Abstract
DNA methylation and chromatin modification operate along a common pathway to repress transcription; accordingly, several experiments demonstrate that the effects of DNA methylation can spread in cis and do not require promoter modification. In order to investigate the molecular details of the inhibitory effect of methylation, we microinjected into Xenopus oocytes a series of constructs containing a human CpG-rich sequence which has been differentially methylated and cloned at different positions relative to a specific promoter. The parameters influencing the diffusion of gene silencing and the importance of histone deacetylation in the spreading effect were analyzed. We demonstrate that a few methylated cytosines can inhibit a flanking promoter but a threshold of modified sites is required to organize a stable, diffusible chromatin structure. Histone deacetylation is the main cause of gene repression only when methylation does not reach levels sufficient to establish this particular structure. Moreover, contrary to the common thought, promoter modification does not lead to the greater repressive effect; the existence of a competition between transactivators and methyl-binding proteins for the establishment of an open conformation justifies the results obtained.
DNA methylation is the major modification of eukaryotic genomes and is known to have a profound effect on gene expression. In mammals, this occurs predominantly at the dinucleotide CpG, and approximately 60 to 90% of the dinucleotides are modified (50). In normal cells, methylation involves mainly CpG-poor regions, while CpG-rich areas (CpG islands), located in regulatory regions of class II genes, seem to be protected from the modification (14). This lack of methylation is likely a prerequisite for active transcription; in fact, methylated CpG islands are found on the inactive X chromosome and on silenced alleles of parentally imprinted genes (41, 47, 48).
Genetic experiments have demonstrated that proper control of DNA methylation is essential for normal mammalian development; accordingly, this epigenetic modification seems to play important roles in X chromosome inactivation, genomic imprinting, senescence, and carcinogenesis (3, 4, 35, 36, 41, 44). The correlation between DNA methylation and gene silencing has been extensively documented by a large body of evidence. In particular, transfection experiments and Xenopus oocyte microinjections, performed with in vitro-methylated DNA, demonstrated that methylation inhibits gene expression (28, 29, 31, 38, 56). Conversely, modified silent genes in cultured cell lines can be activated upon treatment with 5-azacytidine, a demethylating agent (18, 26).
It has been proposed that this modification causes transcriptional repression by directly interfering with the binding of transcription factors to DNA. This hypothesis has been sustained by the identification of a number of transcriptional regulators that cannot bind methylated recognition elements (16). However, the existence of factors indifferent to DNA methylation status and the more recent demonstration that this modification is often capable of repressing transcription only after chromatin has been assembled suggest that this direct effect is not the main mechanism by which modified DNA inhibits gene expression (9, 28, 29).
A second model proposes that methylation attracts proteins that specifically bind modified DNA, thus blocking access to other factors required for gene induction (6, 34). Indeed, several proteins containing a methylated-DNA binding domain (MBD) have recently been described, and four of them have been implicated in transcriptional repression (5, 22). The biochemical characterization of the identified MBD proteins showed that most of them are in complexes containing histone deacetylases, suggesting that methylated CpGs (mCpGs) suppress transcription by establishing a repressive chromatin environment (27, 40, 43, 52, 57). Consistent with this model, histones assembled on methylated DNA are significantly less acetylated than histones assembled on bulk genome, and methylated transfected genes can be reactivated by treatment with trichostatin A (17, 23). Furthermore, a distinctive chromatin structure, insensitive to nuclease digestion, is formed on methylated DNA (20, 28-30). However, it has recently been demonstrated that naturally hypermethylated genes can be transcriptionally reinduced with trichostatin A only after slight demethylation by low doses of 5-azacytidine; the authors proposed that CpG methylation and histone deacetylation act synergistically to silence gene expression (10).
Consistent with the view that DNA methylation inhibits transcription indirectly by means of a particular chromatin structure, it has been reported that transcriptional repression often does not require methylation of promoter sequences and is transmissible in cis (24, 28, 29, 31, 56). Using regionally methylated plasmids, parameters such as position, length, or density of methylated cytosines have been reported as crucial for the efficiency of repression (23, 24, 28). Other authors suggested that transcriptional inhibition relies on methylation at specific critical CpG sites (11, 21, 56). It is possible that the patch-modified templates used in these experiments, characterized by the modification of CpG-rich prokaryotic vector DNAs, and the analyses performed on different promoters in different cell lines have caused these contradictory results.
Injection of a methylated herpes simplex virus (HSV) thymidine kinase (tk) promoter into Xenopus oocytes demonstrated that transcriptional repression occurs by indirect mechanisms and that a repressive nucleoprotein complex, capable of inhibiting transcription more effectively than the nucleosome alone, is assembled (29). Using the same biological system, we wanted to elucidate what parameters influence this repression and its ability to diffuse in cis. To this purpose, a human region containing a physiologically high density of CpG dinucleotides was cloned in different positions relative to the tk promoter. With a systematic approach, the number of methylatable CpG dinucleotides was varied in each construct.
By injecting differentially methylated templates, we demonstrated that a few modified cytosines in front of the tk promoter are enough to drastically inhibit transcription. This repression is not dependent on critical sites and is a nonlinear function of the number of modified dinucleotides. Moreover, there is a clear threshold effect. Transcriptional inhibition can spread for several hundred base pairs only when a sufficient amount of CpGs are methylated; in these conditions, trichostatin A treatment demonstrates that histone acetylation cannot relieve the repression.
A completely different scenario is observed when the number of modified dinucleotides is reduced: silencing occurs only on the regulatory sequences immediately flanking the methylated region, and trichostatin A completely abolishes the repressive effect. We discuss a model that describes two different mechanisms of gene silencing and explains our results. Moreover, we show that, contrary to the common thought, direct methylation of promoter sequences does not have a greater repressive effect than modification of flanking nonregulatory DNA sequences. Binding of basal transcriptional factors or activators to the modified promoter can, in fact, compete with the assembly of methylation-specific repressive nucleoprotein complexes. Our experiments demonstrate that the silencing effect cannot be overcome by a simple DNA-binding domain, but an activating domain is required; the final level of transcriptional activation at the methylated promoter depends on the specific trans-acting factor analyzed.
MATERIALS AND METHODS
Plasmid constructs.
The M13-tkCAT construct was obtained by cloning from pBS-HSV tk DNA (29) a 907-bp BamHI/KpnI fragment containing the chloramphenicol acetyltransferase (CAT) reporter gene under the control of the HSV tk promoter into bacteriophage M13mp19. The aprt-tk-CAT construct was produced by cloning in front of the tk promoter a 275-bp XbaI/BamHI fragment amplified from the CpG island of the human APRT gene (13) with the primers 5′-GCCTTGAAGCTTCTAGACTGTGCAGGCGTCCTTC-3′ and 5′-GCCTTGTCGACGGATCCTGGGCCACCTCCTGG-3′. tkCAT-11aprt was obtained by inserting into KpnI and SstI, downstream of the CAT gene of the vector M13-tkCAT, the fragment amplified from the APRT gene with the primers 5′-GCCTTGTCGACGGTACCTGTGCAGGCGTCCTTC-3′ and 5′-GCCTTGTCTAGAGCTCTGGGCCACCTCTTGG-3′. The aprt-tkCAT+700 vector was constructed inserting into the BamHI site a 776-bp fragment derived from digestion of the V23XHSF-pSP64 plasmid (N. Landsberger and A. P. Wolffe, unpublished data).
To generate the aprt-tkCAT+100 construct, a PCR fragment of 100 bp amplified from V23XHSF-pSP64 DNA with the primers 5′-CAATAAGCTCATCCAGTTCC-3′ and 5′-GCCTTGGATCCTAGGGGGCGCAGGACTCTGC-3′ was subcloned into the BamHI site. A 146-bp fragment containing five Gal4-binding sites was amplified by PCR from pG5-HSPCAT (33) and cloned into the BamHI site of M13-tkCAT (Gal4×5-tkCAT plasmid) or into the BamHI site of the aprt-tkCAT construct (11-aprt-Gal4×5-tkCAT).
The human cytomegalovirus CMVCAT vector has been described previously (32). The GAL4VP16-pSp64 expression vector and the RXR plasmid were described in references 33 and 37, respectively. The GAL4-pSp64 plasmid was obtained by inserting into pSp64polyA a HindIII/SmaI fragment obtained from the GAL4VP16-pSp64 vector. GALTR-pSp64 DNA was kindly provided by Jiemin Wong. The cDNAs coding for Oct1 and Oct2 were excised from pSCTEVGAL(93)OCT1(Q) and pSCTEVGAL(93)OCT2(Q) (15) and inserted into a HindIII/XbaI fragment of pSp64polyA DNA.
All the constructs prepared by inserting a PCR amplification product were confirmed by DNA sequencing.
Generation of differentially methylated constructs.
Large-scale preparation of single-stranded bacteriophage M13 DNA was carried out as described by Sambrook et al. (49).
Different amplified and purified fragments or different oligonucleotides (Table 1) were annealed to 2 μg of single-stranded DNA (ssDNA) at a molar ratio of 3:1 (fragment DNA to ssDNA) in 10 mM Tris (pH 7.5)-10 mM MgCl2-50 mM NaCl-1 mM dithiothreitol by heating for 10 min at 95°C and cooling it from 70 to 30°C in 1 h. To prevent SssI methylase from binding to and methylating the ssDNA region, T4 gene 32 protein (Promega) was added (4.4 μg/μg of ssDNA), and the reaction mixture was incubated on ice for 15 min. The annealed DNA was methylated for 3 h, using the methylase SssI (New England Biolabs) under the conditions recommended by the manufacturer. After proteinase K treatment and phenol extraction, the DNA was filled in and ligated (T4 DNA polymerase and T4 DNA ligase; New England Biolabs) by standard procedures. For each patch-methylated construct, a mock-methylated control was generated following the same protocol but omitting the methylation step.
TABLE 1.
