Abstract
The transcriptional coactivator p300 regulates transcription by binding to proteins involved in transcription and by acetylating histones and other proteins. These transcriptional effects are mainly at promoter and enhancer elements. Regulation of transcription also occurs through scaffold/matrix attachment regions (S/MARs), the chromatin regions that bind the nuclear matrix. Here we show that p300 binds to the S/MAR binding protein scaffold attachment factor A (SAF-A), a major constituent of the nuclear matrix. Using chromatin immunoprecipitations, we established that both p300 and SAF-A bind to S/MAR elements in the transiently silent topoisomerase I gene prior to its activation at G1 during cell cycle. This binding is accompanied by local acetylation of nucleosomes, suggesting that p300-SAF-A interactions at S/MAR elements of nontranscribed genes might poise these genes for transcription.
Activation of gene expression is thought to be a multistep process involving changes in chromatin structure, as well as site-specific events such as the acetylation of histones (20). An important factor in higher-order organization is the nuclear matrix, a proteinaceous structure that consists of a network of RNPs and other nonhistone proteins and that serves as a scaffold for loops of chromatin (reviewed in reference 5). This matrix has been associated with the regulation of transcription, DNA replication, and RNA processing (44). The DNA regions anchoring the chromosomal DNA to the nuclear matrix (called matrix-associated regions [MARs] or scaffold-attachment regions [SARs] [8]) are composed of AT-rich sequences. They have a unique structure characterized by a narrow minor groove, a high unwinding potential and the formation of hairpin structures (5). S/MARs are located in the introns of several large genes (32, 53) and also at borders of transcription units (23, 47). S/MAR elements have often been implicated in the regulation of gene expression. They are frequently found close to enhancers (37, 48), and they can stimulate gene expression of heterologous reporter genes when integrated into the genome (56) and can regulate chromatin accessibility (21, 30).
Several S/MAR binding proteins have been identified and characterized over recent years. One of the first was the scaffold attachment factor-A (SAF-A), initially identified as a component of RNP particles (35) but later also characterized as a S/MAR-binding protein (17, 18, 24, 36, 52). Other S/MAR binding proteins include ubiquitous, abundant proteins such as SAF-B (50), topoisomerase II (1), histone H1 (28), lamin B1 (42), HMGI/Y (66), and nucleolin (11), but also proteins that are expressed in a more cell-type-specific fashion, such as SatB1 (10) and p114 (63). Even though many of these proteins have been thoroughly characterized biochemically, it is still relatively unclear how they are involved in the gene regulating processes of S/MAR elements.
In recent years much progress has been made in the characterization of the events at the site of transcription initiation. Of the factors involved the transcriptional coactivators p300/CBP were originally described as interaction modules that form a bridge between the basal transcription machinery and upstream transcription factors (25). CBP was cloned by virtue of its binding to CREB (7) and p300 as an adenoviral-E1A associating protein (15). Viral oncogenes such as the adenovirus E1A and simian virus 40 (SV40) large T can interact with p300/CBP and disrupt the interaction of p300/CBP with other factors. The homologous proteins p300 and CBP contain three cysteine-histidine (CH)-rich regions, of which CH3 constitutes the major interaction site for adenovirus E1A. An additional role of p300/CBP became clear when it was found that p300/CBP have intrinsic acetyltransferase activity (4, 45) and associate with other proteins with acetyltransferase activity such as p/CAF (64). The acetylation of histone tails is generally related to transcription activation (57). In addition, acetylation of transcription factors by p300/CBP can lead to either activation or repression of transcription (55).
Here we report a strong interaction between p300 and SAF-A. We show that p300 and SAF-A bind to S/MAR elements and that their binding is disrupted when the viral protein E1A or SV40 large T is present. The binding of p300 and SAF-A to S/MAR elements seems to be restricted to the transcriptional inactive state. However, while no transcription occurs, these S/MAR elements are bound by acetylated histones, suggesting that the presence of p300 at S/MAR elements might realize a localized chromatin state ready for transcription.
MATERIALS AND METHODS
Yeast two-hybrid assay.
Yeast two-hybrid screening was performed as described previously (3) with a random-primed human cDNA testis library fused to a VP16 transactivating domain by using the vector pVP16 (61) and the yeast strain Y190 (59). The bait plasmid pMD4-p300 (amino acids [aa] 1570 to 1848) was made by cloning an 834-bp p300 cDNA fragment into pMD4. pMD4 was a gift from M. van Dijk and R. Bernards. Y190, which contains LacZ and HIS3 reporter genes, was transformed with plasmid pMD4-p300 and the pVP16 cDNA library. In the screen 5 × 106 transformants were tested, and the HIS+ positive colonies were subsequently tested for β-galactosidase activity by colony filter assay. Of the double-positive clones (HIS+, LacZ+) one contained the coding sequence for aa 633 to 806 of SAF-A.
Cell lines and cell fractionation.
The cell lines used are the human osteosarcoma cell line U-2 OS, the adenovirus type 5 E1-transformed human embryonic retinoblastoma (HER) cell line 911 (19), human glioblastoma cell line T98G (54), human diploid foreskin fibroblasts VH10, and SV40 early region transformed VH10 cells (38). Cell fractionation of U-2 OS cells was performed essentially as previously described (17, 22). Briefly, cells were washed with phosphate-buffered saline and resuspended in cytoskeleton (CSK) buffer {100 mM NaCl, 300 mM sucrose, 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] [pH 6.8], 3 mM MgCl2, 1 mM EGTA, and 0.5% Triton X-100 containing phenylmethylsulfonyl fluoride [Sigma] and RNasin [Promega]} and incubated at 4°C for 3 min. Skeletal frameworks were pelleted by centrifugation at 650 × g for 5 min, and the supernatant was removed. After an additional wash with CSK buffer, the skeletal framework was treated with RNase A (Roche Diagnostics) for 20 min at room temperature in a CSK buffer with lower NaCl (50 mM) and no RNasin. Digestion was terminated by the addition of ammonium sulfate to a final concentration of 0.25 M. The remaining nuclear skeleton was pelleted at 1,000 × g for 5 min while the RNA-bound fraction stayed in solution. The chromatin-bound fraction was released from the CSK-washed skeletal framework by sonication. After three sonications in a Branson sonifier 250 at 15% output for 20 s, ammonium sulfate was added to a final concentration of 0.15 M, and the remaining nuclear skeleton was pelleted at 1,000 × g for 5 min while the supernatant contained the chromatin-bound fraction.
Antibodies and immunoprecipitation assays.
