Abstract
We present evidence that the inducer-specific regulation of the human tumor necrosis factor alpha (TNF-α) gene in T cells involves the assembly of distinct higher-order transcription enhancer complexes (enhanceosomes), which is dependent upon inducer-specific helical phasing relationships between transcription factor binding sites. While ATF-2, c-Jun, and the coactivator proteins CBP/p300 play a central role in TNF-α gene activation stimulated by virus infection or intracellular calcium flux, different sets of activators including NFATp, Sp1, and Ets/Elk are recruited to a shared set of transcription factor binding sites depending upon the particular stimulus. Thus, these studies demonstrate that the inducer-specific assembly of unique enhanceosomes is a general mechanism by which a single gene is controlled in response to different extracellular stimuli.
Positive and negative control of gene transcription plays a pivotal role in the functional differentiation of cells and in their ability to respond to extracellular signals and environmental stress (18, 28). Transcriptional regulation of eukaryotic genes is accomplished mainly by complex DNA cis-acting promoter elements, called enhancers, which bind to transcription factors that recognize specific DNA sequences. The specific organization of the enhancer sequences in a promoter and the proteins bound to these sequences are involved in the initiation of the assembly of the transcriptional machinery required for the initiation of mRNA synthesis by RNA polymerase II (Pol II) (18, 28). A major question in eukaryotic gene transcription is how specificity in gene activation is achieved; that is, after a particular cellular stress, a variety of transcription factors can be activated and bind to multiple gene promoters, but only a small subset of these genes will be activated.
The tumor necrosis factor alpha (TNF-α) gene provides a unique opportunity in which to study the specificity of transcription, since it is expressed in a variety of cell types stimulated via several different signal transduction pathways and the common end point of these diverse signaling cascades is the binding of a particular set of protein factors to the TNF-α promoter and the induction of TNF-α gene transcription. In most cell types, TNF-α is not expressed prior to stimulation. However, diverse biological processes, which include exposure to virus (12, 14), antigen (15, 16), lipopolysaccharide (LPS) (14, 29), and TNF-α itself (5), can elicit TNF-α gene expression. Recent studies have shown that the activators NFAT, ATF-2/Jun, Ets/Elk, and Sp1 proteins and the CBP/p300 family of coactivator proteins are major regulators of TNF-α gene expression (11, 12, 29-32). In other recent studies, it has been shown that the cell-type- and inducer-specific regulation of the TNF-α gene is accomplished through the recruitment of inducer-specific enhancer complexes to shared promoter regulatory elements (11, 12, 29).
Studies of the T-cell receptor α (TCR-α) and beta interferon (IFN-β) enhancer regions have revealed the details of how combinations of activators bind in a specific three-dimensional arrangement and how these spatial constraints contribute to specific gene expression patterns. Both of these genes are activated in response to a specific cell type or stimulus: the TCR-α enhancer is active only in T cells, and the IFN-β enhancer is activated in response to virus infection. Both enhancers require specific sets of transcription factors and architectural proteins to form an active higher-order nucleoprotein complex, the enhanceosome (reviewed in references 6 and 28). Thus, the IFN-β enhancer and TCR-α enhancer are highly specialized to respond to a certain signal or to a certain cell type.
Here, using the TNF-α gene (which is expressed in multiple cell types in response to a variety of extracellular stimuli) as a model system, we investigated the role of higher-order nucleoprotein complexes in a gene tightly regulated at the transcriptional level by multiple stimuli and in multiple cell types. Employing site-directed mutagenesis, DNase I footprinting analysis with recombinant proteins, and in vivo chromatin immunoprecipitation assays, we show that distinct higher-order transcriptional complexes, or enhanceosomes, are recruited to the TNF-α promoter in response to distinct stimuli (calcium flux or virus infection) in T lymphocytes. After calcium flux or virus infection, NFATp, Sp1, and/or Ets-1 are differentially recruited to shared transcription factor binding sites where precise helical phasing of the DNA between these sites is both absolutely required and inducer specific.
We know of no other example of a gene that is regulated through the recruitment of distinct activators to the same regulatory element depending upon the stimulus. Further, we have examined the overall spatial constraints for activation of the TNF-α promoter in response to different stimuli. The results of these experiments demonstrate the requirement for a distinct and inducer-specific enhanceosome, while previous studies have established the requirement of enhanceosome formation in a one-stimulus-to-one-response fashion or in a single cell type fashion. This work thus represents a unique example of the molecular basis of the specificity of transcription and demonstrates a general mechanism by which a single gene is controlled in response to different extracellular stimuli in T lymphocytes in a tightly controlled and specific fashion.
MATERIALS AND METHODS
Plasmids.
The −200 TNF-α Luc reporter was created by subcloning the SmaI-HincII fragment of −200 TNF-α CAT (14) into the SmaI site of pGL3-Basic (Promega, Madison, Wis.). All point mutations in luciferase reporter constructs (Luc constructs) were created by subcloning the BamHI-XbaI fragment of the corresponding chloramphenicol acetyltransferase constructs (CAT constructs) (15, 30, 31) into pBluescript (Stratagene). Then, the KpnI-XbaI fragment of the resulting vector was subcloned into the KpnI-NheI sites of pGL3-Basic. The −180 M, Up-Sp1M and −149 M luciferase reporters and spacer mutants, where a half-helical turn (5 or 6 bp) or a full-helical turn (10 bp) were introduced between different pairs of activator binding sites, were prepared by using PCR-based mutagenesis methods (circular mutagenesis chain reaction) (CMCR Quick Change; Stratagene).
Cell culture and transfection.
The T-cell hybridoma 68-41 (a gift from Masato Kubo, Science University of Tokyo, Tokyo, Japan) was grown at 37°C, 5% CO2, in RPMI 1640 supplemented with penicillin, streptomycin, 10% fetal bovine serum, and 2 mM l-glutamine. Transfections were performed using DEAE-dextran, as described previously (15). After 6 h, cells were stimulated with 1 μM ionomycin (Calbiochem) or were activated with Sendai virus (Cantell strain; Spafas) at a final concentration of 300 hemagglutinating (HA) units/ml for approximately 16 h and luciferase assays were performed according to the manufacturer's instructions (Dual Luciferase Reporter Assay System; Promega) using a Dynex luminometer, with Renilla luciferase (pRL-TK) as an internal control. The concentrations of ionomycin (from 0.1 to 100 μM) and Sendai virus (from 100 to 1,000 HA units/ml) were titrated, and the optimal concentration of each stimulus for gene induction was utilized.
