Abstract
We have introduced DNA methyltransferase 1 (Dnmt1) mutations into a mouse strain deficient for the Mlh1 protein to study the interaction between DNA mismatch repair deficiency and DNA methylation. Mice harboring hypomorphic Dnmt1 mutations showed diminished RNA expression and DNA hypomethylation but developed normally and were tumor free. When crossed to Mlh1−/− homozygosity, they were less likely to develop the intestinal cancers that normally arise in this tumor-predisposed, mismatch repair-deficient background. However, these same mice developed invasive T- and B-cell lymphomas earlier and at a much higher frequency than their Dnmt1 wild-type littermates. Thus, the reduction of Dnmt1 activity has significant but opposing outcomes in the development of two different tumor types. DNA hypomethylation and mismatch repair deficiency interact to exacerbate lymphomagenesis, while hypomethylation protects against intestinal tumors. The increased lymphomagenesis in Dnmt1 hypomorphic, Mlh1−/− mice may be due to a combination of several mechanisms, including elevated mutation rates, increased expression of proviral sequences or proto-oncogenes, and/or enhanced genomic instability. We show that CpG island hypermethylation occurs in the normal intestinal mucosa, is increased in intestinal tumors in Mlh1−/− mice, and is reduced in the normal mucosa and tumors of Dnmt1 mutant mice, consistent with a role for Dnmt1-mediated CpG island hypermethylation in intestinal tumorigenesis.
Cancer development is thought be driven by the acquisition of both mutations and abnormal methylation patterns (4, 38). Aberrant hypermethylation of promoter CpG islands can lead to transcriptional silencing of key growth-controlling genes (4, 32, 38, 71). DNA methylation is also thought to play a role in the silencing of endogenous repetitive DNA elements such as transposons, retroviruses, LINE-1, and Alu sequences (30, 31, 42, 53, 82). The mechanism driving these methylation events is unclear, but it involves one or more of the DNA methyltransferase enzymes known to methylate genomic DNA at CpG sites.
DNA methyltransferase 1 (Dnmt1), Dnmt3a, and Dnmt3b are three functional DNA methyltransferase enzymes known to contribute to methylation patterns in mammalian cells. These enzymes have differential abilities to catalyze maintenance and de novo methylation (61). In vitro methylation studies have shown that while Dnmt1 has a strong predilection for hemimethylated DNA, Dnmt3a and Dnmt3b have approximately equivalent preferences for hemimethylated and unmethylated DNA substrates (61). This has led to the widely held concept that Dnmt1 is predominantly responsible for maintenance methylation, while Dnmt3a and Dnmt3b are primarily involved in de novo methylation. Some expression studies have suggested that abnormal upregulation of DNA methyltransferase genes might cause aberrant methylation patterns and thus enhance cancer risk, but others have argued against this mechanism (17, 24, 35, 40, 48, 69). The normal activity of methyltransferases may be dependent upon their proper interaction with PCNA through a conserved PCNA-binding motif (11, 83, 84), localization to replication foci during S phase (51), and cell cycle regulation (70). Deregulation of these activities may lead to irregular methylation patterns.
In this study we used a mouse model system to assess the interaction between DNA methylation and DNA mismatch repair deficiency. Cells lacking functional mismatch repair are subjected to 100- to 1,000-fold increases in microsatellite instability (MSI+) and base substitution rates throughout the genome due to the inability to repair replication errors that create base-base and loop mismatched heteroduplex DNA (74). Targeted mutations have been generated for mismatch repair genes Msh2, Msh3, Msh5, Msh6, Mlh1, Pms1, and Pms2 in mice (3, 15, 20, 21, 23, 64, 66). Mutant mice are prone to the development of a spectrum of tumors, in addition to a meiotic defect resulting in infertility (2, 3, 14, 20, 21). However, the mutation spectrum, tumor phenotype, and meiotic phenotype are not always manifest, nor are they the same in all mismatch repair mutant animals, indicating that these genes have distinct yet overlapping functions. Loss-of-function mutations in genes MLH1, MSH2, and MSH6 are the most commonly observed mismatch repair gene mutations in colorectal cancer patients, suggesting that disruption of early steps in the DNA mismatch repair process causes a predisposition to colorectal tumorigenesis.
A role for DNA methylation in the mismatch repair pathway has been proposed (5, 29), possibly involving an interaction between DNA methyltransferase and PCNA (11, 68) and/or the assistance of methylation patterns in mismatch recognition and correction through MBD4 recruitment (5), but direct evidence in support of these mechanisms is lacking. In Escherichia coli, Dam methylation at GATC sites serves as the strand discrimination for the incision and excision of the DNA strand containing the mismatched base (54). However, model organisms such as Saccharomyces cerevisiae and Caenorhabditis elegans have functional mismatch repair despite having no detectable DNA methylation. It is speculated that the orientation of the passing replication fork complex and/or the presence of transient DNA nicks after replication may act as strand discrimination signals in these organisms (77, 78). Whether CpG hemimethylation participates as a signal for strand discrimination in mammalian cells has not been resolved.
Abnormal methylation patterns have been well documented in colorectal cancer (25, 27, 75, 86). Hypermethylation of the MLH1 promoter has been shown to be the basis for the inactivation of the MLH1 mismatch repair gene in the majority of the 10 to 15% of sporadic colorectal cancer cases that exhibit an MSI+ phenotype (33, 81). Several studies have reported that mismatch repair deficiency is associated with a generalized CpG island methylation phenotype (1, 75). It has also been reported that MSI+ colorectal cell lines have a higher de novo methylation activity towards exogenous retroviral sequences than those that are MSI− (50). However, we did not find an increased frequency of CpG island hypermethylation in MSI+ colorectal cell lines (63) or colorectal tumors (86). We have recently suggested that the association between CpG island hypermethylation and mismatch repair deficiency is limited to the association with MLH1 promoter hypermethylation (86). It has been reported that patients with MSI+ cases show loss of imprinting of IGF2 (13, 56). The significance of this observation is poorly understood, but it suggests that DNA methylation and mismatch repair pathways may be intertwined.
In this study, we have devised a genetic strategy based on Dnmt1 hypomorphic alleles to study the effect of DNA methylation on tumor development (45). This system avoids the toxicity associated with the use of 5-aza-2′-deoxycytidine (37). Here, we made use of two hypomorphic alleles which, when introduced in combination, allow for manipulation of the levels of expression of Dnmt1 in vivo without compromising development (52). We applied this experimental system to a mouse Mlh1 knockout strain that is predisposed to the development of multiple tumors, including lymphomas and intestinal adenocarcinomas (3). We show that hypomorphic alleles of Dnmt1 can lead to decreased RNA expression, hypomethylation of centromeric repeat sequences, and reduced hypermethylation of promoter regions containing CpG islands. Paradoxically, a reduction in Dnmt1 expression concomitantly reduced the incidence of intestinal cancers and increased the incidence of aggressive lymphoid tumors in mismatch repair-deficient mice. Our data suggest that in the intestinal epithelium, Dnmt1 may contribute to cancer by causing CpG island hypermethylation. Further, we suggest that the mechanism by which lymphomagenesis is induced by DNA hypomethylation is distinct and possibly could involve a combination of molecular defects, including the lack of mismatch repair, a global change in gene expression patterns with upregulation of repetitive retroelements, and an increased chromosomal instability as a result of a hypomethylated genome.
MATERIALS AND METHODS
Genotyping.
We used PCR to genotype the Mlh1 gene as described previously by Baker et al. (3). The PCR was performed in a 15-μl reaction mixture with 20 mM Tris-HCl (pH 8.4); 50 mM KCl; 1.5 mM MgCl2; 0.2 mM deoxynucleoside triphosphates; 0.3 μM primers MLH1-a, MLH1-U, and MLH1-T5; and 0.35 U of Taq polymerase, using cycling conditions of 94°C for 4 min, followed by 35 cycles of 94°C for 50 s, 58°C for 50 s, and 72°C for 60 s, followed by 72°C for 5 min. The MLH1-a and MLH1-U primers produced a 258-bp amplicon, while the MLH1-a and MLH1-T5 primers produced a 198-bp amplicon, which were diagnostic for the wild-type and targeted alleles, respectively.
Dnmt1 was genotyped by multiplex PCR using primers OL106 (5′-GGGAACTTCCTGACTAGGGG-3′), OL168, (5′CCAACAAACCAGTATGTCTCGT-3′), OL173 (5′-CCCAGTTTCCAGAAAGCTACC-3′), and OL369 (5′-CAATTCCACACAACATACGAGC-3′). Reactions were carried out in a 15-μl volume with 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, a 0.3 μM concentration of each primer, and 0.35 U of Taq polymerase, using cycling conditions of 94°C for 4 min, followed by 35 cycles of 94°C for 50 s, 60°C for 50 s, and 72°C for 1 min 30 s, followed by 72°C for 5 min. OL168 and OL173 produced both a 342-bp wild-type-specific band and a 661-bp Dnmt1R allele-specific band. OL106 and OL173 produced a 430-bp Dnmt1N allele-specific band. OL173 and OL369 produced a second 211-bp Dnmt1R allele-specific band.
Examination of mice.
