Abstract
Hypoxia-inducible factor 1 complex (HIF-1) plays a pivotal role in oxygen homeostasis and adaptation to hypoxia. Its function is controlled by both the protein stability and the transactivation activity of its alpha subunit, HIF-1α. Hydroxylation of at least two prolyl residues in the oxygen-dependent degradation domain of HIF-1α regulates its interaction with the von Hippel-Lindau protein (VHL) that targets HIF-1α for ubiquitination and proteasomal degradation. Several prolyl hydroxylases have been found to specifically hydroxylate HIF-1α. In this report, we investigated possible roles of VHL and hydroxylases in the regulation of the transactivation activity of the C-terminal activating domain (CAD) of HIF-1α. We demonstrate that regulation of the transactivation activity of HIF-1α CAD also involves hydroxylase activity but does not require functional VHL. In addition, stimulation of the CAD activity by a hydoxylase inhibitor, hypoxia, and desferrioxamine was severely blocked by the adenoviral oncoprotein E1A but not by an E1A mutant defective in targeting p300/CBP. We further demonstrate that a hydroxylase inhibitor, hypoxia, and desferrioxamine promote the functional and physical interaction between HIF-1α CAD and p300/CBP in vivo. Taken together, our data provide evidence that hypoxia-regulated stabilization and transcriptional stimulation of HIF-1α function are regulated through partially overlapping but distinguishable pathways.
Lack of oxygen (hypoxia) is a common pathological cause in many human diseases, such as stroke, coronary disease, and pulmonary disorders. Maintenance of oxygen homeostasis and adaptation to hypoxia in higher eukaryotes require hypoxia-inducible factor 1 (HIF-1), a heterodimeric DNA-binding complex that acts as a key regulator of hypoxia-responsive genes. Among its transcriptional targets, HIF-1 activates genes with critical roles in erythropoiesis, angiogenesis, vasomotor function, and intracellular energy metabolism (43). The HIF-1 complex is composed of two subunits (49): HIF-1β (ARNT), which is constitutively expressed, and HIF-1α, which is constantly translated but rapidly degraded under normoxic conditions. The degradation of HIF-1α is mediated by the ubiquitin-proteasome system (14, 22, 38) and requires a highly conserved domain, termed the oxygen-dependent degradation domain (ODD) (15, 33, 45). Lack of oxygen stabilizes HIF-1α, allowing its translocation to the nucleus and its interaction with HIF-1β to form the HIF-1 complex (8, 21). Genetic knockout studies reveal an essential role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation, and tumorigenesis (6, 36, 37). Nonhypoxic stabilization of HIF-1α is observed in response to growth factors and cytokines and also in solid tumors, suggesting a more general role of HIF-1 in tumorigenesis and inflammation (35, 48, 53).
HIF-1α degradation depends on its interaction with the von Hippel-Lindau protein (VHL) (47). VHL is a tumor suppressor whose defect defines a dominantly inherited cancer syndrome, von Hippel-Lindau syndrome (27). Patients with the VHL syndrome are predisposed to develop multiple hemangioblastomas of the central nerve system and retina, renal cell carcinomas, pheochromocytomas, pancreatic carcinoma, and cysts in the kidneys, liver, and pancreas (17, 24, 50). While the functions of VHL have not been completely characterized yet, several studies indicate that VHL is a component of an E3 ubiquitin protein-ligase complex that targets HIF-1α for ubiquitination and proteasomal degradation under normoxic condition (9, 18, 23, 30, 31, 46). Loss of VHL function leads to stabilization of HIF-1α, providing an explanation for the hypervascular nature of VHL-associated neoplasms.
The interaction between VHL and HIF-1α depends on the hydroxylation of proline residues (Pro-402 and Pro-564) in the oxygen-dependent degradation domain of HIF-1α (17, 19, 29, 51). These hydroxylations are mediated by recently identified HIF-specific prolyl hydroxylases (HIF-PHDs) that require oxygen, 2-oxoglutarate, and iron for their full activity (5, 12).
In addition to regulating protein stabilization, hypoxia enhances the transactivation activity of HIF-1α, thus providing a multistep process of regulation (20, 33, 47). Earlier reports have indicated that a region adjacent to the transactivation domains inhibits the transactivation activity of HIF-1α, and this inhibition can be relieved by hypoxia (20). HIF-1α transactivation involves the physical interaction with p300/CBP (1, 10, 21), two coactivators required for the transcriptional activity of a variety of transcription factors, such as MyoD and p53 (2, 34, 41, 52). It has been suggested that hypoxia enhances the recruitment of p300/CBP through a redox-mediated mechanism (7, 11, 13, 21, 26).
In this report, we examine the possible role of VHL and hydroxylases in the regulation of the carboxyl-terminal transactivation activity of HIF-1α. Our data indicate that while both protein stability and the transactivation activity are regulated by the same initial signals and involve hydroxylase activity, two partially overlapping but distinguishable pathways are utilized: one requires VHL, and the other one is VHL independent.
MATERIALS AND METHODS
Plasmids and DNA recombination.
The pM vector encoding the DNA binding domain (DBD) of yeast transcription factor GAL4 and pRL-CMV were purchased from Clontech. pGH1α530-778 and pGH1α740-826 were constructed by inserting a PCR fragment encoding this region into pM as described previously (44, 45). pGH1α786-826 was generated by PCR and subcloned into the pM vector at EcoRI-BamHI sites. pGH1α740-813 was generated by digesting pGH1α740-826 with PstI followed by self-ligation that deletes a small DNA fragment encoding the last 13 amino acids of HIF-1α. Plasmids expressing PHD1, PHD2, and PHD3 were a generous gift from P. Ratcliffe (Oxford University, Oxford, United Kingdom) (12). pFR-luc, a luciferase reporter under the control of a promoter with GAL4 binding sites, was purchased from Stratagene. pEpo-luc was described previously (38). pG4CH1 (amino acids [aa] 300 to 523 of p300 fused with the Gal4 DNA binding domain) and pVP16H1α723-826 (aa 723 to 826 fused with VP16) were kindly provided by D. M. Livingston (Harvard Medical School) (4, 25). pCMVβp300 and pSV40-lacZ were described previously (39).
Antibodies and special chemicals.
Antibodies against HIF-1α (monoclonal; catalog no. 610959), VHL (mouse monoclonal; clone Ig32) and p300 (monoclonal; clone NM11) were purchased from Transduction Lab/Pharmagen. Monoclonal anti-Gal4-DBD antibodies were purchased from Clontech (catalog no. 5399-1) or Santa Cruz (catalog no. SC-510). Monoclonal anti-E1A antibody M73 was a gift from A. Giordano and was described previously (42). Desferrioxamine and cobaltous chloride were purchased from Sigma. Dimethyloxalylglycine (DOG) and anti-HIF-2α were a generous gift from Ratcliffe (Oxford University) (19, 30). Horseradish-peroxidase (HRP)-labeled goat anti-mouse immunoglobulin G (IgG) or goat anti-rabbit IgG antibodies were purchased from GIBCO/Invitrogen. HRP-labeled goat anti-mouse IgG Fc fragments were purchased from Sigma.