Reagents used for the production of patch-methylated constructs
| Differentially methylated template | ssDNA template | Annealed DNA | Primers for PCR amplification |
|---|---|---|---|
| 14-aprt-tkCAT | aprt-tkCAT | 318-bp PCR fragment | 5′GGAAACAGCTATGACCATG3′ + 5′GGGGCCGGATCC3′ |
| 11-aprt-tkCAT | aprt-tkCAT | 248-bp PCR fragment | 5′CTGTGCAGGCGTCCTTCCC3′ + 5′CTGGGCCACCTCTTGGGC3′ |
| 6-aprt-tkCAT | aprt-tkCAT | 142-bp PCR fragment | 5′TGCTTCTTGTTCCTTCTG3′ + 5′GGGGCCGGATCC3′ |
| 4-aprt-tkCAT | aprt-tkCAT | 103-bp PCR fragment | 5′CCACGAGCCAGGCCTTCCCT3′ + 5′GGGGCCGGATCC3′ |
| 3-aprt-tkCAT | aprt-tkCAT | 92-bp PCR fragment | 5′CCACGAGCCAGGCCTTCCCT3′ + 5′CTGGGCCACCTCTTGGGC3′ |
| 1-aprt-tkCAT | aprt-tkCAT | 32-mer oligonucleotide | 5′AGGCCTGGAGGCTCCGGGAG + AGCCCAAGAGGT3′ |
| tkCAT-11aprt | tkCAT-11aprt | 248-bp PCR fragment | 5′CTGTGCAGGCGTCCTTCCC3′ + 5′CTGGGCCACCTCTTGGGC3′ |
| 11-aprt-tkCAT+100 | aprt-tkCAT+100 | 248-bp PCR fragment | 5′CTGTGCAGGCGTCCTTCCC3′ + 5′CTGGGCCACCTCTTGGGC3′ |
| 11-aprt-tkCAT+700 | aprt-tkCAT+700 | 248-bp PCR fragment | 5′CTGTGCAGGCGTCCTTCCC3′ + 5′CTGGGCCACCTCTTGGGC3′ |
| 6-aprt-tkCAT+100 | aprt-tkCAT | 152-bp PCR fragment | 5′CTGTGCAGGCGTCCTTCCC3′ + 5′CTCAATACCAGCTCGCAG3′ |
| 6-aprt-tkCAT+700 | aprt-tkCAT+700 | 132-bp PCR fragment | 5′TGCTTCTTGTTCCTTCTG3′ + 5′CTGGGCCACCTCTTGGGC3′ |
| tkCAT-6aprt | tkCAT-11aprt | 142-bp PCR fragment | 5′CTGTGCAGGCGTCCTTCCC3′ + 5′CAGAAGGAACAAGAAGCAAGG3′ |
| 4-aprt-tkCAT+100 | aprt-tkCAT | 121-bp PCR fragment | 5′GTCCCCAGCCCAGGACAG3′ + 5′CTCAATACCAGCTCGCAG3′ |
| 3-aprt-tkCAT+100 | aprt-tkCAT + 100 | 92-bp PCR fragment | 5′CCACGAGCCAGGCCTTCCCT3′ + 5′CTGGGCCACCTCTTGGGC3′ |
| M13-tkCAT | M13-tkCAT | 174-bp PCR fragment | 5′GGATCCGGCCCCGCCCAG3′ + 5′CTCGAGATCTGCGGCACG3′ |
| 11-aprt-Gal4×5-tkCAT | aprt-tkCAT | 248-bp PCR fragment | 5′CTGTGCAGGCGTCCTTCCC3′ + 5′CTGGGCCACCTCTTGGGC3′ |
| Gal4×5-tkCAT | Gal4×5-tkCAT | 146-bp PCR fragment | 5′GCCTTGGATCCGCGGATACAAGTTTGC3′ + 5′CTCGAGATCTGCGGCACG3′ |
The annealed DNA was loaded on a 0.8% agarose gel, and the supercoiled DNA was purified by GF/C extraction. Briefly, a piece of GF/C and dialysis membrane was inserted in the gel in front of the band that had to be purified. High voltage (150 V) was applied until all the DNA moved on the paper. DNA from the glass microfiber (GF/C) (Whatman) was recovered by centrifugation, phenol extracted, and purified on a spin column.
Analysis of methylation pattern.
The correct differential methylation pattern was analyzed by restriction digestion and/or enzymatic restriction followed by PCR. For the restriction analysis, methylated and mock-methylated DNA was digested with the methylation-sensitive enzyme HpaII, and the restriction fragments were analyzed on an agarose gel. For the PCR approach, the gel-purified templates were restricted with the methylation-sensitive enzyme HpaII or the MspI isoschizomer, which is insensitive to CpG methylation. After extensive digestion, a PCR amplification was carried out using two pairs of primers. The first pair serves as internal control and amplifies a region of the plasmid external to the methylated region and without recognition sites for HpaII. The other set of primers instead permits amplification only if the methylation pattern occurs correctly.
Preparation and microinjection of Xenopus oocytes.
The preparation of Xenopus stage VI oocytes and the microinjection procedure were as previously described (32). A 32-nl amount containing 1.0 ng of methylated or mock-methylated DNA and 0.03 ng of CMVCAT was injected into the oocyte nuclei. The injected oocytes were then incubated at 18°C for 16 h in the absence or presence of trichostatin A (300 nM), and healthy oocytes were collected for DNA and RNA analysis.
Exogenous protein expression (TBP, GAL4, GAL4VP16, GALTR, RXR, GAL4 OCT1, and GAL4 OCT2) was obtained by injecting 32 nl (200 ng/μl) of full-length capped RNA synthesized with the mMessage mMachine kit (Ambion). Oocytes are then incubated for 16 h and subsequently injected with the DNA solution.
RNA and DNA analysis of injected oocytes.
Twenty oocytes were collected and homogenized in 200 μl of 0.25 M Tris (pH 7.5). This homogenate was used for both RNA and DNA analysis. From half of the sample, RNA was isolated using Eurozol (EuroClone), and a primer extension assay was performed as previously described (32). The 30-mer oligonucleotide 5′-GGTGGTATATCCAGTGATTTTTTTCTCCAT-3′ complementary to the CAT gene was used as the primer. Extension products were separated on 6% sequencing gels and visualized by autoradiography. Transcription signals were quantitated with a PhosphorImager (Molecular Dynamics) and normalized to the internal control, CMV.
For DNA analysis, half of the homogenate was incubated at 37°C for 2 h in stop buffer (30 mM EDTA, 20 mM Tris [pH 7.5], 1% sodium dodecyl sulfate [SDS]) and proteinase K (500 μg/ml). DNA was extracted twice with phenol-chloroform and ethanol precipitated. After RNase treatment, DNA was separated by agarose gel electrophoresis, and Southern blotting was performed. Hybridization against a random-primed HSV tk promoter amplification fragment was obtained in Rapid-hyb buffer (Amersham). Quantitation of the signals was performed by PhosphorImager, and the levels of mock-methylated and methylated injected DNA were compared.
To make sure that methylation persisted on the injected templates during the course of the experiment, the extracted DNA was HpaII digested and analyzed by Southern blot using a random primed aprt-tkCAT probe (1,160 bp).
Sequencing of PCR fragments amplified from bisulfite-treated DNA.
A total of 1 ng of methylated or mock-methylated DNA was injected, and oocytes were incubated for 16 h at 18°C. For the Gal4×5tkCAT template, DNA was injected into oocytes that had or had not been previously injected with GAL4VP16 mRNA. Thirty healthy oocytes were collected, and DNA was extracted as previously described. Then 1 μg of salmon sperm DNA was added to each purified DNA before digesting it. The enzymes used were DdeI for the 11-, 6-, and 3-aprt-tkCAT templates and PvuII for the Gal4×5tkCAT DNA. Digested DNA was then phenol extracted, ethanol precipitated, and resuspended in 46.5 μl of water. It was denatured by adding 3.5 μl of 3 M NaOH (freshly prepared) and incubated for 15 min at 37°C. Then 30 μl of freshly prepared 10 mM hydroquinone and 520 μl of 3 M sodium bisulfite (pH 5) were added to the samples. Each reaction was overlaid with mineral oil and incubated in a thermocycler for 16 h using a cycling protocol of 30 s at 95°C and 15 min at 50°C. DNA was desalted using a Wizard purification system (Promega) and desulfonated by addition of NaOH to a final concentration of 0.3 M and incubation at 37°C for 15 min. Ethanol precipitation was then performed.
Resuspended bisulfite-treated DNA was used as the template for subsequent PCR. Primers for the aprt-tkCAT templates were 5′BS11 (5′-TCACTCATTAAACACCCCAAACTTTA), 3′BS11 (5′-TTCAAAACCACACACATCACCTTAA), 11BS-A (5′-TTATTTATTAGGTATTTTAGGTTTTA), and 11BS-B (5′-TTTGAGGTTATATGTGTTATTTTAA). Primers for the Gal4×5tkCAT DNA were 5′BSGALx5 (5′-TTCCAACTCATATATTATATAAAATT), 3′ BSGALx5 (5′-AACTAACTAAAATACCTCAAAAT), GALx5BS-A (5′-TTTTGGTTTGTATGTTGTGTGGATT), and GALx5BS-B (5′-AATTGATTGAAATGTTTTAAAAT). PCR was carried out as follows: 94°C for 30 s, 48°C for 30 s, and 72°C for 45 s for 35 cycles and a final elongation step for 5 min at 72°C. PCR products were gel purified and sequenced using the 11BS-A, 11BS-B, GALx5BS-A, and GALx5BS-B primers.
Analysis of exogenous proteins.
Synthesis of overexpressed proteins was monitored by incubating injected oocytes in [35S]methionine (30 μCi/100 μl of buffer). Whole-cell extracts were prepared by homogenizing the collected oocytes (10 μl per oocyte) in buffer containing 70 mM KCl, 20 mM HEPES (pH 7.5), 1 mM dithiothreitol, and 5% sucrose. Following centrifugation, the supernatant was collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
RESULTS
Experimental strategy.
Previous studies have used protocols involving regional methylation to analyze whether transcriptional repression relies on promoter modification. Following this approach, several works suggested that even partial methylation of the coding region or of the vector can inhibit gene expression (24, 28, 29, 31). However, attempts to define the parameters which affect the efficiency of repression have had contradictory results. Density of CpG dinucleotides, distance of methylation sites from the promoter region, and size of the modified fragments have been proposed to be critical factors influencing gene silencing.
It is possible that some of the discordances are due to the fact that most of these studies have been performed using templates that were unmodified on the region of interest but methylated over the whole CpG-rich prokaryotic vector DNA. Moreover, analyses performed on different promoters and in different cell lines might have contributed to the discrepancies.
We reasoned that by using templates with a well-defined region of methylated DNA, having a physiological density and distribution of modified sites, that is inserted in different positions relative to a tk promoter, we would be able to define which parameters influence this repression and its ability to spread in cis. To this purpose, we used transcription of the CAT gene directed from the HSV tk promoter as the reporter for our silencing assay. We amplified a fragment of 240 bp (aprt) (13), characterized by 11 CpG dinucleotides, from the human aprt CpG island.
