Abstract
Chromatin remodeling by the glucocorticoid receptor (GR) is associated with activation of transcription at the mouse mammary tumor virus (MMTV) promoter. We reconstituted this nucleoprotein transition with chromatin assembled on MMTV DNA. The remodeling event was ATP dependent and required either a nuclear extract from HeLa cells or purified human Swi/Snf. Through the use of a direct interaction assay (magnetic bead pull-down), we demonstrated recruitment of human Swi/Snf to MMTV chromatin by GR. Unexpectedly, we found that GR is actively displaced from the chromatin template during the remodeling process. ATP-dependent GR displacement was reversed by the addition of apyrase and was specific to chromatin templates. The disengagement reaction could also be induced with purified human Swi/Snf. Although GR apparently dissociated during chromatin remodeling by Swi/Snf, it participated in binding of the secondary transcription factor, nuclear factor 1. These results are paralleled by a recent discovery that the hormone-occupied receptor undergoes rapid exchange between chromatin and the nucleoplasmic compartment in living cells. Both the in vitro and in vivo results are consistent with a dynamic model (hit and run) in which GR first binds to chromatin after ligand activation, recruits a remodeling activity, facilitates transcription factor binding, and is simultaneously lost from the template.
The regulation of transcription by nuclear receptors is intimately associated with reorganization of the chromatin structures of target promoters. Receptors interact, either directly or indirectly, with a variety of chromatin-remodeling factors and recruit these activities for chromatin modification in the vicinity of the hormone response elements (HREs). There are two general classes of chromatin-remodeling factors: those that covalently modify components of the nucleoprotein structure (including histone acetylases and deacetylases, methylases, and kinases) (31, 45) and a second group comprised of ATP-dependent nucleosome-remodeling proteins, collectively referred to as the Swi/Snf family of factors (22, 23, 35, 36, 51, 54).
The current paradigm suggests that the purpose of these chromatin-modifying activities is to reorganize the nucleoprotein structures of regulated genes to provide greater access for both site-specific promoter binding proteins and general (or basal) transcription factors, both of which are excluded from their binding sites in nonremodeled chromatin. The nuclear receptors appear to be members of a restricted group of activators (pioneer proteins) with the ability to bind chromatin and initiate the remodeling process (18, 26, 27, 44, 48, 49).
A parallel theme of this model is the concept of a stable initiation complex. Steroid receptors, such as the glucocorticoid (GR), progesterone (PR), and estrogen receptors, require ligand to bind DNA and are believed to assist in the formation of large, multifactor complexes that reside on the template in the continued presence of hormone (55). By using a receptor tagged with a green fluorescent protein and an amplified array of target promoters, the direct interaction of GR with HREs in living cells was recently observed (28). Photobleaching experiments led to the unexpected finding that the GR undergoes rapid exchange with target HREs, in marked contrast to the accepted view of stable template binding in the presence of hormone.
The mouse mammary tumor virus (MMTV) long terminal repeat (LTR) promoter has been a useful system for the study of transcriptional activation by steroid hormone receptors. The MMTV promoter integrated into the cellular genome adopts an organized chromatin structure which restricts transcription factor access (2, 9, 17, 20, 44). Stimulation of transcription by hormone treatment coincides with a chromatin transition (1, 17, 37, 38) and binding of the transcription factor, nuclear factor 1 (NF-1) (6-8, 10, 25, 30, 46, 47).
To characterize factors that affect steroid hormone receptor-chromatin interactions, the MMTV promoter was reconstituted into polynucleosome arrays with a chromatin assembly system (13) and the interaction of purified GR with the nucleoprotein template was monitored under various reaction conditions. A template pull-down assay with bead-immobilized MMTV chromatin or DNA was utilized to directly quantify bound factors. It was found that purified GR binds to specific HREs in the chromatin template, in agreement with current dogma. However, when a nuclear extract containing remodeling proteins and ATP is present in the chromatin binding reaction, both GR and the chromatin-remodeling complex, human Swi/Snf (hSwi/Snf), are displaced from the template. Importantly, GR displacement does not occur on a naked DNA template. This displacement reaction can be replicated with affinity-purified hSwi/Snf. Although GR dissociates during chromatin remodeling, it was found that its presence in the reaction, along with that of hSwi/Snf, is crucial for binding of NF-1 to its site in chromatin.
These findings suggest a more dynamic view of nuclear receptor function. Specifically, we propose that receptors act through a hit-and-run mechanism of action (39, 41). The GR interacts with specific recognition elements in promoter chromatin and recruits coactivators and accessory factors such as Swi/Snf, resulting in increased transcription factor access. Concomitant with chromatin remodeling, however, GR dissociates from the template. This model provides a possible explanation for the unexpected rapid exchange observed for GR on target HREs in living cells (28). We further suggest that the dynamic behavior observed for GR may be a general feature of many transcriptional activators, particularly those that are able to interact productively with unremodeled chromatin.
MATERIALS AND METHODS
Reconstitution of MMTV chromatin.
A 1.1-kb PleI/NcoI fragment of MMTV LTR (positions −437 to +674) was immobilized onto Dynal magnetic beads as described by Fletcher et al. (13). The immobilized fragment was reconstituted into chromatin with Drosophila melanogaster late embryo extract supplemented with mouse histone octamers, as previously described. The reconstituted chromatin was then incubated in embryo extract buffer with Sarkosyl (0.05% final concentration) at room temperature for 5 min and washed twice with cold EX-N buffer (10 mM HEPES [pH 7.6], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, 10 mM β-glycerophosphate, 1 mM dithiothreitol [DTT], 0.05% NP-40, 1 mM aminoethylbenzenesulfonyl fluoride, 1-μg/ml concentrations each of aprotinin, pepstatin, and leupeptin) plus 2 mg of bovine serum albumin (BSA)/ml and 200 mM NaCl to remove proteins from Drosophila embryo extract. Reconstituted chromatin was stored at 4°C in EX-N buffer plus 2 mg of BSA/ml.
Purification of GR.
The GR in all experiments was activated by [6,7-3H]dexamethasone mesylate and then purified and transformed on an anion-exchange column (53). This GR is highly pure according to silver staining results, binds HRE-containing oligonucleotides, activates transcription, and binds to MMTV chromatin promoting a chromatin transition in vitro (13).
For a comparison, experiments were also performed using baculovirus-expressed and ammonium sulfate-enriched GR (Pan Vera). To ensure activation, this GR was incubated for 15 min at room temperature with 100 nM dexamethasone in the pull-down reaction before the addition of template.
Purification of BRG1 and hBRM complexes (hSwi/Snf).
Cell pellets from the clone FL-INI1-11 were obtained by the National Cell Culture Center (Minneapolis, Minn.) with permission from Robert Kingston. Nuclear extracts were incubated with anti-Flag M2 affinity chromatography gels (Kodak) at about 60 mg of protein per 1 ml of affinity gel and washed extensively as described previously (43). Flag-immobilized proteins were eluted by incubation for 1 h in a 10-fold molar excess of Flag peptide in 20 mM HEPES (pH 7.9)-20% glycerol-2 mM EDTA-1 mM DTT-100 mM KCl and then stored at −80°C. The protein concentration was determined by the Bradford assay (Bio-Rad) (5), and proteins were analyzed by silver staining SDS-8% polyacrylamide gels. Proteins were also detected by immunoblotting with anti-BRG1 (J1; a gift from Keji Zhao and G. R. Crabtree), anti-human BRM (anti-hBRM; Signal Transduction Laboratories), and anti-Flag (Kodak).