DNase I footprinting.
DNase I footprinting of the human TNF-α promoter was performed using recombinant NFATp, Ets-1, Elk-1 (generous gifts of D. Thanos, B. Nikolajczyk, and A. Sharrocks, respectively) and Sp1 (Promega) at the concentrations indicated in the figure legends, as described previously (31). The −200 to +87 fragment of the wild-type TNF-α promoter or isogenic constructs bearing the −50 Sp1 mutation and the −50 Sp1 consensus mutation as shown in figures were used as templates.
Formaldehyde cross-linking and chromatin immunoprecipitation.
68-41 T cells (∼2 × 108 cells) and control samples were treated with 1 μM ionomycin (Calbiochem) for 1 h or were activated with Sendai virus (Spafas, Cantell strain) at a final concentration of 300 HA units/ml for 3 h, as indicated in the figure legends. The cells were then treated with formaldehyde (1% final concentration) for 30 min at 4°C. Cells were harvested, and fixed chromatin was sonicated, extracted, and purified as described previously (35), followed by immunoprecipitation with anti-Ets-1 or control normal immunoglobulin G (IgG) antibodies (Santa Cruz Biotechnology). Immunoprecipitated DNA was then amplified by PCR with primers specific to the TNF-α promoter as previously described (12). Titrations of PCR cycles were performed to ensure that experiments were performed in the linear range of amplification.
RESULTS
NFATp and Sp1 bind overlapping TNF-α promoter sequences.
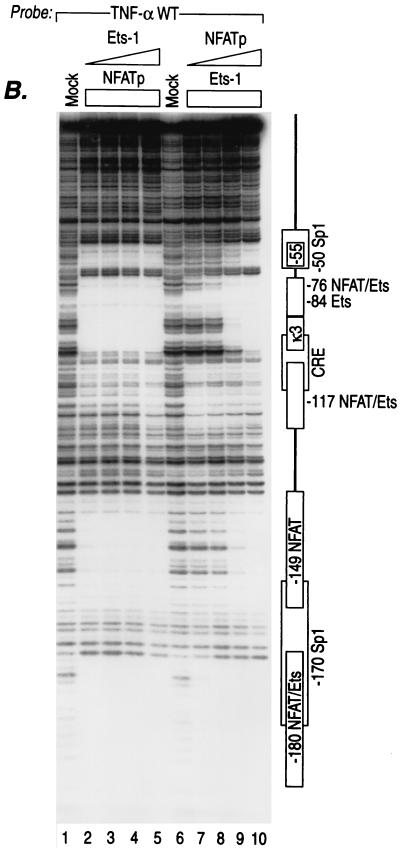
DNase I footprinting using nuclear extracts from T cells stimulated with virus or ionophore showed that the promoter was extensively protected over the previously identified NFAT and Sp1 sites from DNase I digestion (data not shown). Thus, the activator binding sites for NFATp and Sp1 could not be adequately distinguished from each other, and the binding affinities of the cognate sites for these proteins could not be compared using nuclear extracts in the assay. We thus used recombinant NFATp and Sp1 and a human TNF-α promoter fragment spanning from positions −200 to +87 relative to the TNF-α transcription start site and performed quantitative DNase I footprinting to determine the precise patterns of binding of NFATp and Sp1 to the TNF-α promoter in vitro. This analysis revealed two novel NFATp-binding sites located at −55 and −180 nucleotides (nt) relative to the TNF-α cap site (Fig. 1A, lanes 3 to 5) and a high-affinity Sp1 site at −170 nt in addition to the Sp1 site at −50 nt (Fig. 1A, lanes 6 to 9).
FIG. 1.
NFATp, Sp1, and Ets-1 bind to overlapping sites in the TNF-α promoter with different affinities. (A) NFATp binds to six sites and Sp1 binds to two sites in the human TNF-α promoter. The quantitative DNase I footprinting results using the wild-type (WT) human TNF-α promoter (nt −200 to +87 relative to the transcription start site) and increasing concentrations of recombinant NFATp (20 ng, 100 ng, 400 ng, and 2 μg) or Sp1 (0.01, 0.05, 0.25, and 1 footprinting unit [fpu]) (for information on footprinting units, see Promega product information on Sp1 [catalog no. E3391]) are shown. The increasing concentrations are represented in the figure by the height of the triangle over the lanes. The positions of the six NFATp-binding sites, two Sp1-binding sites, and the CRE site are indicated. (B) Sp1 binds to two sites in the TNF-α promoter with different affinities. The quantitative DNase I footprinting results using the wild-type human TNF-α promoter (nt −200 to +87 relative to the transcription start site) and increasing concentrations of recombinant Sp1 (0.5, 1, 2, and 3 fpu) are shown. Sp1 binds with high affinity to the −170 site and with low affinity to the −50 site, which are shown in the figure. (C) Ets-1 binds to the −84 Ets and the −76, −117, and −180 NFAT sites. The quantitative DNase I footprinting results using the wild-type human TNF-α promoter (nt −200 to +87 relative to the transcription start site) and increasing concentrations of recombinant NFATp or Ets-1 (20 ng, 100 ng, 400 ng, and 2 μg) are shown. The positions of the Ets-1, NFATp, Sp1, and CRE sites are indicated.
Intriguingly, the −180 NFAT site binds NFATp with high affinity, whereas the affinity of the −55 site binding with NFATp is lower than for the −180 site (Fig. 1A, lanes 3 to 5). We note that the four previously detected NFAT-binding sites also bind NFATp with different affinities. The −76 NFAT site is a high-affinity site similar to the −180 site, while the −149 and −117 NFAT sites are relatively weak and the κ3-NFAT site binds NFATp with an intermediate affinity (Fig. 1A) (31). Thus, the human TNF-α promoter contains six NFAT-binding sites that bind NFATp with different relative affinities.
Sp1 also binds to two regions of the TNF-α promoter (−50 and −170) with different affinities (Fig. 1A, lanes 6 to 9). The proximal Sp1-binding site at −50 has a much lower binding affinity for Sp1 than that of the upstream −170 Sp1-binding site. As shown in Fig. 1B, even at very high concentrations of Sp1, the −50 site still shows much lower affinity for Sp1 binding than does the −170 site. Furthermore, the regions protected by Sp1 overlap regions that are protected by NFATp at the −55, −180, and −149 NFAT-binding sites (Fig. 1A, compare lanes 5 to 9).