The p300 rabbit polyclonal antibodies 1 and 3 were raised against aa 91 to 328 and 1834 to 2049, respectively, and were as previously described (14). A rabbit polyclonal serum against SAF-A was raised against a purified, bacterially produced His-tagged protein containing aa 1 to 243 of SAF-A. The 3G6 mouse monoclonal antibody against SAF-A and the 4D11 mouse monoclonal antibody against hnRNP L were gifts from G. Dreyfuss (13, 49), and the mouse monoclonal antibody M73 against adenovirus type 5 E1A was a gift from E. Harlow (27). N15 rabbit polyclonal serum against p300, C21 antibody against p90rsk1, and the C19 antibody against ATF-2 were purchased from Santa Cruz. MN11 is a mouse monoclonal antibody against p300/CBP (9) and was purchased from PharMingen. Rabbit polyclonal antibody antiacetylated histone H3, raised against the N-terminal peptide acetylated at lysine 9 and lysine 14, was purchased from Upstate Biotechnology (catalog number 06-599). The antihistone polyclonal antiserum raised against purified Drosophila core histones was a gift from G. E. Chalkley and C. P. Verrijzer (33). Immunoprecipitations were performed as described before (26) in assay buffer (0.1% NP-40-250 mM NaCl-50 mM Tris-HCl [pH 7.5] containing a mixture of protease inhibitors [34]).
DNA constructs.
Glutathione S-transferase (GST)-p300ADA2, GST-p300CH3, and GST-p300BAIT fusion genes were constructed by cloning digested p300 PCR fragments into pGEX-2T (Pharmacia). GST-E1A is a fusion protein of GST and aa 1 to 90 of adenovirus type 5 E1A (34). The pGem4-U21.1 plasmid with full-length SAF-A (35) was a generous gift of G. Dreyfuss. GST-SAF-A was generated by cloning a SAF-A PCR fragment encoding aa 537 to 806 into pGEX-2T.
GST pull-down analysis.
GST fusion protein-coated beads and in vitro-translated proteins were prepared as previously reported (12). In vitro-translated [35S]methionine-labeled proteins were made by a coupled transcription-translation kit (TNT; Promega). For the E1A protein, a plasmid with adenovirus type 5-12S E1A behind a T7 promoter was used; for full-length SAF-A and SAF-A protein fragments a PCR was performed with an upstream oligonucleotide containing the T7 promoter sequence followed by a Kozak sequence (51), and as a downstream primer the sequence at the end of the studied fragment followed by a stop codon. The [35S]methionine-labeled proteins bound to GST fusion proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography. For pull-down assays with whole-cell extracts, the preparation of cell extracts and incubation with beads coated with GST or GST fusion protein were performed as described before (34). Bound proteins were analyzed by SDS-PAGE followed by Western blotting.
Competition assay.
GST-p300CH3 and GST-E1A-N-CR1 (aa 1 to 90) were prepared as previously reported (12). GST-E1A was cleaved with thrombin (Sigma) for 2 h at 4°C. The reaction was terminated by addition of phenylmethylsulfonyl fluoride (Sigma); the sample was then dialyzed in assay buffer (0.1% NP-40-250 mM NaCl-50 mM Tris-HCl [pH 7.5] containing protease inhibitors). The purified protein was analyzed by SDS-PAGE, followed by Coomassie staining. In vitro-translated full-length [35S]methionine-labeled SAF-A was incubated for 1 h in assay buffer with GST-p300CH3, after which different concentrations of E1A or bovine serum albumin (BSA; Sigma) were added. After an additional incubation for 1 h, bound proteins were analyzed by SDS-PAGE followed by autoradiography.
Gel filtration chromatography.
Cell extracts were prepared as described before (34) and loaded onto a preequilibrated Superose 6HR 10/30 gel filtration column (Pharmacia) controlled by an FPLC System (Pharmacia). Chromatography was performed in assay buffer (0.1% NP-40-250 mM NaCl-50 mM Tris-HCl [pH 7.5] containing protease inhibitors), and the flow rate was 0.4 ml/min. Fractions (1.5 ml) were collected and analyzed by SDS-PAGE and Western blotting with 3G6 against SAF-A, MN11 against p300/CBP, and C21 against p90rsk1.
ChIPs.
Cells at 90% confluence were cross-linked with 1% formaldehyde for 10 min at room temperature. Cross-linking was quenched with 125 mM glycine, and whole-cell extracts were prepared for use in the chromatin immunoprecipitations. These chromatin immunoprecipitation assays (ChIPs) were performed as described by Kuras and Struhl (41). The average size of fragmented chromatin was ca. 500 bp as analyzed on agarose gels. For immunoprecipitations, the fragmented chromatin from a 15-cm dish was used, except for p300 immunoprecipitates, for which five times more input was used in parallel with the same amount of input for a nonimmune precipitation. DNA from the immunoprecipitates or from the input (10% of a 15-cm dish) was analyzed either by PCR or by real-time PCR by using the Lightcycler quantification method (Roche Diagnostics). For both PCR analyses, either 1/100 (input) or 1/20 (immunoprecipitates) of the DNA was amplified with 50 pmol of the indicated primers in 40-μl reaction mixtures containing 200 μM concentrations of deoxynucleoside triphosphates, 2.7 mM MgCl2, and 0.25 U of AmpliTaq (Perkin-Elmer). After 4 min at 94°C, 30 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C were performed. PCR products were electrophoresed on 2% agarose gels containing ethidium bromide, analyzed under UV light, and presented in inversed intensity. The real-time PCR results were presented as the amount of template immunoprecipitated relative to the amount precipitated with a nonimmune antibody. All ChIP experiments presented were performed at least two times.
The primers used for amplification were as follows: TopMII, GCTCACTGTGACCTCTGC and CCCAGCACTTTGTGCAGC; pTop, GAGTGGGGACCACCTCCAC and CAATCGGAAATCCGCTTCG; Top exon 13, CAGCAGATAGGTCCACTTGG and CCTTACCTTGATTCGTGAAC; Top exon 20, CACCTTTCTCAGGTGGAGCC and GCTGCACACTTTTCTCTACC; TopMI, CCCTGCTGCTAATGGTATGG and CTTCTGAGGAAACGACTTTGG; c-myc, GCGAACACAACGTCTTGG and GTTCTCGTCGTTTCCGCAAC; c-jun, CTAGGGTGGAGTCTCCATGG and GCTCAACACTTATCTGCTAC; Igκ, CTGCAACAACTTGATAGGAC and GGGTGAATTCTTGATAGCTTTAC; and β-globin, CACAGTCTGCCTAGTACATTAC and GCCCTGAAAGAAAGAGATTAGG.
Reverse transcription-PCR (RT-PCR).
RNA was isolated by using the Promega SV RNA Isolation Kit (Promega). cDNA was made by using random hexamers and the Superscript first-strand synthesis system (Gibco-BRL). PCR was performed as described above, and PCR products were analyzed on ethidium bromide-stained agarose gels. The following primers were used: for GAPDH, AATCCCATCACCATCTTCC and ATGAGTCCTTCCACGATACC; for p300, CGGGATCCGCTGCATCCAGTCTCTG and GCTCTAGATCAAGGGAGGCCCTGTTGCTG; and for topoisomerase I, GAGTGGGGACCACCTCCAC and GGATAGCGCTCTTCTTCCC.
