Abstract
In mouse Sertoli cells, transcription of the Inha gene encoding the α subunit of inhibin, which acts locally as a tumor suppressor, is down-regulated in tumors and in normal cells during aging. Previous studies suggested that regulation of Inha transcription involves the binding of a protein(s) to a repeat of the GGGGC motif in the promoter. Expression screening identified a cDNA encoding a protein that binds this sequence. Of the RING-H2 family, it is the mouse homologue of a human protein of unknown function, RNF6. The mouse gene, Rnf6, is predominantly expressed in two interacting cell types of the testis, Sertoli cells and pachytene spermatocytes. In Sertoli cells, it colocalizes with the PML and Daxx proteins in punctate nuclear bodies. In transient and stable transfectants, Rnf6 expression from a heterologous promoter increased the expression of reporter genes driven by the Inha promoter. In a Sertoli tumor cell line in which expression of both Inha and Rnf6 was reduced, reexpression of the latter restored the level of Inha while, concomitantly, the cells reverted to normal growth control in culture.
Inhibin was initially identified as an endocrine regulator of pituitary function. An important autocrine and/or paracrine function was later demonstrated in the seminiferous epithelium of the testis. Inhibin is a heterodimeric protein composed of a unique subunit, α, associated with either one of two polypeptides βA or βB (reviewed in reference 4). While the β subunits are made in a variety of tissues, to generate homo- and heterodimers (activins), the main site of expression of the α chain (designated α-inhibin, encoded at the Inha locus) is the Sertoli cells in the testis (reviewed in reference 22). In the complex interplay between germ cells and their somatic supporting cells, the Sertoli cells, a role of inhibin was established in the regulation of DNA synthesis (8) and a targeted null mutation in the gene resulted in the development of testicular malignancies (15).
We previously reported (11) that in transgenic mice that express the large T antigen of polyomavirus (PyLT) in Sertoli cells and develop fast-growing tumors at advanced ages (19), Inha levels at the pretumoral stages in the young adult testis were two- to threefold lower than in normal mice. In the tumors that eventually developed in the aging males, Inha expression was further decreased to the point of being, in most cases, undetectable. Cotransfection experiments performed with Sertoli cells in culture directly demonstrated that the Inha promoter is down-regulated by PyLT to an extent comparable to the decrease observed in vivo. A role of inhibin as a tumor suppressor was finally confirmed by studies with cell lines derived from the tumors. Restoration of Inha expression in these cells with a transfected expression vector led to the resumption of normal growth control in culture.
The PyLT and the simian virus 40 large T antigen are regulators of transcription and replication that bind to sites in the early promoters made of direct repeats of a G(A/G)GGC motif (6, 24). We hypothesized that the observed repression of the Inha promoter by PyLT could be due to competition by the viral protein for the binding of a cellular transcriptional activator. The Inha gene includes repeats of the GGGGC motif immediately upstream of the transcription start site. The same sequence is also present in other promoters expressed in Sertoli cells in a region that, in the case of the human urokinase gene, has been shown to be required for expression in Sertoli cells (7). It may also explain our earlier observation of testis-specific expression of the early polyomavirus promoter in transgenic mice (19).
To test the hypothesis that interaction of a positive regulator with GGC boxes is involved in Sertoli cell-specific expression, we screened a testis cDNA expression library for proteins that would bind the Inha promoter. We have identified in this way, cloned, and sequenced a cDNA encoding a protein that is preferentially expressed in the testis in both Sertoli and germ cells. Sequence analysis showed that it is the murine homologue of a human protein of unknown function, RNF6. Cotransfection experiments demonstrated a positive regulatory effect on Inha expression, dependent on the integrity of the GGGGC repeats in the promoter sequence. Furthermore, reexpression of Rnf6 in tumor-derived cell lines was sufficient to restore Inha expression from the endogenous promoter and, consequently, contact inhibition of growth in culture.
MATERIALS AND METHODS
Mice
All experiments were performed with C57BL/6 × DBA/2 F1 hybrids (B6D2F1). Investigations were conducted in accordance with French and European regulations for the care and use of research animals.
Cell culture and fractionation of germ cells.
Cell lines BALB/3T3 (clone A31; ATCC CCL-163), 45T-1, and 15P-1 (20) were cultivated in Dulbecco modified Eagle's medium (Gibco) supplemented with 10% fetal calf serum (Gibco). Fractionation of germ cells by elutriation centrifugation was performed as previously described (26). Primary Sertoli cell cultures from 2- to 3-week-old mice were established in accordance with previously published methods (23). They were maintained for 20 h in Dulbecco modified Eagle's medium with 10% fetal calf serum at 32°C. Attached germ cells were removed by hypotonic treatment (20 mM Tris-HCl, pH 7.4, for 2 to 5 min at 20°C), and the Sertoli cells were washed with culture medium before further treatments.
Transfection and β-galactosidase assays.
Transfection was performed with Fugene 6 in accordance with the supplier's (Boehringer Mannheim) instructions. β-Galactosidase activity was measured in cell extracts by using the Galacto-Light Kit (Tropix, Bedford, Mass.) in accordance with the manufacturer's instructions.
Plasmids and DNA preparation.
Plasmid DNA was extracted from bacteria with a Qiagen Maxiprep kit in accordance with the supplier's instructions. Each construct was checked by sequencing with either a T7 sequencing kit (Pharmacia Biotech) or an Amplicycle PCR sequencing kit (Perkin-Elmer, Foster City, Calif.). Plasmid HSS26 contains the cDNA of the human ribosomal S26 protein (28). Plasmid pPyLT1 codes for PyLT (29). Construction of Inha reporter plasmid pInh-lacZ was done as described previously (11). The full-length cDNA encoding the Rnf6 protein was amplified by reverse transcription-PCR performed on testis mRNA with primers RNF6am (5′CAGGATGGATCTGTCTAGATC3′) and RNF6av (5′AGCAGACCCCACCTCTCAC3′) and cloned into PCR cloning vector pGEM-T easy (Promega). The pNeoRNF6 plasmid was obtained by subcloning the cDNA in the pCDNA3-1 vector (cytomegalovirus promoter; Invitrogen) with NotI restriction sites in both vectors.