Purification of NF-1.
pMT/V5-HisB-NF-1 was generated by amplification of the NF-1 coding sequence from pCTF1 (40) by PCR. The 5′ oligonucleotide (AACTAGTGGTACCCACCATGGATGAGTTCC) introduced a KpnI site (underlined) and a consensus Kozak sequence (bold) at the 5′ end of the NF-1 coding sequence. The 3′ oligonucleotide (AGCCGCGGTCCCAGATACCAGGACTGTGCCTG) introduced a SacII site (underlined) at the 3′ end of NF-1 and removed the stop codon to allow for an in-frame fusion with the V5-His tag of the expression vector. The resulting PCR product was cut with KpnI and SacII and cloned into the similarly cut pMT5/V5-HisB (Invitrogen). Schneider S2 cells (Invitrogen) were transfected, and a stable pool of NF-1 expression cells were obtained according to manufacturer's specifications. Expression of NF-1 in 1.5 liters of cells growing in suspension was induced with 1 μM CuSO4 for 24 h. Cells were lysed with 6 ml of lysis buffer (20 mM Tris HCl [pH 8], 500 mM NaCl, 10% glycerol, 1% NP-40, 2.5 mM imidazole, and the protease inhibitors described above) per 3-ml cell pellet. Lysates were cleared by centrifugation and applied to a Talon spin column (Clontech) equilibrated with lysis buffer plus 1 mg of BSA/ml at 1 ml (10 to 20 mg of protein) of lysate per column. The column was washed three times with lysis buffer and eluted with 1 ml of elution buffer (lysis buffer, 150 mM imidazole) according to the Talon spin protocol. The protein concentration was usually 2 mg/ml, and only NF-1 was detected on a silver-stained 7.5% polyacrylamide gel.
Chromatin and DNA pull-down assay.
Prior to the binding assay, MMTV DNA or chromatin was washed twice with pull-down buffer (20 mM HEPES [pH 7.3], 50 mM NaCl, 10% glycerol, 0.5 mM EDTA, 5 mM MgCl2, 0.1% NP-40, 1 mM DTT, 1 mM aminoethylbenzenesulfonyl fluoride, 1-μg/ml concentrations each of aprotinin, pepstatin, and leupeptin) with 2 mg of BSA/ml and 10 μg of poly(dI-dC)/ml. The binding reaction mixtures (100 μl in pull-down buffer) contained 100 ng of DNA or chromatin template with or without 3 nM purified GR, 0.5 μg of HeLa nuclear extract/μl, and/or 1 mM ATP. When reaction mixtures contained Swi/Snf complexes (0.1 μg of protein), HeLa extract was reduced to a concentration of 25 ng/μl. After incubation at 30°C for 15 min, the template was washed twice with pull-down buffer without BSA or poly(dI-dC) and analyzed by SDS-7.5% polyacrylamide gel electrophoresis (PAGE) and by immunoblotting with the antibodies indicated below. Antibodies used in detection included anti-GR (PAI-512; Affinity Bioreagents), BRG1 (J1 [a gift from Keji Zhao and Gerald Crabtree] or sc-10768 [Santa Cruz, Santa Cruz, Calif.]), hBRM (Signal Transduction Laboratories), and hSNF2h (a gift from Robert Roeder). High-mobility group 1 protein (HMG-1) used in the experiments represented in Fig. 1 was a gift from Michael Bustin.
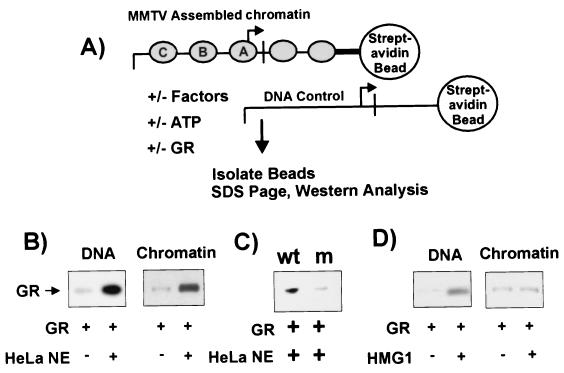
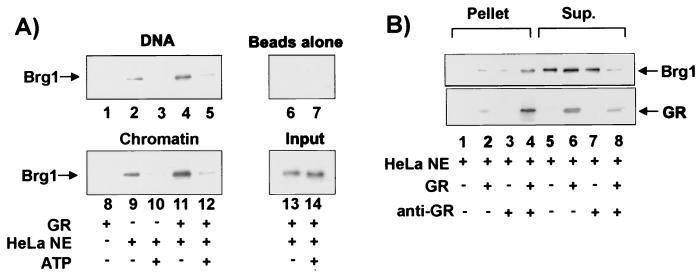
FIG. 1.
Binding of GR to reconstituted MMTV chromatin in vitro. (A) Diagram for chromatin pull-down assay using chromatin reconstituted onto MMTV DNA (positions −437 to +674) attached to streptavidin-coated magnetic beads at +674 (NcoI site). (B) Enhancement of GR binding to MMTV chromatin by HeLa nuclear extract. Purified GR (3 nM) was incubated at 30°C for 15 min with 100 ng of DNA (lanes 1 and 2) or chromatin (lanes 3 and 4) in the presence or absence of 0.5 mg of HeLa nuclear extract/ml as indicated. The associated proteins were analyzed by SDS-PAGE and immunoblotting with PAI-512 anti-GR antibody. (C) Chromatin pull-down assay detecting GR binding specifically to GREs. Purified GR (3 nM) was incubated with wild-type (wt) or GRE-mutated (m) MMTV chromatin and detected as described above. The GRE mutant template was composed of GRE sites 2, 3, 4, 5, and 6 mutated as described by Fletcher et al. (13). (D) Effect of HMG-1 on binding of GR to MMTV promoter. Purified HMG-1 (0.5 μg), instead of HeLa nuclear extract, was added in the pull-down assays described above with either naked DNA or reconstituted chromatin as indicated. HeLa NE, HeLa nuclear extract.
Coimmunoprecipitation.
Reaction mixtures with purified GR and HeLa nuclear extract were identical to the chromatin or DNA pull-down reaction mixtures except that they lacked chromatin and DNA templates. Reaction mixtures were incubated with biotin-labeled anti-GR (MA1.1-510; Affinity Bioreagents) (1 μl/reaction mixture) for 5 min at room temperature, followed by addition of 20 μl of streptavidin-coated beads (200 μg washed once in pull-down buffer plus 1 mg of BSA/ml) and incubation at room temperature with rotation for 2 h. The beads were washed twice with pull-down buffer.