These experiments were conducted with F1 mice derived from two inbred strains. All of the alleles used in this study had been backcrossed to 129/svJae or C57BL/6 mice for at least 10 generations before the experimental crosses were set up. We limited our analysis to homozygous Mlh1−/− and Mlh1+/+ mice. Mlh1 heterozygous offspring were euthanized after genotyping. Experimental mice were monitored closely for 9 months, after which they were euthanized and autopsied. Mice that were moribund before 9 months were also euthanized and autopsied. Tumors that developed in these mice include lymphomas, skin tumors, intestinal adenomas, and adenocarcinomas. Tumor and tissue samples were dissected and snap frozen in liquid N2, as well as paraffin embedded for histology.
DNA isolation.
Genomic DNA was isolated from tumor samples and from tail biopsies of 4-week-old mice as described previously (47). Tissues were lysed in 100 mM Tris-HCl (pH 8.5)-5 mM EDTA-0.2% sodium dodecyl sulfate-200 mM NaCl-100 μg of proteinase K/ml overnight at 55°C with agitation, followed by isopropanol precipitation. The DNA was then spooled from the isopropanol and resuspended in 10 mM Tris-HCl-0.1 mM EDTA (47).
DNA recombination assays.
DNA rearrangement of the immunoglobulin and T-cell receptor loci was analyzed in Mlh1 wild-type and Mlh1−/− thymus, tail, and lymphoma tissues by PCR. We used seminested and nested PCR primers to analyze Dβ1Jβ1, Dβ1Jβ2, and Dβ2Jβ2 recombination at the T-cell receptor beta (TCRβ) locus (85). Immunoglobulin heavy-chain (IgH) DJ recombination was determined using degenerate PCR primers as described previously (73).
Bisulfite treatment and MethyLight analysis.
Genomic DNA was converted with sodium metabisulfite (Sigma) by using a modification of the agarose bead method (62). Denatured genomic DNA was suspended by being mixed with 4% Seakem agarose (FMC Corp.) to form a bead, equilibrated with 5 M bisulfite for 14 h at 50°C, washed, and desulfonated with 0.2 M NaOH. Agarose beads were melted, diluted, and stored at −20 degrees until used. Detection and quantitation of DNA methylation by MethyLight was performed as described previously (16-19). We examined methylation levels at 5 different CpG island sites. For each reaction, we used PCR primer and fluorescent probe combinations that contain CpG sites such that amplification occurs only if those CpG sites are methylated and only if the DNA is completely converted with bisulfite. The fluorescent signal for each reaction was adjusted for DNA input by normalizing to two independent reference reactions (Guca2 and Lhx1), for which the primer and probe sequences do not contain any CpGs and are therefore not dependent on the methylation status of the DNA. Methylation levels are expressed as a ratio of the input DNA-adjusted signal and the signal of a reference methylated DNA sample (16-19). This reference DNA was obtained by the in vitro methylation of genomic DNA derived from 129/sv J1 ES cell line (52) with Sss1 methylase (New England Biolabs) and subsequent bisulfite conversion. The primer and probe sequences used for MethyLight analysis were as follows: for Timp3, forward, 5′-GAGAGGCGGTGGGCGTAG-3′; reverse, 5′-CGAAAATATAAACTAAACGCGTCCT-3′; probe, 6FAM-5′-CGATATACGCTACAACGACGTCCCACGA-3′-TAMRA; for MyoD1, forward, 5′-CGTGTTTCGATTTATTAGATTTGCG-3′; reverse, 5′-CATCCTCACGAACGCCTAAAC-3′; probe, 6FAM-5′-CCACGTACACCAAACGCGAATCCA-3′-TAMRA; for Int4a, forward 5′-CGAGGTGTAGATCGAGGTTTCG-3′; reverse, 5′-AATACCCGATTAACACCCCGA-3′; probe, 6FAM-5′-ACAACATCACCGCTTCCCGAAAAACG-3′-TAMRA; for Mgmt, forward, 5′-CGACACCCTTACGTCACACACT-3′; reverse, 5′-TAGTTCGAGGGTGTAAAGCGG-3′; probe, 6FAM-5′-AACCACGCCCCGCGTACCAA-3′-TAMRA; for Cdkn2a, forward, 5′-CGCGAGGAAAGCGAATTC-3′; reverse, 5′-CGAAACCCCGACTTCCAA-3′; probe, 6FAM-5′-CCCGCACGTCATACACACGACCCT-3′-TAMRA; for Lhx1, forward, 5′-AGAGTGTTTGGAAGTTAGGTGAAGGT-3′; reverse, 5′-CACATTCATAAACACAAATTCACACAAC-3′; probe, 6FAM-5′-CACAATCAACATCCCAAACATATTCACCCA-3′-TAMRA; and for Guca2, forward 5′-GGTGTTGTGGTTTAGAAGGTTATGG-3′; reverse, 5′-ACCTTATCCTCAACTTCCAACATACC-3′; probe, 6FAM-5′-TCTCATCATCTTCTACAAACCAAAAC-3′-TAMRA. (Dyes used were 6-carboxyfluorescein [6 FAM] and 6-carboxy-tetramethylrhodamine [TAMRA]; CpG sites are underlined.)
RNA expression analysis.
We used real-time, fluorescence-based reverse transcription-PCR (RT-PCR) (Taqman) to measure gene expression (17). Total RNA was isolated using the guanidinium isothiocyanate method (Ambion Total RNA). RNA samples were then treated with DNase I to remove contaminating DNA (Ambion DNA-free). Samples were reverse transcribed with random primers and SuperScript II reverse transcriptase (Gibco-BRL) as specified by the manufacturer. Dnmt1 expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh), histone H4, and Pcna control reactions. For the lymphoma analyses, we analyzed 17 different transcripts, including 2 control RNA transcripts, Gapdh and histone H4. Specific gene expression was normalized to both Gapdh and histone H4 expression to correct for RNA input and averaged. The primer and probe sequences were as follows: for Dnmt1, forward, 5′-CCATGGTGCTGAAGCTCACA-3′; reverse, 5′-AGCACACCAAAGGTGCACTG-3′; probe, 6FAM-TAGCCCATGCGGACCAGGCAG-3′-TAMRA; for Bmi-1, forward, 5′-CTGGAGAAGAAATGGCCCAC-3′; reverse, 5′-CCCTCTGGTGACTTCATCTTCAT-3′; probe, 6FAM-5′-CCTTTGAAATACAGAGTTCGGCCAACTTGC-3′-TAMRA; for C-myc, forward 5′-ACAGCAGCTCGCCCAAATC-3′; reverse 5′-CGAGTCCGAGGAAGGAGAGA-3′; probe 6FAM-5′-ACCTCGTCCGATTCCACGGCC-3′-TAMRA; for N-myc, forward 5′-CTGGCGAGCTGATCCTCAA-3′; reverse, 5′-GGTGAGGGTGCAGCATAGTTG-3′; probe, 6FAM-5′-CGCTGTGTTCCCATCCATCAGCA-3′-TAMRA; for Pim-1, forward 5′-TTCGGCTCGGTCTACTCTGG-3′; reverse, 5′-CACGTGCTTAATGGCCACC-3′; probe, 6FAM-5′-ATCCGCGTCGCCGACAACTTG-3′-TAMRA; for Pim-2, forward, 5′-GAACCGTGTGCTAGGCTGGT-3′; reverse, 5′-ACAGCAGCGCAACCTCAAGT-3′; probe, 6FAM-5′-CACCGTGTCAGACTCAGTCACCTGCC-3′-TAMRA; for Frat1, forward, 5′-AGCTCGTGCTCTCGGGAA-3′; reverse, 5′-CTGTAGCTGTCGCGAATGAAGT-3′; probe, 6FAM-5′-TGCGCACCGCCTCCTTGATG-3′-TAMRA; for Frat3, forward, 5′-CTTCATTCGCGAGGGCTACA-3′; reverse 5′-CTGACAAAGGCCCGAGGA-3′; probe, 6FAM-5′-AGGCAAAGCTTCCCGCTCACTCG-3′-TAMRA; for Gfi1, forward, 5′-TGCCGCTCTCTGGACGA-3′; reverse 5′-CCAGCAAGACCGCTCCAT-3′; probe, 6FAM-5′-CCAGCCCTACACGCTGCCTTTCA-3′-TAMRA; for C-abl, forward, 5′-TCTCTTAGGAAGACCCGCCA-3′; reverse, 5′-GTCCAGAACCACACCCTTGG-3′; probe, 6FAM-5′-AGCGCATTGCCAGTGGCACCAT-3′-TAMRA; for Bcl2, forward, 5′-CATCTTCTCCTTCCAGCCTGA-3′; reverse 5′-ACGTCCTGGCAGCCATCTC-3′; probe, 6FAM-5′-AGCAACCCAATGCCCGCTGTG-3′-TAMRA; for C-fos, forward, 5′-CTTCTTGTTTCCGGCATCATC-3′; reverse, 5′-AGGTCCACATCTGGCACAGAG-3′; probe 6FAM-5′-AGGCCCAGTGGCTCAGAGACCTCC-3′-TAMRA; for IAP, forward, 5′-GAGTTTGAAGGCATCCTTGATACC-3′; reverse, 5′-TTGATATCCTAGGCCCTGTAATGAA-3′; probe, 6FAM-5′-CCCAAAGCATGGCCCACCACA-3′-TAMRA; for Line1, forward, 5′-ATAGCGCTGAGTGCCTCCA-3′; reverse, 5′-TTGGGTGAATTTGCTTCCTTTT-3′; probe, 6FAM-5′-TCAAGCTGCTAGTATGTGCTGTCTCCCGT-3′-TAMRA; for Arf, forward, 5′-GGTCGCAGGTTCTTGGTCA-3′; reverse, 5′-AAAACCCTCTCTTGGAGTGGG-3′; probe, 6FAM-5′-CCGCGCGCTGAATCCTCACA-3′-TAMRA; for Cdkn2a, forward, 5′-ACGACTGGGCGATTGGG-3′; reverse, 5′-TCCAGATGGCTCTCCTCGAGT-3′; probe, 6FAM-5′-CACTGAATCTCCGCGAGGAAAGCG-3′-TAMRA; for B1 elements, forward 5′-CACGCCTTTAATCCCAGCAC-3′; reverse, 5′-GCTGGCCTCGAACTCAGAAAT-3′; probe, 6FAM-5′-CGCCTGCCTCTGCCTCCCG-3′-TAMRA; for endogenous long terminal repeat (LTR), forward, 5′-TCTTGCTTCTTACACTCTTGCTCC-3′; reverse 5′-CAGGAAGAACACCACAGACCAG-3′; probe, 6FAM-5′-ATCTTCTGCGGCAAAACTTTATTGCTTACATCTT-3′-TAMRA; for GADPH, forward, 5′-TTGTCAAGCTCATTTCCTGGTATG-3′; reverse, 5′-GCCATGTAGGCCATGAGGTC-3′; probe, 6FAM-5′-CCACCCTGTTGCTGTAGCCGTATTCATT-3′-TAMRA; for histone H4, forward, 5′-TCTCCGGCCTCATCTACGAG-3′; reverse, 5′-CGGATCACGTTCTCCAGGA-3′; probe, 6FAM-5′-CACCTTCAGCACACCGCGGGT-3′-TAMRA; and for PCNA, forward, 5′-GAGCAACTTGGAATCCCAGAAC-3′; reverse, 5′-ATGTGGCTAAGGTCTCGGCATA-3′; probe, 6FAM-5′-TGCAAATTCACCCGACGGCATCTTTATTA-3′-TAMRA.
