Abstract
Rb+/+:Rb−/− chimeric mice are healthy until early in adulthood when they develop lethal pituitary tumors composed solely of Rb−/− cells. In an effort to delineate the minimal structures of the retinoblastoma protein necessary for RB tumor suppression function, chimeric animals derived from stably transfected RB−/− embryonic stem (ES) cells were generated. One such ES cell transfectant expressed a human RB allele encoding a stable, truncated nuclear derivative lacking residues 1 to 378 (Δ1-378). Others encoded either wild-type human RB or an internally deleted derivative of the Δ1-378 mutant. All gave rise to viable chimeric animals with comparable degrees of chimerism. However, unlike control mice derived, in part, from naive Rb−/− ES cells or from ES cells transformed by the double RB mutant, Δ1-378/Δexon22, animals derived from either wild-type RB- or Δ1-378 RB-producing ES cells failed to develop pituitary tumors. Thus, in this setting, a substantial fraction of the RB sequence is unnecessary for RB-mediated tumor suppression.
Considerable knowledge now exists of the functional properties of the retinoblastoma protein (pRB; for reviews, refer to references 6, 11, 45, and 46). pRB is believed to execute its proliferation and differentiation functions, at least in part, by regulating the transcription of selected genes at the hands of two or more distinctive mechanisms. RB interacts with certain members of the E2F transcription factor family. In so doing, it stabilizes these proteins, suppresses their transcription activation function, and engages them in the formation of transcriptional repression complexes (11, 16, 18). The latter contain both a transcriptional corepressor and a histone deacetylase (7, 27, 29, 30, 47). RB also interacts with other proteins, including the oncoprotein MDM2, numerous transcription factors, and at least one member of the SWI/SNF family (10, 43). These biologically important interactions reflect a role for RB-mediated control of gene expression through transcription modulation, including that mediated through influences on the structure and remodeling of chromatin (6).
The results of limited proteolysis and X-ray analysis demonstrate that RB is a globular protein consisting of discrete structural domains (26). One of them is the A/B pocket, consisting of an A domain (residues 379 to 572), a spacer unit (residues 573 to 645), and a B domain (residues 646 to 772), which, together with the C-terminal domain (amino acids [aa] 768 to 928), serve as an interaction center for numerous proteins, including certain E2F species and DNA tumor viral oncoproteins.
Some human tumor-inducing RB mutations map to the pocket domain and encode proteins, the pocket domains of which are incapable of target protein binding. Therefore, the pocket is an important contributor to RB tumor suppression function. When linked to the above-noted C-terminal domain, it is also sufficient for RB-dependent proliferation inhibition, differentiation control, and the stable binding of E2F proteins and transcriptional corepressing elements (7). The C-terminal domain is also essential for interactions with D-type cyclins and MDM2, both important RB binding proteins (12, 15, 50). Naturally occurring, tumor-associated mutations have also been detected in this portion of RB. For example, in a case of acute lymphocytic leukemia, a tumor-associated deletion mutation mapping to exons 24 and 25 was detected (14). This C-terminal region also contains a nuclear localization signal (residues 860 to 876 [53]) and a cyclin A/cdk2 interaction domain from which important elements of the RB phosphorylation process are directed (1, 15). The segment of the protein containing the A/B pocket and C terminus has often been referred to as the large pocket (LP).
By contrast with the pocket and C-terminal domains, the N-terminal 378 residues (N terminus) are dispensable for RB growth suppression function (34). In keeping with this result, overexpression of a truncated RB species lacking this region in RB−/− human tumor cells led to overt suppression of their proliferation (52). Indeed, this particular mutant RB species appeared to be more potent than the full-length protein in these experiments (52).
Other findings relate to the question of whether this N-terminal structural unit plays a role in RB-mediated tumor suppression. First, there are naturally occurring RB mutant alleles in human tumors bearing discrete, internal mutations in the N-terminal segment, including deletion of sequences encoded by exon 4 (aa 127 to 146 [9]) and exon 8 (aa 240 to 287 [17, 25]). Secondly, there is evidence suggesting that mutations in the N terminus in an otherwise intact RB transgene compromise RB tumor suppression function in Rb knockout mice (35). One interpretation of these findings is that the N-terminal segment, while unimportant for RB-mediated proliferation suppression in cultured cells, is, nonetheless, a major contributor to its tumor suppression function. Another is that there is a negative effect of the above-noted N-terminal region mutants on one or more elements of the tumor-suppressing function(s) of the LP segment of RB. In considering which RB functions are material to its tumor-suppressing function and which, if any, are not, the question of the contribution versus noncontribution of the 1-378 segment of RB—approximately 40% of the protein sequence--is a significant one.
Although limited in quantity, there is also evidence pointing to specific biochemical and biological functions of the 1-378 region of RB. For example, RB interacts with Sp1 in vivo, resulting in superactivation of Sp1-mediated transcription (44). In addition to a contribution of the pocket, a segment of the N terminus (residues 140 to 202) is also required for this effect (44). Stimulation of human insulin receptor gene expression by RB also requires an amino-terminal sequence (40). A cell cycle-regulated RB and histone H1 kinase (known as RbK), whose enzymatic and RB association activities are most prevalent in G2/M, also interacts with the RB N-terminal region (42). In addition, there is evidence indicating that a member of a family of proteins dedicated to the initiation of chromosomal DNA replication, MCM7, also interacts with the 1-378 region (41). This region also contains two BRCT motifs, a signature of proteins dedicated, at least in part, to genome integrity and/or proliferation control and, where studied, a major participant in the function of such proteins (4). Therefore, the RB amino-terminal region exhibits multiple biochemical activities. What is less clear is how they relate to the known biological functions of the protein and, in particular, to its tumor suppression function.
