Abstract
The 5′ region of the H19 gene harbors a methylation-sensitive chromatin insulator within an imprinting control region (ICR). Insertional mutagenesis in combination with episomal assays identified nucleosome positioning sequences (NPSs) that set the stage for the remarkably precise distribution of the four target sites for the chromatin insulator protein CTCF to nucleosome linker sequences in the H19 ICR. Changing positions of the NPSs resulted in loss of both CTCF target site occupancy and insulator function, suggesting that the NPSs optimize the fidelity of the insulator function. We propose that the NPSs ensure the fidelity of the repressed status of the maternal Igf2 allele during development by constitutively maintaining availability of the CTCF target sites.
An increasing number of autosomal genes are found to be monoallelically expressed in the soma (1, 5, 18, 20). A subset of these genes are expressed in a parent-of-origin-dependent manner (1, 11). This phenomenon, termed genomic imprinting, provides unique means to assess mechanisms of epigenetically controlled silencing and activation events, since the same somatic cell displays one active and one inactive parental allele. Recent advances suggest that the manifestation of silenced and activated states of imprinted genes involves epigenetically regulated chromatin insulator functions (3, 8, 10, 12-14).
Chromatin insulators organize expression domains by blocking the communication between gene promoters and enhancers or silencers (4). Importantly, insulators generally function only when positioned between the enhancers and promoters and do not directly interfere with enhancer and silencer functions. Most of our knowledge on insulators derives from work on Drosophila melanogaster, although a few vertebrate insulators have been recently uncovered, such as the chicken β-globin FII insulator, the Xenopus repeat organizer elements and the BEAD-A insulator from the T-cell-receptor γ locus (2). One of the most recently identified mammalian insulators maps to the H19 imprinting control region (ICR), which is located approximately 4 to 2 kb upstream of the H19 promoter (3, 8, 10, 12, 13). The H19 ICR directs repression of H19 on the paternal chromosome and repression of Igf2 on the maternal chromosome. This latter feature is perceived to reflect the ability of the H19 ICR to insulate downstream enhancers from the Igf2 promoters on the maternally derived chromosome.
The 11-zinc-finger protein CTCF interacts with the core sequences of all currently known vertebrate insulators (4, 19). Mutational analyses revealed that the CTCF target sites of the H19 ICR are both necessary and sufficient for the insulator function (3, 8, 13). The Igf2/H19 chromatin insulator function is methylation sensitive (10), and chromatin immunopurification analyses revealed a preferential association with the maternal H19 ICR allele (13). Intriguingly, a detailed examination revealed that some of the CTCF target sites map to linker regions which are flanked by positioned nucleosomes on the maternally inherited allele (12, 13). This interpretation has been contended by others who argue that the multiple nuclease hypersensitive sites, some of which depend on CTCF, reflect a nucleosome-free region within the H19 ICR (9, 15).
Given the functional importance of the H19 ICR, we decided to examine the issue of nucleosome positioning more closely by insertional mutagenesis of episomal H19 ICR, followed by transfection and chromatin conformation analyses in stably transfected cells. Our results show that the chromatin insulator function is controlled by multiple nucleosome positioning sites which generate stable nucleosome positioning patterns. The insertion of fragments of various sizes in a strategic point in the H19 ICR upset this balance by repositioning nucleosomes. This led not only to an attenuation of CTCF-target site interaction but also to loss of the insulator function. These observations have implications for a diverse range of topics, such as our understanding of the maintenance of the functional imprint and the emergence of the H19 ICR on the evolutionary stage.
MATERIALS AND METHODS
Cell culture and transfection.
The Hep-3B cell line maintenance and the transfection of plasmid DNAs into these cells followed previously published protocols (12). Stable, episomally transfected Hep3B cells were selected by hygromycin B, as has been described (12).
MNase and DNase I treatments and Southern blot hybridization analysis.