Mutagenesis of the Inha promoter.
PCR amplification was performed on mouse DNA with primers X (5′CGGAATTCCCATAGTTCACTTGCCCGT3′) and Y (5′CCACGCACCACAGTCCCAGTGGCACCTGCC3′) and primers Y′ (5′GGCAGGTGCCACTGGGACTGTGGTGCGTGG3′) and Z (5′CGGAATTCCTCAACCTTAAGCAC3′). The Y and Y′ sequences correspond to an internal region of the promoter (nucleotides [nt] −103 to −73) with the two GGGGC sequences mutated to GGTGC. The amplified products were purified from free oligonucleotides and mixed, and the mixture was further amplified with primers X and Z to generate the mutated 452-bp Inha promoter. The pInhmut-lacZ plasmid was obtained by cloning this fragment at the EcoRI site of pNASSβ (Clontech).
RNA Northern blot analysis.
Twenty micrograms of total RNA from testes or from cells in culture was prepared and hybridized as previously described (11).
Reverse transcription and PCR amplification.
Quantitation of RNA amounts in cell extracts was done by comparison with that of hypoxanthine phosphoribosyltransferase reverse transcripts amplified as an internal standard in the same reaction mixture. Total RNA (1 μg) prepared with a Total RNA Isolation kit (Boehringer Mannheim) was reverse transcribed with murine leukemia virus reverse transcriptase in accordance with the supplier's instructions. PCR amplification was performed with Taq DNA polymerase (Boehringer Mannheim) with oligonucleotide primers for Inha and Hprt as previously described (11). For the Hprt internal controls, a sample was taken after only 5 min of reverse transcription.
Testis cDNA expression library.
A cDNA expression library was generated by starting with 5 μg of poly(A)+ RNA purified from the testes of adult B6D2F1 mice by using the Oligotex direct mRNA kit (Qiagen) in accordance with the manufacturer's instructions. Double-stranded cDNA was obtained by using the ZAP express cDNA synthesis kit, inserted in the correct orientation into the λ-pBK-CMV vector, and encapsidated with the ZAP Express cDNA Gigapack III Gold Cloning kit, both from Stratagene, in accordance with the manufacturer's instructions. The number of independent clones was estimated to be 1 × 106 to 1.2 × 106, with insert sizes between 1.3 and 3 kb. Screening for expression of DNA binding proteins was performed as previously described (25). A 30-bp double-stranded labeled oligonucleotide (NMPI) containing sequences from the Inha promoter (nt −64 to −93) was used as a probe in the screening. The same oligonucleotide with the two GGGGC boxes mutated to GGTGC (DMPI) was used as a negative control. After three rounds of selection, isolated phagemids were selected and the cloned cDNA was obtained in the pBK-CMV vector by rescue with the Ex-assist helper phage in accordance with the manufacturer's (Stratagene) instructions.
In vitro protein synthesis.
Transcription and translation were performed with the TNT kit (Promega) in accordance with the instructions of the manufacturer (Promega).
Gel electrophoresis retardation assay for DNA-protein complexes.
Binding reaction mixtures (25 μl) contained 10 mM Tris-HCl (pH 7.5), 40 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 4% glycerol, 5 μg of poly(dI-dC) heteropolymer, and 0.5 ng (15,000 dpm) of 32P-end-labeled probe. Either total or nuclear protein extracts were prepared as previously described (1) and added last (5 μg of protein in 1 to 5 μl). When indicated, antibodies (final dilution, 1:250) were added to the reaction mixture. After 15 min at 25°C, samples were loaded onto a 4% (29:1 cross-linked) polyacrylamide gel in 40 mM Tris-HCl (pH 7.2)-20 mM Na acetate-1 mM EDTA and electrophoresed at 12.5 V/cm and 4°C. Gels were fixed for 15 min in 5% acetic acid-5% methanol, dried at 80°C, and exposed on Kodak XAR-5 film.
Antibodies.
Two peptides obtained from Eurogentec-France S.A. were used as immunogens. They correspond to the amino-terminal 15 residues of Rnf6 (MDPSRSRSGGSGEESC) and to an internal hydrophilic peptide (FLEQGRERRGTDYTPC; residues 339 to 353). Two rabbits were immunized with each peptide with a boost 4 weeks after the initial injection, and serum was collected after 4 months. Among the four antisera thus obtained, SE2896, directed against the amino-terminal peptide, and SE2893, directed against the internal peptide, were retained for further analysis. Antibodies directed against the PML (sc-9863; goat polyclonal immunoglobulin G [IgG]) and Daxx (sc-8043; mouse monoclonal IgG2a) proteins were obtained from Santa Cruz Biotechnology.
Immunocytochemistry analysis.
Cells (2 × 104/well in four-well Labteck plates [Nalge Nunc International]) were fixed in cold methanol for 10 min before a 30-min blocking step in 0.03% Triton-1% normal goat serum (G9023; Sigma) or fetal bovine serum (Gibco-BRL, Life Technologies) in phosphate-buffered saline (PBS). Either the primary antibodies or the preimmune serum was diluted 100-fold in 0.03% Triton X-100-1% calf serum in PBS and incubated for 2 h at room temperature with the cell preparation. After three washes for 5 min each time in 1% normal serum in PBS, the secondary antibodies (1/400 dilution in PBS) were added and the mixture was incubated for 1 h at room temperature. Three additional washes were performed, and slides were mounted in Vectashield reagent with 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories, Inc., Burlingame, Calif.). For PML and Daxx immunolocalization, negative controls were performed with the secondary antibodies alone.
In situ hybridization.
In situ hybridization experiments were performed as previously described, with digoxigenin-labeled single-stranded RNA probes corresponding to either the sense or antisense Rnf6 messenger (13).
Nucleotide sequence search of data banks.
Sequencing of the cloned cDNAs on both strands was performed with the DNA cycling sequencing kit (Perkin-Elmer) in accordance with the manufacturer's instructions and confirmed by extensive sequencing (Genome Express). The Genetics Computer Group FASTA software was used for comparisons to sequences in libraries.