NFATp and Ets proteins bind to overlapping TNF-α promoter sequences.
Previously, we showed that Ets-1 and Elk-1 bind to two TNF-α NFAT-binding sites, located at −76 and −117 as well as to an Ets/Elk consensus sequence located at −84 (29). Notably, the −76 and −117 NFAT sites are required for calcineurin and NFAT-dependent TNF-α gene expression in lymphocytes (30). In the case of LPS induction of TNF-α in monocytes, Ets/Elk bind to the −117, −84, and −76 sites and are critical in the regulation of the gene (29). Since the −180 NFAT site also contains 5′-GGAA-3′, which is a core element for Ets/Elk binding, next we examined whether Ets-1 and Elk-1 could also bind to this site. As shown in Fig. 1C, Ets-1 (lanes 3 to 6) and Elk-1 (data not shown) bind to the −180 site at high concentrations. Note that Ets/Elk binds with high affinity to the relatively weak −117 NFAT-binding site and with relatively weak affinity to the high-affinity −180 NFAT-binding site (Fig. 1C, compare lanes 6 to 11). Thus, overlapping TNF-α promoter sequences centered at −180, −117, and −76 are the binding targets of the distinct NFAT and Ets families, whereas the sites centered at −180, −170, −149, −55, and −50 are targets of the distinct NFAT and Sp1 families of transcription factors. Furthermore, these transcription factors bind to the sites with different affinities.
Inducer-specific requirements for the NFAT, Sp1, and Ets sites in virus and ionophore regulation of TNF-α.
Next, we transfected human TNF-α luciferase reporter constructs bearing mutations in the novel and previously detected binding sites that were detected in the quantitative DNase I footprinting analyses (Fig. 2A) and transfected these constructs into 68-41 T cells stimulated by ionophore or virus to identify their role in inducer-specific TNF-α gene regulation. Consistent with previous results (12), the CRE, κ3-NFAT, and −76 NFAT sites are all required for both ionophore and virus induction of the TNF-α gene, and the −50 Sp1 site is absolutely required for virus induction of TNF-α. Mutation of the NFAT/Ets-binding sites at −117 and −180 and the −170 Sp1 site also all decrease the transcriptional activation of the gene by ionophore and virus but to a comparatively greater extent by virus (Fig. 2B). Strikingly, however, mutation of the −84 Ets/Elk site abrogates virus induction but does not affect ionophore induction of the gene (Fig. 2B). Thus, the −84 Ets/Elk site plays a novel inducer-specific role in virus induction similar to the inducer-specific role played by the −50 Sp1 site (Fig. 2B) (12), providing further support that unique combinations of regulatory elements are required for activation of the TNF-α gene by virus and by ionophore.
FIG. 2.
Identification of inducer-specific requirements of activator binding sites for the TNF-α promoter activity in response to ionophore and virus. (A) Sequence of the TNF-α promoter from −200 to −20 nt. Activator binding sites are boxed and colored, and sites that are capable of binding two activators contain two colors. The mutations (M's) tested in panel B are shown below the wild-type sequence. The TATA box is shown in a black box. (B) Relative activities of TNF-α-Luc fusion constructs containing mutations (M's) in activator binding sites in ionophore- and virus-stimulated 68-41 T cells. 68-41 T cells were transfected with 1 μg of the wild-type −200 TNF-α promoter-Luc reporter (WT) or with isogenic reporters containing the mutations in different activator binding sites as shown in panel A. A Renilla luciferase control plasmid (1 μg) was cotransfected in all cases. Cells were then stimulated with ionomycin or Sendai virus as described in Materials and Methods, and luciferase assays were performed and normalized to Renilla luciferase activity. The histograms show the results of four independent experiments. Error bars represent the standard errors of the mean.
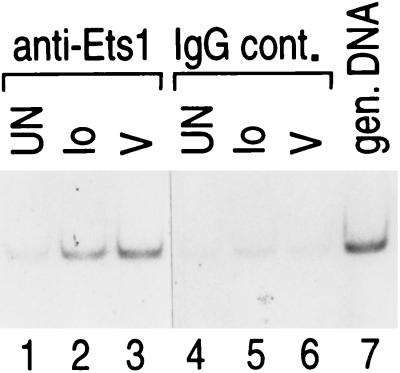
Virus-specific recruitment of Ets to the TNF-α promoter in vivo.
Previous studies using specific antibodies in formaldehyde cross-linking and chromatin immunoprecipitation experiments demonstrated that ATF-2, c-Jun, and NFATp/c were recruited to the TNF-α promoter after both ionophore and virus stimulation of T cells, whereas Sp1 was differentially recruited after virus infection (12). We thus next sought to directly test whether Ets, like Sp1, was also recruited in an inducer-specific manner to the TNF-α promoter in T cells in vivo and whether TNF-α promoter DNA was immunoprecipitated by antibodies to Ets and amplified by PCR. As shown in Fig. 3, although both stimuli result in the in vivo recruitment of Ets-1 to the TNF-α promoter, virus infection resulted in a relatively greater recruitment of Ets-1 binding (lanes 1 to 3), consistent with the virus-specific role of the Ets-binding sites in the transfection analysis.
FIG. 3.
Inducer-specific binding of Ets-1 to the endogenous TNF-α promoter. Formaldehyde cross-linking and chromatin immunoprecipitation of unstimulated (UN) and ionomycin-stimulated (Io)- or virus-stimulated (V) 68-41 T cells are shown. Following stimulation, the cells were treated with formaldehyde to cross-link endogenous protein and DNA. Samples of sonicated and purified chromatin were immunoprecipitated with the indicated antibodies, and DNA isolated from immunoprecipitated material was amplified by PCR with primers specific for the TNF-α gene. An increase in the relative amount of the amplified TNF-α promoter-specific PCR product indicates binding of the protein to the endogenous TNF-α promoter. To demonstrate that the efficiencies of cross-linking and immunoprecipitation of Ets-1 to the TNF-α promoter were equivalent in the mock-, virus-, and ionophore-treated samples, we included isotype-matched IgG antibodies (IgG control [IgG cont.]). gen., genomic.
Activators bind overlapping TNF-α promoter sequences with different affinities.