Nuclei were isolated and subjected to DNase and micrococcal nuclease (MNase) digestions, as has been described (12). The restriction enzyme-digested DNA samples were electrophoresed in 1.7% agarose gels, depurinated, and blotted to Hybond N+ membranes followed by hybridization according to routine protocols. The following probes were used. Probe 1 is a 218-bp fragment (PCR amplified using the 5′ sequence 5′ AAG TCT CGT ACA TGG CAG T 3′ and the 3′ sequence 5′ CCA CAC ATA GTA GCT ATA CTT C 3′) located at −3401 to −3618 bp from the cap site of the H19 gene; probe 2 is a 117-bp AflIII-FokI fragment encompassing sequence from −1253 to −1370 with respect to the cap site of the mouse H19 gene; probe 3 is a 1,406-bp fragment (PCR amplified with the 5′ sequence 5′ AAA GCG GCC GCA GGG CCA TAG TGT GAG TTT AGG 3′ and the 3′ sequence 5′ AAA GCG GCC GCC CAT GAT CAC CAC ACA TAG TAG CT 3′) located at −3389 to −4798 bp from the cap site of H19 gene; probe 4 is a 180-bp fragment (generated by PCR amplification with the 5′ primer sequence 5′-TAG GAT GCC AAG AAG CTA T 3′ and the 3′ primer sequence 5′ CGA ATT CCA GCA CAC TGG CG 3′) located at position +2400 from the cap site of H19 gene; probe 5 is a 292-bp fragment (generated by PCR amplification with forward primer sequence of 5′-TGC AGT ACC ATA ATG CAG ACC 3′ and reverse primer sequence of 5′ AAG ATG ACA GTC ACC AGC GC 3′ followed by BglII digestion) encompassing positions −2366 to −1729 from the cap site of the H19 gene. All probe fragments were radiolabeled by using a multiprime labeling kit and [α-32P]dCTP (Amersham) to a specific activity of more than 108 cpm/μg.
Plasmid cloning strategies.
The construction of the basic pREPH19 vectors has been described previously (12, 13). The insulator function is based on the strategic positioning of the H19 ICR in a 3′ position of the H19 reporter gene, i.e., between the promoter and simian virus 40 (SV40) enhancer. To facilitate chromatin analyses, the 5′ H19 ICR present in the original pREPH19A vector was deleted by XbaI digestion, followed by religation. As previously reported, absence of the H19 ICR in the 5′ position did not affect the insulator assay (12). This modified pREPH19A vector was the basis for a modified pREPH19B (with the wild-type H19 ICR positioned between the SV40 enhancer and the promoter of the H19 reporter gene) and all pREPH19mutB vectors (with a mutated instead of a wild-type H19 ICR).
(i) pREPH19mutB1-4 constructs.
A point mutation was created in the ICR (cloned in the PCR2.1 vector; Pharmacia) at position −2371 with respect to the cap site of the H19 gene (between nuclease-hypersensitive sites [NHSSs] III and IV) to create a new AgeI restriction site by changing CAG to CGG. The primers used for PCR amplification of the pREPH19mutB construct were 5′ GCC GAG CAG CGA ACC AGT GCA GTC CCA CAT AC 3′ and 5′ GTA TGT GGG ACT GCA CTG GTT CGC TGC TCG GC 3′. The neutral fragments of different sizes, i.e., 80, 140, and 280 bp (PCR amplified from the multiple cloning site of the pGEM-T Easy vector [Promega] using the common 5′ primer sequence 5′ ATA CCG GTT GAA GGT CGA CCA TAT GG3′ and different 3′ primer sequences: 5′ ATA CCG GTA CGC CAA GCT ATT TAG GTG 3′ for the 80-bp fragment, 5′ ATA CCG GTT GCA GGT CGA CCA TAT GG 3′ for the 140-bp fragment, and 5′ ATA CCG GTG GAA TTG TGA GCG GAT AAC 3′ for the 280-bp fragment) were cloned into the AgeI site of the H19 ICR. From these constructs, the ICR fragment was excised with KpnI and XhoI and inserted into the modified pREP4H19A plasmid to generate the pREPH19mutB constructs.
(ii) pREP4H19mutB1inv.
The pREP4H19mutB1inv construct was created in the same manner as the pREP4H19mutB1 plasmid but with the orientation of the 80-bp fragment in the pCR2.1 construct reversed prior to insertion into the modified pREPH19AB plasmid.
(iii) pREP4H19mutB1* and pREP4H19mutB2*.
A point mutation was created in NHSS I of the ICR (cloned in the PCR2.1 vector) containing the 80- and 140-bp insertions between NHSS I and II, as has been described (13). This mutated ICR was cloned into the modified pREP4H19A construct as described above.
(iv) pREP4H19mutB4.
The same 80-bp neutral fragment (PCR amplified with the primers mentioned for the pREP4H19mutB1 construct but equipped with BspEI end sites instead of the AgeI site ends) was cloned upstream of NHSS I of the ICR at the BspEI site (−3033 with respect to the cap site of the H19 gene).
(v) pREPH19B5′.
A 1,406-bp fragment, encompassing the sequence from bp −3389 to −4798 from the cap site of H19 gene (PCR amplified with the primer sequences mentioned above for the probe 3 fragment), was cloned into a NotI site in the modified pREP4H19A construct.
