Abstract
We investigated the control of telomere length by the human telomeric proteins TRF1 and TRF2. To this end, we established telomerase-positive cell lines in which the targeting of these telomeric proteins to specific telomeres could be induced. We demonstrate that their targeting leads to telomere shortening. This indicates that these proteins act in cis to repress telomere elongation. Inhibition of telomerase activity by a modified oligonucleotide did not further increase the pace of telomere erosion caused by TRF1 targeting, suggesting that telomerase itself is the target of TRF1 regulation. In contrast, TRF2 targeting and telomerase inhibition have additive effects. The possibility that TRF2 can activate a telomeric degradation pathway was directly tested in human primary cells that do not express telomerase. In these cells, overexpression of full-length TRF2 leads to an increased rate of telomere shortening.
The extremities of eukaryotic chromosomes are protected by telomeres (52, 54, 64). These structures are essential for chromosome stability, and their alteration can lead to genome rearrangements, alterations in cellular programs, and carcinogenesis (8). Telomeres have a specific structure that allows the DNA repair machinery to distinguish them from broken DNA ends (25, 66). Telomeric DNA is a tandem array of repeated sequences, usually with one strand, called the G strand, enriched in guanine nucleotide residues. This G strand ends with a 3′ single-stranded overhang, the G tail. Both the double-stranded telomeric DNA repeats and the G tail are specifically bound by a set of specialized proteins. These proteins constitute the telomeric chromatin and are essential for chromosome capping (40, 58, 63, 74). The number of telomeric DNA repeats is a critical structural and functional determinant. When it falls below a threshold value, checkpoints that lead to growth arrest become activated. If these checkpoints fail, and if telomeric DNA is further shortened, genome instability rapidly increases, ultimately leading to cell death (16). Therefore, the cellular mechanisms that determine the number of telomeric repeats are of primary importance in controlling cell fate and chromosome stability.
The DNA polymerases that replicate the bulk of the chromosomal DNA are unable to fully duplicate the telomeric DNA, and progressive telomeric erosion is an unavoidable consequence of DNA replication. There are both a lagging-strand problem due to the 5′-to-3′ direction of polymerization, which requires an RNA primer for initiation, and a leading-strand problem due to the inability to synthesize the parental G tail (46, 57, 78). Telomere shortening can be exacerbated by the action of processing enzymes, whose activation appears also to be coupled to DNA replication (18). The nature of these enzymes is still elusive, but it is likely that they are associated with nuclease activities.
In certain cells—germ cells, stem cells, cancer cells, and microorganisms, for example—the eroding effect of DNA replication is counterbalanced by the action of an enzyme called telomerase that specifically elongates the G strand (28). Telomerase is a specialized reverse transcriptase that uses an internal RNA molecule as a template (7, 47). The enzymatic activity can be reconstituted in vitro from a catalytic subunit and an RNA moiety that contains the template sequence (3, 79). During telomere replication, telomerase activity is coupled to C-strand synthesis, which is believed to be carried out by the lagging-strand replication machinery (12, 17, 22, 60).
Telomerase is not free to elongate chromosome ends within the cell. In fact, its activity is closely regulated in cis, i.e., at the telomere itself, to prevent the telomeres from becoming too short or too long (29, 42, 49). A model for cis regulation of telomerase activity has emerged from recent intensive studies of budding yeast. Telomerase needs to be recruited or activated at chromosome ends by interacting with a G-tail DNA-binding protein. In Saccharomyces cerevisiae, this protein is Cdc13p (13, 21, 27, 55). The contact between Cdc13p and telomerase is likely to be transient and, presumably, restricted to the period of the cell cycle when telomerase is active (17, 50), which might explain why the interaction has not been detected in cell extracts (35).
Rap1p, a protein that binds along the double-stranded telomeric DNA repeats, enacts an additional level of regulation. An excess of Rap1p molecules bound to the telomere causes cis repression of telomerase activity (39, 40, 49, 51). This creates a negative-feedback loop that contributes to the setting of telomere length. Two factors interacting with Rap1p, Rif1p and Rif2p, are also involved in this regulation (80). How these proteins repress the distally located telomerase in cis at the molecular level is unknown. One can imagine that the gradual folding of the telomeric chromatin into a restrictive higher-order configuration can impair telomerase activity. Interestingly, the components of telomeric chromatin do not impair only the activity of telomerase but also that of telomerase-independent lengthening mechanisms (72), suggesting that similar logics of length setting operate in telomerase-positive and -negative cells. Whether the cis-acting regulations of telomerase mediated by the double-stranded part of the telomeric chromatin and by the G tail are coordinated or have separate outcomes is certainly an exciting question for future investigations.
Strikingly, Cdc13p has no obvious ortholog in species outside budding yeast. Nevertheless, the discovery of other types of G-tail DNA-binding proteins in ciliates, fission yeast, and animal cells suggests the existence of Cdc13p functional homologs (2, 11, 26, 41). Several lines of evidence indicate that the mechanism by which proteins binding to the double-stranded part of telomeres limit telomere elongation might also apply to proteins other than Rap1p and can be well conserved. In fission yeast, a deletion of taz1+, a gene encoding a telomeric DNA-binding protein, dramatically increases telomere length (15). In human cells, the known telomeric DNA-binding proteins are the TTAGGG repeat factors 1 and 2 (TRF1 and TRF2) (5, 6, 9, 14). The C-terminal sequences of TRF1 and TRF2, called teloboxes, specifically recognize a telomeric DNA fragment but require an N-terminal dimerization domain to firmly bind DNA (4, 6, 76). Reminiscent of the properties of the yeast Rap1p protein, an excess of TRF1 and TRF2 inhibits telomere elongation (69, 75). Some additional players in the negative-feedback regulation of telomere length have been identified in human cells: TIN2 (36), PinX1 (81), and tankyrase (67, 68), three TRF1-interacting factors, and hRAP1, a human ortholog of yeast Rap1p, which interacts with TRF2 (44).
A cis repression of telomere elongation by TRF1 or TRF2 has not been demonstrated so far. In order to assay the role in cis of these proteins, we devised a cell system to target multiple molecules of TRF1 or TRF2 to specific telomeres in a human cell line. For that purpose, we adopted the strategy developed by A. Belmont and coworkers in which a chimera containing the Lac repressor (LacI) is directed to an array of Lac operator sequences (LacO) (62). This allowed us to show that TRF1 and TRF2 act in cis to repress telomere elongation. While TRF1 represses the telomerase pathway, TRF2, on the contrary, appears to activate a telomeric degradation pathway, even in cells devoid of telomerase activity.
MATERIALS AND METHODS
Vector construction.
All cloning was carried out according to standard procedures. To construct the seeding vector pLacOTEL, 256 copies of LacO present in pSV2-DHFR-8.32 (a gift from A. Belmont [62]) were inserted into the pCMVT vector (M. P. Zibella, A. Pollice, J. Pulitzer, and E. Gilson, unpublished data), which contains 1.6 kb of TTAGGG sequences and the hygromycin-thymidine kinase resistance gene. Details concerning the exact cloning procedure will be given upon request.
Vectors expressing LacI, TRF1-LacI, and TRF2-LacI, collectively called the LacI-derived proteins, were generated by cloning the corresponding PCR products into the NotI site of pIND (Invitrogen). PCR was carried out on p3′SS (a gift from A. Belmont [62]) for LacI, on the plasmid containing the full-length cDNA of TERF1 (76) for TRF1, and on a plasmid containing the cDNA of TERF2 without its first 87 bp (5) for TRF2 (primers will be communicated on request). The various pIND derivatives were sequenced on both strands (ABI PRISMDNA sequencing system).
The retroviral vector pBloxP was generated by removing the catalytic subunit of telomerase as an EcoRI fragment from the pBabehTERTloxp retroviral vector (a gift from J. Shay [71]), followed by self-ligation of the vector. The cDNA encoding a Myc-tagged version of human full-length TRF2 (a gift from Titia de Lange) was subcloned as an HindIII blunt-ended EcoRI fragment into the SnaBI-EcoRI sites of pBloxP to generate pBTRF2loxP.
Cell culture, transfection, retroviral infection, and cell lines.
EcR-293 cells were purchased from Invitrogen and cultured in 90% Dulbecco's modified Eagle's medium (GIBCO) supplemented with l-glutamine, penicillin-streptomycin, 10% fetal calf serum (HyClone), and zeocin (200 μg/ml). All stable clones were obtained by electroporation, with selection added 2 days after transfection. The selection was decreased by half 2 weeks after the transfection to a final concentration of 40 μg/ml for hygromycin and 150 μg/ml for G418. Induced and noninduced cells were grown in parallel with or without ponasterone A (PonA; Invitrogen).
WI38 normal human diploid fibroblasts (ATCC CCL-75) were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (GIBCO) with 10% fetal calf serum (GIBCO). To create amphotropic retroviruses, PT67 producer cells (Clontech) were transfected with either pBloxP or pBTRF2loxP constructs by calcium phosphate precipitation. Viral supernatants were harvested 48 h after transfection and used to infect WI38 cells with 8 μg of Polybrene (Sigma)/ml. Two days after infection, cells were selected in puromycin (0.5-μg/ml final concentration; Sigma). Infected WI38 cells were grown in medium containing 0.5 μg of puromycin/ml throughout the course of the experiment.