Cell lines, tissue culture, and transfection.
HeLa, Hep3B, 143B, and the VHL-deficient 786-O cell line were purchased from American Type Culture Collection. 786-O#52, expressing wild-type VHL under the Tet-off* control system (Clontech), and 786-O#126, transfected with the backbone vector lacking VHL, were a generous gift from N. Kley (Bristol Myers Squibb). HeLa, 143B, 786-O, and derivative lines were cultured in Dulbecco's modified Eagle medium (DMEM) (Mediatech) supplemented with 10% fetal bovine serum (HyClone), 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Hep3B cells were cultured in minimal essential medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin. Transient transfection was performed with the LipofectAmine Plus reagent (GIBCO/Invitrogen) following the manufacturer's instructions. Serum-free procedures were used for transfection. pRL-CMV (Promega) or pSV40-lacZ were cotransfected for normalization of transfection efficiency. Twenty-four hours after transfection, cells were trypsinized, split into multiwell plates, and exposed to the various experimental conditions.
Hypoxia treatment.
Cell cultures were treated as previously described in a modular incubator (Billups-Rothenburg) and flushed with a gas mixture of 0.5% O2-5% CO2, and balanced with nitrogen (3). In some experiments, cells were directly incubated in a hypoxic workstation (In VIVO2) and the percentage of oxygen in mixed air was described for each experiment.
Luciferase assays.
For single luciferase assays, cells were washed once with 1× phosphate-buffered saline and lysed on plates with 1× reporter lysis buffer (Promega). After incubation at room temperature for 5 min, cells were scraped and collected into Eppendorf tubes. After vigorous vortex and centrifugation, the supernatant was taken for the luciferase assay. For dual luciferase assays, cells were washed and lysed on plates with 1× passive cell lysis buffer (Promega). The cells were scraped, collected, and subjected to three cycles of freezing-thawing. Luciferase assays were performed with the assay reagents purchased from Promega by following the manufacturer's instruction. The luminescence was measured in a TD20/20 luminometer (Promega). When applicable, β-galactosidase activity was assayed in 1× Z-buffer (42) containing 4 mg of o-nitrophenyl β-d-galactopyranoside (Sigma)/ml.
Cell lysate preparation, immunoprecipitation, and Western blot.
For whole-cell lysates, cell pellets were collected and lysed on ice for 1 h with 1× hydroxylase buffer (50 mM Tris-Cl, 250 mM NaCl, 1% Triton-100, 5 mM EDTA, 50 mM NaF, 0.1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 100 μM desferrioxamine, 1× protease inhibitor mix). Immunoprecipitation and Western blot procedures were as previously described except that HRP-labeled goat anti-mouse IgG-Fc was used in immunoblotting (39, 40). The protein concentration was determined by using Bio-Rad reagents.
RESULTS
Common stimuli regulate both protein stability and transactivation activity of HIF-1α.
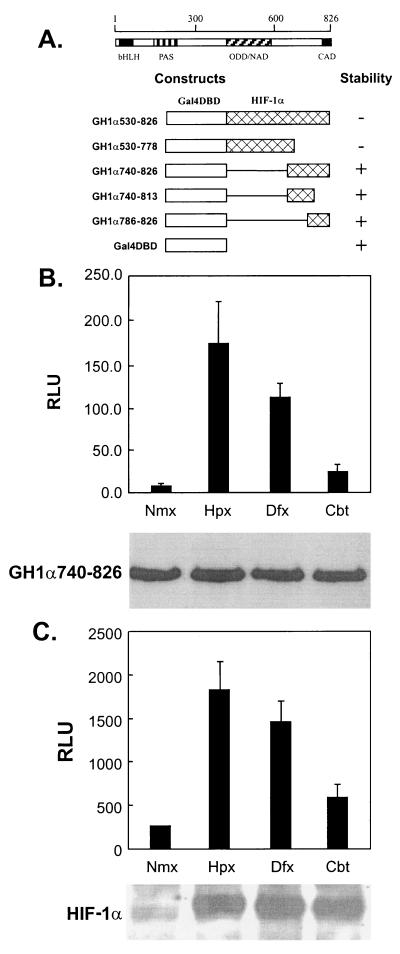
Two transactivation domains of HIF-1α, the N-terminal transactivation domain (NAD) and the carboxyl-terminal transactivation domain (CAD), have been previously described (20, 33) as shown schematically in Fig. 1A. We have reported that the protein levels of both GH1α530-826 and GH1α530-778 were up-regulated by hypoxia and defined an ODD (aa 557 to 571) (45), and the protein stabilities of the different GH1α constructs tested are summarized in Fig. 1A. Since the ODD overlaps NAD, the regulation of NAD activity appears to be determined, at least partially, by variations in protein levels (22, 33). Although the protein levels of GH1α740-826 in transfected HeLa cells were not affected, their transactivation activity was significantly up-regulated by hypoxia and the hypoxia mimics, cobalt and desferrioxamine (Fig. 1B). Similar results were observed in the 143B and Hep3B cell lines (data not shown). Figure 1C shows the increase in endogenous HIF-1α protein levels that accompanies the stimulatory response to hypoxia, desferrioxamine, and cobalt. The finding that the same stimuli that regulate HIF-1α protein stability also affect its transcriptional activity suggests that a similar or identical regulatory pathway is involved in both processes.
FIG. 1.
(A) Schematic structures of HIF-1α and GAL4-H1α constructs used in this study. (B) Effects of hypoxia and hypoxia mimics on the transactivation activity of GH1α740-826. HeLa cells were cotransfected with GH1α740-826 (2.5 μg), pFR-luc reporter (2.5 μg), and pCMVLacZ (0.5 μg). Twenty-four hours later the transfected cells were trypsinized. Fifty percent of the cells were split equally into 12-well culture plates for luciferase assays (top), and the other 50% of the cells were split into 60-mm-diameter dishes to extract proteins for Western blotting with monoclonal antibody against GAL4-DBD (bottom). The cells were cultured under either normoxic (Nmx) or hypoxic (Hpx) (0.5% O2) conditions or with desferrioxamine (Dfx) (130 μM)or cobalt chloride (Cbt) (75 μM), respectively, for 16 h before harvest. (C) Response of endogenous HIF-1 to hypoxia and hypoxic mimics in Hep3B cells. Hep3B cells were transfected with a pEpo-luc reporter (5 μg) and pCMVLacZ (0.5 μg), and the transfected cells were treated as described in panel B. The protein levels of HIF-1α in whole-cell lysates were detected with anti-HIF-1α monoclonal antibody. The luciferase activity was corrected by β-galactosidase activity. RLU, relative luciferase activity.
