Abstract
Histone acetylation and deacetylation have been implicated in the regulation of gene expression. Molecular studies have shown that histone deacetylases (HDACs) function as transcriptional repressors. However, very little is known about their roles during development in multicellular organisms. We previously demonstrated that inhibition of maternal and zygotic expression of histone deacetylase 1 (HDA-1) causes embryonic lethality in Caenorhabditis elegans. Here, we report the identification of an hda-1 genetic mutant which has also been called a gon-10 mutant (for gonadogenesis defective 10) and show that loss of HDA-1 zygotic expression results in specific postembryonic defects in gonadogenesis and vulval development. We provide evidence that the lag-2 gene, which plays a role in gonadogenesis and vulval development and encodes a Notch ligand, is derepressed in gon-10 animals, suggesting that lag-2 may be a target of HDA-1. Our findings reveal a novel and specific function for the ubiquitously expressed HDA-1 in C. elegans gonadogenesis and place hda-1 in the Notch signaling pathway.
Histone acetylation and deacetylation play critical roles in transcriptional regulation. While histone acetylation facilitates chromatin opening and transcriptional activation, histone deacetylation contributes to chromatin condensation and gene silencing (reviewed in references 20, 28, 39, and 41). The histone deacetylation reaction is catalyzed by histone deacetylases (HDACs) that have been conserved and are found in organisms from Saccharomyces cerevisiae to humans. In humans, the list of histone deacetylase genes continues to grow. At the present time, there are 17 hdac genes (reviewed in reference 16) that can be grouped into three classes based on their similarity to the S. cerevisiae HDACs Rpd3p, Hda1p, and Sir2p (13, 19, 27, 33). While class I and II deacetylases share sequence features in the catalytic domain, the class II HDACs contain additional sequences that are absent in class I HDACs. Class II HDACs also display different tissue distributions, while expression of class I HDACs appears to be ubiquitous (13, 19). Class III deacetylases are exemplified by Sir2 which has a unique property in that it requires NAD as a cofactor for its enzymatic activity (27, 33, 44). On the basis of sequence homology, humans are predicted to have seven class III deacetylase genes (reviewed in reference 16). Caenorhabditis elegans has three class I, four class II, and four class III HDACs. Recently, HDA-1, a class I histone deacetylase, has been shown to collaborate with POP-1 and UNC-37, a C. elegans Groucho homolog, in the repression of end-1, an early E-cell fate-determining gene during C. elegans embryogenesis (5).
The developmental roles of HDACs are just beginning to be identified. Generation of germ line clones of a strong hypomorphic allele of Drosophila Rpd3, a class I deacetylase, results in embryonic lethality, highlighting a specific role for RPD3 in segmentation control during embryogenesis (36). Furthermore, in C. elegans, RNA-mediated interference (RNAi) (12) of maternal and zygotic expression of the C. elegans homolog of Rpd3p, HDA-1, results in embryonic lethality. Although cells that form muscle, hypoderm, and intestine are present and appear to be terminally differentiated, the embryos nevertheless die prior to elongation (43; P. Dufourcq and Y. Shi, unpublished results). In addition to its role in embryogenesis, recent RNAi studies have suggested a possible postembryonic function for HDA-1 in C. elegans vulval development (35, 45).
Here, we report the identification and results of analyses of an hda-1 genetic mutant. We provide genetic, molecular, and biochemical evidence that this hda-1 mutant is the previously isolated gon-10 mutant (named for gonadogenesis defective 10). Phenotypic analysis of gon-10 animals revealed multiple developmental defects in gonadogenesis and vulval development. The gonadogenesis defects are characterized by the lack of organized somatic gonad structures, which suggests that these abnormalities may be due to defects in tissue morphogenesis. Hermaphrodite gon-10 animals also display a protruding or everted vulva and often develop multivulvae as a result of hyperinduction of vulval cells. Since gonadogenesis and vulval development are regulated by Notch and Ras, respectively (reviewed in reference 26), our findings suggest that hda-1 may be a component of these two signaling pathways. Consistent with this hypothesis, we have identified lag-2, a gene that encodes a homolog of the Notch ligand Delta, as a potential target for HDA-1. In summary, we provide compelling evidence that a ubiquitous histone deacetylase plays specific roles in a number of critical developmental decision processes in C. elegans.
MATERIALS AND METHODS
Strains and genetics.
C. elegans was cultivated as described previously (3). The following mutant alleles and/or strains were used: gon-10(e1795)/DnT1V; YS40 dpy-11(e224) gon-10(e1795)/unc-76(e911)V; YS47 gon-10(e1795)/unc-76(e911); dpy-11(e224)V; unc-76(e911)V; YS52 dpy-11(e224) gon-10(e1795)/unc-76(e911); bmEx1 [phda-1 (wild-type hda-1 gene) pPD97.93 (myo-3::gfp)]; lin-15(n433); and lin-15(n744) (9). The presence of class A (lin-15A) or B (lin-15B) mutations in the context of the double mutant were confirmed by soaking larval stage 1 (L1) heterozygote gon-10(e1795)/unc-76(e911); lin-15(n433) or gon-10(e1795)/unc-76(e911); lin-15(n744) in lin-15B RNAi and lin-15A RNAi, respectively. The following genes were used in expression studies: egl-26::gfp (23); lag-2::gfp (14), lim-7::gfp (22), flp-8::gfp (K. Kim and C. Li, personal communication.), egl-17::gfp (4), cdh-3::gfp (40), lip-1::gfp (2), myo-3::gfp (pPD97.93) (A. Fire, S. Xu, J. Ahnn, and G. Seydoux, personal communication), sur-5::gfp (21), and let-858::gfp (29).
Isolation of gon-10(e1795).
A mutation induced by ethyl methanesulfonate, originally designated gon-10(e1795), was isolated while screening for abnormal sexual development and mapped to a location on linkage group V, between the egl-41 and unc-76 loci. Hermaphrodites homozygous for e1795 were sterile and maintained as heterozygotes. gon-10(e1795) animals were outcrossed five times; the gon-10(e1795) gene was marked by dpy-11(e224) and balanced by unc-76(e911). In order to score for progeny, larvae, or eggs, adult heterozygote hermaphrodites were allowed to lay eggs for a short period of time, and progeny were examined through time course analysis.
Transformation rescue of gon-10(e1795).
Nested PCR primers were used to isolate the hda-1 gene using cosmid D1027 (canonical form of cosmid C53A5) as a template. The PCR primers were designed on the basis of the sequence information from cosmid C53A5. The outer primer pair is 3361S/6946R (TGCCTCAAAGAGCTTTCCTACG/CATCCAACATCAGATGAAGACAGAC), and the inner primer pair is 3881S/6799R (TTCAACATCGTGAGAGCGTGG/CGACATAAACGATGTCAACTGC). Transgenic lines were established as described previously (37). To perform rescue experiments, we marked gon-10(e1795) with dpy-11. Since dpy-11 is closely linked to gon-10(e1795), we used the Dpy phenotype as an indicator for homozygosity of the gon-10 locus; homozygote dpy-11(e224) gon-10(e1795) animals are phenotypically Dpy, characterized by short and fat physical appearance. If rescue is complete, the sterile homozygous gon-10(e1795) animals with the Dpy phenotype are expected to become fertile. Hermaphrodites were injected with a mixture of test DNA and a green fluorescence protein (GFP) marker pPD97.93 myo-3::gfp at 20 ng/μl. GFP was used to monitor the germ line transmission frequency of the hda-1 transgene. For all transgenic lines, the transmission rate was approximately 60%.
