Abstract
The DNA single-strand break repair (SSBR) protein XRCC1 is required for genetic stability and for embryonic viability. XRCC1 possesses two BRCA1 carboxyl-terminal (BRCT) protein interaction domains, denoted BRCT I and II. BRCT II is required for SSBR during G1 but is dispensable for this process during S/G2 and consequently for cell survival following DNA alkylation. Little is known about BRCT I, but this domain has attracted considerable interest because it is the site of a genetic polymorphism that epidemiological studies have associated with altered cancer risk. We report that the BRCT I domain comprises the evolutionarily conserved core of XRCC1 and that this domain is required for efficient SSBR during both G1 and S/G2 cell cycle phases and for cell survival following treatment with methyl methanesulfonate. However, the naturally occurring human polymorphism in BRCT I supported XRCC1-dependent SSBR and cell survival after DNA alkylation equally well. We conclude that while the BRCT I domain is critical for XRCC1 to maintain genetic integrity and cell survival, the polymorphism does not impact significantly on this function and therefore is unlikely to impact significantly on susceptibility to cancer.
Thousands of DNA single-strand breaks (SSBs) arise in cells each day both directly by the disintegration of damaged sugars and indirectly as intermediates of base excision repair (BER) (4, 29). If not repaired, SSBs pose a threat to both genetic stability and cell survival, resulting in an increased frequency of mutations and chromosome aberrations (15, 23, 38, 50, 53, 56). The threat posed by unrepaired SSBs most likely reflects their ability to become double-strand breaks during chromosome replication (27). The enzymatic pathways adopted by cells for DNA SSB repair (SSBR) can be divided into four basic steps, involving damage detection, end processing, gap filling, and DNA ligation (see reference 8 for a review). For example, in the case of direct SSBs arising from sugar damage or spontaneous cleavage at abasic sites, the breaks are most likely detected by poly(ADP-ribose) polymerase 1 (PARP-1) or poly(ADP-ribose) polymerase 2 (PARP-2) (3, 18-20, 52). Next, damaged 3′ or 5′ termini present at the breaks are converted to conventional 3′-hydroxyl and 5′-phosphate chemistries by AP endonuclease (APE1/HAP1), DNA polymerase β (Polβ), or polynucleotide kinase (PNK). Finally, SSBR is completed by gap filling by Polβ or Polδ/ɛ and DNA ligation by DNA ligase I (Lig1) or DNA ligase IIIα (Lig3α).
A feature of mammalian SSBR appears to be the employment of protein-protein interactions to stimulate individual component steps and/or the overall repair reaction. Arguably the most intriguing SSBR protein in this respect is XRCC1 (X-ray repair cross-complementing group 1), a polypeptide that interacts with PARP-1 (9, 33), PNK (54), Polβ (9, 26), and Lig3α (10, 11) (reviewed in references 8 and 51). The interaction of XRCC1 with PNK stimulates both the 5′-kinase and 3′-phosphatase activities of this enzyme (54), and the interaction with Lig3α increases the intracellular stability of the ligase (11, 46). Studies have demonstrated a role for XRCC1 both in vitro and in vivo during the repair of either direct SSBs or those arising indirectly during BER (13, 14, 16, 17, 21, 48, 49, 54). Consistent with this, rodent cells lacking XRCC1 are hypersensitive to a broad range of genotoxins (7, 13, 15-17, 49, 56). Loss of XRCC1 also results in decreased genetic stability, including increased frequencies of spontaneous and/or induced chromosome translocations and deletions (15, 23, 49, 53, 56).
Given the number of protein-protein interactions involving XRCC1 it is important to identify the contribution made by each to genetic stability. To achieve this we are systematically mutating the individual protein binding domains within XRCC1. A striking feature of XRCC1 is the presence of two BRCA1 carboxyl-terminal (BRCT) domains, denoted BRCT I and BRCT II, that are located centrally and at the C terminus of this polypeptide, respectively (5, 12). The C-terminal domain is responsible for binding and stabilizing Lig3α (31, 37, 47) and is required for SSBR specifically during the G0/G1 phase of the cell cycle (36, 46). In contrast, little is known about the role of the central BRCT I domain. However, this domain has become the focus of considerable interest since it was identified as the site of a common human genetic polymorphism (R399Q) that appears from a large number of epidemiological studies to impact significantly on cancer risk (1, 2, 6, 22, 24, 28, 34, 41-45). This polymorphism has been reported to both increase and decrease cancer frequencies, depending on the type and location of the cancer. In addition, a correlation has been reported between the presence of the 399Q allele and levels of DNA damage, mutation, and ionizing radiation-induced mitotic delay (25, 30, 35). Although provocative, these epidemiological studies are difficult to interpret because little is known about the role and importance of the BRCT I domain and because of the variation in genetic background that is intrinsic to such studies. Here, we have directly examined the importance of the BRCT I domain to cell survival and SSBR in an isogenic cellular background and examined the impact of the common R399Q polymorphism on these processes.
MATERIALS AND METHODS
Expression constructs and Arabidopsis thaliana XRCC1.
The mammalian cell expression vector pcD2E and the derivative pcD2EXH, which encodes C-terminally histidine-tagged XRCC1 (XRCC1-His), have been described previously (10). Derivatives of pcD2EXH harboring mutations within the BRCT I domain were created by site-directed mutagenesis using the QuikChange protocol (Stratagene). The sequence of all resulting open reading frames was confirmed by sequencing. The sequence of the A. thaliana XRCC1 was obtained by direct sequencing of the lambda PRL2 clone, 210J9T7, obtained from the Arabidopsis Biological Research Center (Ohio State University).
Cell culture.
CHO cells were cultured as monolayers or in suspension as appropriate in alpha-minimal essential medium supplemented with 10% fetal bovine serum (GibcoBRL). Expression constructs were introduced into EM9 cells by calcium phosphate coprecipitation, and stable clones were selected in the presence of G418 (1.5 mg/ml) for 10 to 14 days.