Intracellular calcium levels and subsequent NFATp nuclear translocation and levels following treatment with ionophore are much higher than those following virus infection of T and B cells (12). Furthermore, relatively higher levels of Sp1 are inducibly recruited to the promoter after virus infection than after ionophore treatment (12). We thus speculated that a mechanism of inducer-specific regulation of TNF-α might involve the relative levels of NFATp following a specific stimulus combined with the different affinities of NFATp and Sp1 for the overlapping −50 and −55 NFAT/Sp1-binding sites (−50/−55 NFAT/Sp1-binding site). To test this potential mechanism, we performed DNase I footprinting analysis and investigated the ability of NFATp to compete with Sp1 for binding to the overlapping sites to which both proteins bind. As shown in Fig. 4A, although NFATp and Sp1 bind to the same overlapping −50/−55 site, each can be distinguished by their pattern of protection from DNase I digestion (compare lanes 5 and 9 of Fig. 4A). However, incubation of the TNF-α promoter with increasing amounts of both NFATp and Sp1 proteins leads to the NFATp-specific footprint after DNase I digestion (Fig. 4A, compare lanes 5, 9, and 13). Thus, at increasing concentrations of NFATp, Sp1 is displaced from the site.
FIG. 4.
Mutually exclusive binding of different activators to shared sites in the TNF-α promoter. (A) Mutually exclusive binding of NFATp and Sp1 to shared sites in the TNF-α promoter. DNase I footprinting results using the wild-type (WT) human TNF-α promoter (nt −200 to +87 relative to the transcription start site) or isogenic probes bearing mutations in the −50 Sp1 site (Sp1 mut or Sp1 cons mut), as described in the text, and increasing concentrations of recombinant NFATp and/or Sp1 are shown. The positions of the six NFAT-binding sites and two Sp1-binding sites are indicated to the left of the panel. (B) Mutually exclusive binding of NFATp and Ets-1 to shared sites in the TNF-α promoter. DNase I footprinting results using the wild-type human TNF-α promoter and increasing concentrations of either recombinant Ets-1 or NFATp are shown. The concentration of recombinant Ets-1 was increased, while NFATp protein was kept at a constant and maximal concentration (lanes 2 to 5), and vice versa: the concentration of recombinant NFATp was increased, while Ets-1 protein was kept at a constant and maximal concentration (lanes 7 to 10). The concentrations of recombinant proteins used are described in detail in the legend to Fig. 1.
We next tested the effects of mutations that shifted the balance of binding affinities between NFATp and Sp1 within the overlapping −50/−55 NFAT/Sp1 site. Strikingly, the Sp1 mutation which prevents binding of Sp1 to the −50/−55 NFAT/Sp1 site (Fig. 4A, lanes 19 to 21) dramatically increases the affinity of NFATp binding to the site (lanes 16 to 18 and 22 to 24), consistent with the hyperresponsiveness to ionomycin induction of the Sp1 mutant TNF-α reporter construct observed in the transient-transfection analysis (Fig. 2B). In contrast, when the TNF-α Sp1 motif is replaced with a consensus Sp1 binding motif (CCCCGCCCC), the affinity of NFATp for binding to this site is dramatically decreased (Fig. 4A, lanes 27 to 29) and Sp1 binds at the lowest concentration (lane 30). Thus, the relatively weak affinity that the wild-type TNF-α −50 site has for Sp1 allows NFATp to effectively compete for binding under conditions of increased nuclear levels of NFATp such as following calcium flux.
Similarly, the upstream overlapping binding sites for NFATp and Sp1 (−149 NFAT, −170 Sp1, and −180 NFAT) demonstrate the same pattern of mutually exclusive binding: either NFATp or Sp1 and at increasing concentrations of NFATp, Sp1 is displaced from the −170 site (Fig. 4A, lanes 3 to 13). Furthermore, when NFATp and Ets were tested at increasing concentrations, NFATp was able to displace at the −76 and −180 sites as well as at the relatively high-affinity −117 NFAT/Ets sites (Fig. 4B).
Taken together, these data suggest that inducer-specific TNF-α gene regulation is achieved through the presence of overlapping activator binding sites that bind different transcription factors with different affinities combined with the presence of different levels of specific transcription factors in the nucleus following distinct extracellular stimulation. Therefore, in the case of low NFATp levels such as occurs after virus infection, Sp1 and Ets could effectively compete with NFATp for binding to shared binding sites, whereas in the case of high NFATp levels such as occurs following ionophore stimulation, NFATp would bind to these same sites (see the model shown in Fig. 5).
FIG. 5.
Inducer-specific occupancy of the activator binding sites in the TNF-α promoter region. A model of the cis-acting TNF-α promoter elements and transcription factors involved in the inducer-specific regulation of TNF-α by ionophore and virus in T cells is summarized schematically. Activator binding sites that are critical for activation of the promoter are shown in colored boxes. The transcription factors that are recruited to the TNF-α promoter (NFAT, Sp1, Ets, and ATF-2/c-Jun) following the indicated stimuli are shown in different shapes and colors.
Inducer-specific enhanceosomes are recruited to the TNF-α gene enhancer.
A striking characteristic of the TNF-α promoter is the close proximity of the activator binding sites and the fact that all of these sites are required for maximal levels of inducible transcription by virus or ionophore. These findings suggested that protein-protein interactions between the activators might be involved in the inducer-specific regulation of the gene and that the relative positions of the activators on the DNA double helix were critical.
To determine whether precise helical phasing between the activators bound to their recognition elements within the TNF-α promoter was essential for enhancer function, we constructed TNF-α promoters in which half- or full-helical turns (5 or 6 bp or 10 bp, respectively) were inserted between individual transcription factor binding sites (Fig. 6). To avoid the generation of new unexpected binding sites for transcriptional activators or repressors, we used the insertion sequences previously tested, which were selected from the randomized sequence of inserts and were shown not to have transcriptional activity (27).