RNAi.
RNAi experiments were performed as described previously (12). yk109d9 (hda-1 cDNA) served as a template for the production of double-stranded hda-1 RNA using an in vitro transcription kit (Promega). Young adult animals were injected and allowed to lay eggs for 6 to 12 h. Five percent of the animals from eggs laid during the first 6 h survive through adulthood, while 100% of the animals from eggs laid during the second 6 h die during embryogenesis.
RT-PCR analysis.
Reverse transcription-PCR (RT-PCR) analysis was performed to compare the mRNA levels corresponding to either the control sc35 gene (34) or the gfp and lag-2 genes in wild-type N2, lag-2::gfp, and lag-2::gfp; gon-10 transgenic lines. Thirty hermaphrodites (L4 larvae) were collected in Trizol reagent (Gibco BRL Life Technologies). After three cycles of freeze-thawing, total RNA was isolated. RT was performed with 500 ng of total RNA using the Superscript II RNase H− Reverse Transcriptase (Gibco BRL) and oligo(dT) primers (Gibco BRL) following the manufacturer's instructions. Three sets of primers were used for PCR as follows: sc35 F, 5′ CAATGGTCTAACTTCGCTG 3′; sc35 R, 5′ TATCTTGGAGATCTGGAGC 3′; GFP F, 5′ GTAAAGGAGAAGAACTTTTCACTGG 3′; GFP R, 5′ GTATAGTTCATCCATGCCATG 3′; lag-2 F, 5′ CGCTGTGACATCGGATGGATGG 3′; and lag-2 R, 5′ GATGGAGAAGATCACGAAGAGAGC 3′. The optimal number of cycles and amount of RT products used for the PCR were determined in preliminary experiments (not shown). Once the semiquantitative conditions were set up, the RT products were submitted to amplification with the different sets of primers. The samples were then subjected to analysis on an ethidium bromide-stained agarose gel.
Generation of HDA-1 polyclonal antibody and immunofluorescence.
Rabbit polyclonal antiserum was raised against amino acids 374 to 460 of HDA-1 (now available at Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.). The purified antibody recognized a single band at the expected position of 50 kDa (data not shown). In addition, the antibody detected strong immunoreactive signals in wild-type embryos but not in hda-1(RNAi) embryos (not shown).
To synchronize larvae, 20 N2, YS40, or YS47 hermaphrodites were allowed to lay eggs for 4 h before being transferred to new plates; eggs and larvae were subsequently harvested at specific time points as needed. Adult animals were fixed as described previously (10). Anti-HDA-1 antibody was used at a 1/500-fold dilution, and the monoclonal antibody MH27, which recognizes adherens junctions (15), was used at a 1/2,000-fold dilution.
RESULTS
Animals in which zygotic hda-1 expression is inhibited display phenotypes resembling those of gon-10(e1795) animals.
Previous studies have shown that inhibition of both maternal and zygotic hda-1 expression by RNAi results in highly penetrant embryonic lethality (43). Here, we present an investigation of the zygotic function of HDA-1. We focused on the progeny derived from the first batch of eggs laid from the worms injected with double-stranded hda-1 RNA. These embryos contain maternally provided HDA-1 protein, therefore allowing us to analyze the consequence of selective inhibition of zygotic HDA-1 expression by RNAi. The resultant embryos hatched and reached adulthood but were sterile and displayed multiple defects in gonadogenesis. A prominent phenotype is the lack of organized somatic gonad structures. Using the hda-1 zygotic phenotypes as a guide, we searched for an hda-1 mutation candidate among previously isolated gonadogenesis-defective mutations that have been mapped to the region near the hda-1 gene on chromosome V (C. elegans Data Base). This phenotype-based search identified gon-10 (named for gonadogenesis defective 10) as a potential hda-1 mutation. The hda-1(RNAi) and gon-10(e1795) homozygote mutant animals had similar defects in gonadogenesis and vulval development (detailed below). These phenotypic similarities suggest that the gon-10(e1795) mutant may be a hda-1 mutant.
The gon-10(e1795) mutant is an hda-1 mutant.
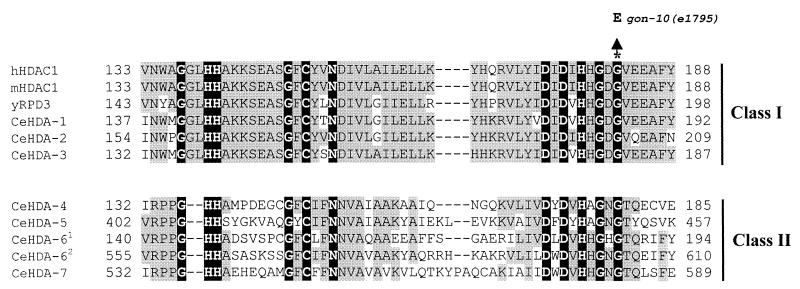
To explore the hypothesis that the gon-10(e1795) mutant is an hda-1 mutant, we sequenced the hda-1 gene isolated from five independent gon-10(e1795) homozygote animals. We found that the hda-1 gene carried a single base change in the coding region which converts a conserved glycine (G) residue to glutamic acid (E) within the catalytic domain of HDA-1 (Fig. 1). No mutations were detected in the promoter region (approximately 1.5 kb) or in the 3′ end of the gene (0.8 kb). This finding suggests the possibility that the hda-1 mutation may underlie the defects observed in gon-10(e1795) animals.
FIG. 1.
A point mutation in the hda-1 gene from gon-10(e1795) mutant animals is located in the conserved region of the protein. The sequences of the predicted C. elegans HDA-1 to HDA-7 proteins (CeHDA-1 to CeHDA-7) and HDA-1 homologs in humans (hHDAC1), mice (mHDAC1), and yeast (yRPD3) were aligned using the ClustalX program (47). Amino acids conserved in all species and C. elegans class I and class II histone deacetylases are shown on a black background. In each class of histone deacetylases, any amino acid that is conserved in more than three deacetylases is shown on a gray background. CeHDA-61 and CeHDA-62 refer to the first and second catalytic domain of HDA-6, respectively. The amino acid positions of the first and last amino acids of the catalytic domain are given to the sides of the sequences. The position of the G186E point mutation found in the gon-10(e1795) mutant and introduced into the human HDAC1 is indicated by the asterisk. Dashes represent gaps introduced to maximize sequence alignment. The HDAs and the open reading frames are as follows: CeHDA-1, C53A5.3; CeHDA-2, C08B11.2; CeHDA-3, R06C1.1; CeHDA-4, Y51H1A.5; CeHDA-5, F43G6.4; CeHDA-61, F41H10.6A; CeHDA-62, F41H10.6B; CeHDA-7, C10E2.3.
