Abstract
The estrogen receptor (ER), a member of the nuclear hormone receptor superfamily important in human physiology and disease, recruits coactivators which modify local chromatin structure. Here we describe effects of ER on large-scale chromatin structure as visualized in live cells. We targeted ER to gene-amplified chromosome arms containing large numbers of lac operator sites either directly, through a lac repressor-ER fusion protein (lac rep-ER), or indirectly, by fusing lac repressor with the ER interaction domain of the coactivator steroid receptor coactivator 1. Significant decondensation of large-scale chromatin structure, comparable to that produced by the ∼150-fold-stronger viral protein 16 (VP16) transcriptional activator, was produced by ER in the absence of estradiol using both approaches. Addition of estradiol induced a partial reversal of this unfolding by green fluorescent protein-lac rep-ER but not by wild-type ER recruited by a lac repressor-SRC570-780 fusion protein. The chromatin decondensation activity did not require transcriptional activation by ER nor did it require ligand-induced coactivator interactions, and unfolding did not correlate with histone hyperacetylation. Ligand-induced coactivator interactions with helix 12 of ER were necessary for the partial refolding of chromatin in response to estradiol using the lac rep-ER tethering system. This work demonstrates that when tethered or recruited to DNA, ER possesses a novel large-scale chromatin unfolding activity.
Nuclear hormone receptors are ligand-inducible transcriptional activators that play a large number of physiological roles (37). These proteins activate transcription through a multistage process involving binding to DNA response elements (26), recruitment of coactivators (18, 25), remodeling of chromatin (11, 32), and assembly of general transcription factors and RNA polymerase II (42). Gene expression, the last step of the process, is typically used as a readout of transcription factor function. Few experimental approaches allow dissection of the chromatin-related activities of a transcriptional activator. In addition, conclusions about how steroid hormone receptors and their associated coregulators function have often been based on studies of transiently expressed reporter genes which lack native chromatin structure.
It has become clear in recent years that the ability of steroid hormone receptors to activate transcription of endogenous genes likely depends upon their ability to affect chromatin structure. Indeed, many steroid hormone receptors interact with coregulator proteins that are implicated in the remodeling of local chromatin structure and the acetylation of histones (reviewed in references 32, 33, and 41). In fact, enhancement of transcription by adding ligand to estrogen receptor (ER) was observed using chromatinized template DNA but not when using naked DNA lacking histones (27). In addition, genes integrated into chromosomes have been shown to be regulated differently from genes located in transiently transfected plasmid DNA (17), often revealing surprising dynamics and regulation (7, 24, 45). This suggests that understanding transcription in the context of native chromosomes will be important for understanding steroid hormone receptor function.
While recent work has revealed changes induced by nuclear receptors at the level of local chromatin structure (i.e., at the level of histones and nucleosomes), it is unclear to what extent steroid hormone receptors might also affect higher levels of chromatin folding. These levels include higher-order chromatin structure (folding which produces the 30-nm chromatin fiber) and large-scale chromatin structure (folding above the level of the 30-nm fiber).
A lac repressor-based system has allowed direct visualization of large-scale chromatin dynamics in mammalian cells (4). By fusing protein domains to the lac repressor, this system was used to demonstrate large-scale chromatin decondensation induced by targeting the acidic activation domain (AAD) of the strong viral transcription factor VP16 to a heterochromatic chromosome arm generated by gene amplification (51). This large-scale chromatin uncoiling occurred even when polymerase II transcription was blocked, suggesting that it was induced through trans factors recruited by the VP16 AAD rather than being the result of transcription per se. While artificial, this lac operator tethering system provides a powerful assay to test the role of specific proteins in chromatin remodeling and to dissect the protein domains required for the observed large-scale chromatin decondensation. Recently this system was used to demonstrate comparable large-scale chromatin decondensation produced by the BRCT domains of BRCA1; moreover, several cancer-predisposing mutations of BRCA1 were shown to increase dramatically the large-scale chromatin decondensation potential of full-length BRCA1 (55).
Here we have applied this system to determine whether the ER could induce comparable changes in large-scale chromatin structure. We were interested in whether the chromatin effects of transcriptional activators would be proportional to their strength and whether different transcriptional activators could unfold chromatin to different levels. Also, would the chromatin unfolding activity of an activator be tightly linked to or clearly separable from transcriptional activation potential? Based on this tethering assay system, our results show a very pronounced chromatin unfolding activity associated with ER which was not dependent on active transcription. Intriguingly, this activity was strong even in the absence of the ligand and was independent of the helix 12 region of the receptor, which mediates ligand-dependent recruitment of several known coactivators. Our results suggest the possibility of novel physiological activities associated with ER, and our approach may provide a new methodology to identify novel proteins involved in ER function.
MATERIALS AND METHODS
Construction of plasmids.
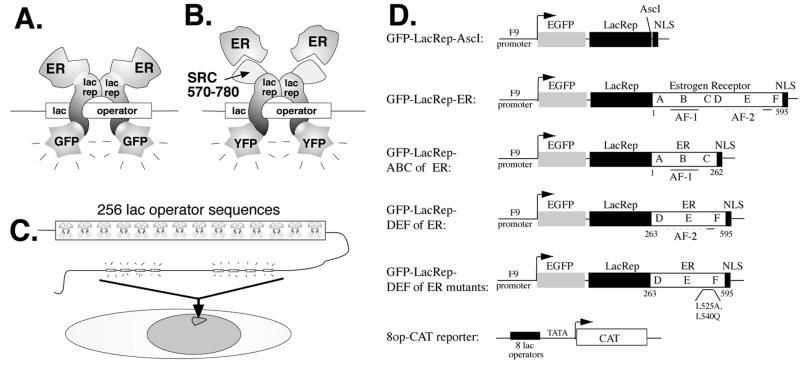
A plasmid expressing the green fluorescent protein (GFP)-dimer lac repressor-simian virus 40 nuclear localization signal (NLS) fusion protein under control of the F9-1 promoter, called p3′SS-EGFP-dimer lac repressor (51), was used as the basis for these studies. This plasmid contains a mutant form of the lac repressor which forms dimers rather than tetramers due to a five-amino-acid deletion at the C terminus (8). The N terminus of the lac repressor was replaced with a proline-3→tyrosine mutant form from pAFS135 (43, 49) to produce a tightly binding lac repressor. This plasmid, p3′SS EGFP-dimer tb lac repressor, was then modified by Quik-change site-directed mutagenesis (Stratagene, La Jolla, Calif.), resulting in the insertion of a 9-bp fragment containing an AscI site into the PvuII site 7 amino acids N-terminal to the linker and NLS. This generated GFP-lac rep-AscI-NLS (NYE4). Selected regions of the ER were amplified by PCR using primers which contain AscI sites in frame. The template for these reactions was either wild-type human ER-alpha from the CMV-ER plasmid (54) or mutant ERs (L525A or L540Q) in the same vector backbone (15, 22, 44). The PCR products were digested with AscI and ligated into the AscI-digested GFP-lac rep-AscI-NLS vector to create in-frame fusions. All regions of constructs which had undergone PCR were sequenced to ensure fidelity. Construction of yellow fluorescent protein (YFP)-steroid receptor coactivator 1 (SRC-1) and cyan fluorescent protein (CFP)-ER has been described (47). The CFP-lac repressor-ER fusion is described elsewhere (48). YFP-lac rep-SRC570-780 was constructed by PCR amplifying lac rep-NLS from NYE4 with primers adding BglII sites and ligation into the BglII site between YFP and SRC570-780 of the YFP-SRC570-780 vector (47).