Nucleotide sequence accession number.
The nucleotide sequence of the Rnf6 gene has been submitted to the GenBank database and assigned accession number AY039004.
RESULTS
Integrity of the GGGGC motifs of the Inha promoter is required for maximal expression in Sertoli cells.
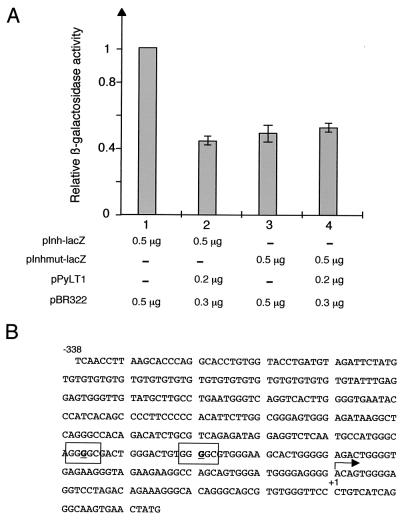
The PyLT protein of polyomavirus is a transcriptional regulator with a binding site in the viral promoter consisting of a repeat of the G(A/G)GGC nucleotide sequence (GGC box). Repeated GGC boxes are also present in the promoter of the α subunit of inhibin (Inha), and we formulated a hypothesis of competitive inhibition by PyLT of the binding of a positive regulatory protein in the same region of the promoter (11). In order to evaluate the role of the GGC boxes in the regulation of Inha expression, we generated a mutated promoter by replacement of the two GGGGC motifs with GGTGC sequences. Reporter constructs with either the wild-type or the mutated Inha promoter driving the lacZ gene were transfected into Sertoli cell primary cultures. Expression of the gene under control of the wild-type promoter was twofold greater than the activity observed with the mutated promoter. Interestingly, coexpression of PyLT repressed the wild-type promoter to the same extent, comparable to the decrease in Inha expression in the PyLT transgenic mouse testis in vivo (11), and it had no effect on the residual expression of the mutant (Fig. 1).
FIG. 1.
GGGGC motifs of the Inha promoter are required for maximal expression in Sertoli cells. (A) Primary cultures of Sertoli cells were transfected with the indicated amounts of plasmid DNA containing the lacZ reporter gene under control of the Inha promoter (pInh-lacZ) either without (bar 1) or with (bar 2) pPyLT1, an expression vector for PyLT (29). The same experiment was performed in parallel with the mutated Inha promoter pInhmut-lacZ, either without (bar 3) or with (bar 4) pPyLT1. Activities measured after transfection of the indicated DNA are presented relative to the activity indicated by bar 1 (mean ± standard error of the mean of triplicate experiments). (B) Sequence of the Inha promoter fragment in pInh plasmids. The G nucleotides underlined are replaced with T nucleotides in the mutated form pInhmut-lacZ.
The murine Rnf6 protein binds the Inha promoter.
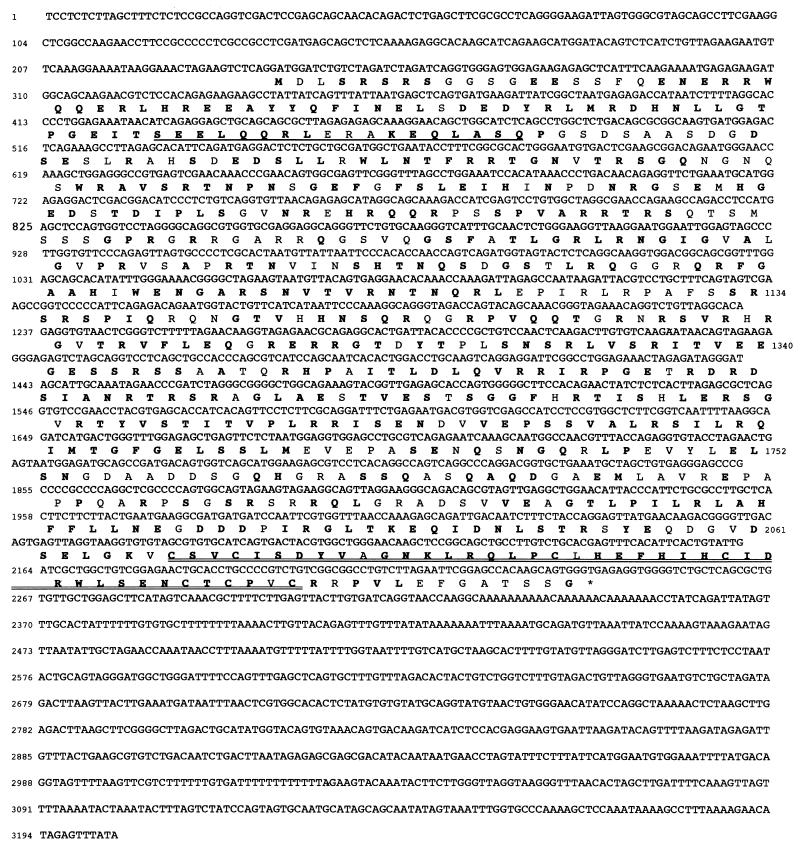
In order to identify a protein(s), that would recognize the region of the Inha promoter containing the GGC box regulatory sequences, we screened a testis expression cDNA library in λ phage. The NMPI double-stranded oligonucleotide probe reproducing the sequence of the promoter from −64 to −93 and the DMPI oligonucleotide mutated in the two GGC boxes were used in parallel to probe the library. After three rounds of selection, several clones positive with NMPI and negative with DMPI were isolated that carried cDNA inserts whose sequences were compared with those in databases. One of them had extensive similarity to a human gene of unknown function designated RNF6, identified on the basis of its proximity to the translocation breakpoint in a myelofibrosis case (14). The sequence of this initial 1.5-kb cDNA fragment was subsequently extended by computer analysis of mouse expressed sequence tag libraries, and a full-length cDNA of 3,204 bp (Rnf6) was finally generated by reverse transcription-PCR amplification of testis RNA. The complete cDNA sequence contains an open reading frame for a 667-amino-acid polypeptide with a predicted molecular mass of 79 kDa. The mouse sequence shows an overall identity of 75% with the human RNF6 amino acid sequence (Fig. 2). The cloned mouse cDNA, in fact, corresponds to a spliced isoform of the human gene containing alternative exon 1A (14). Like the human protein, murine Rnf6 has a zinc finger domain and a coiled-coil domain, two characteristics of the RBCC subgroup of the RING-H2 family (9, 12),
FIG. 2.