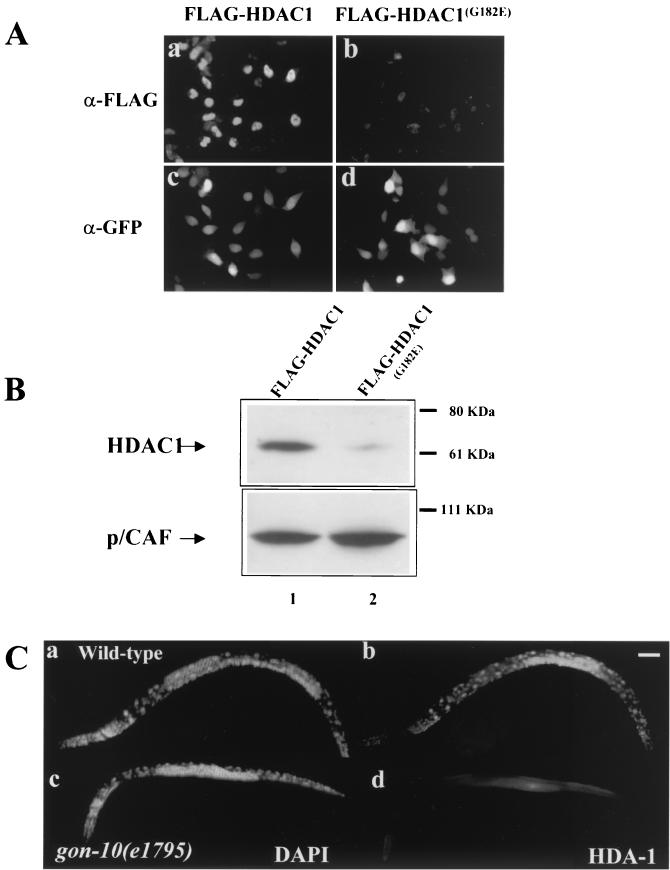
Since the HDA-1G186E mutation is within the predicted catalytic region of HDA-1 (11), it is possible that HDA-1 enzymatic activity is affected. To test this, we asked whether the same mutation would affect the activity of human HDAC1, the homolog of HDA-1, in mammalian cells. We introduced the mutation into human HDAC1 (HDAC1G182E) and transfected the expression plasmid into HeLa cells. Surprisingly, we detected drastically reduced levels of the mutated protein by immunostaining or Western blotting. As shown in Fig. 2A, while the FLAG epitope-tagged wild-type HDAC1 is easily detectable by immunostaining, the mutant protein is detected at a significantly lower level (Fig. 2A, compare panels a and b). To identify transfected cells and to control for transfection efficiency, a plasmid encoding the GFP was cotransfected (Fig. 2A, panels c and d). In parallel, we compared the levels of the wild-type and mutant HDAC1G182E proteins by Western blotting. FLAG-tagged p/CAF was cotransfected and used as a control. As indicated in Fig. 2B, the same result was obtained, i.e., the mutant HDAC1 protein is detected at a significantly reduced level (compare lanes 1 and 2).
FIG. 2.
The G-to-E mutation significantly reduces expression of both human and C. elegans HDAC1. (A) Immunostaining of wild-type and mutant human HDAC1 proteins in transfected HeLa cells. The wild-type and mutant HDAC1G182E proteins were tagged with FLAG at the C terminus, cotransfected with a GFP-expressing plasmid into HeLa cells, and visualized by using an anti-FLAG antibody (α-FLAG). The transfected cells were identified by a GFP polyclonal antibody that recognizes the cotransfected GFP (α-GFP). (B) Western blot analysis of wild-type and mutated human HDAC1 proteins in transfected HeLa cells. Protein extracts from HeLa cells expressing the wild-type (lane 1) or mutated version of human HDAC1 (lane 2) were probed with an anti-FLAG antibody. Cotransfected, FLAG-tagged p/CAF protein was used as controls. The FLAG-tagged HDAC1 and p/CAF proteins can be distinguished by their molecular masses. The positions of molecular mass markers are indicated to the right of the blots. (C) Expression of C. elegans HDA-1 in the wild type and gon-10(e1795) mutants. Wild-type and mutant L4 worms were stained with the HDA-1 polyclonal antibody (b and d) and costained with 4′,6′-diamidino-2-phenylindole (DAPI) (a and c). For a control for immunostaining, the same animals were found to be stained positive for the monoclonal antibody MH27 (data not shown). Bar, 50 μm.
We subsequently examined HDA-1 expression in both wild-type and gon-10(e1795) mutant animals. In the wild-type C. elegans, we found HDA-1 in the germ line, indicating that it is a maternally provided protein (data not shown). We also found HDA-1 expressed ubiquitously throughout embryonic and postembryonic development (Fig. 2C, panel b; also data not shown). HDA-1 immunoreactivity was found in virtually all cell types except sperm (Fig. 2C, panel b). HDA-1 was localized in the nucleus with the exception of the germ line including the oocytes where the signal was also detected in the cytoplasm. Unlike wild-type animals, HDA-1 was virtually undetectable in the gon-10 homozygous mutants (Fig. 2C, panel d). Therefore, the same G-to-E mutation causes a drastically reduced level of HDA-1/HDAC1 expression in both C. elegans and mammalian cells. This finding is consistent with the hypothesis that a severe reduction of HDA-1 protein may be the molecular basis for the phenotypes observed for gon-10(e1795) animals.
We predicted that if gon-10(e1795) is an hda-1 mutant, a wild-type copy of the hda-1 gene would rescue the gonad and sterile phenotype. To accomplish this, three independent transgenic lines were generated using a 3.3-kb hda-1 gene fragment which includes 1.8 kb of the promoter region, the entire coding region, and 0.54 kb of the 3′ sequence. The gon-10 mutation was marked with the recessive marker dpy-11 (see Materials and Methods). We asked whether the transgenic F2 Dpy animals were fertile. In the absence of the hda-1 transgene, 6% (22 of 324) of the Dpy animals laid eggs as a result of recombination between the dpy-11 and gon-10 loci. However, in the presence of the hda-1 transgene, the fertility of Dpy animals rose to 54% (97 of 181). This increase is in line with the germ line transmission frequency of the hda-1 transgene (60%). Significantly, from the gon-10(e1795) homozygous but fertile animals that carry the hda-1 transgene (dpy-11 gon-10; bmEx1 [phda-1 (wild-type hda-1 gene) pPD97.93 (myo-3::gfp)]), we were able to establish three independent lines that are viable and fertile for at least three generations. These results show that a wild-type copy of the hda-1 gene can rescue gon-10(e1795) defects. Interestingly, HDA-1 with a point mutation (HDA-1H145F) predicted to abrogate the catalytic activity (24) failed to rescue gon-10, suggesting that the enzymatic activity of HDA-1 is important for its biological function.
