Abstract
The cytotoxic T-lymphocyte protease granzyme A induces caspase-independent cell death in which DNA single-stranded nicking is observed instead of oligonucleosomal fragmentation. A 270- to 420-kDa endoplasmic reticulum-associated complex (SET complex) containing the nucleosome assembly protein SET, the tumor suppressor pp32, and the base excision repair enzyme APE can induce single-stranded DNA damage in isolated nuclei in a granzyme A-dependent manner. The normal functions of the SET complex are unknown, but the functions of its components suggest that it is involved in activating transcription and DNA repair. We now find that the SET complex contains DNA binding and bending activities mediated by the chromatin-associated protein HMG2. HMG2 facilitates assembly of nucleoprotein higher-order structures by bending and looping DNA or by stabilizing underwound DNA. HMG2 is in the SET complex and coprecipitates with SET. By confocal microscopy, it is observed that cytoplasmic HMG2 colocalizes with SET in association with the endoplasmic reticulum, but most nuclear HMG2 is unassociated with SET. This physical association suggests that HMG2 may facilitate the nucleosome assembly, transcriptional activation, and DNA repair functions of SET and/or APE. HMG2, like SET and APE, is a physiologically relevant granzyme A substrate in targeted cells. HMG1, however, is not a substrate. Granzyme A cleavage after Lys65 in the midst of HMG box A destroys HMG2-mediated DNA binding and bending functions. Granzyme A cleavage and functional disruption of key nuclear substrates, including HMG2, SET, APE, lamins, and histones, are likely to cripple the cellular repair response to promote cell death in this novel caspase-independent death pathway.
Cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells play an essential role in the clearance of virally infected cells and tumor cells, mostly by inducing apoptosis via the release of cytolytic granules (reviewed in reference 49). Cytotoxic granules contain a pore-forming protein perforin (PFP) and a family of serine proteases known as granzymes (Gzm). GzmA and GzmB, the most abundant granzymes in cytotoxic lymphocytes, are delivered to the cytosol of target cells via PFP, concentrate in target cell nuclei, and independently and synergistically induce cell death (26, 33, 38, 58). GzmB activates the ubiquitous apoptotic caspase pathway by initially cleaving caspases 3 and 7 (19, 21, 43, 45, 54, 66) but also activates caspase-independent apoptosis by direct cleavage of key downstream caspase-pathway substrates, such as DFF45/ICAD, Bid, NuMa, DNA-PKcs and lamin B (4, 6, 41, 56, 67). GzmA induces cell death through an alternate caspase-independent mechanism, causing single-stranded DNA nicking and other apoptotic features including nuclear fragmentation, chromatin condensation, loss of mitochondrial potential, and membrane blebbing (9, 47, 67; K. Kaznatcheev and J. Lieberman, unpublished observations).
To understand the GzmA-mediated cell death pathway, affinity chromatography with inactive Ser→Ala mutant GzmA (S-AGzmA) was used to determine potential substrates and other proteins that associate with GzmA (8). A 270- to 420-kDa cytoplasmic complex (SET complex) from K562 cell lysates binds to immobilized S-AGzmA (9). The purified SET complex reconstitutes single-stranded DNA nicking in isolated nuclei in the presence of GzmA (10). The SET complex contains the tumor suppressor protein pp32 (5, 10, 28, 59, 61), the nucleosome assembly protein (NAP) SET (10, 31, 59, 60), and the base excision repair enzyme APE/ref-1 (20; Z. Fan, P. J. Beresford, D. Zhang, Z. Xu, C. D. Novina, A. Yoshida, Y. Pommier, and J. Lieberman, submitted for publication). SET and APE, but not pp32, are substrates of GzmA in CTL attack and after GzmA loading with perforin (8, 10; Fan et al., submitted). pp32 (also known as PHAPI, I1PP2A) is a highly conserved protein phosphatase 2A (PP2A) inhibitor with putative nuclear localization and export signals which shuttles between the nucleus and the cytosol via binding to the nuclear export receptor crm1 (13, 28). SET (also known as PHAPII, TAF-Iβ, I2PP2A) was initially identified as a translocated gene in acute undifferentiated leukemia (60). SET can stimulate adenovirus replication of a viral chromatin template (31). Recent reports show that SET and its homologues bind to the transcriptional coactivators CBP/p300 and the core histones and may serve as a link between transcriptional coactivators and chromatin (40, 46). APE is a multifunctional protein with DNA binding and endonuclease activity associated with its C terminus and redox function in the N-terminal domain (reviewed in reference 22). APE, a major member of the base excision repair pathway, recognizes and is the rate-limiting enzyme in the repair of DNA apurinic/apyrimidinic (AP) sites, the most frequent type of DNA damage in cells (7, 34). It also activates by a redox-based mechanism the DNA binding activity of many transcription factors, including AP-1 (Fos/Jun), NF-κB, and Myb (64).
SET and its homologue TAF-Iα and pp32 and its homologue APRIL were recently shown to coassociate in a smaller ∼150-kDa nuclear complex which has been shown to inhibit histone acetylation and to bind to HuR, a protein that increases the stability of mRNAs with AU-rich regions that are encoded by many early-response genes, protooncogenes, and cytokine genes (13, 40). The size of the nuclear SET complex suggests that SET, pp32, and their homologues may constitute all the components of the nuclear complex, although each of these proteins is so acidic (predicted pIs of ∼4) that it is hard to see how they can stay together without a balancing charge. However, the endoplasmic reticulum (ER)-associated SET complex is substantially larger and not all of its known components (SET, pp32, and APE) account for its apparent molecular weight as estimated by gel filtration (10; Fan et al., submitted). All of the components of the ER-associated SET complex can also be found in the nucleus, where most of their known functions would require them to be. The functions of the cytoplasmic SET complex are unknown, as are the signals, which regulate its nuclear import and export. In this study we show that, despite its nuclear function, the DNA bending protein HMG2 (high mobility group protein 2; reviewed in reference 14) is a component of the ER-associated SET complex. HMG2 binds directly to SET and colocalizes in part with SET to associate with the ER. Just as GzmA destroys the NAP activity of SET and the DNA repair and redox activities of APE, it also cleaves and functionally inactivates the DNA binding and bending activities of HMG2.
MATERIALS AND METHODS
Cell lines and antibodies.
K562 and HeLa cells were obtained from American Type Culture Collection and grown in RPMI 1640 supplemented with 10% fetal calf serum, 2 mM glutamine, 2 mM HEPES, 100 U of penicillin per ml, 100 mg of streptomycin per ml, and 50 μM β-mercaptoethanol. Mouse monoclonal anti-pp32 antibody (RJ1) was produced as previously described (10). Rabbit antiserum to SET peptide amino acids (aa) 3 to 16 was produced following the method of Adachi et al. (1) and protein A purified. Mouse monoclonal anti-SET antibody KM1720 was a kind gift of K. Nagata (32). Mouse polyclonal anti-human GzmA antiserum was generated by immunizing mice with 10 μg of GzmA in complete Freund's adjuvant and boosting three times with GzmA in incomplete Freund's adjuvant. Other commercially available antibodies used are mouse anti-APE monoclonal antibody (Transcription Laboratories), rabbit polyclonal antibodies against HMG1 and HMG2 C-terminal peptides (PharMingen), and glutathione S-transferase (GST) (Clontech), horseradish peroxidase-conjugated sheep anti-mouse immunoglobulin (IgG) and horseradish peroxidase-conjugated monkey anti-rabbit IgG (Amersham), fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Zymed), and tetramethyl rhodamine isocyanate (TRITC)-conjugated goat anti-mouse IgG (Molecular Probes).
Recombinant proteins and purified PFP.
Recombinant GzmA, inactive S-AGzmA, and GzmB were produced and purified as previously reported (8, 65). PFP was purified from the rat RNK-16 cell line and used at a sublytic concentration, titrated to induce <10% cytolysis, as described previously (44). Recombinant pp32 (rpp32) was expressed in BL21-DE3 cells from pET 30a, recombinant SET (rSET) was expressed from pET 26b, recombinant APE was expressed from pET 14b (a kind gift of I. Hickson, University of Oxford), and recombinant GST (rGST) was expressed from pET 30b (Novagen) as described previously (10; Fan et al., submitted). Recombinant proteins with His6 tags were purified sequentially over Novagen nickel and Bio-Rad anion-exchange columns. HMG2 cDNA, a kind gift of P. Sharp, was expressed from pET15b and purified as described previously (48).
Cytoplasmic SET complex isolation and HMG2 identification.