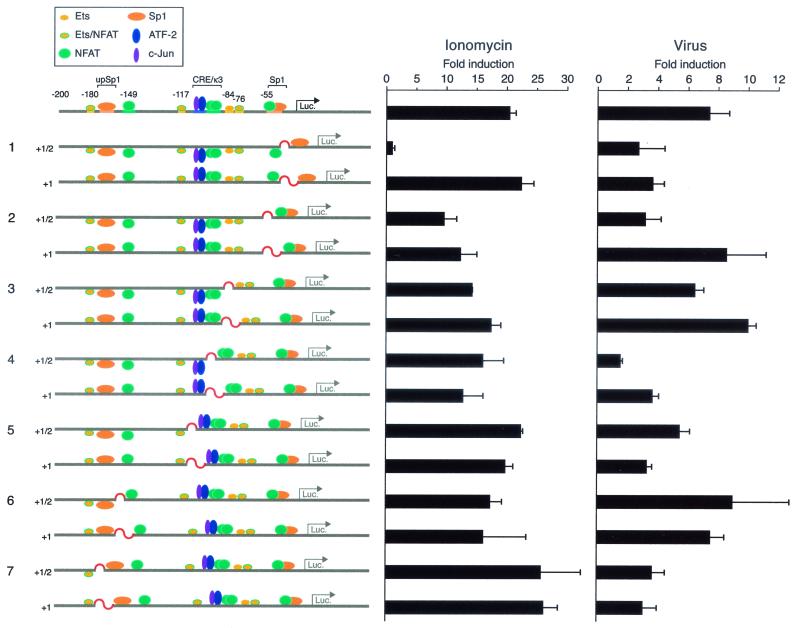
FIG. 6.
TNF-α gene activation requires inducer-specific precise helical phasing between activator binding sites. 68-41 T cells were transfected with the wild-type −200 TNF-α promoter-Luc reporter or with the indicated mutant reporter constructs containing insertions between activator binding sites that disrupt and restore helical phasing as indicated and with the Renilla luciferase (Luc.) control plasmid. After the cells were stimulated with ionomycin or Sendai virus, luciferase activity was measured and divided by Renilla luciferase activity to normalize transfection efficiency and fold induction was calculated. The histograms show the results of four independent experiments. Error bars represent the standard errors of the mean. Note that we observed a strong correlation between the effects of the insertion mutants on basal and induced levels of transcription (data not shown), consistent with the fact that both the basal transcription complex and enhanceosome contribute to the induction of the TNF-α gene.
As shown in Fig. 6, 68-41 cells were transfected with the wild-type −200 TNF-α Luc reporter gene or with constructs with an insertion of 5 or 6 bp or 10 bp and then stimulated with ionophore or Sendai virus. While ionophore induction of the wild-type −200 TNF-α Luc reporter resulted in a >20-fold increase in luciferase activity, the insertion of a half-helical turn between the −50 Sp1 and −55 NFAT-binding sites abrogated ionophore induction (Fig. 6, lane 1). Remarkably, insertion of 10 bp, which reestablished the relative positions of binding sites on the face of the DNA helix, fully restored the activity of the promoter in response to calcium flux (Fig. 6, compare constructs in lane 1). These results thus suggested that after ionophore stimulation, the activators bound to the TNF-α promoter upstream of the −50 Sp1 site, including all NFAT molecules, likely contact the basal transcription complex directly or indirectly through protein-protein interactions with each other or via the coactivator proteins p300/CBP. Flipping all the necessary activators out of the correct helical surface strongly inhibits the formation of the functional enhancer complex on the TNF-α promoter following ionophore stimulation.
In contrast, the insertion of a half-helical turn between the −50/−55 Sp1/NFAT- and −76 NFAT-binding sites inhibits the activity of the reporter gene to approximately 50% of the wild-type activity achieved upon ionophore stimulation, and the insertion of a full-helical turn does not restore the transcriptional activity of the promoter after ionophore stimulation (Fig. 6, lane 2). Thus, the proteins bound to the −76 NFAT-binding site also appear to directly interact with the activators bound to the overlapping −50/−55 Sp1/NFAT site.
T cells transfected with the same helical phasing mutants stimulated with Sendai virus gave us surprisingly different results. Disruption of the DNA phasing at the −50 Sp1 site with half a helical turn of the DNA reduced promoter activity by approximately 50% (Fig. 6, lane 1), and this activity was not restored by correct helical phasing, indicating that in the case of virus-stimulated TNF-α gene expression, proteins bound to the Sp1 site must interact directly with proteins bound to adjacent sites. In contrast, a half DNA turn between the −76 NFAT/Ets-binding site and the −50/−55 Sp1/NFAT composite site decreases the TNF-α promoter response to virus to less than half compared to the wild-type construct, but introduction of a 10-bp spacer between the same sites completely restores the activity of the promoter to wild-type levels (Fig. 6, compare constructs in lane 2). Taken together (and consistent with our mutagenesis data [Fig. 2B]), these results suggest that for virus induction of TNF-α, Sp1 interacts with proteins bound to abutting sequences and that proteins bound to the −76 NFAT/Ets site must be in phase with other activators in order to interact with the basal transcription complex perhaps through protein-protein interactions with the coactivator proteins p300/CBP.
Insertion of nucleotides disrupting the phasing and spacing between other sites also has inducer-specific effects, but none of the effects can be rescued by restoring helical phasing. For example, the introduction of 5 or 10 bp between CRE and κ3 sites decreases the inducibility of the promoter by both ionomycin and virus (Fig. 6, constructs in lane 4). These results are consistent with the CRE-κ3 site functioning as a composite element where ATF-2, c-Jun, and NFATp proteins must directly contact each other or a coactivator protein for gene activation to occur.
Taken together, these results demonstrate that specific protein-protein and protein-DNA interactions are absolutely necessary for the assembly of a functional enhancer complex on the TNF-α promoter. The fact that precise helical phasing requirements are absolutely required and inducer specific is consistent with our observation that different sets of transcriptional activators bind to shared binding sites in the TNF-α promoter in response to different stimuli. We conclude that inducer-specific TNF-α gene activation is achieved through the recruitment of distinct enhanceosomes.
DISCUSSION
We have shown here that inducer-specific TNF-α gene regulation in T cells requires the assembly of distinct and inducer-specific multicomponent higher-order transcription enhancer complexes, or enhanceosomes. In contrast to TNF-α, which is induced by multiple stimuli, the other previously characterized genes that require enhanceosome formation for transcriptional induction are either stimulus specific or cell type specific. For example, the IFN-β gene is tightly regulated in response to a single specific stimulus, virus infection (reviewed in reference 24), whereas the TCR-α enhanceosome is constitutively recruited in a single cell type, T cells (13).
Virus infection of a cell results in the coordinate assembly of a specific set of transcriptional activators on the IFN-β promoter, including NF-κB p50/p65, ATF-2/Jun, interferon regulatory factors 3 and 7, and the architectural protein HMG-I(Y). These activators bind to a fixed set of binding sites in its promoter and associate with the CBP/p300 coactivators (reviewed in reference 24). Thus, the IFN-β enhanceosome has a stringent requirement for a specific set of binding sites and cognate transcription factors, which are coordinately activated by a single stimulus, virus. Similar to the IFN-β enhanceosome, the TCR-α enhanceosome, which is constitutively active in a single cell type, T cells, requires the binding of an architectural protein (LEF-1). Notably, the architectural proteins HMG-I(Y) and LEF-1 induce a bend in the DNA helix when bound to the IFN-β and TCR-α enhancer regions, respectively, and they thus facilitate interactions between factors bound at the flanking sequences of these enhancers (13, 24).
