Abstract
Silencing of gene transcription involves local chromatin modification achieved through the local recruitment of large multiprotein complexes containing histone deacetylase (HDAC) activity. The mammalian corepressors mSin3A and mSin3B have been shown to play a key role in this process by tethering HDACs 1 and 2 to promoter-bound transcription factors. Similar mechanisms appear to be operative in yeast, in which epistasis experiments have established that the mSin3 and HDAC orthologs (SIN3 and RPD3), along with a novel protein, SDS3, function in the same repressor pathway. Here, we report the identification of a component of the mSin3-HDAC complex that bears homology to yeast SDS3, physically associates with mSin3 proteins in vivo, represses transcription in a manner that is partially dependent on HDAC activity, and enables HDAC1 catalytic activity in vivo. That key physical and functional properties are also shared by yeast SDS3 underscores the central role of the Sin3-HDAC-Sds3 complex in eukaryotic cell biology, and the discovery of mSds3 in mammalian cells provides a new avenue for modulating the activity of this complex in human disease.
Complementary genetic and biochemical studies with yeast and mammalian systems have identified key elements that modify chromatin architecture and hence regulate gene expression. Transcriptional activation has been linked to the combined actions of members of the SWI and SNF families, mediated in part through ATP-dependent alterations in chromatin structure that are permissive for transactivating factor accessibility (for a review, see reference 15). In yeast, these positive actions of the SWI/SNF complex are opposed by the repressors SIN3 and RPD3--factors that were first identified in genetic screens designed to uncover gene mutations which alleviate the repressive effects of a swi5 mutation (21). An epistatic relationship between SIN3 and RPD3 is supported by the observation that loss-of-function mutations in either gene lead to derepression of the same set of genes affected by the swi5 mutation (32).
The subsequent identification of the yeast SIN3 mammalian orthologs, Sin3A and Sin3B, and their physical interaction with the Mad family of sequence-specific transcriptional repressors (Mad/Mxi1) supported the view that Mad/Mxi1-mediated transcriptional repression might be executed on the level of chromatin regulation (3, 28). With the identification of the mammalian histone deacetylases HDAC1 and HDAC2 and their significant homology to yeast RPD3 (33, 38), it became apparent that mSin3 functions to tether sequence-specific transcriptional repressors to histone deacetylase activity (1, 12, 13, 16, 20, 40). Biochemical studies with mammalian cells have demonstrated that mSin3 possesses histone deacetylase activity and that this mSin3-associated activity requires physical interaction between HDAC1 and the C-terminal region of mSin3. The mSin3/HDAC1 interaction appears to be indirect as determined by the presence of mSin3 and HDAC1 in reciprocal immunoprecipitations in vivo but not in vitro.
More recent studies with yeast have identified another integral component of the Sin3-Rpd3 complex, the Sds3 transcriptional repressor (6, 17). These studies demonstrated that Sds3 maintains the physical integrity of the Sin3-Rpd3 complex and is required for its histone deacetylase activity (17). These biochemical observations gain added significance in light of the fact that yeast SDS3, SIN3, and RPD3 all emerged from a genetic screen aimed at identifying suppressors of a silencing defective rap1 mutant (34). Further evidence for a genetic and functional link between Sds3 and Sin3-Rpd3 came from finding that sds3 null mutants demonstrated substantial phenotypic overlap with strains mutant for sin3 and/or rpd3 (6, 34, 35).
The C-terminal region of mSin3 that is required for HDAC1 interaction, designated the histone deacetylase interaction domain (HID), plays an essential role in Sin3-mediated repression of both gene expression and Myc-mediated cellular transformation (1, 16). In an effort to understand better the role of the mSin3 HID, we sought to identify factors that may participate in the critical physical and functional interactions between mSin3 and HDAC1. Here, we report the identification of a component of the mSin3-HDAC1 complex that bears striking homology to the previously identified yeast Sds3, designated mSds3. Although mSds3 does not possess intrinsic histone deacetylase activity, mSds3 represses transcription in reporter assays, recruits histone deacetylase activity, and supports the catalytic activity of HDAC1 in a manner analogous to its ortholog in yeast.
MATERIALS AND METHODS
Two-hybrid studies, isolation of mSds3 cDNA, and generation of mSds3 mutants.
A mouse E9.5 and E10.5 cDNA library in the vector pVP16 (kindly provided by S. Hollenberg and described in reference 37) was introduced into the S. cerevisiae L40 reporter strain expressing LexA fused to the C terminus of mSin3A (LexA-mSin3A residues 534 to 1274) in the plasmid pBTM116. Fragment of mSds3 isolated in the above two-hybrid screen was used to probe a mouse newborn brain cDNA phage library (Stratagene). Restriction mapping and sequence analysis of several mSds3 hybridizing clones resulted in the identification of two partial overlapping clones that were used to create a full-length 2.4-kb cDNA. Protein sequence alignments and secondary structure predictions were determined using the BCM Search Launcher (31). For two-hybrid studies, plasmids encoding the VP16 TAD fused to fragments of mSds3 were cointroduced along with plasmids encoding LexA fused to fragments of mSin3A, mSin3B, mSds3, mSAP30 (mSAP30 was isolated in a previous two-hybrid screen for mSin3A-associated proteins [L. Alland and R. A. DePinho, unpublished data]), mSAP18 (kindly provided by D. Reinberg), or MeCP2 (kindly provided by A. Bird) into the L40 yeast strain by small-scale transformation. Deletion mutants of mSds3 for this and other experiments were generated by PCR-based site-directed mutagenesis. Double transformants were assayed for β-galactosidase activity by filter assay as described previously (37).
Immunoprecipitation and Western blot.
