Abstract
The essential Saccharomyces cerevisiae gene BDP1 encodes a subunit of RNA polymerase III (Pol III) transcription factor (TFIIIB); TATA box binding protein (TBP) and Brf1 are the other subunits of this three-protein complex. Deletion analysis defined three segments of Bdp1 that are essential for viability. A central segment, comprising amino acids 327 to 353, was found to be dispensable, and cells making Bdp1 that was split within this segment, at amino acid 352, are viable. Suppression of bdp1 conditional viability by overexpressing SPT15 and BRF1 identified functional interactions of specific Bdp1 segments with TBP and Brf1, respectively. A Bdp1 deletion near essential segment I was synthetically lethal with overexpression of PCF1-1, a dominant gain-of-function mutation in the second tetracopeptide repeat motif (out of 11) of the Tfc4 (τ131) subunit of TFIIIC. The analysis also identifies a connection between Bdp1 and posttranscriptional processing of Pol III transcripts. Yeast genomic library screening identified RPR1 as the specific overexpression suppressor of very slow growth at 37°C due to deletion of Bdp1 amino acids 253 to 269. RPR1 RNA, a Pol III transcript, is the RNA subunit of RNase P, which trims pre-tRNA transcript 5′ ends. Maturation of tRNA was found to be aberrant in bdp1-Δ253-269 cells, and RPR1 transcription with the highly resolved Pol III transcription system in vitro was also diminished when recombinant Bdp1Δ253-269 replaced wild-type Bdp1. Physical interaction of RNase P with Bdp1 was demonstrated by coimmunoprecipitation and pull-down assays.
RNA polymerase III (Pol III) transcribes genes encoding tRNAs, 5S rRNA, U6 snRNA, and other small RNAs. Accurate initiation of this transcription requires basal transcription factor IIIA (TFIIIA), TFIIIB, and TFIIIC. In the yeast Saccharomyces cerevisiae, TFIIIB is required for all Pol III transcription in vitro and in vivo, TFIIIC is required for all Pol III transcription in vivo, and TFIIIA is required only for 5S rRNA gene transcription. TFIIIB is composed of three subunits, TATA-binding protein (TBP), Brf1 (the TFIIB-related factor), and Bdp1; all three are also essential for Pol III transcription in vitro (7, 12, 14, 21, 27, 28, 33, 40, 51, 52, 65). In vivo analysis defines multiple interactions of TFIIIB with the rest of the S. cerevisiae Pol III transcription machinery: Brf1 and Bdp1 interact with Tfc4 (the second largest subunit of TFIIIC), and Brf1 also interacts with the RPC34 and RPC17 subunits of Pol III (3, 5, 19, 20, 39, 47, 52, 64).
In human cells, two TFIIIB-related assemblies have been identified (46, 60). TFIIIBα, which contains TBP, Bdp1 (previously called hTFIIIB150), and Brf2 (a hBrf1 paralogue previously called hTFIIIB50), is required for transcription of Pol III genes with upstream promoter elements, such as 7SK and U6 (53, 61). TFIIIBβ, containing TBP, Bdp1, and Brf1, is required for transcription of genes with internal promoters (53). Alternatively spliced variants of hBrf1 have also been noted (44). Human TFIIIB interacts with a subcomplex of Pol III-specific subunits—hRPC32, hRPC39, and hRPC62 (homologues of yeast Rpc31, Rpc34, and Rpc82, respectively)—through direct interactions hBrf1 and hTBP with hRPC39 (63). The conservation of interactions of yeast and human Brf1 and yRPC34/hRpc39 implies a conservation of TFIIIB functions between yeast and higher eukaryotes. Functional domains of the subunits of yeast TFIIIB have been analyzed by in vitro transcription, gel shift assay and DNA footprinting (3, 13, 24, 26, 29, 30, 36, 55, 56). Although TFIIIB can bind directly to genes with strong TATA boxes (43), most Pol III-transcribed genes of S. cerevisiae, including most tRNA genes, require prior TFIIIC binding to boxA and boxB promoter sites (6, 34). TATA box-directed binding of TFIIIB is mediated by TBP. TFIIIC-dependent recruitment of TFIIIB to Pol III promoters is mediated through interaction with Brf1 and potentially with Bdp1 and TBP (5, 15, 20, 42). The fully assembled TFIIIB-DNA complex is very stable against dissociation by high concentrations of simple electrolytes and polyanions.
The ability to recruit Pol III to the transcription start site is a key property of TFIIIB, but its Brf1 and Bdp1 subunits also play an essential role in postrecruitment steps of transcriptional initiation in vitro (22, 29, 31, 32). Less is known about functions in vivo (22), particularly in regard to Bdp1. The work that is presented here is intended to fill this gap.
Recent studies of RNA polymerase II (Pol II) focused on the relationship between transcription and mRNA maturation. mRNA processing factors, capping enzyme, splicing factors, and polyadenylation factors interact with general transcription initiation factors or with Pol II itself (reviewed in reference 49). Pol III transcripts also undergo processing. In particular, three processing steps are required for tRNA maturation: 5′ end processing by RNase P, 3′ end processing involving La (yeast Lhp1) and intron excision by endonuclease- and ligase-mediated splicing (2, 4, 17, 45, 67, 69). La is not essential for yeast viability (68); all genes encoding subunits of RNase P and splicing endonuclease are essential (8, 62). Direct interactions of transcript-processing enzymes with the Pol III transcription apparatus of yeast have not been examined. Human La, a phosphoprotein, is involved in 5′- and 3′-end processing of tRNA (18, 25), but little is known about its interaction with the relevant endonucleases.
In this work, we present the results of an analysis that identified the parts of Bdp1 that are essential to its functioning in vivo. The effects of deleting segments of Bdp1 on viability were examined, and core essential segments of Bdp1 were identified. We also report on suppressors and enhancers of conditional viability that indicate interactions of particular segments of Bdp1 with other components of Pol III transcription and identify a relationship between Bdp1 and posttranscriptional processing of Pol III transcripts.
MATERIALS AND METHODS
Media and strains.
Cells were grown in synthetic dextrose (SD) medium (2% dextrose, 0.5% ammonium sulfate, and 0.17% yeast nitrogen base without amino acids and ammonium sulfate) containing required amino acids or YPD (1% yeast extract, 2% peptone, and 2% dextrose). 5-FOA plates contained 0.5% 5-fluoroorotic acid in SD medium. The strain used for the genetic analysis is a haploid BDP1 disruptant (MATα bdp1::TRP1 ura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1) carrying a BDP1 expression plasmid (51). The originally resident BDP1-expressing the 2μm plasmid (YEp24; URA+) was replaced by the CEN plasmid pRS316 BDP1WT (constructed as specified below) in two steps. First, the strain was transformed with pRS315 BDP1WT and transformants were grown on 5-FOA medium to eliminate YEp24 BDP1. Second, pRS315 BDP1WT was displaced by freshly introduced pRS316 BDP1WT by selection on SD plates for Leu− Ura+ growth.
Plasmids and primers.
BDP1 expression plasmids with a centromere (CEN6/ARSH4) or 2μm origin were constructed by PCR cloning. The BDP1 expression cassette plasmid pRS315UD was constructed by inserting the flanking segments of the BDP1 open reading frame, 0.5 kbp upstream from its ATG codon and 0.5 kbp downstream from its stop codon, as appropriately cleaved PCR products, using primers AIP003, AIP004, AIP005, and AIP006. The upstream and downstream fragments were inserted between the XhoI and PstI sites and between the BamHI and SacI sites of pRS315 (CEN6/ARSH4 LEU2) (57). The BDP1 wild type and all internal deletion mutants were transferred with the use of primers AIP007 and AIP010 from previously described Escherichia coli expression plasmids (36) between the PstI and BamHI sites of pRS315 UD. Terminally truncated bdp1 mutant expression plasmids were cloned with the use of the following primers: bdp1-(1-487), AIP007-AIP071; bdp1-(1-464), AIP007-AIP072; bdp1-(1-441), AIP007-073; bdp1-(1-352), AIP007-AIP008; bdp1-(40-594), AIP068-AIP010; bdp1-(40-487), AIP068-071; bdp1-(40-464), AIP068-AIP072; bdp1-(40-441), AIP068-AIP073; bdp1-(138-594), AIP013-010; bdp1-(138-487); AIP013-AIP071; bdp1-(138-464), AIP013-AIP072; bdp1-(158-594), AIP069-AIP010; bdp1-(158-487), AIP069-AIP071; bdp1-(186-594), AIP070-AIP010; and bdp1-(352-594), AIP009-AIP010. Wild-type and mutant BDP1 with flanking sequence or the flanking sequence alone was transferred as XhoI/SacI fragments into vectors pRS316(CEN6/ARSH4 URA3) and pRS423(2μm HIS3) (10, 57). Multicopy plasmids pRS423TBP, pRS423B70 (BRF1), and pRS423B90 (BDP1) were gifts from I. Willis (54). pRS423 PCF1 and pRS423 PCF1-1 were derived from pRS313 PCF1 and pRS313 PCF1-1 (50, 66), also from I. Willis, by transfer between the two PvuI sites of pRS423.
