Abstract
Although both RAG-1 and RAG-2 are required for all steps of V(D)J recombination, little is known about the specific contribution of either protein to these steps. RAG-1 contains three acidic active-site amino acids that are thought to coordinate catalytic metal ions. To search for additional catalytic amino acids and to better define the functional anatomy of RAG-1, we mutated all 86 conserved basic amino acids to alanine and evaluated the mutant proteins for DNA binding, nicking, hairpin formation, and joining. We found several amino acids outside of the canonical nonamer-binding domain that are critical for DNA binding, several step arrest mutants with defects in nicking or hairpin formation, and four RAG-1 mutants defective specifically for joining. Analysis of coding joints formed by some of these mutants revealed excessive deletions, frequent use of short sequence homologies, and unusually long palindromic junctional inserts, known as P nucleotides, that result from aberrant hairpin opening. These features characterize junctions found in scid mice, which are deficient for the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs), suggesting that the RAG proteins and DNA-PKcs perform overlapping functions in coding joint formation. Interestingly, the amino acids that are altered in 12 of our mutants are also mutated in human inherited immunodeficiency syndromes. Our analysis of these mutants provides insights into the molecular mechanisms underlying these disorders.
V(D)J recombination generates diversity in the antigen-binding sites of immunoglobulin and T-cell receptor molecules. The RAG-1 and RAG-2 proteins, the only lymphocyte-specific factors required for this reaction, recognize and bind to recombination signal sequences (RSS) that contain conserved heptamer and nonamer motifs separated by either 12 or 23 bp of spacer sequence. Efficient recombination occurs only between two RSS with different spacer lengths (the 12/23 rule) (34). The RAG proteins initiate recombination by introducing a nick in the DNA at the 5′ border of the RSS. This nick is subsequently converted to a double-strand break by a transesterification reaction in which the newly generated 3′-hydroxyl attacks the phosphodiester bond on the opposite strand, leaving a covalently sealed hairpin containing the coding sequence (the coding end) and a blunt 5′-phosphorylated end containing the RSS (the signal end) (39). The broken DNA ends are thought to remain in a postcleavage complex (1, 25) until they are processed and joined, forming two characteristic junctions, coding joints and signal joints. Joining proceeds with the help of several double-strand break repair factors (19, 45). Recent data indicate that RAG-1 and RAG-2 play critical roles in the formation of both signal joints and coding joints in vivo (47, 54).
Both RAG proteins are required for all steps of the recombination reaction, but little is known about the specific role(s) played by each protein. At a single RSS, RAG-1 binds as a dimer, with one or two monomers of RAG-2 (4, 50, 64). Recent work has shown that only one functional active site contained within one RAG-1 monomer is required for both catalytic steps (cleavage and hairpin formation) at a single RSS (32, 62). Site-directed mutagenesis of all evolutionarily conserved acidic amino acids of truncated active core RAG-1 and RAG-2 (the minimal regions of the proteins required for efficient recombination) revealed that RAG-1 contains three acidic active-site amino acids (33) thought to be responsible for coordinating catalytic metal ions (16, 30, 33). Thus, RAG-1 contains at least part of the active site of the recombinase.
Deletion analysis has provided other information about important regions of the RAG-1 protein. The N terminus contains a DNA-binding domain that interacts with the nonamer element (12, 59). RAG-1 also contacts DNA at the border between the heptamer and the adjacent coding sequence (14, 42, 64); recent DNA-protein cross-linking studies have indicated that the region responsible for these contacts resides in the C terminus of RAG-1 (44). Deletion analysis has revealed extensive regions of RAG-1 that interact with RAG-2 (2, 40), but specific amino acids important for this interaction have not been identified.
Basic amino acids are often involved in DNA binding and protein-protein interactions. In transposases and restriction enzymes, basic residues position the substrate for cleavage or stabilize the transition state of the reaction (reviewed in references 22 and 56). Site-directed mutational analyses of all conserved positively charged amino acids in core RAG-2 failed to identify catalytic amino acids (15, 47). To search for such catalytic amino acids in RAG-1 and to better define other functional regions of RAG-1, we mutated all 86 evolutionarily conserved basic amino acids in RAG-1 to alanine (Fig. 1) and evaluated the mutant proteins for their ability to carry out DNA binding, nicking, hairpin formation, and joining.
FIG. 1.
Basic amino acids in RAG-1 targeted for mutagenesis. Eighty-six conserved basic residues (71 absolutely conserved [blue] and 15 with charge conservations [red]) were identified by aligning the RAG-1 sequences from nine species (human, rabbit, mouse, opossum, chicken, Xenopus, bull shark, rainbow trout, and zebra fish). The active-site residues D600, D708, and E962 are shown as white letters on a black background.
This comprehensive mutagenesis approach revealed several different classes of recombination-defective RAG-1 mutants, including 12 with alterations that affect amino acids mutated in human inherited immunodeficiency syndromes. First, we identified several amino acids outside of the canonical nonamer-binding domain that are critical for DNA binding. In the primary sequence, these amino acids are located near active-site metal-coordinating residues, raising the possibility that they may form part of the active-site DNA-binding domain. Second, we found step arrest mutants with specific defects in either nicking or hairpin formation. Interestingly, the mutations that impair hairpin formation are all located between amino acids 855 and 973, a region that includes the active-site glutamate (E962). Finally, we identified four RAG-1 mutants that are specifically defective for the joining phase of the reaction. Two of these constitute a novel class of mutants whose joining defects can be suppressed by certain substrate configurations, suggesting that the mutations may destabilize the postcleavage complex. Analysis of the rare coding joints formed by some of these mutants revealed features characteristic of junctions formed in cells deficient for the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs), suggesting that the RAG proteins and DNA-PKcs perform overlapping functions in coding joint formation.
MATERIALS AND METHODS
Plasmid constructs and mutagenesis.
All 86 conserved basic amino acids in core RAG-1 were mutated to alanine as described previously (33). All mutants (except as noted in Table 1) were constructed with the pMAL-17 expression vector, which is derived from the pMAL-2 expression vector (33); pMAL-17 encodes amino acids 384 to 1008 of murine RAG-1 fused to a nine-histidine tag and three copies of the human c-myc epitope on the carboxyl terminus. The entire open reading frames of all mutants with non-wild-type phenotypes were sequenced to rule out the presence of second-site mutations. Mutants exhibiting a non-wild-type phenotype in vivo were reconstructed with the glutathione S-transferase (GST)-RAG-1 fusion expression vector, pEBG-1ΔN (58), by double-stranded mutagenesis (11) for protein purification. The entire open reading frames of all mutants constructed with pEBG-1ΔN were sequenced. Despite repeated attempts, one mutant, H482/K483, could not be generated with the GST-RAG-1 fusion expression vector.
TABLE 1.
Summary of in vivo recombination analysis
| Defect and mutant |
|---|
| None (wild type) or moderate |
| K388 |
| H395 |
| R409 |
| K412 |
| K416 |
| K421 |
| K428 |
| H445/R446 |
| R458a |
| R486 |
| K489b |
| H501 |
| R504b |
| K555a |
| R556a |
| R579 |
| K618 |
| K645 |
| K652a |
| H665 |
| R679 |
| R696 |
| R703 |
| K710 |
| H744 |
| R756 |
| R761 |
| K777a |
| K782a |
| K806 |
| K823 |
| R826/K827 |
| K844 |
| R848 |
| R875 |
| R927 |
| R931a |
| R949b |
| K997 |
| K1001 |
| K1006a |
| Severe |
| R391c |
| R393c |
| R401/R402d |
| K405/H406/R407 |
| R440d |
| R471 |
| H482/K483 |
| K596 |
| K608 |
| R621 |
| R713d |
| R734a |
| R748/H750d |
| R773/R775 |
| H795 |
| R838/K839/R840d |
| R855/K856 |
| K890a |
| R894 |
| H937/K938 |
| H942d |
| K966 |
| R969/R970 |
| R972/K973d |
| R977 |
| K980d |
| K989/H990/H991 |
This mutant was tested with the pEBG-1ΔN vector (GST-RAG-1 fusion vector) and compared to the appropriate wild-type control.
This mutant exhibited a moderate recombination defect.
This amino acid is in the nonamer-binding domain, and single mutations at this site have been shown to produce a DNA-binding defect (12, 59). This mutant was not further characterized.
This mutant was also tested in the context of full-length, untagged RAG-1.
Transfections.
The pMAL-17 wild-type or mutant RAG-1 expression vector (1.8 μg) and the pMAL-1 wild-type RAG-2 expression vector (33) (2.1 μg), along with a recombination substrate (pJH290, pJH289, or pJH299) (2 μg), were transiently transfected into Chinese hamster ovary fibroblasts (RMP41 cells) by using the Fugene6 transfection reagent (Roche) as previously described (33). After 48 h, the DNA was harvested by the Hirt method as described previously (60).
PCR assays for coding and signal joints.
To detect coding joints and signal joints, 1/30th of the total harvested DNA was assayed by PCR (24 cycles). Coding and signal joints on the excised products of pJH289 and pJH290 were amplified with primers DR55 and ML68 as described previously (61). Coding joints formed on the plasmid product from pJH290 were amplified with primers DR99 and DR100 (21). Coding joints and signal joints formed on the pJH299 substrate were amplified with primers ML68 and DR99 and primers DR55 and DR100, respectively. PCR products (10 μl) were separated on a 6% polyacrylamide gel, transferred to a membrane (Genescreen Plus), and hybridized with a radiolabeled oligonucleotide probe (DR55 or DR99).