We have chosen to assess the question of whether this segment contributes to RB tumor suppression by generating Rb+/+:Rb−/− chimeric mice in which the Rb−/− cells express no RB or synthesize ectopically encoded wild-type (wt) or specifically truncated RB proteins at similar levels. The latter encode stable products devoid of the 1-378 region. The question was whether the absence of this segment from an expressed RB transgene in certain Rb−/− cells affects their tumorigenic fate.
MATERIALS AND METHODS
Construction of plasmids.
The Rb-1 plasmid was obtained from Philip Hinds. It was cleaved with XbaI and SmaI to release a 1.6-kb fragment containing the human RB (hRB) promoter, which was then recloned in an expression vector, pCMV-Rb, to replace the cytomegalovirus promoter. The engineered recombinant encodes hRB under the control of the hRB promoter. This vector was linearized with HindIII and purified for electroporation.
Generation of ES cells and blastocyst injection.
ES2 cells (Rb−/−:LacZ+) were electroporated with a Bio-Rad GenePulser at 600 V and 25 μF. Transfected cells were plated at 106 cells/ml on gelatinized plastic tissue culture plates. Embryonic stem (ES) cells were cultivated in Dulbecco's modified Eagle medium supplemented with 15% fetal bovine serum (ES cell grade; Gibco) and leukemia inhibitory factor (1,000 U/ml; Gibco). Ten to twelve ES cells were injected into each C57BL16 blastocyst to create chimeric animals.
PCR primers.
HAYL1 (5′ GATCTTCCTCATGCTGTT 3′) and HAYR1 (5′ CTCTTCCTTGTTTGAGGT 3′) span exon 22 of the human Rb gene. Their use led to the amplification of fragments from vectors encoding full-length RB, RB-LP, and RB-LPΔ22.
IP and Western blotting.
Tissues were collected and frozen briefly in liquid nitrogen. Upon thawing, they were homogenized in EBC buffer (180 mM NaCl, 10 mM Tris-HCl, pH 8.0, and 0.1% Nonidet P-40) plus protease inhibitors (1 mM phenylmethysulfonyl fluoride, 1 μg of aprotinin/ml, 10 μg of leupeptin/ml, and 1 μg of pepstatin/ml) and phosphatase inhibitors (10 mM NaF and 1 mM sodium orthovanadate). A monoclonal RB antibody, XZ 56, was used in immunoprecipitation (IP) and Western blot analysis, as previously described (19).
Histological analysis of animal tissues.
After sacrifice, animals were perfused with freshly prepared 4% paraformaldehyde (pH 7.4) for 30 to 60 min. Tissues were then removed and dehydrated in alcohol, embedded in paraffin, and sectioned with a microtome at 5 μm and stained with hematoxylin and eosin. For staining with 5-bromo-4-chloro-3-indolylphosphate (X-Gal), tissues were embedded in OCT (Tissue-Tek), frozen in liquid nitrogen, and sectioned at 5 to 10 μm. X-Gal staining was performed in phosphate-buffered saline with 1 mg of X-Gal/ml, 4 mM potassium ferrocyanide, 4 mM potassium ferricyanide, and 2 mM magnesium chloride. Slides were counterstained with nuclear fast red to demonstrate nuclei.
RESULTS
Construction of vectors and generation of ES cell lines.
Rb−/− ES cells that also express a lacZ allele were generated from a cross between ROSA26 mice (8) and Rb+/− mice (22). Rb−/− blastocysts were collected and cultivated on feeder layers to generate Rb−/−:LacZ+ ES cells. Two independent lines, ES2 and ES4, were obtained, and the fates of chimeric mice generated from both lines were essentially indistinguishable from one another and from that of animals analyzed in a previous study of chimeric Rb−/−:Rb+/+ mice (49). Moreover, both ES2 and ES4 contributed extensively to multiple tissues (B. O. Williams and T. Jacks, unpublished results). In this study, we used ES2-derived cells to generate all chimeric mice. Use of similar ES cells has been reported previously (26a).
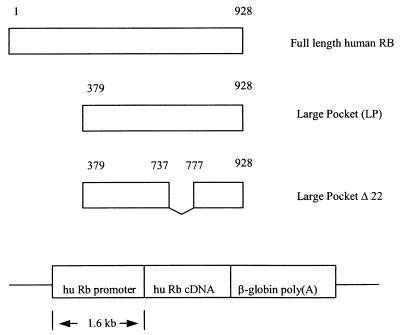
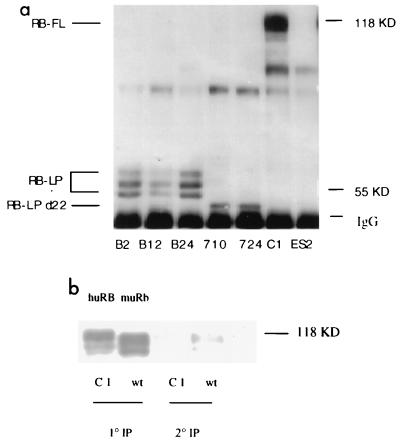
For the preparation of ES2 cells stably transformed by various hRB alleles, expression vectors expressing each of a group of hRB alleles (hpRB) were created. These vectors encoded hpRB (Δ1-378, so-called human LP), hpRB (379-928, Δexon 22, so-called LPΔ22), and full-length hpRB (wt; Fig. 1). A previously characterized hRB promoter was used to direct the expression of these hRB alleles in an effort to approximate a nearly physiologic expression level (Fig. 1). This promoter unit can drive RB transcription in a manner resembling that of the endogenous murine Rb promoter, both spatially and temporally (3). Following their transfection, hygromycin selection of stably transformed clones was undertaken. Drug-resistant clones were first screened by PCR for the relevant human pRB allele and then for its expression by IP and Western blotting (Fig. 2a). Independent clones containing equivalent quantities of RB wt, LP, and LPΔ22 were identified (Fig. 2a). Clones B2, B12, and B24 synthesize RB LP; 710 and 724 synthesize RB LPΔ22; and C1 synthesizes full-length, wt hRB protein.
