Abstract
CLIP-170 is a plus-end tracking protein which may act as an anticatastrophe factor. It has been proposed to mediate the association of dynein/dynactin to microtubule (MT) plus ends, and it also binds to kinetochores in a dynein/dynactin-dependent fashion, both via its C-terminal domain. This domain contains two zinc finger motifs (proximal and distal), which are hypothesized to mediate protein-protein interactions. LIS1, a protein implicated in brain development, acts in several processes mediated by the dynein/dynactin pathway by interacting with dynein and other proteins. Here we demonstrate colocalization and direct interaction between CLIP-170 and LIS1. In mammalian cells, LIS1 recruitment to kinetochores is dynein/dynactin dependent, and recruitment there of CLIP-170 is dependent on its site of binding to LIS1, located in the distal zinc finger motif. Overexpression of CLIP-170 results in a zinc finger-dependent localization of a phospho-LIS1 isoform and dynactin to MT bundles, raising the possibility that CLIP-170 and LIS1 regulate dynein/dynactin binding to MTs. This work suggests that LIS1 is a regulated adapter between CLIP-170 and cytoplasmic dynein at sites involved in cargo-MT loading, and/or in the control of MT dynamics.
Several developmental and cellular processes require tightly controlled polarity signaling to the cytoskeleton. These processes include the migration of neurons during brain development (14), the establishment of apicobasolateral polarity of epithelial cells (83), and the positioning of mitotic spindles before the onset of anaphase (26). Central to this is the role of positional cues assembled at the actin-based cell cortex (65). These positional cues capture microtubules (MTs) and generate pulling forces on the centrioles and attached organelles. The molecular basis for MT capture at the cell cortex is still far from understood. Many proteins are thought to be involved in the process; among them are the MT-binding proteins CLIP-170 and LIS1.
CLIP-170, cortical cues, and dynein-dynactin.
CLIP-170 was identified as an MT-binding protein that localized to a subset of MT distal ends (54). Subsequent experiments demonstrated that it is required for the binding of endocytic carrier vesicles to MTs in vitro (51). In living cells, it is found at MT plus ends (17), but it detaches when the MT stops growing (49). Genetic studies with budding and fission yeasts have also established that the CLIP-170 orthologs, Bik1p and tip1p, may act as an anticatastrophe factor at specific cellular sites, ensuring that the MTs reach the tip of the growing cell (4, 8). CLIP-170 belongs to a unique class of MT-binding proteins, recently termed +TIPS (for plus-end tracking proteins) (64). The closest homolog of CLIP-170 in mammals, CLIP-115 (16), is also a +TIP (1, 32). Like CLIP-170, CLIP-115 contains two MT binding domains at its N terminus and a coiled-coil domain that mediates homodimerization (32, 62), but CLIP-115 lacks the unique C-terminal domain of CLIP-170, which displays two conserved zinc finger motifs (27). It remains unknown whether the latter are functionally important.
In addition to a potential role in the regulation of MT dynamics, the +TIPs may also function to recruit different proteins to the tips of growing MTs. For example, dynein/dynactin and adenomatous polyposis coli have been suggested to be targeted to MT tips by CLIP-170 (74) and EB1 (45), respectively. The positioning of the Saccharomyces cerevisiae mitotic spindle near the bud's neck and its migration into the bud involve interactions of cytoplasmic MTs with positional cues (48). The yeast homolog of EB1, Bim1p, is part of a kinesin-dependent pathway mediating positioning of the spindle near the bud's neck before anaphase (73). Bim1p interacts with Kar9p (44), a protein transported along actin cables to the bud cortex via an interaction with a type V myosin (reviewed in reference 37). This is so far the only elucidated molecular mechanism for tethering MTs to the cell cortex.
Genetic studies have suggested that the minus-end-directed MT motor dynein and its regulator, the dynactin complex, are also involved in MT capture at the cell cortex (7, 12). Dynein's known sites of action include cortical cues involved in centriole/nucleus rotations in Caenorhabditis elegans embryos (33, 69), spindle positioning in fruit fly oogenesis (42, 71), and mammalian epithelial cells (10). So far, the molecular mechanism of the dynein/dynactin-mediated MT tethering at cortical cues remains unknown. CLIP-170 may have an interesting role in this process, since it may tether cortex-associated dynein/dynactin to MTs, but so far attempts to demonstrate a physical interaction between these molecules have been unsuccessful.
Kinetochores as positional cues.
A special example of a positional cue is the kinetochore, a pair of structures assembled during mitosis at the centromeres (34). They contain several large protein complexes that mediate interactions with spindle MTs and are important for fail-safe progression of mitosis (reviewed in references 41, 56, 66, and 68). In vertebrate somatic cells entering mitosis, the first kinetochore movements are mediated by the rapid sliding of one of the sister kinetochores along the matrix of a polar MT that it has captured tangentially (43, 55). This transport is likely to be driven by the dynein/dynactin motor complex (reviewed in reference 2). Kinetochore-localized CLIP-170 (18) could play a role in the MT-capturing process and subsequently aid the motor complex with initiating the transport, but so far, proof for this function is sparse. CLIP-170 colocalizes with the dynein/dynactin complex at nonattached kinetochores in a dynein/dynactin-dependent way (18). Like at MTs (74), this colocalization depends on the carboxy-terminal domain of CLIP-170 (18), indicating a direct or indirect association with the motor complex. Recently, Bik1p was shown to be a +TIP and a component of the kinetochore-MT binding interface in budding yeast. Its binding to the kinetochore is dependent on the C-terminal 40 amino acids (38).
LIS1 and dynein/dynactin.
LIS1 is one of the genes that have a principal role in brain development, since hemizygote mutations in LIS1 result in a severe brain malformation known as lissencephaly (smooth brain) (53). LIS1 contains an N-terminal region with a coiled-coil domain important for dimerizaton (11) and seven WD repeats that mediate protein interactions (52, 53). Recent data have provided compelling evidence for LIS1 function in the dynein/dynactin pathway. nudF, a LIS1 homolog in the fungus Aspergillus nidulans, was identified in a screen for mutants defective in nuclear migration (46, 82). Genetic and biochemical evidence suggested that nudF regulates dynein motor activity and MT dynamics (3, 29, 31, 79-81). These findings are consistent with interaction of mammalian LIS1 with tubulin (58) and are corroborated by its interaction with the first P-loop of dynein heavy chain (60). Dynein, dynactin, and LIS1 also coimmunoprecipitate in cultured cells and in the brain (23, 70). In interphase mammalian cells, LIS1 was localized in a dot-like pattern along MTs but not at MT tips (58, 70), whereas in mitotic cells, it colocalizes at astral MTs, kinetochores, and cortical sites (23). Overexpression of LIS1 results in spindle misorientation and disturbance in mitotic progression (23). LIS1 could therefore be involved in dynein/dynactin-mediated tethering of MTs to cortical sites in a yet-unknown way. Overexpression or reduced expression of LIS1 results in the disturbance of additional dynein-related functions in mammalian cells (60, 70) and in Drosophila melanogaster neurons (39). An additional link of LIS1 to the dynein pathway was demonstrated in yeast. A genetic screen for genes required for viability in budding yeast cells with a kinesin-like motor deleted identified many of the established dynein pathway genes (25). Pac1p and Bik1p, homologs of, respectively, LIS1 and CLIP-170 (4, 51), were identified in that screen as well. Pac1p was also found to be involved in nuclear migration together with other genes in the dynein/dynactin pathway (24, 25).
Taking into consideration the amazing conservation of LIS1 functions from A. nidulans to human, and based on the data from S. cerevisiae, indicating a possible functional relationship between PAC1 and BIK1, we set out to investigate whether LIS1 and CLIP-170 interact in mammalian cells and the role of the CLIP-170 zinc finger motifs in this interaction. Here we show, in addition to the expected localizations at kinetochores and tips of astral MTs, new colocalizations of CLIP-170 and LIS1 at MT tips in interphase cells, nuclei of prophase cells, and cortical sites contacting astral MTs in mitotic cells. We also provide evidence for the existence of a physical interaction between CLIP-170 and LIS1, involving both zinc fingers of CLIP-170 and some of the WD domains of LIS1. We report that the recruitment of LIS1 to kinetochores is dynein/dynactin dependent and that CLIP-170 targeting to these structures is via its LIS1-interacting domain. Although LIS1 has an intrinsic MT binding capacity, we show that association of a phospho-LIS1 isoform (and dynactin) to MT bundles, induced by CLIP-170 overexpression, is dependent on the distal zinc finger of CLIP-170, raising the possibility that CLIP-170 and LIS1 regulate dynein/dynactin binding to MTs. This work suggests that LIS1 is a regulated adapter between CLIP-170 and cytoplasmic dynein at sites involved in cargo MT loading, and/or in the control of MT dynamics.
MATERIALS AND METHODS
Cell culture.
Culturing and transfection of COS-1 cells (32) or HeLa and COS-7 cells (18) was performed as described previously.
Immunological methods.
Antibodies against CLIP-170 were monoclonal antibody 4D3 (54), polyclonal antibody H2A (18), and MT domain antibodies anti-CLIP-115/170 antibody no. 2221 (32). Polyclonal anti-CLIP-170 TA antibodies were produced in rabbit by using a His-tagged fusion protein comprising the last 156 C-terminal amino acids of CLIP-170 (a gift of T. Kreis). CLIP-170 C terminus-specific antibodies (no. 2360) were generated by injecting glutathione S-transferase (GST)-CLIP-170 C terminus (amino acids 1029 to 1320) into rabbits as described previously (32). Antibodies against LIS1 (polyclonal [no. 19, affinity purified] and monoclonal [no. 210]) were described previously (57, 58). Other antibodies were monoclonal anti-p150GLUED (Transduction Laboratories, Lexington, Ky.), anti-cytoplasmic dynein intermediate-chain monoclonal antibody 70.1 (Sigma), human CREST autoimmune antiserum recognizing the centromeric region of chromosomes (a gift from H. Ponstingl, German Cancer Research Center, Heidelberg, Germany), monoclonal anti-β-tubulin (Zymed Laboratories, Inc., South San Francisco, Calif.), and monoclonal anti-myc 9E10 (22). All secondary antibodies used for the immunofluorescence studies were made in goat and conjugated to 6-((7-amino-4-methylcoumarin-3-acetyl)amino) hexonic acid, fluorescein isothiocyanate, Cy3, or Cy5 (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) or ALEXA-488, -594, -350 (Molecular Probes).