Expression and mobility shift analyses.
Assays for RNase protection of the mouse H19 reporter gene and mobility shift assays were performed as has been described (6, 12, 13). The in vitro chromatin reconstitution using rat liver histone extracts (kind gift from ö. Wrange) followed published protocols (16). Recombinant CTCF was expressed in Pichia pastoris and purified by cation-exchange chromatography (21). The 178-bp fragment used for the band shift analyses was PCR amplified with the 5′ primer 5′ TTCAGTCTTGCGCCCTTCA 3′ and the 3′ primer 5′ CTTTGTTGAACCTGGGGTA 3′, encompassing positions −2232 to −2054 with respect to the cap site of the H19 gene.
RESULTS
Nucleosome positioning sites are factor independent and unequally distributed in the H19 ICR.
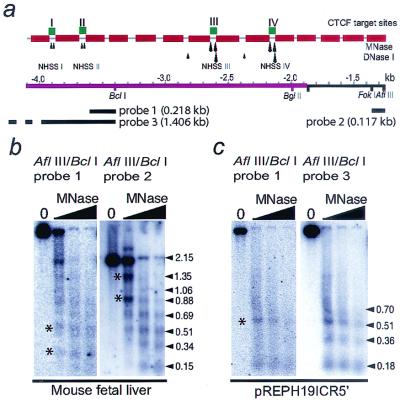
It was previously shown that sequences downstream of an intrinsic BclI site, which is located −3.5 kb upstream of the H19 transcription start site and divides the H19 ICR in halves, are covered by positioned nucleosomes (Fig. 1a) (12). To examine whether this feature extends to the entire ICR, a 218-bp fragment was used to visualize sequences 5′ to the BclI site towards an AflIII site in the 5′ region of the H19 ICR (Fig. 1a). As expected, the unique chromatin conformation with positioned nucleosomes with NHSSs in the linker regions extended throughout the ICR (Fig. 1b). We note with interest that all four CTCF target sites colocalize with NHSSs which map to the linker region flanked by positioned nucleosomes. This conclusion is based on a combination of DNase I footprinting of CTCF target sites (13) and an estimation of nucleosome-linker boundaries (with a precision of ±10 bases [standard error of the mean]) based on seven independent Southern blot analyses (data not shown). While NHSSs I and II were only occasionally observed, NHSSs III and IV, previously termed NHSSs I and II, are much more marked (Fig. 1b) (12).
FIG. 1.
The NPSs map to the 3′ end of the H19 ICR. (a) Schematic map of CTCF target sites (green boxes) and restriction sites and probes used in this study. The nucleosome positions (red boxes) and distribution of NHSSs were identified previously (12) and complemented in this study. Strong NHSSs are shown with larger arrows, and weaker NHSSs are shown with smaller arrows. The four CTCF target sites colocalize with the NHSSs (12), as indicated. The cerise bar depicts the differentially methylated region. The numbers indicate nucleotide positions relative to the transcriptional start site of the H19 gene. (b) Indirect end labeling of MNase-digested chromatin DNA from mouse fetal liver from the BclI and AflIII sites using probes 1 and 2, respectively. The asterisks depict the CTCF target sites and their position in linker regions. (c) Indirect end labeling of MNase-digested chromatin DNA from cells with a stably maintained episome harboring a 1.4-kb 5′ ICR fragment (see Materials and Methods). The blot was hybridized first to a short fragment (probe 1) and then to a full-length fragment (probe 3) to assess the nucleosome positioning pattern of this region.
The nucleosome positioning feature is intrinsic to the ICR, since it has previously been shown that sequences 3′ of the BclI site (including some H19 ICR-H19 promoter spacer sequences) established correct nucleosome positioning pattern in an episomal assay (12). The principles of nucleosome positioning assays are based upon direct or indirect end labeling, which allows the visualization of nucleosomes stably positioned over particular sequences. By inserting portions of the ICR fragment into an episome (pREPH19B5) followed by transfection into cells, propagation of hygromycin-selected transfectants, and MNase digestion of isolated nuclei, we examined if this feature extended to the 5′ H19 ICR. Figure 1c shows that in contrast to the endogenous sequences, the episomal version of the first half of the H19 ICR displayed a smeared signal when a short fragment was used as a probe (probe 1). Since in vivo positioned nucleosomes can be identified only by indirect end labeling, this result suggested that the first half of the H19 ICR has no nucleosome positioning feature. This conclusion is underscored by the demonstration that the nucleosome bands appeared more clearly when the blot was hybridized to a longer fragment (probe 3); i.e., although the sequence is covered by nucleosomes in the transfected cells, they are not positioned. This result also indicates that there is no nucleosome positioning feature imposed by the vector itself. We conclude that nucleosome positioning sites in the H19 ICR are restricted to its 3′ portion.