FIG. 1.
The 1.6-kb hRb promoter directs the expression of hRB cDNAs. A 1.6-kb hRB fragment encoding the hRB promoter has been characterized previously (3). It can faithfully direct the expression of hRb cDNA in mouse tissues. Full-length cDNAs corresponding to human Rb, hRB 379-928 (LP), and hRB 379-928 (Δ738-776, so-called LPΔ22) were cloned downstream of this hRB promoter-containing fragment, and the ensuing recombinant plasmids were transfected or electroporated into Rb−/− ES cells.
FIG. 2.
(a) Expression of various hRB transgenes in ES cell lines. Cell extracts in EBC buffer were immunoprecipitated with the monoclonal hRB antibody XZ 56 (21). Immunoprecipitated proteins were resolved in a sodium dodecyl sulfate-8.5% polyacrylamide gel and Western blotted with the same antibody. An equivalent amount (500 μg) of each cell lysate was used in each IP. Clones B2, B12, and B24 express RB-LP. Clones 710 and 722 express RB-LPΔ22, and clone C1 expresses full-length hRB (RB-FL). ES2 is the parental Rb−/− ES cell line. KD, kilodaltons. (b) Comparison between expression levels of hRB (huRB)-expressing Rb−/− ES cells and endogenous mouse Rb (muRb) in wt ES cells. C1 ES cells (expressing hRB in a Rb−/− background) or wt ES cells were lysed in EBC buffer, and the same amount of ES cell lysate (300 μg) from either cell line was analyzed by serial IP. First we immunoprecipitated with an excess (2 μg) of G3-245 antibody, which was raised against human pRB (recognizes an epitope located between aa 332 and 344) and cross-reacts with murine pRb. After one round of precipitation with antibody-coated beads, the supernatants were reimmunoprecipitated with 2 μg of G3-245 to determine the amount of RB remaining in the supernatant after the first IP. Immunoprecipitated proteins were then analyzed by Western blotting. The blot was probed with G3-245 (Pharmingen).
A wt ES clone that synthesizes only endogenous wt murine Rb and the Rb−/−, stably transfected ES clone that synthesizes only wt hRB were analyzed, in parallel, for RB expression. Specifically, each culture was lysed, and the same amount of total protein in each case was then subjected to IP with an excess of the RB monoclonal antibody, G3-245. The supernatants from the first round of IPs were then subjected to another round of G3-245 IP. This antibody is known to react efficiently with both human and mouse RB, proteins that are 91% identical in primary sequence (2). As shown in Fig. 2b, equivalent quantities of murine Rb and hRB protein were precipitated from each test culture in the above-noted, serial IP protocol. These data indicate that the level of hRB expression was similar to that of endogenous wt murine Rb expression in the relevant ES cell lysates. Since the expression levels of all of the various hRB proteins described in this report were similar (Fig. 2a), this result indicates that the expression levels of the ectopically encoded RB proteins were in the physiologic range.
Generation of chimeric mice expressing various ectopic hpRB alleles.
Chimeras were readily generated with each of the above-noted clones. The degree of chimerism was judged by coat color and by staining tissue sections of various organs with X-Gal and scoring the relative percentage of β-Gal-positive and -negative cells. Twenty-three LP (i.e., hpRB[Δ1-378]), 26 LPΔ22 (i.e., hRB-LPΔ22), 22 wt (i.e., hpRB-FL), and 15 control Rb−/− chimeric animals were created in this study. We also generated nine control Rb+/+:Rb+/+ chimeric animals by introducing BS1 ES cells (murine Rb+/+:lacZ) into wt C57BL/6 blastocysts. LacZ-positive and -negative cells in the liver and kidneys of various chimeric animals were quantitated, and, from these data, the approximate degree of chimerism in this control setting was also determined. On repeated occasions, the animals in each genotype group were approximately 60 to 70% chimeric with respect to coat color.
Expression of hRB transgenes in chimeric tissues.
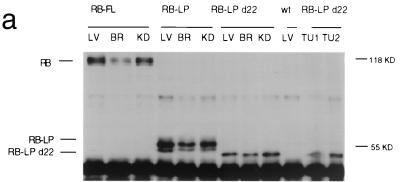
To determine whether hRB proteins were synthesized in the relevant chimeric animals, we analyzed certain tissues by reverse transcriptase PCR (RT-PCR) (Fig. 3) and standardized Western blotting (Fig. 4a). It was previously shown that the hRB promoter fragment used in transgenic animals could direct physiologic RB expression in murine tissues (3). In keeping with this observation, we found that, in all of the tissues examined, including brain, liver and kidney, there were detectable quantities of the relevant hRB protein (Fig. 4a), both wt and mutant, reflecting a common pattern of expression of each transgene under investigation.