The 8op-CAT reporter plasmid (NYE10) was generated by removing a BamHI-HindIII fragment containing 8 lac operator repeats from pPS-8.1 (39). This fragment was ligated into the pATC2 vector (31), whose two estrogen response elements (EREs) had been removed with a BglII/HindIII digestion.
Tissue culture, transfection, and CAT assays.
A03_1 CHO DG44 cells contain a gene-amplified chromosome region containing ∼400-kb blocks of pSV2-DHFR-8.32 vector repeats separated by an estimated 1,000 kb of flanking, coamplified genomic DNA (28). Each vector copy contains the dihydrofolate reductase cDNA transgene and 256 direct repeats of the lac operator. These cells were cultured at 37°C with 5% CO2 in F-12 Ham's media without hypoxanthine or thymidine, with 0.3 μM methotrexate, without phenol red, and with 10% dialyzed fetal bovine serum (HyClone Labs, Logan, Utah) treated with charcoal-dextran. Phenol-red free trypsin was used to passage cells. RRE_B1 cells contain a different gene-amplified chromosome region with lac operator repeats derived from a modified vector (50) and were cultured using the same medium as for A03_1 except with 10 μM methotrexate. Wild-type Chinese hamster ovary CHO-K1 cells (ATCC CRL 9618) were cultured in phenol-red free F-12 Ham's medium with 10% charcoal-dextran-treated fetal bovine serum.
Transfections on coverslips were performed with FuGENE6 reagent (Roche, Indianapolis, Ind.) according to the manufacturer's instructions using 250 ng of DNA and 3 μl of reagent per 35-mm-diameter plate. Fresh medium containing hormone was added 24 h after transfection. Seventy-two hours after transfection, cells were rinsed in calcium-, magnesium-free phosphate-buffered saline (CMF-PBS), fixed in CMF-PBS with 1.6% paraformaldehyde (Polysciences, Warrington, Pa.), and stained with 0.2 μg of DAPI (4′,6′-diamidino-2-phenylindole, dihydrochloride)/ml in CMF-PBS. Transfections for CAT transcription assays used 1.6 μg of 8 op-CAT or pATC4 (4 ERE-TATA-CAT) reporter (31), 0.6 μg of the beta-galactosidase internal reference reporter pCH110 (Pharmacia), and 10 ng of effector plasmid combined with 15 μl of FuGENE6 reagent per 60-mm-diameter plate. Fresh media containing hormone was added 24 h posttransfection, and cells were harvested and lysed 48 h posttransfection. CAT assays were performed and normalized for beta-galactosidase expression as described previously (44).
Immunocytochemistry.
Cells were stained in PBS* solutions (PBS-5 mM MgCl2-0.1 mM EDTA). Cells were rinsed in PBS*, permeabilized for 30 (histone acetylation staining) or 60 s (brahma and BAF170 staining) in 0.1% Triton X-100, and fixed in 1.6% formaldehyde (Polysciences) for 15 to 30 min at room temperature. Coverslips were washed for 5 min in 0.1% Triton X-100 (brahma and BAF170 staining only), three times (5 min each) in PBS*, three times (5 min each) in 20 mM glycine, 5 min in 0.1% Triton X-100 (brahma and BAF170 staining only), and 30 min at room temperature in 5% normal goat serum (histone acetylation staining) or 1 h at room temperature plus 1 h at 4°C in 5% normal donkey serum (brahma and BAF170 staining). Coverslips were washed three times (5 min each) in 0.1% Triton X-100 and incubated overnight at 4°C with a primary antibody diluted in 0.1% Triton X-100 as follows: 1:500 antiacetylated histone H3 (AHP412; Serotec, Raleigh, N.C.), 1:500 antiacetylated histone H4 (AHP418; Serotec), 1:50 anti-brahma N-19 (sc-6450; Santa Cruz Biotechnology, Santa Cruz, Calif.), 1:100 anti-BAF170 C-19 (sc-9744; Santa Cruz Biotechnology). Coverslips were washed three times (5 min each) in 0.1% Triton X-100 and then incubated at 4°C with a secondary antibody diluted in 0.1% Triton X-100 as follows: 6 h at 4°C with a 1:1,000 dilution of Texas Red goat anti-rabbit immunoglobulin G (code 111-075-144; Jackson Immunoresearch, West Grove, Pa.) for histone acetylation staining or 19 h at 4°C with a 1:3,000 dilution of Alexa 594 donkey anti-goat immunoglobulin G (Molecular Probes, Eugene, Ore.) for brahma and BAF170 staining. Coverslips were finally washed three times (5 min each) in 0.1% Triton X-100, three times (5 min each) in PBS*, 5 min in 0.2 μg of DAPI/ml, and three times (5 min each) in PBS*, and mounted in Prolong Antifade solution (Molecular Probes).
Fluorescence microscopy and image analysis.