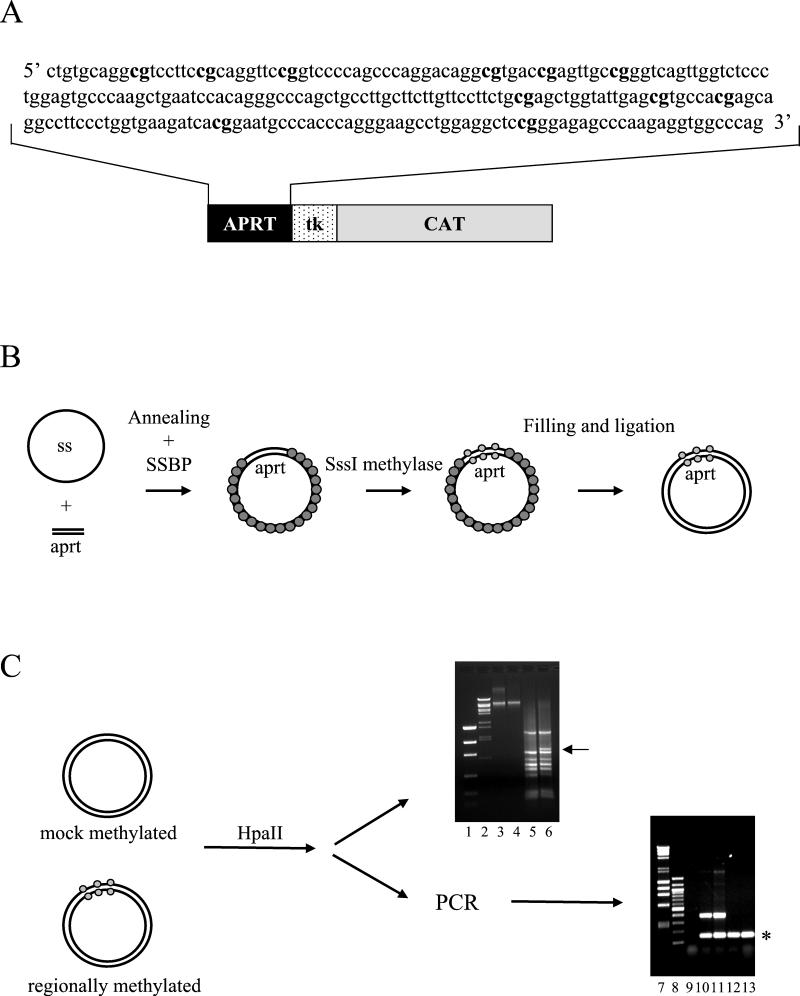
The sequence and distribution of the methylatable sites are shown in Fig. 1A. This region was cloned upstream the HSV tk promoter (Fig. 1A), either immediately before the regulatory elements or separated further by unrelated sequences; we also generated a construct in which the CpG-containing region was located downstream of the CAT gene (see Fig. 4). Following the procedure diagrammed in Fig. 1B (see also Materials and Methods), we obtained plasmids that were methylated precisely only on the cloned aprt fragment; a mock-methylated control was obtained each time by omitting the SssI methylase from the assay. Fully replicated, covalently closed molecules were gel purified (Fig. 1C, lanes 3 and 4), and methylation-sensitive restriction enzymes per se (lanes 5 and 6) or combined with PCR amplification (lanes 10 to 13) were used to confirm that the specific region was completely methylated (51).
FIG. 1.
Schematic representation of the experimental strategy. (A) The 241-bp sequence amplified from the human aprt CpG island is shown. Methylatable CpG dinucleotides are indicated in bold letters. (B) Scheme of the protocol for the generation of differentially methylated templates. (C) Evaluation of the correct pattern of methylation. The methylation pattern of 11-aprt-tkCAT was analyzed by digestion with the methylation-sensitive restriction enzyme HpaII. The digestion profile was directly visualized on a 1% agarose gel (lanes 5 and 6), or the restricted DNA amplified by PCR (lanes 10 to 13). Markers of low-mass DNA (lane 1), λ BstEII-cut DNA (lanes 2), λ HindIII/φχ HaeIII-cut DNA (lane 7), and 100-bp DNA ladder (lane 8) were resolved. The positions of the supercoiled gel-purified mock-methylated and methylated DNAs are shown in lanes 3 and 4, respectively. Lane 5, HpaII-digested mock-methylated DNA. Lane 6, HpaII-digested regionally methylated DNA. The arrow in lane 6 indicates the appearance of a longer DNA fragment indicative of the incapacity of the enzyme to digest the methylated aprt fragment. PCR amplification of the HpaII-restricted DNAs shown in each lane (10 to 13), and the presence of the internal control is indicated with an asterisk. In contrast, the 450-bp fragment, obtainable only if the aprt fragment is methylated, appears only when the digested regionally methylated DNA is used for the amplification (lanes 10 and 11) and not when mock-methylated DNA is analyzed (lanes 12 and 13). In lane 9, a no-DNA control was loaded.
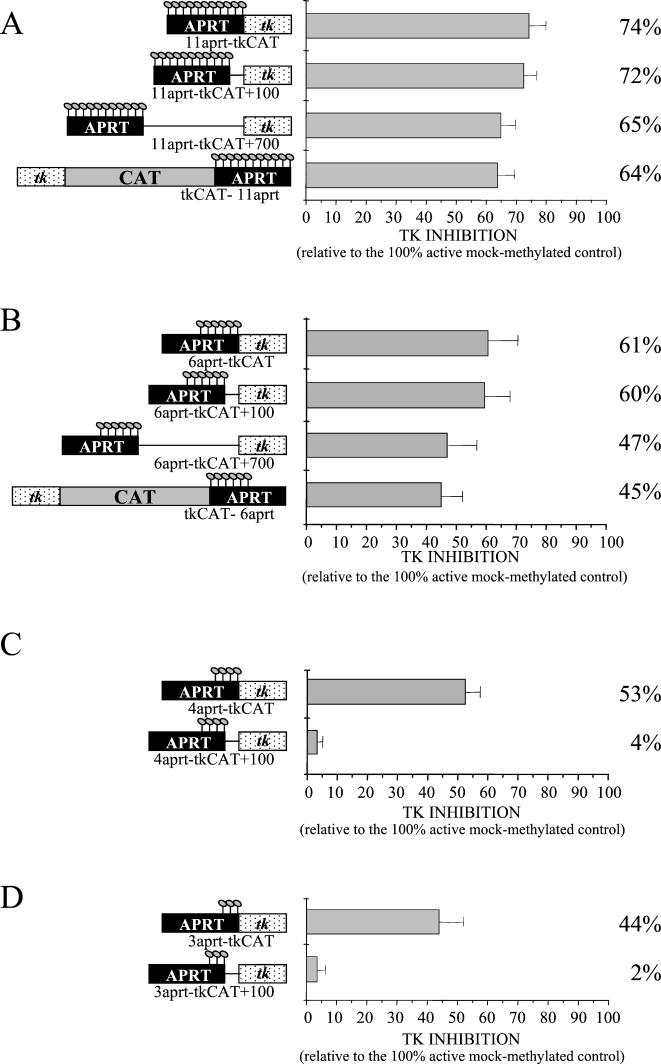
FIG. 4.
Diffusion in cis of the repressive effect is influenced by the number of methylated sites. (A) Repressive activity of the regionally methylated 11-aprt-tkCAT template was compared to that of patch-methylated 11-aprt-tkCAT+100 or 11-aprt-tkCAT+700 or the differentially modified tkCAT-11aprt. Transcription was assayed 16 h after injection, quantified, and standardized against the CMV control compared to the corresponding mock controls. (B) As in panel A, but the inhibitory ability of the modified 6-aprt-tkCAT was compared to that of 6-aprt-tkCAT+100, 6-aprt-tkCAT+700, and tkCAT-6aprt. (C) As in panel A but using the 4-aprt-tkCAT template and the 4-aprt-tkCAT+100 vector. (D) As in panel A but using the 3-aprt-tkCAT DNA and 3-aprt-tkCAT+100. The error bars represent the standard deviations of three independent experiments.
The annealing of different aprt amplification products to ssDNA allowed us to obtain different templates characterized by a variable number of modified dinucleotides (Materials and Methods and Fig. 3); we used this strategy to test their effects on gene silencing.
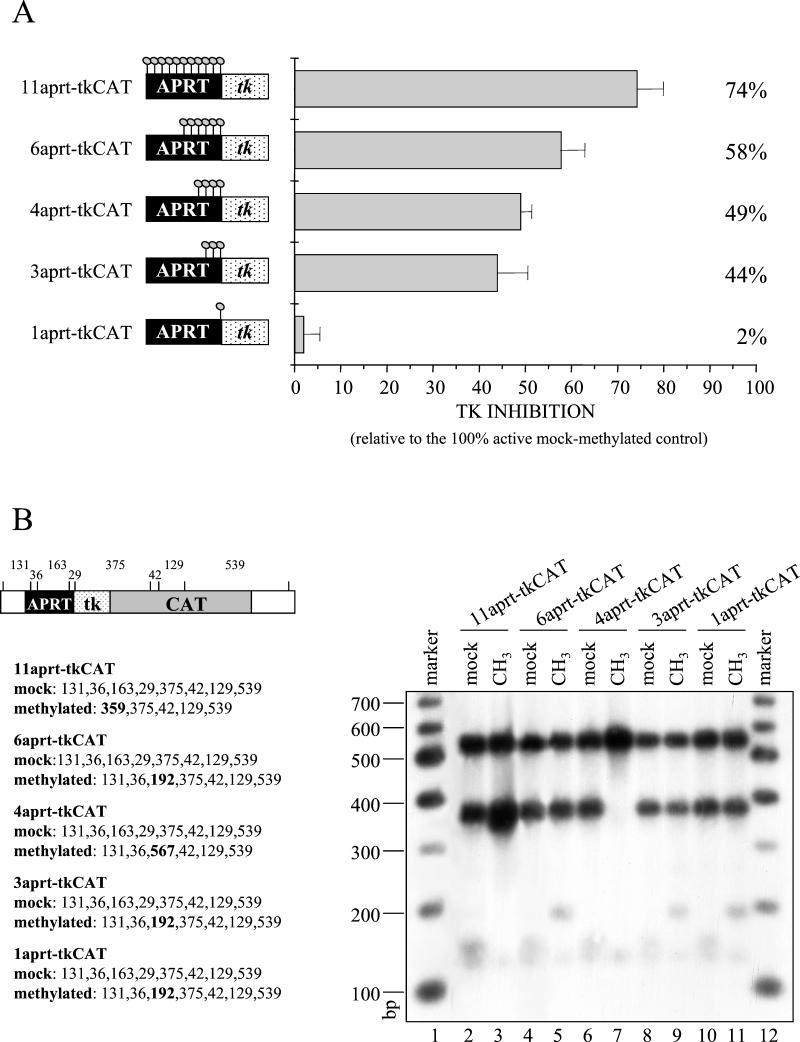
FIG. 3.