RESULTS
Effects of Dnmt1 hypomorphic mutations on expression and DNA methylation levels.
Five independent targeted mutations of the murine Dnmt1 gene have been described so far. These have been designated Dnmt1N (52), Dnmt1S (49), Dnmt1C (49), Dnmt1CHIP (76), and Dnmt11Δo (34), and they all differ in the targeted location within the gene and the degree of disruption of Dnmt1 activity. In this report, we made use of one of these mutations, Dnmt1N, and another Dnmt1 mutation that has not been previously described, which we refer to as Dnmt1R (for insertion to EcoRV site) (Fig. 1A). The Dnmt1N allele is known to be hypomorphic, with very low levels of RNA message and protein (49, 52). The residual protein is slightly reduced in size (49), suggesting that a modified Dnmt1 protein results from an alternative splicing event around the neo cassette insert. Homozygous Dnmt1N/N mouse embryos had 30% of their normal methyl cytosine content and died at mid-gestation (52). Since we had observed that Dnmt1S/R mice show reduced viability (data not shown), while Dnmt1N/R mice are fully viable, we hypothesized that the Dnmt1R allele may be expressed at a higher level than the Dnmt1N allele but at a lower level than the wild-type allele. This was investigated by quantitative RT-PCR analysis (Fig. 1B). The Dnmt1N/R mice show substantially lower expression levels than the Dnmt1N/+ mice, which supports this hypothesis. If the Dnmt1R allele is indeed a hypomorphic allele, then this could potentially result in detectably lower levels of genomic DNA methylation in combination with the other hypomorphic allele, Dnmt1N. Figure 1C shows a Southern blot analysis of genomic DNA obtained from tail biopsies, which was digested with the methylation-sensitive restriction enzyme HpaII and hybridized with a centromeric minor satellite repeat probe (8). Dnmt1R/+ and Dnmt1N/+ genomic DNA samples did not show detectable levels of hypomethylation relative to Dnmt1+/+ DNA. However, Dnmt1N/R DNA showed clear evidence of enhanced digestion of centromeric repeat sequences by HpaII, indicating that these sequences have reduced levels of methylation in these mice. We did not observe an effect of Mlh1 genotype on methylation levels (data not shown).
FIG. 1.
Hypomorphic alleles of the mouse Dnmt1 gene. (A) Schematic map (drawn approximately to scale) showing the positions of the Dnmt1R and Dnmt1N mutations in the 5′ region of the Dnmt1 genomic locus. The first four exons shown are represented as numbered black boxes, and intervening introns are represented as solid lines. The 320-bp insertion (43, 44) containing three copies of the lac operator (lacO) sequence from E. coli is located just upstream of an EcoRV site (R) in intron 3 and is depicted as an open box (O). H, HindIII. The Dnmt1N allele, which has been described previously (49, 52), contains a neomycin cassette that replaces a 900-bp NaeI (N) fragment. This mutation deletes part of exon 4 and causes an almost full disruption of Dnmt1 expression. (B) Dnmt1 expression in mutant mice as determined by real-time RT-PCR analysis. RNA was isolated from two thymus tissues of each genotype, and expression was normalized to expression of Gapdh, histone H4, and Pcna. The bars represent the mean value obtained for each of the normalized measurements shown on a relative scale. The error bars represent the standard error of the mean. The slope of the solid triangle below the graph schematically represents the relative Dnmt1 expression for each genotype. (C) DNA methylation of centromeric minor satellite repeats in Dnmt1 mutant mice. Genomic DNA from tail biopsies of Mlh1−/− mice was digested with methylation-sensitive HpaII (H) or with a methylation-insensitive isoschizomer, MspI (M), as a control and then probed with a fragment derived from pMR150 (8). Results for DNA from a Dnmt1C/C complete-knockout embryonic stem cell line are shown as a control (49). Lower-molecular-weight bands in the HpaII lanes indicate DNA hypomethylation. The slope of the solid triangle above the gels schematically represents the relative Dnmt1 expression for each genotype.
Dnmt1R/R mice were viable and developed normally, in contrast to Dnmt1N/N mice, which show extensive hypomethylation and embryonic lethality (52). Dnmt1N/R mutants were also viable and showed detectable hypomethylation at centromeric repeat sequences, although their DNA was not as severely hypomethylated as complete knockout Dnmt1C/C embryonic stem cell DNA (49) (Fig. 1C). This strongly indicates that expression, enzyme activity, or both are higher for Dnmt1R than Dnmt1N. Additionally, this confirms that while Dnmt1 activity is necessary for viability, mice with reduced expression still develop properly (46, 52). Therefore, we can rank genotypes Dnmt1+/+ > Dnmt1R/+ > Dnmt1N/+ > Dnmt1N/R > Dnmt1N/N in decreasing order of Dnmt1 expression.
Tumor incidence in Mlh1-deficient mice harboring Dnmt1 mutations.
Mice with Dnmt1 hypomorphic alleles were crossed with animals carrying a null mutation in the mismatch repair gene Mlh1 (3). These mismatch repair-deficient mice are prone to the development of a spectrum of tumors, including skin tumors, intestinal adenomas, and adenocarcinomas. 129Sv/Jae Mlh1+/− Dnmt1R/+ females were mated with C57/BL6 Mlh1+/− Dnmt1N/+ males to produce 618 offspring. Of these, we obtained 30% Dnmt1+/+, 26% Dnmt1R/+, 23% Dnmt1N/+, and 20% Dnmt1N/R (P = 0.09 by the chi-square test) and 25% Mlh1+/+, 56% Mlh1+/−, and 19% Mlh1−/− (P = 0.02 by the chi-square test). We observed that Mlh1−/− mice were underrepresented (19% [119 of 618]), in contrast to previous reports (3, 20). The 12 combinations of Mlh1 and Dnmt1 genotypes were obtained at expected frequencies (data not shown); this is consistent with the independent segregation of the two genes and indicates that the simultaneous absence of Mlh1 and reduced Dnmt1 expression does not adversely affect development in mice. We monitored F1 mice for 9 months, after which they were euthanized and assessed for tumors. We also euthanized mice that were moribund before 9 months and screened them for tumors that had developed (see Materials and Methods).
It has been suggested that DNA hypomethylation leads to an elevated rate of gene loss due to mitotic recombination and DNA rearrangement (10). We tested whether reduced Dnmt1 expression by itself would increase tumorigenesis by examining Mlh1+/+ mice with different Dnmt1 genotypes. We euthanized 16 Dnmt1+/+, 17 Dnmt1R/+, 20 Dnmt1N/+, and 22 Dnmt1N/R mice at 9 months, but autopsy failed to reveal tumors in any of these mice. Thus, it appears that the degree of hypomethylation in Dnmt1N/R DNA is insufficient to induce tumor formation in animals with proficient mismatch repair during the first 9 months.