RESULTS
Interaction between p300 and SAF-A.
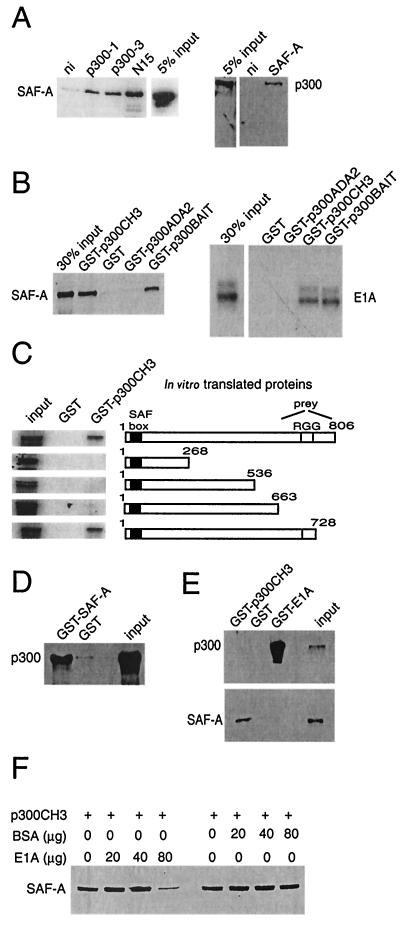
In a search for proteins involved in the functional regulation of p300, a yeast two-hybrid screen was performed with a p300 fragment that comprises the ADA2 homology domain and CH3 (aa 1570 to 1844). This led to the identification of SAF-A as a p300-associating protein. To test whether p300 and SAF-A associate in vivo, we examined whether endogenously expressed p300 and SAF-A could be coimmunoprecipitated from human cells. As shown in Fig. 1A, SAF-A readily coprecipitated when p300 was immunoprecipitated from a U-2 OS cell extract by using three different antibodies against p300. Since p300 also coprecipitated when SAF-A was immunoprecipitated, we conclude that the endogenous p300 and SAF-A proteins interact in cells. To delineate the interaction domain of p300 and SAF-A, in vitro binding studies were performed. Pull-down studies with GST fusion proteins with the p300 bait sequence or its separate ADA2 homology and CH3 domains showed that in vitro-translated SAF-A interacts with the CH3 domain of p300, as does E1A, while neither protein binds to GST or GST-ADA2 (Fig. 1B). The in vitro-translated SAF-A protein used comprises the C-terminal aa 633 to 806, the SAF-A protein domain identified in our yeast two-hybrid screen to interact with p300. GST pull-down studies with in vitro-translated full-length or C-terminally truncated SAF-A protein showed that SAF-A contains a single binding site for p300 localized between aa 663 and 728 (Fig. 1C), a region previously identified as an RNA-binding domain (35). The GST-p300CH3 protein also binds SAF-A from cell extracts (Fig. 1E), while GST-SAF-A (aa 537 to 806) brings down full-length p300 from whole-cell extracts (Fig. 1D).
FIG. 1.
p300 interacts with SAF-A in vivo and in vitro. (A) Endogenous p300 coimmunoprecipitates with SAF-A. U-2 OS whole-cell extracts were immunoprecipitated with antibodies against p300 (p300-1, p300-3, and N15) or SAF-A and tested on Western blot for coimmunoprecipitated SAF-A or p300. (B) GST pull-down assays. In vitro-translated [35S]methionine-labeled 12S-E1A or SAF-A (aa 633 to 806) retained on GST, GST-p300ADA2 (aa 1573 to 1731), GST-p300CH3 (aa 1726 to 1848), or GST-p300BAIT (aa 1573 to 1848) was visualized by autoradiography. (C) Mapping of the SAF-A-p300 interaction by GST pull-down. In vitro-translated SAF-A subdomains retained on GST-p300CH3 were visualized by autoradiography.(D) GST-SAF-A (aa 537 to 806) retains endogenous p300 from U-2 OS whole-cell extracts as visualized on a Western blot with an anti-p300 antibody. (E) GST pull-down assay. GST-E1A (aa 1 to 90) and GST-p300CH3 retain endogenous p300 or SAF-A, respectively, as visualized on Western blots. (F) Competition assay. Increasing amounts of E1A or BSA were used for competing away in vitro-translated SAF-A retained on GST-p300CH3.
The binding of E1A and SAF-A to the same domain of p300 might imply a mutually exclusive binding. Indeed, no indication for a trimeric complex of E1A, p300, and SAF-A is found since a pull-down experiment with GST-E1A (aa 1 to 90) brings down only p300 (Fig. 1E). Further evidence for a mutually exclusive binding is provided by the fact that increasing amounts of E1A can compete for binding of in vitro-translated SAF-A to GST-p300CH3, while increasing amounts of BSA have no effect on the binding of SAF-A to GST-p300CH3 (Fig. 1F). Taken together, these results show that SAF-A interacts, both in vivo and in vitro, with p300. Binding occurs through the CH3 domain of p300, precluding E1A from binding to this site simultaneously.
p300-SAF-A binds S/MAR elements.
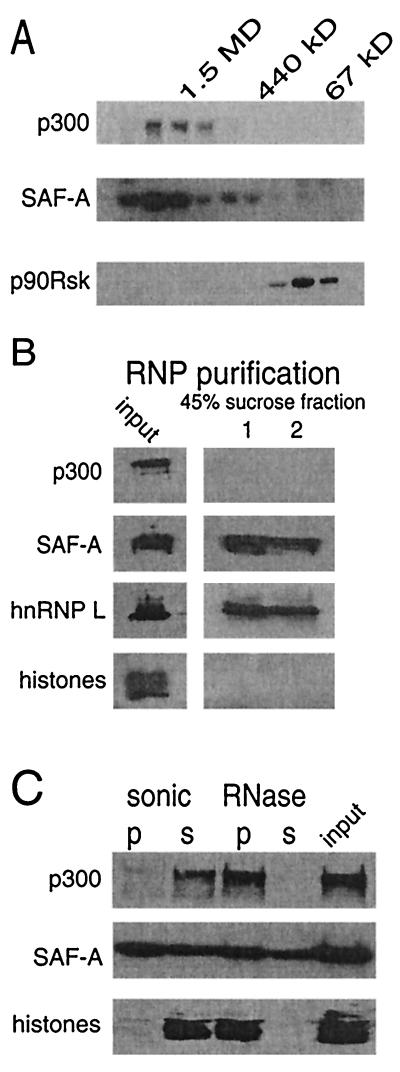
To investigate whether p300 and SAF-A (120 kDa) exist in a multiprotein complex, U-2 OS cell extracts were fractionated on a gel filtration column. Western blotting analysis revealed that the endogenous p300 eluted at ca. 1.5 MDa (Fig. 2A), a finding consistent with previous reports (43). The endogenous SAF-A eluted over a broader range of molecular weights with a large fraction that cofractionated with p300 (Fig. 2A). From these fractions p300 and SAF-A could be readily coimmunoprecipitated (data not shown). As a control, the elution pattern of p90rsk1 kinase was analyzed and shown to elute at ca. 90 kDa, indicating that it is not present in a complex with p300 and/or SAF-A.