Images were collected on an inverted light microscope (IMT-2; Olympus, Success, N.Y.) equipped with a cooled, slow-scan charge-coupled device camera (Photometrics; Tucson, Ariz.) as described previously (19). Optical sections of nuclei were collected and deconvolved as described previously (1). Figures were assembled using Adobe Photoshop. For quantitative measurements, ScionImage software (Scion Corp., Frederick, Md.) was used to measure the area of the lac operator arrays (homogeneously staining regions) in images collected from a large number of cells. A macro based on the “Analyze particles” command was used to measure the area of each lac operator array, which was identified in each image by thresholding. Proper thresholding was verified by visual inspection and defined interactively, if necessary, using the freehand selection tool. Measurements were exported, analyzed, and graphed in Microsoft Excel and SigmaPlot.
RESULTS
Experimental design.
We fused a green fluorescent lac repressor to the ER (Fig. 1A and D). GFP-lac rep-ER binds tightly to lac operator DNA repeats integrated into the genome of CHO cells, making the structure of the chromatin visible by fluorescence microscopy (Fig. 1C) (28). We also fused a yellow fluorescent lac repressor to amino acids 570 to 780 of the coactivator SRC-1, which is normally recruited by ER. In this reciprocal system, wild-type ER or CFP-ER is recruited to lac operator DNA via interaction with YFP-lac rep-SRC570-780 (Fig. 1B).
FIG. 1.
Experimental design. (A) Schematic of the GFP-lac repressor-ER fusion protein used in this study, which binds to lac operator DNA sequences. (B) Schematic of the YFP-lac rep-SRC570-780 fusion protein, which binds to lac operator sequences and can recruit wild-type ER or CFP-ER. (C) Schematic of the A03_1 cell line used in this study, generated by gene amplification of a vector containing the dihydrofolate reductase gene (dhfr) and a 256-copy repeat of the lac operator DNA sequence. The lac operator vector repeats are visible when a fluorescent lac repressor fusion protein is expressed. (D) GFP-lac rep-ER constructs made for this study.
For most of the work in this paper, we used the A03_1 CHO cell line (Fig. 1C), in which lac operator repeats and coamplified genomic DNA form an ∼90-Mbp chromosomal array. This array appears in metaphase as a homogeneously staining region and has properties of heterochromatin, forming a condensed mass roughly 1 μm in diameter through most of interphase. Targeting a control GFP-lac repressor fusion protein produces no obvious conformational changes in the amplified chromosome region (39). Alternatively, we used the RRE_B1 cell line, which contains a large, nonheterochromatic amplified chromosome region forming unusually extended fibrillar structures during interphase in most nuclei.
Nuclear redistribution, transcriptional activation, and coactivator recruitment by ER fusion proteins.
The GFP-lac rep-ER fusion protein retains normal ER activity based on several criteria. First, expression of the GFP-lac rep-ER fusion protein in wild-type CHO cells shows a homogeneous nuclear distribution in the absence of hormone which shifts to a punctate distribution after estradiol addition (data not shown), as previously reported for a GFP-ER fusion (20, 47).
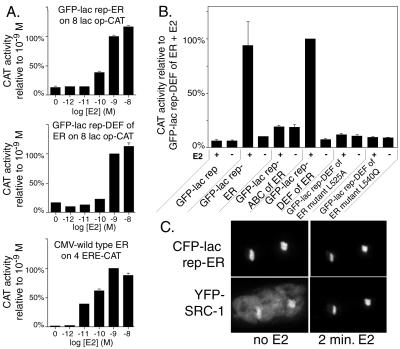
Second, GFP-lac rep-ER fusion proteins activated transcription as expected on a transiently transfected 8 lac op-TATA-CAT reporter plasmid (Fig. 1D). GFP-lac rep-ER and GFP-lac rep-DEF fusion proteins showed hormone-dependent transcriptional activation on the lac operator reporter with a dose response similar to that of wild-type ER on an ERE-based reporter, with activity peaking or reaching a plateau at 10−8 or 10−9 M estradiol (Fig. 2A). Activation by the GFP-lac rep-ER fusion protein, expressed from the F9-1 promoter and using the 8 lac op reporter construct, was at an only 2.5-fold-lower level than activation by wild-type ER, expressed from the stronger cytomegalovirus promoter and using the four-copy ERE reporter construct (direct comparison not shown). We note that ER does not activate transcription in the absence of hormone, despite being tethered to DNA via the lac repressor. The other lac repressor-ER fusion proteins also behaved as expected in the transcription assay (Fig. 2B). GFP-lac rep-ABC contains the weak constitutive transcriptional activation function AF-1 in the A/B domains and ER's DNA binding domain in the C domain. Two mutations in GFP-lac rep-DEF (L525A and L540Q) prevent transcriptional activation; the L525A mutation is known to virtually eliminate estradiol binding (14, 15), and L540Q shows impaired recruitment of coactivators, leading to markedly reduced transcriptional activation (22, 44, 54). The negative control GFP-lac rep did not activate transcription, while the positive control GFP-lac rep-VP16 AAD transcriptional activation was roughly 150 times higher than GFP-lac rep-ER, using the same promoter to drive fusion protein expression (data not shown). We also tested the transcriptional activity of the reciprocal system in which YFP-lac rep-SRC570-780 recruits CFP-ER or wild-type ER to DNA. Recent in vitro experiments have shown that each ER dimer binds only one molecule of SRC-1 (30). Any ER recruited to lac operator DNA via the truncated SRC-1 should not be able to interact with wild-type SRC-1 and possibly other coactivators which bind to the same helix 12 region of ER. Indeed, transient transcription assays indicate that the combination of YFP-lac rep-SRC570-780 and CFP-ER (or wild-type ER) is unable to detectably activate transcription when transfected with a lac operator reporter gene with or without estradiol treatment (data not shown).
FIG. 2.
Transcription and coactivator recruitment by GFP-lac rep-ER fusion proteins. (A) Dose response assays of transcriptional activity of ER fusion proteins in CHO cells in response to various concentrations of 17β estradiol (E2). We placed eight lac operator binding sites upstream of a TATA box followed by the CAT reporter gene (8 lac op-CAT). 4ERE-CAT is identical except that it contains four EREs. The data are normalized to a β-galactosidase internal control reporter plasmid, and results are displayed relative to the response of each construct at 10−9 M E2. (B) Transcriptional activity of constructs, tested with or without 10−9 M 17β estradiol (E2). Results are displayed relative to the sample with the highest average activity, GFP-lac rep-DEF plus E2. The error bars depict the standard error of the mean and in some cases are too small to be visible. (C) CFP-lac rep-ER recruits YFP-SRC-1. A03_1 cells were cotransfected with CFP-lac rep-ER (top row) and YFP-SRC-1 (bottom row) and subjected to live microscopy during hormone addition. Shown are two nuclei side by side, each with one lac operator-containing array. Exposure conditions and image manipulations were the same for each panel to reflect the increase in brightness of YFP-SRC-1 following the recruitment. All images are deconvolved and represent a 3-dimensional reconstruction of a series of z-sections.
