Abstract
Rho family small GTPases are key regulators of the actin cytoskeleton in various cell types. The Rnd proteins, Rnd1, Rnd2, and Rnd3/RhoE, have been recently identified as new members of the Rho family of GTPases, and expression of Rnd1 or Rnd3 in fibroblasts causes the disassembly of actin stress fibers and the retraction of the cell body to produce extensively branching cellular processes. Here we have performed a yeast two-hybrid screening by using Rnd1 as bait and identified a novel protein that specifically binds to Rnd GTPases. We named this protein Socius. Socius directly binds to Rnd GTPases through its COOH-terminal region. When transfected into COS-7 cells, Socius is translocated to the cell periphery in response to Rnd1 and Rnd3 and colocalized with the GTPases. While expression of wild-type Socius in Swiss 3T3 fibroblasts has little effect on the actin cytoskeleton, the expression of a membrane-targeted form of Socius, containing a COOH-terminal farnesylation motif (Socius-CAAX), induces a dramatic loss of stress fibers. The inhibitory effect of Socius-CAAX on stress fiber formation is enhanced by truncation of its NH2 terminus. On the other hand, the expression of Socius-CAAX or its NH2 terminus-truncated form suppresses the Rnd-induced retraction of the cell body and the production of extensively branching cellular processes, although the disassembly of stress fibers is observed. We propose that Socius participates in the Rnd GTPase-induced signal transduction pathways, leading to reorganization of the actin cytoskeleton.
The actin cytoskeleton mediates a variety of essential cellular functions, including motility, cytokinesis, and morphogenesis (17, 26). Much progress has been made in elucidating the molecular mechanisms that control the organization of the actin cytoskeleton, and it is already well known that members of the Rho family of small GTPases are key regulators of the actin cytoskeleton in various cell types (8, 12). Like other GTPases of the Ras superfamily, members of the Rho family of small GTPases serve as molecular switches by cycling between an inactive GDP-bound state and an active GTP-bound state and, once activated, they can interact with their specific effectors, leading to a variety of biological functions. Activation of the Rho family proteins requires GDP-GTP exchange catalyzed by various guanine nucleotide exchange factors (GEFs), and their activation is regulated by GTPase-activating proteins (GAPs), which stimulate the intrinsic GTPase activities of the G proteins. In addition, guanine nucleotide dissociation inhibitors inhibit the exchange of GDP for GTP and might also serve to regulate their localization (25). Presently, at least 14 mammalian Rho family proteins have been identified: Rho (A, B, and C), Rac (1, 2, and 3), Cdc42, RhoD, RhoG, RhoH/TTF, TC10, and Rnd (1, 2, and 3). Among these, the functions of Rho, Rac, and Cdc42 have been extensively characterized. In fibroblasts, the activation of Rho leads to formation of actin stress fibers (22), whereas the activation of Rac and Cdc42 induces formation of lamellipodia and filopodia, respectively (19, 23). These proteins are also involved in other cellular activities such as gene transcription and cell adhesion (12, 27). Furthermore, a number of proteins that specifically interact with them, including their GEFs, GAPs, and effectors, have been already identified (2, 15). However, little is known about binding partners for other Rho family members, especially Rnd proteins, the most recent members to be identified.
The Rnd proteins, Rnd1, Rnd2, and Rnd3 (also known as RhoE), comprise a distinct branch of Rho family GTPases in that they have a low affinity for GDP and very low intrinsic GTPase activities (6, 7, 20). Transient expression of Rnd1 or Rnd3 in fibroblasts inhibits the formation of actin stress fibers and the assembly of focal adhesions induced by lysophosphatidic acid (LPA), a Rho activator and, in addition, leads to the retraction and rounding of the cell body (20). In MDCK epithelial cells, expression of Rnd3/RhoE induces the disappearance of stress fibers and an increase in the speed of cell migration (7), and it is likely that the regulation of Rnd3 expression is involved in the alteration of the actin cytoskeleton associated with oncogenic transformation (9). In neuronal PC12 cells, expression of Rnd1 induces the formation of many neuritic processes from the cell body, with disruption of the cortical actin filaments that is probably due to the inhibition of Rho signaling pathway (1). Furthermore, Xenopus Rnd1 has been recently isolated and shown to be expressed in tissues undergoing extensive morphogenetic changes, such as marginal zone cells, somitogenic mesoderm, and neural crest cells, and overexpression of Xenopus Rnd1 induces the disruption of cell adhesion and is reversed by Xenopus RhoA expression (28). It appears, therefore, that Rnd1 and Rnd3 possess antagonistic effects on the action of RhoA in various cell types. However, the molecular mechanisms underlying Rnd protein-induced cellular functions have not yet been elucidated.
To isolate proteins involved in the signal transduction pathways of Rnd GTPases, we performed a yeast two-hybrid screening of a rat brain cDNA library by using Rnd1 as bait. Here we report the cloning and characterization of Socius, a novel Rnd GTPase-interacting protein (Socius is Latin for “partner”). Socius is the first molecule that specifically interacts with Rnd GTPases and, when transfected into COS-7 cells, Socius is translocated to the cell periphery in response to Rnd1 and Rnd3. While expression of wild-type Socius in fibroblasts causes little effect on the actin cytoskeleton, targeting of Socius to the membrane by the addition of a CAAX motif induces the disappearance of stress fibers. Furthermore, expression of Socius-CAAX or its NH2 terminus-truncated form suppresses the Rnd1- or Rnd3-induced retraction of the cell body and the production of extensively branching processes, although the disassembly of stress fibers is observed. We conclude that Socius participates in Rnd protein-mediated signal transduction pathways involved in the reorganization of actin cytoskeleton.
MATERIALS AND METHODS
Plasmid constructions.
The coding sequences for human Rnd2 and Rnd3 were obtained from human embryonic kidney 293 cells by reverse transcription-PCR (RT-PCR). The PCR products were cloned into pCR2.1 vector (Invitrogen) and sequenced completely. Wild-type Rnd1 and Rnd1N27, as well as wild-type and constitutively active forms of RhoA, Rac1, and Cdc42 (RhoAV14, Rac1V12, and Cdc42V12), were obtained as described previously (1, 13, 14). Rnd1A45 was generated by PCR-mediated mutagenesis (10). For two-hybrid constructs, the CAAX motifs of Rho family proteins were inactivated by a change of Cys to Ser (Rnd1S229, Rnd2S224, Rnd3S241, RhoAV14S190, Rac1V12S189, and Cdc42V12S188) introduced by PCR, and they were fused to the GAL4 DNA-binding domain (DNA-BD) of the pAS2-1 vector (Clontech) by using EcoRI sites. For purification of recombinant proteins, cDNAs encoding wild-type Rnd1, Rnd3, RhoA, Rac1, and Cdc42 with BamHI and EcoRI sites synthesized by PCR were subcloned into the BamHI/EcoRI sites of pGEX-4T-2 (Amersham Pharmacia Biotech). For expression in mammalian cells, cDNAs encoding Rnd1, Rnd2, and Rnd3 were fused in frame with a sequence in pcDNA3 (Invitrogen) encoding an initiating methionine, followed by the hemagglutinin (HA) tag sequence at the NH2 terminus by using KpnI/EcoRI sites. To generate green fluorescent protein (GFP)-fused Rnd GTPases, the cDNAs were subcloned into pEGFP-C1 (Clontech) by using KpnI/BamHI (Rnd1 and Rnd2) or KpnI/ApaI (Rnd3) sites. An expression plasmid encoding GFP-fused Rac1 was generated as described previously (29).
Yeast two-hybrid screening.
A rat brain cDNA library fused to the GAL4 activation domain of the pACT2 vector (Clontech) was screened by using pAS2-1/Rnd1S229 as bait in the yeast strain Y190 according to the manufacturer's instructions. Interaction between the bait and library proteins activates transcription of the reporter gene HIS3 or lacZ. From 106 transformants, ca. 400 His+ colonies appeared, and 48 colonies were also positive for β-galactosidase activity. Plasmids were recovered from positive colonies, and primary positive clones were retransformed into the original yeast strain. Among them, 15 clones were positive for the second round of β-galactosidase activity, and they were further tested for specificity of other Rho family proteins. One of these, clone 83, was positive for β-galactosidase activity in the presence of Rnd1, Rnd2, or Rnd3 but negative in the presence of RhoAV14, Rac1V12, or Cdc42V12. The 977-bp insert of this clone was sequenced and found to encode a novel protein of 300 amino acids. A poly(A) region was present 20 bp downstream of the 3′ stop codon, but it was possible that the clone 83 represented an incomplete open reading frame without an NH2-terminal initiation codon.
