Abstract
The Saccharomyces cerevisiae kinase Ste20 is a member of the p21-activated kinase (PAK) family with several functions, including pheromone-responsive signal transduction. While PAKs are usually activated by small G proteins and Ste20 binds Cdc42, the role of Cdc42-Ste20 binding has been controversial, largely because Ste20 lacking its entire Cdc42-binding (CRIB) domain retains kinase activity and pheromone response. Here we show that, unlike CRIB deletion, point mutations in the Ste20 CRIB domain that disrupt Cdc42 binding also disrupt pheromone signaling. We also found that Ste20 kinase activity is stimulated by GTP-bound Cdc42 in vivo and this effect is blocked by the CRIB point mutations. Moreover, the Ste20 CRIB and kinase domains bind each other, and mutations that disrupt this interaction cause hyperactive kinase activity and bypass the requirement for Cdc42 binding. These observations demonstrate that the Ste20 CRIB domain is autoinhibitory and that this negative effect is antagonized by Cdc42 to promote Ste20 kinase activity and signaling. Parallel results were observed for filamentation pathway signaling, suggesting that the requirement for Cdc42-Ste20 interaction is not qualitatively different between the mating and filamentation pathways. While necessary for pheromone signaling, the role of the Cdc42-Ste20 interaction does not require regulation by pheromone or the pheromone-activated Gβγ complex, because the CRIB point mutations also disrupt signaling by activated forms of the kinase cascade scaffold protein Ste5. In total, our observations indicate that Cdc42 converts Ste20 to an active form, while pathway stimuli regulate the ability of this active Ste20 to trigger signaling through a particular pathway.
The Saccharomyces cerevisiae protein Ste20 is the founding member of the p21-activated kinase (PAK) family of protein kinases. It was originally identified for its signaling role in the yeast mating pathway (2, 26, 49), though subsequently it has been found to function in other signaling pathways that regulate filamentous growth and osmotic stress response (31, 43, 48, 51), as well as in the control of actin organization and polarized growth (13, 16, 22, 55, 63). In the mating pathway, Ste20 mediates activation of a mitogen-activated protein (MAP) kinase cascade in response to extracellular mating pheromones (reviewed in references 15, 17, and 20). These pheromones bind to G protein-coupled receptors and trigger release of Gβγ dimers, which activate the MAP kinase cascade in a manner involving recruitment of the kinase cascade scaffold protein Ste5 to the plasma membrane (18, 36, 47, 62). This is thought to bring the Ste5-associated kinase Ste11 (a MAP kinase kinase kinase) into close proximity with Ste20, which phosphorylates and activates Ste11 (60, 65). Ste20 is enriched at the cell periphery in both growing and mating cells via interaction with the membrane-bound GTPase Cdc42 (27, 39, 45, 47, 64). In addition, Ste20 binds the pheromone-activated Gβγ complex (29), potentially endowing Ste20 with increased kinase activity, increased access to Ste11, or both.
PAKs are commonly activated by small GTPases of the Cdc42/Rac family (4, 14). While Ste20 binds the GTPase Cdc42, there have been conflicting reports regarding the role of this interaction in mating pathway signaling. Early studies suggested that Cdc42 and its guanine nucleotide exchange factor Cdc24 were required for pheromone response (58, 67) and that GTP-bound Cdc42 could stimulate Ste20 kinase activity in vitro (58). Later studies suggested that the apparent requirement for Cdc24 and Cdc42 in pheromone response was an artifact of using conditional mutants that arrest at a nonresponsive position in the cell cycle (42). Furthermore, other studies concluded that while Cdc42 binding was required for proper localization of Ste20 and for its role in the filamentation and osmotic response pathways (27, 45, 48), it was not required for Ste20 kinase activity or for its role in pheromone response (27, 45). In these latter studies, the role of the Cdc42-Ste20 interaction was tested by removal of the entire Cdc42/Rac interaction binding (CRIB) domain from Ste20.
Recent studies of other PAK family members have suggested that these kinases are frequently regulated by an autoinhibitory mechanism that involves the CRIB domain. For some members of this family, including human PAK1/α-PAK and Schizosaccharomyces pombe Pak1/Shk1, it has been observed that the CRIB domain can bind to and inhibit the kinase domain (21), which has been confirmed recently for human PAK1 by crystallography (30). Furthermore, mutations in either domain that disrupt this intramolecular binding can lead to kinase activation (9, 59, 66, 68). In accord with this regulatory mechanism, recent mutational studies of yeast Cdc42 have reasserted its involvement in activating Ste20 for pheromone signaling, because mutant forms of Cdc42 that are impaired for binding Ste20 show defects in pheromone response, and these defects can be rescued by deletion of the Ste20 CRIB domain (39).
In this study, we have reinvestigated the role of the Ste20 CRIB domain in mediating Cdc42 binding and pheromone-responsive signaling. Our results indicate that Ste20 kinase activity and signaling are limited by inhibitory binding between the CRIB and kinase domains. They further suggest that binding of Cdc42 to the Ste20 CRIB domain activates the kinase activity and signaling ability of Ste20 and that this step is required for Gβγ-mediated signaling but is not itself regulated by Gβγ. Removal of inhibitory CRIB residues or of the CRIB domain in its entirety allows Cdc42-independent kinase activity and signaling ability, indicating that Ste20 adheres to the general model for PAK family kinases in which they are activated by small GTPases through relief of autoinhibition.
MATERIALS AND METHODS
Yeast strains.
Yeast strains are listed in Table 1. All two-hybrid assays were performed using strain PPY760 (11), a derivative of strain L40 (5). Filamentation assays used strains in the Σ1278b background, generated by crosses involving strains L5585 (51), L6149 (53), and meiotic segregants of strain L5978 (35). KBY211 (22) was described previously, as were PPY496, PPY640, PPY866, and PT2α (47). PPY913 was derived from YEL206 (27) by integration of a LEU2-marked FUS1-lacZ reporter at the FUS1 locus, using SphI-digested pFC23 (46).
TABLE 1.
Yeast strains used in this study
| Strain background | Strain name | Relevant genotype |
|---|---|---|
| W303a | KBY211 | MATα ste20::ADE2 cla4::LEU2 + YCp TRP1-cla4-75ts |
| PPY398 | MATa | |
| PPY496 | MATaFUS1::FUS1-lacZ::LEU2 ste20-1::TRP1 | |
| PPY640 | MATaFUS1::FUS1-lacZ::LEU2 | |
| PPY866 | MATα FUS1::FUS1-lacZ::LEU2 ste20-1::TRP1 ste5::ADE2 ste4::ura3FOA | |
| PPY913 | MATaFUS1::FUS1-lacZ::LEU2 ste20-3Δ::TRP1 | |
| PPY1203 | MATaFUS1::FUS1-lacZ::LEU2 ste20-Δ334-369 | |
| PPY1205 | MATaFUS1::FUS1-lacZ::LEU2 ste20-S338A/H345G | |
| PPY1234 | MATaFUS1::FUS1-lacZ::LEU2 STE20::myc12::URA3 | |
| PPY1236 | MATaFUS1::FUS1-lacZ::LEU2 ste20-Δ334-369::myc12::URA3 | |
| PPY1238 | MATaFUS1::FUS1-lacZ::LEU2 ste20-S338A/H345G::myc12::URA3 | |
| Σ1278b | PPY966 | MATahis3::hisG leu2::hisG trp1::hisG ura3-52 |
| PPY1200 | MATahis3::hisG leu2::hisG trp1::hisG ura3-52 ste20-Δ334-369 | |
| PPY1202 | MATahis3::hisG leu2::hisG trp1::hisG ura3-52 ste20-S338A/H345G | |
| PPY1209 | MATahis3::hisG leu2::hisG trp1::hisG ura3-52 ste20-1::TRP1 | |
| S288C | PPY760 | MATaade2 his3 leu2 trp1 LYS2::lexAop-HIS3 URA3::lexAop-lacZ far1::ADE2 |
| Other | PT2α | MATα hom3 ilv1 can1 |
W303 is ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1.
Replacement of the genomic STE20 locus with the S338A/H345G or Δ334-369 allele used a pop-in/pop-out strategy, in which cells were first transformed with BamHI-digested pPP1042 or pPP1043, respectively, and selected for uracil prototrophy, and then spontaneous 5-fluoro-orotic acid-resistant clones were screened by PCR (followed by restriction digests) to determine which allele was retained. By this method, PPY1203 and PPY1205 were derived from PPY640, and PPY1200 and PPY1202 were derived from PPY966.
To tag the Ste20 C terminus with 12 tandem copies of the Myc epitope, a tagging cassette was inserted at the genomic STE20 locus by transformation with HindIII-digested pGSMste20::12myc (a gift from J. Moskow and D. Lew), selection for uracil prototrophy, and screening by immunoblotting using anti-Myc antibodies for an appropriate-sized product. By this method, strains PPY1234, PPY1236, and PPY1238 were derived from PPY640, PPY1203, and PPY1205, respectively.
Plasmids.
Plasmids used in this study are listed in Table 2. Signaling properties of Ste20 mutants were mostly studied in two parental constructs, pRL116 and pPP1001. pRL116 (27) is a CEN URA3 plasmid expressing a green fluorescent protein GFP(S65T)-STE20 fusion gene from the native STE20 promoter; it includes native STE20 sequences 480 bp upstream and 30 bp downstream of the open reading frame. pPP1001 (Table 2) is a CEN URA3 plasmid expressing the native, unfused STE20 gene from its own promoter; it includes native STE20 sequences 1,210 bp upstream and 2 bp downstream of the open reading frame and was constructed by replacing the Δ334-369 region in pRS316-STE20Δ334-369 (27) (Table 2) with wild-type sequences from pRL116 as a BamHI-SalI fragment. In theory, pPP1001 should be identical to pRS316-STE20 (27).
