Abstract
Yrr1p is a recently described Zn2Cys6 transcription factor involved in the pleiotropic drug resistance (PDR) phenomenon. It is controlled in a Pdr1p-dependent manner and is autoregulated. We describe here a new genome-wide approach to characterization of the set of genes directly regulated by Yrr1p. We found that the time-course production of an artificial chimera protein containing the DNA-binding domain of Yrr1p activated the 15 genes that are also up-regulated by a gain-of-function mutant of Yrr1p. Gel mobility shift assays showed that the promoters of the genes AZR1, FLR1, SNG1, YLL056C, YLR346C, and YPL088W interacted with Yrr1p. The putative consensus Yrr1p binding site deduced from these experiments, (T/A)CCG(C/T)(G/T)(G/T)(A/T)(A/T), is strikingly similar to the PDR element binding site sequence recognized by Pdr1p and Pdr3p. The minor differences between these sequences are consistent with Yrr1p and Pdr1p and Pdr3p having different sets of target genes. According to these data, some target genes are directly regulated by Pdr1p and Pdr3p or by Yrr1p, whereas some genes are indirectly regulated by the activation of Yrr1p. Some genes, such as YOR1, SNQ2, and FLR1, are clearly directly controlled by both classes of transcription factor, suggesting an important role for the corresponding membrane proteins.
Multidrug resistance (MDR) in tumor cells, resulting in the failure of chemotherapy, remains a major obstacle to the successful treatment of cancer. This phenomenon has been shown to involve mechanisms such as increases in exocytosis, decreases in intracellular pH, cell detoxification, or changes in membrane composition, with the overproduction of molecular pumps (for review, see reference 2). In the yeast Saccharomyces cerevisiae, a complex network of genes, the pleiotropic drug resistance (PDR) network, has developed to mediate MDR. This response is controlled by two major transcription factors, Pdr1p and Pdr3p, which activate the production of various targets, such as the ATP-binding cassette (ABC) transporters encoded by PDR5, PDR10, PDR15, SNQ2, and YOR1; the major facilitator superfamily (MFS) permeases encoded by HXT9 and HXT11; and the lipid metabolism proteins encoded by IPT1 and PDR16 (for review, see reference 15). Other proteins, such as Pdr13p (13) and Yrr1p (28), have also been described as being involved in PDR network regulation and may act directly on the PDR genes themselves or indirectly in general drug resistance.
YRR1 is a member of the diverse Zn2Cys6 zinc finger transcription factor family. It was isolated by reveromycin A resistance screening (5). YRR1 deletion leads to hypersensitivity to 4-nitroquinoline-N-oxide (4-NQO) and oligomycin but not to cycloheximide (5). The SNQ2 gene is involved in YRR1-mediated 4-NQO resistance, as suggested by observations that SNQ2 confers resistance to 4-NQO (6, 23). Promoter deletions have also demonstrated that Yrr1p interacts with the promoter region of SNQ2 (4). Furthermore, chromatin-immunoprecipitation experiments with hemagglutinin (HA)-tagged Yrr1p have shown direct binding to the promoter region of YOR1, which encodes a protein involved in oligomycin resistance (28). Thus, YRR1 is involved in complex PDR network regulation, directly activating SNQ2 and YOR1, both of which are common targets of Pdr1p, Pdr3p, and another regulator of MDR, Yap1p. Finally, as Pdr1p activates Yrr1p (28), it seems highly likely that YRR1 plays a major role in the PDR regulation network.
We used a genomic gain-of-function allele of YRR1 in combination with an approach based on chimeras developed by Devaux et al. (9) to determine the specific regulation network of this gene. We carried out a time-course analysis of the expression of the genes that we found to be regulated by Yrr1p. An electrophoretic mobility shift assay (EMSA) demonstrated specific interactions between parts of the promoter regions of these genes and Yrr1p. Finally, we have identified the region of DNA binding to Yrr1p and propose a consensus sequence for Yrr1p binding. These new data add to existing knowledge, enabling us to define a regulation network involving Yrr1p and the major regulators of the PDR phenomenon.
MATERIALS AND METHODS
Strains.
For the Yrr1p gain-of-function analyses, the S. cerevisiae strains used were isogenic to SEY6210 (MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ190 lys2-801 suc2-Δ9 Mel−). The gain-of-function allele has been described elsewhere (28) and consists of an insertion of three HA tags in the C-terminal region of Yrr1p, after amino acid 730 (YRR1::3X HA-730). The yeast strains transformed with a construct encoding a fusion of the YRR1 binding domain and the GAL4 activating domain (called Yrr1*GAD) were from FY1679-28C (Mata ura3-52 trp1Δ63 leu2Δ1 his3Δ200) and W303 (MATa ura3-52 ade2-1 leu2-3,12 his3-11,15 trp1-289) backgrounds. For the FY strain, the YRR1 deletion mutant was provided by EUROPHAN as FMRN012-01A(A) (MATa ura3-52 LEU2 trp1-289 HIS3 yor162D(4, 2433)::KANMX4). For the W303 strain, the YRR1 deletion mutant was obtained using a His6-URA3-His6 cassette (17).
Plasmid construction.
The pCB-GAD plasmid has been described elsewhere (9). We constructed pCB-Yrr1*GAD by inserting into pCB-GAD either the first 534 nucleotides of the YRR1 open reading frame for the short form (Yrr1S*GAD) or the first 579 nucleotides of the YRR1 open reading frame for the long form (Yrr1L*GAD). These sequences were amplified by PCR with high-fidelity Taq polymerase (Roche) and were inserted into the NotI site of pCB-GAD by homologous recombination in the FY or W303 strain. The 5′ nucleotide used for PCR amplification and homologous recombination was GACGTCCCGGACTATGCAAGGCCTGTTCCATCACACGTGAAAAGAAGAAGCGATGCTTTG. The 3′ oligonucleotides used were CTTTTTTGGAGGCTCGGGAATTAATTCCGCTGCATGTCCGGAATGTTTACTTTGTAGGTA for Yrr1S*GAD and CTTTTTTGGAGGCTCGGGAATTAATTCCGCTGCATGTTGCAATTTGGGTTCTCATAGAAG for Yrr1L*GAD.