We next assessed tumor susceptibility in mismatch repair-deficient Mlh1−/− animals. The Mlh1-deficient mice developed a spectrum of tumors similar to that described previously (22, 64). Lymphomas and intestinal tumors accounted for the vast majority of the observed tumors, but skin tumors were also seen in Mlh1−/− Dnmt1+/+ mice. Of 24 Mlh1−/− Dnmt1+/+ mice, 21% (5 of 24) developed aggressive lymphoid tumors, and 83% (20 of 24) developed either adenomas or adenocarcinomas in the intestinal tract by 9 months of age. One mouse developed a tumor of the uterine epithelium, and another had a skin tumor.
Tumor susceptibility in Mlh1-deficient mice was substantially modified when we crossed them with Dnmt1 mutant mice (Fig. 2A and B). There was a strong effect of Dnmt1 genotype on the frequency of intestinal tumors (Fig. 2A) and of lymphomas (Fig. 2B) in mice lacking Mlh1. In the intestinal epithelium, reduced Dnmt1 expression was associated with a diminished tumor burden (Fig. 2A). Figure 2A shows that at 9 months the cumulative frequency of intestinal tumors was 83% for Dnmt1+/+ mice, but it decreased to 61% for Dnmt1R/+ mice (P = 0.1) and to 41% for Dnmt1N/+ mice (P = 0.004). There were two mice of the Dnmt1N/R genotype that survived to 9 months, and they did not develop any detectable intestinal tumors. These results suggest that mice with reduced Dnmt1 expression are less likely to develop intestinal tumors than Dnmt1 wild-type littermates (P = 0.02 for trend). This reduced frequency of intestinal tumors in Dnmt1 hypomorphic mice is not just an indirect consequence of increased lymphomagenesis (see below). The intestinal tumor incidence in just the surviving mice at 9 months was also reduced (71% for Dnmt1+/+, 62% for Dnmt1R/+ [P = 0.36], 44% for Dnmt1N/+ mice [P = 0.04], and 0% for Dnmt1N/R mice [P = 0.02]). In contrast, lymphoma frequencies were dramatically elevated in these same mismatch repair-deficient Dnmt1 mutant animals. As shown in Fig. 2B, Dnmt1N/R mutants were at highest risk of developing lymphomas (90% by 9 months versus wild type; P < 0.0001 by unpaired t test). Dnmt1R/+ and Dnmt1N/+ mice also developed lymphomas at higher frequencies (P = 0.07 for trend). In addition, Dnmt1 mutant mice presented with lymphoma at a younger age than their Dnmt1+/+ littermates, suggesting an enhanced rate of tumor formation in the lymphoid lineage (Fig. 2C). Consistent with a previous report (64), we found that the majority of Mlh1−/− Dnmt1+/+ animals with lymphoma presented after 6 months. The mean age of onset of lymphomagenesis was significantly reduced in mice with any combination of Dnmt1 hypomorphic alleles. Interestingly, we found that the lymphoma frequency was higher in Dnmt1R/+ than in Dnmt1N/+ mice.
FIG. 2.
Effect of Dnmt1 mutations on the tumor incidence in Mlh1 deficient mice. (A) Effect of Dnmt1 genotype on the cumulative frequency of intestinal tumors in Mlh1-deficient mice by 9 months of age. The number of mice in each cohort is shown at the bottom. The statistical significance of the difference in tumor frequency for each mutant genotype versus the wild type was tested using chi-square analysis, with the P values shown at the bottom. (B) Effect of Dnmt1 genotype on the cumulative frequency of lymphomagenesis in Mlh1-deficient mice by 9 months of age. All F1 Mlh1-deficient and wild-type mice were sacrificed when they were moribund or when they reached9 months. The statistical significance of the difference in tumor frequency for each mutant genotype versus the wild type was tested using chi-square analysis, with the P values shown at the bottom. (C) Effect of Dnmt1 genotype on the age of onset of lymphomas. The horizontal lines represent the mean age at diagnosis for each genotype. The statistical significance of the difference in age of onset of each mutant genotype versus wild-type mice was tested using an unpaired t test, with the P values shown at the bottom.
Since this experiment was conducted in a syngeneic genetic background (F1; 129sv/Jae and C57BL6), the observed tumor predisposition is modified specifically by Dnmt1 genotype and not by strain differences. The effect on tumor frequency is tissue dependent. In the intestinal mucosa, the effect appears to be protective against adenomas and adenocarcinomas. In the lymphoid lineage, the tumor burden is enhanced by Dnmt1 mutations. To our knowledge, this is the first report showing that a reduction in Dnmt1 expression concomitantly exacerbates and diminishes tumor incidence in different tissues.
Effects of Dnmt1 genotype on survival of Mlh1-deficient mice.
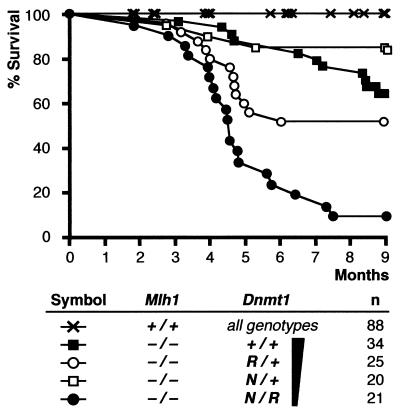
Mlh1-deficient mice are known to have reduced survival (64). We confirmed this in our study as well (Fig. 3). At 9 months, all of the 16 Mlh1+/+ Dnmt1+/+ mice we examined were still alive, but only 65% (22 of 34) of the Mlh1−/− Dnmt1+/+ mice had survived. Dnmt1 hypomorphic alleles showed an interesting interaction with Mlh1−/− homozygosity. Dnmt1N/R mice had the poorest survival due to the accelerated lymphomagenesis. Dnmt1 wild-type mice survived best early on, but they dropped below Dnmt1N/+ mice in survival rates later in life, due to their higher rates of intestinal tumorigenesis (Fig. 3). Our results suggest that lowering Dnmt1 expression levels can substantially affect longevity of Mlh1-deficient mice through modification of tumor burden risks. As Mlh1+/+ Dnmt1 mutant animals did not show reduced survival, a decrease in Dnmt1 expression by itself has little effect on longevity until it is coupled with additional molecular deficiencies such as a loss of mismatch repair.
FIG. 3.
Kaplan-Meier survival curves of Mlh1-deficient, Dnmt1 mutant mice up to 9 months. n, number of mice in each genotype cohort. Each group was monitored closely for 9 months, at which time mice were euthanized and examined on autopsy. Each point represents a death event for a mouse, which is then censored from further survival analysis. Mice removed from cohorts for any other reasons were also censored so that only the survival data for the remaining cohort is monitored.
Methylation analysis of DNA from intestinal tumors.
The reduced frequency of intestinal tumors in Dnmt1 hypomorphic mice suggests that Dnmt1, or its resulting methylation, may contribute to tumorigenesis in the intestinal mucosa. Dnmt1 could potentially participate in oncogenesis by mediating hypermethylation of promoter CpG islands (38). Hypermethylation of some genes, such as tumor suppressors, would be expected to predispose the intestinal mucosa to oncogenic transformation. The occurrence of occasional CpG island hypermethylation in the normal mucosa would thus be expected to predispose some cells to neoplastic transformation. Therefore, we used the sensitive MethyLight assay (16-19) to determine whether CpG island hypermethylation could be detected in the normal intestinal mucosa of Dnmt1 hypomorphic mice (Fig. 4B). The MethyLight assay is well suited for this analysis, since it can detect very low occurrences of fully methylated CpG islands (16-19). Such fully methylated CpG islands are expected most likely to result in transcriptional silencing of the associated gene. We chose a collection of CpG islands that included both genes implicated in oncogenesis (Cdkn2a) and those thought to be neutral to the process (Myod1), so that we could assess whether any observed CpG island hypermethylation was a generalized phenomenon or was restricted to genes providing a selective advantage (Fig. 4A). We isolated genomic DNA from normal mucosa or tumors, performed sodium bisulfite conversion, and analyzed methylation levels for the genes Myod1, Timp3, Itga4, Cdkn2a (p16), and Mgmt. We performed MethyLight analysis of DNA from normal mucosa of Dnmt1 mutant mice to determine if there were significant methylation differences in the normal mucosa between the various Dnmt1 genotypes (Fig. 4B). We found a trend of decreasing methylation in the Dnmt1 mutant mice at genes Myod1, Itga4, Cdkn2a, and Mgmt. CpG island hypermethylation was generally highest in the intestinal mucosa of Dnmt1 wild-type mice. The effect of Dnmt1 genotype on DNA methylation levels was most pronounced for MyoD1. At this locus, there was a strong, statistically significant difference in methylation between Dnmt1+/+ DNA and DNA from other Dnmt1 mutant genotypes. The greatest difference was seen between Dnmt1+/+ and Dnmt1N/R (P = 0.001 by the Mann-Whitney U test; nonparametric statistical tests were used, since the methylation data are not normally distributed). We also observed a significant decrease of methylation levels in the Dnmt1 mutant mice at Cdkn2a (P = 0.02 for Dnmt1+/+ versus Dnmt1R/+). We did not detect an effect of genotype on methylation levels at Timp3, suggesting that this locus is resistant to changes in Dnmt1 activity or that other methyltransferases contribute to methylation at this genomic location. Our results suggest that Dnmt1 contributes to methylation at these CpG islands in the normal mucosa, which could cause a predisposition to neoplastic progression. If this is indeed the case, then one would expect CpG island hypermethylation levels to be higher in the resulting intestinal tumors than in the originating normal mucosa, which consists of many cells without any or with lower levels of CpG island hypermethylation. We therefore investigated whether CpG island hypermethylation was elevated in the intestinal tumors of Mlh1-deficient mice with different Dnmt1 genotypes (Fig. 5).
