Abstract
The mitochondrial proapoptotic protein Smac/DIABLO has recently been shown to potentiate apoptosis by counteracting the antiapoptotic function of the inhibitor of apoptosis proteins (IAPs). In response to apoptotic stimuli, Smac is released into the cytosol and promotes caspase activation by binding to IAPs, thereby blocking their function. These observations have suggested that Smac is a new regulator of apoptosis. To better understand the physiological function of Smac in normal cells, we generated Smac-deficient (Smac−/−) mice by using homologous recombination in embryonic stem (ES) cells. Smac−/− mice were viable, grew, and matured normally and did not show any histological abnormalities. Although the cleavage in vitro of procaspase-3 was inhibited in lysates of Smac−/− cells, all types of cultured Smac−/− cells tested responded normally to all apoptotic stimuli applied. There were also no detectable differences in Fas-mediated apoptosis in the liver in vivo. Our data strongly suggest the existence of a redundant molecule or molecules capable of compensating for a loss of Smac function.
Apoptosis, or programmed cell death (PCD), is a physiological cell suicide program essential for both embryonic development and the maintenance of tissue homeostasis in multicellular organisms (6, 11, 13). The family of cysteine proteases known as caspases are key components of mammalian PCD (1). Caspases are expressed in cells as inactive precursors, which are activated by proteolytic processing (22, 26). Two classes of caspases, initiators and effectors, are involved in mammalian apoptosis (3). Activated initiator caspases, such as caspase 8 and caspase 9, cleave the precursor forms of effector caspases, such as caspases 3, 6, and 7. Activated effector caspases in turn cleave a specific set of cellular substrates resulting in the biochemical and morphological changes associated with the apoptotic phenotype (26).
The activation of initiator caspases is thought to irreversibly trigger the caspase cascade, necessitating that caspase activation be tightly regulated by layered control mechanisms. Among the growing number of cellular proteins that have been shown to regulate caspase activation and activity are the IAPs, including c-IAP1, c-IAP2, XIAP, and survivin. These proteins have been reported to block both death receptor- and mitochondrially-mediated apoptotic pathways by directly inhibiting initiator and effector caspases (4, 28).
Smac/DIABLO, a mitochondrial protein released into the cytosol in response to apoptotic stimuli, was recently found to promote caspase activation by eliminating IAP function (5, 29). Smac binds to most known human IAP family members and relieves their inhibition of caspase activity. The N-terminal 20 amino acids of the mature Smac protein are crucial for Smac-IAP interaction, and removal of this region completely abrogates the ability of Smac to bind to XIAP (2, 33). Since Smac blocks IAP activity, it has been proposed that Smac is a mammalian functional homologue of the Drosophila proapoptotic proteins Reaper, Grim, and Hid (9, 20, 34). This hypothesis is bolstered by the finding that the first four N-terminal residues of Smac, which recognize a surface groove on BIR3, are also conserved in the Drosophila proteins (33).
In this study, we generated gene-targeted Smac-deficient mice and studied the apoptosis of Smac-deficient cells in vitro and in vivo. We demonstrate that several types of Smac-deficient primary cells respond normally to a broad range of apoptotic stimuli. Normal apoptosis is also induced in the liver in vivo . These lines of evidence strongly suggest the existence of molecules and pathways that can circumvent the loss of Smac.
MATERIALS AND METHODS
ES cell culture.
E14K embryonic stem (ES) cells from 129/Ola mice were maintained on a layer of mitomycin C-treated mouse embryonic fibroblasts (MEFs) in Dulbecco's modified Eagle's medium (DMEM) supplemented with leukemia inhibitory factor, 15% heat-inactivated fetal calf serum (HyClone, Logan, Utah), l-glutamine, and β-mercaptoethanol.
Generation of Smac-deficient mice.
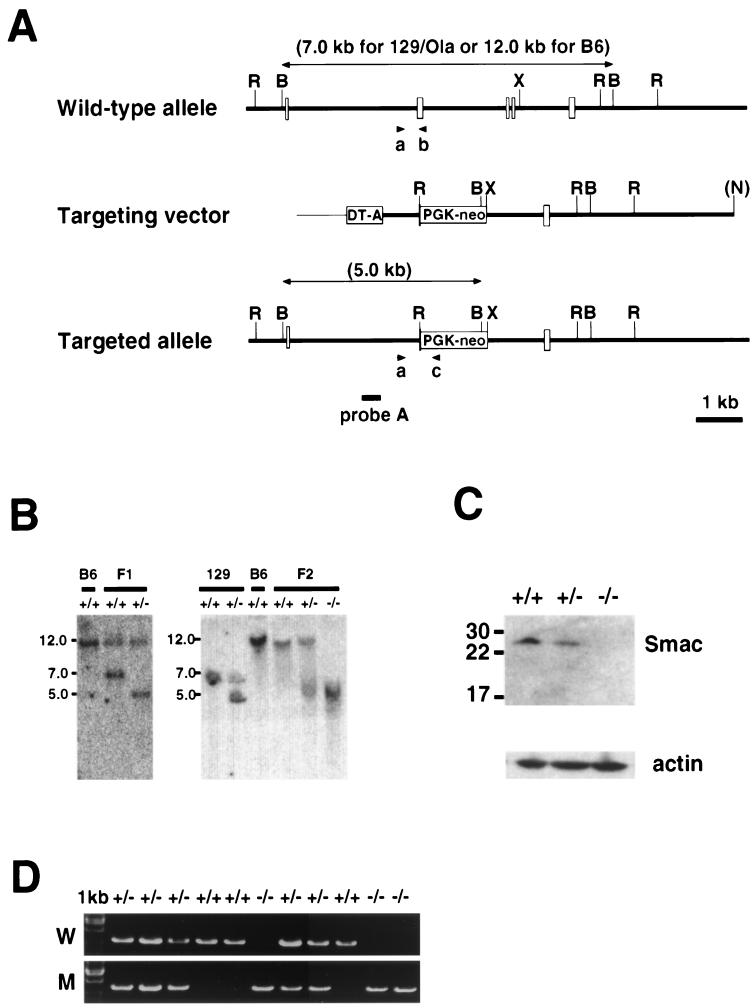
