Abstract
The RNA polymerase II transcription factor TFIID, composed of the TATA-binding protein (TBP) and TBP-associated factors (TAFIIs), nucleates preinitiation complex formation at protein-coding gene promoters. SAGA, a second TAFII-containing multiprotein complex, is involved in transcription regulation in Saccharomyces cerevisiae. One of the essential protein components common to SAGA and TFIID is yTAFII25. We define a minimal evolutionarily conserved 91-amino-acid region of TAFII25 containing a histone fold domain that is necessary and sufficient for growth in vivo. Different temperature-sensitive mutations of yTAFII25 or chimeras with the human homologue TAFII30 arrested cell growth at either the G1 or G2/M cell cycle phase and displayed distinct phenotypic changes and gene expression patterns. Immunoprecipitation studies revealed that TAFII25 mutation-dependent gene expression and phenotypic changes correlated at least partially with the integrity of SAGA and TFIID. Genome-wide expression analysis revealed that the five TAFII25 temperature-sensitive mutant alleles individually affect the expression of between 18 and 33% of genes, whereas taken together they affect 64% of all class II genes. Thus, different yTAFII25 mutations induce distinct phenotypes and affect the regulation of different subsets of genes, demonstrating that no individual TAFII mutant allele reflects the full range of its normal functions.
Initiation of transcription of protein-encoding genes by RNA polymerase II (Pol II) requires the assembly of general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) on promoters to form a preinitiation complex (31). Preinitiation complex assembly on both TATA-containing and TATA-less promoters is often nucleated by transcription factor TFIID, which comprises the TATA-binding protein (TBP) and 14 TBP-associated factors (TAFIIs) (4, 23, 68). To date, most of the TFIID components from Saccharomyces cerevisiae, Drosophila, and humans have been identified, partially characterized, and shown to be well conserved throughout evolution (18, 19, 68).
A large number of recent studies have provided a direct molecular link between histone acetylation and transcriptional activation (reviewed in references 11 and 40). It has been shown that several previously identified coactivators/adapters of transcription possess intrinsic histone acetyltransferase (HAT) activity or are associated in large multiprotein complexes with HATs (71). One of these, the 1.8- to 2-MDa S. cerevisiae SAGA (ySAGA) complex (26), comprises at least four distinct classes of gene products: (i) the Ada proteins (yAda1, yAda2, yAda3, yGcn5 [yAda4], and yAda5 [ySpt20]), which were isolated in a genetic screen for mutations that relieve the toxicity of the strong VP16 activation domain when overexpressed as a fusion with the Gal4 DNA-binding domain in yeast cells (5); (ii) Spt proteins (ySpt3, ySpt7, ySpt8, and ySpt20), initially identified as suppressors of transcription initiation defects caused by promoter insertions of the transposable Ty element (16, 17, 22, 65); (iii) a subset of TBP-associated factors (TAFIIs), including TAFII17, TAFII25, TAFII60, TAFII68/61, and TAFII90 (27); and (iv) the product of the essential gene Tra1 (28, 66), which is also a component of the Esa1-containing HAT complex NuA4 (1).
In addition, several similar human multiprotein complexes have been characterized, such as the TBP-free TAFII-containing complex (10, 81), the PCAF/GCN5 complex (58), and the SPT3-TAFII31-GCN5 acetyltransferase complex (48), all of which contain homologues of the S. cerevisiae GCN5 HAT, ADA proteins, SPTs, TAFIIs, and the human homologue of yTRA1, TRRAP.
Initial sequence comparisons indicated that human TAFII80 (hTAFII80; homologous to Drosophila TAFII60 [dTAFII60] and yTAFII60), hTAFII31 (dTAFII40, yTAFII17), and hTAFII20 (dTAFII30α, yTAFII61/68) have obvious homology to histones H4, H3, and H2B, respectively (33, 38), and X-ray crystallography showed that dTAFII60 and dTAFII40 or hTAFII28 and hTAFII18 interact via their histone fold domains (HFDs) (6, 82). The histone fold is a protein-protein interaction motif originally described in the heterodimerization of the core histones H4 and H3 and histones H2A and H2B and their assembly into a nucleosome (3, 46). The HFD comprises three α-helices linked by two loops. The HFDs of histones H3 and H2B contain an additional α-helix extension at the N- (αN) or C-terminal (αC) end, respectively (46).
In vitro transcription assays using different cell-free systems as well as cell transfection experiments in mammalian cells suggested that TAFIIs are essential for activation of transcription in response to transcriptional activators (13, 25, 35, 50, 63, 73) and important for core promoter recognition (12, 49, 64, 75). Moreover, the largest of the TAFIIs, TAFII250, has been shown to possess enzymatic activities: a kinase activity (14, 57), a HAT activity (53), and a ubiquitin-activating/conjugating activity (59). Thus, TAFIIs seem to be involved in several steps in the regulation of transcription, but their exact role and individual contributions are unknown.
Surprisingly, data from experiments carried out by TAFII depletion or with temperature-sensitive TAFII mutants suggested that some TAFIIs are not generally required for transcription activation and that different yTAFIIs selectively affect the transcription of different subsets of genes (2, 54, 69, 80). Moreover, TAFII-dependent and TAFII-independent promoters have been described in S. cerevisiae, and it has been proposed that following heat shock, TAFII occupancy of TAFII-independent promoters increases (41, 44). Nevertheless, TAFIIs are essential in S. cerevisiae, since full gene deletions are not viable (55, 62).
In contrast to the TAFII class of SAGA subunits, all other subunits, with the exception of Tra1, are not essential for growth (72). The observation that cells containing a temperature-sensitive mutation in certain TAFII alleles arrest at particular points in the cell cycle may further support the idea that individual TAFIIs have selective effects on different subsets of genes required for cell cycle regulation. Arrest of individual yTAFII90 and yTAFII150 mutant strains occurs at the G2/M boundary, whereas yTAFII130 mutant strains arrest in the G1 phase of the cell cycle (2, 79, 80).
Furthermore, genome-wide expression analysis, comparing single conditionally temperature-sensitive mutations in genes encoding components of either TFIID or SAGA complexes (29, 34), indicated that expression of most genes requires one or more of the common TAFII subunits, suggesting that the functions of either TFIID or SAGA or both are widely required for gene expression (42), although the contribution of either complex remains to be investigated. Among the subunits shared by TFIID and SAGA are five histone fold-containing TAFIIs, yTAFII90, yTAFII68/61, yTAFII60, yTAFII25, and yTAFII17, mutants of which have been shown to affect distinct gene expression patterns in S. cerevisiae.
To date, many questions remain concerning the role of TAFIIs in transcription, including whether TAFIIs are required at most or only a small subset of promoters. The interpretation of the published data is complicated by the fact that different temperature-sensitive alleles of a single TAFII can either result in broad effects on Pol II transcription (39, 67) or only affect the transcription of a subset of genes (29, 34, 42). Furthermore, it has not yet been possible to separate the function of a TAFII in TFIID from that in SAGA.
Here we describe a structure-function study of yTAFII25 in S. cerevisiae. We define a minimal 91-amino-acid region of yTAFII25 that is necessary and sufficient for vegetative growth. This region of yTAFII25 contains a putative HFD similar to that in eight other yTAFIIs (18). The HFD of yTAFII25 was shown to mediate selective heterodimerization with TFIID-specific components yTAFII47 and yTAFII65 as well as SAGA-specific component ySPT7 in S. cerevisiae two-hybrid and bacterial coexpression assays (20).
Here we show that yTAFII25 is required for normal cell cycle progression and that different mutations in the minimal region of TAFII25 arrest yeast cell growth at different cell cycle phases with distinct phenotypes, giving evidence for the multiple functions of TAFII25. To investigate the possibility that the different phenotypes of the TAFII25 mutants studied here can be attributed to TFIID- or SAGA-specific functions of TAFII25, we examined the subunit composition of both TFIID and SAGA by immunoprecipitation and tested the genome-wide expression pattern in TAFII25 mutant strains. Our results show that different temperature-sensitive mutations differentially affect the integrity of both complexes and that each TAFII25 mutant allele influences the expression pattern of different and unique sets of genes. However, taken together, our TAFII25 mutations affect the transcription level of about 64% of all class II genes, similar to the number of genes affected by a combination of SAGA and TFIID mutants, as published by Lee et al. (42).
MATERIALS AND METHODS
Strains, medium, and yeast cell transformation.
All strains used in this study (Table 1) are isogenic DKY12 generated from strain PL3(2n) (60). S. cerevisiae strains were propagated according to standard procedures in either rich medium (YPD) or appropriate selective medium (SD) without tryptophan and/or uracil. For temperature shift experiments, cells were grown at 28°C until mid-log phase (optical density at 600 nm [OD600], 0.3 to 0.5). Prior to shifting to 37°C, an equal volume of warm (46°C) medium was added to the cultures, and the cultures were transferred to 37°C for the indicated time. Standard S. cerevisiae genetic manipulations were performed as described previously (30). Yeast cell transformation was carried out as described previously (24).
TABLE 1.
Strains
| Strain | Relevant genotype | Plasmid |
|---|---|---|
| PL3(2n) | ura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 | |
| DKY12 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | PRS316-TAF25+ |
| DKY18 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-ΔC-TAF25 |
| DKY19 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-ΔN-TAF25 |
| DKY21 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Chim. 21 |
| DKY23 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-ΔN-TAF30 |
| DKY24 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-TAF30+ |
| DKY30 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-TAF25+ |
| DKY32 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Del. 32 |
| DKY37 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Chim. 37 |
| DKY38 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Chim.38 |
| DKY41 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Del. 41 |
| DKY42 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Del. 42 |
| DKY43 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Del. 43 |
| DKY71 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Del. 71 |
| DKY72 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Del. 72 |
| DKY92 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Del. 92 |
| DKY96 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Chim. 96 |
| DKY97 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Chim. 97 |
| DKY98 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Chim. 98 |
| DKY105 | MATaura3-Δ1 his3Δ-200 leu2-Δ1 trp1::3ERE-URA3 taf25::KanMX2 | YEP10FL-Del. 105 |
Disruption of genomic TAFII25 locus.
PCR-based homologous recombination was used to replace the entire coding region of one allele of the endogenous TAFII25 gene in strain PL3(2n) by the kanamycin resistance gene (KanMX2) (78). Cells were transformed with the centromeric URA3 plasmid (PRS316) (70) harboring the wild-type yTAFII25 cDNA. The DKY12 strain was obtained from isolated spores by selection for kanamycin resistance and uracil auxotrophy. The proper gene replacement was verified by Southern blot analysis and PCR on the genomic DNA.