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pBTL56 | CEN URA3 GFP-ste20Δ334-369 | 27 |
| pRL116 | CEN URA3 GFP-STE20 | 27 |
| pPP964 | pRL116 ste20-S338A | This study |
| pPP965 | pRL116 ste20-H345G | This study |
| pPP1009 | pRL116 ste20-S338A/H345G | This study |
| pPP1010 | pRL116 ste20-Δ334-369 | This study |
| pPP1109 | pRL116 ste20-S338A/H345G/L369G | This study |
| pPP1117 | pRL116 ste20-L369G | This study |
| pRS316-STE20 (Δ334-369) | CEN URA3 ste20Δ334-369 | 27 |
| pPP1001 | CEN URA3 STE20 | This study |
| pPP1002 | pPP1001 ste20-S338A | This study |
| pPP1003 | pPP1001 ste20-H345G | This study |
| pPP1005 | pPP1001 ste20-S338A/H345G | This study |
| pPP1011 | pPP1001 ste20-Δ334-369 | This study |
| pPP1112 | pPP1001 ste20-S338A/H345G/L369G | This study |
| pPP1118 | pPP1001 ste20-L369G | This study |
| pRS306 | URA3 integrating | 57 |
| pPP1042 | pRS306 ste20-S338A/H345G | This study |
| pPP1043 | pRS306 ste20-Δ334-369 | This study |
| pDH166 | CEN HIS3 GAL1pr-STE20 | 27 |
| pPP1248 | pDH166 ste20-Δ334-369 | This study |
| pPP1272 | pDH166 ste20-L369G | This study |
| pPP1273 | pDH166 ste20-S338A/H345G | This study |
| pPP1274 | pDH166 ste20-S338A/H345G/L369G | This study |
| pBTM116 | 2μm TRP1 lexA DBD vector | 5 |
| pB20N2 | 2μm TRP1 lexA DBD-ste20 1-499 | This study |
| pPP1059 | pB20N2 ste20-S338A | This study |
| pPP1060 | pB20N2 ste20-H345G | This study |
| pPP1061 | pB20N2 ste20-S338A/H345G | This study |
| pPP1062 | pB20N2 ste20-Δ334-369 | This study |
| pPP1115 | pB20N2 ste20-S338A/H345G/L369G | This study |
| pPP1119 | pB20N2 ste20-L369G | This study |
| pPP1309 | 2μm TRP1 lexA DBD-ste20 580-939 (K649M) | This study |
| pGAD424 | 2μm LEU2 GAL4 AD vector | 5 |
| pPP1027 | 2μm LEU2 GAL4 AD-CDC42- G12V/C188S | This study |
| pRL51.1 | 2μm LEU2 GAL4 AD-BEM1 157-551 | 28 |
| pGADXP | 2μm LEU2 ADH1pr (full-length) GAL4 AD vector | 11 |
| pPP1037 | 2μm LEU2 ADH1pr (full-length) GAL4 AD-ste20 1-439 | This study |
| pPP1321 | pPP1037 ste20-L369G | This study |
| pPP1322 | pPP1037 ste20-Δ334-369 | This study |
| pPP1323 | pPP1037 ste20-S338A/H345G | This study |
| pRS316 | CEN URA3 | 57 |
| pRS314 | CEN TRP1 | 57 |
| pRS413 | CEN HIS3 | 57 |
| pPP1219 | CEN HIS3 GAL1pr | This study |
| pPP827 | 2μm LEU2 TyFRE-lacZ | This study |
| pPP1252 | 2μm LEU2 flo11(10/9)-lacZ | This study |
| pPP1253 | 2μm LEU2 FLO11pr-lacZ | This study |
| p2988 | CEN LEU2 YLR042Cpr-lacZ | 50 |
| p2987 | CEN LEU2 KSS1pr-lacZ | 50 |
| pGSM ste20::12myc | STE20 C-term::myc12 URA3 | D. Lew |
| pSTE20-5 | CEN URA3 STE20 | 26 |
| pH-G42-L61 | CEN HIS3 GAL10pr-CDC42O61L | This study |
| pH-GS5-CTM | CEN HIS3 GAL1pr-STE5-CTM | 47 |
| pH-SL2 | CEN HIS3 GAL1pr-His6-myc-STE5 (P44L)-GST | This study |
As mentioned in the text, the results with these plasmids suggest that they may mildly underexpress STE20, possibly due to the absence of native downstream sequences such as transcriptional terminators. This is more evident with the “native” (pPP1001-based) constructs than with the GFP fusion (pRL116-based) constructs. In order to minimize confusion and to facilitate comparison to previous work, Table 3 compares the basic features of several STE20 plasmids and their ability to complement ste20Δ cells for pheromone-induced transcription. The results indicate that the GFP-STE20 fusion construct most closely approximates the level of function supplied by either the genomic STE20 allele or a construct (pSTE20-5) containing STE20 on a large genomic fragment, whereas the native construct (pPP1001) appears to underexpress STE20, in that it provides only 30 to 40% of these levels (see Table 3; also see Fig. 2).
TABLE 3.
Comparison of different STE20 and ste20Δ334-369 plasmids
| Strain | Plasmida | Native STE20 sequence (bp)
|
STE20 allele | Mean FUS1-lacZ expressionb (U) ± SD | |
|---|---|---|---|---|---|
| Upstream | Downstream | ||||
| PPY640 (STE20) | pRS316 | >1,000 | >1,000 | STE20 (genomic) | 80 ± 16 |
| PPY913 (ste20Δ) | pSTE20-5 | >1,000 | >1,000 | STE20 | 68 ± 18 |
| pRL116 | 480 | 30 | GFP-STE20 | 72 ± 15 | |
| pPP1010 | 480 | 30 | GFP-ste20Δ334-369 | 27 ± 13 | |
| pBTL56 | 480 | 0 | GFP-ste20Δ334-369c | 27 ± 8.2 | |
| pPP1001* | 1,210 | 2 | STE20 | 25 ± 9.4 | |
| pPP1011* | 1,210 | 2 | ste20Δ334-369 | 5.2 ± 3.5 | |
| pRS316-STE20Δ334-369* | 1,210 | 2 | ste20Δ334-369 | 4.2 ± 1.7 | |
| pRS316 | Vector | 0.05 ± 0.04 | |||
In some plasmids (∗), STE20 was inserted into the pRS316 vector in an opposite orientation from the others. Conceivably, transcriptional termination is governed by vector sequences in all plasmids except pSTE20-5, and thus different downstream vector sequences in the two orientations may contribute to the different levels of function. A polyadenylation sequence was predicted to lie 149 bp downstream of the open reading frame (26).
Mean β-galactosidase units ± SD (n = 12), measured after 2 h of exposure to 5 μM α-factor.
The STE20 open reading frame in pBTL56 is extended by two codons at the C terminus.
FIG. 2.
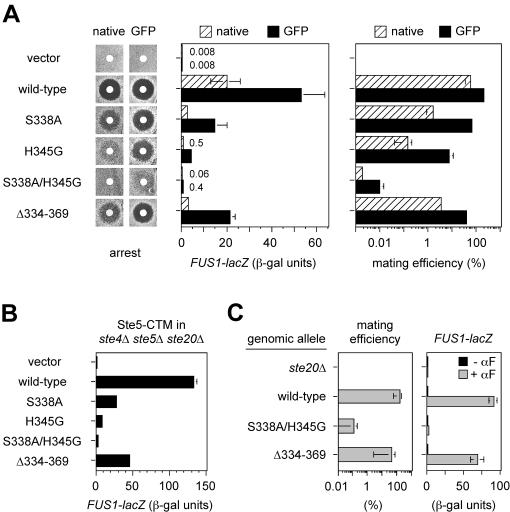
Interaction between the Ste20 CRIB domain and Cdc42 is required for mating pathway function, in a manner that is bypassed by CRIB domain deletion. (A) CRIB domain point mutations reduce pheromone response in proportion to their Cdc42-binding defect. Strain PPY913 harbored a vector (pRS316) or the indicated STE20 plasmids in either native (pPP1001-based) or GFP fusion (pRL116-based) contexts. Tests of pheromone-induced growth arrest (left) and FUS1-lacZ expression (middle) and of mating efficiency (right) were done as described in Materials and Methods. (B) Signaling role of the Cdc42-Ste20 interaction is independent of pheromone and Gβγ. FUS1-lacZ induction was stimulated without addition of pheromone and in the absence of Gβ (Ste4) by galactose-induced synthesis of Ste5-CTM (pH-GS5-CTM) in an ste4Δ ste5Δ ste20Δ strain (PPY866). The strain also harbored a vector (pRS316) or the indicated GFP-STE20 mutant plasmid. (C) Mating and FUS1-lacZ results using alleles integrated at the genomic STE20 locus. Strains were PPY496, PPY640, PPY1205, and PPY1203. αF, α-factor. In A to C, bars indicate mean ± SD for three or more repeats.
Plasmids pPP1010, pPP1011, and pPP1062 contain the ste20Δ334-369 mutation transferred into pRL116, pPP1001, and pB20N2, respectively, by replacement of their BamHI-SalI fragments with the analogous fragment from pBTL56 (27) (Table 2). In theory, plasmid pPP1011 should be identical to pRS316-STE20Δ334-369 (27). Also, pPP1010 (the Δ334-369 derivative of pRL116) is highly similar to pBTL56 (27). See Table 3 for comparisons.
Point mutations were first generated in pRL116 by Pfu polymerase-mediated extension of complementary mutagenic oligonucleotides, according to the Quick-Change site-directed mutagenesis kit (Stratagene). To ensure that the phenotypes did not arise from spurious mutations elsewhere, restriction fragments were transferred back into unmutagenized plasmids, and the transferred fragments were sequenced to confirm the presence of only the desired mutation(s). Mutations S338A, H345G, and L369G were transferred on BamHI-SalI fragments. These fragments were transferred into the GFP-STE20 plasmid pRL116, the non-GFP fusion construct pPP1001, and the two-hybrid constructs pB20N2 and pPP1037 to generate the mutant derivatives listed in Table 2. The S338A/H345G double and S338A/H345G/L369G triple mutants were generated by sequential rounds of mutagenesis, followed by BamHI-SalI fragment transfer as for the single mutants. All point mutations are marked by the gain or loss of a restriction site as follows: S338A, loss of EcoRV; H345G, gain of EcoRI; and L369G, gain of KpnI.