Nucleotide sequence of Rnf6 cDNA and sequence comparison of the human and murine proteins. Nucleotides are numbered from the transcription start. Amino acids in boldface are identical between the murine and human proteins. The coiled-coil and RING-H2 domains are singly and doubly underlined, respectively.
Rnf6 is transcribed in the testis, in both germ cells and Sertoli cells.
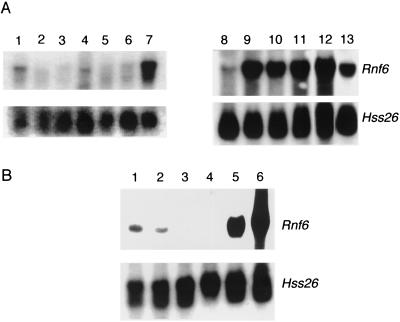
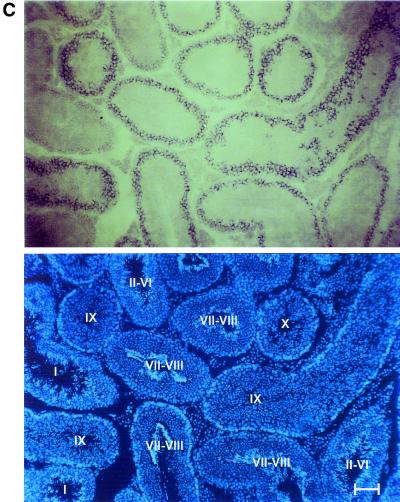
Northern blot analysis of RNA from various tissues demonstrated high levels of a 3.2-kb Rnf6 RNA in the testis (Fig. 3A). Smaller amounts of RNA were detected in somatic tissues, especially in the brain and intestine, but clearly not at the level observed in the testis. Whether or not the doublets of slightly different sizes observed in some of the tissues correspond to splice isoforms remains to be investigated. The same analysis was also performed with prepuberal animals. The first wave of germinal differentiation occurs synchronously, starting after birth from prospermatogonia, which, together with Sertoli cells, constitute the testis cords, precursors of the seminiferous tubules (2). Rnf6 RNA was detected at day 10 postparturition, before the first germ cells enter meiosis, and reached maximal levels between days 10 and 20 postparturition (first meiotic division). Three possible explanations were considered for the increased expression observed during meiosis: increased transcription levels in the maturing Sertoli cells, expression of the gene in spermatocytes, and both events occurring simultaneously. The latter explanation turned out to be the correct one. Robust expression in Sertoli cells was directly demonstrated by Northern blot hybridization of RNA prepared from primary cultures of purified Sertoli cells prepared between 2 and 3 weeks after birth and devoid of germ cells (23) (Fig. 3A). On the other hand, Northern blot analysis of purified fractions of germ cells (Fig. 3B) and in situ hybridization on adult testis sections (Fig. 3C) evidenced Rnf6 RNA in meiotic cells, with a maximal level at stages IX and X of spermatogenesis.
FIG. 3.
Rnf6 messenger is preferentially expressed in testicular cells. (A) Northern blot analysis of total RNA prepared from brain (lane1), heart (lane 2), liver (lane 3), intestine (lane 4), lung (lane 5), spleen (lane 6), adult testis (lane 7), testis of a 10-day-old mouse (lane 8), testis of a 20-day-old mouse (lane 9), testis of a 25-day-old mouse (lane 10), testis of a 30-day-old mouse (lane 11), testis of a 2-month-old mouse (lane 12), and freshly isolated Sertoli cells from 3-week-old mice (lane 13). Hybridization was performed in succession with probes for Rnf6 cDNA and for the ubiquitous ribosomal S26 protein cDNA (28). (B) Northern blot analysis of total RNA prepared from germinal fractions purified (≥90% pure) by elutriation centrifugation. Fractions: 1 and 2, spermatozoa and elongated spermatids; 3 and 4, round spermatids; 5 and 6, spermatocytes. Hybridization was performed in succession with probes for cDNAs of the Rnf6 and S26 proteins. (C) Top, in situ hybridization performed on testis section with Rnf6 antisense riboprobe. Bottom, same section stained with Hoechst 33258. The germinal differentiation stage (2) is indicated for each tubule section in roman numerals. Bar, 50 μm.
The Rnf6 protein is a constituent of the punctate nuclear bodies (PODs) in Sertoli cells.