Cell extracts and affinity purification of histidine-tagged XRCC1 protein complexes.
Whole-cell extracts were prepared from frozen pellets of transfected EM9 cells (1 × 107 to 3 × 107) by resuspension in 1.5 ml of lysis buffer (50 mM Tris-Cl [pH 7.5], 130 mM NaCl, 0.5% Triton X-100, 10% glycerol, 1 mM dithiothreitol, 1/20 dilution of protease inhibitor cocktail [Sigma P8340]) and incubation on ice for 30 min. High-molecular-weight DNA was sheared by passage through a narrow-gauge needle several times, and insoluble material was removed by centrifugation in a microcentrifuge for 5 min at 4°C. To affinity-purify histidine-tagged XRCC1 complexes, clarified protein extracts (1.5 ml) from the different EM9 transfectants were incubated in parallel with 0.5 ml (bed volume) of nickel-nitrilotriacetic acid (NTA)-agarose (Qiagen) prewashed in lysis buffer and incubated with gentle agitation on ice for 30 min. The resulting suspensions were added to disposable 5-ml chromatography columns (Polyprep; Bio-Rad), and the protein-agarose beads were washed with 10 column volumes of lysis buffer containing 20 mM imidazole. Histidine-tagged XRCC1 protein complexes were eluted with 10 column volumes of lysis buffer containing 250 mM imidazole, with 0.25- to 0.5-ml fractions collected dropwise.
Indirect immunofluorescence.
Immunofluorescence was conducted as described previously (46). Images were captured at ×400 magnification.
SSBR assays.
Single cell agarose gel electrophoresis was conducted on asynchronous or synchronized populations of CHO cells as described previously (36, 46).
Survival curves.
Survival curves describing the resistance of transfected CHO cells to methyl methanesulfonate (MMS) were obtained as described previously, for ethyl methanesulfonate (46).
Nucleotide sequence accession number.
The sequence of the open reading frame of A. thaliana XRCC1 was placed in GenBank (accession number AJ276506).
RESULTS
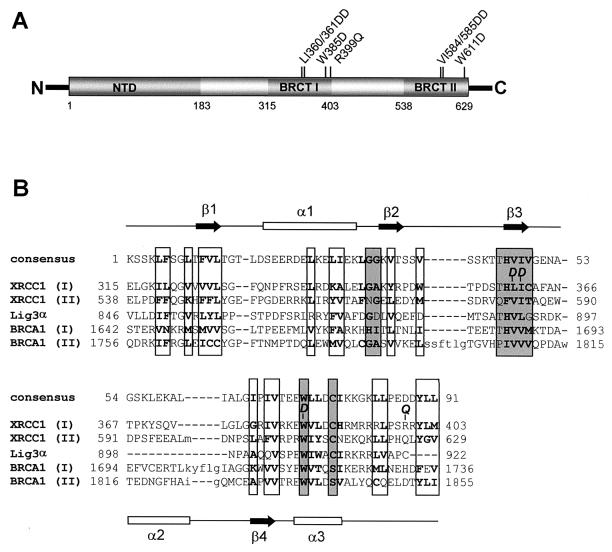
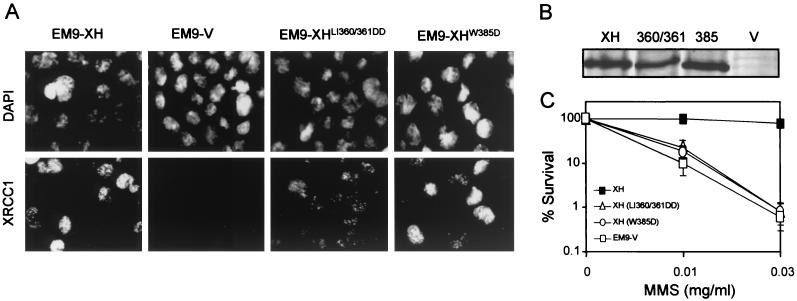
To examine the biological importance of the BRCT I domain, mutations were created that were analogous to those previously shown to disrupt the folding (57) and activity (36, 46, 47) of the BRCT II domain (Fig. 1A). These mutations lie within the β3 sheet (LI360/361DD) and the highly conserved α3 helix (W385D) (Fig. 1B). The mutation in the α3 helix is particularly noteworthy, since it removes the tryptophan residue that is present in almost all BRCT domains (5, 12). Empty pcD2E expression vector or pcD2E constructs encoding wild-type (pcD2EXH) or mutant (pcD2EXHLI360/361DD or pcD2EXHW385D) histidine-tagged XRCC1 were transfected into XRCC1 mutant EM9 cells by calcium phosphate coprecipitation. For further analysis we attempted to isolate single clones that possessed similar levels of wild-type or mutant XRCC1, as measured by indirect immunofluorescence and immunoblotting. In the clones selected, the level of mutant XRCC1W385D was essentially the same as the level of wild-type XRCC1, whereas the level of XRCC1LI360/361DD was approximately half this amount (Fig. 2A and B). A hallmark phenotype of XRCC1 mutant cells is hypersensitivity to MMS (49). Whereas pcD2EXH was able to restore wild-type levels of resistance to MMS, neither pcD2EXHLI360/361DD nor pcD2EXHW385D increased survival much above that observed in EM9 cells expressing empty pcD2E vector (Fig. 2C). In agreement with these data, EM9 cells harboring either pcD2EXHLI360/361DD or pcD2EXHW385D were also unable to survive culture conditions in which 20% of genomic thymine base is replaced with chlorouracil during cell division, conditions that are lethal to cells lacking XRCC1 but which do not affect the survival of cells expressing wild-type XRCC1 (data not shown). The inability of pcD2EXHLI360/361DD and pcD2EXHW385D to complement the MMS sensitivity of EM9 cells did not reflect the use of single clones, since similar results were observed with pooled populations of >100 clones (data not shown). The lack of cellular complementation by pcD2EXHLI360/361DD similarly did not simply reflect the lower level of expression of XRCC1LI360/361DD, since wild-type XRCC1 or XRCC1 harboring the analogous mutation in the BRCT II domain fully restores resistance to the alkylating agent even at protein levels that are undetectable by immunoblotting (36; unpublished observations).These data suggest that mutation of the BRCT I domain disrupts the ability of XRCC1 to promote cell survival following MMS treatment. This contrasts dramatically with the BRCT II domain, which is largely dispensable for MMS resistance in cycling CHO cells (36, 46).