Subconfluent 293T or NIH 3T3 cells were transfected with 3 μg of each of the appropriate expression constructs and 30 μg of Lipofectamine reagent (Gibco). Where indicated, cells were metabolically labeled using [35S]methionine (Amersham) for 6 h prior to harvest. Mouse Sds3 rabbit polyclonal antibody was raised against glutathione-S-transferase (GST)-mSds3 fusion protein encoding residues 1 to 110 of mSds3 and affinity purified (Research Genetics). Immunoprecipitations and Western blotting of overexpressed proteins were performed as described previously (1) using anti-FLAG M2 (Sigma), anti-mSin3B (PAH2) (Santa Cruz Biotechnology), anti-HDAC1 (Upstate Biotechnology), and anti-mSds3 antibodies. For interaction studies of endogenous proteins, newborn (1- to 3-day-old) C57/B6J (Jackson Labs) mouse tissues were isolated and homogenized in lysis buffer, and 1 to 5 mg of protein was used per immunoprecipitation with normal rabbit serum (Santa Cruz Biotechnology), anti-mSin3A (PAH2) (Santa Cruz Biotechnology), anti-HDAC1 (Upstate Biotechnology), or anti-mSds3 antibodies, as described above.
Deacetylase assays.
A peptide encoding for the 24 first amino acids of histone H4 was synthesized, 3H radiolabeled, and purified. Immunoprecipitates were incubated for 2 h at 37°C, and deacetylase activity was determined as described previously (33).
In vitro translation and GST pull-down assays.
The GST fusion constructs were generated by fusing full-length mSds3 or HDAC1 cDNAs in-frame into pGEX (Pharmacia). GST protein was expressed in DH5α or BL21 cells and purified with glutathione-Sepharose beads (Pharmacia). Plasmid encoding full-length SMRT was kindly provided by R. M. Evans, and plasmids encoding full-length RbAp48 and HDAC1 were kindly provided by S. L. Schreiber. Radiolabeled TNT-coupled transcription and translation products (Promega) were incubated with GST protein for 1 h at 4°C in radioimmunoprecipitation buffer.
Reporter assays.
Subconfluent NIH 3T3 cells were transfected with various GAL4-mSds3 fusion constructs or GAL4-mSin3B and the luciferase reporter plasmid containing four GAL4 binding sites upstream of the myelomonocytic growth factor minimal promoter (2), kindly provided by R. N. Eisenman. Trypticase soy agar (TSA) (100 ng/ml) (Sigma) was added for 24 h prior to lysis. Cells were lysed in luciferase lysis buffer (Invitrogen) 48 h posttransfection. Cell extracts were assayed for luciferase activity using luciferase buffer (Invitrogen) and an automated luminometer. Transfection efficiencies were normalized using an internal β-galactosidase control.
RNA interference assays.
RNA oligonucleotides that were 21 bases in length were synthesized by Dharmacon. The first [RNAi(1)] and second [RNAi(2)] sets of complementary primers correspond, respectively, to nucleotides 127 to 145 and 306 to 324 of the mSds3 cDNA sequence relative to the first nucleotide of the start codon. Deoxyribosylthymidines were added 3′ of each primer. Each primer was diluted to 20 μM and annealed with its complementary primer as described (8). Oligofectamine (Life Technology) was used to transfect double-stranded RNA, and Lipofectamine Plus was used to transfect plasmids according to the manufacturer's instructions. The cells were lysed 60 h after transfection, and histone deacetylase assays were performed on 500 μg of protein.
RESULTS
Identification of an mSin3-associated protein with homology to yeast Sds3.
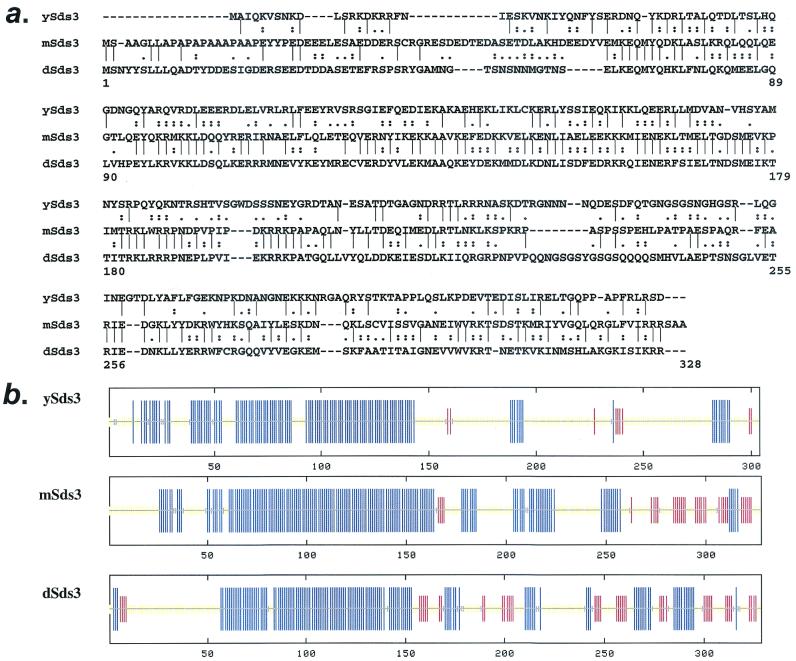
To search for mSin3-interacting proteins, we employed a mSin3A C-terminal bait (residues 534 to 1274, which includes the HID) in the yeast two-hybrid assay. We retrieved 25 His+ LacZ+ clones from a mouse embryo cDNA library (37), all of which encoded overlapping fragments of the same cDNA. Sequence analysis of the full-length 2.4-kb cDNA revealed a putative 328-amino-acid-residue protein bearing significant similarity to the yeast sds3 protein on the levels of primary amino acid sequence (18% identity, 50% similarity) (Fig. 1a) and predicted alpha-helical domains (Fig. 1b). Expression analysis identified a nuclear staining pattern and broad expression of a 2.4-kb transcript, as well as a shorter 1.8-kb transcript arising from an alternative polyadenylation signal in the 3′ untranslated region, in a panel of adult mouse tissues (data not shown). Consistent with the evolutionary conservation of mSin3 and its components, sequences bearing significant homology to mSds3 were identified in the drosophila and human databases. Relative to mSds3, drosophila Sds3 possessed 38% identity and 66% similarity (Fig. 1a), and the human Sds3 possessed 99.6% identity (data not shown). Secondary structure analysis revealed that the N-terminal regions of yeast, mouse, and Drosophila proteins are strongly predicted to form coiled-coil domains (amino acids 59 to 170, 58 to 136, and 64 to 157 of mSds3, ySds3, and dSds3, respectively). We conclude that the above proteins represent orthologs of the yeast Sds3 protein.
