Abstract
The murine p204 protein level is highest in heart and skeletal muscle. During the fusion of cultured myoblasts to myotubes, the p204 level increases due to transcription dependent on the muscle-specific MyoD protein, and p204 is phosphorylated and translocated from the nucleus to the cytoplasm. p204 overexpression accelerates myoblast fusion in differentiation medium and triggers this process even in growth medium. Here we report that p204 is required for the differentiation of C2C12 myoblasts. We propose that it enables the differentiation, at least in part, by overcoming the inhibition of the activities of the MyoD and E47 proteins by the Id proteins: Id1, Id2, and Id3. These are known to inhibit skeletal muscle differentiation by binding and blocking the activity of MyoD, E12/E47, and other myogenic basic helix-loop-helix (bHLH) proteins. Our hypothesis is based on the following findings. (i) A decrease in the p204 level in C2C12 myoblasts by antisense RNA (a) increased the level of the Id2; (b) inhibited the MyoD-, E12/E47-, and other bHLH protein-dependent accumulation of the muscle-specific myosin heavy-chain protein; and (c) inhibited the fusion of myoblasts to myotubes in differentiation medium. (ii) p204 bound to the Id proteins in vitro and in vivo. (iii) In the binding of p204 to Id2, the b segment of p204 and the HLH segment of Id2 were involved. (iv) Addition of p204 overcame the inhibition by the Id proteins of the binding of MyoD and E47 to DNA in vitro. (v) Overexpression of p204 in myoblasts (a) decreased the level of the Id proteins, even in a culture in growth medium, and (b) overcame the inhibition by the Id proteins of MyoD- and E47 dependent transcription and also overcame the inhibition by Id2 of the fusion of myoblasts to myotubes.
The interferons are cytokines of vertebrates with antimicrobial, cell growth-regulatory, and immunomodulatory activities (64, 65). They function by modulating the expression of many genes, including those of the gene 200 cluster (14, 32, 36, 44, 56). This cluster arose by repeated gene duplications and consists of nine or more genes and pseudogenes in mice. In humans only three genes from the cluster are known (8, 21, 67). The 200 cluster genes encode the p200 family proteins (32, 36, 44).
The best-characterized murine p200 family proteins are p202a and p204 (10, 11, 12, 13, 16, 17, 22, 24, 43, 46, 47, 52, 69). p202a and Ifi202a, the gene encoding it, were earlier designated p202 and Ifi202, respectively (68). p202a is inducible by interferon and is primarily nuclear. The overexpression of p202a inhibits cell proliferation (13, 52), and this may be correlated with the binding and inhibition of the activity of several transcription factors by p202a. These include, among others, c-Fos, c-Jun, AP2, E2F1, E2F4, NF-κB, MyoD, myogenin, and c-Myc (10, 13, 17, 52, 69). In most of these cases p202a inhibits the sequence-specific binding of the transcription factor to DNA, whereas in the case of c-Myc it blocks the binding to Max (69). p202a also binds pRb (12) and inhibits the activity of p53 (16). In turn, the transcription of the Ifi202 gene is inhibited by p53 (22). The inhibitory activity of p202a is overcome by the binding to p202a of the p53-binding protein 1 (16) and also of the human adenovirus E1A oncoprotein (74). p202a is also induced during muscle differentiation (17). During this process it can modulate the activity of the myogenic transcription factors MyoD and myogenin, and it also inhibits apoptosis (17, 34, 69). Overexpression of p202 was linked to susceptibility to the autoimmune disease lupus erythematosus in mice (62). p202a has a sister protein, p202b, which is encoded by the Ifi202b gene and differs from p202a in only 7 amino acids out of 445 (68). The disruption of the Ifi202a gene in mice has no obvious phenotype, apparently because of the increase of the p202b level compensating for the loss of p202a.
p204, which is encoded by the Ifi204 gene, is also inducible by interferon (11). Its overexpression is growth inhibitory (11, 43, 46). p204 can inhibit the transcription of rRNA by binding to the ribosomal DNA-specific UBF transcription factor and inhibiting its sequence-specific binding to DNA (46). p204, similarly to p202a, contains the pRb binding motifs LXCXE and can bind to pRb (29, 46). Overexpression of p204 can delay the progression of cells from the G0/G1 phase to the S phase (29). Both of the two LXCXE motifs of p204 were reported to be required for the antiproliferative activity of p204 (29). It was also reported that focus formation of cells transfected with a p204 expression plasmid is inhibited only in the case of Rb+/+ cells and not in that of Rb−/− cells. Transfection of cells with expression plasmids encoding dominant negative p204 mutants (in which a region of 72 amino acids including the LXCXE motif was deleted) inhibited the replication of cytomegalovirus but not of several other viruses (28). On this basis, it was concluded that the replication of cytomegalovirus requires p204.
Among 10 adult mouse tissues tested, the level of p204 is highest in heart and skeletal muscle (47). In cultured C2C12 myoblasts, p204 is nucleoplasmic. During myoblast fusion to myotubes, the p204 level increases manyfold, p204 becomes phosphorylated, and most of it is translocated to the cytoplasm in a nuclear export signal-dependent process. This increase in p204 level is a consequence of MyoD transcription factor binding to E-box (CANNTG) sequences in the Ifi204 gene, followed by transcription. The level of p204 in mouse heart muscle also strongly increases during differentiation, as it is barely detectable in 10.5-day embryos, reaches its peak level in 16.5-day embryos, and remains high thereafter. Overexpression of p204 in C2C12 myoblasts (carrying an inducible p204 expression plasmid) accelerates the fusion of myoblasts in differentiation medium and elicits the fusion even in growth medium (47).
One of the conceivable ways in which p204 might enable C2C12 myoblasts to differentiate is by overcoming the inhibition of this process by the Id proteins (4, 31, 54). Discovered in 1990, the Id (for inhibitor of DNA binding) proteins have a helix-loop-helix (HLH) domain but lack a basic (b) domain (4, 31). They can form nonfunctional heterodimers with bHLH proteins (e.g., MyoD and E12/E47) and thus act as negative regulators of such proteins. Id family proteins (e.g., in mice Id1, Id2, Id3, and Id4) were implicated in the control of differentiation and cell cycle progression in organisms from fly to human (31, 51, 54). Thus, e.g., Id1 and Id3 are required for neurogenesis, angiogenesis, and vascularization of tumor xenografts (48). Id2 binds to pRb and can reverse the inhibition of cell proliferation and cell cycle progression by pRb (30). Id2 transcription is activated by c-Myc or N-Myc, and this accounts for the overexpression of Id2 in neuroblastoma cells carrying extra copies of the N-myc gene (38).
Mouse skeletal muscle differentiation depends on, among others, two classes of bHLH proteins: A (e.g., MyoD and myogenin) and B (e.g., E12 and E47) (18, 23, 39, 73). Their HLH domains mediate homodimerization, or heterodimerization between the class A and class B proteins, thereby allowing the binding of the bHLH proteins to E-box sequences in DNA (49, 55). The overexpression of Id proteins (Id1, Id2, and Id3, but not Id4) in cultured myoblasts inhibits the transactivation by bHLH proteins, (e.g., MyoD and E12/E47), of genes with regulatory E-box sequences, thereby inhibiting (i) the formation of proteins participating in muscle differentiation and, consequently, (ii) the fusion of myoblasts to myotubes (1, 4, 31, 37, 51, 54).
Here we report experiments revealing that (i) decreasing the level of p204 (by antisense RNA) inhibited the differentiation of cultured C2C12 myoblasts to myotubes; (ii) an increase in the level of p204 could overcome the inhibition of myoblast differentiation by Id2; (iii) p204 enabled myoblasts to differentiate by overcoming the inhibition of the activity of MyoD and E47 (as well as presumably other bHLH proteins) by the Id1, Id2, and Id3 proteins; and (iv) p204 accomplished this by binding and sequestering the Id1, Id2, and Id3 proteins, as well as by promoting their disappearance during differentiation.
MATERIALS AND METHODS
Antibodies and cell lines.
The preparation and purification of an antiserum to p204 was reported previously (47). Anti-MyoD, anti-Id1, anti-Id2, and anti-Id3 immunoglobulin Gs (IgGs) were purchased from Santa Cruz Biotechnology. The MF20 monoclonal antibody against light meromyosin (which reacts with all sarcomere myosin), developed by D.A. Fischman, (2), was obtained from the Developmental Studies Hybridoma Bank maintained by the University of Iowa Department of Biological Science, Iowa City. C2C12 (ATCC 1772CRL), a murine thigh muscle myoblast line (75); C3H 10T1/2 (ATCC 266CCL), a cloned murine embryo fibroblast line referred to as 10T1/2 (60); and 293 human embryonic kidney cells (25) were used.
Plasmid constructs.
To obtain the 204 sense and antisense RNA expression plasmids (pCMV204 and pCMV204AS, respectively), 204 cDNA (14) was inserted into the EcoRI site of the pCMVneo vector (27) in the appropriate and inverted orientations, respectively. Glutathione S-transferase (GST)-204 fusion proteins were described previously (46). The mammalian expression plasmids pFlag-Id1, pFlag-Id2, and pFlag-Id3 were generously provided by B. A. Christy; pcDNA3-E47 and pGEX-E47 were provided by A. Cano (59); and GST-Id2 was provided by M. Israel. To obtain bacterial expression plasmids encoding various GST-Id2 segments, three cDNA segments encoding Id2 (amino acids 1 to 29), Id2 (amino acids 30 to 87), and Id2 (amino acids 88 to 134) were cloned in frame into the BamHI-EcoRI sites of pGEX-3X. DNA segments encoding His6-Id1 and His6-Id3 were generated by PCR with appropriate primers. The products were inserted into the BamHI-EcoRI sites of the pGEX-3X vector, resulting in the GST/His-Id1 and GST/His-Id3 expression plasmids. GST-FHF1B (a fibroblast growth factor homologous factor 1B [FHF1B] expression plasmid) was described previously (45), and the His-sodium channel-associated protein 1 (SAP1) expression plasmid was made in our laboratory (C.-J. Liu et al., unpublished). Details of the constructions are available upon request. In the plasmid p4RCAT, chloramphenicol acetyltransferase (CAT) expression is driven by four MyoD binding (R) sites from the muscle-specific creatine kinase gene (71).
Comparison of the levels of Id1, Id2, and Id3 proteins in murine thigh muscle.