GR (3 nM) or β-galactosidase (70 ng/μl; Sigma) was incubated at room temperature for 10 min with or without purified hSwi/Snf complex mixture (0.1 μg of protein) in 100 μl of binding and washing buffer (20 mM HEPES [pH 7.9]), 20% glycerol, 2 mM EDTA, 1 mM DTT, 100 mM KCl, 1 mg of BSA/ml). Anti-Flag M2 affinity beads (50 μl) equilibrated with binding and washing buffer were added to the mixture, followed by overnight incubation at 4°C. The affinity gel was washed twice with binding and washing buffer.
Proteins were eluted with SDS-PAGE loading buffer, run on SDS-7.5% polyacrylamide gels, and detected by immunoblotting with anti-GR, anti-BRG1, anti-hBRM, anti-Flag, or anti-β-galactosidase (Sigma) antibodies.
SacI accessibility assay.
Chromatin (40 ng) was incubated at room temperature for 20 min with different amounts (indicated in Fig. 5) of GR or GR buffer, 0.05 μg of purified hSwi/Snf, and 1 mM ATP in EX-N buffer plus 0.5 mg of BSA/ml (total, 40 μl). Reactions were restricted with 10 U of SacI at 37°C for 15 min and stopped by the addition of 10 μl of 2.5% Sarkosyl-0.1 M EDTA. The reaction products were processed and 32P labeled as described previously (13). DNA fragments were removed from beads by digestion with EcoRI and run on a 1% agarose gel, which was dried and then exposed to a Molecular Dynamics PhosphorImage screen.
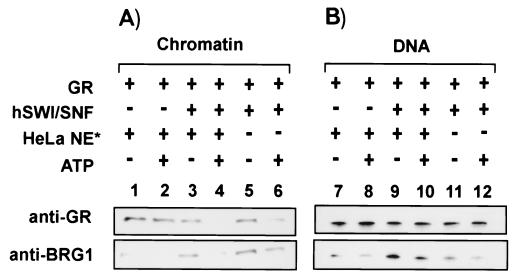
FIG. 5.
hSwi/Snf complexes facilitate displacement of GR from MMTV chromatin. Immobilized MMTV chromatin (A) or DNA (B) (50 ng) was incubated at 30°C for 15 min with hSwi/Snf (2 ng/μl), with or without GR (3 nM), HeLa extract (25 ng/μl), and/or ATP (1 mM) as indicated. An asterisk indicates that the concentration of the HeLa nuclear extract was 1/20 that used for Fig. 1 through 3. GR and BRG1 were detected as indicated next to the panels.
Digested and undigested chromatin were quantitated by using ImageQuant software. SacI access was determined by the fractional cleavage, F(x), calculated by dividing the amount digested by the total amount. The change in fractional cleavage, ΔF(x), for a particular sample was F(x)sample − F(x)control (14).
Footprinting NF-1 with λ exonuclease
Immobilized MMTV DNA (40 ng) with 160 ng of purified NF-1 or Schneider S2 cell extract as indicated in the legend to Fig. 6 was digested simultaneously with 1 U of AlwNI (position −295) and 20 U of λ exonuclease in EX-N buffer (total, 40 μl) at 30°C for 15 min. Samples were deproteinated as described previously for the in vitro SacI assay (13) and then labeled by primer extension with 32P-end-labeled primer 669 (positions +27 to +54 from transcription start site), processed, and run on an 8% sequencing gel as described previously (29).
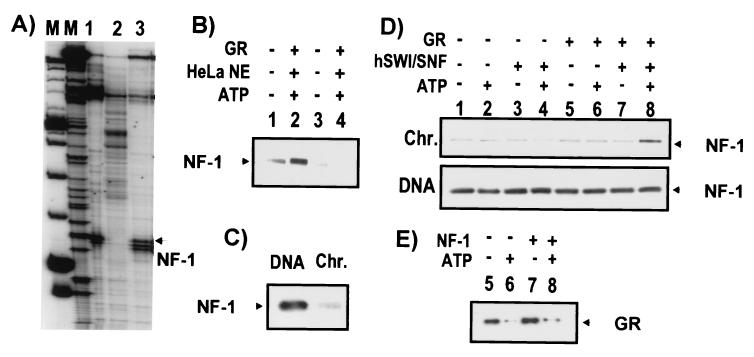
FIG. 6.
GR-dependent interactions of the transcription factor NF-1 with MMTV DNA and chromatin. (A) NF-1 binding detected by λ exonuclease footprinting on 40 ng of immobilized DNA with 10 μg of extract from Schneider S2 cells expressing recombinant NF-1 (lane 1) or lacking NF-1 (lane 2) and 160 ng of purified NF-1 (lane 3). Markers (M) from left to right are a φX174 ladder labeled at the 5′ end with 32P (Life Technologies) and a ddGTP sequencing ladder primer extended with the same primer used in footprinting. The arrow to the right of the gel denotes the λ exonuclease stop at the 5′ edge of the NF-1 site. (B) NF-1 binding to its site detected by chromatin pull-down assay. Immobilized MMTV chromatin (50 ng) with either a wild-type (TTTTGGAATTTATCCAAATCTT) or mutant (TTCTCGAGTTTATCCAGATCTT) NF-1 binding site was incubated at 30°C for 15 min with 160 ng of NF-1, with or without GR (3 nM), HeLa nuclear extract (25 ng/μl), and/or ATP (1 mM) as indicated. V5-epitope-tagged NF-1 was detected by anti-V5 antibody (Invitrogen). HeLa NE, HeLa nuclear extract. (C) Greater binding of NF-1 to MMTV naked DNA than to chromatin. Fifty nanograms of immobilized MMTV DNA or chromatin (Chr.) was incubated at 30°C for 15 min with NF-1 (160 ng of protein). NF-1 was detected as described for panel B. (D) GR and hSwi/Snf facilitation of binding of NF-1 to MMTV chromatin. Chromatin and DNA pull-down assays were performed in the presence of 2 ng of hSwi/Snf per μl and GR, with or without ATP and/or NF-1 as indicated. NF-1 was detected as described for panel B. (E) Absence of effect of NF-1 on ATP-dependent dissociation of GR. Chromatin pull-down assay was performed in the presence of 2 ng of hSwi/Snf per μl and GR, with or without ATP and/or NF-1 as indicated. GR was detected by Western blotting.
RESULTS
Facilitated binding of GR to MMTV LTR chromatin.
To study the interactions between transcription factors and chromatin, we have established an in vitro chromatin pull-down assay using chromatin reconstituted on MMTV LTR DNA. As illustrated in Fig. 1A, a 1-kb DNA fragment containing the MMTV LTR promoter was immobilized onto magnetic beads and reconstituted into a nucleosomal array by using a Drosophila embryo extract supplemented with core histone octamers purified from cultured mouse cells. Chromatin prepared by this method is easily isolated from Drosophila factors in the extract and undergoes a transition similar to that in vivo when supplied with GR and HeLa nuclear extract (13). After being washed in the presence of Sarkosyl (to disrupt Drosophila ISWI-containing complexes), the reconstituted chromatin was then incubated with purified GR for 15 min at 30°C in the presence or absence of HeLa nuclear extract. Naked MMTV DNA attached to magnetic beads was used for comparison. Binding of GR to the template was then analyzed by SDS-PAGE and Western blotting.