Construction of truncations and deletions of yTAFII25 and of chimeras between yTAFII25 and hTAFII30.
The TAFII25 constructs listed in Fig. 1 were generated by PCR, digested with XhoI and BamHI, and cloned in-frame into the corresponding sites of the 2μm expression shuttle vector YEP10-FL (TRP1) (60), containing a Flag epitope tag sequence in-frame and N-terminal to the XhoI site. The inserts in the YEP10-FL plasmid are expressed under the control of the phosphoglycerate kinase (PGK) promoter (60). All the constructs were verified by sequencing. PCR primers used to generate the different fragments are available upon request.
FIG. 1.
Mapping the functional domains of yTAFII25. (A) Schematic representation of the predicted secondary structure of the yTAFII25 and the hTAFII30 proteins. Amino acids important for the generation of the different mutants are indicated for the S. cerevisiae (top) and human (bottom) proteins. The α helices (α) and the loops (L) of the histone fold motif are also shown. (B) Schematic representation of the different deletion and chimeric mutants of yTAFII25 and hTAFII30. yTAFII25 and its derivatives are shown in grey, with the nonconserved insertion indicated by the hatched box (L2). The hTAFII30 sequences are indicated in black. Chimeric constructs between the S. cerevisiae and the human protein are characterized by their color and labeled with their amino acid positions. In the complementation assay, carried out at 28°C, a + indicates the ability of a construct to complement the lethal Δtaf25 phenotype and a − indicates noncomplementation. Asterisks indicate the severity of the slow-growth phenotype of the mutants tested at 28°C. Temperature sensitivity (ts) at 37°C is indicated by a +. When the mutant is viable at the elevated temperature, it is indicated by −, while +/− indicates slow growth at 37°C. Cell cycle arrest phenotypes are indicated by whether they arrest in the G1 or the G2/M (G2) phase of the cell cycle. (C) The noncomplementing yTAFII25 and hTAFII30 mutant proteins expressed before the plasmid shuffle. Whole-cell extracts were made from the indicated strains and analyzed by Western blotting using either the anti-Flag monoclonal antibody (lanes 1 to 4) or the 4G2 anti-hTAFII30 monoclonal antibody (lanes 5 to 8), which recognizes the C-terminal end of hTAFII30 (35). (D) The complementing yTAFII25 mutants and hTAFII30 chimeras are not overexpressed after the plasmid shuffle compared to yTAFII25 levels in wild-type PLa cells (wt). Whole-cell extracts were made from the indicated strains and analyzed by Western blot using polyclonal anti-yTAFII25 antibodies. The same blot was exposed for either 2 (lanes 1 to 3) or 40 s. To facilitate the comparison, Del. 32 is shown on both exposures. The wild-type yTAFII25, the Flag-tagged full-length yTAFII25, and the different TAFII25 mutant proteins are indicated with arrowheads. NS, nonspecific.
Complementation and temperature sensitivity tests.
To determine the abilities of the yTAFII25 mutants to complement the TAFII25 gene, a plasmid shuffle technique was used (8). Strain DKY12 was transformed with plasmid YEP10FL containing either wild-type or mutant alleles of TAFII25, and the transformants were selected for Ura+ and Trp+ prototrophy. Positive colonies were transferred to 5-fluoro-orotic acid plates to screen for cells that had lost the PRS316-TAFII25+ plasmid. Complementing Trp+ mutants were further maintained on medium with URA and without TRP. For the temperature sensitivity tests, the mutants cells were grown in YPD medium to a density of 0.4 × 107 to 1 × 107 cells/ml, and aliquots of different dilutions were spotted on YPD plates and exposed to either 28 or 37°C.
Microscopy.
Cells were grown as described above, and aliquots were taken at the given time points, directly cooled on ice, washed with phosphate-buffered saline (PBS), and mounted on slides. For phase contrast and fluorescence microscopy, a Leica DMLB microscope was used. DAPI (4prime;,6′-diamidino-2-phenylindole) staining was performed at a concentration of 1 μg/ml.
FACS.
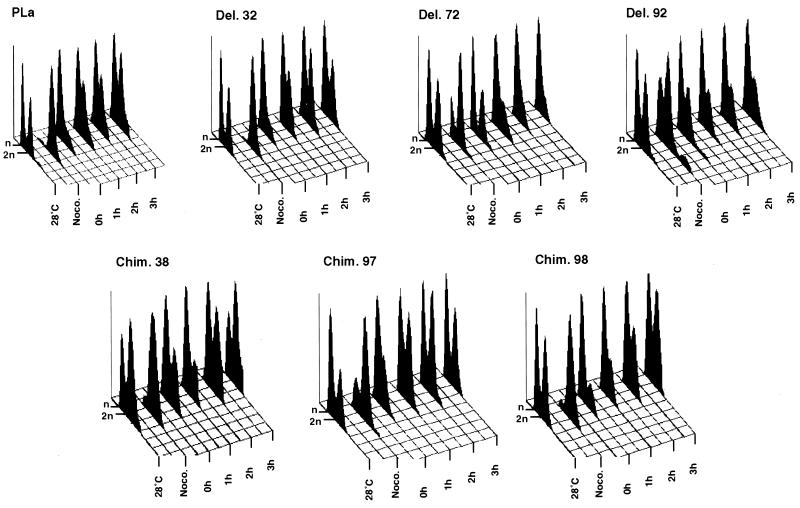
Prior to fluorescence-activated cell sorting (FACS) analysis, exponentially growing cells in full medium were synchronized in G2/M phase by the addition of nocodazole to a final concentration of 20 μg/ml. G2/M arrest of the cells was also monitored microscopically. When 90 to 100% of cells were arrested in the G2/M phase, cells were collected by centrifugation, washed twice with ice-cold YPD medium, resuspended in warm (37°C) YPD for temperature shift, and further incubated at 37°C. Aliquots were taken from exponentially growing cultures (28°C) after synchronization by nocodazole 10 min after the shift at 37°C (t = 0) and then each hour for 3 h (Fig. 4).
FIG. 4.
Distinct yTAFII25 temperature-sensitive mutants accumulate at different cell cycle stages. Cells were grown in YPD medium at 28°C and synchronized with nocodazole. The nocodazole was eliminated, and the cells were shifted to 37°C. Aliquots were taken from exponentially growing cultures (28°C) after synchronization by nocodazole (Noco.), 10 min after the shift at 37°C (t = 0), and then each hour for 3 h. Cells were fixed, stained with propidium iodide, and analyzed by FACS. Wild-type and Del. 32 were used as the reference strains. Strains carrying the Del. 72 and Del. 92 mutations accumulate in G1 phase during the 5 h at 37°C, whereas Del. 38 and Del. 98 strains accumulate in G2/M phase after the shift to 37°C. The x axis indicates the DNA content (1 n or 2 n), the y axis indicates the cell number, and the z axis indicates the time. For each histogram, approximately 20,000 particles were gated.
For FACS analysis, samples were prepared as described previously (43) except that the incubation time for the RNase treatment and pepsin digestion was doubled. The samples were analyzed on a FACScan (Becton Dickinson) equipped with a single argon ion laser. A minimum of 20,000 cells per sample were analyzed using CellQuest software (Becton Dickinson).
Whole-cell extract preparation, Western blot analysis, and antibody production.
Cultures were grown to mid-log phase at 28°C or shifted for 4 h to 37°C as described above, then cells were harvested by centrifugation for 5 min at 3,400 rpm, washed in cold H2O, and recentrifuged, and the pellet was frozen at −80°C and thawed. One volume of buffer A (40 mM Tris-HCl [pH 7.8], 350 mM NaCl, 0.1% Tween, 10% glycerol, 1× protease inhibitor cocktail [1× PIC; 2.5 mg each of leupeptin, pepstatin, chymostatin, antipain, and aprotinin per ml]) was added to the pellet, and cells were broken by addition of acid-washed glass beads (0.5 mm diameter; Sigma). Tubes were vortexed vigorously 6 times for 20 s each with 1 min of cooling on ice. Whole-cell extracts were clarified by centrifugation for 30 min at 4°C, and the protein concentration was determined. Protein extracts were stored at −80°C.
Whole-cell extracts (10 μg) were loaded on a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel, separated by electrophoresis, and electroblotted. The blots were then treated with the indicated antibodies, followed by incubation with peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody (Jackson Immuno Research). Chemiluminescence detection was performed according to the manufacturer's instructions (Amersham). Specific polyclonal rabbit antibodies recognizing TFIID- and SAGA-specific subunits were generated and purified as described previously (22, 67, 68).
Two-hybrid assays.
The L40 S. cerevisiae strain with a lexA-driven β-galactosidase reporter gene was transformed by the polyethylene glycol-lithium acetate method with the plasmid constructs as described in the text and earlier (20, 21). The two-hybrid assay was done as described previously (76).
Immunoprecipitation analysis.
Immunoprecipitations were carried out using anti-Flag M2-agarose beads (Sigma) equilibrated in buffer A. The different TAFII25 mutant-containing whole-cell extracts (5 mg) were mixed with 300 μl of M2 beads each and incubated for 12 h at 4°C by rotation. Beads were washed three times with 10 volumes of buffer A and then three times with 10 volumes of buffer B (40 mM Tris-HCl [pH 7.9], 150 mM NaCl, 0.1% Tween, 10% glycerol, 1× PIC). Bound protein complexes were eluted with 200 μl of buffer B containing 2.5 mg of the Flag peptide per ml for 3 h at 4°C. From these eluates, equal volumes (15 μl) were analyzed by Western blot.
RNA isolation and DNA microarray analysis.
For RNA isolation, two distinct cultures of each mutant were grown in YPD medium at 28°C to an OD600 of 0.25. One volume of YPD medium at 46°C was added to bring each culture to 37°C. The cultures were then incubated for 45 min at 37°C. Following this, the cultures were immediately cooled, pelleted in cooled tubes for 5 min at 3,400 rpm, washed in cold H2O, repelleted, and frozen in liquid nitrogen. Total RNA was isolated by the hot phenol-chloroform method using RNA-Solv reagent (Omega-Blotem) following the manufacturer's instructions and subsequently quantified. Synthesis of cDNA and biotin-labeled cRNA was performed according to the recommendations of Affymetrix Inc. and previously published protocols (45). The fragmented cRNA was hybridized to Affymetrix oligonucleotide arrays (GeneChip YG-S98). Absolute and comparison analyses were conducted using Microarray Suite 4.0 software. The total fluorescence intensity for each array was scaled to a uniform value by normalizing the average intensity of all genes (total intensity/number of genes) to a fixed value of 1,000.