FIG. 1.
mSds3 encodes a mammalian protein that bears homology to yeast and Drosophila Sds3. (a) Amino acid comparisons of ySds3, mSds3, and dSds3. “|” indicates identity, “:” indicates high similarity, and “.” indicates low similarity. (b) Secondary structure predictions of ySds3, mSds3, and dSds3. Blue lines, alpha helix; red lines, beta sheet; yellow lines, indeterminate. Amino acids 59 to 170, 58 to 136, and 64 to 157 of mSds3, ySds3, and dSds3, respectively, correspond to predicted coiled-coil domains.
mSds3 interacts with mSin3 proteins in vitro and in vivo and is capable of homodimerization.
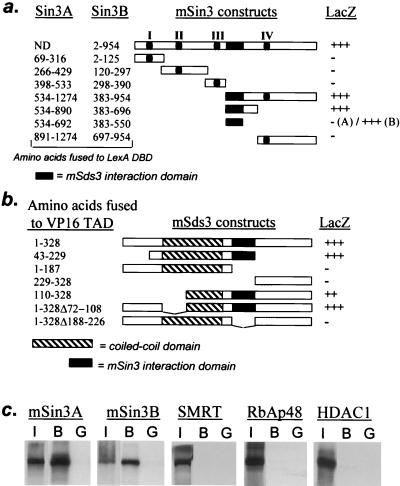
Physical interactions among mSds3 and components of the mSin3 complex were examined in a number of assays. In yeast two-hybrid assays, full-length mSin3B interacted strongly with full-length mSds3, as did various mSin3A and mSin3B fragments that included the HID (Fig. 2a). A portion of the HID within mSin3B was sufficient for interaction with mSds3, delimiting mSin3B residues 383 to 550 as the mSds3 interaction domain (Fig. 2a). The corresponding region within mSin3A (residues 534 to 692) was not sufficient for interaction with mSds3 (Fig. 2a), perhaps reflecting an anomalous presentation of the mSin3A fragment. Employing yeast two-hybrid assays, we found that mSds3 residues 188 to 226 played an essential role in associating with full-length mSin3B (Fig. 2b). Although these results suggest this mSds3 region as the mSin3 interaction domain (SID), a fragment spanning residues 188 to 226 was not sufficient for interaction (data not shown), suggesting either that this fragment is not presented properly to mSin3 or that loss of this region in mSds3 disrupts presentation of a SID elsewhere in the molecule. GST-pulldown assays supported a strong, possibly direct interaction between GST-mSds3 and in vitro-translated mSin3A or mSin3B (Fig. 2c). In vitro interactions were not detected between GST-mSds3 and the known mSin3-associated proteins SMRT, RbAp48, and HDAC1 in GST-pulldown assays (Fig. 2c) and mSAP18, mSAP30, and MeCP2 in yeast two-hybrid assays (data not shown).
FIG. 2.
mSds3 interacts with mSin3A and mSin3B in yeast and in vitro. (a) Yeast two-hybrid studies using various mSin3A and mSin3B baits along with the VP16(TAD)-full-length mSds3 prey to localize mSin3's mSds3 interaction domain. I to IV correspond to the paired amphipatic helix of mSin3A and mSin3B (PAH). (b) Yeast two-hybrid studies using a full-length mSin3B bait along with various mSds3 prey to localize mSds3's mSin3 interaction domain. +++ indicates a LacZ phenotype that is equivalent to that observed for Myc-Max HLH/LZ interaction in yeast. (c) Bacterially expressed GST full-length mSds3 fusion protein was tested for its ability to interact with radiolabeled in vitro-translated mSin3A, mSin3B, or other known components of the mSin3 repression complex. I, input, 10%; B, bound (GST-mSds3); G, GST alone.
Following transient transfection of mSds3 and mSin3B expression constructs into 293T cells, immunoprecipitates of mSds3FLAG contained mSin3B, and similarly, anti-mSin3B immunoprecipitates were found to contain mSds3FLAG, which was consistently visualized as a doublet with a more prominent band of approximately 48 kDa and a less prominent band of 52 kDa when overexpressed in cells (Fig. 3a, lanes 5 and 6). Consistent with previous immunoprecipitations of mSin3 (40), additional polypeptides were coimmunoprecipitated specifically with endogenous and transfected mSin3B that correspond in size to HDAC1 (60 kDa), HDAC2 (58 kDa), and to a lesser extent two polypeptides of 48 and 46 kDa that correspond in size to RbAp48 and RbAp46 (Fig. 3a, lanes 1, 3, and 5). Interestingly, p60 and p58 were also present in mSds3FLAG immunoprecipitates when mSin3B was overexpressed (Fig. 3a, lane 6). Deletion of the N-terminal portion of HID in mSin3B (Δ383-550) abolished interaction with mSds3FLAG (Fig. 3b, lanes 1 and 2) as well as interaction with the polypeptides migrating at 60 and 58 kDa, and at 48 and 46 kDa, that are consistent in size with HDAC1 and HDAC2 and with RbAp48 and RbAp46, respectively (Fig. 3b, compare lanes 1 and 3). Notably, helix-breaking proline substitutions in PAH3 and PAH4 of mSin3B did not affect mSin3B-mSds3 interaction (data not shown). Anti-FLAG immunoprecipitates of mSds3ΔSIDFLAG (Δ188-226) did not possess detectable amounts of mSin3B or of polypeptides corresponding in size to HDAC1/2 or RbAp46/48 (Fig. 3b, lanes 3 and 4). An analysis of various newborn mouse tissues revealed endogenous mSds3-mSin3A complexes as evidenced by the presence of mSin3A in anti-Sds3 immunoprecipates, and correspondingly, mSds3 in the anti-mSin3A immunoprecipitates (Fig. 3c). Together, these results indicate that mSds3 interacts with mSin3 proteins in vitro and in vivo .