Third, a CFP-lac repressor-ER fusion protein (CFP-lac rep-ER) binds to lac operator repeats and shows in vivo, hormone-enhanced recruitment of a YFP-SRC-1 coactivator fusion protein. The construction and functional testing of these fusion proteins is described elsewhere (47, 48). Consistent with recent fluorescence resonance energy transfer results using SRC-1 peptides and truncated ER in living cells (29), there is a partial recruitment of YFP-SRC-1 at the CFP-lac rep-ER-bound lac operator array in the absence of hormone, with a significant amount of YFP-SRC-1 distributed throughout the nucleus (Fig. 2C). Quantitative measurements reveal that within minutes of adding estradiol, absolute levels of YFP-SRC-1 at the lac operator array increase significantly, while nuclear background levels of YFP-SRC-1 show an absolute decrease. Further analysis of the in vivo recruitment of YFP-SRC-1 as well as a YFP-CBP fusion protein to CFP-lac rep-ER is described in a separate report (48).
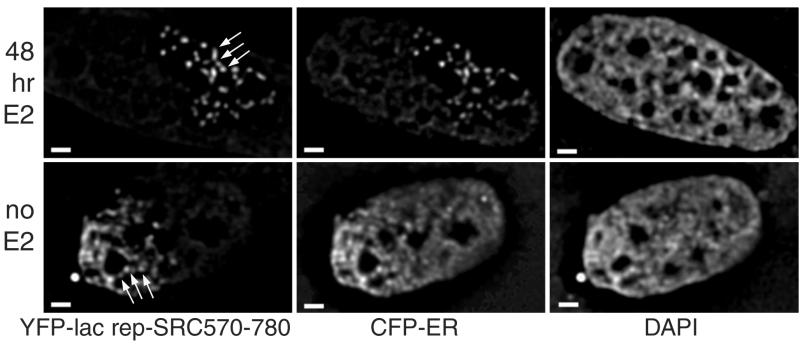
We also found that recruitment of CFP-ER to the YFP-lac rep-SRC570-780-bound lac operator array was as anticipated (see Fig. 5). In the absence of estradiol (n = 50), 6% show strong recruitment of CFP-ER to the YFP-lac rep-SRC570-780-bound lac operator array with only faint nucleoplasmic staining, 52% show moderate recruitment, and 42% show no recruitment. In the presence of estradiol (n = 51), 47% of cells show strong recruitment, 49% of cells show moderate recruitment, and 4% show no recruitment. This significant amount of interaction in the absence of hormone, enhanced by addition of estradiol, is consistent with the observed recruitment of YFP-SRC-1 to CFP-lac rep-ER and with our previous finding that CFP-ER and YFP-SRC570-780 visibly colocalize after addition of estradiol (47).
FIG. 5.
CFP-ER alters large-scale chromatin structure when recruited to YFP-lac rep-SRC570-780. A03_1 cells, which contain a heterochromatic lac operator array, were cotransfected with YFP-lac rep-SRC570-780 (left) and CFP-ER (center) and stained with DAPI (right). Arrows point out regions where fiber segments can be observed. Cells were exposed to 10−9 M estradiol (top) or vehicle (bottom) 24 h after transfection, which is 48 h prior to fixation. Scale bars, 1 μm.
Changes in large-scale chromatin structure after targeting of ER.
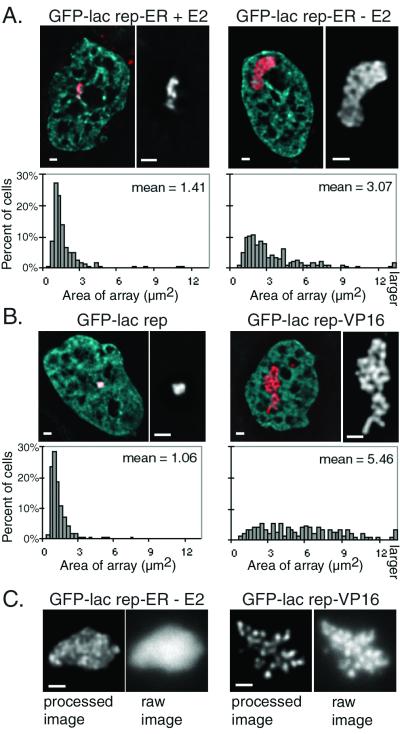
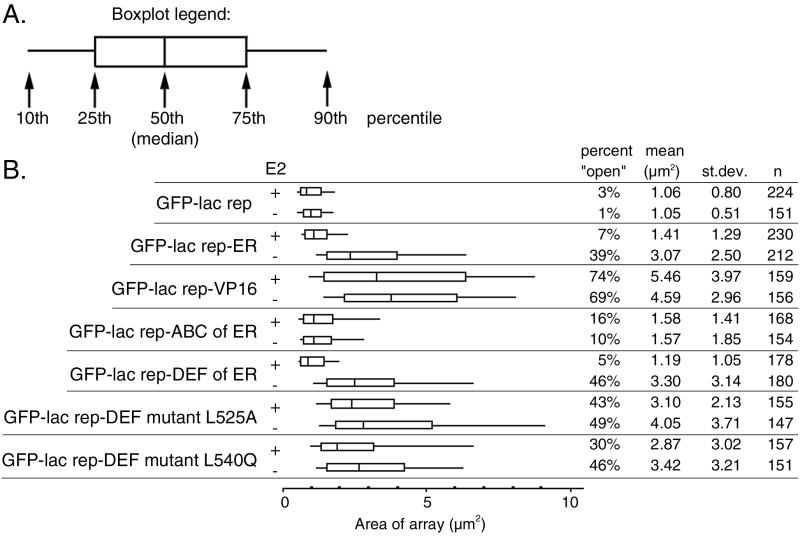
GFP-lac rep-ER fusions were transiently transfected into A03_1 cells, which normally contain a condensed lac operator array. Transfection of GFP-lac rep-ER and 48-h treatment with estradiol caused some cells' GFP-labeled chromosome regions to decondense (Fig. 3A, left) relative to the GFP-lac rep negative control (Fig. 3B, left). To confirm these observations, we collected images of large numbers of transfected cells. Quantitative measurements aided by image analysis software indeed revealed larger average lac operator array sizes in GFP-lac rep-ER-transfected cells (1.41 μm2) relative to GFP-lac repressor-transfected control cells (1.06 μm2) (Fig. 3A and B). lac operator arrays larger than 2.7 μm2 were considered unfolded, or “open.” By this criterion, 7% of cells contained an open lac operator array relative to 1 to 3% in the populations transfected with GFP-lac rep alone.