The quality of total RNA and cRNA (before and after fragmentation) was verified by capillary electrophoresis and on gels. Before application to experimental arrays, cRNA quality was confirmed on test chips. Hybridization quality was controlled with bacterial cRNAs.
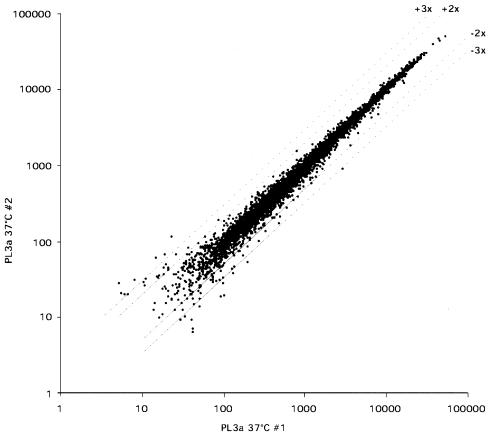
Data analysis was carried out with Microarray Suite 4.0 and Data Mining Tool 2.0 from Affymetrix, Inc. A change in mRNA transcription levels as detected by the gene chips was considered significant if the change was at least twofold up or down when comparing a mutant strain to the wild-type strain at the nonpermissive temperature. The twofold cutoff was chosen after analyzing a duplicate sample preparation of the control TAFII25 wild-type strain PL3a at 37°C. Comparing changes in expression between two independent samples using the same criteria as for comparing mutant samples to wild-type samples gave 40 genes whose expression was more than twofold up or down (0.63%) and only 4 genes that were more than threefold up or down (0.063%) (see Fig. 7 for a representation of the average difference change comparison).
FIG. 7.
Evaluation of microarray data: average difference scatter graph plot of two independent sample preparations analyzed with the Affymetrix GeneChip. The x and y axes show relative hybridization signal intensities for all genes tested. Higher values correspond to stronger expression. Two- and threefold change lines are shown. Only genes that were qualified as present by the Affymetrix software in either of the sample preparations were included (5,734 out of 6,332; see also Materials and Methods and Results). The data show very good reproducibility. Only some of the weakly expressed genes tend to fall out of the two- or threefold change lines.
RESULTS
A 91-amino-acid region of yTAFII25 containing the putative HFD is necessary and sufficient for vegetative growth in S. cerevisiae.
Yeast TAFII25 protein is encoded by a single-copy essential gene (37) containing a putative HFD (20) (see also Fig. 1A). To study the structure-function relationship of TAFII25, an S. cerevisiae Δtaf25 strain complemented with yTAFII25 expressed from a URA/CEN plasmid was generated. Subsequently the plasmid shuffle technique (7) was used to introduce plasmids expressing different Flag epitope-tagged yTAFII25 wild-type and mutant proteins (Fig. 1B). Protein expression was verified either before (noncomplementing mutants; Fig. 1C) or after (complementing mutants; Fig. 1D) the plasmid shuffle.
Although complementing yTAFII25 derivatives were expressed from a 2μm plasmid under the control of the strong PGK promoter, none of the mutant TAFII25 proteins resulting in a temperature-sensitive phenotype were overexpressed compared to yTAFII25 expression in a wild-type strain (Fig. 1D). The proteins that were overexpressed from the 2μm plasmid were the full-length TAFII25 and Del. 32, which did not result in detectable phenotypes (Fig. 1B and D). In complementation tests, the N-terminal nonconserved 73 amino acids of TAFII25 (ΔN-yTAFII25 in Fig. 1B) and the nonconserved extended loop 2 (L2) of the potential HFD (between amino acids 149 and 180; Del 32. in Fig. 1B) (23) could be deleted without any consequence on vegetative growth at 28°C (Fig. 2).
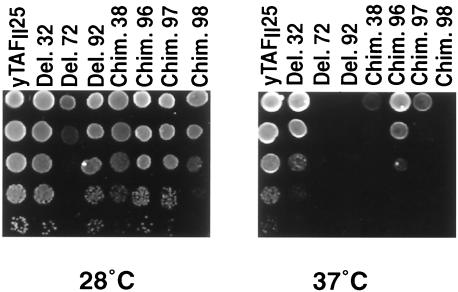
FIG. 2.
Several yTAFII25 mutants show temperature-sensitive phenotypes. The indicated strains were spotted in 10−1 serial dilutions on YPD medium and tested for growth at 28°C (left panel) and at 37°C (right panel) in comparison to the strain expressing full-length yTAFII25.
Interestingly, a well-conserved 10-amino-acid region between amino acids 74 and 83, encoding the putative αN-helix of the HFD (αN in Fig. 2), could also be deleted without affecting growth (Del. 71 in Fig. 1B). When a further 3 amino acids were deleted (amino acids 84 to 86), the growth of this strain was normal at 28°C but slightly impaired at 37°C (Fig. 1B, Del. 100). However, a further 3-amino-acid deletion (amino acids 87 to 89; strain Del. 72) resulted in a temperature-sensitive phenotype characterized by slow growth at 28°C and nonviability at 37°C (Fig. 1B and 2). Further N- or C-terminal deletions were not able to complement the TAF25 null phenotype (Del. 41 to 43 and Del. 105 in Fig. 1B).
Thus, the shortest deletion mutant that is sufficient to support viability with no obvious growth defects at 28°C on full medium and no temperature-sensitive phenotype at 37°C contains less than 50% of the wild-type yTAFII25 protein (Del. 71 in Fig. 1B), i.e., a 91-amino-acid region (from amino acids 84 to 149 and from 180 to 206) including the previously described HFD (20). These results show that the evolutionarily conserved HFD-containing region (including loop N) of yTAFII25 is necessary and sufficient for viability.
In an attempt to dissect the HFD further and determine important structural features, yTAFII25 was analyzed by Ballast (61), a program to predict local maximum segments (i.e., sequence segments conserved relative to their flanking regions). Two short amino acid sequences, the PIIPD and EYGLN motifs (amino acids 86 to 90 and 194 to 198, respectively, in yTAFII25), were identified by Ballast as presenting the highest degree of conservation. As described above, deletion of the PIIPD motif located in the loop N region (N-loop) between the αN and α1 helices of the HFD is important for yTAFII25 function (Del. 72 in Fig. 1B and 2). Deletion of the EYGLN motif C-terminal to the α3 helix did not affect growth under the conditions tested at 28°C but caused temperature sensitivity at 37°C (Del. 92 in Fig. 1B and Fig. 2), indicating a potential role of this region in the stability of the complexes (see below).
Neither full-length human TAFII30 nor its conserved HFD can substitute for yTAFII25 function, but partial substitutions in the HFD support viability and give rise to temperature-sensitive phenotypes
To study the functional conservation between S. cerevisiae TAFII25 and its human homologue hTAFII30, full-length hTAFII30 or its conserved HFD was tested for complementation in the TAF25 null strain. Neither full-length hTAFII30 nor its N-terminally truncated version, which retains the entire HFD (ΔN-hTAFII30), was able to complement yTAFII25 (Fig. 1B), while the presence of these proteins is easily detectable (Fig. 1C). A chimera comprising the N-terminal 73 amino acids of S. cerevisiae TAFII25 fused to the conserved HFD of hTAFII30 was also unable to rescue the TAF25 null phenotype (Chim. 21, Fig. 1B and C).
Thus, despite the relatively high amino acid conservation in the HFD of the S. cerevisiae and human proteins (67% similarity and 43% identity), the human protein cannot functionally replace its S. cerevisiae counterpart. This can be explained by the fact that in two-hybrid interaction assays, the HFD of hTAFII30 interacts with yTAFII47 but not with the two other yTAFII25 interaction partners, the TFIID subunit yTAFII65 and the SAGA component SPT7 (20).
We then analyzed chimeric proteins in which predicted structural motifs within the conserved minimal yTAFII25-complementing region were replaced by the corresponding regions of hTAFII30. To this end, either the first two (αN and α1) or the second two (α2 and α3) α-helices of yTAFII25 were replaced by the corresponding human HFDs (Chim. 37 and 38 in Fig. 1B). The fusion protein containing the αN and α1 helices from the human protein and α2 and α3 from the S. cerevisiae protein did complement the TAF25 null phenotype (Chim. 38 in Fig. 1B and 2), whereas the reverse chimera did not (Chim. 37, Fig. 1B and C, lane 8). This suggests that the α2 and α3 helices of the HFD have species-specific function, probably involved in interaction specificity, whereas the αN and α1 regions do not interfere with the essential functions of the S. cerevisiae protein under normal growth conditions (YPD, 28°C). The fusion protein Chim. 38 showed, however, a temperature-sensitive phenotype at 37°C (Fig. 1B and 2), indicating that the αN and α1 regions of the S. cerevisiae protein are not completely interchangeable with the homologous human sequences and that they may be involved in protein stability and/or protein interaction determination (see two-hybrid data below).
To dissect which region of the αN and α1 helices of the human protein was responsible for the temperature-sensitive phenotype, we made three further chimeras in which only the αN helix, only the N-loop, or only the α1 helix from the human protein were exchanged with the corresponding regions of the S. cerevisiae protein (Chim. 96 to 98 in Fig. 1B). Chim. 96, containing the human αN helix, fully complemented the TAF25 null phenotype, in good agreement with the above deletion studies (Fig. 1B, compare Chim. 96 and Del. 71), further indicating that the αN helix is not contributing to any essential TAFII25 function under the conditions tested. However, the other two chimeras, containing either the human N-loop region (Chim. 97) or the human α1 helix (Chim. 98), grew normally at 28°C, albeit somewhat slower than the wild type, but were temperature sensitive at 37°C, Chim. 97 to a lesser degree (Fig. 1B and 2).
It should be noted that the exchange of the S. cerevisiae for the human sequences in Chim. 97 and 98 reflects only three- and six-amino-acid differences, respectively, compared to wild-type yTAFII25. These results, in addition to the two-hybrid data below, strongly suggest that the N-loop and α1 regions of TAFII25 are important function-determining regions and are involved in interaction specificity.
Distinct TAFII25 mutants have different morphology and cell cycle defects.