A 129/Ola mouse phage genomic library was screened with a murine Smac full-length cDNA probe. Restriction mapping and sequence analysis of subcloned fragments revealed that the murine Smac gene contains five coding exons and four introns spanning a region of at least 11 kb. The targeting vector was designed to replace exons 2 to 4 of the Smac gene (containing the IAP binding region) with a cassette in which the neomycin resistance gene is under the control of the PGK promoter (PGK-neo). The diphtheria toxin A gene (DT-A) driven by the pMC1 promoter was incorporated into the 5′ end of the vector to allow for negative selection (35). The targeting vector was linearized with NotI and electroporated into ES cells (Bio-Rad Gene Pulser; 0.34 kV, 250 μF). We obtained 380 G418-resistant ES cell colonies, which were screened for homologous recombination by PCR. Homologous recombinants were confirmed by Southern blot analysis as described previously (19) with the 5′ external probe A depicted in Fig. 1A. Nineteen correctly targeted clones were identified, and 3 were injected into C57BL/6J blastocysts. Three independent Smac−/− mouse strains were established by standard procedures (16).
FIG. 1.
Targeting of the murine Smac gene by homologous recombination. (A) Schematic representation of the wild-type mouse Smac locus (top), the targeting construct (middle), and the mutated Smac allele (bottom). The coding exons are shown as clear boxes. Exons 2 to 4 were replaced with PGK-neo, and DT-A was added for negative selection. The 5′-flanking probe A used for Southern blot analysis is shown, as are the predicted sizes of the hybridizing fragments. The primer pairs used for PCR (a and b or a and c) are also indicated. R, EcoRI; B, BamHI; X, XbaI; N, NotI. (B) Southern blot analysis of Smac-deficient ES cells and mice. (Left panel) DNA prepared from C57BL6/J (+/+) mice and F1 offspring (+/+ and +/−) of chimeric mice. (Right panel) DNA was from 129/Ola ES cells (+/+), Smac+/− ES cell clones, C57BL6/J (+/+) mice, and F2 offspring (+/+, +/−, and −/−) of intercrosses of Smac mutant F1 mice. In all cases, tail genomic DNA was digested with BamHI and hybridized to probe A. (C) Western blot analysis of Smac protein expression. Protein samples were prepared from Smac+/+, Smac+/−, and Smac−/− MEFs and incubated with anti-Smac antibody. Actin was used as the loading control. (D) Genotypic analysis of F2 littermates by PCR. PCR was performed on genomic tail DNA templates with primer pair a and b to detect the wild-type allele (W) or primers a and c to detect the mutant allele (M).
Genotypes of mutant mice were determined by PCR and confirmed by Southern blot analysis of genomic DNA from tail biopsy samples. As shown in Fig. 1A, the primers a, specific for a Smac-specific intronic sequence, and b, located in deleted exon 2, were used to detect the wild-type allele. Primers a and c specific for neo were used to detect the mutant allele. The sequences were as follows: primer a, 5′-TATAGAGCCCGAATGTCAGAA-3′; primer b, 5′-GAGACAGAAAGGTAGAGGTGC; primer c, 5′-GGTGGATGTGGAATGTGTG-3′. All data presented in this report were confirmed in at least two mutant mouse lines.
Generation of Smac−/− ES cells and MEFs.
To generate Smac−/− ES cells, two independent Smac+/− ES cell lines were cultured in a high concentration (4.0 to 4.5 mg/ml) of G418 for 10 days. Colonies resistant to this level of G418 were expanded, and their genotypes were determined by Southern blot analysis. Two independent clones homozygous for the Smac mutation were selected and used for further studies. To prepare primary MEFs, fibroblasts were established from E14.5 F2 embryos according to standard procedures. The cells were maintained in DMEM supplemented with 10% fetal calf serum (FCS), l-glutamine, and antibiotics.
In vitro assay of caspase 3 activation.
The determination of caspase 3 activation was performed essentially as described previously (5). Briefly, Smac+/+ or Smac−/− MEFs were homogenized in buffer A (20 mM HEPES-KOH [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 1 mM sodium EDTA, 1 mM sodium EGTA, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride) containing 0.5% (wt/vol) CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} and centrifuged at 100,000 × g for 30 min at 4°C. S-100 supernatants were recovered, and samples were stored as aliquots at −80°C. For procaspase 3 cleavage assays, 12 μl of S-100 lysate (50 μg of protein) was incubated with 1 μg of horse heart cytochrome c (Sigma), 1 mM dATP, and 1 mM additional MgCl2 in buffer A at 30°C for 30 or 60 min in a final volume of 20 μl. The reaction mixture was then subjected to Western blotting as described below. Recombinant Smac protein was added to in vitro caspase assays as described previously (5).
Western blot analysis.
