Abstract
The hematopoietic, zinc-finger protein FOG-1 is essential for the development of the erythroid and megakaryocytic lineages. FOG-1's function in hematopoiesis is dependent on its ability to interact with the transcription factor GATA-1. FOG-1 has also been observed to interact with the corepressor molecule C-terminal binding protein (CtBP) through a peptide motif shared by all FOG family members. In this study, we confirmed that FOG-1 and CtBP interact by coimmunoprecipitation. We further demonstrate that a FOG-1 mutant unable to interact with CtBP has increased erythropoietic (but not megakaryocytic) rescue (relative to the wild type) of a FOG-1−/− cell line. To analyze further the physiological role of the FOG-1-CtBP interaction, we generated knock-in mice that express a FOG-1 variant unable to bind CtBP. Surprisingly, these mice are normal and fertile. Furthermore, erythropoiesis at all stages of development is normal in these mice. Erythrocyte production is similar in mutant and wild-type mice even under conditions of erythropoietic stress stimulated by either exogenously added erythropoietin or phenylhydrazine-induced anemia. Thus, despite conservation of the FOG-CtBP interaction site, the in vivo function of FOG-1 in erythroid development is not affected by its inability to interact with the corepressor CtBP.
FOG family proteins are cofactors for GATA transcription factors in diverse developmental contexts. In mice, FOG-1 is coexpressed in hematopoietic lineages with GATA-1, -2, and -3, whereas FOG-2 is coexpressed in the developing heart with GATA-4, -5, and -6 (16, 21, 29, 32). A Xenopus FOG homologue is expressed broadly and overlaps expression of all six Xenopus GATA factors, including in the heart and ventral blood island, where it appears to regulate erythropoiesis (7). In Drosophila, the FOG homologue U-shaped is coexpressed with the GATA factor Pannier in the nervous system, where it participates in control of sensory bristles (6, 11). In Drosophila hemocyte development, U-shaped appears to modulate serpent or other GATA factors (8).
FOG family proteins are related to each other by similar zinc fingers and the presence of a conserved site for interaction with C-terminal binding protein (CtBP). Four of the zinc fingers individually mediate physical interaction with the N-terminal zinc finger of GATA proteins (9). Disruption of this interaction results in phenotypes that mimic GATA factor loss. Hypomorphic alleles of U-shaped or Pannier, or specific mutations of Pannier that prevent its interaction with U-shaped, result in related defects in sensory bristle development (6, 11). In mice, disruption of either FOG-1 or GATA-1 results in embryonic lethality due to anemia with a developmental block at the proerythroblast stage (10, 31). Likewise, disruption of the GATA-1-FOG-1 interaction by a specific mutation of the N-terminal zinc finger of GATA-1 impairs red blood cell development and results in anemia in rare cases in humans (4, 20). Loss of FOG-2 or disruption of the interaction of FOG-2 with GATA-4 by a specific mutation of the N-terminal zinc finger of GATA-4 results in highly similar cardiac defects (5, 30). FOG family members share little sequence similarity in nonfinger regions except for the presence of a CtBP interaction site. Although FOG proteins are required for many aspects of GATA function, the mechanism by which they act to influence transcription is unknown. It has been suggested that FOG family members facilitate gene repression through interaction with the corepressor molecule CtBP (7, 9).
CtBP is a broadly expressed corepressor protein that binds Pro-X-Asp-Leu-Ser (PXDLS) motifs present in diverse nuclear regulatory proteins including Kruppel, Net, ZEB, and the adenoviral E1A oncoprotein (3, 13, 22, 23, 33). Disruption of the latter interaction increases tumorigenicity and transcription activation by E1A (2, 27). Disruption of CtBP's association with the TALE homeodomain protein, TGIF, is associated with a human disorder, familial holoprosencephaly (a disease of craniofacial development) (17). Within the hematopoietic system, CtBP has been suggested to play a role with BKLF, Ikaros, and Evi-1 (12, 15, 33). All FOG family members carry the PIDLS motif.
CtBP mediates repression through several mechanisms. CtBP binds to several histone deacetylases and the polycomb group protein human Polycomb2 (25). These groups of proteins mediate chromatin compaction through histone deacetylase-dependent and -independent mechanisms, respectively (15, 18, 28, 37). Similarly, CtBP is found in association with Ikaros, which itself is found in nucleosome remodeling and deacetylase complexes and silent, centromeric heterochromatin (14, 15). The transcriptional corepressor retinoblastoma protein and BRCA1 have been found in a complex with CtIP, a CtBP-interacting protein (18, 24, 36). Drosophila CtBP mediates repression by interacting with the short-range repressors Hairy, Knirps, and Snail (19). CtBP's ability to homodimerize potentially allows it to serve as a link between these transcriptional silencing complexes and molecules that bear a PXDLS motif (34).
Several studies support a role for CtBP as a transcriptional corepressor with GATA-FOG complexes. FOG-1 represses GATA activation in transient-transfection reporter assays depending on the cell line and promoter used (9). In addition, overexpression of murine FOG-2 in Xenopus embryos or Drosophila results in inhibition of blood formation (7, 8). All of these activities depend, in part, on FOG's ability to interact with CtBP. Amino acid substitutions in FOG-1 that impair interaction with CtBP relieve repression in reporter assays and augment blood formation in both Xenopus and Drosophila assays.