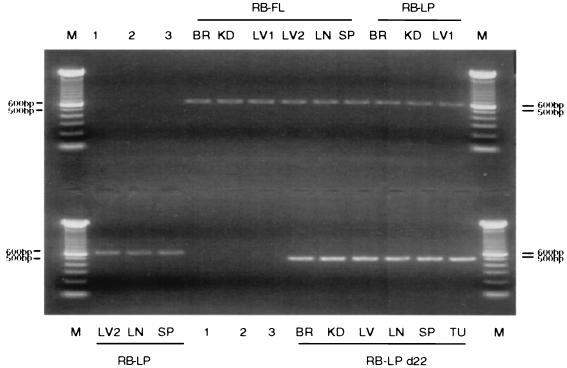
FIG. 3.
Expression of transgene-encoded hRB mRNAs in mouse tissues. Relevant transgenic animals were sacrificed, and their tissues were excised. Poly(A)+ RNA was extracted from each. A standardized quantity of each RNA sample was then subjected to an RT-PCR assay in search of RNA templates that reflect the inherent hRB transgene in the mice of interest. Pituitary tumors were observed in animals that were Rb+/+:LPΔ22 in which the tumor cells arose from ES cells that had been stably transformed with an RB-LPΔ22 vector. The PCR primers spanning exon 22 amplify a 620-bp fragment for RB-FL and RB-LP and a 506-bp fragment for RB-LPΔ22. M denotes the 100-bp ladder marker (the brightest single band is 600 bp). The sources of RNA were as follows: BR, brain; KD, kidney; LV, liver; LN, lung; SP, spleen; and TU, pituitary tumor. Negative controls are as follows: lane 1, no RNA input in RT-PCR assay; lane 2, no RT present in the RT-PCR; and lane 3, no DNA input was present in the PCR.
FIG. 4.
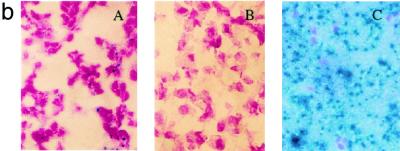
(a) Expression of mutant RB proteins in chimeric murine tissues. Mouse tissues were homogenized in the presence of EBC buffer, as described in Materials and Methods. Standardized quantities of each lysate were then subjected to IP and Western blotting with XZ 56, which recognizes hRB specifically (21). wt refers to wt control animals (which do not carry a transgene). LV, liver; BR, brain; KD, kidney; TU, pituitary tumor; KD, kilodaltons. (b) Staining of pituitary glands from wt and hRB-LP animals. Pituitary glands were fixed with 4% freshly prepared paraformaldehyde solution and stained with X-Gal solution. The tissue segments were then sectioned and counterstained with nuclear fast red, as described in Materials and Methods. Blue-staining intermediate lobe cells were derived from ROSA-26 LacZ-positive ES cells, while, in nonchimeric, wt tissue, there was no detectable X-Gal activity as expected. Photograph A, intermediate lobe tissue from an hRB-LP chimera; photograph B, similar tissue from a C57BL16 control mouse; and photograph C, X-Gal-stained section from a pituitary tumor arising in an hRB-LP Δ22-expressing chimeric mouse. The nuclei stained red. X-Gal staining was dot-like in the relevant normal intermediate lobe tissue and was diffuse in the spongiform pituitary tumor tissue.
Tumor development in transgene- and non-transgene-bearing chimeric animals.
The major phenotypes observed in Rb−/− chimeric mice are cataracts and pituitary tumors (28, 49). The same type of pituitary malignancy has been reproducibly observed in Rb+/− animals with high penetrance (20). The first clinical detection of these tumors in Rb+/− animals occurred between 2 and 10 months (20). With chimeric Rb+/+:Rb−/− mice, clinical tumor detection and death occurred within a rather narrow period (i.e., 4 to 6 months), in keeping with the original reports of such animals (28, 49).
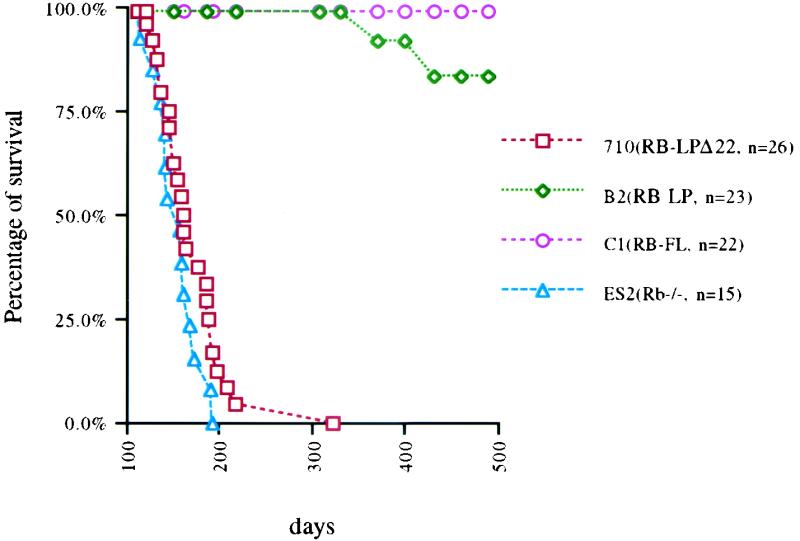
After birth, the various hpRB chimeric mice, described above, were followed closely for tumor development and mortality. Twenty-six hRB-LPΔ22 chimeras and a comparable number of Rb+/+:Rb−/− chimeric animals generated with the same clone of Rb−/− ES cells (untransfected) all developed pituitary tumors with similar kinetics (Fig. 5), implying that this Δ22 allele is defective in its tumor suppression function. Importantly, when analyzed by Western blotting, the LPΔ22 transgene was expressed in all relevant tumors.