FIG. 4.
MethyLight analysis of five CpG islands. (A) CpG density maps of five CpG islands analyzed by MethyLight. Individual CpG sites are indicated by vertical lines. Locations of MethyLight amplicons are indicated by solid boxes. Sequences are aligned relative to their respective transcriptional starts at position 0, indicated by the arrow pointing to the right. GenBank accession numbers are indicated to the right of the CpG density maps. (B) MethyLight analysis of CpG island hypermethylation in normal mucosal tissue. Genomic DNA from jejunum mucosal tissue was bisulfite converted and then used to measure CpG island hypermethylation. CpG island hypermethylation is expressed as percentage of methylated reference (PMR) value (19). The calculation of PMR values is described in Materials and Methods. Geometric means of the PMR values were calculated, since the methylation data are not normally distributed. The error bars represent the standard errors of the means. The statistical significance of the difference in CpG island hypermethylation for each gene of each mutant genotype versus the wild type was tested using the Mann-Whitney U test, with the P values shown at the bottom.
FIG. 5.
MethyLight analysis of CpG island hypermethylation at five sites in normal mucosa and intestinal tumors of Mlh1−/− mice. Genomic DNA from jejunum mucosal tissue or tumor tissue was bisulfite converted and then used to measure CpG island hypermethylation. CpG island hypermethylation is expressed as percentage of methylated reference (PMR) value (19). The calculation of PMR values is described in Materials and Methods. Geometric means of the PMR values were calculated, since the methylation data are not normally distributed. The error bars represent the standard errors of the means. The statistical significance of the difference in CpG island hypermethylation between normal and tumor tissue was tested separately for each gene and for each genotype using the Mann-Whitney U test, with the P values shown at the bottom.
We observed a tendency for tumor DNA to have higher levels of CpG island hypermethylation at three of the five genes analyzed. This is apparent at genes Timp3, Myod1, and Itga4, at which CpG island hypermethylation levels were higher in the tumor DNAs than in their corresponding normal mucosal samples. The difference between tumor and normal tissues was statistically significant for Timp3 and Itga4 for Dnmt1+/+ animals (P = 0.02 at Timp3 and P = 0.01 at Itga4). To our knowledge, this is the first demonstration of increased CpG island hypermethylation of intestinal tumors in mice. CpG island hypermethylation was also observed in tumors derived from Dnmt1R/+ and Dnmt1N/+ mice for the genes Timp3, Itga4, and Cdkn2a, although it was less pronounced than in Dnmt1+/+ tumors. We could not perform a similar test for the mice of the Dnmt1N/R genotype, because these animals did not develop intestinal tumors.
It is interesting that tumor CpG island hypermethylation decreased by Dnmt1 genotype, just as in the normal intestinal mucosa. This suggests that the low rate of tumorigenesis that does occur in Dnmt1 hypomorphic mice may rely less on CpG island hypermethylation. Presumably, other oncogenic mechanisms that are less dependent upon Dnmt1 expression predominate in mice with limiting amounts of Dnmt1. The reduced rates of intestinal tumorigenesis in Dnmt1 hypomorphic mice may be a consequence of the limited capacity for CpG island hypermethylation in these mice, although we do not have direct evidence for this. We observed a correlation between CpG island hypermethylation frequency and rates of intestinal tumorigenesis, but this is not proof of a causal link.
Analysis of lymphoid tumors from Mlh1-deficient mice.
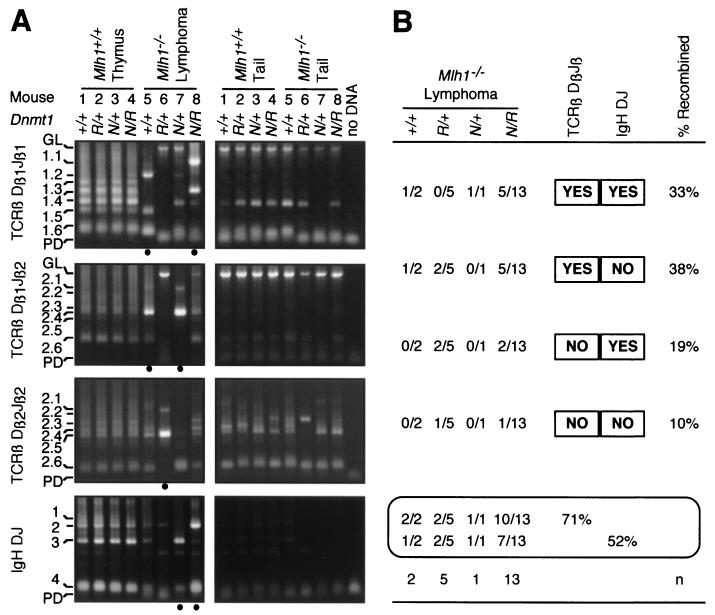
Previous studies have reported that mismatch repair-deficient mice are predisposed to the development of lymphomas, including T-cell lymphomas in Msh2 knockout mice and both B- and T-cell lymphomas in Msh6 mutant mice (15, 20). Mlh1 and Pms2 mutant mice also succumb to lymphomas, but their phenotypes were not reported (3, 20). The vast majority of lymphoid tumors that we observed in our mouse colony presented in the thymus, suggesting that they may be of the T-cell lineage. Splenomegaly was occasionally observed in conjunction with a thymoma. We characterized these lymphomas by using PCR to analyze DJ recombination at the TCRβ locus and at the IgH locus (Fig. 6). Out of 21 lymphoma samples tested, 19 (90%) showed a dominant recombined band (Fig. 6A) when tested for either TCRβ or IgH. DJ rearrangement at TCRβ was detected in 71% (15 of 21) of the tumors, while rearrangement at IgH was seen in 52% (11 of 21) of the tumors. This is consistent with the observation that the majority of these tumors developed in the thymus and would therefore be expected to be of the T-cell lineage. DJ recombination at these two loci was not mutually exclusive, as 33% (7 of 21) of these samples showed simultaneous DNA rearrangements at both TCRβ and IgH. Due to the small sample size, we could not determine if the Dnmt1 genotype had an effect on these recombination frequencies. We did not observe dominant recombined bands in any of the control thymus DNA or matched tail DNA samples (Fig. 6A) when assayed at either TCRβ or IgH.
FIG. 6.
Recombination of lymphoma DNA in Mlh1-deficient, Dnmt1 mutant mice. (A) Dβ1Jβ2 and Dβ1Jβ2 recombinations at the TCRβ locus and DJ recombination at the IgH locus were determined using PCR of genomic DNAs derived from various tissues, and products were separated on an agarose electrophoresis gel (85). Representative DNA samples from four Mlh1 wild-type thymus samples (lanes 1 to 4, left panels) and four lymphoma samples (lanes 5 to 8, left panels) at each Dnmt1 genotype and their matched control tail DNAs are shown. Lanes with thymus DNA show different possible DJ configurations, while some lanes with lymphoma DNA show one or two predominant configurations (indicated by a closed circle below the lane). Recombined product bands seen in the tail DNA lanes (right panel) are due to DNA from blood lymphocytes that contaminated the samples. GL, unrearranged, germ line configuration; PD, primer dimer. (B) Summary of DNA recombination in lymphoma in Mlh1-deficient mice for each Dnmt1 genotype.
We next investigated the mechanism by which reduced levels of Dnmt1 expression might have resulted in enhanced rates of lymphomagenesis in Mlh1-deficient mice. It does not appear that the effects of Dnmt1 hypomorphic alleles on the rates of lymphomagenesis are exerted primarily through CpG island hypermethylation, since lymphomagenesis is increased in these mice, while CpG island hypermethylation is decreased. Since retroviral infection and proviral activation of proto-oncogenes are known to cause lymphomas in mice (6, 79, 80), we hypothesized that Dnmt1 mutant mice might have increased rates of endogenous proviral activation, or increased expression of proto-oncogenes, due to reduced levels of promoter methylation. To test this, we analyzed the expression of 17 different genes and repetitive elements in lymphoma samples by using real-time fluorescence-based Taqman RT-PCR (Fig. 7). The level of expression of each of the 17 transcripts in the lymphomas was calculated relative to the expression level in corresponding Mlh1+/+ thymus control samples. We found that 10 out of 17 transcripts showed a significant difference in expression between lymphoma and thymus samples in at least one of the Dnmt1 genotypes. On average, many of the tested genes and repeat elements showed upregulation in lymphoma tissues. There was a significantly elevated expression of the C-abl, Bmi-1, C-myc, and N-myc genes. These genes (with the exception of C-abl) have been shown to be activated by proviral insertion in Moloney murine leukemia virus-induced lymphoma models (6, 79, 80). Our analysis did not reveal upregulation of some other oncogenes that have been shown to be overexpressed in lymphoma models (e.g., Gfi1 and Frat-1). Interestingly, we found that the tumor suppressor genes p19Arf and Cdkn2a both showed strongly increased expression levels in lymphoid tumors relative to control tissues. Bcl2 was the only gene to show a reduced level of expression in lymphomas compared to normal thymus.
FIG. 7.
