Abstract
Trypanosome RNA editing, the posttranscriptional insertion and deletion of U residues in mitochondrial transcripts, is catalyzed by a protein complex containing seven distinct proteins. In this study, we cloned the gene for band III, a 555-amino-acid protein with two separate zinc finger motifs. We prepared antibodies that showed band III protein cofractionates with the previously characterized band IV protein throughout the purification of the editing complex and is not found free or in other protein associations; therefore, it is a true constituent of the editing complex. Double-stranded RNA interference efficiently depleted band III protein and demonstrated that band III expression is essential for growth of procyclic trypanosomes and for RNA editing. These depleted cell extracts were deficient specifically in guide RNA-directed endonuclease cleavage at both U deletion and U insertion sites and in the activity of the band IV ligase, but they retained the 3′-U-exonuclease and terminal-U-transferase activities as well as band V ligase of the editing complex. Loss of band III protein also resulted in almost complete loss of the band IV ligase protein and altered sedimentation of the band V ligase. These data indicate that band III is either the RNA editing endonuclease or a factor critical for cleavage activity in the editing complex. They also demonstrate that band III is required for proper assembly of the editing complex.
Trypanosomes are early diverging protozoa in which many mitochondrial transcripts undergo posttranscriptional insertion and deletion of U residues at multiple, specific sites (reviewed in references 1, 2, 13, and 42). This RNA editing is directed by short, trans-acting guide RNAs (gRNAs) (4), and both U insertion and U deletion reactions have been reproduced in vitro (19, 38). Once thought to involve coupled transesterification reactions (5, 6), the mechanism of editing has been found to occur through a series of protein-catalyzed reactions (7, 19, 33, 35, 39; also see reference 4). First, the mRNA is cleaved by a gRNA-directed endonuclease, next U residues are added to or removed from the 3′ end of the upstream cleavage product by a terminal-U-transferase (TUTase) or a 3′-U-exonuclease (3′-U-exo), and then the mRNA is rejoined by RNA ligase. Although U insertion and U deletion share a parallel three-step enzymatic mechanism, evidence is accumulating that the two forms of editing utilize distinct activities for all three reaction steps (7, 8, 16, 33; J. Cruz-Reyes et al., submitted for publication).
The editing reactions are catalyzed by a protein complex, or editosome. The simplest complex purified contains seven different potentially equimolar proteins, identified as band I through band VII, based on their relative migration on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (33). This complex actively catalyzes U deletion and U insertion, and it exhibits all of the expected enzymatic activities: gRNA-directed endonuclease, 3′-U-exo, TUTase, and RNA ligase activities (9, 33). The components are stably associated, as they remain together on examined chromatographic matrices, velocity sedimentation, and native gel electrophoresis (33; L. Rusché and B. Sollner-Webb, unpublished observations). Two of the seven proteins of the editing complex are RNA ligases (band IV and band V) (33), but specific functions have not yet been ascribed to any of the other five components.
Even before its purification, the existence of such an editosome had been hypothesized (see references 12 and 31), because the sequential enzymatic reactions would be more efficient if sequestered in a common complex, possibly acting in concert. More recently, it has been suggested that the localization of editing activities within a common complex could increase the specificity and accuracy of the editing process in ways that would not be as feasible if each activity was present as a free protein. For instance, Igo et al. (17) suggest that concerted steps contribute to the specificity of U insertion, with TUTase preferentially adding the number of U's specified by the gRNA, 3′-U-exo trimming any excess added U's not protected by base pairing, and ligase preferentially joining fragments that perfectly pair with the gRNA and therefore have the specified number of added U residues. Huang et al. (16) further show that an additional function of the band IV ligase is to repair RNAs that evidently had been incorrectly cleaved during editing. Such coordinated reactions virtually depend on the editing activities existing in a common complex.
Additional proteins might also interact with the minimal editing complex (for example, see reference 29). Helicases, possibly including the DEAD box containing mHel61p (26); gRNA binding proteins, such as gBP21 (3, 20, 27); and pre-mRNA binding proteins, such as REAP-1 (23, 24), may transiently associate. These have been proposed to stimulate editing by facilitating annealing of the gRNA with the mRNA and by helping the minimal complex associate with its RNA substrate.
We have focused on the proteins comprising the purified seven-polypeptide editing complex, because these alone appear able to catalyze full round U deletion and full round U insertion in vitro; in fact, they support editing more actively than other reported preparations (9, 10, 33). The genes for the band IV and band V RNA ligases are related (33, 34) and have also been isolated by other laboratories, where they are called TbMP52 and TbMP48 (37) or p52 and p48 (25), referring to the size of the cytoplasmic preprotein in Trypanosoma brucei. We have found that band V is specialized for joining in U insertion, while band IV is required for joining in U deletion, yet band IV can also partly substitute for band V in U insertion and serve in RNA repair (16; Cruz-Reyes et al., submitted for publication). Genetic knockout studies have further revealed that band IV's protein-protein interactions are important for maintaining the integrity of the editing complex, since in the absence of this ligase, the complex appears more labile, with the band V ligase and the remaining associated components then sedimenting at ∼7S (16). Conversely, when overproduced, the excess band IV protein beyond that incorporated into the editing complex is rapidly degraded (34).
Except for the ligases, none of the remaining components of the purified seven-polypeptide editing complex have been characterized. We now report the cDNA cloning and initial analysis of band III. This protein derives from a 59-kDa primary translation product, contains two zinc finger motifs, and is evidently the same protein as that just reported by Panigrahi et al. (30). However, through RNA interference (RNAi) studies, we now show that band III expression is essential in procyclic T. brucei. It is required for normal editing, specifically for gRNA-directed cleavage activity and band IV ligase activity. Furthermore, band III protein is needed to maintain the editosome, because in its absence, band IV protein is lost, while the sedimentation of remaining editing components, including band V protein, is disturbed. This indicates that one function of band III may be to serve as a scaffold that allows association of other proteins of the editing complex.
MATERIALS AND METHODS
Oligonucleotides, PCR, and cloning.
Tryptic peptide sequences from band III were generated by the Wistar protein analysis facility (Philadelphia, Pa.) by using this protein from editing complex purified as described previously (33, 41; see reference 34 for details). The degenerate primers designed from this sequence and used for initial cloning of a band III fragment from genomic DNA were III.2C-upstream (5′-CCNATGTTYGGNCARACNAG-3′) and III.3C-downstream (5′-ACYTCNCCRAARCARCA-3′, where R is A or G, Y is C or T, and N is any nucleotide). These primers (256- or 64-fold degeneracy) or primers with reverse 5′-3′ directionality were used in PCRs with 2.5 mM MgCl2, Taq DNA polymerase (Perkin-Elmer) and ∼2 ng of genomic DNA from trypanosome strain TREU 667 basically as described previously (34). Reactions used the manufacturer's buffer and stepped-down annealing temperatures (from 58°C to 48°C) for 40 reaction cycles. PCR products produced from genomic DNA (or reverse transcription-PCR [RT-PCR] products from mRNA [described below]) were cloned into pCR2.1 (Invitrogen) for sequencing. To obtain a nearly full-length clone, a cDNA library from procyclic T. brucei brucei strain TREU 667 in λZAPII (Stratagene; generated by Kenneth Piller) was probed with the initial PCR product, as described previously (34). The three longest clones were each ∼1,600 bp, and one was double strand sequenced. RT-PCR to obtain the 5′-most portion of the cDNA utilized a primer to the miniexon sequence SL-1 (5′-CTCTATTATTAGAACAGTTTCTGTACTATATTG-3′) and a primer to the upstream portion of the ∼1,600-bp clone, band III-reverse (5′-GACTTATCGGTGAGGGCACCGGTGTG-3′) plus rTth polymerase (Perkin-Elmer), as described by Rusché et al. (34). Once complete, the sequence was used to screen the GenBank, Institute for Genomic Research, and Pfam databases.