As shown in Fig. 1B, in the absence of HeLa nuclear extract, we detected a minimal amount of GR bound to MMTV chromatin. Addition of HeLa nuclear extract significantly enhanced the binding of GR to MMTV chromatin. A similar effect was observed on naked MMTV DNA. Binding of GR was specific to DNA or chromatin, since GR did not bind to the magnetic streptavidin beads in spite of the presence of HeLa nuclear factors (Fig. 2A, lanes 1 and 2). Mutation of the HREs abolished the majority of GR binding (13) (Fig. 1C).
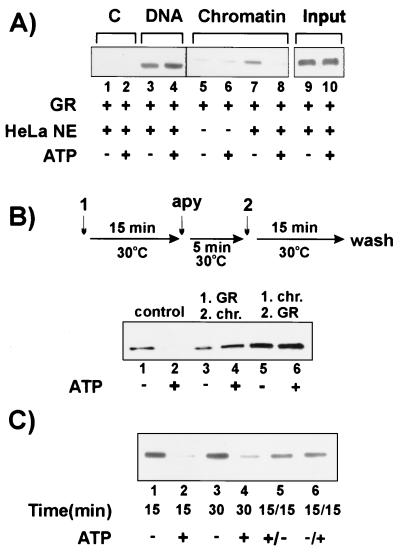
FIG. 2.
Effect of ATP on binding of GR to MMTV chromatin. (A) GR (3 nM) was incubated with reconstituted MMTV chromatin as indicated in the absence (lanes 5 and 6) or presence (lanes 7 and 8) of HeLa nuclear extract (HeLa NE; 0.5 mg/ml). As a control, GR and HeLa nuclear extract were also incubated with beads alone (lanes 1 and 2) or naked DNA (lanes 3 and 4). ATP (1 mM) was added as indicated. GR associated with the beads (lanes 1 through 8) or in the reaction mixtures (lanes 9 and 10) were analyzed by Western blotting with anti-GR antibody. (B) The effect of preincubation of GR or chromatin with ATP on GR binding is shown. The chromatin pull-down assay was done with concentrations of MMTV chromatin, GR, HeLa nuclear extract, and/or ATP as indicated. The top panel illustrates the experimental schedule in which the initial reaction mixture (labeled 1) was incubated as indicated, followed by addition of 1 U of apyrase (apy) to deplete ATP. The second component was added to the ATP-depleted reaction mixture (labeled 2), and the mixture was incubated further, followed by washing (wash) and Western blotting with the anti-GR antibody. The bottom panel shows the Western blot detection of GR. Lanes 1 and 2 (control) contain chromatin template, GR, and HeLa extract, with or without ATP, incubated as described for panel A. Lanes 3 and 4 contain GR incubated first with HeLa extract, with or without ATP, followed by apyrase, and then chromatin was added. Lanes 5 and 6 contain chromatin incubated first with HeLa extract, with or without ATP, followed by apyrase, and then GR was added. (C) GR, HeLa nuclear extract, and ATP (concentrations same as above) were incubated for 15 (lanes 1 and 2) or 30 (lanes 3 and 4) min. A parallel assay was performed in the presence of ATP for 15 min, followed by addition of apyrase (1 U) and further incubation for 15 min (lane 5). Alternatively, the mixture was incubated for 15 min without ATP, followed by 15 min of incubation with ATP (lane 6). The presence of GR was analyzed by Western blotting.
It has been reported previously that HMG proteins, i.e., HMG-1 and -2, increase binding of purified GR to a short stretch of DNA containing an HRE sequence (4). To test if HMGs constitute the activity of HeLa nuclear extract that enhances binding of GR to the MMTV promoter, purified HMG-1 was added in place of the HeLa nuclear extract. Although HMG-1 stimulated binding of GR to MMTV DNA, its effect on GR binding to the chromatin template was minimal (Fig. 1D). Thus, GR exhibits differences in its requirements for binding to MMTV chromatin and DNA templates.
ATP-dependent displacement of GR from reconstituted MMTV chromatin.
It was previously observed that GR blocks access of restriction enzymes to their sites adjacent to or within an HRE. Addition of ATP and HeLa nuclear extract resulted in a significant increase in access, which we attributed to chromatin remodeling (13). The increase in enzyme access in the presence of ATP was also consistent with a loss of GR from the template. Unfortunately, it is impossible to distinguish between chromatin remodeling and displacement of a factor, or both, as a cause of increased nuclease access. To directly study GR promoter interactions with ATP, we included ATP in the chromatin pull-down assay. ATP caused a dramatic decrease in the amount of GR on the chromatin template (Fig. 2A, lanes 7 and 8). The decrease was observed only when HeLa nuclear extract was present (Fig. 2A, lanes 5 and 6), suggesting the involvement of other nuclear factors in an apparent displacement of GR. This activity was not limited to our preparation of GR, since it also occurred with baculovirus-expressed, activated GR (data not shown). ATP-dependent protein degradation did not appear to be involved in the reduction of GR on the template, since the overall level of GR in the reaction mixture was unaffected by ATP (Fig. 2A, lanes 9 and 10). Release of GR from the template was chromatin specific, since this effect did not occur on DNA under similar conditions (Fig. 2A, lanes 3 and 4). Again, these results indicate that interactions of GR with MMTV chromatin and DNA are distinct.
The observed ATP-dependent GR displacement may be a direct modification of either GR or chromatin, resulting in less efficient GR binding. These possible scenarios are illustrated in Fig. 2B. First, GR and HeLa extract were incubated with or without ATP, followed by ATP depletion with apyrase. This premodified mixture was then incubated with the chromatin template. Under these conditions, more GR was bound to chromatin (Fig. 2B, lanes 3 and 4). Preincubation of the chromatin template with ATP followed by apyrase did not affect subsequent binding of GR (Fig. 2B, lanes 5 and 6). These results suggest that an alteration of GR or the chromatin template by ATP is not responsible for the ATP-dependent loss of GR. Moreover, the ATP effect (possibly direct modification) on GR itself resulted in more GR bound to chromatin. To test whether ATP could promote displacement of prebound GR from the template, we allowed GR binding in the absence of ATP, followed by ATP addition. As shown in Fig. 2C, a loss of GR binding was observed under this condition (lanes 1 and 6), suggesting the release of previously bound GR upon the addition of ATP. Conversely, apyrase caused a partial restoration of binding after GR and chromatin were initially incubated with HeLa nuclear extract and ATP (Fig. 2C, lanes 2 and 5). This observation suggests that the constant presence of ATP is required for the dissociation of GR from MMTV chromatin. As a control, we showed that the ATP effect was still maintained when the incubation time was simply extended without changing the ATP conditions (Fig. 2C, lanes 3 and 4). Taken together, these results suggest that GR interactions with MMTV chromatin are dynamic, i.e., the binding-releasing cycle of GR requires the constant presence of ATP and nuclear factors.
Recruitment of chromatin-remodeling factors to the MMTV promoter by GR.