FIG. 3.
mSds3 interacts with mSin3A and mSin3B in vivo. (a and b) 293T cells were metabolically labeled after cotransfection of the various mSin3B and mSds3FLAG expression constructs shown. Radiolabeled whole-cell extracts were immunoprecipitated using anti-mSin3B antibody or anti-FLAG antibody and visualized by autoradiography. Triangles indicate the bands corresponding to the HID deletion mutant of mSin3B (panel b, lane 2) or the SID deletion mutant of mSds3 (panel b, lane 4). (c) Tissue extracts derived from various organs of newborn mice were immunoprecipitated with anti-mSds3, resulting in coimmunoprecipitation of mSin3A detected by Western blotting using mSin3A antibody (top panels), and similarly, extracts were immunoprecipitated with anti-mSin3A antibody, resulting in coimmunoprecipitation of mSds3 detected by Western blot using anti-mSds3 antibody (bottom panels). NRS, normal rabbit serum; HC, heavy chain.
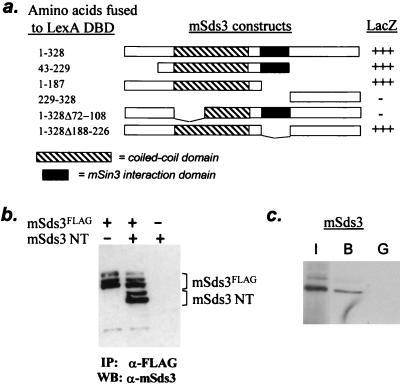
The putative coiled-coil domain of mSds3 was dispensable for mSin3 interaction (Fig. 2b), although the importance of other known coiled-coil domains in protein homodimerization raised the possibility of a similar role for mSds3. Indeed, yeast two-hybrid studies revealed robust interaction between full-length mSds3 and various coiled-coil-containing mSds3 fragments, and abolishment of interaction with deletion of residues 72 to 108 positioned within “helix A” of the coiled-coil domain (see Fig. 4a). The preservation of interaction with mSds3ΔSID indicates that homodimerization is mSin3 independent (Fig. 4a). Consistent with these yeast-based assays, coimmunoprecipitation studies with mammalian cells revealed the capacity of the mSds3 N-terminal region (residues 1 to 187) to engage FLAG-tagged full-length mSds3 (Fig. 4b). Additionally, GST-mSds3 bound in vitro-translated mSds3 (Fig. 4c), indicating that while higher order complexes are likely to form in vivo, homodimerization of mSds3 seems to occur via direct protein-protein interaction.
FIG. 4.
mSds3 forms homodimers. (a) Yeast two-hybrid studies using various mSds3 baits along with VP16 TAD-full-length mSds3 prey to localize mSds3's dimerization domain. Helix A of the putative coiled-coil domain (residues 72 to 108) was defined by secondary structure prediction software. (b) In vivo expression of mSds3FLAG along with mSds3 N-terminal fragment (mSds3NT: residues 1 to 187) in 293T cells followed by immunoprecipitation with anti-FLAG antibody results in coimmunoprecipitation of mSds3NT detected by Western blot using anti-mSds3 antibody. IP, immunoprecipitation; WB, Western blot. (c) Bacterially expressed GST-full-length mSds3 fusion protein was tested for its ability to interact with radiolabeled in vitro translated full-length mSds3. I, input, 10%; B, bound (GST-mSds3); G, GST alone.
mSds3 forms a complex with mSin3B and HDAC1.
Studies with yeast have uncovered an important role for yeast Sds3 in maintaining the Sin3-Rpd3 interaction. The close proximity of mSds3 and HDAC1 interaction sites on mSin3 suggested an analogous role for mSds3 in modulating the integrity of the mSin3-HDAC1 complex. To this end, we first documented that endogenous mSds3 is indeed a component of the mSin3/HDAC1 complex in vivo, by demonstrating that mSds3 was present in anti-HDAC1 immunoprecipitates from various mouse tissues (Fig. 5a). Of note, in contrast to the consistent doublet that is found when mSds3 is overexpressed in transformed cell lines, endogenous mSds3 appeared as a large single band of approximately 46 kDa in mouse tissues. The basis for these differences is not understood.