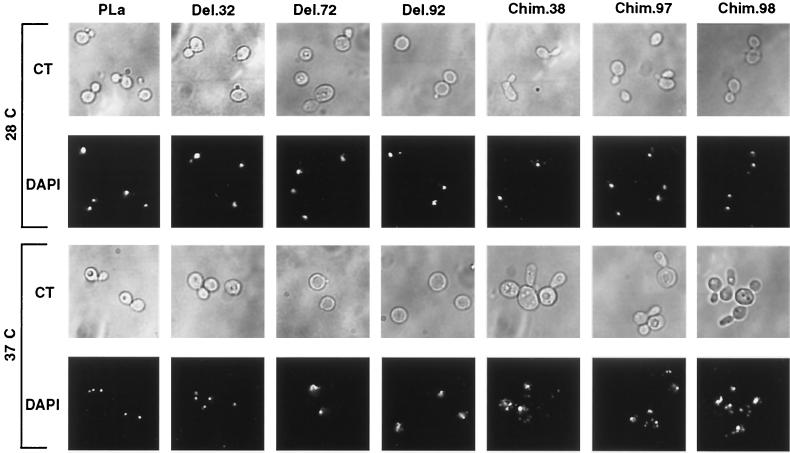
Next, the morphological changes of the above-described TAFII25 temperature-sensitive mutant strains were analyzed at 28 and 37°C by light microscopy, and the nuclei were visualized by using fluorescence microscopy following DAPI staining. At the permissive temperature, only slight differences were found between strains (Fig. 3). For example, cells containing Chim. 38 often had elongated cell and bud morphologies (Fig. 3). At the nonpermissive temperature (37°C), most of the temperature-sensitive strains showed abnormal morphologies that could be divided in two categories: (i) a high proportion of cells with no or “mini” buds together with an aberrant nuclear morphology (Fig. 3, Del. 72 and Del. 92) and (ii) large cells with elongated bud morphologies (10 to 30% of cells with multiple buds) and with several “stained bodies” in the same cell (Fig. 3, Chim. 38, Chim. 97, and Del. 98).
FIG. 3.
Changes in cell morphology and nuclear organization of distinct yTAFII25 mutants upon shift to the nonpermissive temperature. Morphological changes in the yTAFII25 temperature-sensitive mutants were visualized by phase contrast microscopy (CT). Nuclei were visualized by fluorescence microscopy with DAPI staining. In the two upper lines, S. cerevisiae strains are shown at 28°C, whereas in the two bottom lines, strains were shifted for 4 h to 37°C.
Since most of these morphological changes are typical of cell cycle defects and as some other TAFII temperature-sensitive mutants were shown to arrest at different cell cycle stages (see the introduction), we analyzed the DNA content of these mutants by FACS. Cells were grown at 28°C to mid-log phase and synchronized with nocodazole, a drug that blocks cells in the G2/M phase (2 n). Following synchronization, nocodazole was eliminated and cells were shifted to 37°C for 3 h. The DNA content was measured every 60 min (Fig. 4).
Control strains (PLα and Del. 32) grew asynchronously at 28°C (with about 55% of control strain cells having n and 45% having 2 n DNA content), after nocodazole treatment almost all cells were arrested in G2/M phase with 2 n DNA content, and after release the cells rapidly entered G1 phase with 1 n DNA content; after 3 h at 37°C they again became asynchronous (Fig. 4). In contrast, Del. 72 and Del. 92 strains could not be completely arrested with nocodazole in G2/M phase, with 10 to 30% of the cells remaining with 1 n DNA content in G1 phase (Fig. 4). After release from the arrest and shift to the nonpermissive temperature, both strains accumulated with 1 n DNA content in G1 phase. After 3 h at 37°C, about 95% of Del. 72 and 70% of Del. 92 cells had accumulated in G1 (Fig. 4).
In contrast, Chim. 38 cells, which already showed a higher proportion of cells with 2 n DNA content relative to the control strains at 28°C (about 30% of cells have 1 n and 70% have 2 n DNA content), were synchronized efficiently with nocodazole in G2/M phase. After release from the arrest and shift to the nonpermissive temperature Chim. 38 showed a slow progression to G1 phase, and about 70% of the cells arrested in G2/M after 3 h at 37°C (Fig. 4). Chim. 98 behaved like Chim. 38, although the G2/M arrest was less pronounced (Fig. 4).
All these results were confirmed by using another synchronization method (α-factor) and by shifting the cells to the nonpermissive temperature without prior synchronization (data not shown). Finally, mutant Chim. 97 did not show a pronounced arrest at any particular cell cycle phase in any of the cell cycle assays performed (Fig. 4 and data not shown). Thus, our TAFII25 mutants can be classified in three groups: (i) Del. 72 and Del. 92, arresting in G1; (ii) Chim. 38 and Chim. 98, arresting G2/M; and (iii) Chim. 97, for which no specific cell cycle arrest stage could be determined.
These data together demonstrate that S. cerevisiae TAFII25 is involved in proper cell cycle regulation and that distinct TAFII25 mutants display different morphology and cell cycle defects. Moreover, they suggest that mutations Del. 72 and Del. 92 affect TFIID (or TFIID and SAGA) function, which is known to arrest cells in G1 phase, whereas the Chim. 38 and Chim. 98 mutants, which arrest in G2/M phase, might rather compromise SAGA function (see below and Discussion).
Distinct TAFII25 temperature-sensitive mutants display differential interaction profiles with their interaction partners.
The fact that human TAFII30 does not complement the TAF25 deletion and that small sequence exchanges between the S. cerevisiae and human homologues in the HFD protein-protein interaction domain caused severe growth defects or a temperature-sensitive phenotype (Del. 72, Del. 92, Chim. 38, Chim. 97, and Chim. 98 in Fig. 1B and 2) prompted us to test in a two-hybrid assay whether these mutants would interact with the previously described interaction partners, yTAFII47, yTAFII65, and ySPT7 (20).
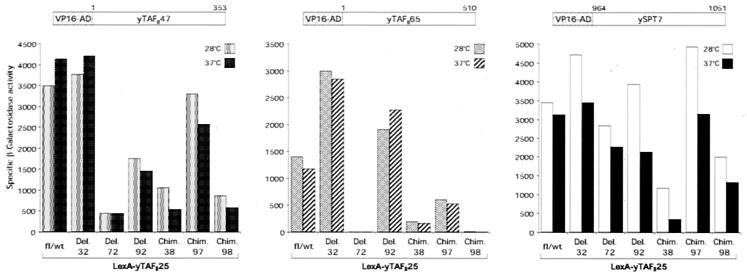
Full-length yTAFII47 and yTAFII65 and the HFD of ySPT7 (amino acids 964 to 1051) were fused to the VP16 activation domain, and the full-length TAFII25 and Del. 32, Del. 72, Del. 92, Chim. 38, Chim. 97, and Chim. 98 mutants were fused to the LexA DNA-binding domain. LexA DNA-binding domain fusion proteins were then tested for interaction with the different VP16 fusions in a series of S. cerevisiae two-hybrid experiments at either 28 or 37°C (Fig. 5). Interactions were assessed by measuring specific β-galactosidase activity in the S. cerevisiae reporter strain L40, which harbors a LexA-responsive lacZ gene (77).
FIG. 5.
Two-hybrid assay for interaction of TAFII25 deletion mutants and chimeras with the TFIID-specific (TAFII47 and TAFII65) and SAGA-specific (SPT7) interaction partners. The indicated TAFII25 mutants were expressed as LexA DNA-binding domain fusions; TAFII25 interaction partners (TAFII47, TAFII65, and SPT7) were expressed as VP16 activation domain (VP16-AD) fusions (amino acid positions are indicated). The reporter gene is β-galactosidase with four LexA-binding sites in the promoter. The results are shown as specific β-galactosidase activity. Interactions were tested at 28 and 37°C (after 4 h of incubation). Shown are the results of at least two qualitatively identical experiments; the error was less than 20%. None of the LexA-TAFII25 fusion constructs activated the reporter on their own or in the presence of the VP16 activation domain only (20) (data not shown).
LexA-TAFII25 in the presence of VP16 alone does not activate the reporter gene (20). Also, with the exception of TAFII47, neither of the unfused TAFII25 interaction partners, TAFII65 and SPT7, activated transcription in the presence of LexA-TAFII25 (data not shown). Activation of the reporter by TAFII47 (coexpressed with LexA-TAFII25) alone was low but detectable (870 U of specific β-galactosidase activity, compared to 3,500 U with the VP16 fusion; see Fig. 5, left panel). Since this activity of TAFII47 is also the result of a specific recruitment by LexA-TAFII25 to the promoter, it does not interfere with our analysis of TAFII25 mutants affecting an interaction with the TAFII25 interaction partners.
Full-length TAFII25 and Del. 32 (Fig. 1B) showed qualitatively similar interactions with yTAFII47, yTAFII65, and ySPT7 (Fig. 5). The effect of the temperature shift to 37°C on the protein interaction is in general (with a few exceptions) not very pronounced. This could be due to a stabilization of the mutant proteins by their fusion partners or a shift in the thermodynamic equilibrium under two-hybrid conditions, where both proteins are highly overexpressed. Thus, a small decrease in interaction strength between two proteins in this assay may reflect a severe functional defect in vivo at endogenous expression levels. Nevertheless, it is possible to qualitatively distinguish among the different mutants for their capacity to interact with the TAFII25 interaction partners.
Chim. 97 showed interactions comparable to the wild type with TAFII47 and SPT7 but a two- to fourfold decrease in interaction with TAFII65. However, mutants Del. 72, Chim. 38, and Chim. 98 all showed severely impaired interaction with both TAFII47 and TAFII65 (Fig. 5). Chim. 38, particularly at the nonpermissive temperature, and Chim. 98 also showed a diminished interaction with ySPT7, with which all the other mutants seemed to interact in a manner comparable to that of wild-type TAFII25. In this assay, Chim. 92 showed interactions with both SPT7 and TAFII65 (similar to the wild type) but a reduced level of interaction with TAFII47.
The two-hybrid interaction results suggest that Chim. 98 and, more specifically, Chim. 38 have a defect in interaction with SPT7 and that the observed phenotypes for these two proteins are at least partially due to an impaired interaction with this protein in the SAGA complex. In contrast, Del. 72, which displayed a slow-growth phenotype already at 28°C, had no obvious defect in SPT7 interaction but was completely impaired for interaction with TAFII65 and, of the mutants tested, had the most severe reduction in interaction with TAFII47. These results indicate that the deletions in Del. 72 most severely affect interactions in TFIID. Surprisingly, Del. 92 did not show any apparent reason in the two-hybrid assay for its functional impairment at the nonpermissive temperature, but showed in the SAGA/TFIID immunoprecipitation experiments (see below) one of the most severe phenotypes at the nonpermissive temperature.
Taken together, the two-hybrid data support the idea that within the HFD of TAFII25, the N-loop and α1 regions are important for determining interaction specificity and that the different observed phenotypes of the mutants tested may be linked to changes in this interaction domain, resulting in differential interaction with their putative partners, affecting different complexes.