Sequence analysis of the TNF-α enhancer region, however, did not reveal any binding motifs for the HMG-domain family members, which includes both HMG-I(Y) and LEF-1 and, furthermore, we were unable to detect binding of recombinant HMG-I(Y) in DNase I footprinting analysis of the TNF-α enhancer (data not shown). However, DNA binding of activators, including members of the AP-1 (8) and Sp1 (19) families of transcription factors, also result in DNA bending, and the AP-1 bending effect can be augmented by the concordant binding of NFATp to the adjacent site (7). Thus, binding of the bZIP heterodimer ATF-2/c-Jun, NFATp, and Sp1 may result in DNA bending and take the role of architectural proteins in TNF-α enhanceosome assembly.
In contrast to TNF-α, which responds to multiple stimuli in a single cell type and is expressed in a variety of cell types, IFN-β and T-cell receptor alpha (TCR-α) are single-switch genes. They require structural proteins to form highly specific and unique enhanceosomes, and none of the activators can be substituted by other proteins, even by closely related family members (13, 24). In the case of the multiswitch TNF-α gene, the inducer-specific enhanceosomes that are formed have different helical phasing requirements, indicating that the ternary structure of these enhanceosomes is distinct. Thus, we imagine that TNF-α promoter DNA adopts an inducer-specific conformation to accommodate different sets of activators and coactivators in response to different stimuli.
Notably, activation of TNF-α gene transcription in T cells by TCR engagement, ionophore stimulation, and virus infection requires the coactivator proteins CBP/p300 (12). The histone acetyltransferases CBP and p300 both may function as transcriptional integrators, interacting with multiple transcription factors, including all the major activators involved in TNF-α gene activation, i.e., NFATp, Ets-1, Sp1, and ATF-2/Jun, and the basal transcription machinery (1, 3, 20, 21, 26, 37). Thus, through protein-protein interactions with the inducer-specific TNF-α enhancer complexes, CBP/p300 may provide a surface for the higher-order enhanceosome structures to be formed and tethered to the target surface within the Pol II transcriptional machinery and, thus, replace the requirement for architectural proteins (see the model shown in Fig. 7).
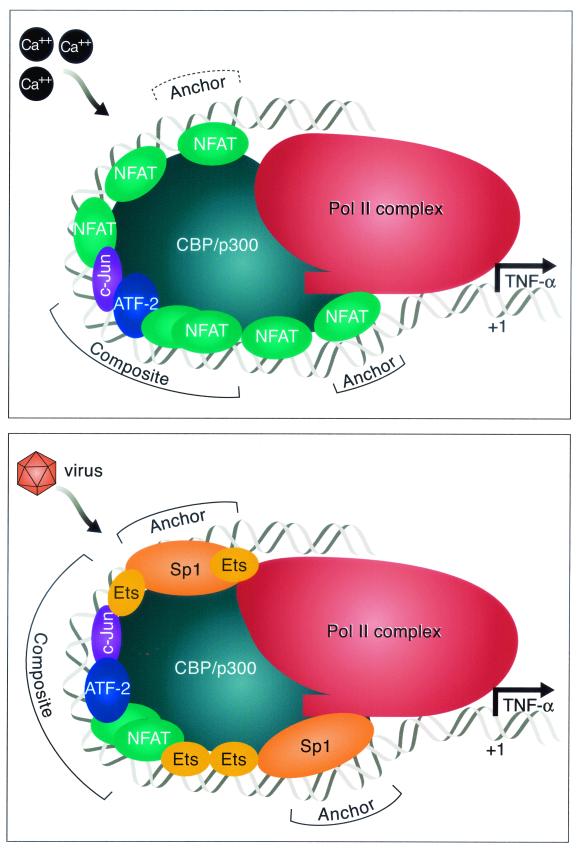
FIG. 7.
Model of the recruitment of inducer-specific TNF-α gene enhanceosomes in T lymphocytes. Distinct enhanceosomes are formed on the TNF-α promoter region in response to ionophore or virus stimulation of T cells. The ionomycin-inducible enhanceosome includes proteins bound to the composite core element CRE/κ3 (ATF-2/c-Jun/NFATp dimer) and two NFAT molecules closest to the basic transcription complex, which we imagine play a major anchoring role in the process of forming the functional transcription-driving complex. We note that phasing mutations upstream of the −76 NFAT site have a relatively modest effect on transcriptional activation by ionomycin, although site-directed mutations in these sites do have a significant impact on gene induction (Fig. 2). The virus-inducible enhanceosome has different components. It has a composite core element ATF-2/c-Jun/NFATp dimer, which also includes Ets protein, and it contains two anchoring Sp1/Ets complexes that, we speculate, are responsible for the recruitment of basic transcription machinery and the activation of transcription. Once assembled, the enhanceosome makes multiple contacts with the basal transcription Pol II complex. The relative sizes of the proteins and the length of the DNA covered are not drawn to scale.
Precise helical phasing of DNA and specific arrangement of protein binding sites on the same surface of duplex DNA are critical for the appropriate binding of transcription factors to form a three-dimensional structure of a functional enhanceosome complex (27). Strikingly, five of the six NFAT-binding sites in the TNF-α enhancer region are positioned on the same phase of the DNA helix (Fig. 2A). Moreover, they are aligned with the ATF-2 and c-Jun proteins bound to the CRE site and with the Sp1- and TATA-binding proteins bound to their respective sites. Only the very low-affinity −149 NFAT-binding site is located on the opposite side of the DNA helix.
In the case of ionomycin induction of TNF-α, the overlapping NFAT/Sp1 site (−50/−55) is occupied by NFAT, which in turn appears to anchor the ionomycin-induced enhanceosome to the basic transcription machinery. This is supported by the demonstration that the −55 NFAT site and the TATA box alone result in moderate levels of ionomycin activation and by the demonstration that when the −55 site is out of phase with the Pol II complex, transcription is abrogated (Fig. 6, construct in lane 1). Using mice deficient in NFATp, we previously showed that NFATp could not be functionally replaced by other NFAT family members in immediate-early TNF-α gene activation in T cells (32). Notably, a partial TFIID complex assembled from recombinant hTBP, hTAFII250, and hTAFII130 supports NFATp-activated transcription, demonstrating the ability of hTAFII130 to serve as a coactivator for NFATp in vitro (22). Thus, NFATp or Sp1 bound to the overlapping NFAT/Sp1 site could be an anchor between the basic transcription machinery and the ionophore- or virus-specific enhanceosome, respectively.
