the dom34Δ strain (Fig. 6). As demonstrated for Hbs1p, the loss of Dom34p served to exacerbate the translation initiation defect of rps30AΔ (Fig. 6). The effects were most apparent for extracts prepared from strains shifted to the nonpermissive temperature. The hbs1Δ dom34Δ rps30AΔ disruption strain demonstrated polyribosome profiles consistent with the hbs1Δ rps30AΔ and dom34Δ rps30AΔ strains (Fig. 6).
FIG. 6.
Polyribosome profiles from a dom34Δ rps30AΔ strain show similar 80S monoribosome increases to those for a hbs1Δ rps30AΔ strain. Wild-type yeast (CP1) or cells lacking dom34Δ (TKY525), hbs1Δ dom34Δ (TKY514), dom34Δ rps30AΔ (TKY522), and dom34Δ hbs1Δ rps30AΔ (TKY535) were grown at 30°C to mid-log phase. Cells were extracted or shifted to 13°C for 2 h prior to extraction. Polyribosome content was analyzed on a 7 to 47% sucrose gradient, and subunits and ribosomes are as indicated in the first panel.
Inhibition of translation initiation with a constitutively active GCN2 mutant allele confers a synthetic growth defect on strains lacking HBS1 or DOM34.
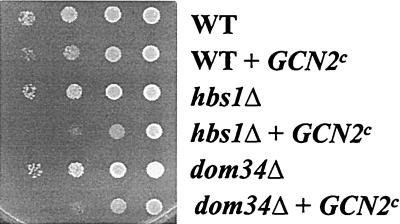
To investigate the model that Hbs1p and Dom34p are required under conditions of limiting translation initiation, we sought to reduce global translation independent of alteration of the concentration of ribosomal subunits within the cell. Constitutively active mutant alleles of GCN2p, the kinase that phosphorylates the α subunit of eIF2, result in slow-growth phenotypes and can serve to reduce cellular mRNA translation (44). To address whether the absence of Hbs1p or Dom34p exacerbates the growth defects associated with reduced eIF2-GTP-initiator-tRNAiMet levels, a constitutively active GCN2p kinase (GCN2p-cE803V) (21) was introduced into wild-type hbs1Δ and dom34Δ strains. Introduction of a plasmid encoding the constitutively active GCN2p but not an empty vector resulted in a slow-growth phenotype in wild-type cells, as previously reported (44) (Fig. 7). Expression of the activated kinase in the hbs1Δ or dom34Δ strain produced a greater growth defect than for the wild-type strain (Fig. 7). In contrast to the effects on the overall rate of protein synthesis, phosphorylation of eIF2 by the GCN2p kinase served to enhance the translation efficiency of GCN4 mRNA (reviewed in reference 24). Strains bearing deletions of HBS or DOM34 or either in combination with an RPS30A null allele did not demonstrate altered derepression of GCN4, as monitored by growth on media lacking histidine and supplemented with 3-AT, relative to the wild-type strain (data not shown), suggesting that Hbs1p or Dom34p does not function to alter eIF2-GTP-initiator-tRNAiMet availability (18). These results suggest that Hbs1p and Dom34p are important under conditions of limiting translation initiation.
FIG. 7.
Expression of a plasmid-borne GCN2c allele confers a synthetic growth defect in hbs1Δ and dom34Δ strains. Exponentially growing cultures of HIS3 strains TKY600 (wild type [WT]), TKY601 (hbs1Δ), and TKY602 (dom34Δ) transformed with a plasmid expressing GCN2p-cE803V were diluted to an A600 of 0.5. Ten microliters of 10-fold serial dilutions was spotted on C-Ura plates and incubated at 30°C for 3 days.
DISCUSSION
HBS1 encodes a previously uncharacterized member of the elongation factor class of G proteins in the yeast S. cerevisiae. The identification of putative HBS1 orthologs from mouse and human (53) and more recently from D. melanogaster, S. pombe, and C. albicans (25) suggests that HBS1 is evolutionarily conserved. As a member of the elongation factor class of G proteins, Hbs1p shares approximate 39% similarity and 28% identity with both eEF1A and eRF3. It is clear that Hbs1p is not functionally redundant with either related protein yet is still related to the protein synthetic apparatus of the cell.