K562 cell lysates (1010 cell equivalents in NP-40 lysis buffer [50 mM Tris HCl, pH 7.5; 0.5% NP-40; 25 mM KCl; 5 mM MgCl2]) were loaded onto an immobilized S-AGzmA column and eluted with 500 mM NaCl in 50 mM Tris-HCl, pH 7.5, as described previously (8). The concentrated S-AGzmA column eluate was applied in Tris-buffered saline to an S400 gel filtration column (Pharmacia). Eluted fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting and compared to the elution profile of Pharmacia gel filtration standards. The S-AGzmA column eluate was also desalted and applied to a Bio-Rad Q column and eluted with an NaCl gradient in 50 mM Tris-HCl, pH 7.5. The dominant 28-kDa band in the flowthrough visualized after Coomassie blue staining was subjected to N-terminal sequencing performed by M. Berne of the Tufts Core facility. A 28-kDa band directly eluted from the S400 column was also analyzed by tryptic digestion and sequencing performed by the Harvard Microchemistry Facility by microcapillary reverse-phase high-pressure liquid chromatography nanoelectrospray tandem mass spectroscopy.
Coimmunoprecipitation and immunoblotting.
Antibodies were preincubated with protein A-Sepharose (Pharmacia) for 1 h at 4°C. Antibody-coated beads were washed twice in phosphate-buffered saline (PBS) before being added to recombinant proteins (50 μg/ml) or postnuclear cytosolic lysates (5 × 106 cell equivalents in 20 μl of NP-40 lysis buffer) that had been preincubated at 4°C for 2 h. After overnight incubation with shaking at 4°C, the beads were washed extensively in 1% NP-40, 0.1% SDS in PBS and boiled in 2× SDS loading buffer before electrophoresis through SDS-PAGE gels. Nuclear lysates were prepared by washing the nuclear pellet, obtained after centrifugation at 1,000 × g of NP-40-lysed K562 cells, twice in NP-40 lysis buffer followed by lysis in NP-40 lysis buffer with 500 mM NaCl. S400 column fractions or nuclear and cytoplasmic fractions were similarly analyzed by SDS-PAGE and then transferred to nitrocellulose before probing with indicated antibodies as described.
Laser-scanning confocal microscopy.
HeLa cells, grown overnight to subconfluency at 37°C in eight-well chamber slides coated with rat collagen I (Becton Dickinson Labware, Bedford, Mass.), were fixed and permeabilized using the Fix-and-Perm kit (Caltag Laboratories, Burlingame, Calif.) according to the manufacturer's instructions and then blocked with permeabilization buffer with 10% goat serum. All antibodies were diluted in permeabilization buffer with 10% goat serum and incubated with the samples at room temperature; washes between steps were with PBS. Primary antibodies were incubated for 2 h, followed by incubation with species-specific FITC- or TRITC-conjugated secondary antibodies for 1 h. Samples were mounted with ProLong Antifade mounting medium (Molecular Probes, Eugene, Oreg.) and dried overnight. Images were acquired with a Bio-Rad Radiance 2000 laser-scanning confocal microscope by focusing on the central plane of each cell.
Granzyme in vitro and in situ cleavage assay.
rHMG2 (1 μM) or postnuclear K562 cell lysates (2 × 105 cell equivalents) were incubated for the indicated times at 37°C with indicated concentrations of GzmA, S-AGzmA, or GzmB in 20 μl of 50 mM Tris-HCl (pH 7.5)-1 mM CaCl2-1 mM MgCl2. Nuclei were pelleted from K562 or HeLa cells lysed in NP-40 lysis buffer with 1 mM phenylmethylsulfonyl fluoride (PMSF) and washed twice in NP-40 lysis buffer and once in NP-40 lysis buffer without NP-40. Nuclei (106 in 50 μl of 50 mM NaCl-0.25 M sucrose-2 mM CaCl2-20 mM Tris-HCl [pH 7.2]) were incubated with the indicated amounts of GzmA or 0.5 μM S-AGzmA at 37°C for 1.5 h. Reaction products were boiled in 5× SDS loading buffer, separated on SDS-12% PAGE gels, and transferred to nitrocellulose for immunoblotting.
Granzyme loading with PFP.
K562 cells (2 × 105) in 100 μl of loading buffer (Hanks' balanced salt solution with 1 mg of bovine serum albumin per ml, 1 mM CaCl2, and 1 mM MgCl2) were incubated for indicated times at 37°C with 1 μM GzmA or 1 μM GzmB and sublytic concentrations of PFP. Cells were then incubated for an additional 15 min in 1 mM PMSF before lysing in 20 μl of 0.5% NP-40 lysis buffer containing PMSF. Nuclear pellets were washed twice in NP-40 lysis buffer and extracted in 20 μl of 1% NP-40 lysis buffer containing PMSF. Samples were boiled in 2× SDS loading buffer before SDS-PAGE and immunoblotting.
DNA binding assay.
pBR322 DNA (0.3 μg) was incubated with indicated amounts of rHMG2, purified SET complex (2 μg/μl), or 1 μM rGST in 10 mM Tris-HCl (pH 7.8)-100 mM NaCl-1 mM EDTA-10 mM MgCl2-1 mM dithiothreitol-10% glycerol-5 μg of bovine serum albumin per ml at 25°C for 1 h. For GzmA pretreatment of HMG2, 2 μM rHMG2 or 2 μl of pooled SET complex (2 μg/μl) was preincubated with 100 or 500 nM GzmA or 500 nM S-AGzmA at 37°C for 1 h. The reaction mixtures were electrophoresed through 1% agarose gels in 40 mM Tris-acetate, pH 7.8, containing 1 mM EDTA, and analyzed after ethidium bromide staining.
DNA bending assay.
DNA fragments (123 bp), prepared with cohesive ends by AvaI digestion of the 123-bp ladder from Life Technologies, Inc., were 5′ end labeled with [γ-32P]ATP using T4 polynucleotide ligase (New England Biolabs) and purified through Sephadex G-50 minicolumns (Pharmacia). Radiolabeled fragments (1 nM) in 10 μl of 30 mM Tris-HCl (pH 7.8)-10 mM MgCl2-10 mM dithiothreitol-0.5 mM ATP were ligated with T4 DNA ligase (0.1 U/reaction) (New England Biolabs) at 30°C for 30 min, followed by termination at 65°C for 15 min. rHMG2 (1 μM) or SET complex (2 μg/μl), pretreated with indicated amounts of GzmA or 1 μM S-AGzmA as above, was then added to the 32P-labeled 123-bp DNA fragments for 30 min at 4°C. Deproteinized samples were electrophoresed through 6% nondenaturing gels in 0.5× Tris-borate-EDTA buffer and visualized by autoradiography after drying.
RESULTS
The cytoplasmic SET complex contains HMG2.