FIG. 5.
Survival curves of chimeric animals of various RB genotypes. Mice were followed for up to 18 months. Mice generated from RB-LP-expressing chimeric animals were labeled B2; those from RB-LPΔ22 animals were labeled 710; and those from wt RB were labeled C1. ES2 (Rb+/+:Rb−/−) chimeric mice were generated by wt blastocyst injection of the parental, Rb−/− ES cell line. n indicates the number of chimeric animals studied in each group. The percentage indicates the mice remaining alive at each time point.
By contrast, we detected no tumors or significant mortality in 22 hpRB-FL or 23 hpRB(Δ1-378) chimeras followed for up to 18 months of age. As shown earlier, there was comparable expression of the hpRB(Δ1-378), LPΔ22, and FL proteins, both in the donor ES cell clones and in the resulting chimeric tissues (Fig. 4a). To measure the degree of pituitary chimerism achieved by the progeny of wt hRB, hRB(Δ1-378), and LPΔ22 transgene-containing ES cells, the intermediate lobe regions of the relevant pituitaries were stained with X-Gal (see examples in Fig. 4b). The results indicated that ∼20% of each stained positively. Thus, lack of pituitary tumors in the hpRB(Δ1-378) animals is not due to the absence of ES-derived pituitary cells. Rather, one can argue that hpRB(Δ1-378) provided specific protection against tumorigenesis in this organ.
Cataracts in transgene-bearing chimeric animals.
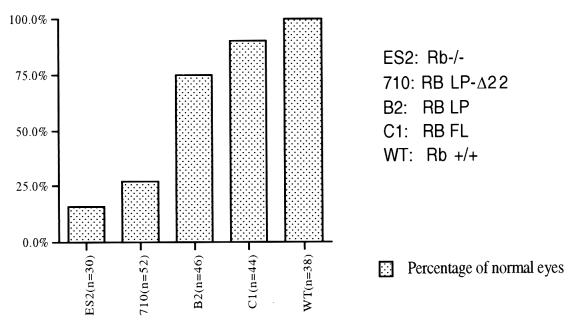
In an effort to detect other clinical effects of a major defect in RB function, we scored for the presence of cataracts, a hallmark of unchecked proliferation of lens cells and ensuing death due to lack of normal RB function (31, 49). In Rb−/− chimeras, cataracts form very early after birth and can be detected as early as 4 weeks of age (Fig. 6). Both the wt hRB and hRB(Δ1-378) transgenes suppressed cataract formation, the former more efficiently than the latter. By contrast, there was dramatic cataract formation in the LPΔ22 animals, with bilateral lesions routinely observed. Thus, a parallel was detected between cataract formation and tumor development in these animals.
FIG. 6.
Fraction of the lenses of RB chimeric mice that were cataract free. Cataracts are hallmarks of unchecked proliferation and apoptosis in Rb−/− embryos and Rb+/+:Rb−/− chimeric animals (31). hRB-LP effectively suppressed the occurrence of cataracts in chimeric animals. The identity labels refer to mice of a defined genotype and were the same used in the experiment reported for Fig. 5. n indicates the total number of lenses examined from each line of chimeric mice.
No other gross anatomical defects in the chimeric animals.
In search of yet other forms of pathology, we examined multiple tissues, including heart, kidney, liver, lung, gastrointestinal tract, and the nervous systems of the hRB(Δ1-378), hRB-LPΔ22, and wt hRB chimeras, and failed to detect any gross anatomical defects, in agreement with previous reports (28, 49). One case of pheochromocytoma was detected in a single hRB-LPΔ22 mouse of the 26 animals generated. Since no such tumors were detected in control Rb+/+:Rb−/− animals, it was likely unrelated to the absence of intact RB protein in the Rb−/− cells.
DISCUSSION
Certain Rb−/− cells of Rb+/+:Rb−/− chimeric mice are at high risk of developing a neoplastic phenotype and emerging as lethal tumors. In particular, with very high penetrance these animals develop malignant pituitary tumors by ∼4 to 6 months of age. Genetic reconstitution of these cells with a wt RB allele or with a mutant RB gene encoding a product lacking residues 1 to 378 effectively suppressed this phenotype. Suppression was pocket domain dependent, since a double mutant lacking both 1-378 and a small, discrete segment of the pocket failed to suppress tumor formation. wt RB and both mutant derivatives were all equivalently expressed in the relevant ES cells and in murine tissues of the ensuing chimeric animals. Therefore, the observed effects are not a product of imbalances in ectopic gene expression. Moreover, the abundance of cells in the intermediate lobe of the pituitary that were derived from Rb−/− ES cells was similar in animals of the relevant chimeric genotypes. Therefore, the differences in clinical outcome—pituitary tumor versus no tumor—cannot be ascribed to differences in pituitary chimerism levels.
Given these observations, it seems reasonable to conclude that the 1-378 segment is dispensable for tumor suppression activity, at least in pituitary cells of 129/Sv origin. It is also dispensable for the normal suppression of unregulated lens cell proliferation, for both RB-LP and the wt protein suppressed cataract formation in the mice analyzed in this study.