We have examined the role of Hbs1p in the process of protein synthesis. Despite homology to known translation factors and a previous genetic interaction between HBS1 to the ribosome-associated Ssb chaperones, we have been unsuccessful in demonstrating a direct role for Hbs1p in protein synthesis. Instead, we have identified a new Hbs1p-interacting factor and identified mutations in additional genes that make HBS1 important for cell growth.
HBS1 and DOM34 share a genetic relationship with genes encoding protein components of the ribosome.
In this work we have identified new genetic relationships between HBS1, DOM34, and genes encoding protein components of the ribosome. Deletion of HBS1 serves to exaggerate the growth and translation defects associated with strains deficient in one of several different ribosomal protein genes. The synthetic growth defects are hypothesized to reflect the requirement for HBS1 under conditions of limiting or inefficient translation initiation. RPS14A and RPS14B genes have previously been demonstrated to be expressed at approximately a 10:1 ratio within the cell (41), and deletion of RPS14A results in increased expression of RPS14B through a posttranscriptional control mechanism where Rps14p binds both its own pre-mRNA and 18S rRNA (22). Even with the coordinate expression, loss of RPS14A but not inactivation of RPS14B creates a 40S subunit deficiency, reduced translation efficiency, and a slow-growth defect. We have demonstrated that the loss of HBS1 results in synthetic slow growth when combined with the loss of RPS14A but not of RPS14B. The proteins derived from RPS14A and RPS14B have been demonstrated to be functionally equivalent in their role in the ribosome, as one copy of either RPS14A or RPS14B is sufficient for viability (34). This result suggests that the genetic relationship between HBS1 and the small-ribosomal-protein genes may not reflect specificity for the ribosomal protein gene product; instead, it may reflect the relative expression of the ribosomal protein gene or the degree to which inactivation of the gene produces a 40S subunit deficiency. Reduction in the availability of eIF2-GTP-initiator-tRNAiMet, by way of the introduction of a constitutively active mutant allele of GCN2, served to confer a synthetic growth defect in the hbs1Δ strain. This provides additional support to the idea that it is the degree to which a 40S subunit deficiency is produced that allows for detection of the synthetic defect with the HBS1 null.
The loss of Hbs1p does not serve to alter rRNA processing or 40S subunit levels within the cell. While delayed processing of the 35S rRNA is observed after hbs1Δ rps30AΔ strains are shifted to the nonpermissive temperature, these effects are subtle and likely result from reduced translational capacity of the cells. Reduced translation can serve to feed back and slow ribosome biogenesis, as the necessary ribosomal proteins and ribosome-processing factors become limiting in the cell. Recently, van Hoof et al. demonstrated that deletion of HBS1 did not result in defects in the processing of the 5.8S rRNA, U4 snRNA, or 5′ externally transcribed spacer. These results suggest that it is unlikely that Hbs1p is involved in rRNA processing events carried out by the exosome (52).
DOM34 shares the same genetic relationship with protein components of the ribosome as HBS1. Deletion of DOM34 serves to exaggerate the growth defects associated with an rps30AΔ strain, and consistent with the hbs1Δ strain, the introduction of a constitutively active GCN2 mutant into the dom34Δ strain also confers a synthetic growth defect. It has been previously reported that a dom34Δ strain conferred growth defects and reduced translation capacity (16), both of which are not observed in our strain background. The yeast strain Σ1287 utilized by Davis and Engebrecht is capable of invasive growth on rich media and demonstrates defective induction of the stress-responsive genes and constitutively transcribed GCN4-responsive genes (50). The growth defects associated with our dom34Δ strain expressing the constitutive active GCN2p mutant allele would be consistent with the phenotypes previously reported (44). An hbs1Δ dom34Δ rps30AΔ strain confers a phenotype similar to those of the hbs1Δ rps30AΔ and dom34Δ rps30AΔ strains, providing genetic support to the hypothesis that the two proteins function together as part of a complex.
Hbs1p and the elongation factor class G proteins of yeast.