Four 15-day-old C129 mice (two males and two females) were sacrificed, and their thigh muscles were quickly excised and stored in liquid nitrogen. To produce muscle extracts, the muscle tissue was cut into small pieces and homogenized in ice-cold lysis buffer (50 mM Tris HCl [pH 7.4], 300 mM NaCl, 5 mM EDTA, 10 mM iodoacetamide, 1 mM phenylmethylsulfonyl fluoride, 1% Triton X-100, 0.02% [wt/vol] sodium azide, 10 μg of aprotinin per ml, and 10 μg of leupeptin per ml). For immunoprecipitation, 30-μl aliquots of protein A-agarose beads (GIBCO) were incubated with 1 μg of affinity-purified polyclonal antibodies to Id1, Id2, and Id3, in 0.5 ml of ice-cold phosphate-buffered saline (PBS) at 4°C for 4 h. The loaded beads were sedimented and washed four times in lysis buffer. One-milliliter aliquots from the muscle extract were precleared by incubation with 30 μl of protein A-agarose at 4°C for 30 min and sedimented by centrifugation at 16,000 × g and 4°C for 5 min. One-milliliter aliquots from the precleared muscle extract (containing 500 μg of protein) were supplemented with 10 μl of 10% bovine serum albumin and incubated at 4°C overnight with 30 μl of protein A-agarose beads loaded with antibodies to Id1, Id2, or Id3. The beads were washed four times with ice-cold washing buffer (50 mM Tris-HCl [pH 7.4], 300 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, 0.02% sodium azide) and once with ice-cold PBS and were sedimented by centrifugation. The beads were then boiled in 30 μl of 2× sodium dodecyl sulfate (SDS) loading buffer for 5 min, and the eluate was subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred to a nitrocellulose membrane and blocked with 8% nonfat milk blocking buffer at 4°C overnight. The membrane was cut into three parts for immunodetection by incubation with Id1, Id2, and Id3 antibodies and for visualization by enhanced chemiluminescence (ECL). We verified that the antibodies to Id1, Id2, and Id3 were specific for their targets and did not cross-react with the other Id proteins.
Generation of stable cell lines. (i) Myoblast lines expressing 204 antisense RNA.
C2C12 myoblasts were transfected with a 204 antisense RNA expression plasmid including the entire 204 cDNA (for generating the p204AS lines) or the pCMV expression vector (for generating the pCMV control lines). Transfectants were selected in the presence of 1.2 mg of G418 per ml. Six stably transfected pCMV204AS clones and a mixture of at least 50 pCMV control clones were amplified, and their p204 levels were examined by immunoblotting using anti-p204 antiserum.
(ii) Two myoblast lines expressing Id2 constitutively, one in which Muristerone induces p204 expression and one that serves as the control line.
Cultures of a C2C12 myoblast line in which Muristerone induces p204 expression (ind.p204) and of the control line (con.) (47) were transfected with a Flag-Id2 expression plasmid (51). Transfectants were selected in a medium including 1 mg of hygromycin per ml, 0.5 mg of G418 per ml, and 0.5 mg of zeocin per ml. The resulting stable clones were amplified. The line in which p204 was induced by Muristerone was designated ind.p204/Id2, and the control line was designated con./Id2. The Id2 mRNA levels were examined by Northern blotting, and the p204 levels were examined by immunoblotting.
Immunofluorescent cell staining.
Cultures of the pCMV control cell line and the pCMV204AS line were plated on glass coverslips coated with polylysine and were grown in growth medium (GM) (Dulbecco modified Eagle medium [DMEM] supplemented with 20% fetal bovine serum [FBS]) in 10% CO2-90% air at 37°C. After reaching confluency, the cultures were shifted to differentiation medium (DM) (DMEM supplemented with 0.5% horse serum) for 2 days. For staining, the cells were fixed with cold methanol for 5 min and air dried. After rehydration in PBS and blocking with 30% goat serum in PBS for 45 min, the cells were incubated with primary antibodies (MF20) against mouse myosin heavy chain at room temperature overnight. After being washed with PBS, the coverslips were incubated with secondary antibodies (against mouse IgG) conjugated with fluorescein isothiocyanate diluted 1:200 (Santa Cruz) for 1 h. The specimens were observed under a fluorescence microscope with appropriate optical filters. Microscopic images were captured using the Image Pro program (Media Cybernetics) and an Olympus microscope. Pictures were arranged using the Adobe Photoshop and Coreldraw program.
Preparation of nuclear extracts.
Nuclear extracts of C2C12 cells were prepared essentially according to the procedure of Kushner and Ricciardi (35) with minor modifications. Briefly, cells were harvested by trypsinization, washed in PBS, pelleted, and resuspended in lysis buffer (10 mM Tris-HCl [pH 8.0], 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, proteinase inhibitors, and 0.3% NP-40). After 5 min on ice, the lysates were centrifuged at 1,000 × g and 4°C for 5 min, and the pelleted nuclei were briefly washed in lysis buffer without NP-40. The nuclear pellet was resuspended in an equal volume of nuclear extraction buffer (20 mM Tris-HCl [pH 8.0], 420 mM NaCl, 1.5 mM MgC12, 0.2 mM EDTA, and 25% glycerol), and 5 M NaCl was added to obtain a final NaCl concentration of 400 mM. After an incubation at 4°C for 10 min, the nuclei were briefly vortexed and centrifuged at 25,000 × g for 5 min. The supernatant fraction was used as nuclear extract.
Expression, purification, and processing of GST fusion proteins.
For expressing GST fusion proteins, the appropriate plasmids, i.e., (i) pGST-204 and its truncated derivatives (46), (ii) pGEX-Id2 and its truncated derivatives, GST-Id2 (amino acids 1 to 29), GST-Id2 (amino acids 30 to 83), and GST-Id2 (amino acids 84 to 134), (iii) GST/His-Id1 and GST/His-Id3, and (iv) GST-FHF1B were transformed into Escherichia coli DH5α (GIBCO/BRL), and His-SAP1 was transformed into E. coli Top-10 (Invitrogen). The fusion proteins synthesized were affinity purified on glutathione-agarose beads as previously described (46). To cleave off and remove the GST moiety, 50 μg of purified GST/His-Id1 (or GST/His-Id3) fusion protein was incubated with 1 μg of Xa factor (New England Biolabs) in 20 μl of 20 mM Tris-HCl [pH 8.0]-100 mM NaCl-2 mM CaCl2 at 23°C for 8 h. The reaction was terminated by the addition of 2 μM dansyl-Glu-Gly-Arg-chloromethyl ketone (New England Biolabs) and incubation at room temperature for 1 min. The completion of the cleavage was established by SDS-PAGE. The resulting His-Id1 and His-Id3 were used in the HisTrap binding assay.
In vitro binding assay.
(i) To examine the binding of p204 to Id2 and Id2 segments, 35S-p204 (IVT-204) was expressed in a rabbit reticulocyte transcription-translation system (Promega). Ten-microliter aliquots of 35S-p204 were incubated at 4°C for 4 h with 20 μl of packed glutathione-Sepharose beads (Pharmacia) loaded with 0.5 μg of GST (control) or 0.5 μg of GST-Id2 or GST-Id2 deletion mutants, as indicated, in 200 μl of buffer AM supplemented with 100 mM KCl and 0.5 mg of bovine serum albumin per ml (46). The bound 35S-p204 was analyzed by SDS-7.5% PAGE and visualized by autoradiography. (ii) To examine the binding of p204 to Id1 and Id3, 0.5 μg of GST-204 was incubated in 150 μl of buffer AM supplemented with 100 mM KCl, 0.5 mg of bovine serum albumin per ml, and 25 μl of HisTrap beads (which were loaded with 0.5 μg of His-Id1, His-Id3, or His-SAP1) at 4°C for 4 h. Thereafter, the beads were washed four times with 1 ml of buffer AM supplemented with 100 mM KCl, sedimented by centrifugation, and eluted by boiling in 20 μl of 1× SDS loading buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol) for 5 min. The eluted proteins were analyzed by SDS-7.5% PAGE and visualized by immunoblotting with anti-p204 antiserum. (iii) To examine the binding of p204 segments to Id2, glutathione-Sepharose beads loaded with GST (0.5 μg) (serving as control), GST-204 (0.5 μg), or GST-204 segments (0.5 μg), as indicated, were incubated with extract (500 μg of protein) from C2C12 myoblasts. The bound proteins were separated by SDS-10% PAGE, and Id2 was detected by immunoblotting with anti-Id2 antibodies.
Coimmunoprecipitation.
C2C12 myoblasts were grown in monolayer in 100-mm-diameter dishes in GM. After reaching 80% confluency, the cultures were shifted to DM for 1 or 2 days. Cell extracts were prepared as described by Min et al. (52). After 1 h of incubation with either anti-p204, anti-Id1, anti-Id2, or anti-Id3 rabbit antiserum, aliquots (200 μl) from the cell extracts were incubated with 30 μl of protein A-agarose (GIBCO/BRL) at 4°C overnight. After washing five times with immunoprecipitation buffer (52), the bound proteins were released by boiling in 20 μl of 2× SDS loading buffer for 3 min (46). The released proteins were examined by Western blotting with anti-Id2 or anti-p204 antiserum. We verified that each of the antisera to Id1, Id2, and Id3 was specific for its target protein and did not immunoprecipitate either of the other two Id proteins (not shown).
Far-Western assay.
An overlay blotting (far-Western) assay was carried out as described previously (42) with minor modifications. Extracts from 293 cells which had been transfected with expression plasmids encoding either green fluorescent protein (GFP) or GFP-204 fusion protein (47) were incubated first with anti-GFP antibodies (Clontech) for 1 h and thereafter with protein A-agarose at 4°C overnight. After washing five times in cell lysis buffer (PBS [pH 7.5], 1% Triton X-100, 5 mM EDTA) and elution with 2× sample buffer, the purified GFP (control) and GFP-204 were subjected to SDS-10% PAGE. The proteins were electrotransferred to a nitrocellulose membrane at 85 mA for 2 h, and the proteins in the blot were denatured and renatured by sequential washings, first in 0.1 M CZ solution (20 mM HEPES [pH 7.9], 20% glycerol, 0.1 M KCl, 5 mM MgC12, 01.mM ZnCl2, 0.1 mM EDTA, 2 mM dithiothreitol) supplemented with 0.02% polyvinylpyrrolidone and 6 M guanidine-HCl for 20 min and then in 0.1 M CZ solution supplemented with 0.02% polyvinylpyrrolidone three times for 2 h each. After blocking with 5% bovine serum albumin, the membranes were incubated with 50 μg of purified GST-Id2 protein, followed by incubation with anti-Id2 antibodies or anti-GFP antibodies as indicated and visualization with the ECL system.