Distinct TAFII25 mutations differentially affect the integrity of the TFIID and SAGA complexes.
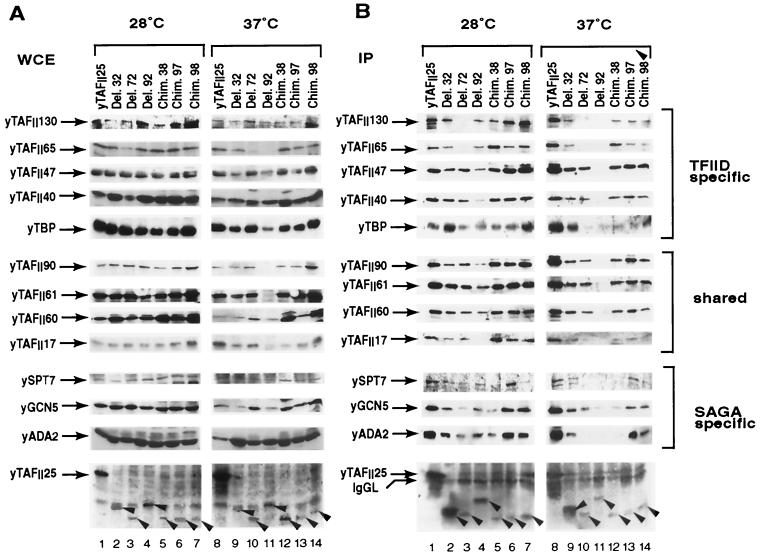
To test whether the loss of pairwise interactions observed between the different TAFII25 mutants and their respective interaction partners in the two-hybrid assay influences the integrity of TFIID and/or SAGA complexes, we analyzed the polypeptide composition of these complexes in the different TAFII25 mutant strains. First, S. cerevisiae whole-cell extracts were prepared from the wild-type and the mutant strains grown either at 28°C or for 4 h at 37°C and analyzed by Western blot for the presence of different TFIID and SAGA components (Fig. 6A).
FIG. 6.
Analysis of TFIID and SAGA complex integrity in yTAFII25 wild-type and mutant strains. (A) Immunoblot of whole-cell extracts (WCE) prepared from yeast cells grown at 28°C or after a 4-h shift to 37°C. Cultures were harvested, whole-cell extracts were prepared, and 10 μg of the respective whole-cell extracts was separated by SDS-PAGE, blotted, and analyzed by Western blot with the indicated antibodies. (B) Whole-cell extracts prepared from the different strains (as indicated) were immunoprecipitated (IP) using a resin-bound anti-Flag antibody which recognizes the Flag epitope localized at the N terminus of each construct. Peptide-eluted complexes were separated by SDS-PAGE, blotted, and analyzed by Western blot with the indicated antibodies. In panels A and B, the bottom panels show the full-length TAFII25 and the different TAFII25 mutants (indicated with arrowheads) revealed with an anti-Flag antibody.
In extracts prepared from strains grown at 28°C, most of the analyzed TAFIIs and SAGA components were comparably expressed (Fig. 6A, lanes 1 to 7). However, in the Del. 72 strain, the yTAFII65 level was lower than in the other strains. Interestingly, in extracts prepared from most of the temperature-sensitive strains grown at the nonpermissive temperature, many of the TFIID and SAGA components were expressed at similar levels (Fig. 6A, lanes 8 to 14). Only two strains had altered TAFII expression levels in the whole-cell extracts at 37°C: strain Del. 72, in which the yTAFII65 level was lower than in the other strains, and strain Del. 92, in which there was no detectable yTAFII90 or yTAFII65 and very low levels of yTAFII61 and yTAFII17.
As all of the different TAFII25 proteins contained a Flag epitope tag on their N-terminal ends, the TAFII25 wild-type and mutant proteins were immunoprecipitated with an anti-Flag antibody from the whole-cell extracts. The presence of the TFIID and SAGA subunits in the immunoprecipitations was analyzed by Western blot (Fig. 6B). The different TAFII25 mutant proteins were completely depleted from the whole-cell extracts by the anti-Flag immunoprecipitation, as a large excess of antibody was used in the immunoprecipitations (data not shown). Importantly, in the immunopurified TAFII25-containing complexes, most of the tested TFIID and SAGA components (i.e., yTAFII90, yTAFII61, yTAFII60, yTAFII47, yTAFII40, and yGCN5) were present at comparable levels, indicating that, independent of the expression level of the different TAFII25 mutants, they incorporated similarly into endogenous TFIID and SAGA complexes (Fig. 6B). Note that in these experiments, the Del. 32 strain is a better control because the temperature-sensitive mutants were made in the Del. 32 mutant context (Fig. 1B).
Surprisingly, in complexes prepared from the Del. 72 strain, already at 28°C two of the TFIID-specific TAFIIs, yTAFII65 and yTAFII130, were missing, and after the shift to 37°C two additional SAGA-specific components (SPT7 and ADA2) were also lost (Fig. 6B, lanes 3 and 10). The lack of yTAFII65 from the Del. 72-containing complexes is in good agreement with the two-hybrid results. Moreover, the Del. 72-containing complexes contained only trace amounts of TBP at 37°C (Fig. 6B, lane 10). In the TAFII25-containing complexes prepared from the Del. 92 strain at 28°C, most of the tested subunits were present (except for yTAFII17), whereas at 37°C almost all of the tested TFIID and SAGA components were missing (Fig. 6B, lanes 4 and 11), although most of these components were easily detectable in Del. 92 whole-cell extract at 37°C (Fig. 6A, lane 11). This indicates a loss of Del. 92 from both complexes rather than the disintegration of SAGA and TFIID.
In immunopurified complexes prepared from the Chim. 38, Chim. 97, and Chim. 98 strains at 37°C, all the tested components of TFIID were comparable to those present in the Del. 32 control complexes (Fig. 6B, lanes 12 to 14). However, the Chim. 38-containing complexes clearly lacked the SAGA-specific components SPT7 and ADA2 (lane 12), and the Chim. 98-containing complexes lacked SPT7 (lane 14), in agreement with the above-observed impaired interaction of these two mutants with SPT7 (see Fig. 5).
These results demonstrate that the temperature-sensitive phenotype of the tested strains (Del. 72 and 92 and Chim. 38, 97, and 98) at 37°C (see Fig. 2) is not the consequence of a general disintegration of the respective TAFII25-containing complexes. Instead, the studied TAFII25 mutations affect the TAFII25-containing complexes in several different ways: (i) in the Del. 92 strain at 37°C, the corresponding TAFII25 mutation is not able to incorporate in either TFIID or SAGA; (ii) in the slow-growing Del. 72 strain, two TFIID-specific TAFIIs (yTAFII130 and yTAFII65) are absent from TFIID already at 28°C, while at 37°C additional subunits dissociated from the corresponding complexes without triggering a total disintegration of TFIID; (iii) complexes from the Chim. 38 and 98 strains lost only SAGA-specific subunits at 37°C, while TFIID was unaffected; and (iv) complexes from the Chim. 97 strain have a TFIID and SAGA composition similar to that of those prepared from the Del. 32 reference strain, which shows no growth defect. Thus, the distinct TAFII25 mutations differentially affect the composition of the corresponding TFIID and SAGA complexes: Del. 72 seems to affect TFIID (at 28°C) or TFIID and SAGA composition (at 37°C), whereas Chim. 38 and 98 seem to affect SAGA composition specifically (at 37°C). These results appear to be in good agreement with the two-hybrid interaction assay.
Moreover, our results, together with the study of Natarajan et al. (56), demonstrate that conclusions based on yTAFII and other SAGA subunit expression in whole-cell extracts prepared from mutant strains can be misleading and that the corresponding multiprotein complexes have to be immunopurified to analyze the weakened subunit interactions.
Distinct mutations in TAFII25 differentially affect the expression of class II genes.
To investigate how the different TAFII25 mutants influence Pol II transcription, Affymetrix DNA microarray analysis was performed (see Materials and Methods). Total RNA was prepared from the above-described temperature-sensitive strains (Del. 72, Del. 92, Chim. 38, Chim. 97, and Chim 98) grown at 28°C or transferred for 45 min to 37°C. As a reference, we used the wild-type strain PL3a. To test the reproducibility of the results and to determine a cutoff for changes in gene expression, we compared the results of two independent RNA preparations, each carried out on the wild-type strain at 37°C and tested on Affymetrix DNA microarrays. The results obtained indicate a very high reproducibility (Fig. 7). Only about 0.6% of mostly weakly expressed genes changed twofold up or down when the two independent preparations were compared. We therefore chose a twofold cutoff as being significant for changes in gene expression in our mutant analysis, with the possibility of a 0.6% error (see also Materials and Methods).
A total of 6,332 Pol II genes with a systematic locus name were included in the analysis (Table 2). Of these genes, 64% (4,073 genes) were either up- or downregulated more than twofold at the nonpermissive temperature compared to the wild-type strain. These genes include the nonredundant affected genes (up- and downregulated) in the five mutant strains together. In any individual mutant, the number of affected genes was between 1,121 and 2,110 (or 18 and 33% of all genes analyzed) (see Table 2). Interestingly, in the different mutants, more genes showed increased expression than a decrease in transcription, 40 and 30%, respectively.
TABLE 2.
Total gene expression analysisa
| TAFII25 mutant | Change, no. of genes (% of 6,332 analyzed)
|
||
|---|---|---|---|
| Downregulated >2-fold | Upregulated >2-fold | Up- or downregulated >2-fold | |
| Del. 72 | 1,124 (18) | 447 (7) | 1,571 (25) |
| Del. 92 | 554 (9) | 700 (11) | 1,254 (20) |
| Chim. 38 | 516 (8) | 1,106 (17) | 1,622 (26) |
| Chim. 97 | 524 (8) | 1,586 (25) | 2,110 (33) |
| Chim. 98 | 483 (8) | 638 (10) | 1,121 (18) |
| Total | 1,925 (30) | 2,512 (40) | 4,073 (64) |
Affymetrix DNA microarray analysis was performed. Total RNA was prepared from the Del. 72, Del. 92, Chim. 38, Chim. 97, and Chim 98 strains grown at 28°C and transferred for 45 min to 37°C. As a reference, we used the wild-type strain PL3a. Numbers and percentages of genes affected by different TAFII25 mutation at the nonpermissive temperature compared to the wild-type strain PL3a are indicated. Total reflects the nonredundant number of genes up- and downregulated by the mutants. Genes were considered affected when their expression level was at least twofold above or below that of the wild-type control. The error was estimated to be about 0.6%.