FIG. 3.
The ER alters the appearance of large-scale chromatin fibers. A03_1 cells which contain a heterochromatic lac operator array were transfected. (A and B) Deconvolved optical sections of fixed cells. DAPI staining is shown as blue and GFP as red in the left panel of each pair. A black-and-white close-up of each GFP-labeled chromosomal region is shown in the right panel of each pair. Quantitative measurements on large numbers of cells are shown below the images as histograms. The sizes of lac operator arrays were measured as described in the text and Materials and Methods. (C) Optical sections of live cells. The left panel of each pair shows a deconvolved section, and the right panel shows the raw image. Scale bars, 1 μm.
We hypothesized that GFP-lac rep-ER in the absence of estradiol would have no effect on large-scale chromatin structure. Unexpectedly, a significant fraction of the cell population transfected with GFP-lac rep-ER in the absence of estradiol contained lac operator arrays which are even more unfolded than those produced in the presence of estradiol (Fig. 3A, right). These structures were often similar in size (mean = 3.07 μm2) to those seen with the GFP-lac rep-VP16 AAD fusion protein (mean = 5.46 μm2) with or without estradiol (Fig. 3B, right). Thirty-nine percent of lac operator arrays with GFP-lac rep-ER minus estradiol were open, compared to ∼70% of lac operator arrays using GFP-lac rep-VP16 AAD and 3% of lac operator arrays with the control GFP-lac repressor constructs.
The fibers produced by GFP-lac rep-ER in the absence of hormone appear significantly less distinct than is typically seen after decondensation by GFP-lac rep-VP16 AAD, suggesting a qualitative difference in the decondensed structures. To ensure that these differences were not due to fixation conditions, we confirmed this finding by performing microscopy on living cells (Fig. 3C). Indeed, the GFP-lac rep-ER-produced structures appear much less distinct than those produced by GFP-lac rep-VP16 AAD, suggesting a qualitatively different type of unfolding.
Unfolding of heterochromatic array induced by ER fusion proteins is partially reversed by estradiol.
The unfolding observed with GFP-lac rep-ER in the absence of estradiol was much larger than that seen in the presence of estradiol. Direct observation of living cells revealed a striking recondensation of large-scale chromatin structure beginning within minutes after estradiol addition (Fig. 4). Data sets with more frequent time points can be viewed as Quicktime movies at www.life.uiuc.edu/belmont. Exposure to UV light in the process of collecting these four-dimensional images appeared to inhibit this condensation (data not shown). We therefore limited UV exposure by taking individual optical sections at each time point and found condensation to occur in 15 out of 16 cells, producing an average decrease in lac operator array area of ∼50% (Table 1). The antiestrogens, trans-hydroxytamoxifen and ICI182,780, produced comparable changes in lac operator array area (Table 1).
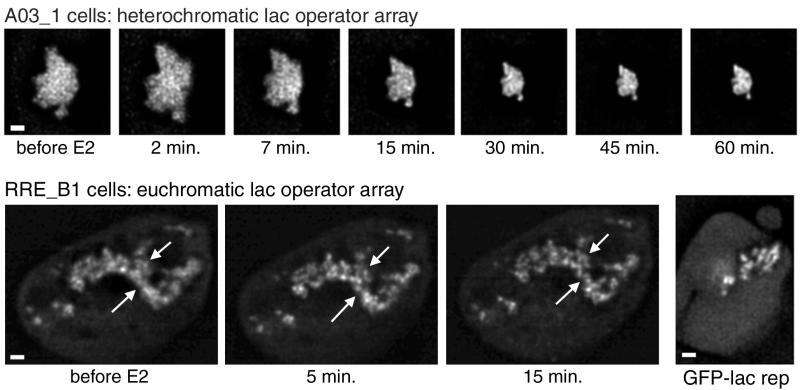
FIG. 4.
Effects of estradiol observed in living cells. Cells were transfected with GFP-lac rep-ER, cultured in the absence of estradiol, and observed in a live cell chamber on the fluorescence microscope. Optical sections were taken before adding 17β-estradiol at 10−9 M and at each time point afterwards. The top series of images shows a deconvolved section for each time point of an A03_1 cell which contains a heterochromatic lac operator array. The bottom series shows a projection of a stack of deconvolved images for each time point of an RRE_B1 cell which contains a euchromatin-like lac operator array. For comparison, an example of an RRE_B1 cell transfected with the negative control GFP-lac repressor is shown to the right, although this cell line shows great variability. Arrows emphasize examples of fibers which condense locally. Scale bars, 1 μm.
TABLE 1.
Live recondensation of GFP-lac rep-ER-bound heterochromatic lac operator arrays induced by estradiol and antiestrogens in A03_1 cells
| Treatment | % Original sizea | % SD | nb |
|---|---|---|---|
| 10−9 M E2 (17β-estradiol) | 52 | 22 | 16 |
| 2 × 10−6 M trans-hydroxytamoxifen | 40 | 17 | 23 |
| 2 × 10−6 M ICI182,780 | 39 | 12 | 31 |
| None | 116 | 27 | 13 |
Sizes of arrays were measured as in Fig. 3, and the size after 30 min of treatment was divided by the original size to determine the % original size.
n, number of cells examined.