In contrast, the virus-specific enhanceosome includes Sp1 and Ets family members, and intriguingly, Sp1 and Ets factors can interact and promote transcription through protein-protein interactions (4, 9, 10, 23). Insertion of a half-helical turn of DNA between the −50/−55 Sp1/NFAT site and the −76/−84 Ets site significantly inhibits virus-inducible TNF-α promoter activity, and insertion of a full-helical turn, which restores the normal helical phasing, restores the activity of the promoter in response to virus (Fig. 6, constructs in lane 2). These results thus suggest that an interaction between the activators bound to the Sp1 and Ets sites is necessary for the formation of the virus-inducible TNF-α enhanceosome.
We note that four different glutamine-rich regions of hTAFII130, a component of the Pol II complex, can interact with Sp1 and are required for Sp1-mediated transcriptional enhancement in mammalian cells, and furthermore, different activation domains of the Sp1 protein target distinct subdomains of hTAFII130 (25). Taken together with our observations that both Sp1 sites are critical for virus induction and that Sp1 is inducibly recruited by virus to the TNF-α promoter in vivo, we speculate that Sp1 anchors the virus-inducible enhanceosome to the basic transcription machinery, likely through an interaction with coactivators and the Pol II complex (Fig. 7).
Adjacent Ets- and AP-1-binding sites occur in a large number of promoter/enhancer elements (8, 34, 36), and functional cooperation between Ets and AP-1 is critical for the controlled expression of many genes, including cytokines (17, 33). The physical association between the DNA-binding domain of Ets family proteins and AP-1 was demonstrated both in vitro and in vivo in activated human T cells (2). In the TNF-α enhanceosome, ATF-2/c-Jun bind to the CRE site, which abuts the high-affinity Ets-binding site at −117. All three proteins bound to their sequences are positioned on the same helical surface of DNA that would allow them to interact with each other. Indeed, mutation of either the −117 Ets site or the CRE site leads to complete unresponsiveness of the reporter gene to virus (Fig. 2B). Moreover, insertion of a half-helical turn of DNA between these two sites reduces the activity of the promoter in response to virus, and correct phasing that displaces the sites away from each other further does not restore activity (Fig. 6, constructs in lane 5).
These data support the speculation that Ets/ATF-2/c-Jun protein-protein interactions are functionally important for the activation of TNF-α gene transcription by virus. They also demonstrate that the −117 Ets site is part of a composite site that includes the CRE/κ3 element and that it must be intact for virus activation of TNF-α gene expression. Thus, the core composite element required for virus and ionophore activation is distinct. In the case of virus activation, this core composite site includes the −117 Ets site along with the CRE/κ3 element, which binds ATF-2/c-Jun and NFATp (30), while the core composite element of the ionomycin-inducible enhanceosome does not include Ets (Fig. 7).
Thus, our results demonstrate that stimulus-specific TNF-α gene expression depends on the precise arrangement of activator recognition sites in the promoter and inducer-specific combinations of bound activators, which together generate a unique network of protein-protein and protein-DNA interactions. We imagine that after the stimulus-specific enhanceosome is formed, it presents a particular activation surface that is complementary to a target surface within the Pol II transcriptional machinery, thereby recruiting it to the TNF-α promoter, resulting in high levels of transcriptional synergy. Thus, these data demonstrate a novel mechanism by which a high level of transcriptional specificity is achieved in response to distinct extracellular signals and that may be common to genes that are activated by multiple extracellular stimuli.
Acknowledgments
We thank Robert Barthel and James Falvo for helpful discussions and comments on the manuscript. For the generous gifts of recombinant proteins, we thank Dimitris Thanos (NFATp protein), Andrew Sharrocks (Elk-1 protein), and Barbara Nikolajczyk (Ets-1).
This work was supported in part by grants from the NIH (GM-56492) and the American Heart Association to A.E.G. and from the Arthritis National Research Foundation to A.V.T.
REFERENCES
- 1.Arias, J., A. S. Alberts, P. Brindle, F. X. Claret, T. Smeal, M. Karin, J. Feramisco, and M. Montminy. 1994. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature 370:226-229. [DOI] [PubMed] [Google Scholar]
- 2.Bassuk, A. G., and J. M. Leiden. 1995. A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity 3:223-237. [DOI] [PubMed] [Google Scholar]
- 3.Billon, N., D. Carlisi, M. B. Datto, L. A. van Grunsven, A. Watt, X.-F. Wang, and B. B. Rudkin. 1999. Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene 18:2872-2882. [DOI] [PubMed] [Google Scholar]
- 4.Block, K. L., Y. Shou, and M. Poncz. 1996. An Ets/Sp1 interaction in the 5′-flanking region of the megakaryocyte-specific alpha IIb gene appears to stabilize Sp1 binding and is essential for expression of this TATA-less gene. Blood 88:2071-2080. [PubMed] [Google Scholar]
- 5.Brinkman, B. M., J. B. Telliez, A. R. Schievella, L. L. Lin, and A. E. Goldfeld. 1999. Engagement of tumor necrosis factor (TNF) receptor 1 leads to ATF-2- and p38 mitogen-activated protein kinase-dependent TNF-alpha gene expression. J. Biol. Chem. 274:30882-30886. [DOI] [PubMed] [Google Scholar]
- 6.Carey, M. 1998. The enhanceosome and transcriptional synergy. Cell 92:5-8. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L., J. N. Glover, P. G. Hogan, A. Rao, and S. C. Harrison. 1998. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature 392:42-48. [DOI] [PubMed] [Google Scholar]
- 8.Chinenov, Y., and T. K. Kerppola. 2001. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 20:2438-2452. [DOI] [PubMed] [Google Scholar]
- 9.Dittmer, J., C. A. Pise-Masison, K. E. Clemens, K. S. Choi, and J. N. Brady. 1997. Interaction of human T-cell lymphotropic virus type I Tax, Ets1, and Sp1 in transactivation of the PTHrP P2 promoter. J. Biol. Chem. 272:4953-4958. [DOI] [PubMed] [Google Scholar]