In this study, we examined the functional significance of FOG-1-CtBP interaction in mice in vivo. We tested this first in a cellular assay in which retrovirally expressed FOG-1 rescues erythroid and megakaryocytic maturation of a murine FOG-1 null cell line. We found that a mutant FOG-1 molecule with reduced binding affinity for CtBP rescues erythropoiesis considerably better than the wild-type molecule, consistent with CtBP acting as a corepressor in concert with FOG-1. We then tested the significance of this finding in vivo by creating a non-CtBP-interacting FOG-1 allele in mice. Surprisingly, such mutant mice develop normally and have no demonstrable changes in erythropoiesis. These results suggest that despite conservation of the CtBP binding site in all FOG family members, FOG-1's role in erythropoiesis does not require interaction with CtBP.
MATERIALS AND METHODS
Construction of the mutCtBP-FOG-1 targeting vector and generation of mutant mice.
A 10-kb KpnI-NotI fragment of murine Fog-1 genomic DNA was subcloned into a polylinker-modified Litmus 28 plasmid (New England Biolabs). A floxed neomycin expression cassette was blunt-end cloned into an SfiI site approximately 500 bp 3′ of the last FOG-1 exon. The PIDLSK sequence motif was mutated by PCR to PIASSK (ΔDL) in a FOG-1 cDNA clone, and a novel XhoI site was inserted to facilitate genotyping. This mutation was designed in a manner similar to that described and analyzed by Schaeper et al. (23) for E1A. An XbaI-to-NotI fragment containing the mutation was substituted in the targeting vector. The herpes simplex virus-thymidine kinase (HSV-TK) cassette was cloned into a SalI site 5′ of the homology region. The targeting construct was linearized with SspI and electroporated into CJ7 ES cells. Five positive clones were identified with a probe generated from a 3′ external SpeI-to-HindIII fragment. Two of these carried the mutation, as confirmed by restriction digestion with XhoI of genomic PCR products. A clone with a normal karyotype was injected into C57BL/6 blastocysts to generate chimeras. Genotyping for heterozygous mice was performed by Southern blot analysis. The neomycin resistance cassette was removed by mating to a ubiquitous Cre-expressing transgenic mouse, and subsequent genotyping was performed by PCR over the remaining LoxP site.
Sequencing of mutated locus.
Genomic DNA was isolated from tails of homozygous mutCtBP-FOG-1 or wild-type littermate control mice by standard procedures. The region encompassing the mutations was amplified by PCR using the oligonucleotides 5′-GCGTCGCAAGCTGTACGAGC-3′ and 5′-CAGTGGTGCGCAAAGGCGCG-3′ and cloned into the vector pGEM-T Easy (Promega, Inc., Madison, Wis.). Representative clones were subjected to automated fluorescent sequencing with an SP6 primer.
Retroviral infections of FOG-1−/− cells.
A hematopoietic cell line was derived from FOG-1−/− embryonic stem cells by in vitro differentiation followed by immortalization with HOX-11 (A. B. Cantor, S. G. Katz, and S. H. Orkin, submitted for publication). cDNAs encoding FOG-1 or mutCtBP-FOG-1 (FOG-1 ΔCtBP) were cloned into the retroviral expression vector MMP-IRES-GFP, and the resultant constructs were packaged into virus and used to infect the FOG-1−/− cells as described elsewhere (Cantor et al., submitted). After green fluorescent protein (GFP) selection and incubation for 6 days in the presence of erythropoietin (EPO; 2 U/ml) and 1% (vol/vol) thrombopoietin (recombinant human thrombopoietin tissue culture supernatant) (35), cells were washed three times in phosphate-buffered saline and used for either histocytochemistry, Western blot analysis, or semiquantitative reverse transcriptase PCR (RT-PCR) as described elsewhere (Cantor et al., submitted).
Hematological blood parameters.
Periorbital bleeds were performed on adult mice into EDTA tubes (catalog no. 365973; Becton Dickinson). Blood was diluted in citrate dextrose (catalog no. c-3821; Sigma, St. Louis, Mo.) and analyzed on an ADVI A120B hematology system blood cell analyzer. In addition, hematocrits were determined by capillary tube centrifugation.
Coimmunoprecipitation and Western blot analysis.