Protein samples were prepared in buffer A containing 0.5% CHAPS and supplemented with protease inhibitor cocktail (Amersham-Pharmacia, Piscataway, N.J.). A total of 20 μg of protein per lane was fractionated on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10 to 20% polyacrylamide) gradient gel (Invitrogen, Carlsbad, Calif.), and the gel was transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, Mass.). The blot was subsequently incubated with a rabbit polyclonal antibody recognizing Smac (5), an antibody specifically recognizing the cleaved, active form of caspase 3 (Cell Signaling), an antibody recognizing procaspase 3 (Transduction Laboratories), anti-cIAP-1 antibody (R&D Systems), anti-cIAP-2 antibody (R&D Systems), anti-XIAP antibody (Transduction Laboratories), antisurvivin antibody (Santa Cruz Biotechnology), or anti-Omi antibody (a kind gift from E. Alnemri, Kimmel Cancer Institute, Thomas Jefferson University, Philadelphia, Pa.). Immunoblot analysis was performed with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody or goat anti-mouse secondary antibody, and proteins were visualized with the ECL Plus enhanced chemiluminescence system (Amersham-Pharmacia) according to the manufacturer's protocol.
Apoptosis in ES cells, MEFs, and thymocytes.
For ES cell apoptosis, ES cells (105 per well) were seeded on 1% gelatinized 24-well plates in ES cell medium. After 24 h, the cells were treated with the following stimuli to induce cell death: UV at 60 J/m2, anisomycin (Sigma) at 30 μM, adriamycin (Sigma) at 1 μM, and etoposide (Sigma) at 100 μM. ES cells were harvested 8 or 24 h after the induction of cell death. Cell viability was determined by staining with annexin-propidium iodide (PI) (R&D Systems apoptosis detection kit; ) according to the manufacturer's protocol followed by flow cytometric analysis. For MEF apoptosis, MEFs (5 × 104 per well) were plated in 12-well plates and treated 24 h later with the following apoptotic stimuli: UV light at 30, 60, 90, and 120 mJ/cm2; staurosporine (Sigma) at 0.03, 0.1, 0.3, and 1.0 μM; adriamycin at 0.3, 1, and 3 μM; etoposide at 3, 10, 30, and 100 μM; anisomycin at 3, 10, 30, and 100 μM; tumor necrosis factor-cycloheximide (TNF/CHX) (Sigma) at 0.1, 0.3, 1, and 3 μg of TNF per ml in the presence of 5 μg of CHX per ml. For thymocyte apoptosis, thymocytes prepared from Smac+/+ or Smac−/− mice by standard procedures (21) were plated at 5 × 106/ml in 24-well plates in RPMI containing 10% FCS. Thymocytes were cultured for 24 h at 37°C in 5% CO2 in the presence or absence of etoposide at 0.3, 1.0, 3, and 10 μM; dexamethasone (Sigma) at 1, 3, 10, and 30 nM; or anti-Fas antibody (Jo2; BD-Pharmingen)-CHX at 0.3 μg/ml, and 1 μg of anti-Fas antibody per ml in the presence of 10 μg of CHX per ml. Apoptosis of MEFs and thymocytes was determined as for ES cells.
Apoptosis in MEFs overexpressing E1A or c-Myc.
To establish MEFs overexpressing E1A, we first transfected LPC-12S (23) into the Phoenix ecotropic retrovirus packaging cell line to produce retrovirus carrying the E1A 12S gene and conferring resistance to puromycin. This retrovirus was used to infect Smac+/+ and Smac−/− MEFs. Cells that had integrated the virus were selected in medium containing 2.5 μg of puromycin per ml (Sigma). To establish MEFs overexpressing c-Myc, we transfected mouse cMyc/MarXII-hygro (30) into the Phoenix ecotropic retrovirus packaging cell line to produce retrovirus carrying the c-Myc gene and conferring resistance to hygromycin. Cells that integrated this virus were selected in medium containing 100 μg of hygromycin B per ml (Roche, Mannheim, Germany). Selected MEFs of both retroviral types and both genotypes were cultured in the presence or absence of adriamycin at 0.1, 0.25, and 0.5 μg/ml; TRAIL at 20 μg/ml; or paclitaxel (Sigma) at 0.1, 0.5, and 1 μg/ml to induce apoptosis. After 24 h, cell viability was evaluated by trypan blue exclusion.
Proliferation and survival of T and B cells.
For activation-induced cell death (AICD) of T cells, lymph node cells were purified with magnetic beads (21) from Smac+/+ and Smac−/− mice, and then incubated in six-well tissue culture plates (6 × 106 cells/well) in the presence of 10 μg of plate-bound anti-CD3ɛ (clone 145-2C11; BD-Pharmingen) per ml and interleukin-2 (50 U/ml) (Genzyme) for 48 h to induce activation. The activated cells (0.5 × 106) were replated in 24-well plates precoated with 1 μg of anti-CD3ɛ per ml and harvested 24, 48, or 72 h after restimulation. The number of viable cells was determined by trypan blue exclusion. For T-cell proliferation, purified Smac+/+ and Smac−/− T cells were plated into round-bottom 96-well plates (105 cells per well) in freshly prepared RPMI 1640 (10% FCS, 10 μM β-mercaptoethanol). The cells were stimulated with 10 ng of phorbol myristate acetate per ml (Sigma) plus 100 ng of Ca2+ ionophore A23617 per ml, 0.1 to 100 μg of soluble anti-CD3ɛ per ml, 0.02 μg of soluble anti-CD28 per ml (clone 37.51; BD-PharMingen), or 2 μg of the mitogen concanavalin A per ml (Amersham-Pharmacia). For B-cell proliferation, purified B cells (105 cells per well) were stimulated with 20 μg of anti-immunoglobulin M (IgM) (61-6800; Zymed), 10 μg of anti-IgM F(ab′)2 fragment per ml (61-5900; Zymed), 1 μg of anti-CD40 per ml (clone HM40; BD-PharMingen), or 2 μg of lipopolysaccharide per ml (Sigma). Cells were stimulated in triplicate for different time periods and pulsed for the last 12 to 18 h with 1 μCi of [3H]thymidine per well (NEN, Boston, Mass.). Labeled cells were harvested with a Filtermate-196 harvester (Canberra Packard, Mississauga, Canada), and thymidine incorporation was determined with a Matrix-996 direct counter (Canberra Packard).