Genomic blotting and telomeric DNA analysis.
Genomic DNA was extracted from subconfluent cells at various mean population doublings (MPDs) using the Nucleon Bacc2 kit (Amersham Pharmacia Biotech). For measuring the lengths of the terminal restriction fragments of the total population of telomeres, we followed a Southern blotting procedure with a HinfI/RsaI digestion and a TTAGGG probe as described previously (16). For telomere length analysis of the tagged telomeres, we released the terminal restriction fragment containing the LacO sequences by HindIII restriction and analyzed the lengths of these fragments by Southern blotting using the LacO probe (a 10-kb LacO256 fragment excised from psv2-dhfr-8.32). The Southern gels were scanned with a PhosphorImager and analyzed by ImageQuant software.
The length corresponding to the apex of the distribution of terminal restriction fragments is called TRF for the total population of telomeres and OTRF for the LacO-containing telomeres. The location of the apex on the densitometric scan was determined empirically, after smoothing the profile by hand if necessary. In the case of TRF analysis, no correction was made to account for the dependence of signal strength on telomere length. When indicated in the text and in the figures, the mean OTRF was calculated as the center of mass of the densitometric profile as described previously (59). Each measurement of telomere length was made at least twice; the standard error between the two measures has been estimated to be smaller than 10%.
FISH and immunofluorescence of metaphase spreads.
Preparation of metaphase chromosomes, harvesting, and fixation were performed following standard cytogenetic methods (20). Fluorescence in situ hybridization (FISH) with plasmids was done as described earlier (43). The LacO probe was labeled with digoxigenin using a nick translation kit (Boehringer). Chromosome painting (Oncor) was performed according to the manufacturer's instructions. Chromosome instability was scored from centromere (Cambio-Vysis) and telomere [PNA-Cy3(C3TA2)3; Perseptive Biosystems] labeled slides.
For immunofluorescence analysis, cells were treated with Colcemid (0.4 μg/ml; 60 min), harvested by trypsinization, and sedimented onto glass slides (Shandon). Chromosomes were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) and permeabilized with 0.2% Triton. Samples were blocked with 2% bovine serum albumin in PBS containing 0.05% Tween 20. Antibody incubations were performed at 37°C for 1 h with anti-LacI and then with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit antibody (Jackson Laboratories). Chromosomes were counterstained with 4,6-diamino-2-phenylindole (DAPI) or propidium iodide (PI). The slides were embedded in Vectashield (Vector Laboratories). Digital fluorescence analysis was performed using a Leica DM-RBE epifluorescence microscope equipped with a Cohu camera. Image acquisition and analysis were accomplished on a Cytovision workstation (Applied Imaging).
Nuclear extracts and Western blotting.
Cells at the indicated MPDs were harvested in cold PBS and treated for nuclear extracts as described previously (65). The protein concentration was determined using the Bradford assay (Bio-Rad) with bovine serum albumin as a standard. Proteins (25 μg) were analyzed by Western blotting following standard procedures. Analyses were performed using polyclonal antiserum to LacI (Stratagene), anti-topoisomerase II clone KiS1 (Dako), or polyclonal antibodies raised against the endogenous telomeric proteins TRF1 and TRF2 (5).
In vivo cross-linking, immunoprecipitation, and quantitative PCR analysis.
Cell cross-linking was performed exactly as described by Hsu et al. (34). Extracts were immunoprecipitated with anti-LacI serum or with rabbit serum (preimmune) for 2 h at 4°C. An aliquot (called the input) was not subjected to immunoprecipitation. Protein A-coupled beads (Amersham Pharmacia Biotech) were then added for 1 h and washed four times with NET buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 0.5% NP-40). To isolate coimmunoprecipitated DNA, elution was done first in 100 μl of 1% sodium dodecyl sulfate-0.1 M NaHCO3 and then in 150 μl of TE (10 mM Tris·HCl [pH 7.5], 1 mM EDTA) buffer. The pooled eluates and the input fraction were heated at 65°C overnight to reverse the formaldehyde cross-links and then treated with proteinase K solution (0.1 μg of glycogen/ml and 0.4 μg of proteinase K/ml in TE). They both were equilibrated with 0.2 mM LiCl, extracted in phenol-chloroform-isoamyl alcohol, and finally ethanol precipitated. The immunoprecipitates and the input fractions were each resuspended in 50 μl of TE buffer. Chromatin immunoprecipitation analyses were done by real-time detection and quantitative PCR using the LightCycler system (Roche) according to the manufacturer's instructions. In this way, we measured the recovery of the LacO sequences in the induced and uninduced conditions. Alternatively, the immunoprecipitates were analyzed by a standard slot blot procedure.
In vivo inhibition of telomerase activity.
In vivo inhibition of telomerase activity was done as described previously (32) with LacO 293 cells expressing either TRF1-LacI or TRF2-LacI. Match, a modified oligonucleotide recognizing the telomerase RNA template, was purchased from Eurogentec. Its sequence is 5′-CAGUUAGGGUUAG, where the underlined nucleotides possess phosphothioate linkages. Cells (3 × 106) were plated on day 1 in a 100-mm-diameter dish. They were transfected on day 2 with 10 μg of the Match oligonucleotide, using the Max1 transfection reagent (Eurogentec). The cells were harvested on day 4 and counted; 3 × 106 cells were replated, and the remaining cells were used for telomere length analysis (as described above) and the semiquantitative telomerase activity assay (TRAPeze telomerase detection kit; Intergen).
RESULTS
In this study, we investigated the molecular nature of telomere length regulation by TRF1 and TRF2. We designed an inducible LacI-based system for targeting TRF1 and TRF2 to a particular subtelomeric position in an immortalized human cell line. This strategy has proved to be a valuable tool in yeast to study the cis effect of telomeric factors and to dissect the domains of Rap1 involved in silencing and length regulation (48, 51).
Stable cell lines with subtelomeric LacO and an inducible expression of LacI-based hybrid proteins.
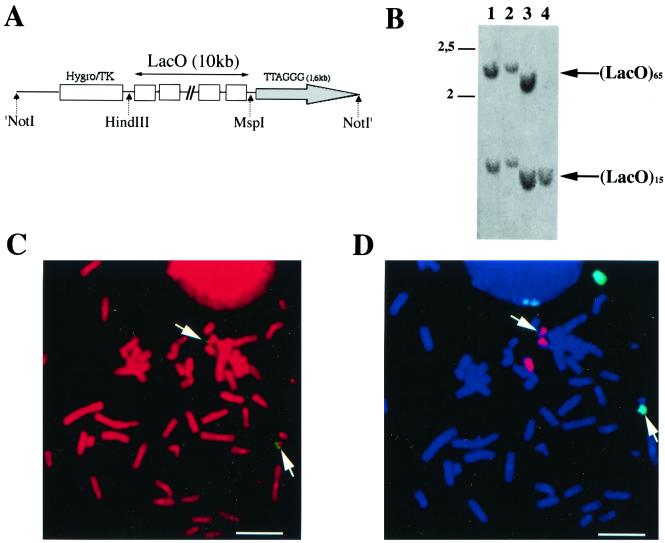
We sought to engineer a tagged telomere carrying an array of binding sites for the LacI protein (LacO) juxtaposed to the telomeric repeats. To this end, we used a seeding procedure with the linearized pLacOTEL plasmid (Fig. 1A) (23, 31). This was carried out in EcR-293 cells, which are adenovirus-transformed p53-inactive embryonic kidney cells that constitutively express the Drosophila RXR and VgEcR proteins and permit the inducible expression of proteins upon addition of PonA, an ecdysone analog. Hygromycin-resistant stable clones were selected. In roughly 50% of them, the DNA fragments hybridizing with the LacO probe exhibited a smear by Southern analysis, reflecting the length polymorphism of telomeric restriction fragments and successful telomere seeding (data not shown). We chose one clone, 182E6, in which the lengths of the LacO-containing terminal restriction fragments remain unchanged during long-term cell culture (data not shown). When genomic DNA from clone 182E6 was digested with HindIII and MspI and hybridized with the LacO probe, two bands were obtained, suggesting that two telomeres contain a LacO insert in this clone (Fig. 1B, lane 1). This was confirmed by FISH using the LacO probe, followed by chromosome painting: LacO sequences are present on one tip of chromosome 18 and on one tip of chromosome 19 (Fig. 1C and D). According to the lengths of the two LacO-containing HindIII-MspI fragments (Fig. 1B, lane 1), one apparently harbors roughly 15 LacO repeats (LacO15) and the other harbors 65 repeats (LacO65). The telomeres containing LacO sequences will subsequently be referred to as the tagged telomeres.
FIG. 1.