FIG. 5.
mSds3 forms a complex with mSin3B and HDAC1 in mammalian cells and mouse tissues. (a) Tissue extracts were derived as for Fig. 3c and immunoprecipitated with anti-HDAC1 antibody, resulting in coprecipitation of mSds3, detected by Western blot using mSds3 antibody. Anti-mSds3 immunoprecipitates were performed as a positive control for mSds3. NRS, normal rabbit serum; HC, heavy chain. (b) 293T cells were metabolically labeled after transfection of the various mSin3B, mSds3FLAG, and HDAC1 expression constructs shown. Whole-cell extracts were immunoprecipitated using anti-mSin3B antibody and visualized by autoradiography. The triangle indicates the band corresponding to the HID deletion mutants of mSin3B (lane 4). (c) NIH 3T3 cells were transfected with mSin3BFLAG and/or HDAC1FLAG expression constructs as shown. Whole-cell extracts were immunoprecipitated with an anti-mSds3 antibody, and the presence of mSin3BFLAG and/or HDAC1FLAG was detected by Western blotting using anti-FLAG antibody (right panel, lanes 4 to 6). The left panel (lanes 1 to 3) represents a Western blot performed with an anti-FLAG antibody on 10% of the extracts before immunoprecipitation.
To better define the physical relationship among these three proteins, coimmunoprecipitation assays were performed following transfection of various combinations of mSin3B, mSds3, and HDAC1. Anti-mSin3B immunoprecipitates efficiently captured HDAC1 and mSds3FLAG in 293T cells cotransfected with all three expression constructs (Fig. 5b, lane 3). As shown in Fig. 3b, mSin3B was able to coprecipitate with two polypeptides at 60 and 58 kDa, and this coprecipitation was dependent on the HID domain (Fig. 5b, compare lanes 1 to 3 to lane 4). The amount of HDAC1 in anti-mSin3B immunoprecipitates did not appear to change in the presence of overexpressed mSds3FLAG (compare lanes 1 and 3), and similarly, the amount of mSin3B-associated mSds3FLAG was not altered significantly in the presence of overexpressed HDAC1 (compare lanes 2 and 3). The mSin3B deletion mutation (Δ383-550) that is unable to bind mSds3 also failed to bind HDAC1 (lane 4), indicating that the integrity of this region of mSin3B is essential for interaction with both mSds3 and HDAC1. Together, these results suggest that despite the use of operationally defined HID of mSin3B, mSds3 and HDAC1 do not compete for the same site within the HID.
Given that mSin3B can associate with both HDAC1 and mSds3, the role of mSin3B in the interaction between mSds3 and HDAC1 was investigated. To this end, 293T cells were transfected with mSin3BFLAG and/or HDAC1FLAG, and their presence in endogenous anti-mSds3 immunoprecipitates was analyzed. In separate transfections, anti-mSds3 immunoprecipitates contained mSin3BFLAG, as expected (Fig. 5c, lane 5), but only modest amounts of HDAC1FLAG. Interestingly, the amount of HDAC1FLAG in anti-mSds3 immunoprecipitates was significantly increased by overexpression of mSin3BFLAG (Fig. 5c, compare lane 4 to lane 6). Together, these results strongly suggest that mSin3B, HDAC1, and mSds3 are part of the same complex in vivo and that mSin3B and mSds3 cooperate to stabilize the interaction with HDAC1.
mSds3 represses transcription and augments histone deacetylase activity of HDAC1 in vivo.
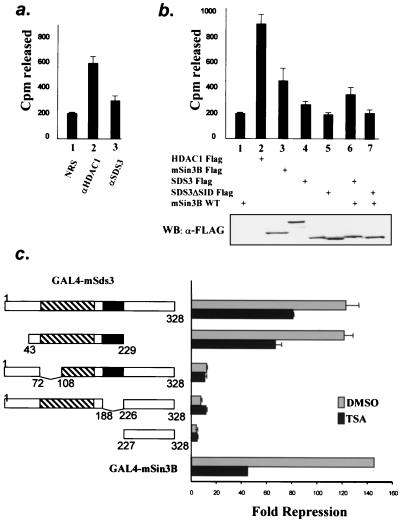
To determine whether mSds3 recruits histone deacetylase activity, histone deacetylase assays were performed on anti-mSds3 immunoprecipitates in three independent experiments. mSds3 immunoprecipitates from untransfected NIH 3T3 cells contained a modest but reproducible deacetylase activity above background levels (Fig. 6a, lane 3; P = 0.005). Similarly, reproducible deacetylase activity was detected in anti-FLAG immunoprecipitates of 293T cells transfected with mSds3FLAG, and this activity was enhanced by mSin3B overexpression (Fig. 6b, compare lanes 4 and 6), consistent with the physical interaction data presented in Fig. 5c above. Deacetylase activity above background levels was not detected in immunoprecipitates of the mSds3 mutant that is unable to bind mSin3B (lane 5), and the activity of this mSds3 mutant was not altered by mSin3B overexpression (lane 7). Taken together, these results are consistent with the view that mSds3 does not possess intrinsic deacetylase activity but instead functions to recruit HDAC catalytic activity in a manner that is augmented by mSin3B.
FIG. 6.
mSds3 recruits histone deacetylase and represses basal transcription. (a and b) Deacetylase activity assays on immunoprecipitates from untransfected NIH 3T3 cells (a) or 293T cells transfected with the various expression constructs shown (b). Shown are the means from three experiments performed in duplicate and 95% confidence intervals. (a) endogenous histone deacetylase activity immunoprecipitated using normal rabbit serum (NRS) (lane 1), anti-HDAC1 antibody (lane 2), or anti-mSds3 antibody (lane 3). (b) Histone deacetylase assays on anti-FLAG immunoprecipitates from cells transfected with the indicated expression constructs (upper panel). The level of expression of transfected constructs was assessed by an anti-FLAG Western blot (bottom panel). (c) Reporter assays using various GAL4-mSds3 fusions and GAL4-mSin3B to repress a luciferase reporter gene containing GAL4 binding sites upstream of the myelomonocytic minimal promoter. The constructs and their levels of repression relative to GAL4 are shown. Shown are the means of three experiments performed in triplicate and 95% confidence intervals, except for GAL-mSin3B, for which the experiment was done once in triplicate. Gray bars, untreated cells; solid bars, cells treated with TSA for 24 h. DMSO, dimethyl sulfoxide.