The H19 ICR harbors multiple nucleosome positioning sequences (NPSs).
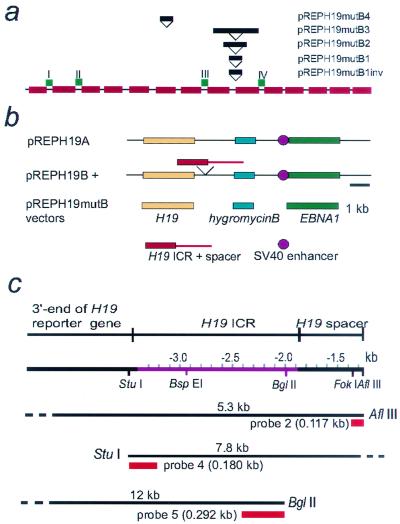
To examine more closely the origin of the positioning feature in the 3′ portion of the H19 ICR, DNA fragments of 80, 140, and 280 bp encompassing almost one half, almost one, and almost two turns, respectively, of the average length of nucleosomal DNA were inserted into different positions followed by structural and functional analyses of the chromatin. The different fragments were parts of a multiple cloning site. Figure 2a depicts the insertion sites in relation to the chromatin map of the H19 ICR. The mutated H19 ICRs were inserted between the promoter of the mouse H19 reporter gene and the SV40 enhancer (Fig. 2b). The nucleosome positioning assays were performed by indirect end labeling of MNase-treated episomal DNA, which was stably propagated in Hep3B cells. To facilitate interpretation of Southern chromatin blot analyses, we have also indicated restriction enzymes, probes, and the resulting fragments (Fig. 2c).
FIG. 2.
Schematic map of the episomal pREPH19 displacement vectors. (a) Positions of the insertion of the neutral fragment (part of a multiple cloning site and containing an AflIII site). The red rectangles depict nucleosomes positions. (b) Schematic drawing of the episomes used in this study. Although their general design is as previously described, all episomes of this study lacked the H19 ICR in the 5′ position, i.e., upstream of the H19 reporter gene, to facilitate chromatin conformation analyses. (c) Restriction enzymes, probes, and restriction fragment lengths, described in this report, on the chromatin map of the H19 ICR.
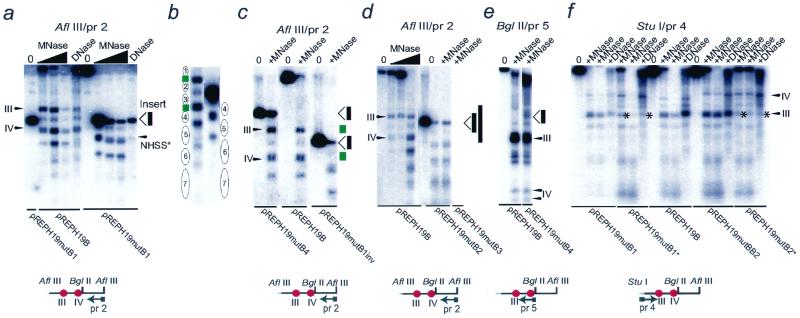
When the 80-bp fragment was inserted between NHSSs III and IV to generate the pREPH19mutB1 construct, the nucleosomes downstream of this insert were found at a new location (Fig. 3a and b). This observation suggested the existence of a nucleosome positioning domain upstream of the insertion site between NHSSs III and IV. Since both orientations of the insert in the position between NHSSs III and IV generated an identical result (Fig. 3c), this vector effect could not be due to any direction-driven property of the fragment. Strikingly, when the longer versions of this fragment (140- and 280-bp insertions to generate the pREPH19mutB2 and -3 plasmids) were inserted in the same position of the H19 ICR, the patterns of nucleosome displacements 3′ of the insertion site were identical to that observed in the pREPH19mutB1 construct, in stably transfected cells (Fig. 3d). In all of these instances, the nucleosome positioning patterns were identical upstream of NHSS III (Fig. 3f). Other changes were observed when the 80-bp fragment was inserted instead at a position upstream of NHSS III to generate the pREPH19mutB4 construct: nucleosomes flanking the insertion site in the pREPH19mutB4 construct by and large retained their positioning pattern (Fig. 3c and e), suggesting that the flanking nucleosomes could not be displaced due to NPSs.