Ability of a methylated DNA to influence an adjacent promoter is a function of the number of modified sites. The 11-aprt-tkCAT, 6-aprt-tkCAT, 4-aprt-tkCAT, 3-aprt-tkCAT, and 1-aprt-tkCAT templates (see Materials and Methods) were microinjected and transcription(A) and methylation stability (B) were assayed 16 h after injection. (A) Transcription signals obtained from primer extensions were quantitated with a PhosphorImager, and tk inhibition, expressed as a percentage of repression related to the mock-methylated control, is reported. The graph plots the means of triplicate determinations, and the error bars show the standard deviation. (B) Total DNA was isolated from injected oocytes, and the methylation pattern was analyzed by digestion with the methylation-sensitive restriction enzyme HpaII and subsequent Southern blot analysis. A fragment spanning from the aprt sequence to the end of the CAT gene was used as the probe. The fragments expected from the digestion of the mock-methylated and methylated templates are indicated in the scheme. In bold are predicted fragments expected only if the DNAs are correctly methylated.
A short region of methylated DNA can influence the expression of a flanking unmodified promoter.
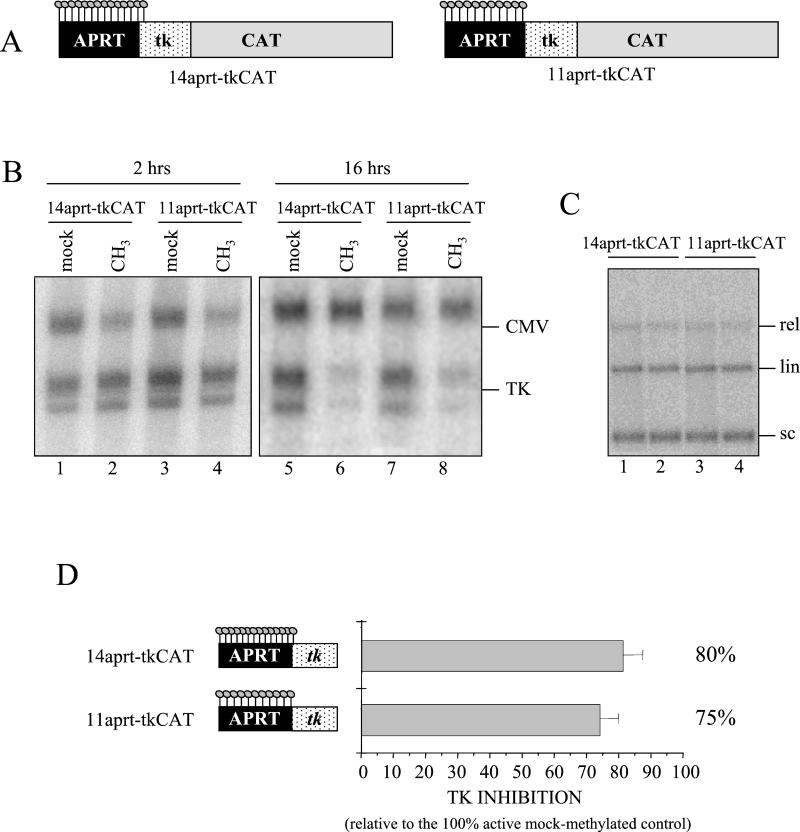
In order to examine the parameters influencing inhibition of transcription by DNA methylation, we started to evaluate whether a methylated aprt region, cloned immediately upstream of the HSV tk promoter, affects gene expression. Regionally methylated templates, characterized by 11 or 14 modified dinucleotides (Fig. 2A and Materials and Methods), or mock-methylated templates were mixed with a plasmid containing an unmodified CMV promoter as an internal control and microinjected into oocyte nuclei. We checked by Southern blot that the amounts of tk vectors injected into the oocytes were comparable (Fig. 2C), and digestion of the reisolated DNA with the methylation-sensitive restriction enzyme HpaII confirmed the stability of regional methylation in vivo (Fig. 3B).
FIG. 2.
A short region of methylated DNA severely affects tk expression. (A) Schematic representation of the regionally methylated 14-aprt-tkCAT and 11-aprt-tkCAT plasmids. The methylated sites are depicted by lollipops. (B) Methylated (CH3; lanes 2, 4, 6, and 8) or mock-methylated (mock; lanes 1, 3, 5, and 7) plasmids 14-aprt-tkCAT (lanes 1, 2, 5, and 6) and 11-aprt-tkCAT (lanes 3, 4, 7, and 8) were injected into oocyte nuclei, and transcription from the HSV tk promoter was assayed by primer extension at 2 h (lanes 1 to 4) and 16 h (lanes 5 to 8) after injection. Coinjection of pCMVCAT served as an internal standard. The positions of correctly initiated transcripts from the tk promoter (tk) and the CMV promoter (CMV) are indicated. (C) Total DNA was isolated from oocytes 16 h after injection. To evaluate if comparable amounts of mock-methylated (lanes 1 and 3) or patch-methylated (lanes 2 and 4) DNAs were injected, purified DNA was analyzed by Southern blot using the tk promoter fragment as a probe. Positions of supercoiled (sc), linear (lin), and relaxed (rel) DNA are indicated. (D) Methylated or mock-methylated 14-aprt-tkCAT or 11-aprt-tkCAT DNAs were injected together with the CMV internal standard, and primer extension was performed 16 h later. Transcription signals were quantitated with a PhosphorImager, and tkCAT transcription was standardized against the internal standard. tkCAT inhibition is expressed as a percentage of the activity of the 100% active mock-methylated control and is indicated as the mean value calculated from three independent experiments. Vertical bars represent standard deviations.
At the time indicated (Fig. 2B), total RNA was extracted from oocytes, and the CAT transcription level was evaluated by primer extension. At 16 h after injection, when nucleosomes are fully assembled over the templates (2), both methylated constructs revealed a dramatic inhibition of CAT gene transcription compared to the mock-methylated vectors (Fig. 2B, compare lanes 6 to 5 and 8 to 7). In contrast, methylated and unmethylated tk templates showed equal transcriptional activity 2 h after injection (Fig. 2B, lanes 1 to 4), when chromatin assembly is very inefficient (2).
To determine more precisely the level of inhibition, the same experiment was repeated three times, and the level of tk transcription relative to that of the CMV control was quantitated. Figure 2D shows that modification of 14 and 11 dinucleotides upstream of a promoter results in a transcriptional inhibition of about 80 and 75%, respectively. These values do not agree with a previous report, in which 11 mCpGs upstream of the simian virus 40 promoter reduced CAT reporter expression by only 4% (28). These discrepancies may be explained by considering that the repressive effect of DNA methylation depends on promoter strength and protein environment in a given cell type (7).
The results obtained demonstrate that a short region of methylated DNA not covering regulatory sequences and characterized by few modified dinucleotides at a physiological density can severely affect tk expression. As shown previously, methylation alone is not sufficient to cause the inactivation, which requires some time to be established (Fig. 2) (29).
The number of methylated sites influences the diffusion in cis of transcriptional repression.
Having defined that a limited region of modified DNA can silence an adjacent tk promoter, we wished to determine the minimum number of methylated dinucleotides necessary to establish this inhibitory effect. We produced differentially methylated templates, characterized by a similar density but a reduced number of modified dinucleotides, by annealing progressively shorter PCR fragments of the aprt sequence to single-stranded DNA. As described above, we microinjected mixtures of patch-methylated or mock-methylated aprt-tk templates together with the CMV control, analyzed transcription 16 h after injection, and checked methylation stability.
Persistence of regional methylation in vivo was assessed by two different approaches. In the first (Fig. 3B), microinjected DNA was isolated, digested with the methylation-sensitive restriction enzyme HpaII, and analyzed by Southern blot. The comparison of the HpaII fragments obtained with the injected mock templates (lanes 2, 4, 6, 8, and 10) to the ones obtained with the regionally methylated plasmids (lanes 3, 5, 7, 9, and 11) clearly demonstrates that both light and heavy methylation was stably maintained during the course of the experiment. Moreover, since the probe used in the Southern blot covers the tk promoter and the CAT reporter gene, which are not modified in any injected template, the same experiment demonstrates that spreading of methylation does not occur in these conditions.
Methylation stability was also verified by sequencing PCR fragments amplified from bisulfite-treated 11-, 6-, and 3-aprt-tkCAT DNAs (data not shown) (19, 53).
To analyze the silencing effects, three independent sets of injections were performed for each template. After verifying the presence of comparable amounts of the tk construct in the injected oocytes (Fig. 2C and 3B), transcription levels of methylated DNA relative to CMV were compared to those of unmethylated DNA. Moreover, we were able to change the specific methylated sites, holding the number constant by annealing different amplification products to ssDNA. With this strategy, we verified that the results we obtained do not depend on modification of key sites (11).
Figure 3 shows that all regionally methylated templates, with the exception of the construct containing a single methylated CpG, inhibit transcription. As already observed in Fig. 2, the activity of the tk promoter decreased about 75% in the presence of 11 modified dinucleotides. Transcriptional levels increased when the methylated sites were reduced to six, but a significant repression of nearly 45% was observed even in the presence of only three methylated CpGs.
In conclusion, our data indicate that the ability of methylated DNA to influence an adjacent promoter is a function of the number of modified sites. We observed a nonlinear relationship and a clear threshold effect, since three modified cytosines can reduce transcription by 45%, whereas a single methylated dinucleotide has no effect.
In our next set of experiments, we wished to examine whether the methylated aprt sequence had to be located immediately upstream of the tk promoter or if the repressive effect could spread far away on flanking DNA. To this purpose, 100 or 700 bp of unrelated DNA was inserted between the aprt CpG island and the promoter. We also produced a construct in which the aprt fragment was cloned downstream of the CAT reporter gene, approximately 800 bp 3′ of the tk sequences (Materials and Methods). We prepared regionally methylated templates, characterized by methylation of the 11 aprt CpGs, and assayed the effect on transcription 16 h after injection.