Characterization of VHL-deficient and VHL-expressing cell lines.
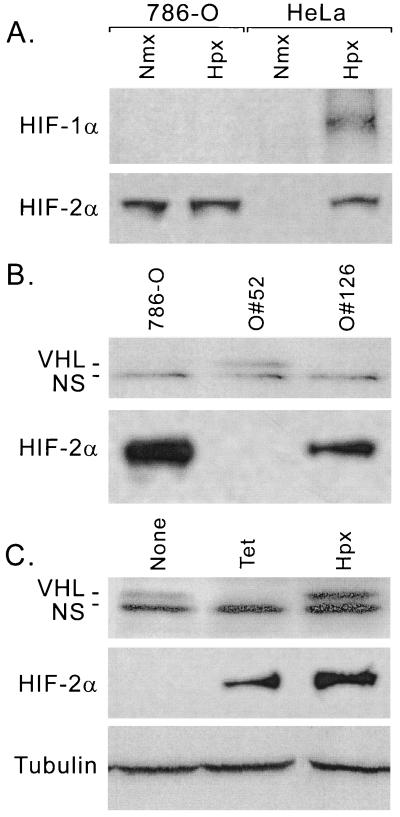
Since VHL is involved in the regulation of HIF-1α stability, we investigated the role of VHL in the regulation of the CAD transactivation activity. A renal carcinoma cell line, 786-O, which lacks one VHL allele and expresses a truncated nonfunctional protein from the second allele (16), was stably transfected with a vector expressing a tetracycline-regulatable (Tet-off*) wild-type VHL protein (786-O#52) or with the vector backbone (786-O#126). The expression of HIF-α and VHL in 786-O cells was examined by immunoblot analysis. As shown in Fig. 2A, HIF-2α but not HIF-1α is detectable in 780-O cells, confirming the observations made by others (30). In addition, HIF-2α levels were constitutively expressed and were not affected by hypoxia in the VHL-deficient cells. Figure 2B shows the lack of VHL expression in 786-O cell or 786-O#126, whereas it was detected in the 786-O#52 cells. Treatment of 786-O#52 cells with tetracycline for 36 h increased HIF-2α levels, while VHL expression became undetectable (Fig. 2C). Hypoxia, desferrioxamine, and cobalt did not repress the expression of VHL but stabilized HIF-2α (Fig. 2C and data not shown). These data confirmed that tetracycline efficiently represses the expression of VHL in 786-O#52 cells and that the expressed VHL functions normally.
FIG. 2.
(A) Hypoxia-regulated expression of HIF-α proteins in VHL-deficient cells (786-O). 786-O cells and HeLa cells were cultured under either normoxic (Nmx) (21% O2) or hypoxic (Hpx) (0.5% O2) conditions for 6 h. Whole-cell lysates were prepared and examined for the expression levels of HIF-1α and HIF-2α. (B) Expression of VHL and HIF-2α in 786-O-derived cell lines. Whole-cell lysates prepared from 786-O, 786-O#52, and 786-O#126 cells were fractionated on an SDS-10% polyacrylamide gel and transferred onto a PVDF membrane, and the expression of VHL and HIF-2α was detected by immunoblotting. (C) Regulated expression of VHL in 786-O#52 cells by tetracycline. Cells were seeded and cultured in DMEM with or without 3 μg of tetracycline/ml for a total of 36 h, and the media were changed every 12 h to maintain the tetracycline concentration. Immediately before harvest, cells were exposed to normoxia (21% O2) (lanes 1 and 2) or hypoxia (2% O2) (lane 3) for 12 h. The expression status of VHL and HIF-2α was detected by immunoblot. The same membrane was stripped and detected by a monoclonal antibody against β-tubulin (bottom). NS, nonspecific band.
VHL is not required for the up-regulation of HIF-1α transactivation activity in response to hypoxia or hypoxia mimics.
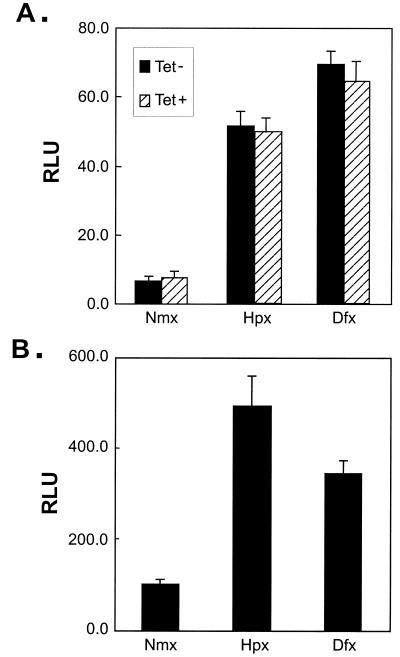
Next, we studied whether VHL was required for the stimulation of HIF CAD transactivation activity in response to hypoxia and hypoxia mimics. A GAL4 fusion construct, GH1α740-826 (Fig. 1A), was cotransfected into 786-O#52 cells with a luciferase reporter under the control of five GAL4 binding sites, and the cells were maintained with or without tetracycline. As shown in Fig. 3A, in response to hypoxia or desferrioxamine, the transactivation activity of GH1α740-826 increased regardless of the treatment of tetracycline. In addition, hypoxia and hypoxia mimics increased the transactivation activity of GH1α740-826 in the 786-O#126 cell line, which lacks functional VHL (Fig. 3B). These data demonstrate that expression of a functional VHL is not necessary for hypoxia-induced up-regulation of HIF CAD activity.
FIG. 3.
(A) Regulation of CAD activity in 786-O#52 (Tet-off VHL). Cells were seeded and cotransfected with GH1α740-826 and pFR-Luc reporter. After transfection, cells were cultured in DMEM with or without tetracycline (3 μg/ml) for 36 h. Twelve hours before harvest and luciferase assay, cells were treated with hypoxia (Hpx) (2% O2) or desferrioxamine (Dfx) (130 μM). The activity was corrected by cotransfected pRL-CMV. (B) Regulation of CAD activity in 786#126 (VHL−) cells. Cells were transfected, treated, and assayed as for panel A, except for the addition of tetracycline. Nmx, normoxic conditions.
Effect of hydroxylase inhibitor on CAD activity.
