Abstract
Focal adhesion kinase (FAK) is a nonreceptor tyrosine kinase whose focal adhesion targeting (FAT) domain interacts with other focal adhesion molecules in integrin-mediated signaling. Localization of activated FAK to focal adhesions is indispensable for its function. Here we describe a solution structure of the FAT domain bound to a peptide derived from paxillin, a FAK-binding partner. The FAT domain is composed of four helices that form a “right-turn” elongated bundle; the globular fold is mainly maintained by hydrophobic interactions. The bound peptide further stabilizes the structure. Certain signaling events such as phosphorylation and molecule interplay may induce opening of the helix bundle. Such conformational change is proposed to precede departure of FAK from focal adhesions, which starts focal adhesion turnover.
Adhesion of cells to the extracellular matrix (ECM) allows ECM proteins to interact with cell membrane-bound receptors. Such interactions generate intracellular signals that are important for cell growth, survival, and migration (11, 12, 35, 48). Integrins are a large family of transmembrane receptors that have long been recognized for their roles in linking ECM proteins to the actin cytoskeleton and in regulating cell shape and tissue architecture (14, 15, 44). The binding of integrins to ECM proteins elicits signals that are transmitted into the cell and cause actin and associated cytoskeletal proteins, including paxillin, talin, vinculin, and tensin, to gather at cell substratum sites termed focal adhesions. Although integrins have no enzymatic activity per se, one of the most prominent alterations observed upon integrin clustering is the phosphorylation of tyrosine residues within a variety of proteins in focal adhesions (40).
Focal adhesion kinase (FAK) is a nonreceptor kinase that can be activated by integrin signaling. Since the discovery of FAK in the early 1990s, this protein and its related signaling pathways have been studied in some detail (6, 36, 39, 54). Analysis of Fak knockout mice has shown that the null mutation results in an embryonic lethal phenotype that is very similar to that of the fibronectin knockout mice (24). It is now clear that FAK plays an important role in relaying the signals that are generated by the attachment of cells to the ECM and are transmitted through integrins to cytoplasmic and nuclear targets. In this way, FAK regulates cellular processes such as migration, survival, and proliferation (6, 39).
The central role of FAK in integrin signaling and the involvement of integrins in tumor progression and metastasis (15) suggest that FAK is a key player in the multistep progression toward a malignant phenotype. Indeed, mounting evidence shows that the expression as well as activity of FAK is upregulated in many cancer cells and that FAK may be involved in the development of invasive cancer (27). Furthermore, it has been demonstrated that plasma membrane-associated pY397FAK is a marker of cytotrophoblast invasion in vivo and in vitro (25). Thus, although FAK does not appear to function as a classic oncoprotein, its role in transducing survival signals suggests that it is a potential target for inhibiting tumor growth (28). This proposal is also attractive because the disruption of the function of FAK is known to induce cell apoptosis (23).
FAK and a second nonreceptor tyrosine kinase that has been called proline-rich tyrosine kinase 2 (PYK2) (also known as cell adhesion kinase β [CAKβ] and calcium-dependent protein-tyrosine kinase [CADTK]) compose a subfamily of nonreceptor tyrosine kinases. The overall amino acid sequence similarity of the two kinases is approximately 45%, and the two do not contain SH2 or SH3 domains (39). The highest sequence conservation is within the central region, which contains the kinase domain, and within the N- and C-terminal regions. The C-terminal part of FAK is rich with protein-protein interaction sites. In certain cells, this part of FAK is autonomously expressed (20) and termed FAK-related nonkinase (FRNK). FRNK contains a C-terminal focal adhesion targeting (FAT) domain and several proline-rich regions that serve as docking sites for many SH3-containing proteins. It acts as an endogenous inhibitor of FAK signals (47).
Upon activation, FAK colocalizes at focal adhesions (6, 39), and mutation studies have shown that the FAT domain is responsible for this localization. It has been proposed that this localization plays a key role in FAK signaling (23), and this is supported by the recent observation that a truncated FAK isoform that contains the FAT domain and the rest of the C-terminal portion of full-length FAK inhibits cell spreading and migration (36, 47). The function of the FAT domain is further demonstrated by studies which showed that overexpression of the FAT domain in certain cell lines can induce cell death (27, 53). Interestingly, although adenoviral transduction of a DNA segment encoding the FAT domain had no effect on adhesion and viability in normal mammary cells, this transduction resulted in the loss of adhesion and in apoptosis of breast cancer cells (52).
Much is known about the role of FAK in integrin signaling. However, the mechanism by which activated FAK localizes to focal adhesions remains unclear. Although extensive genetic and biochemical studies of this area have been performed (6), the lack of detailed structural information on FAK and the proteins with which it interacts has prevented investigators from fully understanding the results of such studies. It has been proposed that FAT results from multiple protein-protein interactions and that the well-defined interaction between the FAT domain and paxillin is necessary but not sufficient for localization of FAK to focal adhesions (8). Indeed, another focal adhesion molecule, talin, can bind to the FAT domain (4, 55). Furthermore, it has been clearly demonstrated that the ability of FAK to bind to several proteins within focal adhesions, including growth factor receptors, allows FAK to play a key role in regulating cross talk between various receptor tyrosine kinases and integrin (43). However, the details of the mechanism by which the FAT domain is released from a dispersing focal adhesion remain to be elucidated.
To help understand the mechanisms of FAT of the FAT domain, we have elucidated the solution structure of the FAT domain of FAK in the context of the complex formed by the FAT domain bound to a peptide corresponding to the LD2 sequence of paxillin. Our structure of the FAT domain is consistent with findings from genetic studies and allows us to interpret results of mutagenesis studies and to clarify some apparently controversial findings (8, 21, 46). We have also identified putative binding sites of other focal adhesion molecules on the surface of the FAT domain and propose a mechanism by which the interaction between the focal adhesion and the FAT domain is disrupted by the binding of the SH2 domain of Grb2 to the phosphorylated FAT domain. Our structural studies will potentially be helpful in the rational design of inhibitors that disrupt the function of the FAT domain of endogenous FAK. Such inhibitors may well lead to novel therapies for cancer and other diseases.
MATERIALS AND METHODS
Expression and purification of the FAT domain of FAK.
The chicken FRNK cDNA was kindly provided by J. Thomas Parsons (University of Virginia). The cDNA encoding the FAT domain (residues 916 to 1053) was subcloned into a PET28a vector. The N-terminal His-tagged FAT domains were subsequently expressed in Escherichia coli. The methods of protein induction, harvest, and purification have been described previously. To isotope label protein, we used morpholinepropanesulfonic acid-buffered medium that contained 15NH4Cl (1 g/liter) and 13C6-glucose (2.5 g/liter).
LD2 peptide and spin labeling.