The results shown in Fig. 4A demonstrate that the presence of a short region of 100 bp of unmodified DNA (11-aprt-tk+100) separating the methylated CpG from the promoter does not influence transcriptional repression and the insertion of 700 bp of unmethylated DNA (11-aprt-tk+700) determines only a weak reduction of the repressive effects. Moreover, since the template containing the methylated sequence 3′ of the CAT gene (tkCAT-11aprt) shows the same repressive ability as the 11-aprt-tk+700 vector, we can state that this mechanism of silencing can spread for several hundred base pairs in both directions.
Since we have previously demonstrated that the inhibitory effect is a function of the number of methylated sites (Fig. 3), we proceeded analyzing if templates characterized by reduced methylation are still able to influence gene expression at a long distance. Injection of plasmids methylated on only six residues demonstrated a behavior similar to the 11-aprt-tk construct; in fact, six modified CpGs inhibited transcription of a tk promoter located either immediately adjacent or 100 bp away from the modified fragment (Fig. 4B). Transcriptional repression decreased from 60% to about 45% when 700 to 800 bp of unmodified DNA separated the promoter either 5′ or 3′ from the aprt sequence.
A different result was observed when a template containing only four methylated sites (Fig. 4C) or three methylated sites (Fig. 4D) was analyzed. In fact, even though four or three methylated dinucleotides can inhibit a flanking promoter, the repressive effect cannot diffuse over 100 bp, suggesting the existence of a threshold effect for the mechanism that controls spreading of the inhibitory structure as well.
Contribution of histone deacetylation to gene silencing is a function of the number of modified CpGs.
Recently, it has been demonstrated that many polypeptides that preferentially bind to methylcytosines (MBDs) participate in protein complexes, which recruit histone deacetylases (5, 27, 40, 43, 52, 57). Therefore, it has been proposed that DNA methylation and histone deacetylation might work in concert to establish a repressive chromatin environment and silence gene expression. However, the effects of trichostatin A on naturally methylated genes indicated that histone deacetylation is not the only mechanism contributing to gene silencing (10, 12).
Since our experiments revealed that the number (Fig. 3 and 4) of methylated CpGs affects the repressive strength and the capability of diffusing in cis, we assessed the contribute of histone deacetylation to the repression mediated by the different regionally methylated templates. The results obtained from microinjected oocytes incubated for 16 h in the absence or presence of trichostatin A are shown in Fig. 5A. Since both the tk promoter and the CMV control resulted in sensitivity to drug treatment, we performed these experiments omitting the CMVCAT construct and comparing the transcriptional levels of mock-methylated HSV promoters to the methylated ones; the quality of the injections, as well as the presence in the injected oocytes of comparable amounts of DNA, was verified each time (Fig. 5B). Moreover, we assessed the stability of regional methylation in the presence of trichostatin A by isolating microinjected DNAs, digesting with HpaII, and subsequent Southern blot analysis (data not shown).
FIG. 5.
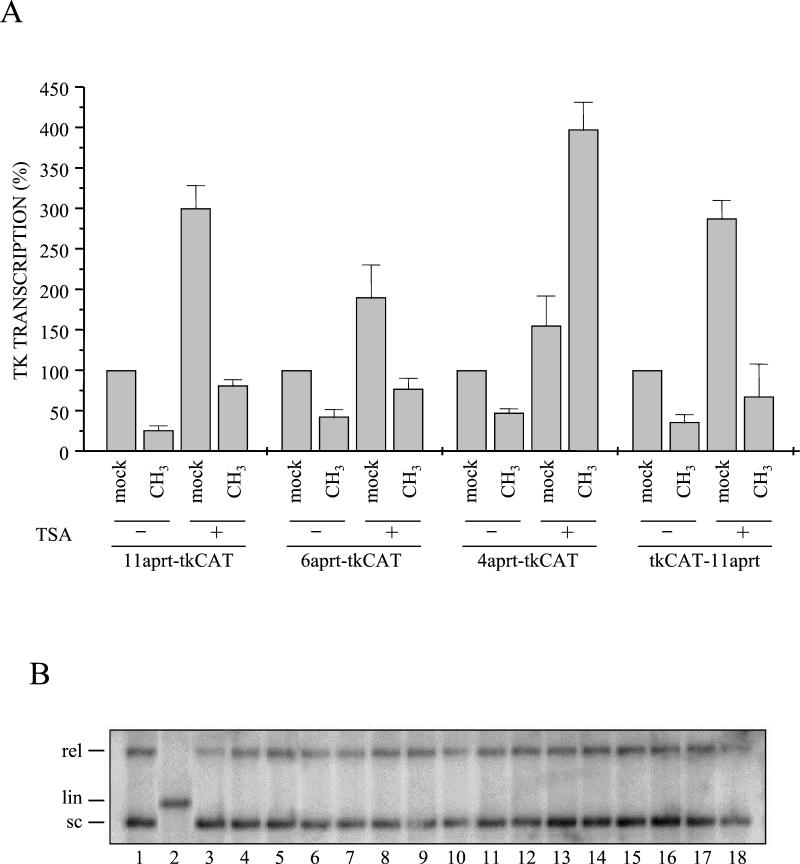
Trichostatin A relieves transcriptional inhibition only when a limited number of CpGs are modified. Xenopus oocytes were injected with mock-methylated or regionally methylated DNAs (11-aprt-tkCAT, 6-aprt-tkCAT, 4-aprt-tkCAT, and tkCAT-11aprt) and incubated in the absence (−) or presence (+) of trichostatin A. (A) Transcription was evaluated by primer extension 16 h after injection. Transcriptional signals were quantified with a PhosphorImager, and the levels of transcription relative to DNA mass injected (estimated from Southern blotting, panel B) are indicated in the graph. Data represent the mean values obtained from three independent experiments; vertical bars represent standard deviations. (B) DNA was extracted from half of the collected oocytes and analyzed by electrophoresis on a 1% agarose gel before transfer to a membrane and hybridization with an aprt-tk-CAT probe. Lanes 1 and 2 indicate positions of supercoiled (sc) and linear (lin) DNA. In lanes 3 to 18, purified DNAs were loaded following the same order as in panel A.
Figure 5A indicates that the expression level of unmethylated templates increases between 2- and 4-fold when the oocytes are incubated with trichostatin A. This variation is not dependent on the particular construct analyzed but seems to depend on the oocyte batch. The analysis of trichostatin A effects on patch-methylated vectors shows that transcription is influenced by histone deacetylation; in fact, gene expression from all methylated DNAs increased when the inhibitor was present. However, comparing the transcriptional level obtained with the 11-aprt-tk mock-methylated templates to the equivalent regionally methylated construct, it is clear that, even in the presence of the drug, the modified DNA is inhibited almost 70% with respect to its control.
An identical behavior was observed analyzing the templates characterized by six modified CpGs (6aprt-tkCAT) or 11 methylated dinucleotides positioned downstream of the CAT gene (tkCAT-11aprt). A completely different expression profile was instead obtained when the injected templates contained four (4-aprt-tkCAT) or three (3-aprt-tkCAT; data not shown) methylated sites upstream of the promoter; in fact, whereas methylated DNAs were inhibited, with respect to the mock control, in the absence of the drug (Fig. 5 and 3), trichostatin A reverted the situation, bringing the patch-modified templates to a higher transcription level than their controls.
These results indicate that histone deacetylation has a repressive effect both on the unmethylated tk vector and on regionally methylated DNAs. However, the contribution of deacetylation to this mechanism of gene silencing seems to depend on the number of modified sites. In particular, when methylated dinucleotides reach the threshold necessary for spreading the repressive effect a long distance (Fig. 4), histone deacetylation has only a general inhibitory effect and transcriptional repression is maintained with respect to the unmethylated control. On the contrary, when the methylated sites do not allow the formation of a strong repressive structure, capable of diffusing in cis (Fig. 4), histone deacetylation seems to be the most important factor involved in methylation-dependent transcriptional inhibition.
Even though we do not know why trichostatin A produces a higher transcription from the methylated 4-aprt-tkCAT construct than from its mock-methylated counterpart, we can speculate that binding of MBD proteins and their associated factors to the sequences immediately upstream of the promoter could compete with nucleosome deposition, thus participating in maintaining this promoter structure more accessible to transcription factors. Therefore, the removal of the transcriptional block conferred by deacetylation might lead to a higher transcription from the modified vector than from the control. We believe that the analysis of the chromatin structure organized on the injected templates and chromatin immunoprecipitation experiments will help elucidate if our hypothesis is correct.
Methylation-mediated repression is weaker when regulative sequences are modified.
It is generally assumed that repression is greater if the promoter itself is modified (39, 42). In accordance with this, changes in promoter methylation are usually investigated in losses of gene function mediated by epigenetic events (4).
To verify this idea, we injected vectors modified only on the tk promoter, containing 23 CpGs in 174 bp. The result of the primer extension (Fig. 6A) indicates a repressive effect due to the methylated promoter of about 40%, much weaker than what might be anticipated from the number of methylated dinucleotides. This slight repression could suggest that the presence of transcription factors binding to the promoter could destabilize the formation of the methylation-specific repressive structure, thus reducing its inhibitory strength.
FIG. 6.