It has been proposed that steroid receptors and other transcription factors act by recruiting other accessory proteins or complexes, including chromatin-remodeling complexes, to promoters. Here, we directly tested recruitment of hSwi/Snf present in the HeLa nuclear extract to reconstituted MMTV chromatin by GR in vitro with the chromatin pull-down assay. Moderate levels of BRG1 (Fig. 3A) and hBRM (data not shown), catalytic subunits of hSwi/Snf complexes, associated with MMTV DNA and chromatin even in the absence of GR. Addition of GR to these reaction mixtures increased the amount of BRG1 and hBRM bound to both the DNA and chromatin templates. BRG1 also coimmunoprecipitated with purified GR, indicating that they can form a complex in solution under our reaction conditions (Fig. 3B, lane 4). Components of Swi/Snf in either yeast or mammalian cells have been shown to be involved in transcriptional activation by GR (4, 32, 56). Hormone-dependent association of GR and BRG1 in solution has also been observed (15). Our results provide direct evidence of recruitment of BRG1 and hBRM to a chromatin template by the intact GR.
FIG. 3.
Recruitment of hBRG1 to MMTV promoter by GR. (A) Reaction mixtures contained MMTV DNA (lanes 1 through 5), chromatin (lanes 8 through 12), or beads alone (lanes 6 and 7). The presence of hBRG1 on chromatin (lanes 1 through 12) or in one-fifth of the total protein in the mixtures (lanes 13 and 14) was analyzed by Western blotting with the J1 anti-BRG1 antibody. (B) Reaction mixtures containing 3 nM GR and 0.5 mg of HeLa nuclear extract/ml were coimmunoprecipitated with MA1.1-510 (biotin-labeled BuGR2) anti-GR antibody. Lanes 1 and 2 contain streptavidin beads alone as negative controls. Lanes 3 and 4 contain biotin-labeled anti-GR antibody with streptavidin beads. Lanes 5 and 6 contain aliquots of supernatants from lanes 1 and 2. Lanes 7 and 8 contain aliquots of supernatants from lanes 3 and 4. Both the pellets and the supernatants were analyzed for the presence of GR and hBRG1 as described above. HeLa NE, HeLa nuclear extract.
ATP is required for the remodeling of chromatin by Swi/Snf and may play an important role in the interaction between BRG1 and hBRM and a chromatin template. Here, we directly tested the effect of ATP on BRG1 (Fig. 3A) and hBRM (data not shown) binding to the MMTV promoter. The majority of BRG1 and hBRM, either constitutively bound or GR recruited to MMTV, was released from the MMTV template in the presence of ATP. Thus, dynamic interactions with chromatin may not be exclusive to steroid receptors. However, unlike GR, BRG1 and hBRM dissociate from both chromatin and naked DNA in the presence of ATP.
Although the results in Fig. 3 demonstrate that GR can recruit BRG1 and hBRM to MMTV, these results do not demonstrate the ability of GR to interact directly with hSwi/Snf complexes in solution and recruit them to the MMTV template. To address this, hSwi/Snf complexes were affinity purified from a HeLa cell line expressing Flag-tagged INI1 as described by Sif and colleagues (43). These complexes contained either BRG1 or hBRM plus p250 (BAF250), p170 (BAF170), p155 (SWI3), p60 (BAF60), p50 (BAF57), p47 (BAF53), or p43 (INI1). Since BRG1 and hBRM exist in distinct hSwi/Snf complexes containing INI1, this preparation provided both types of hSwi/Snf complexes (Fig. 4A and B). This mixture is able to disrupt mononucleosomes containing the nucleosome B region of MMTV in an ATP-dependent manner, as judged by changes in DNase I pattern and restriction enzyme access (data not shown).
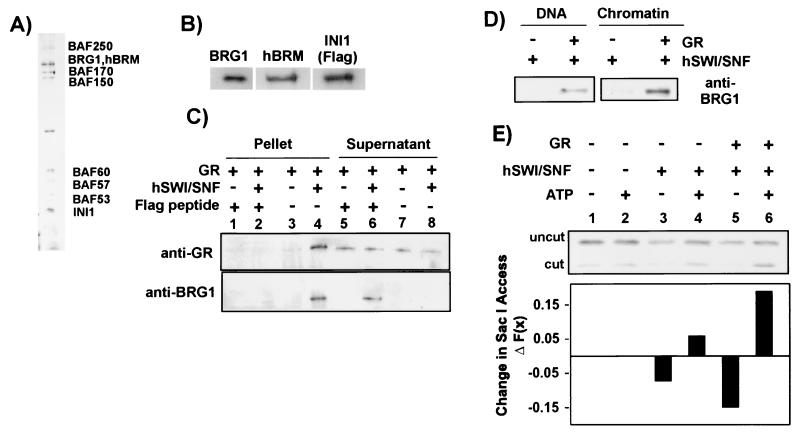
FIG. 4.
Recruitment of purified hSwi/Snf to MMTV DNA and chromatin by GR. (A) Silver staining of an SDS-7.5% PAGE gel containing Flag affinity-purified hSwi/Snf (0.675 ng of protein). (B) Western blot demonstrating the presence of hBRM, BRG1, and INI in affinity-purified hSwi/Snf. (C) Coimmunoprecipitation of GR with hSwi/Snf. Reaction mixtures containing 3 nM GR with or without 0.1 μg of hSwi/Snf were immunoprecipitated with an anti-Flag affinity gel. Lanes 1 and 2 contain 0.5 mg of Flag peptide/ml added to the immunoprecipitation reaction mixture as negative controls. Lanes 5 and 6 contain supernatants of the reaction mixtures in lanes 1 and 2. Lanes 7 and 8 contain supernatants of the reaction mixtures shown in lanes 3 and 4. GR and BRG1 were detected in both pellets and an aliquot of the supernatants by the antibodies, as indicated next to the panels. (D) Chromatin and DNA pull-down assay. Immobilized MMTV DNA or chromatin (50 ng) was incubated at 30°C for 15 min with hSwi/Snf (2 ng/μl) with or without GR (3 nM). GR and BRG1 were detected by the antibodies, as indicated in the panels. (E) Change in SacI cleavage [ΔF(x)] of immobilized MMTV chromatin (50 ng) with hSwi/Snf (25 ng), with or without GR (1 nM) and/or ATP (1 mM) as indicated. The ΔF(x) values were obtained by subtracting the fractional cleavage of the control [F(x)control] from the fractional cleavage with hSwi/Snf, GR, and/or ATP [F(x)sample].
An interaction of GR with hSwi/Snf was demonstrated by coimmunoprecipitation with anti-Flag (Fig. 4C). Purified GR was immobilized minimally by the anti-Flag affinity gel (Fig. 4C, lane 3). However, addition of hSwi/Snf containing the Flag-tagged INI1 resulted in an increase of GR in the pellet (Fig. 4C, lane 4). Addition of Flag peptide to the mixture disrupted interactions of hSwi/Snf with anti-Flag, preventing immobilization of either hSwi/Snf or GR (Fig. 4C, lanes 1 and 2). β-Galactosidase (negative control) did not associate with the purified hSwi/Snf (data not shown).