For immunofluorescence of MT tips, cells were fixed in 100% methanol-1 mM EGTA for 10 min at −20°C, followed by 15 min of fixation in 4% paraformaldehyde in phosphate-buffered saline (PBS). Cells were washed for 5 min in 0.15% Triton X-100-PBS, blocked in PBS-1% bovine serum albumin (BSA)-0.1% Tween, and labeled with different antibodies for 1 h at room temperature. After washing in 0.05% Tween 20-PBS sections were incubated with secondary antibodies. Slides were mounted using Vectashield mounting medium (Vector laboratories) with DAPI (4′,6′-diamidino-2-phenylindole) (Sigma) and analyzed on a Leica DMRBE fluorescence microscope equipped with a Hamamatsu C4880 charge-coupled device camera.
For the colocalization experiment, HeLa cells were lysed for 2 min in 0.5% Triton X-100 and fixed in 3% paraformaldehyde in PHEM buffer (18). They were washed three times for 5 min each in PBS and further permeabilized for 25 min in 0.1% Triton X-100 in PBS. The cells were treated with 50 mM NH4Cl in PBS for 10 min, washed three times for 5 min each in PBS, blocked in PBS-BSA (PBS containing 0.1% BSA), and labeled with a polyclonal anti-LIS1 antibody and the monoclonal 4D3 antibodies for 1 h at 37°C. After being washed in PBS-BSA, the cells were incubated with secondary antibodies for 45 min. The cells were postfixed in 4% formaldehyde in PBS for 16 min and treated with 50 mM NH4Cl in PBS for 10 min. The chromosomes were stained with DAPI (Sigma) for 5 min. Coverslips were mounted using PBS-15% glycerol containing the antifading agent 1.4-diazabicyclo-(2.2.2) octane (DABCO) (Sigma) at 100 mg/ml (36). All antibodies were diluted in PBS-BSA.
For the kinetochore-targeting experiment, transfected COS-7 cells were treated with 33 μM nocodazole for 1 h just before the fixation step. The cells were lysed for 30 s in 0.5% Triton X-100 in PHEM to remove excess soluble expressed protein, fixed for 20 min in 3% paraformaldehyde in PHEM at room temperature, washed three times for 5 min each in PBS, and further permeabilized for 25 min in 0.5% Triton X-100 in PBS. The immunofluorescences were processed as described above using CREST human serum and a polyclonal anti-GFP antibody.
For enhanced green fluorescent protein (EGFP)-p50 overexpression experiments, transfected HeLa cells were treated with 10 μM nocodazole for 4 h before lysis and fixation, as described above. Cells were immunolabeled as previously described, using either anti-p150, polyclonal anti-LIS1 and CREST serum, or anti-p150, TA, and CREST.
For recruitment on MTs, transfected COS-7 cells were lysed for 30 s in 0.5% Triton X-100 in PHEM to remove excess soluble expressed protein and fixed for 20 min in 3% paraformaldehyde. The immunofluorescences were processed as described above using a polyclonal or monoclonal anti-c-myc antibody with a polyclonal anti-LIS1 antibody, the 210 monoclonal antibody, or the monoclonal anti-p150 antibody. For COS-7 cells transfected with CLIP-ΔRΔC-DsRed, the immunofluorescence was processed with a monoclonal anti-EB1 antibody or the TA antibody.
Wide-field optical sectioning fluorescence microscopy.
Pictures of fixed cells were collected using a three-dimensional deconvolution imaging system, the detailed description and validation of which will be published elsewhere (J.-B. Sibarita and J. R. De Mey, unpublished data). A brief description has been published previously (61).
Protein methods.
GST pull-down experiments with brain extracts and reticulocyte lysates were done as described previously (59). HeLa cells were extracted as described previously (15). For GST pull-down experiments, 100 μg of HeLa cells proteins and 10 μg of recombinant proteins on glutathione beads were used in a final volume of 500 μl. Beads were processed as described previously (59). Immunoprecipitations were done as described previously (13). Yeast two-hybrid assays were done according to the instructions of the manufacturer (Origene, Rockville, Md.).
Expression constructs.
The mammalian expression vectors pEGFP-C (Clontech, Palo Alto, Calif.) were used to construct GFP-CLIP-115 and GFP-CLIP-170-br (32). These vectors were also used to construct GFP-CLIP-170(−tail), in which the last 79 amino acids of CLIP-170-br are deleted, and GPF-CLIP-115(+tail), which contains the C-terminal 159 amino acids of CLIP-170 linked in frame to CLIP-115 in front of its stop codon. GFP-LIS1 (mouse LIS1 cDNA was obtained by reverse transcription-PCR from total mouse brain RNA), GFP-NudC (mouse NudC cDNA is IMAGE clone 2646247 [accession number AW322862]), GFP-NudE (human NudE cDNA is IMAGE clone 2820974 [accession number AW249237]), and GFP-NudEL (mouse NudEL cDNA is IMAGE clone 2646029 [accession number AW322683]) were cloned by PCR into pEGFP vector. To generate HA-CLIP-115 and HA-CLIP-170-br fusions, the pMT2SM-HA vector was used; HA-LIS1 was produced by substituting GFP for the hemagglutinin (HA) tag in the corresponding fusion construct.
The plasmids encoding wild-type CLIP-170 and CLIP-170 Δ55-346 (lacking the MT-binding regions) are referred to here as wt-CLIP-170 and ΔN-CLIP-170, respectively, and have been described elsewhere (50, 51). These fusion proteins have been tagged with a c-myc epitope at the N terminus. These cDNA constructs were a gift of T. Kreis and P. Pierre (University of Geneva, Geneva, Switzerland). The myc-tagged wt-CLIP-170 cDNA was inserted into the EcoRI site of the pSG5 vector. The expression of the fusion protein is under control of the simian virus 40 promoter. The C-terminal XhoI/BamHI fragment of CLIP-170 (corresponding to the last 156 amino acids) was inserted in the pBluescript II KS(−), pEGFPC3, pEG202 (with a modified multiple cloning site [MCS]), and pJG4-5 (with a modified MCS) vectors in phase with the GFP (Clontech) and LEXA and AD (Origene) genes, respectively. These constructs were named, respectively, pBS-Cter, pEGFP-Cter, pEG202-Cter, and pJG4-5-Cter. pBS-Cter was digested with XhoI and NotI enzymes, and the resulting XhoI/NotI fragment was inserted in pGEX4T3 at the SalI and NotI sites to give pGEX-Cter. A C-terminal fragment corresponding to the last 64 amino acids of the CLIP-170 was generated by PCR using ΔN-CLIP170 as a template and the primers 5′TCCAAGCTCGAGCCTCGCCTCTTCTGTGACATT-3′ and 5′AGATCTGGATCCGAATTCCCTAGTCTGTTATGT3′. This fragment was inserted in the pEGFPC3, pEG202 (with a modified MCS), and pJG4-5 (with a modified MCS) vectors to give pEGFP-Z1 + 2, pEG202-Z1 + 2, and pJG4-5-Z1 + 2.
Fusion proteins lacking the C-terminal 20 amino acids of CLIP-170 were generated as follows. ΔN-CLIP170 was used as a template to amplify by PCR the cDNA sequence of CLIP-170 and introduce a stop codon and a BamHI restriction site by using the primers 5′-CTGAGAAATGAGGTCACAGT-3′ and 5′TATTAGGATCCTTAGTATGGGCGTTCCTCACC-3′. The PCR product was digested with XhoI and BamHI enzymes and was exchanged with the XhoI/BamHI fragment of pSG5-wt-CLIP-170, pSG5-ΔN-CLIP-170, pEGFPC3, pEG202 (modified MCS), pJG4-5 (modified MCS), and pBluescript II KS(−). These constructs were named, respectively, ΔC2, ΔNΔC2, pEGFP-ΔC2, pEG202-ΔC2, pJG4-5-ΔC2, and pBS-ΔC2. The pBluescript vector containing the XhoI/BamHI PCR product was digested with XhoI and NotI, and the resulting XhoI/NotI fragment was inserted in pGEX4T3 at the SalI and NotI sites to give pGEX-ΔC2.
Fusion proteins lacking the C-terminal 14 and 6 amino acids were generated as described above using the primers 5′TATTAGGATCCTTACATCTCACAGATTTCACAGTATGG-3′ and 5′TATTAGGATCCTTAGCAGTTGGTGGCCCAGTGTCC-3′ to introduce the modifications. They were named, respectively, ΔC3, ΔNΔC3, pEGFP-ΔC3, pEG202-ΔC3, pJG4-5-ΔC3, pBS-ΔC3, pGEX-ΔC3, ΔC4, ΔNΔC4, pEGFP-ΔC4, pBS-ΔC4, and pGEX-ΔC4.
Fusion proteins with mutations C1333M and C1336M (mutations in the first zinc finger motif) were generated using the Quick Change Kit (Stratagene, La Jolla, Calif.). The C-terminal XhoI/BamHI fragment of CLIP-170 cDNA subcloned in pBluescript KS(−) was used as a template with the primers 5′TGGAGATCAAAGCAGTCCATAATGTCCATGAAGAGGCGAGGTTTCTT-3′ and 5′AAGAAACCTCGCCTCTTCATGGACATTATGGACTGCTTTGATCTCCA-3′. The mutated XhoI/BamHI fragment was exchanged with that of pSG5-wt-CLIP-170 and was inserted in pEGFPC3, pEG202 (modified MCS), and pJG4-5 (modified MCS) to give Z1m, pEGFPC3-Z1m, pEG202-Z1m, and pJG4-5-Z1m, respectively. The pBluescript vector containing the XhoI/BamHI mutated fragment was digested with XhoI and NotI, and the resulting XhoI/NotI fragment was inserted in pGEX4T3 at the SalI and NotI sites. The resulting construct was named pGEX-Z1m.