Next, we examined the capacity of mSds3 to repress transcription in reporter assays. On the basis of its physical interactions with mSin3 and HDAC1, it is anticipated that mSds3 would possess potent repression activity; however, it is also possible that mSds3 could exert repressive activity through Sin3/HDAC1-independent mechanisms. mSds3 and various fragments were fused to the GAL4 DNA binding domain and tested for their ability to modulate transcription of luciferase reporters driven by one of several minimal promoters bearing GAL4 binding sites. In this assay, mSds3 exhibited strong repression of basal transcription on a level comparable to that of mSin3B (Fig. 6c). A fragment of mSds3 that is sufficient for homodimerization and binding to mSin3 (amino acids 43 to 229) also showed strong repression activity. Disruption of the homodimerization domain or the putative SID strongly diminished the repression activity of mSds3, indicating that both of these functions contribute significantly to the mSds3-associated repression activity.
The addition of TSA, a potent inhibitor of histone deacetylase activity, only partly reduced the repressive activity driven by mSds3 (Fig. 6c). This reduction was only 35% for GAL-mSds3 compared to 70% for GAL-mSin3B. As expected, TSA affected the repression activity of the mutants of mSds3 which were able to bind to mSin3, and therefore HDAC1, whereas repression was unchanged for those mutants that did not bind to mSin3/HDAC1. Western blot analysis of cell lysates following transient transfection of the various GAL4-mSds3 fusions demonstrated equivalent expression levels (data not shown). Together, these results suggest that mSds3 brings both HDAC-dependent and -independent repressive activity to the mSin3/HDAC complex.
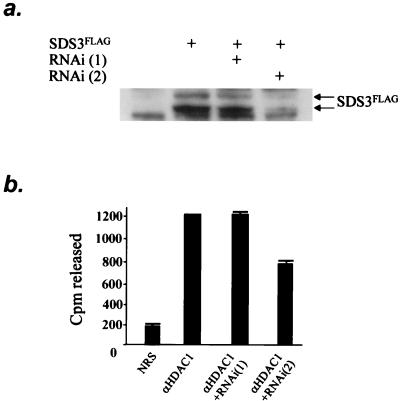
Next, we sought to determine whether the mSds3, like its yeast counterpart, played a role in supporting the catalytic activity of the histone deacetylase molecule. The RNA interference method was used to assess the impact of reduced mSds3 levels on HDAC1 enzymatic activity. We generated two sets of cRNA primers corresponding to nucleotides 127 to 145 [RNAi(1)] or to nucleotides 306 to 324 [RNAi(2)] of the mSds3 cDNA. The efficiency of these double-stranded RNAs in down-regulating the level of mSds3 protein was tested on lysates of NIH 3T3 cells expressing a small amount of mSds3FLAG protein. Indeed, transfection of RNAi(2) efficiently down-regulated the level of exogenous mSds3, while mock transfection or transfection with RNAi(1) did not affect the level of exogenously expressed mSds3 (Fig. 7a). This result correlates with previous observations showing that RNAi are far more efficient when the 5′ nucleotide sequence is AA, as is the case for RNAi(2) (T. Tuschl, personal communication).
FIG. 7.
mSds3 contributes to HDAC1 enzymatic activity. (a) Western blot performed with an anti-FLAG antibody on NIH 3T3 cell extracts after transfection with mSds3-FLAG construct with or without RNA primers. (b) Histone deacetylase activity assays were performed on anti-HDAC1 immunoprecipitates from NIH 3T3 cells after transfection with RNA primers. Shown are the means from three experiments. SDS3FLAG, mSds3FLAG.
We then tested the effect of these cRNA primers on HDAC1 enzymatic activity. In three independent experiments with each point conducted in triplicate, NIH 3T3 cells were mock transfected or transfected with RNAi(1) or RNAi(2). The histone deacetylase activity of endogenous HDAC1 immunoprecipitates was then determined. As expected, deacetylase activity was not affected by transfection of RNAi(1) (Fig. 7b, lanes 1 and 2). In contrast, transfection of RNAi(2) reproducibly reduced deacetylase activity of HDAC1 by 40% (Fig. 7b, lane 3), though the HDAC1 protein level was not affected (data not shown). Together, these results strongly indicate that mSds3 enables HDAC1 enzymatic activity.
DISCUSSION
In this study, we report the identification of mammalian Sds3 as a key component of the mSin3 corepressor complex, sharing physical and functional properties of its yeast ortholog. These observations underscore the importance of the Sin3 corepressor complex from yeast to mammals. Indeed, all known components of the mammalian Sin3 corepressor complex, mSin3A/B, HDAC1/2, RbAP46/48, SAP18, SAP30, and mSds3, appear to be represented in yeast, thus reinforcing the central and essential role of this complex in eukaryotic cell biology (24, 36, 40, 42).
Beyond its role in promoting a functional HDAC1 activity in the mSin3 complex, the precise mechanism(s) through which mSds3 operates in not yet clear. When tethered to DNA, mSds3 exhibits potent transcriptional repression activity in reporter assays, and this activity is only partly neutralized by the HDAC inhibitor, TSA, or by deletion of the putative SID of mSds3. Thus, it is possible that mSds3 might recruit activities other than histone deacetylase. Within the context of the mSin3 complex, the additional finding that mSds3 can homodimerize raises the intriguing possibility that mSds3 might allows for the assembly of a large complex composed of two Sin3 molecules, each with distinct repressor activities. Although this supposition remains unproven, it is notable that yeast Sin3p is present in a large multiprotein complex with an apparent molecular mass of greater than 2 million Da (14).