Taken together, our findings show that HDA-1 protein is present at nearly undetectable levels in the gon-10(e1795) animals due to a point mutation in the hda-1 gene. This suggests that the gon-10(e1795) mutation causes a severe loss of function. In support of this interpretation, heterozygote animals carrying a copy of the gon-10 mutation in trans with a deficiency covering the hda-1 locus display phenotypes identical to those of the gon-10 homozygotes or animals in which zygotic hda-1 expression is selectively inhibited by RNAi. Thus, the result is consistent with the hypothesis that gon-10(e1795) is likely a genetic null hda-1 mutation.
The lack of zygotically expressed gon-10 HDA-1 causes defects in gonadogenesis and vulval development.
To investigate the function of HDA-1 in C. elegans development, we analyzed the development of mutant hermaphrodites. Surprisingly, the gon-10(e1795) mutant exhibited a restricted role for HDA-1 in specific developmental processes. gon-10(e1795) homozygote animals complete embryogenesis and mature to adulthood presumably due to the maternal supply of the HDA-1 protein. However, adult animals are sterile with a number of interesting gonadal and vulval phenotypes. The main phenotypes are summarized in Table 1 and described below.
TABLE 1.
Summary of developmental defects in gon-10(e1795) animals
| Tissue | Defect | % of animals with defectsa |
|---|---|---|
| Gonad | ||
| Somatic | Shorter proximal gonad armb | 99 (163/164) |
| Shorter distal gonad armc | 100 (164/164) | |
| No spermatheca | 100 (164/164) | |
| Germ line | Fewer oocytes | 82 (56/68) |
| No oogenesis | 16 (11/68) | |
| Vulva | Protruding vulva | 100 (>300/>300) |
| Multivulvae | 20 (16/82) |
The number of animals with defects/total number of animals studied is shown in parentheses.
Abnormal early turn of gonadal arms.
Abnormal distal parts of the gonadal arms not properly elongated.
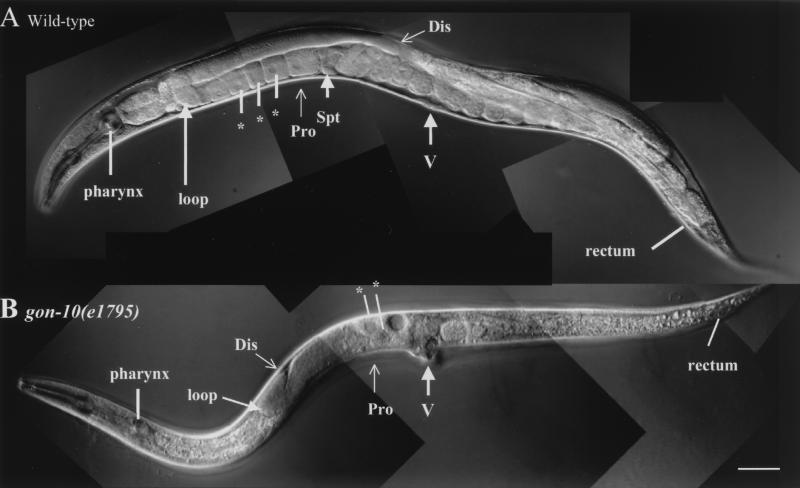
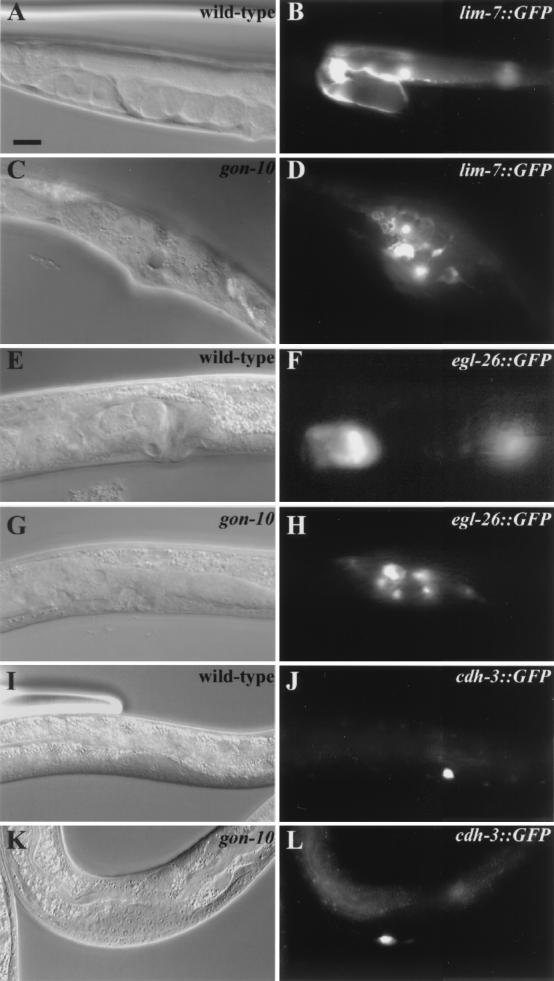
We found that gonadogenesis is severely affected in the gon-10(e1795) homozygote animals (Fig. 3). Although cells necessary for the formation of the somatic gonad appear to be present and differentiated, the corresponding tissues are nevertheless severely disorganized (Fig. 4 and data not shown). This is best illustrated through our analysis of GFP reporter expression in somatic gonad tissues. The lim-7::gfp reporter marks 16 of the 20 terminally differentiated sheath cells in a wild-type gonad. We found that all 16 sheath cells were present in the gon-10(e1795) homozygotes. However, instead of being located along the sheath, as in the wild-type animals, they remain located centrally in the region of the vulva (Fig. 4, compare panels B and D). Similarly, improper localization of the spermatheca cells, as seen with reporter egl-26::gfp, was observed (Fig. 4, compare panels F and H). Compared with the somatic gonad tissues, germ cell development does not appear to be as grossly affected. We detected both mitotic and meiotic populations of the germ cells in gon-10(e1795) animals (data not shown). However, the number of germ cells was reduced by a third (380 versus 500; n = 3), fertilized eggs were rarely found (for eggs with fewer than 100 cells, data not shown), and none of the homozygous mutants were fertile (n > 500). It is unclear whether the germ cell defects are direct or indirect consequences of the gon-10(e1795) mutation.
FIG. 3.
HDA-1 is necessary for hermaphrodite gonadogenesis and vulval development. Differential interference-contrast Nomarski images of wild-type and gon-10(e1795) adult hermaphrodites. The distal (Dis) and proximal (Pro) parts of the gonad, oocytes in the proximal half of the gonad (asterisks), spermatheca (Spt), and vulva (V) are indicated. The distal part of the gonad is not fully elongated and fewer developing oocytes are located in the region of the vulva in the gon-10 (e1795) animals compared to the wild-type animals. No visible gonadal somatic structure, such as the spermatheca and uterus, are detected in the mutant animals. Note the protruding vulva in the gon-10(e1795) animal. The anterior part of the animal is on the left, while the posterior part is on the right. Bar, 50 μm.