Ligation-mediated PCR for detection of signal ends.
One-thirtieth of the harvested DNA was assayed by ligation-mediated PCR as described previously (60). For PCR assays, 1 μl of a 1:100 dilution of the ligation mixture was amplified with primers DR55 and DR20. PCR products were separated on a 6% polyacrylamide gel, transferred to a membrane, and hybridized with a radiolabeled oligonucleotide probe (DR69) (60).
Transformation assay for detection of coding and signal joints.
Coding joints and signal joints formed on the plasmid products from the pJH290 and pJH289 substrates were detected as chloramphenicol-resistant colonies as described previously (23).
Analysis of coding and signal joint sequences.
PCR products containing the coding joint from pJH289 were gel purified and cloned by using a Topo TA cloning kit (Invitrogen). Positive colonies were identified by colony PCR screening, and the PCR products were sequenced by using a BigDye terminator cycle sequencing kit (version 2.0; PE Biosystems). Sequences for coding joints for RAG-1 mutants were obtained from three independent transfections. The structures of signal joints were determined by digesting PCR products containing the signal joint from pJH290 with 20 U of ApaLI at 37°C for 3 h.
Purification of GST fusion proteins.
The wild-type or mutant GST-RAG-1 expression vector (pEBG-1ΔN) and the wild-type GST-RAG-2 expression vector (pEBG-2ΔC) (58) were cotransfected into RMP41 cells. The GST-RAG-1 and GST-RAG-2 proteins were copurified as described previously (47, 52, 54). Protein concentrations were determined by Coomassie blue staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Multiple preparations of mutant R471 yielded only very low quantities of protein, so we were unable to assay this mutant in vitro. Note that for all biochemical assays, estimations of the relative activities of the mutant proteins were made based on comparisons to the activities of wild-type control protein preparations assayed at the same time and run on the same gel.
DNA-binding assays.
DNA-binding assays with a single radiolabeled 12-RSS substrate (DAR39/40) (39) and the copurified GST-RAG-1 and GST-RAG-2 proteins were performed as described previously (25). DNA binding was analyzed by nondenaturing electrophoresis through a 4 to 20% polyacrylamide gel in Tris-borate-EDTA and visualized by using a PhosphorImager (Molecular Dynamics).
Oligonucleotide cleavage assays.
Coupled cleavage of oligonucleotide substrates was performed as described previously (25) but with some modifications. Briefly, the purified RAG proteins (2 μl) were incubated with 25 fmol of radiolabeled 12-RSS oligonucleotide substrate (DAR39/40) and 250 fmol of unlabeled 23-RSS (or unlabeled 12-RSS, when needed) substrate (DG61/62) (39) in a 10-μl volume containing 25 mM morpholinepropanesulfonic acid (MOPS) (pH 7.0), 2 mM dithiothreitol, 100 μg of bovine serum albumin/ml, 5 mM CaCl2, 19 mM potassium acetate, and 200 ng of high-mobility group protein 1 (HMG-1). Histidine-tagged, full-length recombinant human HMG-1 was purified from bacteria containing the plasmid pET-HMG (18). Reaction mixtures were incubated for 10 min at 37°C. MgCl2 was added to a final concentration of 5 mM, and the reaction mixtures were incubated for an additional 20 min at 37°C. Reactions were terminated by the addition of an equal volume of formamide loading dye. Products were resolved on a 10% acrylamide gel containing 30% formamide, 0.67× Tris-borate-EDTA, and 12.5 mM HEPES (pH 7.5) and visualized by using a PhosphorImager. Oligonucleotide substrates with nonpermissive coding flank sequences were created by annealing SK5/SK6 (12 RSS) and SK38/SK39 (23 RSS) (28). All oligonucleotides were gel purified prior to labeling and annealing.
Cleavage at a single RSS and cleavage of a prenicked substrate were performed as described for coupled cleavage but with the following modifications: The purified RAG proteins were incubated with the 12-RSS oligonucleotide substrate or the prenicked substrate, which was made by annealing DAR42 (32P end labeled), DG10, and DAR40 (39). MnCl2 was substituted for MgCl2, and HMG storage buffer (25 mM Tris-Cl [pH 8.0], 1 mM EDTA, 1 mM dithiothreitol, 150 mM KCl, 10% glycerol) replaced HMG-1.
RESULTS
In vivo analysis reveals RAG-1 mutants with specific defects in cleavage or in joining.
To determine whether basic amino acids in RAG-1 are essential for catalysis and to further define regions of RAG-1 that are important in DNA binding and protein-protein interactions, we constructed 70 RAG-1 mutants in which all 86 evolutionarily conserved arginines, lysines, and histidines (71 absolutely conserved; 15 with positive charges conserved) in the truncated active core domain of RAG-1 were changed to alanine (Fig. 1). We analyzed the in vivo phenotypes of all mutants by using an established transient transfection system that allows examination of protein levels (by Western blotting) and V(D)J recombination of plasmid substrates (33). Mutant proteins were generated in the pMAL-17 vector, which encodes the truncated active core domain of RAG-1 fused to His and myc epitope tags (33). All but two mutants produced wild-type levels of RAG-1 protein (R494 and K508 were poorly expressed and were not analyzed further). The remaining 68 mutants were analyzed for their ability to form coding and signal joints in vivo by using three different recombination substrates, pJH290, pJH289, and pJH299. These substrates assay for coding joint formation (with the retention of the coding joint on the plasmid), signal joint formation (with the retention of the signal joint on the plasmid), and inversional recombination (with the retention of both the coding joint and the signal joint on the plasmid), respectively. The results are summarized in Table 1.
Thirty-eight mutants were proficient for recombination, forming signal and coding joints at ≥10% wild-type levels, and 3 mutants consistently exhibited moderate recombination defects, giving between 2 and 10% wild-type levels of recombination. The 27 remaining mutants were severely defective, giving ≤1% recombination in multiple experiments. Eight of these mutants were reconstructed in the context of full-length, untagged RAG-1 and tested in combination with full-length RAG-2. No decrease in the severity of the in vivo recombination defects was observed (Table 1). Two mutants, R391 and R393, were studied previously; they affect critical amino acids in a domain with homology to hin recombinase, which has been implicated in binding to the nonamer (12, 59). As expected, we found these two mutants severely defective for recombination (Table 1) and did not study them further.
We determined the effects of the 25 remaining mutants on cleavage in vivo by using an established semiquantitative ligation-mediated PCR assay for signal ends (60). Twenty-one mutants were severely (at least 100-fold) defective for cleavage (Table 2). Four mutants produced levels of signal ends within 10-fold wild-type levels (Table 2) (see below) and thus were specifically defective for the joining phase of the reaction.
TABLE 2.
In vivo cleavage analysis
| Type of deficiency | Mutant | Signal endsa |
|---|---|---|
| Cleavage | K405/H406/R407 | − |
| R471 | − | |
| H482/K483 | − | |
| K596 | − | |
| K608 | − | |
| R621 | − | |
| R713 | − | |
| R734 | − | |
| R748/H750 | − | |
| R773/R775 | − | |
| H795 | − | |
| R855/K856 | − | |
| K890 | − | |
| R894 | − | |
| H937/K938 | − | |
| H942 | − | |
| K966 | − | |
| R969/R970 | − | |
| R972/K973 | − | |
| R977 | − | |
| K989/H990/H991 | − | |
| Joiningb | R401/R402 | + |
| R440 | + | |
| R838/K839/R840 | + | |
| K980 | + |
Presence (+) or absence (−).
Cleavage within 10-fold that of the wild type.
Analyses of purified proteins.
The 25 mutants with severe in vivo recombination defects were coexpressed as GST fusion proteins along with GST-tagged truncated active core RAG-2 and were purified from mammalian cells. Despite repeated attempts, two mutant proteins, R471 and H482/K483, could not be purified and were not studied further. Two independent protein preparations of each of the 23 remaining mutants were subjected to biochemical analyses. These studies, in combination with the in vivo results described above, allowed us to group mutants into five categories (Table 3). Group 1 mutants have severe or moderate DNA-binding defects, group 2 mutants have catalytic defects and are unable to nick or form hairpins, group 3 mutants can nick but cannot form hairpins, the ability of group 4 mutants to cleave is dependent on coding flank sequences, and group 5 mutants are defective in joining.
TABLE 3.
Functional categorization of recombination-defective mutants
| Group | Mutant | Defect |
|---|---|---|
| 1 | K405/H406/R407 | Impaired DNA binding, nicking, and hairpin formation |
| R748/H750 | ||
| R773/R775 | ||
| H937/K938 | ||
| H942 | ||
| R969/R970 | ||
| K966 | ||
| R977 | ||
| 2 | K596 | Impaired nicking and hairpin formation |
| R621 | ||
| R713a | ||
| R734 | ||
| H795a | ||
| K989/H990/H991 | ||
| 3 | R855/K856b | Impaired hairpin formation |
| K890b | ||
| R894b | ||
| R972/K973 | ||
| 4 | K608 | Conditionally defective in hairpin formation (sensitive to coding flank sequence) |
| 5 | R401/R402c | Impaired joining |
| R440c | ||
| R838/K839/R840d | ||
| K980d |
Cannot form hairpins even on a prenicked substrate.