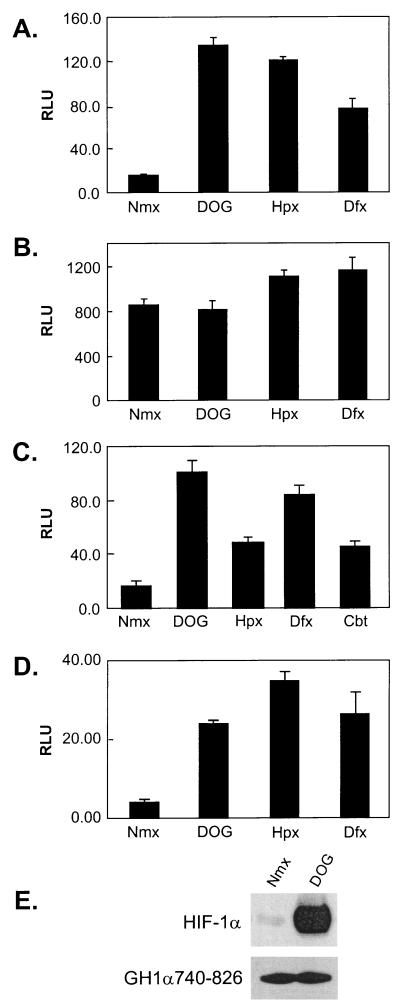
PHD enzymes are involved in the regulation of oxygen-dependent degradation of HIF-1α (5, 12). These enzymes require Fe2+, O2, and 2-oxoglutarate to hydroxylate prolyl residues at the ODD of HIF-1α, thus promoting its interaction with VHL. Since the carboxyl-terminal transactivation domain is responsive to hypoxia, iron chelators, and transition metals as well, we hypothesized that the regulation of CAD activity might involve a similar enzymatic process. To test this hypothesis, we treated cells with a 2-oxoglutarate analogue, DOG, which serves as a cell-permeative competitive inhibitor of hydroxylases (19). As shown in Fig. 4A, DOG stimulated the transactivation activity of CAD in HeLa cells under normoxic conditions as efficiently as desferrioxamine or hypoxia. This regulation requires the negative regulatory region (NRR) (aa 740 to 785) of HIF-1α because another Gal4 fusion construct, GH1α786-826, is constitutively active and is not stimulated by DOG or desferrioxamine (Fig. 4B). To examine if VHL participated in this hydroxylation-dependent modulation of CAD activity, we utilized the VHL− 786-O#126 or VHL+ 786-O#52 cells. Figure 4C and D shows that both cell lines responded normally to DOG, indicating that the responsiveness of HIF CAD to hydroxylase inhibition is also independent of the expression of functional VHL. To examine the effect of a hydroxylase inhibitor on protein levels of GH1α740-826, we transfected a plasmid expressing GH1α740-826 into HeLa cells. Western blot analysis revealed that while treatment of cells with DOG significantly increased the protein levels of HIF-1α, the protein levels of GH1α740-826 did not change (Fig. 4E), confirming that inhibition of hydroxylation enhanced the transactivation activity, not the stability, of GH1α740-826.
FIG. 4.
PHD inhibitor induces CAD activity under normoxic conditions. (A and B) HeLa cells were transfected with GH1α740-826 and pFR-luc reporter (A) or GH1α786-826 and pFR-luc (B). Luciferase assays were performed after the cells were treated with DOG (1 mM), hypoxia (Hpx) (2% O2), and desferrioxamine (Dfx) (130 μM) for 12 h. (C and D) Effects of hydroxylase inhibitor on GH1α740-826 in 786-O#52 (VHL+) and 786-O#126 (VHL−) cells. (E) Effect of hydroxylase inhibitor on protein stability of GH1α740-826. HeLa cells were transfected with GH1α740-826, pooled, and reseeded in two dishes that were treated with or without DOG. Whole-cell lysates were prepared, and the expression levels of HIF-1α and GH1α740-826 were examined by Western blotting by using anti-HIF-1α and anti-GAL4-DBD monoclonal antibodies, respectively. Cbt, cobalt chloride.
Effect of overexpression of HIF-PHDs on HIF CAD activity.
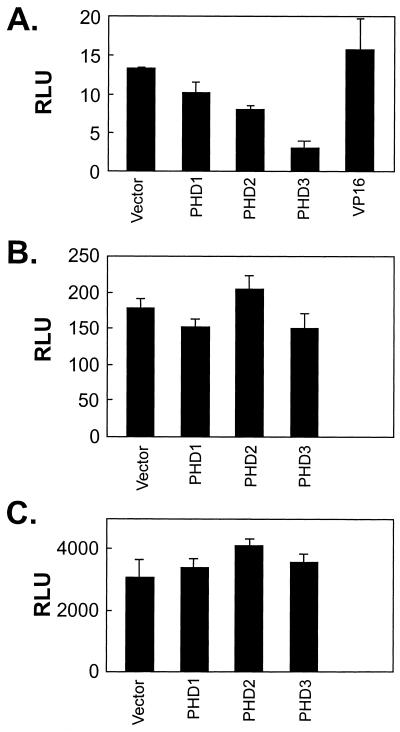
The recently identified HIF-PHD family includes three structurally and functionally related enzymes (5, 12). Since overexpression of these enzymes enhanced the degradation of HIF-1α, hydroxylase activity was considered limiting for the normoxic degradation of HIF-1α (5). If hydroxylase activity is a limiting factor for the repression of the transactivation activity of HIF-1α, and any of these PHDs is involved in this repression, overexpression of that hydroxylase would repress the basal transactivation activity of GH1α740-826. We cotransfected plasmids expressing PHD1, PHD2, and PHD3 with GH 1α740-826 in HeLa cells and observed that at least PHD3 repressed the transactivation activity of GH1α740-826 under normoxic conditions, while VP16 showed no effect (Fig. 5A). In contrast, none of the enzymes inhibited the constitutive transactivation activity of GH1α786-826 (Fig. 5B) or a control Gal4-VP16 protein (Fig. 5C). As expected, none of these enzymes inhibited hypoxia or desferrioxamine-stimulated activity of CAD (data not shown). It should be pointed out that the protein levels of PHD1, PHD2, and PHD3 were not examined because of unavailability of antibodies. Therefore, these data do not rule out the involvement of PHD1 in normoxic repression. Nevertheless, these results suggest that a hydroxylase activity is involved in the normoxic repression of HIF CAD and that this activity is limiting.
FIG. 5.
Effects of overexpression of PHDs on CAD activity. HeLa cells were cotransfected with GH1α740-826 (1 μg), pFR-Luc (1 μg), and pRL-CMV (0.1 μg). In addition, either empty vector (3 μg) or plasmids coding for PHD1, PHD2, PHD3, or VP16 (3 μg) was cotransfected (A). Similarly, the effects of PHDs on GH1α786-826 and GVP16 were examined (B and C). RLU, relative luciferase activity.
Hydroxylase inhibitor enhances the functional interaction between HIF-1α and p300/CBP.