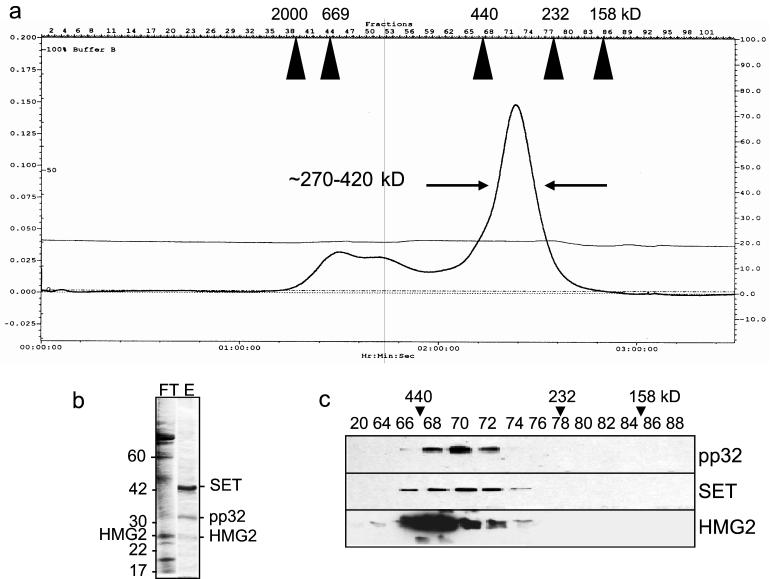
We previously showed that a 270- to 420-kDa multimeric complex (termed the SET complex) elutes with 500 mM NaCl from immobilized S-AGzmA (10). The concentrated S-AGzmA column eluate migrates as a single broad peak through an S400 gel filtration column (Fig. 1a). The complex was previously found to contain the 44-kDa GzmA substrate SET, the tumor suppressor protein pp32, and the 38-kDa GzmA substrate APE as well as an unidentified 25-kDa nuclease (10; Fan et al., submitted). The purified SET complex incubated with isolated nuclei initiates DNA nicking in the presence of active GzmA (10). Neither SET complex nor GzmA alone induce DNA breaks. To identify other SET complex proteins, the S-AGzmA eluate was applied to an anion-exchange column, which disrupts the complex and binds SET and pp32 (Fig. 1b). The highly acidic SET and pp32 proteins (calculated pI, 4.1 and 4.0, respectively) elute at approximately 0.75 M NaCl. The flowthrough contains a prominent 28-kDa band which was analyzed by N-terminal sequencing and found to be identical to the N-terminal nine amino acids of HMG-2. Although HMG2, like SET and pp32, has an acidic C-terminal tail, the calculated pI of HMG2 is 7.8, which is consistent with its primarily fractionating with the flowthrough of the anion-exchange column. (A faint 28-kDa band in the anion column eluate may represent a small amount of HMG-2 that remains stuck to pp32 or SET and separates in the eluate despite its presumed positive charge at the buffer pH of 7.5.) A 28-kDa band directly eluted from the S400 column was also digested with trypsin and analyzed by microcapillary reverse-phase high-pressure liquid chromatography nanoelectrospray tandem mass spectroscopy. Twelve distinct peptide sequences are identical to predicted tryptic fragments of HMG2. The presence of HMG2 in the SET complex was confirmed by immunoblotting (Fig. 1c). The elution profile of HMG-2 in fractions 66 to 76 coincides with the fractions that contain pp32 and SET. However, the peak fractions containing HMG-2 (66-68) do not coincide with the peak of SET and pp32 immunoreactivity (fraction ∼70). This, taken together with the breadth of the SET complex peak from the sizing column, may indicate some heterogeneity in the complex.
FIG. 1.
HMG2 coelutes with the SET complex proteins SET and pp32. (a) A 270- to 420-kDa complex elutes from K562 cell lysates applied sequentially to immobilized S-AGzmA and S400 gel filtration columns. Migration of Pharmacia gel filtration standards is indicated. (b) The SET complex, isolated from the S-AGzmA affinity column, is disrupted by purification on an anion-exchange column. The acidic SET and pp32 proteins stick to the anion-exchange column (eluate, E), but most proteins are in the flowthrough (FT). HMG2 was identified by N-terminal sequencing of a prominent 28-kDa band in the flowthrough. The identification of the indicated bands was verified by immunoblotting (not shown). (c) Immunoblots confirm the comigration of HMG2, SET, and pp32 in the S400 column fractions.
HMG2 associates with the nucleosome assembly protein SET.
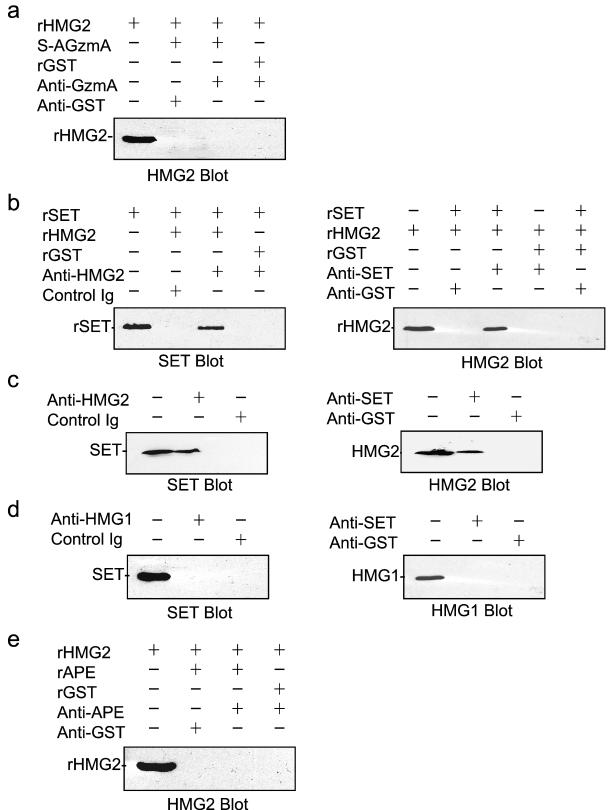
To determine the interactions between SET complex proteins, we did coimmunoprecipitation experiments with recombinant proteins. rSET, but not rpp32, coprecipitates with S-AGzmA (10). However, rHMG2 does not coprecipitate with S-AGzmA (Fig. 2a). Therefore, HMG2 is not binding to immobilized S-AGzmA by a direct interaction. However, rHMG2 coprecipitates with rSET when antibodies to either HMG2 or SET are used (Fig. 2b). Similar results are found when K562 cell lysates (Fig. 2c) or purified SET complex proteins (not shown) are used. HMG2 in cytoplasmic or nuclear lysates also binds specifically to immobilized SET and elutes with 0.24 M NaCl (data not shown). Therefore, HMG2 directly binds to SET in vitro and associates with SET in vivo.
FIG. 2.
HMG2 binds directly to SET and associates with SET in K562 cell cytosol. (a) rHMG2 does not bind directly to mutant GzmA. The positive control of S-AGzmA binding to rSET is not shown (10). (b) rHMG2 binds directly to rSET. rHMG2 and rSET were coincubated and immunoprecipitated with anti-HMG2 antisera (left) or anti-SET monoclonal antibody (right) or control antibody. (c) The experiment shown in panel b was repeated with K562 postnuclear lysates to coprecipitate native SET and HMG2. (d) However, despite the close homology between HMG1 and HMG2, HMG1 and SET do not coprecipitate from K562 cell lysates. (e) rHMG2 does not bind directly to rAPE, another SET complex protein (Fan et al., submitted).
HMG1 is highly homologous to HMG2 (78% identical and 83% similar). The major differences are in Ser or Thr residues (potential phosphorylation sites) and a longer acidic tail on HMG1. The functional distinctions between these two molecules are unclear. We therefore tested whether HMG1 coprecipitates with SET in K562 lysates (Fig. 2d). However, HMG1 does not coprecipitate with SET. HMG1 (which migrates with an apparent Mr of 29 kDa, compared to 28 kDa for HMG2) was also not detected by silver staining in the SET complex, although HMG2 was clearly visible by Coomassie blue staining (not shown). These two results suggest that HMG1 is not a component of the SET complex. The lack of binding of HMG1 is a good specificity control for the HMG2 coprecipitation experiments.
Experiments analogous to those for the interaction of HMG2 with SET were performed for the interaction with pp32. However, neither rHMG2 nor native HMG2 in the SET complex or in cytoplasmic lysates coprecipitates with pp32, except in the presence of S-AGzmA. This is similar to what we found for the interaction of SET and pp32. The recombinant and native proteins interact only weakly in the absence of S-AGzmA, but they strongly associate in the presence of S-AGzmA. rHMG2 also does not coprecipitate with rAPE (Fig. 2e).
HMG2 distributes both in the cytoplasm and the nucleus and colocalizes with SET in a perinuclear rim.
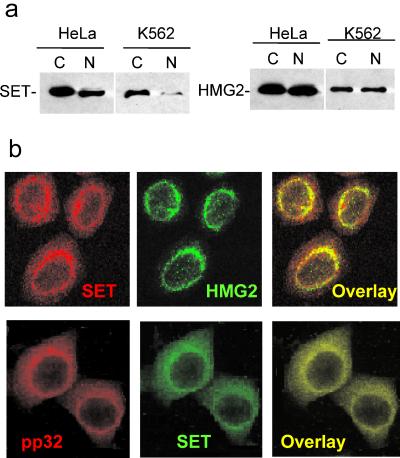
HMG2 is an abundant nonhistone chromosomal protein that has been implicated in the maintenance and establishment of chromatin structure. A major role of HMG box DNA-binding proteins is to bend DNA to facilitate the formation of complex nucleoprotein assemblies. The known functions of HMG2 suggest that it should localize to the nucleus. However, we isolated it from cytoplasmic cell lysates in association with SET, which we previously found to stain in a perinuclear rim with the ER. When postnuclear cytoplasmic supernatants and washed nuclear pellets obtained after NP-40 lysis are analyzed by SDS-PAGE and immunoblotting for SET, most of the protein is found in the cytoplasmic fraction (Fig. 3a). When the same cell fractions are analyzed for HMG2, HMG2 is equally distributed between the nucleus and cytoplasm. These results were confirmed by laser-scanning confocal microscopy using the Caltag Fix-and-Perm kit (Fig. 3b). In the cytoplasm of HeLa cells, HMG2 and SET concentrate in perinuclear regions and colocalize with the ER marker calreticulin and BiP (not shown). HMG2 cytoplasmic staining also coincides with pp32 and APE (10; Fan et al., submitted; and data not shown). However, a signal for HMG2 is clearly detected in the nucleus. This agrees well with the cell fractionation data. Similar results are found when 4% paraformaldehyde is used for fixation in place of the Caltag reagent (not shown). However, if harsher fixation conditions, such as methanol or 1% SDS, are used, the perinuclear staining for SET, pp32, and HMG2 is no longer visible and the signal is limited to the nucleus (reference 10 and data not shown). Differences in staining procedure may explain why some published immunohistochemical studies identify all of these as exclusively nuclear proteins.