Phenotype analyses.
Cells were grown in rich medium (1% Bacto-peptone and 1% yeast extract) containing glucose (YPGlu; 2% glucose) or galactose (YPGal; 2% galactose) to an optical density at 600 nm (OD600) of 0.6. Four serial 1:10 dilutions of this stock culture were prepared and plated on rich medium containing either glucose or galactose with 2% bacterial agar and various drug dilutions (see Fig. 1 legend for details). For oligomycin resistance analyses, cells were plated on rich respiration medium (2% Bacto-peptone, 1% yeast extract, 2% glycerol, and 2% ethanol). The plates were photographed 3 to 5 days after plating.
FIG. 1.
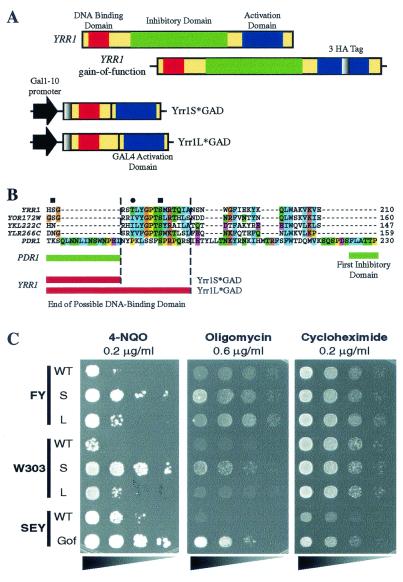
Experimental strategy and physiological validation for construction of a chimeric transcription factor consisting of the DNA-binding domain of Yrr1p and the activating domain of Gal4p. (A) Schematic representation of the various YRR1 constructs used in this study. At the top, the YRR1 gene is shown with its three main regions (DNA-binding domain, inhibitory region, and activating domain). Below is the YRR1 gain-of-function mutant with a 3-HA tag insertion in the activating domain. The two chimeric constructs encoding the N-terminal amino acids of Yrr1p and the activating domain of GAL4 transcription factor are also represented. The short (Yrr1S*GAD) and long (Yrr1L*GAD) chimeras differ by 16 amino acids, as indicated in panel B. (B) Detail of an amino acid alignment including the closest homologs of YRR1 and PDR1 previously used in similar experimental approaches (9). The limit of the YRR1 region chosen for the two constructs is shown for the short (Yrr1S*GAD) and the long (Yrr1L*GAD) forms. The end of the Pdr1p chimera is shown with the beginning of the first inhibitory region for comparison. Using the PhosphoBase online analysis tool (16), we identified three sites in Yrr1p that could be phosphorylated by protein kinase C (square) and protein kinase A (circle). (C) Phenotypes observed for the Yrr1*GAD chimeras. Serial cell dilutions of various strains, from the most concentrated (on the left) to the least concentrated (on the right), were plated after induction with galactose for 20 h in the liquid phase. For the FY and W303 strains, we compared Yrr1S*GAD (S) and Yrr1L*GAD (L) with their corresponding wild-type (WT) strains. We also compared the phenotype of the gain-of-function (Gof) mutant with that of the SEY wild-type strains. Three different drugs were tested: 4-NQO at a concentration of 2 μg/ml, oligomycin at 6 μg/ml, and cycloheximide at 2 μg/ml. The plates were photographed after 72 h for cycloheximide, after 120 h for 4-NQO, and after 144 h for oligomycin. More complete drug resistance analysis is available via the Internet (http://www.biologie.ens.fr/yeast-publi.html).
Microarray experiments.
Microarrays containing all the open reading frames of yeast were obtained from Hitachi Software and MWG-Biotech. For YRR1 gain-of-function activation analyses, we performed six independent microarray experiments. For Yrr1S*GAD in the FY background, the experiment was performed three times. The detailed microarray protocols are available via the Internet (http://www.biologie.ens.fr/yeast-publi.html). The arrays were read with an Axon Genepix 4000A scanner and analyzed with Genepix 3.0 software.
Northern blot analyses.
Northern blot analyses were performed with total RNA extracted from the strains used for the microarray experiments. Cells transformed with Yrr1S*GAD were grown in YPGlu to an OD600 of 0.8 and were then transferred to YPGal for various lengths of time, from 30 min to 16 h. Total RNA was extracted, using the same protocol as for microarray experiments. Northern blotting was performed as previously described (9). Specific PCR products of each of the genes were labeled with the NonaPrimer kit (Quantum-Appligene) and used as probes.
EMSA.
The promoter region of each gene was amplified with the Expand High-Fidelity PCR System (Roche), using 100 ng of genomic DNA and 40 pmol of each oligonucleotide. PCR products were purified on a NucleoSpin column (Machery-Nagel). DNA (10 pmol) was labeled with T4 polynucleotide kinase (BioLabs) and [γ-32P]ATP (Amersham). Unincorporated nucleotides were removed on ProbeQuant G-50 Micro Columns (Amersham). The oligonucleotide pairs used to amplify 200 bp of the gene promoters are described in Table 1.
TABLE 1.