The LD2 peptide of chicken paxillin (residues 139 to 162) and the four mutant peptides (Ser143Cys, Arg147Cys, Asn153Cys, and Gln156Cys) were chemically synthesized by the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital. The cysteine-specific spin label (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate (MTSSL) was purchased from Toronto Research Chemicals (Toronto, Canada). MTSSL was attached to the modified LD2 peptides as described earlier (26). In brief, 1.1 mM LD2 peptide and a 10-fold excess of MTSSL were mixed and stirred for 12 h in a 4:1 (vol/vol) solution of 130 mM NaCl, 20 mM sodium phosphate buffer (pH 7.2), and acetonitrile. Spin-labeled LD2 was purified by reverse-phase high-performance liquid chromatography.
Physical biochemistry studies.
Circular dichroism (CD) spectra were obtained with an Aviv 62DS CD spectrometer (Aviv, Lakewood, N.J.). Light-scattering experiments were performed with a DynaPro-801TC dynamic light-scattering instrument (ProteinSolution, Charlottesville, Va.). The interaction between the LD2 peptide and the FAT domain was measured by a microcalorimetry system isothermal titration calorimeter (Microcal, Northampton, Mass.).
NMR samples.
LD2 peptide samples were prepared by dissolving the lyophilized peptide in 550 μl of either 99.996% D2O or 90% H2O-10% D2O; the final concentration of peptide was about 4.0 mM. The pH of the peptide solution was adjusted to 6.5. The samples of the FAT domain:LD2 peptide complex were generated by titrating increasing amounts of unlabeled LD2 peptide with 15N-labeled or 15N/13C-labeled FAT domain. The progress of the titration was monitored by recording two-dimensional (2D) 1H-15N correlated spectra. Sample concentrations for nuclear magnetic resonance (NMR) experiments were typically 0.5 to 1.6 mM in 10 mM potassium phosphate buffer (pH 6.5) and 0.1% NaN3.
NMR spectroscopy.
All NMR data were acquired with Varian Inova 600-MHz spectrometers at 37°C. Data were processed and displayed by the program packages NMRpipe and NMRDraw (9) on an SGI Octane workstation. The programs XEASY (T.-H. Xia, C. Bartels, and K. Wuthrich, user manual for XEASY ETH automated spectroscopy for X Windows system, ETH-Honggerberg, Zurich, Switzerland) and CSI (51) were used for data analysis and semiautomatic assignments. For the LD2-bound FAT domain, backbone resonances were assigned on the basis of 3D HNCA, HNCACB, CBCA(CO)NH, HNCO, and HNCOCA, whereas aliphatic side-chain resonances were assigned on the basis of 3D 15N-edited TOCSY, HCCH-COSY, and HCCH-TOCSY spectra. Aromatic side-chain resonances were assigned on the basis of 2D ROESY, NOESY, and TOCSY of the D2O sample. The nuclear Overhauser effect (NOE) connections were assigned on the basis of 3D 15N-edited NOESY and 13C-edited NOESY, with the help of 4D 15N/13C NOESY and 4D 13C/13C HMQC-NOESY-HSQC. For the LD2 peptide, proton resonances were assigned on the basis of 2D NOESY, ROESY, COSY, and TOCSY either in H2O or in D2O; NOEs were mainly obtained from 2D ROESY and NOESY spectra.
Structural calculation.
For the LD2 peptide, a total of 476 distance constraints were derived from 2D ROESY (mixing time [τm] = 100 ms) and NOESY (τm = 100 ms). For the LD2-bound FAT domain, a total of 2,743 meaningful distance constraints were derived from 3D 15N-edited NOESY (τm = 100 ms), 3D aliphatic 13C-edited NOESY (τm = 100 ms), and 2D D2O NOESY. NOEs were assigned manually. Integrated NOE peaks were calibrated and converted to distance constraints with the program CALIBA (16). Dihedral constraints for the LD2 peptide were derived from DQF-COSY; those for the FAT domain were derived from 3D HNHA. The program DYANA (17) was used to calculate a family of 200 structures starting from randomly generated conformers in 10,000 annealing steps. Torsion angle dynamics combined with a simulated annealing algorithm were employed in the calculation. With preliminary DYANA-calculated structures available, the scaling factors for the volume-to-distance conversion for each class of distance constraints were evaluated with CALIBA by plotting volumes of peaks arising from pairs of protons at a fixed distance. Several cycles of the structure calculations were carried out to recalibrate the NOE distance constraints. The 25 structures with the lowest target functions were included in the final families.
The program DYANA was also used to build the model of the complex of the FAT domain and the LD2 peptide. The distance constraints were derived from paramagnetic relaxation effect. A 12-Å upper limit in distance was set between the paramagnetic center of each spin-labeled peptide and those amide protons of the FAT domain which their resonance missed in the 1H-15N correlation spectra of 15N-labeled FAT bound to the spin-labeled peptide. A family of 100 structures were calculated starting from the randomly oriented FAT domain and the LD2 peptide. During the calculation, both the backbone conformations of the FAT domain and the LD2 peptide were fixed. The structure of the complex was determined by averaging the 20 best structures with lowest target functions.
Nucleotide sequence accession number.
The coordinates of the FAT domain reported in this paper have been deposited in the Protein Data Bank with accession code 1KTM.
RESULTS
Structure of the FAT domain.
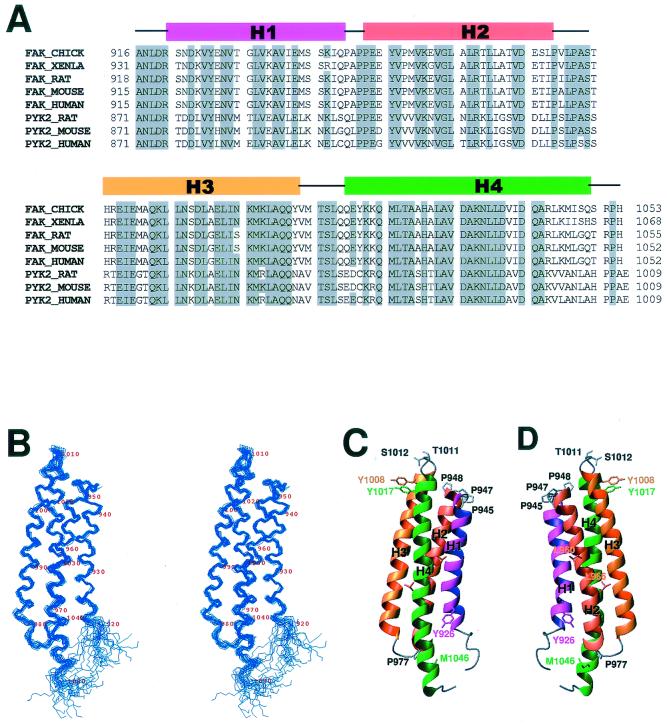
The FAT domain of FAK consists of approximately 140 amino acids and is well conserved among different species. Its amino acid sequence is highly similar to the C-terminal domain of PYK2, which also binds to the focal adhesion molecule paxillin (Fig. 1A). Using protein NMR spectroscopy, we determined the solution structure of the FAT domain of chicken FAK. The FAT domain extends from residues 916 through 1053 of chicken FAK and was expressed as a recombinant protein in E. coli. Our initial measurements of biophysical characteristics, including light scattering, gel filtration, and molecular diffusion properties, indicated that the FAT domain nonspecifically aggregates when its concentration is approximately 1 mM, the concentration required for NMR studies of protein structure. The aggregation state was completely disrupted when the FAT domain bound to a chemically synthesized, 24-amino-acid peptide that was identical to the LD2 motif (residues 139 to 162) of chicken paxillin. Our measurements, including those of light scattering, gel filtration, and molecular diffusion properties, indicated that, when excess LD2 peptide was added to the solution of the FAT domain, the LD2-bound form of the FAT domain existed as a monomer. Therefore, we used a sample of 13C/15N-labeled peptide representing the FAT domain bound to unlabeled LD2 peptide for NMR measurements. Because of the relatively low binding affinity, the molar ratio between the FAT domain and the LD2 peptide in the NMR samples was 1:5. Under such conditions, the chemical-shift perturbation indicated that all of the FAT domain was bound to the LD2 peptide.