Fusion proteins with mutations C1373M and C1376M (mutations in the second zinc finger motif) were generated as described above using the primers 5′GGTGAGGAACGCCCATACATGGAAATCATGGAGATGTTTGGACAC-3′ and 5′-GTGTCCAAACATCTCCATGATTTCCATGTATGGGCGTTCCTCACC-3′. The resulting constructs were named, respectively, Z2m, pEGFPC3-Z2m, pEG202-Z2m, pJG4-5-Z2m, and pGEX-Z2m.
Fusion proteins with mutations C1333M, C1336M, C1373M, and C1376 M (mutations in the two zinc finger motifs) were generated as described above using the mutated C1333M and C1336M C-terminal XhoI/BamHI fragment of CLIP-170 cDNA subcloned in pBluescript II KS(−) as a template with the primers 5′GGTGAGGAACGCCCATACATGGAAATCATGGAGATGTTTGGACAC-3′ and 5′-GTGTCCAAACATCTCCATGATTTCCATGTATGGGCGTTCCTCACC-3′. The resulting constructs were named, respectively, dblZm, pEGFPC3-dblZm, pEG202-dblZm, pJG4-5-dblZm, and pGEX-dblZm.
A fusion protein corresponding to the first 361 amino acids of c-mycCLIP-170 was generated as follows. The DsRed gene was subcloned in the pVNC7 vector (modified from pVM116 [35]) at the BamHI and NotI sites to give pVNC7-DsRed. The EcoRI/BglII fragment of CLIP-170 was inserted into the EcoRI and BglII sites of pVNC7-DsRed. This construct was digested with BglII and AgeI enzymes and treated with Klenow polymerase. The final construct was named CLIP-ΔRΔC-DsRed.
All PCR-amplified sequences and all translation phases of the fusion proteins have been verified by sequencing.
The expression vector pDsRed-FLAG-LIS1 was generated by site-directed mutagenesis of the stop codon, introducing an NcoI site, this was cloned in frame with a FLAG epitope and then into pDsRed (Clontech). Point mutations were generated using the Quick Change protocol (Stratagene). The LIS1 yeast two-hybrid construct was described previously (67).
RESULTS
Colocalization of LIS1 and CLIP-170 at MT-dependent and -independent sites.
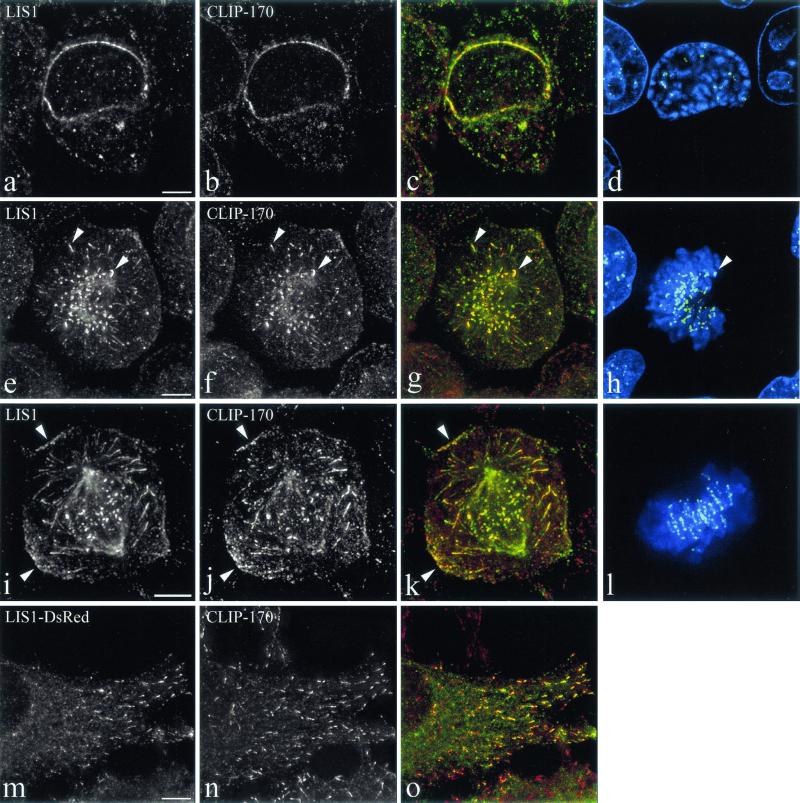
Published data indicate that LIS1 and CLIP-170 may each colocalize at sites also known to accumulate the dynein/dynactin complex (10, 18, 23, 77). In order to assess this, we used mainly HeLa cells, but comparable results were obtained in COS-7 and MDCK cells (not shown). During mitosis, colocalization of LIS1 and CLIP-170 was seen at the nuclear envelopes of prophase cells (Fig. 1a to d). In early prometaphase cells, colocalization was observed at unattached kinetochores (Fig. 1e to h). The position of the kinetochores is marked by the autoimmune CREST serum (Fig. 1h and l). The proteins localize to kinetochores in dots that are adjacent to CREST but do not completely overlap, since anti-CREST autoimmune serum labels the centromeres (19, 20, 41) while CLIP-170 and dynein localize to the fibrous corona (18; reviewed in reference 41). In early prometaphase cells, LIS1 and CLIP-170 also colocalize at the tips of astral MTs (Fig. 1e to h) but also at the cell cortex, where sites accumulating LIS1 and CLIP-170 can be seen (Fig. 1i to l). At all of these sites, dynactin and dynein were detected (not shown). We described earlier such cortical sites accumulating dynein and dynactin in polarized MDCK cells (10). In view of their restricted topology (the belt of adherens junctions, just beneath the tight junctions) and the fact that they interact with astral MTs, we have proposed that these cortical sites play a role in spindle positioning and movements. This is the first demonstration of colocalization of LIS1 and CLIP-170 in these structures. LIS1 was shown to interact with MTs before (58, 70), but it was never seen at MT tips in interphase cells. Since some of the results described below pointed to the possibility of LIS1 binding to MT tips in a CLIP-170-dependent way, we expressed a LIS1-FLAG-DsRed-tagged form and studied its colocalization with CLIP-170. We first ascertained that this form was distributed in a manner similar to all of the known localizations of LIS1, and we also observed that under the conditions used here, it did not cause aberrant mitosis (data not shown), as described by others (23). In interphase cells, LIS1-FLAG-DsRed colocalized with endogenous CLIP-170 at MT plus ends, even in cells expressing very moderate levels (Fig. 1m to o). LIS1 was also distributed as a series of linearly arranged spots, in agreement with the usual immunostaining along MTs (58). We conclude that in mammalian cells too, LIS1 is associated with growing MT plus ends. This localization pattern resembles the dynein labeling at MT tips (77) and is consistent with LIS1 being attached to dynein (23, 60, 70). These results are also consistent with the possibility that LIS1 is being attached to any of a variety of other proteins such as dynactin or EB1; however, at present we do not have evidence supporting these possibilities. The colocalization of CLIP-170 and LIS1 prompted us to investigate further the potential physical interactions between LIS1 and CLIP-170.
FIG. 1.
Colocalization of LIS1 and CLIP-170 at MT-dependent and MT-independent sites. HeLa cells were labeled with a polyclonal anti-LIS1 antibody (a, e, and i), the monoclonal 4D3 anti-CLIP-170 antibody (b, f, and j), and CREST autoimmune serum. Panels c, g, and k are superimpositions of CLIP-170 (red) and LIS1 (green) signals. Panels d, h, and l are superimpositions of CREST (green) and DAPI signals. LIS1 and CLIP-170 colocalize on the nuclear envelope in prophase cells (a to d). In prometaphase, they colocalize on the kinetochores and MT tips (early prometaphase) (e to h, arrows) and on the cortical sites (late prometaphase) (i to l, arrows). HeLa cells were transiently transfected with LIS1-DsRed (m) and fixed under conditions preserving the MT tips (see Materials and Methods). They were labeled with polyclonal anti-CLIP-170 antibody TA (n). Panel o is a superimposition of LIS1-DsRed (green) and CLIP-170 (red). All images are maximal-intensity projections of x/y optical section stacks, acquired by three-dimensional deconvolution microscopy. Bars, 5 μm.
Physical interactions and mapping of interaction domains.