A multicopy S. cerevisiae genomic library constructed with 4- to 5-kbp inserts from a partial Sau3AI digest inserted into the BamHI site of YEp352 (58) was a gift from S. Emr (University of California, San Diego). The RPR1 gene was lifted out of suppressor plasmid pDm1SR#14 by PCR amplification using primers AIP095 and AIP096 and inserted between the SalI and EcoRI sites of YEp352 (CEN4/ARS1, URA3) and pBluescript KS(+), respectively, to generate YEp352 RPR1 and p RPR1.
The following primers were used: AIP003 (5′-AACTCGAGCGAACACTGGCTCACTGCTAATTCTTTCG-3′), AIP004 (5′-AACTGCAGCTGGTAATCAGTGGCTCCTTGCCGC-3′), AIP005 (5′-AAGGATCCTTTATATGCATACATAATGGATAAATAGC-3′), AIP006 (5′-AAGAGCTCTTATAATTAAGATCTGGAAACTTCGC-3′), AIP007 (5′-AACTGCAGATGAGTAGTATTGTTAATAAAAGTGG-3′), AIP008 (5′-AAGGATCCTTAAGGAATGTCAGCGTTGAGTAGTTTATC-3′), AIP009 (5′-AACTGCAGATGCCTGAGTCAGACCGCAAAGCACATACG-3′), AIP010 (5′-AAGGATCCTTATTGATCAATCTCAGGCTCTTC-3′), AIP013 (5′-AACTGCAGATGGACAACGAAAGTCGTCCAAGTTTTAA-3′), AIP068 (5′-AAACTGCAGATGGAAAGTAAAGAAATAGAAGAAGAT-3′), AIP069 (5′-AAACTGCAGATGGGGACTGCGCGACGTTTATCTAC-3′), AIP070 (5′-AAACTGCAGATGCTGAAAACTTTCAAAAGACATAGG-3′), AIP071 (5′-AAGGATCCTTATTCGCAGCAATACTCATCAAAATT-3′), AIP072 (5′-AAGGATCCTTATTTCTTCTCCTCATTAACGAATTTG-3′), AIP073 (5′-AAGGATCCTTACAAATTGAAATCTGTCCCCCACAT-3′), AIP095 (5′-AAAGAATTCTAAAAATCAATCAATCATCGTGTG-3′), AIP096 (5′-AAAGTCGACGCCGATAAGGTGTACTGGCG-3′).
In vitro transcription.
TFIIIC, Pol III, and the recombinant proteins TBP, Brf1, and Bdp1 were purified as previously described (32). Preinitiation complexes were formed in 20 μl of transcription buffer (40 mM Tris-HCl [pH 8.0], 70 mM NaCl, 7 mM MgCl2, 3 mM dithiothreitol [DTT], 100 μg of bovine serum albumin [BSA] per ml) with 100 ng of template plasmid (pTZ1 [SUP4] or p RPR1) at 21°C for 40 min. Transcription was started by adding 5 μl of transcription buffer containing 1 mM (each) GTP, ATP, and CTP and 125 μM [α-32P]UTP (10,000 cpm/pmol) and continued at 21°C for 30 min (30). Transcripts were separated by 8 M urea-8% polyacrylamide gel electrophoresis (PAGE) and analyzed with phosphorimager plates.
Immunoprecipitation and pull-down assays.
Yeast total cell extracts were prepared from 50 g of S. cerevisiae (strain YBS334; MATα pep4-3 prb1-1122 ura3-52 leu2-3,112 reg1-501 gal) (23) grown in YPD medium. Cells were broken in 100 ml of disruption buffer (75 mM Tris HCl [pH 8.0], 6% [vol/vol] glycerol, 200 mM ammonium sulfate, 1.5 mM DTT, 0.15 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 0.5% benzamidine HCl) with glass beads. After centrifugation (10,000 × g, 1 h), the supernatant was dialyzed against buffer containing 50 mM Tris HCl (pH 8.0), 10% (vol/vol) glycerol, 100 mM NaCl, 0.5 mM DTT, 0.1 mM EDTA, and 0.01% Tween 20. Rabbit antibodies to E. coli-produced recombinant yeast TBP (full length), Brf1 (full length), and Bdp1 (amino acids 40 to 487) were prepared. Protein A affinity beads (protein A-Sepharose CL-4B; 25 μl) were saturated by incubation with each antiserum (250 μl) and used for immunoprecipitation. Recombinant Bdp1, Bdp1Δ253-269, or BSA (50 μg each) was immobilized on cyanogen bromide-activated Sepharose 4B beads (10 μl) in 1 ml of buffer containing 50 mM HEPES (pH 7.8), 10% (vol/vol) glycerol, 100 mM NaCl, 0.5 mM DTT, 0.1 mM EDTA, and 0.01% (vol/vol) Tween 20 overnight at 4°C and blocked with the same buffer containing BSA (100 μg/ml). These beads were incubated with yeast cell extract (5 mg of protein) in 1 ml of binding buffer (50 mM Tris HCl [pH 8.0], 10% [vol/vol] glycerol, 100, 200, or 300 mM NaCl, 0.5 mM DTT, 0.1 mM EDTA, 0.01% [vol/vol] Tween 20, 1 mM PMSF) for 3 h at 4°C. After two washes with 1 ml of the corresponding binding buffer, ligands were eluted in 200 μl of binding buffer with 100 mM NaCl and 1% (wt/vol) sodium dodecyl sulfate (SDS) (see Fig. 7A) or in 200 μl of 50 mM Tris HCl (pH 8.0)-10% (vol/vol) glycerol-1 M NaCl-0.5 mM DTT-0.1 mM EDTA-0.1% Tween 20-1 mM PMSF (Fig. 7B). These samples were extracted with phenol-chloroform-isoamyl alcohol (25:24:1), ethanol precipitated with carrier sonicated salmon sperm DNA (6 μg), separated by 6 M urea-6% PAGE, and analyzed by Northern blotting.
FIG. 7.
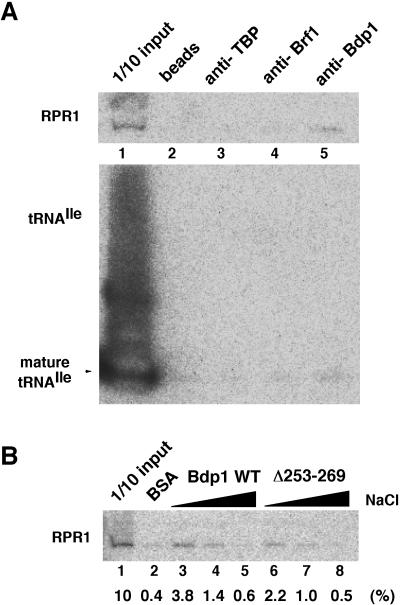
Affinity purification of RPR1 RNA with Bdp1. (A) Immunoprecipitation by TFIIIB-specific antibodies. Yeast total cell extracts were mixed with anti-TBP, anti-Brf1, and anti-Bdp1 immobilized on protein A affinity beads. Protein A beads without antibody were used as a control (lane 2). Material eluting from washed beads with buffer containing SDS was separated by PAGE, and RPR1 RNA was detected by Northern blotting for RPR1 RNA (top) and for tRNAIle (bottom). Three percent of input RPR1 RNA is pulled down by immobilized Bdp1 while the greatly overexposed lower panel shows less than 0.01% of input tRNAIle binding to immobilized Bdp1 (compare lanes 5 and 1). (B) Pull-down assay using immobilized recombinant Bdp1. RNase P was pulled down from yeast total cell extract by these beads in buffer containing 100 (lanes 2, 3, and 6), 200 (lanes 4 and 7), or 300 (lanes 5 and 8) mM NaCl. Immobilized BSA was used as a negative control (lane 2; buffer with 100 mM NaCl). Material eluting from washed beads with buffer containing 1 M NaCl was resolved and detected by Northern blotting as in panel A. The averages of three experiments are given at the bottom of the panel as a percentage of input RPR1 RNA. Input controls (lanes 1 of all panels) were loaded with 10% of the total nuclear extract used for each assay.
Northern blotting
BDP1 wild-type and bdp1-Δ253-269 mutant cells were grown at 30°C in YPD containing 20 μg of adenine, 15 μg of l-lysine, 10 μg of l-histidine, and 10 μg of uracil per ml and harvested just before and 30, 60, 120, and 240 min after a temperature shift to 37°C. Harvested cells were frozen, disrupted with glass beads in disruption buffer (100 mM NaCl, 10 mM EDTA, 50 mM Tris-HCl [pH 7.5], 5% SDS), and extracted with an equal amount of phenol-chloroform-isoamyl alcohol (50:50:1). After precipitation with ethanol, aliquots of extracted total RNA (10 μg) were separated by 7 M urea-PAGE (6 or 8%) and transferred to nylon membranes in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Membranes were incubated in hybridization buffer (40% formamide, 180 mM NaCl, 20 mM Na cacodylate [pH 7.5], 1 mM EDTA, 0.1% SDS, 120 μg sonicated salmon sperm DNA/ml, and 1% dextran sulfate) for 2 h and then overnight at 45°C in the same buffer containing each radioactively labeled probe (1 μCi/ml). Membranes were washed with 2× SSC at room temperature and again with 0.2× SSC-0.1% SDS at 60°C to dissociate nonspecifically binding hybridization probe. Signals were analyzed and quantified by phosphorimager plate analysis.