Summary of genes induced (white boxes), repressed (black boxes), or unchanged (gray boxes) in lymphoid tumors from Mlh1-deficient mice. Gene expression levels of 17 transcripts were analyzed using real-time RT-PCR in lymphomas (the number is indicated at the bottom of the chart) and compared to the mean values for eight age-matched normal thymus tissues from Mlh1 wild-type mice. Gene expression was normalized to both Gapdh and histone H4 expression to correct for RNA input and averaged. The values shown in each box represent the relative increase or decrease in expression level in the lymphomas versus the control tissues.
Although the expression levels of IAP and endogenous LTR elements were modestly enhanced in Dnmt1R/+ and Dnmt1N/R mice, we did not observe a strong and convincing difference in gene upregulation between the different Dnmt1 genotypes. This implies that similar lymphomagenic pathways are utilized in mice with different Dnmt1 genotypes. Since the rate of lymphomagenesis is higher in Dnmt1 hypomorphic mice, this may indicate that the oncogenic pathways shared by these mice are activated more readily in animals with reduced levels of Dnmt1 expression but that the relative contribution of each of the pathways does not shift in the Dnmt1 hypomorphs. In other words, the nature of the lymphomagenic process has not been altered by reduced Dnmt1 expression, but the rate at which these processes are activated or proceed has increased.
DISCUSSION
Mice carrying germ line mutations of the Mlh1 mismatch repair gene have both an enhanced mutational burden (100- to 1,000-fold increase) and a greater tumor incidence (22, 64). Our results clearly demonstrate that hypomorphic mutations of Dnmt1 differentially affect neoplastic development in two separate tissue compartments, as was illustrated by an exacerbated tumor frequency in lymphoid tissues and a diminished tumor frequency in the intestinal epithelium in Dnmt1 mutant, Mlh1-deficient mice. The protection from intestinal tumors in Dnmt1 mutant animals may be a consequence of reduced CpG island-associated promoter hypermethylation, while the enhanced lymphomagenesis in the same animals may be the result of overexpression of endogenous retroviruses and/or proto-oncogenes or a consequence of an increase in genomic rearrangements.
It is generally thought that global DNA hypomethylation and silencing of growth control genes by promoter hypermethylation contribute to cancer progression (38). However, the causal role of these abnormal epigenetic changes in cancer development is sometimes unclear, since they could either directly contribute to neoplastic transformation or simply be a consequence of the malignant transformation. Promoter CpG island hypermethylation has been shown to interfere with transcription by causing the local chromatin to form a compact, transcriptionally repressed state through the recruitment of methyl-binding proteins, histone deacetylase, and corepressors (26, 39, 57-59, 67). Some studies involving mouse embryonic stem cells with reduced Dnmt1 expression and patients with immunodeficiency, centromeric region instability, and facial anomaly syndrome support the hypothesis that DNA hypomethylation can increase cancer risk by enhancing chromosomal instability (10, 28, 60, 87). However, we and others have also reported that DNA hypomethylation can suppress tumorigenesis and lead to reduced rates of gene inactivation (7, 12, 46, 55, 65). In light of our findings, one could argue that the effects of reduced methylation levels depend on the tissue origin of the tumor. Indeed, our results suggest that DNA hypomethylation can concomitantly enhance and diminish tumor development.
This study relies on the use of two Dnmt1 hypomorphic alleles. Although the molecular and phenotypic studies indicate that the expression from these alleles is below wild-type levels, the precise mechanism by which this is achieved is not fully clear. It appears that both of these hypomorphic mutations cause a reduced expression at the RNA level by interfering with efficient and correct splicing, leading to an accompanying decrease in protein levels. The transcript generated from the Dnmt1R allele appears to be normal in length and sequence composition but present at lower-than-wild-type levels.
Although we observed clear effects of Dnmt1 hypomorphic alleles on tumorigenesis in mismatch repair-deficient animals, we cannot conclude that this effect is a direct consequence of reduced levels of DNA methylation. It is now known that the Dnmt1 protein itself is associated with activities that are distinct from the enzymatic ability of Dnmt1 to methylate DNA (11, 26, 41, 67, 72). Therefore, the effect of reduced levels of Dnmt1 on tumorigenesis may be a consequence of alterations in the transcriptional repressive complexes associated with Dnmt1, rather than being due to changes in genomic DNA methylation levels.
We found a good correlation between predicted Dnmt1 expression levels and intestinal tumorigenesis (Fig. 2A). These results are consistent with previous studies documenting suppression of polyp multiplicity in ApcMin mice by Dnmt1 mutations and 5-azadeoxycytidine treatment (12, 46). On the other hand, we did not observe a perfect correlation between predicted Dnmt1 expression levels and lymphomagenesis, since the lymphoma incidence in Dnmt1N/+ mice was lower than that in the Dnmt1R/+ mice (Fig. 2B). This result could be due to a statistical aberration, or it may reflect a dominant activity by a truncated form of the Dnmt1 protein expressed by one of the hypomorphic alleles. Further experiments will be needed to investigate whether this result is reproducible, and if so, what the molecular mechanism is that is responsible for this finding.
We suggest that the mechanism of intestinal tumor suppression by Dnmt1 hypomorphic alleles is through a reduced frequency of CpG island hypermethylation in the normal mucosa and during intestinal tumor formation. We provide evidence that Dnmt1 mutant mice have a reduced frequency of CpG island hypermethylation in both normal mucosal and tumor DNAs, as determined for a limited number of CpG islands. We do not believe that the five loci that we analyzed are necessarily involved in intestinal tumor formation, but we suggest that the process of CpG island hypermethylation does occur in the mouse intestinal mucosa, that it is increased in intestinal tumors, and that it is suppressed in Dnmt1 hypomorphic mice.
Since Mlh1-deficient mice are already subjected to greatly elevated mutation rates, it seems unlikely that Dnmt1 insufficiency would contribute substantially to lymphomagenesis by further increasing the mutation rate. Rather, the observed acceleration of lymphomagenesis in Mlh1−/− mice by Dnmt1 hypomorphic alleles is likely to be due to an additive or synergistic effect of mismatch repair deficiency and DNA methyltransferase insufficiency through different mechanisms. In particular, our data do not provide evidence for a direct interaction between the mismatch repair process and DNA methylation, since functional mismatch repair is presumed to be absent in Mlh1−/− mice. It is possible that Dnmt1 mutations lead to an increased MSI by enhancing DNA polymerase slippage or misincorporation rates, but there is no evidence to support this. It seems more likely that the enhanced lymphomagenesis is a consequence of an increased rate of the same type of endogenous retroviral or proto-oncogene activation or chromosomal instability that occurs normally in Mlh1−/−-induced lymphomas. We did find evidence for increase expression of some proto-oncogenes and also of endogenous LTR elements and IAP in our lymphoid tumors. Endogenous IAP and related repetitive viral elements are normally silenced by dense methylation (82) but can be induced to reactivate by hypomethylation and thereby could afford higher rates of de novo integration which then could either disrupt genes or cause constitutive gene activation. However, since only a few lymphoid tumors per genotype were available for analysis, we could not definitively show that hypomethylation induces lymphomagenesis through an endogenous retroviral reactivation mechanism. An alternative view is that hypomethylation enhances lymphomagenesis by directly allowing proto-oncogenes to be expressed at higher levels. A recent study showed that up to 10% of genes are abnormally expressed in Dnmt1-deficient cells (36). Thus, the risk of developing lymphoma could be greatly exacerbated by a combination of mismatch repair deficiency, aberrant gene expression and the inherent chromosomal instability that is proposed to be associated with a hypomethylated genome (10, 28, 60, 87).
Finally, it is yet unclear if our observations can be readily extended to human cancer. The opposing effects of DNA hypomethylation on tumorigenesis are interesting and potentially relevant to human clinical trials of methyltransferase inhibitors. Inhibition of Dnmt1 activity could be effective in the prevention of colorectal cancer in at-risk individuals with familial adenomatous polyposis or hereditary nonpolyposis colorectal cancer. We had previously shown that reduced Dnmt1 expression can suppress polyp formation in a mouse model of familial adenomatous polyposis (46). In this study, we now also provide evidence that reduced Dnmt1 expression can inhibit intestinal tumor formation in a mouse model of hereditary nonpolyposis colorectal cancer. Drugs such as 5-azacytidine and 5-aza-2′-deoxycytidine are in clinical trials for the treatment of various hematopoietic and solid malignancies but are considered too toxic and mutagenic for long-term preventive therapy (37). Thus, development of less toxic inhibitors of Dnmt1 may be beneficial in the treatment and prevention of colorectal cancer. However, our finding of increased rates of lymphomagenesis in Dnmt1 hypomorphic mice suggests that long-term inhibition of DNA methyltransferases in humans may have an associated increased lymphoma or leukemia risk. Obviously, there are important differences between the mouse and human tumor systems. Nevertheless, our results offer new insights into the tissue-dependent balance of oncogenic effects of Dnmt1 expression levels.
Acknowledgments
This work was supported by NIH/NCI grant R01 CA 75090 to P.W.L.
We thank Michael Liskay for providing Mlh1 knockout mice.