The cDNAs for murine CtBP1 and human CtBP2 were cloned into the mammalian expression vector pEF1αFLAG-pgkpuro, incorporating a FLAG epitope tag fused to the amino terminus of the expressed products. COS-7 cells (10-cm dish at 50 to 80% confluence) were transfected with 5 μg of either pEF1αFLAG-pgkpuro (empty vector), pEF1αFLAG-CtBP1-pgkpuro, pEF1αFLAG-CtBP2-pgkpuro, pEF1αHA-FOG-1-pgkpuro, or pEF1αHA-FOG-1mutCtBP-pgkpuro by using Fugene 6 (Roche) following the manufacturer's instructions. After 48 h, nuclear extracts were prepared (1) and diluted to reduce the final NaCl concentration to 150 mM. Diluted extracts were immunoprecipitated at 4°C for 12 h with 4.2 μg of a monoclonal anti-FLAG antibody (Sigma) in immunoprecipitation buffer (150 mM NaCl, 20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 mM dithiothreitol, 0.1% NP-40) containing protease inhibitors. Protein G-agarose (Roche) (50 μl of a 1:1 slurry) was added, and the samples were incubated for 1.5 h at 4°C with constant agitation. The agarose beads were washed three times with cold immunoprecipitation buffer, and proteins were eluted by heating at 95 to 100°C in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Eluted proteins were separated by SDS-PAGE on 7.5% acrylamide gels and transferred to nitrocellulose (Schleicher & Schuell). Blots were probed with either antihemagglutinin (anti-HA) antibody (Y-11; Santa Cruz Biotechnology) at a 1:1,000 dilution or anti-FLAG antibody (M2) at a 1:1,000 dilution followed by appropriate horseradish peroxidase (HPR)-conjugated secondary antibodies, washed, and developed with enhanced chemiluminescence detection (Amersham). Blots were exposed to Kodak X-Omat AR film for approximately 5 s and developed. Nuclear extract (5 μl, representing 6.25% of the immunoprecipitated input material) was also separated by SDS-PAGE and immunoblotted as a control.
In vivo stimulation of erythropoiesis.
Mice were injected subcutaneously with 20 U of EPO on days 2, 4, 6, 8, 10, 12, and 14, as previously described (26). Hematological parameters were measured before and after EPO treatment by phlebotomizing on days 1 and 15.
Mice were injected intraperitoneally with 60 mg of phenylhydrazine (PHZ) per kg of body weight on days 1 and 2. Periorbital bleeds for hematocrits were performed on days 1, 4, 6, 9, 13, and 15.
All mouse protocols complied with federal and Children's Hospital policies.
RESULTS
CtBP binds to FOG-1 through a conserved PIDLS motif.
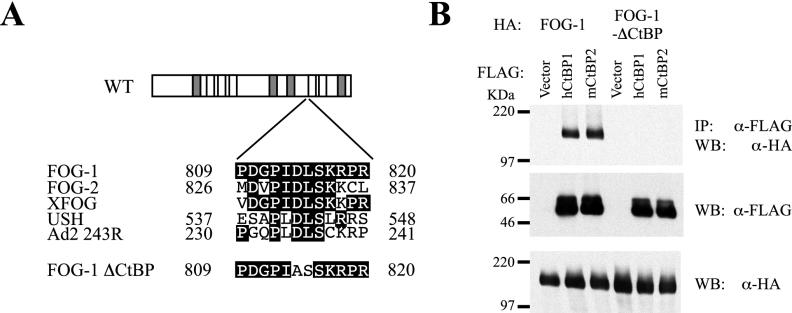
FOG-1 interacts with CtBP through the PIDLS motif located between zinc fingers 6 and 7 (9). This motif is conserved in FOG family members from Drosophila to humans despite low sequence conservation beyond the zinc fingers (Fig. 1A). Mutation of the PLDLS motif to PLASS in the adenoviral E1A oncoprotein disrupts its binding to CtBP (23). A coimmunoprecipitation experiment was performed to test if full-length FOG-1 interacts with CtBP1 and/or CtBP2 in mammalian cells and if this interaction is disrupted by similar mutations (change of aspartic acid 814 to alanine and leucine 815 to serine [PIDLS to PIASS]) (Fig. 1B). Expression vectors encoding either wild-type or mutated HA-tagged FOG-1 and FLAG-tagged human CtBP1 or murine CtBP2 were transfected into COS cells. Nuclear extracts were immunoprecipitated with an anti-FLAG antibody and immunoblotted with an anti-HA antibody. HA-tagged FOG-1 was detected in both CtBP1- and CtBP2-transfected extracts but not when the PIDLS motif was mutated. Therefore, wild-type FOG-1 is capable of interacting with either CtBP1 or CtBP2, and this interaction is abolished by mutating the PIDLS motif to PIASS.
FIG. 1.
Interaction of FOG-1 with CtBP family members in mammalian cells. (A) Schematic depiction of conserved PIDLS motif from adenovirus E1A in FOG family members and the mutation in FOG-1 that disrupts CtBP binding. Boxes represent zinc fingers; shaded boxes represent GATA-binding zinc fingers. XFOG, Xenopus FOG; USH, Drosophila U-shaped; Ad2 243R, adenovirus E1A. (B) Coimmunoprecipitation of HA-FOG-1 and HA-mutCtBP-FOG-1 (FOG-1 ΔCtBP) with FLAG-tagged human CtBP2 or murine CtB2 in transfected COS cells. Cells were transfected with plasmids expressing the indicated proteins, and nuclear lysates were immunoprecipitated (IP) with anti-FLAG antibody. Western blot (WB) analysis was performed with anti-HA antibody or anti-FLAG antibody.
Mutation of the conserved CtBP binding motif in FOG-1 results in enhanced erythroid rescue of a FOG-1 null cell line.