FIG. 3.
The identification of NPSs in the H19 ICR. (a to d) Indirect end labeling from the 3′ AflIII site, as described previously (12). The presence of an additional AflIII site in the neutral fragment uncovered its insertion site. NHSS* (a) marks a new NHSS which is not identical to NHSS IV, since that CTCF target site (green box) has not been shifted with respect to the position of the probe. The ovals depict nucleosomes, which have been numbered to emphasize the change in nucleosome phasing. (e) Positioned nucleosomes and NHSSs upstream of the BglII site. (f) Indirect end labeling from the 5′ end of the ICR insert. Asterisks depict loss of nuclease hypersensitivity of NHSS III, which was mutated in the pREPH19mutB1 and -B2* constructs. The gray line depicts the 3′ end of the mouse H19 reporter gene, which is separated from the H19 ICR by a multiple cloning site in the episomal constructs. The red circles and roman numerals in the restriction and probe map underneath each panel depict NHSSs III and IV.
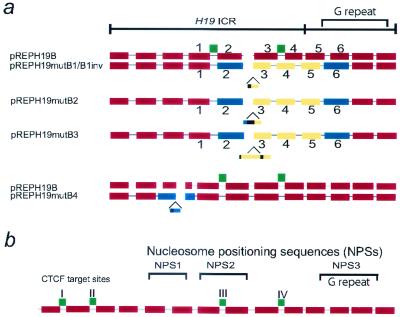
Figure 4 summarizes these results by marking the displaced nucleosomes and nucleosomes on wild-type H19 ICR. Several conclusions can be made. Firstly, the nucleosomes nr 2 and 6 (Fig. 4a; see also Fig. 3b) are not displaced in any of the insertion mutants. Secondly, the DNA sequences covered by nucleosome nr 6 are extended in the insertion mutant ICRs. We interpret this to mean that the insertion mutants upset a balance between at least two different and antagonistic NPSs. These sites map to or in the vicinity of nucleosomes nr 2 and 6. Interestingly, the extended domain covered by nucleosome nr 6 maps to a repeat sequence (TATAG[T/C]GGGG) which is similar to a previously discovered TATA tetrad repeat element with strong nucleosome positioning properties (22).
FIG. 4.
Summary of the effect of insertion mutagenesis on chromatin conformations. (a) Interpretation of data in Fig. 3 to show local or directional displacements of nucleosomes by the neutral inserts depending on their position within the H19 ICR. The displaced nucleosomes (including those on the inserted sequences) are depicted as yellow boxes or blue if demarcating borders of change. Nucleosomes on the wild-type H19 ICR are in red. (b) The H19 ICR chromatin map is based on the maternal H19 ICR allele of mouse fetal liver and interprets the results in Fig. 3. The insertional mutagenesis strategy identified several NPSs.
The nucleosome positioning pattern is essential for the insulator function.
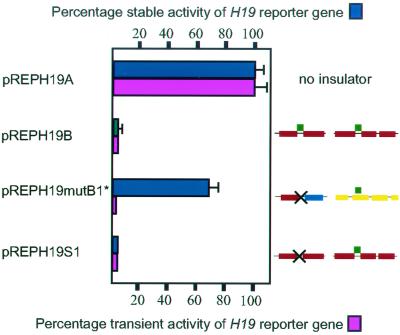
We have proposed previously that nucleosome positioning features are intimately linked with the insulator function of the H19 ICR (12). To put this contention to the test, we analyzed the ability of the H19 ICR (with an 80-bp fragment inserted between NHSSs III and IV) in the pREPH19mutB1 construct to insulate the reporter gene from the strong SV40 enhancer in stably maintained episomes in transfected Hep3B cells. In this construct, the CTCF target site IV is covered by a nucleosome, whereas the CTCF target site III remained available (see above). Since it has been shown (see also vector pREPH19S1 in Fig. 5) that each of the CTCF target sites III and IV has insulator function (13), we mutated the CTCF target site III in pREPH19B1 to generate pREPH19B1*. This and other control vectors were propagated in Hep3B cells for at least 2 months in the presence of hygromycin prior to analysis of reporter gene expression and copy numbers, as reported earlier (12). Figure 5 (blue bars) shows that the insulator function was strongly reduced when CTCF target site III was mutated while CTCF target site IV was covered by a nucleosome. Strikingly, when the performance of these plasmids was examined by transient-transfection analyses, all H19 ICR constructs, including pREPmutB1*, displayed potent insulator activity (Fig. 5, pink bars). Collectively, these results show that the Igf2/H19 insulator function depends on the superimposition mode of the nucleosome positioning and CTCF site distribution patterns.