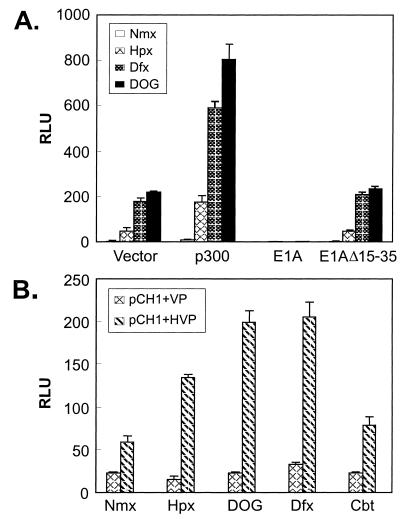
Since the transactivation activity of HIF-1α involves its interaction with p300/CBP, we next tested if inhibition of the PHDs enhances HIF-1α activity by promoting the functional interaction between HIF-1α and p300/CBP. As shown in Fig. 6A, exogenous expression of p300 mildly increased the activity of GH1α740-826 under normoxic condition. Treatment of cells with DOG, hypoxia, and desferrioxamine substantially enhanced the activity. The adenoviral oncoprotein E1A, which sequesters endogenous p300/CBP (41), blocked the response of GH1α740-826 to DOG, hypoxia, and desferrioxamine. An E1A mutant that is defective in binding p300/CBP is defective in blocking the response, demonstrating that recruitment of p300/CBP is required for GH1α740-826 responses. Since DOG, hypoxia, and desferrioxamine do not directly increase the protein levels or transactivation activity of p300/CBP (N. Sang and J. Caro, data not shown), the results presented suggest that these stimuli are affecting the interaction between HIF CAD and p300/CBP.
FIG. 6.
(A) Hydroxylase inhibitor enhances the ability of p300 to potentiate CAD activity. HeLa cells were cotransfected with GH1α740-826 and pFR-Luc. In addition, empty vector, p300, E1A, or an E1A mutant was cotransfected to test its effects on CAD activity. The transfected cells were split and exposed to various conditions as indicated. (B) Hydroxylase inhibitor enhances functional interaction between HIF-1α and the CH1 domain of p300. Indicated combinations of mammalian two-hybrid partners (2 μg of each) were cotransfected with pFR-luc reporter (1.5 μg). Twenty-four hours after transfection, cells were split and subjected to various conditions as indicated. The relative luciferase activity in pDBD-, pHVP-transfected cells was too low to be seen in the chart. Nmx, normoxia; Hpx, hypoxia; Dfx, desferrioxamine; Cbt, cobalt chloride.
Using the mammalian two-hybrid system, we further tested the hypothesis that DOG, hypoxia, and desferrioxamine enhance HIF CAD-p300 interactions. In this system, the CH1 domain (aa 300 to 528) of p300, which is responsible for the interaction with HIF-1α, was fused with the DBD of GAL4 (GCH1) (4). A DNA fragment encoding aa 723 to 826 of HIF-1α was fused in frame with the VP16 activating domain (VP16H1α723-826) (4, 25). G4CH1 gives rise to basal transactivation activity because CH1 is capable of interacting with the basal transcription complex (TBP) directly (52). Enhanced transactivation activity would be detected if VP16H1α723-826 interacts with G4CH1, because VP16 is a much stronger transactivator than CH1 (4). As shown in Fig. 6B, cotransfection with VP16H1α723-826 increased the transactivation activity around twofold under normoxic conditions. Treatment with DOG, hypoxia, and desferrioxamine further enhanced the transactivation activity, confirming that they promoted the functional interaction between p300-CH1 and HIF CAD.
Hydroxylase inhibitor and hypoxia enhance the physical interaction of HIF-1α-p300 in vivo.
We next examined the effects of hydroxylase inhibitor, hypoxia, and desferrioxamine on the physical interaction between HIF CAD and p300/CBP in vivo. Plasmids expressing GH1α740-826 and GH1α786-826 were transfected into HeLa cells. Protein complexes were immunoprecipitated by an anti-p300 monoclonal antibody and were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes. An anti-GAL4 (DBD) antibody was used to detect whether GAL4-HIF CAD copurified with p300 under different experimental conditions. GH1α786-826 was found to copurify with p300 under all conditions tested (Fig. 7A). However, the interaction between p300 and GH1α740-826 was only detectable when cells were exposed to DOG, hypoxia, or desferrioxamine (Fig. 7B). These results suggest that the NRR (aa 740 to 785) inhibits the interaction between p300 and HIF CAD and that this inhibition can be overridden by a hydroxylase inhibitor, hypoxia, and desferrioxamine.
FIG. 7.
Hydroxylase inhibitor enhances the physical interaction between HIF-1α and p300 in vivo. HeLa cells were transfected with GH1α786-826 (A) or GH1α740-826 (B). Twenty-four hours after transfection, transfected dishes were trypsinized, pooled, and reseeded to fresh dishes to normalize the transfection efficiency. The dishes then were treated overnight with various conditions as indicated (samples 1 to 5). As controls, none-transfected HeLa cells were exposed to normoxia (Nmx) or hypoxia (Hpx) (samples 6 and 7). Whole-cell lysates (WCL) were prepared. WCL (5%) were separated directly through an SDS-10% polyacrylamide gel (top) to check the expression of transgenes. The remaining lysates (95%) were immunoprecipitated (IMP) with a monoclonal antibody against human p300 (bottom, (lanes 1 to 4, 6, and 7) or an anti-E1A monoclonal antibody as the control (bottom, lane 5). The precipitated protein complexes were resolved through an SDS-10% polyacrylamide gel and transferred onto PVDF membranes. The membranes were detected with a monoclonal antibody specific for GAL4-DBD and developed by an ECL kit. Dfx, desferrioxamine.
Overexpression of the negative regulatory region up-regulates HIF CAD activity in vivo.
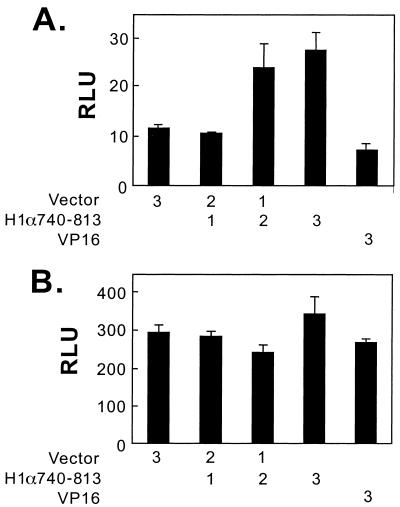
The NRR (aa 740 to 813) may directly inhibit the interaction between p300 and HIF-1α by itself or indirectly by recruitment of an inhibitory factor. If recruitment of an inhibitory factor is required for the inhibition, overexpression of NRR in trans should be able to sequester the inhibitory factor and thus alleviate the inhibition. For this purpose, we constructed a plasmid expressing the NRR fused in frame with a fragment of VP16 that provides a nuclear localization signal. As shown in Fig. 8, transfection of the plasmid expressing H1α740-813 increased the transactivation activity of GH1α740-826 in normoxic Hep3B cells, while it had no stimulatory effect on the activity of GH1α786-826. Transfection with the VP16 backbone plasmid had no effect. These data support a model in which the NRR interacts with a cellular inhibitory factor.
FIG. 8.