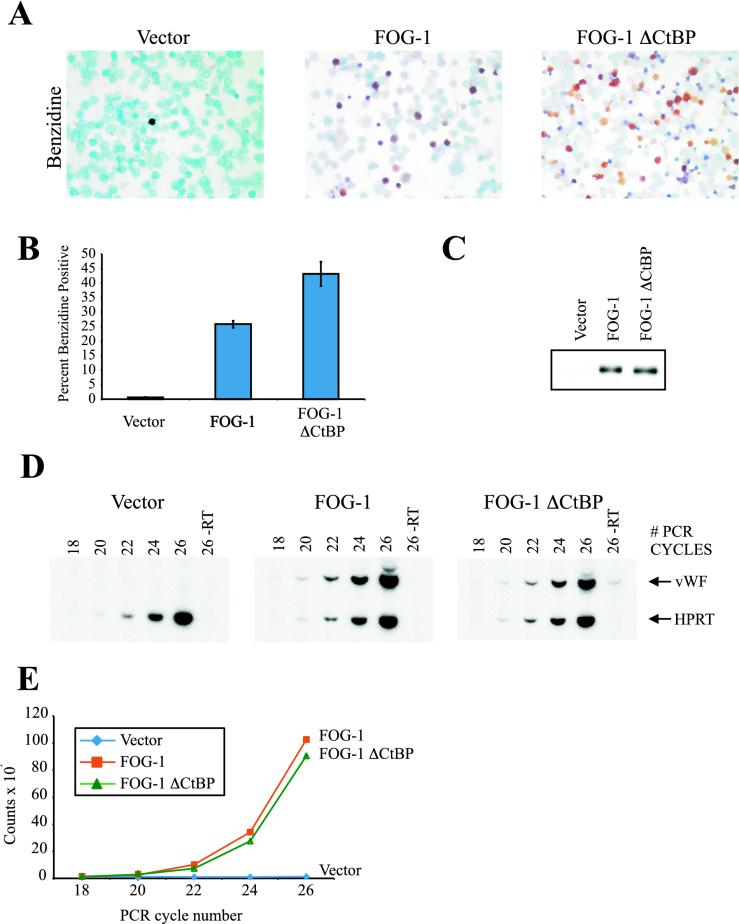
We initially tested the functional significance of FOG-1-CtBP interaction using a cell-based assay of FOG-1 activity. This system is based on the rescue of terminal erythroid and megakaryocytic maturation of a Hox-11 immortalized FOG-1−/− cell line by retrovirally expressed FOG-1 (Cantor et al., submitted). We infected FOG-1−/− cells with an MMP-IRES-GFP retrovirus expressing either the wild-type or CtBP-binding motif mutant FOG-1 molecule (mutCtBP-FOG-1). GFP-positive cells were isolated by fluorescence-activated cell sorting and cultured in the presence of EPO and thrombopoietin for 6 days. Compared to wild-type-rescued cells, the cells rescued with mutCtBP-FOG-1 exhibited marked enhancement of erythroid rescue (Fig. 2A and B), despite equivalent expression of the transgenes (Fig. 2C). In contrast to the enhanced erythroid rescue of these cells, there was no significant difference in rescue of megakaryocyte development based on RT-PCR of vWF (Fig. 2D and E), on the percentage of acetylcholinesterase-positive cells (vector, 0.6% ± 0.2%; FOG-1, 13.2% ± 1.6%; mutCtBP-FOG-1, 18.5% ± 4.0%), or on total acetylcholinesterase activity of rescued cells (1.3 ± 0.2, total acetylcholinesterase activity of mutCtBP-FOG-1-rescued cells relative to wild-type-rescued cells).
FIG. 2.
Mutation of the FOG-1-CtBP interaction motif enhances FOG-1-mediated rescue of erythropoiesis but not megakaryopoiesis of a FOG-1−/− hematopoietic cell line. (A) Benzidine stains of FOG-1−/− cells rescued with each of the constructs. Magnification, ×400. Positive (hemoglobinized) cells appear black or brown. (B) Percent benzidine-positive cells obtained from at least 10 independent rescue experiments, given as means ± standard errors of the means. (C) Western blot analysis of rescued cells for expression of each construct. Nuclear extracts were prepared from a portion of the rescued cells 1 day after sorting for GFP expression. Equivalent amounts of total protein were separated by SDS-PAGE and immunoblotted with an anti-HA antibody. (D) Semiquantitative RT-PCR analysis of cells rescued with each construct. RT-PCR using vWF- and HPRT-specific primers was performed with [32P]dCTP for radioisotope incorporation, and the products were separated by PAGE. (E) Bands from panel D were quantified with a PhosphorImager (Molecular Dynamics, Inc.). Data are shown as the vWF signal normalized to the HPRT control signal.
Generation of mutCtBP-FOG-1 knock-in mice.