The following 5′ 32P-labeled DNA probes were used: RPR1 (5′-GGCTCCACTGTGTTCCACCGAATTTCCCAC-3′), U4 snRNA (5′-CACCGAATTGACCATGAGGAGACGGTCTGG-3′), 5S rRNA (5′-CAGTTGATCGGACGGGAAACGGTG-3′), U6 snRNA (5′-CGGTTCATCCTTATGCAGGGGAACTGC-3′), and tRNAIle(UAU) (5′-GCTCGAGGTGGGGTTTGAACCCACGACGG-3′).
RESULTS
Regions of Bdp1 that are required in vivo.
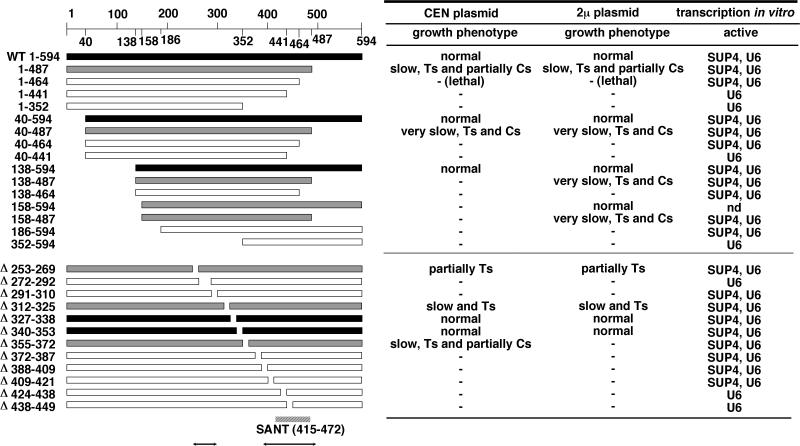
In order to identify the essential core of Bdp1, yeast expression plasmids carrying 16 terminally truncated and internal deletion mutants of BDP1 were constructed in centromeric and multicopy (2μm) versions. All plasmids were introduced into a haploid strain with a disrupted chromosomal copy of BDP1 (bdp1::TRP1) and resident wild-type BDP1 expression plasmid pRS316 BDP1WT (CEN URA3). Viability was tested after selection for plasmid shuffling on 5-FOA-containing plates. Viable mutants were maintained on 5-FOA plates and checked for elimination of pRS316 BDP1WT by their uracil requirement. High temperature sensitivity (Ts) was monitored at 37°C and cold sensitivity (Cs) at 18°C. In the CEN plasmid series, nine mutants [bdp1-(1-487), bdp1-(40-594), bdp1-(40-487), bdp1-(138-594), bdp1-Δ253-269, bdp1-Δ312-325, bdp1-Δ327-338, bdp1-Δ340-353, and bdp1-Δ355-372 mutants] were viable and five of these [bdp1-(1-487), bdp1-(40-487), bdp1-Δ253-269, bdp1-Δ312-325, and bdp1-Δ355-372 mutants] showed different growth phenotypes (Fig. 1). In the multicopy (2μm) plasmid series, three additional mutants [bdp1-(138-487), bdp1-(158-594), and bdp1-(158-487) mutants] were viable, and only one mutant that was viable in the CEN series, the bdp1-Δ355-372 strain, was inviable in the multicopy series (Fig. 1). These results identified three separate regions of Bdp1 (amino acids 158 to 252, 269 to 312, and 372 to 487; regions III, II, and I, respectively) that are required in vivo. Regions II and I overlap with domains of Bdp1 that interact with other components of the TFIIIB-DNA complex, as determined by protein footprinting (36), and region I overlaps with the relatively conserved SANT structural domain (amino acids 415 to 472) of Bdp1 (Fig. 1). In general, the requirements for Bdp1 in vivo are more restrictive than for function in the resolved Pol III transcription system in vitro (36; unpublished results) (Fig. 1, right).
FIG. 1.
Viability of cells producing truncated and internal-deletion Bdp1. Bars show the extent of the retained BDP1 open reading frame (594 amino acids) in expression cassette plasmids pRS315UD (CEN LEU2) and pRS423UD (2μm HIS3), both of which contain flanking 0.5-kbp promoter- and terminator-proximal segments of BDP1. Shading indicates growth at 30°C after 5 days. Open bars, completely defective; black bars, viable; gray bars, viable but showing Ts and/or Cs conditional growth with a CEN or multicopy plasmid, as indicated on the right. Growth of strains in the CEN series was observed on SD plates lacking tryptophan and leucine but containing 5-FOA. Growth of strains in the multicopy plasmid series was observed on SD plates lacking histidine and leucine but containing 5-FOA. The abilities of the corresponding proteins to support TFIIIC-dependent transcription of the SUP4 tRNATyr gene and TFIIIC-independent transcription of the U6 snRNA gene are shown on the right (data from reference 36 and unpublished data). The locations of the SANT domain and of two segments of Bdp1(↔) that are protected from hydroxyl radical-mediated cleavage upon assembly into a TFIIIB-DNA complex (↔) are indicated at the bottom. nd, not determined.
Multicopy suppression.
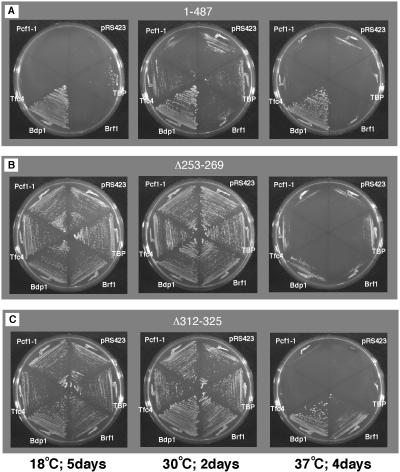
It was anticipated that the viability or temperature sensitivity (Ts or Cs) of some bdp1 mutants might be affected by overproduction of proteins that interact with Bdp1. Recent reports show that TBP, Brf1, and Tfc4 (also called τ131, TFIIIC131, and Pcf1) interact directly with Bdp1 (5, 9, 13, 35, 47, 52). To detect suppression by plasmid shuffling, strains carrying pRS316 BDP1WT and mutant genes on CEN plasmids were transformed by multicopy pRS423 plasmids carrying the SPT15 (encoding TBP), BRF1, or TFC4 gene or the TFC4 dominant mutant PCF1-1 gene with a mutation in the second tetracopeptide repeat (originally isolated as a suppressor of negative effect of a tRNA gene boxA promoter mutation [50]), and also the wild-type BDP1 gene as a control. None of the bdp1 deletion mutants that were inviable when harbored on CEN plasmids in Fig. 1 were rescued by TBP, Brf1, or Tfc4 overexpression. One bdp1 deletion strain, bdp1(40-487), which was Ts and Cs in the initial screen (Fig. 1), failed to grow in the somewhat more complex multicopy suppression screen in attempts with two different CEN vectors (data not shown). The remaining eight bdp1 mutant strains that retained viability after loss of pRS316 BDP1WT (by 5-FOA counterselection followed by verification of uracil auxotrophy) were examined for growth phenotype at 18, 30, and 37°C in the multicopy suppression strains. The amino acid 355 to 372 deletion proved to be lethal at 30°C in cells overexpressing PCF1-1, the dominant gain-of-function variant of Tfc4 (data not shown), and was not examined at other temperatures. The Cs phenotype of bdp1-(1-487) was weakly suppressed by SPT15 (TBP) overexpression (Table 1; Fig. 2A, left). The completely Ts phenotype of bdp1-Δ312-325 was suppressed by overexpression of BRF1 (Table 1; Fig. 2C, right). The Ts phenotype of another internal deletion mutant, the bdp1-Δ253-269 mutant, was evidently dominant, as it was not suppressed by wild-type BDP1 (Fig. 2B, right). The normal growth phenotype of the bdp1-(40-594) strain at 37°C was somewhat impaired by overexpression of PCF1-1 both at 18°C and at 37°C (Table 1). Curiously, the bdp1-(138-594) strain, with the more extensive N-terminal deletion, did not show any influence of PCF1-1 overexpression.
TABLE 1.