Also tends to form aberrant hairpins.
Defective for both coding joint and signal joint formation.
More severely defective for coding joint than for signal joint formation.
(i) Group 1 mutants showed impaired DNA binding.
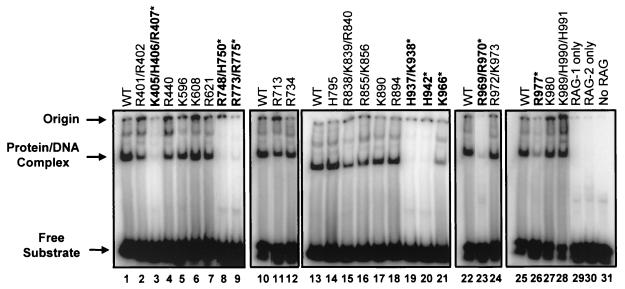
DNA binding was tested by using a standard electrophoretic mobility shift assay (Fig. 2). Most mutants bound to a radiolabeled 12-RSS substrate at or near wild-type levels. However, six mutants (K405/H406/R407, R748/H750, R773/R775, H937/K938, H942, and R969/R970) exhibited consistent, severe (15- to 50-fold) binding defects in multiple experiments (representative data are shown in Fig. 2, lanes 3, 8, 9, 19, 20, and 23), and two mutants (K966 and R977) exhibited consistent but less severe (8- to 10-fold) binding defects (lanes 21 and 26). As would be expected, these eight mutants also showed defects in nicking and hairpin formation (Fig. 3A, lanes 3 to 10). Finally, five mutants showed very mild (≤3-fold) binding defects (R401/R402, R440, R838/K839/R840, R855/K856, and K890) (Fig. 2, lanes 2, 4, 15, 16, and 17), which did not substantially affect their ability to nick the DNA or, in some cases, to form hairpins (see below).
FIG. 2.
Mutations outside of the nonamer-binding domain in RAG-1 affect DNA binding. DNA binding was evaluated by electrophoretic mobility shift assays in which GST-tagged RAG-1 mutants coexpressed with GST-tagged RAG-2 were incubated with a radiolabeled 12-RSS oligonucleotide substrate. The positions of the free substrate, the protein-DNA complex, and the origin are indicated. Mutants with severe DNA-binding defects are in boldface type and are indicated by an asterisk. All lanes within each panel are from the same gel. Quantification was done by PhosphorImager analysis of the results of binding experiments with two independent protein preparations. WT, wild type. (Mutant K966 was previously shown to be proficient for RSS binding [30].)
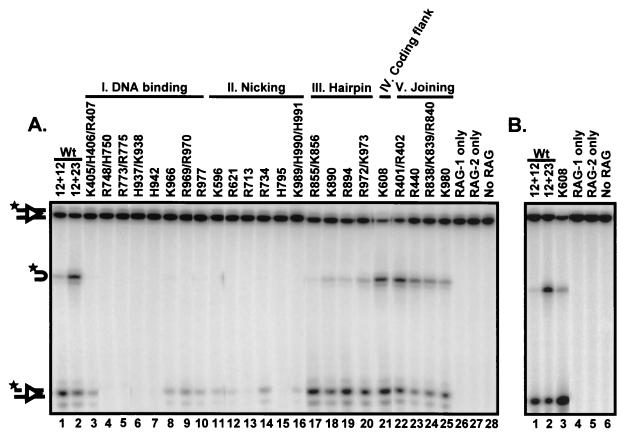
FIG. 3.
Analysis of coupled cleavage of oligonucleotide substrates in Mg2+ allows classification of RAG-1 mutants. GST-tagged RAG-1 mutants coexpressed with GST-tagged RAG-2 were incubated with a radiolabeled 12-RSS oligonucleotide substrate, an unlabeled 23-RSS substrate, and HMG-1 in Mg2+. (A) Coupled cleavage with oligonucleotide substrates. Mutants that were identified as having a DNA-binding defect (panel I; DNA binding) were also severely defective for both nicking and hairpin formation (lanes 3 to 10). The second mutant class (panel II; nicking) was defective for nicking (lanes 11 to 16). The third mutant class (panel III; hairpin) was specifically defective for hairpin formation (lanes 17 to 20). The fourth class of mutants (panel IV; coding flank) contained one mutant, K608, which exhibited wild-type (Wt) levels of nicking and hairpin formation on substrates with permissive coding flanks (5′-TCTTA-heptamer) (lane 21). The fifth class of mutants (panel V; joining) was capable of both nicking and hairpin formation (lanes 22 to 25). Representative data are shown. All lanes are from the same exposure of the same gel but have been rearranged for clarity. The positions of the uncleaved substrate and the nick and hairpin products are indicated by diagrams on the left. The star indicates the position of the radioactive label. (B) Coupled cleavage with nonpermissive coding flank (5′-TCGAC-heptamer) oligonucleotide substrates. Compared to the wild type, mutant K608 exhibited a decrease in hairpin formation with a concurrent accumulation of nicks on substrates with nonpermissive coding flanks (compare lanes 2 and 3), indicating that this mutant is sensitive to the sequence of the coding flank.
Mutations that cause defective DNA binding could directly hinder interactions between RAG-1 and the DNA substrate. Indeed, one mutant that failed to bind to the RSS, K405/H406/R407, has alterations in conserved amino acids that are part of the previously identified nonamer-binding domain (12, 59). Impaired binding to RAG-2, however, could also cause defective DNA binding, since stable RSS binding requires both RAG proteins (24). To test this possibility, we used a GST pull-down assay to test for the interaction of the GST-tagged RAG-1 mutants with myc-tagged RAG-2. None of the GST-tagged RAG-1 DNA-binding mutants showed measurable defects in binding to myc-tagged RAG-2 (data not shown). In reciprocal experiments, GST-tagged RAG-2 was also able to pull down myc-tagged versions of the RAG-1 DNA-binding mutants (data not shown). These data demonstrate that the mutations do not substantially affect the ability of RAG-1 to interact with RAG-2 and also indicate that the mutant proteins are not grossly misfolded. The affected amino acids lie outside the known nonamer-binding domain and cluster in the vicinity of two active-site amino acids, D708 and E962, in the primary sequence. Based on these observations and on additional data (see Discussion), we suggest that some or all of these residues may reside in one or more DNA-binding domains that contact the cleavage site.
(ii) Group 2 mutants showed impaired nicking.
We next examined the ability of the purified mutant proteins to perform coupled cleavage at an RSS pair. Mutant proteins were incubated in Mg2+ with radiolabeled 12-RSS substrate and an unlabeled 23-RSS oligonucleotide. Under these conditions, efficient hairpin formation requires a 12-RSS-23-RSS pair, but nicking does not (25) (Fig. 3A, compare lanes 1 and 2). Six mutants that were capable of RSS binding displayed catalytic defects and were unable to efficiently nick the DNA substrate (Fig. 3A, lanes 11 to 16): K596, R621, R713, R734, H795, and K989/H990/H991.
To determine whether these mutants are capable of catalysis under relaxed conditions, we examined cleavage in Mn2+, which allows nicking and hairpin formation at a single RSS. Mn2+ partially restored nicking and/or hairpin formation by K596, R734, and K989/H990/H991 (Fig. 4, compare lane 1 with lanes 5 to 7), indicating that these amino acids are not absolutely required for catalysis. Three mutants (R621, R713, and H795) remained unable to nick or form hairpins (Fig. 4, lanes 2 to 4). Because nicking is a prerequisite for hairpin formation, these three mutants were tested for their ability to form hairpins with a prenicked oligonucleotide substrate in Mn2+. Under these conditions, R621 was able to form hairpins at nearly wild-type levels (Fig. 5, lane 2); its defect, therefore, is specifically in the nicking step. R713 and H795, however, remained defective for both nicking and hairpin formation (Fig. 5, lanes 3 and 4). Thus, these two mutants, while proficient for DNA binding, are defective for cleavage. These data suggest that R713 and H795, like the previously identified acidic triad (D600, D708, and E962), play critical roles in the catalysis of both cleavage steps.
FIG. 4.
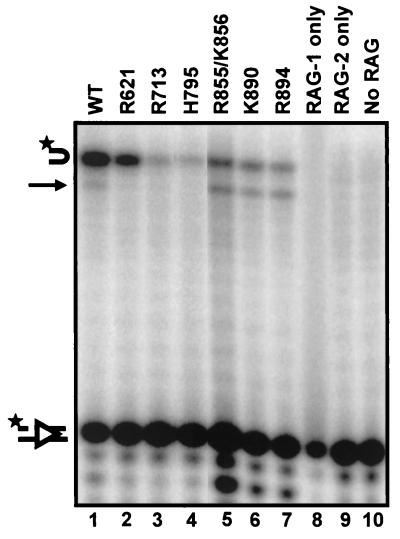
Analysis of nick-defective and hairpin-defective mutants under relaxed RSS cleavage conditions in Mn2+. The 10 mutants showing defective nicking and/or hairpin formation in the coupled cleavage assay were tested for cleavage of a radiolabeled 12-RSS substrate under relaxed cleavage conditions (in Mn2+). Mutants are grouped and labeled according to their phenotypes in Mg2+. Three mutants (R621, R713, and H795) remained defective for nicking (lanes 2 to 4), and three mutants remained specifically defective for hairpin formation (lanes 8 to 10). Interestingly, these hairpin-defective mutants made low levels of aberrant hairpins, indicated by arrows. Representative data are shown. All lanes are from the same exposure of the same gel but have been rearranged for clarity. The positions of the uncleaved substrate, the nick product, and the normal hairpin product are indicated by diagrams on the left. The star indicates the position of the radiolabel. WT, wild type.