Oligonucleotides used for EMSAsa
| Expt. and gene | Promoter region (5′/3′) | 5′ Oligonucleotide | 3′ Oligonucleotide |
|---|---|---|---|
| Analysis of 200-bp region | |||
| APD1 | −344/−174 | GGCAATTTGAACTTTTTAGG | TAGAAGATATCATCAGCGCC |
| AZR1 | −271/−93 | TTACCAATTTGCATTATTTC | CTCTAGCATTATATATAATC |
| FLR1 | −471/−299 | CCTCTGATGCTGACACACGC | CTTCCGGAAAAACTAGTGATG |
| SNG1 | −434/−248 | ATAGGTGGATCTCGAAAAGGA | TCACGGCTTTCGCTTTTCTT |
| SNQ2 | −693/−503 | CCACGGATCACCCCATTTGG | GATTTAAACGGAAATGGGCGG |
| YGR035C | −430/−242 | GGGGCCGATTGTTCCAC | CTAAATCCGCGGCTTTCC |
| YKL051W | −639/−469 | GAAAAGAACGGAAACAAACG | AGTGGAAATGCGATATGTCC |
| YLL056C | −348/−182 | ACATAAGTCTTCCGTGGAAG | ATATCTTCAGGAAAGCCAAG |
| YLR046C | −442/−267 | TTACTGTCAAAAAAGGGCCC | TGGAAATGTGCTTATTCCCG |
| YLR179C | −397/−200 | GAAAACTTTTTACTCTTCCC | ACGGAAATAGAATTTGAAG |
| YLR346C | −388/−221 | GCTGAAAGTGAAAAGGCAC | ATTGATAGGAATCAGCCGC |
| YMR102C | −770/−577 | CGCTTATACCCGCGGATG | CGCGCAAGGAAATCCAG |
| YOR1 | −500/−318 | CCGTGGAAATAGCCGG | ACCTTCGCTAGCTACCTCTG |
| YPL088W | −357/−212 | GACAGACTTCGTCACCGTGG | GCCGAGTTATCTCCCGTTCC |
| Reduction of specific Yrr1p consensus sequence | |||
| AZR1 5′ | −271/−182 | GACACTTTTTATTGTATTTCC | |
| AZR1 3′ | −193/−93 | AAAAAGTGTCAAGCAGAACC | |
| AZR1 5′/5′ | −271/−225 | GGAAATACAGCGGATATC | |
| AZR1 5′/3′ | −233/−182 | TGTATTTCCGCGCTTC | |
| SNG1 5′ | −434/−332 | ACGCATATACACGGAAATAGG | |
| SNG1 3′ | −336/−248 | TGCGTGGTTATTCGTTTTCG | |
| SNG1 5′/5′ | −434/−382 | TTTCTCAACATTTCTTTTTCCTG | |
| SNG1 5′/3′ | −386/−332 | GAAAAAAATAAACCGATTCCC |
This table lists all the oligonucleotides used for the amplification by PCR of the selected 200-bp region of each regulated gene promoter. For each regulated gene identified in microarray experiments, the exact location of the amplified promoter region is indicated with the complete sequence of both the 5′ and 3′ oligonucleotides. Each oligonucleotide sequence is given in the 5′ to 3′ sense. This table also contains the oligonucleotides used for the reduction experiments carried out with the selected 200-bp regions of the AZR1 and SNG1 promoters to delineate more precisely the consensus sequence for Yrr1p. Only one oligonucleotide is shown in each row because the complementary oligonucleotide was described for the previous experimental step.
Protein extracts were prepared from 100 ml of cell cultures grown to an OD600 of 1.5. The cells were washed and broken by vortexing with glass beads in extraction buffer (100 mM Tris-HCl [pH 8.0], 1 mM dithiothreitol [DTT], 20% glycerol, 1 mM phenylmethylsulfonyl fluoride, and 1× protein inhibitor cocktail [Roche]). The cell debris and glass beads were removed by centrifugation for 15 min at 10,000 × g at 4°C, with the final supernatant corresponding to the crude extract. Binding reactions were carried out for 20 min at 30°C in a mixture of 7.5% glycerol, 90 mM KCl, 0.1× salmon sperm, 100 fmol of labeled DNA, and 1 to 5 μl of crude extract in binding buffer (20 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 5 mM DTT, 5 mM CaCl2, 0.1 mM MgSO4, and 0.1 mM EDTA). The binding reaction mixture was loaded onto a 5% acrylamide-biacrylamide (29.1:0.9) gel in 1× Tris-borate-EDTA and run at 5 W and 4°C.
The promoter regions of AZR1 and SNG1 were shortened by PCR amplification of half of the 200-bp region selected from the 800 upstream nucleotides. The oligonucleotides used are described in Table 1.
Bioinformatic analyses.
For microarray experiments, we filtered our data, excluding artifactual spots, saturated spots, and spots with only weak signals. Assuming that most genes displayed no change in expression, Cy3/Cy5 ratios were normalized using the median of all the ratios for each experiment. We clustered data from six independent experiments for the gain-of-function mutant and three independent experiments for the chimera, using the principal component analysis (PCA) module of J-express (11). The cluster shown below in Fig. 2 was generated with Treeview software (12). The profiles of genes belonging to up- and down-regulated clusters were visually checked, and genes with irregular profiles were discarded. We used the Consensus module of RSA tools (27) to select a 200-bp region within the promoters (between −800 and +1) of the up-regulated genes. We used Motif Sampler online software (25) to search for a consensus sequence in the regulated genes displaying specific binding. The consensus sequences identified were aligned and represented using the Sequence Logo (22). The final global PDR network was determined from data provided by the Yeast Proteome Database (3) and Yeast Microarray Global Viewer (18).
FIG. 2.
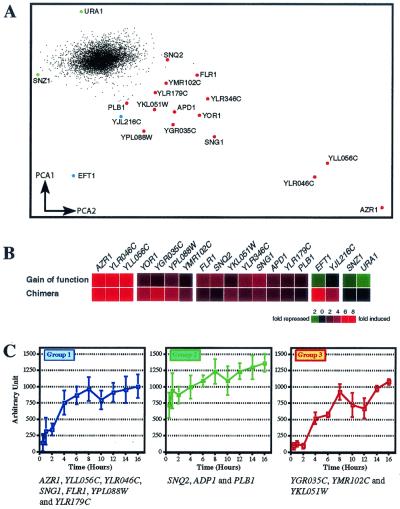
Genes upregulated by YRR1. (A) Microarray results for the gain-of-function mutant and chimera experiments were analyzed by PCA (11) to determine the two key variables plotted on the two axes (PCA1 versus PCA2). This method clearly distinguished repressed genes (left, green) from activated genes (right, red) and the majority of genes not regulated by YRR1 (central, black). Genes specifically activated in the chimera experiments are marked in blue. The upregulated genes are distributed along a gradient, which may indicate the existence of different subgroups of genes. (B) Cluster analyses have been performed (12) and are shown here. The upper part of the diagram presents the results of six independent experiments in which a strain constitutively producing the gain-of-function mutant Yrr1p protein was compared with the SEY strain used as a control. The lower part of the diagram shows the results of three independent experiments in which the strain with the Yrr1*GAD construct was subjected to galactose induction for 4 h and compared with the wild-type FY strain, used as a control. Genes were identified as described in the text. The color scale is indicated in the lower part of the figure. The complete set of data is available at http://www.biologie.ens.fr/yeast-publi.html. (C) Time course of the expression of Yrr1p-regulated genes under galactose induction. Northern blot analyses were performed for each regulated gene identified by microarray experiments. Total RNA (10 μg) was deposited on the gel after induction in galactose medium for various lengths of time, from 30 min to 16 h. Three groups were defined according to the quantification profile obtained from Northern blot analysis (see the related website http://www.biologie.ens.fr/yeast-publi.html). For these three groups, mean RNA levels (arbitrary units) are plotted against the duration of galactose induction (in hours). Group 1 contains YLR046C, YLL056C, AZR1, SNG1, FLR1, YPL088W, and YLR179C, the most strongly induced genes. Group 2 comprises APD1, SNQ2, and PLB1, which display high levels of basal expression. Finally, group 3 consists of YGR035C, YMR102C, and YKL051W, which show an induction only after 2 h in galactose. Error bars are standard deviations.