Effects of overexpression of Bdp1-interacting proteins on temperature phenotypes of Bdp1 mutant strains
| Growth conditions | Bdp1 mutation | Growth of strain overexpressing:
|
|||||
|---|---|---|---|---|---|---|---|
| None (vector) | TBP | Brf1 | Bdp1 | Tfc4 | Pcf1-1 | ||
| 18°C, 5 days | 1-594 | +++++ | +++++ | +++++ | ++++ | +++++ | +++++ |
| 1-487 | − | ++ | − | +++++ | − | − | |
| 40-594 | +++++ | +++++ | +++ | ++++ | +++++ | +++ | |
| 138-594 | ++++ | +++++ | ++++ | +++++ | ++++ | ++++ | |
| Δ253-269 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | |
| Δ312-325 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | |
| Δ327-338 | +++++ | +++++ | +++++ | +++++ | +++++ | ++++ | |
| Δ340-353 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | |
| Δ355-372 | +/− | +/− | +/− | +++++ | +/− | NDa | |
| 30°C, 2 days | 1-594 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
| 1-487 | +++ | +++ | ++ | +++++ | +++ | ++ | |
| 40-594 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | |
| 138-594 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | |
| Δ253-269 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | |
| Δ312-325 | ++++ | ++++ | ++++ | +++++ | ++++ | ++++ | |
| Δ327-338 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | |
| Δ340-353 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | |
| Δ355-372 | ++++ | +++ | ++ | +++++ | +++++ | ND | |
| 37°C, 4 days | 1-594 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
| 1-487 | − | − | − | +++++ | − | − | |
| 40-594 | +++++ | ++++ | ++++ | ++++ | +++++ | ++ | |
| 138-594 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | |
| Δ253-269 | + | + | + | + | + | + | |
| Δ312-325 | − | − | ++ | +++++ | − | − | |
| Δ327-338 | +++++ | +++++ | +++++ | +++++ | +++++ | ++++ | |
| Δ340-353 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | |
| Δ355-372 | − | − | − | +++++ | − | ND | |
ND, not determined.
FIG. 2.
Multicopy suppression by Bdp1-interacting proteins. 5-FOA-resistant strains (Table 1) were tested for growth at 18, 30, and 37°C. (A) The bdp1-(1-487) partial Cs phenotype (very slow growth at 18°C) is partly suppressed by TBP overexpression. (B) The partial Ts (very slow growth) at 37°C phenotype of the bdp1-Δ253-269 internal deletion mutant is not suppressed, even by BDP1. (C) Brf1 overexpression suppresses the Ts phenotype of bdp1-Δ312-325.
A split Bdp1 is functional.
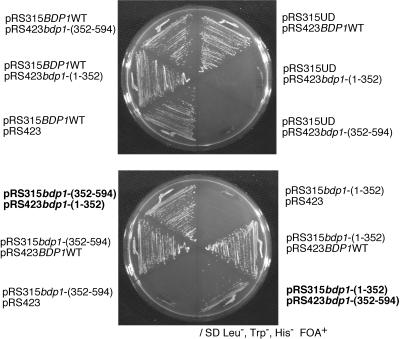
Because plasmid shuffling experiments identified amino acids 327 to 353 of Bdp1 as dispensable (Fig. 1), one might anticipate that severing the N- and C-proximal halves of Bdp1 in this region would not destroy function in vivo. Split Bdp1 functions in vitro (55). To examine the in vivo counterpart of this result, the BDP1-disruptant strain carrying pRS316 BDP1WT was transformed with plasmid pair pRS315 bdp1-(1-352)/pRS423 bdp1-(352-594) or pRS315 bdp1-(352-594)/pRS423 bdp1-(1-352), and viability was tested on 5-FOA plates (Fig. 3). As controls, the empty cassette plasmids and wild-type Bdp1 plasmids were also substituted for the four plasmids encoding Bdp1(1-352) and Bdp1(352-594) in all screenable combinations (Fig. 3). The strain carrying the pRS315 bdp1-(1-352)/pRS423 bdp1-(352-594) combination was indistinguishable from the wild type, and the pRS315 bdp1-(352-594)/pRS423 bdp1-(1-352) combination resulted in a slower-growth phenotype. No high or low temperature sensitivity was noted (data not shown). Bdp1(1-352) is stable, and large amounts accumulate in vivo (data not shown). We have not tried to identify localization. However, these results suggest that the C-terminal fragment Bdp1(352-594) may be less stable or not normally distributed in the cell.
FIG. 3.
Viability of cells producing split Bdp1. Twelve combinations of plasmids were tested by plasmid shuffling. Three-plasmid transformant cell lines carrying pRS315 BDP1WT and two other plasmids in various pairwise combinations were constructed. Neither of the terminally truncated bdp1-(1-352) nor bdp1-(352-594) mutants confers viability separately after elimination of the wild-type BDP1 gene, but they do so in combinations of plasmids pRS315 bdp1-(1-352) with pRS423 bdp1-(352-594) and pRS315 bdp1-(352-594) with pRS423 bdp1-(1-352) after elimination of the wild-type gene (bold).
Suppressor gene of Bdp1Δ253-269.
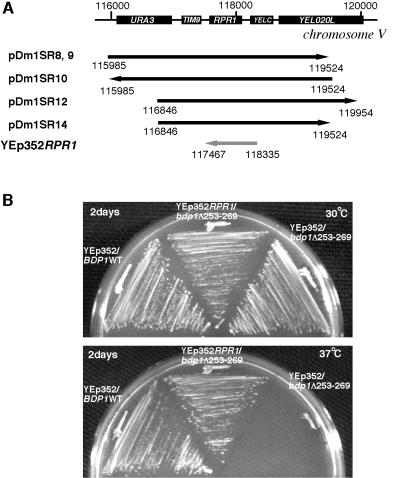
Cells producing Bdp1Δ253-269 in place of wild-type Bdp1 grow very slowly at 37°C but normally at 30 and 18°C. The Ts phenotype conferred by this deletion was not suppressed by overexpression of TBP, Brf1, or Tfc4 (Table 1). As noted above, even the BDP1 wild-type gene on plasmid pRS423 did not rescue temperature sensitivity (Fig. 2B). In order to isolate suppressors, a yeast genomic library in the multicopy plasmid YEp352(URA3) was screened. From 10,000 transformants, 16 candidates were isolated (Dm1SR#1 to -16) at 37°C as cells that grew well. Seven of these were 5-FOA sensitive at 37°C (Dm1SR#3, -4, -8, -9, -10, -12, and -14). The corresponding candidate plasmids were isolated and used to retransform the bdp1-(Δ253-269) strain. All retained the normal growth phenotype at 37°C, and their inserts were sequenced. Five plasmids (pDm1SR#8, -9, -10, -12, and -14) contained DNA from the same location on chromosome V (bp 116,000 to 120,000) (Fig. 4A); two plasmids (pDm1SR#3 and -4) contained BDP1.
FIG. 4.
Identification of a suppressor gene of bdp1-Δ253-269. Arrows show inserted yeast genomic DNA fragments (from a partial Sau3AI digest genomic library) and orientations in the BamHI site of YEp352. (A) Five of seven suppressor plasmids cover the same region of chromosome V. YEp352 RPR1 has the indicated RPR1 fragment cloned between the EcoRI and SalI sites of YEp352 by PCR cloning (gray arrow). (B) Suppression of bdp1-Δ253-269 by YEp352 RPR1. Extremely slow growth at 37°C is fully restored by RPR1 overexpression.
It is surprising that BDP1 was isolated as a suppressor of bdp1-Δ253-269, because this mutation was found to be dominant relative to coexpression of BDP1 in the pRS423 multicopy plasmid (Table 1). It is conceivable that expression efficiency differs between pRS423B90 (BDP1) and the genomic library BDP1 suppressors in YEp352 (Fig. 2B and Table 1). In fact, Western blotting indicated lower levels of Bdp1 accumulation when BDP1 was expressed in pRS423B90 relative to the genomic BDP1 clone pDm1SR#3 (in YEp352) (data not shown).
The suppressor is RPR1.
All five non-BDP1 suppressor clones harbor three genes: TIM9 (a mitochondrial inner membrane protein), YELC (a transposon Ty4 long terminal repeat), and RPR1 (the RNA subunit of RNase P). The suppressor gene was anticipated to be RPR1, for three reasons. First, RNase P is one of the tRNA maturation enzymes, processing the 5′ ends of pre-tRNAs. Yeast RNase P is composed of one RNA subunit (RPR1) and nine protein subunits; eight of the nine proteins are common to RNase P and MRP (the mitochondrial rRNA processing enzyme, which also processes nuclear rRNAs). Although the function of RNase P has been characterized, the possibility of a relationship between tRNA recognition and Pol III transcription has not been examined. Second, RPR1 has been isolated as a suppressor gene of a mutation in TFC3, the gene encoding τ138, the largest subunit of TFIIIC (39). Third, RPR1 appears to be transcribed by RNA Pol III in vivo, as its transcription is impaired by both a temperature-sensitive lesion in RNA Pol III and point mutations in putative boxA and boxB TFIIIC recognition elements (37). To confirm this expectation, RPR1 with its promoter region (see below) was cloned into the multicopy vector YEp352 (2μm, URA3) and used to transform cells producing Bdp1Δ253-269 (off pRS315). YEp352 RPR1 (Fig. 4A) restored normal growth at 37°C (Fig. 4B).
The Ts phenotype of cells producing Bdp1Δ253-269 and its suppression by overexpression of RPR1 might reflect three possible situations. (i) Assembly of TFIIIB with Bdp1Δ253-269 in vivo may lead to defective transcription of RPR1, which has a tRNA-like promoter with a boxA and a suboptimally placed, imperfect (nonconsensus) boxB (38). (ii) Interaction between RNase P and Bdp1 in the Pol III initiation complex may be required for effective RNA processing; the deletion of amino acids 253 to 269 may weaken that interaction. (iii) Globally lower Pol III transcription levels due to Bdp1Δ253-269 may generate a deficit of RPR1 RNA that is critical at high temperatures. However, TFIIIBs assembled with Bdp1Δ253-269 and wild-type Bdp1 have approximately the same in vitro transcription activity, DNA-binding activity, and DNase I footprint as TFIIIB assembled with wild-type Bdp1 (36), arguing against the probability of a global effect on all Pol III transcription in vivo.