For estradiol-treated cells, the most dramatic condensation occurred within 30 min and the lac operator arrays showed minimal further change in structure through the end of the experiment, up to 3 h later. After 30 min, the lac operator arrays had not condensed completely to sizes typical of a population treated with estradiol for 48 h; in these cells, the average lac operator array area decreased from 8.4 to 4.3 μm2 in 30 min, which is still significantly larger than the 1.4 μm2 seen after long-term estradiol exposure. Therefore, estradiol induces rapid recondensation within 30 min followed by further gradual condensation over a period of hours or days.
Recondensation occurred with minimal changes in nuclear position or shape; the lac operator arrays did not appear to fold over long distances as they condensed but rather decreased in size uniformly, perhaps due to local condensation of individual large-scale chromatin fibers. In addition to the global recondensation caused by hormone, we also noticed a local condensation of large-scale chromatin structure and the appearance of distinct, large-scale chromatin fibers within several minutes of adding estradiol to cells expressing GFP-lac rep-ER. At light microscopy resolution, the diffuse fibrillar appearance of these lac operator arrays became more well defined, becoming very similar to the distinct fibers normally observed with the GFP-lac rep-VP16 AAD protein.
A nonheterochromatic, decondensed lac operator array shows no significant changes in array area and only local changes in large-scale chromatin structure after estradiol treatment.
The lac operator array in A03_1 cells is normally condensed and late replicating. By these criteria, the array behaves as heterochromatin. The extensive, hormone-induced reversal of the array decondensation produced by GFP-lac rep-ER could be explained in several ways. First, dimerization of ER in response to ligand might cause GFP-lac rep-ER fusions to aggregate and therefore appear to condense chromatin. Second, addition of hormone might induce an active, chromatin-condensing activity of the ER itself. Third, addition of hormone might simply reverse or down-regulate whatever large-scale chromatin decondensation activity is produced by ER targeting, with the global condensation of the lac operator array simply representing a reversion to the array's normal, heterochromatic state.
To distinguish between these possibilities, we examined changes in lac operator array morphology occurring in cells expressing GFP-lac rep-ER after hormone treatment using a different cell line, RRE_B1. The lac operator array conformation in this cell line is variable but has a very high percentage of cells with unusually extended and fibrillar arrays, relative to the bulk large-scale chromatin structure seen throughout most of the nucleus (3). For the euchromatin-like arrays in RRE_B1 cells expressing GFP-lac rep-ER, addition of estradiol does not lead to significant changes in array area, for either the more highly extended or the more compact arrays within this cell line (Fig. 4). Thus, it is unlikely that the condensation seen in the other cell line, A03_1, is due to dimerization or aggregation of GFP-lac rep-ER, nor is it due to a chromatin-condensing activity of ER. We therefore conclude that the global condensation of lac operator arrays in the presence of hormone in the A03_1 cell line represents a reversal or down-regulation of ER's unfolding activity, allowing the array to revert to its normal heterochromatin state.
While the overall size of the lac operator array in RRE_B1 cells remains unchanged, the appearance of the fibers does change after exposure to estradiol. As shown in Fig. 4, RRE_B1 cells expressing GFP-lac rep-ER show open lac operator arrays with a more diffuse appearance. Because of the high variability of the structures in control cells expressing GFP-lac repressor, we cannot easily determine whether this structure differs from that seen with GFP-lac repressor alone. Yet, upon addition of estradiol to GFP-lac rep-ER expressing RRE_B1 cells, we see a local condensation in which the diffuse fibers change to more sharply demarcated, distinct fibers (Fig. 4). These local changes in array structure are quite similar to those described above for the A03_1 array, but they occur without the reversal of global decondensation.
Targeting wild-type ER via a lac repressor-SRC570-780 fusion protein also produces dramatic large-scale chromatin decondensation.
Observing maximal chromatin decondensation in the absence of ligand using the GFP-lac rep-ER was unexpected. We therefore considered the possibility of a problem with the GFP-lac rep-ER fusion proteins despite the fact that they properly localize, activate transcription from a transiently transfected reporter gene, and recruit SRC-1. For example, the lac repressor DNA binding domain may bind lac operator DNA more tightly than ER binds to EREs, possibly preventing GFP-lac rep-ER from diffusing throughout the nucleus and obtaining posttranslational modifications or protein interactions which might be important for ER function. We therefore used an alternative mechanism to target unmodified, wild-type ER to the lac operator array by fusing the lac repressor to amino acids 570 to 780 of the coactivator SRC-1 (Fig. 1B). This region of SRC-1 contains three LXXLL motifs, known as nuclear receptor boxes, which interact with the ER as well as other nuclear receptors. As noted above, transcription is not activated upon estradiol addition because any ER recruited to YFP-lac rep-SRC570-780 is unable to bind endogenous SRC-1 and possibly other coactivators which bind the same domain of ER. This feature is an advantage because it allows examination of the effect of ER on large-scale chromatin structure in the absence of transcriptional activation.
In roughly half of the A03_1 cells transfected with YFP-lac rep-SRC570-780 alone, the lac operator array is significantly increased in size relative to the control. This opening in the absence of cotransfected ER is difficult to interpret, as YFP-lac rep-SRC570-780 could recruit any number of transcription factors endogenous to this CHO-derived cell line. However, a quite distinct unfolded structure is observed when CFP-ER is cotransfected with YFP-lac rep-SRC570-780, with or without estradiol (Fig. 5). These unusually decondensed lac operator arrays are qualitatively different from any array conformations seen with either lac repressor-VP16 or lac repressor-ER. The fibers produced are often very numerous and thin, protruding from a region where fibers cannot be individually traced (seen best in the “no E2” example in Fig. 5). Frequently, there are gaps in the fibers which could indicate unfolding to the extent that non-lac operator intervening DNA is detectable as a visible gap in lac repressor staining (seen best in the “48 hr E2” example in Fig. 5). In the absence of estradiol, 20% of cells examined (n = 49) display this unusual structure while the remainder are comparable to YFP-lac rep-SRC570-780 alone. In the presence of estradiol, 36% of cells examined (n = 50) display this unusual structure. By contrast, no such structures were seen when YFP-lac rep-SRC570-780 was expressed with estradiol but without cotransfected ER (n = 83). We also observed these unusual decondensed structures when YFP-lac rep-SRC570-780 was cotransfected with completely wild-type ER (data not shown). This confirms that the large-scale chromatin decondensing activity observed is due to an intrinsic property of wild-type ER and is not an artificial property of the ER fusion proteins per se.