In RNAi-mediated knock-down experiments, HDAC1 activity was markedly impaired in vivo. This observation, coupled with mSin3B-driven enhancement of the mSds3/HDAC1 interaction, suggests that these three proteins act cooperatively to maintain the catalytic activity of the complex. Correspondingly, it is possible that the basis for the catalytically inactive nature of HDAC1 generated in bacterial or insect cells (41; G. David and R. A. DePinho, unpublished results) relates to the absence of mSds3-like factors. These observations are reminiscent of several studies emphasizing an essential role of SANT domain-containing proteins in histone deacetylase activity (the SANT domain is a putative DNA-binding domain in SWI3, ADA2, NCoR, and TFIIB). Specifically, Zhang et al. (41) reported that the formation of a catalytically active NuRD complex requires the presence of the SANT domain protein MTA2. Similarly, SMRT and NCoR, which also contain SANT domains, are necessary for catalytically active HDAC3 (10). Finally, the CoREST protein, which is a core component of an mSin3-independent HDAC1/2 complex, possesses two SANT domains (39). With regard to the mSin3/HDAC complex, SANT domain-containing proteins have not yet been identified and it is plausible that mSds3 may function to recruit such a protein to the Sin3/HDAC complex.
Current database searches have not identified highly related mSds3 homologs, although two mammalian proteins share some relatedness. Brms1, identified previously as a breast cancer metastatic suppressor gene (30), shares 24% identity and 50% similarity with more than half of the mSds3 protein sequence (mSds3 residues 62 to 232), including the putative SID domain. A second mammalian protein (human MGC11296) whose function has not yet been characterized also shares homology to mSds3 and to a higher extent to Brms1. This protein contains an additional C-terminal portion that is absent from brms1 but that shares homology to mSds3 (residues 232 to 323 in mSds3). Together, these findings raise the possibility of the existence of a family of mammalian mSds3-related proteins, which, like mSds3, might play a role in mSin3-related complexes.
Several lines of evidence point to a potential role for mSds3 in tumor suppressor pathways. First, Mxi1, a member of the Mad family of transcriptional repressors, functions as a bona fide tumor suppressor as evidenced by a cancer predisposition in mice null for Mxi1 (29) and a possible linkage to the pathogenesis of prostate cancer in humans (7, 25). Similarly, cell culture-based transformation assays have established that recruitment of the mSin3 complex is essential to Mxi1's antioncogenic actions (1, 26). Second, there is an emerging role of histone deacetylases in the development of hematopoietic malignancies (4, 5, 9, 11, 18, 19), and HDAC inhibitors have exhibited therapeutic activity in the clinic (for a review, see reference 23). Third, as noted above, mSds3 bears structural relatedness to a putative metastases suppressor gene, Brms1 (30). Finally, using computational analysis, we localized the gene encoding human SDS3 on chromosome 12q24.2, a region known to be deleted in 25% (4 of 16) of cases of human prostate cancer (27). Given the requirement of an active mSin3/HDAC complex for Mxi1 to function as a tumor suppressor, it is tempting to speculate that mSds3 may be an important component of the Mxi1-mSin3-HDAC1 tumor suppressor pathway. An improved understanding of the regulation of histone deacetylase activity by mSds3 might allow the identification of novel avenues for modulation of HDAC activity. Histone deacetylase inhibitors have shown promise in the treatment of hematologic malignancies, but their utility has been limited by toxicity and short-term efficacy (22). Future studies into the mechanisms by which mSds3 regulates the catalytic activity of HDAC might augment the therapeutic impact of these deacetylase inhibitor programs.
Acknowledgments
Leila Alland and Gregory David contributed equally to this work.
We thank Thomas Graf, Nicole Schreiber-Agus, and Nabeel Bardeesy for helpful discussions, Maria Dudas and Carlos Cordon-Cardo for providing mouse embryo sections, and Annick Harel-Bellan for advice on histone deacetylase assays. We also thank Jim DeCaprio and Richard Maser for critical reading of the manuscript.
This work was supported by NIH grant K11CA68266 (L.A. and R.A.D.), the Ruth Estrin Goldberg Memorial Award (L.A.), an American Cancer Society Young Investigator Award (L.A.), and Human Frontier Science Program Organization (G.D.). R.A.D. is an American Cancer Society Research Professor.