Stable introduction of LacO at chromosome ends. (A) schematic representation of the NotI-linearized form of the pLacOTEL plasmid. (B) Southern blot after digestion of genomic DNA with HindIII and MspI and hybridization with the LacO probe. Lane 1, 182E6 cells; lane 2, LacI-expressing cells; lane 3, TRF1-LacI-expressing cells; lane 4, TRF2-LacI-expressing cells. Due to a local gel effect, the LacO bands in lanes 3 and 4 do not appear to exactly comigrate with the bands seen in lanes 1 and 2. We repeatedly noticed that these bands are identical and do not change over time (data not shown). (C) FISH of the integrated LacO sequences on a metaphase spread of the 182E6 clone with a LacO probe stained with FITC. The LacO sequence staining is indicated by open arrows. The DNA was visualized with PI in red. (D) The same metaphase shown in panel C was hybridized with probes specific for either chromosome 18 (FITC; green) or chromosome 19 (rhodamine; red) for chromosome painting. The DNA was stained with DAPI in blue. Bar = 5 μm.
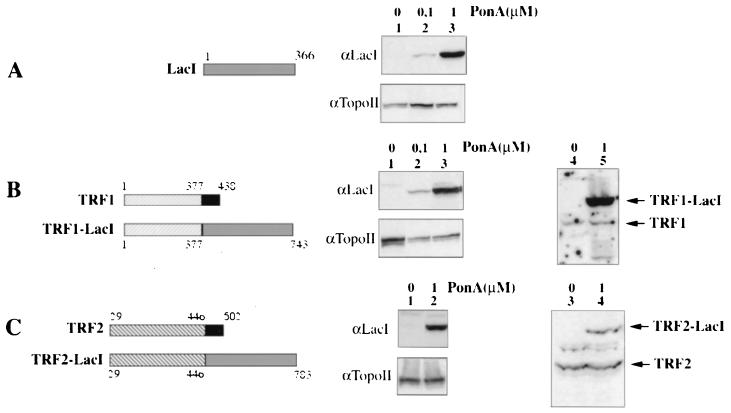
We then generated cell lines that allow the inducible expression of either the bacterial lactose repressor LacI or chimeras between LacI and TRF1 or TRF2 in which LacI replaces the telobox domain (Fig. 2). This domain is required for telomeric DNA binding by TRF1 and TRF2. After transfection of 182E6 cells with the corresponding plasmid DNAs, we selected one clone for each construction in which expression of the LacI-derived proteins was dependent on the addition of PonA. Typical results of an induction experiment, 10 MPDs after the addition of PonA, are shown in Fig. 2A and B, lanes 1 to 3, and C, lanes 1 and 2. Western blotting using antibodies directed against either TRF1 or TRF2 showed that induction of the hybrid proteins does not affect the expression of the corresponding endogenous proteins (Fig. 2B, lanes 4 to 5, and C, lanes 3 to 4). Upon induction by 0.1 μM PonA, the amounts of LacI and TRF1-LacI increased over more than 20 MPDs (data not shown). In cells treated with 1 μM PonA, LacI expression also increased with successive passages, while the amount of TRF1-LacI or TRF2-LacI progressively decreased after roughly five MPDs (data not shown). The reasons for this decline in the expression of TRF1-LacI and TRF2-LacI are unknown. It may be due to selection against cells expressing high levels of the hybrid proteins, although no gross defects in growth rate were apparent (data not shown). Alternatively, the induction by 1 μM PonA may be unable to maintain an active chromatin state of the hybrid genes. However, a reduced concentration of PonA does not lead to a decrease in TRF1-LacI expression, arguing against this possibility.
FIG. 2.
Inducible expression of LacI fusion proteins. On the left, schematic structures of LacI (A), TRF1 (B), and TRF2 (C) are presented (the telobox domain is solid), as well as fusion proteins between LacI (shaded) and the nontelobox portion of TRF1 or TRF2 (hatched): TRF1-LacI or TRF2-LacI. Western analysis of inducible expression of each LacI protein in 182E6-derived cells is presented in the middle of each panel. Nuclear extracts prepared at 10 MPDs from uninduced cells (lanes 1) or clones induced with 0.1 (A and B, lanes 2) or 1 (A and B, lanes 3; C, lane 2) μM PonA were analyzed with an anti-LacI antibody (αLacI) or an anti-topoisomerase II antibody (αTopoII). Clones expressing TRF1- or TRF2-LacI under 1 μM induction (B, lane 5, and C, lane 4) or uninduced (B, lane 4, and C, lane 3) were also analyzed using an anti-TRF1 antibody (B, right) or an anti-TRF2 antibody (C, right)
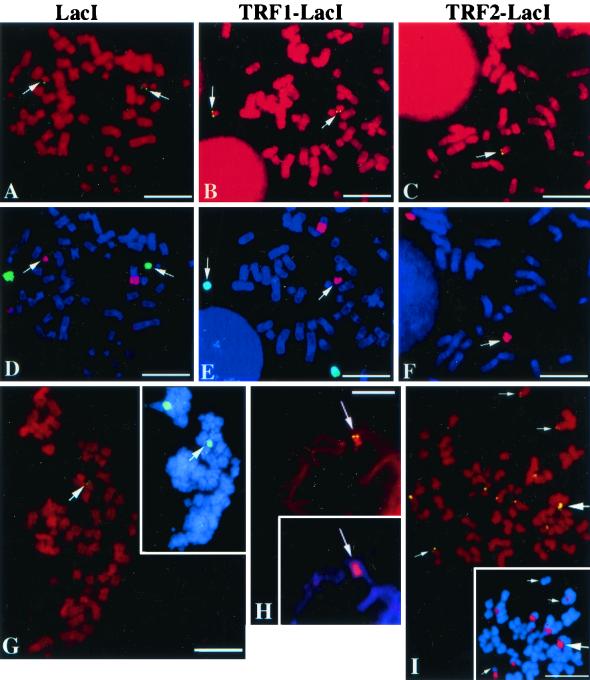
In the LacI- and TRF1-LacI-expressing clones, the two tagged telomeres are indistinguishable from those of the parental cell line, 182E6. This was confirmed both by Southern blotting (Fig. 1B, lanes 1 to 3) and by FISH analysis (compare Fig. 1C and D with Fig. 3A, B, D, and E). However, in the TRF2-LacI-expressing clone, only the LacO15 fragment (Fig. 1B, lane 4) can be detected by Southern blotting. Consistently in that clone, chromosome 19 is the only one showing a signal after FISH with a LacO probe (Fig. 3C and F). Therefore, the tagged telomere of chromosome 18 has been rearranged in the TRF2-LacI clone. Parallel cytogenetic studies showed that the LacO sequences of chromosome 18 have become interstitial in this clone, probably due to fusion with another chromosome (data not shown). One can also infer from these data that in 182E6, the LacO65 fragment corresponds to the tagged telomere of chromosome 18, while the LacO15 fragment corresponds to the tagged telomere of chromosome 19. The difference in the number of LacO repeats between the two chromosomes probably accounts for our observation that in all cells, the LacO FISH signal on chromosome 18 was recovered in most of the metaphases analyzed while the signal on chromosome 19 was usually fainter and more difficult to detect (data not shown). We conclude that the 182E6, LacI, and TRF1-LacI cells contain two tagged telomeres, one at chromosome 18 with 65 LacO sequences and one on chromosome 19 with 15 LacO sequences, and that the TRF2-LacI cells contain only the chromosome 19 LacO15 telomere.
FIG. 3.
Cytological analysis of three clones derived from 182E6. Metaphase chromosomes were analyzed for LacO sequence localization. (A to C) First, in situ hybridization using a LacO FITC probe was done. (D to F) Then chromosome painting was performed, as for Fig. 1D. The DNA was visualized with PI (top panels) or DAPI (bottom panels). Metaphase chromosomes of clones expressing LacI (G), TRF1-LacI (H), and TRF2-LacI (I) were subjected to immunofluorescence analysis with an anti-LacI antibody reacting with a FITC-conjugated secondary antibody (green). The DNA was stained with PI (red). Within the insets, the same metaphase was reanalyzed using specific chromosome 18 (FITC; green) or 19 (rhodamine; red) painting probes and counterstained with DAPI. The majority of observed doublets are at, or close to, the telomeres of chromosome 18 or 19 (large arrows); strikingly, however, in TRF1-LacI and TRF2-LacI cells, but not in LacI cells, some telomeric doublets were detected that did not colocalize with either chromosome 18 or 19 (small arrows). Bars, 5 μm.
LacI-derived proteins are bound in vivo to tagged telomeres.
We verified by a gel shift assay that in vitro-translated LacI, TRF1-LacI, and TRF2-LacI proteins bind to LacO sequences in vitro (data not shown). We next analyzed their in vivo binding to the tagged telomeres by immunofluorescence and by chromatin immunoprecipitation.