To express C-terminal six-histidine-tagged protein in Escherichia coli for antibody productions (described below), the entire band III open reading frame (ORF) was amplified with 5′-GGGAATTCCATATGTGCAGGAATGCTAGCAGC-3′ and 5′-GCATAAGCTTGATCACAGCTAGAGTCCCTG TG-3′ and genomic DNA template. The product was cloned into the NdeI and HindIII sites of pET29 (Invitrogen). A similar cloning of the band IV ORF used 5′-GGGAATTCCATATGCAACTCCAAAGGTTGGG-3′ and 5′-GTATAAGCTTTTCGCCCTTTGTGGGGGCAGC-3′.
For double-stranded RNA (dsRNA) production, a 754-bp fragment of band III was amplified with 5′-GATCTCGAGAAATGGCGGCTAAGTTGCACC-3′ and 5′-TCGAAGCTTAGCTGCTGCTGCAAATGAGGC-3′ primers and directionally cloned into the XhoI and HindIII sites of pZJM vector (43). This vector has two opposing tetracycline-responsive T7 promoters, which drive expression of both strands of interest, producing dsRNA. It also has a constitutive T7 promoter driving the phleomycin resistance gene (BLE).
Trypanosome propagation, transfection, and induction.
Our initial studies of the minimal editing complex used T. brucei brucei strain TREU 667 (33). These cells, the extracts of which were used for the experiments shown in Fig. 2, have two allelic isoforms of band IV, so they run as a doublet (34). The RNAi studies presented here used T. brucei brucei strain 427, where band IV appears as a single band (34). Cell line 29-13, a 427 derivative that expresses T7 RNA polymerase and tetracycline repressor protein (45), was grown with 15 μg of G418 per ml (Gibco BRL) and 50 μg of hygromycin per ml (Sigma) to ensure retention of those transgenes. This cell line was transfected with the linearized pZJM construct bearing the inserted band III PCR fragment as described by Wirtz et al. (44), except that following addition of phleomycin (2 μg/ml; Sigma), the culture was not plated, but was allowed to grow until nontransformed cells had died and phleomycin-resistant cells were doubling normally (approximately 2 weeks). Transformants were then cloned by extreme dilution (M. Klingbeil, personal communication), by diluting approximately 10 live cells into 20 ml of fresh culture medium, aliquotting this mixture into a 96-well dish, and incubating it in a 5% CO2 incubator at 27°C for approximately 10 days, when positive wells were identified. This single-cell cloning was necessary because there is variation in expression level of the dsRNA, and mixed cultures are quickly overtaken by cells that are nonresponsive or express low levels of dsRNA upon induction. Some commercially prepared media exacerbated an apparent partial expression of dsRNA prior to induction (S. F. O’Hearn, unpublished observations); use of Cunningham's medium (JRH Biosciences) along with 15% “Tet system approved” fetal bovine serum (Clontech) minimized this problem. However, long-term culture or recovery from cryo-storage, which could provide a selection against properly expressing clones, was avoided. For induction of the RNAi construct, tetracycline was added to 1 μg/ml, and cultures were protected from light. Cells were counted with an automated cell counter (Coulter) and observed with a Zeiss Axioskop microscope (Carl Zeiss, Inc.) and a 100×, 1.3-numerical-aperture plan-NEOFLUAR oil immersion objective.
FIG. 2.
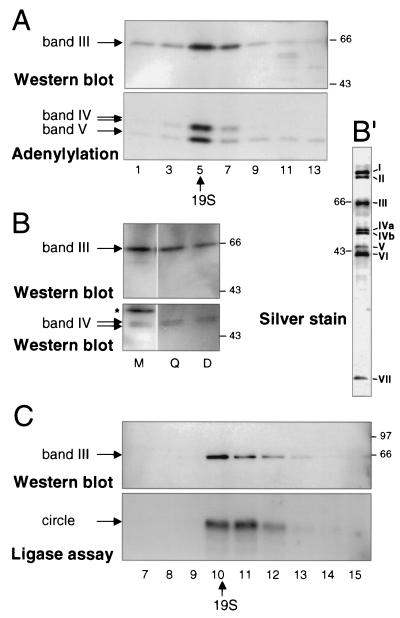
Band III copurifies with band IV. (A) Glycerol gradient sedimentation analysis of band III and bands IV and V. Fractions from mitochondrial extract (strain TREU 667; the two band IV isoforms are described in Materials and Methods) were resolved and analyzed by Western blotting for band III and by adenylylation for bands IV and V. Fraction 1 was at the bottom of the gradient, and fraction 14 was at the top; 19S was identified in a parallel glycerol gradient of thyroglobulin. There are inhibitors of band IV adenylylation high in the gradient (25). The same SDS gel, transferred to a membrane, was first exposed for adenylylation analysis and then used for Western analysis. The adenylylated proteins are not seen in the Western blot, because film was exposed for ∼10 s to score the chemiluminescence generated in the Western analysis, but 18 h was required to score the adenylylation. (B) SDS-PAGE gel analysis of band III and band IV in crude extract and partly and highly purified editing complex. The gel was loaded with 0.1 μl of crude mitochondrial extract (∼0.25 μg), 0.5 μl of the peak fraction after this extract had been put over a Q-Sepharose column (Q), and 5 μl of the peak fraction after this partially purified extract had been further purified over a DNA cellulose column (D) to yield the seven-polypeptide editing complex. The blot was sequentially probed with polyclonal antibodies against band IV (lower panel) and band III (upper panel). In crude mitochondrial extract, some cross-reacting proteins are observed (∗). Silver staining of proteins from a Q-Sepharose- and DNA cellulose-purified editing complex is shown in panel B′. (C) Such a Q-Sepharose- and DNA cellulose-purified seven-polypeptide complex had been subjected to glycerol gradient sedimentation by Rusché et al. (33), where the editing activities were shown to sediment at ∼20S; the assay of RNA ligase activity from these fractions is reproduced in the lower panel. The same glycerol gradient fractions were here reassayed for band III protein (upper panel). Sedimentation time for panel C was considerably shorter than that for panel A.
Northern blotting, antibody preparation, and Western blotting.
Total cell RNA was prepared with Trizol reagent (Gibco BRL) according to the manufacturer's instructions. For Northern blotting, 15 μg was run on a 1% agarose-formaldehyde gel and transferred to GeneScreen Plus membrane (NEN Research Products). After prehybridization in ULTRAhyb (Ambion), membranes were hybridized at 42°C in this buffer with a denatured, random-primed labeled probe encompassing the region used for dsRNA production and then extensively washed and exposed to film.
Antibodies were prepared in accordance with standard procedures (14). Specifically, 50 μg of full-length, six-His-tagged recombinant band III or band IV protein, purified under denaturing conditions on Probond Nickel columns according to the manufacturer's instructions (Invitrogen) and dialyzed into phosphate-buffered saline, was mixed with an equal volume of complete Freund's adjuvant (Sigma) and injected into the peritoneal cavity of 6-week-old BALB/c or CD1/Swiss female mice. Mice were boosted with 50 μg of antigen solubilized in incomplete Freund's adjuvant (Sigma) approximately every 2 weeks for 20 weeks. Mice were exsanguinated, and the total blood volume was collected and centrifuged for the serum, which was stored at −70°C with 0.02% sodium azide. Tail test bleeds were assayed by Western blotting of recombinant protein and whole-cell trypanosome lysate.