These conclusions stand in contrast to those of Riley et al. (35), who found that, unlike their wt counterpart, human pRB transgenes bearing certain internal deletion mutations within the N-terminal region failed to suppress pituitary tumor formation in Rb+/− mice. In considering the significance of these findings, it should be noted that one of the N-terminal mutations reported (35) negatively affects pocket domain function (Δ76-181 [P. W. Hinds, unpublished results]). Moreover, others have reported the existence of cis-dominant negative effects of other mutations within the N-terminal 378 residues on the function of the pocket domain. Specifically, certain small, internal deletions within this 378-residue region clearly compromised RB proliferation suppression and E1A binding activities, both pocket-dependent functions (33). Furthermore, deletion of exon 4 (encodes residues 127 to 146) abolished E2F binding activity by the pocket domain, while complete ablation of the amino-terminal region (residues 1 to 378) did not affect this activity (38). Given these observations, one wonders whether certain N-terminal region mutants that fail to rescue RB tumor suppression function have acquired an ability to compromise RB pocket function in cis and, therefore, RB tumor suppression activity. In that light, it might be difficult to conclude from a genetic study in which selected N-terminal region mutations are found to be defective in pituitary tumor suppression that the N-terminal region is necessary for this RB function.
From the data described in this report, it is clear that total elimination of the N-terminal region did not affect the proliferation suppression function of the pocket domain. Similarly, it did not compromise the ability of RB(Δ1-378) to act as a transcriptional repressor, to interact with E2F:DP complexes and suppress their transactivation function, or to be phosphorylated in a cell cycle-dependent manner (1, 7, 29). The N-terminal region is also dispensable for MDM2 and c-Abl binding (48, 50). The former effect can now be linked to the ability of RB to enhance p53-mediated apoptosis, potentially a tumor-suppressing activity (19). The N-terminal region is also dispensable for the performance of at least one aspect of the differentiation regulation function of RB (13). RB mutants lacking N-terminal region sequences were competent to elicit differentiating effects in RB−/− osteogenic sarcoma cells and in myoblasts, the latter in the presence of myoD (13, 39).
What, then, is the significance of the observation that tumor suppression does not depend upon a functional contribution by a large segment of the protein? First, none of the studies reported to date have tested a full range of RB tumor suppression activities, and it is conceivable that the N-terminal region operates in this regard in certain nonpituitary cells. For example, RB/p107 double-knockout chimeric mice develop retinoblastomas (36). Whether N-terminal region deletion would affect the emergence of these lesions, which closely reflect the analogous human condition, is unknown. Similarly, there is no model, as yet, for RB-mediated sarcoma development, postpuberal sarcomas being another established component of the hRB syndrome. On the other hand, considering all of the evidence, one can argue that the pituitary tumor-suppressive effects of both wt and RB(Δ1-378) alleles, observed in the chimeric mice, correlate closely with the ability of RB (and RB[Δ1-378]) to suppress proliferation. In this regard, RB-mediated suppression of proliferation has long been considered an N-terminal-region-independent process (51). The anticataract effect of these alleles would also support this hypothesis.
Like the remainder of the protein, the N-terminal region, too, has been well conserved through evolution. While its function is as yet incompletely understood, there are data showing that it can interact with at least one MCM protein (41). MCM proteins are essential participants in the operation of eukaryotic replication origins. Moreover, recent work indicates a strong genetic connection between Drosophila Rb action and origin recognition complex (ORC) function or control of replication origin firing in ovarian follicle cells undergoing a normal state of chorion gene amplification (5, 37). In addition, when overproduced in S phase, un(der)phosphorylated RB can direct cessation of DNA synthesis (23, 24), and it has been reported that complexes of RB and replication factor C contribute to the regulation of cell death following DNA damage (32). These findings, taken together, strongly suggest a link between pocket protein function and the normal regulation of DNA replication, perhaps putting into a physiologic context the aforementioned observations of N-terminal region/MCM binding and RB control of S-phase activity.
Whatever the case, it now appears that contributions by all elements of its primary sequence are nonessential for RB-mediated pituitary tumor suppression function. This is, perhaps, not surprising, given that RB, the structure of which has been widely conserved among metazoans and plants, is a protein likely selected to play an embryonic survival/development function(s). Therefore, tumor suppression, which results in effects largely manifest after birth, likely plays a secondary role to embryonic survival and may have emerged as a by-product of certain RB survival functions. In that context, given that RB is a multidomain protein, it would not be surprising to find that tumor suppression depends upon some but not all of its multiple biochemical functions.
Acknowledgments
We thank our colleagues Fabio Martelli and Zhiyan Wang for numerous helpful discussions. We are also grateful to Kim Mercer and Denise Crowley for their expert assistance with histological analyses, to John Mkandawire for help with chimeric animal care, and to Shizuo Mukai for assistance in the characterization of murine cataracts. We thank Steve Grossman most enthusiastically for his expert advice during the preparation of the manuscript.
This work was supported by grants from the National Cancer Institute and the Sharf-Green family fund. Hong Yang is the Sharf-Green Fellow of the Dana-Farber Cancer Institute.