FIG. 2.
SAF-A and p300 cofractionate. (A) U-2 OS cell extract was fractionated on a Superose 6 column, and fractions were collected and tested on Western blot for the presence of SAF-A, p300, and the protein kinase p90rsk. At the top the elution of markers is indicated by their molecular sizes in kilodaltons. (B) RNP particles were isolated from whole-cell extracts in the 45% sucrose fractions (lanes 1 and 2 represent two independent gradients). The presence of SAF-A, hnRNP L, histones, and p300 was tested by Western blot. (C) U-2 OS nuclear skeleton was separated by two procedures into a chromatin and an RNP-associating fraction. In the sonication procedure, the pellet (p) represents the RNP-associated fraction and the supernatant (s) represent the chromatin-associated fraction. In the RNase procedure, the pellet represents the chromatin-associated fraction and the supernatant represents the RNP-associated fraction. The presence of p300, SAF-A (120 kDa), and histones was detected by Western blotting.
SAF-A has been identified as both chromatin and RNP bound. To investigate whether p300 binds SAF-A in both locations, we analyzed the presence of p300 and SAF-A via various cellular fractionation methods (17, 22). First, we purified RNP particles in the 45% sucrose fraction by centrifugation of whole-cell extracts over a sucrose gradient (22). In these RNP particles, SAF-A and hnRNP L, another RNP protein, were clearly present, but neither p300 nor histones were detected (Fig. 2B), indicating that p300 does not bind to SAF-A present in hnRNP particles. To determine the presence of p300 and SAF-A in chromatin, we prepared distinct nuclear fractions (17). For this, cells were lysed in an isotonic buffer in the presence of 1% Triton X-100. This treatment released only a small amount of SAF-A, whereas the majority of the protein remained bound to nuclear structures. From this nuclear fraction, which still contained p300, chromatin can be released by sonication and, alternatively, RNA can be released by an RNase treatment. SAF-A was found equally distributed in the nuclear fractions, as described by Fackelmayer et al. (17). However, p300 was mainly found in the chromatin fractions, which were identified by the presence of histone proteins (Fig. 2C). These data indicate that p300 and SAF-A most likely associate in a chromatin-related context.
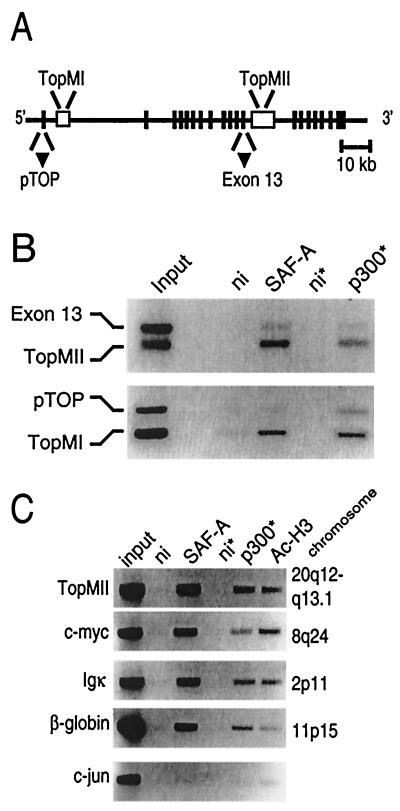
The binding of SAF-A to S/MAR elements has been intensively studied with the MII S/MAR element of topoisomerase I. In order to study whether both SAF-A and p300 are present at S/MARs, ChIPs on the S/MAR elements of the topoisomerase I gene were performed in U-2 OS cells. Immunoprecipitation of SAF-A brought down both the MI and the MII S/MAR element as detected with primer sets in these elements (Fig. 3A and B). More interestingly, an antibody against p300 also brought down these elements. A nonimmune control serum was used to show the specificity of the immunoprecipitations. The antibodies to SAF-A or to p300 do not immunoprecipitate other parts of the topoisomerase I gene, as shown by the use of two different primer sets, one in the promoter and one in exon 13, that were added as internal controls for the PCR (Fig. 3A and B). Therefore, these data indicate a specific interaction of the p300-SAF-A complex with S/MAR elements in U-2 OS cells.
FIG. 3.
SAF-A/p300 bind S/MAR elements. (A) Schematic representation of the topoisomerase I gene (39, 53) showing the MI and MII S/MAR elements. Vertical bars represent the exons, open boxes represent the S/MAR elements, and triangles represent the positions of the internal controls in panel B. (B and C) ChIP assays with U-2 OS cells with antibodies against SAF-A, p300, and acetylated histone H3. Nonimmune serum (ni) was used as a control. The DNAs of the immunoprecipitated fractions were isolated, and topoisomerase I sequences in the immunoprecipitated DNA were amplified by PCR. Asterisks indicate that five times more input was used in these assays. TopMI and TopMII are fragments within the S/MAR elements of topoisomerase I; pTOP is a fragment within the promoter region, and exon 13 is a fragment within exon 13 of the topoisomerase I gene. In panel C, additional S/MAR sequences were tested: MII of the topoisomerase I gene (TopMII) and the S/MAR sequences of the c-myc, Igκ, and β-globin genes, respectively. As a control for specific amplification, a primer set in the c-jun promoter was used. The chromosomal location of the various genes is shown on the right.
To test whether the association of p300 and SAF-A with S/MAR elements was gene specific, we used ChIP assays to analyze their binding to several other well-defined S/MAR elements. Binding of SAF-A and p300 was found to occur at the S/MARs of the topoisomerase I (MI and MII [40]), c-myc (6), β-globin (29), and Igκ (62) genes (Fig. 3B and C). Since p300 is known to have acetyltransferase activity, we also examined the acetylation status of histone H3 at the different S/MARs. Interestingly, we found the presence of acetylated histone H3 at the S/MAR elements (Fig. 3C). Apart from nonimmune controls, we studied the c-jun promoter and found no binding of p300 and SAF-A to this site. The S/MAR elements we tested were located on various chromosomes, and we conclude that the presence of a p300-SAF-A complex on S/MAR elements seems to be a general phenomenon.
Viral oncogenes disrupt p300-SAF-A-S/MAR complexes.