Western blots were performed with precut polyvinylidene difluoride membranes (Novex), electrotransferring proteins from SDS gels with the mini-Protean II system (Bio-Rad) and 25 mM Tris-0.2 M glycine-20% methanol buffer. Membrane blocking and incubation with antibodies were performed at room temperature. Blocking was done in TBS-T (20 mM Tris, 137 mM NaCl, 0.25% Tween 20 [pH 7.6]) with 0.5% bovine serum albumin fraction V (ICN) for at least 1 h. Then membranes were incubated with a 1:1,000 dilution of primary antibody (in TBS-T with 0.5% bovine serum albumin fraction V) for 1 h, washed repeatedly with TBS-T, incubated with a 1:12,500 dilution of secondary antibody (horseradish peroxidase conjugated anti-mouse immunoglobulin G [Santa Cruz] in TBS-T with 0.5% bovine serum albumin fraction V) for 1 h, washed repeatedly in TBS-T, and developed with the ECL-Plus chemiluminescent detection kit following the manufacturer's instructions (Amersham Pharmacia).
Extract preparation, adenylylation, and in vitro editing reactions.
Small-scale mitochondrial extract preparations were as described previously (34), based on the method of Sabatini et al. (36). Large-scale mitochondrial extract preparation and its further purification by column chromatography or glycerol gradient sedimentation were described elsewhere (16, 33, 41). Extracts were prepared from the same number of cells and yielded very similar protein concentrations.
Adenylylation assays were carried out following deadenylylation as described previously (16) with ∼1 μg of trypanosome mitochondrial extract or 4 μl of glycerol gradient fraction.
In vitro editing reactions, PCR amplification of template DNA, RNA production, and its 3′-end labeling were done as described previously (7). Reactions were assembled from preannealed mRNA-gRNA, mitochondrial extract, and the appropriate buffer, described as follows. Natural A6 pre-mRNA, called m[0,4], contains four U's at editing site 1 (ES1) and none at ES2 (39). Optimized gRNA D33′ directs removal of 3 U's at ES1, is much more efficient than natural A6 gRNA, and forms no chimeras (10). Optimized gRNA I47G directs insertion of 2 U's at ES2 and is a few fold more efficient than natural A6 gRNA (Cruz-Reyes et al., unpublished data; see also reference 16). Twenty-five to 50 fmol of 3′-end-labeled mRNA (m[0,4]) and 1.25 pmol of gRNA per reaction were preannealed in TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) at 42°C for 5 min, followed by cooling to 22°C over ∼15 min. Reactions used equal protein amounts of mitochondrial extract (∼0.45 μg) made in parallel from noninduced or tetracycline-induced cells. U deletion reactions were in 10 mM KCl-MRB (25 mM Tris-HCl [pH 7.5], 10 mM magnesium acetate, 10 mM KCl, 1mM EDTA, 0.5 mM dithiothreitol [DTT], 5% glycerol) supplemented with 0.15 mM ATP, 3 mM ADP, 5 mM CaCl2, 4.5 ng of torula RNA per μl, 5 mM DTT, 0.8 U of RNasin per ml (Promega). Total U deletional cleavage was assayed in such reactions supplemented with 1 mM PPi to inactivate ligase and with no ATP or CaCl2 (8); total U insertional cleavage was assayed analogously, except with omission of ADP and with 0.045 ng of torula RNA per μl (8). The 20-μl reaction mixtures were incubated for 40 min at 22°C, and recovered RNAs were resolved on 9% polyacrylamide-7.5 M urea gels and visualized by autoradiography.
Assays of the other individual editing steps also used equal protein amounts of mitochondrial extract (∼0.2 μg) made from noninduced or tetracycline-induced cells in parallel. Analysis of 3′-U-exo activity used a 5′ kinase-labeled (New England Biolabs, following the manufacturer's recommendations) gRNA with a long U tail (Anc + U16) (10) and was performed in MRB for 30 min at 22°C. TUTase activity was assessed with RNAs designed to mimic a U insertion site following its cleavage, specifically the mRNA and gPCA6-2A gRNA of Igo et al. (17), resolving the products as described by these authors; we included 1.5 mM PPi (Sigma) to inhibit ligation and performed the reaction in MRB for 1 h at 27°C. RNA ligase activity was assessed by using 5′ kinase-labeled (New England Biolabs) gPCA6-2A RNA (17) in MRB with 0.3 mM ATP for 30 min at 22°C, assessing formation of dimers, trimers, and circles.
RESULTS
Band III cDNA encodes a novel protein with zinc finger motifs.
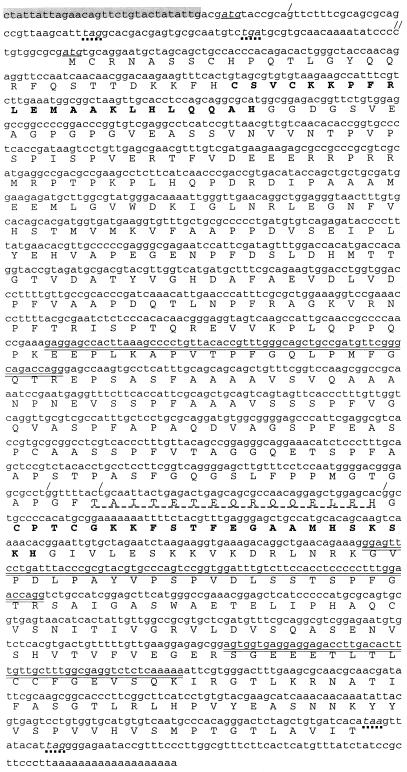
Three tryptic peptides of the band III protein isolated from purified T. brucei editing complex were subjected to Edman degradation, yielding sequences of 24, 21, and 18 amino acids (the double-overlined amino acids in Fig. 1). PCR with trypanosome genomic DNA and degenerate primers designed to portions of two of these peptides generated an ∼800-bp fragment that encodes these two tryptic peptides in phase, including the adjacent 8 amino acids not used for primer design, plus the entire 24 amino acids of the third tryptic peptide. Therefore, the cloned fragment does indeed correspond to the isolated band III protein.
FIG. 1.
Nucleotide and amino acid sequences of T. brucei band III. Nucleotide sequence and the predicted amino acid sequence of the cloned trypanosome cDNA are shown. The three sequenced tryptic peptides are indicated by the double-overlined amino acids. The trypanosome spliced leader sequence is highlighted in gray, and the initiating AUG, as well as an alternate AUG (4 residues downstream of the spliced leader) evidently utilized in some strains, is underlined and in italics. In-frame termination codons are in italics with dotted underlining. Differences from the TbMP63 T. brucei sequence in the BLAST database are shown: the slash on the first line indicates the position of an additional G residue found in the TbMP63 sequence, but not in this sequence, making the indicated termination codons out of frame, and the double slash indicates a C residue in this sequence, but not in the TbMP63 sequence, returning the two sequences to be in frame. Similarly, each of the three slashes two-thirds of the way through the gene indicates an additional base that changes the reading frame in this region (dashed underlining). Two zinc finger motifs discussed in the text are shown in boldface.
From a trypanosome cDNA library, this PCR product identified three clones of ∼1,600 bp. One was double strand sequenced, confirming that it contained the entire PCR product and encoded the remaining 14 amino acids of known tryptic peptide sequence. However, this cDNA was shorter than the ∼2,000-nucleotide (nt) mature transcript (see Fig. 3A), and it lacked the 39-nt miniexon sequence that is trans-spliced onto all mature nucleus-encoded trypanosome mRNAs. Therefore, to obtain the complete 5′ end, RT-PCR was performed with poly(A)+ trypanosome RNA, with primers to the trypanosome miniexon sequence and band III gene sequence.