- 10.Eichbaum, Q., D. Heney, D. Raveh, M. Chung, M. Davidson, J. Epstein, and R. A. Ezekowitz. 1997. Murine macrophage mannose receptor promoter is regulated by the transcription factors PU.1 and SP1. Blood 90:4135-4143. [PubMed] [Google Scholar]
- 11.Falvo, J. V., B. M. N. Brinkman, A. V. Tsytsykova, E. Y. Tsai, T.-P. Yao, A. L. Kung, and A. E. Goldfeld. 2000. A stimulus-specific role for CREB-binding protein (CBP) in T-cell receptor-activated tumor necrosis factor α gene expression. Proc. Natl. Acad. Sci. USA 97:3925-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falvo, J. V., A. M. Uglialoro, B. M. Brinkman, M. Merika, B. S. Parekh, E. Y. Tsai, H. C. King, A. D. Morielli, E. G. Peralta, T. Maniatis, D. Thanos, and A. E. Goldfeld. 2000. Stimulus-specific assembly of enhancer complexes on the tumor necrosis factor alpha gene promoter. Mol. Cell. Biol. 20:2239-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giese, K., C. Kingsley, J. R. Kirshner, and R. Grosschedl. 1995. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 9:995-1008. [DOI] [PubMed] [Google Scholar]
- 14.Goldfeld, A. E., C. Doyle, and T. Maniatis. 1990. Human tumor necrosis factor alpha gene regulation by virus and lipopolysaccharide. Proc. Natl. Acad. Sci. USA 87:9769-9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldfeld, A. E., P. G. McCaffrey, J. L. Strominger, and A. Rao. 1993. Identification of a novel cyclosporin-sensitive element in the human tumor necrosis factor α gene promoter. J. Exp. Med. 178:1365-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldfeld, A. E., J. L. Strominger, and C. Doyle. 1991. Human tumor necrosis factor α gene regulation in phorbol ester stimulated T and B cell lines. J. Exp. Med. 174:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottschalk, L. R., D. M. Giannola, and S. G. Emerson. 1993. Molecular regulation of the human IL-3 gene: inducible T cell-restricted expression requires intact AP-1 and Elf-1 nuclear protein binding sites. J. Exp. Med. 178:1681-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill, C. S., and R. Treisman. 1995. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell 80:199-211. [DOI] [PubMed] [Google Scholar]
- 19.Imanishi, M., Y. Hori, M. Nagaoka, and Y. Sugiura. 2000. DNA-bending finger: artificial design of 6-zinc finger peptides with polyglycine linker and induction of DNA bending. Biochemistry 39:4383-4390. [DOI] [PubMed] [Google Scholar]
- 20.Janknecht, R., and A. Nordheim. 1996. MAP kinase-dependent transcriptional coactivation by Elk-1 and its cofactor CBP. Biochem. Biophys. Res. Commun. 228:831-837. [DOI] [PubMed] [Google Scholar]
- 21.Kawasaki, H., J. Song, R. Eckner, H. Ugai, R. Chiu, K. Taira, Y. Shi, N. Jones, and K. K. Yokoyama. 1998. p300 and ATF-2 are components of the DRF complex, which regulates retinoic acid- and E1A-mediated transcription of the c-jun gene in F9 cells. Genes Dev. 12:233-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, L. J., A. G. Seto, T. N. Nguyen, and J. A. Goodrich. 2001. Human TafII130 is a coactivator for NFATp. Mol. Cell. Biol. 21:3503-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krehan, A., H. Ansuini, O. Bocher, S. Grein, U. Wirkner, and W. Pyerin. 2000. Transcription factors ets1, NF-kappaB, and Sp1 are major determinants of the promoter activity of the human protein kinase CK2alpha gene. J. Biol. Chem. 275:18327-18336. [DOI] [PubMed] [Google Scholar]
- 24.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon-β enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 25.Saluja, D., M. F. Vassallo, and N. Tanese. 1998. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol. Cell. Biol. 18:5734-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shikama, N., J. Lyon, and N. B. La Thangue. 1997. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 7:230-236. [DOI] [PubMed] [Google Scholar]
- 27.Thanos, D., and T. Maniatis. 1995. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell 83:1091-1100. [DOI] [PubMed] [Google Scholar]
- 28.Tjian, R., and T. Maniatis. 1994. Transcriptional activation: a complex puzzle with few easy pieces. Cell 77:5-8. [DOI] [PubMed] [Google Scholar]
- 29.Tsai, E. Y., J. V. Falvo, A. V. Tsytsykova, A. K. Barczak, A. M. Reimold, L. H. Glimcher, M. J. Fenton, D. C. Gordon, I. F. Dunn, and A. E. Goldfeld. 2000. A lipopolysaccharide-specific enhancer complex involving ets, elk-1, sp1, and CREB binding protein and p300 is recruited to the tumor necrosis factor alpha promoter in vivo. Mol. Cell. Biol. 20:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai, E. Y., J. Jain, P. A. Pesavento, A. Rao, and A. E. Goldfeld. 1996. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol. Cell. Biol. 16:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai, E. Y., J. Yie, D. Thanos, and A. E. Goldfeld. 1996. Cell-type-specific regulation of the human tumor necrosis factor alpha gene in B cells and T cells by NFATp and ATF-2/Jun. Mol. Cell. Biol. 16:5232-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsytsykova, A. V., and A. E. Goldfeld. 2000. Nuclear factor of activated T cell transcription factor NFATp controls superantigen-induced lethal shock. J. Exp. Med. 192:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, C. Y., A. G. Bassuk, L. H. Boise, C. B. Thompson, R. Bravo, and J. M. Leiden. 1994. Activation of the granulocyte-macrophage colony-stimulating factor promoter in T cells requires cooperative binding of Elf-1 and AP-1 transcription factors. Mol. Cell. Biol. 14:1153-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wasylyk, B., S. L. Hahn, and A. Giovane. 1993. The Ets family of transcription factors. Eur. J. Biochem. 211:7-18. [DOI] [PubMed] [Google Scholar]
- 35.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 36.Westermarck, J., and V. M. Kahari. 1999. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 13:781-792. [PubMed] [Google Scholar]
- 37.Yang, C., L. H. Shapiro, M. Rivera, A. Kumar, and P. K. Brindle. 1998. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol. Cell. Biol. 18:2218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]