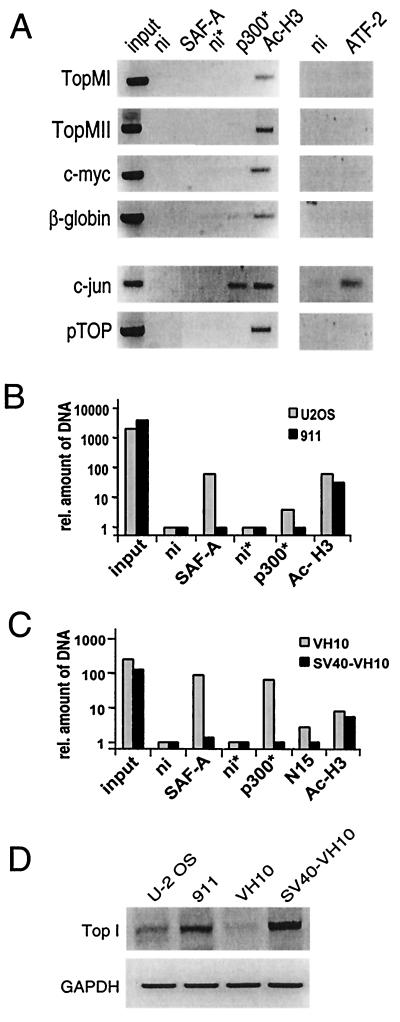
Binding of SAF-A to p300 occurs via the p300CH3 domain that also interacts with the adenovirus E1A protein (15). This interaction is mutually exclusive (Fig. 1E and F), suggesting that the binding of SAF-A and p300 may be disrupted in adenovirus-transformed cell lines. To test whether the presence of E1A interfered with the formation of a p300-SAF-A-S/MAR complex, ChIP assays were performed in the adenovirus E1-transformed HER cell line 911 (19). As shown in Fig. 4A, no binding of either p300 or SAF-A to S/MAR elements could be detected in 911 cells, suggesting that disruption of the p300-SAF-A interaction by E1A also disrupts the SAF-A binding to the S/MAR element. The mechanism of this disruption is not established but might include the presence of other histone acetyltransferase (HAT)-containing complexes at S/MAR elements in virally transformed cells, since we found at all S/MAR elements studied the presence of acetylated histone H3 (see also Discussion). As a control, ChIP assays were performed on the c-jun promoter, which is active in adenovirus-transformed cells (14). Both p300 and the transcription factor ATF-2 could be detected. Similar results were obtained by using 293 cells, an adenovirus-transformed human embryonic kidney cell line (data not shown). A quantitative comparison of these assays in U-2 OS and 911 cells, based on real-time PCR and with the TopMII S/MAR primer set, is shown in Fig. 4B. Quantification shows that in U-2 OS cells immunoprecipitation of p300 and SAF-A brings down almost 5 and 75 times more MII S/MAR DNA, respectively, than a control immunoprecipitation, whereas in adenovirus-transformed cells in both cases levels comparable to the control are immunoprecipitated.
FIG. 4.
Absence of binding of SAF-A or p300 to S/MAR elements in virally transformed cells. (A) ChIP assays with adenovirus-transformed 911 cells with a control antibody (ni) and antibodies against SAF-A, p300, acetylated histone H3, and ATF-2. Primer sets in the topoisomerase I promoter (pTOP) and the c-jun promoters were used as a control. Asterisks indicate that five times more input was used. (B) Quantification of ChIP results. The relative difference in the amount of precipitated DNA as determined by real-time PCR is presented for the ChIP results found for the MII S/MAR element of topoisomerase I in Fig. 4A (911 cells) and Fig. 3B (U-2 OS cells) on a log scale. Nonimmune values are set at 1. (C) Absence of binding of SAF-A or p300 to S/MAR elements in an SV40-transformed cell line compared to nontransformed VH10 cells. The relative difference inamount of precipitated DNA as determined by real-time PCR is presented for the ChIP results found for the MII S/MAR element of topoisomerase I (TopMII). (D) RT-PCR for the mRNA expression of the topoisomerase I and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) genes in U-2 OS, 911, VH10, and SV40-transformed VH10 cells.
Like E1A, SV40 large T antigen has also been shown to interact with the CH3 domain of p300 (2, 16) and similarly might reduce the binding of p300 and SAF-A to S/MAR elements. Therefore, we compared VH10 primary human foreskin fibroblasts with their SV40-transformed counterpart (38). The quantified ChIP assays using the TopMII S/MAR primer set with VH10 cells show a strong binding of SAF-A and p300 to the S/MAR element of topoisomerase I (Fig. 4C), while in the SV40-transformed VH10 cells binding of SAF-A and p300 to the S/MAR element is strongly reduced (Fig. 4C). From these results we conclude that the binding of p300 and SAF-A to the MII S/MAR element, as observed in U-2 OS and VH10 cells, is absent in SV40- and adenovirus-transformed cells.
Several lines of evidence indicate that S/MAR elements are involved in the regulation of transcription. In order to investigate whether the binding of a p300-SAF-A complex to S/MAR correlates with active transcription, we studied the expression of the S/MAR-containing gene topoisomerase I. RT-PCR of topoisomerase I mRNA showed a higher level of expression in the adenovirus-transformed cell line compared to the U-2 OS and VH10 cells (Fig. 4D), as described earlier (54). Also, for the SV40-transformed cell line we found increased topoisomerase I expression. These data show expression of topoisomerase I while no p300-SAF-A is bound to the S/MAR elements of topoisomerase I and indicate a possible role for p300-SAF-A complexes at S/MAR elements of genes that are not transcribed.
Cell cycle-dependent disruption of p300-SAF-A-S/MAR complexes.