FIG. 3.
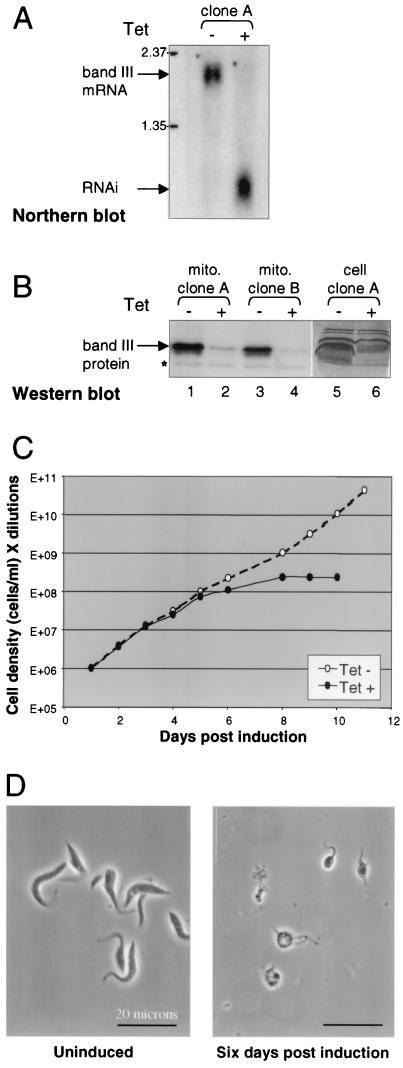
RNAi depletes band III mRNA and protein and halts cell growth. (A) Northern blot analysis of mRNA levels in cells expressing the band III dsRNA construct, noninduced (−) and 48 h postinduction with 1 μg of tetracycline (Tet) per ml (+). The full-length band III mRNA is ∼2 kb (upper arrow), while RNA generated from the pZJM construct is ∼800 nt (lower arrow). (B) Western blot with anti-band III polyclonal antibody of either mitochondrial extract (lanes 1 to 4) or total cell protein (lanes 5 and 6) from cells expressing the band III RNAi construct noninduced (−) and 6 days postinduction (+). The antibody reaction to nonspecific proteins serves as an internal loading control; a nonspecific band present in the blot of mitochondrial extract is marked (∗). The band III band is distorted in whole-cell extract, because a very abundant protein runs just below it. (C) Cell growth profile of one clonal population of band III RNAi cells, either noninduced (−) or induced with 1 μg of tetracycline per ml (+). From these cells, aliquots were taken to perform the experiments shown in panels A and B, lanes 5 and 6. The cells were diluted throughout the experiment to make sure their densities remained compatible with exponential growth, and cell density times dilution is represented on the y axis. Similar experiments with different band III RNAi clones gave very similar results. The cell growth profile of the parental 29-13 cell line was not affected by addition of tetracycline. (D) Phase-contrast microscopy images of band III RNAi cell line either noninduced (left panel) or following 6 days of tetracycline treatment (right panel).
The complete band III cDNA encodes a protein of 555 amino acids with a predicted molecular mass of 59.3 kDa that contains two Cys2His2 zinc finger motifs (Fig. 1). This band III sequence is almost identical to one in the BLAST database (TbMP63, accession no. AF382334), except for five single-base insertions or deletions, which make for two interesting differences. The band III sequence has two stop codons in frame with the first AUG, so the subsequent AUG, 96 nt beyond, is most likely utilized. Relative to this band III sequence, TbMP63 has a 1-nt insertion just beyond the first AUG and a 1-nt deletion 77 nt later, making these stop codons out of phase, so TbMP63 likely utilizes this first AUG (Fig. 1). Additionally, in the central portion of the gene, TbMP63 has inserted 2 nt that change the reading frame, and 43 nt later another inserted nucleotide that corrects the reading frame, so 15 amino acids of band III are replaced by 16 different amino acids in TbMP63 (Fig. 1). These differences likely represent allelic variations of the same protein. In fact, strain TREU 667, from which the protein was isolated, may well be heterozygous, because the purified band III protein did not run as a single band (33), yet the complexity of the tryptic profile for the combined bands suggested only a single protein (D. Reim, personal communication).
A C-terminal segment of the band III protein shows the greatest homology with band VII protein, extending over much of the length of band VII (E value, 8 × 10−5). However, it also shows marked homology with the equivalent C-terminal regions of band II and band VI (E values, 2 × 10−3 and 7 × 10−3, respectively). Furthermore, the region encompassing the N-terminal zinc finger motif of band III shows homology to regions encompassing a zinc finger of band II and band VI. Thus, out of the entire sequence database, band III has the most similarity to three other proteins of the T. brucei seven-protein editing complex (33). TbMP81, -42, and -18, recently reported to copurify by the Stuart laboratory (30), correspond to band II, band VI, and band VII, respectively.
Band III is a component of the minimal RNA editing complex.
Band III was initially identified as part of the minimal T. brucei RNA editing complex, because it purified along with the RNA ligase proteins and the other editing activities. At the final DNA cellulose column, these seven coeluting proteins were the only bound species of appreciable abundance (Fig. 2B′), and they stayed associated upon subsequent Q-Sepharose chromatography and native gel electrophoresis (33). However, even after several sequential column purifications, contaminant proteins can still be present (11, 29), and surreptitious protein associations could occur within the most purified fractions. To provide additional lines of evidence that band III is a constituent of the minimal editing complex, we prepared polyclonal antibodies to the six-His-tagged translation product of the cloned band III gene synthesized in E. coli and isolated on a nickel column. These antibodies enabled comparison of the sedimentation profile and purification characteristics of the band III protein with those of the band IV RNA ligase, the best-characterized member of the editing complex (33, 34, 37). The band III antibody indeed identifies a trypanosome protein of the expected size (Fig. 2A and B′). Analysis of glycerol gradient fractions of crude mitochondrial extract showed that band III protein cosediments with the band IV and band V RNA ligases (Fig. 2A; ligases identified by adenylylation with [α-32P]ATP, which donates AMP for covalent attachment to activate the ligases). Therefore, the vast majority of band III protein is in a complex that sediments with the RNA editing ligases at ∼20S, and virtually none is present as free protein. Furthermore, throughout purification of the editing complex, the ratio of band III protein to band IV protein is quite constant from crude mitochondrial extract to Q-Sepharose and DNA cellulose (Fig. 2B), demonstrating that band III does not purify away from the editing complex. Finally, we made use of our earlier observation (33) that when the Q-Sepharose- and DNA cellulose-purified RNA editing complex containing only seven major polypeptides was subjected to velocity centrifugation, the editing activities still cosedimented at ∼20S (33) (Fig. 2C, lower panel), as before purification. Reassaying these glycerol gradient fractions, we now find that band III protein continues to cosediment with the highly purified editing complex (Fig. 2C, upper panel), confirming its intimate association. Therefore, band III is a bona fide constituent of this editing complex.
Band III protein is essential for viability in procyclic trypanosomes.