FIG. 4.
Abnormal somatic tissue organization in gon-10(e1795) animals. Paired differential interference-contrast Nomarski and fluorescence images of the wild-type and gon-10(e1795) mutant animals expressing GFP reporter transgenes are shown. lim-7:: GFP was used as a sheath cell marker (A and B versus C and D), egl-26:: GFP was used as a marker of the spermatheca (E and F versus G and H), and cdh-3:: GFP was used to visualize the anchor cell (I and J versus K and L). Animals are oriented so that the anterior part is to the left. Bar, 10 μm.
gon-10(e1795) affects vulval induction.
In addition to gonadogenesis defects, vulval development is affected by the gon-10(e1795) mutation. For instance, 20% of gon-10(e1795) animals display a multivulva (Muv) phenotype (Table 1). In wild-type animals, vulva formation is induced by the anchor cell. Thus, one possible explanation for the observed Muv phenotype could be the presence of supernumerary anchor cells. However, this does not appear to be the case, as the wild-type number of anchor cells (1 cell, n = 50) is present in the mutant (1 cell, n = 68), as observed using the anchor cell reporter cdh-3::gfp (Fig. 4J and L).
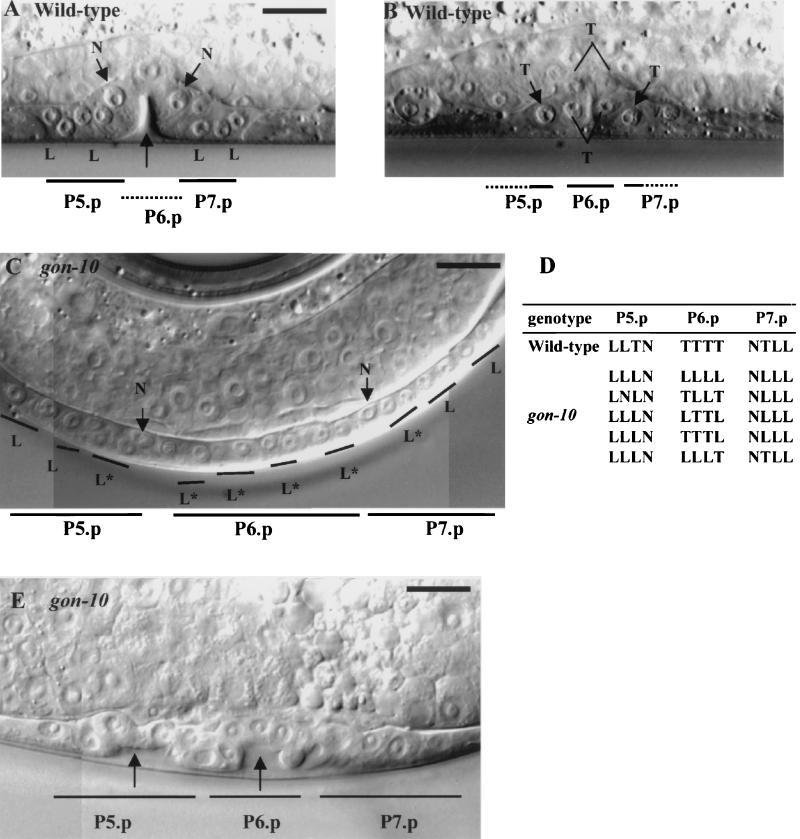
The vulva is derived from three of six potential vulval precursor cells (VPCs) (P5.p to P7.p cells) through a series of stereotypically oriented cell divisions. Whereas these VPCs undergo two longitudinal divisions, the third division is either longitudinal or transversal; 2 of the 12 cells resulting from the second division do not divide further (42). All gon-10(e1795) hermaphrodites display what appears to be a protruding vulva (Fig. 3B). Lineage analyses in gon-10(e1795) revealed that the first two rounds of cell division are oriented longitudinally, as in the wild type. However, cells that divide transversally during the third division in the wild type sometimes divide instead along the longitudinal axis in the mutant (Fig. 5C and D). Therefore, HDA-1 activity appears to be specifically required either for preventing the longitudinal division or promoting the transverse division during the final round of vulval cell division. In gon-10(e1795) animals, the abnormal division orientation is often accompanied by ectopic invagination in the region of the descendants of P5.p to P7.p cells (Fig. 5A and E).
FIG. 5.
Abnormal orientation of cell division during vulval development in gon-10(e1795) animals. The orientation of the division is longitudinal (L) or transversal (T); cells which do not divide (N) are indicated. (A and B) Stereotypical divisions of P5.p to P7.p cells result in a three-dimensional vulval structure with cells in two focal planes in wild-type animals. The unlabeled, black arrow in panel A points to an invagination. (C) Defective transverse divisions in gon-10(e1795) animals leave all descendants of P5.p to P7.p cells in the same focal plane. Abnormal transverse divisions are indicated by asterisks. (D) Orientation of the third division of P5.p to P7.p cells in wild-type and gon-10(e1795) animals. (E) Abnormal invagination in gon-10(e1795). Two invaginations are indicated by the unlabeled, black arrows. Bar, 10 μm.
In addition to the lineage defect, approximately 20% of the gon-10(e1795) animals have multivulvae, suggesting a role for HDA-1 in vulval induction. Vulval development in C. elegans includes positive regulation by Ras signaling and negative regulation by a set of genes termed the synMuv genes (for synthetic multivulva) (8). Worms carrying double mutations of a synMuvA gene and a synMuvB gene result in synthetic Muv phenotypes (9). Previous studies using RNAi suggested that HDA-1 is a member of the synMuv family, either solely as a synMuvB gene or as both a synMuvA and synMuvB gene (35, 45). We tested the role of HDA-1 in the synMuv pathway by constructing double mutants carrying gon-10(e1795) and either a synMuvA lin-15A(n433) or synMuvB lin-15B(n744) gene. As indicated in Table 2, we failed to observe an increase in the induction of VPCs in the double mutants over that for gon-10(e1795) single mutants. This observation together with the finding that 20% of the gon-10(e1795) animals already displayed a Muv phenotype suggest that hda-1 is not a classical synMuv gene.
TABLE 2.
Vulval induction phenotype caused by gon-10(e1795) and genetic interactions between gon-10 (e1795) and synMuv mutanta
| Genotype | No. of induced VPCs | No. of animals examined |
|---|---|---|
| Wild-type | 3.0 | 15 |
| lin-15A(n433) | 3.0 | 15 |
| lin-15B(n744) | 3.0 | 15 |
| gon-10(e1795)b | 3.2 | 39 |
| gon-10(e1795); lin-15A(n433)b | 3.2 | 28 |
| gon-10(e1795); lin-15B(n744)b | 3.2 | 24 |
The average number of induced VPCs for each genotype is shown. The state of morphogenesis was assessed by counting the number of induced VPCs (normally three in the wild type) at the L4 stage.