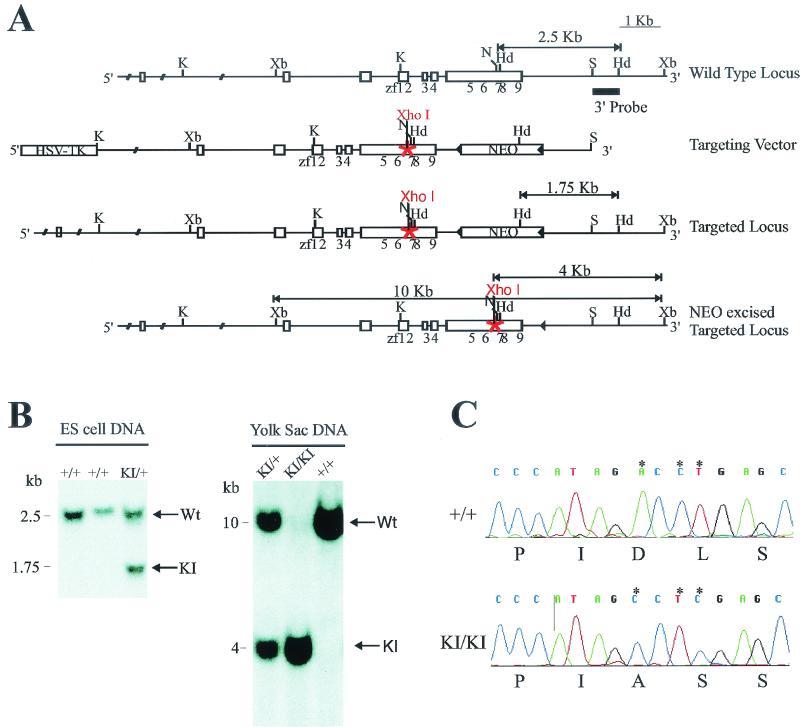
To assess the in vivo significance of these findings (Fig. 2A), we generated mice harboring a knock-in mutation at the FOG-1 locus that prevents interaction with CtBP. A targeting vector designed to introduce the PIDLS-to-PIASS mutation in the FOG-1 genomic locus was electroporated into ES cells (Fig. 3A). Properly targeted ES cells with both the knock-in mutation and a floxed neomycin cassette 3′ to FOG-1 were injected into host blastocysts to generate chimeric mice. Chimeras were bred to mice ubiquitously expressing Cre recombinase to remove the neomycin cassette and generate FOG-1 knock-in heterozygotes (+/KI). The presence of the mutation was identified by Southern blot analysis utilizing a unique XhoI restriction site introduced as part of the mutation (Fig. 3B). In addition, we confirmed the presence of the mutations by direct DNA sequencing from PCR fragments generated from the genomic DNA (Fig. 3C). Upon interbreeding of heterozygous mice, the expected genotypes were generated at a Mendelian ratio (Table 1). Thus, the FOG-1-CtBP interaction is not essential for survival.
FIG. 3.
Targeting the FOG-1-CtBP interaction in mice. (A) Partial restriction map of the murine Fog-1 wild-type locus, the Fog-1 knock-in targeting vector, and targeted homologous recombination before and after Cre-mediated excision of the selection cassette. The targeting construct contains the HSV-TK and neomycin resistance (NEO) cassettes under the control of the mouse phosphoglycerate kinase promoter. Homologous recombination results in replacement of wild-type Fog-1 with genomic DNA harboring a mutation of aspartic acid 814 to alanine and leucine 815 to serine (PIDLS to PIASS), as well as the incorporation of the neomycin resistance cassette. Fog-1 coding exons are shown as empty boxes, with positions of zinc fingers (zf) underneath. Solid triangles are LoxP sites. The probe used in Southern blotting is shown as a black line. K, KpnI; Xb, XbaI; N, NotI; Hd, HindIII; S, SpeI. (B) Southern blot analysis of ES cell DNA or yolk sac DNA showing the presence of wild-type (+/+), heterozygous (KI/+), and homozygous mutant (KI/KI) genotypes. (C) Sequencing chromatograms obtained with PCR fragments generated from genomic DNA from a homozygous mutCtBP-FOG-1 mouse and from a wild-type littermate.
TABLE 1.
Survival of mutCtBP-FOG-1 knock-in mice
| Type of value | Survivala
|
||
|---|---|---|---|
| +/+ | +/KI | KI/KI | |
| Observed | 11 | 33 | 17 |
| Expected | 15 | 31 | 15 |
Number of pups of each genotype produced by mating of heterozygous (+/KI) parents.
Normal red cell function in mutCtBP-FOG-1 knock-in mice.
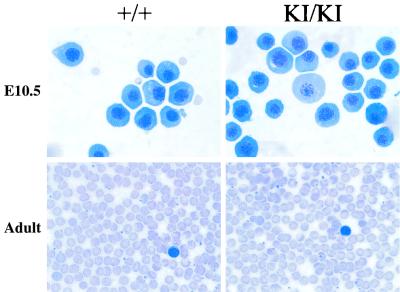
Blood counts were performed on FOG-1 KI/KI adult mice to assess the functional significance of FOG-1-CtBP interactions during steady-state hematopoiesis. In contrast to the results obtained from the in vitro cellular assay, the KI/KI mice exhibited no increase in hematocrit or hemoglobin compared to heterozygous or wild-type mice (Table 2). Furthermore, other blood cell parameters were unremarkable (Table 2). Blood smears exhibited no changes in red blood cell morphology (Fig. 4).
TABLE 2.