Surprisingly, only 132 genes were similarly affected in all five mutant strains analyzed. This is either 3.2% of all genes affected by any one of the mutations (4,073 genes) or 2.1% of all genes analyzed (6,332 genes). In addition, when mutant strains were compared pairwise for genes that are commonly decreased twofold or more, the overlap was limited to 11% to 28% or to 5% to 13% when comparing any group of three mutants. For upregulated genes, comparable results were obtained. These data show that, based on their affected global gene expression pattern, our TAFII25 temperature-sensitive mutants cannot be put in well-defined categories. Thus, any given individual TAFII25 mutant affects (directly or indirectly) a very specific and only partially overlapping subset of Pol II genes.
To further test our mutants for SAGA and/or TFIID specificity, we compared our data to a list of genes showing differential requirements for TFIID and SAGA as published by Lee et al. (42). This list contains four categories of 10 genes: TFIID dependent, SAGA dependent, TFIID and SAGA dependent, and genes whose transcription is affected only in a SAGA TFIID double mutant. Surprisingly, out of 10 genes in each category, we found between one and four genes unaffected by all five of our TAFII25 mutants. However, in mutant Del. 72, four genes of the first category (CAM1, WTM2, GNA1, and MSH6), five genes of the second (PHO84, PHO5, SPL2, YHB1, and YJL012C), four genes in the third (AAH1, FAR1, YOR095C, and PHO3), and two genes of the fourth category (HEM4 and TOM7) are downregulated by more than twofold, suggesting that this TAFII25 mutant may affect both SAGA and TFIID activity.
In contrast, Chim. 38 and Chim. 98 do not downregulate the expression of any gene in the first and fourth categories (with the exception of one gene downregulated by Chim. 38 in the first category), but the expression of five genes is decreased more then twofold in the second (PHO84, PHO5, SPL2, YHB1, and YDL124W), SAGA-specific category, and four genes in the third category (AAH1, FAR1, YOR095C, and YOR306C), suggesting that these two mutants influence gene transcription by affecting SAGA function rather than that of TFIID. The results for the two other mutants, Del. 92 and Chim. 97, are much less clear when compared to these published lists of genes. The data suggest, however, that different mutations of the same TAFII shared by SAGA and TFIID can specifically affect the function of one or the other complex.
DISCUSSION
The results described in this study indicate that a minimal 91-amino-acid region containing the HFD of the yTAFII25 protein is necessary and sufficient for vegetative growth. This minimal region may even be shorter, as the last seven amino acids of yTAFII25 (C-terminal to the HFD) can be deleted without influencing important HFD-HFD interactions between yTAFII25 and yTAFII47 or yTAFII25 and yTAFII47 (20; unpublished data). We also show that the distinct temperature-sensitive mutations (truncations, deletions, and multiple point mutations) in the minimal HFD-containing region of yTAFII25 do not result in a single well-defined phenotype but in different morphological and cell cycle phenotypes as well as in surprisingly distinct gene expression patterns.
These findings are in agreement with other recent studies (15, 36, 52, 74), arguing that most individual temperature-sensitive mutants of a given TAFII cannot be used to evaluate all functions of that TAFII in gene regulation. Depending on the nature of the temperature-sensitive mutation, it can result in very different phenotypes and thus reflect many different functions of the same protein. Moreover, our observations also show that the effects of a TAFII mutation on its function should be studied in the biochemical context of the corresponding multiprotein complexes. Our data indicate that effects on complex integrity need to be determined and that characterization of SAGA- and TFIID-specific functions of shared TAFIIs should be possible by using appropriate mutations in the histone fold domain.
Different observed phenotypes of tested mutants may be linked to changes in TFIID and/or SAGA complexes.
Comparison of the different results obtained with our yTAFII25 mutants suggests that the effect of some mutations can be correlated with the integrity of the TAFII25-containing complexes. Strains in which the TAFII25 gene is deleted are not viable (37), suggesting that TFIID and SAGA complexes are not functional without TAFII25. It is thus conceivable that the TAFII25 mutant-containing complexes represent TFIID and SAGA complexes which are sufficient for viability at 28°C.
In strains where the mutated yTAFII25 protein is able to enter the TAFII25-containing complexes but prevents the cointegration of other TFIID- and SAGA-specific components (Del. 72 at 37°C) or is unable to integrate into TFIID and SAGA complexes (Del. 92 at 37°C), cells arrest in the G1 phase, suggesting that TFIID function is crucial for entry into the S-phase of the cell cycle. Interestingly, the immunopurified TFIID and SAGA complexes from the Del. 72 strain were very similar when purified from cells grown at 28 or 37°C; however, the Del. 72 cells were able to grow (though extremely slowly) at 28°C but not at 37°C (Fig. 2). One reason for this could be that in the Del. 72-containing complexes, TBP incorporation is seriously impaired at 37°C. Moreover, it is possible that weakened interactions between the Del. 72 mutant and yTAFII65 and/or the Del. 72 mutant and yTAFII130 are still functional in the cells at 28°C, allowing them to survive, but these interactions may be disrupted during biochemical purification and TAFII130 as well as TAFII65 may be lost from the immunoprecipitation.
The fact that strains containing the Del. 72 and Del. 92 mutant proteins arrest in the G1 stage of the cell cycle, together with the observation that in mouse cells, in which the two alleles of the vertebrate homologue of the yeast taf25 gene, TAFII30, have been conditionally disrupted, also arrest in G1 phase (51), suggests that TAFII25 and its metazoan homologues are required (directly or indirectly) for progression through the G1 phase.
On the other hand, yeast cells containing the Chim. 38 and Chim. 98 mutant TAFII25 proteins, in which solely the polypeptide composition of the SAGA complex is compromised, accumulate at the G2/M stage. These TAFII25 mutants seem to affect the interaction mainly with the SAGA-specific HFD-containing interaction partner SPT7 in both immunoprecipitation and two-hybrid experiments. Moreover, an ada1 null mutation (another SAGA component) results in morphological features (large elongated cells with several elongated buds; E. vom Baur, unpublished observation) very similar to those of the Chim. 38 and Chim. 98 mutants at 37°C (Fig. 3). Thus, there seems to be a good correlation between the TAFII25 mutations that compromise SAGA integrity and mutations of other SAGA-specific components. These results together suggest that when cells are impaired in TFIID (and SAGA) function, they arrest at the G1 phase of the cell cycle, and that when mostly SAGA-specific functions are compromizsed, cells accumulate in the G2/M phase of the cell cycle.
Interestingly, deleting the αN and loop N regions of the HFD in strain Del. 72 resulted in cells accumulating in G1 phase at 37°C; in contrast, replacing the αN, loop N, and α1 regions with the corresponding human sequences (strain Chim. 38) resulted in cells accumulating in G2/M phase at 37°C. The compositions of TFIID and SAGA complexes at 37°C are also different in these strains: TAFII25-containing TFIID and SAGA complexes from strain Del. 72 seem to be partially dissociated, whereas in the Chim. 38 strain, the SAGA complex is affected, but not TFIID (Fig. 6). In agreement with the biochemical data, the gene expression pattern in these two mutants differs significantly (see below). Together, these observations show that temperature-sensitive mutations that are generated in the same region of yTAFII25 can result in a different composition of the corresponding multiprotein complexes, different phenotypic changes, and different gene expression patterns.
Moreover, the fact that in the Chim. 97 temperature-sensitive strain the protein-protein interactions within the TAFII25-containing complexes are not seriously affected (Fig. 6) suggests that other key interactions between the TAFII-containing complexes and transcription factors and/or the promoter DNA are impaired, and this may lead to the observed phenotypic and gene expression changes.
How do the different TAFII25 mutations influence TAFII-TAFII or TAFII-SAGA component interactions?
Our observations suggest that the loop N region of the HFD is crucial for the interaction between yTAFII25 and yTAFII65, because a deletion of this region (mutant Del. 72) disrupts this interaction (Fig. 5) and, in good agreement, yTAFII65 (together with yTAFII130) is lost from the Del. 72-containing TFIID complex (Fig. 6B). Interestingly, when the loop N region is present but the S. cerevisiae α1 helix is replaced with the corresponding human sequences (mutant Chim. 98), yTAFII25 fails to interact with yTAFII65 in the two-hybrid assay (Fig. 5), but yTAFII65 stays associated with the Chim. 98-containing TFIID complexes (together with yTAFII130; Fig. 6B). These data suggest that in Chim. 98-containing complexes, other additional TAFII-TAFII interactions are able to stabilize the weakened Chim. 98-yTAFII65 interaction in TFIID and that the loop N region is important in this stabilization effect. The intact HFD of human TAFII30 does not interact with yTAFII65 (20), further underlining the importance of the additional TAFII-TAFII interactions in our mutants.
Mutants Chim. 38, which has the entire human αN-N-loop-α1 region, and Chim. 97, which has only the human loop N region (with three amino acid differences compared to the S. cerevisiae sequences) and the remaining S. cerevisiae sequences (Fig. 1B), retain about 10 to 40% of the two-hybrid interactions with yTAFII65 and have an unchanged TFIID composition, suggesting that the loop N region is important for the interaction between yTAFII25 and yTAFII65. Moreover, our study predicts important interactions among TAFII25, yTAFII65, and TAFII130, as the results obtained with the Del. 72-containing TFIID complex indicate that the loop N regions of TAFII25 and yTAFII65 are both necessary for the stable association of yTAFII130 with TFIID. These interactions are consistent with immunoelectron microscopy data which showed that TAFII25, yTAFII65, and TAFII130 are all present in close proximity in the same lobe of the yTFIID (9; C. Leurent, S. L. Sanders, V. Mallouh, P. A. Weil, D. B. Kirschner, L. Tora, and P. Schultz, submitted for publication). In agreement, an interaction between hTAFII250, the human homologue of yTAFII130, and hTAFII30 has also been reported (35).
The HFD of yTAFII25 and that of the human TAFII30 have also been shown to interact both physically and genetically with that of the yTFIID component TAFII47 (20). Interestingly, none of the TAFII25 mutants tested completely abolished the interaction between yTAFII47 and yTAFII25 in the two-hybrid assay (however, some of them weakened the interactions by five- to sevenfold at 37°C [Fig. 5]), and the presence of yTAFII47 in the immunopurified complexes was not affected (Fig. 6B). Even the Del. 72 mutant, which showed the weakest interaction with TAFII47, did not impair the cointegration of TAFII47 into the TFIID complex, suggesting that additional interactions are able to stabilize the weakened Del. 72-yTAFII47 interaction or that yTAFII47 makes additional contacts with other proteins in TFIID.