FIG. 1.
Sequence and structure of the FAT domain. (A) Structure-based amino acid sequence alignment of the FAT domains of FAKs and the C-terminal domains of PYK2s. The alignment was produced by ClustalW and manually modified. (B) Stereo view of the peptide backbone (N, Cα, C') of 20 superimposed structures of the FAT domain with lowest target functions. (C and D) Ribbon diagrams of the structure of the FAT domain. The images were generated by the MOLMOL program (29).
The structure of the LD2-bound FAT domain was based on 2,848 NMR-derived constraints, which include 2,743 distance constraints derived from NOE data, 34 backbone dihedral angle restraints obtained from measurements of three-bond scalar coupling constants, and 71 hydrogen bond constraints attained from hydrogen-deuterium exchange experiments. All the NOE assignments were verified by the results of 4D 13C/15N- and 13C/13C-edited NOE experiments. Twenty-five structures that best fit the experimental constraints in the structural calculation were selected for detailed analysis (Fig. 1B). Most of the backbone conformations of the final structures are in favorable regions of φ-ψ space (Table 1).
TABLE 1.
NMR structure determination statistics
| Parameter | Value for:
|
|
|---|---|---|
| FAT | LD2 | |
| No. of NOE distance restraints | ||
| Intraresidue | 666 | 98 |
| Interresidue | ||
| Sequential | 669 | 162 |
| Medium range ([I, I+2] to [I, I+5]) | 958 | 195 |
| Long range | 450 | 1 |
| Total | 2,743 | 456 |
| No. of hydrogen bonds | 71 | 0 |
| No. of dihedral angle restraints | 34 | 20 |
| Root mean square deviations from the mean (Å) | ||
| Overall structure, backbone | 0.78a | 0.65b |
| Overall structure, heavy atoms | 1.35a | 1.21b |
| Helix region, backbone | 0.49c | 0.38d |
| Helix region, heavy atoms | 0.93c | 1.03d |
| No. of residues in Ramachandran plot (%) | ||
| Most favorable regions | 81 | 68 |
| Additionally allowed region | 17 | 32 |
| Generously allowed regions | 2 | 0 |
| Disallowed regions | 0 | 0 |
Residues 922 to 1050.
Residues 922 to 944, 948 to 975, 981 to 1008, and 1013 to 1049.
Residues 141 to 160.
Residues 142 to 195.
The FAT domain has four α-helices and comprises an elongated “right-turn” helical bundle (Fig. 1C and D). Its topology is square, as defined by Harris and coworkers (18). In antiparallel helix packing, all four helices (H1 to H4) are connected by short “underhand” loops. Apart from helix H4, all helices are relatively straight. The lengths of the four helices differ: H1 and H2 are the shortest, and H4 is the longest and contains a kink in the middle (around Leu1035). Like typical functional domains (30), the FAT domain has N and C termini that are near one another.
Almost all the residues sequestered inside the bundle are hydrophobic, and the helix bundle is therefore mainly stabilized by the hydrophobic core. We observed extensive interhelical interactions among these hydrophobic residues, not only between neighbor helices but also between cross-diagonal helices (H1 to H3 and H2 to H4). However, our examination of the side chains of charged residues revealed that few interhelical salt bridges are present. Most residues which are exposed to solvent are hydrophilic. However, there are two leucines (Leu960 and Leu966) on the surface of helix H2 that form a perfect unpaired leucine zipper. The hydrophobic residues that are involved in these core contacts are also present in a comparable region of PYK2 (Fig. 1A), and this suggests that the structure of the C-terminal domain of PYK2 is similar to that of the FAT domain.
Each of the three underhand loops that connect the four helices in the FAT domain is short and highly structured. Loop 1, which connects helices H1 and H2, is proline rich. Helix H1 ends with Pro945, helix H2 starts with Pro948, and the intervening tight turn contains Ala946 and Pro947. Loop 2, which connects helices H2 and H3, is also proline rich. Helix H2 ends at Pro974, and Pro977 sits in the middle of the loop. Both Pro947 and Pro977 exist in the trans conformation. Pro947 is exposed to the solvent, whereas Pro977 has a hydrophobic contact with the side chain of Met1046 in H4. Loop 3, which connects helices H3 and H4, contains Thr1011 and Ser1012. However, the most noteworthy feature of this loop is the two tyrosine residues, Tyr1008 and Tyr1017, that sit at the two ends of the loop. The side chains of the two tyrosines are in perfect ring-stacking contact. This contact, which was clearly indicated by the NOE spectra, undoubtedly plays a role in stabilizing the loop.
Although the four-helix bundle is a common structure in proteins, elongated helix bundles, such as that found in the FAT domain, are rare. Searching the Protein Data Bank with the DALI program (22), we found that the structure of the FAT domain resembles that of exchangeable apolipoproteins (50) and also that of the C-terminal tail domain of the focal adhesion protein vinculin (2). However, apolipoproteins are left-turn helix bundles. Only the tail domain of vinculin has the same topology as that of the FAT domain, which is a right-turn helix bundle. The structures of helices H1 to H4 of the FAT domain are similar to the C-terminal four helices (H2, H3, H4, and H5) of the tail domain of vinculin. The homology to vinculin is particularly interesting; vinculin is also colocalized in the focal adhesions, and its tail domain also binds to paxillin. Like helix H4 of the FAT domain, helix H3 of the tail domain of vinculin contains a kink, and this may be significant because helix H4 of the FAT domain and helix H3 of the tail domain of vinculin interact with paxillin (see Discussion).
Structure of the LD2 motif of paxillin.