For the β-galactosidase filter assay, colonies of yeast transformants were transferred onto Hybond-N filter papers (Amersham Pharmacia Biotech) and permeabilized in liquid nitrogen. Each filter was placed on a Whatman no. 2 filter paper that had been presoaked in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 37.5 mM β-mercaptoethanol) containing 0.33 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml and was then incubated at 30°C for 8 h.
Cloning of full-length Socius.
When the expressed sequence tag (EST) databases were searched by using the basic local alignment search tool (BLAST; i.e., the BLAST network service at the National Center for Biotechnology Information) with the clone 83 sequence as the query, a partial human sequence highly similar to that of clone 83 was obtained from EST AW583166. When we further searched a human EST database by using the sequence of EST AW583166 as the query, the putative NH2-terminal sequence of human clone 83 was obtained from EST R18647. Two primers were synthesized: 5′-CTAGAATTCCGAGCTCACCTCTGGCCTCTC-3′, derived from EST R18647 and corresponding to the 5′ end of the putative coding sequence along with an EcoRI site, and 5′-TTTGTCAGGGCGGCCGCTTTATTCAGGGGA-3′, derived from the original yeast clone and corresponding to the minus strand of the 3′ end of the putative coding sequence along with a NotI site. These two primers were used in RT-PCR with rat testis RNA as a template, and an ∼1.4-kb PCR fragment was obtained. The PCR product was cloned into pCR2.1 vector and sequenced completely. Five independent clones of the PCR product were analyzed to eliminate any errors due to PCR amplification. Longer 5′ regions of rat Socius were isolated with the FirstChoice RLM-RACE kit (Ambion) from rat testis RNA by using a primer corresponding to the minus strand of 64 to 85 bp of Socius (5′-TTATCCCTCGCCTCCCGGGAT-3′) and the Ambion RNA adapter primer. For expression in mammalian cells, the cDNA for full-length Socius with EcoRI/NotI sites synthesized by PCR was fused in frame with a sequence in pcDNA3 encoding an initiating methionine, followed by the Myc tag sequence at the NH2 terminus by using EcoRI/NotI sites. To generate glutathione S-transferase (GST) fusion proteins, cDNAs encoding full-length and various truncated forms of Socius were generated by PCR amplification with primers that incorporated EcoRI/NotI sites and were subcloned into pGEX-4T-2, except for the UBX domain of Socius (amino acids 362 to 485), which was subcloned into pGEX-5X-2 by using BamHI/XhoI sites. The COOH-terminal 17-amino-acid polybasic and farnesylation sequence of K-Ras (KDGKKKKKKSKTKCVIM) was generated by PCR amplification from embryonic kidney 293 cells and fused in frame with a sequence in pcDNA3 encoding full-length or truncated forms of Socius without a stop codon by using XhoI/XbaI sites.
Northern blot analysis.
Total RNA (20 μg) isolated from various rat tissues by using an Isogen RNA isolation kit (Nippon-Gene, Tokyo, Japan) was separated by electrophoresis on a 1.5% agarose gel and transferred onto a nylon membrane (Biodyne [Pall Biosupport Division]). The membrane was hybridized at 65°C for 15 h in a mixture containing 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.5% sodium dodecyl sulfate (SDS), 5× Denhardt solution, 100 μg of denatured salmon sperm DNA/ml, and a 32P-labeled probe (2 × 106 cpm/ml) encoding the original two-hybrid fragment of Socius. The membrane was then washed twice in 2× SSC and twice in 2× SSC-1% SDS at 65°C. The membrane was dried, and the radioactivity was visualized by BAS 2000 image analyzer (Fuji).
Recombinant proteins.
All GST-fused proteins were purified from Escherichia coli as described previously (13). Nonfused Rnd1 and Rnd3 were recovered by incubation with 10 U of thrombin/ml (Sigma) for 4 h at 4°C, and then thrombin was removed by absorption to p-amino-benzamidine-agarose beads (Sigma). Protein concentration was determined by comparing with bovine serum albumin standards after SDS-polyacrylamide gel electrophoresis (PAGE) and staining with Coomassie brilliant blue. For in vitro binding assays, recombinant GTPases except Rnd1 were loaded with guanine nucleotides by incubation with 100 μM GTPγS or GDP in loading buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 0.1 mM dithiothreitol [DTT]; 5 mM EDTA) at 30°C for 30 min. The reaction was stopped by addition of MgCl2 to a final concentration of 10 mM. Since Rnd1 was unstable in the presence of EDTA as described previously (20), it was loaded with GTPγS in the buffer containing 2 mM MgCl2 without EDTA at 30°C for 10 min.
Immunoblotting.
Proteins were separated by SDS-10 or 12.5% PAGE and were electrophoretically transferred onto a polyvinylidene difluoride membrane (Millipore Corp.). The membrane was blocked with 3% low-fat milk in Tris-buffered saline and then incubated with primary antibodies. The primary antibodies were detected by using horseradish peroxidase-conjugated secondary antibodies (Dako) and an ECL detection kit (Amersham Pharmacia Biotech). Mouse monoclonal anti-Myc antibody 9E10 (Santa Cruz Biotechnology, Inc.) and anti-HA antibody 12CA5 (Boehringer Mannheim Corp.) were used at 1:100 and 1:200 dilutions, respectively.
In vitro binding assays.
COS-7 cells were cultured in Dulbecco modified Eagle medium containing 10% fetal bovine serum, 4 mM glutamine, 100 U of penicillin/ml, and 0.2 mg of streptomycin/ml under humidified conditions in 95% air and 5% CO2 at 37°C. Transient transfections were carried out by using LipofectAMINE 2000 (Life Technologies, Inc.) according to the manufacturer's instructions. Cells transfected with Myc-tagged Socius or HA-tagged Rnd GTPases were rinsed once with phosphate-buffered saline (PBS) and lysed with the ice-cold cell lysis buffer (20 mM Tris-HCl, pH 7.4; 2 mM MgCl2; 1 mM DTT; 0.2% Triton X-100; 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin/ml; 10 μg of leupeptin/ml). Cell lysates were then centrifuged for 10 min at 18,000 × g at 4°C. The supernatants were incubated for 5 min at 4°C with 10 μg of GST fusion proteins and subsequently incubated with glutathione-Sepharose beads for 2 h at 4°C. After the beads were washed with the ice-cold cell lysis buffer, the bound proteins were eluted in Laemmli sample buffer and analyzed by SDS-PAGE and immunoblotting with anti-Myc or anti-HA antibody.
To examine direct interaction between Socius and Rnd proteins, an overlay assay was performed according to the modified method of Manser et al. (16). GST fusion proteins (2 μg of each) were subjected to SDS-PAGE and transferred onto nitrocellulose membrane. The membrane was soaked for 5 min in 6 M guanidinium hydrochloride dissolved in buffer A (25 mM HEPES-NaOH, pH 7.0; 0.5 mM MgCl2; 0.05% Triton X-100) at 4°C, and the buffer was diluted with an equal volume of buffer A and agitated for a further 5 min. This process was repeated four times. The membrane was then agitated for 10 min five times in buffer A; transferred to PBS containing 1% bovine serum albumin, 0.1% Triton X-100, 0.5 mM MgCl2, and 5 mM DTT; and incubated in GAP buffer (25 mM HEPES-NaOH, pH 7.0; 5 mM MgCl2; 0.05% Triton X-100; 2.5 mM DTT; 100 μM GTP) containing [γ-32P]GTP (6,000 Ci/mmol; NEN Life Science Products, Inc.)-loaded Rnd1 or Rnd3. After being washed with wash buffer (25 mM HEPES-NaOH, pH 7.0; 5 mM MgCl2; 0.05% Triton X-100), the membrane was dried, and the bound radioactivity was visualized by FLA-3000 image analyzer (Fuji). Nonfused Rnd1 and Rnd3 (1 μg) were loaded with [γ-32P]GTP as described above.
Immunoprecipitation.