REFERENCES
- 1.Adams, P. D., X. Li, W. R. Sellers, K. B. Baker, X. Leng, J. W. Harper, Y. Taya, and W. G. Kaelin, Jr. 1999. Retinoblastoma protein contains a C-terminal motif that targets it for phosphorylation by cyclin-cdk complexes. Mol. Cell. Biol. 19:1068-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernards, R., G. M. Schackleford, M. R. Gerber, J. M. Horowitz, S. H. Friend, M. Schartl, E. Bogenmann, J. M. Rapaport, T. McGee, T. P. Dryja, et al. 1989. Structure and expression of the murine retinoblastoma gene and characterization of its encoded protein. Proc. Natl. Acad. Sci. USA 86:6474-6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bignon, Y. J., Y. Chen, C. Y. Chang, D. J. Riley, J. J. Windle, P. L. Mellon, and W. H. Lee. 1993. Expression of a retinoblastoma transgene results in dwarf mice. Genes Dev. 7:1654-1662. [DOI] [PubMed] [Google Scholar]
- 4.Bork, P., K. Hofmann, P. Bucher, A. F. Neuwald, S. F. Altschul, and E. V. Koonin. 1997. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11:68-76. [PubMed] [Google Scholar]
- 5.Bosco, G., W. Du, and T. L. Orr-Weaver. 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3:289-295. [DOI] [PubMed] [Google Scholar]
- 6.Brehm, A., and T. Kouzarides. 1999. Retinoblastoma protein meets chromatin. Trends Biochem. Sci. 24:142-145. [DOI] [PubMed] [Google Scholar]
- 7.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Z., G. A. Friedrich, and P. Soriano. 1994. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 8:2293-2301. [DOI] [PubMed] [Google Scholar]
- 9.Dryja, T. P., J. Rapaport, T. L. McGee, T. M. Nork, and T. L. Schwartz. 1993. Molecular etiology of low-penetrance retinoblastoma in two pedigrees. Am. J. Hum. Genet. 52:1122-1128. [PMC free article] [PubMed] [Google Scholar]
- 10.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 11.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 12.Ewen, M. E., H. K. Sluss, C. J. Sherr, H. Matsushime, J. Kato, and D. M. Livingston. 1993. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell 73:487-497. [DOI] [PubMed] [Google Scholar]
- 13.Gu, W., J. W. Schneider, G. Condorelli, S. Kaushal, V. Mahdavi, and B. Nadal-Ginard. 1993. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell 72:309-324. [DOI] [PubMed] [Google Scholar]
- 14.Hansen, M. F., R. Morgan, A. A. Sandberg, and W. K. Cavenee. 1990. Structural alterations at the putative retinoblastoma locus in some human leukemias and preleukemia. Cancer Genet. Cytogenet. 49:15-23. [DOI] [PubMed] [Google Scholar]
- 15.Harbour, J. W., R. X. Luo, A. Dei Santi, A. A. Postigo, and D. C. Dean. 1999. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98:859-869. [DOI] [PubMed] [Google Scholar]
- 16.Hateboer, G., R. M. Kerkhoven, A. Shvarts, R. Bernards, and R. L. Beijersbergen. 1996. Degradation of E2F by the ubiquitin-proteasome pathway: regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes Dev. 10:2960-2970. [DOI] [PubMed] [Google Scholar]
- 17.Henson, J. W., B. L. Schnitker, K. M. Correa, A. von Deimling, F. Fassbender, H. J. Xu, W. F. Benedict, D. W. Yandell, and D. N. Louis. 1994. The retinoblastoma gene is involved in malignant progression of astrocytomas. Ann. Neurol. 36:714-721. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann, F., F. Martelli, D. M. Livingston, and Z. Y. Wang. 1996. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 10:2949-2959. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh, J. K., F. S. Chan, D. J. O'Connor, S. Mittnacht, S. Zhong, and X. Lu. 1999. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol. Cell 3:181-193. [DOI] [PubMed] [Google Scholar]
- 20.Hu, N., A. Gutsmann, D. C. Herbert, A. Bradley, W. H. Lee, and E. Y. Lee. 1994. Heterozygous Rb-1 delta 20/+mice are predisposed to tumors of the pituitary gland with a nearly complete penetrance. Oncogene 9:1021-1027. [PubMed] [Google Scholar]
- 21.Hu, Q. J., C. Bautista, G. M. Edwards, D. Defeo-Jones, R. E. Jones, and E. Harlow. 1991. Antibodies specific for the human retinoblastoma protein identify a family of related polypeptides. Mol. Cell. Biol. 11:5792-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacks, T., A. Fazeli, E. M. Schmitt, R. T. Bronson, M. A. Goodell, and R. A. Weinberg. 1992. Effects of an Rb mutation in the mouse. Nature 359:295-300. [DOI] [PubMed] [Google Scholar]
- 23.Karantza, V., A. Maroo, D. Fay, and J. M. Sedivy. 1993. Overproduction of Rb protein after the G1/S boundary causes G2 arrest. Mol. Cell. Biol. 13:6640-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knudsen, E. S., C. Buckmaster, T. T. Chen, J. R. Feramisco, and J. Y. Wang. 1998. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 12:2278-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubota, Y., K. Fujinami, H. Uemura, Y. Dobashi, H. Miyamoto, Y. Iwasaki, H. Kitamura, and T. Shuin. 1995. Retinoblastoma gene mutations in primary human prostate cancer. Prostate 27:314-320. [DOI] [PubMed] [Google Scholar]
- 26.Lee, J. O., A. A. Russo, and N. P. Pavletich. 1998. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature 391:859-865. [DOI] [PubMed] [Google Scholar]