RESULTS
Experimental strategy.
The transcription factors of the Zn2Cys6 zinc finger family are all similar in general structure and are composed of three main regions (21). The N-terminal DNA-binding domain and the C-terminal activation domain are linked by a central regulatory domain which, in the case of Pdr1p and Pdr3p, constitutively inhibits the activity of the protein. We recently developed a general genome-wide strategy for the systematic analysis of regulatory networks under the control of Zn2Cys6 transcription factors (9). This approach is based on the conditional expression of a chimeric gene encoding the DNA-binding region of the transcription factor studied fused to the transcription-activating domain of GAL4. We used a similar strategy with YRR1 to create a chimera called Yrr1*GAD (Fig. 1A). The key point in this approach is that the chimera must contain a specific DNA-binding domain devoid of inhibitory activity due to the central domain. We reasoned that the central regulatory domain should be highly divergent among members of the Zn2Cys6 zinc finger family and should be easily distinguished from the DNA-binding domain. We therefore aligned the sequences of the 54 transcription factors of this family, using ClustalW software (26). We used this alignment to generate a phylogenetic tree (available from the website http://www.biologie.ens.fr/yeast-publi.html). YRR1 belongs to a subfamily of four genes, the others being YKL222C, YOR172W, and YLR266C. We aligned the sequences of these four genes with that of PDR1 (Fig. 1B) and identified a highly conserved region, from amino acids 175 to 190 in Yrr1p, which was included in the long form, Yrr1L*GAD, but absent from the short form, Yrr1S*GAD, of the artificial chimera protein. This conserved region contained a protein kinase C phosphorylation site (Fig. 1B), as predicted by PhosphoBase (16).
We also analyzed the properties of a gain-of-function form of Yrr1p characterized by Zhang and coworkers (28) and obtained by insertion screening with a transposon (1). This gain-of-function form results from the insertion of a triple HA tag in the C-terminal region of the Yrr1p protein.
Physiological characterization of Yrr1*GAD chimeras.
We checked that the production of each chimera followed the normal temporal pattern by performing Northern and Western blotting (data not shown). The phenotype conferred by the production of both chimera proteins was compared with the known properties of the Yrr1p gain-of-function mutant (28). We observed that only the overproduction of Yrr1S*GAD resulted in a phenotype similar to that of the gain-of-function mutant (Fig. 1C). This was especially clear with 4-NQO and oligomycin in the case of the W303 strain overproducing Yrr1S*GAD. However, oligomycin resistance was not observed in the FY background, probably because of the respiratory deficiencies of this strain (20). Finally, if cycloheximide were added to the medium, no resistance was observed after induction of the chimera, whatever the construct and the genetic background used.
All these results are consistent with previous observations made for YRR1 drug resistance (5). Thus, for further analyses we considered that Yrr1S*GAD production followed a similar pattern to production of the natural Yrr1p protein. In Western blot analysis, the chimeric protein produced in the FY background was clearly visible after 4 h of galactose induction in rich medium. Growth rates were not affected by production of the chimeric protein (data not shown), and the corresponding wild-type FY strain was used as a control.
A genome-wide search of Yrr1p target genes.
We investigated the YRR1 regulation network by microarray experiments to compare the Yrr1S*GAD chimera and the genomic gain-of-function mutant with a wild-type strain. We performed six analyses with two different types of commercial slides, testing the Yrr1p gain-of-function mutant against the SEY6210 wild-type strain, and three microarray experiments in which the strain producing Yrr1S*GAD, induced for 4 h, was tested against the wild-type strain. All these data were analyzed by PCA (11) and scored with specific weighting according to expression ratio and image quality (Fig. 2A). PCA clearly distinguished Yrr1p-regulated genes from the large number of genes not regulated by this protein. Figure 2B presents the expression ratios of the genes displaying significant induction. The complete analysis criteria and data are available via the Internet (http://www.biologie.ens.fr/yeast-publi.html).
We identified 15 genes that were similarly activated by the artificial chimera Yrr1S*GAD and by the gain-of-function mutant (Table 2). The FLR1, SNG1, SNQ2, YOR1, YLR346C, YLL056C, YGR035C, YMR102C, and YPL088W genes were up-regulated by both proteins, but the recorded expression ratios differed slightly between the gain-of-function mutant and the chimeric form of Yrr1p (Fig. 2B and Table 2). However, these differences are unlikely to reflect real differences between the two forms of the transcription factor but are instead likely to reflect difficulties in controlling biological reproducibility in these experiments. Only EFT1, SNZ1, URA1, and YJL216C appeared to be repressed by the gain-of-function mutant, but further studies are required to ensure that the observed repression was really caused directly by the activity of the gain-of-function mutant.
TABLE 2.