FIG. 5.
Prenicked oligonucleotide substrates specifically assay for the hairpin formation step. Mutants that exhibited a nicking defect (lanes 2 to 4) or aberrant hairpin formation (lanes 5 to 7) under relaxed cleavage conditions were tested for hairpin formation with a prenicked substrate in Mn2+, a strategy which bypasses the requirement for the generation of a nick at the correct position. The position of the aberrant hairpin is indicated by an arrow. Representative data are shown. All lanes are from the same exposure of the same gel but have been rearranged for clarity. Lane 5 is overloaded, but quantification reveals ratios of products and substrate similar to those in other lanes. See the legend to Fig. 3 for explanations of symbols. WT, wild type.
(iii) Group 3 mutants showed impaired hairpin formation.
Group 3 mutants (R855/K856, K890, R894, and R972/K973) were capable of nicking but showed specific defects in hairpin formation (Fig. 3A, lanes 17 to 20). These mutants could be unable to perform the chemistry of transesterification or could be defective for other steps required for hairpin formation. One such step is the introduction of DNA distortion at the cleavage site. Several lines of evidence indicate that the RAG proteins must distort DNA in the vicinity of the cleavage site in order to form hairpins (3, 8, 28, 48, 63). Previous work showed that substrates with two mismatched bases at the cleavage site facilitate hairpin formation (8, 48) and can restore activity to a RAG-1 mutant with a specific defect in this reaction (28). We therefore tested the ability of such a mispaired substrate to rescue hairpin formation by the four hairpin-defective mutants. No rescue was observed (data not shown), suggesting that the defect in cleavage is not simply due to the inability of the mutant proteins to induce distortion at the cleavage site.
Mn2+ completely restored catalytic activity to one hairpin-defective mutant, R972/K973 (Fig. 4, lane 11), indicating that it is not completely defective for the ability to perform transesterification. The remaining three hairpin-defective mutants (R855/K856, K890, and R894) formed aberrant hairpins in Mn2+ (Fig. 4, compare lane 1 with lanes 8 to 10). Although the wild-type protein forms some abnormal hairpins in Mn2+, the mutants produce a higher proportion of aberrant hairpins. These mutants may have difficulty positioning the 3′ OH for hairpin formation; alternatively, aberrant hairpins may result from aberrant nicking followed by correct hairpin formation. When a correctly positioned nick was provided by a prenicked substrate, R855/K856, K890, and R894 formed approximately equal proportions of perfect and aberrant hairpins (Fig. 5, lanes 5 to 7), although the levels of hairpins remained lower than those seen with the wild type. These results support the hypothesis that these mutants have difficulty positioning the 3′ OH of the nicked strand for transesterification.
(iv) A group 4 mutant showed a conditional defect in hairpin formation.
One mutant, K608, was severely defective for cleavage in vivo. When tested in vitro, however, it cleaved proficiently (Fig. 3A, compare lanes 2 and 21). We considered the possibility that this difference might result from differences in the substrates used in the two assays. The in vivo substrates contain nonpermissive coding flank sequences that block hairpin formation by two other mutants with mutations located in the same region of RAG-1 (between amino acids 606 and 611), whereas the in vitro experiments were done with oligonucleotides with permissive coding flank sequences that allow hairpin formation by these mutants (28).
We therefore tested the ability of K608 to cleave an oligonucleotide substrate containing the same nonpermissive coding flank sequence as that found in the plasmid (5′-TCGAC-heptamer) in vitro. With this substrate, K608 exhibited defective hairpin formation (Fig. 3B, compare lanes 2 and 3). In agreement with these results, the purified K608 protein was able to cleave plasmid substrates with permissive but not nonpermissive coding flank sequences (data not shown). Mutant K608 thus exhibits a conditional defect in hairpin formation with certain coding flank sequences. It is not clear why this defect was not observed in earlier in vivo studies of this mutant (51).
(v) Group 5 mutants showed joining defects.
The fifth group contains four joining-deficient mutants (R401/R402, R440, R838/K839/R840, and K980). We tested these mutants by using three types of recombination substrates: those that form coding and signal joints by inversion, those that retain coding joints on the plasmid, and those that retain signal joints on the plasmid (diagrammed in Fig. 6A). All four mutants displayed mild in vivo cleavage defects, with approximately 10-fold decreases in the levels of signal ends (Fig. 6F, Table 2, and data not shown).
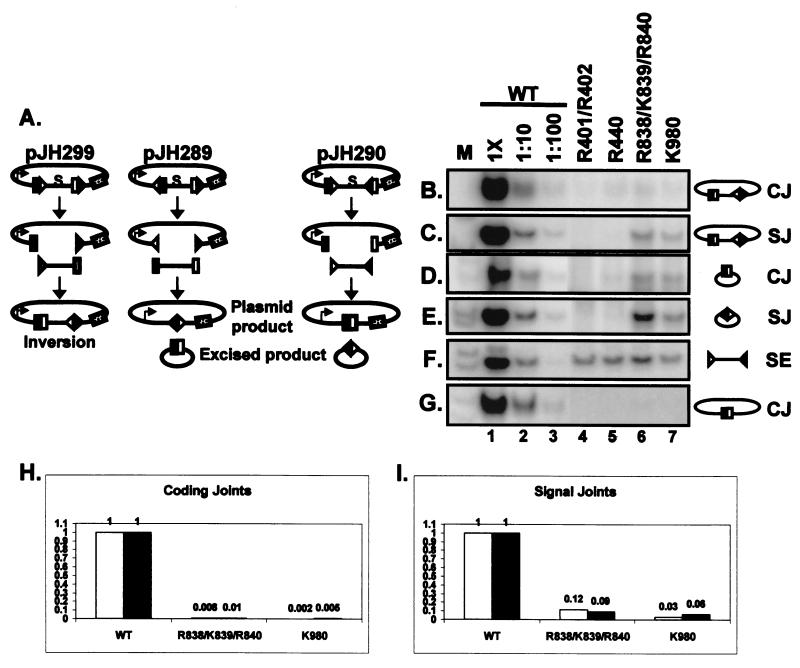
FIG. 6.
Identification of joining-deficient RAG-1 mutants. (A) Diagram illustrating the three recombination substrates used for in vivo analysis of the mutants. pJH299 assays for coding joints and signal joints formed by inversion on the plasmid. pJH289 assays for signal and coding joints formed by deletion, with the formation of signal joints on the plasmid product and coding joints on the excised product. pJH290 assays for coding joints and signal joints formed by deletion, with the formation of coding joints on the plasmid product and signal joints on the excised product. Coding flanks are represented by open and filled rectangles. The 12- and 23-RSS are indicated by open and filled triangles, respectively. The arrow on the plasmid products indicates the promoter for the chloramphenicol acetyltransferase (cat) gene, represented by the stippled rectangle. cat is expressed upon recombination, removing the transcription terminator (represented by S). Joints formed on the plasmid products can be identified by a bacterial transformation assay (23). (B to G) PCR assays (24 cycles) to detect coding joints (CJ) (B, E, and G), signal joints (SJ) (C and E), and signal ends (SE) (F). Diagrams of the PCR substrates are shown on the right. Wild-type (WT)-transfected DNA was assayed at 1× and 1:10 and 1:100 dilutions. The mutants were assayed at 1× concentrations. M, radiolabeled markers. (B) PCR for inversional coding joints on pJH299. (C) PCR for inversional signal joints on pJH299. (D) PCR for coding joints on the excised product of pJH289. (E) PCR for signal joints on the excised product of pJH290. (F) Signal ends (12 RSS) from the pJH290 substrate were detected by ligation-mediated PCR. Analysis of signal ends from the pJH299 substrate gave similar results (data not shown). (G) PCR for coding joints on the plasmid product of pJH290. (H and I) Analysis of coding joints on the plasmid product of pJH290 (H) and signal joints on the plasmid product of pJH289 (I) by a transformation assay. The wild-type result is normalized to 1, and recombination by the mutants is shown relative to that of the wild type. Data from two independent experiments are shown (white and black bars).
In contrast to the mild cleavage defect, R401/R402 and R440 exhibited severe (≥100-fold) joining defects in coding and signal joint formation with all three substrates (Fig. 6B to E and G, lanes 4 and 5). R838/K839/R840 and K980, however, were only mildly defective (approximately 10-fold) for signal joint formation (Fig. 6C and E, lanes 6 and 7). This defect in signal joint formation can be largely accounted for by the partial (10-fold) cleavage defect exhibited by these mutants (Fig. 6F, lanes 4 to 7). These two mutants were severely defective (≥100-fold) for the formation of coding joints with the pJH299 substrate, which recombines by inversion (Fig. 6B, lanes 6 and 7), and the pJH290 substrate, which generates coding joints on the plasmid product (Fig. 6G, lanes 6 and 7). Interestingly, these mutants were only mildly defective (approximately 10-fold) for coding joint formation on the small excised product of pJH289 (Fig. 6D, lanes 6 and 7), suggesting that the defect in coding joint formation can be modified by the configuration of the substrate. A smaller but reproducible increase in signal joint formation was also observed on the excised product.