REFERENCES
- 1.Alland, L., R. Muhle, H. Hou, Jr., J. Potes, L. Chin, N. Schreiber-Agus, and R. A. DePinho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49-55. [DOI] [PubMed] [Google Scholar]
- 2.Ayer, D. E., C. D. Laherty, Q. A. Lawrence, A. P. Armstrong, and R. N. Eisenman. 1996. Mad proteins contain a dominant transcription repression domain. Mol. Cell. Biol. 16:5772-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayer, D. E., Q. A. Lawrence, and R. N. Eisenman. 1995. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80:767-776. [DOI] [PubMed] [Google Scholar]
- 4.David, G., L. Alland, S. H. Hong, C. W. Wong, R. A. DePinho, and A. Dejean. 1998. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene 16:2549-2556. [DOI] [PubMed] [Google Scholar]
- 5.Dhordain, P., O. Albagli, R. J. Lin, S. Ansieau, S. Quief, A. Leutz, J. P. Kerckaert, R. M. Evans, and D. Leprince. 1997. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. USA 94:10762-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorland, S., M. L. Deegenaars, and D. J. Stillman. 2000. Roles for the Saccharomyces cerevisiae SDS3, CBK1 and HYM1 genes in transcriptional repression by SIN3. Genetics 154:573-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eagle, L. R., X. Yin, A. R. Brothman, B. J. Williams, N. B. Atkin, and E. V. Prochownik. 1995. Mutation of the MXI1 gene in prostate cancer. Nat. Genet. 9:249-255. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 9.Grignani, F., S. De Matteis, C. Nervi, L. Tomassoni, V. Gelmetti, M. Cioce, M. Fanelli, M. Ruthardt, F. F. Ferrara, I. Zamir, C. Seiser, M. A. Lazar, S. Minucci, and P. G. Pelicci. 1998. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 391:815-818. [DOI] [PubMed] [Google Scholar]
- 10.Guenther, M. G., O. Barak, and M. A. Lazar. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guidez, F., S. Ivins, J. Zhu, M. Soderstrom, S. Waxman, and A. Zelent. 1998. Reduced retinoic acid-sensitivities of nuclear receptor corepressor binding to PML- and PLZF-RARalpha underlie molecular pathogenesis and treatment of acute promyelocytic leukemia. Blood 91:2634-2642. [PubMed] [Google Scholar]
- 12.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 13.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 14.Kasten, M. M., S. Dorland, and D. J. Stillman. 1997. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol. Cell. Biol. 17:4852-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 16.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 17.Lechner, T., M. J. Carrozza, Y. Yu, P. A. Grant, A. Eberharter, D. Vannier, G. Brosch, D. J. Stillman, D. Shore, and J. L. Workman. 2000. Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3·Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J. Biol. Chem. 275:40961-40966. [DOI] [PubMed] [Google Scholar]
- 18.Lin, R. J., L. Nagy, S. Inoue, W. Shao, W. H. Miller, Jr., and R. M. Evans. 1998. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 391:811-814. [DOI] [PubMed] [Google Scholar]
- 19.Melnick, A. M., J. J. Westendorf, A. Polinger, G. W. Carlile, S. Arai, H. J. Ball, B. Lutterbach, S. W. Hiebert, and J. D. Licht. 2000. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol. Cell. Biol. 20:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 21.Nasmyth, K., D. Stillman, and D. Kipling. 1987. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell 48:579-587. [DOI] [PubMed] [Google Scholar]
- 22.Newmark, H. L., J. R. Lupton, and C. W. Young. 1994. Butyrate as a differentiating agent: pharmacokinetics, analogues and current status. Cancer Lett. 78:1-5. [DOI] [PubMed] [Google Scholar]
- 23.Pandolfi, P. P. 2001. Histone deacetylases and transcriptional therapy with their inhibitors. Cancer Chemother. Pharmacol. 48(Suppl. 1):S17-S19. [DOI] [PubMed] [Google Scholar]
- 24.Parthun, M. R., J. Widom, and D. E. Gottschling. 1996. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell 87:85-94. [DOI] [PubMed] [Google Scholar]
- 25.Prochownik, E. V., L. Eagle Grove, D. Deubler, X. L. Zhu, R. A. Stephenson, L. R. Rohr, X. Yin, and A. R. Brothman. 1998. Commonly occurring loss and mutation of the MXI1 gene in prostate cancer. Genes Chromosomes Cancer 22:295-304. [PubMed] [Google Scholar]
- 26.Rao, G., L. Alland, P. Guida, N. Schreiber-Agus, K. Chen, L. Chin, J. M. Rochelle, M. F. Seldin, A. I. Skoultchi, and R. A. DePinho. 1996. Mouse Sin3A interacts with and can functionally substitute for the amino-terminal repression of the Myc antagonist Mxi1. Oncogene 12:1165-1172. [PubMed] [Google Scholar]
- 27.Sattler, H. P., V. Rohde, H. Bonkhoff, T. Zwergel, and B. Wullich. 1999. Comparative genomic hybridization reveals DNA copy number gains to frequently occur in human prostate cancer. Prostate 39:79-86. [DOI] [PubMed] [Google Scholar]
- 28.Schreiber-Agus, N., L. Chin, K. Chen, R. Torres, G. Rao, P. Guida, A. I. Skoultchi, and R. A. DePinho. 1995. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell 80:777-786. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber-Agus, N., Y. Meng, T. Hoang, H. Hou, Jr., K. Chen, R. Greenberg, C. Cordon-Cardo, H. W. Lee, and R. A. DePinho. 1998. Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature 393:483-487. [DOI] [PubMed] [Google Scholar]
- 30.Seraj, M. J., R. S. Samant, M. F. Verderame, and D. R. Welch. 2000. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 60:2764-2769. [PubMed] [Google Scholar]
- 31.Smith, R. F., B. A. Wiese, M. K. Wojzynski, D. B. Davison, and K. C. Worley. 1996. BCM Search Launcher-an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 6:454-462. [DOI] [PubMed] [Google Scholar]
- 32.Stillman, D. J., S. Dorland, and Y. Yu. 1994. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SW15 transcriptional activator. Genetics 136:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 34.Vannier, D., D. Balderes, and D. Shore. 1996. Evidence that the transcriptional regulators SIN3 and RPD3, and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in S. cerevisiae. Genetics 144:1343-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vannier, D., P. Damay, and D. Shore. 2001. A role for Sds3p, a component of the Rpd3p/Sin3p deacetylase complex, in maintaining cellular integrity in Saccharomyces cerevisiae. Mol. Genet. Genomics 265:560-568. [DOI] [PubMed] [Google Scholar]
- 36.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87:95-104. [DOI] [PubMed] [Google Scholar]
- 37.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205-214. [DOI] [PubMed] [Google Scholar]
- 38.Yang, W. M., C. Inouye, Y. Zeng, D. Bearss, and E. Seto. 1996. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. USA 93:12845-12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You, A., J. K. Tong, C. M. Grozinger, and S. L. Schreiber. 2001. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 98:1454-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, Y., Z. W. Sun, R. Iratni, H. Erdjument-Bromage, P. Tempst, M. Hampsey, and D. Reinberg. 1998. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell 1:1021-1031. [DOI] [PubMed] [Google Scholar]