The GAL1pr-ste20 mutant constructs pPP1248, pPP1272, pPP1273, and pPP1274 contain the SpeI-SgrAI fragments from pPP1010, pPP1117, pPP1009, and pPP1109, respectively, transferred into pDH166 (27). Plasmids pPP1042 and pPP1043 contain the NotI-XhoI fragments from pPP1005 and pPP1011, respectively, transferred into pRS306.
Plasmid pB20N2 was constructed by PCR of sequences encoding Ste20 residues 1 to 499, using pSTE20-5 as the template, and insertion into pBTM116 (5) as an EcoRI-PstI fragment; a silent mutation from the downstream PCR primer disrupts the EcoRI site normally upstream of STE20 codon 499. pPP1309 was constructed by PCR of sequences encoding Ste20 residues 580 to 939, using pRD56-K649M (16) as the template to incorporate the K649M mutation, and insertion into pBTM116 as a BamHI-PstI fragment. pPP1037 was created by PCR of sequences encoding Ste20 residues 1 to 439, using pSTE20-5 as the template, transferred first as an EcoRI-PstI fragment into pGAD424 and then as an EcoRI-BglII fragment into pGADXP.
Plasmid pPP1027 was constructed by PCR amplification of CDC42G12V from pGAL-CDC42-Val12 (69), with the C188S mutation introduced by the downstream primer, and insertion into pGAD424 as an EcoRI-PstI fragment. pH-G42-L61 contains the GAL10pr-CDC42Q61L cassette from pGAL-CDC42-Leu61 (69) transferred as a BamHI-XhoI fragment into pRS413. Plasmids pPP827, pPP1252, and pPP1253 are LEU2-marked derivatives of pFRE(Ty1)::lacZ (33), p3941, and p3782 (53), respectively, created by homologous recombination in S. cerevisiae using the marker swap construct pUC4-ura3::LEU2 (38) digested with HindIII. pPP1219 contains the GAL1 promoter from pPP450 transferred into pRS413 as a SacI-ApaI fragment. Plasmid pH-SL2 was created by transferring the BamHI-BamHI insert from pSL2 (54) into the BamHI site of pPP1219.
β-Galactosidase assays.
For liquid assays, cells (usually 1 ml of culture) were harvested by centrifugation and then resuspended in 0.5 ml of Z-buffer (5) and processed as described before (46).
Pheromone response assays.
Halo assays of growth arrest were performed by spreading 3 × 105 cells/plate on −Ura plates, then applying sterile filter disks containing 20 μl of 1 mM α-factor, and incubating for 3 days at 30°C.
For FUS1-lacZ transcriptional induction assays, cells were grown overnight at 30°C to mid-exponential phase (optical density at 660 nm [OD660] = 0.3 to 0.6) in selective (plasmid alleles) or complete (integrated alleles) glucose medium and then incubated with and without 5 μM α-factor for 2 h before harvesting 1 ml for the β-galactosidase assay. For FUS1-lacZ activated by galactose-inducible constructs, cells were grown overnight in selective raffinose medium to the mid-exponential phase and then induced with 2% galactose for 3 h before harvest.
For quantitative mating assays, cells were grown overnight to the mid-exponential phase in selective or complete glucose medium, as appropriate, and then 5 × 106 a cells were mixed with 107 PT2α partner cells and collected onto filters, and the filters were placed onto complete glucose plates. After 4 h of mating at 30°C, cells on the filters were suspended in liquid, and serial dilutions were plated on medium selective for diploids. Mating efficiency was calculated as the percentage of input a cells that formed diploids; the number of input cells was determined by plating a dilution of the a cell cultures on selective (plasmid) or complete (integrated) plates at the start of the mating assay.
Patch mating assays were performed by patching a cell transformants directly onto a lawn of PT2α partner cells, incubating overnight at 30°C, replicating to minimal medium to select for diploids, and then incubating for 2 days at 30°C.
Yeast cell lysates and kinase assays.
Transformants were grown in selective medium overnight at 30°C to an OD660 of 0.6 to 1.0. For kinase assays involving Cdc42Q61L, transformants were first grown in −His/raffinose medium and then induced with galactose (2% final) for 90 min. Equivalent numbers of cells (usually 1 × 108 to 4 × 108) were harvested by centrifugation at 4°C, suspended in residual supernatant, transferred to an ice-cold microcentrifuge tube, and recentrifuged for 30 s in the cold; the supernatant was aspirated, and the pellets were stored at −80°C. Pellets were thawed by addition of 1 ml of ice-cold buffer A (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM dithiothreitol [DTT], 2 mM EDTA, 10% glycerol, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF]), washed once with 1 ml of buffer A, and then resuspended in 350 μl of buffer B (buffer A containing 0.1% Triton X-100 and 1 mM Na3VO4 plus protease inhibitors [PIs: 20 μg of 4-(2-aminoethyl)-benzenesulfonyl fluoride, 2 μg of leupeptin, 1 μg of pepstatin, and 1 μg of aprotinin per ml]), kept cold throughout. Approximately 0.4 ml of glass beads was added, followed by three 1-min pulses at full speed in a multitube vortexer (VWR Scientific Products Corp.), with 30 s on ice between pulses. Additional Triton X-100 was added (40 μl of a 10% solution), and the lysate was mixed on a nutator for 10 min. Following 2 min of centrifugation, the supernatant was transferred to a fresh tube and centrifuged for 4 min, and the new supernatant containing the clarified lysate was transferred to a fresh tube.
Epitope-tagged Ste20 was immunoprecipitated by adding 2 μg of anti-GFP monoclonal (clones 7.1 and 13.1; Boehringer Mannheim) or 0.4 μg of anti-Myc monoclonal (9E10; Santa Cruz Biotech.) to the clarified lysate and mixing for 2 h at 4°C. Protein G beads (Sigma; 40 μl of a 50% slurry in buffer A) were added and mixed for 1 h. Beads were pelleted at 2,400 × g, washed three times with 200 μl of cold buffer B and once with 200 μl of kinase buffer (KB; 10 mM Tris-HCl [pH 7.4], 10 mM MgCl2). Residual supernatant was removed using a 26-gauge needle, and beads were resuspended in 20 μl of room-temperature KB plus PIs. Then 10 μl of kinase assay cocktail (KB containing 1 mg of myelin basic protein per ml, 50 μM ATP, and 0.8 μM [γ-32P]ATP [3,000 Ci/mmol]) was added, and incubation proceeded for 30 min in a 30°C bath with occasional gentle mixing. Reactions were stopped by adding 20 μl of 2.5× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, boiling for 2 min, and then centrifuging for 5 min.
To analyze phosphorylation, 15 μl was run on an SDS-15% PAGE gel, and the gel was dried and exposed to a Phosphorimager screen (Molecular Dynamics). To determine Ste20 levels, 15 μl was run on an SDS-7% PAGE gel, followed by semidry transfer to an Immobilon-P membrane (Amersham); epitope-tagged Ste20 was detected by using rabbit anti-GFP polyclonal antibody (K. Gould laboratory, Vanderbilt University; a gift of D. McCollum) or mouse anti-Myc monoclonal antibody (9E10; Santa Cruz Biotechnology) at a 1:1,000 dilution, followed by alkaline phosphatase-conjugated secondary antibody diluted 1:3,000 (goat anti-rabbit or anti-mouse immunoglobulin; Bio-Rad) and chemiluminescence with Immun-Star substrate (Bio-Rad).
Filamentation assays.
Agar invasion assays were performed as previously described (51). Cells were patched onto YPD plates and incubated for 3 days at 30°C and then for 2 days at room temperature. Plates were photographed before and after being rinsed under a gentle stream of deionized water. Filamentation reporter expression was measured in exponentially growing cultures, prepared by diluting overnight cultures into fresh glucose medium (lacking appropriate nutrients to maintain plasmid selection) to an OD660 of 0.2 to 0.3 and incubating at 30°C for at least 3 h before harvest.
Microscopy.
Transformants were grown at 30°C in −Ura/glucose medium containing extra adenine (0.008%) to inhibit accumulation of fluorescent pigment in ade2 cells. Cells were examined without fixation under a Nikon E600 epifluorescence microscope equipped with a 50× Plan oil-immersion objective, and images were captured using a cooled black and white charge-coupled device camera (DAGE-MTI, Inc.).
RESULTS
Pheromone-responsive signaling requires Cdc42-Ste20 interaction.
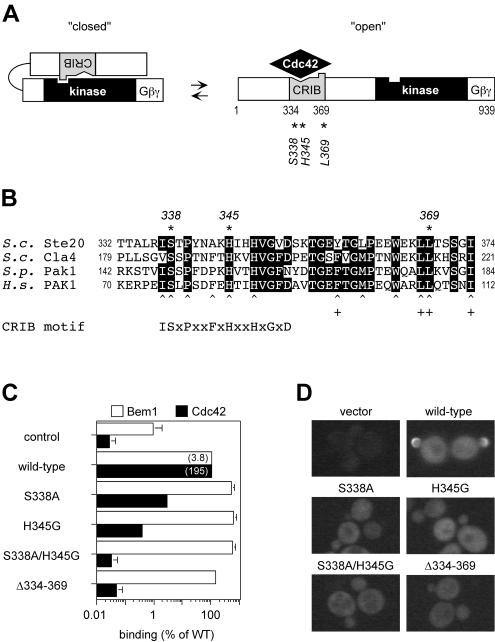
Previous studies tested the role of the Cdc42-Ste20 interaction in pheromone response by deleting the entire Ste20 CRIB domain (27, 45). To reexamine this issue, we made point mutations at two highly conserved residues in the Ste20 CRIB domain (Fig. 1A and B), changing Ser338 to Ala (S338A) or His345 to Gly (H345G). Alteration of the homologous residues in human and S. pombe PAKs disrupts Cdc42 binding (59, 66, 68). The Ste20 mutations were also combined to make an S338A/H345G double mutant.
FIG. 1.