FIG. 1.
(A) XRCC1 protein-protein interaction domains. The location of the NTD and the two BRCT domains within XRCC1 are indicated. The location of mutations in BRCT II previously employed (36, 46, 47) to disrupt activity of this domain, and the analogous BRCT I mutations employed in this study, are shown. Also shown is the location of the common human genetic polymorphism in BRCT I, R399Q. (B) CD alignment (NCBI) of the pfam00533 consensus BRCT domain with BRCT domains present in XRCC1, Lig3α, and BRCA1. Boxed residues denote regions of hydrophobicity (with hydrophobic residues in boldface) that are conserved in the BRCT family, with those in dark grey denoting those that are most conserved. The position of secondary structure, based on the crystal structure of the BRCT II domain of XRCC1 (57), is shown at the top and bottom. The amino acids mutated to aspartate (D) in this study, and the position of the common human polymorphism (Q) is also indicated.
FIG. 2.
Characterization of EM9 cells expressing wild-type or mutant XRCC1. (A) XRCC1 protein was examined in the indicated cell lines by indirect immunofluorescence, using the primary anti-XRCC1 MAb 33-2-5 and a fluorescein isothiocyanate-conjugated rabbit antimouse secondary antibody (DAKO). Nuclei were visualized with the DNA stain 4′,6′-diamidino-2-phenylindole. Photographs were taken under ×400 magnification. (B) XRCC1 protein levels were examined in cell extracts (20 μg of total protein) from the indicated cell lines (XH, EM9-XH; 360/361, EM9-XHLI360/361DD; 385, EM9-XHW385D; V, EM9-V) by immunoblotting, using the anti-XRCC1 MAb 33-2-5. (C) The cell lines indicated (see panel A for an explanation of symbols) were plated in six-well dishes (200 cells/well) and treated with the indicated concentrations of MMS for 1 h. After a wash, cells were incubated in drug-free medium for 7 to 10 days to allow formation of macroscopic colonies. The fraction of cells surviving MMS treatment was calculated by dividing the number of colonies in treated wells by the number in untreated wells. Results are the mean ± 1 standard deviation of three independent experiments.
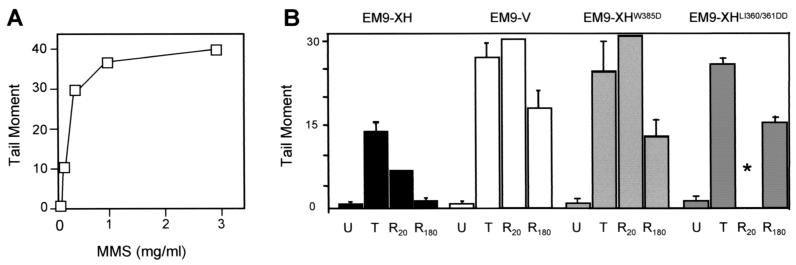
To examine whether the mutated BRCT I domain disrupts SSBR, this process was measured in transfected EM9 cell using alkaline agarose single-cell gel electrophoresis. This technique, also known as the comet assay, measures DNA strand breaks by their ability to increase the amount of DNA that exits the cell nucleus, and the distance migrated by this DNA, under an electric field. DNA damage is expressed graphically as the tail moment, an arbitrary unit reflecting the product of these two parameters. In EM9-V cells lacking XRCC1-dependent SSBR, tail moment increased linearly with doses of up to ∼0.5 mg of MMS/ml (Fig. 3A), and so a concentration of 0.3 mg/ml was chosen for subsequent experiments. Whereas the level of SSBs in EM9-XH cells rose ∼15-fold during MMS treatment, the level in EM9-V, EM9-XHLI360/361DD, and EM9-XHW385D cells rose ∼30-fold (Fig. 3B). A difference between repair-proficient and mutant EM9 cells in the level of SSBs accumulated during DNA alkylation has been noted previously and is due to a differential ability to repair SSBs during drug treatment (36, 46, 49). Thus, the observation that EM9-XHLI360/361DD and EM9-XHW385D accumulated levels of SSBs similar to those of EM9-V rather than EM9-XH suggests that the former cell lines are SSBR defective. This was confirmed during a subsequent repair incubation in drug-free medium, because the level of MMS-induced SSBs in EM9-XH cells dropped by ∼40% within 20 min whereas the level in EM9-V and EM9-XHW385D cells appeared to increase by ∼20% (Fig. 3B). This increase in the last two cell lines most likely reflects continued excision of damaged bases in the absence of efficient SSB rejoining. The number of MMS-induced SSBs removed by EM9-XHLI360/361DD and EM9-XHW385Dcells was also no greater than that removed by EM9-V cells after a repair incubation of 180 min (Fig. 3B), further suggesting that the mutated BRCT I domain ablated XRCC1-dependent SSBR. It was noted that some residual SSBR was present in EM9-V, EM9-XHLI360/361DD, and EM9-XHW385Dcells, however, since after the initial increase the level of MMS-induced SSBs dropped by 40 to 60% by the end of the experiment (Fig. 3B). Residual SSBR in XRCC1-deficient cell lines has been noted previously (49) and is consistent with the activity of an XRCC1-independent SSBR process such as that occurring during BER mediated by PCNA, DNA polymerase δ/ɛ, and Lig1 (reviewed in reference 8).