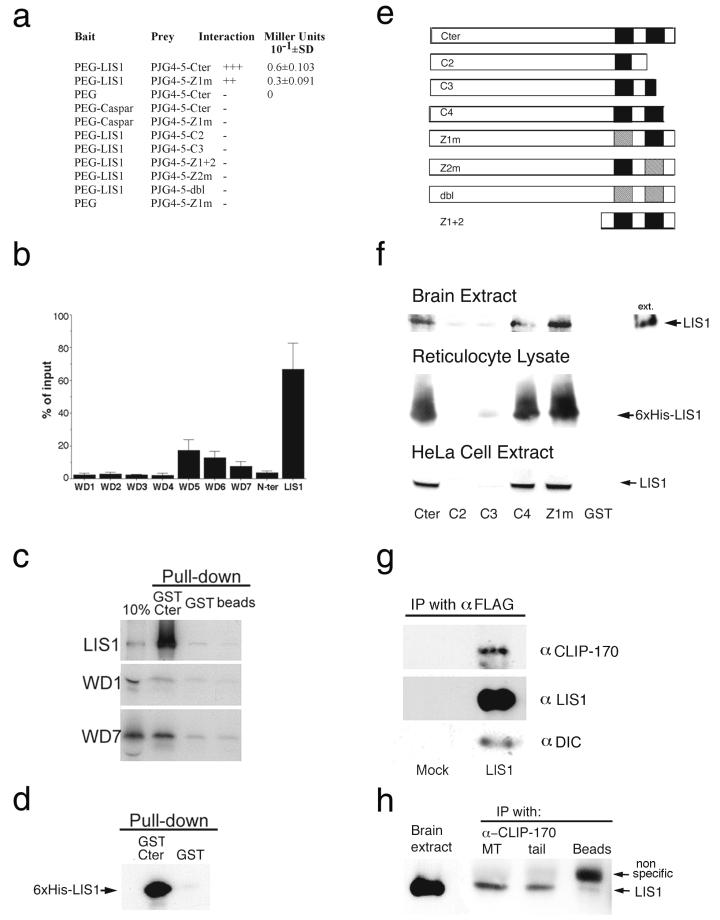
Several assays allowed us to demonstrate an interaction between LIS1 and CLIP-170 and also to map the domains of interaction in both proteins. The yeast two-hybrid system (28) revealed LIS1 interaction with the C-terminal domain of CLIP-170 (Cter in Fig. 2e). A point mutation in the first zinc domain (Z1m in Fig. 2e) weakened the interaction as measured by a liquid β-galactosidase assay (Fig. 2a). A pull-down experiment using the purified recombinant proteins GST-Cter and 6-His-LIS1 demonstrated a direct interaction without the presence of other cellular proteins (Fig. 2d). Several deletions and mutations (C2, C3, and Z2m in Fig. 2e) abolished the interaction. Mapping of the precise interacting domain in CLIP-170 was done using GST pull-down assays with in vitro-translated LIS1 in a reticulocyte lysate extract or endogenous LIS1 from brain or HeLa cell extract (Fig. 2f). The distal zinc finger domain of CLIP-170 was shown to be necessary for LIS1 binding. Deletion of part or all of this domain (C2 and C3) (Fig. 2f) and point mutations in the second or both zinc finger motifs abolished the interaction. However, deleting the last six amino acids that are not part of the zinc finger (C4) (Fig. 2f) did not affect LIS1 binding. Consistent with the results obtained in the yeast two-hybrid experiments, point mutations in the first zinc finger (Z1m) did not disrupt the interaction. Mapping of the interaction domain in LIS1 indicated that the full-length protein is most efficient in binding CLIP-170; nevertheless, WD5, -6, and -7, in reducing order of importance, play a role in the interaction (Fig. 2b and c). In vivo experiments also support an interaction between LIS1 and CLIP-170, as these proteins can be coimmunoprecipitated from LIS1-FLAG-DsRed-transfected cells (Fig. 2g) and from mouse embryonic brain extract (Fig. 2h). We could coprecipitate the dynein intermediate chain by using anti-FLAG antibodies from LIS1-FLAG-DsRed-transfected cells (Fig. 2g). However, dynein intermediate chain was not immunoprecipitated with anti-CLIP-170 antibodies (data not shown).
FIG. 2.
Physical interaction between LIS1 and CLIP-170. (a) LIS1 interacts with the C-terminal region of CLIP-170 in a yeast two-hybrid system. Positive interactions were demonstrated between PEG-LIS1 and pJG4-5-Cter or Z1m. Other controls with empty baits or an unrelated bait (Caspar) resulted in no blue colonies. (b) CLIP-170 interaction with LIS1 requires the presence of the WD5 domain. GST pull-down assays using GST-Cter or GST (negative control) with individual domains of LIS1 that were in vitro translated in a reticulocyte lysate using [35S]methionine were performed. The percentage of input protein that was pulled down specifically by GST-Cter is indicated. The results presented are averages from four independent experiments ± standard errors of the means. (c) Representative autoradiogram used for quantification of the different LIS1 domains in panel b. (d) Recombinant six-histidine LIS1 and GST-Cter-CLIP-170 interact in a GST pull-down assay. The negative control GST does not interact with LIS1. (e) Schematic presentation of the different C-terminal CLIP-170 constructs used in this study. (f) LIS1 interacts with the C-terminal domain of CLIP-170 and requires the second zinc finger domain (results of GST pull-down assays with brain extract, reticulocyte lysate, and HeLa cell extract) are shown. (g) LIS1, CLIP-170, and dynein intermediate chain coimmunoprecipitate (IP) from cells transfected with DsRed-LIS1-FLAG using anti-FLAG antibodies. (h) LIS1 and CLIP-170 coimmunoprecipitate from E15.5 mouse brain extract with two different anti-CLIP-170 antibodies. The antibodies used were the MT domain antibodies anti-CLIP-115/170 antibody no. 2221 (32) and CLIP-170 C terminus (or tail)-specific antibodies (no. 2360), as described in Materials and Methods.
LIS1 and CLIP-170 at kinetochores.
To unravel the functional importance of the LIS1-CLIP-170 interaction we have carefully examined kinetochores, where both proteins colocalize at the fibrous corona.
(i) LIS1 recruitment to kinetochores is dynein/dynactin dependent.
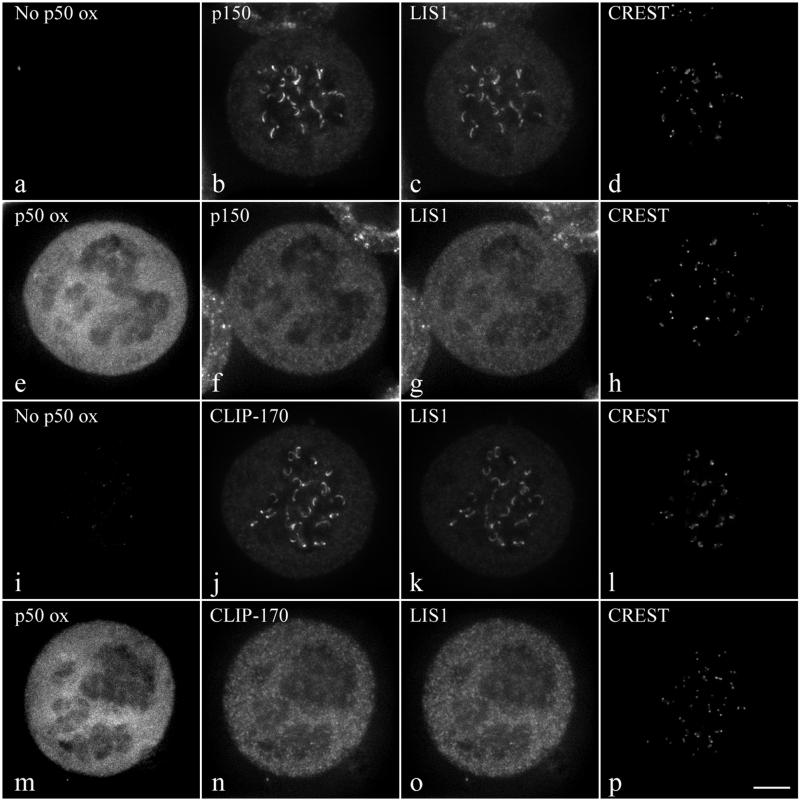
Disruption of the dynein/dynactin protein complex by overexpression of the dynactin subunit p50 causes dissociation of cytoplasmic dynein (9, 21) and CLIP-170 (18) from prometaphase kinetochores. We demonstrate here that overexpression of p50 also results in a significant reduction of LIS1 kinetochore localization (Fig. 3g versus c and o versus k), similar to the reduction in the dynactin subunit p150 (Fig. 3f versus b) and CLIP-170 (Fig. 3n versus j). We conclude that LIS1 targeting to kinetochores is dependent upon the dynein/dynactin complex.
FIG. 3.
Effect of p50 overexpression on CLIP-170 and LIS1 targeting to kinetochores. Triple immunostaining of mitotic control (a to d and i to l) or EGFP-p50-overexpressing (e to h and m to p) HeLa cells for either p150 (b and f), LIS1 (c and g), and CREST (d and h) or CLIP-170 (j and n), LIS1 (k and o), and CREST (l and p). Cells were treated with 10 μM nocodazole for 4 h prior to fixation. As expected, p150 (b and f) and CLIP-170 (j and h) staining from kinetochores decreased under EGFP-p50 expression. Under these conditions LIS1 staining is also dramatically diminished (c, g, k, and o). The images in panels a and i demonstrate that there is no p50 overexpression (ox) while those in panels e and m show eGFP-p50 overexpression. All images are maximal-intensity projections of 10 consecutive planes acquired by three-dimensional deconvolution microscopy. Bar, 5 μm.
(ii) Recruitment of CLIP-170 at kinetochores depends on its LIS1-interacting domain.
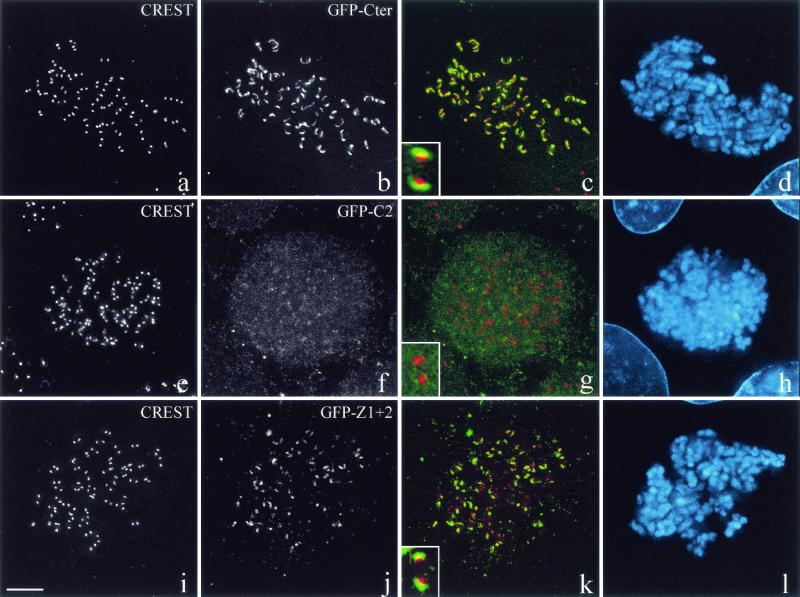
A GFP-tagged wild-type C-terminal fragment of CLIP-170 localizes to kinetochores (Fig. 4a to d), but deletion of the second zinc finger results in a diffuse GFP signal (Fig. 4e to h). Mutations in the first zinc finger motif reduce the CLIP-170 kinetochore signal significantly (data not shown), thus mirroring our results with yeast two-hybrid assays. Nevertheless, expression of the two zinc finger domains by themselves (Z1 + 2) is sufficient to successfully target CLIP-170 to the kinetochores (Fig. 4i to l). We cannot exclude the possibility that the second zinc finger domain interacts directly with additional proteins located at the kinetochore; however, our interpretation is based on the available data. We conclude that the LIS1 binding domain is necessary and sufficient for the recruitment of CLIP-170 to kinetochores.