RPR1 was also tested for suppression of other Bdp1 deletion Ts and Cs phenotypes (Table 1). Introducing pDm1SR#14 (multicopy expression of RPR1) into strains producing Bdp1(1-487), Bdp1Δ312-325, and Bdp1Δ355-372 did not restore normal growth at 37°C or, in the case of Bdp1(1-487) and Bdp1Δ355-372, growth at 18°C (data not shown). We conclude that RPR1 is uniquely a suppressor of bdp1-Δ253-269.
Pol III transcripts of cells producing Bdp1Δ253-269.
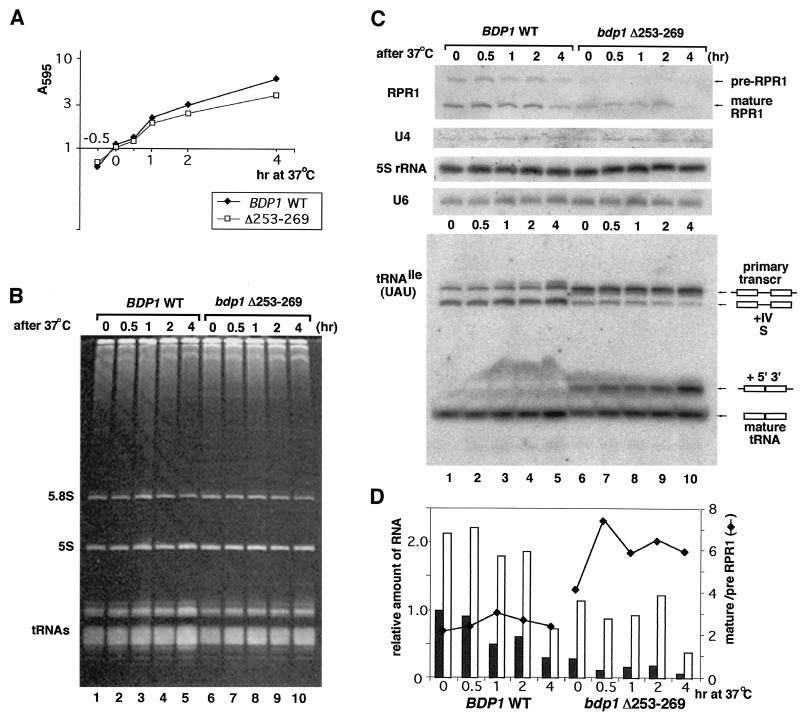
In order to understand the Bdp1-RPR1 connection, Pol III transcripts were analyzed by Northern blotting. Cells producing wild-type Bdp1 and Bdp1Δ253-269 (off the appropriate pRS315 plasmid) were grown in YPD medium at 30°C and collected before temperature shift to 37°C as well as 0.5, 1, 2, or 4 h later (Fig. 5A). Aliquots of total RNA (prepared as described in Materials and Methods) were separated on 7 M urea 6 or 8% polyacrylamide gels (Fig. 5B and C) and transferred to nylon membranes for hybridization to detect Pol III transcripts. Specific probes for RPR1 RNA, 5S rRNA, U6 snRNA, and tRNAIle(UAU) were used, and a U4 snRNA (a Pol II transcript) probe served as an internal control. Levels of 5S rRNA, U6 snRNA, and U4 RNA in cells producing wild-type Bdp1 and Bdp1Δ253-269 were comparable at 30°C and after a shift to 37°C (Fig. 5C). RPR1 RNA and tRNAIle precursors were abnormal in cells producing Bdp1Δ253-269 at both temperatures (Fig. 5C, lanes 1 and 6). The accumulation of RPR1 RNA was also deficient in mutant cells and the proportion of mature to pre-RPR1 RNA was very different (Fig. 5D). Although tRNAIle levels were approximately normal, the pools of tRNAIle processing intermediates were elevated. Maturation of tRNA can proceed along two pathways: either the 5′ and 3′ ends of the primary transcript are processed first and the intron is excised subsequently, or both termini are processed after splicing. In wild-type Bdp1-producing cells, intron-retaining intermediates of tRNAIle maturation accumulated to the same levels at 30 and 37°C and the +5′/3′ form was not detected (Fig. 5C, bottom). In mutant cells, transcripts retaining the 5′ leader (reflecting absence of cleavage by RNase P) were much more abundant than in wild-type cells. Thus, very clearly, 5′-end processing of the tRNAIle Pol III transcript is defective in cells producing Bdp1Δ253-269. These results eliminate globally defective Pol III transcription in vivo as a primary cause of temperature sensitivity in cells producing Bdp1Δ253-269 and argue in favor of an RPR1-specific effect, possibly involving a direct interaction of a Bdp1 and RNase P.
FIG. 5.
RNA analysis of cells producing Bdp1Δ253-269 and wild-type Bdp1. (A) Growth in YPD medium. Cells were harvested at 30°C and 30, 60, 120, and 240 min after a shift to 37°C. (B) Total RNA (10 μg) was separated by 7 M urea-PAGE. The gel is stained with ethidium bromide. (C) Northern blotting of Pol III transcripts (transcr): RPR1 RNA, 5S rRNA, U6 snRNA, and tRNAIle(UAU). U4 snRNA, a Pol II transcript, was used as the internal standard. Two forms of RPR1 RNA were detected; their abundances differ in cells producing Bdp1Δ253-269. There are distinct differences of tRNA processing at 30 and 37°C. (D) Data from panel C, quantified and compared. Shaded bars, pre-RPR1 primary transcript; open bars, mature RPR1 RNA; diamonds, ratio of mature RPR1 to pre-RPR1 RNA.
RPR1 transcription in vitro.
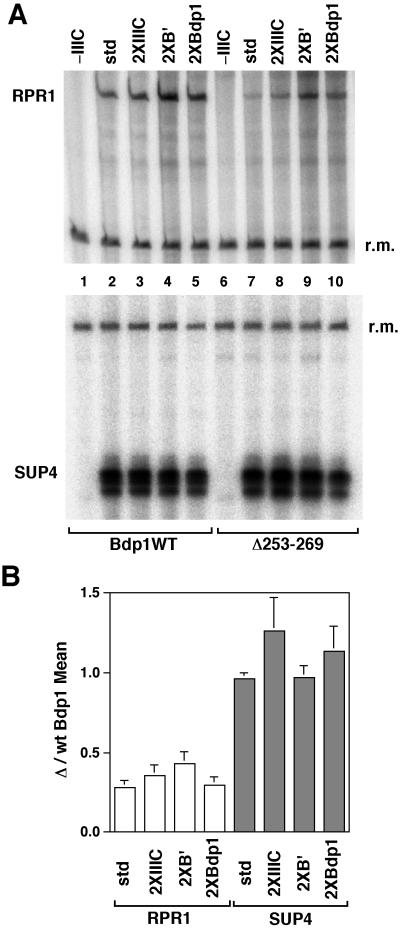
RPR1 transcription with recombinant TFIIIB containing wild-type and mutant Bdp1 was analyzed in vitro (Fig. 6). Accurately initiating transcription of RPR1, like tRNA gene transcription (SUP4), was TFIIIC dependent (Fig. 6A, lanes 1 and 6) and also required Bdp1 and Brf1 (data not shown). Surprisingly, although there were no apparent differences of SUP4 transcription between wild-type Bdp1 and Bdp1Δ253-269 (Fig. 6A, bottom, compare lanes 2 to 5 with lanes 7 to 10) the yield of RPR1 transcripts with Bdp1Δ253-269 was reduced to approximately one-half to one-third of the wild-type level (Fig. 6A, top, compare lanes 2 to 5 with lanes 7 to 10, and 6B), comparable with what was observed in vivo (Fig. 5C and 5D). The transcriptional defect of the RPR1 promoter with Bdp1Δ253-269 was not compensated for by doubling its concentration or the concentrations of TFIIIC (Fig. 6B). Doubling the concentrations of both Brf1 and TBP approximately doubled RPR1 transcription with both wild-type Bdp1 and Bdp1Δ253-269 (Fig. 6A and data not shown) and did not compensate for the Bdp1Δ253-269 defect (Fig. 6B, 2×B′). These results indicate that the RPR1 and SUP4 tRNA promoters require the same complement of initiation factors but suggest a greater sensitivity of transcription of the RPR1 gene, with its variant promoter, to deletion of amino acids 253 to 269 in Bdp1.
FIG. 6.
Transcription. (A) In vitro transcription of the RPR1 (top) and SUP4 (bottom) genes with highly purified Pol III (5 fmol), TFIIIC (1×, 26 fmol), TBP (1×, 50 fmol), Brf1 (1×, 35 fmol), Bdp1 (1×, 150 fmol; wild type or Bdp1Δ253-269), and template DNA (50 fmol). Lanes 1 to 5, full-length Bdp1; lanes 6 to 10, Bdp1Δ253-269; lanes 1 and 6, no TFIIIC; lanes 2 and 7, standard quantities of TFIIIC and components of TFIIIB; lanes 3 and 8, twofold-higher quantity of TFIIIC; lanes 4 and 9, twofold-higher quantities of TBP and Brf (B′); lanes 5 and 10, twofold-higher quantity of wild-type or mutant Bdp1. r.m., recovery marker. (B). Ratios of transcript yields with Bdp1Δ253-269 relative to wild-type Bdp1 for transcription of the SUP4 (closed bars) and RPR1 genes. std, 1× quantities of all components. The averages and standard errors of the means for three experiments are shown.