We have demonstrated that Hbs1p is a bona fide G protein, with the capacity to bind GTP in vitro in a nucleotide-specific manner. Truncations and mutational analysis of the Hbs1p in vivo suggest that the eEF1A-like C terminus of Hbs1p is the functional domain with regard to the genetic relationship that it exhibits with rp gene deficiencies. Hbs1p also shares sequence similarity with Ski7p, a member of this family recently demonstrated to play a role in cytoplasmic mRNA degradation mediated by the exosome (52). Hbs1 is not required for exosome-mediated RNA processing (52); however, it has been that speculated Hbs1p may play a role in nuclear mRNA degradation (3, 52). It is interesting that recent work has demonstrated that the eEF1A domain of the Ski7p protein is not required for its function in 3′-to-5′ mRNA degradation; instead, the N-terminal domain mediates interaction with the exosome (3). The identification of factors that physically interact with the N-terminal domain of Hbs1p may certainly provide additional insight into its function.
Dom34p and the RNA binding motif.
Dom34p, like Hbs1p, shares sequence similarity factors linked to protein synthesis. Dom34p is a member of a group of proteins, including eRF1, Rpl30p, and SBP2, that share a putative RNA binding motif initially identified by Koonin et al. (31), referred to as the L30 RNA binding motif (14). Ribosomal protein L30 functions in the regulation of the splicing and translation of its own mRNA, and nuclear magnetic resonance analysis has established the role of the L30 RNA binding motif in L30 interaction with its own pre-mRNA (36). The novel mammalian factor SBP2 binds the unique selenocysteine insertion sequence present within the 3′ untranslated region of the mRNA and functions with the specialized translation elongation factor eEFSec in the cotranslational incorporation of this rare amino acid in mammalian cells (14). Additionally, Dom34p and eRF1 share an additional, approximately 60-aa span of weak homology that includes the putative RNA binding motif. The additional sequence similarity between eRF1 and Dom34p that extends beyond the RNA binding motif may reflect RNA targets for the two proteins related in sequence or structure.
An attractive hypothesis for the function of Dom34p and Hbs1p is in the translation regulation of a subset of mRNAs. Many examples of translation regulation involving RNA binding proteins that function through interaction with specific sequence in the 3′ untranslated region of the mRNA have been identified. Binding of Dom34p to such an RNA structure and interaction with Hbs1p could possibly serve to alter translation of a particular mRNA or set of mRNAs. Alternatively, this complex could function to facilitate the translation of a subset of mRNAs uniquely sensitive to alterations in the initiation of protein synthesis.
There are several lines of evidence indicating that the processes of mRNA stability and translation are linked (reviewed in reference 51). Mutations in factors involved in mRNA stability, such as Pat1p, affect translation initiation (8). Additionally, mutations in translation initiation factors such as eIF4E and eIF4G have also been demonstrated to affect mRNA turnover (48). In the absence of direct evidence implicating Hbs1p in the process of protein synthesis, an alternative interpretation of our results is that Hbs1p may indirectly affect translation as a result of affects on mRNA turnover. It has been hypothesized that Hbs1p may play a role in mRNA degradation, based on its similarity to Ski7p (52). While the loss of HBS1 fails to affect rRNA processing mediated by the exosome, a role in nuclear mRNA decay has not been ruled out.
Acknowledgments
We thank Raj Munshi for technical assistance, John Woolford for plasmids, and Thomas Dever for helpful comments.
This work was supported by NIH-RO1 GM28301 to T.G.K. and by NIH-5RO1 GM17139 to E.A.C. A.C.-S. was supported by NIH Predoctoral Training Grant T32-AI07403 and C.P. was supported by T32-GM07215.