FIG. 3.
Measurement of MMS-induced SSBs in asynchronous populations of transfected EM9 cells. (A) EM9-V cells were treated for 15 min with the indicated concentrations of MMS, and the level of SSBs was quantified by alkaline agarose gel electrophoresis (comet assay). SSB levels are expressed as the tail moment, an arbitrary unit reflecting the product of the amount of DNA present in the “comet” tail and the tail length, after electrophoresis. (B) SSBs were quantified in the indicated EM9 transfectants before treatment for 15 min with 0.3 mg of MMS/ml (U, untreated), immediately after MMS treatment (T, treated), or after a subsequent repair incubation in drug-free medium for 20 min (R20) or 3 h (R180). Results are the average from four independent experiments (±1 standard deviation). The asterisk denotes the absence of a data point for a 20-min repair incubation in EM9-XHLI360/361DD cells.
Mutation of the BRCT II domain selectively ablates XRCC1-dependent SSBR in G0/G1 phases of the cell cycle (8, 36, 46). To examine whether mutation of the BRCT I domain similarly ablates SSBR in a cell cycle-specific manner, SSBR was measured in cells that were synchronized in G1 or S/G2 phase prior to MMS treatment (Fig. 4a). Synchronization was achieved as previously described (36, 39, 46) by a combination of serum starvation and mimosine to arrest the cells at the G1/S boundary (39). We have shown previously by BrdUrd pulse-labeling that populations of CHO cells synchronized in this way do not possess replicating cells (36). The steady-state level of SSBs increased ∼20-fold in each of the cell lines during treatment with MMS in G1 (Fig. 4B, top panel). Once again, neither EM9-XHLI360/361DD nor EM9-XHW385D cells removed significantly more MMS-induced SSBs during a 3-h repair incubation than did EM9-V cells, with ∼30% removed in these cell lines compared to >90% in EM9-XH cells (Fig. 4B, top panel). Similar results were observed in cells synchronized in the S/G2 phase of the cell cycle, with the level of SSBs remaining in EM9-XHLI360/361DD and EM9-XHW385D cells after 3 h similar to those present in EM9-V cells (Fig. 4B, bottom panel). These data indicate that in contrast to the BRCT II domain, mutation of the BRCT I domain ablates XRCC1-dependent SSBR in both G1 and S/G2 phases of the cell cycle.
FIG. 4.
Measurement of MMS-induced SSBs in synchronized populations of EM9 transfectants. (A) DNA content of EM9-V cells synchronized in G1 (top panel) or S/G2 (bottom panel) by serum starvation and mimosine as described previously (36, 39, 46). Similar synchrony was observed with other EM9 transfectants (data not shown). The respective arrows denote peak positions of cells with G1 and G2 DNA content. (B) SSBs were quantified in the indicated EM9 transfectants in G1 (top panel) or S/G2 (bottom panel) before treatment for 15 min with 0.3 mg of MMS/ml (U, untreated), immediately after MMS treatment (T, treated), or after a subsequent repair incubation in drug-free medium for 3 h (R180). Results are the mean (±1 standard deviation) from three independent experiments.
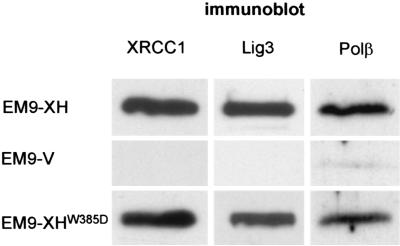
XRCC1 contains two other known protein-protein interaction domains in addition to the BRCT I domain. These are the N-terminal domain (NTD) and the BRCT II domain, which together flank the central BRCT I domain (see Fig. 1A) and which mediate interactions with Polβ and Lig3α, respectively (26, 31, 32, 37, 47). It was considered possible that mutation of the BRCT I domain may have ablated XRCC1-dependent SSBR indirectly, by interfering with the function of one or both of these domains. To examine this possibility, the presence of Polβ and Lig3α in XRCC1 protein complexes affinity purified from EM9-XH and EM9-XHW385D cells was measured by immunoblotting. These experiments revealed that similar amounts of Polβ and Lig3α copurified with XRCC1 complexes from EM9-XH and EM9-XHW385D cell extract (Fig. 5), suggesting that the inability of XRCC1W385D to support SSBR was not due to the W385D mutation interfering with the domains that bind these proteins. Rather, these experiments suggest that the inability of XRCC1W385D to support SSBR reflects a direct requirement for the BRCT I domain for this process.
FIG. 5.
Presence of Lig3 and Polβ in affinity-purified XRCC1 protein complexes. Total cellular protein (20 μg) from the cell lines indicated on the left was subjected to metal chelate affinity chromatography (nickel agarose; Qiagen) to purify histidine-tagged XRCC1 protein complexes. Aliquots of the recovered complexes were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with anti-XRCC1 MAb (33-2-5), anti-Lig3 Pab (TL-25), or anti-Polβ MAb (Clone 18S; Lab Vision).
Several common polymorphisms have been described in XRCC1 that are overrepresented or underrepresented in individuals with certain types of cancer (1, 2, 6, 22, 24, 28, 34, 41-45). Intriguingly, the polymorphism most commonly reported to affect cancer risk is R399Q, which is located at the C terminus of the BRCT I domain (see Fig. 1). The allele frequency of 399Q is 15 to 30%, based on studies conducted so far, resulting in a high frequency of individuals (5 to 15%) that are homozygous for this allele. In addition, a correlation has been reported between the presence of the 399Q allele and levels of DNA damage, mutation, and ionizing radiation-induced mitotic delay (25, 30, 35). We therefore examined directly whether the two isoforms of XRCC1 differed in their ability to support XRCC1-dependent cell survival and SSBR following exposure to MMS. Analysis of pooled populations of EM9 transfectants harboring pcD2E, pcD2EXH, or pcD2EXHR399Q failed to reveal any difference in the sensitivity of these cell lines to MMS (Fig. 6A). The lack of quantitative difference between the MMS sensitivities of EM9XH and EM9XHR399Q cell populations (in both of which >90% of individual clones expressed recombinant XRCC1) did not reflect differences in steady-state levels of XRCC1, since similar levels of this protein were observed in these cell lines (Fig. 6B). We also examined the 399Q allele for the ability to support SSBR following treatment with MMS. XRCC1R399Q removed an amount of single-strand breakage similar to that removed by wild-type XRCC1 during a 3-h repair-incubation, suggesting that the two isoforms are not significantly different in SSBR capacity (Fig. 6C and D).