COS-7 cells cotransfected with Myc-tagged Socius and HA-tagged Rnd1 were lysed with ice-cold cell lysis buffer (20 mM Tris-HCl, pH 7.4; 100 mM NaCl; 2 mM MgCl2; 1 mM DTT; 0.2% Triton X-100; 1 mM phenylmethylsulfonyl fluoride; 10 μg of aprotinin/ml; 10 μg of leupeptin/ml). After centrifugation, the supernatants were incubated with anti-HA polyclonal antibody (MBL) for 1 h and then with protein A-Sepharose (Amersham Pharmacia Biotech) for 1 h. The beads were washed with the lysis buffer, and bound proteins were analyzed by SDS-PAGE and immunoblotting.
Separation of membrane and cytosolic fractions.
COS-7 cells transiently transfected with various Socius constructs were suspended in homogenization buffer (20 mM Tris HCl, pH 7.4; 100 mM NaCl; 1 mM EDTA; 5 mM MgCl2; 1 mM DTT; 5% glycerol). The cell suspensions were then lysed by homogenizing them with a Potter-Elvehjem homogenizer, and the homogenates were centrifuged at 9,300 × g for 5 min to remove the unbroken cells and nuclear fractions. The supernatants were further fractionated at 100,000 × g for 1 h into particulate and cytosolic fractions. The particulate pellet was resuspended in the same volume as the cytosol fraction, and equal volumes of them were analyzed by SDS-PAGE and immunoblotting.
Microinjection.
Swiss 3T3 fibroblasts were cultured in the same condition as described above for the COS-7 cells. For microinjection, cells were seeded onto round 13-mm glass coverslips in 35-mm culture dishes at a density of 104 cells. Then cells were serum starved for 24 h, and the indicated plasmids were microinjected into the nucleus at a concentration of 100 μg/ml in reversed PBS (3.7 mM NaCl, 137 mM KCl, 1 mM KH2PO4, 8 mM Na2HPO4). Microinjection was performed by using an IMM-188 microinjection apparatus (Narishige, Tokyo, Japan). During microinjection, cells were maintained in HEPES-buffered Dulbecco modified Eagle medium (pH 7.4) at 37°C. After microinjection, cells were returned to the incubator for a further 2 h before fixation. For quantification analysis of the effect of Socius expression on stress fiber formation, microinjected cells were stimulated with 1 μM LPA (Sigma) for 30 min before fixation. A proteasome inhibitor, MG132 (10 μM; Sigma), was added 2 h before microinjection.
Immunofluorescence microscopy.
All steps were carried out at room temperature, and cells were rinsed with PBS between steps. At the indicated times, cells on coverslips were fixed with 4% paraformaldehyde in PBS for 15 min. After residual formaldehyde had been quenched with 50 mM NH4Cl in PBS for 10 min, cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min and incubated with 10% fetal bovine serum in PBS for 30 min to block nonspecific antibody binding. For the detection of cells expressing Myc-tagged Socius, cells were incubated with anti-Myc monoclonal antibody 9E10 (0.5 μg/ml) in PBS for 1 h, followed by incubation with a fluorescein isothiocyanate (FITC) or rhodamine-conjugated goat anti-mouse immunoglobulin G (IgG) antibody (Chemicon International, Inc.) in PBS (1:500 dilution) for 1 h. Filamentous actin (F-actin) was stained with rhodamine-conjugated phalloidin (Molecular Probes) in PBS (0.5 U/ml) for 1 h. Cells on coverslips were mounted in 90% glycerol containing 0.1% p-phenylenediamine dihydrochloride in PBS. Confocal microscopy was performed by using an MRC-1024 laser scanning confocal imaging system (Bio-Rad Laboratories) equipped with a Nikon Eclipse E800 microscope and a Nikon Plan Apo 40-by-1.0 or 60-by-1.4 oil immersion objective lens.
Nucleotide sequence accession number.
The nucleotide sequence of rat Socius is available from GenBank/EMBL/DDBJ under accession number AB072920.
RESULTS
Isolation of a novel Rnd1-interacting protein.
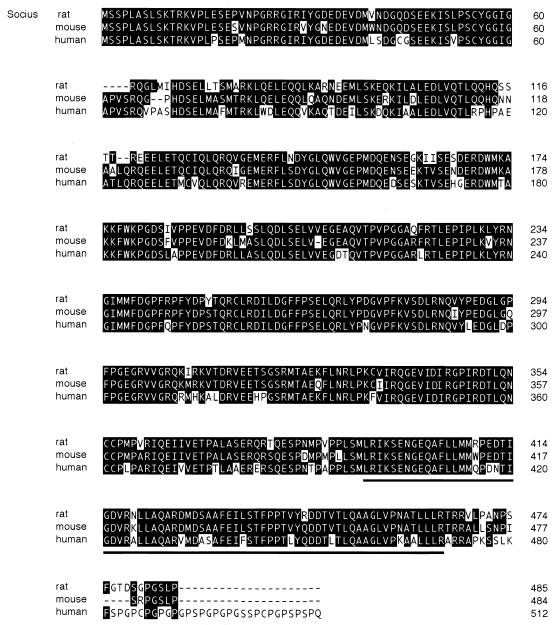
To isolate Rnd1-interacting proteins, we employed a yeast two-hybrid screening of a rat brain cDNA library with Rnd1S229, a mutant of Rnd1 lacking the COOH-terminal CAAX motif, as bait. The CAAX motif of Ras-related small GTPases targets them to the membrane and would interfere with the localization into the nucleus required for transcriptional activation of the reporter genes in this assay (11, 24). Approximately 106 clones were screened as described in Materials and Methods; 15 positive clones were obtained, and one of these, clone 83, was positive for β-galactosidase activity in the presence of Rnd1, Rnd2, and Rnd3 but was negative in the presence of RhoAV14, Rac1V12, and Cdc42V12, as well as a negative control lamin C and GAL4 DNA-BD alone (Table 1). Sequence analysis revealed that clone 83 encoded a protein of 300 amino acids, but it was possible that clone 83 represented an incomplete open reading frame without the NH2-terminal initiation codon. The predicted amino acid sequence of clone 83 was used to search a protein database by BLAST, but no homologous protein was found. We further searched EST databases by BLAST by using the nucleotide sequence of clone 83 as the query, and a partial human sequence encoding the putative NH2-terminal sequence of clone 83 was found. By RT-PCR with two primers derived from the EST clone corresponding to the 5′ end of the putative coding sequence and the original yeast clone corresponding to the 3′ end of the putative coding sequence, one major PCR product was obtained (1.5 kb in length), and sequence analysis of this product indicated that the original two-hybrid clone 83 represented an incomplete open reading frame. Longer 5′ regions of rat clone 83 were isolated, by using a 5′ rapid amplification of cDNA ends technique, from rat testis. The full open reading frame encodes a protein of 485 amino acids, and the predicted molecular mass is 54.7 kDa (Fig. 1). We found no additional upstream in-frame ATG in the rat clone 83 sequence. Furthermore, a human NH2-terminal partial sequence of clone 83 from the EST database containing the MSSPLA sequence is preceded by an in-frame stop codon, and there is no homology upstream of this sequence between human and rat clone 83, strongly suggesting that the MSSPLA sequence is the NH2 terminus of clone 83. We named this protein Socius. While the present study was in preparation, a mouse sequence highly similar to Socius (87% identity in amino acid sequence; GenBank accession number AK015720) and partial human EST clones that were longer than those obtained in our first BLAST search (EST AL555488 and AY007163) were reported (Fig. 1).
TABLE 1.
Yeast two-hybrid interaction assay between clone 83 and various Rho family GTPases
| GAL4 DNA-BD fusion protein | β-Galactosidase filter assaya |
|---|---|
| GAL4 DNA-BD | White |
| Rnd1S229 | Blue |
| Rnd2S224 | Blue |
| Rnd3S241 | Blue |
| RhoAV14S190 | White |
| Rac1V12S189 | White |
| Cdc42V12S188 | White |
| Lamin C | White |
White, color development was not observed after an 8-h incubation; blue, dark blue color was observed within 8 h of incubation in all three samples.
FIG. 1.
Deduced amino acid sequences of Socius. The amino acid sequence of rat Socius protein was aligned with mouse and human sequences highly similar to Socius obtained by a BLAST search (mouse sequence, GenBank AK051720; human sequences, EST AL555488 and AY007163). The original clone isolated by a yeast two-hybrid screening contains amino acids 186 to 485. Identical amino acids are boxed. A UBX domain is indicated by an underline.