REFERENCES
- 1.Ahuja, N., A. L. Mohan, Q. Li, J. M. Stolker, J. G. Herman, S. R. Hamilton, S. B. Baylin, and J. P. Issa. 1997. Association between CpG island methylation and microsatellite instability in colorectal cancer. Cancer Res. 57:3370-3374. [PubMed] [Google Scholar]
- 2.Baker, S. M., C. E. Bronner, L. Zhang, A. W. Plug, M. Robatzek, G. Warren, E. A. Elliott, J. Yu, T. Ashley, N. Arnheim, et al. 1995. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell 82:309-319. [DOI] [PubMed] [Google Scholar]
- 3.Baker, S. M., A. W. Plug, T. A. Prolla, C. E. Bronner, A. C. Harris, X. Yao, D. M. Christie, C. Monell, N. Arnheim, A. Bradley, T. Ashley, and R. M. Liskay. 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13:336-342. [DOI] [PubMed] [Google Scholar]
- 4.Baylin, S. B., and J. G. Herman. 2000. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 16:168-174. [DOI] [PubMed] [Google Scholar]
- 5.Bellacosa, A., L. Cicchillitti, F. Schepis, A. Riccio, A. T. Yeung, Y. Matsumoto, E. A. Golemis, M. Genuardi, and G. Neri. 1999. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc. Natl. Acad. Sci. USA 96:3969-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berns, A., N. van der Lugt, M. Alkema, M. van Lohuizen, J. Domen, D. Acton, J. Allen, P. W. Laird, and J. Jonkers. 1994. Mouse model systems to study multistep tumorigenesis. Cold Spring Harbor Symp. Quant. Biol. 59:435-447. [DOI] [PubMed] [Google Scholar]
- 7.Chan, M. F., R. van Amerongen, T. Nijjar, E. Cuppen, P. A. Jones, and P. W. Laird. 2001. Reduced rates of gene loss, gene silencing, and gene mutation in dnmt1-deficient embryonic stem cells. Mol. Cell. Biol. 21:7587-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman, V., L. Forrester, J. Sanford, N. Hastie, and J. Rossant. 1984. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature 307:284-286. [DOI] [PubMed] [Google Scholar]
- 9.Chen, D., C. L. Luongo, M. L. Nibert, and J. T. Patton. 1999. Rotavirus open cores catalyze 5′-capping and methylation of exogenous RNA: evidence that VP3 is a methyltransferase. Virology 265:120-130. [DOI] [PubMed] [Google Scholar]
- 10.Chen, R. Z., U. Pettersson, C. Beard, L. Jackson-Grusby, and R. Jaenisch. 1998. DNA hypomethylation leads to elevated mutation rates. Nature 395:89-93. [DOI] [PubMed] [Google Scholar]
- 11.Chuang, L. S., H. I. Ian, T. W. Koh, H. H. Ng, G. Xu, and B. F. Li. 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277:1996-2000. [DOI] [PubMed] [Google Scholar]
- 12.Cormier, R. T., and W. F. Dove. 2000. Dnmt1N/+ reduces the net growth rate and multiplicity of intestinal adenomas in C57BL/6-multiple intestinal neoplasia (Min)/+ mice independently of p53 but demonstrates strong synergy with the modifier of Min 1(AKR) resistance allele. Cancer Res. 60:3965-3970. [PubMed] [Google Scholar]
- 13.Cui, H., I. L. Horon, R. Ohlsson, S. R. Hamilton, and A. P. Feinberg. 1998. Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nat. Med. 4:1276-1280. [DOI] [PubMed] [Google Scholar]
- 14.de Vries, S. S., E. B. Baart, M. Dekker, A. Siezen, D. G. de Rooij, P. de Boer, and H. te Riele. 1999. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13:523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Wind, N., M. Dekker, A. Berns, M. Radman, and H. te Riele. 1995. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell 82:321-330. [DOI] [PubMed] [Google Scholar]
- 16.Eads, C. A., K. D. Danenberg, K. Kawakami, L. B. Saltz, C. Blake, D. Shibata, P. V. Danenberg, and P. W. Laird. 2000. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 28:E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eads, C. A., K. D. Danenberg, K. Kawakami, L. B. Saltz, P. V. Danenberg, and P. W. Laird. 1999. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 59:2302-2306. [PubMed] [Google Scholar]
- 18.Eads, C. A., R. V. Lord, S. K. Kurumboor, K. Wickramasinghe, M. L. Skinner, T. I. Long, J. H. Peters, T. R. DeMeester, K. D. Danenberg, P. V. Danenberg, P. W. Laird, and K. A. Skinner. 2000. Fields of aberrant CpG island hypermethylation in Barrett's esophagus and associated adenocarcinoma. Cancer Res. 60:5021-5026. [PubMed] [Google Scholar]
- 19.Eads, C. A., R. V. Lord, K. Wickramasinghe, T. I. Long, S. K. Kurumboor, L. Bernstein, J. H. Peters, S. R. DeMeester, T. R. DeMeester, K. A. Skinner, and P. W. Laird. 2001. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 61:3410-3418. [PubMed] [Google Scholar]
- 20.Edelmann, W., P. E. Cohen, M. Kane, K. Lau, B. Morrow, S. Bennett, A. Umar, T. Kunkel, G. Cattoretti, R. Chaganti, J. W. Pollard, R. D. Kolodner, and R. Kucherlapati. 1996. Meiotic pachytene arrest in MLH1-deficient mice. Cell 85:1125-1134. [DOI] [PubMed] [Google Scholar]
- 21.Edelmann, W., P. E. Cohen, B. Kneitz, N. Winand, M. Lia, J. Heyer, R. Kolodner, J. W. Pollard, and R. Kucherlapati. 1999. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21:123-127. [DOI] [PubMed] [Google Scholar]
- 22.Edelmann, W., K. Yang, M. Kuraguchi, J. Heyer, M. Lia, B. Kneitz, K. Fan, A. M. Brown, M. Lipkin, and R. Kucherlapati. 1999. Tumorigenesis in Mlh1 and Mlh1/Apc1638N mutant mice. Cancer Res. 59:1301-1307. [PubMed] [Google Scholar]
- 23.Edelmann, W., K. Yang, A. Umar, J. Heyer, K. Lau, K. Fan, W. Liedtke, P. E. Cohen, M. F. Kane, J. R. Lipford, N. Yu, G. F. Crouse, J. W. Pollard, T. Kunkel, M. Lipkin, R. Kolodner, and R. Kucherlapati. 1997. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell 91:467-477. [DOI] [PubMed] [Google Scholar]
- 24.El-Deiry, W. S., B. D. Nelkin, P. Celano, R. W. Yen, J. P. Falco, S. R. Hamilton, and S. B. Baylin. 1991. High expression of the DNA methyltransferase gene characterizes human neoplastic cells and progression stages of colon cancer. Proc. Natl. Acad. Sci. USA 88:3470-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feinberg, A. P., C. W. Gehrke, K. C. Kuo, and M. Ehrlich. 1988. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 48:1159-1161. [PubMed] [Google Scholar]
- 26.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes-Davies, and T. Kouzarides. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88-91. [DOI] [PubMed] [Google Scholar]
- 27.Goelz, S. E., B. Vogelstein, S. R. Hamilton, and A. P. Feinberg. 1985. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science 228:187-190. [DOI] [PubMed] [Google Scholar]
- 28.Hansen, R. S., C. Wijmenga, P. Luo, A. M. Stanek, T. K. Canfield, C. M. Weemaes, and S. M. Gartler. 1999. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. USA 96:14412-14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hare, J. T., and J. H. Taylor. 1985. One role for DNA methylation in vertebrate cells is strand discrimination in mismatch repair. Proc. Natl. Acad. Sci. USA 82:7350-7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hata, K., and Y. Sakaki. 1997. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene 189:227-234. [DOI] [PubMed] [Google Scholar]
- 31.Hellmann-Blumberg, U., M. F. Hintz, J. M. Gatewood, and C. W. Schmid. 1993. Developmental differences in methylation of human Alu repeats. Mol. Cell. Biol. 13:4523-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herman, J. G., and S. B. Baylin. 2000. Promoter-region hypermethylation and gene silencing in human cancer. Curr. Top. Microbiol. Immunol. 249:35-54. [DOI] [PubMed] [Google Scholar]
- 33.Herman, J. G., A. Umar, K. Polyak, J. R. Graff, N. Ahuja, J. P. Issa, S. Markowitz, J. K. Willson, S. R. Hamilton, K. W. Kinzler, M. F. Kane, R. D. Kolodner, B. Vogelstein, T. A. Kunkel, and S. B. Baylin. 1998. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc. Natl. Acad. Sci. USA 95:6870-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howell, C. Y., T. H. Bestor, F. Ding, K. E. Latham, C. Mertineit, J. M. Trasler, and J. R. Chaillet. 2001. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104:829-838. [DOI] [PubMed] [Google Scholar]
- 35.Issa, J. P., P. M. Vertino, J. Wu, S. Sazawal, P. Celano, B. D. Nelkin, S. R. Hamilton, and S. B. Baylin. 1993. Increased cytosine DNA-methyltransferase activity during colon cancer progression. J. Natl. Cancer Inst. 85:1235-1240. [DOI] [PubMed] [Google Scholar]
- 36.Jackson-Grusby, L., C. Beard, R. Possemato, M. Tudor, D. Fambrough, G. Csankovszki, J. Dausman, P. Lee, C. Wilson, E. Lander, and R. Jaenisch. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27:31-39. [DOI] [PubMed] [Google Scholar]
- 37.Jackson-Grusby, L., P. W. Laird, S. N. Magge, B. J. Moeller, and R. Jaenisch. 1997. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc. Natl. Acad. Sci. USA 94:4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones, P. A., and P. W. Laird. 1999. Cancer epigenetics comes of age. Nat. Genet. 21:163-167. [DOI] [PubMed] [Google Scholar]