Recruitment of hSwi/Snf by GR to an MMTV template was also observed. In contrast to the results with HeLa extract, very little purified hSwi/Snf associated with MMTV chromatin and DNA in the absence of GR under the experimental conditions described for Fig. 4D. However, addition of GR caused a significant increase in hSwi/Snf binding. Recruitment of hSwi/Snf activity by GR could also be detected by changes in restriction enzyme access [change in fractional cleavage, ΔF(x)] in the B nucleosome region of MMTV. An increase in SacI access has been observed in vivo in the presence of dexamethasone (14, 19, 34) and in vitro with GR, extracts, and ATP (13). hSwi/Snf remodeling in the presence of ATP resulted in a slight increase in SacI access (Fig. 4E, lane 4). Although GR blocked SacI access by binding to chromatin (13) (Fig. 4E, lane 5), recruitment of hSwi/Snf activity to MMTV by GR resulted in a significant increase in access with the addition of ATP (lane 6).
hSwi/Snf facilitates release of GR from MMTV chromatin.
The dynamic behavior of GR in the presence of ATP could be minimized by reducing the amount of HeLa extract in the reaction mixture (Fig. 5A, lanes 1 and 2). Interestingly, the ATP-dependent loss of GR from chromatin was greatly enhanced by the addition of hSwi/Snf (Fig. 5A, lanes 3 and 4). An ATP-dependent GR release from chromatin also occurred with hSwi/Snf alone (Fig. 5A, lanes 5 and 6). Importantly, GR was not displaced from a naked DNA template under the reaction conditions in which it was displaced from chromatin (Fig. 5B, lanes 9 through 12). Since ISWI complexes present in the Drosophila assembly extracts were attributed to a role in the binding of retinoic acid receptor and retinoid X receptor to chromatin (11), we also performed our experiments on non-detergent-washed chromatin. We found that GR displacement still occurs and no longer requires HeLa nuclear extract, which suggested that the abundant ISWI complexes from the Drosophila assembly extracts are also capable of this activity (data not shown) and that this activity is removed from chromatin by washing (Fig. 2A, lanes 5 and 6). As shown in Fig. 3, hSwi/Snf has binding properties distinct from those of GR under the same reaction conditions. ATP-dependent displacement was observed on both chromatin and naked DNA templates (Fig. 5). However, both GR and hSwi/Snf displacement appeared to be slightly reduced when HeLa extract was absent (Fig. 5A, lanes 5 and 6, and B, lanes 11 and 12).
The restriction enzyme access results in Fig. 4 and the results in Fig. 5 demonstrate that the nuclease hypersensitivity previously observed (13) is due to both recruitment of chromatin-remodeling machinery by GR and GR dissociation during the remodeling reaction.
Both GR and hSwi/Snf are required for NF-1 binding to MMTV chromatin.
NF-1 expressed in Schneider S2 cells was capable of interacting with its site on MMTV DNA, as judged by λ exonuclease footprinting (Fig. 6A). The pull-down assay also detected site-specific binding of NF-1 on wild-type MMTV chromatin but not on chromatin with a mutated NF-1 site (Fig. 6B). Furthermore, addition of GR, HeLa nuclear extract, and ATP caused an increase in NF-1 binding. Previously, it has been shown that the binding affinity of NF-1 is much greater on naked DNA than on mononucleosomes (3). NF-1 is also excluded from reconstituted MMTV minichromosomes (50). Using the pull-down assay, we also demonstrated substantially less binding of NF-1 to its site on MMTV chromatin than on naked DNA (Fig. 6C), suggesting that chromatin remodeling may be required. Surprisingly, remodeling by hSwi/Snf alone did not promote NF-1 binding to chromatin (Fig. 6D, lane 4). Neither did GR alone facilitate NF-1 binding (Fig. 6D, lanes 5 and 6). However, a considerable increase in NF-1 binding to MMTV chromatin required GR, hSwi/Snf, and ATP (Fig. 6D, lane 8). None of these factors, either alone or in combination, had a significant effect on NF-1 binding to naked DNA (Fig. 6D). Although GR affected NF-1 binding, NF-1 had little influence on the ATP-dependent displacement of GR (Fig. 6E).
DISCUSSION
The formation of stable initiation complexes on regulatory elements is a central paradigm of transcriptional regulation. As a class, steroid receptors have been argued to bind statically to response elements in the presence of ligand. The DNA-receptor complex would then serve as a platform for the assembly of secondary factors, including coactivators and general transcription factors. These stable complexes would attract the wide variety of activities required for local chromatin transitions and the eventual assembly of a productive polymerase II initiation event. Under this model, the DNA-receptor complex would have a long lifetime in the presence of hormone, with removal of the ligand from the cellular environment, leading to the breakup of the static complex.
While the results of many studies have been interpreted to be in support of this general model, several puzzling findings are contrary to the central thesis. In an analysis of the GR response element (GRE) 2.6 kb upstream in the tyrosine aminotransferase promoter, Rigaud and colleagues reported that activation of GR led to the recruitment of hepatocyte nuclear factor 5 (39). Despite this ordered recruitment, GR and hepatocyte nuclear factor 5 cannot cooccupy their respective sites on deproteinized DNA. Thus, it appears in this case that the pioneer protein that initiates a local nucleoprotein transition cannot remain bound to the template after recruitment of the secondary protein. A simple and straightforward explanation for the tyrosine aminotransferase conundrum is that GR initiates the nucleosome transition but is immediately ejected from the template during chromatin remodeling, leaving behind a region of reorganized chromatin. It is this reorganization event that is in turn responsible for secondary transcription factor recruitment.
Mechanism of GR displacement.
Chromatin remodeling is a critical step for transcriptional activation on nucleoprotein templates. Although the ability to remodel chromatin is intrinsic to chromatin-remodeling machineries such as Swi/Snf, several lines of evidence from in vitro studies demonstrate that the remodeling activity is targeted to specific promoters by interaction with transcriptional activators. In particular, yeast Swi/Snf can be recruited to a chromatin-containing GAL4 site in vitro by acidic activation domains fused to GAL4 (57). Yeast Swi/Snf can also stimulate transcription in vitro when targeted to Gal4 sites by a Gal4 DNA binding domain fused to the τ1 domain of GR (52). In vitro-translated GR has been found to interact specifically with the hSwi/Snf subunit BAF 250 (33). Evidence of in vivo recruitment of hSwi/Snf by estrogen receptors was demonstrated by DiRenzo and colleagues using chromatin immunoprecipitation assays (12). In this report, we have shown recruitment of hSwi/Snf complexes to a nucleosomal array assembled on a natural promoter, most likely through direct interactions with intact GR. We found, however, that the recruitment reaction is highly transient.
One mechanism for GR displacement is suggested by our findings. Displacement may occur as a direct result of the nucleosome reorganization event. That is, GR binding is destabilized as a direct consequence of the remodeling process. An interesting possibility in this regard is suggested by reports of local changes in DNA topology (16, 21, 24) during remodeling. In the first two studies mentioned (16, 21), localized changes in DNA topology were found to require constant ATP hydrolysis reminiscent of the GR reassociation that we observed with the apyrase addition (Fig. 2C).