Analysis of the predicted amino acids sequence of Socius revealed that Socius contained a UBX domain at the COOH terminus (amino acids 393 to 464; Fig. 1). The UBX domain is an ∼80-amino-acid residue module of unknown function present typically at the COOH terminus of a variety of eukaryotic proteins, including Fas-associated factor-1 and p47, a cofactor for p97-mediated membrane fusion (3). The other region of Socius showed no significant sequence homology to any other known proteins. Northern blot analysis of RNA from various rat tissues demonstrated that a transcript of Socius was strongly detected in testis (Fig. 2A). However, Socius was expressed as two transcripts in various tissues examined upon longer exposure and, among them, higher levels of expression were observed in lung, brain, and thymus tissue. The major smaller size of the transcript correlates well with the isolated 1.5-kb cDNA of Socius, and this result supports the fact that the isolated cDNA clone is close to full length.
FIG. 2.
Expression of Socius mRNA in rat tissues. (A) Northern blot analysis. A membrane blotted with total RNAs from rat various tissues was hybridized with an α-32P-labeled cDNA probe encoding the COOH-terminal fragment of Socius originally isolated by yeast two-hybrid screening (amino acids 186 to 485). The results other than testis are shown by a 10-fold-longer exposure. (B) RT-PCR amplification of Socius NH2-terminal fragments from rat testis, lung, thymus, and brain tissues. Total RNA (2.5 μg) was reverse transcribed with random primers, followed by amplification of the NH2-terminal fragment of Socius with the primers Socius-F (5′-ATGAGCTCACCCCTGGCCTCT-3′, corresponding to amino acids 1 to 7 of Socius) and Socius-R2 (5′-AACATCATGATTCCATTCCG-3′, corresponding to amino acids 408 to 413). For the PCR product of rat testis, a 10-fold-smaller amount of the RT-PCR sample was analyzed. Deduced spliced variants (products a, b, and c) of Socius are shown schematically. FL, full-length form of Socius.
While the isolation of longer 5′ regions of Socius was in process, we obtained clones containing the internal insertion or deletion at the NH2-terminal half of Socius from rat brain by RT-PCR (Fig. 2B). PCR product a, shown in Fig. 2B, had a 205-bp insert, creating a new short coding region downstream after amino acid 36 of Socius, which contained a new stop codon located 60 bp downstream from the junction (Fig. 2B, product a). PCR product b contained a 126-bp deletion of the coding region corresponding to amino acids 97 to 137 of Socius but would generate a protein with the same reading frame as that of full-length of Socius beyond the deletion point (Fig. 2B, product b). PCR product c contained a 253-bp deletion, resulting in a frameshift at the deletion point, and would generate a protein lacking the main part of Socius (amino acids 97 to 485) but instead gaining another 20-residue COOH terminus (Fig. 2B, product c). These splice variants of Socius were abundant in the brain, whereas full-length Socius was the main transcript in other tissues expressing higher levels of Socius mRNA (Fig. 2B). On the other hand, no splice variant containing an insertion or deletion at the COOH-terminal half of Socius was detected by RT-PCR (data not shown).
Characterization of the interaction between Socius and Rnd GTPases.
In a yeast two-hybrid assay, the isolated COOH-terminal fragment of Socius (amino acids 186 to 485) interacted specifically with Rnd GTPases but not with other Rho family GTPases (Table 1). To confirm the specificity of the interaction by in vitro binding assay, Myc-tagged full-length Socius was expressed in COS-7 cells and pull-down assays were performed by using purified GST-fused Rnd1, Rnd3, and other Rho family GTPases. GST-fused GTPases preloaded with GTP-γS or GDP were incubated with lysates of COS-7 cells expressing Myc-tagged full-length Socius, immobilized on glutathione-Sepharose beads, and bound Socius was detected by immunoblotting with anti-Myc antibody (Fig. 3A). Consistent with the result of the yeast two-hybrid assay, GTP-γS-loaded, GST-fused Rnd1 and Rnd3 strongly interacted with the full length of Socius. However, GST alone or GST-fused RhoA, Rac1, or Cdc42 loaded with either GTP-γS or GDP did not bind to Myc-tagged Socius. Since Rnd proteins were very poorly bound to GDP (6, 7, 20), we could not examine the interaction between Socius and GDP-loaded Rnd1 and Rnd3. On the other hand, we could not obtain recombinant Rnd2 protein from E. coli as described previously (20). Therefore, Socius was expressed as a GST fusion protein, and the interaction with HA-tagged Rnd2 expressed in COS-7 cells was examined by pull-down assays. As shown in Fig. 3B, HA-tagged Rnd2 was strongly precipitated by GST-fused Socius (amino acids 186 to 485, corresponding to the original isolated yeast clone) but was not precipitated by nonfused GST. HA-tagged Rnd1 and Rnd3 were also bound to GST-fused Socius. These results demonstrate that Socius specifically interacts with Rnd subfamily GTPases.
FIG. 3.
In vitro interaction of Socius with Rnd GTPases. (A) COS-7 cells were transiently transfected with an expression vector encoding Myc-tagged full-length Socius, and cell lysates were incubated with GST (lane 2) or GST-fused Rnd1 (lane 3), Rnd3 (lane 4), RhoA (lanes 5 and 6), Rac1 (lanes 7 and 8), or Cdc42 (lanes 9 and 10) preloaded with GTP-γS (lanes 2, 3, 4, 5, 7, and 9) or GDP (lanes 6, 8, and 10). Then they were immobilized on glutathione-Sepharose beads, and bound proteins and lysate input (Lysate, lane 1) were analyzed by immunoblotting with anti-Myc monoclonal antibody. (B) COS-7 cells were transiently transfected with an expression vector encoding HA-tagged Rnd1 (top), Rnd2 (middle) or Rnd3 (bottom), and cell lysates were incubated with GST (lane 2) or GST-fused Socius (amino acids 186 to 485, lane 3). Then they were immobilized on glutathione-Sepharose beads, and bound proteins and lysate input (Lysate, lane 1) were analyzed by immunoblotting with anti-HA monoclonal antibody. (C) GST and GST-Socius (amino acids 186 to 485) were subjected to SDS-PAGE, transferred onto nitrocellulose membrane, and then probed with [γ-32P]GTP-Rnd1, as described in Materials and Methods (left). GST and GST-fused Socius used in this experiment were shown by staining with Coomassie brilliant blue (right).
To determine whether the interaction between Socius and Rnd GTPases was direct, an overlay assay was performed with recombinant nonfused Rnd1 preloaded with [γ-32P]GTP as a probe. GST-fused Socius showed strong binding to [γ-32P]GTP-loaded Rnd1 on a nitrocellulose membrane, but nonfused GST did not (Fig. 3C). Similar results were obtained with [γ-32P]GTP-loaded Rnd3 as a probe (data not shown).
To identify the region of Socius required for the interaction with Rnd GTPases, various truncated forms of Socius were expressed as GST fusion proteins, and their binding to HA-tagged Rnd3 expressed in COS-7 cells was examined by the pull-down assay. HA-tagged Rnd3 was precipitated by the GST-fused, COOH-terminal half of Socius (Socius CT, amino acids 275 to 485), as well as the full-length protein (Socius FL) or the fragment corresponding to the original yeast clone (Socius CT*, amino acids 186 to 485), whereas it was not precipitated by the GST-fused NH2-terminal half of Socius (Socius NT, amino acids 2 to 238) or GST alone (Fig. 4A). This result indicates that the binding site of Rnd proteins is located at the COOH-terminal half of Socius. We further divided Socius CT into two fragments, the UBX domain (amino acids 362 to 485) and the other region of Socius CT (amino acids 275 to 362), and their interaction with HA-tagged Rnd3 was examined by the pull-down assay. However, both fragments did not interact with HA-tagged Rnd3 (data not shown), suggesting that the entire COOH-terminal region of Socius is required for the interaction with Rnd GTPases.
FIG. 4.