FIG. 4.
CLIP-170 localization at the kinetochore is dependent on its LIS1-binding site. COS-7 cells were transiently transfected with GFP-Cter, GFP-C2, or GFP-Z1 + 2 and blocked in prometaphase by nocodazole treatment. The fixation was preceded by extraction in Triton X-100 in order to render kinetochore-bound CLIP-170 detectable. The cells were double labeled with CREST autoimmune serum (a, e, and i) and anti-GFP (b, f, and j). Panels c, g, and k are superimpositions of CREST (red) and GFP (green) signals. Chromosomes were stained with DAPI (d, h, and l). The insets in panels c, g, and k show the relative locations of GFP chimera proteins (in green) and the CREST antigens (in red). All images are maximal-intensity projections of x/y optical section stacks acquired by three-dimensional deconvolution microscopy. Bar, 5 μm.
Recruitment of LIS1 to MT ends in interphase cells overexpressing CLIP-170.
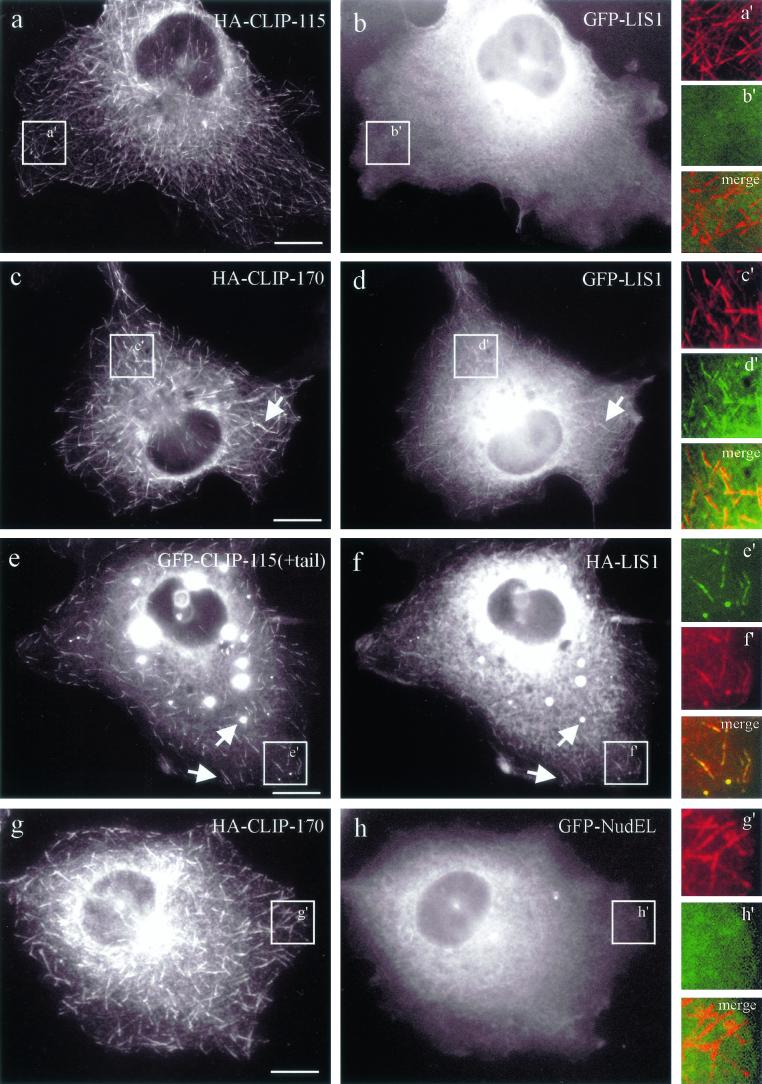
Overexpression of LIS1 has been shown to abolish dynactin accumulation at the MT plus ends (23). We wanted to test if overexpression of CLIP-170 or its closest mammalian homolog, CLIP-115, can influence the MT plus-end targeting of LIS1 and other proteins (NudE, NudEL, and NudC) which are implicated in the dynein/dynactin pathway as well (47, 63, 76). Overexpression of HA-CLIP-170 induced accumulation of coexpressed GFP-tagged LIS1 at the tips of MTs (Fig. 5c and d). This was not the case when HA-CLIP-115 was instead expressed at medium overexpression levels (Fig. 5a and b), suggesting that its MT binding and coiled-coil domains are not interacting with LIS1. Based on these observations, a chimeric protein consisting of GFP-CLIP-115 and the C terminus of CLIP-170 [GFP-CLIP-115(+tail)] was tested. At low levels of expression, GFP-CLIP-115(+tail) is present at MT plus ends, and at higher levels it accumulates in patchy (data not shown) or spherical (Fig. 5e) aggregates, which also recruit HA-tagged LIS1 (Fig. 5f). This demonstrates that the last 159 amino acids of CLIP-170, which include the two zinc finger motifs, are sufficient to relocalize LIS1. GFP fusion proteins of NudE, NudEL, and NudC were not recruited to MTs by any of the CLIPs (either in double transfections with HA-CLIP fusions or in triple transfections with an untagged CLIP and an HA-LIS1-expressing construct) (Fig. 5g and h and data not shown). In addition, none of the NudE/EL/C constructs had an apparent influence on the localization of dynactin to MT tips (data not shown). We conclude that the LIS1-associated proteins NudE, NudEL, and NudC are not involved in CLIP-170-mediated recruitment of LIS1 to MT plus ends.
FIG. 5.
CLIP-170, but not CLIP-115, recruits epitope-tagged LIS1 to MT distal ends. COS-1 cells were cotransfected with HA-CLIP-115 and GFP-LIS1 (a and b), HA-CLIP-170 and GFP-LIS1 (c and d), GFP-CLIP-115(+tail) and HA-LIS1 (e and f), and HA-CLIP-170 and GFP-NudEL (g and h). In the GFP-CLIP-115(+tail) construct, the zinc “knuckle”-containing domain of CLIP-170 was transferred to CLIP-115. LIS1, recruited to CLIP-positive structures, is indicated by arrows. Panels a′ to h′ represent colored enlargements of the areas indicated in panels a to h. The merged images of panels a′ to h′ demonstrate overlap (or the lack of it) at MT distal ends of the different tagged proteins. Bar, 10 μm.
A phospho-LIS1 isoform binds to MT bundles in interphase cells in a CLIP-170-dependent way.
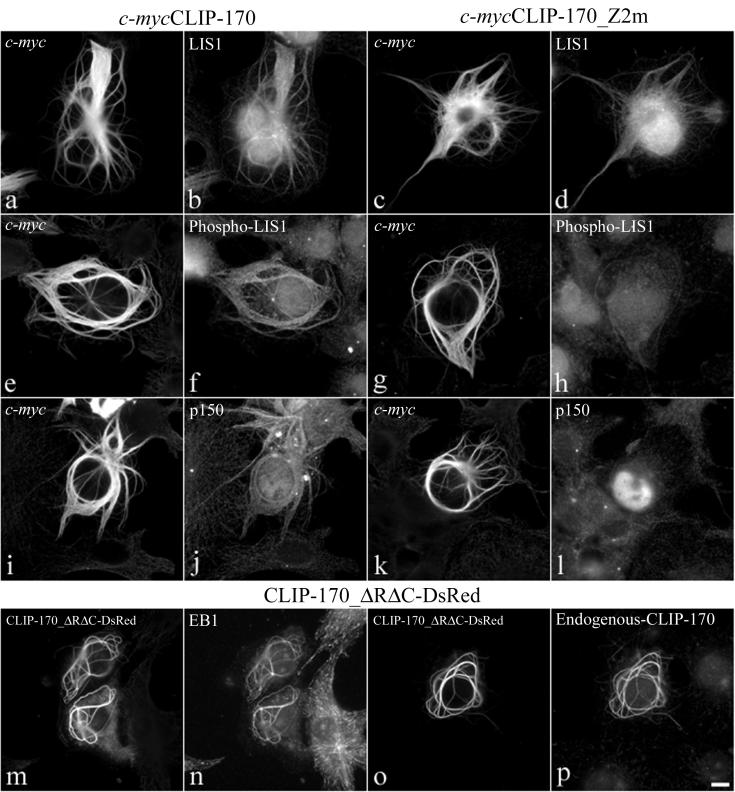
Cells transfected with CLIP-170 constructs containing the MT binding domains form dense MT bundles (50) (Fig. 6a, c, e, g, i, and k). These bundles are formed also when CLIP-170 lacks the coiled-coil dimerization domain (CLIP-170-ΔRΔC) (Fig. 6m and o). LIS1 is recruited to these bundles (Fig. 6b) also when the second zinc finger motif of CLIP-170 is mutated (Fig. 6d), or when the bundles are induced by monomeric CLIP-170-ΔRΔC (data not shown). However, the second zinc finger appears to be crucial in the recruitment of a phospho-LIS1 isoform that is specifically detected by the monoclonal anti-LIS1 antiserum, clone 210 (57). Mutations in the distal zinc finger motif of CLIP-170 reduced the recruitment of this phospho-LIS1 isoform to the bundles (Fig. 6f versus h). Interestingly, the dynactin subunit p150 also is not recruited to the MT bundles formed by CLIP-170-Z2m (Fig. 6l versus h) or by CLIP-170-ΔRΔC (data not shown). On the other hand, both EB1 and endogenous CLIP-170 are recruited to the MT bundles induced by all of the MT-binding CLIP-170 forms tested (shown here is CLIP-170-ΔRΔC-DsRed [Fig. 6o and p]). Taken together, these results suggest that LIS1 targeting to specific intracellular structures is regulated both by phosphorylation and by CLIP-170. These results are supported by our previous observations that another monoclonal antibody, clone 338, and clone 210 recognize distinct, but not all, phospho-epitopes of LIS1 (reference 57 and data not shown), suggesting that LIS1 exists in the cell as several pools that are differentially regulated.