FIG. 6.
MMS sensitivity and SSBR capacity of EM9 cells harboring the human genetic polymorphism XRCC1R399Q. (A) The indicated cell lines (pooled populations of >100 independent transfectants) were plated in six-well dishes (200 cells/plate) and treated with the indicated concentration of MMS for 1 h. After a wash, cells were incubated in drug-free medium for 7 to 10 days to allow formation of macroscopic colonies, and the fraction of surviving cells was calculated as described in the legend for Fig. 2. (B) Total cellular protein (20 μg) from the indicated cell lines was fractionated by SDS-PAGE and immunoblotted with the anti-XRCC1 MAb, 33-2-5. (C) SSBs (expressed as tail moments) were quantified in the indicated EM9 transfectants before (−MMS) or immediately after (+MMS) treatment with 0.3 mg of MMS/ml. (D) The percentage of the MMS-induced SSBs shown in panel C remaining after a 3-h repair incubation in drug-free medium (calculated from the tail moment present after the repair incubation). Symbols are as in panel C. Results are the mean ± 1 standard deviation for three independent experiments.
DISCUSSION
The BRCT I domain is required for XRCC1 function during SSBR and for resistance to MMS.
XRCC1 is important for genetic stability and for embryonic viability (23, 48, 49). This polypeptide appears to function as a scaffold protein that mediates the assembly of SSBR protein complexes (8, 54). To examine the contribution of individual protein interactions to SSBR, we have been systematically mutating individual proteinbinding domains within XRCC1. Two such domains are BRCT I and BRCT II, which are located in the center and C terminus of the polypeptide, respectively (5, 12). We reported previously that the BRCT II domain mediates interaction with Lig3α and is required for XRCC1-dependent SSBR during G0/G1 phases of the cell cycle (36, 46). However, this domain is largely dispensable for XRCC1-dependent SSBR during S/G2 phase and consequently for cellular resistance to alkylating agents (46). In contrast to BRCT II, we report here that mutation of the BRCT I domain ablates XRCC1-dependent SSBR in both G1 and S/G2 and also prevents XRCC1 from maintaining cell survival after DNA alkylation.
It seems unlikely that the BRCT I mutations ablate XRCC1-dependent SSBR through an indirect effect on regions located outside of this domain. This is because these mutations did not disrupt binding of XRCC1 to either Polβ or Lig3α, two interactions known to occur within XRCC1 domains that flank the BRCT I domain. Rather, these data suggest a direct requirement for the BRCT I domain for SSBR in G1 and S/G2 and for cellular resistance to MMS.
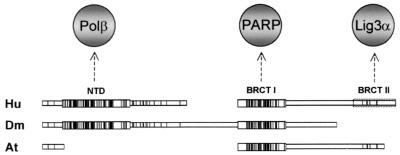
The suggestion that the BRCT I domain is a fundamental component of XRCC1 function is supported by the comparison of XRCC1 homologues in different species. Whereas the BRCT II domain is present in human and other mammalian species, it is absent from both Drosophila melanogaster and A. thaliana XRCC1 (Fig. 7). In contrast, the BRCT I domain is present in all XRCC1 homologues so far identified.
FIG. 7.
Evolutionary conservation of XRCC1 protein domains. Human (Hu), D. melanogaster (Dm), and A. thaliana (At) XRCC1 homologues were aligned using MACAW software, with vertical bars denoting conserved residues and enlarged boxes denoting regions of extensive conservation. The dotted box denotes the position of the BRCT II domain present in mammalian XRCC1 (represented here by the human protein). The domains identified by this alignment are the NTD, which contains the binding site for Polβ, and the two BRCT domains, which contain the binding sites for PARP-1, poly(ADP-ribose), and Lig3α. The Hu and Dm BRCT I domains, and the Hu and At BRCT I domains, share 54.5 and 52.8% identity, respectively.
Biochemical role of the BRCT I domain.
In contrast to the BRCT II domain, the BRCT I domain appears to be indispensable for XRCC1-dependent SSBR in both G1 and S/G2. What biochemical role does the BRCT I domain fulfill? This domain does not appear to be required for XRCC1 to assemble into discrete subnuclei foci (see Fig. 2A), structures previously shown to assemble primarily during S phase and in response to DNA damage (46). However, in addition to Polβ and Lig3α, XRCC1 also interacts with PARP-1 (9, 33), and the site of this interaction is located within the BRCT I domain (33). It is likely that the interaction with PARP-1 may serve to recruit XRCC1 protein complexes to sites of single-strand breakage, since XRCC1 preferentially binds the activated form of PARP-1 that arises once the latter polypeptide has bound an SSB (33, 40). This ability to discriminate between active and inactive PARP-1 most likely reflects the ability of XRCC1 to bind poly(ADP-ribose), the polymeric product of PARP-1 activity (40). Further work is required to determine whether or not binding by the XRCC1 BRCT I domain to PARP-1 and poly(ADP-ribose) can account for the importance of this domain to SSBR and cell survival.