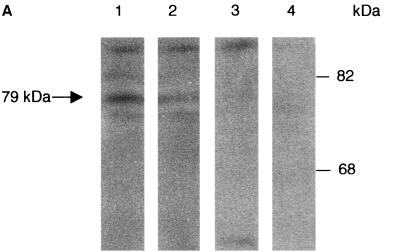
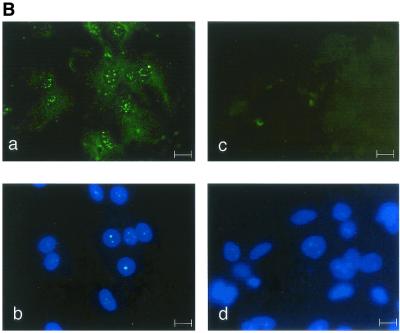
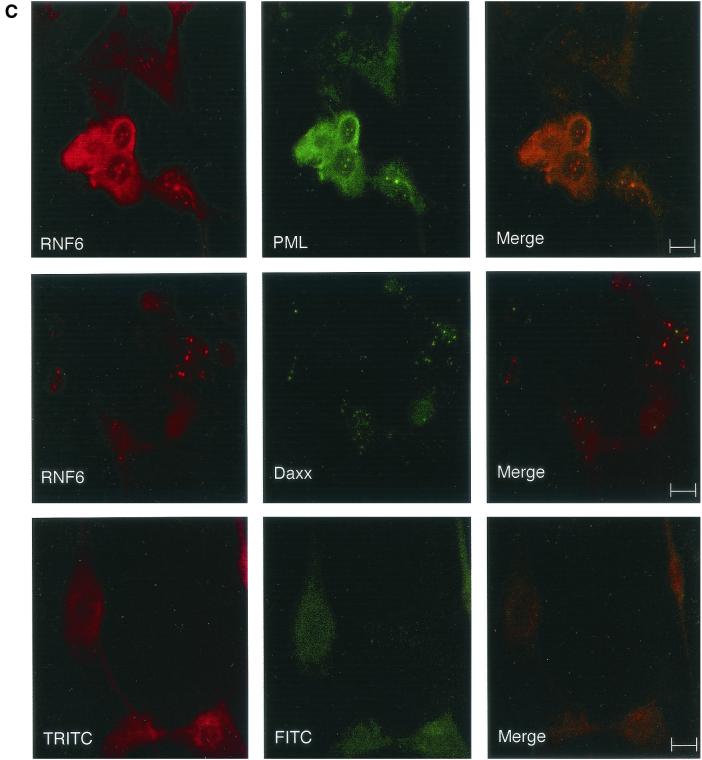
Protein localization was performed by immunocytochemical analysis with antipeptide antibodies SE2896 and SE2893, directed, respectively, against the amino-terminal and internal regions of Rnf6. Identical results were, in every instance, generated by the two types of antibodies, in agreement with the observation that they both revealed a polypeptide with the same apparent molecular mass (79 kDa) on Western blots prepared from testicular protein extracts (Fig. 4A). Immunocytochemical localization assays with similar results were performed with freshly prepared Sertoli cells in primary cultures (Fig. 4B) and the established line of Sertoli origin 15P-1 (20) (data not shown). A punctate distribution was evidenced in the interphasic Sertoli cell nuclei. This localization was evocative of the nuclear structures designated ND10, PML bodies, or PML oncogenic domains (PODs; reviewed in reference 16). That Rnf6 was indeed present in these structures was ascertained by double immunostaining of the same preparations with antibodies against two of the known components of nuclear bodies, the PML and Daxx proteins. Results presented in Fig. 4C show colocalization of Rnf6 with PML and Daxx immunostaining in interphasic nuclei. As expected (16), this punctate pattern was not seen in mitotic cells and, similarly, was also not seen in pachytene spermatocytes engaged in the first meiotic division.
FIG. 4.
Immunolocalization of the Rnf6 protein in the nuclei of Sertoli cells. (A) Western blot analysis of total testicular protein extracts with antibodies against the Rnf6 protein. Lanes: 1 and 3, immunization against the amino-terminal 15-mer (SE2896); 2 and 4, immunization against an internal peptide (SE2893; see Materials and Methods); 1 and 2, immune serum; 3 and 4, preimmune serum. MW (kDa), molecular mass in kilodaltons. (B) Immunofluorescence staining (a and c) and Hoechst 33258 staining (b and d) of Sertoli cell primary cultures with SE2896 antibodies (a) and the corresponding preimmune serum (c). Bar, 10 μm. (C) Double immunofluorescence staining of Sertoli cell primary cultures with antibodies directed against the Rnf6 protein (SE2896) and either the PML or the Daxx protein. The Rnf6 protein was revealed with tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit IgG and staining of PML or Daxx protein with fluorescein isothiocyanate-coupled anti-goat or anti-mouse IgG, respectively. The images at the bottom show staining with the preimmune serum and the corresponding mixed secondary antibodies. Bars, 10 μm.
Rnf6 binding to the Inha promoter: a role of the GGGGC repeats.
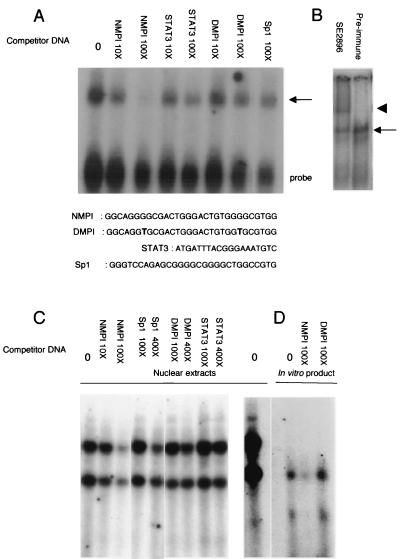
Gel electrophoresis retardation assays were performed to monitor complex formation with a labeled NMPI probe by first using the in vitro transcription-translation product of the complete Rnf6 cDNA. As shown in Fig. 5A, a protein-DNA complex was detected. Specificity of complex formation was ascertained by its sensitivity to competition by an excess of the unlabeled probe and by the lack of effect of the oligonucleotides DMPI (mutated in the two GGC boxes) and STAT3 (an unrelated sequence). It is of interest that the Sp1 oligonucleotide, which includes a direct repeat of GGGGC without a spacer, did not compete to a significant extent with binding of the protein to NMPI. Addition of anti-Rnf6 antibodies resulted in the appearance of a slowly migrating complex, thereby confirming that the protein was associated with the DNA probe.
FIG. 5.