Blood indices of mutCtBP-FOG-1 knock-in mice
| Mouse genotype (n) | Mean ± SEa
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RBC (103/mm3) | Hemoglobin (g/dl) | Hematocrit (%) | MCV (fl) | MCHC (g/dl) | RBC distribution width (%) | Reticulocytes (%) | CHr (pg) | Platelets (103/mm3) | WBC (103/mm3) | |
| +/+ (6) | 9.3 ± 0.5 | 15.1 ± 0.6 | 51.1 ± 1.8 | 55.8 ± 3.0 | 29.8 ± 0.8 | 14.6 ± 1.5 | 2.9 ± 0.6 | 15.2 ± 0.6 | 1,145.8 ± 46.5 | 4.7 ± 0.6 |
| KI/+ (21) | 9.3 ± 0.2 | 15.0 ± 0.3 | 51.1 ± 1.4 | 54.7 ± 1.1 | 29.6 ± 0.3 | 13.5 ± 0.2 | 3.6 ± 0.2 | 16.0 ± 0.2 | 1,099.8 ± 67.7 | 5.3 ± 0.4 |
| KI/KI (17) | 9.3 ± 0.2 | 14.2 ± 0.9 | 50.0 ± 1.4 | 53.7 ± 1.0 | 28.5 ± 1.7 | 13.5 ± 0.3 | 2.9 ± 0.3 | 15.8 ± 0.3 | 856.6 ± 67.4 | 5.2 ± 0.4 |
RBC, red blood cells; MCV, mean corpuscular volume; MCHC, mean corpuscular hemoglobin concentration; CHr, concentration of hemoglobin in reticulocytes; WBC, white blood cells.
FIG. 4.
Normal primitive and definitive erythropoiesis in mice lacking FOG-1-CtBP interaction. (Top) May-Grunwald-Giemsa staining of blood cells from wild-type and mutant yolk sacs at embryonic day 10.5 (E10.5). Magnification, ×1,000. (Bottom) May-Grunwald-Giemsa staining of blood cells from wild-type and mutant adult mice. Magnification, ×400.
To exclude selection against abnormal red cells as an explanation for these findings, we examined mice in the early perinatal period. Hematocrits were normal at this time (Table 3). Blood smears of primitive erythrocytes at the yolk sac stage of development were also normal (Fig. 4). These data show that a FOG-1-CtBP interaction is not essential for red cell function during development or in the adult at steady state.
TABLE 3.
Hematocrits of neonatal mutCtBP-FOG-1 knock-in mice
| Agea | Mean hematocrit ± SE (n)
|
||
|---|---|---|---|
| +/+ | +/KI | KI/KI | |
| P1 | 44 ± 0.9 (3) | 37 ± 4.2 (7) | 42 ± 1.5 (6) |
| P3 | 25 ± 0.9 (10) | 23 ± 1.0 (16) | 29 ± 1.5 (2) |
| P6 | 32 ± 1.2 (11) | 32 ± 2.5 (3) | 35 ± 1.9 (4) |
P, postnatal day of life, where birth is day 1.
Normal response to stress in mutCtBP-FOG-1 knock-in mice.
A second possibility to account for differences between our in vitro and in vivo studies relates to in vivo compensation for increased erythropoietic drive. For instance, reduced levels of EPO might lead to normal red cell formation in the face of increased intrinsic drive. That is, red blood cell precursors in KI/KI mutant mice might be more sensitive to EPO. No reliable methods are available to measure murine EPO levels, especially below normal. As an alternative means of testing this possibility, recombinant EPO or an equivalent amount of saline was injected intraperitoneally into wild-type and mutant mice, and differences in red blood cell production were analyzed. Mice were injected every other day for a total of seven injections. Complete blood counts were measured before and after treatment. As anticipated, mice treated with EPO demonstrated a rise in both hematocrit and hemoglobin compared to mice injected with saline. In KI/KI mice, no significant difference in EPO responsiveness was seen. The absence of an altered response in the KI/KI mice suggests that the EPO pathway is not a means of compensation within the mutant mice.
FIG. 5.
Percent change in hematocrit in wild-type and mutant mice due to EPO administration. Hematocrits measured before and after EPO treatment were subtracted, divided by the initial value, and multiplied by 100. Data are means ± standard errors of the means.
We also examined the recovery of KI/KI mice from acute anemia induced by PHZ administration (Fig. 6). PHZ (60 mg/kg) was injected on two consecutive days, and mice were phlebotomized on days 1, 4, 6, 9, 13, and 15. PHZ administration induced anemia by day 4. Recovery occurred between days 9 and 13. KI/KI mice became anemic and recovered at the same rate as wild-type mice.
FIG. 6.
Hematocrits of wild-type and mutant mice during recovery from anemia induced by PHZ (two consecutive injections, indicated by arrows). Results are means ± standard errors of the means.
DISCUSSION
The development of specific cell lineages requires the interplay of multiple transcription factors. The direct interaction of GATA-1 with FOG-1 is essential for differentiation of mature erythroid and megakaryocytic lineages (4, 20). In this study we assessed the in vivo significance of the potential interaction between FOG-1 and the corepressor molecule CtBP. As all known members of the FOG family of proteins share a conserved CtBP binding motif, it seemed plausible that this interaction plays a critical role in FOG's function. We disrupted the interaction by mutating the PIDLS motif in FOG-1, which mediates interaction with CtBP. In vitro, mutCtBP-FOG-1 was able to rescue erythropoiesis of a FOG-1 null, bipotential cell line somewhat better than the wild type. However, mice harboring the same mutation in FOG-1 exhibited entirely normal erythropoiesis. Furthermore, no differences were observed under conditions of increased erythropoietic drive.