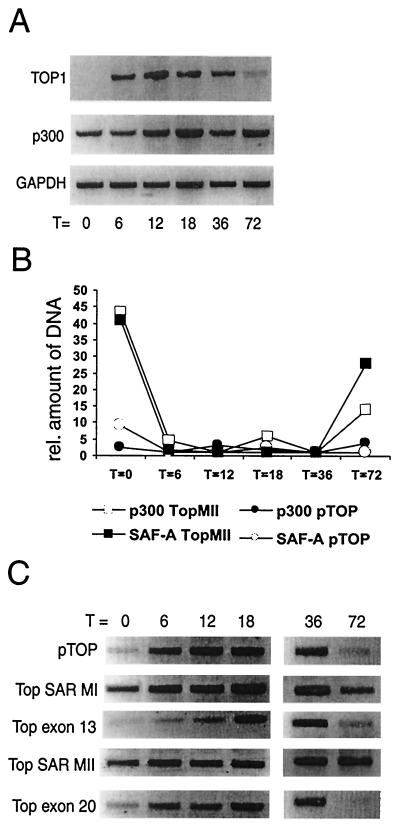
In order to further substantiate the hypothesis that binding of p300-SAF-A to S/MAR elements is restricted to nontranscribed genes, we studied the cell cycle-dependent induction of the topoisomerase I expression in parallel with binding of SAF-A and p300 to the MII S/MAR element in the topoisomerase I gene. For this we used the glioblastoma cell line T98G (see, e.g., reference 58) since these cells can be arrested by serum deprivation and can then be efficiently stimulated to reenter the cell cycle by addition of serum. Moreover, these cells can be density arrested. The mRNA expression of topoisomerase I was studied by RT-PCR and was found to be absent after serum deprivation, while increased expression was found during the first 36 h after serum stimulation (Fig. 5A). After growth for 72 h (when the cells are density arrested), topoisomerase I expression again decreased. In parallel with the expression, the binding of p300 and SAF-A to the S/MAR elements of topoisomerase I was determined by ChIP assays. The quantitative results from real-time PCR are depicted in Fig. 5B. Before serum stimulation a relatively strong binding of the SAF-A and p300 proteins to the MII S/MAR element could be observed. After serum stimulation, the binding of both p300 and SAF-A to the MII S/MAR element was decreased at all time points tested, except at 72 h when again the binding of both p300 and SAF-A to the MII S/MAR element was detected. As a control, a primer set in the topoisomerase I promoter was used, which showed no binding of SAF-A or p300 at any time point. These results clearly show that p300 and SAF-A are not bound to S/MAR elements when the topoisomerase I gene is expressed. To investigate the possible consequence of the presence of the acetyltransferase p300 at S/MAR elements, we assessed the acetylation of histone H3 during the same time course. As shown in Fig. 5C, acetylated histone H3 was present at the S/MAR elements of the topoisomerase I gene, irrespective of its expression. In contrast, acetylated histone H3 could only be detected in other parts (e.g., the promoter) when the gene was actively transcribed. The presence of acetylated H3 during transcription is most likely due to HAT activities associated with the transcription machinery. Hyperacetylation in the absence of transcription, however, was found to be restricted to S/MAR elements. These findings show that, although SAF-A-p300 may somehow be involved in keeping H3 acetylated at S/MAR elements in the absence of transcription (T = 0 h, T = 72 h), the complex is dispensable for keeping H3 acetylated.
FIG. 5.
Inactive topoisomerase I gene has SAF-A, p300, and acetylated histone H3 bound to its S/MAR element. (A) RT-PCR for the RNA expression of the topoisomerase I, p300, and GAPDH genes of serum-arrested T98G cells and after subsequent stimulation by serum for 6, 12, 18, 36, and 72 h, representing growing and subsequently (72 h) density-arrested cells. (B) ChIP assays with the serum-stimulated cells were analyzed with real-time PCR with primers within the topoisomerase I S/MAR element and in the topoisomerase I promoter. Depicted is the relative level of precipitated DNA with antibodies to SAF-A or p300 in comparison to a nonimmune control antibody. (C) ChIP assays with an antibody against acetylated histone H3 show high levels of histone H3 acetylation at S/MAR elements of topoisomerase I when the gene is inactive, in contrast to promoter and exon sequences that only show hyperacetylated histone H3 when the gene is expressed.
DISCUSSION
The transcriptional coactivators p300 and CBP were originally described as bridging factors between various transcription factors and the basal transcription machinery. They were later found to contain intrinsic acetyltransferase activity and to associate with other proteins containing acetyltransferase activity, such as p/CAF. Here we identified a novel function of p300, namely, its association with SAF-A, a protein found as a S/MAR-interacting component of the nuclear matrix and as a component of RNP particles (13). Our studies suggest a role for p300 bound to SAF-A at S/MAR elements. SAF-A interacts with S/MARs via its N-terminal SAF box (36) and via its C-terminal domain with p300, allowing binding of SAF-A to both p300 and a S/MAR element. The C-terminal domain of SAF-A also contains the RGG-box, involved in binding to RNA (35), suggesting that RNA and p300 binding to SAF-A are mutually exclusive. Our results showing that p300 is absent from RNP particles support this view.
For the topoisomerase I gene, release of p300-SAF-A from the S/MAR elements by stimulation of the S phase or by viral oncoproteins is accompanied by enhanced transcription. This suggests that the p300-SAF-A complex functions at the S/MAR elements of nontranscribed genes. The presence of p300, together with SAF-A, at S/MAR elements of inactive genes is unlikely to contribute to the inactive state of the gene. Perhaps, p300 at S/MAR elements contributes to the enigmatic feature of S/MAR elements to stimulate transcription of nearby genes (5). One possibility would be that p300 forms a bridge between SAF-A at S/MAR elements and transcription factors already bound to the promoter, enabling a quick complete activation of the gene. However, this would implicate that p300 should be present at the promoter elements of S/MAR- containing genes, which is in contrast to our findings.
Alternatively, the main contribution of p300 or associated proteins to the transcriptionally competent state through S/MAR elements might be by acetylation of histone tails and, indeed, we found acetylated histone H3 at all S/MAR elements studied. An analysis of the topoisomerase I gene during activation showed that the S/MAR elements of the inactive gene are bound by p300-SAF-A and hyperacetylated histones are present at the S/MAR elements but not at other parts of the gene. When transcription occurs, hyperacetylated histones are present throughout the gene, probably due to histone acetylation activities accompanying the elongation by RNA polymerase II (46). The S/MAR-restricted occurrence of acetylation observed here has recently also been shown for the chromosomal integrated immunoglobulin μ enhancer (21). Together, these findings suggest that one of the mechanisms by which S/MAR elements are able to affect transcription of nearby genes is via the generation of an extended domain of histone acetylation.
The presence of viral oncoproteins such as E1A and SV40 large T can disrupt the binding of p300 to the S/MAR elements. Surprisingly, they also disrupt the binding of SAF-A to these elements. However, no effect of the viral oncogenes was observed on the acetylation state of histone H3 at the S/MAR elements. The release of p300-SAF-A from S/MAR elements is unlikely to be a consequence of transcription, since the c-myc gene is barely expressed in adenovirus-transformed cells (60; data not shown) and there is hardly any expression of β-globin in the nonerythroid cells used here (data not shown). Another option is that the presence of viral proteins results in SAF-A protein modifications leading to an inability to bind S/MAR elements. Finally, it is also possible that the viral oncogenes induce an altered protein composition at the S/MAR elements localizing acetyltransferases other than p300 to these elements. An altered protein composition at the S/MARs might explain both the absence of binding of SAF-A and the presence of acetylated histones without detectable levels of p300 at S/MAR elements in these cells. However, at present we cannot exclude that other virus-induced mechanisms are involved in releasing p300 and SAF-A from S/MAR elements and in keeping the nucleosomes at S/MAR elements acetylated.
Taken together, our results suggest that p300 not only functions at promoter and enhancer sequences but also at S/MAR elements via association with SAF-A. In non-virally transformed cells and in primary cells S/MAR elements of nontranscribed genes bind p300-SAF-A, while there is only localized histone H3 hyperacetylation at the S/MAR elements. This strongly suggests that in this situation the presence of the p300-SAF-A complex is responsible for the acetylation. The most obvious candidate is p300, but it is certainly also possible that the complex contains other p300-associated HATs such as p/CAF (64) and SRC-1 (31, 65). Association of p300 and SAF-A with S/MAR elements of nontranscribed genes might ensure the presence of localized histone acetylation and poise these chromatin regions for transcription.