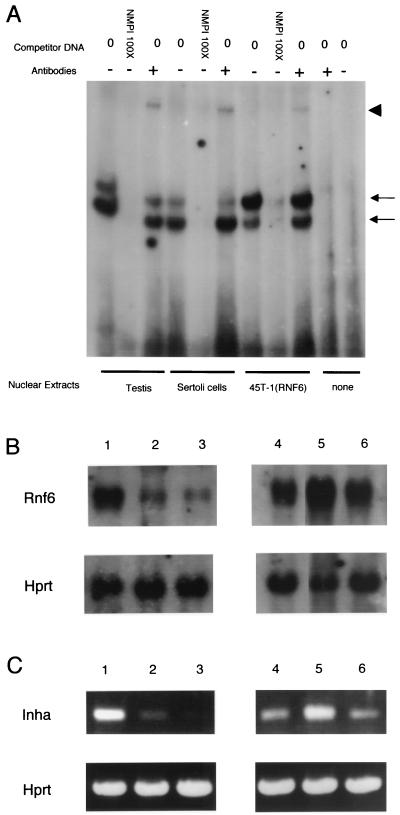
Binding of the Rnf6 protein to the Inha promoter and requirement for the GGGGC repeat. (A) Electrophoretic retardation assay performed with the double-stranded oligonucleotide NMPI corresponding to part of the Inha promoter and the in vitro transcription-translation product of Rnf6 cDNA. The competitors used were oligonucleotides STAT3 (unrelated); Sp1, with two immediately adjacent GGGGC boxes; and DMPI, with the same sequence as the probe except for GGC-to-TGC mutations in the two boxes. (B) Same experiment as in panel A but with the addition of anti-Rnf6 serum (SE2896, diluted 1:250). The arrow indicates the protein complex bound to the NMPI probe, and the arrowhead indicates the position of the higher-molecular-weight complex formed in the presence of antibodies. (C) Same assay performed with nuclear protein extracts from a primary culture of Sertoli cells (5 μg per assay). (D) Comparison in the same experiments of the migration rates of the complexes generated by the nuclear extract (left lane) and the in vitro translation product (right lanes).
When nuclear protein extracts prepared from primary-culture Sertoli cells were used as a source of protein, two complexes with distinct migration rates were evidenced (Fig. 5B), with the faster-migrating complex at the same position as the one generated by the in vitro translation products (Fig. 5C). Although DNA-protein interactions in a complex nuclear extract cannot be directly compared with those allowed by the simplified transcription-translation system, the presence of the protein was ascertained in both cases by the changes in mobility seen in all cases in the presence of antibodies (see Fig. 7).
FIG. 7.
Rnf6 overexpression increases Inha messenger level. (A) Complex formation between the NMPI probe and nuclear protein extracts from total testis cells, primary-culture Sertoli cells, and one of the 45T-1(RNF6) clones. The probe was incubated with the protein extracts either with or without an excess (100-fold) of unlabeled NMPI oligonucleotide. In the indicated lanes, the binding reaction was performed in the presence of anti-Rnf6 antibodies (SE2896). The arrows correspond to the protein complexes bound to the NMPI probe, and the arrowhead indicates the position of the higher-molecular-weight complex in the presence of antibodies. (B) Northern blot analysis of Rnf6 expression in primary-culture Sertoli cells (lane 1), in the 45T-1 cell line (lane 2), in 45T-1 cells transfected with the empty vector (lane 3), and in three independent 45T-1(RNF6) stable transformants (lanes 4, 5, and 6). The same membrane was hybridized in succession with probes for the Rnf6 (top) and Hprt (bottom) mRNAs. (C) The same RNA preparations as in panel B were reverse transcribed with oligo(dT) primers and tested by PCR for the presence of Inha (top) and Hprt (bottom) messengers.
Rnf6 up-regulates the Inha promoter.
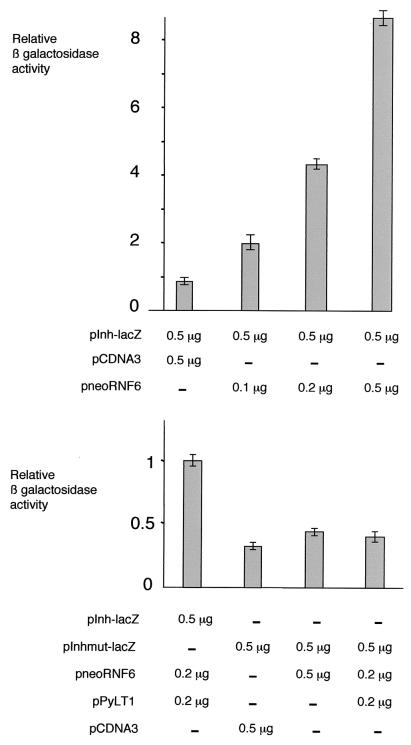
Following up from our initial hypothesis that the PyLT protein competes for the binding of a transcriptional activator to the Inha promoter, we sought evidence of an activating function of Rnf6 by transient-cotransfection experiments with BALB/3T3 cells. The pNeoRNF6 expression vector for the complete cDNA was transfected together with pInh-lacZ. As shown in Fig.6, expression of Rnf6 increased the activity of the Inha promoter in a dose-dependent manner. This increase in expression was abolished in the presence of the PyLT protein (cotransfection of the pPyLT1 expression vector). As predicted, Rnf6 had no effect on the activity of the Inha promoter mutated in the two GGC boxes.
We previously reported that expression of the Inha tumor suppressor was greatly reduced in 45T-1, a cell line derived from a Sertoli tumor (11). Rnf6 expression was, in fact, also reduced to very low levels in the same cells (Fig. 7). To confirm directly an effect of Rnf6 on the expression of the endogenous Inha promoter, 45T-1 cells were transfected with pNeoRNF6 DNA and, as a control, with the pCDNA3-1 empty vector. Three independent transformants were cloned in each series after selection in G418-containing medium. As shown in Fig. 7, increased levels of Rnf6 transcripts were observed by Northern blot hybridization in all three of them. Protein extracts prepared from these clones generated complexes in gel electrophoresis retardation assays comparable to those produced by extracts from native Sertoli cells, and the presence of the Rnf6 protein in these complexes was demonstrated by a shift in electrophoretic mobility upon addition of anti-Rnf6 antibodies. Positive regulation of endogenous Inha transcription was demonstrated by the increased levels of Inha RNA in the three clones in which Rnf6 was reexpressed from the transfected expression vector (Fig. 7C). In agreement with previous findings on the effect of Inha expression, these clones recovered the contact inhibition of growth in culture characteristic of the nontransformed Sertoli line (data not shown).