Mutations to test whether the Ste20 CRIB domain behaves as a Cdc42-regulated autoinhibitory domain. (A) Model for activation of Ste20, supported by this study, involving transition between low-activity (closed) and high-activity (open) conformations. The CRIB domain (grey) is indicated as having regions that bind Cdc42 (notch) or the kinase domain (bump). Asterisks denote residues mutated in this study. The Gβγ-binding domain was defined previously (29). (B) Alignment of CRIB domains in PAKs from S. cerevisiae (S.c.), S. pombe (S.p.) and Homo sapiens (H.s.). Residues identical in three or more proteins are boxed in black, and mutated residues are indicated (∗). Also indicated are human PAK1 residues (30, 37, 66, 68) involved in binding Cdc42 (caret) or the PAK1 kinase domain (+). A consensus CRIB motif is shown at the bottom; sequences C-terminal to this motif are well conserved among PAKs (68) but not between PAKs and other Cdc42 targets, such as mammalian WASP and ACK or yeast Gic1 and Gic2 (8, 37). (C) Effects of CRIB domain mutations on binding to Cdc42 and Bem1, measured by two-hybrid assay. Wild-type (pB20N2) and mutant (pPP1059, pPP1060, pPP1061, and pPP1062) derivatives of a DNA-binding-domain fusion to Ste201-499 were coexpressed in PPY760 with activation domain fusions to Bem1157-551 (pRL51.1) or Cdc42G12V/C188S (pPP1027), or with vector (pGAD424). β-Galactosidase measurements (mean ± standard deviation [SD]; n = 4) were normalized to the wild-type (WT) Ste20 allele (= 100%) for each binding partner; values in parentheses are the mean units for the wild-type interactions. The negative control (control) gives the basal signal of pB20N2 with pGAD424 (mean units = 0.04); the mutant derivatives of pB20N2 gave similar basal signals (not shown). (D) Effects on subcellular localization. Representative fluorescence images of cells (PPY913) expressing the indicated GFP-Ste20 mutants or carrying only a vector (pRS316).
To test for effects on binding, the mutations were introduced into a two-hybrid fusion construct containing Ste20 residues 1 to 499. To assay function, they were introduced into low-copy-number vectors encoding either native Ste20 or a GFP-Ste20 fusion protein, each expressed from the STE20 promoter. We found it useful to study the mutations in both expression contexts, as they appeared to confer different levels of function—e.g., the native constructs gave lower signaling and thus revealed the subtle defects of mild mutations, whereas the GFP constructs gave higher signaling and thus revealed the residual function of severe mutations (see Table 3, Materials and Methods, and also Fig. 2 below). For comparison to the point mutations, a CRIB domain deletion (Δ334-369) studied previously (27, 45) was introduced into all of the same vectors.
Cdc42 binding, measured by a two-hybrid assay (Fig. 1C), was disrupted by the point mutations to various degrees: S338A showed the mildest defect, H345G showed a stronger defect, and the S338A/H345G double mutant showed the strongest defect, which approximated that observed with the complete CRIB domain deletion (Δ334-369). These effects were specific to Cdc42, as the mutations did not interfere with binding to Bem1 in a similar assay (Fig. 1C). The subcellular localization of GFP-Ste20 harboring these mutations was consistent with loss of Cdc42 binding (27, 39, 45), as all mutants showed loss of the sharp peripheral fluorescence at bud tips (Fig. 1D), including that with the weakest Cdc42-binding defect, S338A.
Importantly, we found that the point mutants showed defects in mating pathway function that paralleled their Cdc42-binding defects (Fig. 2A), with the S338A mutant showing mild defects, H345G showing stronger defects, and the S338A/H345G mutant showing very strong defects that approached the null phenotype. This pattern was observed in all assays of mating pathway function, including growth arrest, transcriptional induction (FUS1-lacZ), and mating ability. In stark contrast to these point mutants, the mutant lacking the entire CRIB domain (Δ334-369) was largely signaling competent (e.g., roughly 50-fold-higher FUS1-lacZ induction and 1,000-fold-higher mating efficiency than the S338A/H345G mutant) despite negligible Cdc42 binding. These results suggest that interaction with Cdc42 is normally critical for Ste20 to function in the mating pathway, whereas deletion of the CRIB domain bypasses this requirement.
In order to rule out the possibility that these pheromone response defects result from disruption of binding between Ste20 and Gβγ (29), we assayed signaling that was initiated by a membrane-targeted derivative (Ste5-CTM) of the kinase cascade scaffold protein Ste5, which bypasses the requirement for Gβγ but still depends on Ste20 (47). In this setting (Fig. 2B), the CRIB domain mutants showed a spectrum of defects similar to those found when signaling was initiated by pheromone; in the most extreme case, FUS1-lacZ induction in the S338A/H345G double mutant was reduced to roughly 1% of the wild-type level. These data are consistent with the pheromone response assays and show that the signaling defects are not caused by defective Ste20-Gβγ interaction. In addition, because of the absence of Ste4 (Gβ) and pheromone in these experiments, they demonstrate that the signaling role of the Cdc42-Ste20 interaction operates independently of receptor activation and does not require regulatory input from pheromone or Gβγ.
We also integrated into the genomic STE20 locus the two alleles with the strongest Cdc42-binding defects, S338A/H345G and Δ334-369. In both mating and transcriptional induction assays, these alleles showed opposing phenotypes: the S338A/H345G mutant was nearly null, while the Δ334-369 mutant was nearly wild type (Fig. 2C). These results largely confirm the results with plasmid-borne alleles, though one difference was notable: the ability of the Δ334-369 allele to approach wild-type levels of function was greatest when integrated (e.g., 76% of wild-type FUS1-lacZ), and when it was plasmid-borne, function was greater for the GFP fusion (40% of wild-type level) than for the native, non-GFP derivative (14% of wild-type level). Thus, whether the CRIB deletion form behaves as largely competent or as significantly defective can depend on the expression context (see also Table 3). This pattern is important when comparing the effects of CRIB domain deletion on different pathways (such as mating versus filamentation pathways; see below). For example, the conclusion that Ste20Δ334-369 is defective at diploid pseudohyphal growth (27) derived from a comparison of plasmids (pSTE20-5 versus pRS316-STE20Δ334-369) in which wild-type and mutant alleles were expressed in different contexts, and which we found also differed by roughly 15-fold for FUS1-lacZ induction (see Table 3). Overall, our observations indicate that deletion of the CRIB domain creates a hypomorphic form of Ste20 that is mildly compromised for mating pathway signaling, to a degree that may be enhanced in contexts of reduced expression.
Cdc42 binding is required to antagonize autoinhibitory interaction between Ste20 CRIB and kinase domains.
To address the possibility that something special about the extensive size of the Δ334-369 deletion allows it to bypass the requirement for Cdc42 binding or that something detrimental about the point mutations makes them defective for reasons unrelated to Cdc42 binding, we tested whether the S338A/H345G double mutant could be rescued by a third point mutation at Ste20 residue Leu369 (L369G), at the downstream end of the CRIB domain (see Fig. 1A and B). In other PAKs, mutation at the analogous residue interferes with intramolecular interaction between the CRIB and kinase domains (9, 59, 66). Thus, we predicted that if Ste20 can adopt an autoinhibited conformation, the L369G mutation might disrupt this conformation and render Ste20 Cdc42 independent.
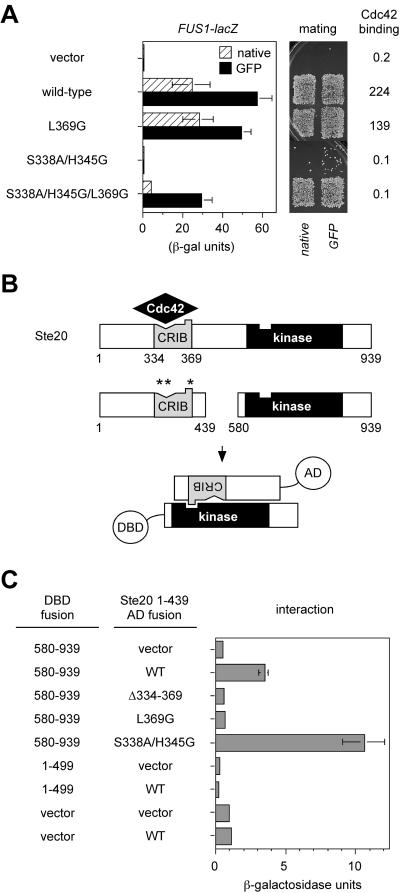
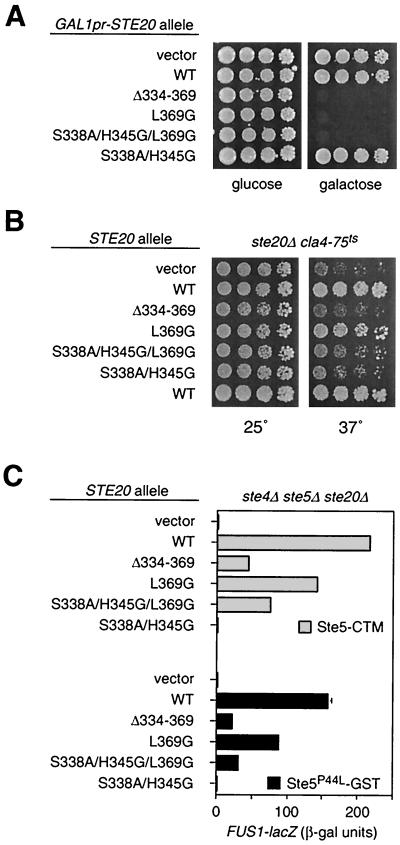
Indeed, the L369G mutation on its own had little effect on either Cdc42 binding or signaling by Ste20, but it showed a dramatic ability to suppress the signaling defects of the S338A/H345G mutation, and did so without restoring Cdc42 binding (Fig. 3A). This indicates that the Cdc42-Ste20 interaction is primarily required to relieve the negative influence of Leu369 and that loss of Leu369 is sufficient to explain the Cdc42-independent behavior of the larger Δ334-369 deletion.