It is also possible that the BRCT I domain mediates the recently identified interaction of XRCC1 with PNK (54). This interaction stimulates the activity of PNK at SSB termini, thereby enabling restoration of the conventional 3′-hydroxyl and 5′-phosphate chemistry required for completion of SSBR by gap filling and DNA ligation. Finally, it is possible that BRCT I also interacts with an as yet unidentified polypeptide or that it fulfills some other role. For example, it has been reported that BRCT domains can bind DNA directly, though the physiological significance of this is unclear (55). Clearly, given the importance of the BRCT I domain to XRCC1 function and genetic integrity, it is important to identify the role/s fulfilled by this structure.
A human polymorphism within BRCT I.
One class of genetic factors that are likely to contribute to cancer predisposition is genetic polymorphisms. The XRCC1 BRCT I domain has recently attracted considerable interest because of the identification of a common genetic polymorphism (R399Q) at the C terminus of the BRCT I domain (43). Intriguingly, a large number of studies have reported an overrepresentation of one allele over the other among groups of individuals with a variety of cancers, including those of the lung, breast, bladder, and esophagus (1, 2, 6, 22, 24, 28, 34, 41-45). The explanation suggested for overrepresentation of one or another of the XRCC1 alleles among cancer groupings has been that the polymorphism impacts upon the activity of XRCC1 and consequently on SSBR efficiency and genetic stability. Consistent with this, a correlation has also been reported between the presence of the 399Q allele and levels of DNA damage, mutation, and ionizing radiation-induced mitotic delay (25, 30, 35).
Although provocative, such epidemiological studies are difficult to interpret in lieu of functional data concerning the impact of the polymorphism on protein function. Moreover, there is currently no clear consensus among the epidemiological studies as to which of the alleles is detrimental. A further complication with respect to XRCC1 is the additional location of several other DNA repair genes within the region of chromosome 19q13.2-13.3, such as ERCC1, ERCC2, DNA ligase I, and polynucleotide kinase, that could harbor polymorphisms that contribute to the apparent impact of the XRCC1 polymorphism. To circumvent these problems, in this study we compared directly for the first time the two XRCC1 isoforms for their ability to conduct SSBR and mediate cell survival in an isogenic background. In these experiments the two alleles were equally able to complement both the SSBR defect and sensitivity to MMS observed in XRCC1-deficient EM9 cells. Given that the BRCT I domain is critical to these functions, these experiments suggest that the polymorphism has little effect on the BRCT I domain or XRCC1 function.
In summary, we report that the BRCT I domain comprises the evolutionarily conserved core motif of XRCC1 and that this domain is critical for efficient SSBR and cell survival. However, the common human polymorphism located within this domain does not appear to significantly affect the ability of the BRCT I domain to support either of these processes following DNA alkylation, suggesting that it is unlikely to impact significantly on susceptibility to cancer.
Acknowledgments
R.M.T. was funded by Medical Research Council grants to K.W.C. (grants G9809326, G0001259, and G9821041).
REFERENCES
- 1.Abdel-Rahman, S. Z., and R. A. El Zein. 2000. The 399Gln polymorphism in the DNA repair gene XRCC1 modulates the genotoxic response induced in human lymphocytes by the tobacco-specific nitrosamine NNK. Cancer Lett. 159:63-71. [DOI] [PubMed] [Google Scholar]
- 2.Abdel-Rahman, S. Z., A. S. Soliman, M. L. Bondy, S. Omar, S. A. El Badawy, H. M. Khaled, I. A. Seifeldin, and B. Levin. 2000. Inheritance of the 194Trp and the 399Gln variant alleles of the DNA repair gene XRCC1 are associated with increased risk of early-onset colorectal carcinoma in Egypt. Cancer Lett. 159:79-86. [DOI] [PubMed] [Google Scholar]
- 3.Ame, J. C., V. Rolli, V. Schreiber, C. Niedergang, F. Apiou, P. Decker, S. Muller, T. Hoger, J. Menissier-de Murcia, and G. de Murcia. 1999. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274:17860-17868. [DOI] [PubMed] [Google Scholar]
- 4.Beckman, K. B., and B. N. Ames. 1997. Oxidative decay of DNA. J. Biol. Chem. 272:19633-19636. [DOI] [PubMed] [Google Scholar]
- 5.Bork, P., K. Hofmann, P. Bucher, A. F. Neuwald, S. F. Altschul, and E. V. Koonin. 1997. A superfamily of conserved domains in DNA damage responsive cell cycle checkpoint proteins. FASEB J. 11:68-76. [PubMed] [Google Scholar]
- 6.Butkiewicz, D., M. Rusin, L. Enewold, P. G. Shields, M. Chorazy, and C. C. Harris. 2001. Genetic polymorphisms in DNA repair genes and risk of lung cancer. Carcinogenesis 22:593-597. [DOI] [PubMed] [Google Scholar]
- 7.Caldecott, K., and P. Jeggo. 1991. Cross-sensitivity of gamma-ray-sensitive hamster mutants to cross-linking agents. Mutat. Res. 255:111-121. [DOI] [PubMed] [Google Scholar]
- 8.Caldecott, K. W. 2001. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays 23:447-455. [DOI] [PubMed] [Google Scholar]