RNAi was then used to specifically degrade band III mRNA (28, 40) and to determine the phenotype when band III protein is depleted. dsRNA corresponding to an ∼750-bp segment from the central portion of the band III gene was inducibly expressed in procyclic trypanosomes. This expression was driven from opposing T7 promoters controlled by tetracycline operators by using a construct stably integrated into the ribosomal DNA locus (43). When independent clonal cultures were induced with tetracycline for 48 h, the interfering RNA was abundant and the full-length band III mRNA was undetectable by Northern blotting, so evidently it was degraded (Fig. 3A). As induction proceeded, morphological abnormalities and cell debris became apparent (data not shown). By 6 days postinduction, Western blotting revealed that virtually all band III protein is absent from the mitochondria (Fig. 3B, lanes 1 to 4), also indeed from the whole cell (Fig. 3B, lanes 5 and 6). Upon band III depletion, cell growth ceased (Fig. 3C), and almost all of the remaining cells had highly abnormal morphologies (Fig. 3D). Frequently, the cell growth crisis was so severe that the culture could not be revived by tetracycline removal (data not shown). Control cells, including analogously constructed lines inducibly expressing heterologous dsRNA of similar lengths and the 29-13 parental line, were grown in parallel, and exhibited neither cell growth nor morphological defects upon tetracycline treatment (data not shown). Therefore, we conclude that band III is an essential protein in procyclic trypanosomes.
Band III is required for in vitro editing.
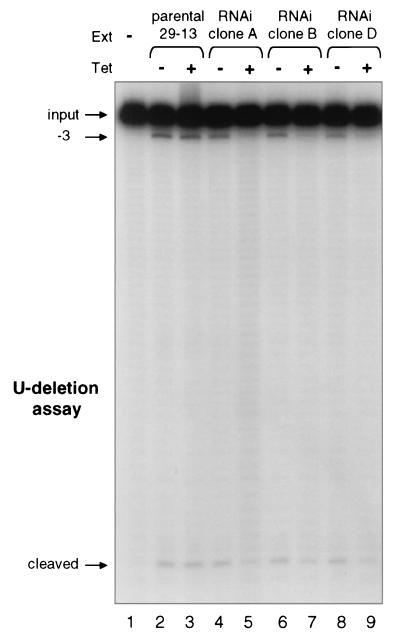
To look more specifically at the effect of RNAi-mediated band III depletion, we assessed RNA editing in vitro. Mitochondrial extracts were prepared from control cell lines and multiple independent clonal band III RNAi cell lines either noninduced or induced with tetracycline for 6 days. For comparison of editing activities, the same numbers of noninduced and induced cells were used to prepare extracts in parallel, yielding similar protein concentrations. We first assessed full round U deletion reactions, using an active gRNA that directs removal of 3 U residues at the first editing site of ATPase 6 (A6) pre-mRNA (D33′) (10). The noninduced and induced 29-13 control cell extracts showed almost identical levels of the −3 U deletion product (Fig. 4, lanes 2 and 3), demonstrating that tetracycline treatment alone does not have adverse effects (16). Furthermore, extracts from noninduced band III RNAi clones were similarly active (Fig. 4, lanes 4, 6, and 8). However, extracts from induced band III RNAi clones were severely inhibited in U deletion, as only very low levels of the −3 product were produced (Fig. 4, lanes 5, 7, and 9). Therefore, since RNA editing is essential in procyclic trypanosomes (32), the cell death observed in Fig. 3C may arise because depletion of band III protein inhibits proper pre-mRNA editing.
FIG. 4.
Depletion of band III through RNAi inhibits in vitro RNA editing. Full round U deletion reaction at the first editing site of A6 pre-mRNA is shown. Reaction mixtures used equal amounts of protein (∼0.45 μg) from mitochondrial extracts (Ext) of the parental 29-13 cell line or independent clones of band III RNAi cells, either noninduced (−) or induced with tetracycline (Tet) for 6 days (+). The final −3 deletion product and the remaining 3′ cleavage fragment are seen.
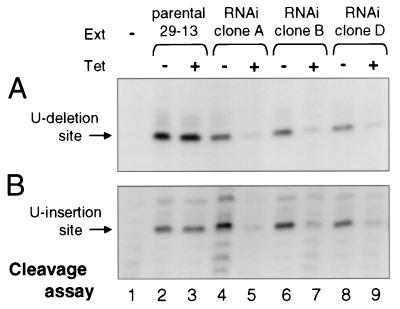
In order to gain insight into the role of band III in RNA editing, we dissected the in vitro editing process into its three component reactions: gRNA-directed cleavage, U removal or addition, and mRNA religation. Total cleavage was scored in reactions where the ligases are inhibited with pyrophosphate (8). This analysis was performed for both U deletional and U insertional cleavage (8), using a gRNA directing U deletion at the first editing site of A6 pre-mRNA (Fig. 5A) or a gRNA directing U insertion at the second editing site of this pre-mRNA (Fig. 5B). Both forms of cleavage are markedly reduced when band III is depleted by the RNAi induction (Fig. 5A and B). This explains why, in the full round reaction (Fig. 4), extracts of the induced band III RNAi cells generate less total cleaved plus edited RNA. In contrast to recent studies indicating that the ligases of the minimal editing complex act predominantly in either U deletion or U insertion (16; Cruz-Reyes et al., submitted), here we find that depletion of band III affects steps of both the in vitro U deletion and U insertion cycles (Fig. 5A and B). This is also the first report of a protein needed for either of the first two enzymatic steps in the U deletion or U insertion pathway.
FIG. 5.
Depletion of band III inhibits U deletional and U insertional endonuclease activity. (A) Total U deletional cleavage reactions were performed as described in the legend to Fig. 4, except omitting ATP and including PPi to inhibit the ligases. Ext, extract; Tet, tetracycline. (B) Total U insertional cleavage reactions were much as in panel A (see Materials and Methods), except with gRNA I47G (Cruz Reyes et al., unpublished data; see Materials and Methods) to assess the second editing site.
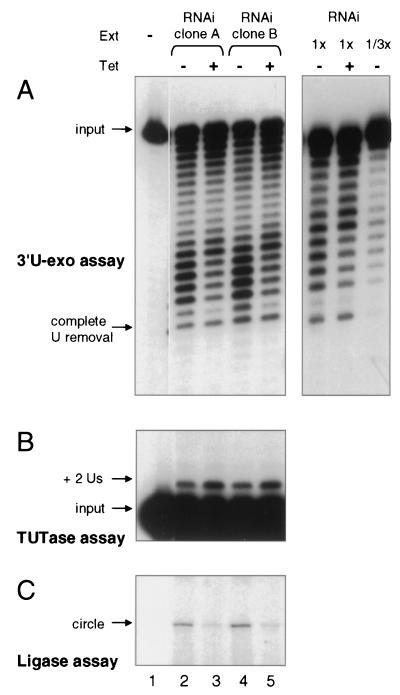
Because the cleavage steps of both U deletion and U insertion are greatly reduced in extracts of induced band III RNAi cells, these extracts would generate negligible substrate with which to analyze the subsequent steps of editing in the naturally coupled reaction pathway. However, these enzymatic activities can also be scored individually, independent of the prior editing steps, using heterologous RNAs (33, 39). The 3′-U-exo activity was assessed by shortening of a 5′-labeled RNA that ends in 19 U residues (33). It remained active in the induced band III RNAi cell extracts (Fig. 6A), demonstrating that band III is not needed for 3′-U-exo activity. Both the noninduced and induced extracts were able to catalyze extensive U removal, but the profile of U removal products was slightly different. Titration of the extract amount using a more purified preparation (Fig. 6A, right panel, and data not shown) indicated this was not due to marked differences in the amount of the enzyme. It thus may be that 3′-U-exo, while present and active, is subtly altered in the absence of band III.
FIG. 6.