FIG. 3.
The Ste20 CRIB domain behaves as an autoinhibitory domain. (A) Mutation of Leu369 is sufficient to allow Ste20 to signal without binding Cdc42. FUS1-lacZ and mating assays used strain PPY913 harboring the vector (pRS316) or plasmids encoding the indicated native Ste20 or GFP-Ste20 derivatives, similar to Fig. 2. Bars, mean ± SD (n = 4). Cdc42 binding two-hybrid results show mean β-galactosidase units (n = 3) from PPY760 cells cotransformed with pPP1027 and either pBTM116, pB20N2, pPP1119, pPP1061, or pPP1115. (B) Schematic diagram of interaction between CRIB and kinase domains, as detected by two-hybrid analysis (in panel C) using activation domain (AD) and DNA-binding domain (DBD) fusions to the indicated Ste20 fragments. Positions of point mutations are denoted by asterisks as in Fig. 1A. (C) CRIB-kinase interaction results. Strain PPY760 harbored plasmids encoding the indicated DNA-binding domain fusions (pPP1309, pB20N2, and pBTM116) in combination with the vector (pGADXP) or activation domain fusions to wild-type (WT) and mutant derivatives of Ste20 residues 1 to 439 (pPP1037, pPP1321, pPP1322, and pPP1323). The kinase domain fragment (580 to 939) bears a kinase-inactivating mutation, K649M, in order to prevent toxicity. Values are means ± SD (n = 5).
Consistent with these observations, we were able to detect a specific interaction between the Ste20 C-terminal kinase domain and an N-terminal fragment containing the CRIB domain in a two-hybrid assay (Fig. 3B and C). Importantly, this interaction was eliminated by each of the CRIB domain mutations that conferred Cdc42-independent signaling—i.e., the large deletion (Δ334-369) and the L369G point mutation. In contrast, the interaction was not disrupted by the double mutation (S338A/H345G), which produced the most severe pheromone response defect (and which actually increased the CRIB-kinase interaction signal, perhaps by reduced interference from endogenous Cdc42). Therefore, the Ste20 CRIB domain interacts both with Cdc42 (via S338 and H345) and with the kinase domain (via L369), and the role of the former (Cdc42-CRIB) interaction is to antagonize the negative effect of the latter (CRIB-kinase) interaction.
Cdc42 binding stimulates Ste20 kinase activity by relief of autoinhibition.
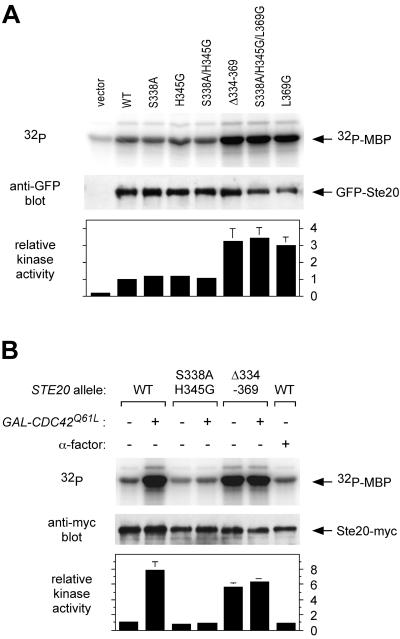
We tested the kinase activity of all of the Ste20 mutants upon immunoprecipitating the GFP fusion derivatives from cell lysates using anti-GFP antibodies (Fig. 4A). We found that kinase activity was elevated three- to fourfold for the three mutants whose signaling properties suggested they are Cdc42 independent, namely, Ste20Δ334-369, Ste20S338A/H345G/L369G, and Ste20L369G. This hyperactivity supports the contention that these mutations disrupt an autoinhibited conformation of Ste20, although it was not observed in previous studies of Ste20 deleted for the CRIB domain (see Discussion).
FIG. 4.
Ste20 kinase activity and stimulation by Cdc42-GTP. (A) Hyperactive kinase activity displayed by Cdc42-independent Ste20 mutants. Strain PPY913 was transformed with the indicated GFP-STE20 plasmids or pRS316 (vector). Ste20 mutant proteins were immunoprecipitated with anti-GFP antibodies and tested for kinase activity by using myelin basic protein (MBP) as the substrate. Portions of the kinase reactions were also analyzed by anti-GFP immunoblot to determine protein levels. Bottom, quantification of two independent experiments (mean ± range), normalized in each experiment to the level of [32P]MBP in the wild-type (WT) sample (= 1). (B) Cdc42-GTP stimulates kinase activity of wild-type but not mutant Ste20 derivatives. Ste20 kinase activity was assayed using strains PPY1234, PPY1238, and PPY1236, expressing Myc12-tagged wild-type, S338A/H345G, and Δ334-369 derivatives of Ste20, respectively. The strains harbored a vector (pRS413) or a galactose-inducible CDC42Q61L construct (pH-G42-L61), as denoted by − and + GAL-CDC42Q61L, respectively. Transformants were induced with 2% galactose for 90 min in the absence (−) or presence (+) of 10 μM α-factor as indicated. The Myc12-tagged proteins were immunoprecipitated, assayed for kinase activity, and tested for Ste20-Myc protein levels, as for panel A. Bottom, quantification of two independent experiments as in panel A.
Surprisingly, we did not observe reduced kinase activity with the signaling-deficient mutants (Fig. 4A), even for the mutant showing the strongest signaling defect in vivo, S338A/H345G. We considered the possibility that only a small fraction of wild-type molecules are fully active at any one time in the cell, and thus any reduction in this fraction may be masked by an excess of low-activity molecules or perhaps by molecules that become activated by unfolding during cell lysis. Therefore, we tested whether the level of active Ste20 in vivo could be boosted by increasing the amount of GTP-bound Cdc42, using the GTPase-deficient mutant Cdc42Q61L (Fig. 4B). Indeed, galactose-induced expression of Cdc42Q61L stimulated the kinase activity of wild-type Ste20 (Ste20WT). In contrast, Ste20S338A/H345G could not be stimulated, whereas Ste20Δ334-369 activity was constitutively elevated and was not further increased by Cdc42Q61L. These results show clearly that the kinase activity of Ste20 can be stimulated by Cdc42 in vivo. Furthermore, they suggest that the signaling defects of Ste20S338A/H345G result from an inability to become stimulated by Cdc42, whereas the Cdc42-independent kinase activity of Ste20Δ334-369 (or Ste20S338A/H345G/L369G) allows signaling without binding Cdc42.
We also took advantage of our ability to detect Cdc42-mediated stimulation of Ste20 to revisit the issue of whether pheromone stimulates Ste20 kinase activity, but we found no effect (Fig. 4B, far right lane). This is consistent with a previous report (65), and hence there remains no evidence that pheromone can stimulate Ste20 kinase activity. Moreover, because it did not mimic expression of Cdc42Q61L, pheromone treatment also does not produce an obvious increase in the levels of GTP-bound Cdc42. While it remains possible that such effects do occur (see Discussion), the alternative scenario raised by these observations is that Cdc42 and Ste20 activities per se may be uninfluenced by pheromone.
L369G highlights separable roles for Cdc42 in Ste20 kinase activity and function.
It has been observed previously that overexpression of a fragment encoding only the kinase domain of Ste20 (Ste20ΔN) is toxic due to disruption of actin organization (27, 49). Consistent with their hyperactive kinase activity, we found that Ste20Δ334-369, Ste20L369G, and Ste20S338A/H345G/L369G were also toxic when overexpressed from the GAL1 promoter, whereas wild-type Ste20 and the signaling-deficient mutant Ste20S338A/H345G were not (Fig. 5A).
FIG. 5.
Additional contribution of Cdc42 to Ste20 function, separable from kinase activity. (A) Hyperactive kinase mutants are toxic when overexpressed. Strain PPY398 was transformed with pPP1219, pDH166, pPP1248, pPP1272, pPP1274, or pPP1273, and then fivefold serial dilutions were spotted onto −His/glucose or −His/raffinose/galactose plates, as indicated, and incubated for 3 days at 30°C. (B) Loss of Cdc42-binding, not deregulated Ste20 activity, disrupts the Cla4-redundant essential function of Ste20. Strain KBY211 (ste20Δ cla4Δ YCp-TRP1-cla4-75ts) was transformed with the indicated GFP-STE20 plasmids, and then fivefold serial dilutions were spotted onto −Ura plates and incubated for 3 days at 25 or 37°C, as indicated. (C) Loss of Cdc42 binding decreases Ste20 signaling activity even when kinase activity is already made Cdc42 independent by the L369G mutation. Strain PPY866 harbored the indicated GFP-STE20 plasmids plus either pH-GS5-CTM (Ste5-CTM) or pH-SL2 (Ste5P44L-GST). Bars show FUS1-lacZ induction (mean ± SD, n = 3) after galactose-induced synthesis of the indicated Ste5 derivative.
Despite their similar kinase activities and toxicities, however, the L369G mutant differs from the other two hyperactive mutants in that it retains the ability to bind Cdc42. This prompted us to ask whether Cdc42 binding might still contribute to Ste20 function—e.g., by localization—even when kinase activity is deregulated. Two phenotypes indicate that it does. First, we compared the CRIB domain mutants for their ability to perform an essential cell polarization function (13, 22) that is redundant with a related kinase, Cla4 (Fig. 5B). Previous work showed that this function was impaired by removal of the Ste20 CRIB domain (27, 45), which, in light of our results, could indicate that cells lacking Cla4 are sensitized to either delocalization or hyperactivity of Ste20. We found that the Ste20L369G mutant was distinguishable from the other two hyperactive mutants in that it remained capable of supporting growth when Cla4 was inactivated. This suggests that it is delocalization, not deregulated kinase activity alone, that causes loss of the essential (Cla4-redundant) function in the Ste20Δ334-369 and Ste20S338A/H345G/L369G mutants.