A dynamic view of interactions among GR, hSwi/Snf, NF-1, and the MMTV promoter.
In this study, we demonstrated that all three factors—GR, hSwi/Snf, and NF-1—have distinct interactions with both DNA and chromatin. While displacement of GR occurs only on chromatin, the ATP-dependent release of Swi/Snf is observed on both chromatin and DNA templates. This does not necessarily suggest that hSwi/Snf interacts identically with DNA and chromatin. Yeast Swi/Snf binds to nucleosomal and naked DNA through different subunits (42). Interestingly, contacts between particular subunits in these complexes and nucleosomal DNA are altered during remodeling. Our observations may be a reflection of Swi/Snf conformational changes with ATP hydrolysis which modifies its interactions with nucleosomal arrays and naked DNA.
Like GR, ATP-dependent changes in NF-1 binding are exclusive to chromatin. However, an increase in NF-1 binding which requires the presence of GR during chromatin remodeling by Swi/Snf occurs. The PR also induces MMTV transcription and recruits NF-1 to the promoter. PR action is facilitated by remodeling complexes; Di Croce and colleagues have argued that direct protein-protein interactions contribute to a stable PR/NF-1/template complex (10). However, NF-1 binding does not appear to prevent ATP-dependent GR dissociation from MMTV chromatin in our system. This demonstrates either a potential distinction between the two receptors or the ability of the template pull-down assay to monitor dynamic interactions between transcription factors and chromatin during remodeling. In light of our findings that GR participates in NF-1 binding to chromatin, it is conceivable that NF-1 and GR (or PR) can interact, albeit transiently, on a chromatin template.
The findings presented here represent a significant departure from the static view of nuclear receptor function that has dominated the literature for more than 2 decades. Many secondary implications arise from the dynamic view of receptor behavior. As one example, the receptors would be immediately and constantly available for interaction with other signaling pathways, rather than requiring a separate (slow) disengagement process that in turn might require the removal of ligand from the cellular environment. Thus, coordination of the receptor response with alternative regulatory pathways could in principle be integrated much more effectively under the dynamic model. We anticipate that future studies may lead to similar findings for other members of the large nuclear receptor superfamily.
Finally, the dynamic view of receptor function described here will have important implications for mechanisms of transcriptional control by other regulatory pathways. In particular, we suggest that the exchange process may be a general feature of the initial remodeling events required for gene activation and that other pioneer proteins may function through a similar mechanism.
Acknowledgments
T.M.F. and N.X. contributed equally to this work.
We thank Michael Bustin for generously providing HMG-1. We are also grateful for the assistance of Carl Wu, Raphael Sandaltopoulus, and Ju-Gyeong Kang (NIH) for providing the Drosophila embryos for chromatin assembly extract preparation. We thank Catharine Smith and Denise Thurber (NIH) for the MMTV construct containing the mutant NF-1 site, Robert Kingston (Harvard Medical School) for permission to use the FL-INI1-11 cell line, and the National Cell Culture Center for providing cell pellets. The anti-BRG1 antibodies were generously provided by Keji Zhou and Gerald Crabtree (Stanford University), and anti-Snf2 h was provided by Robert Roeder (Rockefeller University). We thank Matthias Becker and Sam John for critical reading of the manuscript.
REFERENCES
- 1.Archer, T. K., M. G. Cordingley, R. G. Wolford, and G. L. Hager. 1991. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol. Cell. Biol. 11:688-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beato, M. 1996. Chromatin structure and the regulation of gene expression: remodeling at the MMTV promoter. J. Mol. Med. 74:711-724. [DOI] [PubMed] [Google Scholar]
- 3.Blomquist, P., Q. Li, and O. Wrange. 1996. The affinity of nuclear factor 1 for its DNA site is drastically reduced by nucleosome organization irrespective of its rotational or translational position. J. Biol. Chem. 271:153-159. [DOI] [PubMed] [Google Scholar]
- 4.Boonyaratanakornkit, V., V. Melvin, P. Prendergast, M. Altmann, L. Ronfani, M. E. Bianchi, L. Taraseviciene, S. K. Nordeen, E. A. Allegretto, and D. P. Edwards. 1998. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol. Cell. Biol. 18:4471-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourachot, B., M. Yaniv, and C. Muchardt. 1999. The activity of mammalian brm/SNF2α is dependent on a high-mobility-group protein I/Y-like DNA binding domain. Mol. Cell. Biol. 19:3931-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Buetti, E., B. Kuhnel, and H. Diggelmann. 1989. Dual function of a nuclear factor I binding site in MMTV transcription regulation. Nucleic Acids Res. 17:3065-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cato, A. C., P. Skroch, J. Weinmann, P. Butkeraitis, and H. Ponta. 1988. DNA sequences outside the receptor-binding sites differently modulate the responsiveness of the mouse mammary tumor virus to various steroid hormones. EMBO J. 7:1403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordingley, M. G., A. T. Riegel, and G. L. Hager. 1987. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell 48:261-270. [DOI] [PubMed] [Google Scholar]
- 9.Deroo, B. J., and T. K. Archer. 2001. Glucocorticoid receptor-mediated chromatin remodeling in vivo. Oncogene 20:3039-3046. [DOI] [PubMed] [Google Scholar]
- 10.Di Croce, L., R. Koop, P. Venditti, H. M. Westphal, K. P. Nightingale, D. F. Corona, P. B. Becker, and M. Beato. 1999. Two-step synergism between the progesterone receptor and the DNA-binding domain of nuclear factor 1 on MMTV minichromosomes. Mol. Cell 4:45-54. [DOI] [PubMed] [Google Scholar]
- 11.Dilworth, F. J., C. Fromental-Ramain, K. Yamamoto, and P. Chambon. 2000. ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR in vitro. Mol. Cell 6:1049-1058. [DOI] [PubMed] [Google Scholar]
- 12.DiRenzo, J., Y. Shang, M. Phelan, S. Sif, M. Myers, R. Kingston, and M. Brown. 2000. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol. Cell. Biol. 20:7541-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher, T. M., B.-W. Ryu, C. T. Baumann, B. S. Warren, G. Fragoso, S. John, and G. L. Hager. 2000. Structure and dynamic properties of a glucocorticoid receptor-induced chromatin transition. Mol. Cell. Biol. 20:6466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fragoso, G., W. D. Pennie, S. John, and G. L. Hager. 1998. The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames. Mol. Cell. Biol. 18:3633-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fryer, C. J., and T. K. Archer. 1998. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88-91. [DOI] [PubMed] [Google Scholar]
- 16.Gavin, I. M., P. J. Horn, and C. L. Peterson. 2001. SWI/SNF chromatin remodeling requires changes in DNA topology. Mol. Cell 7:97-104. [DOI] [PubMed] [Google Scholar]
- 17.Hager, G. L. 2001. Understanding nuclear receptor function: from DNA to chromatin to the interphase nucleus. Prog. Nucleic Acid Res. Mol. Biol. 66:279-303. [DOI] [PubMed] [Google Scholar]
- 18.Hager, G. L., and T. K. Archer. 1991. The interaction of steroid receptors with chromatin, p. 217-234. In M. G. Parker (ed.), Structure and function of nuclear hormone receptors. Academic Press, London, England.