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed essentially as described previously (47) except that the MEF-1 consensus oligodeoxynucleotide (Santa Cruz) was used as a probe. Briefly, the binding assay was performed in a 20-μl volume containing 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, 1 μg of poly(dI-dC), 3 ng of the labeled DNA probe, and various combinations of GST, GST-MyoD, GST-204, anti-MyoD IgG, the wild-type or mutant MEF-1 oligonucleotide (Santa Cruz), and Id1, Id2, and Id3 translated in vitro (IVT-Id1, IVT-Id2, and IVT-Id3, respectively), as indicated. In the case of EMSA involving nuclear extracts, 25-μl reaction volumes including 4-μl aliquots of nuclear extract were used. After incubation at room temperature for 35 min, the samples were subjected to 4% PAGE in 0.5% TBE (45 mM Tris-borate, 1 mM EDTA) at 15 V/cm and room temperature for 3 h. The gel was dried, and autoradiography was performed at −70°C.
Assay of the effects of decreasing the p204 level on the Id2 level in myoblasts.
Each line of C2C12 myoblasts stably transfected with pCMV204AS (expressing 204AS RNA) and each line of C2C12 myoblasts stably transfected with pCMV (serving as a control) was plated in two 100-mm-diameter culture dishes in DMEM-10% FBS. When the cells reached 40 to 50% confluency, one of the two plates was further incubated in DMEM-10% FBS and the other plate was shifted to DM. After 48 h, cell extracts were prepared in 1 ml of ice-cold lysis buffer. To aliquots of the cell lysates containing 500 μg of protein, 30 μl of protein A beads was added. After incubation at 4°C for 30 min, the reaction mixtures were centrifuged at 16,000 × g and 4°C for 5 min. The supernatant fraction was incubated with protein A beads (coated with 1 μg of anti-Id2 antibody protein) at room temperature for 2 h. Thereafter the beads were washed twice with washing buffer and once with PBS. The bound protein was released by boiling in 30 μl of 2× SDS loading buffer for 3 min, subjected to SDS-12% PAGE, and transferred to a polyvinylidene difluoride membrane for Western blotting with anti-Id2 antibodies (diluted 1:1,000). The level of p204 in the cell lysates was determined by immunoblotting.
Assay of the effects of increasing the p204 level on the Id, Id2, and Id3 levels in myoblasts.
Stable cell lines in which Muristerone induces p204, together with a control line (con.), were generated from C2C12 myoblasts by transfection of appropriate constructs and selection (47) The cells were grown in DMEM-10% FBS without or with 2.5 μM Muristerone to 40% confluency in approximately 48 h. The medium was replaced with fresh DMEM-10%FBS (without or with Muristerone as in the previous incubation), and after a 12-h incubation the cells were lysed in 0.5 ml of lysis buffer (Pierce) per 10-cm-diameter plate and the lysate was centrifuged at 16,000 × g and 4°C for 10 min. The protein concentration in the supernatant fraction was adjusted to 1 mg/ml. A 0.5-ml portion of the solution was incubated with 0.1 mg of antibodies to Id1, Id2, or Id3 (Santa Cruz at) 4°C overnight, supplemented with 0.4 ml of immobilized protein G (Pierce), further incubated at room temperature for 2 h, and sedimented by centrifugation. The immune complexes in the precipitate were washed four times with 0.14 M NaCl-0.008 M sodium phosphate-0.002 M potassium phosphate-0.001 M KCl (pH 7.4) and eluted with Immunopure elution buffer (Pierce). The samples were concentrated using Microcon centrifugal filters (Millipore). Aliquots were subjected to SDS-12% PAGE and immunoblotted with antibodies to Id1, Id2, or Id3. The levels of p204 in the cell lysates were determined by immunoblotting.
Transient-transfection assay.
10T1/2 fibroblasts or C2C12 myoblasts grown to around 50% confluency in GM in six-well plates were transfected with 1 μg of the reporter construct p4RCAT together with 1 μg of pSVGal internal control plasmid and 1 μg of the expression plasmids indicated. At 48 h after transfection, the cultures were harvested and lysed, and CAT and β-galactosidase activities were determined using kits from Promega.
RESULTS
A decrease in the p204 level inhibits myoblast differentiation.
The effect of a decrease in the p204 level on myoblast differentiation was tested in six individually picked, stable C2C12 myoblast lines (p204AS). These lines had been generated by transfection of an expression plasmid (pCMV204AS) encoding 204 antisense RNA and selection with G418. The outcomes of the tests with each of the six lines were similar to that shown in Fig. 1 for one of the six lines. The expression of the antisense 204 RNA in cultures in GM decreased the level of p204 three- to fourfold (Fig. 1C) compared to that in a control culture transfected with the pCMV vector. Both the control and the p204AS cultures grown to confluency in GM remained as undifferentiated cells (Fig. 1A, panels a and b). Shifting the cultures to DM for 4 days resulted in the differentiation, i.e., fusion to myotubes, of the control culture (Fig. 1A, panel c), whereas the differentiation of the p204AS culture was strongly inhibited (Fig. 1A, panel d).
FIG. 1.
Lowering the level of p204 inhibits the fusion of C2C12 myoblasts to myotubes and the formation of myosin heavy-chain protein. (A) Stable lines of C2C12 myoblasts carrying the expression vector pCMV (control) or the antisense expression plasmid pCMV204AS (p204AS) were grown in GM for 2 days and then were shifted to DM for 4 days. The cultures were observed with a phase-contrast microscope. Bar, 150 μm. (B) The control and p204AS cultures were treated as described for panel A except that they were kept in DM for only 2 days. Thereafter the cultures were fixed, blocked with goat serum, incubated with mouse antibodies against myosin heavy chain as well as secondary antibodies against mouse IgG conjugated with fluorescein isothiocyanate, and photographed using a fluorescence microscope. (C) Assay of the p204 level in stable C2C12 lines expressing pCMV (control) or pCMV204AS (p204AS). The control (lane 1) and p204AS (lane 2) cultures were grown to confluency in GM and lysed, and 40-μg protein samples from the lysates were assayed for p204 by Western blotting with anti-p204 antiserum. The relative levels of p204 protein and the p204 band are indicated. For further details, see Materials and Methods.
We performed immunofluorescent cell staining to examine whether the decrease in p204 level inhibited the biosynthesis of myosin heavy chain, a protein present in differentiated myotubes but not in the precursor myoblasts. The results (Fig. 1B, panels e and f) revealed that it did: myosin heavy chain accumulated in the myotubes formed after the control culture was kept in DM, whereas it was undetectable in the p204AS culture also incubated in DM.
These observations indicate that an appropriate level of p204 was essential for the differentiation of C2C12 myoblasts under our conditions.
p204 binds to the Id1, Id2, and Id3 proteins.
Among the four murine Id proteins, Id1, Id2, Id3, and Id4, only the first three were shown to inhibit the differentiation of cultured skeletal myoblasts (1, 31, 37, 51, 54). The presence of these three proteins in thigh muscle from 15-day-old C129 mice is shown in Fig. 2A.
FIG. 2.
Interaction of p204 with Id1, Id2, and Id3. (A) Comparison of the levels of the Id1, Id2, and Id3 proteins in murine thigh muscle. Id1, Id2, and Id3 were immunoprecipitated from a thigh muscle extract of 15-day-old C129 mice by using polyclonal antibodies to the Id1, Id2, or Id3 proteins immobilized on protein A-agarose beads. The proteins were eluted from the washed beads, and the eluate was subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and blocked with nonfat milk. The Id1, Id2, and Id3 proteins were detected by using antibodies to Id1, Id2, and Id3, respectively, and the ECL system. (B) Binding of p204 to GST-Id2 determined by pull-down assay in vitro. [35S]methionine-labeled p204 translated in a reticulocyte lysate (IVT204) (lane 1) bound to GST-Id2 (lane 4) but not to GST (lane 2) or GST-FHF1B (lane 3) immobilized on glutathione-Sepharose beads. The amount of p204 used in the experiment in lane 1 was 1/10 of that in the experiments in lanes 2 and 3. The bound proteins were analyzed by gel electrophoresis and fluorography. The position of the p204 band is indicated. The GST-FHF1B protein was used as a negative control. (C) Left panel, purified GFP (lane 1) or GFP-204 fusion protein (lane 2) was subjected to SDS-10% PAGE, electrotransferred to a nitrocellulose membrane, denatured, and renatured. After blocking with bovine serum albumin, the membrane was incubated with purified GST-Id2 protein, followed by incubation with anti-Id2 antibodies and visualization. Right panel, the same membrane was stripped and reprobed with anti-GFP antibodies. The GST-Id2 band (as retained by GFP-204) and the IgG band are indicated by arrows. (D) Interaction of p204 with Id2 in vivo, assayed by coimmunoprecipitation with anti-p204 antiserum. Extracts prepared from C2C12 myoblasts in GM or in DM for 1 day (DM1) or 2 days (DM2) were immunoprecipitated (IP) with anti-p204 antiserum (lanes 2 to 4). The immunoprecipitates and the cell extract (lane 1) were examined by immunoblotting with anti-Id2 antibodies. The amount of lysate used in the experiment in lane 1 was 10% of that used in the experiments in lanes 2 to 4. The Id2 and IgG bands are indicated. (E) Binding of p204 to His-Id1 and His-Id3 determined by pull-down assay in vitro. GST-204 was incubated with HisTrap beads alone (lane1) or with HisTrap beads loaded with His-SAP1 (lane 2) His-Id1 (lane 3), or His-Id3 (lane 4). The beads were washed and eluted, and the proteins released were analyzed by immunoblotting with anti-p204 antibodies. The GST-204 band is indicated. (F) Interaction of p204 with Id1, Id2, and Id3 in vivo, assayed by coimmunoprecipitation with antisera to Id1, Id2, and Id3. Extracts prepared from C2C12 myoblasts in DM for 1 day were immunoprecipitated with control IgG (lane 3) or antiserum to Id2 (lane 2), Id1 (lane 4), or Id3 (lane 5). The immunoprecipitates, together with cell lysate (lane 1) (20% of the amount used for immunoprecipitation), were examined by immunoblotting with an anti-p204 antiserum. The p204 and IgG bands are indicated. For further details, see Materials and Methods.