Histological analysis.
Tissues were fixed in freshly prepared 4% paraformaldehyde (Sigma) overnight at 4°C. Samples were dehydrated in an ethanol series and embedded in wax. Sections were stained with either hematoxylin and eosin (H&E) or by terminal deoxynucleotide transferase nick-end labeling (TUNEL) staining. TUNEL staining was performed with the Roche In Situ Cell Death Detection kit according to the manufacturer's instructions.
Induction of apoptosis in the liver.
Young adult mice (8 to 10 weeks old) were injected intraperitoneally with anti-Fas antibody (Jo2; BD-Pharmingen) at a dose of 10 or 100 μg per animal. Some animals were monitored for survival, while others were killed 3 h after injection for histological analysis of their tissues. Sections of liver were stained with H&E or TUNEL as described above. Numbers of TUNEL-positive cells were counted in at least five fields per liver.
RESULTS
Targeted disruption of the Smac gene in mice.
To determine the physiological role of Smac in vivo, we used homologous recombination in ES cells to disrupt the Smac gene and generate Smac knockout mice (Fig. 1A). The targeting vector was designed to replace most of the Smac coding region, including exons 2 to 4, with the PGK-neo cassette. A polymorphism was discovered in a BamHI restriction site in the Smac locus when DNA from the C57BL6/J and 129/Ola genetic backgrounds was compared. Probe A indicated in Fig. 1A recognizes a 7-kb fragment from the 129/Ola wild-type allele, a 12.0-kb fragment from the C57BL6/J wild-type allele, and a 5.0-kb fragment from the targeted allele. We obtained 19 independent ES cell clones carrying the mutated Smac allele, and 3 (sm-7, sm-24, and sm-33) were injected into C57BL/6J blastocysts to generate chimeric mice. F1 heterozygotes originating from the sm-24 or sm-33 clones were intercrossed to produce the F2 progeny analyzed in this report. Figure 1B shows Southern blot analyses of DNA from C57BL6/J wild-type mice, C57BL6/J-129/Ola F1 offspring, 129/Ola ES cells, and F2 offspring from F1 intercrosses.
To confirm that the knockout allele was a null mutation, MEFs were prepared from Smac+/+, Smac+/−, and Smac−/− littermates and lysates were subjected to Western blotting to detect Smac expression. As shown in Fig. 1C, Smac−/− MEFs do not express Smac. Genotypic analysis of F2 offspring was confirmed by PCR (Fig. 1D). Of 243 F2 pups, 65 were wild type (26.7%), 117 were heterozygous for the mutation (48.2%), and 61 were homozygous mutants (25.1%), consistent with the ratio expected from Mendelian inheritance. Both male and female Smac−/− mice were healthy and fertile, and F3 Smac−/− offspring obtained by homozygous intercrosses were also healthy. Both Smac+/− and Smac−/− mice developed normally and did not exhibit any obvious macroscopic or microscopic abnormalities (data not shown). Aged mice (more than 12 months of age) did not show any sign of anomalies, such as autoimmune disease or tumor formation (data not shown).
Caspase 8 activation in vitro is impaired in Smac−/− cell lysates.
The Smac protein is localized in the soluble membrane fraction of cell lysates, which promotes caspase 3 activation in a manner dependent on caspase 9, Apaf-1, and cytochrome c (5). We therefore compared in vitro cleavage of procaspase 3 in cell lysates prepared from Smac−/− or Smac+/+ MEFs. Caspase 3 activation was induced by the addition of dATP and cytochrome c to cell lysates, and the presence of cleaved, endogenous caspase 3 was detected by immunoblotting with antibody specifically recognizing active caspase 3. While procaspase 3 cleavage could be detected within 30 min in Smac+/+ lysates, activated caspase 3 was not observed in Smac−/− lysates even after 60 min (Fig. 2A). To confirm that this phenotype was caused by the loss of Smac, we reconstituted the Smac−/− lysate with recombinant Smac protein (5). Procaspase 3 cleavage was restored in the Smac−/− lysate in a dose-dependent manner (Fig. 2B). To exclude the possibility that the loss of Smac affected the protein levels of the IAPs, we compared the levels of several IAPs in Smac+/+ and Smac−/− lysates by Western blotting. Equivalent amounts of c-IAP1, c-IAP2, XIAP, and survivin were detected in the presence and absence of Smac (data not shown). We also investigated the expression of Omi/HtrA2, a recently reported functional homologue of Smac (10, 14, 25, 27). Equivalent amounts of Omi protein were detected in Smac−/− and Smac+/+ lysates (data not shown). From these observations, we conclude that we have introduced a functionally null mutation into the Smac gene and confirm that Smac potentiates procaspase 3 cleavage in vitro as previously reported (5).
FIG. 2.