Characterization of the interaction between Socius and Rnd GTPases. (A) COS-7 cells were transiently transfected with an expression vector encoding HA-tagged Rnd3, and cell lysates were incubated with GST (lane 2) or various GST-Socius fusion proteins (FL, full-length, amino acids 2 to 485, lane 3; NT, amino acids 2 to 238, lane 4; CT*, amino acids 186 to 485, lane 5; CT, amino acids 275 to 485, lane 6). Then they were immobilized on glutathione-Sepharose beads, and bound proteins and lysate input (Lysate, lane 1) were analyzed by immunoblotting with anti-HA monoclonal antibody (top). Coomassie brilliant blue staining of various GST fusion proteins used in this experiment was also shown (bottom, indicated by arrowheads). (B) COS-7 cells were transiently transfected with an expression vector encoding HA-tagged wild-type Rnd1 (WT, lanes 1 and 2), HA-tagged Rnd1N27 (N27, lane 3), or HA-tagged Rnd1A45 (A45, lane 4), and cell lysates were incubated with GST (lane 1) or GST-fused Socius CT (lanes 2 to 4). Then they were immobilized on glutathione-Sepharose beads, and bound proteins were analyzed by immunoblotting with anti-HA monoclonal antibody (top). Expression of Rnd1 proteins in cell lysates are also shown (bottom).
We next examined the interaction of Socius with various mutants of Rnd1 by the pull-down assay. HA-tagged wild-type Rnd1, Rnd1N27 (equivalent to a dominant-negative mutation in Ras), and Rnd1A45 (equivalent to a mutation in the effector region of Ras) were expressed in COS-7 cells, and the cell lysates were incubated with GST-fused Socius CT. The interaction between HA-tagged Rnd1A45 and GST-fused Socius CT was detected but weak compared to that of wild-type Rnd1, whereas no interaction between HA-tagged Rnd1N27 and GST-fused Socius CT was observed (Fig. 4B).
Previous studies have shown that Rnd1 and Rnd3 have very low intrinsic GTPase activities (6, 7, 20). To determine whether Socius had GAP activities for Rnd GTPases, recombinant nonfused Rnd1 and Rnd3 loaded with [γ-32P]GTP were incubated with GST-fused full-length of Socius, and the hydrolysis rates of GTP were measured. However, Socius did not change the GTPase activity of Rnd1 or Rnd3 (data not shown).
Rnd-dependent translocation of Socius to the cell periphery.
The effect of the expression of Rnd1 and Rnd3 on the subcellular localization of transiently expressed Socius was examined in COS-7 cells. The morphologies of COS-7 cells were not significantly affected by the expression of Rnd1 or Rnd3, probably because the morphological effects of Rnd1 or Rnd3 are cell type specific, as described in previous studies (1, 7, 20). Immunofluorescence confocal microscopy with anti-Myc monoclonal antibody showed that Socius was observed diffusely throughout the cytoplasm and the nucleus in the cells coexpressing Myc-tagged Socius and GFP (Fig. 5Aa and b) or in the cells expressing Myc-tagged Socius alone (data not shown). In contrast, when cells were cotransfected with Myc-tagged Socius and GFP-fused Rnd1 (Fig. 5Ac and d) or GFP-fused Rnd3 (Fig. 5Ae and f), a part of Socius was translocated to the cell periphery where Socius and Rnd proteins were colocalized. The translocation of wild-type Socius was observed in more than 80% of GFP-Rnd1- or GFP-Rnd3-expressing cells, whereas <5% of GFP-expressing cells showed the translocation of Socius. On the other hand, in the cells coexpressing Myc-tagged Socius and GFP-fused Rac1, another Rho family GTPase that was also localized to the cell periphery, the distribution of Socius did not change (Fig. 5Ag and h). The translocation of Socius to the cell periphery was weakly detected in the cells cotransfected with GFP-Rnd1A45, a mutant which can weakly bind to Socius (data not shown). We also examined the effect of Rnd2 expression on the distribution of Socius. However, when cells were cotransfected with Socius and GFP-fused Rnd2, they were diffusely localized in the cells but not localized to the cell periphery (data not shown).
FIG. 5.
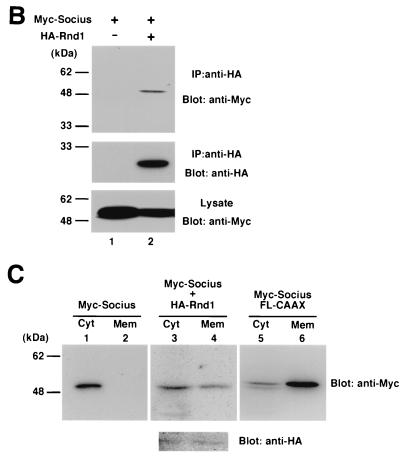
Subcellular localization of Socius in COS-7 cells. (A) Immunofluorescence confocal microscopy. COS-7 cells were transiently transfected with an expression vector encoding Myc-tagged Socius along with a vector encoding GFP (a and b) or GFP-fused Rnd1 (c and d), Rnd3 (e and f), or Rac1 (g and h). At 2 h after transfection, cells were fixed and stained with anti-Myc monoclonal antibody (b, d, f, and h). Localization of GFP fusion proteins was also shown (a, c, e, and g). The scale bar represents 10 μm. (B) Immunoprecipitation. COS-7 cells transfected with an expression vector encoding Myc-tagged Socius alone (lane 1) or along with HA-tagged Rnd1 (lane 2), and cell lysates were immunoprecipitated with anti-HA antibody. The immunoprecipitates (top and middle panels) and the total cell lysates (bottom panel) were analyzed by immunoblotting. (C) Biochemical fractionation. Cellular homogenates from COS-7 cells transfected with a vector encoding Myc-tagged Socius (lanes 1 and 2) or Myc-tagged Socius FL-CAAX (lanes 5 and 6) or cotransfected with expression vectors encoding Myc-tagged Socius and HA-tagged Rnd1 (lanes 3 and 4) were separated into cytosolic (Cyt, lanes 1, 3, and 5) and membrane (Mem, lanes 2, 4, and 6) fractions as described in Materials and Methods. The fractionated samples were analyzed by immunoblotting with anti-myc monoclonal antibody. Expression of HA-tagged Rnd1 was also shown (lanes 3 and 4, bottom panel).
Colocalization of Socius and Rnd1 was also confirmed by coimmunoprecipitation of two proteins from COS-7 cells when they were cotransfected (Fig. 5B). Furthermore, we prepared crude membrane and cytosolic fractions from cellular homogenates of the cells transfected with Myc-tagged Socius alone or cotransfected with Myc-tagged Socius and HA-tagged Rnd1, and the distribution of Socius was analyzed by immunoblotting. In the cells transfected with Socius alone, it was exclusively localized in the cytosolic fraction. In contrast, when cells were cotransfected with Socius and Rnd1, Socius was partly translocated to the membrane fraction (Fig. 5C), a finding consistent with the immunofluorescence confocal microscopy data.
Effect of Socius expression on the actin cytoskeleton in Swiss 3T3 fibroblasts.