Effect of overexpression of NRR on CAD activity. In Hep3B cells, pFR-Luc (1 μg) and pRL-CMV (0.1 μg) were cotransfected with either GH1α740-826 (1 μg) (A) or GH1α786-826 (B). In addition, the indicated amount (in micrograms) of empty vector, plasmids expressing HIF-1α740-813, or VP16 was cotransfected. Luciferase assays were performed 48 h after transfection, and the luciferase activity (RLU) was corrected by cotransfected pRL-CMV.
DISCUSSION
Since HIF-1α stability and the transactivation activity of its CAD domain are responsive to the same stimuli (Fig. 1), it seems reasonable to suggest that a common signaling pathway controls both protein degradation and transactivation activity. HIF-1α degradation is determined by its interaction with VHL, which targets it for ubiquitination and proteasomal destruction. In turn, HIF-VHL interaction depends on the activity of oxygen-dependent PHD enzymes that hydroxylate specific residues in the ODD of HIF-1α. In this study, we examined the possible involvement of VHL and hydroxylase in the regulation of the CAD activity. We provide evidence that up-regulation of HIF CAD activity in response to hypoxia can be mimicked by hydroxylase inhibitors but does not depend on the presence of VHL. Thus, it appears that while both regulatory pathways overlap partially, the pathway regulating HIF-1α stability is distinguishable from the one that regulates the CAD activity.
Three structurally and functionally related PHDs (PHD1 to PHD3) have been cloned and proven to be highly conserved in distant species recently (5, 12). Our data show that the regulation of CAD also involves hydroxylase activity. It is not clear whether the different hydroxylases have equivalent activities on HIF-1α degradation and transcriptional activation. During our studies, we observed that while cobalt is a very effective inducer of HIF-1α accumulation, it is not an effective stimulator of HIF CAD transactivation activity (Fig. 1B and C). The involvement of different hydroxylases in the regulation of protein degradation and transactivation may provide an explanation for that discrepancy. It is assumed that cobalt replaces iron in the hydroxylase enzymatic core, thus inhibiting its activity. Since cobalt has limited effects on CAD activity, it is possible that the hydroxylase involved in regulation of CAD may have a higher affinity for Fe2+ binding. In addition, while there are already three members in the PHD family, the number may still grow, and functionally they may not be redundant. The existence of multiple hydroxylases in human cells reflects complexity in oxygen sensing and hypoxia adaptation.
The role of p300/CBP in hypoxic response was first suggested by its interaction with HIF-1α in a protein-protein interaction screening (1). Since then, several studies have linked the recruitment of p300/CBP to the hypoxia-induced up-regulation of HIF-1α transactivation activity (7, 10, 11, 13, 21). Mutations in the CH1 domain of p300 disrupting the interaction between p300/CBP and HIF CAD completely abolish the transactivation activity of HIF-1α (13, 25). Furthermore, blocking the interactions between p300 and HIF-1α in vivo by overexpressing polypeptides corresponding to the p300-CH1 or HIF-1α786-826 domain results in down-regulation of HIF-1α transactivation activity and a significant inhibition of downstream gene expression (25). These observations indicate that the interaction with p300/CBP is indispensable for the transactivation activity of HIF-1α CAD and that HIF-1α is not able to interact directly with the basic transcriptional machinery.
Previous studies have shown that the HIF-1α contains an internal domain that inhibits HIF-1α transcriptional activity, and mutations of aa 781 to 783 (RLL to AAA) within this domain inactivate its repressive activity (20, 32, 33). Consistent with previous observations, we find that GH1α786-826 is constitutively active and is hardly regulated by hypoxia or hypoxia mimics. Correspondingly, a HIF-1α fragment encompassing aa 786 to 826 interacts with p300 in vivo under normoxia conditions. In contrast, GH1α740-826 shows a striking transcriptional response to hypoxia, desferrioxamine, and hydroxylase inhibitors. Therefore, the region of aa 740 to 785 serves as an NRR that prevents p300/CBP from binding to the active 786-826 region in vivo under normoxic conditions. This hypothesis is confirmed by our finding that coimmunoprecipitation of GH1α740-826 and p300 only becomes detectable under hypoxic conditions or following treatment with desferrioxamine or hydroxylase inhibitors. How NRR blocks p300/CBP interaction with HIF-1α in vivo is not clear. The repressive effect appears not to be caused by intramolecular interactions between the NRR and the 786-826 region, since studies utilizing a mammalian two-hybrid system failed to show any interaction between these two fragments (Sang and Caro, data not shown). Since HIF-1α723-826 is able to interact with p300 efficiently in vitro (4), the repressive effect of the NRR appears to depend on posttranslational modification. One possibility is that this region serves as a destabilizing factor that represses the interaction between HIF CAD and p300. In this context, normoxic hydroxylation would affect the properties of either p300 or HIF-1α, making it more difficult for p300 to access CAD. Although there are two proline residues within the region of aa 740 to 785, neither of them conforms to the suggested consensus hydroxylation motif LXXLAP (12, 29). Similarly, while multiple prolyl residues exist, no prolyl hydroxylation consensus is found in the CH1 domain of p300. Another possibility is that the NRR interacts with protein factors that function as hydroxylation-dependent repressors that prevent the association between p300/CBP and CAD under normoxic conditions. Inhibition of hydroxylation destabilizes the interaction of these factors and the NRR, thus allowing p300/CBP to access the CAD domain. Our finding that overexpression of the NRR relieves the normoxic repression of CAD supports the notion that NRR interacts with a negative regulatory factor.
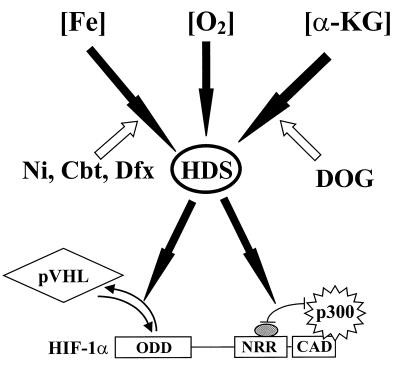
Based on previous reports and the data presented here, we propose a model for the regulation of the protein stability and transactivation activity of HIF-1α (Fig. 9) in which hydroxylases are the key factors. Since hydroxylase activity depends on iron, oxygen, and 2-oxoglutarate, a decrease in the concentration of any of these three factors leads to a hypoxia-like response. While the signaling pathways upstream of the hydroxylases are identical, the mechanism underlying the regulation of protein stability and the modulation of the transactivation activity are differentially controlled. The former involves direct hydroxylation of HIF-1α at its prolyl residues in the ODD, thus promoting the interaction with VHL and its subsequent targeting for degradation. The latter involves the hydroxylation of protein factor(s) that could be p300, HIF-1α, a putative inhibitory factor, or any combination thereof. This hydroxylation would most likely promote the interaction between HIF-1α and the inhibitory factor, thus disrupting the interaction between HIF-1α and p300/CBP. Conversely, hypoxia and hypoxia mimics directly or indirectly inhibit hydroxylase activity, thus resulting in activation of the CAD.