Homozygote gon-10(e1795) animals are easily distinguished from heterozygotes by their abnormal gonad.
lag-2 is derepressed in gon-10(e1795) homozygotes.
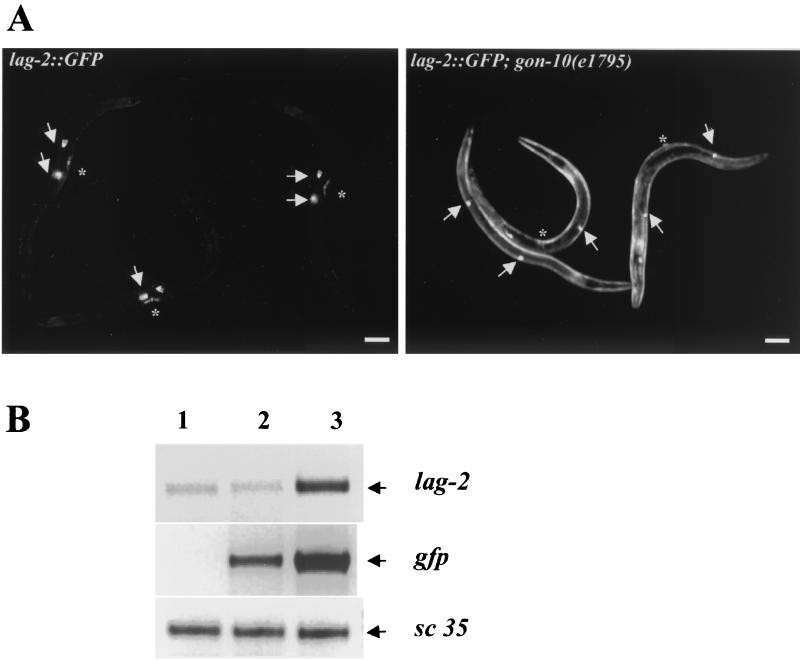
To begin to address the molecular mechanisms underlying the gon-10(e1795) phenotypes, we screened for potential target genes of HDA-1 by looking for the ectopic activation of various cell type-specific promoters fused to gfp reporter transgenes in the gon-10(e1795) background. The expression of the majority of these reporter transgenes was not detected outside the cells normally expressing the transgenes (data not shown). A notable exception is the lag-2 promoter which is misregulated in the mutant background (Fig. 6). lag-2 encodes a Delta/Serrate homologue, a ligand of the Notch receptor family which has previously been implicated in somatic gonad cell fate decisions (49) and in germ cell (25, 46) and vulval development (31) in C. elegans. As shown in Fig. 6A, in wild-type animals the expression of the lag-2 promoter-driven GFP is restricted to the distal tip cells and a few cells of the vulva. However, this promoter was globally derepressed in gon-10(e1795) animals (Fig. 6A, right panel). Derepression of the lag-2::gfp gene was observed using three independent lines of transgenic animals where the reporter gene was either integrated on different chromosomes (two lines) or not integrated.
FIG. 6.
lag-2 is derepressed in the gon-10(e1795) mutant. (A) The lag-2:: GFP transgene is derepressed in gon-10 mutants. lag-2:: GFP transgene expression in wild-type animals (left panel) and gon-10(e1795) animals (right panel). Arrows indicate the locations of the distal tip cell, and the asterisks mark the position of the vulva. Bar, 50 μm. (B) The endogenous lag-2 promoter is derepressed in gon-10 animals. Total RNA was isolated from wild-type N2, lag-2::gfp, and lag-2::gfp; gon-10(e1795) transgenic lines. RT-PCR experiments were performed to determine the levels of the endogenous lag-2 mRNA and gfp mRNA in the wild-type N2 (lane 1), lag-2::gfp (lane 2), and lag-2::gfp; gon-10 transgenic (lane 3) animals. Both the endogenous and ectopic lag-2 promoters were derepressed (compare lanes 2 and 3). The expression of sc35, a ubiquitous mRNA, was used as an internal control.
Using RT-PCR, we also compared the levels of gfp transcripts in lag-2::gfp transgenic animals with and without the mutation. As expected, gfp expression was significantly increased in mutant animals (Fig. 6B, compare lanes 2 and 3), which is consistent with the results for GFP activity in the lag2::gfp transgenic animals. We next asked whether the endogenous lag-2 promoter was similarly regulated by HDA-1 by comparing the levels of lag-2 transcripts in wild-type and gon-10(e1795) animals. As shown in the top panel of Fig. 6B, a low level of endogenous lag-2 was detected in wild-type N2 animals (lane 1) and in the lag2::gfp transgenic animals (lane 2), probably due to the limited number of cells expressing lag-2. However, the level of lag-2 transcripts was significantly elevated in gon-10(e1795) animals (lane 3), suggesting that this gene is derepressed in the absence of HDA-1 function. For a control, sc35, a ubiquitously expressed constitutive splicing factor (34), was found to be expressed at comparable levels in both wild-type and mutant animals (Fig. 6B). The finding that lag-2 is derepressed in gon-10(e1795) suggests that lag-2 may be a direct target of HDA-1 repression. Furthermore, these results implicate deregulation of Notch/Delta signaling in the gonad phenotype of gon-10(e1795) mutant animals.
DISCUSSION
HDA-1 has previously been shown to play a critical role during early development, as animals depleted of the maternal and zygotic HDA-1 die as embryos (43). In this study, we provide evidence that zygotic HDA-1 is required during gonadogenesis and vulval development. We have identified a mutation in the hda-1 gene as the cause of the gonadogenesis and vulval defects of the previously isolated C. elegans mutant gon-10(e1795) animals. This conclusion is based on the following observations. (i) gon-10(e1795) worms carry an hda-1 gene with a point mutation that results in a change of a conserved amino acid within the catalytic domain of HDA-1. (ii) This point mutation is correlated with nearly undetectable levels of HDA-1 expression in gon-10(e1795) homozygotes. (iii) A wild-type copy of the hda-1 gene can fully rescue the developmental defects associated with the gon-10 mutant. Although gon-10(e1795) animals lack zygotic HDA-1, they express the two other highly related class I HDACs, HDA-2 and HDA-3 (unpublished result). Interestingly, the presence of HDA-2 and -3 in the reproductive system does not substitute for the loss of HDA-1, indicating that zygotic HDA-1 plays a highly specific role in gonadogenesis. We also provide evidence that HDA-1 plays a role in the control of cell division orientation of the vulval precursor cells during vulval development. Taken together, these findings identify a surprisingly specific developmental requirement for an ubiquitously expressed histone deacetylase.
HDA-1 and C. elegans gonadogenesis.