We established that p204 translated in vitro bound to GST-Id2 but neither to GST nor to another GST fusion protein (GST-FHF1B) (45) (Fig. 2B). Far-Western blotting revealed that the interaction between purified Id2 (i.e., GST-Id2) and purified p204 (i.e., GFP-204) was direct; moreover, GST-Id2 did not interact with GFP (Fig. 2C). An antiserum to p204 (but not preimmune serum [data not shown]) coimmunoprecipitated Id2 from extracts of C2C12 myoblasts in GM or DM for 1 day (Fig. 2D, lanes 2 and 3). No Id2, however, was coprecipitated by the antiserum from the extract of a myoblast culture kept in DM for 2 days (lane 4). This is in accord with the finding that the level of Id2 strongly decreases during differentiation (31, 51) (see also Fig. 8A). Purified GST-204 also bound to purified His-Id1 or His-Id3 immobilized on HisTrap beads, but it bound neither to the beads themselves nor to another His-fusion protein (His-SAP1) (Liu et al. unpublished) (Fig. 2E, lanes 2, 3, and 4). Furthermore, antisera to Id2, Id1, or Id3 (but not control IgG) coimmunoprecipitated p204 from an extract of C2C12 myoblasts in DM for 1 day (Fig. 2F, lanes 1, 2, 3, and 4). These results indicate that p204 bound to Id1, Id2, or Id3 in vivo and in vitro and that the interaction between the proteins in vitro was direct.
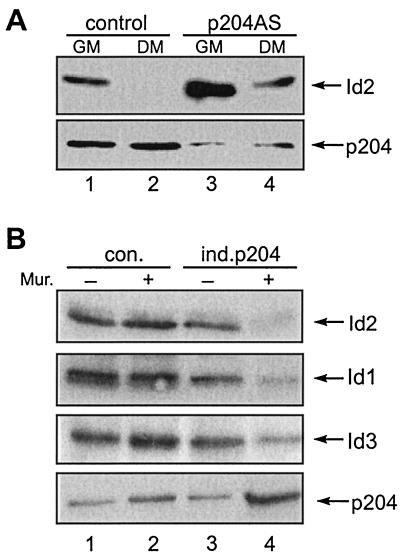
FIG. 8.
Effects of decreasing or increasing the level of p204 on the levels of Id1, Id2, and Id3 in myoblasts. (A) Effect of decreasing the level of p204 on the level of Id2. Two cultures from each of three C2C12 lines expressing 204 antisense RNA (p204AS lines) and from the appropriate control line were grown to 40 to 50% confluency in GM. Thereafter, one of the two cultures from each line was shifted to DM and the other was kept in GM. After 48 h the cultures were lysed. The levels of p204 in the lysates were determined by immunoblotting. To increase the sensitivity of the Id2 assay, Id2 was immunoprecipitated from aliquots of the lysates, and the levels of Id2 in the immunoprecipitates were determined by immunoblotting. Results with only one of three p204As lines are shown. The results with the other two lines were similar. (B) Effect of increasing the level of p204 on the levels of Id2, Id1, and Id3. Cultures of stable C2C12 lines in which p204 can be induced by Muristerone (ind.p204) and the appropriate control (con.) line were grown to 40% confluency, with or without Muristerone (Mur.) as indicated, during approximately 48 h. Thereafter the medium was replaced with fresh GM, with or without Muristerone, and after a 12-h incubation the cells were lysed. The levels of p204 in the lysates were determined by immunoblotting, and the levels of Id2, Id1, and Id3 were determined by immunoprecipitation followed by immunoblotting. For further details, see Materials and Methods.
To identify the p204 segment(s) to which one of the Id proteins, Id2, binds, various such segments were linked to GST (Fig. 3). Pull-down assays involving the immobilized p204 segments, an extract from C2C12 cells, and Western blotting with antiserum to Id2 were performed. These assays revealed a strong binding of Id2 to the p204 b segment, with somewhat more to the b1 than to the b2 segment, and no or only very weak binding to the N-terminal and a segments (Fig. 3A and B).
FIG. 3.
Id2 binds to the b1 and b2 segments in p204. (A) Schematic diagrams of GST-204 fusion proteins used to map the sites of Id2 binding on p204. The numbers refer to amino acid residues in p204. N, N terminal; a1, a2, b1, and b2, segments encoded by single exons. The a1 and a2 segments constitute the a segment, and the b1 and b2 segments constitute the b segment. The strengths of the binding of Id2 to the various GST-204 segments, as shown in panel B, are indicated. (B) Binding of Id2 by p204 and its segments. Glutathione-Sepharose beads carrying GST, GST-204, or its segments, as indicated, were incubated with extracts prepared from C2C12 myoblasts, and the bound Id2 was detected by immunoblotting with anti-Id2 antibodies. The Id2 band is indicated. (C) Expression of free GST or of GST linked to p204 or its segments. Samples (0.5 μg) of affinity-purified GST, GST-204, or its segments, as indicated, were examined by SDS-PAGE and Coomassie blue staining. The positions of size markers are indicated. For further details, see Materials and Methods.
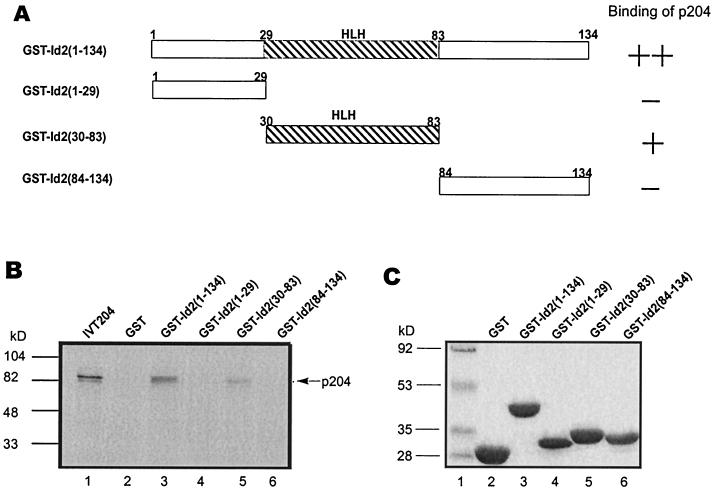
To identify the segment(s) in Id2 to which p204 binds, Id2 and three of its segments were linked to GST (Fig. 4A). Pull-down assays involving immobilized Id2 and its segments and 35S-p204 translated in vitro were performed (Fig. 4B and C). These assays revealed binding of p204 to the complete GST-Id2 and its middle segment (amino acids 30 to 83) but not to its N- and C-terminal segments. The middle segment includes the HLH domain (amino acids 35 to 75) of Id2 (49, 54). The HLH domain constitutes the longest sequence that is partially conserved among the four murine Id proteins. This makes it probable that the Id proteins (other than Id2) might also bind p204 by their HLH domains. This is also the domain in the Id proteins which can bind to the HLH domains of MyoD and some other HLH proteins. The binding of p204 to the HLH domain in the middle fragment (amino acids 30 to 83) from Id2 suggests that the HLH might fold into its proper conformation even in the fragment. The lack of binding of p204 to the N-terminal and C-terminal Id2 segments in our experiments does not necessarily reflect the situation involving the complete Id2 protein. Thus, despite the fact that equal weights (and not moles) of GST-Id2(1-134) and GST-Id2(30-83) were used, more p204 was retained on the full-size GST-Id2 than on its middle segment. Both the N- and C-terminal Id2 segments are short, and their conformations do not necessarily correspond to those within intact Id2.
FIG. 4.
p204 binds to the HLH segment of Id2, as determined by GST pull-down assay in vitro. (A) Schematic diagrams of GST-Id2 and GST-Id2 segment fusion proteins used to identify an Id2 segment binding to p204. The numbers refer to amino acid residues in Id2. The strength of binding or lack of binding of the various GST-Id2 segments to p204, as shown in panel B, is indicated. (B) 35S-labeled p204 translated in a reticulocyte lysate (IVT204) was incubated with glutathione-Sepharose beads carrying GST, GST-Id2, or GST-Id2 segment fusion proteins as indicated. The beads were washed and eluted, and the released proteins were analyzed by SDS-PAGE and autoradiography (lanes 2 to 6). As a control, p204 translated in vitro was also run (lane 1). The p204 band is indicated by an arrow. (C) Aliquots (0.5 μg) of free GST or GST linked to Id2 or to the indicated Id2 segments expressed in E. coli and purified on glutathione-Sepharose were examined by SDS-PAGE and Coomassie blue staining. For further details, see Materials and Methods.
p204 overcomes the inhibition by the Id proteins of the binding to DNA of the MyoD and E47 homodimers and the MyoD-E47 heterodimer.
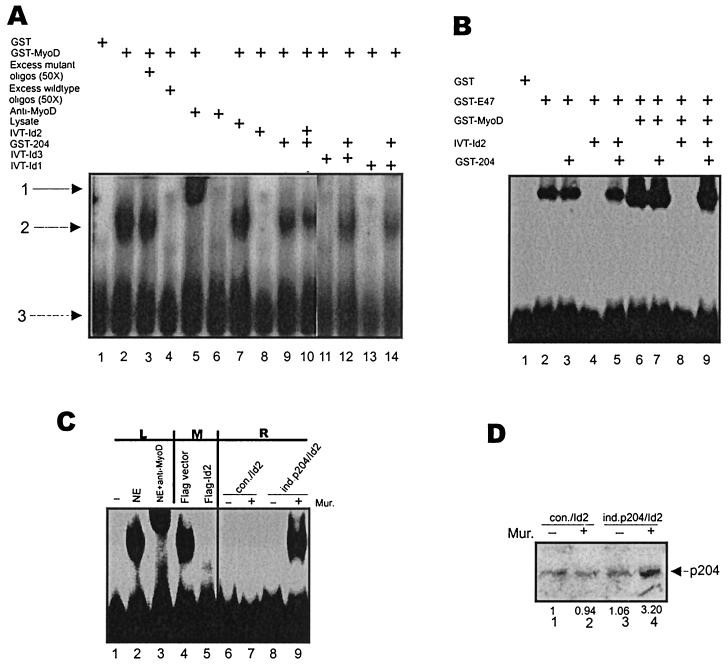
The MyoD and E47 homodimers and also the MyoD-E47 heterodimer can bind to the E-box sequences in DNA. This binding is inhibited by the Id proteins, which can form heterodimers with MyoD and also with E47. We first tested in an EMSA whether purified p204 (i.e., GST-204) can overcome the inhibition by the Id proteins of the binding of purified MyoD (i.e., GST-MyoD) to an E-box sequence. The binding of MyoD to an E-box sequence in vitro (Fig. 5A, lane 2) was competed by excess wild-type (lane 4) but not mutant (lane 3) oligodeoxynucleotide, and the MyoD-E-box sequence band was supershifted by antibodies to MyoD (lane 5). As expected, Id2, Id3, or Id1 (translated in a reticulocyte lysate) inhibited the binding of MyoD to the E-box sequence (Fig. 5A, lanes 8, 11, and 13), whereas the lysate did not (lane 7). GST-204, which did not affect the binding of MyoD to an E-box sequence (lane 9), clearly did overcome the inhibition of the binding by any one of the three Id proteins (lanes 10, 12, and 14). Thereafter we tested whether purified p204 can also overcome the inhibition by an Id protein of the binding of purified E47 (i.e., GST-E47) and of MyoD-E47 (i.e., GST-MyoD-GST-E47) heterodimers to an E-box sequence. The data in Fig. 5B indicate that it can. The binding of E47 to an E-box sequence (lane 2) was unaffected by p204 (lane 3); it was inhibited by Id2 (lane 4), and this inhibition was overcome by p204 (lane 5). Moreover, the strong binding to an E-box sequence by a mixture of MyoD and E47 (i.e., presumably mainly heterodimer) (Fig. 5B, lane 6) was unaffected by p204 (lane 7); it was inhibited by Id2 (lane 8), and this inhibition was overcome by p204 (lane 9). Proteins in the nuclear extract from a C2C12 myoblast culture also bound to an E-box sequence (Fig. 5C, lanes 1 and 2). The proteins binding included MyoD, as revealed by the supershift of the bulk of the band by antibodies to MyoD (lane 3). As expected, the transfection of a construct encoding Flag-Id2 (but not that of a construct encoding Flag vector) into the cultured C2C12 myoblasts inhibited the binding to E-box sequences of proteins (including MyoD) in the nuclear extract from the culture (Fig. 5C, lanes 4 and 5).