To determine which chromosomes the LacI-derived proteins were bound to, we successively performed immunofluorescence assays with LacI antibodies and chromosome painting on metaphase spreads. We considered as bona fide immunofluorescence signals only the doublets, which likely reflect the proteins bound to paired sister chromatids. In LacI-, TRF1-LacI-, and TRF2-LacI-induced cells, the majority of observed doublets are at, or close to, the telomeres of chromosome 18 or 19 (Fig. 3G, H, and I). This strongly suggests that the LacO sequences inserted in these chromosomes are in vivo binding sites for the LacI-derived proteins. In LacI and TRF1-LacI cells, we were unable to observe the telomeric doublets on both chromosome 18 and 19 on the same metaphase as expected from FISH analysis. We believe that the poor sensitivity of detection of the metaphasic chromosome-associated LacI proteins is due to the high level of condensation of metaphasic chromatin and/or to the small number of copies of the LacO sequence inserted at chromosome tips (discussed in reference 62). Strikingly, in TRF1-LacI and in TRF2-LacI cells, but not in LacI cells, some telomeric doublets were detected that did not colocalize with either chromosome 18 or 19 (Fig. 3I and data not shown). Their intensity is systematically lower than that of the chromosome 18 or 19 telomeric doublets (Fig. 3I, compare the doublets marked by the small arrows and the one marked by the large arrow). These observations suggest that the TRF1 and TRF2 moieties have the ability to lead to the association of the chimeras with nontagged telomeres but to a lesser extent and less frequently than at the tagged telomere.
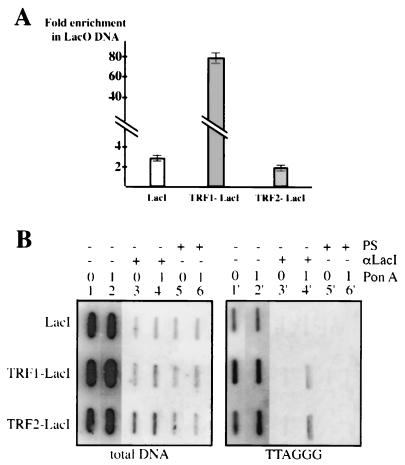
To further analyze the in vivo localization of the LacI-derived proteins, we next performed immunoprecipitation assays of formaldehyde-cross-linked chromatin. First, we estimated by quantitative PCR the amount of LacO sequence in the chromatin immunoprecipitated by LacI antibodies (see Materials and Methods) (Fig. 4A). The fraction of LacO DNA was significantly and reproductively increased upon PonA induction, by a factor of 2 or 3 for the LacI- and TRF2-LacI-expressing cells and roughly 80 times for TRF1-LacI cells. Therefore, the recovery of the LacO DNA sequences is dependent upon the dosage of the fusion proteins in the cells, confirming the immunofluorescence studies (see above). We were unable to detect any significant difference in the in vitro binding activities of the two chimeras (data not shown) or in their expression levels (Fig. 2A and B) that could account for the greater LacO DNA enrichment upon TRF1-LacI induction. Consequently, we favor the hypothesis of a better accessibility of the TRF1-LacI-containing chromatin to LacI antibodies. In agreement with this, TRF1 is less tightly bound to telomeric chromatin than TRF2 (5, 9), suggesting that the TRF1-containing chromatin is a more “open” configuration.
FIG. 4.
Analysis of LacI-derived protein localization by in vivo cross-linked chromatin immunoprecipitation. (A) Quantification of the immunoprecipitation of LacO DNA with anti-LacI antibody. Total and immunoprecipitated LacO DNA from cellular extracts of each LacI clone were quantified by quantitative PCR (LightCycler). Different amounts of LacO DNA associated with immunoprecipitated LacI, TRF1-LacI, or TRF2-LacI were normalized to the levels of total LacO DNA. Then, the fraction of LacO DNA associated with LacI proteins under induced conditions was compared to that under uninduced conditions. This ratio is reported on the y axis as the fold enrichment in LacO sequences. (B) Immunoprecipitation of human telomeric DNA with anti-LacI antibodies in cross-linked chromatin from 182E6-derived clones. DNA from input cellular extracts (lanes 1, 2, 1′, and 2′) and DNA immunoprecipitated (+) with anti-LacI antibodies (lanes 3, 4, 3′, and 4′) or preimmune serum (PS) (lanes 5, 6, 5′, and 6′) were blotted onto a membrane using a slot blot apparatus. The membrane was successively hybridized with either a total human genomic DNA probe (total DNA) or a TTAGGG probe. Lanes 1, 3, 5, 1′, 3′, and 5′, uninduced (0); lanes 2, 4, 6, 2′, 4′, and 6′), 1 μM PonA (1).
We next investigated by slot blot analysis whether the DNA immunoprecipitated by LacI antibodies was also enriched in TTAGGG DNA repeats (Fig. 4B). The binding of LacI to the centromeric side of the two LacO telomeres does not correlate with a detectable enrichment in the immunoprecipitate of the distal TTAGGG DNA repeats in this assay. By contrast, telomeric DNA was immunoprecipitated from the chromatin of induced TRF1-LacI and TRF2-LacI cells (Fig. 4B). This can be simply explained by an association of TRF1-LacI and TRF2-LacI, but not LacI, to nontagged telomeres, as revealed by immunostaining (see above). Additionally, the TRF1-LacI or TRF2-LacI molecules bound to the centromeric side of the tagged telomere could specifically interact with the distal part of this chromosome end, in a configuration reminiscent of t loops (30).
The targeting of TRF2-LacI could create a local dominant-negative effect on the endogenous TRF2. Therefore, we explored the telomere-capping functions in cells expressing TRF2-LacI. First, we scored chromosome dicentrics, with special attention to end-to-end fusions, in 170 metaphases of both uninduced and induced TRF2-LacI cells. Only one sporadic dicentric was detected in uninduced cells. We then estimated the stability of the LacO chromosome 19 by chromosome painting. This chromosome is stable in both cell lines, even after more than 20 MPDs postinduction. Since the telomeres, including the tagged one, are efficiently capped in induced cells, TRF2-LacI binding does not appear to provoke a dysfunction of the endogenous TRF2.
Overall, we conclude that upon PonA induction, the LacI-derived proteins are specifically bound to the tagged telomeres without altering telomere-capping activities. It is noteworthy that a small amount of TRF1-LacI and TRF2-LacI, but not LacI, can occasionally be associated with nontagged telomeres.
Effects of LacI-derived proteins on length distribution of the telomere population.
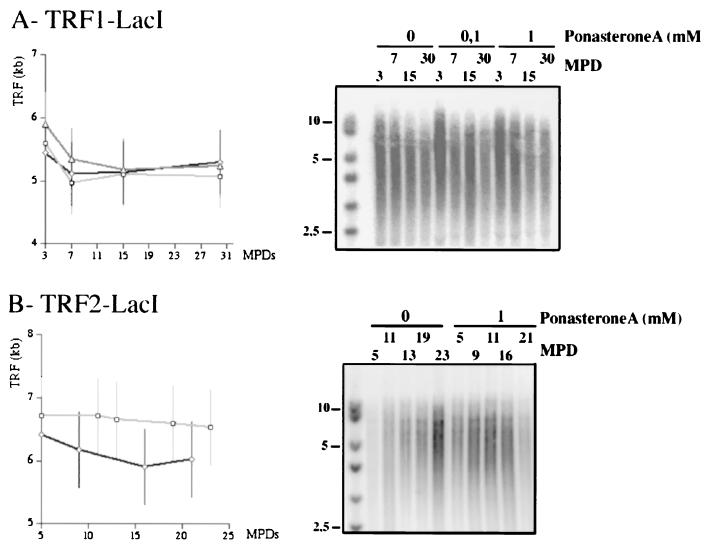
We then sought to examine the effects of the LacI-derived proteins on the lengths of the telomere population. We estimated the length corresponding to the apex of the distribution of terminal restriction fragments (TRF; see Materials and Methods). Treatment of LacI or TRF1-LacI cells with PonA did not change TRF over more than 30 MPDs (Fig. 5A and data not shown). Therefore, LacI or TRF1-LacI has no apparent trans effect on telomere length regulation. For the TRF2-LacI cells, a slight but detectable decrease in TRF can be detected after PonA treatment, with a maximal difference between uninduced and induced cells of 0.6 kb after 15 MPDs postinduction (Fig. 5B). This result suggests that the occasional incorporation of TRF2-LacI in nontagged telomeres (Fig. 4B) can be sufficient to exert a negative effect on the elongation of the whole population of telomeres.
FIG. 5.
TRF1-LacI and TRF2-LacI effects on the lengths of the total population of telomeres. Shown are the results of TRF analysis (see Materials and Methods) of TRF1-LacI-expressing cells (A) and of TRF2-LacI-expressing cells (B). Squares, uninduced; triangles, 0.1 μM PonA; diamonds, 1 μM PonA.
Expression of TRF1-LacI and TRF2-LacI leads to a specific shortening of LacO telomeres.
We next estimated the lengths of the tagged telomeres over more than 20 MPDs after induction of the expression of LacI, TRF1-LacI, or TRF2-LacI (Fig. 6). The length corresponding to the apex of the distribution of LacO-containing terminal restriction fragments (OTRF) was manually estimated from the densitometry profile (see Materials and Methods). The OTRF values are given in kilobases of (TTAGGG)n DNA, since we subtracted 2.5 (65 LacO repeats) or 0.6 (15 LacO repeats) kb from the apex length. In uninduced cells, OTRF is between 4 and 5 kb in the three cell lines. The change in size of OTRF over time is solely due to variations in the number of telomeric repeats, since the number of LacO repeats present at the telomere remained constant over the course of our experiment (Fig. 1B and data not shown).