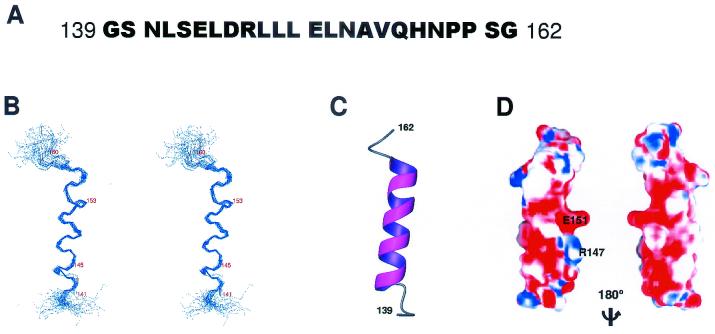
Although the LD2 peptide of paxillin consists of only 24 residues, CD spectra indicated that it has a predominantly α-helical structure when dissolved in aqueous solution at pH 6.5. NMR proton spectra also showed that the proton resonances of the amide groups within the LD2 backbone are well dispersed, which is characteristic of a folded protein. Furthermore, in the NOE spectra (mainly the ROESY spectra) of the peptide, some NOEs between protons of the amide groups within the backbone were observed, which is consistent with the helical structures revealed by the CD spectra.
The solution structure of the LD2 peptide was elucidated by using conventional 2D NMR experiments. All the proton resonances were assigned, and the structure was based on approximately 476 NMR-derived constraints. These included 456 structurally meaningful NOE distance constraints (mostly from ROESY spectra) and 20 backbone dihedral angle restraints attained from measurements of three-bond scalar coupling constants. The relatively high number of constraints per residues permitted characterization of a high-resolution structure. For 20 selected NMR conformers that had the lowest target function, the global root mean square deviations for the folded region (residues 3 to 23) were approximately 0.6 Å. The backbone conformations of all the final structures are in the most-favored and additionally allowed regions of φ-ψ space (Table 1).
The collection of the 20 best-calculated structures of the paxillin LD2 peptide is shown in Fig. 2. We observed a well-ordered, four-turn helix that is relatively straight. The N terminus of the helix starts at Leu142 and is N capped by Asn141. The helix ends at His157 and is C capped by Asn158 and Pro159. This typical capping at both ends of the helix suggests that the capping residues play a significant role in stabilizing the helical structure of the peptide in solution (1).
FIG. 2.
Structure of paxillin LD2 peptide. (A) Amino acid sequence of the LD2 peptide of paxillin. (B) Stereo view of the peptide backbone (N, Cα, C') of 20 superimposed solution structures of the LD2 peptide with lowest target functions. (C) Ribbon diagram of the structure of the LD2 peptide. (D) The molecular surface of the LD2 peptide. Red represents negative electrostatic potential, blue represents positive, and white represents natural. The image was generated by the GRASP program (33).
An examination of the electrostatic potential of the surface of the LD2 peptide in solution revealed that Arg147 and Glu151 form a strong electric dipole (Fig. 2). This dipolar nature of the LD2 peptide suggests that the surface of LD2 may be involved in protein-protein recognition through an electrostatic interaction (42).
Interaction between the FAT domain and the LD2 motif of paxillin.
The chemical-shift pertubation that occurred when the LD2 peptide was added to the FAT domain clearly revealed the interaction between the two. By comparing the series of 1H-15N correlated NMR spectra of the 15N-labeled FAT domain in the presence of different concentrations of unlabeled LD2 peptide, we found that many but not all resonances of the backbone amide groups differed from those that were observed in the absence of LD2 binding. These differences clearly demonstrate that the LD2 peptide interacts with a defined region of the FAT domain. On the basis of chemical-shift values that we obtained during the titration of the LD2 peptide used, we found that the binding affinity was approximately 10 μM. This binding affinity was confirmed by measurements from isothermal titration calorimetry experiments. Furthermore, gel filtration chromatography indicated that the self-aggregation of the FAT domain was completely abolished when the FAT domain was bound to the LD2 peptide. This result has been verified by light-scattering measurement and diffusion measurement by NMR spectroscopy.
Examining a series of 1H-15N correlation spectra of the FAT domain in the presence of different concentrations of LD2 peptide, we found it difficult to define the interaction surface between the FAT domain and the LD2 peptide by solely analyzing the chemical-shift perturbations. All four helices of the FAT domain contained residues whose amide-proton resonances changed upon binding of the LD2 peptide. Clearly, some of these changes in resonances are due to the ligand-binding effect and to the disruption of FAT domain aggregation and the possible conformational changes associated with it. Because of the lack of feasible methods, it was not possible to distinguish the sources that caused the chemical-shift perturbations. Moreover, because the binding of the LD2 peptide to the FAT domain is relatively weak, the exchange rate between FAT domain-bound LD2 peptide and free LD2 peptide is high. Therefore, the typical isotope edited-filtered NMR experiments were unable to yield intermolecular NOEs between the FAT domain and the LD2 peptide.
To overcome this problem, we used an approach that involves spin labeling. Paramagnetic relaxation caused by site-directed spin labeling can be employed as a powerful tool for determining the solution structure of a molecule because the paramagnetic center can enhance the spin lattice relaxation of magnetic nuclei in a distance-dependent manner (3, 13). Oxidized nitroxide has been commonly used as such a paramagnetic center, and previous reports showed that spin labeling can be achieved by the MTSSL method, which covalently links nitroxide to a solvent-exposed cysteine residue through methanethiosulfonate (26). Using a similar method, we performed site-directed labeling of the LD2 peptide.
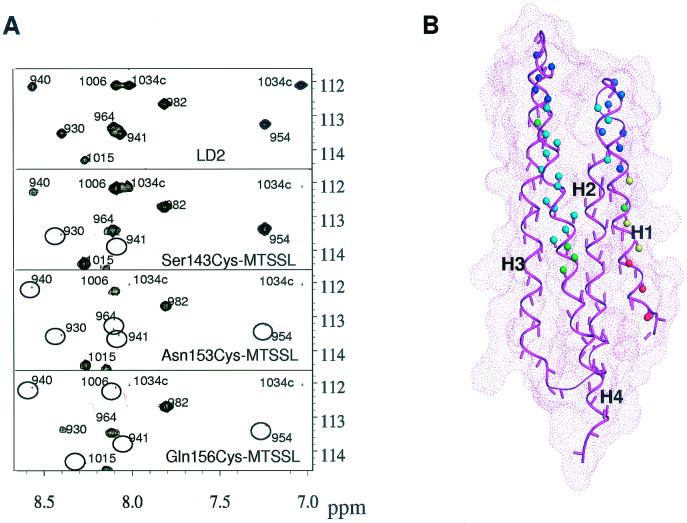
We chemically synthesized four LD2 peptides that each contained a cysteine mutation, Ser143Cys, Arg147Cys, Asn153Cys, and Gln156Cys. These peptides were then subjected to MTSSL (26). In our initial screening, we found that three of the four spin-labeled peptides bound to the FAT domain with a binding affinity similar to that of the wild-type peptide. Only the spin-labeled Arg147Cys peptide, which we designated Arg147Cys-MTSSL, bound to the FAT domain very weakly (data not shown). We compared the 1H-15N correlation spectra of the 15N-labeled FAT domain bound to the LD2 peptide with that of 15N-labeled FAT domain bound to the three spin-labeled LD2 peptides. Our comparison showed that the positions of the peaks of most amide protons were unchanged, and this indicates that spin labeling does not interfere in the binding of the LD2 peptide to the FAT domain. However, the paramagnetic effects on the relaxation resonances of amide protons within the backbone were clearly observed. The peak intensities of those residues in the proximity of spin labeling were markedly reduced (Fig. 3) by the relaxation enhancement and line broadening.