FIG. 5.
Nucleosome positioning critically contributes to the Igf2/H19 insulator function. The H19 reporter gene activity in stable, episomally transfected Hep3B cells was assessed by RNase protection analysis. The expression levels were normalized not only for RNA input but also for episome copy number. The mean deviation of at least three different experiments is indicated for each vector construct, unless the differences were too small to allow visualization. The SV40 enhancer-driven expression of the modified pREPH19A construct was, for convenience, assigned a value of 100, and all other values were related to this. The central portion of the ICR and its chromatin map are visualized for each of the constructs. Green boxes depict CTCF target sites with the point-mutated site crossed out in the pREPH19mutB1* and pREPH19S1 constructs. The S1 construct is described elsewhere (13). The CTCF target sites are indicated by green boxes. See Fig. 1 and 4 for additional information.
Nucleosomes regulate the interaction between CTCF and its target sites.
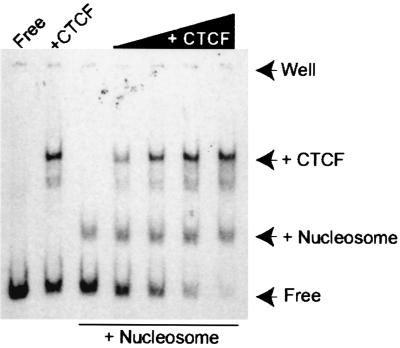
To further examine the nucleosome-CTCF link, we generated a 178-bp fragment with NHSS IV positioned in the middle. The central position of the CTCF target site ensured that it would always be covered by a nucleosome upon in vitro chromatin reconstitution. Figure 6 shows that this fragment interacted with either recombinant CTCF or nucleosomes. When nucleosomes were first reconstituted on the fragment followed by CTCF incubation, however, there was no indication of any supershift of the nucleosome-fragment complex despite the fact that essentially all of the remaining free fragment interacted with the recombinant CTCF. The sensitivity of CTCF to the chromatin reconstitution protocol precluded a reversal of the order of nucleosome-CTCF incubations. Nonetheless, the results collectively underscore our deduction that nucleosomes regulate the availability of target sites for CTCF in the H19 ICR.
FIG. 6.
Nucleosomes regulate interaction of CTCF with target sites. The band-shifted PCR fragment contained the NHSS IV site in its central portion. The arrows depict the position of the fragment when this was complexed with rat liver histone extracts and/or recombinant CTCF. The position of the sample well is indicated to show the absence of any band shift that is larger than the CTCF-DNA complex.
DISCUSSION
The chromatin conformation of the H19 ICR is essential to our understanding of the establishment and manifestation of the epigenetic marks associated with this region. Whereas one report has claimed that both paternal H19 ICR alleles display positioned nucleosomes (12), two other groups claim that the H19 ICR is nonnucleosomal (9, 15). However, neither of those two studies exploited the indirect end-labeling technique in association with MNase digestions that is absolutely essential for uncovering patterns of positioned nucleosomes. We resolve this controversy by showing here that insertion of DNA fragments of various lengths displaced the nucleosome positioning in a pattern that depended on where they were inserted within the ICR. Importantly, displacement of nucleosomes over a CTCF target site led to occlusion of CTCF and loss of insulator function in episomes that were stably propagated in transfected cells. We propose that the nucleosome positioning feature presumably has facilitated and optimized the insulator function. Our results also suggest that CTCF reads the parental mark in a postreplicative manner.
The H19 ICR displays a unique chromatin conformation.
Strikingly, all four CTCF target sites map to linker regions between positioned nucleosomes. By insertional mutagenesis we were also able to show that the nucleosome positioning feature depends on at least two NPSs within the H19 ICR as well at least one NPS site in the 3′ region of the H19 ICR. The inserted sequences are unlikely to provide any nucleosome positioning signal themselves for the following reasons: (i) the nucleosome displacement was independent of the orientation or the length of the insert, and (ii) the nucleosome displacement depended on the position and not the sequence of the insert, as documented by using pREPH19mutB1-3 and pREPH19mutB4 constructs. We cannot rule out the existence of additional NPSs in the 5′ region of the H19 ICR. Such NPSs might jointly with the H19 ICR NPSs ensure that the nucleosome positioning feature extends throughout the H19 ICR.