To confirm these results, we used a standard bacterial transformation assay to detect the plasmid products of V(D)J recombination (23). The measurement of signal joints formed by R838/K839/R840 and K980 from pJH289 revealed that both mutants form signal joints at 5 to 10% wild-type levels (Fig. 6I), consistent with the magnitude of the cleavage defect (Fig. 6F) and the results of PCR assays for signal joints on the inversion substrate (Fig. 6C). Coding joint formation on the pJH290 substrate, however, was severely defective (1% wild-type levels or less) (Fig. 6H), in agreement with the PCR results. Thus, these mutants displayed more severe defects in coding joint formation on the plasmid product (≤1% recombination) than on the excised product (∼10% recombination). This result could be related to the relatively short distance between the ends of the excised product (∼250 nucleotides [nt]), which may allow partial rescue of the joining defect by promoting end-to-end associations (see Discussion).
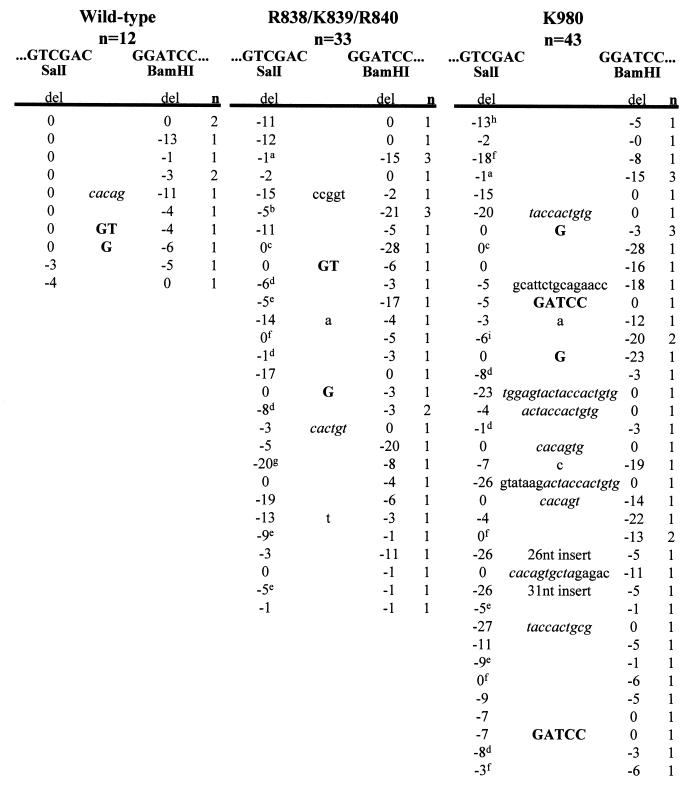
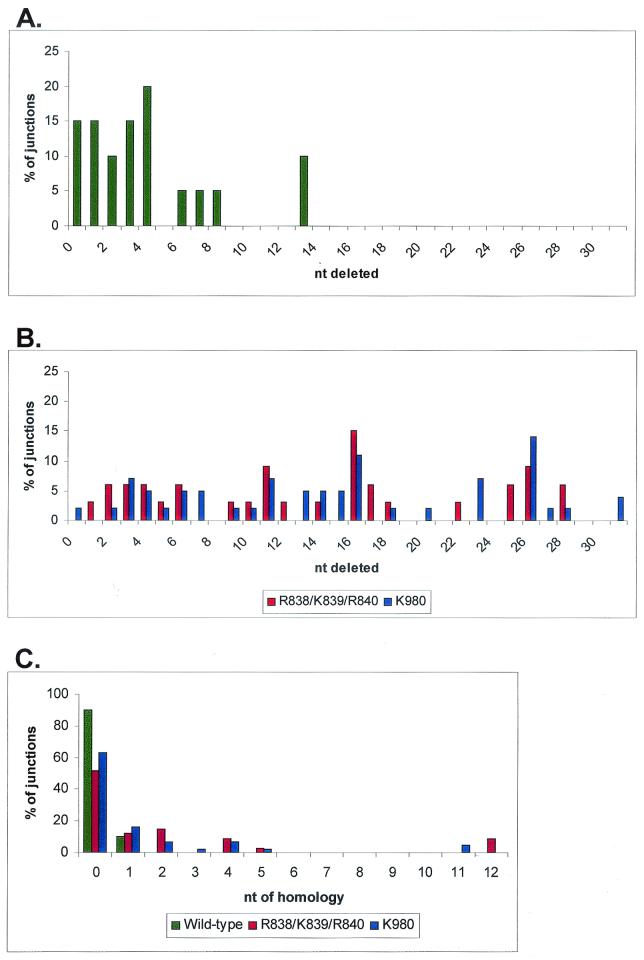
Another interesting feature of coding joints formed on the excised product by these mutants is the smearing of the coding joint products (Fig. 6D, lanes 6 and 7), which was observed in multiple experiments and suggested the presence of junctions with excessive deletions. To test this hypothesis, we performed nucleotide sequence analysis of cloned coding joints derived from the excised product. Whereas 90% of wild-type coding joints lose less than 10 nt (Fig. 7 and 8A) (65), analysis of 76 coding joints from R838/K839/R840 and K980 revealed that 60 to 70% of the junctions showed deletions of greater than 10 nt, with many junctions losing more than 20 nt (Fig. 7 and 8B). These data suggest that certain RAG-1 mutations cause the coding ends to be abnormally exposed to nucleolytic degradation. Notably, the signal joints formed in the presence of these mutants were generally perfect, without addition or loss of nucleotides (data not shown), indicating that excessive deletions were specific to the coding joints.
FIG. 7.
Nucleotide sequence analysis of coding joints from R838/K839/R840 and K980. PCR products containing coding joints were cloned and sequenced. The total numbers of unique sequences obtained for the wild type, R838/K839/R840, and K980 are indicated. The first 6 nt on either side of a perfect junction are shown. A perfect coding joint is GGTCGTTGATCCCCCATCGATGAGAGTCGAC-GGATCCTCTCATCGATGAGAGGATCGACGACGACATGGC. n, number of unique junctions with the indicated sequence. The number of deleted (del) nucleotides from each end is indicated beneath the corresponding end sequences. Letters in boldface type between the two ends indicate presumptive P nucleotides. Lowercase letters between the two ends indicate nucleotide insertions. Italics denote nucleotides derived from imprecise cleavage events. The sequence of the 26-nt insert is CTAGAGTCGATCCGTCCCCGGGCGAG. The sequence of the 31-nt insert is CAAAACCCTCTGTAACTCTAGATCCAGGAAT. Superscripts designate junctions that exhibit short sequence homologies, as follows: a, TCGA; b, CATCGATGAGAG; c, TCGAC; d, GA; e, G; f, C; g, TC; h, ATC; I, CATCGATGAGA.
FIG. 8.
Coding joint deletions and short sequence homologies. Distribution of the number of nucleotides deleted from coding joints from the wild type (A) (includes sequences from this study and reference 47) and R838/K839/R840 and K980 (B). The x axis represents the total number of nucleotides lost from both coding ends. Data are from Fig. 7. (C) Mutant coding joints exhibit short sequence homologies at the coding junction. The x axis represents the number of homologous nucleotides at the junction. Data are from Fig. 7.
The coding joints formed by these two RAG-1 mutants are reminiscent of the coding joints seen in cells with defects in nonhomologous end-joining factors (such as DNA-PKcs), which also show frequent excessive deletions. Therefore, we searched for another feature characteristic of coding joints formed in DNA-PKcs mutants: excessive P nucleotide insertions. P nucleotide insertions result from opening of the hairpin a few nucleotides away from the tip and are almost always <3 nt (31, 41). In DNA-PKcs-deficient cells, however, longer P nucleotide insertions are observed (29, 53, 65), indicating that in addition to impairing hairpin opening, this mutation also causes opening at abnormal positions. Indeed, two different coding joint sequences isolated from mutant K980 exhibited abnormally long (5-nt) P nucleotide additions, both derived from the same coding end (Fig. 7). A large survey of P nucleotides generated by wild-type RAG proteins from substrates with the same coding end as that used in our study failed to detect P nucleotides >3 nt in length associated with this end (41). Thus, the presence of 5-nt P nucleotide insertions at ∼5% of the junctions produced by the K980 mutant provides strong evidence that the mutation in this RAG-1 mutant causes abnormal hairpin opening.
Another feature characteristic of coding joints formed in double-strand break repair-deficient cells, including cells lacking DNA-PKcs, is the frequent presence of short sequence homologies (5, 37). Although only 10% of coding joints made by wild-type RAG proteins exhibit junctional homologies (Fig. 8C), 40 to 50% of the junctions made by R838/K839/R840 and K980 contain short sequence homologies (Fig. 8C). These data suggest that coding joint formation by these mutants may be facilitated by base-pairing interactions. Finally, a large fraction of the coding joints made by mutant K980 contain large insertions of random DNA, another feature characteristic of coding joints made in Ku- and DNA-PKcs-deficient cells (5, 65).