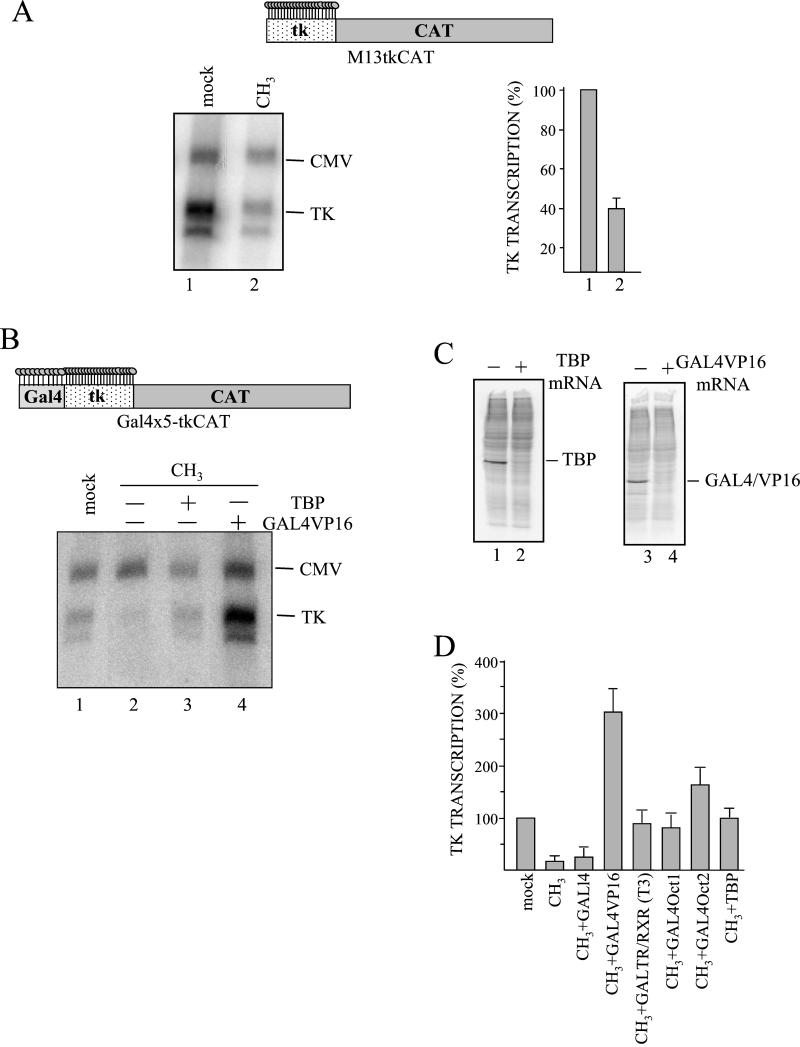
Transcriptional activators compete with methylation-specific repressors in vivo. (A) M13-tkCAT vector methylated only on the promoter (lane 2) or the corresponding unmethylated DNA (lane 1) was microinjected. Transcription was evaluated by primer extension and quantitated with a PhosphorImager. Transcriptional levels, relative to the CMV internal standard, were calculated and are reported in the graph as percentages of the 100% active mock DNA. The mean value and standard deviation of three independent experiments are reported. In the upper part of the panel, the regionally methylated M13-tkCAT DNA, characterized by 23 mCpGs distributed over the HSV tk sequence, is represented. (B) TBP (lane 3) or GAL4VP16 (lane 4) mRNA was injected into stage VI oocytes. After incubation for 16 h, a mixture of templates containing unmodified Gal4×5-tkCAT DNA (lane 1) or the same regionally methylated template (lanes 2 to 4) and the CMVCAT internal control was microinjected. Oocytes were collected 16 h after DNA injection, and transcription was analyzed. In the upper part, as above, is a scheme of the partially methylated Gal4×5-tkCAT DNA. (C) A total of 6 ng of TBP (lane 1) or GAL4VP16 (lane 3) mRNAs was injected, and oocytes were incubated for 16 h in the presence of [35S]methionine (100 μCi/ml). Batches of 10 oocytes were collected, and a whole-cell extract (1 oocyte equivalent) from uninjected (lanes 2 and 4) or injected (lanes 1 and 3) oocytes was loaded on SDS-10% PAGE followed by fluorography. The correct positions of exogenously expressed TBP and GAL4VP16 proteins are indicated. (D) A total of 6 ng of a specific mRNA (GAL4, GAL4VP16, a mixture of GALTR and RXR, GAL4OCT1, GAL4OCT2, and TBP) was injected, oocytes were incubated for 16 h and subsequently reinjected with a mixture of regionally methylated Gal4×5-tkCAT and CMV DNAs. A mixture of unmethylated Gal4×5-tkCAT and CMV DNA was also injected as a control (mock). Thyroid hormone (T3, 50 nM) was added to the oocytes synthesizing GALTR/RXR, and all samples were incubated overnight. Transcription was analyzed by primer extension and quantified with a PhosphorImager. The histogram reports the percentage of transcription relative to CMV, calculated from two independent experiments.
We tested this hypothesis by examining whether an increase in the abundance of transcriptional activators that might compete with MBDs can relieve the repression conferred by promoter methylation. To this purpose we microinjected tkCAT templates modified with five GAL4 binding sites immediately upstream of the tk sequence. Regionally methylated DNA was prepared, modifying the whole regulatory DNA fragment (Fig. 6B); note that in this case there are 33methylated dinucleotides, 10 more than in Fig. 6A.
Under physiological conditions, in the absence of exogenous transcriptional factors, methylation of the GAL4×5-tk sequences results in dramatic repression (Fig. 6B, lane 2). When TBP is expressed in oocytes by microinjection of its specific mRNA (Fig. 6C, lane 1), expression of the regionally methylated promoter is significantly increased and reaches levels comparable to those in the unmethylated control (Fig. 6B, lane 3). The relief of inhibition is further augmented when high levels of the GAL4VP16 transcriptional activator (Fig. 6C, lane 3) are present (Fig. 6B, compare lanes 3 and 4).
In order to evaluate if the ability of TBP and GAL4VP16 to overcome repression was limited to these activators, the same experiment was repeated, examining also the influence of the thyroid hormone receptor, OCT1, and OCT2 fused to the GAL4 DNA-binding domain (15, 54, 55). Moreover, the importance of the presence of an activating domain was assayed by synthesizing only an exogenous GAL4 binding domain (GAL4). Once again, DNA was injected after exogenous protein expression, and oocytes were collected 16 h later.
Figure 6D shows that in the absence of activators, transcription from the Gal4×5tk promoter was repressed by regional methylation. Quantitation of transcriptional activities calculated from two independent experiments indicates a repression of about 90%. In the presence of exogenous activators (GALTR/RXR, GAL4VP16, GAL4OCT1, GAL4OCT2, or TBP), repression was relieved and the level of activation was a function of the specific factor analyzed. As expected, GAL4VP16 conferred the highest transcriptional ability. On the contrary, an increase in the abundance of a simple DNA-binding domain (GAL4) did not overcome the silencing effect.
These experiments demonstrate that methylation of promoter sequences does not have a greater repressive effect than the modification of flanking, nonregulatory DNA sequences. The presence of a transcription factor can compete for the establishment of an inactive promoter conformation, thus reducing the silencing effect. Relief of transcriptional inhibition requires an activating domain, and the final level of activation depends on the specific trans-acting factor. It is important to note that this model seems to be confirmed by the fact that the increase from 23 to 33 modified dinucleotides, in the absence of factors able to bind the GAL4 binding sites, brings the inhibition level from 40 to 90%, with an evident synergistic effect.
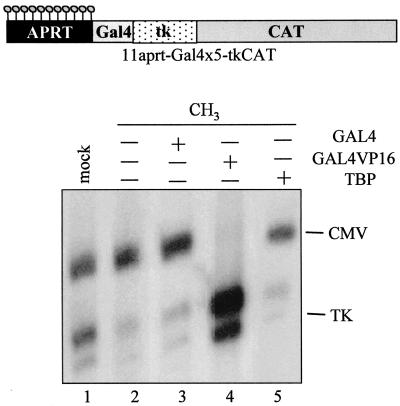
Finally, we examined whether an increase in the abundance of transcriptional activators might impede diffusion of methylation-dependent gene silencing. To approach this issue, we modified the 11-aprt-tkCAT vector to contain five binding sites for GAL4 immediately upstream of the tk promoter (Fig. 7A). Mock-methylated DNA or regionally modified templates methylated only on the aprt sequence were prepared and microinjected.
FIG. 7.
Diffusion of methylation-dependent silencing is blocked by exogenous GAL4VP16 but not TBP. GAL4 (lane 3), GAL4VP16 (lane 4), or TBP (lane 5) mRNA was injected into stage VI oocytes. After 16 h, a mixture of templates containing the regionally methylated 11-aprtGal4×5-tkCAT and CMVCAT DNAs was injected (lanes 2 to 5). In lane 1, the injected DNA mixture contained the mock-methylated vector together with CMVCAT. Oocytes were incubated for 16 h, and RNA was extracted and analyzed by primer extension. The upper part of the figure shows a scheme of the regionally methylated 11-aprtGal4×5-tkCAT template.
The presence of GAL4 sites did not influence the repressive effect of the 11 mCpGs contained in the aprt region; in fact, under physiological conditions, transcription from the methylated template was repressed almost 70% (Fig. 7, compare lane 1 to lane 2, and see Fig. 3A). Expression of high quantities of TBP (lane 5) or of a simple DNA-binding domain (GAL4, lane 3) did not lead to any derepression. On the contrary, exogenous GAL4VP16 sustained vigorous transcriptional activity from the regionally methylated DNA (lane 4). In this experiment, the disappearance of the CMV control suggests that, in the presence of large quantities of GAL4VP16, transcription factors shared between the two promoters can be limiting. To our knowledge, this effect is only obtained with the artificial activator GAL4VP16 and seems to vary from experiment to experiment, depending on the oocyte batch.
These data show that the nucleoprotein structure formed at the methylated aprt sequence exerts a dominant, transmissible repression, capable of overcoming not only physiological concentrations of transcription factors (Fig. 7, lane 2, and Fig. 2 to 4), but also the presence of high concentrations of TBP. However, as previously demonstrated (29), a strong activator such as GAL4VP16 abolishes the silencing effect of methylated DNA.
DISCUSSION
It is widely recognized that DNA methylation inhibits transcription mainly by recruiting methyl-binding proteins that, associating with histone deacetylase and chromatin remodeling complexes, stabilize a condensed chromatin conformation (5). In accordance with this, transfection and microinjection experiments of differentially methylated templates have demonstrated that transcriptional silencing often does not require modification of promoter DNA, suggesting that methylation effects are transmissible in cis (21, 24, 28, 29, 31, 56). In this work, microinjections of regionally methylated vectors into Xenopus oocytes were used to address some basic yet unanswered questions regarding CpG methylation and to analyze the parameters influencing diffusion of methylation-mediated gene silencing and the importance of histone deacetylation in this spreading effect.
The human aprt CpG island is characterized by a natural distribution and density of methylatable dinucleotides, and the HSV thymidine kinase promoter has been previously demonstrated to be inhibited by DNA methylation by an indirect mechanism (29). Using a protocol that we optimized, we were able to obtain patch-methylated constructs characterized by having only a portion of this CpG island as the methylated region and the HSV tk promoter.