Regulated genes identified in microarray experimentsa
| ORF | Gene | Gof | Yrr1S*GAD | Function | Promoter region analyzed | Group | EMSA | PDRE | YRRE | Drug resistance |
|---|---|---|---|---|---|---|---|---|---|---|
| YGR224W | AZR1 | 29 | 16.2 | Protein involved in resistance to azoles and acetic acid; member of the MFS MDR protein family | −271/−93 | 1 | 1 | 1 | Acetic acid, ketoconazole, and fluconazole | |
| YLR046C | YLR046C | 12.6 | 8.8 | Protein with similarity to Rtm1p | −442/−267 | 1 | NS | 0 | ||
| YLL056C | YLL056C | 11.2 | 10.5 | Protein with weak similarity to Yersinia pseudotuberculosis CDP-3, 6-dideoxy-d-glycero-l-glycero-4-hexulose-5-epimerase | −348/−182 | 1 | 1 | 1 | 1 | |
| YBR008C | FLR1 | 3.7 | 2 | Member of the MDR 12-span (DHA12) family of MFS-MDR | −471/−299 | 1 | 1 | 1 | 2 | Fluconazole, 4-NQO, benomyl, methotrexate, fluconazole, and cycloheximide |
| YPL088W | YPL088W | 2.5 | 5.2 | Putative aryl alcohol dehydrogenase, may participate in late steps of degradation of aromatic compounds that arise from the degradation of lignocellulose | −357/−212 | 1 | 1 | 1 | 3 | |
| YLR179C | YLR179C | 2 | 1.7 | Protein with similarity to Tfs1p Cdc25p-dependent nutrient- and ammonia-response protein | −397/−200 | 1 | NS | 0 | ||
| YGR197C | SNG1 | 5 | 3.1 | Probable transport protein, confers resistance to MNNG and nitrosoguanidine | −434/−248 | 1 | 1 | 1 | MNNG | |
| YDR011W | SNQ2 | 3.2 | 1.5 | Drug-efflux pump involved in resistance to multiple drugs, member of the ABC-transporter superfamily | −693/−503 | 2 | 1 | 4 | 4 | 4-NQO, fluphenazine, azole antifungal agents, staurosporin, cercosporin, and clotimazol Copper |
| YBR151W | APD1 | 2.6 | 2.1 | Protein required for normal cellular structure, localization of actin patches, and resistance to copper | −344/−174 | 2 | 1 | 2 | ||
| YMR008C | PLB1 | 1.8 | 1.7 | Phospholipase B, preferentially deacylates phosphatidylcholine and phosphatidylethanolamine | ? | 2 | ? | 2 | ||
| YGR035C | YGR035C | 1.9 | 7.1 | Protein of unknown function | −430/−242 | 3 | NS | 2 | 3 | |
| YMR102C | YMR102C | 1.4 | 2.9 | Protein of unknown function, contains WD (WD-40) repeats | −770/−577 | 3 | NF | 2 | 2 | |
| YKL051W | YKL051W | 1.9 | 2.9 | Protein of unknown function | −639/−469 | 3 | NF | 2 | ||
| YGR281W | YOR1 | 2.3 | 5.7 | Oligomycin resistance factor; member of the ABC superfamily | −500/−318 | ? | 1 | 1 | 2 | Oligomycin, staurosporin, azole antifungal agents, tetracycline, and erythromycin |
| YLR346C | YLR346C | 3.7 | 2.8 | Protein of unknown function | −388/−221 | ? | 2 | 1 | 2 |
This table provides information concerning the 15 regulated genes identified in experiments with the gain-of-function mutant and the Yrr1S∗GAD chimera. The mean expression ratio was determined by microarray analysis for the gain-of-function (Gof) mutant and the short chimera (Yrr1S∗GAD). Each value is the mean for six independent arrays. Gene function is given according to YPD definition (3). The results of promoter analysis are also given. We also provide details of the location of the promoter region used for EMSA analysis. The group number according to Northern blot kinetic analysis is indicated. We also indicate the number of significant shifts seen in Fig. 3, nonspecific mobility shifts (NS), and the region for which no shift was seen (NF). We also give the number of PDR elements (PDRE) and Yrr1p response elements (YRRE) present in the promoter region of each target. Drug resistance is included for each gene for which it has been characterized. MNNG, N-methyl-N′-nitro-N-nitrosoguanidine ORF, open reading frame.
Time course of Yrr1p-regulated gene induction.
Northern blot analyses of the transcription products of these 15 genes were carried out to analyze the kinetics of Yrr1p-dependent activation (see supplementary data at http://www.biologie.ens.fr/yeast-publi.html). The results of these analyses were entirely consistent with those of microarray experiments. Quantification of the level of each transcript led to the definition of three classes of genes on the basis of behavior in the presence of the activated form of Yrr1p (Fig. 2C). Group 1 comprised AZR1, FLR1, SNG1, YOR1, YLL056C, YLR046C, YLR179C, YLR346C, and YPL088W. All these genes are induced early and display large changes in expression level, with low levels of mRNA detected before galactose induction. Group 2 consisted of SNQ2, APD1, and PLB1, which display high levels of constitutive expression but only a small, linear increase in expression ratio after galactose induction. Finally, group 3 corresponded to late-induced genes. The expression ratios of YGR035C, YKL051W, and YMR102C only began to increase 2 to 4 h after galactose induction. This suggests that Yrr1p does not directly control the expression of these genes. This is consistent with the lack of induction of a specific band shift with the promoter of YGR035C in the presence of Yrr1p (Fig. 3A). See the following website for information about quantification and complete results: http://www.biologie.ens.fr/yeast-publi.html.
FIG. 3.