FIG. 3.
HMG2 distributes between the cytosol and the nucleus and colocalizes with SET in the perinuclear regions. (a) HMG2 immunoblots of postnuclear cytosoplasmic lysate (C) and nuclear pellet (N) fractions of NP-40-lysed K562 and HeLa cells show that HMG2 is present in comparable amounts in the cytosol and the nucleus but most SET is in the cytoplasm. (b) Cytoplasmic HMG2 colocalizes with SET in HeLa cells in a perinuclear distribution. The top row shows confocal laser-scanning microscopy images of cells indirectly labeled with TRITC for SET and FITC for HMG2; the bottom row cells are stained for TRITC to pp32 and FITC to SET. All three proteins costain with the ER marker calreticulin (reference 10 and data not shown).
GzmA cleaves recombinant HMG2 at Lys65.
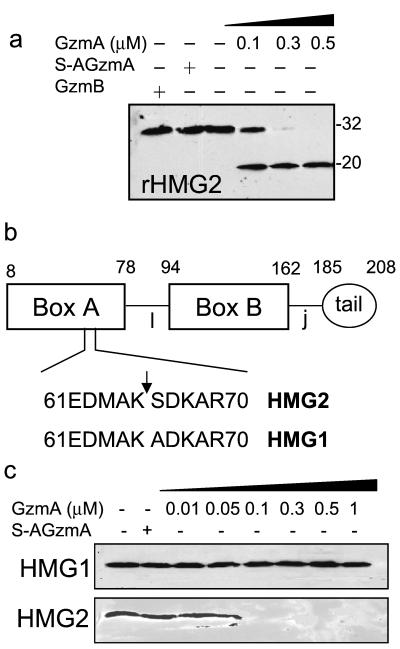
SET and APE, but not pp32, are GzmA substrates (8, 10; Fan et al., submitted). To determine whether HMG2 is a GzmA substrate, rHMG2 (1 μM) was incubated for 2 h at 37°C with 100 to 500 nM GzmA. On immunoblots probed with an antibody to a C-terminal peptide of HMG2, a 20-kDa cleavage product is clearly identified with the lowest concentration of GzmA tested and no intact full-length HMG2 is detected at the highest concentration (Fig. 4a). Neither S-AGzmA nor GzmB cuts HMG2 even when present at a 1 μM concentration. N-terminal sequencing of the 20-kDa cleavage product indicates cleavage after Lys65 in the sequence 61EDMAK SDKAR70. This cleavage site in the midst of HMG box A (aa 8 to 78) (Fig. 4b) is consistent with the tryptase activity of GzmA.
FIG. 4.
GzmA cleaves recombinant HMG2 to produce a 20-kDa product in vitro. (a) rHMG2 is cleaved by GzmA but not GzmB. rHMG2 (1 μM) was incubated for 2 h at 37°C with the indicated amounts of GzmA or 1 μM S-AGzmA or GzmB, and the reaction products were analyzed by HMG2 immunoblotting. (b) Schematic of HMG2 showing the GzmA cleavage site in HMG box A, one of the DNA binding domains of HMG2. HMG2 contains HMG boxes A and B, a linker region (l), a joining region (j), and an acidic tail. The cleavage site was identified by N-terminal sequencing of the 20-kDa cleavage fragment shown in panel a. (c) Although HMG2 is degraded after adding GzmA to cytoplasmic lysates, HMG1 in cell lysates is not cleaved by GzmA. This is despite the fact that the region around the HMG2 cleavage site differs only by a single amino acid change in the P1′ site.
HMG2, but not HMG1, in cytoplasmic lysates is cleaved by GzmA.
In the region of HMG2 cleavage, HMG1 differs by a single amino acid change in the P1 site Ser->Ala (HMG2: 61EDMAKSDKAR70 versus HMG1: 61EDMAKADKAR70; amino acid change underlined) (Fig. 4b). Although HMG1 is not in the SET complex, it might nonetheless be a substrate for GzmA. We therefore incubated cytoplasmic lysates for 2 h at 37°C with GzmA. However, when the reacted lysates are probed for HMG1 and HMG2, HMG2 is completely degraded in the presence of 100 nM GzmA, but there is no evidence of HMG1 cleavage, even with 1 μM GzmA (Fig. 4c). The difference in the cleavages of HMG1 and HMG2 suggests that GzmA proteolytic activity is highly specific and that the P′ region may be important in GzmA activity, as was previously noted (10).
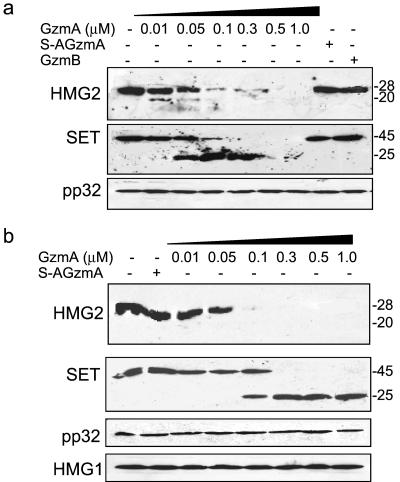
GzmA, but not GzmB, degrades HMG2 in cell cytosol and isolated nuclei.
Because GzmA directly cleaves rHMG2 in vitro and HMG2 in cell lysates, we next compared cleavage of HMG2 with cleavage of SET and determined whether GzmB might also cleave HMG2. K562 cell lysates were incubated with various amounts of GzmA, S-AGzmA, or GzmB and analyzed by immunoblotting. HMG2 was degraded after treatment of GzmA beginning at nanomolar concentrations (Fig. 5a). The same blots were stripped and reprobed for SET and pp32. SET is processed by GzmA to produce the previously reported 25-kDa fragment (9, 10) (Fig. 5), while pp32 is unchanged. Cleavage of HMG2 is detected at 10 nM GzmA, but clear cleavage of SET is not seen until a concentration of 50 nM is reached. GzmB and inactive mutant GzmA do not cleave HMG2 in cell lysates.
FIG. 5.
GzmA, but not GzmB, cleaves native HMG2 in K562 cell lysates and isolated nuclei. (a) K562 postnuclear cytoplasmic lysates (2 × 105 equivalents) were incubated with the indicated concentrations of GzmA or 1 μM S-AGzmA or GzmB at 37°C for 2 h and analyzed by immunoblotting for HMG2, SET, and pp32. HMG2 and SET are cleaved at nanomolar concentrations of GzmA, but pp32 is unchanged. (b) Increasing concentrations of GzmA or 1 μM inactive S-AGzmA was incubated with 106 K562 nuclei and nuclear lysates were analyzed 1.5 h later for cleavage of HMG2, SET, pp32, and HMG1. Full-length SET and HMG2 are completely cleaved at nanomolar concentrations of GzmA.
GzmA enters isolated nuclei in the absence of PFP (26, 68). We therefore tested whether GzmA also degrades nuclear HMG2 or SET in situ. Isolated K562 nuclei were treated with GzmA and analyzed by immunoblotting with probes for SET, pp32, APE, and HMG2. GzmA also cleaves nuclear HMG2, SET, and APE, but not HMG1 or pp32, in a dose-dependent fashion (Fig. 5b).