- 9.Caldecott, K. W., S. Aoufouchi, P. Johnson, and S. Shall. 1996. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular "nick-sensor' in vitro. Nucleic Acids Res. 24:4387-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caldecott, K. W., C. K. Mckeown, J. D. Tucker, S. Ljungquist, and L. H. Thompson. 1994. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 14:68-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caldecott, K. W., J. D. Tucker, L. H. Stanker, and L. H. Thompson. 1995. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 23:4836-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callebaut, I., and J. P. Mornon. 1997. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 400:25-30. [DOI] [PubMed] [Google Scholar]
- 13.Cantoni, O., D. Murray, and R. E. Meyn. 1987. Induction and repair of DNA single-strand breaks in EM9 mutant CHO cells treated with hydrogen-peroxide. Chem.-Biol. Interact. 63:29-38. [DOI] [PubMed] [Google Scholar]
- 14.Cappelli, E., R. Taylor, M. Cevasco, A. Abbondandolo, K. Caldecott, and G. Frosina. 1997. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J. Biol. Chem. 272:23970-23975. [DOI] [PubMed] [Google Scholar]
- 15.Carrano, A. V., J. L. Minkler, L. E. Dillehay, and L. H. Thompson. 1986. Incorporated bromodeoxyuridine enhances the sister-chromatid exchange and chromosomal aberration frequencies in an EMS-sensitive Chinese hamster cell line. Mutat. Res. 162:233-239. [DOI] [PubMed] [Google Scholar]
- 16.Churchill, M. E., J. G. Peak, and M. J. Peak. 1991. Correlation between cell-survival and DNA single-strand break repair proficiency in the Chinese-hamster ovary cell-lines AA8 and EM9 irradiated with 365-nm ultraviolet-a radiation. Photochem. Photobiol. 53:229-236. [DOI] [PubMed] [Google Scholar]
- 17.Churchill, M. E., J. G. Peak, and M. J. Peak. 1991. Repair of near-visible-light-induced and blue-light-induced DNA single-strand breaks by the CHO cell-lines AA8 and EM9. Photochem. Photobiol. 54:639-644. [DOI] [PubMed] [Google Scholar]
- 18.Dantzer, F., G. de la Rubia, J. Menissier-de Murcia, Z. Hostomsky, G. de Murcia, and V. Schreiber. 2000. Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry 39:7559-7569. [DOI] [PubMed] [Google Scholar]
- 19.Dantzer, F., V. Schreiber, C. Niedergang, C. Trucco, E. Flatter, G. de la Rubia, J. Oliver, V. Rolli, J. Menissier-de Murcia, and G. de Murcia. 1999. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie 81:69-75. [DOI] [PubMed] [Google Scholar]
- 20.de Murcia, G., and D. M. Menissier. 1994. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem. Sci. 19:172-176. [DOI] [PubMed] [Google Scholar]
- 21.Dillehay, L. E., L. H. Thompson, and A. V. Carrano. 1984. DNA-strand breaks associated with halogenated pyrimidine incorporation. Mutat. Res. 131:129-136. [DOI] [PubMed] [Google Scholar]
- 22.Divine, K. K., F. D. Gilliland, R. E. Crowell, C. A. Stidley, T. J. Bocklage, D. L. Cook, and S. A. Belinsky. 2001. The XRCC1 399 glutamine allele is a risk factor for adenocarcinoma of the lung. Mutat. Res. 461:273-278. [DOI] [PubMed] [Google Scholar]
- 23.Dominguez, I., P. Daza, A. T. Natarajan, and F. Cortes. 1998. A high yield of translocations parallels the high yield of sister chromatid exchanges in the CHO mutant EM9. Mutat. Res. 398:67-73. [DOI] [PubMed] [Google Scholar]
- 24.Duell, E. J., R. C. Millikan, G. S. Pittman, S. Winkel, R. M. Lunn, C. K. Tse, A. Eaton, H. W. Mohrenweiser, B. Newman, and D. A. Bell. 2001. Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiol. Biomarkers Prev. 10:217-222. [PubMed] [Google Scholar]
- 25.Hu, J. J., T. R. Smith, M. S. Miller, H. W. Mohrenweiser, A. Golden, and L. D. Case. 2001. Amino acid substitution variants of APE1 and XRCC1 genes associated with ionizing radiation sensitivity. Carcinogenesis 22:917-922. [DOI] [PubMed] [Google Scholar]
- 26.Kubota, Y., R. A. Nash, A. Klungland, P. Schar, D. E. Barnes, and T. Lindahl. 1996. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 15:6662-6670. [PMC free article] [PubMed] [Google Scholar]
- 27.Kuzminov, A. 2001. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl. Acad. Sci. USA 98:8241-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J. M., Y. C. Lee, S. Y. Yang, P. W. Yang, S. P. Luh, C. J. Lee, C. J. Chen, and M. T. Wu. 2001. Genetic polymorphisms of XRCC1 and risk of the esophageal cancer. Int. J. Cancer 95:240-246. [DOI] [PubMed] [Google Scholar]
- 29.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 30.Lunn, R. M., R. G. Langlois, L. L. Hsieh, C. L. Thompson, and D. A. Bell. 1999. XRCC1 polymorphisms: effects on aflatoxin B-1-DNA adducts and glycophorin A variant frequency. Cancer Res. 59:2557-2561. [PubMed] [Google Scholar]
- 31.Mackey, Z. B., W. Ramos, D. S. Levin, C. A. Walter, J. R. Mccarrey, and A. E. Tomkinson. 1997. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol. Cell. Biol. 17:989-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marintchev, A., M. A. Mullen, M. W. Maciejewski, B. Pan, M. R. Gryk, and G. P. Mullen. 1999. Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nat. Struct. Biol. 6:884-893. [DOI] [PubMed] [Google Scholar]
- 33.Masson, M., C. Niedergang, V. Schreiber, S. Muller, J. Menissier-de Murcia, and G. de Murcia. 1998. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 18:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matullo, G., S. Guarrera, S. Carturan, M. Peluso, C. Malaveille, L. Davico, A. Piazza, and P. Vineis. 2001. DNA repair gene polymorphisms, bulky DNA adducts in white blood cells and bladder cancer in a case-control study. Int. J. Cancer 92:562-567. [DOI] [PubMed] [Google Scholar]