FIG. 6.
cis repression of telomere elongation by TRF1-LacI or TRF2-LacI targeting. (A and D) LacI; (B and E) TRF1-LacI; (C and F) TRF2-LacI. (A to C) Examples of Southern blots. (D to F) Graphic representation of the variations of the OTRF during the experiments (rectangles, uninduced; triangles, 0.1 μM PonA; diamonds, 1 μM PonA). The dotted lines (F) correspond to values of the mean OTRF (see Materials and Methods).
In LacI and TRF1-LacI cells, the distribution of OTRF appears unimodal without induction (Fig. 6A and B and data not shown). This might seem surprising, given the presence of two telomeres with different numbers of LacO repeats in these cells (Fig. 1 and 3). We believe that the signal generated by the telomere containing 15 LacO repeats is masked by the signal of the other tagged telomere, which contains almost five times as many repeats. Consequently, the length profiles of LacO-containing fragments are expected to characterize mostly the chromosome 18 telomere containing 65 LacO repeats.
When the expression of TRF1-LacI or TRF2-LacI was induced with 1 μM PonA, OTRF progressively decreased (Fig. 6B and E and 6C and F, respectively). For TRF1-LacI-expressing cells, OTRF shortened from 4.3 to 2.7 kb in 20 MPDs in a linear fashion, corresponding to a shortening rate of roughly 80 nucleotides (nt) per MPD. An intermediate rate was observed upon the induction of TRF1-LacI with 0.1 μM PonA (Fig. 6B and E). The kinetics of telomere shortening in the TRF2-LacI-induced cells appeared more rapid, with an initial loss of 1.9 kb after five MPDs postinduction, corresponding to a shortening rate of 380 nt per MPD, followed by an apparent stabilization of OTRF at 3 kb (Fig. 6F). The tagged-telomere shortening rate (380 nt/MPD) is much higher than that of the telomere population (40 nt/MPD), confirming that TRF2-LacI is mainly acting at the tagged telomere (compare Fig. 6C and F to 5B). In the uninduced culture, a progressive shortening of OTRF can also be detected, albeit at a lower rate (Fig. 6F). Leaky expression of TRF2-LacI in the uninduced cells might explain this effect. Finally, the fact that the induction of the control protein LacI does not significantly affect OTRF demonstrates that the effect is specific to the TRF1 or TRF2 portion of the chimeras (Fig. 6A and D).
In the TRF2-LacI cell line, only the telomere of chromosome 19 with 15 repeats is present, explaining why the signal is less intense and corresponds to a broader distribution than with the other cell lines (compare Fig. 6A or B with C; data not shown). This can render an accurate determination of the apex value difficult. Therefore, in order to rule out any possible artifact, we also calculated the center of mass of the length distribution (mean OTRF [Fig. 6F]; see Materials and Methods). The values obtained by this method are in nice agreement with those obtained by determining the apex (Fig. 6F).
Since we observed a progressive decline in the detectable expression of TRF1-LacI, and TRF2-LacI is observed with increasing population doublings at 1 μM of PonA (see above), there is no apparent correlation between the hybrid expression level and the extent of telomere shortening. An obvious explanation is that a low dosage of the hybrids within the cells is expected to lead to a rapid occupancy of the LacO sequences due to the high-affinity binding of LacI to these sequences. Importantly, the data in Fig. 6 show that there is a clear correlation between the shortening and the induction of the TRF1 and TRF2 hybrids, showing that the effects are due to their presence in the cells.
Overall, the induction of the expression of either TRF1-LacI or TRF2-LacI leads to shortening of the tagged telomere. This effect is restricted to the tagged telomere in TRF1-LacI-induced cells and is more pronounced at the tagged telomere than at nontagged telomeres in TRF2-LacI-induced cells. Therefore, we conclude that the TRF1 and TRF2 hybrids exert a cis-acting inhibition of telomere elongation of the LacO telomere. It is noteworthy that the effect of TRF2-LacI appears to be much stronger than that of TRF1-LacI.
Evidence for distinct mechanisms of action for TRF1-LacI and TRF2-LacI.
The ability of TRF1-LacI or TRF2-LacI to shorten telomeres in cis can be explained a priori either by telomerase inhibition or by increased telomere degradation. To investigate this point, we inhibited telomerase activity in cells expressing TRF1-LacI or TRF2-LacI by following a protocol involving the transfection of a 2′-O-MeRNA oligonucleotide complementary to the template region of the hTR (OM-RNA) (32). We performed a series of six successive transfections 4 days apart either without an oligonucleotide (mock experiment) or with OM-RNA (Fig. 7). To verify the efficiency of this treatment, we assayed telomerase activity in a semiquantitative manner by a TRAPeze assay (see Materials and Methods). We found telomerase activity to be significantly reduced 3 days after the OM-RNA transfection compared to the mock experiments (data not shown). This indicates that the OM-RNA transfections were successful in inhibiting the enzyme. However, it is not known to what extent this in vitro activity reflects the level of in vivo inhibition in OM-RNA-transfected cells.
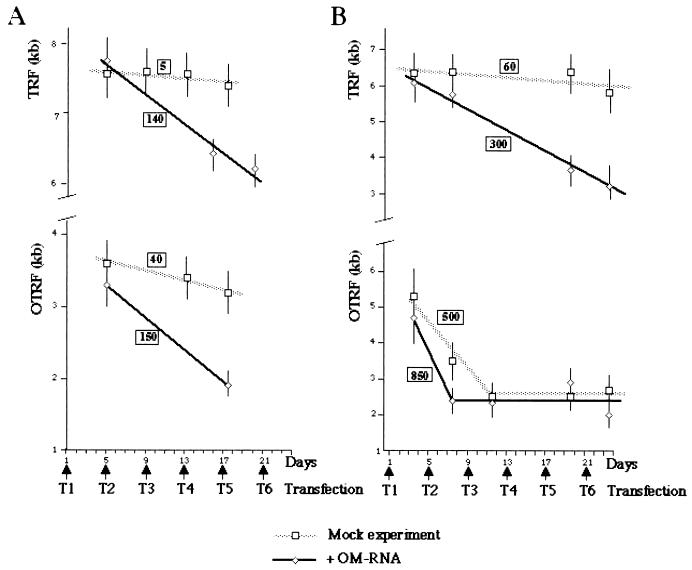
FIG. 7.
OM-RNA inhibition of telomerase uncovers different mechanisms for TRF1-LacI and TRF2-LacI effects. Graphical representation of TRF and ORTF variations in TRF1-LacI (A) and TRF2-LacI (B) cells treated with 1 μM PonA during successive transfections either with no oligonucleotide (Mock experiment) or with OM-RNA (+OM-RNA). The curves correspond to the linear regressions that were used to estimate the degradation rate of telomeric DNA (the boxed values are expressed in nucleotides/MPD).
During mock transfections, OTRF decreased both in TRF1-LacI- and in TRF2-LacI-expressing cells, while TRF remained approximately unchanged (Fig. 7A and B). The rate of OTRF shortening is much higher in cells expressing TRF2-LacI, rapidly leading to a plateau of telomere length (Fig. 7B). These data confirm the results shown in Fig. 5 and 6. Upon successive OM-RNA transfections, TRF decreases as expected from the drop in the in vitro telomerase activity (Fig. 7A and B, top, and data not shown).
No clear difference can be observed in the rate of shortening in TRF1-LacI-expressing cells between TRF and OTRF, both corresponding to roughly 140 to 150 nt/MPD (Fig. 7A). The lack of specific effect of TRF1-LacI at the tagged telomere indicates that TRF1-LacI loses its cis-acting effects when the telomerase activity is inhibited. This strongly suggests that TRF1-LacI acts in the pathway of telomerase regulation.
Upon telomerase inhibition, the rate of telomere shortening of the population of telomeres is higher in cells expressing TRF2-LacI (300 nt/MPD [Fig. 7B, top]) than in cells expressing TRF1-LacI (140 nt/MPD [Fig. 7A, top]). The difference between these two rates is significantly higher than the simple effect of TRF2-LacI on TRF when telomerase is not inhibited by OM-RNA (roughly 40 to 60 nt/MPD [Fig. 5B and 7B]). Since TRF2-LacI has been shown to interact slightly with nontagged telomeres, it seems that TRF2-LacI and telomerase inhibition have additive effects. At the tagged telomere, in contrast to TRF1-LacI, the expression of TRF2-LacI together with telomerase inhibition resulted in an accentuated shortening rate of OTRF compared to the TRF rate (Fig. 7B). This rate has been estimated at 850 nt/MPD within the first cell divisions (Fig. 7B, bottom). This is higher than the rate corresponding to the expression of TRF2-LacI in the absence of telomerase inhibition by OM-RNA (500 nt/MPD [Fig. 6C and F and Fig. 7B, bottom]). Remarkably, it corresponds to an additive effect with telomerase inhibition, as estimated for the telomere population in the same kinetics experiment (300 nt/MPD [Fig. 7B, top]). The simplest explanation is that the telomere shortening due to TRF2-LacI occurs as a consequence of an alteration of a telomere length pathway different from telomerase.
TRF2 overexpression leads to an increased rate of telomere shortening in telomerase-negative cells.