FIG. 6.
Binding of a specific phospho-LIS1 isoform to CLIP-170-induced MT bundles is dependent on the second zinc finger domain of CLIP-170. COS-7 cells were transiently transfected with c-mycCLIP-170 (a, b, e, f, i, and j), with c-mycCLIP-170-Z2m (c, d, g, h, k, and l), or with CLIP-ΔRΔC-DsRed (m, n, o, and p). The fixation was preceded by a brief extraction in Triton X-100. (a to d) The cells were double labeled with a polyclonal anti-c-myc antibody (a and c) and a polyclonal anti-LIS1 (b and d). (e to l) The cells are double labeled with the monoclonal anti-c-myc 9E10 antibody (e, g, i, and k) and the monoclonal anti-phospho-LIS1 210 antibody (f and h) or a monoclonal anti-p150 antibody (j and l). (m to o) The cells were labeled with a monoclonal anti-EB1 antibody (n) or the polyclonal antibody TA against the C-terminal portion of CLIP-170 (p). All of the CLIP-170 constructs give rise to the formation of thick MT bundles. LIS1 is always recruited to the bundles (b and d). A mutation in the second zinc finger domain (Z2m) diminishes the recruitment of a phospho-LIS1 isoform and p150 to the bundles (f and j versus h and l). Bar, 10 μm.
DISCUSSION
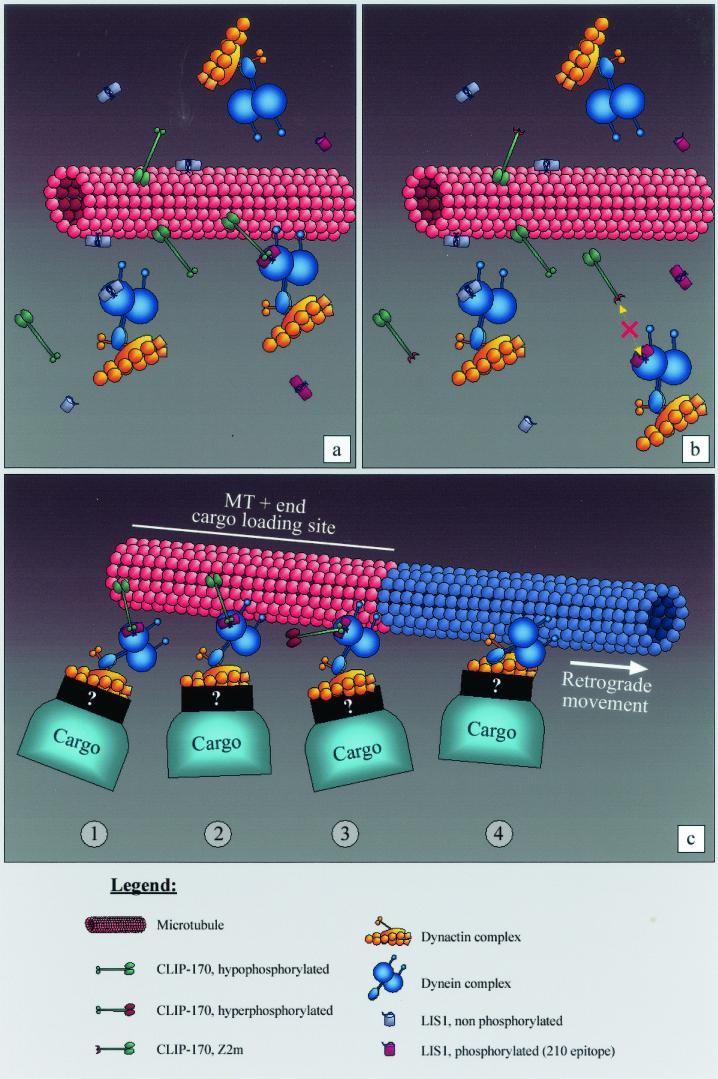
Orchestrating cellular functions requires modular and dynamic interactions between proteins. Our data indicate a direct physical interaction between LIS1 and CLIP-170 in vitro; in vivo the two proteins are found in the same complex in embryonic brain and coimmunoprecipitate. GST pull-down assays and yeast two-hybrid experiments demonstrated that the CLIP-170 second zinc finger is necessary for this interaction. Overexpressed GFP-LIS1 is recruited by epitope-tagged CLIP-170 to MT tips during interphase. The experiments indicated clearly that the MT binding domains and the coiled-coil region are not mediating the interaction between CLIP-170 and LIS1, since CLIP-115 does not relocalize LIS1 to MT tips in the absence of the CLIP-170 zinc finger motifs. Yeast two-hybrid results and kinetochore targeting indicated that point mutations in the first zinc finger domain reduce the strength of interaction, suggesting a functional role for this domain as well. The regulatory role of the first zinc finger domain most likely developed during evolution, since budding and fission yeasts contain only one zinc finger domain (4, 8, 51). Within LIS1, the WD 5, -6, and -7 domains contribute to the interaction; however, it is clear that the protein as a whole interacts with CLIP-170 much better than its subdomains. Furthermore, our results suggest that specific LIS1 phosphorylation might be an in vivo mode of regulating its interaction with CLIP-170. Our previous results indeed indicated that LIS1 exists in the cell as several pools that are differentially regulated on several phosphorylation sites, some of which are recognized by different monoclonal antibodies (57). Endogenous LIS1 molecules bind to MTs along their length in a CLIP-170-independent manner, and therefore, the same can be assumed to hold for the LIS1 pool recognized on these bundles by our affinity-purified polyclonal antibodies. The phospho-LIS1 isoform recognized by clone 210 (57) is strongly recruited to kinetochores (not shown), and interestingly, its recruitment to MT bundles induced by overexpressed MT-binding forms of CLIP-170 is dependent on CLIP-170's distal zinc finger. Our proposed model in Fig. 7 illustrates our assumptions.
FIG. 7.
A model of LIS1 mediating CLIP-170 interactions with the dynein/dynactin pathway. (a) LIS1 can bind to MT bundles; however, phospho-LIS1 (recognized by the 210 antibody) binding to MT bundles is mediated through the distal zinc finger motif of CLIP-170. (b) When the distal zinc finger motif of CLIP-170 is mutated, the phospho-LIS1 isoform can no longer bind to MT bundles. This can also explain why p150/dynactin is not recruited to the MT bundles. (c) Possible kinetics of the interactions. CLIP-170 targets the dynein/dynactin complex to the MT, using phopho-LIS1 as an adapter (step 1); the motor complex, being in the proximity of the MT, associates with it (step 2); the initial static link to the MT provided by CLIP-170 is released by its hyperphosphorylation (step 3); and the retrograde dynein/dynactin-dependent movement of the cargo (endocytic vesicle, kinetochore, etc.) takes place (step 4). Tip1p, the CLIP-170 ortholog in S. pombe, has been reported by Brunner and Nurse (8) to act as an anticatastrophe factor. Based on the model depicted here, it also could be that CLIP-170 plays a similar role in eukaryotic cells by targeting MT-stabilizing factors as cargo to the plus tips. The model is based on that of Rickard and Kreis (54; reviewed in references 1a, 54a, and 75).
One important cellular colocalization of LIS1 and CLIP-170 is at the kinetochores, probably the best-characterized structure tethering MTs. The dynein/dynactin motor complex is present at unattached kinetochores (75), and proteins like CLIP-170 and LIS1 are thought to contribute to its functioning, in yet-unresolved ways (18, 23). The recruitment of dynein/dynactin/LIS1/CLIP-170 to the kinetochores is not dependent upon MTs, as following nocodazole treatment these proteins still accumulate at these sites 18, 30, 72; this study). In this work we demonstrate a hierarchy of interactions at the kinetochores. The initial targeting of LIS1 to kinetochores is dynein/dynactin dependent, since overexpression of p50 results in reduction of LIS1 at these sites. Recruitment of CLIP-170 to kinetochores appears to be LIS1 dependent, since deletions or mutations in its LIS1 binding domain abolish CLIP-170 kinetochore targeting. The CLIP-170 MT binding domains play a role in chromosome congression to the equatorial plate (18). We propose that the recruitment of CLIP-170 to kinetochores via LIS1/dynein/dynactin is an important step in this process and facilitates the initial coupling of kinetochores to MTs prior to their first poleward movement.
We show here that CLIP-170 and LIS1 also colocalize at cortical sites which capture astral MTs. This further illustrates the analogy between kinetochores and the cortical sites (34). It may be hypothesized that LIS1 is involved there in MT capturing since its over- or underexpression perturbs spindle positioning in both D. melanogaster oogenesis and early embryos (40) and in mammalian epithelial cells (23). The regulated physical link with CLIP-170 that we report here may be involved in the molecular mechanism for tethering MTs to positional cortical cues.
Another new finding is that in interphase mammalian cells, LIS1 colocalizes with CLIP-170 at the tips of polymerizing MTs. Dynein/dynactin, which is known to regulate MT dynamics, also associates with MT tips (77). This motor complex could function via a highly regulated linker function of LIS1 connecting it to CLIP-170, but our data and those of others (74) do not yet provide direct evidence that this is indeed the case.