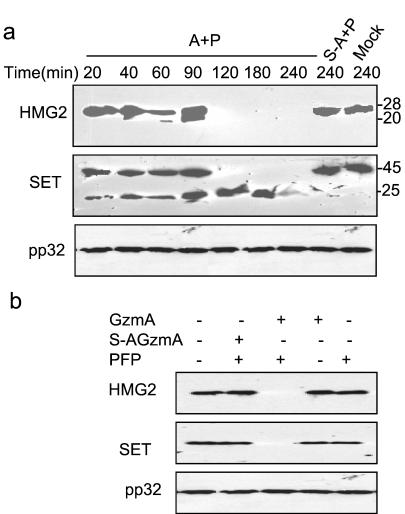
HMG2 is cleaved in cells after GzmA loading with PFP.
To investigate whether HMG2 cleavage is physiologically relevant, we looked at HMG2 degradation in the cytosol and nuclei of K562 cells loaded with GzmA and PFP (Fig. 6). Treating cells with either GzmA or PFP alone (not shown) or with mutant GzmA with PFP does not result in changes in cytosolic SET or HMG2 (Fig. 6a). Cytosolic HMG2 cleavage in loaded cells occurs within 1 h, and neither uncleaved HMG2 nor the C-terminal cleavage product can be detected within 2 h. SET cleavage, however, is evident within 20 min of loading. pp32, which is not a GzmA substrate, is unchanged and provides a good control for loading. Nuclear fractions were also analyzed 4 h after PFP-mediated GzmA loading (Fig. 6b). Nuclear HMG2 and SET were completely proteolyzed in cells loaded with active GzmA, but not in cells loaded with inactive S-AGzmA or treated with either PFP or GzmA alone. The GzmA concentration required to cleave HMG2 and SET in vivo is comparable to that required to induce cell death and DNA damage (9). Therefore, HMG2 is a direct physiological substrate of GzmA in vivo.
FIG. 6.
HMG2 is cleaved in cells perforin loaded with GzmA. (a) K562 cells were treated for the indicated times at 37°C with 1 μM GzmA (A) or 1 μM S-AGzmA (S-A) and sublytic concentrations of perforin (P). Cell lysates were analyzed by immunoblotting for HMG2, SET, and pp32. Neither full-length HMG2 nor SET are detected after 2 h, but pp32 is unchanged. (b) Nuclear fractions were isolated 4 h after GzmA loading with perforin and analyzed by immunoblotting. Nuclear SET and HMG2 are completely degraded by active, but not inactive, enzyme in a perforin-dependent manner.
The DNA binding and bending activities of HMG2 are disrupted by GzmA treatment.
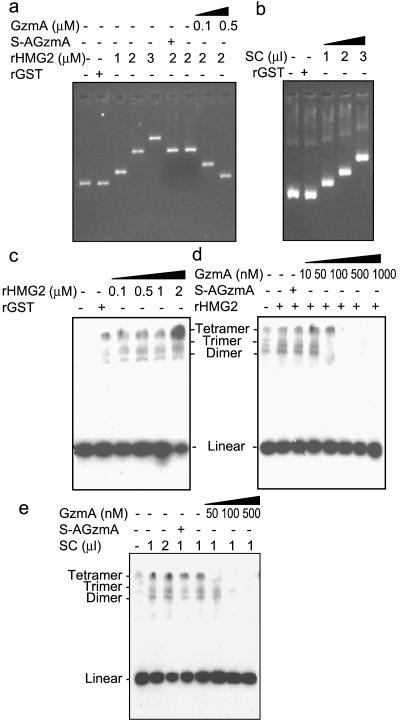
HMG2 plays an architectural role facilitating the formation of nucleoprotein assemblies and has been implicated in almost all DNA processes, including DNA repair, replication, recombination, transcription, and nucleosome assembly and disassembly (14). HMG1 and HMG2 bind double- and single-stranded DNA without sequence specificity but preferentially bind to distorted DNA, such as cis-platinum DNA adducts and supercoiled and cruciform structures, and mediate DNA bending and unwinding (23, 36, 42). Although box A or box B fragments of HMG2 can independently bind DNA to some extent, truncational mutational analysis has shown that DNA bending, binding, and unwinding are synergistically enhanced when both boxes are joined (35). Since GzmA cleavage towards the C-terminal end of box A disrupts box A and separates it from box B, we would predict that DNA binding and bending by HMG2 would be impaired after GzmA treatment. DNA gel retardation was carried out to analyze the effect of GzmA on binding of recombinant HMG2 to DNA. With increasing amounts of rHMG2, but not with the control protein rGST, DNA mobility was progressively retarded (Fig. 7a). The purified SET complex also possesses DNA binding activity (Fig. 7b). Preincubation of rHMG2 with GzmA inhibits its DNA binding in a dose-dependent manner. As expected, inactive S-AGzmA has no such effect (Fig. 7a).
FIG. 7.
GzmA disrupts HMG2 DNA binding and bending. (a) rHMG2-mediated DNA binding activity is destroyed by GzmA treatment. pBR322 DNA was incubated with the indicated amount of rHMG2 or 1 μM rGST at 25°C for 1 h. HMG2 samples were either mock treated or incubated at 37°C for 1 h with 0.1 or 0.5 μM GzmA or 0.5 μM S-AGzmA before adding to plasmid DNA. The reaction mixture was analyzed by ethidium bromide staining. (b) The SET complex (SC) contains DNA binding activity. pBR322 DNA (0.3 μg) was incubated with the indicated amount of the purified SET complex (2 μg/μl) or 1 μM rGST and analyzed as indicated in the legend to panel a. (c) rHMG2 induces DNA bending activity. 32P-labeled 123-bp DNA fragments were preincubated with different concentrations of rHMG2 or 1 μM rGST, treated with T4 DNA ligase, deproteinized, and electrophoresed through 6% nondenaturing gels. The first lane contains no ligase. (d) GzmA pretreatment of rHMG2 abolishes its DNA bending activity in a dose-dependent manner. rHMG2 (1 μM) was preincubated with the indicated amount of GzmA or 1 μM S-AGzmA at 37°C for 1 h prior to adding 32P-labeled 123-bp DNA fragments. Samples were analyzed as for panel c. (e) Similar results were obtained when the SET complex was used in place of rHMG2 in experiments described in the legends to panels c and d.
One of the most direct methods to analyze DNA bending by non-sequence-specific proteins is the ligase-mediated circularization assay, which measures the efficiency with which T4-DNA ligase forms circles from fragments of DNA that are shorter than 150 bp (36). In the absence of internal curvature, the stiffness of a short DNA fragment (<150 bp) prevents intramolecular alignment of its ends to form circles. Circles are detected only in the presence of a DNA bending protein. 32P-labeled 123-bp DNA fragments with cohesive ends were preincubated with rHMG2 and treated with T4 DNA ligase before proteinase K digestion and electrophoresis. The formation of DNA circles induced by rHMG2 was dose dependent (Fig. 7c). The control protein rGST has no DNA bending activity. Pretreatment of rHMG2 with GzmA, at GzmA concentrations as low as 100 nM, blocks DNA circularization (Fig. 7d). Similar results were obtained by using the SET complex in place of rHMG2 (Fig. 7e). The SET complex facilitates ligase-mediated circularization and this effect is abolished by nanomolar concentrations of GzmA.
DISCUSSION
HMG2 is an abundant nonhistone sequence-independent DNA binding protein involved in bending DNA for critical steps in DNA replication, transcription, and recombination (reviewed in reference 14). HMG2 binds preferentially to AT-rich regions in the minor groove in the linker regions of DNA between nucleosomes and to bent or altered DNA, such as occurs in cis-platinum adducts or four-way junctions. We now show that HMG2 associates with the ER in a 270- to 420-kDa complex containing the nucleosome assembly protein SET, the tumor suppressor protein pp32, and the base excision repair enzyme APE (10; Fan et al, submitted). HMG2 binds directly to SET. HMG2, like SET and APE, is targeted for cleavage by the CTL death-inducing protease GzmA, which induces a novel caspase-independent cell death pathway (9, 47). Both nuclear and ER-associated HMG2 are physiologically relevant targets during cytolysis induced by loading GzmA into target cells. GzmA cleaves HMG2 after Lys65 within the HMG box A, which is required for efficient DNA binding, bending, and unwinding (35). In fact, GzmA treatment of HMG2 disrupts DNA binding and circularization of small linear oligonucleotides. We previously showed that GzmA similarly disrupts the nucleosome assembly functions of SET and the apurinic endonuclease and redox functions of APE, which are important for base excision repair and activating key transcription factors (10; Fan et al., submitted). GzmA also targets important nuclear targets outside the SET complex, including histones and lamins (67, 68).