- 26a.Lipinski, M. M., K. F. Macleod, B. O. Williams, T. L. Mullaney, D. Crowley, and T. Jacks. 2001. Cell-autonomous and non-cell-autonomous functions of the Rb tumor suppressor in developing central nervous system. EMBO J. 20:3402-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 28.Maandag, E. C., M. van der Valk, M. Vlaar, C. Feltkamp, J. O'Brien, M. van Roon, N. van der Lugt, A. Berns, and H. te Riele. 1994. Developmental rescue of an embryonic-lethal mutation in the retinoblastoma gene in chimeric mice. EMBO J. 13:4260-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnaghi, J. L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, V. J. Le, F. Troalen, D. Trouche, and B. A. Harel. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 30.Meloni, A. R., E. J. Smith, and J. R. Nevins. 1999. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc. Natl. Acad. Sci. USA 96:9574-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgenbesser, S. D., B. O. Williams, T. Jacks, and R. A. DePinho. 1994. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature 371:72-74. [DOI] [PubMed] [Google Scholar]
- 32.Pennaneach, V., I. Salles-Passador, A. Munshi, H. Brickner, K. Regazzoni, F. Dick, N. Dyson, T. Chen, J. Y. Wang, R. Fotedar, and A. Fotedar. 2001. The large subunit of replication factor C promotes cell survival after DNA damage in an LxCxE motif- and Rb-dependent manner. Mol. Cell 7:715-727. [DOI] [PubMed] [Google Scholar]
- 33.Qian, Y., C. Luckey, L. Horton, M. Esser, and D. J. Templeton. 1992. Biological function of the retinoblastoma protein requires distinct domains for hyperphosphorylation and transcription factor binding. Mol. Cell. Biol. 12:5363-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin, X. Q., T. Chittenden, D. M. Livingston, and W. Kaelin, Jr. 1992. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 6:953-964. [DOI] [PubMed] [Google Scholar]
- 35.Riley, D. J., C. Y. Liu, and W. H. Lee. 1997. Mutations of N-terminal regions render the retinoblastoma protein insufficient for functions in development and tumor suppression. Mol. Cell. Biol. 17:7342-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robanus-Maandag, E., M. Dekker, M. van der Valk, M. L. Carrozza, J. C. Jeanny, J. H. Dannenberg, A. Berns, and H. te Riele. 1998. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 12:1599-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Royzman, I., R. J. Austin, G. Bosco, S. P. Bell, and T. L. Orr-Weaver. 1999. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 13:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sellers, W. R., B. G. Novitch, S. Miyake, A. Heith, G. A. Otterson, F. J. Kaye, A. B. Lassar, and W. G. Kaelin, Jr. 1998. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 12:95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sellers, W. R., J. W. Rodgers, and W. G. Kaelin, Jr. 1995. A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc. Natl. Acad. Sci. USA 92:11544-11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen, W. J., H. S. Kim, and S. Y. Tsai. 1995. Stimulation of human insulin receptor gene expression by retinoblastoma gene product. J. Biol. Chem. 270:20525-20529. [DOI] [PubMed] [Google Scholar]
- 41.Sterner, J. M., S. Dew-Knight, C. Musahl, S. Kornbluth, and J. M. Horowitz. 1998. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol. Cell. Biol. 18:2748-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sterner, J. M., Y. Murata, H. G. Kim, S. B. Kennett, D. J. Templeton, and J. M. Horowitz. 1995. Detection of a novel cell cycle-regulated kinase activity that associates with the amino terminus of the retinoblastoma protein in G2/M phases. J. Biol. Chem. 270:9281-9288. [DOI] [PubMed] [Google Scholar]
- 43.Trouche, D., C. C. Le, C. Muchardt, M. Yaniv, and T. Kouzarides. 1997. RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl. Acad. Sci. USA 94:11268-11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Udvadia, A. J., D. J. Templeton, and J. M. Horowitz. 1995. Functional interactions between the retinoblastoma (Rb) protein and Sp-family members: superactivation by Rb requires amino acids necessary for growth suppression. Proc. Natl. Acad. Sci. USA 92:3953-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, J. Y., E. S. Knudsen, and P. J. Welch. 1994. The retinoblastoma tumor suppressor protein. Adv. Cancer Res. 64:25-85. [DOI] [PubMed] [Google Scholar]
- 46.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 47.Weintraub, S. J., K. N. Chow, R. X. Luo, S. H. Zhang, S. He, and D. C. Dean. 1995. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature 375:812-815. [DOI] [PubMed] [Google Scholar]
- 48.Welch, P. J., and J. Y. Wang. 1993. A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-Abl tyrosine kinase in the cell cycle. Cell 75:779-790. [DOI] [PubMed] [Google Scholar]
- 49.Williams, B. O., E. M. Schmitt, L. Remington, R. T. Bronson, D. M. Albert, R. A. Weinberg, and T. Jacks. 1994. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J. 13:4251-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao, Z. X., J. Chen, A. J. Levine, N. Modjtahedi, J. Xing, W. R. Sellers, and D. M. Livingston. 1995. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature 375:694-698. [DOI] [PubMed] [Google Scholar]
- 51.Xu, H. J., K. Xu, Y. Zhou, J. Li, W. F. Benedict, and S. X. Hu. 1994. Enhanced tumor cell growth suppression by an N-terminal truncated retinoblastoma protein. Proc. Natl. Acad. Sci. USA 91:9837-9841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, H. J., Y. Zhou, J. Seigne, G. S. Perng, M. Mixon, C. Zhang, J. Li, W. F. Benedict, and S. X. Hu. 1996. Enhanced tumor suppressor gene therapy via replication-deficient adenovirus vectors expressing an N-terminal truncated retinoblastoma protein. Cancer Res. 56:2245-2249. [PubMed] [Google Scholar]
- 53.Zacksenhaus, E., R. Bremner, R. A. Phillips, and B. L. Gallie. 1993. A bipartite nuclear localization signal in the retinoblastoma gene product and its importance for biological activity. Mol. Cell. Biol. 13:4588-4599. [DOI] [PMC free article] [PubMed] [Google Scholar]