These results are unexpected in light of observations regarding FOG-1 and CtBP in other experimental systems. Transient-transfection experiments in NIH 3T3 fibroblasts demonstrated a repressive role for FOG-1 at the mouse α-globin and EKLF promoters (9). Repression was partially relieved by mutating FOG-1's CtBP interaction motif. Injection of mouse FOG-1 or FOG-2 or Xenopus FOG into Xenopus embryos inhibited red blood cell formation. However, overexpression of FOG-2 containing a mutation in its CtBP motif resulted in an increase in red blood cell production (7). Finally, overexpression of U-shaped, FOG-1, or FOG-2 in Drosophila inhibited formation of crystal cells (i.e., blood cells of Drosophila) (8). Inhibition was not observed when a non-CtBP-interacting FOG-2 mutant molecule was tested. However, mutant CtBP and wild-type FOG constructs had similar effects on heart and eye development. Taken together, these results suggest that FOG-1 might inhibit red blood cell formation through transcriptional repression mediated by CtBP, at least under certain conditions.
Several explanations might account for the differences between previous observations and the absence of an increased production of erythroid cells in the knock-in mice reported here. First, overexpression of FOG proteins might lead to nonphysiological effects, whereas in knock-in mice, FOG-1 protein is expressed at an appropriate level. Alternatively, the mutant mouse might be able to compensate for changes in erythroid cell function resulting from expression of mutant FOG-1. Such compensation would presumably occur through a pathway independent of CtBP. Finally, FOG-1-CtBP interaction might be critical in development in a setting other than erythropoiesis. Our study illustrates once again the limitations of in vitro assay systems for assessing consequences of specific protein interactions for development and demonstrates the need to validate findings in vivo.
Based on our in vivo observations, we conclude that repression by FOG-1 mediated through interaction with CtBP is dispensable for normal erythropoiesis. Efforts are under way to determine what other molecules, coactivators or corepressors, might be associated with GATA-FOG-1 complexes in hematopoiesis. It will also be of interest to determine if FOG-1 or FOG-2 molecules fulfill their predicted repressive roles in other contexts.
Acknowledgments
The first two authors contributed equally to this work.
We thank S. Tevosian for generously providing us with the mutCtBP-FOG-1 cDNA. We thank G. Chinnadurai and M. Crossley for providing cDNAs for murine CtBP1 and human CtBP2, respectively. We also thank Y. Fujiwara, C. Browne, A. Chapdelaine, and S. Galusha for their expertise and technical assistance.
A.B.C. is supported by National Institutes of Health grant 5K08CA82175. S.H.O. is an Investigator of the Howard Hughes Medical Institute. Partial support for these studies was derived from an NIH grant to S.H.O.
REFERENCES
- 1.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, J. M., T. Subramanian, U. Schaeper, M. La Regina, S. Bayley, and G. Chinnadurai. 1993. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 12:469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Criqui-Filipe, P., C. Ducret, S. M. Maira, and B. Wasylyk. 1999. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 18:3392-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crispino, J. D., M. B. Lodish, J. P. Mackay, and S. H. Orkin. 1999. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell 3:219-228. [DOI] [PubMed] [Google Scholar]
- 5.Crispino, J. D., M. B. Lodish, B. L. Thurberg, S. H. Litovsky, T. Collins, J. D. Molkentin, and S. H. Orkin. 2001. Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG factors. Genes Dev. 15:839-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cubadda, Y., P. Heitzler, R. P. Ray, M. Bourouis, P. Ramain, W. Gelbart, P. Simpson, and M. Haenlin. 1997. u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila. Genes Dev. 11:3083-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deconinck, A. E., P. E. Mead, S. G. Tevosian, J. D. Crispino, S. G. Katz, L. I. Zon, and S. H. Orkin. 2000. FOG acts as a repressor of red blood cell development in Xenopus. Development 127:2031-2040. [DOI] [PubMed] [Google Scholar]
- 8.Fossett, N., S. G. Tevosian, K. Gajewski, Q. Zhang, S. H. Orkin, and R. A. Schulz. 2001. The Friend of GATA proteins U-shaped, FOG-1, and FOG-2 function as negative regulators of blood, heart, and eye development in Drosophila. Proc. Natl. Acad. Sci. USA 98:7342-7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox, A. H., C. Liew, M. Holmes, K. Kowalski, J. Mackay, and M. Crossley. 1999. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 18:2812-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiwara, Y., C. P. Browne, K. Cunniff, S. C. Goff, and S. H. Orkin. 1996. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 93:12355-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haenlin, M., Y. Cubadda, F. Blondeau, P. Heitzler, Y. Lutz, P. Simpson, and P. Ramain. 1997. Transcriptional activity of Pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev. 11:3096-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izutsu, K., M. Kurokawa, Y. Imai, K. Maki, K. Mitani, and H. Hirai. 2001. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor beta signaling. Blood 97:2815-2822. [DOI] [PubMed] [Google Scholar]