FIG. 3.
Interaction between the FAT domain and LD2 peptide. (A) H-15N correlation maps of 15N-labeled FAT domain bound to LD2 peptides. The top panel shows a partial spectrum of the FAT domain bound to LD2 peptide, and the lower three panels show the same region in the spectrum of which the FAT domain is bound to one of the spin-labeled LD2 peptides. The missing peaks due to the paramagnetic relaxation enhancement are denoted by circles. (B) The amide protons of the FAT domain for which the resonance peaks vanished in the complex with spin-labeled LD2 from the 1H-15N correlation spectra are indicated by small balls in the structure of the FAT domain. The effects of Ser143Cys-MTSSL, Asn153Cys-MTSSL, and Gln156Cys-MTSSL are represented by red, blue, and green balls, respectively, with the structure of the FAT domain. Yellow (red + blue) represents the amide protons that are affected by both Ser143Cys-MTSSL and Asn153Cys-MTSSL, and cyan (blue + green) is for both Asn153Cys-MTSSL and Gln156Cys-MTSSL.
Structure of the complex of the FAT domain and the LD2 motif.
Analysis of the paramagnetic effects of site-directed spin labeling allowed us to identify the binding sites of the LD2 peptide on the surface of the FAT domain. Resonances belonging to amide protons that were within 10 Å of the paramagnetic center were usually broadened beyond detection at a reasonable signal-to-noise ratio, which agrees with previously documented results (13). These “lost peaks” in the 1H-15N correlation spectra of the FAT indicate that the surface of the FAT domain to which LD2 peptide binds spans two helices, H1 and H4. When the spin-labeled peptide Ser143Cys-MTSSL bound to the FAT domain, the lost peaks were probably those representing the residues located in the first helix H1 of the FAT domain. When Asn153Cys-MTSSL and Gln156Cys-MTSSL bound to the FAT domain, most of the residues that were affected by spin labeling were located in helix H4.
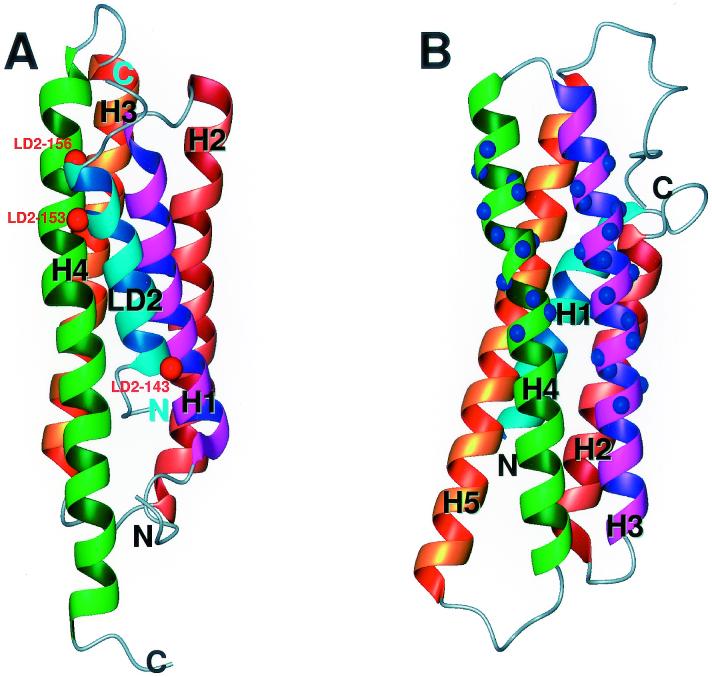
Using the distance information derived from spin-labeling experiments and assuming that residues Ser143, Asn153, and Gln156 in the FAT-bound LD2 peptide are exposed to solvent, we constructed a model of the complex of the FAT domain bound to the LD2 peptide (Fig. 4A). In the model, the LD2 peptide contacts the FAT domain through charge-charge and hydrophobic interactions, and the chief residues of the LD2 peptide that are involved in the interaction are Arg147, Leu148, Glu151, and Leu152. Another noticeable feature of the complex is that the bound LD2 peptide holds helices H1 and H4 together and appears to further stabilize the globular structure of the FAT domain. Indeed, when the LD2 peptide was titrated into the solution of the FAT domain, we observed small chemical-shift perturbations throughout all four helices of the FAT domain. This result indicates that, upon binding of the LD2 peptide, all four helices of the FAT domain undergo some degree of conformational change.
FIG. 4.
Structure comparison of LD2 bound to the FAT domain with the tail domain of vinculin. (A) Ribbon diagram of the structure of the FAT domain-LD2 peptide (in cyan) complex. The red balls on the ribbon of LD2 peptide represent the positions of site-directed spin labels. (B) Ribbon diagram of the structure of the tail domain of vinculin. The blue balls on the ribbon represent a paxillin-binding site (46).
DISCUSSION
Earlier mutagenesis studies showed that the LD2 motif of paxillin interacts with FAK (5), and a detailed analysis identified the key residues in the FAT domain that are involved in binding to paxillin (8, 46). The solution structure of the FAT domain is fully consistent with the findings of these mutagenesis studies and provides a comprehensive view of the interaction at the atomic level. The binding site of LD2 intersects helices H1 and H4 on the surface of the FAT domain. Although the two previously identified binding sites are separated in sequence, in the 3D structure the sites are so close together that they form one binding patch. It had been previously determined by biochemical studies that the sequences of two separate paxillin-binding sites in the FAT domain are similar to that of a contiguous stretch in the tail domain of a focal adhesion protein, vinculin, which also binds to paxillin (46). Comparison of the structures of the two proteins revealed that the paxillin-binding sites are very similar and encompass two structurally adjacent helices. However, the prospective paxillin-binding site of vinculin is formed by two contiguous helices, H3 and H4, whereas the paxillin-binding site of the FAT domain, involving the helices H1 and H4, is not sequentially connected. This result explains why all the residues in the paxillin-binding site of vinculin are within a consecutive stretch of sequence (46).
A previous report had also shown that Tyr926 (Tyr925 in human and mouse sequences) in the FAT domain can be phosphorylated after FAK has localized to the focal adhesion (41). Although the solution structure of the FAT domain showed that Tyr926 is partially exposed to the solvent, Tyr926 is partially covered by the bound LD2 peptide in the structure of the FAT domain:LD2 peptide complex. Therefore, for Tyr926 to be accessible to the kinase, the interaction between the LD2 peptide and the FAT domain has to be disrupted. The weak interaction between the FAT domain and the LD2 peptide ensures an easy breakdown of such interaction so that Tyr926 can be phosphorylated.