RNA ligation, but not 3′-U-exo or TUTase, is inhibited by band III depletion. (A) 3′-U-exo assay, scoring removal of a long U-tail from a 5′-end-labeled RNA. The position of a fragment lacking all of the terminal U's is indicated. The left panel assays the same small-scale extract (Ext) preparations as those in Fig. 3 to 7A on an RNA with 19 terminal U's. The right panel assays two amounts of large-scale extract preparations of clone B (as were also used for the glycerol gradient in Fig. 7C) on an RNA with 13 terminal U's, verifying that the assays are responsive to the amount of 3-U-exo. Tet, tetracycline. (B) TUTase assay using the precleaved editing system (17), which calls for addition of two U's onto the 3′ end of the 5′-end-labeled upstream mRNA fragment (17). Ligation of this fragment was inhibited by addition of PPi. (C) RNA ligase assay, showing circularization. The lack of a bridging oligonucleotide should favor scoring of band IV but not band V activity (see text).
TUTase activity was assessed in a precleaved editing system with a 5′-labeled artificial upstream mRNA fragment, a downstream fragment, and an artificial gRNA specifying insertion of two U residues (17), but also including pyrophosphate to inhibit ligation. It showed that this U addition activity was not reduced in the induced band III RNAi cell extracts (Fig. 6B). Thus, the activities catalyzing the second steps of both the U deletion and U insertion pathways, 3′-U-exo and TUTase, remain active in the absence of band III.
RNA ligation activity was also assessed with a heterologous substrate RNA. Because the ends being joined do not have a base-pairing ligation bridge, this assay should score band IV but not band V RNA ligase (10, 17, 34; C. E. Huang, unpublished observations). It showed joining was severely reduced in the band III-depleted extracts (Fig. 6C).
From these assays of editing activities, we conclude that depletion of band III diminishes U deletional and U insertional cleavage and ligation by band IV. We also conclude that the 3′-U-exo and TUTase activities do not depend on band III protein. However, we cannot rigorously assign a specific enzymatic activity to this protein. In fact, we hypothesize that the loss of band III exerts at least some of its effects by disrupting the coordinated actions of editing complex.
Band III is required to maintain the editing complex.
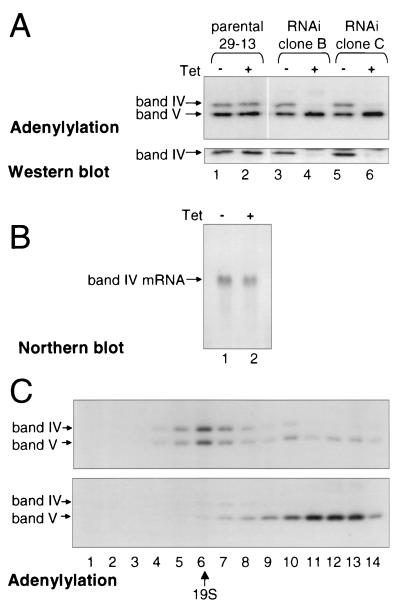
To address how band III loss could affect several different steps in the editing process, we analyzed other proteins of the minimal editing complex, specifically the ligases, which can be labeled by adenylylation with [α-32P]ATP (Fig. 7A, upper panel). When band III protein is depleted by RNAi induction, band IV protein is coordinately depleted, while band V protein is not depleted. Indeed, in multiple independent cell lines, band V ligase protein appears present in at least control levels. The loss of band IV protein was also confirmed by Western blotting with an anti-band IV mouse polyclonal antibody (Fig. 7A, lower panel), loading equal concentrations of extract protein. Because band IV mRNA does not contain sequences in common with the band III region used for RNAi and band IV mRNA remains at control levels upon induction of band III RNAi (Fig. 7B), band IV protein depletion is most likely a secondary consequence of the band III protein depletion. Thus, loss of band III causes loss of some but not other components of the editing complex. That band IV protein is lost from the entire mitochondrion, not just the editing complex, reinforces the previous finding (34) that band IV protein is quickly degraded when not in an editing complex. These results also explain why the extracts of induced band III RNAi cells showed minimal ligation activity of the kind characteristic of band IV.
FIG. 7.
Depletion of band III through RNAi causes loss of band IV and disrupts the editing complex. (A) Equal amounts of protein (∼1.35 μg) from mitochondrial extracts of the parental 29-13 cell line or two independent clones of band III RNAi cells, either noninduced (−) or induced with tetracycline (Tet) for 6 days (+), were assessed by adenylylation (upper panel) and by Western blotting (lower panel), showing that the effect on band IV is on protein abundance, not adenylylation activity. (B) Northern blot analysis demonstrating that band IV mRNA levels are unaffected by band III RNAi expression. Total RNA from a control cell line (lane 1) and a band III RNAi cell line (lane 2), both induced with tetracycline for 2 days, was probed for band IV coding sequences. (C) The parental 29-13 cell line (top panel) and band III RNAi clone B cell line (lower panel) were induced with 1 μg of tetracycline per ml for 6 days; 460 μg of mitochondrial extract protein was sedimented over glycerol gradients, and adenylylation reactions were performed with the collected fractions. Fraction 1 was at the bottom of the gradient, and fraction 14 was at the top; 19S thyroglobulin was identified in a parallel glycerol gradient.
Because band III protein appears important for the assembly of band IV protein into the editing complex (Fig. 7A) and band IV RNA ligase is essential for editing (16, 34, 37), the inhibition of editing by band III RNAi could be due to depletion of band IV alone. However, band IV ligase activity is not needed for either the U deletional or U insertional cleavage steps (16), which are lost upon band III RNAi (Fig. 5), suggesting that the observed effects on cleavage are due to the loss or inactivation of another component of the editing complex. The simplest explanation would be that this deficient cleavage component is band III itself, but it could also be yet a third component. In fact, it would not be surprising if the absence of two members (band III and band IV) of such a small complex would disrupt the configuration or function of a remaining component.
To learn more about the residual editing complex that remains after the loss of band III and band IV, mitochondrial extract from induced band III RNAi cells and control cells was resolved by glycerol gradient sedimentation, and ligases were detected by adenylylation. The editing complexes in the induced band III RNAi extract indeed appear markedly disrupted (Fig. 7C, lower panel). Not only is band IV protein almost completely lost, but band V sediments at <5S, indicative of substantially disaggregated associations. The small remaining amount of band IV protein, plus a corresponding amount of band V, sediment at the position of the normal ∼20S editing complex. Parallel glycerol gradient sedimentation of control extract from 29-13 parental cells similarly treated with tetracycline (Fig. 7C, upper panel) showed that the altered sedimentation is not an unanticipated consequence of tetracycline administration, but is due to the loss of band III protein. Since loss of band III disrupts the editing complex, the data do not clarify whether band III loss prevents cleavage because band III is the endonuclease or because of secondary structural effects on a distinct endonuclease protein. However, the data clearly demonstrate that band III protein is required for the assembly or maintenance of a complete and intact editing complex and is needed for the proper function of other editing components.