HMG1 and HMG2 are highly homologous with a similar structure consisting of two homologous DNA-binding domains (HMG boxes) A and B, each of ∼70 amino acid residues, and a long acidic C-terminal tail of ∼30 (HMG1) or 20 (HMG2) acidic residues (14) (Fig. 4b). The acidic tail may be involved in interactions with other basic proteins such as histones or in regulating DNA-binding affinity (27, 37, 51). SET and pp32 in the same complex have even longer C-terminal acidic regions (55 and 82 aa, respectively), which make them both highly acidic (calculated pIs, ∼4). These acidic regions may bind to as-yet-unidentified basic proteins in the SET complex, which would be required to balance the charge of the complex. Despite their high homology and ability to substitute for one another in most in vitro assays, HMG1 and HMG2 may not be completely redundant. For example, antisense suppression of HMG2 blocks cell cycle progression, which is not compensated for by HMG1 (27). Similarly, mice genetically deficient only in HMG1 suffer from lethal hypoglycemia (17). Many of the amino acid differences between the two proteins are in potential Ser or Thr phosphorylation sites, which suggests that the two proteins may be differentially regulated and have distinct functions. We did not identify a clear 29-kDa HMG1 band in the SET complex, and unlike HMG2, HMG1 does not bind to SET. Therefore HMG2 is preferentially involved in the functions of the SET complex.
Although HMG1 differs by only one amino acid residue at the P1′ site from HMG2 (Ala66 in HMG1 versus Ser66 in HMG2), HMG1 is not a substrate of GzmA. This difference in susceptibility to GzmA demonstrates the enzyme's exquisite specificity. At present eight distinct physiologically validated cleavage sites have been identified (Table 1). No obvious similarities among the substrates N terminal to the cleavage site are evident, and cleavage after either Lys or Arg appears to be equally favored. Specificity may come from tertiary structure or from P′ sites as has been previously noted (10); P′ sites are also important for GzmB substrates (53). Common (but not universal) features in the P′ region are Ser at P1′ or P2′ and a basic residue at P5′. Lack of cleavage of HMG1 supports the importance of the Ser residue in the P′ motif.
TABLE 1.
GzmA cleavage sites
| Substrate | Cleavage site | Sequencea | Reference or source |
|---|---|---|---|
| pIL-1β | R120 | DAPVR SLNCT | 25 |
| Thrombin receptor | R41 | TLDPR SFLLR | 52 |
| Histone H1 | K85 | KLGLK SLVSK | 68 |
| Histone H2b | K12 | APAPK KGSKK | 68 |
| SET | K176 | QTQNK ASRKR | 10 |
| Lamin B | R392 | VTVSR ASSSR | 67 |
| APE | K31 | KTAAK KNDKE | Fan et al., submitted |
| HMG2 | K65 | EDMAK SDKAR |
Residue in bold indicates the P1 site for GzmA cleavage.
Since both SET and APE are known to interact with DNA, it is not surprising to find HMG2 in the same complex. The coassociation of HMG2 with SET and APE may shed light on the function(s) of the SET complex. Since SET, but not APE, directly interacts with HMG2, an intriguing hypothesis is that HMG2 facilitates the nucleosome assembly function of SET. Both HMG2 and SET act in a sequence-independent manner. SET facilitation of transcription has been attributed to its postulated ability to reverse nucleosome assembly and increase DNA accessibility (30). SET and other NAP family members physically associate with the p300/CREB-binding protein family of transcriptional coactivators and with core histones (29, 46). In fact SET binding to histones blocks their acetylation (40). SET may therefore provide a link between transcriptional activators and chromatin (46). Just as HMG proteins bend DNA around an enhanceosome, HMG2 may be involved in bending DNA as part of nucleosome assembly/disassembly (55). SET was also identified as TAF-1β because of its ability to stimulate replication of an adenoviral chromatin template (31). HMG2 has also been implicated in enhancing viral DNA replication (18).
Since HMG2 binds preferentially to distorted DNA, it may help target SET and APE to sites of DNA damage where SET unwinds the damaged DNA from the nucleosome and APE participates in its repair. Little is known about the effect of chromatin structure on DNA repair. However, damaged DNA in actively transcribed genes (with more open chromatin) is preferentially repaired (12). Not surprisingly, a recent study showed that a DNA oligonucleotide containing a T(6-4)T photoproduct was repaired 10 times less efficiently in nucleosomal DNA than in naked DNA (24). Therefore, it makes sense for the rate-limiting base excision repair enzyme to have a mechanism for detecting damage within packed chromatin and for unwinding the damaged DNA for easy access for repair. Since SET and APE both facilitate transcription, it is also possible that much of the APE base excision repair occurs during transcription as the integrity of DNA is examined for bends secondary to ongoing oxidative DNA damage. The SET complex also contains an as-yet-unidentified Mg2+-dependent DNase, which induces single-stranded DNA nicks when the SET complex is incubated with GzmA and isolated nuclei (10). This DNase might also be involved in repair.
Because of the nuclear functions of the SET complex proteins, it is perhaps surprising to find that a large proportion of these proteins are in the cytoplasm associated with the ER. They may be stored there for regulated nuclear entry. In fact pp32 has both a canonical nuclear localization signal and leucine-rich regions for binding to crm1 for nuclear export. pp32 binds to crm1 and its nuclear export is inhibited by leptomycin B (13). SET and pp32 also associate in a smaller, ∼150-kDa complex in the nucleus that binds to HuR, a protein that stabilizes mRNAs with AU-rich regions (13). SET and pp32 might be exported from the nucleus with HuR-bound mRNAs. It is possible that the ER-associated SET complex might also have a posttranscriptional function.
Although most studies have identified HMG2 in the nucleus, in some cells a substantial fraction of HMG2 is cytoplasmic (16). In fact, staining for HMG-2 is highly dependent on the fixation protocol (data not shown). Interestingly, a perinuclear staining pattern of HMG1/HMG2 autoantibodies that develop in patients with refractory ulcerative colitis has been previously reported (50). Autoantibodies to HMG1 and HMG2 have also been found in patients with systemic lupus erythematosis and juvenile rheumatoid arthritis (15, 63). One theory is that autoantibodies arise to proteins and other compounds modified during apoptosis. Since HMG2 is cleaved by GzmA in T-cell-mediated cytolysis, the finding of autoantibodies to HMG2 is consistent with this theory about the origins of autoantibodies. However, HMG2 is not cleaved by GzmB or in GzmB-loaded cells, so it is unlikely to be cleaved during caspase activation.
Most GzmA substrates so far identified (histone H1, core histones, SET, APE, and HMG2) modify chromatin structure. We previously found that GzmA loading of isolated nuclei enhances the access and DNA cleavage by exogenous DNases (68). We have now described several mechanisms used by GzmA which open up chromatin for digestion by apoptotic nucleases. GzmA degrades lamins A, B, and C, disrupting the integrity of the nuclear envelope and possibly interfering with chromatin attachment at matrix attachment regions (67). GzmA also completely degrades the linker histone H1 and cuts the tails from the core histones (68). Chromatin stripped of linker histones is released from a compacted configuration to an extended state (3, 62). Trypsinization of polynucleosomal DNA, which also cuts the tails from core histones (but at somewhat different sites), further opens up compacted chromatin (2). HMG2 is a nonhistone protein that also binds to the internucleosomal linker region of DNA and to core histones (39). The interaction of HMG2 with H2AH2B histone dimers is via the acidic C terminus, whereas the N terminus is involved in the interactions with (H3H4)2 tetramers (11). GzmA cleavage of HMG2 is likely to open up chromatin and contribute to blocking de novo transcription required for the cellular repair response.
Opening up chromatin may also contribute to the observed synergy of GzmA and GzmB in inducing oligonucleosomal DNA fragmentation during CTL cytolysis (9, 33, 45). HMG2, however, has been shown to enhance the nuclease activity of the GzmB- and caspase-activated DNase DFF40 or CAD (57), and HMG2 cleavage would likely counter that effect. However, perhaps one of the homologous HMG proteins such as uncleaved HMG1 could also perform the same function of maintaining the high activity of CAD/DFF40 during CTL-mediated apoptosis.
Acknowledgments
This work was supported by Public Health Service grant AI-45587 from the National Institute of Allergy and Infectious Diseases (J.L.).
We thank Zhan Xu and David Oh for technical support. We also thank K. Nagata for supernatant from hybridoma KM1720, Z. Damuni for pp32 plasmid, I. Hickson for APE plasmid, and P. Sharp for HMG2 plasmid.