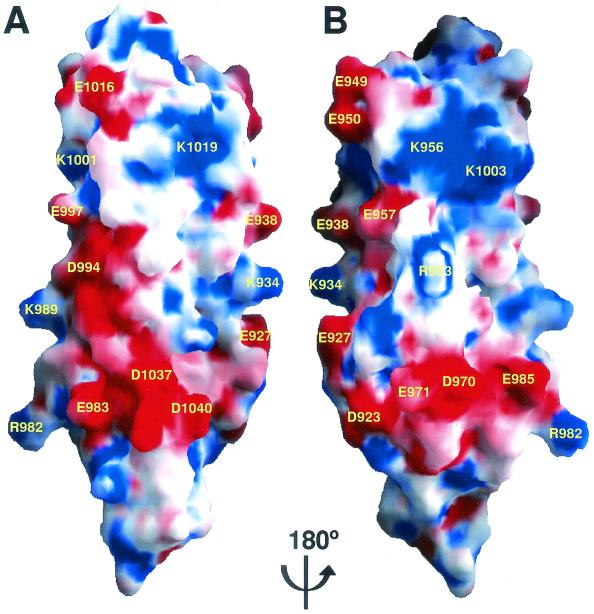
One feature of the surface of the FAT domain is the many unpaired, charged residues that are exposed to solvent and which form several well-defined electrostatic polar patches (Fig. 5A and B). In addition, helix H2 contains an unpaired leucine zipper on its surface. Our NMR data indicate that the two solvent-exposed leucine residues are involved in the aggregation of the FAT domain at high concentrations (data not shown). Although the biological relevance of such polymerization in vivo is not clear, the two leucine residues may provide an additional site for protein-protein interaction. Thus, it is plausible that through electrostatic and hydrophobic interactions, the FAT domain interacts with proteins in addition to paxillin and talin in the focal adhesion complex.
FIG. 5.
Surface of the FAT domain. (A and B) Two views of molecular surface of the FAT domain generated by GRASP (33). Red represents negative electrostatic potential, blue represents positive, and white represents natural.
Another feature of the FAT domain is that hydrophobic interaction is the chief force that holds the four helices together. Few salt bridges between the helices are present. Therefore, interrupting the core hydrophobic interactions by replacing hydrophobic residues with hydrophilic residues will probably disrupt the 3D structure completely. Indeed, the structure of the dominant-negative mutant of chicken FAK is substantially altered by the mutation Leu1035Ser (Leu1034 in human and mouse sequences) (19), and this dominant-negative protein cannot localize to focal adhesions. The 1H-15N-correlated NMR spectrum of the 15N-labeled Leu1035Ser mutant is very different from the spectrum of the wild-type FAT domain (data not shown), and the Leu1035Ser mutant appears to have a completely different structure. In the structure of the FAT domain, Leu1035 is located in helix H4, and the side chain engages in intensive hydrophobic interactions with side chains of other core hydrophobic residues, such as Ala987 in helix H3 and Val869 in helix H2. Furthermore, within helix H4 is a kink that is located to the right of Leu1035. We hypothesize that the hydrophobic core will collapse and that the structure of the FAT domain will be completely altered if Leu1035 is replaced by a hydrophilic residue, e.g., serine. In addition, we speculate that another previously identified dominant-negative mutant, ΔC14 (7), also has a folding pattern that differs from that of wild-type FAT. This mutant, which lacks the last 14 residues and is unable to bind to focal adhesions, inhibits the activity of endogenous FAK. Most of the last 14 residues of the FAT domain reside in helix H4, and many of the residues, such as Ala1042, Leu1044, and Met1046, participate in interhelical hydrophobic interactions. Therefore, deletion of these residues would reduce the stability of the hydrophobic core and would probably disturb the structure of the FAT domain.
Because the four-helix bundle of the FAT domain is mainly maintained by hydrophobic interactions, the FAT domain may exist in an “open” form under certain physiologic conditions. Indeed, the two proteins whose structures are most similar to that of the FAT domain, exchangeable apolipoproteins and the tail domain of vinculin, are present in an open form under certain conditions (2, 50). In the lipid-bound state, exchangeable apolipoproteins are in an open conformation, and each helix stably interacts with lipids (32). It has also been proposed that, when vinculin is located near the membrane, the helix bundle of the tail domain of vinculin is opened and that the hydrophobic sites of the helix interact with lipids (2). Although there is no direct evidence, the results of our structural studies and earlier findings of other investigators support our hypothesis that the FAT domain is in an open form in certain biological surroundings. For example, our solution structure of the FAT domain strongly suggests that interaction between the FAT domain and SH2 domain of Grb2 requires the FAT domain to be in an open conformation.
The phosphorylation of Tyr926 results in the creation of a consensus Grb2 SH2 recognition motif, pYNQV. However, the solution structure of the FAT domain showed that, although Tyr926 is partially accessible to the solvent, this residue and its neighboring residues are located in a well-folded region of helix H1. The structures of the Grb2 SH2 domain bound to two phosphopeptides, both of which have a pYXQV motif, have been elucidated (34, 37). According to those structural studies, the phosphotyrosine residues and its neighboring residues, including Glu927, Asn928 and Val929, have to be pulled out from the helical structure of the FAT domain for phosphorylated Tyr926 to bind to the SH2 domain of Grb2. In the solution structure of the FAT domain, both Asn928 and Val929 participate in the core hydrophobic interaction that stabilizes the four-helix bundle. The binding of the SH2 domain of Grb2 to phosphorylated Tyr926 will certainly break these hydrophobic contacts. As a result, the disruption of these contacts probably causes the helix bundle to collapse. In addition, our solution structure of the FAT domain indicates that it is not possible for the SH2 domain of Grb2 and the LD2 peptide of paxillin to simultaneously bind to the FAT domain, because the SH2-binding motif overlaps the binding site of the LD2 peptide. Therefore, the binding of the SH2 domain of Grb2 to phosphorylated Tyr926 of FAK is likely to cause the release of bound paxillin from FAK.
In a migrating cell, the dynamics of focal adhesions promote cell migration by alternately forming and disassembling. The roles of FAK in the dynamics of focal adhesions (38) remain unclear. At a certain point, integrin-activated FAK needs to be deactivated. The dissociation of FAK from the focal adhesion complex may be the first step in terminating FAK signaling, and it is possible that certain FAK phosphorylation events initiate such action (10). Indeed, it has been shown that during mitosis, the interaction between FAK and an adapter, p130Cas, is mediated by FAK serine phosphorylation (31). In this work, our solution structure strongly suggests that it is not possible for phosphorylated FAK to bind to the SH2 domain of Grb2 while it is still associated with the focal adhesion complex. This notion is supported by a recent study. Volberg et al. (49) reported that, in the Src−/− cells, FAK could not be phosphorylated. However, the FAT of FAK was not affected in the Src−/− cells. Furthermore, they showed that FAK-containing adhesions were considerably larger and more intense in the Src−/− cells. Tyr926 is a major Src phosphorylation site (41). According to the model that we proposed, disrupting the phosphorylation at Tyr926 would stop (or at least slow down) the FAK-involved focal adhesion turnover. This matches perfectly with the observation made in the Src−/− cells. Indeed, it has been recognized that multiple tyrosine residues in FAK play important and in many cases different roles in FAK function (45). No doubt, an exact structural and biochemical definition of the manner in which FAK acts in concert with other molecules to achieve the turnover of focal adhesion will be a fascinating target of future study.