DISCUSSION
To date, characterization of the proteins involved in the catalysis of RNA editing has been quite limited. Several proteins that appear to contribute to the efficiency of the overall in vivo editing process have been identified (3, 20, 23, 24, 26, 27), but the band IV and band V RNA ligases are the only members of the seven-polypeptide editing complex (33) that have been studied (16, 25, 29, 34, 37; Cruz-Reyes et al., submitted). Therefore, we now focus on another approximately stoichiometric component of this minimal editing complex, band III. Its gene was cloned using tryptic fragment sequencing data and encodes a 555-amino-acid protein with two separate zinc finger motifs (Fig. 1). An antibody prepared to the recombinant protein showed that trypanosome band III has the same sedimentation profile as the ligase markers of the editing complex (Fig. 2A), that it and the ligase proteins are similarly enriched when the editing complex is purified from crude mitochondrial extract through Q-Sepharose and DNA cellulose chromatography (Fig. 2B), and that they also remain together upon glycerol gradient sedimentation of that DNA cellulose fraction (Fig. 2C). dsRNA-mediated interference showed that band III is essential for cell growth (Fig. 3C and D) and for in vitro editing (Fig. 4). In the absence of band III protein, band IV protein of the editing complex is no longer present (Fig. 7A) and the ∼20S editing complex is disrupted into smaller associations (Fig. 7C). These findings indisputably demonstrate that band III is a critical member of the T. brucei RNA-editing complex.
Analysis of the individual steps of the editing cycles showed that the 3′-U-exo (Fig. 6A) and the TUTase activities (Fig. 6B), which catalyze the second steps in U deletion and U insertion, remained active upon depletion of band III protein. However, the gRNA-directed endonuclease activities for both U deletion and U insertion was severely diminished (Fig. 5A and B). The RNA ligation activity of band IV, which is required for sealing in U deletion (10, 16; Cruz-Reyes et al., submitted), was similarly diminished in the induced band III RNAi cell extracts (Fig. 6C). The deficiency of the ligation step in U deletion explains why the relative amount of cleaved mRNA from induced versus noninduced extracts was more different when assaying total cleavage (Fig. 5A and B) than when assaying the full round editing reaction (Fig. 4): the few molecules that are cleaved are less efficiently sealed in the ligation step of the full round reaction, so relatively more cleaved molecules remain. Accordingly, the RNAi-induced depletion of band III protein results in depletion of band IV protein; however, band V protein remains (Fig. 7A and B).
Our results show that RNAi-induced extracts are lacking band III protein and gRNA-directed endonuclease activity, as well as band IV and its ligation activity (Fig. 3B, 5, 6C, and 7). One obvious possible explanation is that band III is the gRNA-directed endonuclease for both U deletion and U insertion sites. Although band III recombinant proteins made in multiple different expression systems did not exhibit such endonuclease activity or any component editing activity (C.E.H. and S.F.O., unpublished observations), enzymatic function could require additional components of the editing complex or posttranslational modifications. Furthermore, while no sequences or motifs predictive of an endonuclease were detected in the band III protein sequence by using Pfam, not all nucleases are identified by this search, and the novel features of the trypanosome editing endonuclease (41) suggest it could be highly diverged. One unprecedented aspect of the editing endonuclease activities is in the response to adenosine nucleotides: U deletional cleavage is stimulated by ADP or ATP, while U insertional cleavage is inhibited by these nucleotides (9). If band III provides the endonuclease activity, this adenosine nucleotide effect would likely be exerted through an allosteric interaction, with the single endonuclease adopting alternative states for U deletion and U insertion. Alternatively, it is possible that the band III protein does not contain the endonuclease active site, but instead provides a scaffold needed for proper assembly of other proteins of the editing complex that provide the endonuclease catalytic center.
Clearly, one critical function of the band III protein is as a scaffold to allow proper assembly of other components into the editing complex. The seven-component polypeptides are separately encoded in the nuclear genome and likely imported into the mitochondria as individual proteins, where they are then assembled into an ordered ∼20S complex. Band III appears essential for this orderly assembly; when it is depleted, band IV is also absent from the complex. Band IV is also absent from mitochondrial extract (Fig 7A), providing additional support for previous studies (34), which showed that band IV protein not assembled into the editing complex is quickly degraded. Another member of the complex, band V, remains present but assembles only into small associations (Fig. 7C) Once antibodies for the other proteins of the complex are developed, their fate upon band III depletion can be assessed. Note, however, that those proteins providing the 3′-U-exo activity and the TUTase activity remain present and active upon band III depletion. Their uncompromised activities suggest that the observed effects of band III depletion on editing are unlikely to be due to global disruption of the cell but are specifically due to the loss of band III.
Band III seems well suited to a role in protein-protein interactions and protein assembly, since it, along with band II and band VI, are putative zinc finger proteins. Zinc finger motifs confer diverse functions, including RNA binding and packaging and protein binding, folding, and assembly (reviewed in references 18 and 21). Intriguingly, band III has two presumptive zinc fingers that are at opposite ends of the polypeptide, an unusual zinc finger configuration that suggests structural bridging. One other polypeptide showing two distinct zinc-binding domains is RPB9, a component of the yeast RNA polymerase II complex, which appears to aid in conformational changes of that complex (15). It is currently unknown whether the two putative zinc fingers in band III do actually function, much less whether they mediate binding to proteins within the complex or to substrate RNA.
Our studies depleting band III protein underscore the importance of maintaining protein interactions within the editing complex. Previous experiments reducing band IV protein to approximately one-half, through single allele gene knockout, demonstrated that editing complexes lacking band IV are somewhat more labile; approximately half the band V protein sedimented at ∼7S, indicative of broken complexes (16). The current study extends this analysis. Depletion of band III causes almost complete loss of band IV, and almost all of band V sediments much more slowly than 20S (Fig. 7C). These residual complexes are smaller than the ∼7S observed upon band IV depletion, consistent with band IV, band III, and possibly additional components being lost in the induced band III RNAi cells. The sequestration of editing enzymes in an editing complex extends back in evolution to at least the split between the trypanosoma and leishmania clades, ∼108 years ago (22). Therefore, instead of merely a collection of physically attached but independently acting proteins, the editing constituents have had ample time to develop strong interdependence for proper assembly, regulation of abundance, structural configuration, and function.
Acknowledgments
We greatly thank Laura Rusché for preparing the band III protein for tryptic peptide analysis and for making the fractions of partially and fully purified editing complex used in Fig. 2 as well as for critically reading the manuscript. We acknowledge David Reim at the Wistar Institute for sequencing of the tryptic peptides. We also thank Ken Piller for preparing the procyclic strain 667 cDNA library; Alevtina Zhelonkina and Jorge Cruz-Reyes for providing the I47G gRNA; Zefeng Wang, Jim Morris, and Mark Drew for providing the pZJM vector and advice on its use; and Paul Englund and Michele Klingbeil for helpful discussions.
This work was supported by NIH grant GM34231.
C.E.H. and S.F.O. contributed equally to this work.