Association of Bdp1 with RNase P.
Physical interactions between Bdp1 (or TFIIIB) and RNase P have also been explored by coimmunoprecipitation and pull-down assays. A total cell extract (yeast strain YBS334; BDP1 wild type) was reacted with polyclonal antibody to TBP, Brf1, and Bdp1, and immune complexes were adsorbed to protein A affinity beads. Coimmobilizing material was eluted with detergent (SDS)-containing buffer and probed for RPR1 RNA (Fig. 7A, top), which was seen to have been captured with anti-Bdp1 antibody (lane 5) but not significantly (above background) with anti-TBP or anti-Brf1 antibody (lanes 3 and 4). An essentially identical result was obtained when coimmobilizing material was eluted with high-salt buffer (data not shown). A control probing for tRNAIle (cf. Fig. 5C) showed essentially no signal above background (Fig. 7A, bottom). Anti-Bdp1 pulled down 3% of the input RPR1 RNA (upper panel) but, in contrast, less than 0.01% of input tRNAIle (lower panel) under conditions of direct competition by all RNA species in the cell extract for binding to Bdp1. An interaction with Bdp1 was also detected by direct pull-down assay, using recombinant wild-type Bdp1 immobilized on beads (Fig. 7B, lane 3). RPR1 RNA was also bound by immobilized Bdp1Δ253-269 with somewhat lower efficiency (compare lanes 3 and 6), but the dependence of RPR1 capture by wild-type Bdp1 and Bdp1Δ253-269 on electrolyte concentration in the binding buffer was comparable (lanes 3 to 8). These experiments point to a physical interaction (though not necessarily a direct one) between Bdp1 and RNase P, although they do not account directly or entirely for the multicopy suppression by RPR1 of the temperature sensitivity conferred by bdp1-Δ253-269. The interaction that associates RPR1 RNA with Bdp1 is not especially strong, since it can be dissociated with neutral salt.
DISCUSSION
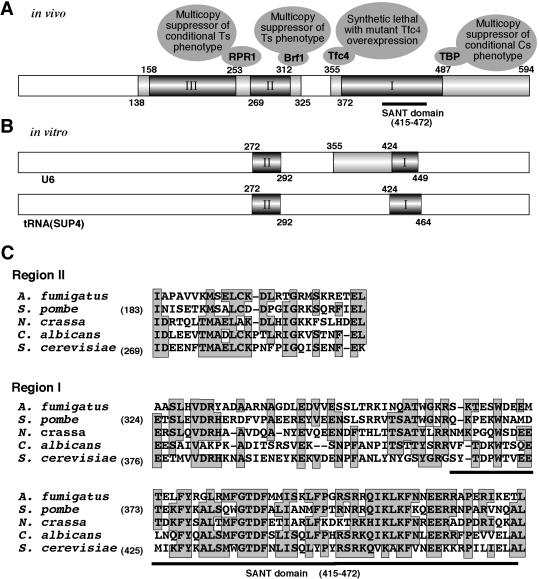
We have identified three separate segments of Bdp1 that are required for viability (Fig. 8). Regions I, II, and III correspond to parts of Bdp1 that have already been assigned some significance in the context of other analysis. Regions I and II overlap with segments of Bdp1 that were found to become less reactive to cleavage by hydroxyl radicals upon assembly of Bdp1 into a TFIIIB-DNA complex (36), implying at least partial burial as a consequence of changing protein-protein and/or protein-DNA interactions. Region III overlaps with a segment of Bdp1 that was found to become more reactive to hydroxyl radicals, implying possible displacement from an internal protein-protein interaction upon formation of the TFIIIB-DNA complex. Region I is also associated with other features of Bdp1: (i) it encompasses the SANT domain, a motif associated with protein-protein interaction (1) and is the segment that is most conserved among Bdp1 genes (Fig. 8), and (ii) it also comes to notice in connection with analysis of in vitro transcription. Deletions between amino acids 424 and 464 in region I make Bdp1 defective for TFIIIC-dependent transcription of supercoiled DNA (36). Deletions of 13- to 22-amino-acid segments between amino acids 355 and 421 of Bdp1 make TFIIIB defective for TFIIIC-independent transcription of linear DNA. Further analysis of this defect has revealed that TFIIIB plays a role in Pol III transcription extending beyond polymerase recruitment to the promoter (29). Region II encompasses a segment of Bdp1 that cross-links efficiently to DNA in a TFIIIB-DNA complex ∼10 bp upstream of the TATA box (55). Deleting amino acids 272 to 292 in region II also abolishes TFIIIC-dependent transcription in vitro (36). However, in general, the global requirements for Bdp1 function in vivo are more demanding and restrictive than requirements for function in the defined Pol III in vitro transcription system (36).
FIG. 8.
Functional map of Bdp1. (A) Summary of this work. Solid bars show absolutely required regions I, II, and III. Lightly shaded segments cover abnormal but viable alleles. Open segments are not necessary for normal growth at 18, 30, or 37°C. Sites associated with suppression by RPR1, Brf1, and TBP and synthetic lethality with Tfc4 (Pcf1-1) are marked. (B) Regions of Bdp1 required in vitro (29, 36). U6 gene transcription requires one of the two darkly shaded segments, and both segments are required for tRNA (SUP4) gene transcription. The lightly shaded segment is required for transcription of linear but not supercoiled DNA. (C) Alignment of fungal Bdp1 homologues. Region I and II consensus residues in at least three species are boxed and shaded. Region I is the most conserved and includes the SANT domain. Region II is also conserved. Region III is divergent and not required for in vitro transcription. The preliminary sequences of Bdp1 from C. albicans, N. crassa, and A. fumigatus were obtained from Stanford Genome Technology Center Contig6-2237 (http://www-sequence.stanford.edu/group/candida), WIMCGR Contig2.668 assembly 2 (http://www-genome.wi.mit.edu), and TIGR_5085 (http://www.tigr.org), respectively. The GenBank accession numbers for Bdp1 from S. cerevisiae and Schizosaccharomyces pombe are AAC49073 and CAA22645, respectively.
Deletions of several segments of Bdp1 generate high- or low-temperature sensitivity. Some of these defects are partly rescued by overexpression of other components of Pol III transcription. Thus, bdp1-(1-487) partial Cs and bdp1-Δ312-325 Ts phenotypes were suppressed by overexpression of TBP and Brf1. In contrast, the bdp1-Δ355-372 deletion, which confers Ts phenotype on its own, was seen to be synthetically lethal with overexpression of the dominant gain-of-function variant PCF1-1 of the TFC4 gene. These effects suggest that specific segments of Bdp1 are required for (normal) interactions with other components of TFIIIB and with TFIIIC. Although the lethality of deletions covering regions I, II, and III was not rescued by overproducing Brf1, TBP, or Tfc4, this does not exclude these regions as potential sites of such interaction. In fact, the rescue of conditional phenotypes generated by deletions at the boundaries of these Bdp1 segments suggests their involvement also. On the other hand, the ability to split Bdp1 at amino acid 352 is consistent with the nonessential nature of the surrounding 325-to-355 segment of Bdp1.
Relationships between transcription and RNA processing have remained largely unexplored for yeast Pol III. The finding that yeast genomic library screening captures RPR1 as a suppressor of the partial temperature sensitivity of the bdp1-Δ253-269 strain therefore holds particular interest. The product of the RPR1 gene is the RNA subunit of RNase P. RNase P trims 5′ ends of pre-tRNAs to generate mature tRNA, and this function is universally conserved. Bacterial, archaeal, and eucaryal RNase Ps are composed of one RNA subunit and several protein subunits, and their catalytic activity is contained in the RNA subunit (67). Yeast RNase P is also composed of one RNA subunit (RPR1 RNA) and nine protein subunits that are all essential for cell viability (8, 11, 16, 38, 41, 59). Bacterial RNase P recognizes pre-tRNA structure directly (48). This is evidently not the case for the yeast enzyme (67), and how it is brought into contact with its substrate is not known. Finding RPR1 as a suppressor of the temperature sensitivity of the bdp1-Δ253-269 strain suggests a possible involvement of TFIIIB in posttranscriptional processing of tRNA but could also reflect defective transcription of RPR1 itself, due to the bdp1-Δ253-269 deletion. RPR1 has a relatively weak promoter with a nonconsensus and suboptimally placed boxB, and RPR1 transcription in vitro by Pol III in conjunction with wild-type TFIIIC and TFIIIB is relatively weak compared to that of the standard SUP4 tRNATyr gene with its near-consensus and optimally configured boxA and boxB promoter sites (Fig. 6A). Under the conditions of analysis, transcription of RPR1 with TFIIIB(Bdp1Δ253-269) is reduced ∼2- to 3-fold relative to transcription with wild-type TFIIIB when activities are normalized to SUP4 transcription (Fig. 6). When DNA templates are transcribed under favorable conditions in vitro (using genes with strong promoters, and with transcription factors in excess), Bdp1Δ253-269 does not generate quantitative defects of transcript yield or qualitative changes of DNA footprint (36, 55), but RPR1 and other genes with weaker promoters might be more sensitive indicators of defects in the transcription apparatus, particularly when transcribed in competition with all Pol III genes in the cell (Fig. 5B and C).