FIG. 5.
p204 can overcome the inhibition of the sequence-specific binding to DNA of MyoD, E47, and the MyoD-E47 heterodimer by the Id proteins. (A) Id2, Id1, and Id3 inhibit the sequence-specific binding of MyoD to the MEF-1 oligodeoxynucleotide in vitro, and p204 overcomes the inhibition, as determined by EMSA. The proteins indicated, i.e., GST (0.5 μg); GST-MyoD (0.5 μg); Id2, Id1, or Id3 translated in a reticulocyte lysate (IVT-Id2, IVT-Id1, and IVT-Id3, respectively) (0.5 μg); and GST-204 (1 μg), were mixed in the reaction buffer (20 μl). For competition experiments, a 50-fold excess of wild-type or mutant MEF-1 oligodeoxynucleotide was added. For supershift assays, anti-MyoD IgG (0.5 μg) was included. After 15 min of incubation the 32P-MEF-1 probe was added, and the reaction mixture was incubated for a further 20 min and analyzed by gel electrophoresis. The positions of the supershifted MyoD-DNA complex (arrow 1), the MyoD-DNA complex (arrow 2), and the free DNA probe (arrow 3) are indicated. (B) Id2 inhibits the sequence-specific binding of E47 homodimers and MyoD-E47 heterodimers to the MEF-1 oligodeoxynucleotide in vitro, and p204 overcomes the inhibition. The proteins indicated, i.e., GST (0.5 μg), GST-MyoD (0.5 μg), and Id2 translated in a reticulocyte lysate (0.5 μg), were mixed in the reaction buffer (20 μl) and incubated with the 32P-MEF-1 probe for 15 min. Thereafter GST-204 (1 μg) was added to the reaction mixture, and it was incubated for another 20 min and analyzed by gel electrophoresis. (C) Transfection of a Flag-Id2 expression plasmid inhibits the binding of C2C12 nuclear proteins to the MEF-1 oligodeoxynucleotide, and induced p204 overcomes the inhibition. EMSA was performed using 10 μg of nuclear extract proteins from the indicated cells cultured in growth medium and the 32P-MEF-1 oligodeoxynucleotide. L, EMSA with no cell extract (lane 1), with nuclear extract (NE) from C2C12 control cells (lane 2), and with nuclear extract from C2C12 control cells but supplemented with anti MyoD antiserum (lane 3). M, nuclear extract from control cells which had been transfected with Flag vector (lane 4) and nuclear extract from control cells which had been transfected with Flag-Id2 plasmid (lane 5). R, the Muristerone (Mur.)-inducible line (ind.p204/Id2) and the control line (con./Id2) were incubated in growth medium in the absence or presence of 2.5 μM Muristerone for 48 h. (The effects of this incubation on the p204 levels in the two cell lines are shown in panel D.) EMSA was performed using 10 μg of nuclear extract proteins and the 32P-MEF-1 oligodeoxynucleotide. EMSA with nuclear extracts from con./Id2 cells (lanes 6 and 7) and ind.p204/Id2 cells (lanes 8 and 9) is shown. (D) Induction of p204 expression by Muristerone in the ind.p204/Id2 line but not in the con./Id2 line. Both lines were incubated in GM in the presence (+) or absence (−) of 2.5 μM Muristerone for 48 h. The effects of this incubation on the p204 level were determined by Western blotting. The p204 band is indicated by an arrow. The fold induction of p204 is shown. For further details, see Materials and Methods.
We generated two cell lines for establishing whether p204, when overexpressed in cells, can overcome this inhibition by Id2. We transfected the Flag-Id2 plasmid (i) into a cell line derived from C2C12 cells in which p204 expression could be increased approximately threefold upon incubation with Muristerone (47) (thereby obtaining, after cloning, the ind.p204/Id2 line) and (ii) into the appropriate control line in which Muristerone did not affect the expression of p204 (thereby obtaining, after cloning, the con./Id2 line). As expected, treatment with Muristerone increased the p204 level (approximately threefold) in the ind.p204/Id2 line without affecting this level in the con./Id2 line (Fig. 5D). After exposure to Muristerone, the inhibition of the binding of nuclear proteins to the E-box sequence by Id2 was overcome in the case of the ind.p204/Id2 line (Fig. 5C, lanes 8 and 9) but not in the case of the con./Id2 line (lanes 6 and 7).
p204 overcomes the inhibition by Id1, Id2, and Id3 of MyoD-dependent transcription.
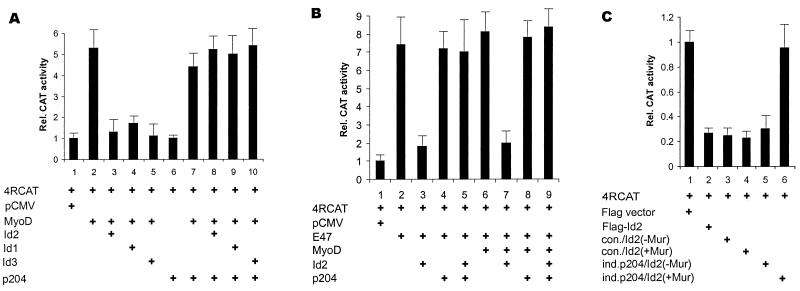
The findings in the previous section prompted us to explore the effect of overexpressing p204 on the inhibition of the MyoD- and E47-dependent expression of a reporter gene by the Id proteins. The reporter gene used, p4RCAT, contained E-box sequences (71). These sequences are binding sites, for, e.g., MyoD, myogenin, E47, and some other bHLH proteins (9, 23, 39, 40). Transfection of a Flag-Id2 plasmid into CV1 cells was shown to result in the inhibition of the activity of the pMoCAT reporter construct including such E-box sequences (51). We established that in 10T1/2 murine fibroblasts the expression of transfected MyoD boosted the p4RCAT reporter activity approximately fivefold (Fig. 6A, bars 1 and 2). Cotransfection of a plasmid encoding Id2, Id1, or Id3 with a MyoD plasmid inhibited the reporter activity over threefold (bars 3, 4, and 5). The cotransfection of a p204 plasmid with the MyoD and Id2, Id3, or Id1 plasmids overcame the inhibition by any one of the three Id proteins (bars 8, 9, and 10). Transfection of a p204 plasmid without transfection of an Id2 plasmid and with or without transfection of a MyoD plasmid affected the reporter activity barely, if at all (bars 6 and 7). A further control experiment revealed that the cotransfection of a p204 plasmid with the MyoD and Flag-Id2 plasmids did not appreciably affect the levels of MyoD and Flag-Id2 RNAs (not shown).
FIG. 6.
p204 can overcome the inhibition of the MyoD-, E47-, or MyoD-E47 heterodimer-dependent transcription of a reporter gene by the Id proteins. (A) Experiments with a 10T1/2 line. One microgram of p4RCAT reporter construct was transfected into 10T1/2 cells together with 1 μg of pSVGal internal control plasmid and 1 μg each of the expression plasmid(s) indicated: pCMV, MyoD, Id2, Id1, Id3, or p204 (standing for pCSA-MyoD, pFlag-Id2, pFlag-Id1, pFlag-Id3, and pCMV-204, respectively). At 48 h after transfection, the cultures were harvested and lysed, and the β-galactosidase and CAT activities were determined. The CAT activities were normalized to the β-galactosidase activities. This normalized activity was taken as 1 in the case of cells transfected with the pCMV vector. The standard deviations based on three experiments are indicated. (B) Experiments with a 10T1/2 line. One microgram of p4RCAT reporter construct was transfected into 10T1/2 cells together with 1 μg of pSVGal internal control plasmid and 1 μg each of the expression plasmid(s) indicated: pCMV, E47, MyoD, Id2, and p204 (standing for pCMV-E47, pCSA-MyoD, pFlag-Id2, and pCMV-204, respectively). However, in the case of cotransfection with E47 and MyoD, 0.5 μg of each plasmid was used. At 48 h after transfection, the cultures were processed as described for panel A. The standard deviations based on three experiments are indicated. (C) Experiments with C2C12 cell lines. The same four cell lines were used as in Fig. 5C. One microgram of p4RCAT reporter construct was cotransfected with 1 μg of pSVGal internal control plasmid into each of the stable lines: C2C12 transfected with the Flag vector, C2C12 transfected with the Flag-Id2 plasmid, con./Id2, and ind.p204/Id2. Two dishes were used for each of the con./Id2 and ind.p204/Id2 cultures. One of these two dishes was incubated without Muristerone (−Mur), and the other was incubated with 2.5 μM Muristerone (+Mur). The cultures were incubated in GM for 48 h and processed as described for panel A. The normalized activity in cells transfected with the Flag vector was taken as 1. The standard deviations based on three experiments are indicated. For further details, see Materials and Methods.
In the 10T1/2 murine fibroblasts the expression of transfected E47 boosted the reporter activity over sevenfold (Fig. 6B, bars 1 and 2). Cotransfection of a plasmid encoding Id2 inhibited the reporter activity over threefold. The cotransfection of a p204 plasmid with the E47 and Id2 plasmids overcame the inhibition by the Id protein (bar 5). Furthermore, transfection of a p204 plasmid with the E47 plasmid affected the reporter activity barely if at all (bar 4). Cotransfection of plasmids encoding MyoD and E47 boosted the reporter activity about eightfold (bar 6). Cotransfection also of a plasmid encoding Id2 inhibited the reporter activity about fourfold (bar 7). The cotransfection of a p204 plasmid with the E47, MyoD, and Id2 plasmids overcame the inhibition by the Id2 protein (bar 9). Furthermore, cotransfection of a p204 plasmid with MyoD and E47 plasmids affected the reporter activity barely, if at all (bar 8).