The second possibility is consistent with immunoelectron microscopy data, which indicated the presence of at least two molecules of yTAFII47 in the same TFIID particle (C. Leurent, S. L. Sanders, V. Mallouh, P. A. Weil, D. B. Kirschner, L. Tora, and P. Schultz, submitted for publication). Moreover, based on two-hybrid data, the Del. 92 mutation does not completely destroy the interaction between TAFII47 and TAFII25 even at 37°C, and thus a lack of interaction cannot be the reason for the failure of this TAFII25 mutant to incorporate into the TFIID complex at 37°C. Since the Del. 92 mutation does not show any obvious reason for its functional impairment, we speculate that this mutation may affect other interactions than those of its known binding partners, and the lack of these would then result in a rapid degradation of many other TAFIIs (see TAFII90, TAFII65, TAFII61, and TAFII17 in the whole-cell extracts at 37°C; Fig. 6A).
It has been shown that the HFD of the SAGA component SPT7 can interact with yTAFII25 but not with human TAFII30, both in two-hybrid assays and in vitro (20). Comparison of the functional complementation experiment (Fig. 1B) with our two-hybrid interaction studies (Fig. 5) and with the SAGA/TFIID immunoprecipitation experiment (Fig. 6B) suggests that the human loop N region does not influence the TAFII25-SPT7 interaction (Chim. 97), whereas the human α1 region does (Chim. 98). However, when the corresponding mutant contains both of these human regions (Chim. 38), this results in a severe defect in the two-hybrid TAFII25-SPT7 interaction and in the parallel disintegration of the SAGA complex. In contrast, the amino acid changes in the Chim. 38 and Chim. 98 mutants do prevent yTAFII47 and yTAFII65 from associating with the corresponding TFIID complexes (Fig. 6B).
Thus, introducing either the human αN-N-loop-α1 region of the HFD or the human α1 region of the HFD into the S. cerevisiae yTAFII25 backbone creates mutants that seem to mainly impair SAGA composition and function. Importantly, both the Chim. 38 and Chim. 98 mutants arrest in the G2/M cell cycle phase, suggesting that in both mutants similar, possibly SAGA-specific functions are affected at the nonpermissive temperature. The human α1 region contains six amino acid substitutions, and thus in the future it will be important to test which of these six amino acids is crucial for the yTAFII25-SPT7 interaction.
Individual TAFII25 mutants influence different allele-specific gene expression patterns.
The main suggestion that followed from the above observations is that distinct conditionally temperature-sensitive mutations in the same TAFII may affect transcription in different ways, partially due to specific effects on TFIID and/or SAGA function. To test this hypothesis, a genome-wide transcription analysis was carried out with DNA microarrays. We found that only 2.1% of the genome is affected in the same way in all five mutants, whereas about 64% of global gene expression is affected when all of the nonredundant transcription effects on any gene caused by the mutations are added. Our gene array data, together with other mutant TAFII allele analyses (15, 36, 52, 74), argue that results obtained with any single temperature-sensitive mutation in a TAFII does not necessarily reveal all of the functions of this protein.
It is clear that our yTAFII25 mutants individually did not affect the expression of all genes (between 18 and 33%); however, the sum of the individual contributions revealed a global role for yTAFII25 (64% of total genes analyzed). Moreover, it is conceivable that by testing more or other yTAFII25 mutants, it would be possible to show a combined effect on a larger portion of the genome or maybe on the whole genome. In this respect, it is interesting that in a recent genome-wide analysis, the combined effect of single mutations in several shared TFIID and SAGA subunits can account for changes in more than 70% of the genome (42). Unexpectedly, here we found that expression of most genes was affected by multiple individual mutations in an HFD-containing TAFII shared by TFIID and SAGA.
As the distinct mutations in the HFD of yTAFII25 differently impair TAFII-TAFII, TAFII-SAGA, and TAFII-other transcription interactions (see above) we speculate that there are different requirements for the specific regions of the TAFII25 HFD in global gene expression. As our TAFII25 mutants do not result in a total disintegration of TFIID and/or SAGA complexes (with the possible exception of Del. 92), we think that the differential gene expression patterns observed reflect distinct functional defects caused by the given mutation in TFIID and/or SAGA complexes and that partial complexes can still play a role in gene regulation at certain promoters. Moreover, the fact that there were only 132 genes whose transcription was affected in all five mutants in the same way suggests that these genes are the ones which are the most sensitive to changes in TAFII25.
Interestingly, when looking for the expression of the genes which were proposed to be TFIID, SAGA, and TFIID and SAGA dependent (42), we found that Del. 72 affected most of the genes in these categories, suggesting that Del. 72 may affect both TFIID- and SAGA-specific functions. This observation is also in good agreement with our biochemical purification data, showing that in Del. 72-containing complexes both TFIID- and SAGA-specific subunits are lost at 37°C. Moreover, the Chim. 38 and Chim. 98 mutants did influence the expression of genes in the SAGA-dependent but not in the TFIID- or TFIID- plus SAGA-dependent categories, further supporting a mostly SAGA-specific defect for these mutants. Thus, it seems possible to create mutations in shared TAFIIs that affect mainly TFIID (and SAGA) or SAGA functions. To identify such SAGA- and TFIID-specific mutants, it seems to be necessary to combine expression profiles and biochemical analysis of the complexes involved.
The fact, however, that almost every TAFII25 mutant (36; this study) shows an individual phenotype with rather limited congruity with any other mutant argues against a very specific role of TAFII25 (and probably the other HFD-TAFIIs) in mediating specific interactions with distinct activators or promoter elements. Instead, these observations favor a view in which the histone fold domain TAFIIs would play instead a structural role in which numerous interactions, both between the HFD-TAFIIs and with promoter sequences, activators, and/or (general) transcription factors, would contribute to transcriptional regulation by the proposed HFD-TAFII nucleosome-like structure(s). Likewise, a great number of histone mutations were generated, giving rise to a plethora of phenotypes and transcription defects (32). The crystal structure of the nucleosome bound to DNA showed how numerous the histone-DNA contacts are and how little a single or even a few contacts influence the overall nucleosome structure (47).
It is clear that the HFD is for most of the HFD-TAFIIs the only essential—usually evolutionary conserved—part of the protein, whereas N- and C-terminal sequences can be deleted without apparent phenotype. Nevertheless, these sequences may play very important and specific roles at particular promoters, but they may not contribute to the general structural organization of the TAFII-containing complexes. Thus, to understand the role of HFD-TAFIIs, it will be of paramount importance to understand the organization and three-dimensional structure of TAFIIs in both TFIID and SAGA. If HFD-TAFIIs really form a nucleosome-like structure(s), it will be important to identify mutations (or combinations of mutations) that effectively destroy the whole structure in one or the other complex to understand their real importance in transcription.
Acknowledgments
We are grateful to F. Winston for antibodies; C. Waltzinger for help in the flow cytometry studies; R. Martin, C. Gaudon, and R. Losson for advice and reagents; U. Kothe for help at certain stages of this study; A. Collet for help in the figure preparations; and B. Bell and W. Mohan for critically reading the manuscript. We also thank F. Ruffenach for oligonucleotide synthesis.
D.B.K. was supported by fellowships from the European Community (Marie Curie) and the Fondation pour la Recherche Médicale (FRM), and E.v.B. was supported by a European Community RTN grant (HPRN-CT-2000-00087). This work was supported by funds from the Institut National de la Santé et de la Recherche Médicale, the CNRS, the Hôpital Universitaire de Strasbourg, the Association pour la Recherche sur le Cancer, the FRM, the Ligue Nationale contre le Cancer, European Community grants HPRN-CT-2000-00087 and HPRN-CT-2000-00088, the Human Frontier Science Program (RG 196/98) to L.T., and NIH grant GM 52461 to P.A.W.
REFERENCES
- 1.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Cote. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apone, L. M., C. M. Virbasius, J. C. Reese, and M. R. Green. 1996. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 10:2368-2380. [DOI] [PubMed] [Google Scholar]
- 3.Arents, G., R. W. Burlingame, B. C. Wang, W. E. Love, and E. N. Moudrianakis. 1991. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA 88:10148-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, B., and L. Tora. 1999. Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp. Cell Res. 246:11-19. [DOI] [PubMed] [Google Scholar]
- 5.Berger, S. L., B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251-265. [DOI] [PubMed] [Google Scholar]
- 6.Birck, C., O. Poch, C. Romier, M. Ruff, G. Mengus, A.-C. Lavigne, I. Davidson, and D. Moras. 1998. Human TAFII28 and TAFII18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell 94:239-249. [DOI] [PubMed] [Google Scholar]
- 7.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 8.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 9.Brand, M., C. Leurent, V. Mallouh, L. Tora, and P. Schultz. 1999. Three-dimensional structures of the TAFII-containing complexes TFIID and TBP-free TAFII-containing complex. Science 286:2151-2153. [DOI] [PubMed] [Google Scholar]
- 10.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 274:18285-18289. [DOI] [PubMed] [Google Scholar]
- 11.Brown, C. E., I. Lechner, I. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15-19. [DOI] [PubMed] [Google Scholar]
- 12.Burke, T. W., and J. T. Kadonaga. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, J. L., L. D. Attardi, C. P. Verrijzer, K. Yokomori, and R. Tjian. 1994. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell 79:93-105. [DOI] [PubMed] [Google Scholar]
- 14.Dikstein, R., S. Ruppert, and R. Tjian. 1996. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell 84:781-790. [DOI] [PubMed] [Google Scholar]
- 15.Durso, R. J., A. K. Fisher, T. J. Albright-Frey, and J. C. Reese. 2001. Analysis of taf90 mutants displaying allele-specific and broad defects in transcription. Mol. Cell. Biol. 21:7331-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eizenmann, D. M., K. M. Arndt, S. L. Ricupero, J. W. Rooney, and F. Winston. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6:1319-1331. [DOI] [PubMed] [Google Scholar]
- 17.Eizenmann, D. M., C. Chapon, S. M. Roberts, C. Dollard, and F. Winston. 1994. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics 137:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gangloff, Y., C. Romier, S. Thuault, S. Werten, and I. Davidson. 2001. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26:250-257. [DOI] [PubMed] [Google Scholar]
- 19.Gangloff, Y. G., J. C. Pointud, S. Thuault, L. Carre, C. Romier, S. Muratoglu, M. Brand, L. Tora, J. L. Couderc, and I. Davidson. 2001. The TFIID components human TAFII140 and Drosophila BIP2 [TAFII155] are novel metazoan homologues of yeast TAFII47 containing a histone fold and a PHD finger. Mol. Cell. Biol. 21:5109-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gangloff, Y. G., S. L. Sanders, C. Romier, D. Kirschner, P. A. Weil, L. Tora, and I. Davidson. 2001. Histone folds mediate selective heterodimerization of yeast TAFII25 with TFIID components yTAFII47 and yTAFII65 and with SAGA component ySPT7. Mol. Cell. Biol. 21:1841-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gangloff, Y. G., S. Werten, C. Romier, L. Carre, O. Poch, D. Moras, and I. Davidson. 2000. The human TFIID components TAFII135 and TAFII20 and the yeast SAGA components ADA1 and TAFII68 heterodimerize to form histone-like pairs. Mol. Cell. Biol. 20:340-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gansheroff, L. J., C. Dollard, P. Tan, and F. Winston. 1995. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics 139:523-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgieva, S., D. B. Kirschner, T. Jagla, E. Nabirochkina, S. Hanke, H. Schenkel, C. de Lorenzo, P. Sinha, K. Jagla, B. Mechler, and L. Tora. 2000. Two novel drosophila TAFIIs have homology with human TAFII30 and are differentially regulated during development. Mol. Cell. Biol. 20:1639-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodrich, J. A., T. Hoey, C. J. Thut, A. Admon, and R. Tjian. 1993. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75:519-530. [DOI] [PubMed] [Google Scholar]
- 26.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 27.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates, and J. L. Workman. 1998. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 28.Grant, P. A., D. Schieltz, M. G. Pray-Grant, J. R. Yates, and J. L. Workman. 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2:863-867. [DOI] [PubMed] [Google Scholar]
- 29.Green, M. R. 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25:59-63. [DOI] [PubMed] [Google Scholar]
- 30.Guthrie, C., and G. F. Fink. 1991. Guide to yeast genetics and molecular biology, vol. 194. Academic Press, Inc., New York, N.Y.