Second, we tested mating pathway signaling initiated by targeting Ste5 to the plasma membrane (47), using Ste5-CTM (Fig. 5C, top). Of the three hyperactive kinase mutants, signaling was demonstrably stronger for Ste20L369G than for the other two (Ste20Δ334-369 and Ste20S338A/H345G/L369G), suggesting that retention of Cdc42 binding allows Ste20L369G to signal more efficiently. Curiously, similar results were observed with a Ste5 reagent (54) that is not explicitly targeted to the periphery, but instead is activated by a combination of mutation and fusion to GST (Ste5P44L-GST; Fig. 5C, bottom). This suggests either that the function of Cdc42 binding which is disrupted in the Ste20Δ334-369 and Ste20S338A/H345G/L369G mutants can affect Ste5 signaling that is unlocalized or that signaling by Ste5P44L-GST is still initiated primarily at the cell periphery (see Discussion); relevant to this latter possibility, signaling by this form of Ste5 is considerably stronger when Gβγ is present (R. E. Lamson and P. M. Pryciak, unpublished observations).
Filamentation pathway signaling.
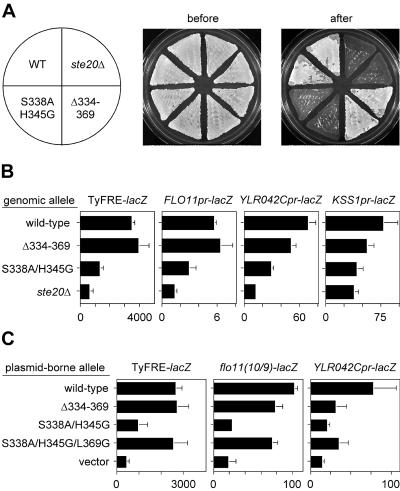
Ste20 also functions in a signal transduction pathway that governs filamentous growth (31, 51). To analyze the role of Cdc42-Ste20 binding in this pathway, we replaced the genomic STE20 gene with the S338A/H345G and Δ334-369 alleles in cells of the Σ1278b strain background, which is necessary for tests of filamentous growth (19, 51). The relative ability of these two alleles to function in the filamentation pathway paralleled that for the mating pathway—i.e., the S338A/H345G double mutant resembled a ste20Δ null mutant, whereas the Δ334-369 mutant was nearly wild type for agar invasion ability (Fig. 6A). This was confirmed quantitatively (Fig. 6B) by assaying transcriptional expression from several different Tec1- and Kss1-dependent reporter constructs (33, 34, 50). All reporters were strongly reduced by the S338A/H345G double mutant. The Δ334-369 mutant, on the other hand, was indistinguishable from the wild type for two reporters (TyFRE and FLO11) and mildly reduced for two other reporters (YLR042C and KSS1), reminiscent of its mild deficiency in the mating pathway.
FIG. 6.
Effects of Ste20 CRIB mutants on the filamentation pathway. (A) Agar invasion in haploid strains PPY966, PPY1209, PPY1200, and PPY1202. Duplicate patches of each strain were grown on a rich medium (YPD) plate, which is shown before and after being rinsed under a stream of water. Note that the Δ334-369 allele retains agar invasion activity, which differs from previous reports of agar invasion or pseudohyphal growth assays when this allele was tested in plasmid-borne form (27, 45). In contrast, the S338A/H345G allele is strongly defective. These results are representative of multiple assays and were confirmed by using independently derived genomic replacements with the Δ334-369 and S338A/H345G alleles. (B) Transcriptional expression of filamentation reporters. The strains tested in panel A harbored the indicated lacZ reporter constructs (plasmids pPP827, pPP1253, p2988, and p2987). Bars show β-galactosidase units (mean ± SD; n = 6 to 8). Note that the Δ334-369 allele activates the TyFRE and FLO11 reporters to a degree indistinguishable from the wild-type (WT) level but is detectably reduced for the other two reporters. (C) Effects of plasmid-borne alleles on filamentation reporter expression. β-Galactosidase activity was measured in strain PPY1209 harboring the indicated lacZ reporter (pPP827, pPP1252, or p2988) plus plasmids expressing native Ste20 derivatives as follows, from top to bottom: pPP1001, pPP1011, pPP1005, pPP1112, and pRS316. Bars show β-galactosidase units (mean + SD, n = 9). The flo11(10/9)-lacZ reporter contains a 440-bp segment of the 3-kb promoter present in the full-length FLO11pr-lacZ construct (53); it is included here to illustrate the range of transcriptional effects caused by removal of the Ste20 CRIB domain, from no effect (TyFRE and FLO11), to mild effect (flo11[10/9]), to moderate effect (YLR042C and KSS1).
Our results with the CRIB deletion mutant were surprising in light of prior reports that concluded it was defective for filamentation pathway function (27, 45). Previous tests used plasmid-borne STE20 alleles, which may enhance mild signaling defects (see above). We were unable to observe robust agar invasion using plasmid-borne alleles (data not shown), but we did compare them for transcriptional reporter expression (Fig. 6C). These tests yielded a pattern similar to that found for the integrated alleles, with the S338A/H345G double mutant approximating the null phenotype and the Δ334-369 mutant showing an intermediate phenotype in which the severity was dependent on the reporter—i.e., either no defect (TyFRE), a mild defect (flo11[10/9]), or a stronger defect (YLR042C).
As seen with mating pathway signaling, defects in expression of filamentation reporters (e.g., YLR042C) were more pronounced for the Δ334-369 mutant when plasmid borne (39% of wild-type level) than when integrated (72% of wild-type level). More importantly, the strong defects in filamentation pathway signaling observed for the S338A/H345G double mutant were suppressed by the L369G mutation, with the resulting triple mutant matching the Δ334-369 derivative for all reporters (Fig. 6C). This shows that for filamentation, as for mating, Cdc42-Ste20 binding is primarily required to antagonize inhibitory CRIB sequences. In total, our results indicate that rather than being qualitatively distinct (27, 45), the role of the Cdc42-Ste20 interaction is highly similar between the mating and filamentation pathways.
DISCUSSION
In this study we have readdressed the signal transduction role of the Cdc42-binding (CRIB) domain in Ste20, a yeast PAK family kinase. We provide genetic and biochemical evidence that Cdc42-Ste20 binding regulates Ste20 kinase activity and signaling competence. Point mutations in the CRIB domain decrease pheromone response to a degree proportional to their Cdc42-binding defect (Fig. 2), suggesting that the Cdc42-Ste20 interaction is normally critical for pheromone signaling. This conclusion contrasts with that made previously from studies in which larger deletions were used to remove the Cdc42-binding site (27, 45). In retrospect, those studies appear to have bypassed the Cdc42 requirement (39) rather than show it is unnecessary, because the deletions also removed inhibitory residues immediately downstream of the CRIB consensus motif.
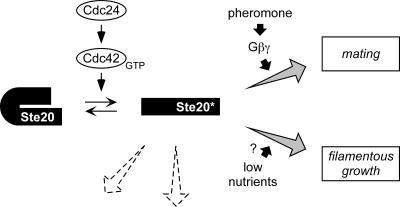
Indeed, we found here that the Ste20 CRIB domain interacts with the kinase domain and disruption of this interaction by mutation relieves the requirement for Cdc42 binding (Fig. 3). Our results overwhelmingly support a model in which Cdc42 activates Ste20 by antagonizing the negative influence of sequences within the CRIB domain (see Fig. 1A and 7). They also complement the recent data for Cdc42 mutants that show defects in both Ste20 binding and pheromone response (39) and are consistent with other recent studies on PAKs from other organisms (9, 30, 59, 66). We suggest that the basic mechanism proposed for GTPase-mediated regulation of PAKs (reviewed in reference 21) is shared by Ste20, indicating that Ste20 continues to serve as a model for PAK function. Our results with Ste20 are equally compatible with models in which autoinhibition occurs intramolecularly (in cis) or intermolecularly between members of a homodimer (in trans), as shown recently for mammalian PAK1 (44).
FIG. 7.
General model for the production and utilization of activated Ste20. Based on results in this study, the model proposes that GTP-bound Cdc42 (presumably activated by its exchange factor, Cdc24) converts Ste20 to an active form (asterisk) by antagonizing the autoinhibitory influence of CRIB domain sequences in the Ste20 N terminus. As discussed in the text, this conversion is required for pheromone-dependent signaling, but there is no evidence that mating pheromones stimulate this conversion, and instead Cdc42-activated Ste20 is competent to signal even without prior exposure to pheromone. Therefore, pheromone-mediated activation of the Gβγ complex may promote signaling through the mating pathway by an existing pool of activated Ste20 kinase. The lifetime of activated Ste20 is unknown, as is the degree to which Ste20 activity requires continual interaction with Cdc42; evidence from other systems suggests that PAK autophosphorylation stabilizes the open conformation, allowing kinase activity to persist after dissociation of the GTPase (10, 66). The pathway leading to filamentous growth also requires activation of Ste20 by Cdc42. The question mark implies that while nutritional cues can trigger filamentous growth, they do not necessarily stimulate the Ste20-dependent filamentation MAP kinase cascade, as discussed previously (35); instead, nutritional cues may primarily modulate a separate (protein kinase A) pathway that is required in parallel for filamentous growth (12, 32). Dashed-line arrows denote that Ste20 also functions in other cellular processes (e.g., regulation of actin, cell polarity, and osmoregulatory signaling); these likely also depend on activation by Cdc42, but little is known about Ste20 regulation during these events.
Interestingly, PAKs are not alone among targets of Rho family GTPases in being regulated by conversion between autoinhibited and uninhibited forms, generically termed intrasteric regulation (25). Instead, this mechanism appears to be common among many kinase and nonkinase targets of Rho, Rac, and Cdc42 (7, 21, 24, 30). It is also notable that the yeast pheromone response pathway (for a review, see reference 15) repeatedly uses a strategy in which positive activation is accomplished by antagonism of a negative regulator: (i) ligand-bound receptor activates Gβγ by inhibiting the negative effect of the Gα subunit; (ii) Cdc42 activates Ste20 by inhibiting the negative effect of the Ste20 CRIB domain; (iii) Ste20 activates Ste11 by inhibiting the negative effect of the Ste11 N terminus; (iv) Fus3 and Kss1 activate Ste12-mediated transcription by inhibiting repressors of Ste12, Dig1/Rst1, and Dig2/Rst2; and (v) release of an interaction between the N and C termini of Ste5 may also contribute to activation of the MAP kinase cascade (54).