One important prediction of the above-mentioned experiments with telomerase inhibition is that TRF2 should be able to shorten telomere length in telomerase-negative cells. To test this, we determined the rate of telomere shortening in telomerase-negative primary human cells upon overexpression of the full-length TRF2.
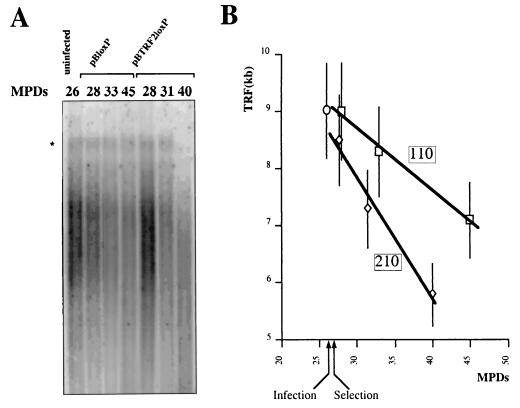
We infected normal human fibroblasts (WI38) at MPD 26 with Myc-TRF2-expressing retrovirus (pBTRF2loxP) and the parental empty vector (pBloxP). After 2 days, the cells were placed under puromycin selection to purify the polyclonal-infected population. By analyzing the lengths of the telomeres (TRF) of these cells, we noticed a band of 14 kb located above the characteristic smear of the telomeric DNA fragment (Fig. 8A). The origin of this band is unclear, but we consider that it might represent either a subpopulation of very long telomeres or telomeric fusions, as was previously detected by a similar Southern analysis of tumor cells expressing a mutant form of TRF2 (74). Cells infected with the empty vector pBloxP lose telomeric DNA at a rate of roughly 110 nt/MPD (Fig. 8), similar to the value reported previously for WI38 cells (77). However, the rate of telomere shortening is significantly higher in cells infected with pBTRF2loxp, reaching 210 nt/MPD (Fig. 8). These cells overexpressed TRF2 during the time course of the experiment, as revealed by immunoblotting of total cellular protein extracts using the c-Myc 9E10 antibody and TRF2 antibodies (data not shown). Interestingly, the high-molecular-weight band disappeared in the TRF2-overexpressing cells at late MPDs (Fig. 8A, pBTRF2loxP-infected cells at 40 MPDs), indicating that these telomere-hybridizing DNA species can be shortened by an excess of TRF2, like the bulk of telomeres. The effects of TRF2 overexpression on the life span of these cells will be described elsewhere (M. Brunori, A. Roborel de Climens, and E. Gilson, unpublished data).
FIG. 8.
TRF2 overexpression increases the rate of telomere erosion in telomerase-negative fibroblasts. We infected WI38 cells at MPD 26 with pBloxP control or TRF2-expressing pBTRF2loxP. Genomic DNA was isolated at the indicated MPDs and analyzed for TRF length. (A) Example of HinfI-RsaI Southern blot used to calculate the TRF of infected WI38 cells. The asterisk indicates the location of a high-molecular-weight band (see the text). (B) Quantitative analysis of TRF during the time course of the experiment. The curves correspond to the linear regressions that were used to estimate the shortening rate of telomeric DNA (the boxed values are expressed in nucleotides per MPD).
We conclude that an excess of full-length TRF2 can lead to telomere shortening in the absence of telomerase, which is in full agreement with the TRF2-LacI experiments showing that at least part of the shortening due to TRF2 tethering is not the consequence of telomerase inhibition.
DISCUSSION
We have presented evidence that telomere length in human cells is regulated by different mechanisms involving the cis actions of TRF1 and TRF2. In addition, we have demonstrated the feasibility of a LacI-targeting approach (62) to study the functions of human telomeric proteins.
We generated derivatives of the 293 cell line in which the expression of LacI-derived proteins could be induced by addition of PonA to the culture medium. We demonstrated a correlation between the amount of induced TRF1-LacI or TRF2-LacI, their association with distinct telomeres containing LacO repeats, and their ability to specifically decrease the lengths of such telomeres. No telomere shortening was observed in LacI-expressing cells, which proves that the TRF1 and TRF2 moieties are responsible for the effects of the chimeras. The specific targeting of amino acids 1 to 377 of TRF1 or amino acids 29 to 446 of TRF2 is sufficient to negatively regulate telomere elongation in cis.
The forms of TRF1 and TRF2 hybrid proteins that we used in this study lack the C-terminal telobox sequence required for specific telomeric DNA binding (5, 6, 9, 14, 38, 76). It was previously reported that similar, but not identical, truncated forms of TRF1 or TRF2 lacking the telobox exhibit negative trans-dominant effects, including telomere lengthening and telomere fusion (74, 75). These TRF1 and TRF2 mutant proteins fail to bind DNA and act as trans-dominant inhibitors, probably because they can still dimerize with the endogenous full-length proteins. The situation appears quite different with our LacI hybrids. First, we observed a faint but significant binding of TRF1 or TRF2 to telomeres by immunofluorescence (Fig. 3). This LacO-independent binding is far less efficient than the specific targeting to the LacO tagged telomeres and is more pronounced for TRF2 than for TRF1 hybrids (Fig. 3). It is not observed for LacI alone, indicating that the nontelobox part of TRF1 and TRF2 has the ability to target the LacI hybrids to telomeres. This is in contrast to the absence of telomeric localization of the trans-dominant forms previously described (74, 75). The association of our hybrids with telomeres could reflect their ability to dimerize with the endogenous proteins or with other components of the telomeric chromatin. A difference from the previously published truncated forms of TRF1 and TRF2 could be that the telomeric association is stabilized by the DNA-binding activities of the LacI portion of the chimeras. Second, our hybrids do not exhibit the hallmarks of the previously described negative trans-dominant effects of TRF1 and TRF2. Indeed, their expression did not lead to telomere lengthening or to chromosome instability, as revealed by an unchanged rate of dicentric formation involving the tagged telomere upon TRF2-LacI expression.
Upon induction of the TRF1-LacI protein, no apparent shortening could be detected for the total population of telomeres (Fig. 5A and 7A), while a significant decrease in length occurred for the tagged telomere, at a rate of roughly 50 to 70 nt per MPD (Fig. 6E and 7A). This clearly shows that TRF1-LacI exerts a cis repression on telomere elongation at the tagged telomere. The induction of the expression of TRF2-LacI protein induces a shortening of about 50 to 70 nt per MPD of the total population of telomeres compared to noninduced cells (Fig. 5B and 7B) and a dramatic decrease of the tagged-telomere length, initially at a rate of roughly 500 nt per MPD before the length is maintained at a plateau of 3 kb over time (Fig. 6F and 7B). Importantly, in the induced cells, one can observe an occasional faint association of TRF2-LacI with the population of telomeres and an important tethering of the hybrids to the LacO telomere (Fig. 3I). The fact that the number of telomere-associated hybrids nicely correlates with the extent of telomeric DNA shortening shows a specific effect of TRF2-LacI at associated versus unassociated telomeres. This strongly argues that the TRF2-LacI hybrid specifically represses telomeric DNA elongation of the telomeres with which it is associated, i.e., in cis.
It has been reported that an overexpression of full-length TRF1 and TRF2 leads to a progressive shortening of telomere length in telomerase-positive cells (69, 75). Our results are consistent with these findings and further indicate that telomere shortening is a direct consequence of an increased amount of TRF1 and TRF2 at chromosome ends and not of trans effects promoted by the overexpressed proteins. Overall, we conclude that the TRF1 and TRF2 chimeras analyzed here mimic part of the normal activity of their native (full-length) counterparts with respect to telomere length control.
It was previously reported that in yeast, mammalian cells, and Trypanosoma, an inverse relation between telomere elongation and telomere length exists, providing a negative-feedback loop that maintains telomeres at a defined mean length (19, 33, 49, 51, 61, 70). If this regulation involves the binding of TRF1 and TRF2, we expect that the tethering of a constant amount of these proteins would lead to a new setting of the telomeric DNA length specifically at the tagged telomere. Indeed, upon TRF2-LacI expression, the lengths of the telomere population are only moderately reduced (Fig. 5B) while the length of the tagged telomere is rapidly scaled down to a lower limit of about 2.5 to 3 kb of TTAGGG repeats (Fig. 6C and F and 7B, bottom). We propose that this plateau reflects a new length setting, where degradation and elongation are balanced, for the following two reasons. First, the plateau does not seem to be due to selection against cells that have a critically short tagged telomere, because we were unable to detect, at least over 25 MPDs postinduction, any increased chromosome instability or alteration in growth rate (data not shown). Second, since the barely detectable amount of TRF2-LacI present in uninduced cells seems sufficient to cause a significant erosion of the tagged telomere (Fig. 6F), we do not think that the cessation of shortening is due to the reduction in the amount of TRF2-LacI observed upon long-term growth of the induced cells. For instance, the absence of further shortening of the tagged telomere starts at 10 MPDs (Fig. 6F), i.e., at the time when TRF2-LacI is still expressed much more than in uninduced cells (Fig. 2C). It is noteworthy that after 7.5 MPDs, the additive effects of TRF2-LacI induction and telomerase inhibition by OM-RNA transfection also lead to an apparent stabilization of telomere length, but at a value slightly lower than that of the plateau reached after TRF2-LacI induction alone (Fig. 7B, bottom). This new equilibrium likely depends upon the residual telomerase activity present in OM-RNA-transfected cells.