Acknowledgments
We thank G. Dreyfuss for the 3G6 and 4D11 antibodies and the pGem-SAF-A vector; S. Elledge for Y190; R. Bernards for the pMD4 plasmid; E. Harlow for M73; and R. G. J. Vries, A. J. van der Eb, and C. P. Verrijzer for helpful discussions.
This work was supported by the Counsel Chemical Sciences of The Netherlands Organization for Scientific Research (NWO-CW) and EC grants TMR-CT96-0044 and Biomed CT97-2567. E.K. and J.C.D. were supported by grants from the Dutch Cancer Society (KWF).
REFERENCES
- 1.Adachi, Y., E. Kas, and U. K. Laemmli. 1989. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 8:3997-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avantaggiati, M. L., M. Carbone, A. Graessmann, Y. Nakatani, B. Howard, and A. S. Levine. 1996. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional coactivator, p300. EMBO J. 15:2236-2248. [PMC free article] [PubMed] [Google Scholar]
- 3.Bai, C., and S. J. Elledge. 1997. Gene identification using the yeast two-hybrid system. Methods Enzymol. 283:141-156. [DOI] [PubMed] [Google Scholar]
- 4.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 5.Bode, J., C. Benham, A. Knopp, and C. Mielke. 2000. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements). Crit. Rev. Eukaryot. Gene Expr. 10:73-90. [PubMed] [Google Scholar]
- 6.Chou, R. H., J. R. Churchill, M. M. Flubacher, D. E. Mapstone, and J. Jones. 1990. Identification of a nuclear matrix-associated region of the c-myc protooncogene and its recognition by a nuclear protein in the human leukemia HL-60 cell line. Cancer Res. 50:3199-3206. [PubMed] [Google Scholar]
- 7.Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy, and R. H. Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855-859. [DOI] [PubMed] [Google Scholar]
- 8.Cockerill, P. N., and W. T. Garrard. 1986. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell 44:273-282. [DOI] [PubMed] [Google Scholar]
- 9.Dallas, P. B., P. Yaciuk, and E. Moran. 1997. Monoclonal antibody NM11 recognizes a C-terminal epitope shared by p300 and CBP. Hybridoma 16:273-275. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson, L. A., T. Joh, Y. Kohwi, and T. Kohwi-Shigematsu. 1992. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70:631-645. [DOI] [PubMed] [Google Scholar]
- 11.Dickinson, L. A., and T. Kohwi-Shigematsu. 1995. Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol. Cell. Biol. 15:456-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorsman, J. C., A. F. Teunisse, A. Zantema, and, A. J. van der Eb. 1997. The adenovirus 12 E1A proteins can bind directly to proteins of the p300 transcription co-activator family, including the CREB-binding protein CBP and p300. J. Gen. Virol. 78:423-426. [DOI] [PubMed] [Google Scholar]
- 13.Dreyfuss, G., Y. D. Choi, and S. A. Adam. 1984. Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol. Cell. Biol. 4:1104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duyndam, M. C., H. van Dam, P. H. Smits, M. Verlaan, A. J. van der Eb, and A. Zantema. 1999. The N-terminal transactivation domain of ATF2 is a target for the co-operative activation of the c-jun promoter by p300 and 12S E1A. Oncogene 18:2311-2321. [DOI] [PubMed] [Google Scholar]
- 15.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 16.Eckner, R., J. W. Ludlow, N. L. Lill, E. Oldread, Z. Arany, N. Modjtahedi, J. A. DeCaprio, D. M. Livingston, and J. A. Morgan. 1996. Association of p300 and CBP with simian virus 40 large T antigen. Mol. Cell. Biol. 16:3454-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fackelmayer, F. O., K. Dahm, A. Renz, U. Ramsperger, and A. Richter. 1994. Nucleic-acid-binding properties of hnRNP-U/SAF-A, a nuclear-matrix protein which binds DNA and RNA in vivo and in vitro. Eur. J. Biochem. 221:749-757. [DOI] [PubMed] [Google Scholar]
- 18.Fackelmayer, F. O., and A. Richter. 1994. Purification of two isoforms of hnRNP-U and characterization of their nucleic acid binding activity. Biochemistry 33:10416-10422. [DOI] [PubMed] [Google Scholar]
- 19.Fallaux, F. J., O. Kranenburg, S. J. Cramer, A. Houweling, H. Van Ormondt, R. C. Hoeben, and, A. J. van der Eb. 1996. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 7:215-222. [DOI] [PubMed] [Google Scholar]
- 20.Felsenfeld, G., J. Boyes, J. Chung, D. Clark, and V. Studitsky. 1996. Chromatin structure and gene expression. Proc. Natl. Acad. Sci. USA 93:9384-9388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez, L. A., M. Winkler, and R. Grosschedl. 2001. Matrix attachment region-dependent function of the immunoglobulin μ enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol. Cell. Biol. 21:196-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fey, E. G., G. Krochmalnic, and S. Penman. 1986. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J. Cell Biol. 102:1654-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasser, S. M., and U. K. Laemmli. 1986. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell 46:521-530. [DOI] [PubMed] [Google Scholar]
- 24.Gohring, F., and F. O. Fackelmayer. 1997. The scaffold/matrix attachment region binding protein hnRNP-U (SAF-A) is directly bound to chromosomal DNA in vivo: a chemical cross-linking study. Biochemistry 36:8276-8283. [DOI] [PubMed] [Google Scholar]
- 25.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 26.Hagmeyer, B. M., M. C. Duyndam, P. Angel, R. P. de Groot, M. Verlaan, P. Elfferich, A. van der Eb, and A. Zantema. 1996. Altered AP-1/ATF complexes in adenovirus-E1-transformed cells due to EIA-dependent induction of ATF3. Oncogene 12:1025-1032. [PubMed] [Google Scholar]
- 27.Harlow, E., B. R. Franza, and C. Schley. 1985. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 55:533-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izaurralde, E., E. Kas, and U. K. Laemmli. 1989. Highly preferential nucleation of histone H1 assembly on scaffold-associated regions. J. Mol. Biol. 210:573-585. [DOI] [PubMed] [Google Scholar]
- 29.Jarman, A. P., and D. R. Higgs. 1988. Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J. 7:3337-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenuwein, T., W. C. Forrester, L. A. Fernandez-Herrero, G. Laible, M. Dull, and R. Grosschedl. 1997. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature 385:269-272. [DOI] [PubMed] [Google Scholar]
- 31.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Kurokawa, B. Gloss, S. C. Lin, R. A. Heyman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403-414. [DOI] [PubMed] [Google Scholar]
- 32.Kas, E., and L. A. Chasin. 1987. Anchorage of the Chinese hamster dihydrofolate reductase gene to the nuclear scaffold occurs in an intragenic region. J. Mol. Biol. 198:677-692. [DOI] [PubMed] [Google Scholar]