We also examined the effect of p204 on the inhibition of the activity of the same reporter gene by Id2 in our two C2C12 cell lines (Fig. 6C). The transfection of a Flag-Id2 plasmid decreased the reporter activity to less than one-third of that in the control culture (bars 1 and 2). Incubation with Muristerone, which in the case of the ind.p204/Id2 cell line resulted in a threefold increase in the p204 level (Fig. 5C, bars 3 and 4), increased the reporter gene activity more than threefold (Fig. 6C, bars 5 and 6), thus overcoming the inhibition by Id2. In the case of con./Id2 cells, in which the treatment with Muristerone did not affect the p204 level (Fig. 5C, bars 1 and 2), the reporter gene activity did not increase upon incubation with Muristerone (actually, it slightly decreased) (Fig. 6B, bars 3 and 4). The results in Fig. 6A and B clearly indicate that the increase in p204 expression can overcome the inhibition of the activity of a reporter gene with E-box enhancers by Id1, Id2, or Id3.
p204 overcomes the inhibition by Id2 of the fusion of myoblasts to myotubes.
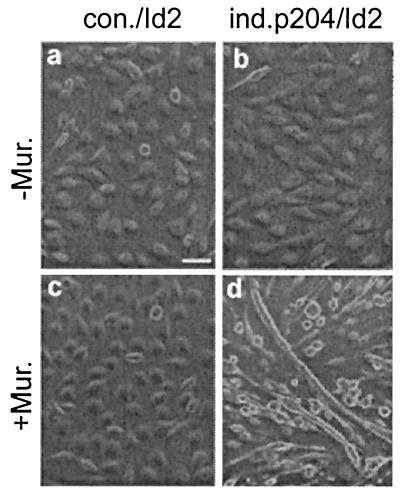
These findings prompted us to explore whether p204 could also overcome the demonstrated inhibition by Id2 of the fusion of cultured myoblasts to myotubes (51). The data in Fig. 7 reveal that indeed it could. Myoblasts from the ind.p204/Id2 line expressing Flag-Id2 did fuse to myotubes after incubation with Muristerone in GM and a shift to DM (Fig. 7d). If incubated without Muristerone, they did not fuse (Fig. 7b). The myoblasts from the control con./Id2 line did not fuse irrespective of whether they were incubated in GM without (Fig. 7a) or with (Fig. 7c) Muristerone prior to being shifted to DM.
FIG. 7.
p204 can overcome the inhibition of myoblast fusion to myotubes by Id2. Two dishes were used from each of the con./Id2 and ind.p204/Id2 cultures. One of the two dishes was incubated without Muristerone (−Mur.) and the other was incubated with 2.5 μM Muristerone (+Mur.) in GM for 48 h. All four dishes were shifted to differentiation medium (without Muristerone) for 4 days prior to observation under a phase-contrast microscope. Bar, 150 μm. For further details, see Materials and Methods.
Involvement of p204 in control of the Id1, Id2, and Id3 levels in myoblasts.
The data presented so far revealed that p204 (i) is required in myoblast differentiation, (ii) can overcome the inhibition of this process by Id2 (and presumably by the other Id proteins), and (iii) binds to the Id proteins and overcomes the inhibition of MyoD activity by Id1, Id2, or Id3. We wished to examine whether, in addition to binding and sequestering Id proteins, p204 can also affect their level during proliferation and differentiation.
This is of interest since in the course of the fusion of C2C12 myoblasts to myotubes, as triggered by shifting the culture from GM to DM, the level of Id1, Id2, or Id3 strongly decreases (5). This is a consequence of (i) the dependence of the transcription of the Id genes on mitogenic growth factors (3) and (ii) the translocation of the Id proteins from the nucleus to the cytoplasm (70), followed by their degradation in the cytoplasm by the ubiquitin pathway (7). To explore whether p204 affects the level of the Id proteins during myoblast proliferation and differentiation, we used two types of derivatives of the C2C12 line: (i) cell lines in which the level of p204 was decreased by the expression of 204 antisense RNA (p204AS) and (ii) cell lines in which p204 was inducible by Muristerone (ind.p204).
Three cell lines expressing 204 antisense RNA were used: p204AS1, p204AS6, and p204AS8. These three were among the six lines noted earlier to be unable to fuse to myotubes after shifting their culture to DM. A comparison of the p204 and Id2 levels in the control line with those in p204AS8, one of the lines expressing 204 antisense RNA, is shown in Fig. 8A. In the cases of the other two cell lines expressing 204AS RNA (not shown), the patterns are similar to the one illustrated. As expected, the p204 level was lower in the 204AS line than in the control line. This was the case both for cultures in GM and for cultures shifted to DM for 48 h. At that time myoblast fusion had started in the control culture but not in the cultures expressing 204AS RNA, in which the fusion was inhibited. In cultures in GM the Id2 level was higher in the 204AS line than in the control line. The shift from GM to DM resulted in a decrease in the Id2 level in both cultures. However, Id2 disappeared (its level decreased below detectability under our conditions) in the control line, whereas Id2 persisted at a diminished level in the 204AS line (Fig. 8A).
The effect of increasing the p204 level by incubation with Muristerone on the levels of Id2, Id1, and Id3 in cultures grown in GM are shown in Fig. 8B. Muristerone had no significant effect on either the p204 or the Id1, Id2, or Id3 levels in the control line. However, in the line (ind.p204) in which Muristerone increased the p204 level (approximately 2.8-fold), it strongly decreased the levels of the Id proteins (for Id2, approximately 6.3-fold; for Id1, approximately 3.8-fold; and for Id3, approximately 3.6-fold). This line (but not the control line) started to fuse 60 h after the addition of Muristerone. In a second Muristerone-inducible line, in which the increase of p204 was approximately 2.0-fold, the decrease in the Id2 level was approximately 5.6-fold (not shown). These data reveal the important role of p204 in the control of the levels of the three Id proteins: an increase in the p204 level can result in a strong decrease in the level of Id proteins, whereas a decrease in the p204 level interferes with the disappearance of Id2 after a culture is shifted from GM to DM. Moreover, significantly, p204 was shown also to affect the Id protein level in a culture kept in GM. Thus, it is reasonable to assume that the myoblast fusion, elicited by the overexpression of p204 as induced by Muristerone in a culture in GM (47), depends on the strong decrease in the levels of the Id proteins (as demonstrated here in Fig. 8B).
DISCUSSION
This project concerned the interactions between the interferon- and MyoD-inducible p204 protein and the Id proteins and the role of these interactions in skeletal muscle differentiation. Most of the experiments involved cultured C2C12 myoblasts. The inclusion of the Id1, Id2, and Id3 proteins in the study was based on their detection in a murine thigh muscle extract by immunoprecipitation and immunoblotting (Fig. 2A). Thigh muscle contains, in addition to multinucleated myotubes, also smaller amounts of mononucleated precursors termed satellite cells. These cells are quiescent, but they can become activated and, after a limited number of divisions, differentiate to myotubes (6, 63). No efforts were made to determine the distribution of Id1, Id2, and Id3 between myotubes and satellite cells. It was reported that Id2 RNA was not detected in a (commercial) poly(A)+ RNA preparation from murine skeletal muscle by Northern blotting (38). The basis of this possibly apparent discrepancy remains to be established.
Our study revealed that (i) the lowering of the p204 level in C2C12 myoblasts by 204 antisense RNA resulted in an increase in their levels of Id1, Id2, and Id3 and in the inhibition of their fusion to myotubes; (ii) p204 bound Id1, Id2, and Id3 in vitro and in vivo; (iii) in the binding of p204 to Id2, the b segment of p204 and the HLH segment of Id2 were involved; and (iv) an increase in the p204 level in the myoblasts resulted in a decrease in the levels of Id1, Id2, and Id3 and also could overcome the inhibition by Id1, Id2, and Id3 of (a) the sequence-specific binding of MyoD, E47, and the MyoD-E47 heterodimer to E-box sequences in vitro, (b) the MyoD-, E47- and MyoD-E47 heterodimer-dependent expression of reporter genes with E-box sequences in myoblasts, and (c) the inhibition by Id2 of the fusion of myoblasts to myotubes. These and earlier results suggest that the increase in the p204 level may enable myoblast differentiation by decreasing the level of Id proteins and by overcoming the inhibition of the activities of MyoD, E47, and the MyoD-E47 heterodimer (and conceivably of other relevant bHLH proteins) by Id1, Id2, and Id3. Other experiments established that p204 is required for skeletal muscle differentiation also in the case of P19 undifferentiated stem cells derived from murine teratocarcinoma (B. Ding et al., unpublished data).
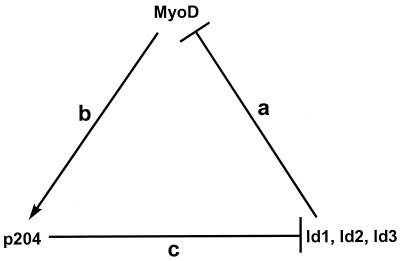
There is a regulatory circuit involving the Id1, Id2, Id3, MyoD, and p204 proteins (Fig. 9): the Id proteins can bind MyoD and E47 and inhibit their various activities (51, 54, 66) (Fig. 9, step a), MyoD can transactivate the expression of p204 during myoblast differentiation (47) (step b), and p204 can bind the Id proteins and prevent the inhibition of MyoD (and E47) activity by the Id proteins (this study) (step c). The interactions among MyoD, p204, and the Id proteins result in a positive feedback loop: p204, whose expression is triggered by MyoD, can boost the transcriptional activity of MyoD towards its various target genes (26, 58, 72), including Ifi204, the gene encoding p204. This is a consequence of the binding to Id proteins of p204, which can overcome the inhibition of MyoD activity by the Id proteins.
FIG. 9.