The new equilibrium upon TRF2-LacI expression was reached after a loss of 2 kb of telomeric DNA. Assuming that the LacI binding sites are fully occupied, 15 sites are vacant for the binding of LacI dimers, and therefore 30 molecules of TRF2-LacI are expected to be present at telomere 19. If we hypothesize that the tethering of 30 TRF2-LacI molecules on the centromeric side of the tagged telomere results in an equivalent loss of TRF2 molecules on the telomeric-DNA side, we can expect that 2 kb of telomeric DNA is bound to 30 TRF2 molecules. This is far below the maximal binding capacity for TRF2 on telomeric DNA, taking 10 nt as a binding site for one TRF2 monomer (4). This is also below the expected amount of TRF2 present per kilobase of (TTAGGG)n (78 molecules/kb in 293 cells) (56). In fact, it is highly likely that the telomeric DNA is not completely covered by TRF2 because of the competition with other telomeric-DNA-binding factors and because TRF1 and TRF2 appear to be present only in limiting amounts at human telomeres (69, 75).
The stabilization of the length of the tagged telomere was not observed in the time course of the TRF1-LacI experiment. It is likely that more population doublings are required to reach a new length equilibrium, because TRF1-LacI shortens telomeres at a lower rate than TRF2-LacI. In agreement with this prediction, the maintenance of short telomeres was previously observed upon long-term overexpression of TRF1 (69).
Transfections of an antisense telomerase oligonucleotide lead to telomere shortening, and this occurs at a rate that is not further increased by targeting of TRF1-LacI (Fig. 7A). This strongly argues that the main effect of TRF1-LacI is to repress telomerase activity in cis, explaining why this effect is no longer observable under conditions where telomerase is inhibited by other means. We do not know at present whether this inhibitory effect is mediated by TRF1 itself or by associated proteins. The latter hypothesis should be seriously considered in light of the capacity of the TRF1-interacting factors TIN2 and PinX1 to inhibit telomerase-mediated telomere elongation (36, 81). A tempting model suggested by the capacity of PinX1 to interact with the catalytic subunit of telomerase (81) is that PinX1 serves as a bridge between TRF1 and telomerase, leading to the inactivation of the catalytic activity. This would explain how an excess of TRF1 molecules artificially tethered on the centromeric side of a telomere can exert a repressive effect on the distally located telomerase molecules.
By contrast, targeting TRF2-LacI (Fig. 7B) can exacerbate the rate of telomere shortening attained by telomerase inhibition. This indicates that the cis-acting effects of TRF2-LacI involve, at least in part, a pathway different from simple telomerase repression. This prediction was directly confirmed in human primary cells that do not express telomerase: overexpression of full-length TRF2 in WI38 fibroblasts causes an increase in the rate of telomere shortening (Fig. 8). In addition, this result argues that the behavior of the targeted TRF2-LacI mimics at least part of the activities of the normal TRF2. Since EcR-293 cells are p53 inactive and WI38 cells are wild type for p53, the telomerase-independent telomere shortening induced by TRF2 appears to be p53 independent. An implication of these data is that TRF2 activates a telomeric DNA degradation pathway. Interestingly, the expression of a trans-negative form of TRF2 is associated with a significantly decreased amount of the G-rich single-stranded part of telomeric DNA detected, which likely reflects either a trimmed or a masked G tail (74). Therefore, TRF2 may be directly involved in the processing of chromosomal DNA ends. For example, the targeting to telomeres of the RAD50-MRE11 complex via TRF2 (82) can be involved in specific telomeric DNA degradation events by virtue of the nuclease activities of the complex (73). Alternatively, TRF2-mediated t loops (30) could serve as intermediates for an intrachromatid deletion event. A related excision process has already been demonstrated in yeast, leading to a rapid shortening of overelongated telomeres (45). Interestingly, the intrachromatid deletion event in yeast is dependent upon the Rad50-Mre11 complex (10). Therefore, it is tempting to speculate that the dramatic rate of telomere shortening induced by an excess of TRF2 (Fig. 6F and 7B) reflects an increased number of intrachromatid deletion events mediated by t-loop formation and Rad50-Mre11 telomeric targeting.
In conclusion, TRF1 and TRF2 appear to regulate telomeric length by different mechanisms, TRF1 as a cis inhibitor of telomerase activity and TRF2 as an activator of telomere degradation. Importantly, our results indicate that TRF2 can contribute to telomere length regulation in telomerase-negative cells. The mechanisms that control the rate of telomere shortening in human cells are still unknown (53), and interestingly enough, they appear to be developmentally regulated (24). The studies presented here raise the possibility that variations in TRF2 expression and/or activity modulate the rate of telomere shortening both in normal cells and in cells with alterations in telomere length dynamics, as in progeroid syndromes (37) and cancer (1).
Acknowledgments
We thank A. Belmont, T. de Lange, A. Pollice, J. Pulitzer, J. Shay, and M. P. Zibella for plasmids. We are grateful to P. A. Defossez and S. Marcand for helpful comments on the manuscript.
The work in the Gilson laboratory is supported by La Ligue Nationale contre le Cancer and the Région Rhône-Alps (contract 00816045). The work in the Sabatier laboratory is supported by CEC funding (FIGH-CT-1999-0009). K.A. is a Ph.D. fellow from La Ligue National contre le Cancer, M.B. is supported by the CEC (HPRN-CT-2000-00089), and S.B. is supported by a postdoctoral fellowship from the Centre Léon Bérard (Lyon, France).
REFERENCES
- 1.Artandi, S. E., and R. A. DePinho. 2000. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr. Opin. Genet. Dev. 10:39-46. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. [DOI] [PubMed] [Google Scholar]
- 3.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 1998. Reconstitution of human telomerase activity in vitro. Curr. Biol. 8:177-180. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi, A., R. M. Stansel, L. Fairall, J. D. Griffith, D. Rhodes, and T. de Lange. 1999. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 18:5735-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilaud, T., C. Brun, K. Ancelin, C. E. Koering, T. Laroche, and E. Gilson. 1997. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17:236-239. [DOI] [PubMed] [Google Scholar]
- 6.Bilaud, T., C. E. Koering, E. Binet-Brasselet, K. Ancelin, A. Pollice, S. M. Gasser, and E. Gilson. 1996. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 24:1294-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackburn, E. H. 2000. The end of the (DNA) line. Nat. Struct. Biol. 7:847-850. [DOI] [PubMed] [Google Scholar]
- 8.Blackburn, E. H. 2000. Telomere states and cell fates. Nature 408:53-56. [DOI] [PubMed] [Google Scholar]
- 9.Broccoli, D., A. Smogorzewska, L. Chong, and T. de Lange. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17:231-235. [DOI] [PubMed] [Google Scholar]
- 10.Bucholc, M., Y. Park, and A. J. Lustig. 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson, D. L., R. Skopp, and C. M. Price. 1997. DNA-binding properties of the replication telomere protein. Biochemistry 36:15900-15908. [DOI] [PubMed] [Google Scholar]
- 12.Carson, M. J., and L. Hartwell. 1985. CDC17: an essential gene that prevents telomere elongation in yeast. Cell 42:249-257. [DOI] [PubMed] [Google Scholar]
- 13.Chandra, A., T. R. Hughes, C. I. Nugent, and V. Lundblad. 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chong, L., B. van Steensel, D. Broccoli, H. Erdjument-Bromage, J. Hanish, P. Tempst, and T. de Lange. 1995. A human telomeric protein. Science 270:1663-1667. [DOI] [PubMed] [Google Scholar]
- 15.Cooper, J. P., E. R. Nimmo, R. C. Allshire, and T. R. Cech. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385:744-747. [DOI] [PubMed] [Google Scholar]
- 16.Counter, C. M., A. A. Avilion, C. E. Le Feuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diede, S. J., and D. E. Gottschling. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99:723-733. [DOI] [PubMed] [Google Scholar]
- 18.Dionne, I., and R. J. Wellinger. 1998. Processing of telomeric DNA ends requires the passage of a replication fork. Nucleic Acids Res. 26:5365-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ducray, C., J. P. Pommier, L. Martins, F. D. Boussin, and L. Sabatier. 1999. Telomere dynamics, end-to-end fusions and telomerase activation during the human fibroblast immortalization process. Oncogene 18:4211-4223. [DOI] [PubMed] [Google Scholar]
- 20.Dutrillaux, B., and J. Lejeune. 1975. New techniques in the study of human chromosomes: methods and applications. Adv. Hum. Genet. 5:119-156. [DOI] [PubMed] [Google Scholar]
- 21.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 22.Fan, X., and C. M. Price. 1997. Coordinate regulation of G- and C strand length during new telomere synthesis. Mol. Biol. Cell 8:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farr, C., J. Fantes, P. Goodfellow, and H. Cooke. 1991. Functional reintroduction of human telomeres into mammalian cells. Proc. Natl. Acad. Sci. USA 88:7006-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frenck, R. W., Jr., E. H. Blackburn, and K. M. Shannon. 1998. The rate of telomere sequence loss in human leukocytes varies with age. Proc. Natl. Acad. Sci. USA 95:5607-5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasser, S. M. 2000. A sense of the end. Science 288:1377-1379. [DOI] [PubMed] [Google Scholar]