- 33.Katsani, K. R., J. J. Arredondo, A. J. Kal, and C. P. Verrijzer. 2001. A homeotic mutation in the trithorax SET domain impedes histone binding. Genes Dev. 15:2197-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keblusek, P., J. C. Dorsman, A. F. Teunisse, H. Teunissen, A. J. van der Eb, and A. Zantema. 1999. The adenoviral E1A oncoproteins interfere with the growth-inhibiting effect of the cdk-inhibitor p21(CIP1/WAF1). J. Gen. Virol. 80:381-390. [DOI] [PubMed] [Google Scholar]
- 35.Kiledjian, M., and G. Dreyfuss. 1992. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 11:2655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kipp, M., F. Gohring, T. Ostendorp, C. M. van Drunen, R. van Driel, M. Przybylski, and F. O. Fackelmayer. 2000. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 20:7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klehr, D., K. Maass, and J. Bode. 1991. Scaffold-attached regions from the human interferon beta domain can be used to enhance the stable expression of genes under the control of various promoters. Biochemistry 30:1264-1270. [DOI] [PubMed] [Google Scholar]
- 38.Klein, B., A. Pastink, H. Odijk, A. Westerveld, and A. J. van der Eb. 1990. Transformation and immortalization of diploid xeroderma pigmentosum fibroblasts. Exp. Cell Res. 191:256-262. [DOI] [PubMed] [Google Scholar]
- 39.Kunze, N., G. C. Yang, M. Dolberg, R. Sundarp, R. Knippers, and A. Richter. 1991. Structure of the human type I DNA topoisomerase gene. J. Biol. Chem. 266:9610-9616. [PubMed] [Google Scholar]
- 40.Kunze, N., G. C. Yang, Z. Y. Jiang, H. Hameister, S. Adolph, K. H. Wiedorn, A. Richter, and R. Knippers. 1989. Localization of the active type I DNA topoisomerase gene on human chromosome 20q11.2-13.1 and two pseudogenes on chromosomes 1q23-24 and 22q11.2-13. 1. Hum. Genet. 84:6-10. [DOI] [PubMed] [Google Scholar]
- 41.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 42.Luderus, M. E., A. de Graaf, E. Mattia, J. L. den Blaauwen, M. A. Grande, L. de Jong, and R. van Driel. 1992. Binding of matrix attachment regions to lamin B1. Cell 70:949-959. [DOI] [PubMed] [Google Scholar]
- 43.McKenna, N. J., Z. Nawaz, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1998. Distinct steady-state nuclear receptor coregulator complexes exist in vivo. Proc. Natl. Acad. Sci. USA 95:11697-11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nickerson, J. A., B. J. Blencowe, and S. Penman. 1995. The architectural organization of nuclear metabolism. Int. Rev. Cytol. 162A:67-123. [DOI] [PubMed]
- 45.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 46.Orphanides, G., and D. Reinberg. 2000. RNA polymerase II elongation through chromatin. Nature 407:471-475. [DOI] [PubMed] [Google Scholar]
- 47.Phi-Van, L., and W. H. Stratling. 1996. Dissection of the ability of the chicken lysozyme gene 5′ matrix attachment region to stimulate transgene expression and to dampen position effects. Biochemistry 35:10735-10742. [DOI] [PubMed] [Google Scholar]
- 48.Phi-Van, L., J. P. von Kries, W. Ostertag, and W. H. Stratling. 1990. The chicken lysozyme 5′ matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol. Cell. Biol. 10:2302-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinol-Roma, S., M. S. Swanson, J. G. Gall, and G. Dreyfuss. 1989. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J. Cell Biol. 109:2575-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Renz, A., and F. O. Fackelmayer. 1996. Purification and molecular cloning of the scaffold attachment factor B (SAF-B), a novel human nuclear protein that specifically binds to S/MAR-DNA. Nucleic Acids Res. 24:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roest, P. A., R. G. Roberts, S. Sugino, G. J. van Ommen, and J. T. den Dunnen. 1993. Protein truncation test (PTT) for rapid detection of translation-terminating mutations. Hum. Mol. Genet. 2:1719-1721. [DOI] [PubMed] [Google Scholar]
- 52.Romig, H., F. O. Fackelmayer, A. Renz, U. Ramsperger, and A. Richter. 1992. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 11:3431-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romig, H., J. Ruff, F. O. Fackelmayer, M. S. Patil, and A. Richter. 1994. Characterisation of two intronic nuclear-matrix-attachment regions in the human DNA topoisomerase I gene. Eur. J. Biochem. 221:411-419. [DOI] [PubMed] [Google Scholar]
- 54.Stein, G. H. 1979. T98G: an anchorage-independent human tumor cell line that exhibits stationary phase G1 arrest in vitro. J. Cell Physiol. 99:43-54. [DOI] [PubMed] [Google Scholar]
- 55.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stief, A., D. M. Winter, W. H. Stratling, and A. E. Sippel. 1989. A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature 341:343-345. [DOI] [PubMed] [Google Scholar]
- 57.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 58.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 59.Toyoshima, H., and T. Hunter. 1994. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 78:67-74. [DOI] [PubMed] [Google Scholar]
- 60.van Dam, H., R. Offringa, A. M. Smits, J. L. Bos, N. C. Jones, and A. J. van der Eb. 1989. The repression of the growth factor-inducible genes JE, c-myc and stromelysin by adenovirus E1A is mediated by conserved region 1. Oncogene 4:1207-1212. [PubMed] [Google Scholar]
- 61.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205-214. [DOI] [PubMed] [Google Scholar]
- 62.Whitehurst, C., H. R. Henney, E. E. Max, H. W. Schroeder, Jr., F. Stuber, K. A. Siminovitch, and W. T. Garrard. 1992. Nucleotide sequence of the intron of the germline human kappa immunoglobulin gene connecting the J and C regions reveals a matrix association region (MAR) next to the enhancer. Nucleic Acids Res. 20:4929-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yanagisawa, J., J. Ando, J. Nakayama, Y. Kohwi, and T. Kohwi-Shigematsu. 1996. A matrix attachment region (MAR)-binding activity due to a p114 kilodalton protein is found only in human breast carcinomas and not in normal and benign breast disease tissues. Cancer Res. 56:457-462. [PubMed] [Google Scholar]
- 64.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 65.Yao, T. P., G. Ku, N. Zhou, R. Scully, and D. M. Livingston. 1996. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl. Acad. Sci. USA 93:10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao, K., E. Kas, E. Gonzalez, and U. K. Laemmli. 1993. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 12:3237-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]