- 39.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 40.Kautiainen, T. L., and P. A. Jones. 1986. DNA methyltransferase levels in tumorigenic and nontumorigenic cells in culture. J. Biol. Chem. 261:1594-1598. [PubMed] [Google Scholar]
- 41.Knox, J. D., F. D. Araujo, P. Bigey, A. D. Slack, G. B. Price, M. Zannis-Hadjopoulos, and M. Szyf. 2000. Inhibition of DNA methyltransferase inhibits DNA replication. J. Biol. Chem. 275:17986-17990. [DOI] [PubMed] [Google Scholar]
- 42.Kochanek, S., D. Renz, and W. Doerfler. 1995. Transcriptional silencing of human Alu sequences and inhibition of protein binding in the box B regulatory elements by 5′-CG-3′ methylation. FEBS Lett. 360:115-120. [DOI] [PubMed] [Google Scholar]
- 43.Labow, M. A. 1995. Use of lac activator proteins for regulated expression of oncogenes. Methods Enzymol. 254:375-387. [DOI] [PubMed] [Google Scholar]
- 44.Labow, M. A., S. B. Baim, T. Shenk, and A. J. Levine. 1990. Conversion of the lac repressor into an allosterically regulated transcriptional activator for mammalian cells. Mol. Cell. Biol. 10:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laird, P. W. 2000. Mouse models in DNA-methylation research. Curr. Top. Microbiol. Immunol. 249:119-134. [DOI] [PubMed] [Google Scholar]
- 46.Laird, P. W., L. Jackson-Grusby, A. Fazeli, S. L. Dickinson, W. E. Jung, E. Li, R. A. Weinberg, and R. Jaenisch. 1995. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 81:197-205. [DOI] [PubMed] [Google Scholar]
- 47.Laird, P. W., A. Zijderveld, K. Linders, M. A. Rudnicki, R. Jaenisch, and A. Berns. 1991. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 19:4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee, P. J., L. L. Washer, D. J. Law, C. R. Boland, I. L. Horon, and A. P. Feinberg. 1996. Limited up-regulation of DNA methyltransferase in human colon cancer reflecting increased cell proliferation. Proc. Natl. Acad. Sci. USA 93:10366-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lei, H., S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122:3195-3205. [DOI] [PubMed] [Google Scholar]
- 50.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1997. DNA methylation and genetic instability in colorectal cancer cells. Proc. Natl. Acad. Sci. USA 94:2545-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leonhardt, H., A. W. Page, H. U. Weier, and T. H. Bestor. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865-873. [DOI] [PubMed] [Google Scholar]
- 52.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 53.Liu, W. M., and C. W. Schmid. 1993. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res. 21:1351-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu, A. L., S. Clark, and P. Modrich. 1983. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc. Natl. Acad. Sci. USA 80:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacLeod, A. R., and M. Szyf. 1995. Expression of antisense to DNA methyltransferase mRNA induces DNA demethylation and inhibits tumorigenesis. J. Biol. Chem. 270:8037-8043. [DOI] [PubMed] [Google Scholar]
- 56.Nakagawa, H., R. B. Chadwick, P. Peltomaki, C. Plass, Y. Nakamura, and A. de La Chapelle. 2000. Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc. Natl. Acad. Sci. USA 19:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nan, X., F. J. Campoy, and A. Bird. 1997. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88:471-481. [DOI] [PubMed] [Google Scholar]
- 58.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 59.Ng, H. H., and A. Bird. 1999. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 9:158-163. [DOI] [PubMed] [Google Scholar]
- 60.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 61.Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19:219-220. [DOI] [PubMed] [Google Scholar]
- 62.Olek, A., J. Oswald, and J. Walter. 1996. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 24:5064-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pao, M. M., G. Liang, Y. C. Tsai, Z. Xiong, P. W. Laird, and P. A. Jones. 2000. DNA methylator and mismatch repair phenotypes are not mutually exclusive in colorectal cancer cell lines. Oncogene 19:943-952. [DOI] [PubMed] [Google Scholar]
- 64.Prolla, T. A., S. M. Baker, A. C. Harris, J. L. Tsao, X. Yao, C. E. Bronner, B. Zheng, M. Gordon, J. Reneker, N. Arnheim, D. Shibata, A. Bradley, and R. M. Liskay. 1998. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat. Genet. 18:276-279. [DOI] [PubMed] [Google Scholar]
- 65.Ramchandani, S., A. R. MacLeod, M. Pinard, E. von Hofe, and M. Szyf. 1997. Inhibition of tumorigenesis by a cytosine-DNA, methyltransferase, antisense oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA 94:684-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reitmair, A. H., R. Schmits, A. Ewel, B. Bapat, M. Redston, A. Mitri, P. Waterhouse, H. W. Mittrucker, A. Wakeham, B. Liu, et al. 1995. MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nat. Genet. 11:64-70. [DOI] [PubMed] [Google Scholar]
- 67.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 68.Robertson, K. D., and P. A. Jones. 1997. Dynamic interrelationships between DNA replication, methylation, and repair. Am. J. Hum. Genet. 61:1220-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robertson, K. D., K. Keyomarsi, F. A. Gonzales, M. Velicescu, and P. A. Jones. 2000. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res. 28:2108-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robertson, K. D., E. Uzvolgyi, G. Liang, C. Talmadge, J. Sumegi, F. A. Gonzales, and P. A. Jones. 1999. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 27:2291-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robertson, K. D., and A. P. Wolffe. 2000. DNA methylation in health and disease. Nat. Rev. Genet. 1:11. [DOI] [PubMed] [Google Scholar]
- 72.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 73.Sollbach, A. E., and G. E. Wu. 1995. Inversions produced during V(D)J rearrangement at IgH, the immunoglobulin heavy-chain locus. Mol. Cell. Biol. 15:671-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strand, M., T. A. Prolla, R. M. Liskay, and T. D. Petes. 1993. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365:274-276. [DOI] [PubMed] [Google Scholar]
- 75.Toyota, M., N. Ahuja, M. Ohe-Toyota, J. G. Herman, S. B. Baylin, and J. P. Issa. 1999. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA 96:8681-8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tucker, K. L., C. Beard, J. Dausmann, L. Jackson-Grusby, P. W. Laird, H. Lei, E. Li, and R. Jaenisch. 1996. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 10:1008-1020. [DOI] [PubMed] [Google Scholar]
- 77.Umar, A., J. C. Boyer, and T. A. Kunkel. 1994. DNA loop repair by human cell extracts. Science 266:814-816. [DOI] [PubMed] [Google Scholar]
- 78.Umar, A., A. B. Buermeyer, J. A. Simon, D. C. Thomas, A. B. Clark, R. M. Liskay, and T. A. Kunkel. 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87:65-73. [DOI] [PubMed] [Google Scholar]
- 79.van Lohuizen, M., S. Verbeek, P. Krimpenfort, J. Domen, C. Saris, T. Radaszkiewicz, and A. Berns. 1989. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell 56:673-682. [DOI] [PubMed] [Google Scholar]
- 80.van Lohuizen, M., S. Verbeek, B. Scheijen, E. Wientjens, H. van der Gulden, and A. Berns. 1991. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 65:737-752. [DOI] [PubMed] [Google Scholar]
- 81.Veigl, M. L., L. Kasturi, J. Olechnowicz, A. H. Ma, J. D. Lutterbaugh, S. Periyasamy, G. M. Li, J. Drummond, P. L. Modrich, W. D. Sedwick, and S. D. Markowitz. 1998. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc. Natl. Acad. Sci. USA 95:8698-8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed] [Google Scholar]
- 83.Warbrick, E. 1998. PCNA binding through a conserved motif. Bioessays 20:195-199. [DOI] [PubMed] [Google Scholar]
- 84.Warbrick, E. 2000. The puzzle of PCNA's many partners. Bioessays 22:997-1006. [DOI] [PubMed] [Google Scholar]
- 85.Whitehurst, C. E., S. Chattopadhyay, and J. Chen. 1999. Control of V(D)J recombinational accessibility of the D beta 1 gene segment at the TCR beta locus by a germline promoter. Immunity 10:313-322. [DOI] [PubMed] [Google Scholar]
- 86.Xiong, Z., A. H. Wu, C. M. Bender, J. L. Tsao, C. Blake, D. Shibata, P. A. Jones, M. C. Yu, R. K. Ross, and P. W. Laird. 2001. Mismatch repair deficiency and CpG island hypermethylation in sporadic colon adenocarcinomas. Cancer Epidemiol. Biomarkers Prev. 10:799-803. [PubMed] [Google Scholar]
- 87.Xu, G. L., T. H. Bestor, D. Bourc'his, C. L. Hsieh, N. Tommerup, M. Bugge, M. Hulten, X. Qu, J. J. Russo, and E. Viegas-Pequignot. 1999. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402:187-191. [DOI] [PubMed] [Google Scholar]