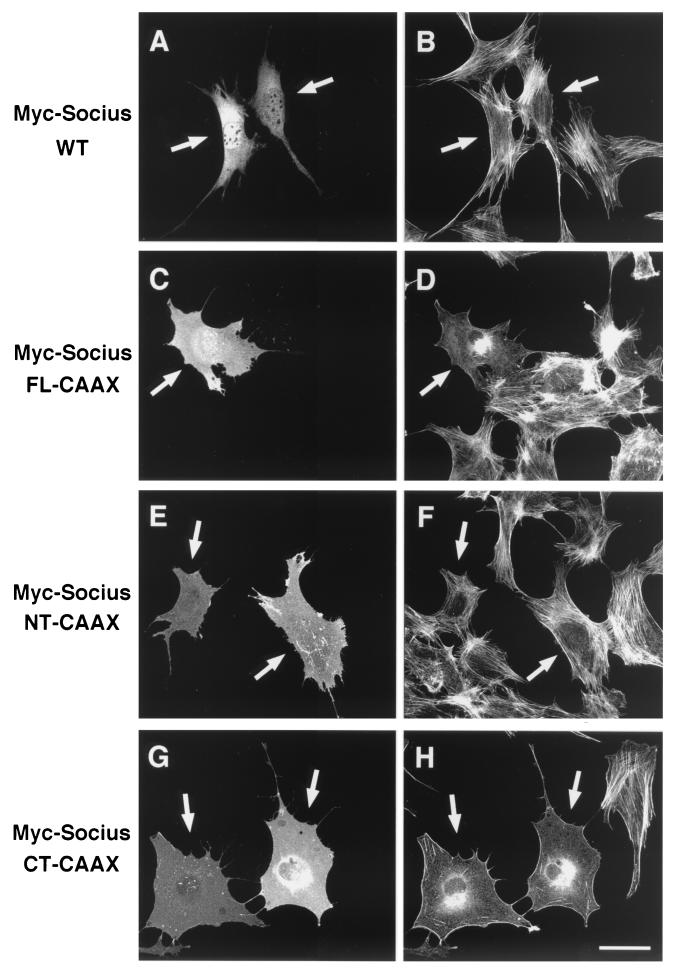
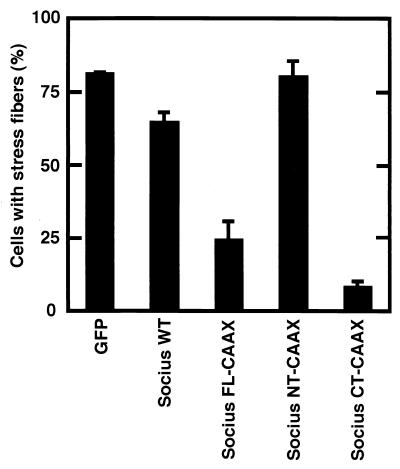
Previous studies have shown that expression of Rnd1 or Rnd3 induces the disappearance of actin stress fibers in Swiss 3T3 fibroblasts and MDCK epithelial cells and that Swiss 3T3 cells expressing Rnd1 or Rnd3 subsequently retract and severely round up to produce a phenotype like that of dendritic cells (7, 20). RT-PCR analysis with Socius-specific primers revealed that Socius was expressed in these cells (data not shown). Therefore, to test the ability of Socius to induce reorganization of the actin cytoskeleton, a plasmid encoding Myc-tagged wild-type Socius (Myc-Socius WT) was microinjected into the nuclei of serum-starved Swiss 3T3 fibroblasts, and their F-actin structures were examined. However, expression of Myc-Socius WT did not significantly affect the actin cytoskeleton at 2 h (Fig. 6A and B) or at 24 h (data not shown) after microinjection. Considering that Socius was translocated to the cell periphery by Rnd1 and Rnd3 (Fig. 5), the membrane localization of Socius may be required for its action. To examine whether artificially targeting Socius to the plasma membrane would affect the F-actin structure in Swiss 3T3 fibroblasts, we generated Myc-tagged full-length of Socius with the 17-amino-acid COOH-terminal membrane-targeting sequence from K-Ras (Myc-Socius FL-CAAX) (4, 5). We verified that Myc-Socius FL-CAAX was actually localized to the membrane by the biochemical fractionation study (Fig. 5C). Therefore, this construct was microinjected into the nuclei of serum-starved Swiss 3T3 fibroblasts, and their F-actin structures were examined. In contrast to the expression of Myc-Socius WT, the expression of Myc-Socius FL-CAAX induced the complete disappearance of stress fibers and a dramatic loss of F-actin staining (Fig. 6C and D). These effects were observed in >90% of the microinjected cells. When microinjected cells were stimulated with LPA, a strong Rho activator, expression of Myc-Socius FL-CAAX was also able to suppress LPA-induced stress fiber formation (Fig. 7).
FIG. 6.
Effect of Socius expression on the actin cytoskeleton in Swiss 3T3 fibroblasts. Serum-starved Swiss 3T3 fibroblasts were microinjected with an expression vector encoding Myc-Socius WT (A and B), Myc-Socius FL-CAAX (C and D), Myc-Socius NT-CAAX (E and F), or Myc-Socius CT-CAAX (G and H). At 2 h after microinjection, cells were fixed and stained with rhodamine-conjugated phalloidin to visualize F-actin (B, D, F, and H) and with anti-Myc monoclonal antibody, followed by staining with FITC-conjugated anti-mouse IgG antibody (A, C, E, and G). The arrows indicate injected cells. Scale bar, 50 μm.
FIG. 7.
Effect of Socius expression on LPA-induced stress fiber formation. Serum-starved Swiss 3T3 fibroblasts were microinjected with an expression vector encoding Myc-Socius WT, Myc-Socius FL-CAAX, Myc-Socius NT-CAAX, or Myc-Socius CT-CAAX. At 2 h after microinjection, cells were stimulated with 1 μM LPA for 30 min. After fixation, cells were double stained with rhodamine-conjugated phalloidin and with anti-Myc monoclonal antibody, followed by staining with FITC-conjugated anti-mouse IgG antibody, and positively stained cells were assessed. Cells with well-defined stress fibers were scored as a percentage of the total number of microinjected cells. At least 50 cells were assessed in one experiment, and data are the means ± the standard error of triplicate experiments.
We next assessed the ability of truncated forms of Socius to affect the actin cytoskeleton in Swiss 3T3 fibroblasts. Expression of an NH2-terminal region of Socius with the CAAX motif (Myc-Socius NT-CAAX) had no effect on the F-actin structure in serum-starved cells and on LPA-induced stress fiber formation. (Fig. 6E and F; Fig. 7). In contrast, expression of a COOH-terminal region of Socius with the CAAX motif (Myc-Socius CT-CAAX), containing the Rnd-binding domain, induced the disassembly of stress fibers similar to that induced by expression of Myc-Socius FL-CAAX (Fig. 6G and H). The inhibitory effect of Socius CT-CAAX on the LPA-induced stress fiber formation was stronger than that of Myc-Socius FL-CAAX (Fig. 7). In these experiments, expression of various Socius constructs was almost at the same levels (data not shown). In contrast, the expression of Myc-Socius FL- or Myc-Socius CT-CAAX failed to induce the rounding of the cell body and the extensive branching, another Rnd-induced phenotype. The membrane localization of Myc-tagged Socius NT-CAAX and CT-CAAX was also confirmed by the biochemical fractionation. Furthermore, the CAAX motif did not affect the interaction of Socius with Rnd GTPases by the immunoprecipitation study in COS-7 cells (data not shown).
Effect of Socius expression on the Rnd-induced morphological change.
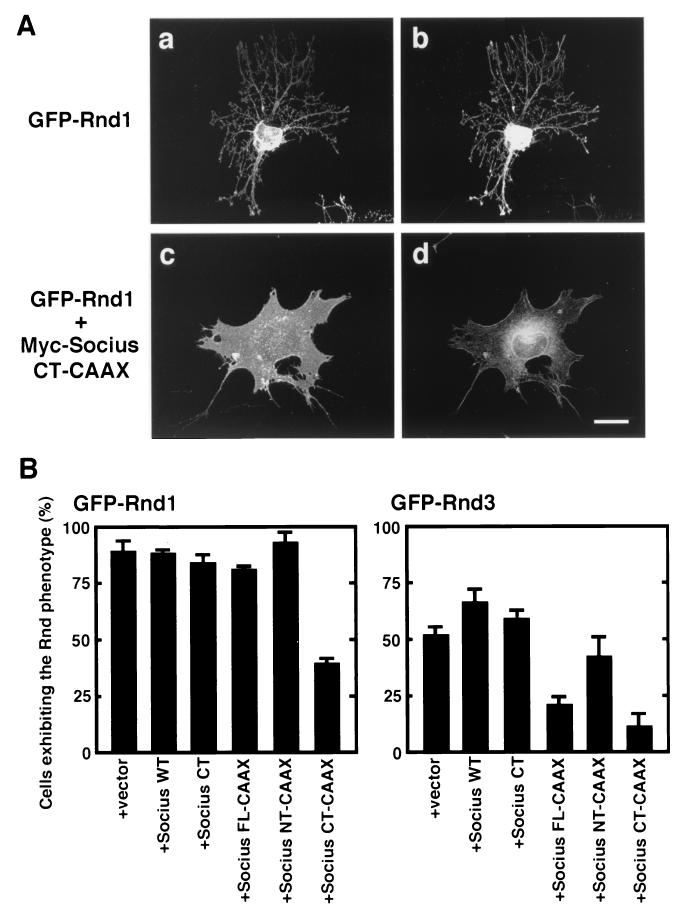
To examine whether expression of Socius would affect the Rnd1- or Rnd3-induced morphological change, plasmids encoding wild-type or truncated forms of Socius were comicroinjected with a plasmid encoding GFP-tagged Rnd1 or Rnd3 into the nuclei of serum-starved Swiss 3T3 fibroblasts, and the morphology of microinjected cells was observed at 2 h after microinjection. Expression of GFP-Rnd1 alone induced the retraction and rounding of the cell body to produce extensively branching cellular processes (Fig. 8Aa and b). In contrast, when cells were comicroinjected with plasmids encoding GFP-Rnd1 and Myc-Socius CT-CAAX, the Rnd1-induced rounding of the cell body and the extensive branching were suppressed, but the disappearance of stress fibers was observed (Fig. 8Ac and d and 8B). Expression of Myc-Socius FL-CAAX slightly inhibited the Rnd1-induced branching phenotype, whereas expression of Myc-Socius WT, Myc-Socius CT, or Myc-Socius NT-CAAX had no effect (Fig. 8B). We also examined the effects of various Socius constructs on the Rnd3-induced morphological change. Expression of GFP-Rnd3 also induced the branching phenotype, as did that of GFP-Rnd1 (data not shown), but it was less effective than the expression of GFP-Rnd1 (Fig. 8B). Comicroinjection of GFP-Rnd3 with Myc-Socius FL-CAAX or Myc-Socius CT-CAAX suppressed the Rnd3-induced branching phenotype, but comicroinjection of GFP-Rnd3 with Myc-Socius CT or Myc-Socius NT-CAAX had no effect. In contrast, comicroinjection of GFP-Rnd3 with Myc-Socius WT slightly enhanced the Rnd3-induced phenotype. A previous study has shown that expression of Rnd1A45 does not cause the loss of stress fibers and the branching phenotype (20), but it can weakly bind to Socius (Fig. 4B). Coexpression of GFP-Rnd1A45 with Socius WT in Swiss 3T3 fibroblasts induced the weak disruption of stress fibers but did not cause the branching phenotype (data not shown). In these experiments, the expression levels of GFP-Rnd1, GFP-Rnd3, and various Socius constructs in the comicroinjected cells were similar (data not shown). Furthermore, we also obtained similar results with HA-tagged Rnd1 or Rnd3 (data not shown). Finally, Socius has a UBX domain at the COOH terminus, which is known to be structurally homologous to ubiquitin (3). However, pretreatment of the cells with the proteasome inhibitor MG132 had no effect on the Socius FL-CAAX-induced loss of stress fibers or on the Rnd1-induced branching phenotype (data not shown), suggesting that the ubiquitin/proteasome system is not involved in the Socius-induced actions.