REFERENCES
- 1.Adachi, Y., G. N. Pavlakis, and T. D. Copeland. 1994. Identification and characterization of SET, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. J. Biol. Chem. 269:2258-2262. [PubMed] [Google Scholar]
- 2.Allan, J., N. Harborne, D. C. Rau, and H. Gould. 1982. Participation of core histone “tails” in the stabilization of the chromatin solenoid. J. Cell Biol. 93:285-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allan, J., P. G. Hartman, C. Crane-Robinson, and F. X. Aviles. 1980. The structure of histone H1 and its location in chromatin. Nature 288:675-679. [DOI] [PubMed] [Google Scholar]
- 4.Andrade, F., S. Roy, D. Nicholson, N. Thornberry, A. Rosen, and L. Casciola-Rosen. 1998. Granzyme B directly and efficiently cleaves several downstream caspase substrates: implications for CTL-induced apoptosis. Immunity 8:451-460. [DOI] [PubMed] [Google Scholar]
- 5.Bai, J., J. R. Brody, S. S. Kadkol, and G. R. Pasternack. 2001. Tumor suppression and potentiation by manipulation of pp32 expression. Oncogene 20:2153-2160. [DOI] [PubMed] [Google Scholar]
- 6.Barry, M., J. A. Heibein, M. J. Pinkoski, S. F. Lee, R. W. Moyer, D. R. Green, and R. C. Bleackley. 2000. Granzyme B short-circuits the need for caspase 8 activity during granule-mediated cytotoxic T-lymphocyte killing by directly cleaving Bid. Mol. Cell. Biol. 20:3781-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett, R. A., D. M. R. Wilson, D. Wong, and B. Demple. 1997. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc. Natl. Acad. Sci. USA 94:7166-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beresford, P. J., C. M. Kam, J. C. Powers, and J. Lieberman. 1997. Recombinant human granzyme A binds to two putative HLA-associated proteins and cleaves one of them. Proc. Natl. Acad. Sci. USA 94:9285-9290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beresford, P. J., Z. Xia, A. H. Greenberg, and J. Lieberman. 1999. Granzyme A loading induces rapid cytolysis and a novel form of DNA damage independently of caspase activation. Immunity 10:585-594. [DOI] [PubMed] [Google Scholar]
- 10.Beresford, P. J., D. Zhang, D. Oh, Z. Fan, E. L. Greer, M. Jaju, and J. Lieberman. 2001. Granzyme A activates an endoplasmic reticulum-associated caspase-independent nuclease to induce single-stranded DNA nicks. J. Biol. Chem. 276:43285-43293. [DOI] [PubMed] [Google Scholar]
- 11.Bernues, J., E. Espel, and E. Querol. 1986. Identification of the core-histone-binding domains of HMG1 and HMG2. Biochim. Biophys. Acta 866:242-251. [DOI] [PubMed] [Google Scholar]
- 12.Bohr, V. A., C. A. Smith, D. S. Okumoto, and P. C. Hanawalt. 1985. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40:359-369. [DOI] [PubMed] [Google Scholar]
- 13.Brennan, C. M., I. E. Gallouzi, and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bustin, M., B. Dunn, R. Gillette, E. Mendelsohn, and N. Soares. 1982. Antigenic determinants of high mobility group chromosomal proteins 1 and 2. Biochemistry 21:6773-6777. [DOI] [PubMed] [Google Scholar]
- 16.Bustin, M., and N. K. Neihart. 1979. Antibodies against chromosomal HMG proteins stain the cytoplasm of mammalian cells. Cell 16:181-189. [DOI] [PubMed] [Google Scholar]
- 17.Calogero, S., F. Grassi, A. Aguzzi, T. Voigtlander, P. Ferrier, S. Ferrari, and M. E. Bianchi. 1999. The lack of chromosomal protein HMG1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 22:276-280. [DOI] [PubMed] [Google Scholar]
- 18.Cotmore, S. F., and P. Tattersall. 1998. High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J. Virol. 72:8477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darmon, A. J., D. W. Nicholson, and R. C. Bleackley. 1995. Activation of the apoptotic protease CPP32 by cytotoxic T-cell-derived granzyme B. Nature 377:446-448. [DOI] [PubMed] [Google Scholar]
- 20.Demple, B., T. Herman, and D. S. Chen. 1991. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl. Acad. Sci. USA 88:11450-11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan, H., K. Orth, A. M. Chinnaiyan, G. G. Poirier, C. J. Froelich, W. W. He, and V. M. Dixit. 1996. ICE-LAP6, a novel member of the ICE/Ced-3 gene family, is activated by the cytotoxic T cell protease granzyme B. J. Biol. Chem. 271:16720-16724. [DOI] [PubMed] [Google Scholar]
- 22.Evans, A. R., M. Limp-Foster, and M. R. Kelley. 2000. Going APE over ref-1. Mutat. Res. 461:83-108. [DOI] [PubMed] [Google Scholar]
- 23.Farid, R. S., M. E. Bianchi, L. Falciola, B. N. Engelsberg, and P. C. Billings. 1996. Differential binding of HMG1, HMG2, and a single HMG box to cisplatin-damaged DNA. Toxicol. Appl. Pharmacol. 141:532-539. [DOI] [PubMed] [Google Scholar]
- 24.Hara, R., J. Mo, and A. Sancar. 2000. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol. 20:9173-9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irmler, M., S. Hertig, H. R. MacDonald, R. Sadoul, J. D. Becherer, A. Proudfoot, R. Solari, and J. Tschopp. 1995. Granzyme A is an interleukin 1 beta-converting enzyme. J. Exp. Med. 181:1917-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jans, D. A., L. J. Briggs, P. Jans, C. J. Froelich, G. Parasivam, S. Kumar, V. R. Sutton, and J. A. Trapani. 1998. Nuclear targeting of the serine protease granzyme A (fragmentin-1). J. Cell Sci. 111:2645-2654. [DOI] [PubMed] [Google Scholar]
- 27.Lee, K. B., and J. O. Thomas. 2000. The effect of the acidic tail on the DNA-binding properties of the HMG1,2 class of proteins: insights from tail switching and tail removal. J. Mol. Biol. 304:135-149. [DOI] [PubMed] [Google Scholar]
- 28.Li, M., A. Makkinje, and Z. Damuni. 1996. Molecular identification of I1PP2A, a novel potent heat-stable inhibitor protein of protein phosphatase 2A. Biochemistry 35:6998-7002. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto, K., K. Nagata, M. Okuwaki, and M. Tsujimoto. 1999. Histone- and chromatin-binding activity of template activating factor-I. FEBS Lett. 463:285-288. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto, K., M. Okuwaki, H. Kawase, H. Handa, F. Hanaoka, and K. Nagata. 1995. Stimulation of DNA transcription by the replication factor from the adenovirus genome in a chromatin-like structure. J. Biol. Chem. 270:9645-9650. [DOI] [PubMed] [Google Scholar]
- 31.Nagata, K., H. Kawase, H. Handa, K. Yano, M. Yamasaki, Y. Ishimi, A. Okuda, A. Kikuchi, and K. Matsumoto. 1995. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc. Natl. Acad. Sci. USA 92:4279-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagata, K., S. Saito, M. Okuwaki, H. Kawase, A. Furuya, A. Kusano, N. Hanai, A. Okuda, and A. Kikuchi. 1998. Cellular localization and expression of template-activating factor I in different cell types. Exp. Cell Res. 240:274-281. [DOI] [PubMed] [Google Scholar]
- 33.Nakajima, H., H. L. Park, and P. A. Henkart. 1995. Synergistic roles of granzymes A and B in mediating target cell death by rat basophilic leukemia mast cell tumors also expressing cytolysin/perforin. J. Exp. Med. 181:1037-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura, J., and J. A. Swenberg. 1999. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 59:2522-2526. [PubMed] [Google Scholar]
- 35.Nakamura, Y., K. Yoshioka, H. Shirakawa, and M. Yoshida. 2001. HMG box A in HMG2 protein functions as a mediator of DNA structural alteration together with box B. J. Biochem. (Tokyo) 129:643-651. [DOI] [PubMed] [Google Scholar]
- 36.Paull, T. T., M. J. Haykinson, and R. C. Johnson. 1993. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 7:1521-1534. [DOI] [PubMed] [Google Scholar]
- 37.Payet, D., and A. Travers. 1997. The acidic tail of the high mobility group protein HMG-D modulates the structural selectivity of DNA binding. J. Mol. Biol. 266:66-75. [DOI] [PubMed] [Google Scholar]