Gonadogenesis in C. elegans involves the development of both germ cells and somatic gonad tissues. Somatic gonadogenesis involves two morphogenic processes, the extension of tissue buds that elongate and form the C. elegans bilobal gonad long arms and the formation of complex, differentiated epithelial tubes composed of distinct modular units, i.e., the uterus, spermatheca, and sheaths in hermaphrodites and the vesicle and vas deferens in males (30). One salient feature of the gonadogenesis phenotype in gon-10(e1795) animals is the lack of organized somatic gonad tissues. Using cell type-specific promoter-driven GFP genes as markers, we were able to visualize the presence of the various differentiated cells necessary for the formation of these tissues. Thus, the lack of organized somatic tissue is probably due to defects in tissue morphogenesis. Taken together, these findings suggest that genes whose products are important for cell-cell communication and cell polarity may be targets of HDA-1. It will be interesting to identify HDA-1 target genes during gonadogenesis to test this hypothesis.
Compared with the somatic gonad tissue defects, the germ line development appeared to be affected to a lesser extent in gon-10(e1795) mutants. Both mitosis and meiosis appear to take place, and as a result we could identify sperm and oocytes by Nomarski analysis. However, a reduction in the number of meiotic cells in the mutants was observed. At present, it is unclear whether germ line defects are direct or indirect consequences of the mutation. In our rescue experiments, we introduced the wild-type gene into gon-10(e1795) animals as simple tandem arrays, which are susceptible to germ line silencing (29). We were nonetheless able to achieve efficient rescue in which the rescued lines were stable for several generations. Since a low level of germ line expression might be sufficient for rescue, these findings do not rule out the possibility that the germ line defects may also be a direct consequence of the gon-10(e1795) mutation.
HDA-1 and C. elegans vulval development.
Our analysis of gon-10(e1795) indicates that HDA-1 plays a crucial role in vulval development, a process in C. elegans which is known to be regulated positively by Ras signaling and negatively by the synMuv genes (named for synthetic multivulva) (8). Worms carrying combinations of two mutations of the synMuvA and synMuvB genes result in the synthetic multivulval phenotype (9). A number of transcription factors have been identified as synMuv genes, including the C. elegans homologs of E2F, DP1, and Rb (6, 35). Previous studies suggested that hda-1 acts as a synMuv gene (35, 45). However, we were unable to observe an increase in vulval cell induction when gon-10(e1795) was placed either in a synMuvA or synMuvB background. This discrepancy could be due to the fact that previous studies used RNAi, while this study analyzed a genetic mutant. We also cannot rule out the presence of a persisting maternal HDA-1 component or other histone deacetylase activities in the gon-10(e1795) mutant which might mask a synMuv phenotype. Interestingly, the lineage defect or morphogenic phenotype observed for gon-10(e1795) animals has also been reported for lin-40 metastasis-associated factor 1 (MTA) (7), which has been identified along with HDAC1 as a member of the NURD complex (32, 38). Further analyses are necessary to understand the precise molecular role of HDA-1 in vulval development.
Vulval development in C. elegans is also regulated by the LIN-12/Notch signaling pathways (reviewed in references 17 and 31). Through a mechanism of lateral inhibition, Notch signaling prevents certain VPCs from adopting the primary vulval cell fate. We show here that transcription of one of the Notch ligand-encoding genes, lag-2, is derepressed in gon-10(e1795) mutants, resulting in widespread expression of LAG-2. It is interesting to speculate that the abnormal expression of LAG-2 may contribute to the Muv phenotypes seen in gon-10(e1795) animals. It is possible that derepression of the expression of LAG-2 leads to overactivation of LIN-12/Notch signaling and mimics gain-of-function alleles of lin-12 which have already been shown to result in multivulval phenotypes (17, 18).
Corepressor complexes and development of the reproductive systems.
In mammals, class I HDACs such as HDAC1 and HDAC2 (which are both homologs of HDA-1) are components of multiple corepressor complexes. HDAC1 and -2 have been found to be present in at least two distinct biochemical complexes, i.e., the SIN3 and NURD/Mi-2 complexes (32, 38). Members of the NURD/Mi-2 complex, with the exception of MBD3, are conserved in C. elegans. Interestingly, in addition to HDA-1, two other members of the NURD/Mi-2 complex, the C. elegans MTA1 homolog LIN-40, and Mi-2 homologs LET-418 and CHD-3 have recently been shown to play a role in vulval development (7, 45, 48). The C. elegans SIN3 complex is less well understood, but at least two components of this complex, SIN3 (encoded by open reading frame F02E9.4) and SAP18 (encoded by open reading frame C16C10.4), are present in C. elegans (1). Preliminary experiments suggest that inhibition of SIN3 expression results in sterile animals (unpublished result) and therefore may play a role in either one of these two processes.
In addition, HDAC1 and -2 also interact with Rb and Groucho, both of which are corepressors and can be targeted to promoters via interactions with DNA-binding transcription factors (reviewed in reference 32). The C. elegans Rb homolog LIN-35 has been shown to play a role in vulval development (35): the role of the C. elegans Groucho homolog UNC-37 in gonadogenesis and vulval development is unknown. However, on the basis of biochemical results, we predict that the SIN3 complex and UNC-37 probably play a role in either one or both of these processes. Since HDA-1 is a component of a number of the different corepressor complexes discussed above, it is not surprising that the hda-1 mutation can affect both gonadogenesis and vulval development in postembryonic C. elegans, perhaps acting through distinct corepressor complexes.
In summary, we have provided compelling evidence that the gon-10(e1795) mutant is an hda-1 mutant. We have shown that mutation in this ubiquitous histone deacetylase causes surprisingly specific defects during C. elegans development, compromising the development of somatic gonad tissues (germ cells and the vulva). Our findings highlight the essential and specific roles ubiquitously expressed histone deacetylases play in a multicellular organisms and suggest possible important functions for HDAC-containing corepressor complexes in the development of reproductive systems of other organisms as well.
Acknowledgments
We thank Keith Blackwell, Patrick Blader, and Grace Gill for critical reading of the manuscript. We gratefully acknowledge Zhe Chen for help with the analyses of vulval development. We thank Yuji Kohara for providing the many expressed sequence tag clones used in this study, and we thank the C. elegans Data Base for providing information. Some nematode strains used in this work were provided by the Caenorhabditis Genetic Center.
This work was supported in part by a grant from the National Institutes of Health (GM58012) to Y.S. P.D. was supported by a fellowship from the Lalor Foundation. M.V. was supported by a fellowship from the DAAD (German Scientific Exchange Service) and the Taplin Foundation. The Caenorhabditis Genetic Center is supported in part by the National Institutes of Health's National Center for Research Resources.