Overall, LIS1 phosphorylation may affect its binding to MTs, as previously suggested (57). It is likely that several phosphorylation sites regulate LIS1 activity, its cellular localization, and its binding to other interacting proteins. The monoclonal antibody 210 has provided us with unique opportunities to investigate this question. A hierarchy of interactions was exemplified by examining the MT bundles induced by CLIP-170 mutated in the distal zinc finger domain or lacking the coiled-coil and C-terminal domains, where no specific phospho-LIS1 isoform accumulated. On these bundles we detected EB1 and endogenous CLIP-170; however, p150 was not recruited there. This result is interesting in view of the finding that EB1 can associate with p150, the intermediate chain of cytoplasmic dynein, and with dynamitin (p50) (5). None of the interactions demonstrated were necessarily direct; nevertheless, within the context of these MT bundles, our results suggest that EB1 and LIS1 interact directly with MTs. The specific phospho-LIS1 isoform, however, is recruited to the bundles only via CLIP-170, and we suggest that p150 is recruited to the bundles via this phospho-LIS1 form, probably mediated by dynein. This somewhat resembles the dynactin interaction with the kinesin-related motor HsEg5, which is regulated by phosphorylation (6). In addition, very similar to the case for LIS1 and CLIP-170, the interaction of the cytoplasmic dynein intermediate chain with dynactin is regulated by serine 84 phosphorylation (78). The dynein intermediate chain is also a WD repeat protein, similar to LIS1, and contains a phosphorylation site within its N-terminal region following a coiled-coil domain (78). It is amazing to reveal such a similar pattern of regulation of interaction by phosphorylation of residues within the same protein module. This work suggests that LIS1 is a phosphorylation-dependent linker between CLIP-170 and cytoplasmic dynein at sites involved in positional cue-MT tethering and/or in control of MT dynamics.
Acknowledgments
The first two authors contributed equally to this work.
We thank Anne Moreau for technical assistance. Jean-Baptiste Sibarita is acknowledged for his expertise with image deconvolution.
This work was supported in part by the Fritz Thyssen Stiftung Foundation, HFSP grant no. RG283199 9, March of Dimes grant no. 6-FY01-5, and the Minerva Foundation (to O.R.) and by the Association pour la Recherche contre le Cancer (Villejuif, France) (to J.R.D.M.). O.R. is an Incumbent of the Aser Rothstein Career Development Chair in Genetic Diseases.
REFERENCES
- 1.Akhmanova, A., C. C. Hoogenraad, K. Drabek, T. Stepanova, B. Dortland, T. Verkerk, W. Vermeulen, B. M. Burgering, C. I. De Zeeuw, F. Grosveld, and N. Galjart. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104:923-935. [DOI] [PubMed] [Google Scholar]
- 1a.Allan, V. 1996. Motor proteins: a dynamic duo. Curr. Biol. 6:630-633. [DOI] [PubMed] [Google Scholar]
- 2.Banks, J. D., and R. Heald. 2001. Chromosome movement: dynein-out at the kinetochore. Curr. Biol. 11:R128-R31. [DOI] [PubMed] [Google Scholar]
- 3.Beckwith, S. M., C. H. Roghi, B. Liu, and N. Ronald Morris. 1998. The “8-kD” cytoplasmic dynein light chain is required for nuclear migration and for dynein heavy chain localization in Aspergillus nidulans. J. Cell Biol. 143:1239-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlin, V., C. A. Styles, and G. R. Fink. 1990. BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiae, colocalizes with tubulin. J. Cell Biol. 111:2573-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berrueta, L., J. S. Tirnauer, S. C. Schuyler, D. Pellman, and B. E. Bierer. 1999. The APC-associated protein EB1 associates with components of the dynactin complex and cytoplasmic dynein intermediate chain. Curr. Biol. 9:425-428. [DOI] [PubMed] [Google Scholar]
- 6.Blangy, A., L. Arnaud, and E. A. Nigg. 1997. Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J. Biol. Chem. 272:19418-19424. [DOI] [PubMed] [Google Scholar]
- 7.Bloom, K. 2001. Nuclear migration: cortical anchors for cytoplasmic dynein. Curr. Biol. 11:326-329. [DOI] [PubMed] [Google Scholar]
- 8.Brunner, D., and P. Nurse. 2000. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102:695-704. [DOI] [PubMed] [Google Scholar]
- 9.Burkhardt, J. K., C. J. Echeverri, T. Nilsson, and R. B. Vallee. 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 139:469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busson, S., D. Dujardin, A. Moreau, J. Dompierre, and J. R. De Mey. 1998. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr. Biol. 8:541-544. [DOI] [PubMed] [Google Scholar]
- 11.Cahana, A., T. Escamez, R. S. Nowakowski, N. L. Hayes, M. Giacobini, A. von Holst, O. Shmueli, T. Sapir, S. K. McConnell, W. Wurst, S. Martinez, and O. Reiner. 2001. Targeted mutagenesis of Lis1 disrupts cortical development and LIS1 homodimerization. Proc. Natl. Acad. Sci. USA 98:6429-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carminati, J. L., and T. Stearns. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caspi, M., R. Atlas, A. Kantor, T. Sapir, and O. Reiner. 2000. Interaction between LIS1 and doublecortin, two lissencephaly gene products. Hum. Mol. Genet. 9:2205-2213. [DOI] [PubMed] [Google Scholar]
- 14.Chenn, A., J. E. Braisted, S. K. McConnell, and D. D. M. O'Leary. 1997. Development of the cerebral cortex: mechanisms controlling cell fate, laminar and areal patterning, and axonal connectivity, p. 440-473. In W. M. Cowan, T. M. Jessell, and S. L. Zipursky (ed.), Molecular and cellular approaches to neural development. Oxford University Press, New York, N.Y.
- 15.Clark, I. B., and D. I. Meyer. 1999. Overexpression of normal and mutant Arp1alpha (centractin) differentially affects microtubule organization during mitosis and interphase. J. Cell Sci 112:3507-3518. [DOI] [PubMed] [Google Scholar]
- 16.De Zeeuw, C. I., C. C. Hoogenraad, E. Goedknegt, E. Hertzberg, A. Neubauer, F. Grosveld, and N. Galjart. 1997. CLIP-115, a novel brain-specific cytoplasmic linker protein, mediates the localization of dendritic lamellar bodies. Neuron 19:1187-1199. [DOI] [PubMed] [Google Scholar]
- 17.Diamantopoulos, G. S., F. Perez, H. V. Goodson, G. Batelier, R. Melki, T. E. Kreis, and J. E. Rickard. 1999. Dynamic localization of CLIP-170 to microtubule plus ends is coupled to microtubule assembly. J. Cell Biol. 144:99-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dujardin, D., U. I. Wacker, A. Moreau, T. A. Schroer, J. E. Rickard, and J. R. De Mey. 1998. Evidence for a role of CLIP-170 in the establishment of metaphase chromosome alignment. J. Cell Biol. 141:849-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earnshaw, W. C., H. Ratrie III, and G. Stetten. 1989. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma 98:1-12. [DOI] [PubMed] [Google Scholar]
- 20.Earnshaw, W. C., and N. Rothfield. 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91:313-321. [DOI] [PubMed] [Google Scholar]
- 21.Echeverri, C. J., B. M. Paschal, K. T. Vaughan, and R. B. Vallee. 1996. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 132:617-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faulkner, N. E., D. L. Dujardin, C. Y. Tai, K. T. Vaughan, C. B. O'Connell, Y. Wang, and R. B. Vallee. 2000. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2:784-791. [DOI] [PubMed] [Google Scholar]
- 24.Fujiwara, T., K. Tanaka, E. Inoue, M. Kikyo, and Y. Takai. 1999. Bni1p regulates microtubule-dependent nuclear migration through the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8016-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geiser, J. R., E. J. Schott, T. J. Kingsbury, N. B. Cole, L. J. Totis, G. Bhattacharyya, L. He, and M. A. Hoyt. 1997. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol. Biol. Cell 8:1035-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goode, B. L., D. G. Drubin, and G. Barnes. 2000. Functional cooperation between the microtubule and actin cytoskeletons. Curr. Opin. Cell Biol. 12:63-71. [DOI] [PubMed] [Google Scholar]
- 27.Griparic, L., J. M. Volosky, and T. C. Keller III. 1998. Cloning and expression of chicken CLIP-170 and restin isoforms. Gene 206:195-208. [DOI] [PubMed] [Google Scholar]
- 28.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 29.Han, G., B. Liu, J. Zhang, W. Zuo, N. R. Morris, and X. Xiang. 2001. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biol. 11:1-20. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman, D. B., C. G. Pearson, T. J. Yen, B. J. Howell, and E. D. Salmon. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at ptk1 kinetochores. Mol. Biol. Cell 12:1995-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann, B., W. Zuo, A. Liu, and N. R. Morris. 2001. The LIS1 related protein NUDF of Aspergillus nidulans and its interaction partner NUDE bind directly to specific subunits of dynein and dynactin and to α- and γ-tubulin. J. Biol. Chem. 16:16. [DOI] [PubMed] [Google Scholar]
- 32.Hoogenraad, C. C., A. Akhmanova, F. Grosveld, C. I. De Zeeuw, and N. Galjart. 2000. Functional analysis of CLIP-115 and its binding to microtubules. J. Cell Sci. 113:2285-2297. [DOI] [PubMed] [Google Scholar]
- 33.Hyman, A. 1989. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J. Cell Biol. 109:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirschner, M., and T. Mitchison. 1986. Beyond self-assembly: from microtubules to morphogenesis. Cell 45:329-342. [DOI] [PubMed] [Google Scholar]