Activation function 1 and helix 12 are not required for chromatin unfolding; helix 12 is required for reversal of unfolding.
The fact that estradiol does not reverse unfolding by lac rep-SRC-tethered ER implies that recruitment of SRC-1, or another coactivator which binds the same domain of ER as SRC-1, is required along with ligand binding to reverse the unfolding. We tested several truncations and mutations of ER to determine which domains of ER are capable of producing unfolding and the reversal of unfolding. Domains A, B, and C are not required for chromatin unfolding, since a GFP-lac rep-DEF construct produced results similar to those with full-length ER (Fig. 6A and 7). A truncation at amino acid 534 and the mutation L540Q both eliminate coactivator recruitment via helix 12 of ER. Both constructs are able to unfold chromatin as well as wild-type ER (Fig. 6C and 7) (48). This indicates that the well-studied coactivators recruited via LXXLL domains to helix 12 of ER (25) are not responsible for the dramatic unfolding seen in the absence of hormone. Instead, a region from amino acids 262 to 534 is able to recruit the proteins required for chromosome unfolding. ER truncated at amino acid 534 or with the L540Q mutation produces unfolded lac operator array chromatin even in the presence of hormone. Moreover, estradiol does not reverse unfolding by lac rep-SRC-tethered ER. These results indicate that a functional AF-2/helix 12, and possibly SRC-1 recruitment, is required for recondensation.
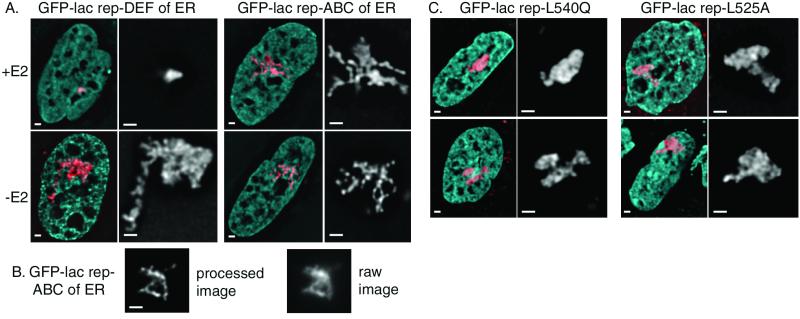
FIG. 6.
Domain and mutational analysis of chromatin unfolding by ER. (A) A03_1 cells, which contain a heterochromatic lac operator array, were transfected, and images were collected as with Fig. 3. GFP-lac rep-DEF of ER resembles the full-length ER, whereas GFP-lac rep-ABC of ER displays an unusual structure of thin fibers in a small percentage of cells. (B) An image of a live cell was collected as with Fig. 3. (C) Images were collected from A03_1 cells transfected with GFP-lac rep-DEF with an L540Q mutation (left) and GFP-lac rep-DEF with an L525A mutation (right). Scale bars, 1 μm.
FIG. 7.
Measurements of all GFP-lac rep-ER fusion proteins. (A) The box plot shown is a simplified version of a histogram. Ends of the lines show the 10th and 90th percentiles, ends of the boxes show the 25th and 75th percentiles, and the center line shows the 50th percentile (median). (B) Quantitative measurements of lac operator arrays were made using images collected from A03_1 cells transfected with each GFP-lac rep fusion protein. The data for GFP-lac rep-ER, GFP-lac rep, and GFP-lac rep-VP16 is the same as that shown in the histograms in Fig. 3. For the percent “open” column, lac operator arrays were counted as “open” if they measured larger than 2.7 μm2. st.dev., standard deviation; n, number of cells examined.
The GFP-lac rep-ABC construct, containing the weak, constitutive AF-1 transcriptional activation domain, produced dramatic unfolding in about 10% of the cells which was qualitatively different from that with any other construct (Fig. 6A and B and 7). These cells showed thin, extended fibers protruding several microns out from a more densely folded core. This unfolding was not altered by estradiol, consistent with the construct's lack of a ligand binding domain.
Finally, we tested whether the unfolding of chromatin in the absence of estradiol was due to trace amounts of estrogens in the medium, despite our using charcoal-dextran-treated serum and phenol red-free medium. We found that structures produced by ER with an L525A mutation, which does not bind ligand, produced unfolded structures whether or not estradiol was present, just as its wild-type counterpart does in the absence of hormone (Fig. 6C and 7). Therefore, the large-scale chromatin decondensation observed cannot be attributed to residual estrogens in the cell medium but reflects a true activity of the ER in the absence of hormone, which may be dependent on a hormone-independent signaling pathway.
Chromatin remodeling and histone acetylation.
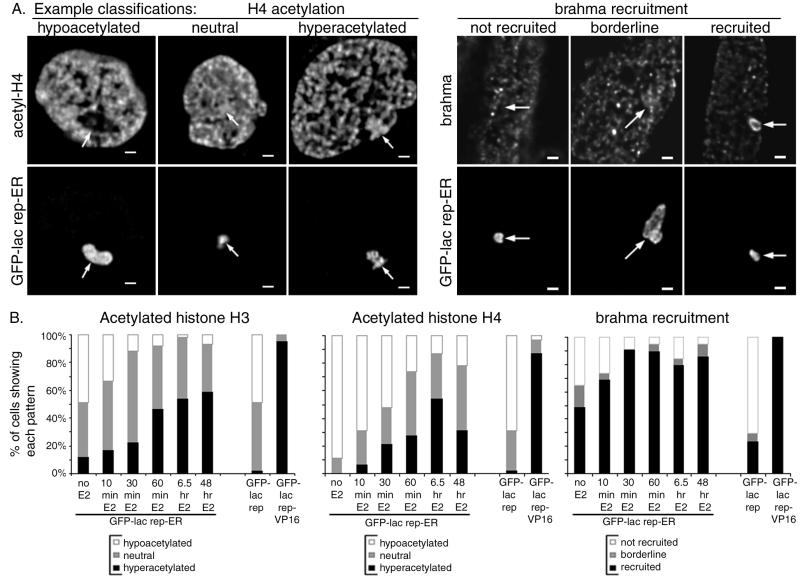
Changes in chromatin structure at the nucleosomal level produced by chromatin remodeling complexes (52) and histone acetyltransferases (23) have been proposed to affect higher levels of chromatin folding. To pursue the potential link between these two levels of structure, cells were immunostained for components of chromatin remodeling complexes. Human brahma (hbrm/hSnf2α), a human homologue of the yeast SWI2/SNF2 chromatin remodeling complex component (35), has been shown to enhance estrogen receptor transcriptional activity (9). Yeast two-hybrid experiments previously revealed a potential ligand-independent interaction between ER and brahma, enhanced by estradiol (21). Immunostaining of A03_1 cells revealed strong recruitment of brahma to GFP-lac rep-ER in the absence of estradiol in ∼50% of cells (Fig. 8, right). After estradiol treatment, ∼85% of cells showed strong recruitment. BAF170 (BRG1-associated factor 170) is a component of the human BRG1-containing chromatin remodeling complex (53), which has not previously been tested for interaction with ER. BAF170 was not detectably recruited to GFP-lac rep-ER with or without estradiol (data not shown), whereas GFP-lac rep-VP16 strongly recruited both brahma and BAF170 in nearly every cell (data not shown).