We observed hyperactive kinase activity in all Ste20 mutants with Cdc42-independent signaling ability (Fig. 4). The level of hyperactivity (three- to eightfold) is similar to that observed with another yeast PAK, Cla4 (6), whereas each of these is relatively modest compared to mammalian PAKs bearing analogous mutations (9, 66). While our results are consistent with those for other PAKs, they were not observed in previous studies of Ste20 mutants lacking the CRIB domain (27, 45). It is not clear why our results were different, though they were consistent regardless of whether an N-terminal GFP tag or a C-terminal Myc tag was used to purify Ste20. It is possible that in prior studies, the wild-type protein was artificially activated by unfolding to the “open” conformation, either during preparation of cell lysates or by binding of polyclonal anti-Ste20 antibodies (27); alternatively, overexpression may have made Ste20 resistant to negative regulation (45). It is also conceivable that in our experiments the wild-type protein became inactivated during lysate preparation by a mechanism to which the Δ334-369 mutant is insensitive (e.g., refolding); if so, this may have simultaneously obscured differences between the wild type and the signaling-deficient mutants (e.g., S338A/H345G).
Regardless of the explanation, our results indicate that there are measurable differences in kinase properties between wild-type Ste20, signaling-deficient mutants, and Cdc42-independent mutants which are likely to be of fundamental importance to signaling. The toxicity resulting from overexpression of the hyperactive mutants (Fig. 5A) independently suggests that the kinase hyperactivity observed in vitro reflects real changes in kinase properties in vivo.
We also found that Ste20 kinase activity is stimulated by expression of GTP-bound Cdc42 in vivo. This has not been reported previously for S. cerevisiae; stimulation in vitro was observed using baculovirus-produced proteins (58), but this was not reproduced using Ste20 purified from yeast extracts (27). Most importantly, we found that the signaling defect of the S338A/H345G mutant correlated with an inability to respond to Cdc42 stimulation, while the ability of L369G or Δ334-369 mutations to confer Cdc42-independent signaling correlated with deregulated, Cdc42-independent kinase activity. Thus, our observations link the Cdc42 dependence of Ste20 kinase activity to the in vivo signaling behavior and argue that signaling by wild-type Ste20 requires that its kinase activity be stimulated by Cdc42.
How is Ste20 kinase activity harnessed by the mating pathway? It is informative that, despite their hyperactive kinase activity, the Cdc42-independent alleles Δ334-369, L369G, and S338A/H345G/L369G do not show constitutive signaling—i.e., there is no increase in either the basal or induced levels of FUS1-lacZ (Fig. 2C and data not shown; see also reference 27). While this may appear counterintuitive, it is in fact consistent with our previous arguments (39, 47) that the rate-limiting step in pheromone signaling is unlikely to be the production of active Ste20, but rather the access of Ste20 to its substrate—namely, the MAP kinase kinase kinase Ste11 associated with the scaffold protein Ste5 (18, 47)—perhaps along with other events, such as conformational changes in Ste5 (54). Consistent with this view, there is no indication that pheromone can stimulate Ste20 kinase activity, as noted previously (65) and confirmed here in parallel with experiments in which Ste20 kinase activity clearly could be stimulated by another method, expression of Cdc42Q61L (Fig. 4B). The fact that pheromone did not mimic Cdc42Q61L expression may also indicate that the levels of GTP-bound Cdc42 are not increased by pheromone, despite the known ability of Gβγ to assemble with the Cdc42 exchange factor, Cdc24. While association of Gβγ with Cdc24 helps guide cell polarization along pheromone gradients (11, 40, 41), it is unresolved whether Gβγ alters only the localization or also the activity levels of Cdc24 and Cdc42, but currently there is no evidence for the latter.
It remains conceivable that pheromone effects on Cdc24, Cdc42, and Ste20 activities do occur but are difficult to detect because they either are extremely labile (e.g., GTP hydrolysis) or involve only a small fraction of molecules. Nevertheless, it seems clear that assembly of complexes involving Gβγ and Cdc24 is not necessary for pheromone to induce Ste20-dependent signaling, as mutants in which these complexes are disrupted are fully competent at pheromone response (41). Furthermore, we show here (Fig. 2B and 5C) that the effects of disrupting the Cdc42-Ste20 interaction are apparent without pheromone stimulation and in the absence of Gβ (Ste4), which, along with other observations (39, 47), suggests that the Cdc42-dependent step normally precedes pathway activation rather than requiring regulation by pheromone or Gβγ. Therefore, in total these findings indicate that although both the Cdc42-Ste20 interaction and Ste20 kinase activity are required for pheromone-dependent signaling, pheromone stimulation of these states is neither required nor evident.
The simplest model would seem to be that pheromone regulates the ability of an existing pool of active Ste20 to activate the downstream MAP kinase cascade (Fig. 7). It is noteworthy in this regard that overexpression of Ste20ΔN, an “activated” form of Ste20 lacking its N-terminal 495 residues, induces FUS1-lacZ to relatively miniscule levels compared to the level when pheromone is added (27); this suggests that pheromone still triggers a critical rate-limiting event, such as phosphorylation of Ste11 by Ste20 (60, 65) or expedited signal transmission from activated Ste11 (18).
Related to the issue of how pathway stimuli harness Ste20 activity, our observations suggest that the role of the Cdc42-Ste20 interaction is not qualitatively distinct between the mating and filamentation pathways, in contrast to previous conclusions (27, 45). Instead, we found that the two pathways are impacted similarly by CRIB domain mutations: (i) precise disruption of the Cdc42-Ste20 interaction by point mutation causes a severe reduction in function, and (ii) these defects are suppressed by either the L369G point mutation or the complete CRIB domain deletion, though not to wild-type efficacy for either pathway. These observations indicate that both pathways require Cdc42 to bind Ste20 and that in each pathway a primary role for this binding is to antagonize the negative effect of the Ste20 CRIB domain (Fig. 7). In addition, when the CRIB domain is completely removed from Ste20, producing a hyperactive kinase, signaling in both pathways is not hyperactive but instead is demonstrably less efficient (to a degree that is exaggerated by reduced expression). It seems likely that this handicap is due to delocalization from the plasma membrane, where activation of the mating pathway MAP kinase cascade is thought to be initiated (36, 47, 61). Whether activation of the filamentation pathway MAP kinase cascade is also initiated at the membrane is less clear, though interactions of Ste11 and Ste7 kinases with the polarity proteins Spa2 and Sph1 (52, 56) could potentially play a role in restricting filamentation pathway signaling to the cell periphery. Proper localization is also critical to the essential function shared by Ste20 and Cla4, as removal of the Cdc42-binding site disrupts this function in either kinase (6, 27, 45) (Fig. 5B). Relatedly, excess levels of hyperactive Ste20 are lethal (27, 49) (Fig. 5A) and produce a depolarized actin phenotype (27), similar to when Ste20 and Cla4 are absent (22) or when Cdc42 is inactivated (1). This requirement for not only PAK activity per se but also its proper spatial regulation is consistent with the fundamental asymmetry of cytoskeletal reorganization events regulated by Cdc42 during cell growth and division (23).
Although Ste20Δ334-369 and Ste20S338A/H345G/L369G are detectably impaired for signaling (e.g., in comparison to the Ste20L369G form, which retains Cdc42-binding; cf. Fig. 2A versus 3A and see Fig. 5), it is somewhat surprising how well they do signal, given their delocalization. It is conceivable that the kinase hyperactivity of these forms compensates for their delocalization to an extent that is purely coincidental. It is also possible that these forms are still restricted to signaling in specialized subcellular locales, in a manner assisted by interactions with other proteins. Indeed, the signaling efficiency of Ste20Δ334-369 appears to depend on interaction with the SH3 domain protein Bem1, as it is reduced by deletion of BEM1 (39) and by mutation of a Bem1-binding domain within Ste20 (M. J. Winters and P. M. Pryciak, unpublished data). This may indicate that while Ste20Δ334-369 is delocalized, it might still signal predominantly at the cell periphery, where Bem1 is abundant (3). Ste20 also binds the pheromone-activated Gβγ complex (29), potentially allowing recruitment of Ste20 to its mating pathway substrates in a manner that compensates to some degree for defects in localization via Cdc42.
Further work will be required to determine the full spectrum of mechanisms involved in colocalizing activated Ste20 with its substrates and to describe the complete set of events triggered by a specific pathway stimulus that are rate limiting for Ste20-dependent signal transduction.
Acknowledgments
We are grateful to K. Blumer, C. Boone, D. Drubin, G. Fink, E. Leberer, D. Lew, H. Madhani, A. Neiman, S. O'Rourke, M. Peter, J. Thorner, and M. Whiteway for plasmids, strains, and antibodies. We thank D. Guertin, D. McCollum, and J. Moskow for advice on kinase assays and A. Breitkreutz and C. Goutte for comments on the manuscript.
This work was supported by a grant from the NIH (GM57769) to P.M.P.