However, this is not the sole defect generated by bdp1-Δ253-269. Maturation of tRNA [assayed with tRNAIle(UAU)] is also aberrant at 30 and 37°C (Fig. 5C), and the relative accumulation of pre-RPR1 and mature RPR1 RNA is strongly affected (Fig. 5D). It remains conceivable that Bdp1, separately or in conjunction with promoter-bound TFIIIB, interacts directly with RNase P. Pol II processing factors, capping enzyme, and splicing and polyadenylation complexes interact with Pol II transcription through the C-terminal domain of the largest Pol II subunit (49). It is plausible to consider a comparable coupling between Pol III transcription and posttranscriptional processing. Because Pol III transcription units are characteristically short, it is also reasonable to consider the possibility that this coupling could be effected by a transcription initiation factor instead of the elongating RNA polymerase complex.
It might be thought that RPR1 suppression of temperature sensitivity conferred by the bdp1-Δ253-269 deletion is merely due to nonspecific relief of a quantitative defect of Pol III transcription. If this were the case it would point to RNA processing by RNase P as the Pol III transcript-supported cellular process that is quantitatively most limiting. A similar inference was made when RPR1, along with the genes encoding all three subunits of TFIIIB, the TFC1 and TFC4 subunits of TFIIIC, and the RPC10 subunit of Pol III turned up as multicopy suppressors of a temperature-sensitive mutation in TFC3, the gene encoding the largest subunit of TFIIIC (39, 52). However, we surmise that the connection between RNase P and Bdp1/TFIIIB has a wider significance. First, the temperature sensitivity of the bdp1-Δ253-269 mutant was not suppressed by overproduction of Brf1, TBP, or Tfc4 (Fig. 2; Table 1). Second, RPR1 multicopy suppression is specific to the 253-to-269 deletion in BDP1. If a mere two- to threefold reduction of RPR1 transcription in vivo were responsible for temperature sensitivity, then RPR1 multicopy suppression should be general to conditions and mutations that depress Pol III transcription. Instead, the RPR1 multicopy suppression is specific to a single BDP1 deletion. Other BDP1 mutations, which presumably also lower transcription in vivo, are specifically suppressed by overproducing TBP and Brf1, for example, but not RPR1 RNA. Third, the specificity of action of yeast RNase P in pre-tRNA processing evidently does not reside in direct substrate recognition. A mediator of specificity is called for; presumably, this additional component is required for efficient catalysis. Finding RPR1 multicopy suppression has, in fact, led to experiments demonstrating that Bdp1 interacts with RNase P/RPR1 RNA with a high degree of specificity and moderate affinity.
Acknowledgments
We are grateful to S. Hahn for providing a yeast strain, to I. M. Willis for yeast expression vectors, and to S. Emr for a yeast genomic library and helpful advice.
We are grateful to the National Institute of General Medical Sciences for support of this work. We appreciate access to the following information. Candida albicans sequence data was obtained from the Stanford Genome Technology Center. Sequencing of C. albicans was accomplished with support of the NIDR and the Burroughs Wellcome Fund. Aspergillus fumigatus sequence data was obtained from The Institute for Genomic Research. Sequencing of A. fumigatus was accomplished with support from NIH, NIAID, and the Wellcome Trust. Neurospora crassa sequence data were obtained from the Neurospora Sequencing Project; Whitehead Institute/MIT Center for Genome research. Sequencing of N. crassa was accomplished with support from NSF.
REFERENCES
- 1.Aasland, R., A. F. Stewart, and T. Gibson. 1996. The SANT domain--a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional corepressor N-CoR and TFIIIB. Trends Biochem. Sci. 21:87-88. [PubMed] [Google Scholar]
- 2.Abelson, J., C. R. Trotta, and H. Li. 1998. tRNA splicing. J. Biol. Chem. 273:12685-12688. [DOI] [PubMed] [Google Scholar]
- 3.Andrau, J. C., A. Sentenac, and M. Werner. 1999. Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol. 288:511-520. [DOI] [PubMed] [Google Scholar]
- 4.Baldi, M. I., E. Mattoccia, S. Ciafrè, D. G. Attardi, and G. P. Tocchini-Valentini. 1986. Binding and cleavage of pre-tRNA by the Xenopus splicing endonuclease: two separable steps of the intron excision reaction. Cell 47:965-971. [DOI] [PubMed] [Google Scholar]
- 5.Bartholomew, B., G. A. Kassavetis, and E. P. Geiduschek. 1991. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol. Cell. Biol. 11:5181-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, B. R., D. L. Riggs, G. A. Kassavetis, and E. P. Geiduschek. 1989. Multiple states of protein-DNA interaction in the assembly of transcription complexes on Saccharomyces cerevisiae 5S ribosomal RNA genes. Proc. Natl. Acad. Sci. USA 86:2530-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buratowski, S., and H. Zhou. 1992. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell 71:221-230. [DOI] [PubMed] [Google Scholar]
- 8.Chamberlain, J. R., Y. Lee, W. S. Lane, and D. R. Engelke. 1998. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 12:1678-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaussivert, N., C. Conesa, S. Shaaban, and A. Sentenac. 1995. Complex interactions between yeast TFIIIB and TFIIIC. J. Biol. Chem. 270:15353-15358. [DOI] [PubMed] [Google Scholar]
- 10.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 11.Chu, S., J. M. Zengel, and L. Lindahl. 1997. A novel protein shared by RNase MRP and RNase P. RNA 3:382-391. [PMC free article] [PubMed] [Google Scholar]
- 12.Colbert, T., and S. Hahn. 1992. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 6:1940-1949. [DOI] [PubMed] [Google Scholar]
- 13.Colbert, T., S. Lee, G. Schimmack, and S. Hahn. 1998. Architecture of protein and DNA contacts within the TFIIIB-DNA complex. Mol. Cell. Biol. 18:1682-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cormack, B. P., and K. Struhl. 1992. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell 69:685-696. [DOI] [PubMed] [Google Scholar]
- 15.Deprez, E., R. Arrebola, W. Conesa, and A. Sentenac. 1999. A subunit of yeast TFIIIC participates in the recruitment of TATA-binding protein. Mol. Cell. Biol. 19:8042-8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dichtl, B., and D. Tollervey. 1997. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 16:417-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabbri, S., P. Fruscoloni, E. Bufardeci, E. D. N. Negri, M. I. Baldi, D. G. Attardi, E. Mattoccia, and G. P. Tocchini-Valentini. 1998. Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science 280:284-286. [DOI] [PubMed] [Google Scholar]
- 18.Fan, H., J. L. Goodier, J. R. Chamberlain, D. R. Engelke, and R. J. Maraia. 1998. Processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol. Cell. Biol. 18:3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferri, M. L., G. Peyroche, M. Siaut, O. Lefèbvre, C. Carles, C. Conesa, and A. Sentenac. 2000. A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol. Cell. Biol. 20:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flores, A., J.-F. Briand, O. Gadal, J.-C. Andrau, L. Rubbi, V. Van Mullem, C. Boschiero, M. Goussot, C. Marck, C. Carles, P. Thuriaux, A. Sentenac, and M. Werner. 1999. A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl. Acad. Sci. USA 96:7815-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiduschek, E. P., and G. A. Kassavetis. 2001. The RNA polymerase III transcription apparatus. J. Mol. Biol. 310:1-26. [DOI] [PubMed] [Google Scholar]
- 22.Hahn, S., and S. Roberts. 2000. The zinc ribbon domains of the general transcription factors TFIIB and Brf: conserved functional surfaces but different roles in transcription initiation. Genes Dev. 14:719-730. [PMC free article] [PubMed] [Google Scholar]
- 23.Hovland, P., J. Flick, M. Johnston, and R. A. Sclafani. 1989. Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene 83:57-64. [DOI] [PubMed] [Google Scholar]
- 24.Huet, J., C. Conesa, C. Carles, and A. Sentenac. 1997. A cryptic DNA binding domain at the COOH terminus of TFIIIB70 affects formation, stability, and function of preinitiation complexes. J. Biol. Chem. 272:18341-18349. [DOI] [PubMed] [Google Scholar]
- 25.Intine, R. V. A., A. L. Sakulich, S. B. Koduru, Y. Huang, E. Pierstorff, J. L. Goodier, L. Phan, and R. J. Maraia. 2000. Control of transfer RNA maturation by phosphorylation of the human La antigen on serine 366. Mol. Cell 6:339-348. [DOI] [PubMed] [Google Scholar]
- 26.Kassavetis, G. A., C. Bardeleben, A. Kumar, E. Ramirez, and E. P. Geiduschek. 1997. Domains of the Brf component of RNA polymerase III transcription factor IIIB (TFIIIB): functions in assembly of TFIIIB-DNA complexes and recruitment of RNA polymerase to the promoter. Mol. Cell. Biol. 17:5299-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kassavetis, G. A., B. Bartholomew, J. A. Blanco, T. E. Johnson, and E. P. Geiduschek. 1991. Two essential components of the Saccharomyces cerevisiae transcription factor TFIIIB: transcription and DNA-binding properties. Proc. Natl. Acad. Sci. USA 88:7308-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kassavetis, G. A., C. A. Joazeiro, M. Pisano, E. P. Geiduschek, T. Colbert, S. Hahn, and J. A. Blanco. 1992. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell 71:1055-1064. [DOI] [PubMed] [Google Scholar]