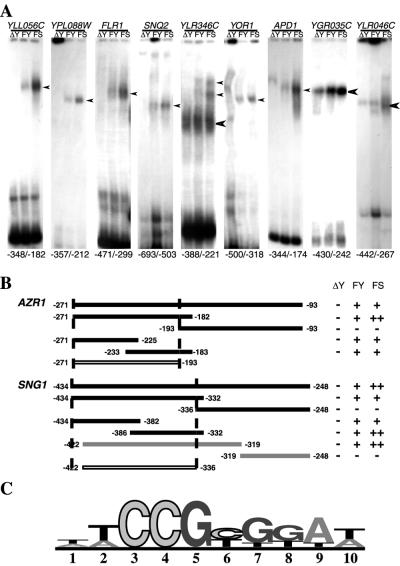
Specific interactions between Yrr1p and the promoters of target genes. (A) EMSA of the main YRR1-regulated genes. Bioinformatic analysis (27) of the promoters of the 15 genes up-regulated by Yrr1p revealed a 200-bp segment with similar features in each case. These similar fragments were amplified by PCR, and EMSAs were performed with the total protein extract from an FY strain in which YRR1 was deleted (ΔY), a wild-type strain (FY), and an FY strain producing Yrr1S*GAD (FS). The precise location of the promoter region selected for each gene is indicated below the autoradiograph. Specific (narrow arrowhead) and nonspecific (thick arrowhead) DNA-binding sites are indicated. (B) Demarcation of the Yrr1p binding sites of the AZR1 and SNG1 promoters. A set of oligonucleotides was used to construct subregions of the 200-bp region of the promoter by PCR. These subregions were delimited by bioinformatic prediction for the AZR1 and SNG1 upstream sequences (Table 1). For each subpromoter region, an EMSA was done with whole-cell extract from an FY strain in which YRR1 was deleted (ΔY), the wild-type FY strain (FY), or an FY strain producing Yrr1S*GAD (FS). The intensity of the mobility shift observed is indicated, from − (no band) to ++ (intense band). The complete gels from EMSAs can be viewed at the related website http://www.biologie.ens.fr/yeast-publi.html. (C) Sequence motif depicting the DNA-binding site preferences of Yrr1p. Promoter sequence analysis of the seven genes up-regulated and involved in a specific shift revealed a core consensus sequence: (T/A)CCG(C/T)(G/T)(G/T)(A/T)(A/T). Only half a dyad seems to be conserved in this consensus sequence. The numbers underneath the sequence motif indicate the sequence of the primers from 5′ to 3′. The height of the nucleotide at each position of the DNA-binding site is based on all the information available for that position, with taller nucleotides more likely to give correct binding than shorter nucleotides.
Identification of specific interactions between Yrr1p and the promoter region of the regulated genes.
We searched for genes directly regulated by Yrr1p by bioinformatic identification of putative DNA-binding regions in the 800 bp upstream from the transcription start site, using online RSA tools (27). Oligonucleotide and dyad analyses were conducted to identify 200-bp regions with a high probability of Zn2Cys6 zinc finger binding within the promoter of each candidate gene. The complete strategy is described in detail at the website http://www.biologie.ens.fr/yeast-publi.html. We performed EMSAs with these selected fragments (Fig. 3A). For each promoter analyzed, we assessed the band shift properties of whole-cell extracts of the strain producing Yrr1S*GAD and of the wild-type strain (FY) expressing the endogenous version of YRR1. The negative controls were conducted with extracts from the FY strain in which YRR1 had been deleted. The selected 200-bp promoter regions of YLL056C, YPL088W, FLR1, and SNQ2 displayed specific binding in the presence of Yrr1p or Yrr1S*GAD (Fig. 3A). EMSA analysis of the YLR346C promoter region revealed two bands, which may reflect the presence of several Yrr1p binding sites. For the YOR1 promoter, different band shift patterns were observed in the presence and absence of Yrr1p, suggesting that other proteins interact with this region if Yrr1p is not present. For YGR035C and YLR046C, the band shifts observed in the three lanes were qualitatively similar, but a larger amount of the putative protein-DNA complex was detected in the presence of activated Yrr1p. Further experiments are required to demonstrate definitively that the quantitative increase in amount of the retarded fragments observed does indeed result from interaction with Yrr1p.
Search for a Yrr1p consensus binding site.
The EMSA and microarray analyses strongly suggested that at least one Yrr1p binding site (Fig. 3B) is present in the selected 200 bp of the promoter regions of AZR1 and SNG1 (see the website http://www.biologie.ens.fr/yeast.publi.html). We used these two promoter regions to analyze more precisely the region interacting with Yrr1p. Specific oligonucleotides were designed to amplify fragments of the relevant regions. The promoter regions −271 to −182 and −434 to −332 of AZR1 and SNG1, respectively, contained sufficient information for binding to Yrr1p (Fig. 3B). We attempted to define the interacting region more precisely by splitting these two fragments into smaller units. We identified two smaller fragments, both of which interacted with Yrr1p. We were therefore able to reduce the binding-competent promoter regions to −271 to −193 for AZR1 and −422 to −336 for SNG1. We used this new information to carry out a search of the sequences common to the AZR1 and SNG1 putative Yrr1p binding sites and then of all seven promoters shown to be involved in specific Yrr1p binding (Fig. 3C). The consensus sequence identified, (T/A)CCG(C/T)(G/T)(G/T)(A/T)(A/T), contains a highly conserved CCG triplet followed by a similar but degenerate triplet. This association mimics the PDR element binding site (PDRE), although the second half of the consensus sequence is less well conserved.
DISCUSSION
We used several complementary approaches to analyze the gene regulatory network directly controlled by the newly discovered transcription factor Yrr1p. Only two genes, YOR1 and SNQ2, have previously been shown to be under the control of Yrr1p. We demonstrate here that 15 genes are actually controlled by Yrr1p. This considerably increases our knowledge of the PDR phenomenon and establishes new connections with other yeast regulatory networks.
Complete genome-wide studies of regulatory networks have suggested that each transcription factor involved plays a precise role. Several conditions are required to determine the complete set of target genes under the control of a given transcription factor. First, it is necessary to activate this transcription factor in such a way that its behavior is similar to that under natural conditions. In this study, the two activated forms of Yrr1p investigated gave similar results. One was a gain-of-function mutant in which negative regulation has been overcome by the insertion of a short peptide in the activating domain (28). The second form, Yrr1S*GAD, is an artificial factor consisting of the Yrr1p DNA-binding domain fused to the heterologous GAL4 activation domain. This is the second example, following similar studies on Pdr1p (9), to demonstrate that the DNA-binding properties of a Zn2Cys6 zinc transcription factor are sufficient to guide the activated protein to the appropriate promoters. This observation is important, both for determination of the regulatory properties of new unknown putative transcription factors discovered in genome sequencing programs and for the design of new transcription factors to control genome expression. The accurate definition of the DNA-binding domain seems to be critical for this sort of approach. A form with a DNA-binding domain 15 amino acids longer, Yrr1L*GAD, displayed only weak activation. We observed posttranslational modification of this DNA-binding extension, which seemed to inhibit the activity of this transcription factor (S. Le Crom et al., unpublished data). This is consistent with previous suggestions that the central domain of yeast Zn2Cys6 zinc transcription factors plays an important role in inhibition (21).