REFERENCES
- 1.Abovich, N., L. Gritz, L. Tung, and M. Rosbash. 1985. Effect of RP51 gene dosage alterations on ribosome synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 5:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abovich, N., and M. Rosbash. 1984. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosome. Mol. Cell. Biol. 4:1871-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araki, Y., S. Takahashi, T. Kobayashi, H. Kajiho, S. Hoshino, and T. Katada. 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 20:4684-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baim, S. B., D. F. Pietras, D. C. Eustice, and F. Sherman. 1985. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol. Cell. Biol. 5:1839-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baniahmad, C., A. Baniahmad, and B. W. O'Malley. 1994. A rapid method combining a functional test of fusion proteins in vivo and their purification. BioTechniques 16:194-196. [PubMed] [Google Scholar]
- 6.Bender, A., and J. R. Pringle. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Sacchraomyces cerevisiae. Mol. Cell. Biol. 11:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 8.Bonnerot, C., R. Boeck, and B. Lapeyre. 2000. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 20:5939-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourne, H. R., D. A. Sanders, and F. McCormick. 1990. The GTPase superfamily: a conserved switch for diverse cellular functions. Nature 348:125-132. [DOI] [PubMed] [Google Scholar]
- 10.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117-127. [DOI] [PubMed] [Google Scholar]
- 11.Carr-Schmid, A., N. Durko, J. Cavallius, W. C. Merrick, and T. G. Kinzy. 1999. Mutations in a GTP-binding motif of eEF1A reduce both translational fidelity and the requirement for nucleotide exchange. J. Biol. Chem. 274:30297-30302. [DOI] [PubMed] [Google Scholar]
- 12.Carr-Schmid, A., L. Valente, V. I. Loik, T. Williams, L. M. Starita, and T. G. Kinzy. 1999. Mutations in elongation factor 1β, a guanine nucleotide exchange factor, enhance translational fidelity. Mol. Cell. Biol. 19:5257-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvalho, M. G., J. F. Carvalho, and W. C. Merrick. 1984. Biological characterization of various forms of elongation factor 1 from rabbit reticulocytes. Arch. Biochem. Biophys. 234:603-611. [DOI] [PubMed] [Google Scholar]
- 14.Copeland, P. R., J. E. Fletcher, B. A. Carlson, D. L. Hatfield, and D. M. Driscoll. 2000. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 19:306-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czaplinski, K., M. J. Ruiz-Echevarria, C. I. Gonzalez, and S. W. Peltz. 1999. Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. Bioessays 21:685-696. [DOI] [PubMed] [Google Scholar]
- 16.Davis, L., and J. Engebrecht. 1998. Yeast dom34 mutants are defective in multiple developmental pathways and exhibit decreased levels of polyribosomes. Genetics 149:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dever, T. 1997. Using GCN4 as a report of eIF2a phosphorylation and translational regulation in yeast. Methods 11:403-417. [DOI] [PubMed] [Google Scholar]
- 18.Dever, T. E., L. Feng, R. C. Wek, A. M. Cigan, T. F. Donahue, and A. G. Hinnebusch. 1992. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68:585-596. [DOI] [PubMed] [Google Scholar]
- 19.Dever, T. E., M. J. Glynias, and W. C. Merrick. 1987. GTP-binding domain: three consensus sequence elements with distinct spacing. Proc. Natl. Acad. Sci. USA 84:1814-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinman, J. D., and T. G. Kinzy. 1997. Translational misreading: mutations in translation elongation factor 1α differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA 3:870-881. [PMC free article] [PubMed] [Google Scholar]
- 21.Dong, J., H. Qui, M. Garcia-Barrio, J. Anderson, and A. G. Hinnebusch. 2000. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 6:269-279. [DOI] [PubMed] [Google Scholar]
- 22.Fewell, S. W., and J. L. Woolford. 1999. Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18S rRNA. Mol. Cell. Biol. 19:826-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 24.Hinnebusch, A. G. 1997. Translational regulation of yeast GCN4. J. Biol. Chem. 272:21661-21664. [DOI] [PubMed] [Google Scholar]
- 25.Inagaki, Y., and W. F. Doolittle. 2000. Evolution of the eukaryotic translation termination system: origins of release factors. Mol. Biol. Evol. 17:882-889. [DOI] [PubMed] [Google Scholar]
- 26.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James, P., C. Pfund, and E. A. Craig. 1997. Functional specificity among hsp70 molecular chaperones. Science 275:387-389. [DOI] [PubMed] [Google Scholar]
- 29.Jones, J., and L. Prakash. 1990. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast 6:363-366. [DOI] [PubMed] [Google Scholar]