Smac deficiency impairs caspase 3 cleavage in vitro. (A) Impaired in vitro caspase 3 cleavage in Smac−/− MEFs. Lysates of Smac+/+ or Smac−/− MEFs were incubated in vitro with the indicated assay reagents, and the cleavage of procaspase 3 was detected by Western blotting with antibodies recognizing either the cleaved (active) form of caspase 3 or procaspase 3. One of the representative results from three independent samples is shown. cyt C, cytochrome c. (B) Restoration of procaspase 3 cleavage by addition of recombinant Smac protein. Lysates of Smac−/− MEFs were incubated with the indicated reagents plus 1 nM to 1 μM recombinant Smac protein. Detection of activated caspase 3 was as for panel A.
Induction of apoptosis is normal in primary and transformed Smac−/− cells.
Since procaspase 3 cleavage is a defining event in most instances of apoptosis, we investigated whether the loss of Smac had any effect on apoptosis induced in MEFs subjected to UV irradiation. It has previously been shown that overexpression of Smac itself does not induce apoptosis in cells, but rather sensitizes them to UV-induced cell death (5). We subjected Smac−/− and Smac+/+ primary MEFs to UV irradiation and assessed viability. As shown in Fig. 3A, similar levels of PCD were induced in Smac−/− and Smac+/+ MEFs at 24 h after UV irradiation. It has been reported that Smac starts to be released from the mitochondria into the cytosol within the first few hours after UV irradiation (5). To examine the possibility that Smac might inhibit procaspase 3 cleavage at early time points, we compared the amount of active caspase 3 accumulated in Smac−/− and Smac+/+ MEFs at 0, 4, 8, 12, and 16 h after UV irradiation. No difference in the level of active caspase 3 was detected at any time point (Fig. 3B), indicating that UV-induced apoptosis of primary MEFs is not affected by the loss of Smac.
FIG. 3.
Smac-deficient cells respond normally to apoptotic stimuli. (A) PCD induced by UV irradiation in primary MEFs. Smac+/+ or Smac−/− MEFs were subjected to UV irradiation, and cell viability was determined by annexin-PI staining at 24 h. (B) Procaspase 3 cleavage in UV-treated MEFs. Protein samples were prepared from Smac+/+ or Smac−/− MEFs at 0, 4, 8, 12, or 16 h after UV irradiation. The amount of active caspase 3 was determined by Western blot analysis as for Fig. 2A. (C to F) Effect of Smac deficiency on PCD induced by various other stimuli in MEFs and other cell types. MEFs (C), ES cells (D), thymocytes (E), and E1A- or c-Myc-overexpressing MEFs (F) were treated with the indicated reagents for 24 h, and cell viability was determined by annexin-PI staining. The data shown are means ± standard deviations of triplicate measurements and are representative of three experiments with similar results.
We next examined a range of chemical agents to determine whether Smac is associated with PCD induced only in certain cell types or only by certain apoptotic stimuli. Staurosporine, adriamycin, anisomycin, etoposide, and TNF/CHX all induced PCD equally in Smac−/− and Smac+/+ MEFs (Fig. 3C) and also in Smac−/− and Smac+/+ ES cells (Fig. 3D [TNF/CHX not tested]). Similarly, Smac−/− thymocytes treated with etoposide, dexamethasone, or Fas/CHX underwent normal PCD (Fig. 3E). PCD induced by staurosporine or by UV or gamma irradiation was also indistinguishable in thymocytes from Smac+/+ and Smac−/− mice (data not shown). Thus, despite the involvement of IAPs in apoptosis, loss of Smac has no effect on PCD induced in these cell types under these circumstances.
Oncogenes can sensitize cells to apoptosis mediated by the mitochondrial pathway (7, 8). We therefore infected Smac−/− and Smac+/+ MEFs with retroviruses causing overexpression of the oncogene c-Myc or E1A. These cells were then treated with adriamycin, TRAIL, or paclitaxel to induce PCD. Cells of both genotypes became equally hypersensitive to apoptotic stimuli (Fig. 3F). This result was somewhat unexpected in view of the fact that transformed caspase 3-deficient MEFs are resistant to PCD induced by these stimuli (32). Our data suggest that other molecules must be able to substitute for Smac in the induced PCD of both primary and oncogene-overexpressing cells.
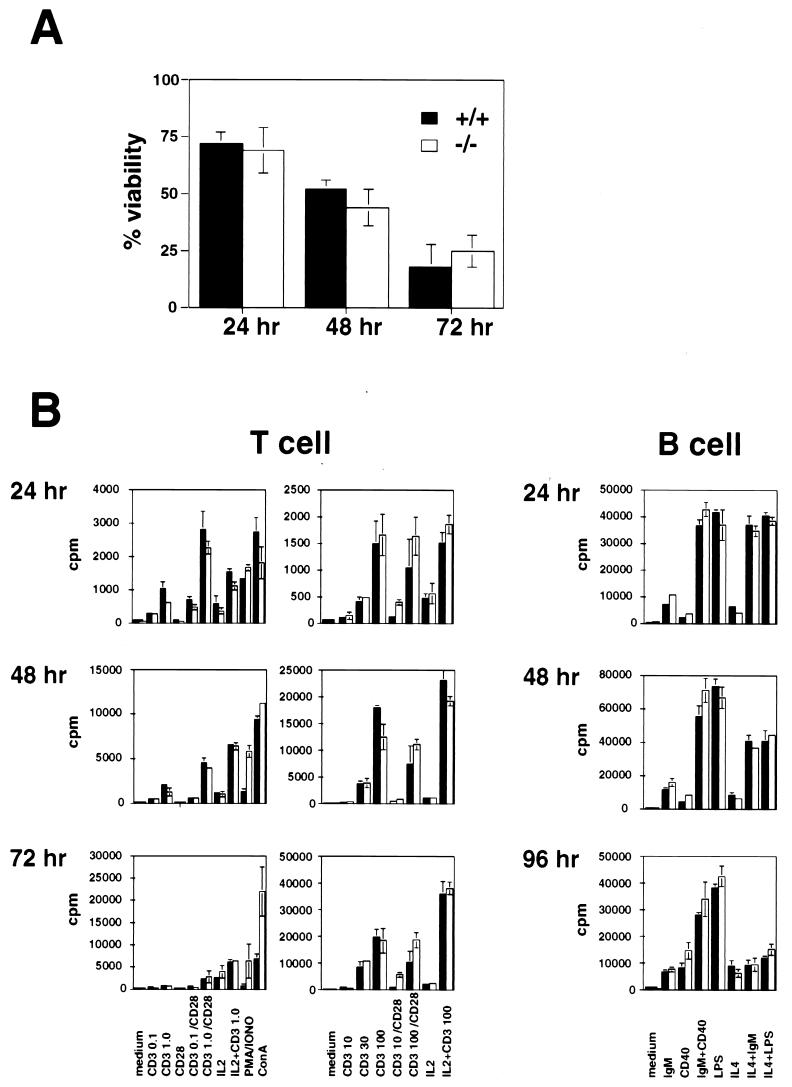
Smac-deficient T and B cells show normal proliferation and survival.