- 31.Hampsey, M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62:465-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartzog, G. A., and F. Winston. 1997. Nucleosomes and transcription: recent lessons from genetics. Curr. Opin. Genet. Dev. 7:192-198. [DOI] [PubMed] [Google Scholar]
- 33.Hoffmann, A., C. M. Chiang, T. Oelgeschlager, X. Xie, S. K. Burley, Y. Nakatani, and R. G. Roeder. 1996. A histone octamer-like structure within TFIID. Nature 380:356-359. [DOI] [PubMed] [Google Scholar]
- 34.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 35.Jacq, X., C. Brou, Y. Lutz, I. Davidson, P. Chambon, and L. Tora. 1994. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell 79:107-117. [DOI] [PubMed] [Google Scholar]
- 36.Kirchner, J., S. L. Sanders, E. Klebanow, and P. A. Weil. 2001. Molecular genetic dissection of TAF25, an essential yeast gene encoding a subunit shared by TFIID and SAGA multiprotein transcription factors. Mol. Cell. Biol. 21:6668-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klebanow, E. R., D. Poon, S. Zhou, and P. A. Weil. 1996. Isolation and characterization of TAF25, an essential yeast gene that encodes an RNA polymerase II-specific TATA-binding protein-associated factor. J. Biol. Chem. 271:13706-13715. [DOI] [PubMed] [Google Scholar]
- 38.Kokubo, T., D.-W. Gong, J. C. Wootton, M. Horikoshi, R. G. Roeder, and Y. Nakatani. 1994. Molecular cloning of Drosophila TFIID subunits. Nature 367:484-487. [DOI] [PubMed] [Google Scholar]
- 39.Komarnitsky, P. B., B. Michel, and S. Buratowski. 1999. TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev. 13:2484-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 41.Kuras, L., P. Kosa, M. Mencia, and K. Struhl. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 42.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405:701-704. [DOI] [PubMed] [Google Scholar]
- 43.Lew, D. J., N. J. Marini, and S. I. Reed. 1992. Different G1 cyclins control the timing of cell cycle commitment in mother and daughter cells of the budding yeast S. cerevisiae. Cell 69:317-327. [DOI] [PubMed] [Google Scholar]
- 44.Li, X. Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 45.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 46.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 47.Luger, K., and T. J. Richmond. 1998. DNA binding within the nucleosome core. Curr. Opin. Struct. Biol. 8:33-40. [DOI] [PubMed] [Google Scholar]
- 48.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. 1998. A human SPT3-TAFII31-GCN5-l-acetylase complex distinct from transcription factor IID. J. Biol. Chem. 273:23781-23785. [DOI] [PubMed] [Google Scholar]
- 49.Martinez, E., Q. Zhou, N. D. L'Etoile, T. Oelgeschlager, A. J. Berk, and R. G. Roeder. 1995. Core promoter-specific function of a mutant transcription factor TFIID defective in TATA-box binding. Proc. Natl. Acad. Sci. USA 92:11864-11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.May, M., G. Mengus, A. C. Lavigne, P. Chambon, and I. Davidson. 1996. Human TAFII28 promotes transcriptional stimulation by activation function 2 of the retinoid X receptors. EMBO J. 15:3093-3104. [PMC free article] [PubMed] [Google Scholar]
- 51.Metzger, D., E. Scheer, A. Soldatov, and L. Tora. 1999. Mammalian TAFII30 is required for cell cycle progression and specific cellular differentiation programmes. EMBO J. 18:4823-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michel, B., P. Komarnitsky, and S. Buratowski. 1998. Histone-like TAFs are essential for transcription in vivo. Mol. Cell 2:663-673. [DOI] [PubMed] [Google Scholar]
- 53.Mizzen, C. A., X. J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell 87:1261-1270. [DOI] [PubMed] [Google Scholar]
- 54.Moqtaderi, Z., Y. Bai, D. Poon, P. A. Weil, and K. Struhl. 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383:188-191. [DOI] [PubMed] [Google Scholar]
- 55.Moqtaderi, Z., J. D. Yale, K. Struhl, and S. Buratowski. 1996. Yeast homologues of higher eukaryotic TFIID subunits. Proc. Natl. Acad. Sci. USA 93:14654-14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Natarajan, K., B. M. Jackson, E. Rhee, and A. G. Hinnebusch. 1998. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol. Cell 2:683-692. [DOI] [PubMed] [Google Scholar]
- 57.O'Brien, T., and R. Tjian. 1998. Functional analysis of the human TAFII250 N-terminal kinase domain. Mol. Cell 1:905-911. [DOI] [PubMed] [Google Scholar]
- 58.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schlitz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 59.Pham, A. D., and F. Sauer. 2000. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 289:2357-2360. [DOI] [PubMed] [Google Scholar]
- 60.Pierrat, B., D. M. Heery, Y. Lemoine, and R. Losson. 1992. Functional analysis of the human estrogen receptor using a phenotypic transactivation assay in yeast. Gene 119:237-245. [DOI] [PubMed] [Google Scholar]
- 61.Plewniak, F., J. D. Thompson, and O. Poch. 2000. Ballast: Blast postprocessing based on locally conserved segments. Bioinformatics 16:750-759. [DOI] [PubMed] [Google Scholar]
- 62.Poon, D., Y. Bai, A. M. Campbell, S. Bjorklund, Y. J. Kim, S. Zhou, R. D. Kornberg, and P. A. Weil. 1995. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92:8224-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pugh, B. F., and R. Tjian. 1992. Diverse transcriptional functions of the multisubunit eukaryotic TFIID complex. J. Biol. Chem. 267:679-682. [PubMed] [Google Scholar]
- 64.Pugh, B. F., and R. Tjian. 1991. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev. 5:1935-1945. [DOI] [PubMed] [Google Scholar]
- 65.Roberts, S. M., and F. Winston. 1996. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:3206-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saleh, A., D. Schieltz, N. Ting, S. B. McMahon, D. W. Litchfield, J. R. Yates, S. P. Lees-Miller, M. D. Cole, and C. J. Brandl. 1998. Tra1p is a component of the yeast Ada. Spt transcriptional regulatory complexes. J. Biol. Chem. 273:26559-26565. [DOI] [PubMed] [Google Scholar]
- 67.Sanders, S. L., E. R. Klebanow, and P. A. Weil. 1999. TAF25p, a nonhistone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem. 274:18847-18850. [DOI] [PubMed] [Google Scholar]
- 68.Sanders, S. L., and P. A. Weil. 2000. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem. 275:13895-13900. [DOI] [PubMed] [Google Scholar]
- 69.Shen, W. C., and M. R. Green. 1997. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90:615-624. [DOI] [PubMed] [Google Scholar]
- 70.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanese, N., B. F. Pugh, and R. Tjian. 1991. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 5:2212-2224. [DOI] [PubMed] [Google Scholar]
- 74.Tsukihashi, Y., T. Miyake, M. Kawaichi, and T. Kokubo. 2000. Impaired core promoter recognition caused by novel yeast TAF145 mutations can be restored by creating a canonical TATA element within the promoter region of the TUB2 gene. Mol. Cell. Biol. 20:2385-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verrijzer, C. P., J. L. Chen, K. Yokomori, and R. Tjian. 1995. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell 81:1115-1125. [DOI] [PubMed] [Google Scholar]
- 76.vom Baur, E., M. Harbers, S. J. Um, A. Benecke, P. Chambon, and R. Losson. 1998. The yeast Ada complex mediates the ligand-dependent activation function AF-2 of retinoid X and estrogen receptors. Genes Dev. 12:1278-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.vom Baur, E., C. Zechel, D. Heery, M. J. Heine, J. M. Garnier, V. Vivat, B. Le Douarin, H. Gronemeyer, P. Chambon, and R. Losson. 1996. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 15:110-124. [PMC free article] [PubMed] [Google Scholar]
- 78.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 79.Walker, S. S., J. C. Reese, L. M. Apone, and M. R. Green. 1996. Transcription activation in cells lacking TAFIIs. Nature 383:185-188. [DOI] [PubMed] [Google Scholar]
- 80.Walker, S. S., W. C. Shen, J. C. Reese, L. M. Apone, and M. R. Green. 1997. Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell 90:607-614. [DOI] [PubMed] [Google Scholar]
- 81.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of TAFII-containing complex without TBP in transcription by RNA polymerase II. Nature 393:187-191. [DOI] [PubMed] [Google Scholar]
- 82.Xie, X., T. Kokubo, S. L. Cohen, U. A. Mirza, A. Hoffmann, B. T. Chait, R. G. Roeder, Y. Nakatani, and S. K. Burley. 1996. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature 380:316-322. [DOI] [PubMed] [Google Scholar]