- 35.Matullo, G., D. Palli, M. Peluso, S. Guarrera, S. Carturan, E. Celentano, V. Krogh, A. Munnia, R. Tumino, S. Polidoro, A. Piazza, and P. Vineis. 2001. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis 22:1437-1445. [DOI] [PubMed] [Google Scholar]
- 36.Moore, D. J., R. M. Taylor, P. Clements, and K. W. Caldecott. 2000. Mutation of a BRCT domain selectively disrupts DNA single-strand break repair in noncycling Chinese hamster ovary cells. Proc. Natl. Acad. Sci. USA 97:13649-13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nash, R. A., K. W. Caldecott, D. E. Barnes, and T. Lindahl. 1997. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry 36:5207-5211. [DOI] [PubMed] [Google Scholar]
- 38.Ochs, K., R. W. Sobol, S. H. Wilson, and B. Kaina. 1999. Cells deficient in DNA polymerase beta are hypersensitive to alkylating agent-induced apoptosis and chromosomal breakage. Cancer Res. 59:1544-1551. [PubMed] [Google Scholar]
- 39.Orren, D. K., L. N. Petersen, and V. A. Bohr. 1995. A UV-responsive G2 checkpoint in rodent cells. Mol. Cell. Biol. 15:3722-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pleschke, J. M., H. E. Kleczkowska, M. Strohm, and F. R. Althaus. 2000. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275:40974-40980. [DOI] [PubMed] [Google Scholar]
- 41.Ratnasinghe, D., S. X. Yao, J. A. Tangrea, Y. L. Qiao, M. R. Andersen, M. J. Barrett, C. A. Giffen, Y. Erozan, M. S. Tockman, and P. R. Taylor. 2001. Polymorphisms of the DNA repair gene XRCC1 and lung cancer risk. Cancer Epidemiol. Biomarkers Prev. 10:119-123. [PubMed] [Google Scholar]
- 42.Shen, H., Y. Xu, Y. Qian, R. Yu, Y. Qin, L. Zhou, X. Wang, M. R. Spitz, and Q. Wei. 2000. Polymorphisms of the DNA repair gene XRCC1 and risk of gastric cancer in a Chinese population. Int. J. Cancer 88:601-606. [DOI] [PubMed] [Google Scholar]
- 43.Shen, M. R., I. M. Jones, and H. Mohrenweiser. 1998. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 58:604-608. [PubMed] [Google Scholar]
- 44.Stern, M. C., D. M. Umbach, C. H. van Gils, R. M. Lunn, and J. A. Taylor. 2001. DNA repair gene XRCC1 polymorphisms, smoking, and bladder cancer risk. Cancer Epidemiol. Biomarkers Prev. 10:125-131. [PubMed] [Google Scholar]
- 45.Sturgis, E. M., E. J. Castillo, L. Li, R. Zheng, S. A. Eicher, G. L. Clayman, S. S. Strom, M. R. Spitz, and Q. Wei. 1999. Polymorphisms of DNA repair gene XRCC1 in squamous cell carcinoma of the head and neck. Carcinogenesis 20:2125-2129. [DOI] [PubMed] [Google Scholar]
- 46.Taylor, R. M., D. J. Moore, J. Whitehouse, P. Johnson, and K. W. Caldecott. 2000. A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol. Cell. Biol. 20:735-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, R. M., B. Wickstead, S. Cronin, and K. W. Caldecott. 1998. Role of a BRCT domain in the interaction of DNA ligase III-alpha with the DNA repair protein XRCC1. Curr. Biol. 8:877-880. [DOI] [PubMed] [Google Scholar]
- 48.Tebbs, R. S., M. L. Flannery, J. J. Meneses, A. Hartmann, J. D. Tucker, L. H. Thompson, J. E. Cleaver, and R. A. Pedersen. 1999. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol. 208:513-529. [DOI] [PubMed] [Google Scholar]
- 49.Thompson, L. H., K. W. Brookman, L. E. Dillehay, A. V. Carrano, J. A. Mazrimas, C. L. Mooney, and J. L. Minkler. 1982. A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutat. Res. 95:427-440. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, L. H., K. W. Brookman, L. E. Dillehay, C. L. Mooney, and A. V. Carrano. 1982. Hypersensitivity to mutation and sister-chromatid-exchange induction in CHO cell mutants defective in incising DNA containing UV lesions. Somatic Cell Genet. 8:759-773. [DOI] [PubMed] [Google Scholar]
- 51.Thompson, L. H., and M. G. West. 2000. XRCC1 keeps DNA from getting stranded. Mutat. Res. 459:1-18. [DOI] [PubMed] [Google Scholar]
- 52.Trucco, C., F. J. Oliver, G. de Murcia, and J. Menissier-de Murcia. 1998. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 26:2644-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veld, C. W. O. H., J. Jansen, M. Z. Zdzienicka, H. Vrieling, and A. A. vanZeeland. 1998. Methyl methanesulfonate-induced hprt mutation spectra in the Chinese hamster cell line CH09 and its xrccl-deficient derivative EM-C11. Mutat. Res.-Fundam. Mol. Mech. Mutagenes. 398:83-92. [DOI] [PubMed] [Google Scholar]
- 54.Whitehouse, C. J., R. M. Taylor, A. Thistlethwaite, H. Zhang, F. Karimi-Busheri, D. D. Lasko, M. Weinfeld, and K. W. Caldecott. 2001. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 104:1-11. [DOI] [PubMed] [Google Scholar]
- 55.Yamane, K., E. Katayama, and T. Tsuruo. 2000. The BRCT regions of tumor suppressor BRCA1 and of XRCC1 show DNA end binding activity with a multimerizing feature. Biochem. Biophys. Res. Commun. 279:678-684. [DOI] [PubMed] [Google Scholar]
- 56.Zdzienicka, M. Z., G. P. Vanderschans, A. T. Natarajan, L. H. Thompson, I. Neuteboom, and J. W. I. M. Simons. 1992. A Chinese-hamster ovary cell mutant (EM-C11) with sensitivity to simple alkylating agents and a very high level of sister chromatid exchanges. Mutagenesis 7:265-269. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, X. D., S. Morera, P. A. Bates, P. C. Whitehead, A. I. Coffer, K. Hainbucher, R. A. Nash, M. J. E. Sternberg, T. Lindahl, and P. S. Freemont. 1998. Structure of an XRCC1 BRCT domain: a new protein-protein interaction module. EMBO J. 17:6404-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]