In the absence of caspase 3, T cells are less susceptible to AICD (30), and activated B cells hyperproliferate in response to mitogenic stimulation (M. Woo, C. Furlonger, R. Hakem, C. Paige, and T. W. Mak, submitted for publication). Since Smac deficiency decreased the amount of activated caspase 3 in MEFs, we examined AICD of T cells and proliferation of both activated T and B cells. T cells were activated in vitro as described in Materials and Methods, and cell viability at 24, 48, and 72 h was evaluated by trypan blue exclusion. No significant differences in AICD were observed between Smac+/+ and Smac−/− T cells (Fig. 4A). Proliferation of T and B cells activated as described in Materials and Methods was measured by [3H]thymidine incorporation at 24, 48, or 72 h after activation for T cells and at 24, 48, or 96 h after activation for B cells. There was no difference between Smac+/+ and Smac−/− cells in [3H]thymidine incorporation by either activated T or B cells (Fig. 4B). These results show that Smac does not affect either cell proliferation or survival of activated lymphocytes.
FIG. 4.
Activated Smac-deficient T and B cells show normal proliferation and survival in culture. (A) Activation-induced cell death in T cells. T cells prepared from either Smac+/+ or Smac−/− mice were activated in vitro and restimulated with anti-CD3ɛ. The number of viable cells remaining was determined at the indicated time points (solid bars, +/+; open bars, −/−). (B) Proliferation of T and B cells. T or B cells prepared from either Smac+/+ or Smac−/− mice were stimulated in vitro with the indicated stimuli. Cell proliferation was determined by measuring [3H]thymidine incorporation (cpm) at the indicated time points (solid bars,+/+; open bars, −/−). The data shown are means ± standard deviations of triplicate measurements. Experiments were repeated three times with similar results.
Fas-mediated apoptosis occurs normally in Smac−/− hepatocytes.
It has been reported that Fas-mediated hepatocyte cell death is delayed in caspase 3-deficient mice (31). Since caspase 3 activation was impaired in vitro in the absence of Smac, we investigated the role of Smac in Fas-mediated PCD in vivo by injecting Smac−/− mice with agonistic anti-Fas antibody. Both Smac+/+ and Smac−/− mice died within 3 or 10 h after injection of 100 or 10 μg of anti-Fas antibody, respectively, with similar kinetics (Fig. 5A). Hepatocyte apoptosis was verified by staining liver sections with H&E (Fig. a, c, and e) or TUNEL (Fig. b, d, and f). No statistically significant differences in numbers of TUNEL-positive cells were observed between Smac+/+ and Smac−/− mice livers (data not shown). Similar results were obtained when apoptosis was induced in heart or kidney by injecting adriamycin (12) or tunicamycin (15), respectively (data not shown).
FIG. 5.
Fas-mediated apoptosis is intact in the liver of Smac-deficient mice. (A) Survival curve of mice injected with anti-Fas antibody. Either 10 or 100 μg of anti-Fas antibody was injected into the intraperitoneal cavity of Smac+/+ or Smac−/− mice (n = 3 per group per dose). One experiment representative of five independent trials is shown (solid symbols, 100 μg; open symbols, 10 μg) (B) Histological analysis of livers of mice injected with anti-Fas antibody. Liver sections were prepared 3 h after Smac+/+ (a and b), Smac+/− (c and d), and Smac−/− (e and f) mice were injected with 100 μg of anti-Fas antibody. Sections were stained with H&E (a, c, and e) or by TUNEL (b, d, and f).
DISCUSSION
Our finding in vivo that targeted disruption of the Smac gene has no effect on apoptosis stands in sharp contrast to previous evidence established by biochemical, cell biological and structural analyses that favor a role for Smac/DIABLO in PCD (5, 29). One of the best examples of this apparent contradiction in our report is the discrepancy between the results of the in vitro caspase 3 activation assay and the phenotype of Smac−/− cells. We were able to demonstrate differences in procaspase 3 cleavage in Smac−/− and Smac+/+ MEF lysates, but could not detect any differences in induced PCD in MEFs. The simplest explanation for this discrepancy is that a functional homologue of Smac exists in vivo, but was not present or functional in cell lysates. We found equivalent protein expression of Omi/HtrA2, a functional homologue of Smac, in wild-type and Smac-deficient cell lysates, but the possibility remains that Omi may have been inactive or only weakly active under the in vitro assay conditions used. In this context, it is noteworthy that some activated caspase 3 was detected in Smac−/− samples after prolonged incubation (data not shown); however, this amount of activated caspase 3 was still significantly less than that in the wild type.