REFERENCES
- 1.Adams, A. E., D. I. Johnson, R. M. Longnecker, B. F. Sloat, and J. R. Pringle. 1990. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111:131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akada, R., L. Kallal, D. I. Johnson, and J. Kurjan. 1996. Genetic relationships between the G protein βγ complex, Ste5p, Ste20p and Cdc42p: investigation of effector roles in the yeast pheromone response pathway. Genetics 143:103-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayscough, K. R., J. Stryker, N. Pokala, M. Sanders, P. Crews, and D. G. Drubin. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137:399-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagrodia, S., and R. A. Cerione. 1999. Pak to the future. Trends Cell Biol. 9:350-355. [DOI] [PubMed] [Google Scholar]
- 5.Bartel, P. L., and S. Fields. 1995. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol 254:241-263. [DOI] [PubMed] [Google Scholar]
- 6.Benton, B. K., A. Tinkelenberg, I. Gonzalez, and F. R. Cross. 1997. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol. Cell. Biol. 17:5067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop, A. L., and A. Hall. 2000. Rho GTPases and their effector proteins. Biochem. J. 348:241-255. [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, J. L., M. Jaquenoud, M. P. Gulli, J. Chant, and M. Peter. 1997. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 11:2972-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, J. L., L. Stowers, M. Baer, J. Trejo, S. Coughlin, and J. Chant. 1996. Human Ste20 homologue hPAK1 links GTPases to the JNK MAP kinase pathway. Curr. Biol. 6:598-605. [DOI] [PubMed] [Google Scholar]
- 10.Buchwald, G., E. Hostinova, M. G. Rudolph, A. Kraemer, A. Sickmann, H. E. Meyer, K. Scheffzek, and A. Wittinghofer. 2001. Conformational switch and role of phosphorylation in PAK activation. Mol. Cell. Biol. 21:5179-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butty, A. C., P. M. Pryciak, L. S. Huang, I. Herskowitz, and M. Peter. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282:1511-1516. [DOI] [PubMed] [Google Scholar]
- 12.Cullen, P. J., and G. F. Sprague, Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cvrckova, F., C. De Virgilio, E. Manser, J. R. Pringle, and K. Nasmyth. 1995. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9:1817-1830. [DOI] [PubMed] [Google Scholar]
- 14.Dan, I., N. M. Watanabe, and A. Kusumi. 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11:220-230. [DOI] [PubMed] [Google Scholar]
- 15.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703-754. [DOI] [PubMed] [Google Scholar]
- 16.Eby, J. J., S. P. Holly, F. van Drogen, A. V. Grishin, M. Peter, D. G. Drubin, and K. J. Blumer. 1998. Actin cytoskeleton organization regulated by the PAK family of protein kinases. Curr. Biol. 8:967-970. [DOI] [PubMed] [Google Scholar]
- 17.Elion, E. A. 2000. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3:573-581. [DOI] [PubMed] [Google Scholar]
- 18.Feng, Y., L. Y. Song, E. Kincaid, S. K. Mahanty, and E. A. Elion. 1998. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr. Biol. 8:267-278. [DOI] [PubMed] [Google Scholar]
- 19.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 20.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman, G. R., and R. A. Cerione. 2000. Flipping the switch: the structural basis for signaling through the CRIB motif. Cell 102:403-406. [DOI] [PubMed] [Google Scholar]
- 22.Holly, S. P., and K. J. Blumer. 1999. PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 147:845-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, D. I. 1999. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63:54-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, A. S., L. T. Kakalis, N. Abdul-Manan, G. A. Liu, and M. K. Rosen. 2000. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature 404:151-158. [DOI] [PubMed] [Google Scholar]
- 25.Kobe, B., and B. E. Kemp. 1999. Active site-directed protein regulation. Nature 402:373-376. [DOI] [PubMed] [Google Scholar]
- 26.Leberer, E., D. Dignard, D. Harcus, D. Y. Thomas, and M. Whiteway. 1992. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 11:4815-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leberer, E., C. Wu, T. Leeuw, A. Fourest-Lieuvin, J. E. Segall, and D. Y. Thomas. 1997. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 16:83-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leeuw, T., A. Fourest-Lieuvin, C. Wu, J. Chenevert, K. Clark, M. Whiteway, D. Y. Thomas, and E. Leberer. 1995. Pheromone response in yeast: association of Bem1p with proteins of the MAP kinase cascade and actin. Science 270:1210-1213. [DOI] [PubMed] [Google Scholar]
- 29.Leeuw, T., C. Wu, J. D. Schrag, M. Whiteway, D. Y. Thomas, and E. Leberer. 1998. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 391:191-195. [DOI] [PubMed] [Google Scholar]
- 30.Lei, M., W. Lu, W. Meng, M. C. Parrini, M. J. Eck, B. J. Mayer, and S. C. Harrison. 2000. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell 102:387-397. [DOI] [PubMed] [Google Scholar]
- 31.Liu, H., C. A. Styles, and G. R. Fink. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741-1744. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 34.Madhani, H. D., T. Galitski, E. S. Lander, and G. R. Fink. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA 96:12530-12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madhani, H. D., C. A. Styles, and G. R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91:673-684. [DOI] [PubMed] [Google Scholar]
- 36.Mahanty, S. K., Y. Wang, F. W. Farley, and E. A. Elion. 1999. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell 98:501-512. [DOI] [PubMed] [Google Scholar]
- 37.Morreale, A., M. Venkatesan, H. R. Mott, D. Owen, D. Nietlispach, P. N. Lowe, and E. D. Laue. 2000. Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat. Struct. Biol. 7:384-388. [DOI] [PubMed] [Google Scholar]
- 38.Mosch, H. U., R. L. Roberts, and G. R. Fink. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5352-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moskow, J. J., A. S. Gladfelter, R. E. Lamson, P. M. Pryciak, and D. J. Lew. 2000. Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:7559-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nern, A., and R. A. Arkowitz. 1999. A Cdc24p-Far1p-Gβγ protein complex required for yeast orientation during mating. J. Cell Biol. 144:1187-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nern, A., and R. A. Arkowitz. 1998. A GTP-exchange factor required for cell orientation. Nature 391:195-198. [DOI] [PubMed] [Google Scholar]
- 42.Oehlen, L. J., and F. R. Cross. 1998. The role of Cdc42 in signal transduction and mating of the budding yeast Saccharomyces cerevisiae. J. Biol. Chem. 273:8556-8559. [DOI] [PubMed] [Google Scholar]
- 43.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parrini, M. C., M. Lei, S. C. Harrison, and B. J. Mayer. 2002. Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol. Cell 9:73-83. [DOI] [PubMed] [Google Scholar]
- 45.Peter, M., A. M. Neiman, H. O. Park, M. van Lohuizen, and I. Herskowitz. 1996. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 15:7046-7059. [PMC free article] [PubMed] [Google Scholar]
- 46.Pryciak, P. M., and L. H. Hartwell. 1996. AKR1 encodes a candidate effector of the Gβγ complex in the Saccharomyces cerevisiae pheromone response pathway and contributes to control of both cell shape and signal transduction. Mol. Cell. Biol. 16:2614-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pryciak, P. M., and F. A. Huntress. 1998. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 12:2684-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raitt, D. C., F. Posas, and H. Saito. 2000. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19:4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramer, S. W., and R. W. Davis. 1993. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 51.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 52.Roemer, T., L. Vallier, Y. J. Sheu, and M. Snyder. 1998. The Spa2-related protein, Sph1p, is important for polarized growth in yeast. J. Cell Sci. 111:479-494. [DOI] [PubMed] [Google Scholar]
- 53.Rupp, S., E. Summers, H. J. Lo, H. Madhani, and G. Fink. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sette, C., C. J. Inouye, S. L. Stroschein, P. J. Iaquinta, and J. Thorner. 2000. Mutational analysis suggests that activation of the yeast pheromone response mitogen-activated protein kinase pathway involves conformational changes in the Ste5 scaffold protein. Mol. Biol. Cell 11:4033-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheu, Y. J., Y. Barral, and M. Snyder. 2000. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:5235-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheu, Y. J., B. Santos, N. Fortin, C. Costigan, and M. Snyder. 1998. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18:4053-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon, M. N., C. De Virgilio, B. Souza, J. R. Pringle, A. Abo, and S. I. Reed. 1995. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature 376:702-705. [DOI] [PubMed] [Google Scholar]
- 59.Tu, H., and M. Wigler. 1999. Genetic evidence for Pak1 autoinhibition and its release by Cdc42. Mol. Cell. Biol. 19:602-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Drogen, F., S. M. O'Rourke, V. M. Stucke, M. Jaquenoud, A. M. Neiman, and M. Peter. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10:630-639. [DOI] [PubMed] [Google Scholar]
- 61.van Drogen, F., V. M. Stucke, G. Jorritsma, and M. Peter. 2001. MAP kinase dynamics in response to pheromones in budding yeast. Nat. Cell Biol. 3:1051-1059. [DOI] [PubMed] [Google Scholar]
- 62.Whiteway, M. S., C. Wu, T. Leeuw, K. Clark, A. Fourest-Lieuvin, D. Y. Thomas, and E. Leberer. 1995. Association of the yeast pheromone response G protein βγ subunits with the MAP kinase scaffold Ste5p. Science 269:1572-1575. [DOI] [PubMed] [Google Scholar]
- 63.Wu, C., S. F. Lee, E. Furmaniak-Kazmierczak, G. P. Cote, D. Y. Thomas, and E. Leberer. 1996. Activation of myosin-I by members of the Ste20p protein kinase family. J. Biol. Chem. 271:31787-31790. [DOI] [PubMed] [Google Scholar]
- 64.Wu, C., T. Leeuw, E. Leberer, D. Y. Thomas, and M. Whiteway. 1998. Cell cycle- and Cln2p-Cdc28p-dependent phosphorylation of the yeast Ste20p protein kinase. J. Biol. Chem. 273:28107-28115. [DOI] [PubMed] [Google Scholar]
- 65.Wu, C., M. Whiteway, D. Y. Thomas, and E. Leberer. 1995. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 270:15984-15992. [DOI] [PubMed] [Google Scholar]
- 66.Zenke, F. T., C. C. King, B. P. Bohl, and G. M. Bokoch. 1999. Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J. Biol. Chem. 274:32565-32573. [DOI] [PubMed] [Google Scholar]
- 67.Zhao, Z. S., T. Leung, E. Manser, and L. Lim. 1995. Pheromone signaling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol. Cell. Biol. 15:5246-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao, Z. S., E. Manser, X. Q. Chen, C. Chong, T. Leung, and L. Lim. 1998. A conserved negative regulatory region in αPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol. Cell. Biol. 18:2153-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ziman, M., J. M. O'Brien, L. A. Ouellette, W. R. Church, and D. I. Johnson. 1991. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol. Cell. Biol. 11:3537-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]