- 35.Klempnauer, K. H., H. Arnold, and H. Biedenkapp. 1989. Activation of transcription by v-myb: evidence for two different mechanisms. Genes Dev. 3:1582-1589. [DOI] [PubMed] [Google Scholar]
- 36.Langanger, G., J. De Mey, and H. Adam. 1983. 1,4-Diazobicyclo-(2,2,2)-octane (DABCO) retards the fading of immunofluorescence preparations. Mikroskopie 40:237-241. [PubMed] [Google Scholar]
- 37.Lee, L., J. S. Tirnauer, J. Li, S. C. Schuyler, J. Y. Liu, and D. Pellman. 2000. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science 287:2260-2262. [DOI] [PubMed] [Google Scholar]
- 38.Lin, H., P. de Carvalho, D. Kho, C.-Y. Tai, P. Pierre, G. R. Fink and D. Pellman. 2001. Polyploids require Bik1 for kinetochore-microtubule attachment. J. Cell Biol. 155:1173-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, Z., R. Steward, and L. Luo. 2000. Drosophila Lis1 is required for neuroblast proliferation, dendritic elaboration and axonal transport. Nat. Cell Biol. 2:776-783. [DOI] [PubMed] [Google Scholar]
- 40.Liu, Z., T. Xie, and R. Steward. 1999. Lis1, the Drosophila homolog of a human lissencephaly disease gene, is required for germline cell division and oocyte differentiation. Development 126:4477-4488. [DOI] [PubMed] [Google Scholar]
- 41.Maney, T., L. M. Ginkel, A. W. Hunter, and L. Wordeman. 2000. The kinetochore of higher eucaryotes: a molecular view. Int. Rev. Cytol. 194:67-131. [DOI] [PubMed] [Google Scholar]
- 42.McGrail, M., and T. S. Hays. 1997. The microtubule motor cytoplasmic dynein is required for spindle orientation during germline cell divisions and oocyte differentiation in Drosophila. Development 124:2409-2419. [DOI] [PubMed] [Google Scholar]
- 43.Merdes, A., and J. De Mey. 1990. The mechanism of kinetochore-spindle attachment and polewards movement analyzed in PtK2 cells at the prophase-prometaphase transition. Eur. J. Cell Biol. 53:313-325. [PubMed] [Google Scholar]
- 44.Miller, R. K., K. K. Heller, L. Frisen, D. L. Wallack, D. Loayza, A. E. Gammie, and M. D. Rose. 1998. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell 9:2051-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mimori-Kiyosue, Y., N. Shiina, and S. Tsukita. 2000. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10:865-868. [DOI] [PubMed] [Google Scholar]
- 46.Morris, N. R., V. P. Efimov, and X. Xiang. 1998. Nuclear migration, nucleokinesis and lissencephaly. Trends Cell Biol. 8:467-470. [DOI] [PubMed] [Google Scholar]
- 47.Morris, S. M., U. Albrecht, O. Reiner, G. Eichele, and L.-Y. Yu-Lee. 1998. The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein, NudC. Curr. Biol. 8:603-606. [DOI] [PubMed] [Google Scholar]
- 48.Ohkura, H., I. M. Hagan, and D. M. Glover. 1995. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 9:1059-1073. [DOI] [PubMed] [Google Scholar]
- 49.Perez, F., G. S. Diamantopoulos, R. Stalder, and T. E. Kreis. 1999. CLIP-170 highlights growing microtubule ends in vivo. Cell 96:517-527. [DOI] [PubMed] [Google Scholar]
- 50.Pierre, P., R. Pepperkok, and T. E. Kreis. 1994. Molecular characterization of two functional domains of CLIP-170 in vivo. J. Cell Sci. 107:1909-1920. [DOI] [PubMed] [Google Scholar]
- 51.Pierre, P., J. Scheel, J. E. Rickard, and T. E. Kreis. 1992. CLIP-170 links endocytic vesicles to microtubules. Cell 70:887-900. [DOI] [PubMed] [Google Scholar]
- 52.Reiner, O. 2000. LIS1: let's interact sometimes…(part 1). Neuron 28:633-636. [DOI] [PubMed] [Google Scholar]
- 53.Reiner, O., R. Carrozzo, Y. Shen, M. Whenert, F. Faustinella, W. B. Dobyns, C. T. Caskey, and D. H. Ledbetter. 1993. Isolation of a Miller-Dieker lissencephaly gene containing G protein β-subunit-like repeats. Nature 364:717-721. [DOI] [PubMed] [Google Scholar]
- 54.Rickard, J. E., and T. E. Kreis. 1991. Binding of pp170 to microtubules is regulated by phosphorylation. J. Biol. Chem. 266:17597-17605. [PubMed] [Google Scholar]
- 54a.Rickard, J. E., and T. E. Kreis. 1996. CLIPs for organelle-microtubule interactions. Trends Cell Biol. 6:178-183. [DOI] [PubMed] [Google Scholar]
- 55.Rieder, C. L., and S. P. Alexander. 1990. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J. Cell Biol. 110:81-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rieder, C. L., and E. D. Salmon. 1998. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8:310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sapir, T., A. Cahana, R. Seger, S. Nekhai, and O. Reiner. 1999. LIS1 is a microtubule-associated phosphoprotein. Eur. J. Biochem. 265:181-188. [DOI] [PubMed] [Google Scholar]
- 58.Sapir, T., M. Elbaum, and O. Reiner. 1997. Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 16:6977-6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sapir, T., D. Horesh, M. Caspi, R. Atlas, H. A. Burgess, S. Grayer Wolf, F. Francis, J. Chelly, M. Elbaum, S. Pietrokovski, and O. Reiner. 2000. Doublecortin mutations cluster in evolutionary conserved functional domains. Hum. Mol. Genet. 5:703-712. [DOI] [PubMed] [Google Scholar]
- 60.Sasaki, S., A. Shionoya, M. Ishida, M. Gambello, J. Yingling, A. Wynshaw-Boris, and S. Hirotsune. 2000. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron 28:681-696. [DOI] [PubMed] [Google Scholar]
- 61.Savino, T. M., J. Gebrane-Younes, J. De Mey, J. B. Sibarita, and D. Hernandez-Verdun. 2001. Nucleolar assembly of the rRNA processing machinery in living cells. J. Cell Biol. 153:1097-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheel, J., P. Pierre, J. E. Rickard, G. S. Diamantopoulos, C. Valetti, F. G. van der Goot, M. Haner, U. Aebi, and T. E. Kreis. 1999. Purification and analysis of authentic CLIP-170 and recombinant fragments. J. Biol. Chem. 274:25883-25891. [DOI] [PubMed] [Google Scholar]
- 63.Schroer, T. A. 2001. Microtubules don and doff their caps: dynamic attachments at plus and minus ends. Curr. Opin. Cell Biol. 13:92-96. [DOI] [PubMed] [Google Scholar]
- 64.Schuyler, S. C., and D. Pellman. 2001. Microtubule “plus-end-tracking proteins.” The end is just the beginning. Cell 105:421-424. [DOI] [PubMed] [Google Scholar]
- 65.Schuyler, S. C., and D. Pellman. 2001. Search, capture and signal: games microtubules and centrosomes play. J. Cell Sci. 114:247-255. [DOI] [PubMed] [Google Scholar]
- 66.Shah, J. V., and D. W. Cleveland. 2000. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell 103:997-1000. [DOI] [PubMed] [Google Scholar]
- 67.Sheffield, P., S. Garrard, M. Caspi, J. Aoki, K. Inoue, U. Derewenda, B. Suter, O. Reiner, and Z. S. Derewenda. 2000. Homologues of the α- and β-subunits of mammalian brain platelet-activating factor acetylhydrolase Ib in the Drosophila melanogaster genome. Proteins 39:1-8. [DOI] [PubMed] [Google Scholar]
- 68.Skibbens, R. V., and P. Hieter. 1998. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu. Rev. Genet. 32:307-337. [DOI] [PubMed] [Google Scholar]
- 69.Skop, A. R., and J. G. White. 1998. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr. Biol. 8:1110-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith, D. S., M. Niethammer, R. Ayala, Y. Zhou, M. J. Gambello, A. Wynshaw-Boris, and L. H. Tsai. 2000. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol. 2:767-775. [DOI] [PubMed] [Google Scholar]
- 71.Theurkauf, W. E. 1997. Oocyte differentiation: a motor makes a difference. Curr. Biol. 7:548-551. [DOI] [PubMed] [Google Scholar]
- 72.Thrower, D. A., M. A. Jordan, and L. Wilson. 1996. Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell. Motil. Cytoskeleton 35:121-133. [DOI] [PubMed] [Google Scholar]
- 73.Tirnauer, J. S., and B. E. Bierer. 2000. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J. Cell Biol. 149:761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valetti, C., D. M. Wetzel, M. Schrader, M. J. Hasbani, S. R. Gill, T. E. Kreis, and T. A. Schroer. 1999. Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol. Biol. Cell 10:4107-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vallee, R. B., and M. P. Sheetz. 1996. Targeting of motor proteins. Science 271:1539-1544. [DOI] [PubMed] [Google Scholar]
- 76.Vallee, R. B., C. Tai, and N. E. Faulkner. 2001. LIS1: cellular function of a disease-causing gene. Trends Cell Biol. 11:155-160. [DOI] [PubMed] [Google Scholar]
- 77.Vaughan, K. T., S. H. Tynan, N. E. Faulkner, C. J. Echeverri, and R. B. Vallee. 1999. Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J. Cell Sci. 112:1437-1447. [DOI] [PubMed] [Google Scholar]
- 78.Vaughan, P. S., J. D. Leszyk, and K. T. Vaughan. 2001. Cytoplasmic dynein intermediate chain phosphorylation regulates binding to dynactin. J. Biol. Chem. 276:26171-26179. [DOI] [PubMed] [Google Scholar]
- 79.Willins, D. A., B. Liu, X. Xiang, and N. R. Morris. 1997. Mutations in the heavy chain of cytoplasmic dynein suppress the nudF nuclear migration mutation of Aspergillus nidulans. Mol. Gen. Genet. 255:194-200. [DOI] [PubMed] [Google Scholar]
- 80.Xiang, X., S. M. Beckwith, and N. R. Morris. 1994. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 91:2100-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiang, X., G. Han, D. A. Winkelmann, W. Zuo, and N. R. Morris. 2000. Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein Arp1. Curr. Biol. 10:603-606. [DOI] [PubMed] [Google Scholar]
- 82.Xiang, X., A. H. Osmani, S. A. Osmani, M. Xin, and N. R. Morris. 1995. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol. Biol. Cell 6:297-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yeaman, C., K. K. Grindstaff, and W. J. Nelson. 1999. New perspectives on mechanisms involved in generating epithelial cell polarity. Physiol. Rev. 79:73-98. [DOI] [PubMed] [Google Scholar]