FIG. 9.
Proposed roles of VHL and hydroxylases (HDS) in HIF-1α stabilization and transactivation. Filled arrows indicate stimulation or enhancement, and empty arrows stand for inhibition or repression.
During the preparation of this report, it was reported by others that an inhibitory factor, FIH-1 (factor inhibiting HIF), interacts with HIF-1α and inhibits the transcriptional activity of CAD (28). It was shown that the transactivation activity of HIF-1α is regulated by VHL but not through protein stabilization. Instead, FIH-1 binds to VHL wherein VHL functions as a transcriptional corepressor by recruiting histone deacetylases. In addition, these interactions between HIF-1α, FIH-1, and VHL were suggested to be independent of hydroxylation (28). While their findings support the involvement of a cellular inhibitory factor, our data demonstrate that the regulation of the CAD activity of HIF-1α involves a hydroxylase activity and is, at least partly, a VHL-independent event.
ADDENDUM IN PROOF
While this paper was in press, Lando et al. published results that are consistent with our experimental findings and conclusions (D. Lando, D. J. Peet, D. A. Whelan, J. J. Gorman, and M. L. Whitelow, Science 295:858-861, 2002).
Acknowledgments
We thank P. Ratcliffe, D. Livingston, N. Kley, and A. Giordano for kindly providing expression plasmids, cell lines, and other reagents necessary for this study. We thank R. Silvano for professional proofreading and editing of the manuscript. N.S. and J.F. thank Y. Zhang (Huashan Hospital of Fudan University Medical Center, Shanghai) for his support.
N.S. is supported by a training grant from the National Institutes of Health (T32HL007821). This work was supported in part by a grant from American Heart Association (9950122N) and by a grant from National Institutes of Health (RO1CA89212-02) to J.C. V.S. is supported by a grant from American Heart Association (0060194U). J.F. is a visiting researcher from Huashan Hospital of Fudan University Medical Center, Shanghai, China.
REFERENCES
- 1.Arany, Z., L. E. Huang, R. Eckner, S. Bhattacharya, C. Jiang, M. A. Goldberg, H. F. Bunn, and D. M. Livingston. 1996. An essential role for p300/CBP in the cellular response to hypoxia. Proc. Natl. Acad. Sci. USA 93:12969-12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avantaggiati, M. L., V. Ogryzko, K. Gardner, A. Giordano, A. S. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89:1175-1184. [DOI] [PubMed] [Google Scholar]
- 3.Beck, I., S. Ramirez, R. Weinmann, and J. Caro. 1991. Enhancer element at the 3′ flanking region controls transcriptional response to hypoxia in the human erythropoietin gene. J. Biol. Chem. 266:15563-15566. [PubMed] [Google Scholar]
- 4.Bhattacharya, S. M., C. L. Michels, M. K. Leung, Z. P. Arany, A. L. Kung, and D. M. Livingston. 1999. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 13:64-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294:1337-1340. [DOI] [PubMed] [Google Scholar]
- 6.Carmeliet, P., Y. Dor, J. M. Herbert, D. Fukumura, K. Brusselmans, M. Dewerchin, M. Neeman, F. Bono, R. Abramovitch, P. Maxwell, C. J. Koch, P. Ratcliffe, L. Moons, R. K. Jain, D. Collen, and E. Keshet. 1998. Role of HIF1α in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394:485-490. [DOI] [PubMed] [Google Scholar]
- 7.Carrero, P., K. Okamoto, R. Coumailleau, S. O'Brien, H. Tanaka, and L. Poellinger. 2000. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1α. Mol. Cell. Biol. 20:402-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chilov, D., G. Camenisch, I. Kvietikova, U. Ziegler, M. Gassmann, and R. H. Wenger. 1999. Induction and nuclear translocation of hypoxia-inducible factor-1 (HIF-1): heterodimerization with ARNT is not necessary for nuclear accumulation of HIF1α. J. Cell Sci. 112:1203-1212. [DOI] [PubMed] [Google Scholar]
- 9.Cockman, M. E., N. Masson, D. R. Mole, P. Jaakkola, G. W. Chang, S. C. Clifford, E. R. Maher, C. W. Pugh, P. J. Ratcliffe, and P. H. Maxwell. 2000. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 275:25733-25741. [DOI] [PubMed] [Google Scholar]
- 10.Ebert, B. L., and H. F. Bunn. 1998. Regulation of transcription by hypoxia requires a multiprotein complex that includes hypoxia-inducible factor 1, an adjacent transcription factor, and p300/CREB binding protein. Mol. Cell. Biol. 18:4089-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ema, M., K. Hitota, J. Mimura, H. Abe, J. Yodoi, K. Sogawa, L. Poellinger, and Y. Fujii-Kuriyama. 1999. Molecular mechanisms of transcription activation by HLF and HIF1α in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 18:1905-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epstein, A. C., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43-54. [DOI] [PubMed] [Google Scholar]
- 13.Gu, J., J. Milligan, and L. E. Huang. 2001. Molecular mechanism of hypoxia-inducible factor 1-p300 interaction. A leucine-rich interface regulated by a single cysteine. J. Biol. Chem. 276:3550-3554. [DOI] [PubMed] [Google Scholar]
- 14.Huang, L. E., Z. Arany, D. M. Livingston, and H. F. Bunn. 1996. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem. 271:32253-32259. [DOI] [PubMed] [Google Scholar]
- 15.Huang, L. E., J. Gu, M. Schau, and H. F. Bunn. 1998. Regulatin of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 95:7987-7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iliopoulos, O., A. P. Levy, C. Jiang, W. G. Kaelin, Jr., and M. A. Goldberg. 1996. Negative regulation of hypoxia-inducible genes by the von Hippel Lindau protein. Proc. Natl. Acad. Sci. USA 93:10595-10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 18.Iwai, K., K. Yamanaka, T. Kamura, N. Minato, R. C. Conaway, J. W. Conaway, R. D. Klausner, and A. Pause. 1999. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA 96:12436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. V. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, B. H., J. Z. Zheng, S. W. Leung, R. Roe, and G. L. Semenza. 1997. Transactivation and inhibitory domains of hypoxia-inducible factor 1alpha. J. Biol. Chem. 272:19253-19260. [DOI] [PubMed] [Google Scholar]
- 21.Kallio, P. J., K. Okamoto, S. O'Brein, P. Carrero, Y. Mkino, H. Tanaka, and L. Poellinger. 1998. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1α. EMBO J. 17:6573-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kallio, P. J., W. J. Wilson, S. O'Brien, Y. Makino, and L. Poellinger. 1999. Regulation of the hypoxia inducible transcription factor 1α by the ubiquitin-proteasome pathway. J. Biol. Chem. 274:6519-6525. [DOI] [PubMed] [Google Scholar]