Our results raise several novel questions. How did the NPSs and insulator function evolve? Did the NPS functions adapt to the insulator function or vice versa? Or did these two functions coevolve? Preliminary results (M. Kanduri et al., unpublished observation) show that most of the nucleosome positioning features can be established in vitro with pure histone octamers, suggesting that the NPSs constitute factor-independent nucleosome-attracting sequences. Such sequences are not expected to display evolutionarily conserved sequence composition, since a wide variety of sequences are able to attract or repel nucleosomes. Indeed, although there are no detectable sequence similarities between mouse and human H19 ICRs beyond the CTCF target sites, the NPS feature is also present in the human H19 ICR (Kanduri et al., unpublished). Although these considerations imply that the NPSs evolved subsequent to the emergence of CTCF target sites, they do not explain the precise distribution of the four CTCF target sites to linker regions flanked by positioned nucleosomes. We envisage, therefore, that the sequential duplication of CTCF target sites and the subsequent adjustment of distances between these elements coevolved with the NPSs in order to increase the stringency of the insulator function and hence the repressed status of the maternal Igf2 allele. These considerations are supported by our observation that modifying the distance between NPSs 2 and 3 upset the nucleosome phasing, resulting in loss of insulator function.
CTCF is not a nucleosome positioning factor.
The H19 ICR constitutes the first demonstration that CTCF target sites map to linker regions which are flanked by positioned nucleosomes (12, 13). However, additional examples with similar features are now emerging in the DM1 locus (7), the Myc locus (V. Lobanenkov et al., unpublished observation) and an ICR in the Kcnq1 locus (Kanduri et al., unpublished). It is unexpected, therefore, that CTCF is not a nucleosome positioning factor. This conclusion is based on the following results and arguments: (i) the NPS function did not include the CTCF target site in NHSS IV; (ii) the nucleosomes were also positioned on the paternal ICR allele, which does not interact with CTCF in vivo; (iii) the chromatin of the pREPH19mutB1* construct (which contains an 80-bp insert between NHSSs III and IV and has the NHSS III site point mutated) did not display nuclease hypersensitivity for either the NHSS III (Fig. 3c) or IV site but nevertheless displayed positioned nucleosomes; (iv) the 5′ portion of the H19 ICR contains two CTCF target sites, although this region is not able to maintain positioned nucleosomes in episomes and in vitro-reconstituted chromatin; and (v) the correct nucleosome positioning pattern could be recapitulated in Drosophila conceptuses which were transgenic for the mouse H19 ICR (Kanduri et al., unpublished), despite the fact that Drosophila CTCF does not interact with the H19 ICR (E. Pugacheva and V. Lobanenkov, unpublished observations). We propose that the regulation of CTCF function by chromatin conformation has necessitated the emergence of a general theme: the coevolution of NPSs to ensure constitutive availability of CTCF target sites.
The CTCF-nucleosome connection.
Due to exceptionally high-affinity binding and stability of CTCF-DNA complexes on the ICR targets (13), they might be expected to outcompete nucleosomes and to establish the insulator function in close association with DNA replication. However, the different outcomes of the transient- and stable-transfection analyses of Fig. 5 presumably reflected a differential competition between CTCF and nucleosomes. The exclusion of CTCF from NHSS IV in the stably propagated pREPmutB1* vector indicates that CTCF is absent from the replicase complex and that it cannot bind once the target site becomes covered by a nucleosome. This deduction was further supported by our observation of nucleosomes preventing the interaction between CTCF and its target site when the nucleosomes were allowed to interact with H19 ICR sequences before the addition of CTCF in band shift assays. Conversely, in the transient-transfection experiments, CTCF apparently outcompeted the pool of histone octamers in a replication-independent process (17). When these data are taken together, there is strong support for the possibility that CTCF target site interaction is regulated by chromatin and that CTCF cannot be a DNA-bound part of the replication complex that copies the maternal H19 ICR allele.
Acknowledgments
We gratefully acknowledge valuable advice during the course of this work from Andreas Reik, Andy P. Feinberg, Gary Franklin, Sandy Morse, Rainer Renkawitz, Alan Wolffe, and Örjan Wrange.
This work was supported by grants from the Natural Science Research Council (NFR), the Swedish Cancer Research Foundation (RmC/CF), the Swedish Pediatric Cancer Foundation (BCF), and the Lundberg Foundation to R.O. and intramural NIAID NIH Research Funding to V.L.