REFERENCES
- 1.Ahringer, J. 2000. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16:351-356. [DOI] [PubMed] [Google Scholar]
- 2.Berset, T., E. F. Hoier, G. Battu, S. Canevascini, and A. Hajnal. 2001. Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science 291:1055-1058. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burdine, R. D., C. S. Branda, and M. J. Stern. 1998. EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development 125:1083-1093. [DOI] [PubMed] [Google Scholar]
- 5.Calvo, D., M. Victor, F. Gay, G. Sui, M. P. Luke, P. Dufourcq, G. Wen, M. Maduro, J. Rothman, and Y. Shi. 2001. A POP-1 repressor complex restricts inappropriate cell type-specific gene transcription during Caenorhabditis elegans embryogenesis. EMBO J. 20:7197-7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceol, C. J., and H. R. Horvitz. 2001. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol. Cell 7:461-473. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Z., and M. Han. 2001. Role of C. elegans lin-40 MTA in vulval fate specification and morphogenesis. Development 128:4911-4921. [DOI] [PubMed] [Google Scholar]
- 8.Fay, D. S., and M. Han. 2000. The synthetic multivulval genes of C. elegans: functional redundancy, Ras-antagonism, and cell fate determination. Genesis 26:279-284. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson, E. L., and H. R. Horvitz. 1989. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123:109-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finney, M., and G. Ruvkun. 1990. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63:895-905. [DOI] [PubMed] [Google Scholar]
- 11.Finnin, M. S., J. R. Donigian, A. Cohen, V. M. Richon, R. A. Rifkind, P. A. Marks, R. Breslow, and N. P. Pavletich. 1999. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401:188-193. [DOI] [PubMed] [Google Scholar]
- 12.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 13.Fischle, W., S. Emiliani, M. J. Hendzel, T. Nagase, N. Nomura, W. Voelter, and E. Verdin. 1999. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J. Biol. Chem. 274:11713-11720. [DOI] [PubMed] [Google Scholar]
- 14.Fitzgerald, K., and I. Greenwald. 1995. Interchangeability of Caenorhabditis elegans DSL proteins and intrinsic signaling activity of their extracellular domains in vivo. Development 121:4275-4282. [DOI] [PubMed] [Google Scholar]
- 15.Francis, R., and R. H. Waterston. 1991. Muscle cell attachment in Caenorhabditis elegans. J. Cell Biol. 114:465-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, S. G., and T. J. Ekstrom. 2001. The human histone deacetylase family. Exp. Cell Res. 262:75-83. [DOI] [PubMed] [Google Scholar]
- 17.Greenwald, I. 1998. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 12:1751-1762. [DOI] [PubMed] [Google Scholar]
- 18.Greenwald, I. S., P. W. Sternberg, and H. R. Horvitz. 1983. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34:435-444. [DOI] [PubMed] [Google Scholar]
- 19.Grozinger, C. M., C. A. Hassig, and S. L. Schreiber. 1999. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl. Acad. Sci. USA 96:4868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 21.Gu, T., S. Orita, and M. Han. 1998. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell. Biol. 18:4556-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall, D. H., V. P. Winfrey, G. Blaeuer, L. H. Hoffman, T. Furuta, K. L. Rose, O. Hobert, and D. Greenstein. 1999. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev. Biol. 212:101-123. [DOI] [PubMed] [Google Scholar]
- 23.Hanna-Rose, W., and M. Han. 2002. The Caenorhabditis elegans EGL-26 protein mediates vulval cell morphogenesis. Dev. Biol. 241:247-258. [DOI] [PubMed] [Google Scholar]
- 24.Hassig, C. A., J. K. Tong, T. C. Fleischer, T. Owa, P. G. Grable, D. E. Ayer, and S. L. Schreiber. 1998. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl. Acad. Sci. USA 95:3519-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson, S. T., D. Gao, E. J. Lambie, and J. Kimble. 1994. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120:2913-2924. [DOI] [PubMed] [Google Scholar]
- 26.Herman, T., and H. R. Horvitz. 1997. Mutations that perturb vulval invagination in C. elegans. Cold Spring Harbor Symp. Quant. Biol. 62:353-359. [PubMed] [Google Scholar]
- 27.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 28.Imhof, A., X. J. Yang, V. V. Ogryzko, Y. Nakatani, A. P. Wolffe, and H. Ge. 1997. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 7:689-692. [DOI] [PubMed] [Google Scholar]
- 29.Kelly, W. G., S. Xu, M. K. Montgomery, and A. Fire. 1997. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics 146:227-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimble, J., and D. Hirsh. 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70:396-417. [DOI] [PubMed] [Google Scholar]
- 31.Kimble, J., and P. Simpson. 1997. The LIN-12/Notch signaling pathway and its regulation. Annu. Rev. Cell Dev. Biol. 13:333-361. [DOI] [PubMed] [Google Scholar]
- 32.Knoepfler, P. S., and R. N. Eisenman. 1999. Sin meets NuRD and other tails of repression. Cell 99:447-450. [DOI] [PubMed] [Google Scholar]
- 33.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Sternglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longman, D., I. L. Johnstone, and J. F. Caceres. 2000. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19:1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, X., and H. R. Horvitz. 1998. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95:981-991. [DOI] [PubMed] [Google Scholar]
- 36.Mannervik, M., and M. Levine. 1999. The Rpd3 histone deacetylase is required for segmentation of the Drosophila embryo. Proc. Natl. Acad. Sci. USA 96:6797-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mello, C. C., J. M. Kramer, D. Stinchcomb, and V. Ambros. 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10:3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng, H. H., and A. Bird. 2000. Histone deacetylases: silencers for hire. Trends Biochem. Sci. 25:121-126. [DOI] [PubMed] [Google Scholar]
- 39.Pazin, M. J., and J. T. Kadonaga. 1997. What's up and down with histone deacetylation and transcription? Cell 89:325-328. [DOI] [PubMed] [Google Scholar]
- 40.Pettitt, J., W. B. Wood, and R. H. Plasterk. 1996. cdh-3, a gene encoding a member of the cadherin superfamily, functions in epithelial cell morphogenesis in Caenorhabditis elegans. Development 122:4149-4157. [DOI] [PubMed] [Google Scholar]
- 41.Roth, S. Y., and C. D. Allis. 1996. Histone acetylation and chromatin assembly: a single escort, multiple dances? Cell 87:5-8. [DOI] [PubMed] [Google Scholar]
- 42.Sharma-Kishore, R., J. G. White, E. Southgate, and B. Podbilewicz. 1999. Formation of the vulva in Caenorhabditis elegans: a paradigm for organogenesis. Development 126:691-699. [DOI] [PubMed] [Google Scholar]
- 43.Shi, Y., and C. Mello. 1998. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev. 12:943-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solari, F., and J. Ahringer. 2000. NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr. Biol. 10:223-226. [DOI] [PubMed] [Google Scholar]
- 46.Tax, F. E., J. J. Yeargers, and J. H. Thomas. 1994. Sequence of C. elegans lag-2 reveals a cell-signaling domain shared with Delta and Serrate of Drosophila. Nature 368:150-154. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Zelewsky, T., F. Palladino, K. Brunschwig, H. Tobler, A. Hajnal, and F. Muller. 2000. The C. elegans Mi-2 chromatin-remodeling proteins function in vulval cell fate determination. Development 127:5277-5284. [DOI] [PubMed] [Google Scholar]
- 49.Wilkinson, H. A., K. Fitzgerald, and I. Greenwald. 1994. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell 79:1187-1198. [DOI] [PubMed] [Google Scholar]