- 29.Kassavetis, G. A., A. Kumar, G. A. Letts, and E. P. Geiduschek. 1998. A post-recruitment function for the RNA polymerase III transcription initiation factor TFIIIB. Proc. Natl. Acad. Sci. USA 95:9196-9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kassavetis, G. A., A. Kumar, E. Ramirez, and E. P. Geiduschek. 1998. The functional and structural organization of Brf, the TFIIB-related component of the RNA polymerase III transcription initiation complex. Mol. Cell. Biol. 18:5587-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kassavetis, G. A., G. A. Letts, and E. P. Geiduschek. 1999. A minimal RNA polymerase III transcription system. EMBO J. 18:5042-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kassavetis, G. A., G. A. Letts, and E. P. Geiduschek. 2001. The RNA polymerase III transcription initiation factor TFIIIB participates in two steps of promoter opening. EMBO J. 20:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kassavetis, G. A., S. T. Nguyen, R. Kobayashi, A. Kumar, E. P. Geiduschek, and M. Pisano. 1995. Cloning, expression, and function of TFC5, the gene encoding the B" component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc. Natl. Acad. Sci. USA 92:9786-9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kassavetis, G. A., D. L. Riggs, R. Negri, L. H. Nguyen, and E. P. Geiduschek. 1989. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol. Cell. Biol. 9:2551-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khoo, B., B. Brophy, and S. P. Jackson. 1994. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 8:2879-2890. [DOI] [PubMed] [Google Scholar]
- 36.Kumar, A., G. A. Kassavetis, E. P. Geiduschek, M. Hambalko, and C. J. Brent. 1997. Functional dissection of the B" component of RNA polymerase III transcription factor IIIB: a scaffolding protein with multiple roles in assembly and initiation of transcription. Mol. Cell. Biol. 17:1868-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, J. Y., C. F. Evans, and D. R. Engelke. 1991. Expression of RNase P RNA in Saccharomyces cerevisiae is controlled by an unusual RNA polymerase III promoter. Proc. Natl. Acad. Sci. USA 88:6986-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, J. Y., C. E. Rohlman, L. A. Molony, and D. R. Engelke. 1991. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol. Cell. Biol. 11:721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lefèbvre, O., J. Rüth, and A. Sentenac. 1994. A mutation in the largest subunit of yeast TFIIIC affects tRNA and 5 S RNA synthesis. Identification of two classes of suppressors. J. Biol. Chem. 269:23374-23381. [PubMed] [Google Scholar]
- 40.López-de-León, A., M. Librizzi, K. Puglia, and I. M. Willis. 1992. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell 71:211-220. [DOI] [PubMed] [Google Scholar]
- 41.Lygerou, Z., P. Mitchell, E. Petfalski, B. Seraphin, and D. Tollervey. 1994. The Pop1 gene encodes protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 8:1423-1433. [DOI] [PubMed] [Google Scholar]
- 42.Marck, C., O. Lefèbvre, C. Carles, M. Riva, N. Chaussivert, A. Ruet, and A. Sentenac. 1993. The TFIIIB-assembling subunit of yeast transcription factor TFIIIC has both tetratricopeptide repeats and basic helix-loop-helix motifs. Proc. Natl. Acad. Sci. USA 90:4027-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Margottin, F., G. Dujardin, M. Gerard, J. M. Egly, J. Huet, and A. Sentenac. 1991. Participation of the TATA factor in transcription of the yeast U6 gene by RNA polymerase C. Science 251:424-426. [DOI] [PubMed] [Google Scholar]
- 44.McCulloch, V., P. Hardin, W. C. Peng, J. M. Ruppert, and S. M. Lobo-Ruppert. 2000. Alternatively spliced hBRF variants function at different RNA polymerase III promoters. EMBO J. 19:4134-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miao, F., and J. Abelson. 1993. Yeast tRNA-splicing endonuclease cleaves precursor tRNA in a random pathway. J. Biol. Chem. 268:672-677. [PubMed] [Google Scholar]
- 46.Mital, R., R. Kobayashi, and N. Hernandez. 1996. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol. Cell. Biol. 16:7031-7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moir, R. D., I. Sethy-Coraci, K. Puglia, M. D. Librizzi, and I. M. Willis. 1997. A tetratricopeptide repeat mutation in yeast transcription factor IIIC131 (TFIIIC131) facilitates recruitment of TFIIB-related factor TFIIIB70. Mol. Cell. Biol. 17:7119-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niranjanakumari, S., T. Stams, S. M. Crary, D. W. Christianson, and C. A. Fierke. 1998. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc. Natl. Acad. Sci. USA 95:15212-15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Proudfoot, N. 2000. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 25:290-293. [DOI] [PubMed] [Google Scholar]
- 50.Rameau, G., K. Puglia, A. Crowe, I. Sethy, and I. Willis. 1994. A mutation in the second largest subunit of TFIIIC increases a rate-limiting step in transcription by RNA polymerase III. Mol. Cell. Biol. 14:822-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts, S., S. J. Miller, W. S. Lane, S. Lee, and S. Hahn. 1996. Cloning and functional characterization of the gene encoding the TFIIIB90 subunit of RNA polymerase III transcription factor TFIIIB. J. Biol. Chem. 271:14903-14909. [DOI] [PubMed] [Google Scholar]
- 52.Rüth, J., C. Conesa, G. Dieci, O. Lefèbvre, A. Düsterhoft, S. Ottonello, and A. Sentenac. 1996. A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 15:1941-1949. [PMC free article] [PubMed] [Google Scholar]
- 53.Schramm, L., P. S. Pendergrast, Y. L. Sun, and N. Hernandez. 2000. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev. 14:2650-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sethy-Coraci, I., R. D. Moir, A. López-de-León, and I. M. Willis. 1998. A differential response of wild type and mutant promoters to TFIIIB70 overexpression in vivo and in vitro. Nucleic Acids Res. 26:2344-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shah, S. M., A. Kumar, E. P. Geiduschek, and G. A. Kassavetis. 1999. Alignment of the B" subunit of RNA polymerase III transcription factor IIIB in its promoter complex. J. Biol. Chem. 274:28736-28744. [DOI] [PubMed] [Google Scholar]
- 56.Shen, Y., G. A. Kassavetis, G. O. Bryant, and A. J. Berk. 1998. Polymerase (Pol) III TATA box-binding protein (TBP)-associated factor Brf binds to a surface on TBP also required for activated Pol II transcription. Mol. Cell. Biol. 18:1692-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stagljar, I., S. te Heesen, and M. Aebi. 1994. New phenotype of mutations deficient in glucosylation of the lipid-linked oligosaccharide: cloning of the ALG8 locus. Proc. Natl. Acad. Sci. USA 91:5977-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stolc, V., and S. Altman. 1997. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes Dev. 11:2414-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teichmann, M., and K. H. Seifart. 1995. Physical separation of two different forms of human TFIIIB active in the transcription of the U6 or the VAI gene in vitro. EMBO J. 14:5974-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teichmann, M., Z. Wang, and R. G. Roeder. 2000. A stable complex of a novel transcription factor IIB-related factor, human TFIIIB50, and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc. Natl. Acad. Sci. USA 97:14200-14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trotta, C. R., F. Miao, E. A. Arn, S. W. Stevens, C. K. Ho, R. Rauhut, and J. N. Abelson. 1997. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89:849-858. [DOI] [PubMed] [Google Scholar]
- 63.Wang, Z., and R. G. Roeder. 1997. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 11:1315-1326. [DOI] [PubMed] [Google Scholar]
- 64.Werner, M., N. Chaussivert, I. M. Willis, and A. Sentenac. 1993. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J. Biol. Chem. 268:20721-20724. [PubMed] [Google Scholar]
- 65.White, R. J. 1998. RNA polymerase III transcription, 2nd ed., vol. XIV. Springer-Verlag & Landes Bioscience, New York, N.Y.
- 66.Willis, I., P. Schmidt, and D. Söll. 1989. A selection for mutants of the RNA polymerase III transcription apparatus: PCF1 stimulates transcription of tRNA and 5S RNA genes. EMBO J. 8:4281-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiao, S. H., F. Houser-Scott, and D. R. Engelke. 2001. Eukaryotic ribonuclease P: increased complexity to cope with the nuclear pre-tRNA pathway. J. Cell. Physiol. 187:11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoo, C. J., and S. L. Wolin. 1994. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol. Cell. Biol. 14:5412-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoo, C. J., and S. L. Wolin. 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89:393-402. [DOI] [PubMed] [Google Scholar]