To gain further insights into the mechanisms of the joining defects, all four mutants were examined biochemically as purified proteins. As expected, all were capable of both nicking and hairpin formation in vitro (Fig. 3A, lanes 22 to 25), with only mild defects that reflect the modest decrease in signal ends observed in vivo. All mutants transposed signal ends at wild-type levels in vitro (data not shown), indicating that the mutant RAG proteins remain associated with the signal ends after cleavage in a postcleavage complex. Furthermore, as observed previously with some coding joint-deficient RAG mutants (47, 54), all four joining mutants opened oligonucleotide hairpins in a standard hairpin opening assay performed in Mn2+ (data not shown). Nevertheless, the specific defect in coding joint formation, along with the presence of excessive P nucleotide insertions, strongly suggests that mutant K980 is defective for hairpin opening in vivo. Clearly, while the in vitro assay (which tests the opening of artificial hairpins in Mn2+) is capable of detecting mutants that lack the catalytic capability to open hairpins (47, 54), it does not recapitulate all physiologically relevant aspects of this reaction (such as its dependence upon DNA-PK in vivo). Thus, we suggest that the K980 mutation impairs a regulatory aspect of the hairpin opening reaction, such as the ability of the postcleavage complex to interact with DNA-PK.
DISCUSSION
Basic residues in the C terminus of RAG-1 that are important for DNA binding.
DNA footprinting and DNA-protein cross-linking studies have shown that RAG-1 alone contacts the nonamer region of the RSS (3, 36, 43, 46). In the presence of RAG-2, the RAG-1 footprint extends into the heptamer and the coding flank (14, 42, 43, 64). Although there is extensive information about the footprint of RAG-1 on DNA, very little is known about the specific regions of RAG-1 that are involved in making those contacts. A nonamer-binding domain (amino acids 376 to 477) in the N terminus of RAG-1, which has homology to the homeodomain of the hin recombinase (59), is the only well-characterized DNA-binding domain in RAG-1. Deletions and point mutations in this region disrupt DNA binding (12, 59). Consistent with these observations, we found that the K405/H406/R407 mutation, located in the hin homology region, is defective for DNA binding.
Besides binding to the nonamer, the RAG complex must also contact the heptamer and the DNA at the border of the cleavage site. Recent DNA-protein cross-linking studies have suggested that the C terminus of RAG-1, specifically a region between methionine residues 889 and 974, contacts the DNA at the coding flank (44). Our data show that several basic residues within this region (H937/K938, H942, K966, and R969/R970) are essential for DNA binding. One additional mutation, R977, which lies just outside this region, also causes a moderate binding defect. Importantly, these mutants retain the ability to interact with RAG-2, suggesting that they are specifically defective for DNA-protein interactions (although we cannot completely rule out the possibility that some of these mutants may have defects in RAG-1 dimerization). Together, these data support a role for the C terminus of RAG-1 in DNA binding and identify specific basic residues that contribute to binding.
The region of RAG-1 encompassing the C2H2 zinc-binding domain (amino acids 723 to 754), termed ZFB, is involved in nonspecific DNA binding (49) and RAG-2 interactions (2). Residues R748 and H750 are located in the zinc finger region, and H750 is predicted to help coordinate zinc. Mutation of the cysteine or histidine ligands in a zinc finger most often results in a loss of function (68). We found that the R748/H750 mutant is defective for DNA binding but maintains the ability to interact with RAG-2, suggesting that the zinc finger is essential for DNA binding but perhaps not for RAG-2 interaction.
Step arrest mutants that specifically block nicking or hairpin formation.
Although three active-site residues, D600, D708, and E962, are required for both nicking and hairpin formation (16, 30, 33), each of these steps is likely to also involve some distinct active-site amino acids. Our R621A mutant is severely and specifically defective for nicking, in agreement with recent studies of R621H, which was found in patients with B-cell-negative SCID (35). We also identified two classes of mutants with specific defects in hairpin formation. The first class consists of the K608A mutant, which is defective for hairpin formation in the presence of certain nonpermissive coding flank sequences. Two other mutations located between amino acids 606 to 611 share similar properties and, like K608A, have been implicated in generating a distorted DNA intermediate required for hairpin formation (28).
The second class of hairpin-defective mutants contains four mutants (R855/K856, K890, R894, and R972/K973) with novel phenotypes. Unlike the RAG-1 mutants described above, these mutants are not sensitive to the coding flank sequence and are not rescued by coding flank sequences containing mismatches; their defect in hairpin formation is not obviously related to problems with distortion of the DNA at the cleavage site. Three of these mutants (R855/K856, K890, and R894) remain defective for accurate hairpin formation even under relaxed cleavage conditions (in Mn2+) that allow efficient hairpin formation at a single RSS. Interestingly, all three mutants form hairpins at aberrant locations in Mn2+, even with a prenicked substrate. These data suggest that they are unable to correctly position the attacking nucleophile (the 3′ OH) in the transesterification reaction. Other related systems, such as human immunodeficiency virus type 1 integrase and Tn5, use basic residues to position the DNA close to the active site (10, 26). Perhaps R855/K856, K890, and R894 serve a similar role in RAG-1.
Catalytic residues important for both nicking and hairpin formation.
Structural analyses of the active sites of several transposases and integrases have revealed the presence of basic amino acids (6, 10, 22). Mutational analysis of these residues has shown that they are essential for both cleavage and strand transfer (6, 22, 26). In addition to orienting the DNA in the active site, basic residues in Tn5 play a role in catalysis by stabilizing a DNA bend (10). Our analysis revealed two residues in RAG-1 (R713 and H795) that, when mutated, produce severe defects in both nicking and hairpin formation without affecting DNA binding (a defect in nicking by the R713 mutant was reported earlier [30]). Our data indicate that R713 and H795 are essential for the catalysis of both cleavage steps.
Basic residues in RAG-1 that are required for joining.
We identified four RAG-1 mutants (R401/R402, R440, R838/K839/R840, and K980) that specifically affect the joining phase of V(D)J recombination. All four mutants are severely impaired for coding joint formation; two mutants (R401/402 and R440) also have a block in signal joint formation. Like other recently described RAG-1 and RAG-2 joining-deficient mutants which are specifically defective for both coding and signal joint formation, R401/R402 and R440 retain the ability to open artificial hairpins in vitro (47, 54). These mutants may be defective for recruiting joining factors, as their joining phenotype is similar to that observed in Ku- and XRCC4-deficient cells.
The remaining two joining mutants identified in this study (R838/K839/R840 and K980) share two novel phenotypes. The first of these is the differential effect of substrate length on coding joint formation (and, to a lesser degree, signal joint formation). A salient feature of the excised product, which permits coding joint formation, is the short distance between the coding ends (∼250 nt). This short distance may facilitate joining by increasing the probability of end-end interactions. Support for this hypothesis is provided by studies of intramolecular ligation, which have shown that recircularization is favored at substrate lengths of between a few hundred to approximately 1,000 bp; above these lengths, the ends begin to behave as if they are on separate DNA molecules (57).
The second novel phenotype of these mutants is their effect on the structure of the coding joints that can be recovered. The sequences of these joints are distinctly abnormal, with features characteristic of the rare junctions recovered from cells bearing double-strand break repair mutations, including excessive deletions and short sequence homologies at the junctions. Previous RAG joining mutations had not been found to affect the structure of these junctions (47, 54). The observation that mutations in RAG-1 and double-strand break repair factors affect the structure of coding joints in similar ways provides strong support for the idea that the RAG proteins are intimately involved in the joining process. These RAG-1 mutants may release coding ends prematurely or exhibit defects in recruiting one or more of the nonhomologous end-joining factors, leaving the coding ends to be joined by alternative repair pathways that allow more exonucleolytic degradation. The observation that the only coding joints that could be recovered from these two RAG-1 mutants are formed on the short excised product and frequently use short sequence homologies suggests that the alternative joining pathways require assistance in aligning the ends for joining. Thus, one important function of the RAG proteins in the postcleavage complex may be to serve as a scaffold to assist end joining.
The phenotypes of these two RAG-1 mutants share many striking parallels with the effects of DNA-PKcs deficiency on V(D)J recombination, including the effects on the junction structures noted above and also the fact that coding joints are more severely affected than signal joints. As noted above, the abnormally long P nucleotide inserts observed in coding joints formed by K980 are characteristic of coding joints recovered from cells lacking DNA-PKcs and suggest a defect in hairpin opening. These observations provide strong evidence that the RAG proteins play a role in hairpin opening in vivo and suggest that the RAG proteins and DNA-PKcs collaborate in the joining process.
Refining the functional map of RAG-1.