FIG. 8.
Effect of Socius expression on the Rnd-induced morphological change. (A) Serum-starved Swiss 3T3 fibroblasts were microinjected with an expression vector encoding GFP-Rnd1, along with a vector encoding Myc-Socius CT-CAAX (c and d) or a vector alone (a and b). At 2 h after microinjection, the cells were fixed and stained with rhodamine-conjugated phalloidin to visualize F-actin (b and d). Rnd1-expressing cells were identified by the fluorescence of GFP (a and c). Scale bar, 50 μm. (B) Quantification of the effect of various forms of Socius expression on Rnd1-induced morphological change. Serum-starved Swiss 3T3 fibroblasts were microinjected with an expression vector encoding GFP-Rnd1 (left) or GFP-Rnd3 (right), along with a vector encoding various forms of Socius. At 2 h after microinjection, cells were fixed and stained with rhodamine-conjugated phalloidin, and cells with the fluorescence of GFP were assessed. Cells exhibiting the Rnd phenotype were defined as cells comprising <1,000 μm2 of the cell body and with extensive branching of the cellular processes, as shown in panels a and b, and were scored as a percentage of the total number of microinjected cells. At least 50 cells were assessed in one experiment, and the data are the means ± the standard error of triplicate experiments.
DISCUSSION
Rnd GTPases, especially Rnd1 and Rnd3, have been shown to possess antagonistic effects on the actions of RhoA, such as the disassembly of actin stress fibers and the loss of cell adhesion (1, 7, 20, 28). However, the mechanisms underlying Rnd GTPase-induced morphological effects and molecules that specifically interact with Rnd GTPases are unknown. Here, we have described identification and characterization of Socius, a novel Rnd GTPase-interacting protein that may play an important role in Rnd-mediated regulation of the actin cytoskeleton. Transiently expressed Socius is localized diffusely in cells but, when it is coexpressed with Rnd1 or Rnd3, it is translocated to the cell periphery. While expression of Socius into Swiss 3T3 fibroblasts has little effect on the actin cytoskeleton, targeting of Socius to the membrane by the addition of a CAAX motif causes a dramatic loss of stress fibers and regulates Rnd-induced morphological change. Taken together, these results suggest that Socius is an important mediator for Rnd protein regulation of the actin cytoskeleton.
Socius is the first molecule that specifically binds to Rnd GTPases. The domain of Socius binding to Rnd GTPases is located at its COOH-terminal half, but its UBX domain alone or a region just before the UBX domain of Socius is not sufficient for its binding to Rnd proteins. These results suggest that the entire COOH-terminal region of Socius is required for the interaction with Rnd GTPases. Searching of EST and known protein databases showed no significant sequence homology to this region. Therefore, the Rnd-binding domain in Socius is unique. On the other hand, Socius does not bind to Rnd1N27, which is equivalent to a dominant-negative mutant of Ras. It is known that introducing a T17N mutation in Ras increases its affinity for Ras GEFs (21). It is likely, therefore, that Socius does not function as a Rnd GEF. We have also shown that Socius does not have GAP activities for Rnd1 and Rnd3 by using recombinant proteins, thereby indicating that Socius is not a Rnd GAP.
The biochemical mechanisms underlying the Socius-mediated regulation of Rnd signaling are not known yet. Considering that Rnd1 and Rnd3 cause the translocation of Socius to the cell periphery, it is likely that Socius acts at the plasma membrane downstream of Rnd1 and Rnd3. Indeed, the membrane-targeted form of Socius has a strong inhibitory effect on LPA-induced stress fiber formation compared to wild-type Socius. Furthermore, a mutation in the effector domain of Rnd1, Rnd1A45, loses the ability to induce the Rnd1 phenotype (20) and decreases the binding affinity for Socius, speculating that Socius is a downstream regulator. In Swiss 3T3 fibroblasts, expression of Rnd1 or Rnd3 causes the retraction and subsequent rounding up of the cell body with extensive branching of cellular processes and inhibits LPA-induced stress fiber formation. In the present study, we have shown that the membrane-targeted form of Socius in Swiss 3T3 fibroblasts induces the disassembly of stress fibers but does not cause the branching phenotype. On the other hand, the membrane-targeted form of Socius suppresses the Rnd-induced branching phenotype, but the disappearance of stress fibers is observed in the cells. One possible explanation for these results is that the Rnd GTPase-induced disassembly of stress fibers and branching phenotype are differentially regulated by at least two distinct signaling pathways and that Socius is involved in the pathway for the disassembly of stress fibers while the branching phenotype is regulated by another, unknown, pathway. Overexpression of the membrane-targeted forms of Socius may tether Rnd to the pathway for stress fiber disassembly and perturb linking of Rnd to the other pathway for the branching phenotype. Identification of another downstream target of Rnd GTPases will provide more information about the mechanisms of the Rnd-induced morphological effects. However, wild-type Socius, which is recruited to the cell periphery by Rnd1 or Rnd3, cannot inhibit the Rnd-induced branching phenotype. This may be because the inhibition of the Rnd-induced branching phenotype occurs when a large amount of Socius is continuously expressed at the plasma membrane, and only a part of wild-type Socius translocated to the cell periphery by Rnd GTPases is insufficient. On the other hand, the Rnd3-induced branching phenotype is strongly inhibited by Socius FL-CAAX, whereas the effect of Rnd1 is not. This is probably due to the stronger activity of Rnd1 than Rnd3 in induction of branching morphology (Fig. 8B, compare the bars labeled “+vector”). Our results also demonstrate that the COOH-terminal fragment of Socius is sufficient for the ability to induce the disassembly of stress fiber formation. Considering that this COOH-terminal fragment of Socius contains the Rnd-binding domain and the UBX domain, Socius may serve as an adapter or scaffold protein, linking Rnd GTPases to cytoskeletal regulatory molecules that would bind to the UBX domain of Socius. The UBX domain is an ∼80-amino-acid residue module that is present typically at the COOH terminus of a variety of eukaryotic proteins and is structurally homologous to ubiquitin (3). However, a number of proteins containing the UBX domain so far appear to be unrelated to ubiquitination processes, and the function of the UBX domain remains unknown. Indeed, a proteasome inhibitor has no effect on the loss of stress fibers by Socius. On the other hand, the NH2-terminal region of Socius may negatively regulate the activity of the COOH-terminal region because Socius CT-CAAX is stronger than Socius FL-CAAX in the ability to suppress LPA-induced stress fiber formation. Furthermore, the NH2-terminal region of Socius may regulate the interaction of its COOH-terminal domain with Rnd GTPases, since the full length of Socius more efficiently binds to Rnd3 compared to the NH2-terminally truncated forms in vitro. However, Socius CT-CAAX is a more potent inhibitor of the Rnd-induced branching phenotype compared to Socius FL-CAAX. We do not know the reason for this, but it is possible that the binding activity of Socius with Rnd GTPases in Swiss 3T3 fibroblasts is distinct from that obtained by the in vitro assay.