The phenotype of Smac-deficient mice could easily be accounted for if Omi was fully active and sufficient to compensate for the loss of Smac in vivo. It will therefore be useful to generate double mutant mice lacking both Smac and Omi to clarify the precise physiological role of the Smac-mediated pathway. Alternatively, other mitochondrial proapoptotic molecules, such as AIF (24), NOXA (17), and p53AIP (18), may be able to compensate for a loss of Smac function in certain contexts in vivo. Finally, the phenotype of Smac−/− mice could result if Smac does not play an essential role in the common apoptotic machinery, but instead participates in regulating PCD only in a specific situation or in tissues yet to be identified.
A role for Smac in apoptosis is not evident from our gene disruption study. However, no obvious abnormalities were found in Smac−/− mice, meaning that investigation of alternative functions for the Smac protein will require extensive efforts to find specific conditions under which loss of Smac results in pathology. The generation of combined mutants by crossing Smac-deficient mice with transgenic mice prone to cancer formation or autoimmune disease might help to reveal covert phenotypes. On the other hand, the phenotype of Smac-deficient mice presented in this study strongly suggests the existence of a redundant molecule or molecules. Research to identify such molecules may bring to light new regulatory mechanisms of apoptosis, which could ultimately lead to novel therapeutic targets for diseases caused by the deregulation of apoptosis.
Acknowledgments
We thank Takeshi Yagi and Tetsuo Noda for the DT-A plasmid, and Emad S. Alnemri for the anti-Omi/HtrA2 antibody. We are grateful to numerous members of the Mak laboratory for helpful comments and discussion and to Mary Saunders for scientific editing.
This work was supported by National Cancer Institute of Canada. H.O. was partially supported by Japanese Foundation for Clinical Pharmacology. J.J. is supported by a DOD Breast Cancer Research Program postdoctoral fellowship. S.W.L. is supported by the Rita Allen Foundation and grant CA13106 from the NIH.
REFERENCES
- 1.Alnemri, E. S., D. J. Livingston, D. W. Nicholson, G. Salvesen, N. A. Thornberry, W. W. Wong, and J. Yuan. 1996. Human ICE/CED-3 protease nomenclature. Cell 87:171.. [DOI] [PubMed] [Google Scholar]
- 2.Chai, J., C. Du, J. W. Wu, S. Kyin, X. Wang, and Y. Shi. 2000. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature 406:855-862. [DOI] [PubMed] [Google Scholar]
- 3.Cryns, V., and J. Yuan. 1998. Proteases to die for. Genes Dev. 12:1551-1570. [DOI] [PubMed] [Google Scholar]
- 4.Deveraux, Q. L., and J. C. Reed. 1999. IAP family proteins—suppressors of apoptosis. Genes Dev. 13:239-252. [DOI] [PubMed] [Google Scholar]
- 5.Du, C., M. Fang, Y. Li, L. Li, and X. Wang. 2000. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102:33-42. [DOI] [PubMed] [Google Scholar]
- 6.Ellis, R. E., J. Y. Yuan, and H. R. Horvitz. 1991. Mechanisms and functions of cell death. Annu. Rev. Cell Biol. 7:663-698. [DOI] [PubMed] [Google Scholar]
- 7.Fearnhead, H. O., M. E. McCurrach, J. O'Neill, K. Zhang, S. W. Lowe, and Y. A. Lazebnik. 1997. Oncogene-dependent apoptosis in extracts from drug-resistant cells. Genes Dev. 11:1266-1276. [DOI] [PubMed] [Google Scholar]
- 8.Fearnhead, H. O., J. Rodriguez, E. E. Govek, W. Guo, R. Kobayashi, G. Hannon, and Y. A. Lazebnik. 1998. Oncogene-dependent apoptosis is mediated by caspase 9. Proc. Natl. Acad. Sci. USA 95:13664-13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goyal, L., K. McCall, J. Agapite, E. Hartwieg, and H. Steller. 2000. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19:589-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegde, R., S. M. Srinivasula, Z. Zhang, R. Wassell, R. Mukattash, L. Cilenti, G. DuBois, Y. Lazebnik, A. S. Zervos, T. Fernandes-Alnemri, and E. S. Alnemri. 2002. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts IAP-caspase interaction. J. Biol. Chem. 277:432-438. [DOI] [PubMed] [Google Scholar]
- 11.Jacobson, M. D., M. Weil, and M. C. Raff. 1997. Programmed cell death in animal development. Cell 88:347-354. [DOI] [PubMed] [Google Scholar]
- 12.Kang, Y. J., Y. Chen, and P. N. Epstein. 1996. Suppression of doxorubicin cardiotoxicity by overexpression of catalase in the heart of transgenic mice. J. Biol. Chem. 271:12610-12616. [DOI] [PubMed] [Google Scholar]
- 13.Kerr, J. F., A. H. Wyllie, and A. R. Currie. 1972. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J. Cancer 26:239-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martins, L. M., I. Iaccarino, T. Tenev, S. Gschmeissner, N. F. Totty, N. R. Lemoine, J. Savopoulos, C. W. Gray, C. L. Creasy, C. Dingwall, and J. Downward. 2002. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a Reaper-like motif. J. Biol. Chem. 277:439-444. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa, T., H. Zhu, N. Morishima, E. Li, J. Xu, B. A. Yankner, and J. Yuan. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 403:98-103. [DOI] [PubMed] [Google Scholar]