- 23.Kamura, T., S. Sato, K. Iwai, M. Czyzyk-Krzeska, R. C. Conaway, and J. W. Conaway. 2000. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. USA 97:10430-10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo, K., and W. G. Kaelin, Jr. 2001. The von Hippel-Lindau tumor suppressor gene. Exp. Cell Res. 264:117-125. [DOI] [PubMed] [Google Scholar]
- 25.Kung, A. L., S. Wang, J. M. Klco, W. G. Kaelin, and D. M. Livingston. 2000. Suppression of tumor growth through disruption of hypoxia-inducible transcription. Nat. Med. 6:1335-1340. [DOI] [PubMed] [Google Scholar]
- 26.Lando, D., I. Pongratz, L. Poellinger, and M. L. Whitelaw. 2000. A redox mechanism controls differential DNA binding activities of hypoxia-inducible factor (HIF) 1α and the HIF like factor. J. Biol. Chem. 275:4618-4627. [DOI] [PubMed] [Google Scholar]
- 27.Latif, F., K. Tory, J. Gnarra, M. Yao, F. M. Duh, M. L. Orcutt, T. Stackhouse, I. Kuzmin, W. Modi, and L. Geil. 1993. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 260:1317-1320. [DOI] [PubMed] [Google Scholar]
- 28.Mahon, P. C., K. Hirota, and G. L. Semenza. 2001. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15:2675-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masson, N., C. William, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J. 20:5197-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 31.Ohh, M., C. W. Park, M. Ivan, M. A. Hoffman, T. Y. Kim, L. E. Huang, V. Chau, and W. G. Kaelin, Jr. 2000. Ubiquitination of HIF requires direct binding to the von Hippel-Lindau protein beta domain. Nat. Cell Biol. 2:423-427. [DOI] [PubMed] [Google Scholar]
- 32.O'Rourke, J. F., Y-M. Tian, P. J. Rateliffe, and C. W. Pugh. 1999. Oxygen-regulated and transactivating domains in endothelial PAS protein 1: Comparison with hypoxia-inducible factor-1alpha. J. Biol. Chem. 274:2060-2071. [DOI] [PubMed] [Google Scholar]
- 33.Pugh, C. W., J. F. O'Rourke, M. Nagao, J. M. Gleadle, and P. J. Ratcliffe. 1997. Activation of hypoxia-inducible factor-1, definition of regulatory domains within the α subunit. J. Biol. Chem. 272:11205-11214. [DOI] [PubMed] [Google Scholar]
- 34.Puri, P. L., M. L. Avantaggiati, C. Balsano, N. Sang, A. Graessmann, A. Giordano, and M. Levrero. 1997. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 16:369-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richard, D. E., E. Berra, and J. Pouyssegur. 2000. Nonhypoxic pathway mediates the induction of hypoxia inducible factor 1 alpha (HIF-1α) in vascular smooth muscle cells. J. Biol. Chem. 275:26765-26771. [DOI] [PubMed] [Google Scholar]
- 36.Ryan, H., M. Poloni, W. McNulty, D. Elson, M. Gassmann, J. M. Arbeit, and R. S. Johnson. 2000. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 60:4010-4015. [PubMed] [Google Scholar]
- 37.Ryan, H. E., J. Lo, and R. S. Johnson. 1998. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 17:3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salceda, S., and J. Caro. 1997. Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. J. Biol. Chem. 272:22642-22647. [DOI] [PubMed] [Google Scholar]
- 39.Sang, N., M. L. Avantaggiati, and A. Giordano. 1997. Roles of p300, pocket proteins, and hTBP in E1A-mediated transcriptional regulation and inhibition of p53 transactivation activity. J. Cell. Biochem. 66:277-285. [DOI] [PubMed] [Google Scholar]
- 40.Sang, N., and A. Giordano. 1997. Extreme N-terminus of E1A oncoprotein specifically associates with a new set of cellular proteins. J. Cell. Physiol. 170:182-191. [DOI] [PubMed] [Google Scholar]
- 41.Sang, N., J. Caro, and A. Giordano. 2002. Adenoviral E1A: everlasting tool, versatile applications, continuous contributions and new hypotheses. Frontiers Biosci. 7:d407-d413. [DOI] [PubMed] [Google Scholar]
- 42.Sang, N., P. P. Claudio, Y. Fu, N. Horikoshi, U. Graeven, R. Weinmann, and A. Giordano. 1997. Transforming region of 243R E1A contains two overlapping but distinct transactivation domains. DNA Cell Biol. 16:1321-1333. [DOI] [PubMed] [Google Scholar]
- 43.Semenza, G. L. 2000. Expression of hypoxia-inducible factor 1: mechanisms and consequences. Biochem. Pharmacol. 59:47-53. [DOI] [PubMed] [Google Scholar]
- 44.Srinivas, V., I. Leshchinsky, N. Sang, M. P. King, A. Minchenko, and J. Caro. 2001. Oxygen sensing and HIF-1 activation does not require an active mitochondrial respiratory chain electron-transfer pathway. J. Biol. Chem. 276:21995-22002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srinivas, V., L-P. Zhang, X-H. Zhu, and J. Caro. 1999. Characterization of an oxygen/redox-dependent degradation domain of hypoxia-inducible factor α (HIF-α) proteins. Biochem. Biophys. Res. Commun. 260:557-561. [DOI] [PubMed] [Google Scholar]
- 46.Stebbins, C. E., W. G. Kaelin, Jr., and N. P. Pavletich. 1999. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science 284:455-461. [DOI] [PubMed] [Google Scholar]
- 47.Tanimoto, K., Y. Makino, T. Pereira, and L. Poellinger. 2000. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO. J. 19:4298-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thornton, R. D., P. Lane, R. C. Borghaei, E. A. Pease, J. Caro, and E. Mochan. 2000. Interleukin-1 induces hypoxia inducible factor-1 in human gingival and synovial fibroblasts. Biochem. J. 350:307-312. [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, G. L., and G. L. Semenza. 1995. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270:1230-1237. [DOI] [PubMed] [Google Scholar]
- 50.Yang, H., and W. G. Kaelin, Jr. 2001. Molecular pathogenesis of the von Hippel-Lindau hereditary cancer syndrome: implications for oxygen sensing. Cell Growth Differ. 12:447-455. [PubMed] [Google Scholar]
- 51.Yu, F., S. B. White, Q. Zhao, and F. S. Lee. 2001. HIF-1 binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. USA 98:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan, W., G. Condorelli, M. Caruso, S. Felsani, and A. Giordano. 1996. Human p300 protein is a coactivator for the transcription factor MyoD. J. Biol. Chem. 271:9009-9013. [DOI] [PubMed] [Google Scholar]
- 53.Zhong, H., K. Chiles, D. Feldser, E. Laughner, C. Hanrahan, M.-M. Georgescu, J. W. Simons, and G. L. Semenza. 2000. Modulation of hypoxia-inducible factor 1α expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 60:1541-1545. [PubMed] [Google Scholar]