- 13.Katsanis, N., and E. M. Fisher. 1998. A novel C-terminal binding protein (CTBP2) is closely related to CTBP1, an adenovirus E1A-binding protein, and maps to human chromosome 21q21.3. Genomics 47:294-299. [DOI] [PubMed] [Google Scholar]
- 14.Kim, J., S. Sif, B. Jones, A. Jackson, J. Koipally, E. Heller, S. Winandy, A. Viel, A. Sawyer, T. Ikeda, R. Kingston, and K. Georgopoulos. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10:345-355. [DOI] [PubMed] [Google Scholar]
- 15.Koipally, J., and K. Georgopoulos. 2000. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem. 275:19594-19602. [DOI] [PubMed] [Google Scholar]
- 16.Laverriere, A. C., C. MacNeill, C. Mueller, R. E. Poelmann, J. B. Burch, and T. Evans. 1994. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J. Biol. Chem. 269:23177-23184. [PubMed] [Google Scholar]
- 17.Melhuish, T. A., and D. Wotton. 2000. The interaction of the carboxyl terminus-binding protein with the Smad corepressor TGIF is disrupted by a holoprosencephaly mutation in TGIF. J. Biol. Chem. 275:39762-39766. [DOI] [PubMed] [Google Scholar]
- 18.Meloni, A. R., E. J. Smith, and J. R. Nevins. 1999. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc. Natl. Acad. Sci. USA 96:9574-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nibu, Y., H. Zhang, and M. Levine. 1998. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science 280:101-104. [DOI] [PubMed] [Google Scholar]
- 20.Nichols, K. E., J. D. Crispino, M. Poncz, J. G. White, S. H. Orkin, J. M. Maris, and M. J. Weiss. 2000. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA-1. Nat. Genet. 24:266-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orkin, S. H. 1992. GATA-binding transcription factors in hematopoietic cells. Blood 80:575-581. [PubMed] [Google Scholar]
- 22.Postigo, A. A., and D. C. Dean. 1999. ZEB represses transcription through interaction with the corepressor CtBP. Proc. Natl. Acad. Sci. USA 96:6683-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA 92:10467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaeper, U., T. Subramanian., L. Lim, J. M. Boyd, and G. Chinnadurai. 1998. Interaction between a cellular protein that binds to the c-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J. Biol. Chem. 273:8549-8552. [DOI] [PubMed] [Google Scholar]
- 25.Sewalt, R. G., M. J. Gunster, J. van der Vlag, D. P. Satijn, and A. P. Otte. 1999. C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol. 19:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silver, D. F., and M. S. Piver. 1999. Effects of recombinant human erythropoietin on the antitumor effect of cisplatin I SCID mice bearing human ovarian cancer: a possible oxygen effect. Gynecol. Oncol. 73:280-284. [DOI] [PubMed] [Google Scholar]
- 27.Sollerbrant, K., G. Chinnadurai, and C. Svensson. 1996. The CtBP binding domain in the adenovirus E1A protein controls CR1-dependent transactivation. Nucleic Acids Res. 24:2578-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sundqvist, A., K. Sollerbrant, and C. Svensson. 1998. The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein-histone deacetylase complex. FEBS Lett. 429:183-188. [DOI] [PubMed] [Google Scholar]
- 29.Tevosian, S. G., A. E. Deconinck, A. B. Cantor, H. I. Reiff, Y. Fujiwara, G. Corfas, and S. H. Orkin. 1999. FOG-2: a novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc. Natl. Acad. Sci. USA 96:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tevosian, S. G., A. E. Deconinck, M. Tanaka, M. Schinke, L. H. Silvio, S. Izumo, Y. Fujiwara, and S. H. Orkin. 2000. FOG-2, a cofactor of GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell 101:729-739. [DOI] [PubMed] [Google Scholar]
- 31.Tsang, A. P., Y. Fujiwara, D. B. Hom, and S. H. Orkin. 1998. Failure of megakaryocytopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcription factor cofactor FOG. Genes Dev. 12:1176-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsang, A. P., J. E. Visvader, A. C. Turner, Y. Fujiwara, C. Yu, M. Weiss, M. Crossley, and S. H. Orkin. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109-119. [DOI] [PubMed] [Google Scholar]
- 33.Turner, J., and M. Crossley. 1998. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 17:5129-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner, J., and M. Crossley. 2001. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. Bioessays 23:683-690. [DOI] [PubMed] [Google Scholar]
- 35.Villeval, J.-L., K. Cohen-Solal, M. Tulliez, S. Giraudier, J. Guichard, S. A. Burstein, E. M. Cramer, W. Vainchenker, and F. Wendling. 1997. High thrombopoietin production by hematopoietic cells induces a fatal myeloproliferative syndrome in mice. Blood 90:4369. [PubMed] [Google Scholar]
- 36.Yu, X., L. C. Wu, A. M. Bowcock, A. Aronheim, and R. Baer. 1998. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 273:25388-25392. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, C. L., T. A. McKinsey, J. R. Lu, and E. N. Olson. 2001. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem. 276:35-39. [DOI] [PubMed] [Google Scholar]