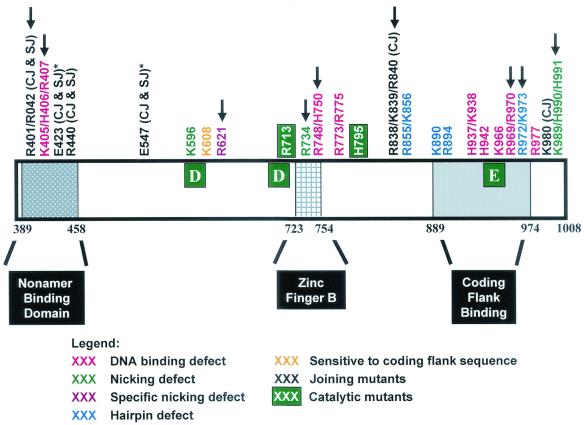
Mapping of mutations along the RAG-1 primary sequence revealed several phenotypic classes that cluster in specific regions of the sequence (Fig. 9). Hairpin-defective mutants implicated in DNA distortion lie in the region of amino acids 606 to 611, which is near the active-site residue, D600. Two joining-deficient mutants with defects in both coding and signal joint formation (R401/R402 and R440) are located in the N terminus, where two previously identified RAG-1 joining mutants (E423Q and E547Q) are also found (54). Furthermore, we found a cluster of joining mutants in the C terminus of the protein, including the R838/K839/R840 and K980 mutants. Preliminary analyses of other mutants have revealed three additional joining mutations in the same region of RAG-1 (unpublished observations). These data suggest that RAG-1 contains two domains important for joining: an N-terminal domain that contacts the nonamer and a C-terminal domain that contacts the coding flank. Interestingly, all five C-terminal joining mutants have specific defects in coding joint formation without a significant impairment in signal joints. Thus, the C-terminal domain, which contacts the coding flank prior to cleavage (44), may also play a special role in coding joint formation, perhaps by binding to the hairpin coding ends.
FIG. 9.
Schematic diagram of the core domain of RAG-1 (amino acids 384 to 1008) mapping the phenotypic classes of mutants identified in this study. The active-site residues, D600, D708, and E962, are shown as large white letters in green boxes. The nonamer-binding domain, zinc finger B, and coding flank region are also shown. Asterisks indicate two acidic residues previously identified as joining mutants (54). Arrows indicate mutants with corresponding mutants found in T− B− Scid and Omenn syndrome patients. The human mutations are R396C, R396L, or R396H; R404W; R410Q; R474H; R507W; R624H or R624C; R737H; H753L; R841W; R973H; R975Q; and K992E (7, 55, 66, 67).
Two additional groups of mutants cluster in the C terminus of the protein near the active-site residue, E962, further emphasizing the importance of this residue. One cluster in this region includes mutants with a specific defect in hairpin formation, which we suggest is due to the inappropriate positioning of the attacking nucleophile (3′-OH) for the transesterification reaction. Several of the DNA-binding mutants that we identified (H937/K938, H942, K966, R969/R970, and R977) are also located in this region. These DNA-binding residues, which are in or near a region of the protein that contacts the coding flank, may participate in one or more DNA-binding domains that are responsible for bringing the active-site residues into close proximity with the cleavage site.
Clinical implications.
It is interesting that 12 of our recombination-deficient RAG-1 mutations alter residues mutated in patients with inherited immunodeficiency syndromes (7, 55, 66, 67). Clearly, these amino acids are critical for recombination of the endogenous antigen receptor loci in developing lymphocytes. The phenotypes of our alanine substitution mutants suggest that patient mutations may fall into the following functional classes: DNA binding (R393, R407, H750, and R970), nicking (R621, R734, and K989), hairpin-forming (R972), and joining (R401 and R838).
Perhaps even more interesting is the similarity between the effects of our RAG joining mutants and mutants with mutations in DNA-PKcs or other nonhomologous end-joining (NHEJ) factors (defects in coding and signal joint formation or structurally aberrant coding joints). The NHEJ factors were recently designated “genome guardians” because mice lacking DNA-PKcs, Ku70, XRCC4, or Ku80 suffer genomic instability and an increased incidence of lymphomas (9, 13, 17, 20, 27, 38). These lymphomas, in fact, bear characteristic chromosome translocations indicative of mistakes in V(D)J recombination. Like mutations in DNA-PKcs and other NHEJ proteins, RAG joining mutations may increase the frequency of oncogenic DNA rearrangements catalyzed by the V(D)J recombinase.
Acknowledgments
We thank Magdalena Walkiewicz for technical support; Heather Yarnall Schultz, Sam Kale, Matt Neiditch, Greg Lee, and Jian-Xia Qiu for critical input; Barbara Corneo for helpful discussions; and Vicky Brandt for helpful suggestions on the manuscript.
This work was supported by a grant from the National Institutes of Health (AI-36420). L.E.H was supported by a National Institutes of Health predoctoral fellowship (T32-AI07495). M.M.P. was supported by a fellowship from the Cancer Research Institute. D.B.R. is an Assistant Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Agrawal, A., and D. G. Schatz. 1997. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell 89:43-53. [DOI] [PubMed] [Google Scholar]
- 2.Aidinis, V., D. C. Dias, C. A. Gomez, D. Bhattacharyya, E. Spanopoulou, and S. Santagata. 2000. Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J. Immunol. 164:5826-5832. [DOI] [PubMed] [Google Scholar]
- 3.Akamatsu, Y., and M. A. Oettinger. 1998. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol. Cell. Biol. 18:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailin, T., X. Mo, and M. J. Sadofsky. 1999. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol. Cell. Biol. 19:4664-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogue, M. A., C. Wang, C. Zhu, and D. B. Roth. 1997. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity 7:37-47. [DOI] [PubMed] [Google Scholar]
- 6.Bolland, S., and N. Kleckner. 1996. The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell 84:223-233. [DOI] [PubMed] [Google Scholar]
- 7.Corneo, B., D. Moshous, T. Gungor, N. Wulffraat, P. Philippet, F. L. Le Deist, A. Fischer, and J. P. de Villartay. 2001. Identical mutations in RAG1 or RAG2 genes leading to defective V(D)J recombinase activity can cause either T-B-severe combined immune deficiency or Omenn syndrome. Blood 97:2772-2776. [DOI] [PubMed] [Google Scholar]
- 8.Cuomo, C. A., C. L. Mundy, and M. A. Oettinger. 1996. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol. 16:5683-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Custer, R. P., G. C. Bosma, and M. J. Bosma. 1985. Severe combined immunodeficiency in the mouse: pathology, reconstitution, neoplasms. Am. J. Pathol. 120:464-477. [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, D. R., I. Y. Goryshin, W. S. Reznikoff, and I. Rayment. 2000. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289:77-85. [DOI] [PubMed] [Google Scholar]
- 11.Deng, W. P., and J. A. Nickoloff. 1992. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem. 200:81-88. [DOI] [PubMed] [Google Scholar]
- 12.Difilippantonio, M. J., C. J. McMahan, Q. M. Eastman, E. Spanopoulou, and D. G. Schatz. 1996. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell 87:253-262. [DOI] [PubMed] [Google Scholar]
- 13.Difilippantonio, M. J., J. Zhu, H. T. Chen, E. Meffre, M. C. Nussenzweig, E. E. Max, T. Ried, and A. Nussenzweig. 2000. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eastman, Q. M., I. J. Villey, and D. G. Schatz. 1999. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol. Cell. Biol. 19:3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fugmann, S., and D. G. Schatz. 2001. Identification of basic residues in RAG2 critical for DNA binding by the RAG1-RAG2 complex. Mol. Cell 8:899-910. [DOI] [PubMed] [Google Scholar]
- 16.Fugmann, S. D., I. J. Villey, L. M. Ptaszek, and D. G. Schatz. 2000. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell 5:97-107. [DOI] [PubMed] [Google Scholar]
- 17.Gao, Y., D. O. Ferguson, W. Xie, J. P. Manis, J. Sekiguchi, K. M. Frank, J. Chaudhuri, J. Horner, R. A. DePinho, and F. W. Alt. 2000. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404:897-900. [DOI] [PubMed] [Google Scholar]
- 18.Ge, H., and R. Roeder. 1994. The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J. Biol. Chem. 269:17136-17140. [PubMed] [Google Scholar]
- 19.Grawunder, U., R. B. West, and M. R. Lieber. 1998. Antigen receptor gene rearrangement. Curr. Opin. Immunol. 10:172-180. [DOI] [PubMed] [Google Scholar]
- 20.Gu, Y., K. J. Seidl, G. A. Rathbun, C. Zhu, J. P. Manis, N. van der Stoep, L. Davidson, H.-L. Cheng, J. M. Sekiguchi, K. Frank, P. Stanhope-Baker, M. S. Schlissel, D. B. Roth, and F. W. Alt. 1997. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7:653-665. [DOI] [PubMed] [Google Scholar]
- 21.Han, J.-O., S. B. Steen, and D. B. Roth. 1997. Ku86 is not required for protection of signal ends or for formation of nonstandard V(D)J recombination products. Mol. Cell. Biol. 17:2226-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. [DOI] [PubMed] [Google Scholar]
- 23.Hesse, J. E., M. R. Lieber, M. Gellert, and K. Mizuuchi. 1987. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V(D)J joining signals. Cell 49:775-783. [DOI] [PubMed] [Google Scholar]
- 24.Hiom, K., and M. Gellert. 1997. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell 88:65-72. [DOI] [PubMed] [Google Scholar]
- 25.Hiom, K., and M. Gellert. 1998. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell 1:1011-1019. [DOI] [PubMed] [Google Scholar]