Regulatory circuit among the Id1, Id2, Id3, MyoD, and p204 proteins. a, binding and inhibition of activity; b, activation of transcription; c, binding, inhibition of activity, and downregulation of the level. Myogenin can substitute for MyoD in the transactivation of the gene encoding p204 (47). The Id proteins also inhibit MyoD activity indirectly, e.g., by binding the E12 and E47 proteins, with which MyoD forms heterodimers (66). p204 also binds MyoD but weakly (this study [data not shown]). In addition to binding and sequestering Id1, Id2, and Id3, p204 is also involved in controlling the levels of these proteins (this Fig. 8 and the text).
p204 can affect the Id proteins not only by binding, and thereby sequestering them but also by triggering a decrease in their level. The mechanism(s) of this effect remains to be explored. It is possible that p204, which binds the Id proteins, mediates the translocation of the bound Id proteins from the nucleus to the cytoplasm, thereby accelerating their degradation by the ubiquitin pathway (7, 19, 41, 61). The probability of such a boost of the translocation of the Id proteins is increased by the fact that p204 has (whereas Id proteins lack) a nuclear export signal and that much of p204 is phosphorylated and translocated from the nucleus to the cytoplasm during myoblast fusion (47). Such a hypothetical removal of Id proteins from the nucleus to the cytoplasm by p204 during differentiation would resemble the stimulation of the translocation (i) of Id proteins from the cytoplasm to the nucleus by an E protein (19) and (ii) of HDAC5, an inhibitor of differentiation, from the nucleus to the cytoplasm by the 14-3-3 protein during myoblast fusion (50). However, it is conceivable that p204 also decreases the Id protein levels by other mechanisms (e.g., by affecting the synthesis, turnover, or translation of the Id mRNAs).
The differentiation of cultured myoblasts to myotubes in vitro is triggered by shifting them from high-serum medium (GM) to low-serum medium (DM) (75). The trigger(s) of myoblast differentiation in vivo has not been identified. The ability of p204 to overcome the Id block of differentiation makes it a possible candidate for serving as a component of the trigger. This is especially the case since overexpression of p204 results in a strong decrease in the Id protein levels (this study) and triggers the fusion of cultured myoblasts even in high-serum medium (47).
The level of p202a, another member of the p200 protein family, also increases during myoblast differentiation, though after a delay. The multiple activities of p202a make its role in myoblast fusion incompletely understood (17, 47). The antiproliferative and antiapoptotic activities of p202a, together with its inhibition of the transcriptional activity of c-Myc (34, 52, 69) (which may result in a decrease of the expression of Id2 [38]), could all support differentiation. Furthermore, p202a binds MyoD and inhibits its sequence-specific binding to DNA, as well as its synthesis (17). This might account for the decrease in the level of MyoD during differentiation. The inhibition of the activity and synthesis of MyoD by p202a might also account for the apparently paradoxical finding that whereas an increase in p204 level prior to induction of differentiation accelerates myoblast differentiation (47), an increase in the p202a level prior to induction of differentiation inhibits this process (17).
p204, and also p202a, can bind to pRb and other pocket proteins (12, 29, 46). The significance of these interactions in the process of muscle differentiation remains to be explored.
Id proteins are involved in modulating the differentiation of many tissues besides skeletal muscle (e.g., in the cases of hematopoiesis or neurogenesis, etc.) (15, 48, 53, 57, 70, 76). Thus, it is likely that p204, and possibly other p200 family proteins, may also be involved in regulating these processes. It is conceivable that an increase in the p204 level might facilitate differentiation in the cases of some malignancies based on impaired differentiation, especially those in which an Id protein is overexpressed (33). The finding that Notch signaling elicits the transcription of the 204 gene in murine thymocytes in the process of maturation may indicate the participation of p204 in this process (20).
Acknowledgments
We thank A. Cano, B. A. Christy, M. A. Israel, A. Lassar, and N. Speck for plasmids and E. Vellali for preparing the manuscript for publication. We thank the Department of Biological Science, University of Iowa, Iowa City, for the MF20 monoclonal antibodies.
These studies were supported by a research grant from the NIH (2 R21 AI12320) and a postdoctoral fellowship to H.W. from the Cancer Research Foundation.
REFERENCES
- 1.Atherton, G. T., H. Travers, R. Deed, and J. D. Norton. 1996. Regulation of cell differentiation in C2C12 myoblasts by the Id3 helix-loop-helix protein. Cell Growth Differ. 7:1059-1066. [PubMed] [Google Scholar]
- 2.Bader, D., T. Masaki, and D. A. Fischman. 1982. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J. Cell Biol. 96:763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belletti, B., M. Prisco, A. Morrione, B. Valentinis, M. Navarro, and R. Baserga. 2001. Regulation of Id2 gene expression by the insulin-like growth factor I receptor requires signaling by phosphatidylinositol 3-kinase. J. Biol. Chem. 276:13867-13874. [DOI] [PubMed] [Google Scholar]
- 4.Benezra, R., R. Davis, D. Lockshon, D. Turner, and H. Weintraub. 1990. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61:49-59. [DOI] [PubMed] [Google Scholar]
- 5.Biggs, J. R., Y. Zhang, and E. V. Murphy. 1995. Repression of the Id2 (inhibitor of differentiation) gene promoter during exit from the cell cycle. J. Cell Physiol. 164:249-258. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff, R. 1994. The satellite cell and muscle regeneration, p. 97-118. In A. G. Engel and C. Franzini-Armstrong (ed.), Myology, vol. 1. McGraw-Hill, Inc., New York, N.Y. [Google Scholar]
- 7.Bounpheng, M. A., J. J. Dimas, S. G. Dodds, and B. A. Christy. 1999. Degradation of Id proteins by the ubiquitin-proteasome pathway. FASEB J. 13:2257-2264. [PubMed] [Google Scholar]
- 8.Briggs, J. A., G. R. Burrus, B. D. Stickney, and R. C. Briggs. 1992. Cloning and expression of the human myeloid cell nuclear differentiation antigen: regulation by interferon alpha. J. Cell. Biochem. 49:82-92. [DOI] [PubMed] [Google Scholar]
- 9.Buskin, J. N., and S. D. Hauschka. 1989. Identification of a myocyte nuclear factor that binds to the muscle-specific enhancer of the mouse muscle creatine kinase gene. Mol. Cell. Biol. 9:2627-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choubey, D., and J. U. Gutterman. 1997. Inhibition of E2F-4/DP-1-stimulated transcription by p202. Oncogene 15:291-301. [DOI] [PubMed] [Google Scholar]
- 11.Choubey, D., and P. Lengyel. 1992. Interferon action: nucleolar and nucleoplasmic localization of the interferon-inducible 72-kD protein that is encoded by the Ifi204 gene from the gene 200 cluster. J. Cell Biol. 116:1333-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choubey, D., and P. Lengyel. 1995. Binding of an interferon-inducible protein (p202) to the retinoblastoma protein. J. Biol. Chem. 270:6134-6140. [DOI] [PubMed] [Google Scholar]
- 13.Choubey, D., S.-J. Li, B. Datta, J. U. Gutterman, and P. Lengyel. 1996. Inhibition of E2F-mediated transcription by p202. EMBO J. 15:5668-5678. [PMC free article] [PubMed] [Google Scholar]
- 14.Choubey, D., J. Snoddy, V. Chaturvedi, E. Toniato, G. Opdenakker, A. Thakur, H. Samanta, D. A. Engel, and P. Lengyel. 1989. Interferons as gene activators. Indications for repeated gene duplication during the evolution of a cluster of interferon-activatable genes on murine chromosome 1. J. Biol. Chem. 264:17182-17189. [PubMed] [Google Scholar]
- 15.Cooper, C. L., G. Brady, F. Bilia, N. N. Iscove, and P. J. Quesenberry. 1997. Expression of the Id family helix-loop-helix regulators during growth and development in the hematopoietic system. Blood 89:3155-3165. [PubMed] [Google Scholar]
- 16.Datta, B., B. Li, D. Choubey, G. Nallur, and P. Lengyel. 1996. p202, an interferon-inducible modulator of transcription, inhibits transcriptional activation by the p53 tumor suppressor protein, and a segment from the p53-binding protein 1 that binds to p202 overcomes this inhibition. J. Biol. Chem. 271:27544-27555. [DOI] [PubMed] [Google Scholar]
- 17.Datta, B., W. Min, S. Burma, and P. Lengyel. 1998. Increase in p202 expression during skeletal muscle differentiation: inhibition of MyoD protein expression and activity by p202. Mol. Cell. Biol. 18:1074-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis, R. H., H. Weintraub, and A. B. Lassar. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51:987-1000. [DOI] [PubMed] [Google Scholar]
- 19.Deed, R. W., S. Armitage, and J. D. Norton. 1996. Nuclear localization and regulation of Id protein through an E protein-mediated chaperone mechanism. J. Biol. Chem. 271:23603-23606. [DOI] [PubMed] [Google Scholar]
- 20.Deftos, M. L., E. Huang, E. W. Ojala, K. A. Forbush, and M. J. Bevan. 2000. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity 13:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeYoung, K. I., M. E. Ray, Y. A. Su, S. L. Anzick, R. W. Johnstone, J. A. Trapani, P. S. Meltzer, and J. M. Trent. 1997. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene 15:453-457. [DOI] [PubMed] [Google Scholar]
- 22.D'Souza, S., H. Xin, S. Walter, and D. Choubey. 2001. The gene encoding p202, an interferon-inducible negative regulator of the p53 tumor suppressor, is a target of p53-mediated transcriptional repression. J. Biol. Chem. 276:298-305. [DOI] [PubMed] [Google Scholar]
- 23.Edmondson, D. G., and E. N. Olson. 1989. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 3:628-640. [DOI] [PubMed] [Google Scholar]
- 24.Geng, Y., S. D'Souza, H. Xin, S. Walter, and D. Choubey. 2000. p202 levels are negatively regulated by serum growth factors. Cell Growth Differ. 11:475-483. [PubMed] [Google Scholar]
- 25.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 26.Halevy, O., B. G. Novitch, D. B. Spicer, S. X. Skapek, J. Rhee, G. J. Hannon, D. Beach, and A. B. Lassar. 1995. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 267:1018-1021. [DOI] [PubMed] [Google Scholar]
- 27.Helin, K., J. A. Lees, M. Vidal, N. Dyson, E. Harlow, and A. Fattaey. 1992. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell 70:337-350. [DOI] [PubMed] [Google Scholar]