The second condition required for determination of the complete set of target genes of a transcription factor is the possibility of distinguishing between direct and indirect effects. This relies on the use of an appropriate design for the biological experiments. It is particularly important to carry out time-course experiments, which make it possible to distinguish between different classes of target genes. Thus, the early activated genes AZR1, FLR1, SNG1, SNQ2, YOR1, YLL056C, YLR046C, YLR179C, and YPL088W may be direct targets of Yrr1p (Fig. 2C), whereas further evidence is required before such a conclusion can be drawn for YGR035C, YMR102C, and YKL051W. However, it should be stressed that such time-course experiments are not sufficient in themselves to conclude that the early activated genes are direct targets of the transcription activator; more direct biochemical evidence is required. EMSA analyses showed that the promoters of AZR1, SNG1, FLR1, SNQ2, YOR1, YLR346C, YLL056C, and YPL088W displayed different electrophoretic properties in the presence and absence of Yrr1p. This was not the case for YLR046C and YLR179C. The promoter region responsible for these band shift properties was restricted to smaller fragments for the AZR1 and SNG1 promoters. Although these observations do not prove that Yrr1p directly interacts with these promoters, they provide independent information concerning the role of Yrr1p. We therefore tried to identify a putative DNA-binding sequence common to all the relevant promoter fragments. The proposed sequence (Fig. 3C) differs from the sequence AAATxxCCGG(C)xxAATTT previously proposed based on analysis of the SNQ2 promoter (4) but is consistent with deletion analysis of the YOR1 promoter (28). It contains an invariant CCG triplet in all the sequences. The Uga3 factor has also been shown to recognize the perfect triplet as a monomer for subsequent dimerization and interaction with the rest of the signal (14). This would be consistent with a split signal composed of two repeats of the proposed YRRE (Yrr1p response element) sequence.
The putative YRRE upstream activation signal is strikingly similar to the PDRE signal described for the Pdr1p and Pdr3p transcription factors (8, 9). This similarity may be of biological importance, especially for promoters recognized by both transcription factors, such as that of YRR1 itself (28). This YRRE sequence is not present in the promoters of YLR046C and YLR179C, which is consistent with the absence of band shift in EMSAs (Fig. 3A). This strongly suggests that these two genes are regulated indirectly, despite their early activation in the presence of Yrr1p. This highlights the importance of using several complementary approaches to directly characterize target genes.
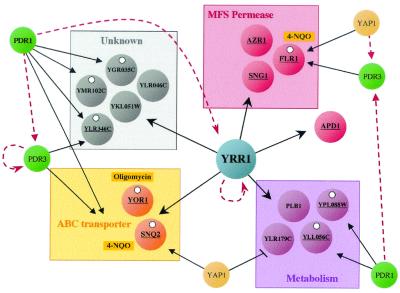
Our complete view of the regulatory network controlled by Yrr1p can be integrated into a more general view of the expression properties of the yeast genome. Even in yeast, in which transcription regulation has been extensively studied, few complete regulatory networks associated with specific transcription factors have been described previously (for a review, see reference 10). The PDR1/PDR3, and YRR1 regulation networks are one of the best-described cases of overlapping networks. The overlap between these two networks results partly from the control exerted by Pdr1p over the expression of YRR1 (28). Thus, genes such as YLL056C, YLR346C, and YPL088W, which are regulated by both classes of factor, may be the only direct targets of Yrr1p. We showed that the promoters of these genes are indeed recognized by Yrr1p (Fig. 3). In addition, all these genes have perfect PDREs in their promoters, and time-course induction of an activated form of Pdr1p has strongly suggested that they are also direct targets of Pdr1p (9). Several important target genes appear to be under the tight control of an intricate interplay of transcription factors; in that respect the case of FLR1 is worth noting. The transcription factor Yap1p can activate FLR1 either directly (19) or via PDR3 (24). We show here that FLR1 can also be activated by YRR1, which is itself controlled by PDR1 (28). Taking into account that PDR1 also activates PDR3 and that PDR3 and YRR1 positively autoregulate their own transcription (7, 28), this provides an interesting example of complex transcription factor regulation (Fig. 4). FLR1 encodes a plasma membrane transporter of the MFS conferring resistance to multiple drugs, in particular to benomyl. The apparent redundancy in the factors controlling FLR1 expression probably reflects the multiplicity of pathways that can be followed to produce this functionally important protein. More generally, the PDR network, in which PDR1 and PDR3 exerts a major control, appears to overlap with satellite networks such as that controlled by YRR1 or other new genes encoding transcription factors, such as YLR266C (I. Hikkel et al., unpublished data). The complete description of such related networks is a prerequisite if we are to control the complex PDR phenomenon and analyze similar networks operating in pathogenic strains to develop new antifungal strategies.
FIG. 4.
The YRR1 regulation network is linked to the PDR network. All the regulated genes found to be activated by Yrr1p fall into the three main characterized functional groups: MFS permease, ABC transporter, and metabolic genes. For all these genes, the consensus PDRE sequence is shown as an open circle. The promoters of the underlined genes gave positive results in the EMSAs presented above. Most of these genes are also regulated by Pdr1p, Pdr3p, and Yap1p. These transcription factors control the expression of other transcription factors in this series (dotted arrows) or, in some cases, their own expression (YRR1 and PDR3). Finally, the most common drugs used (4-NQO and oligomycin) are marked next to the genes associated with specific resistance.
Acknowledgments
W. Scott Moye-Rowley is supported by an NIH grant (GM49825). This work was supported by a grant from the Association pour la Recherche contre le Cancer (ARC 5691). The microarray facilities used in this work are part of the Genopole Ile de France.