- 26.Gottschling, D. E., and V. A. Zakian. 1986. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell 47:195-205. [DOI] [PubMed] [Google Scholar]
- 27.Grandin, N., C. Damon, and M. Charbonneau. 2000. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol. 20:8397-8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greider, C. W., and E. H. Blakburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 29.Greider, C. W. 1996. Telomere length regulation. Annu. Rev. Biochem. 65:337-365. [DOI] [PubMed] [Google Scholar]
- 30.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 31.Hanish, J. P., J. L. Yanowitz, and T. de Lange. 1994. Stringent sequence requirements for the formation of human telomeres. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 32.Herbert, B., A. E. Pitts, S. I. Baker, S. E. Hamilton, W. E. Wright, J. W. Shay, and D. R. Corey. 1999. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl. Acad. Sci. USA 96:14276-14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horn, D., C. Spence, and A. K. Ingram. 2000. Telomere maintenance and length regulation in Trypanosoma brucei. EMBO J. 19:2332-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu, H. L., D. Gilley, E. H. Blackburn, and D. J. Chen. 1999. Ku is associated with the telomere in mammals. Proc. Natl. Acad. Sci. USA 96:12454-12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes, T. R., S. K. Evans, R. G. Weilbaecher, and V. Lundblad. 2000. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 10:809-812. [DOI] [PubMed] [Google Scholar]
- 36.Kim, S. H., P. Kaminker, and J. Campisi. 1999. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23:405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kipling, D., and R. G. Faragher. 1997. Progeroid syndromes: probing the molecular basis of aging? Mol. Pathol. 50:234-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konig, P., L. Fairall, and D. Rhodes. 1998. Sequence-specific DNA recognition by the myb-like domain of the human telomere binding protein TRF1: a model for the protein-DNA complex. Nucleic Acids Res. 26:1731-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krauskopf, A., and E. H. Blackburn. 1996. Control of telomere growth by interaction of RAP1 with the most distal telomeric repeats. Nature 383:354-357. [DOI] [PubMed] [Google Scholar]
- 40.Kyrion, G., K. A. Boakye, and A. J. Lustig. 1992. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:5159-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaBranche, H., S. Dupuis, Y. Ben-David, M. R. Bani, R. J. Wellinger, and B. Chabot. 1998. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 19:199-202. [DOI] [PubMed] [Google Scholar]
- 42.Larson, D. D., E. A. Spangler, and E. H. Blackburn. 1987. Dynamics of telomere length variation in Tetrahymena thermophila. Cell 50:477-483. [DOI] [PubMed] [Google Scholar]
- 43.Lemieux, N., B. Malfoy, R. Fetni, M. Muleris, N. Vogt, C. L. Richer, and B. Dutrillaux. 1994. In situ hybridization approach at infragenic level on metaphase chromosomes. Cytogenet. Cell Genet. 66:107-112. [DOI] [PubMed] [Google Scholar]
- 44.Li, B., S. Oestreich, and T. de Lange. 2000. Identification of human Rap1: implications for telomere evolution. Cell 101:471-483. [DOI] [PubMed] [Google Scholar]
- 45.Li, B., and A. J. Lustig. 1996. A novel mechanism for telomere size control in S. cerevisiae. Genes Dev. 10:1310-1326. [DOI] [PubMed] [Google Scholar]
- 46.Lingner, J., J. P. Cooper, and T. R. Cech. 1995. Telomerase and DNA end replication: no longer a lagging strand problem? Science 269:1533-1534. [DOI] [PubMed] [Google Scholar]
- 47.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lunblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 48.Lustig, A. J. C., C. Liu, C. Zhang, and J. P. Hanish. 1996. Tethered Sir3p nucleate silencing at telomeres and internal loci in S. cerevisiae. Mol. Cell. Biol. 16:2483-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marcand, S., V. Brevet, and E. Gilson. 1999. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marcand, S., V. Brevet, C. Mann, and E. Gilson. 2000. Cell cycle restriction of telomere elongation. Curr. Biol. 10:487-490. [DOI] [PubMed] [Google Scholar]
- 51.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986-990. [DOI] [PubMed] [Google Scholar]
- 52.McClintock, B. 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26:234-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331-358. [DOI] [PubMed] [Google Scholar]
- 54.Müller, H. J. 1938. The remaking of chromosomes. Collect. Net 13:182-198. [Google Scholar]
- 55.Nugent, C. I., T. R. Hughes, N. F. Lue, and V. Lundblad. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249-252. [DOI] [PubMed] [Google Scholar]
- 56.Okabe, J., A. Eguchi, A. Masago, T. Hayakawa, and M. Nakanishi. 2000. TRF1 is a critical trans-acting factor required for de novo telomere formation in human cells. Hum. Mol. Genet. 9:2639-2650. [DOI] [PubMed] [Google Scholar]
- 57.Olovnikov, A. M. 1971. Principles of marginotomy in template synthesis of polynucleotides. Dokl. Akad. Nauk SSSR 201:1496-1499. [PubMed] [Google Scholar]
- 58.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 59.Pommier, J. P., L. Gauthier, J. Livartowski, P. Galanaud, F. Boue, A. Dulioust, D. Marce, C. Ducray, L. Sabatier, J. Lebeau, and F. D. Boussin. 1997. Immunosenescence in HIV pathogenesis. Virology 231:148-154. [DOI] [PubMed] [Google Scholar]
- 60.Qi, H., and V. A. Zakian. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 14:1777-1788. [PMC free article] [PubMed] [Google Scholar]
- 61.Ray, A., and K. W. Runge. 1999. The yeast telomere length counting machinery is sensitive to sequences at the telomere-nontelomere junction. Mol. Cell. Biol 19:31-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinett, C. C., A. Straight, G. Li, C. Willelm, G. Sudlow, A. Murray, and A. S. Belmont. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Samper, E., F. A. Goytisolo, P. Slijepcevic, P. P. van Buul, and M. A. Blasco. 2000. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1:244-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sandell, L. L., and V. A. Zakian. 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75:729-739. [DOI] [PubMed] [Google Scholar]
- 65.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shore, D. 2001. Telomeric chromatin: replicating and wrapping up chromosome ends. Curr. Opin. Genet. Dev. 11:189-198. [DOI] [PubMed] [Google Scholar]
- 67.Smith, S., and T. de Lange. 2000. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10:1299-1302. [DOI] [PubMed] [Google Scholar]
- 68.Smith, S., I. Giriat, A. Schmitt, and T. de Lange. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282:1484-1487. [DOI] [PubMed] [Google Scholar]
- 69.Smogorzewska, A., B. van Steensel, A. Bianchi, S. Oelmann, M. R. Schaefer, G. Schnapp, and T. de Lange. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol 20:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sprung, C. N., G. E. Reynolds, M. Jasin, and J. P. Murnane. 1999. Chromosome healing in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 96:6781-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steinert, S., J. W. Shay, and W. E. Wright. 2000. Transient expression of human telomerase extends the life span of normal human fibroblasts. Biochem. Biophys. Res. Commun. 273:1095-1098. [DOI] [PubMed] [Google Scholar]
- 72.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 73.Trujillo, K. M., S. S. Yuan, E. Y. Lee, and P. Sung. 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 273:21447-21450. [DOI] [PubMed] [Google Scholar]
- 74.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 75.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 76.Vassetzky, N. S., F. Gaden, C. Brun, S. M. Gasser, and E. Gilson. 1999. Taz1p and Teb1p, two telobox proteins in Schizosaccharomyces pombe, recognize different telomere-related DNA sequences. Nucleic Acids Res. 27:4687-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.von Zglinicki, T., G. Saretzki, W. Docke, and C. Lotze. 1995. Mild hyperoxia shortens telomeres and inhibits proliferation of fibroblasts: a model for senescence? Exp. Cell Res. 220:186-193. [DOI] [PubMed] [Google Scholar]
- 78.Watson, J. D. 1972. Origin of concatemeric DNA. Nat. New Biol. 239:197-201. [DOI] [PubMed] [Google Scholar]
- 79.Weinrich, S. L., R. Pruzan, L. Ma, M. Ouelette, V. M. Tesmer, S. E. Holt, A. G. Bodnar, S. Lichtsteiner, N. W. Kim, J. B. Trager, R. D. Taylor, R. Carlos, W. H. Andrews, W. E. Wright, J. W. Shay, C. B. Harley, and G. B. Morin. 1997. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17:498-502. [DOI] [PubMed] [Google Scholar]
- 80.Wotton, D., and D. Shore. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in S. cerevisiae. Genes Dev. 11:748-760. [DOI] [PubMed] [Google Scholar]
- 81.Zhou, X. Z., and K. P. Lu. 2001. The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 107:347-359. [DOI] [PubMed] [Google Scholar]
- 82.Zhu, X. D., B. Kuster, M. Mann, J. H. Petrini, and T. Lange. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25:347-352. [DOI] [PubMed] [Google Scholar]