- 30.Kisselev, L. L., and L. Frolova. 1995. Termination of translation in eukaryotes. Biochem. Cell Biol. 73:1079-1086. [DOI] [PubMed] [Google Scholar]
- 31.Koonin, E. V., P. Bork, and C. Sander. 1994. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 22:2166-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kressler, D., M. Doère, M. Rojo, and P. Linder. 1999. Synthetic lethality with conditional dbp6 alleles identifies Rsa1p, a nucleoplasmic protein involved in the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 19:8633-8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.la Cour, T. F. M., J. Nyborg, S. Thirup, and B. F. C. Clark. 1985. Structural details of the binding of guanosine diphosphate to elongation factor Tu from E. coli as studied by X-ray crystallography. EMBO J. 4:2385-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larkin, J. C., J. R. Thompson, and J. L. Woolford, Jr. 1987. Structure and expression of the Saccharomyces cerevisiae CRY1 gene: a highly conserved ribosomal protein gene. Mol. Cell. Biol. 7:1764-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, J. H., S. K. Choi, A. Roll-Mecak, S. K. Burley, and T. E. Dever. 1999. Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. Proc. Natl. Acad. Sci. USA 96:4342-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao, H., S. A. White, and J. R. Williamson. 1999. A novel loop-loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nat. Struct. Biol. 6:1139-1147. [DOI] [PubMed] [Google Scholar]
- 37.Morikawa, K., T. F. M. la Cour, J. Nyborg, K. M. Rasmussen, D. L. Miller, and B. F. C. Clark. 1978. High resolution X-ray crystallographic analysis of a modified form of the elongation factor Tu: guanosine triphosphate complex. J. Mol. Biol. 125:325-338. [DOI] [PubMed] [Google Scholar]
- 38.Nelson, R. J., T. Ziegelhoffer, C. Nicolet, M. Werner-Washburne, and E. A. Craig. 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71:97-105. [DOI] [PubMed] [Google Scholar]
- 39.Pai, E. F., W. Kabsch, U. Krengel, K. C. Holmes, J. John, and A. Wittinghofer. 1989. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature 341:209-214. [DOI] [PubMed] [Google Scholar]
- 40.Pai, E. F., U. Krengel, G. A. Petsko, R. S. Goody, W. Kabsch, and A. Wittinghofer. 1990. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 9:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulovich, A. G., J. R. Thompson, J. C. Larkin, Z. Li, and J. L. J. Woolford. 1993. Molecular genetics of cryptopleurine resistance in Saccharomyces cerevisiae: expression of a ribosomal protein gene family. Genetics 135:719-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pestova, T. V., I. B. Lamokin, J. H. Lee, S. K. Choi, T. E. Dever, and C. U. T. Hellen. 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403:332-335. [DOI] [PubMed] [Google Scholar]
- 43.Pfund, C., N. Lopez-Hoyo, T. Ziegelhoffer, B. A. Schilke, P. Lopez-Buesa, W. A. Walter, M. Wiedmann, and E. A. Craig. 1998. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 17:3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramirez, M., R. C. Wek, C. R. Bazquez de Aldana, B. M. Jackson, B. A. Freeman, and A. G. Hinnebusch. 1992. Mutations activating the yeast eIF2α kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol. Cell. Biol. 12:5801-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose, M. D., P. Novick, J. H. Thomas, D. Botstein, and G. R. Fink. 1987. A Saccharomyces cerevisiae genomic plasmid based on a centromere containing shuttle vector. Gene 60:237-243. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz, D. C., and R. Parker. 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:5247-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Stanhill, A., N. Schick, and D. Engelberg. 1999. The yeast Ras/cyclic AMP pathway induces invasive growth by suppressing the cellular stress response. Mol. Cell. Biol. 19:7529-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tucker, M., and R. Parker. 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 69:571-595. [DOI] [PubMed] [Google Scholar]
- 52.van Hoof, A., R. R. Staples, R. E. Baker, and R. Parker. 2000. Function of the Ski4p(Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol. Cell. Biol. 20:8230-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallrapp, C., S. Verrier, G. Zhouravleva, H. Philippe, M. Philippe, T. M. Gress, and O. Jean-Jean. 1998. The product of the mammalian orthologue of the Saccharomyces cerevisiae HBS1 gene is phylogenetically related to eukaryotic release factor 3 (eRF3) but does not carry eRF3-like activity. FEBS Lett. 440:387-392. [DOI] [PubMed] [Google Scholar]
- 54.Warner, J. R. 1989. The synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol. Rev. 53:256.. [DOI] [PMC free article] [PubMed] [Google Scholar]