- 16.Niki, M., H. Okada, H. Takano, J. Kuno, K. Tani, H. Hibino, S. Asano, Y. Ito, M. Satake, and T. Noda. 1997. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc. Natl. Acad. Sci. USA 94:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oda, E., R. Ohki, H. Murasawa, J. Nemoto, T. Shibue, T. Yamashita, T. Tokino, T. Taniguchi, and N. Tanaka. 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288:1053-1058. [DOI] [PubMed] [Google Scholar]
- 18.Oda, K., H. Arakawa, T. Tanaka, K. Matsuda, C. Tanikawa, T. Mori, H. Nishimori, K. Tamai, T. Tokino, Y. Nakamura, and Y. Taya. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102:849-862. [DOI] [PubMed] [Google Scholar]
- 19.Okada, H., T. Watanabe, M. Niki, H. Takano, N. Chiba, N. Yanai, K. Tani, H. Hibino, S. Asano, M. L. Mucenski, Y. Ito, T. Noda, and M. Satake. 1998. AML1(−/−) embryos do not express certain hematopoiesis-related gene transcripts including those of the PU.1 gene. Oncogene 17:2287-2293. [DOI] [PubMed] [Google Scholar]
- 20.Ranger, A. M., B. A. Malynn, and S. J. Korsmeyer. 2001. Mouse models of cell death. Nat. Genet. 28:113-118. [DOI] [PubMed] [Google Scholar]
- 21.Ruland, J., G. S. Duncan, A. Elia, I. del Barco Barrantes, L. Nguyen, S. Plyte, D. G. Millar, D. Bouchard, A. Wakeham, P. S. Ohashi, and T. W. Mak. 2001. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-kappaB and neural tube closure. Cell 104:33-42. [DOI] [PubMed] [Google Scholar]
- 22.Salvesen, G. S., and V. M. Dixit. 1997. Caspases: intracellular signaling by proteolysis. Cell 91:443-446. [DOI] [PubMed] [Google Scholar]
- 23.Samuelson, A. V., and S. W. Lowe. 1997. Selective induction of p53 and chemosensitivity in RB-deficient cells by E1A mutants unable to bind the RB-related proteins. Proc. Natl. Acad. Sci. USA 94:12094-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Susin, S. A., H. K. Lorenzo, N. Zamzami, I. Marzo, B. E. Snow, G. M. Brothers, J. Mangion, E. Jacotot, P. Costantini, M. Loeffler, N. Larochette, D. R. Goodlett, R. Aebersold, D. P. Siderovski, J. M. Penninger, and G. Kroemer. 1999. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397:441-446. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki, Y., Y. Imai, H. Nakayama, K. Takahashi, K. Takio, and R. Takahashi. 2001. A serine protease, htra2, is released from the mitochondria and interacts with xiap, inducing cell death. Mol. Cell 8:613-621. [DOI] [PubMed] [Google Scholar]
- 26.Thornberry, N. A., and Y. Lazebnik. 1998. Caspases: enemies within. Science 281:1312-1316. [DOI] [PubMed] [Google Scholar]
- 27.Verhagen, A. M., J. Silke, P. G. Ekert, M. Pakusch, H. Kaufmann, L. M. Connolly, C. L. Day, A. Tikoo, R. Burke, C. Wrobel, R. L. Moritz, R. J. Simpson, and D. L. Vaux. 2002. HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins. J. Biol. Chem. 277:445-454. [DOI] [PubMed] [Google Scholar]
- 28.Verhagen, A. M., E. J. Coulson, and D. L. Vaux. 2001. Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol. 2:3009.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verhagen, A. M., P. G. Ekert, M. Pakusch, J. Silke, L. M. Connolly, G. E. Reid, R. L. Moritz, R. J. Simpson, and D. L. Vaux. 2000. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102:43-53. [DOI] [PubMed] [Google Scholar]
- 30.Wang, J., L. Y. Xie, S. Allan, D. Beach, and G. J. Hannon. 1998. Myc activates telomerase. Genes Dev. 12:1769-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woo, M., A. Hakem, A. J. Elia, R. Hakem, G. S. Duncan, B. J. Patterson, and T. W. Mak. 1999. In vivo evidence that caspase 3 is required for Fas-mediated apoptosis of hepatocytes. J. Immunol. 163:4909-4916. [PubMed] [Google Scholar]
- 32.Woo, M., R. Hakem, M. S. Soengas, G. S. Duncan, A. Shahinian, D. Kagi, A. Hakem, M. McCurrach, W. Khoo, S. A. Kaufman, G. Senaldi, T. Howard, S. W. Lowe, and T. W. Mak. 1998. Essential contribution of caspase 3/CPP32 to apoptosis and its associated nuclear changes. Genes Dev. 12:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, G., J. Chai, T. L. Suber, J. W. Wu, C. Du, X. Wang, and Y. Shi. 2000. Structural basis of IAP recognition by Smac/DIABLO. Nature 408:1008-1012. [DOI] [PubMed] [Google Scholar]
- 34.Wu, J. W., A. E. Cocina, J. Chai, B. A. Hay, and Y. Shi. 2001. Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides. Mol. Cell 8:95-104. [DOI] [PubMed] [Google Scholar]
- 35.Yagi, T., S. Nada, N. Watanabe, H. Tamemoto, N. Kohmura, Y. Ikawa, and S. Aizawa. 1993. A novel negative selection for homologous recombinants using diphtheria toxin A fragment gene. Anal. Biochem. 214:77-86. [DOI] [PubMed] [Google Scholar]