Socius can bind to Rnd2, as well as to Rnd1 and Rnd3. However, the expression of Rnd2 in Swiss 3T3 fibroblasts has no observable effect on the actin cytoskeleton and does not inhibit the formation of actin stress fibers (20). Since Rnd2 does not change the localization of Socius in COS-7 cells, in contrast to Rnd1 or Rnd3, which causes the translocation of Socius to the cell periphery, the loss of actin stress fibers induced by Rnd1 or Rnd3 may require at least the translocation of Socius to the cell periphery. However, in light of the predominant expression of Rnd2 in neurons (18), it is possible that Socius mediates unknown functions of Rnd2 in neurons.
We have found at least three splice variants of Socius in rat tissues, especially in brain. Two of these variants encode proteins lacking the functional COOH-terminal part of Socius, and the other would generate a short form of Socius with a 40-amino-acid deletion in its NH2-terminal side. We have not tried to examine whether the expression of this short variant can affect the structure of the actin cytoskeleton or the action of full-length Socius in fibroblasts. The significance of these splice variants of Socius, especially in the brain, is currently unknown, but they might modulate the function of full-length Socius. We have also found several sequences highly similar to Socius with deletions of various lengths in the NH2-terminal region in a human EST database.
In conclusion, we have identified a novel Rnd GTPase-interacting protein, Socius, involved in the Rnd-induced reorganization of the actin cytoskeleton. Socius is the first molecule that specifically binds to Rnd GTPases, but many questions about Rnd proteins have yet to be answered. For example, what molecules participate in the upstream signaling of Rnd proteins? Further studies focusing on the identification of Rnd-interacting proteins will contribute to our understanding of Rnd signaling and the regulation of cell morphologies.
Acknowledgments
This work was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan (13041029, 13210068, and 13780571).
REFERENCES
- 1.Aoki, J., H. Katoh, K. Mori, and M. Negishi. 2000. Rnd1, a novel Rho family GTPase, induces the formation of neuritic processes in PC12 cells. Biochem. Biophys. Res. Commun. 278:604-608. [DOI] [PubMed] [Google Scholar]
- 2.Aspenström, P. 1999. Effectors for the Rho GTPases. Curr. Opin. Cell Biol. 11:95-102. [DOI] [PubMed] [Google Scholar]
- 3.Buchberger, A., M. J. Howard, M. Proctor, and M. Bycroft. 2001. The UBX domain: a widespread ubiquitin-like module. J. Mol. Biol. 307:17-24. [DOI] [PubMed] [Google Scholar]
- 4.Choy, E., V. K. Chiu, J. Silletti, M. Feoktistov, T. Morimoto, D. Michaelson, I. E. Ivanov, and M. R. Philips. 1999. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69-80. [DOI] [PubMed] [Google Scholar]
- 5.Daniels, R. H., P. S. Hall, and G. M. Bokoch. 1998. Membrane targeting of p21-activated kinase 1 (PAK1) induces neurite outgrowth from PC12 cells. EMBO J. 17:754-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster, R., K.-Q. Hu, Y. Lu, K. M. Nolan, J. Thissen, and J. Settleman. 1996. Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in vivo farnesylation. Mol. Cell. Biol. 16:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guasch, R. M., P. Scambler, G. E. Jones, and A. J. Ridley. 1998. RhoE regulates actin cytoskeleton organization and cell migration. Mol. Cell. Biol. 18:4761-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 9.Hansen, S. H., M. M. P. Zegers, M. Woodrow, P. Rodriguez-Viciana, P. Chardin, K. E. Mostov, and M. Mcmahon. 2000. Induced expression of Rnd3 is associated with transformation of polarized epithelial cells by the Raf-MEK-extracellular signal-regulated kinase pathway. Mol. Cell. Biol. 20:9364-9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito, W., H. Ishiguro, and K. Kurosawa. 1991. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene 102:67-70. [DOI] [PubMed] [Google Scholar]
- 11.Joberty, G., R. R. Perlungher, and I. G. Macara. 1999. The Borgs, a new family of Cdc42 and TC10 GTPase-interacting proteins. Mol. Cell. Biol. 19:6585-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaibuchi, K., S. Kuroda, and M. Amano. 1999. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu. Rev. Biochem. 68:459-486. [DOI] [PubMed] [Google Scholar]
- 13.Katoh, H., J. Aoki, A. Ichikawa, and M. Negishi. 1998. p160 RhoA-binding kinase ROKα induces neurite retraction. J. Biol. Chem. 273:2489-2492. [DOI] [PubMed] [Google Scholar]
- 14.Katoh, H., H. Yasui, Y. Yamaguchi, J. Aoki, H. Fujita, K. Mori, and M. Negishi. 2000. Small GTPase RhoG is a key regulator for neurite outgrowth in PC12 cells. Mol. Cell. Biol. 20:7378-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kjøller, L., and A. Hall. 1999. Signaling to Rho GTPases. Exp. Cell Res. 253:166-179. [DOI] [PubMed] [Google Scholar]
- 16.Manser, E., T. Leung, and L. Lim. 1995. Purification and assay of kinases that interact with Rac/Cdc42. Methods Enzymol. 256:215-227. [DOI] [PubMed] [Google Scholar]
- 17.Mitchison, T. J., and L. P. Cramer. 1996. Actin-based cell motility and cell locomotion. Cell 84:371-379. [DOI] [PubMed] [Google Scholar]
- 18.Nishi, M., H. Takeshima, T. Houtani, K. Nakagawa, T. Noda, and T. Sugimoto. 1999. RhoN, a novel small GTP-binding protein expressed predominantly in neurons and hepatic stellate cells. Mol. Brain Res. 67:74-81. [DOI] [PubMed] [Google Scholar]
- 19.Nobes, C. D., and A. Hall. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81:53-62. [DOI] [PubMed] [Google Scholar]
- 20.Nobes, C. D., I. Lauritzen, M.-G. Mattei, S. Paris, A. Hall, and P. Chardin. 1998. A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J. Cell Biol. 141:187-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quilliam, L. A., R. Khosravi-Far, S. Y. Huff, and C. J. Der. 1995. Guanine nucleotide exchange factors: activators of the Ras superfamily of proteins. Bioessays 17:395-404. [DOI] [PubMed] [Google Scholar]
- 22.Ridley, A. J., and A. Hall. 1992. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70:389-399. [DOI] [PubMed] [Google Scholar]
- 23.Ridley, A. J., H. F. Paterson, C. L. Johnston, D. Diekmann, and A. Hall. 1992. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70:401-410. [DOI] [PubMed] [Google Scholar]
- 24.Sahai, E., A. S. Alberts, and R. Treisman. 1998. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J. 17:1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki, T., and Y. Takai. 1998. The Rho small G protein family-Rho GDI system as a temporal and spatial determinant for cytoskeletal control. Biochem. Biophys. Res. Commun. 245:641-645. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt, A., and M. N. Hall. 1998. Signaling to the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 14:305-338. [DOI] [PubMed] [Google Scholar]
- 27.Van Aelst, L., and C. D'Souza-Schorey. 1997. Rho GTPases and signaling networks. Genes Dev. 11:2295-2322. [DOI] [PubMed] [Google Scholar]
- 28.Wünnenberg-Stapleton, K., I. L. Blitz, C. Hashimoto, and K. W. Y. Cho. 1999. Involvement of the small GTPases XRhoA and XRnd1 in cell adhesion and head formation in early Xenopus development. Development 126:5339-5351. [DOI] [PubMed] [Google Scholar]
- 29.Yasui, H., H. Katoh, Y. Yamaguchi, J. Aoki, H. Fujita, K. Mori, and M. Negishi. 2001. Differential response to NGF and EGF in neurite outgrowth of PC12 cells is determined by Rac1 activation systems. J. Biol. Chem. 276:15298-15305. [DOI] [PubMed] [Google Scholar]