The major conclusions from this work are that (i) a certain number of modified cytosines are required to organize a stable, diffusible chromatin structure (Fig. 2 to 4), (ii) histone deacetylation contributes significantly to gene repression only when the number of modified sites is insufficient to exert repression over a long distance (Fig. 5); and (iii) because of competition between transcriptional factors and methyl-binding proteins, promoter modification does not lead to a greater repressive effect (Fig. 6).
DNA methylation can inhibit gene expression at different levels. Even though direct interference of modified CpG can block binding of transcriptional factors, this level of repression does not represent the main mechanism by which methylation-mediated gene silencing is exerted. In most cases, DNA methylation seems to repress gene expression by recruiting binding proteins specific for methylated DNA (5). MBD proteins can, per se, block transcriptional factor association or mediate the formation of a repressive, inaccessible chromatin structure. Since it has been demonstrated that promoter methylation is often not required for inhibition, it is possible to foresee that a particular chromatin structure is seeded at methylated DNA and diffuses on flanking, unmodified sequences. Different parameters, such as the number or density of modified dinucleotides, might influence the composition of chromatin structure, its stability, and, therefore, its capability of spreading on the fiber.
The results shown in Fig. 2 demonstrate that 11 modified CpGs upstream of the tk promoter can silence its expression. Since, as demonstrated previously, this repression requires time to occur (Fig. 2) and correlates with chromatin assembly (29) (data not shown), we suggest that inhibition is not mediated simply by recruitment of a deacetylase activity which modifies transcription factors and/or the transcriptional apparatus. Therefore, nucleation of a repressive chromatin structure at the methylated DNA and its diffusion on the unmodified tk sequences must occur. Decreasing the number of modified dinucleotides (Fig. 3) reveals that even three methyl groups are able to modulate tk expression, and a nonlinear relationship between the number of methylated sites and repression level is observed.
However, the importance of the number of methylcytosines shows up when the ability of the repressive effect to spread over a long distance is investigated (Fig. 4). In fact, our results demonstrate that inhibition can propagate for several hundreds of base pairs 3′ or 5′ from the modified DNA only when a sufficient number of CpGs are methylated. In contrast, a short region of methylated DNA silences only an immediately adjacent tk promoter, suggesting that a few methylated sites cannot seed the formation of the repressive chromatin structure, nor can they guarantee its stability.
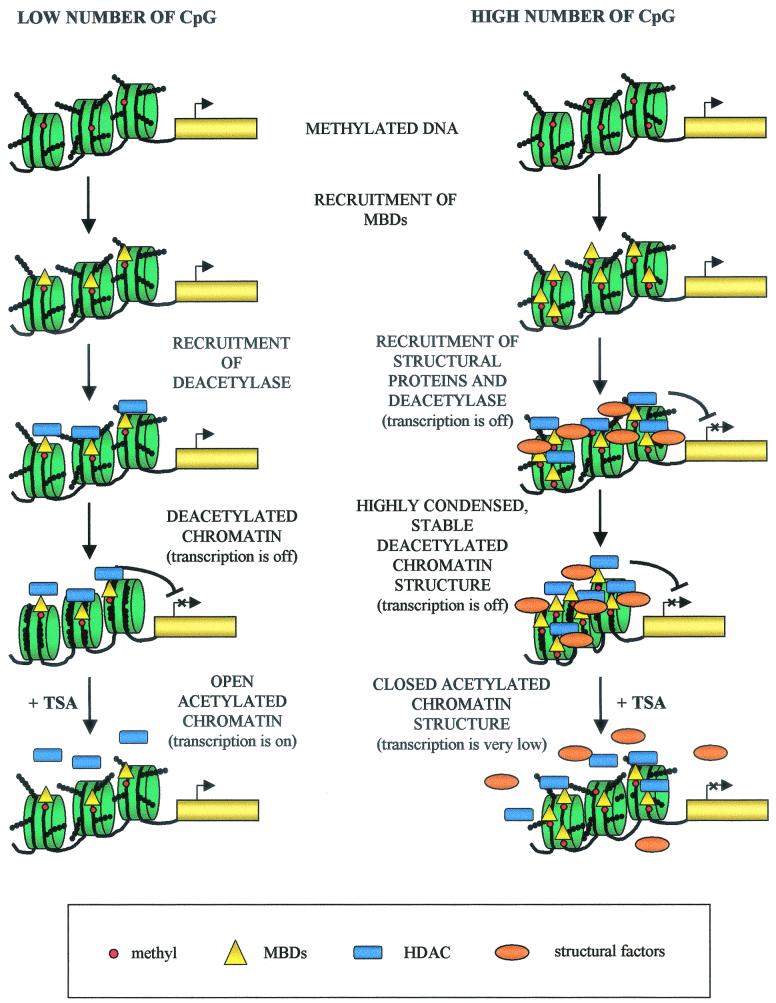
The analysis of the contribution of histone deacetylation to this repression (Fig. 5) seems to reinforce these observations. In fact, our experiments show that histone deacetylation is of fundamental importance to the silencing mechanism only when the number of modified sites does not reach the threshold sufficient for an effect over a long distance. We propose (see the model in Fig. 8) that when only a limited number of modified dinucleotides are close to a promoter, they recruit MBD proteins and their associated histone deacetylation activity; histone deacetylation occurs, remaining localized to a small number of nucleosomes, and transcriptional repression is observed. In this situation, trichostatin A treatment allows bypassing the main mechanism by which methylated DNA silences gene expression, and therefore inhibition is relieved.
FIG. 8.
Model for the molecular mechanisms of gene silencing mediated by DNA methylation. The left side of the model represents the repressive mechanism determined by a small number of modified dinucleotides; on the right, the hypothetical mechanism by which a large number of methylated dinucleotides exert a long-distance inhibitory effect is reported. In this cartoon, DNA is depicted as a black line wrapped around nucleosomes (green), methylcytosines are indicated with red circles, histone tails with a chain of black circles, methyl-binding proteins with yellow triangles, histone deacetylases (HDAC) with blue rectangles, and remodeling factors with orange ovals.
The repression mechanism is significantly different when the number of methylated sites is increased and reaches the threshold that leads to diffusion of gene silencing on the DNA fiber. We propose that in these conditions, a specialized chromatin structure, formed not only by MBD proteins but also by other structural and remodeling activities, is organized on the modified DNA. Afterward, nucleation of this chromatin conformation and propagation on the flanking, unmodified DNA occur (Fig. 8, right panel). The contribution of histone deacetylation to transcriptional inhibition in these conditions is of secondary importance; in fact, even in the presence of trichostatin A, transcriptional levels remain significantly lower than in the unmethylated controls (Fig. 5).
Obviously, we cannot exclude that even a limited number of methylated CpGs can recruit on modified DNA proteins other than histone deacetylases; however, if this is the case, the chromatin structure nucleated is not strong enough to propagate itself, and transcriptional repression is mainly due to histone deacetylation anyway (Fig. 4 and 5).
We believe that our data and the proposed model can explain the apparently contradictory data existing in the literature about the response of methylated DNA to trichostatin A treatment. In fact, while methylated transfected genes can be reactivated by tricosthatin A (reviewed in reference 46), naturally densely methylated endogenous genes cannot be reinduced with trichostatin A alone. Remarkably, this drug does lead to a strong reexpression of several hypermethylated tumor suppressors only following minimal demethylation by 5-aza-2′deoxycytidine treatment (10).
Even though the model was obtained from experiments performed with the tk promoter injected into Xenopus oocytes, we think that it might be considered a general mechanism of methylation-dependent gene silencing. In fact, the oocyte system has proven generally useful in defining transcriptional regulation in a chromatin context, permitting confirmations of earlier results obtained with different experimental systems (1, 37, 45, 54, 55). In particular, in studying DNA methylation, Xenopus oocytes were used to confirm and extend existing data deduced from microinjections of methylated templates into mammalian cells (8, 20). Molecular characterization of Xenopus MeCP2 and other MBD proteins demonstrated almost identical behavior in mammalian and frog cells (5). Moreover, since, as already mentioned, it has been reported that DNA methylation inhibits gene expression mainly by an indirect mechanism and that methylation effects are transmissible in cis, our data seem to be in perfect accord with previous reports.
The response of differentially methylated templates to trichostatin A also seems to confirm previous observations obtained with different promoters (10). Therefore, we believe that this model of methylation-dependent gene silencing can be applied to other promoters; the final level of repression as well as the number of mCpGs useful to establish a stable repressive structure will vary depending on the promoter strength and transcription factor concentration present in the particular cell type.
It is generally assumed that methylation effects are more effective when the regulative gene sequences are modified (42); therefore, changes in promoter methylation are usually investigated as causes of loss of gene function in epigenetic events (4). The results shown in Fig. 6 seem to contradict this assumption; in fact, injection of regionally methylated constructs clearly demonstrates that when the tk promoter is modified, despite the high number of methylated CpGs, transcriptional inhibition is much weaker than that seen on templates characterized by a smaller number of methylated sites positioned upstream of the promoter. Moreover, the ability of exogenous activators to overcome these methylation effects (Fig. 6) demonstrates the existence of competition between transcription factors and MDB proteins in vivo. This effect is not restricted to a particular transcriptional factor; in fact, any activator analyzed (TBP, GAL4VP16, TR, OCT1, and OCT2) is able to reinduce tk expression. However, the final transcriptional levels are a function of the factor examined.
It is important to note that in these experiments, exogenous proteins are highly expressed; we believe that in a more physiological context, transcription from a methylated promoter is a function not only of the strength but also of the concentration of the specific activator. The experiment performed using only the GAL4 DNA-binding domain indicates that an increase in the abundance of a DNA-binding protein does not eliminate the silencing effect; an activation domain seems to be required. In the future, it will be interesting to compare the nucleoprotein structure organized over the methylated promoter in the presence of GAL4 or GAL4VP16.
Finally, the experiment shown in Fig. 7 demonstrates that an increase in the concentration of a general transcription factor, such as TBP, cannot block the silencing effect imposed by the methylation of a nonregulatory region. This result confirms all previous experiments (Fig. 2 to 4), in which it was evident that even though the general transcription machinery is present in oocytes and able to sustain transcription from a tk promoter, it cannot impede spreading of the silencing effect. Therefore, in this context, DNA methylation exerts a dominant transmissible repression. As already demonstrated, expression of a strong activator, such as GAL4VP16, can, however, reinduce expression.
In conclusion, when a promoter is methylated, its expression will depend mainly on transcriptional factor abundance and, of course, on the ability to bind to methylated recognition elements. On the contrary, when DNA sequences lacking transcription factor binding sites are modified, MBD proteins can easily bind and seed a repressive chromatin structure involving nonmethylated flanking regulatory sequences. In this situation, only specific activators can overcome the inhibitory effect.