REFERENCES
- 1.Alfonzo, J., O. Thiemann, and L. Simpson. 1997. The mechanism of U insertion/deletion RNA editing in kinetoplastid mitochondria. Nucleic Acids Res. 25:3751-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benne, R., and D. Speijer. 1998. Appendix 2: RNA editing types and characteristics, p. 551-554. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 3.Blom, D., J. Burg, C. Breek, D. Speijer, A. Muijsers, and R. Benne. 2001. Cloning and characterization of two guide RNA-binding proteins from mitochondria of Crithidia fasciculata: gBP27, a novel protein, and gBP29, the orthologue of Trypanosoma brucei gBP21. Nucleic Acids Res. 29:2950-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blum, B., N. Bakalara, and L. Simpson. 1990. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60:189-198. [DOI] [PubMed] [Google Scholar]
- 5.Blum, B., N. Sturm, A. Simpson, and L. Simpson. 1991. Chimeric gRNA-mRNA molecules with oligo(U) tails covalently linked at sites of RNA editing suggests that U addition occurs by transesterification. Cell 65:543-550. [DOI] [PubMed] [Google Scholar]
- 6.Cech, T. 1991. RNA editing: world's smallest intron? Cell 64:667-669. [DOI] [PubMed] [Google Scholar]
- 7.Cruz-Reyes, J., and B. Sollner-Webb. 1996. Trypanosome U deletional RNA editing involves gRNA-directed endonuclease cleavage, terminal U-exonuclease, and RNA ligase activities. Proc. Natl. Acad. Sci. USA 93:8901-8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz-Reyes, J., L. Rusché, K. Piller, and B. Sollner-Webb. 1998. T. brucei RNA editing: adenosine nucleotides inversely affect U deletion and U insertion reactions at mRNA cleavage. Mol. Cell 1:401-409. [DOI] [PubMed] [Google Scholar]
- 9.Cruz-Reyes, J., L. Rusché, and B. Sollner-Webb. 1998. Trypanosoma brucei U insertion and U deletion activities co-purify with an enzymatic editing complex but are differentially optimized. Nucleic Acids Res. 26:3634-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz-Reyes, J., A. Zhelonkina, L. Rusché, and B. Sollner-Webb. 2001. Trypanosome RNA editing: simple guide RNA features enhance U deletion 100-fold. Mol. Cell. Biol. 21:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estevez, A., F. Kierszenbaum, E. Wirtz, F. Bringaud, J. Grunstein, and L. Simpson. 1999. Knockout of the glutamate dehydrogenase gene in bloodstream Trypanosoma brucei in culture has no effect on editing of mitochondrial mRNAs. Mol. Biochem. Parasitol. 100:5-17. [DOI] [PubMed] [Google Scholar]
- 12.Göringer, U., J. Koller, and H. Shu. 1995. Multicomponent complexes involved in kinetoplastid RNA editing. Parasitol. Today 11:265-267. [Google Scholar]
- 13.Gott, J., and R. Emeson. 2000. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34:499-531. [DOI] [PubMed] [Google Scholar]
- 14.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Hemming, S., and A. Edwards. 2000. Yeast RNA polymerase II subunit RPB9. J. Biol. Chem. 275:2288-2294. [DOI] [PubMed] [Google Scholar]
- 16.Huang, C., J. Cruz-Reyes, A. Zhelonkina, S. O'Hearn, E. Wirtz, and B. Sollner-Webb. 2001. Roles for ligases in the Trypanosoma brucei RNA editing complex: band IV is needed for U-deletion and RNA repair. EMBO J. 20:4694-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Igo, R. P., Jr., S. S. Palazzo, M. L. Burgess, A. K. Panigrahi, and K. Stuart. 2000. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol. Cell. Biol. 20:8447-8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iuchi, S. 2001. Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci. 58:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kable, M. S. Seiwert, S. Heidmann, and K. Stuart. 1996. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science 273:1189-1195. [DOI] [PubMed] [Google Scholar]
- 20.Köller, J., U. Müller, B. Schmid, A. Missel, V. Kruft, K. Stuart, and U. Göringer. 1997. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem. 272:3749-3757. [DOI] [PubMed] [Google Scholar]
- 21.Laity, J., B. Lee, and P. Wright. 2001. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opinion Struct. Biol. 11:39-46. [DOI] [PubMed] [Google Scholar]
- 22.Lake, J., V. de la Cruz, P. Ferrierira, C. Morel, and L. Simpson. 1988. Evolution of parasitism: kinetoplastid protozoan history reconstructed from mitochondrial rRNA gene sequences. Proc. Natl. Acad. Sci. USA 85:4779-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madison-Antenucci, S., R. Sabatini, V. Pollard, and S. Hajduk. 1998. Kinetoplastid RNA-editing associated protein 1 (REAP-1): a novel editing complex protein with repetitive domains. EMBO J. 17:6368-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madison Antenucci, S., and S. Hajduk. 2001. RNA editing-associated protein 1 is an RNA binding protein with specificity for preedited mRNA. Mol. Cell 7:879-886. [DOI] [PubMed] [Google Scholar]
- 25.McManus, M., M. Shimamura, J. Grams, and S. Hajduk. 2001. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA 7:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Missel, A., A. E. Souza, G. Nörskau, and H. U. Göringer. 1997. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol. 17:4895-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller, U., L. Lambert, and U. Göringer. 2001. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 6:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ngo, H., C. Tschudi, K. Gull, and E. Ullu. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95:14687-14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panigrahi, A. K., S. P. Gygi, N. L. Ernst, R. P. Igo, Jr., S. S. Palazzo, A. Schnaufer, D. S. Weston, N. Carmean, R. Salavati, R. Aebersold, and K. D. Stuart. 2001. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21:380-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panigrahi, A. K., A. Schnaufer, N. Carmean, R. P. Igo, Jr., S. P. Gygi, N. L. Ernst, S. S. Palazzo, D. S. Weston, R. Aebersold, R. Salavati, and K. D. Stuart. 2001. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21:6833-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard, V., M. Harris, and S. Hajduk. 1992. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 11:4429-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Priest, J., and S. Hajduk. 1994. Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J. Bioenerg. Biomembr. 26:179-191. [DOI] [PubMed] [Google Scholar]
- 33.Rusché, L., J. Cruz-Reyes, K. Piller, and B. Sollner-Webb. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16:4069-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rusché, L. N., C. E. Huang, K. J. Piller, M. Hemann, E. Wirtz, and B. Sollner-Webb. 2001. The two RNA ligases of the Trypanosoma brucei RNA editing complex: cloning the essential band IV gene and identifying the band V gene. Mol. Cell. Biol. 21:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rusché, L. N., K. J. Piller, and B. Sollner-Webb. 1995. Guide RNA-mRNA chimeras, which are potential RNA editing intermediates, are formed by endonuclease and RNA ligase in a trypanosome mitochondrial extract. Mol. Cell. Biol. 15:2933-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabatini, R., B. Adler, S. Madison-Antenucci, M. McManus, and S. Hajduk. 1998. Biochemical methods for analysis of kinetoplastid RNA editing. Methods (Orlando) 15:15-26. [DOI] [PubMed] [Google Scholar]
- 37.Schnaufer, A., A. Panigrahi, B. Panicucci, R. Igo, R. Salavati, and K. Stuart. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291:2159-2162. [DOI] [PubMed] [Google Scholar]
- 38.Seiwert, S., and K. Stuart. 1994. RNA editing: transfer of genetic information from gRNA to precursor mRNA in vitro. Science 266:114-117. [DOI] [PubMed] [Google Scholar]
- 39.Seiwert, S., S. Heidmann, and K. Stuart. 1996. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84:831-841. [DOI] [PubMed] [Google Scholar]
- 40.Shi, H., A. Djikeng, T. Mark, E. Wirtz, C. Tschudi, and E. Ullu. 2000. Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA 6:1069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sollner-Webb, B., L. Rusché, and J. Cruz-Reyes. 2001. Generally useful novel nuclease activities of the trypanosome RNA editing complex: endonucleases specific for (5′) single strand-(3′) double strand RNA junctions and an exonuclease specific for 3′ U residues. Methods Enzymol. 341:154-174. [DOI] [PubMed] [Google Scholar]
- 42.Stuart, K., T. E. Allen, S. Heidmann, and S. D. Seiwert. 1997. RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev. 61:105-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Z., J. Morris, M. Drew, and P. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
- 44.Wirtz, E., M. Hoek, and G. Cross. 1998. Regulated processive transcription of chromatin by T7 RNA polymerase in Trypanosoma brucei. Nucleic Acids Res. 26:4626-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wirtz, E., S. Leal, C. Ochatt, and G. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in T. brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]