REFERENCES
- 1.Bartolomei, M. S., and S. M. Tilghman. 1997. Genomic imprinting in mammals. Annu. Rev. Genet. 31:493-525. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A., A. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 3.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. [DOI] [PubMed] [Google Scholar]
- 4.Bell, A. C., A. G. West, et al. 2001. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science 291:447-450. [DOI] [PubMed] [Google Scholar]
- 5.Constancia, M., W. Dean, et al. 2000. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat. Genet. 26:203-206. [DOI] [PubMed] [Google Scholar]
- 6.Filippova, G., S. Fagerlie, E. Klenova, C. Myers, Y. Dehner, G. Goodwin, P. Neiman, S. Collins, and V. Lobanenkov. 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippova, G. N., C. P. Thienes, B. H. Penn, D. H. Cho, J. J. Hu, J. M. Moore, T. R. Klesert, V. V. Lobanenkov, and S. J. Tapscott. 2001. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet. 28:335-343. [DOI] [PubMed] [Google Scholar]
- 8.Hark, A. T., C. J. Schoenherr, D. J. Katz, R.S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486-489. [DOI] [PubMed] [Google Scholar]
- 9.Hark, A. T., and S. M. Tilghman. 1998. Chromatin conformation of the H19 epigenetic mark. Hum. Mol. Genet. 7:1979-1985. [DOI] [PubMed] [Google Scholar]
- 10.Holmgren, C., C. Kanduri, G. Dell, A. Ward, R. Mukhopadhya, M. Kanduri, V. Lobanenkov, and R. Ohlsson. 2001. CpG methylation regulates the Igf2/H19 insulator. Curr. Biol. 11:1128-1130. [DOI] [PubMed] [Google Scholar]
- 11.Horsthemke, B., M. A. Surani, T. C. James, and R. Ohlsson. 1999. The mechanisms of genomic imprinting, p. 91-118. In R. Ohlsson (ed.), Genomic imprinting: an interdisciplinary approach. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 12.Kanduri, C., C. Holmgren, G. Franklin, M. Pilartz, E. Ullerås, M. Kanduri, L. Liu, V. Ginjala, E. Ulleras, R. Mattsson, and R. Ohlsson. 2000. The 5′-flank of the murine H19 gene in an unusual chromatin conformation unidirectionally blocks enhancer-promoter communication. Curr. Biol. 10:449-457. [DOI] [PubMed] [Google Scholar]
- 13.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C.-F. Qi, A. Wolffe, R. Ohlsson, and A. Lobanenkov. 2000. Functional interaction of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10:853-856. [DOI] [PubMed] [Google Scholar]
- 14.Kanduri, C., G. Fitzpatrick, R. Mukhopadhyay, M. Kanduri, V. Lobanenkov, M. Higgins, and R. Ohlsson. A differentially methylated imprinting control region within the Kcnq1 locus harbours a methylation-sensitive chromatin insulator. J. Biol. Chem., in press. [DOI] [PubMed]
- 15.Khosla, S., A. Aitchison, R. Gregory, N. D. Allen, and R. Feil. 1999. Parental allele-specific chromatin configuration in a boundary/imprinting-control element upstream of the mouse H19 gene. Mol. Cell. Biol. 19:2556-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Q., U. Bjork, and O. Wrange. 1999. Assays for interaction of transcription factor with nucleosome. Methods Enzymol. 304:313-332. [DOI] [PubMed] [Google Scholar]
- 17.Mello, J. A., and G. Almouzni. 2001. The ins and out of nucleosome assembly. Curr. Opin. Genet. Dev. 11:136-141. [DOI] [PubMed] [Google Scholar]
- 18.Ohlsson, R., A. Paldi, and J. M. Graves. 2001. Did genomic imprinting and X inactivation arise from stochastic expression? Trends Genet. 17:136-141. [DOI] [PubMed] [Google Scholar]
- 19.Ohlsson, R., R. Renkawitz, and V. Lobanenkov. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17:520-527. [DOI] [PubMed] [Google Scholar]
- 20.Ohlsson, R., B. Tycko, and C. Sapienza. 1998. Monoallelic expression: ‘there can only be one.’ Trends Genet. 14:435-438. [DOI] [PubMed] [Google Scholar]
- 21.Quitschke, W. W., M. J. Taheny, L. J. Fochtmann, and A. A. Vostrov. 2000. Differential effect of zinc finger deletions on the binding of CTCF to the promoter of the amyloid precursor protein gene. Nucleic Acids Res. 28:3370-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Widlund, H., H. Cao, S. Simonsson, E. Magnusson, T. Simonsson, P. Nielsen, J. Kahn, D. Crothers, and M. Kubista. 1997. Identification and characterization of genomic nucleosome-positioning sequences. J. Mol. Biol. 267:807-817. [DOI] [PubMed] [Google Scholar]