DISCUSSION
Previous results (11) led us to a hypothesis of competition between PyLT and a still undefined cellular transcriptional activator for binding to a critical site in the Inha promoter. A similar hypothesis had been proposed previously by Grimaldi et al. (7) for a different promoter also expressed in Sertoli cells (human urokinase) and simian virus 40 T antigen, which has the same binding sites (repeated G[G/A]GGC boxes) as the polyomavirus oncoprotein (23). In both instances, the identity of the putative transcriptional activator had remained unknown. In order to identify a candidate protein(s), we screened a testis cDNA expression library for the binding of a double-stranded oligonucleotide containing a fragment of the Inha promoter region with the characteristic repeat of a GGGGC motif. A negative criterion was introduced into the screening procedure by retaining only cDNA clones that would bind the probe (NMPI) but would not recognize the mutated oligonucleotide DMPI, in which the GGGGC motifs were replaced with GGTGC.
Sequence analysis of one of the clones initially selected showed that it corresponds to the murine homologue of the RNF6 protein identified in humans (14). It belongs to a subfamily of the zinc finger proteins designated RING-H2 (12). Its function is unknown, but expression in the testis of several proteins belonging to this family and their possible implication in spermatogenesis were recently reported (5, 17, 21). Northern blotting and in situ hybridization revealed that Rnf6 is preferentially expressed in the testis, in both Sertoli cells and pachytene spermatocytes. The protein belongs to the RBCC subfamily of RING finger proteins that contain B-box and coiled-coil domains (9). The discrete nuclear speckles revealed by immunostaining with anti-Rnf6 antibodies were evocative of the nuclear bodies known as PODs or ND10 (reviewed in reference 3), and the Rnf6, PML, and Daxx proteins were indeed found to colocalize in the interphase nuclei of primary-culture Sertoli cells and of cells of the Sertoli-derived 15P-1 line. The POD structures have been proposed to represent a nuclear depot for storage, degradation, and/or titration of nuclear proteins involved in regulatory functions. The dot structure is known to disappear during mitosis and to be replaced by a diffuse intranuclear distribution, and it was, similarly, not seen during meiosis.
The complexity of the observed gel shift patterns suggests that a protein(s) other than Rnf6 participates in the complexes controlling Inha expression. The modulatory role of Rnf6 is exerted within a two- to threefold range, a moderate extent but one that may be critical in physiological control mechanisms, as suggested by in vivo observations on tumor development in transgenic animals (11). The occurrence in other Sertoli cell-specific promoters of sequences similar to the Rnf6 binding sites of Inha (reference 7 and our unpublished results) suggests that the protein may regulate a number of target genes during spermatogenesis that remain to be characterized. A detailed analysis of the features of the promoter sequence critical for protein binding is in progress. Preliminary results indicate that one GGGGC box alone is not sufficient for RNF6 binding and that spacing between two of them may be critical. This is already indicated by the low efficiency as a competitor for Rnf6 binding of the Sp1 oligonucleotide, which contains two adjacent GGGGC motifs (Fig. 5A).
On the other hand, recent reports from several laboratories (reviewed in reference 10) have assigned to the RING finger proteins a critical role as E3 ubiquitin ligases that are able to transfer ubiquitin to heterologous substrates, as well as to the RING proteins themselves. These reports point to a possible function of these proteins in the regulation of protein stability, a feature of germinal differentiation that has yet to be examined in detail but is likely to be important. Establishment of whether or not Rnf6 could act as an E3 ligase in Sertoli and/or germ cells may provide a first clue in this direction, as well as the identification of potential targets and cell-specific partners of the protein in the somatic and germinal cells of the testis.
Because of the lack of a culture system for meiotic cells, mutagenesis by targeted meiotic recombination (27) appears to be the most hopeful approach to elucidation of the function of Rnf6 in the germinal component of the testis. Analysis of the function of the protein and its RING domain in Sertoli cells may, on the other hand, be performed by a genetic analysis of primary-culture cells and the established line 15P-1 (20). Another possible outcome of these studies may be to gather information on the mechanism responsible for the decrease in Inha expression associated with the aging process (11, 18). Possible changes in Rnf6 expression during that period of life, either at the transcriptional level or in the properties of the protein, are under investigation.
FIG. 6.
Rnf6 expression upregulates the activity of the Inha promoter. β-Galactosidase activity was measured in BALB/3T3 cells 72 h after transfection of the indicated plasmid DNA mixtures. Relative values are indicated as the mean ± the standard error of the mean in triplicate measurements. The experiment shown is representative of a set of four independent experiments.
Acknowledgments
The expert technical assistance of F. Ranc, E. Couchi, K. Laurent, M. Radjkumar, M. Cutajar, and Y. Fanteï is gratefully acknowledged.
This work was supported by a grant from the Association pour la Recherche sur le Cancer.