Acknowledgments
This work was supported by the American Lebanese Syrian Associated Charities and a grant-in-aid from the American Heart Association (0051073B).
We thank J. Thomas Parsons for kindly providing the chicken FRNK cDNA. We thank J. Thomas Parsons, Jun-Lin Guan, and Yi Zheng for critical reading and valuable discussions of the manuscript and Julie Cay Jones for editing the manuscript. We thank Weixing Zhang for technical support. We are particularly grateful to Stephen White for his help, advice, and encouragement during the course of the studies.
REFERENCES
- 1.Aurora, R., and G. D. Rose. 1998. Helix capping. Protein Sci. 7:21-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakolitsa, C., J. M. de Pereda, C. R. Bagshaw, D. R. Critchley, and R. C. Liddington. 1999. Crystal structure of the vinculin tail suggests a pathway for activation. Cell 99:603-613. [DOI] [PubMed] [Google Scholar]
- 3.Bertini, I., A. Donaire, C. Luchinat, and A. Rosato. 1997. Paramagnetic relaxation as a tool for solution structure determination: Clostridium pasteurianum ferredoxin as an example. Proteins 29:348-358. [PubMed] [Google Scholar]
- 4.Borowsky, M. L., and R. O. Hynes. 1998. Layilin, a novel talin-binding transmembrane protein homologous with C-type lectins, is localized in membrane ruffles. J. Cell Biol. 143:429-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, M. C., J. A. Perrotta, and C. E. Turner. 1996. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J. Cell Biol. 135:1109-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cary, L. A., and J. L. Guan. 1999. Focal adhesion kinase in integrin-mediated signaling. Front. Biosci. 4:D102-D113. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H. C., P. A. Appeddu, J. T. Parsons, J. D. Hildebrand, M. D. Schaller, and J. L. Guan. 1995. Interaction of focal adhesion kinase with cytoskeletal protein talin. J. Biol. Chem. 270:16995-16999. [DOI] [PubMed] [Google Scholar]
- 8.Cooley, M. A., J. M. Broome, C. Ohngemach, L. H. Romer, and M. D. Schaller. 2000. Paxillin binding is not the sole determinant of focal adhesion localization or dominant-negative activity of focal adhesion kinase/focal adhesion kinase-related nonkinase. Mol. Biol. Cell 11:3247-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 10.Fincham, V. J., and M. C. Frame. 1998. The catalytic activity of Src is dispensable for translocation to focal adhesions but controls the turnover of these structures during cell motility. EMBO J. 17:81-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedl, P., and E. B. Brocker. 2000. The biology of cell locomotion within three-dimensional extracellular matrix. Cell. Mol. Life Sci. 57:41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedl, P., K. S. Zanker, and E. B. Brocker. 1998. Cell migration strategies in 3-D extracellular matrix: differences in morphology, cell matrix interactions, and integrin function. Microsc. Res. Tech. 43:369-378. [DOI] [PubMed] [Google Scholar]
- 13.Gaponenko, V., J. W. Howarth, L. Columbus, G. Gasmi-Seabrook, J. Yuan, W. L. Hubbell, and P. R. Rosevear. 2000. Protein global fold determination using site-directed spin and isotope labeling. Protein Sci. 9:302-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giancotti, F. G. 2000. Complexity and specificity of integrin signalling. Nat. Cell Biol. 2:E13-E14. [DOI] [PubMed] [Google Scholar]
- 15.Giancotti, F. G., and E. Ruoslahti. 1999. Integrin signaling. Science 285:1028-1032. [DOI] [PubMed] [Google Scholar]
- 16.Guntert, P., W. Braun, and K. Wuthrich. 1991. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J. Mol. Biol. 217:517-530. [DOI] [PubMed] [Google Scholar]
- 17.Guntert, P., C. Mumenthaler, and K. Wuthrich. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273:283-298. [DOI] [PubMed] [Google Scholar]
- 18.Harris, N. L., S. R. Presnell, and F. E. Cohen. 1994. Four helix bundle diversity in globular proteins. J. Mol. Biol. 236:1356-1368. [DOI] [PubMed] [Google Scholar]
- 19.Hauck, C. R., C. K. Klingbeil, and D. D. Schlaepfer. 2000. Focal adhesion kinase functions as a receptor-proximal signaling component required for directed cell migration. Immunol. Res. 21:293-303. [DOI] [PubMed] [Google Scholar]
- 20.Hildebrand, J. D., M. D. Schaller, and J. T. Parsons. 1993. Identification of sequences required for the efficient localization of the focal adhesion kinase, pp125FAK, to cellular focal adhesions. J. Cell Biol. 123:993-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hildebrand, J. D., M. D. Schaller, and J. T. Parsons. 1995. Paxillin, a tyrosine phosphorylated focal adhesion-associated protein binds to the carboxyl terminal domain of focal adhesion kinase. Mol. Biol. Cell 6:637-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 23.Ilic, D., E. A. Almeida, D. D. Schlaepfer, P. Dazin, S. Aizawa, and C. H. Damsky. 1998. Extracellular matrix survival signals transduced by focal adhesion kinase suppress p53-mediated apoptosis. J. Cell Biol. 143:547-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ilic, D., Y. Furuta, S. Kanazawa, N. Takeda, K. Sobue, N. Nakatsuji, S. Nomura, J. Fujimoto, M. Okada, and T. Yamamoto. 1995. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature 377:539-544. [DOI] [PubMed] [Google Scholar]
- 25.Ilic, D., O. Genbacev, F. Jin, E. Caceres, E. A. Almeida, V. Bellingard-Dubouchaud, E. M. Schaefer, C. H. Damsky, and S. J. Fisher. 2001. Plasma membrane-associated pY397FAK is a marker of cytotrophoblast invasion in vivo and in vitro. Am. J. Pathol. 159:93-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson, J. S., B. R. Gibney, F. Rabanal, K. S. Reddy, and P. L. Dutton. 1998. A designed cavity in the hydrophobic core of a four-alpha-helix bundle improves volatile anesthetic binding affinity. Biochemistry 37:1421-1429. [DOI] [PubMed] [Google Scholar]
- 27.Jones, G., J. Machado, Jr., and A. Merlo. 2001. Loss of focal adhesion kinase (FAK) inhibits epidermal growth factor receptor-dependent migration and induces aggregation of nh(2)-terminal FAK in the nuclei of apoptotic glioblastoma cells. Cancer Res. 61:4978-4981. [PubMed] [Google Scholar]