REFERENCES
- 1.Burns, N., B. Grimwade, P. B. Ross-Macdonald, E.-Y. Choi, K. Finberg, G. S. Roeder, and M. Snyder. 1994. Large-scale characterization of gene expression, protein localization and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8:1087-1105. [DOI] [PubMed] [Google Scholar]
- 2.Chauncey, T. R. 2001. Drug resistance mechanisms in acute leukemia. Curr. Opin. Oncol. 13:21-26. [DOI] [PubMed] [Google Scholar]
- 3.Costanzo, M. C., M. E. Crawford, J. E. Hirschman, J. E. Kranz, P. Olsen, L. S. Robertson, M. S. Skrzypek, B. R. Braun, K. L. Hopkins, P. Kondu, C. Lengieza, J. E. Lew-Smith, M. Tillberg, and J. I. Garrels. 2001. YPD, PombePD and WormPD: model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res. 29:75-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui, Z., D. Hirata, and T. Miyakawa. 1999. Functional analysis of the promoter of the yeast SNQ2 gene encoding a multidrug resistance transporter that confers the resistance to 4-nitroquinoline N-oxide. Biosci. Biotechnol. Biochem. 63:162-167. [DOI] [PubMed] [Google Scholar]
- 5.Cui, Z., T. Shiraki, D. Hirata, and T. Miyakawa. 1998. Yeast gene YRR1, which is required for resistance to 4-nitroquinoline N-oxide, mediates transcriptional activation of the multidrug resistance transporter gene SNQ2. Mol. Microbiol. 29:1307-1315. [DOI] [PubMed] [Google Scholar]
- 6.Decottignies, A., A. M. Grant, J. W. Nichols, H. de Wet, D. B. McIntosh, and A. Goffeau. 1998. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 273:12612-12622. [DOI] [PubMed] [Google Scholar]
- 7.Delahodde, A., T. Delaveau, and C. Jacq. 1995. Positive autoregulation of the yeast transcription factor Pdr3p, which is involved in control of drug resistance. Mol. Cell. Biol. 15:4043-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 9.Devaux, F., P. Marc, C. Bouchoux, T. Delaveau, I. Hikkel, M.-C. Potier, and C. Jacq. 2001. An artificial transcription activator mimics the genome-wide properties of the yeast Pdr1 transcription factor. EMBO Rep. 2:493-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devaux, F., P. Marc, and C. Jacq. 2001. Transcriptomes, transcription activators and microarrays. FEBS Lett. 498:140-144. [DOI] [PubMed] [Google Scholar]
- 11.Dysvik, B., and I. Jonassen. 2001. J-Express: exploring gene expression data using Java. Bioinformatics 17:369-370. [DOI] [PubMed] [Google Scholar]
- 12.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallstrom, T. C., D. J. Katzmann, R. J. Torres, W. J. Sharp, and W. S. Moye-Rowley. 1998. Regulation of transcription factor Pdr1p function by an Hsp70 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1147-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Idicula, A. M., and R. A. Dorrington. 2001. The binding sites in the upstream activation sequence (UAS GABA) of Uga3p is different from others of the Cys 6 zinc finger family of transcription factors in Saccharomyces cerevisiae. Yeast 18:S56. [Google Scholar]
- 15.Kolaczkowska, A., and A. Goffeau. 1999. Regulation of pleiotropic drug resistance in yeast. Drug Resist. Updates 2:403-414. [DOI] [PubMed] [Google Scholar]
- 16.Kreegipuu, A., N. Blom, and S. Brunak. 1999. PhosphoBase, a database of phosphorylation sites: release 2.0. Nucleic Acids Res. 27:237-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, P., B. P. Davis, J. Hendrick, and T. W. Jeffries. 1998. Cloning and disruption of the beta-isopropylmalate dehydrogenase gene (LEU2) of Pichia stipitis with URA3 and recovery of the double auxotroph. Appl. Microbiol. Biotechnol. 49:141-146. [DOI] [PubMed] [Google Scholar]
- 18.Marc, P., F. Devaux, and C. Jacq. 2001. yMGV: a database for visualization and data mining of published genome-wide yeast expression data. Nucleic Acids Res. 29:E63-E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen, D. T., A. M. Alarco, and M. Raymond. 2001. Multiple Yap1p-binding sites mediate induction of the yeast major facilitator FLR1 gene in response to drugs, oxidants, and alkylating agents. J. Biol. Chem. 276:1138-1145. [DOI] [PubMed] [Google Scholar]
- 20.Rieger, K. J., A. Kaniak, J.-Y. Coppée, G. Aljinovic, A. Baudin-Baillieu, G. Orlowska, R. Gromadka, O. Groudinski, J.-P. di Rago, and P. P. Slonimski. 1997. Large scale phenotypic analysis. The pilot project on yeast chromosome III. Yeast 13:1547-1562. [DOI] [PubMed] [Google Scholar]
- 21.Schjerling, P., and S. Holmberg. 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24:4599-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Servos, J., E. Haase, and M. Brendel. 1993. Gene SNQ2 of Saccharomyces cerevisiae, which confers resistance to 4-nitroquinoline-N-oxide and other chemicals, encodes a 169 kDa protein homologous to ATP-dependent permeases. Mol. Gen. Genet. 236:214-218. [DOI] [PubMed] [Google Scholar]
- 24.Tenreiro, S., A. R. Fernandes, and I. Sa-Correia. 2001. Transcriptional activation of FLR1 gene during Saccharomyces cerevisiae adaptation to growth with benomyl: role of Yap1p and Pdr3p. Biochem. Biophys. Res. Commun. 280:216-222. [DOI] [PubMed] [Google Scholar]
- 25.Thijs, G., M. Lescot, K. Marchal, S. Rombauts, B. De Moor, P. Rouzé, and Y. Moreau. 2001. A higher order background model improves the detection of regulatory elements by Gibbs Sampling. Bioinformatics 17:1113-1122. [DOI] [PubMed] [Google Scholar]
- 26.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequences alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Helden, J., B. Andre, and J. Collado-Vides. 2000. A web site for the computational analysis of yeast regulatory sequences. Yeast 16:177-187. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, X., Z. Cui, T. Miyakawa, and W. S. Moye-Rowley. 2001. Cross-talk between transcriptional regulators of multidrug resistance in Saccharomyces cerevisiae. J. Biol. Chem. 276:8812-8819. [DOI] [PubMed] [Google Scholar]