REFERENCES
- 1.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellve, A. R. 1998. The male germ cell: origin, migration, proliferation and differentiation. Semin. Cell Dev. Biol. 9:379-391. [DOI] [PubMed] [Google Scholar]
- 3.Bloch, D. B., J. D. Chiche, D. Orth, S. M. de la Monte, A. Rosenzweig, and K. D. Bloch. 1999. Structural and functional heterogeneity of nuclear bodies. Mol. Cell. Biol. 19:4423-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C.-L. C. 1993. Inhibin and activin as paracrine/autocrine factors. Endocrinology 132:4-5. [DOI] [PubMed] [Google Scholar]
- 5.Fujii, T., K. Tamura, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, K. Yomogida, H. Tanaka, Y. Nishimune, H. Nojima, and Y. Abiko. 1999. Sperizin is a murine RING zinc-finger protein specifically expressed in haploid germ cells. Genomics 57:94-101. [DOI] [PubMed] [Google Scholar]
- 6.Gaudray, P., C. Tyndall, R. Kamen, and F. Cuzin. 1981. The high affinity binding site on polyoma virus DNA for the viral large-T protein. Nucleic Acids Res. 9:5697-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimaldi, P., R. Geremia, C. Albanesi, and P. Rossi. 1996. The same sequence mediates activation of the human urokinase promoter by cAMP in mouse Sertoli cells and by SV40 large T antigen in COS cells. Mol. Cell. Endocrinol. 117:167-173. [DOI] [PubMed] [Google Scholar]
- 8.Hakovirta, H., A. Kaipia, O. Soder, and M. Parvinen. 1993. Effects of activin-A, inhibin-A, and transforming growth factor-beta 1 on stage-specific deoxyribonucleic acid synthesis during rat seminiferous epithelial cycle. Endocrinology 133:1664-1668. [DOI] [PubMed] [Google Scholar]
- 9.Henry, J., I. H. Mather, M. F. McDermott, and P. Pontarotti. 1998. B30.2-like domain proteins: update and new insights into a rapidly expanding family of proteins. Mol. Biol. Evol. 15:1696-1705. [DOI] [PubMed] [Google Scholar]
- 10.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 11.Lopez, P., F. Vidal, M. Rassoulzadegan, and F. Cuzin. 1999. A role of inhibin as a tumor suppressor in Sertoli cells: down-regulation upon aging and repression by a viral oncogene. Oncogene 18:7303-7309. [DOI] [PubMed] [Google Scholar]
- 12.Lovering, R., I. M. Hanson, K. L. Borden, S. Martin, N. J. O'Reilly, G. I. Evan, D. Rahman, D. J. Pappin, J. Trowsdale, and P. S. Freemont. 1993. Identification and preliminary characterization of a protein motif related to the zinc finger. Proc. Natl. Acad. Sci. USA 90:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luis, O., L. Lopez-Fernandez, and J. Del Mazo. 1999. Tex 27, a gene containing a zinc-finger domain, is up regulated during the haploid stages of spermatogenesis. Exp. Cell Res. 249:320-326. [DOI] [PubMed] [Google Scholar]
- 14.Macdonald, D. H., D. Lahiri, A. Sampath, A. Chase, J. Sohal, and N. C. Cross. 1999. Cloning and characterization of RNF6, a novel RING finger gene mapping to 13q12. Genomics 58:94-97. [DOI] [PubMed] [Google Scholar]
- 15.Matzuk, M. M., M. J. Finegold, J.-G. J. Su, A. J. W. Hsueh, and A. Bradley. 1992. α-Inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature 360:313-319. [DOI] [PubMed] [Google Scholar]
- 16.Maul, G. G., D. Negorev, P. Bell, and A. M. Ishov. 2000. Properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129:278-287. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa, S., W. Goto, A. Orimo, T. Hosoi, Y. Ouchi, M. Muramatsu, and S. Inoue. 1998. Molecular cloning of a novel RING finger B box-coiled coil (RBCC) protein, terf, expressed in the testis. Biochem. Biophys. Res. Commun. 251:515-519. [DOI] [PubMed] [Google Scholar]
- 18.Pal, D., T. J. Collins, and T. A. Parkening. 1991. Levels of alpha-inhibin in aging female mice. Biol. Reprod. 45:869-875. [DOI] [PubMed] [Google Scholar]
- 19.Paquis-Flucklinger, V., J. Michiels, F. Vidal, C. Alquier, G. Pointis, V. Bourdon, F. Cuzin, and M. Rassoulzadegan. 1993. Expression in transgenic mice of the large T antigen of polyomavirus induces Sertoli cell tumours and allows the establishment of differentiated cell lines. Oncogene 8:2087-2094. [PubMed] [Google Scholar]
- 20.Rassoulzadegan, M., V. Paquis-Flucklinger, B. Bertino, J. Sage, M. Jasin, K. Miyagawa, V. van Heyningen, P. Besmer, and F. Cuzin. 1993. Transmeiotic differentiation of male germ cells in culture. Cell 75:997-1006. [DOI] [PubMed] [Google Scholar]
- 21.Shyu, H., S. Hsu, H. Hsieh-Li, and H. Li. 2001. A novel member of the RBCC family, Trif, expressed specifically in the spermatids of mouse testis. Mech. Dev. 108:213-216. [DOI] [PubMed] [Google Scholar]
- 22.Skinner, M. K. 1993. Secretion of growth factors and other regulatory factors, p. 237-247. In L. D. Russell and M. D. Griswold (ed.), The Sertoli cell. Cache River Press, Clearwater, Fla.
- 23.Steinberger, A., and A. Jakubowiak. 1993. Sertoli cell culture: historical perspective and review of methods, p. 155-180. In L. D. Russell and M. D. Griswold (ed.), The Sertoli cell. Cache River Press, Clearwater, Fla.
- 24.Tjian, R. 1978. The binding site on SV40 DNA for a T antigen-related protein. Cell 13:165-179. [DOI] [PubMed] [Google Scholar]
- 25.Vidal, F., A. Blangy, M. Rassoulzadegan, and F. Cuzin. 1992. A murine sequence-specific DNA binding protein shows extensive local similarities to the amyloid precursor protein. Biochem. Biophys. Res. Commun. 189:1336-1341. [DOI] [PubMed] [Google Scholar]
- 26.Vidal, F., P. Lopez, L. A. Lopez-Fernandez, F. Ranc, J. C. Scimeca, F. Cuzin, and M. Rassoulzadegan. 2001. Gene trap analysis of germ cell signaling to Sertoli cells: NGF-TrkA mediated induction of Fra1 and Fos by post-meiotic germ cells. J. Cell Sci. 114:435-443. [DOI] [PubMed] [Google Scholar]
- 27.Vidal, F., J. Sage, F. Cuzin, and M. Rassoulzadegan. 1998. Cre expression in primary spermatocytes: a tool for genetic engineering of the germ line. Mol. Reprod. Dev. 51:274-280. [DOI] [PubMed] [Google Scholar]
- 28.Vincent, S., L. Marty, and P. Fort. 1993. S26 ribosomal protein RNA: an invariant control for gene regulation experiments in eucaryotic cells and tissues. Nucleic Acids Res. 21:1498.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu, Z., G. M. Veldman, A. Cowie, A. Carr, B. Schaffhausen, and R. Kamen. 1984. Construction and functional characterization of polyomavirus genomes that separately encode the three early proteins. J. Virol. 51:170-180. [DOI] [PMC free article] [PubMed] [Google Scholar]