- 28.Hertel, L., M. DeAndrea, B. Azzimonti, A. Rolle, M. Gariglio, and S. Landolfo. 1999. The interferon-inducible 204 gene, a member of the Ifi200 family, is not involved in the antiviral state induction by IFN-alpha, but is required by the mouse cytomegalovirus for its replication. Virology 262:1-8. [DOI] [PubMed] [Google Scholar]
- 29.Hertel, L., S. Rolle, M. DeAndrea, B. Azzimonti, R. Osello, G. Gribaudo, M. Gariglio, and S. Landolfo. 2000. The retinoblastoma protein is an essential mediator that links the interferon-inducible 204 gene to cell-cycle regulation. Oncogene 19:3598-3608. [DOI] [PubMed] [Google Scholar]
- 30.Iavarone, A., P. Garg, A. Lasorella, J. Hsu, and M. A. Israel. 1994. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 8:1270-1284. [DOI] [PubMed] [Google Scholar]
- 31.Jen, Y., H. Weintraub, and R. Benezra. 1992. Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes Dev. 6:1466-1479. [DOI] [PubMed] [Google Scholar]
- 32.Johnstone, R. W., and J. A. Trapani. 1999. Transcription and growth regulatory functions of the HIN-200 family of proteins. Mol. Cell. Biol. 19:5833-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleeff, J., T. Ishiwata, H. Friess, M. W. Buchler, M. A. Israel, and M. Korc. 1998. The helix-loop-helix protein Id2 is overexpressed in human pancreatic cancer. Cancer Res. 58:3769-3772. [PubMed] [Google Scholar]
- 34.Koul, D., R. Lapushin, H. J. Xu, G. B. Mills, J. U. Gutterman, and D. Choubey. 1998. p202 prevents apoptosis in murine AKR-2B fibroblasts. Biochem. Biophys. Res. Commun. 247:379-382. [DOI] [PubMed] [Google Scholar]
- 35.Kushner, D. B., and R. P. Ricciardi. 1999. Reduced phosphorylation of p50 is responsible for diminished NF-kappaB binding to the major histocompatibility complex class I enhancer in adenovirus type 12-transformed cells. Mol. Cell. Biol. 19:2169-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landolfo, S., G. Gribaudo, and D. Lembo. 1998. The Ifi 200 genes: an emerging family of IFN-inducible genes. Biochimie 80:721-728. [DOI] [PubMed] [Google Scholar]
- 37.Langlands, K., X. Yin, G. Anand, and E. V. Prochownik. 1997. Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. J. Biol. Chem. 272:19785-19793. [DOI] [PubMed] [Google Scholar]
- 38.Lasorella, A., M. Noseda, M. Beyna, and A. Iavarone. 2000. Id2 is a retinoblastoma protein target and mediates signaling by Myc oncoprotein. Nature 407:592-598. [DOI] [PubMed] [Google Scholar]
- 39.Lassar, A. B., R. L. Davis, W. E. Wright, T. Kadesch, C. Murre, A. Voronova, D. Baltimore, and H. Weintraub. 1991. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66:305-315. [DOI] [PubMed] [Google Scholar]
- 40.Lassar, A. B., S. X. Skapek, and B. Novitch. 1994. Regulatory mechanisms that coordinate skeletal muscle differentiation and cell cycle withdrawal. Curr. Opin. Cell Biol. 6:788-794. [DOI] [PubMed] [Google Scholar]
- 41.Lau, J. F., J. P. Parisien, and C. M. Horvath. 2000. Interferon regulatory factor subcellular localization is determined by a bipartite nuclear localization signal in the DNA-binding domain and interaction with cytoplasmic retention factors. Proc. Natl. Acad. Sci. USA 97:7278-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, W. S., C. C. Kao, G. O. Bryant, X. Liu, and A. J. Berk. 1991. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell 67:365-376. [DOI] [PubMed] [Google Scholar]
- 43.Lembo, M., C. Sacchi, C. Zappador, G. Bellomo, M. Gaboli, P. P. Pandolfi, M. Gariglio, and S. Landolfo. 1998. Inhibition of cell proliferation by the interferon-inducible 204 gene, a member of the Ifi 200 cluster. Oncogene 16:1543-1551. [DOI] [PubMed] [Google Scholar]
- 44.Lengyel, P., D. Choubey, S.-J. Li, and B. Datta. 1995. The interferon-activatable gene 200 cluster: from structure toward function. Semin. Virol. 6:203-213. [Google Scholar]
- 45.Liu, C. J., S. D. Dib-Hajj, and S. G. Waxman. 2001. Fibroblast growth factor homologous factor 1B binds to the C terminus of the tetrodotoxin-resistant sodium channel rNa(v)1.9a (NaN) J. Biol. Chem. 276:18925-18933. [DOI] [PubMed] [Google Scholar]
- 46.Liu, C. J., H. Wang, and P. Lengyel. 1999. The interferon-inducible nucleolar p204 protein binds the ribosomal RNA-specific UBF1 transcription factor and inhibits ribosomal RNA transcription. EMBO J. 18:2845-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, C. J., H. Wang, Z. Zhao, S. Yu, Y. Lu, J. Meyer, G. Chatterjee, S. Deschamps, B. A. Roe, and P. Lengyel. 2000. MyoD-dependent induction during myoblast differentiation of p204, a protein also inducible by interferon. Mol. Cell. Biol. 20:7024-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyden, D., A. Z. Young, D. Zagzag, W. Yan, W. Gerald, R. O'Reilly, B. L. Bader, R. O. Hynes, Y. Zhuang, K. Manova, and R. Benezra. 1999. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature 401:670-677. [DOI] [PubMed] [Google Scholar]
- 49.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulation of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2000. Activation of the myocyte enhance factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97:14400-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melnikova, I. N., M. Bounpheng, G. C. Schatteman, D. Gilliam, and B. A. Christy. 1999. Differential biological activities of mammalian Id proteins in muscle cells. Exp. Cell Res. 247:94-104. [DOI] [PubMed] [Google Scholar]
- 52.Min, W., S. Ghosh, and P. Lengyel. 1996. The interferon-inducible p202 protein as a modulator of transcription: inhibition of NF-κB, c-Fos, and c-Jun activities. Mol. Cell. Biol. 16:359-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mori, S., S. I. Nishikawa, and Y. Yokota. 2000. Lactation defect in mice lacking the helix-loop-helix inhibitor Id2. EMBO, J. 19:5772-5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norton, J. D., R. W. Deed, G. Craggs, and F. Sablitzky. 1998. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 8:58-65. [PubMed] [Google Scholar]
- 55.Olson, E. N., and W. H. Klein. 1994. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 8:1-8. [DOI] [PubMed] [Google Scholar]
- 56.Opdenakker, G., J. Snoddy, D. Choubey, E. Toniato, D. D. Pravtcheva, M. F. Seldin, F. Ruddle, and P. Lengyel. 1989. Interferons as gene activators: a cluster of six interferon-activatable genes is linked to the erythroid alpha-spectrin locus on murine chromosome 1. Virol. 171:568-578. [DOI] [PubMed] [Google Scholar]
- 57.Pan, L., S. Sato, J. P. Frederick, X. H. Sun, and Y. Zhuang. 1999. Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol. Cell. Biol. 19:5969-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parker, S. B., G. Eichele, P. Zhang, A. Rawls, A. T. Sands, A. Bradley, E. N. Olson, J. W. Harper, and S. J. Elledge. 1995. p-53-independent expression of p21 Cip1 in muscle and other terminally differentiating cells. Science 267:1024-1027. [DOI] [PubMed] [Google Scholar]
- 59.Perez-Moreno, M. A., A. Locascio, I. Rodrigo, G. Dhondt, F. Portillo, M. A. Nieto, and A. Cano. 2001. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J. Biol. Chem. 276:27424-27431. [DOI] [PubMed] [Google Scholar]
- 60.Reznikoff, C. A., D. W. Brankow, and C. Heidelberger. 1973. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res. 33:3231-3238. [PubMed] [Google Scholar]
- 61.Rosin-Arbesfeld, R., F. Townsley, and M. Bienz. 2000. The APC tumour suppressor has a nuclear export function. Nature 406:1009-1012. [DOI] [PubMed] [Google Scholar]
- 62.Rozzo, S. J., J. D. Allard, D. Choubey, T. J. Vyse, S. Izui, G. Peltz, and B. L. Kotzin. 2001. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity 15:435-443. [DOI] [PubMed] [Google Scholar]
- 63.Schultz, E. 1996. Satellite cell proliferative compartments in growing skeletal muscles. Dev. Biol. 175:84-94. [DOI] [PubMed] [Google Scholar]
- 64.Sen, G. C., and R. M. Ransohoff. 1998. Transcriptional regulation in the interferon system. Chapman and Hall, New York, N.Y.
- 65.Stark, G. R., B. R. G. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-262. [DOI] [PubMed] [Google Scholar]
- 66.Sun, X. H., N. G. Copeland, N. A. Jenkins, and D. Baltimore. 1991. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol. Cell. Biol. 11:5603-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trapani, J. A., M. Dawson, V. A. Apostolidis, and K. A. Browne. 1994. Genomic organization of IFI16, an interferon-inducible gene whose expression is associated with human myeloid cell differentiation: correlation of predicted protein domains with exon organization. Immunogenetics 40:415-424. [DOI] [PubMed] [Google Scholar]
- 68.Wang, H., G. Chatterjee, J. J. Meyer, C. J. Liu, N. A. Manjunath, P. Bray-Ward, and P. Lengyel. 1999. Characterics of three homologous 202 genes (Ifi202a, Ifi202b, and Ifi202c) from the murine interferon-activatable gene 200 cluster. Genomics 60:281-294. [DOI] [PubMed] [Google Scholar]
- 69.Wang, H., C. J. Liu, Y. Lu, G. Chatterjee, X. Ma, R. N. Eisenman, and P. Lengyel. 2000. The interferon- and differentiation-inducible p202a protein inhibits the transcriptional activity of c-Myc by blocking its association with Max. J. Biol. Chem. 275:27377-27385. [DOI] [PubMed] [Google Scholar]
- 70.Wang, S., A. Sdrulla, J. E. Johnson, Y. Yokota, and B. A. Barres. 2001. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron 29:603-614. [DOI] [PubMed] [Google Scholar]
- 71.Weintraub, H., S. D. Hauschka, and S. J. Tapscott. 1991. The MCK enhancer contains a p53 responsive element. Proc. Natl. Acad. Sci. USA 88:4570-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weintraub, H., S. J. Tapscott, R. L. Davis, M. J. Thayer, M. A. Adam, A. B. Lassar, and A. D. Miller. 1989. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. USA 86:5434-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wright, W. E., D. A. Sassoon, and V. K. Lin. 1989. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell 56:607-617. [DOI] [PubMed] [Google Scholar]
- 74.Xin, H., S. D'Souza, L. Fang, P. Lengyel, and D. Choubey. 2001. p202, an interferon-inducible negative regulator of cell growth, is a target of the adenovirus E1A protein. Oncogene 20:6828-6839. [DOI] [PubMed] [Google Scholar]
- 75.Yaffe, D., and O. Saxel. 1977. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270:725-727. [DOI] [PubMed] [Google Scholar]
- 76.Yokota, Y., A. Mansouri, S. Mori, S. Sugawara, S. Adachi, S. Nishikawa, and P. Gruss. 1999. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature 397:702-706. [DOI] [PubMed] [Google Scholar]