- 19.Hager, G. L., T. K. Archer, G. Fragoso, E. H. Bresnick, Y. Tsukagoshi, S. John, and C. L. Smith. 1993. Influence of chromatin structure on the binding of transcription factors to DNA. Cold Spring Harbor Symp. Quant. Biol. 58:63-71. [DOI] [PubMed] [Google Scholar]
- 20.Hager, G. L., T. M. Fletcher, N. Xiao, C. T. Baumann, W. G. Muller, and J. G. McNally. 2000. Dynamics of gene targeting and chromatin remodeling by nuclear receptors. Biochem. Soc. Trans. 28:405-410. [PubMed] [Google Scholar]
- 21.Havas, K., A. Flaus, M. Phelan, R. E. Kingston, P. A. Wade, D. M. Lilley, and T. Owen-Hughes. 2000. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 103:1133-1142. [DOI] [PubMed] [Google Scholar]
- 22.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 23.Krebs, J. E., and C. L. Peterson. 2000. Understanding “active” chromatin: a historical perspective of chromatin remodeling. Crit. Rev. Eukaryot. Gene Expr. 10:1-12. [PubMed] [Google Scholar]
- 24.Langst, G., and P. B. Becker. 2001. ISWI induces nucleosome sliding on nicked DNA. Mol. Cell 8:1085-1092. [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre, P., D. S. Berard, M. G. Cordingley, and G. L. Hager. 1991. Two regions of the mouse mammary tumor virus long terminal repeat regulate the activity of its promoter in mammary cell lines. Mol. Cell. Biol. 11:2529-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemon, B. D., and L. P. Freedman. 1999. Nuclear receptor cofactors as chromatin remodelers. Curr. Opin. Genet. Dev. 9:499-504. [DOI] [PubMed] [Google Scholar]
- 27.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 28.McNally, J. G., W. G. Mueller, D. Walker, R. G. Wolford, and G. L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262-1265. [DOI] [PubMed] [Google Scholar]
- 29.Mellentin-Michelotti, J., S. John, W. D. Pennie, T. Williams, and G. L. Hager. 1994. The 5′ enhancer of the mouse mammary tumor virus long terminal repeat contains a functional AP-2 element. J. Biol. Chem. 269:31983-31990. [PubMed] [Google Scholar]
- 30.Mink, S., E. Hartig, P. Jennewein, W. Doppler, and A. C. B. Cato. 1992. A mammary cell-specific enhancer in mouse mammary tumor virus DNA is composed of multiple regulatory elements including binding sites for CTF/NFI and a novel transcription factor, mammary cell-activating factor. Mol. Cell. Biol. 12:4906-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizzen, C. A., and C. D. Allis. 2000. Transcription. New insights into an old modification. Science 289:2290-2291. [DOI] [PubMed] [Google Scholar]
- 32.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie, Z., Y. Xue, D. Yang, S. Zhou, B. J. Deroo, T. K. Archer, and W. Wang. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20:8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pennie, W. D., G. L. Hager, and C. L. Smith. 1995. Nucleoprotein structure influences the response of the mouse mammary tumor virus promoter to activation of the cAMP signalling pathway. Mol. Cell. Biol. 15:2125-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson, C. L. 2000. ATP-dependent chromatin remodeling: going mobile. FEBS Lett. 476:68-72. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 37.Richard-Foy, H., and G. L. Hager. 1987. Sequence specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 6:2321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richard-Foy, H., F. D. Sistare, A. T. Riegel, S. S. Simons, Jr., and G. L. Hager. 1987. Mechanism of dexamethasone 21-mesylate antiglucocorticoid action. II. Receptor-antiglucocorticoid complexes are unable to interact productively with mouse mammary tumor virus long terminal repeat chromatin. Mol. Endocrinol. 1:659-665. [DOI] [PubMed] [Google Scholar]
- 39.Rigaud, G., J. Roux, R. Pictet, and T. Grange. 1991. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell 67:977-986. [DOI] [PubMed] [Google Scholar]
- 40.Santoro, C., N. Mermod, P. C. Andrews, and R. Tjian. 1988. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature 334:218-224. [DOI] [PubMed] [Google Scholar]
- 41.Schaffner, W. 1988. Gene regulation. A hit-and-run mechanism for transcriptional activation? Nature 336:427-428. [DOI] [PubMed] [Google Scholar]
- 42.Sengupta, S. M., M. VanKanegan, J. Persinger, C. Logie, B. R. Cairns, C. L. Peterson, and B. Bartholomew. 2001. The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J. Biol. Chem. 276:12636-12644. [DOI] [PubMed] [Google Scholar]
- 43.Sif, S., P. T. Stukenberg, M. W. Kirschner, and R. E. Kingston. 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 12:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, C. L., and G. L. Hager. 1997. Transcriptional regulation of mammalian genes in vivo: a tale of two templates. J. Biol. Chem. 272:27493-27496. [DOI] [PubMed] [Google Scholar]
- 45.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toohey, M. G., J. W. Lee, M. Huang, and D. O. Peterson. 1990. Functional elements of the steroid hormone-responsive promoter of mouse mammary tumor virus. J. Virol. 64:4477-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Truss, M., J. Bartsch, A. Schelbert, R. J. Hache, and M. Beato. 1995. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 14:1737-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urnov, F. D., and A. P. Wolffe. 2001. A necessary good: nuclear hormone receptors and their chromatin templates. Mol. Endocrinol. 15:1-16. [DOI] [PubMed] [Google Scholar]
- 49.Urnov, F. D., and A. P. Wolffe. 2001. An array of positioned nucleosomes potentiates thyroid hormone receptor action in vivo. J. Biol. Chem. 276:19753-19761. [DOI] [PubMed] [Google Scholar]
- 50.Venditti, P., L. Di Croce, M. Kauer, T. Blank, P. B. Becker, and M. Beato. 1998. Assembly of MMTV promoter minichromosomes with positioned nucleosomes precludes NF1 access but not restriction enzyme cleavage. Nucleic Acids Res. 26:3657-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallberg, A. E., K. E. Neely, A. H. Hassan, J.-Å. Gustafsson, J. L. Workman, and A. P. H. Wright. 2000. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor τ1 activation domain. Mol. Cell. Biol. 20:2004-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warren, B. S., P. Kusk, R. G. Wolford, and G. L. Hager. 1996. Purification and stabilization of transcriptionally active glucocorticoid receptor. J. Biol. Chem. 271:11434-11440. [DOI] [PubMed] [Google Scholar]
- 54.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 55.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 56.Yoshinaga, S. K., C. L. Peterson, I. Herskowitz, and K. R. Yamamoto. 1992. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258:1598-1604. [DOI] [PubMed] [Google Scholar]
- 57.Yudkovsky, N., C. Logie, S. Hahn, and C. L. Peterson. 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 13:2369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]