Acknowledgments
We are grateful to Sonia Castiglioni and Zoe Christoni for preparing reagents and to all the members of the lab for stimulating discussions. We are indebted to Jiemin Wong and Roberto Mantovani for the generous gifts of the GAL4 fusion expression vectors. Moreover, we thank Marco Muzi Falconi and Roberto Mantovani for reading the manuscript.
This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro, Cofinanziamento MURST-Universita' dell'Insubria.
REFERENCES
- 1.Almouzni, G., and A. P. Wolffe. 1993. Replication coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes Dev. 7:2033-2047. [DOI] [PubMed] [Google Scholar]
- 2.Almouzni, G., and A. P. Wolffe. 1993. Nuclear assembly, structure and function: the use of Xenopus in vitro systems. Exp. Cell. Res. 205:1-15. [DOI] [PubMed] [Google Scholar]
- 3.Baylin, S. B., and J. G. Herman. 2000. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 16:168-174. [DOI] [PubMed] [Google Scholar]
- 4.Baylin, S. B., M. Esteller, M. R. Rountree, K. Bachman, K. Schuebel, and J. G. Herman. 2001. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 10:687-692. [DOI] [PubMed] [Google Scholar]
- 5.Bird, A. P., and A. P. Wolffe. 1999. Methylation-induced repression-belts, braces and chromatin. Cell 99:451-454. [DOI] [PubMed] [Google Scholar]
- 6.Boyes, J., and A. P. Bird. 1991. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64:1123-1134. [DOI] [PubMed] [Google Scholar]
- 7.Boyes, J., and A. P. Bird. 1992. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 11:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buschausen, G., B. Wittig, M. Graessmann, and A. Graessmann. 1987. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc. Natl. Acad. Sci. USA 84:1177-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buschausen, G., B. Wittig, M. Graessmann, and A. Graessmann. 1987. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc. Natl. Acad. Sci. USA 84:1177-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cameron, E. E., K. E. Bachman, S. Myohanen, J. Herman, and S. B. Baylin. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21:103-107. [DOI] [PubMed] [Google Scholar]
- 11.Chien, C., M. C. K. Yang, and T. P. Yang. 2001. Evidence that silencing of the HPRT promoter by DNA methylation is mediated by critical CpG sites. J. Biol. Chem. 276:320-328. [DOI] [PubMed] [Google Scholar]
- 12.Coffee, B., F. Zhang, S. T. Warren, and D. Reines. 1999. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat. Genet. 22:98-101. [DOI] [PubMed] [Google Scholar]
- 13.Cross, S. H., J. A. Charlton, X. Nan, and A. P. Bird. 1994. Purification of CpG islands using a methylated DNA binding column. Nat. Genet. 6:236-244. [DOI] [PubMed] [Google Scholar]
- 14.Cross, S. H., and A. P. Bird. 1995. CpG islands and genes. Curr. Opin. Genet. Dev. 5:309-314. [DOI] [PubMed] [Google Scholar]
- 15.Di Silvio, A., C. Imbriano, and R. Mantovani. 1999. Dissection of the NF-Y transcriptional activation potential. Nucleic Acids Res. 27:2578-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eden, S., and H. Cedar. 1994. Role of DNA methylation in the regulation of transcription. Curr. Opin. Genet. Dev. 4:255-259. [DOI] [PubMed] [Google Scholar]
- 17.Eden, S., T. Hashimshony, I. Keshet, H. Cedar, and A. W. Thorne. 1998. DNA methylation models histone acetylation. Nature 394:842. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson, A. T., R. G. Lapidus, S. B. Baylin, and N. E. Davidson. 1995. Demethylation of the estrogen receptor gene in estrogen receptor-negative breast cancer cells can reactivate estrogen receptor gene expression. Cancer Res. 55:2279-2283. [PubMed] [Google Scholar]
- 19.Frommer, M., L. E. McDonald, D. S. Milllar, C. M. Collis, F. Watt, G. W. Grigg, P. L. Monoy, and C. L. Paul. 1992. A genomic sequencing protocol that yields a positive display of 5-methyl cytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 89:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graessmann, A., and M. Graessmann. 1988. DNA methylation, chromatin structure and regulation of herpes simplex virus tk gene expression. Gene 74:135-137. [DOI] [PubMed] [Google Scholar]
- 21.Graessmann, A., G. Sandberg, E. Guhl, and M. Graessmann. 1994. Methylation of single sites within the herpes simplex virus tk coding region and the simian virus 40 T-antigen intron causes gene inactivation. Mol. Cell. Biol. 14:2004-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrich, B., and A. P. Bird. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18:6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh, C. L. 1994. Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol. 14:5487-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh, C. L. 1997. Stability of patch methylation and its impact in regions of transcriptional initiation and elongation. Mol. Cell. Biol. 17:5897-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaenisch, R. 1997. DNA methylation and imprinting: why bother? Trends Genet. 13:323-329. [DOI] [PubMed] [Google Scholar]
- 26.Jones, P. A. 1985. Altering gene expression 5-azacytidine. Cell 40:485-486. [DOI] [PubMed] [Google Scholar]
- 27.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 28.Kass, S. U., Goddard, J. P., and R. L. Adams. 1993. Inactive chromatin spreads from a focus of methylation. Mol. Cell. Biol. 13:7372-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kass, S. U., Landsberger, N., and A. P. Wolffe. 1997. DNA methylation directs a time-dependent repression of transcription initiation. Curr. Biol. 7:157-165. [DOI] [PubMed] [Google Scholar]
- 30.Keshet, I., Lieman-Hurwitz, J., and H. Cedar. 1986. DNA methylation affects the formation of active chromatin. Cell 44:535-543. [DOI] [PubMed] [Google Scholar]
- 31.Keshet, I., Yisraeli, J., and H. Cedar. 1985. Effect of regional DNA methylation on gene expression. Proc. Natl. Acad. Sci. USA 82:2560-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landsberger, N., and A. P. Wolffe. 1995. Role of chromatin and Xenopus heat shock transcription factor (XHSF1) in the regulation of Xenopus hsp70 promoter in vivo. Mol. Cell Biol. 15:6013-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landsberger, N., and A. P. Wolffe. 1997. Remodeling of regulatory nucleoproteins complexes on the Xenopus hsp70 promoter during meiotic of the Xenopus oocyte. EMBO J. 16:4361-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis, J. D., R. R. Meehan, W. J. Henzel, I. Maurer-Fogy, P, Jeppesen, F. Klein, and A. P. Bird. 1992. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69:905-914. [DOI] [PubMed] [Google Scholar]
- 35.Li, E., Beard, C., and R. Jaenisch. 1993. Role for DNA methylation in genomic imprinting. Nature 366:362-365. [DOI] [PubMed] [Google Scholar]
- 36.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 37.Minucci, S., Wong, J., Blanco, J. C. G., Shi, Y-B., Wolffe, A. P., and Ozato, K. 1998. Retinoid receptor-induced alteration of the chromatin assembled on a ligand-responsive promoter in Xenopus oocytes. Mol. Endocrinol. 12:315-324. [DOI] [PubMed] [Google Scholar]
- 38.Murray, E. J., and F. Grosveld. 1987. Site specific demethylation in the promoter of human globin gene does not alleviate methylation mediated suppression. EMBO J. 6:2329-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nan, X., Campoy, F. S., and A. P. Bird. 1997. MeCP2 is a trascriptional repressor with abundant sites in genomic chromatin. Cell 88:471-481. [DOI] [PubMed] [Google Scholar]
- 40.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. P. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 41.Neumann, B., and D. P. Barlow. 1996. Multiple roles for DNA methylation in gametic imprinting. Curr. Opin. Genet. Dev. 6:159-163. [DOI] [PubMed] [Google Scholar]
- 42.Newell-Price, J., Clark, A. J. L., and P. King. 2000. DNA methylation and silencing of gene expression. TEM 11:142-148. [DOI] [PubMed] [Google Scholar]
- 43.Ng, H. H., Y. Zhang, B. Hendrich, C. A. Johnson, B. M. Turner, and H. Erdjument-Bromage. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23:58-61. [DOI] [PubMed] [Google Scholar]
- 44.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferase Dnmt3A and Dnmt3B are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 45.Perlmann, T., and O. Wrange. 1991. Inhibition of chromatin assembly in Xenopus oocytes correlates with derepression of the mouse mammary tumor virus promoter. Mol. Cell. Biol. 11:5259-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Razin, A. 1998. CpG methylation, chromatin structure and gene silencing a three way connection. EMBO J. 17:4905-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Razin, A., and H. Cedar. 1994. DNA methylation and genomic imprinting. Cell 77:473-476. [DOI] [PubMed] [Google Scholar]
- 48.Riggs, A. D., and G. P. Pfeifer. 1992. X-chromosome inactivation and cell memory. Trends Genet. 8:169-174. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Siegfried, Z., and H. Cedar. 1997. DNA methylation: a molecular lock. Curr. Biol. 7:305-307. [DOI] [PubMed] [Google Scholar]
- 51.Singer-sam, J., M. Grant, J. M. LeBon, K. Okuyama, V. Chapman, M. Monk, and A. D. Riggs. 1990. Use of HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-I CpG island of mouse embryos at the time of X-chromosome inactivation. Mol. Cell. Biol. 10:4987-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wade, P., A. Gegonne, P. L. Jones, E. Ballestar, F. Aubry, and A. P. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 23:62-66. [DOI] [PubMed] [Google Scholar]
- 53.Wang, R. Y.-H., C. W. Gehrke, and M. Ehrlich. 1980. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res. 8:4777-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong, J., Shi, Y.-B. and A. P. Wolffe. 1995. A role for nucleosome assembly in both silencing and activation of the Xenopus TRβA gene by the thyroid hormone receptor. Genes Dev. 9:2696-2711. [DOI] [PubMed] [Google Scholar]
- 55.Wong, J., Y.-B. Shi, and A. P. Wolffe. 1997. Determinants of chromatin disruption and transcriptional regulation instigated by the thyroid hormone receptor: hormone regulated chromatin disruption is not sufficient for transcriptional activation. EMBO J. 16:3158-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yisraeli, J., D. Frank, A. Razin, and H. Cedar. 1988. Effect of in vitro DNA methylation on β-globin gene expression. Proc. Natl. Acad. Sci. USA 85:4638-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. P. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]