- 26.Jenkins, T. M., D. Esposito, A. Engelman, and R. Craigie. 1997. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 16:6849-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jhappan, C., H. C. Morse, R. D. Fleischmann, M. M. Gottesman, and G. Merlino. 1997. DNA-PKcs: a T-cell tumour suppressor encoded at the mouse scid locus. Nat. Genet. 17:483-486. [DOI] [PubMed] [Google Scholar]
- 28.Kale, S. B., M. A. Landree, and D. B. Roth. 2001. Conditional RAG-1 mutants block the hairpin formation step of V(D)J recombination. Mol. Cell. Biol. 21:459-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kienker, L. J., W. A. Kuziel, and P. W. Tucker. 1991. T cell receptor γ and δ gene junctional sequences in scid mice: excessive P nucleotide insertion. J. Exp. Med. 174:769-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, D. R., Y. Dai, C. L. Mundy, W. Yang, and M. A. Oettinger. 1999. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 13:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lafaille, J. J., A. DeCloux, M. Bonneville, Y. Takagaki, and S. Tonegawa. 1989. Junctional sequences of T cell receptor γδ genes: implications for γδ T cell lineages and for a novel intermediate of V(D)J joining. Cell 59:859-870. [DOI] [PubMed] [Google Scholar]
- 32.Landree, M. A., S. B. Kale, and D. B. Roth. 2001. Functional organization of single and paired V(D)J cleavage complexes. Mol. Cell. Biol. 21:4256-4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landree, M. A., J. A. Wibbenmeyer, and D. B. Roth. 1999. Mutational analysis of RAG-1 and RAG-2 identifies three active site amino acids in RAG-1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 13:3059-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis, S. M. 1994. The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv. Immunol. 56:27-150. [DOI] [PubMed] [Google Scholar]
- 35.Li, W., F. C. Chang, and S. Desiderio. 2001. Rag-1 mutations associated with B-cell-negative scid dissociate the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 21:3935-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, W., P. Swanson, and S. Desiderio. 1997. RAG-1- and RAG-2-dependent assembly of functional complexes with V(D)J recombination substrates in solution. Mol. Cell. Biol. 17:6932-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, Z., T. Otevrel, Y. Gao, H.-L. Cheng, B. Seed, T. D. Stamato, G. E. Taccioli, and F. W. Alt. 1995. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell 83:1079-1089. [DOI] [PubMed] [Google Scholar]
- 38.Lim, D. S., H. Vogel, D. M. Willerford, A. T. Sands, K. A. Platt, and P. Hasty. 2000. Analysis of Ku80-mutant mice and cells with deficient levels of p53. Mol. Cell. Biol. 20:3772-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBlane, J. F., D. C. van Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 40.McMahan, C. J., M. J. Sadofsky, and D. G. Schatz. 1997. Definition of a large region of RAG1 that is important for coimmunoprecipitation of RAG2. J. Immunol. 158:2202-2210. [PubMed] [Google Scholar]
- 41.Meier, J. T., and S. M. Lewis. 1993. P nucleotides in V(D)J recombination: a fine-structure analysis. Mol. Cell. Biol. 13:1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mo, X., T. Bailin, S. Noggle, and M. J. Sadofsky. 2000. A highly ordered structure in V(D)J recombination cleavage complexes is facilitated by HMG1. Nucleic Acids Res. 28:1228-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mo, X., T. Bailin, and M. J. Sadofsky. 1999. RAG1 and RAG2 cooperate in specific binding to the recombination signal sequence in vitro. J. Biol. Chem. 274:7025-7031. [DOI] [PubMed] [Google Scholar]
- 44.Mo, X., T. Bailin, and M. J. Sadofsky. 2001. A C-terminal region of RAG1 contacts the coding DNA during V(D)J recombination. Mol. Cell. Biol. 21:2038-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moshous, D., I. Callebaut, R. de Chasseval, B. Corneo, M. Cavazzana-Calvo, F. Le Deist, I. Tezcan, O. Sanal, Y. Bertrand, N. Philippe, A. Fischer, and J. P. de Villartay. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 20:177-186. [DOI] [PubMed] [Google Scholar]
- 46.Nagawa, F., K.-I. Ishiguro, A. Tsuboi, T. Yoshida, A. Ishikawa, T. Takemori, A. J. Otsuka, and H. Sakano. 1998. Footprint analysis of the RAG protein recombination signal sequence complex for V(D)J type recombination. Mol. Cell. Biol. 18:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu, J., S. B. Kale, H. Y. Schultz, and D. B. Roth. 2001. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol. Cell 7:77-87. [DOI] [PubMed] [Google Scholar]
- 48.Ramsden, D. A., J. F. McBlane, D. C. van Gent, and M. Gellert. 1996. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 15:3197-3206. [PMC free article] [PubMed] [Google Scholar]
- 49.Rodgers, K. K., Z. Bu, K. G. Fleming, D. G. Schatz, D. M. Engelman, and J. E. Coleman. 1996. A zinc-binding domain involved in the dimerization of RAG1. J. Mol. Biol. 260:70-84. [DOI] [PubMed] [Google Scholar]
- 50.Rodgers, K. K., I. J. Villey, L. Ptaszek, E. Corbett, D. G. Schatz, and J. E. Coleman. 1999. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 27:2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roman, C. A. J., and D. Baltimore. 1996. Genetic evidence that the RAG1 protein directly participates in V(D)J recombination through substrate recognition. Proc. Natl. Acad. Sci. USA 93:2333-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawchuk, D. J., F. Weis-Garcia, S. Malik, E. Besmer, M. Bustin, M. C. Nussenzweig, and P. Cortes. 1997. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J. Exp. Med. 185:2025-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuler, W., N. R. Ruetsch, M. Amsler, and M. J. Bosma. 1991. Coding joint formation of endogenous T cell receptor genes in lymphoid cells from scid mice: unusual P-nucleotide additions in VJ-coding joints. Eur. J. Immunol. 21:589-596. [DOI] [PubMed] [Google Scholar]
- 54.Schultz, H. Y., M. A. Landree, J. Qiu, S. B. Kale, and D. B. Roth. 2001. Joining-Deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Mol. Cell 7:65-75. [DOI] [PubMed] [Google Scholar]
- 55.Schwarz, K., G. H. Gauss, L. Ludwig, U. Pannicke, Z. Li, D. Lindner, W. Friedrich, R. A. Seger, T. E. Hansen-Hagge, S. Desiderio, M. R. Lieber, and C. R. Bartram. 1998. RAG mutations in human B cell-negative SCID. Science 274:97-99. [DOI] [PubMed] [Google Scholar]
- 56.Selent, U., T. Ruter, E. Kohler, M. Liedtke, V. Thielking, J. Alves, T. Oelgeschlager, H. Wolfes, F. Peters, and A. Pingoud. 1992. A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry 31:4808-4815. [DOI] [PubMed] [Google Scholar]
- 57.Shore, D., J. Langowski, and R. L. Baldwin. 1981. DNA flexibility studies by covalent closure of short fragments into circles. Proc. Natl. Acad. Sci. USA 78:4833-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spanopoulou, E., P. Cortes, C. Shih, C.-M. Huang, D. P. Silver, P. Svec, and D. Baltimore. 1995. Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity 3:715-726. [DOI] [PubMed] [Google Scholar]
- 59.Spanopoulou, E., F. Zaitseva, F.-H. Wang, S. Santagata, D. Baltimore, and G. Panayotou. 1996. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell 87:263-276. [DOI] [PubMed] [Google Scholar]
- 60.Steen, S. B., L. Gomelsky, and D. B. Roth. 1996. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells 1:543-553. [DOI] [PubMed] [Google Scholar]
- 61.Steen, S. B., L. Gomelsky, S. L. Speidel, and D. B. Roth. 1997. Initiation of V(D)J recombination in vivo: role of recombination signal sequences in formation of single and paired double-strand breaks. EMBO J. 16:2656-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swanson, P. C. 2001. The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 21:449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swanson, P. C., and S. Desiderio. 1998. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity 9:115-125. [DOI] [PubMed] [Google Scholar]
- 64.Swanson, P. C., and S. Desiderio. 1999. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 19:3674-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taccioli, G. E., H.-L. Cheng, A. J. Varghese, G. Whitmore, and F. W. Alt. 1994. A DNA repair defect in Chinese hamster ovary cells affects V(D)J recombination similarly to the murine scid mutation. J. Biol. Chem. 269:7439-7442. [PubMed] [Google Scholar]
- 66.Villa, A., S. Santagata, F. Bozzi, S. Giliani, A. Frattini, L. Imberti, L. B. Gatta, H. D. Ochs, K. Schwarz, L. D. Notarangelo, P. Vezzoni, and E. Spanopoulou. 1998. Partial V(D)J recombination activity leads to Omenn syndrome. Cell 93:885-896. [DOI] [PubMed] [Google Scholar]
- 67.Villa, A., C. Sobacchi, L. D. Notarangelo, F. Bozzi, M. Abinun, T. G. Abrahamsen, P. D. Arkwright, M. Baniyash, E. G. Brooks, M. E. Conley, P. Cortes, M. Duse, A. Fasth, A. M. Filipovich, A. J. Infante, A. Jones, E. Mazzolari, S. M. Muller, S. Pasic, G. Rechavi, M. G. Sacco, S. Santagata, M. L. Schroeder, R. Seger, D. Strina, A. Ugazio, J. Valiaho, M. Vihinen, L. B. Vogler, H. Ochs, P. Vezzoni, W. Friedrich, and K. Schwarz. 2001. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood 97:81-88. [DOI] [PubMed] [Google Scholar]
- 68.Wolfe, S. A., L. Nekludova, and C. O. Pabo. 2000. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 29:183-212. [DOI] [PubMed] [Google Scholar]