- 38.Pinkoski, M. J., U. Winkler, D. Hudig, and R. C. Bleackley. 1996. Binding of granzyme B in the nucleus of target cells. Recognition of an 80-kilodalton protein. J. Biol. Chem. 271:10225-10229. [DOI] [PubMed] [Google Scholar]
- 39.Schroter, H., and J. Bode. 1982. The binding sites for large and small high-mobility-group (HMG) proteins. Studies on HMG-nucleosome interactions in vitro. Eur. J. Biochem. 127:429-436. [DOI] [PubMed] [Google Scholar]
- 40.Seo, S., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104:119-130. [DOI] [PubMed] [Google Scholar]
- 41.Sharif-Askari, E., A. Alam, E. Rheaume, P. J. Beresford, C. Scotto, K. Sharma, D. Lee, W. E. DeWolf, M. E. Nuttall, J. Lieberman, and R. P. Sekaly. 2001. Direct cleavage of the human DNA fragmentation factor-45 by granzyme B induces caspase-activated DNase release and DNA fragmentation. EMBO J. 20:3101-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheflin, L. G., N. W. Fucile, and S. W. Spaulding. 1993. The specific interactions of HMG 1 and 2 with negatively supercoiled DNA are modulated by their acidic C-terminal domains and involve cysteine residues in their HMG 1/2 boxes. Biochemistry 32:3238-3248. [DOI] [PubMed] [Google Scholar]
- 43.Shi, L., G. Chen, G. MacDonald, L. Bergeron, H. Li, M. Miura, R. J. Rotello, D. K. Miller, P. Li, T. Seshadri, J. Yuan, and A. H. Greenberg. 1996. Activation of an interleukin 1 converting enzyme-dependent apoptosis pathway by granzyme B. Proc. Natl. Acad. Sci. USA 93:11002-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi, L., C. M. Kam, J. C. Powers, R. Aebersold, and A. H. Greenberg. 1992. Purification of three cytotoxic lymphocyte granule serine proteases that induce apoptosis through distinct substrate and target cell interactions. J. Exp. Med. 176:1521-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi, L., R. P. Kraut, R. Aebersold, and A. H. Greenberg. 1992. A natural killer cell granule protein that induces DNA fragmentation and apoptosis. J. Exp. Med. 175:553-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shikama, N., H. M. Chan, M. Krstic-Demonacos, L. Smith, C. W. Lee, W. Cairns, and N. B. La Thangue. 2000. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell. Biol. 20:8933-8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shresta, S., T. A. Graubert, D. A. Thomas, S. Z. Raptis, and T. J. Ley. 1999. Granzyme A initiates an alternative pathway for granule-mediated apoptosis. Immunity 10:595-605. [DOI] [PubMed] [Google Scholar]
- 48.Shykind, B. M., J. Kim, and P. A. Sharp. 1995. Activation of the TFIID-TFIIA complex with HMG-2. Genes Dev. 9:1354-1365. [DOI] [PubMed] [Google Scholar]
- 49.Smyth, M. J., J. M. Kelly, V. R. Sutton, J. E. Davis, K. A. Browne, T. J. Sayers, and J. A. Trapani. 2001. Unlocking the secrets of cytotoxic granule proteins. J. Leukoc. Biol. 70:18-29. [PubMed] [Google Scholar]
- 50.Sobajima, J., S. Ozaki, F. Osakada, H. Uesugi, H. Shirakawa, M. Yoshida, and K. Nakao. 1997. Novel autoantigens of perinuclear anti-neutrophil cytoplasmic antibodies (P-ANCA) in ulcerative colitis: non-histone chromosomal proteins, HMG1 and HMG2. Clin. Exp. Immunol. 107:135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stros, M., J. Stokrova, and J. O. Thomas. 1994. DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res. 22:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suidan, H. S., J. Bouvier, E. Schaerer, S. R. Stone, D. Monard, and J. Tschopp. 1994. Granzyme A released upon stimulation of cytotoxic T lymphocytes activates the thrombin receptor on neuronal cells and astrocytes. Proc. Natl. Acad. Sci. USA 91:8112-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun, J., J. C. Whisstock, P. Harriott, B. Walker, A. Novak, P. E. Thompson, A. I. Smith, and P. I. Bird. 2001. Importance of the P4′ residue in human granzyme B inhibitors and substrates revealed by scanning mutagenesis of the PI-9 reactive center loop. J. Biol. Chem. 267:15177-15184. [DOI] [PubMed] [Google Scholar]
- 54.Talanian, R. V., X. H. Yang, J. Turbov, P. Seth, T. Ghayur, C. A. Casiano, K. Orth, and C. J. Froelich. 1997. Granule-mediated killing: pathways for granzyme B-initiated apoptosis. J. Exp. Med. 186:1323-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thanos, D., W. Du, and T. Maniatis. 1993. The high mobility group protein HMG I(Y) is an essential structural component of a virus-inducible enhancer complex. Cold Spring Harbor Symp. Quant. Biol. 58:73-81. [DOI] [PubMed] [Google Scholar]
- 56.Thomas, D. A., C. Du, M. Xu, X. Wang, and T. J. Ley. 2000. DFF45/ICAD can be directly processed by granzyme B during the induction of apoptosis. Immunity 12:621-632. [DOI] [PubMed] [Google Scholar]
- 57.Toh, S. Y., X. Wang, and P. Li. 1998. Identification of the nuclear factor HMG2 as an activator for DFF nuclease activity. Biochem. Biophys. Res. Commun. 250:598-601. [DOI] [PubMed] [Google Scholar]
- 58.Trapani, J. A., K. A. Browne, M. J. Smyth, and D. A. Jans. 1996. Localization of granzyme B in the nucleus. A putative role in the mechanism of cytotoxic lymphocyte-mediated apoptosis. J. Biol. Chem. 271:4127-4133. [DOI] [PubMed] [Google Scholar]
- 59.Vaesen, M., S. Barnikol-Watanabe, H. Gotz, L. A. Awni, T. Cole, B. Zimmermann, H. D. Kratzin, and N. Hilschmann. 1994. Purification and characterization of two putative HLA class II associated proteins: PHAPI and PHAPII. Biol. Chem. Hoppe-Seyler 375:113-126. [DOI] [PubMed] [Google Scholar]
- 60.von Lindern, M., S. van Baal, J. Wiegant, A. Raap, A. Hagemeijer, and G. Grosveld. 1992. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol. Cell. Biol. 12:3346-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walensky, L. D., D. S. Coffey, T. H. Chen, T. C. Wu, and G. R. Pasternack. 1993. A novel M(r) 32,000 nuclear phosphoprotein is selectively expressed in cells competent for self-renewal. Cancer Res. 53:4720-4726. [PubMed] [Google Scholar]
- 62.Weintraub, H., and F. Van Lente. 1974. Dissection of chromosome structure with trypsin and nucleases. Proc. Natl. Acad. Sci. USA 71:4249-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wittemann, B., G. Neuer, H. Michels, H. Truckenbrodt, and F. A. Bautz. 1990. Autoantibodies to nonhistone chromosomal proteins HMG-1 and HMG-2 in sera of patients with juvenile rheumatoid arthritis. Arthritis Rheum. 33:1378-1383. [DOI] [PubMed] [Google Scholar]
- 64.Xanthoudakis, S., and T. Curran. 1992. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 11:653-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia, Z., C. M. Kam, C. Huang, J. C. Powers, R. J. Mandle, R. L. Stevens, and J. Lieberman. 1998. Expression and purification of enzymatically active recombinant granzyme B in a baculovirus system. Biochem. Biophys. Res. Commun. 243:384-389. [DOI] [PubMed] [Google Scholar]
- 66.Yang, X., H. R. Stennicke, B. Wang, D. R. Green, R. U. Janicke, A. Srinivasan, P. Seth, G. S. Salvesen, and C. J. Froelich. 1998. Granzyme B mimics apical caspases. Description of a unified pathway for trans-activation of executioner caspase-3 and -7. J. Biol. Chem. 273:34278-34283. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, D., P. J. Beresford, A. H. Greenberg, and J. Lieberman. 2001. Granzymes A and B directly cleave lamins and disrupt the nuclear lamina during granule-mediated cytolysis. Proc. Natl. Acad. Sci. USA 98:5746-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, D., M. S. Pasternack, P. J. Beresford, L. Wagner, A. H. Greenberg, and J. Lieberman. 2001. Induction of rapid histone degradation by the cytotoxic T lymphocyte protease granzyme A. J. Biol. Chem. 276:3683-3690. [DOI] [PubMed] [Google Scholar]