- 28.Jones, R. J., V. G. Brunton, and M. C. Frame. 2000. Adhesion-linked kinases in cancer; emphasis on src, focal adhesion kinase and PI 3-kinase. Eur. J. Cancer 36:1595-1606. [DOI] [PubMed] [Google Scholar]
- 29.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:29-32, 51-55. [DOI] [PubMed]
- 30.Kuriyan, J., and D. Cowburn. 1997. Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct. 26:259-288. [DOI] [PubMed] [Google Scholar]
- 31.Ma, A., A. Richardson, E. M. Schaefer, and J. T. Parsons. 2001. Serine phosphorylation of focal adhesion kinase in interphase and mitosis: a possible role in modulating binding to p130(Cas). Mol. Biol. Cell 12:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayanaswami, V., and R. O. Ryan. 2000. Molecular basis of exchangeable apolipoprotein function. Biochim. Biophys. Acta 1483:15-36. [DOI] [PubMed] [Google Scholar]
- 33.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 34.Ogura, K., S. Tsuchiya, H. Terasawa, S. Yuzawa, H. Hatanaka, V. Mandiyan, J. Schlessinger, and F. Inagaki. 1999. Solution structure of the SH2 domain of Grb2 complexed with the Shc-derived phosphotyrosine-containing peptide. J. Mol. Biol. 289:439-445. [DOI] [PubMed] [Google Scholar]
- 35.Ohene-Abuakwa, Y., and M. Pignatelli. 2000. Adhesion molecules in cancer biology. Adv. Exp. Med. Biol. 465:115-126. [DOI] [PubMed] [Google Scholar]
- 36.Parsons, J. T., K. H. Martin, J. K. Slack, J. M. Taylor, and S. A. Weed. 2000. Focal adhesion kinase: a regulator of focal adhesion dynamics and cell movement. Oncogene 19:5606-5613. [DOI] [PubMed] [Google Scholar]
- 37.Rahuel, J., B. Gay, D. Erdmann, A. Strauss, C. Garcia-Echeverria, P. Furet, G. Caravatti, H. Fretz, J. Schoepfer, and M. G. Grutter. 1996. Structural basis for specificity of Grb2-SH2 revealed by a novel ligand binding mode. Nat. Struct. Biol. 3:586-589. [DOI] [PubMed] [Google Scholar]
- 38.Sastry, S. K., and K. Burridge. 2000. Focal adhesions: a nexus for intracellular signaling and cytoskeletal dynamics. Exp. Cell Res. 261:25-36. [DOI] [PubMed] [Google Scholar]
- 39.Schlaepfer, D. D., C. R. Hauck, and D. J. Sieg. 1999. Signaling through focal adhesion kinase. Prog. Biophys. Mol. Biol. 71:435-478. [DOI] [PubMed] [Google Scholar]
- 40.Schlaepfer, D. D., and T. Hunter. 1998. Integrin signalling and tyrosine phosphorylation: just the FAKs? Trends Cell Biol. 8:151-157. [DOI] [PubMed] [Google Scholar]
- 41.Schlaepfer, D. D., K. C. Jones, and T. Hunter. 1998. Multiple Grb2-mediated integrin-stimulated signaling pathways to ERK2/mitogen-activated protein kinase: summation of both c-Src- and focal adhesion kinase-initiated tyrosine phosphorylation events. Mol. Cell. Biol. 18:2571-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheinerman, F. B., R. Norel, and B. Honig. 2000. Electrostatic aspects of protein-protein interactions. Curr. Opin. Struct. Biol. 10:153-159. [DOI] [PubMed] [Google Scholar]
- 43.Sieg, D. J., C. R. Hauck, D. Ilic, C. K. Klingbeil, E. Schaefer, C. H. Damsky, and D. D. Schlaepfer. 2000. FAK integrates growth-factor and integrin signals to promote cell migration. Nat. Cell Biol. 2:249-256. [DOI] [PubMed] [Google Scholar]
- 44.Simon-Assmann, P., M. Kedinger, A. De Arcangelis, V. Rousseau, and P. Simo. 1995. Extracellular matrix components in intestinal development. Experientia 51:883-900. [DOI] [PubMed] [Google Scholar]
- 45.Slack, J. K., R. B. Adams, J. D. Rovin, E. A. Bissonette, C. E. Stoker, and J. T. Parsons. 2001. Alterations in the focal adhesion kinase/Src signal transduction pathway correlate with increased migratory capacity of prostate carcinoma cells. Oncogene 20:1152-1163. [DOI] [PubMed] [Google Scholar]
- 46.Tachibana, K., T. Sato, N. D'Avirro, and C. Morimoto. 1995. Direct association of pp125FAK with paxillin, the focal adhesion-targeting mechanism of pp125FAK. J. Exp. Med. 182:1089-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, J. M., C. P. Mack, K. Nolan, C. P. Regan, G. K. Owens, and J. T. Parsons. 2001. Selective expression of an endogenous inhibitor of FAK regulates proliferation and migration of vascular smooth muscle cells. Mol. Cell. Biol. 21:1565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Troyanovsky, S. M. 1999. Mechanism of cell-cell adhesion complex assembly. Curr. Opin. Cell Biol. 11:561-566. [DOI] [PubMed] [Google Scholar]
- 49.Volberg, T., L. Romer, E. Zamir, and B. Geiger. 2001. pp60(c-src) and related tyrosine kinases: a role in the assembly and reorganization of matrix adhesions. J. Cell Sci. 114:2279-2289. [DOI] [PubMed] [Google Scholar]
- 50.Wilson, C., M. R. Wardell, K. H. Weisgraber, R. W. Mahley, and D. A. Agard. 1991. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science 252:1817-1822. [DOI] [PubMed] [Google Scholar]
- 51.Wishart, D. S., and B. D. Sykes. 1994. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4:171-180. [DOI] [PubMed] [Google Scholar]
- 52.Xu, L. H., X. Yang, C. A. Bradham, D. A. Brenner, A. S. Baldwin, Jr., R. J. Craven, and W. G. Cance. 2000. The focal adhesion kinase suppresses transformation-associated, anchorage-independent apoptosis in human breast cancer cells. Involvement of death receptor-related signaling pathways. J. Biol. Chem. 275:30597-30604. [DOI] [PubMed] [Google Scholar]
- 53.Xu, L. H., X. Yang, R. J. Craven, and W. G. Cance. 1998. The COOH-terminal domain of the focal adhesion kinase induces loss of adhesion and cell death in human tumor cells. Cell Growth Differ. 9:999-1005. [PubMed] [Google Scholar]
- 54.Zachary, I. 1997. Focal adhesion kinase. Int. J. Biochem. Cell Biol. 29:929-934. [DOI] [PubMed] [Google Scholar]
- 55.Zheng, C., Z. Xing, Z. C. Bian, C. Guo, A. Akbay, L. Warner, and J. L. Guan. 1998. Differential regulation of Pyk2 and focal adhesion kinase (FAK). The C-terminal domain of FAK confers response to cell adhesion. J. Biol. Chem. 273:2384-2389. [DOI] [PubMed] [Google Scholar]