FIG. 8.
Histone acetylation and brahma recruitment by GFP-lac rep-ER fusion proteins. A03_1 cells were transfected with GFP-lac rep-ER, GFP-lac rep, or GFP-lac rep-VP16 and treated with 10−9 M estradiol for various time points prior to fixation and staining with antibodies against the acetylated forms of histone H3 and H4 or against brahma. (A) For histone acetylation (left set of images), each cell's lac operator array was scored as having staining at a lower level than (hypoacetylated), comparable to (neutral), or higher than (hyperacetylated) that for the rest of the chromatin in the nucleus. For brahma recruitment (right set of panels), each cell's lac operator array was scored as having staining comparable to that for the rest of the nucleus (not recruited), slightly brighter staining relative to the rest of the nucleus (borderline), or brighter staining than the rest of the nucleus (recruited). (B) For each time point, cells were scored as in panel A for histone H3 acetylation (left) and histone H4 acetylation (middle) and brahma recruitment (right).
Several histone acetyltransferases are known to be strongly recruited to ER by estradiol, including SRC-1 (46) and CBP (2). We have confirmed recruitment of SRC-1 (Fig. 2C) (48) and CBP (48) to GFP-lac rep-ER. To determine whether this recruitment is accompanied by increased histone acetylation, GFP-lac rep-ER-transfected cells were fixed at various time points after estradiol treatment and stained with antibodies against the acetylated forms of histone H3 and H4 (Fig. 8). In the absence of estradiol, lac operator arrays with GFP-lac rep-ER typically showed histone H3 and H4 hypoacetylation. Histone H3 and H4 acetylation gradually increased after hormone addition, with acetylation of histone H4 possibly decreasing after long-term estradiol treatment. Hyperacetylation was much less dramatic than that produced by GFP-lac rep-VP16 in nearly every cell (data not shown) (51). These results are generally consistent with previous chromatin immunoprecipitation studies, which revealed an increase in histone acetylation in the vicinity of estrogen-responsive promoters after treatment with estradiol, although the dynamics of such changes from study to study have not been resolved (7, 45). Using the alternate tethering approach, cells transfected with YFP-lac rep-SRC570-780 and CFP-ER or wild-type ER did not show detectable increases in acetylation at the lac operator array compared to YFP-lac rep-SRC570-780 alone (data not shown), although the large-scale chromatin decondensation produced was even greater than that seen with GFP-lac rep-VP16. The lack of detectable changes in histone acetylation may be due to a dominant-negative interaction between ER and SRC570-780. However, it may also result from a lowered detection sensitivity for histone acetylation due to the unusually extended conformation of the lac operator array. What is clear is that the chromatin unfolding produced by YFP-lac rep-SRC570-780-recruited ER is not accompanied by a high degree of histone hyperacetylation.
DISCUSSION
Transcriptional activators and large-scale chromatin unfolding.
Using the lac operator/repressor tethering system, we observed the capability of the estrogen receptor to decondense large-scale chromatin structure. We also addressed several fundamental questions about the relationship between large-scale chromatin structure and transcription. First, different transcriptional activators are capable of unfolding chromatin to qualitatively different levels. That is, the fibers produced by ER appeared less distinct than those produced by VP16. In addition, unusually thin fibers were produced in A03_1 cells by GFP-lac rep-ABC and by full-length CFP-ER recruited via a YFP-SRC570-780 fusion. Second, stronger activators do not necessarily produce more extensive unfolding, since GFP-lac rep-ER is 150 times weaker by transient transcription assay than GFP-lac rep-VP16, yet the global unfolding produced by either construct is close in magnitude. Third, we found that there is not a simple correlation between unfolded chromatin and transcriptional potential. Our hypothesis that chromatin would be condensed in the absence of ligand and unfolded upon ligand binding was too simplistic. GFP-lac rep-ER unfolded chromatin by about 35% after long-term estradiol treatment. A more dramatic unfolding was observed by GFP-lac repressor-ER in the absence of ligand, and the largest degree of unfolding, exceeding that previously observed with the VP16 acidic activation domain, was observed using the transcriptionally dead, lac repressor-SRC1 tethering system to recruit wild-type ER.
Unliganded ER is capable of unfolding large-scale chromatin structure.
While the decondensing capability of ER in the absence of ligand was surprising, results from several different lines of investigation yielded similar, internally consistent results. Specifically, the same conclusion was reached using two different methods for ER recruitment and using various ER mutations and truncations. Our results suggest that a region encompassing receptor domains D and E, from amino acids 262 to 534, is responsible for the major ligand-independent decondensation activity observed with ER. This decondensation is insufficient for transcriptional activation, because the mutants and the YFP-lac rep-SRC570-780 plus ER recruitment system lack transcriptional activity yet are still able to unfold chromatin in the absence of hormone.
While we have shown in this work that unliganded ER is capable of unfolding chromatin, this does not prove that it actually does unfold chromatin at endogenous ER-regulated promoters. Interpretation of our findings depends on whether the estrogen receptor is recruited to chromatin in its unliganded state, and there is considerable evidence that this is the case.
While numerous studies have demonstrated the binding of unliganded ER to ERE-containing DNA in vitro (5), the situation in vivo has been less clear (26). Several independent, quantitative experimental approaches have demonstrated that significant ER is bound at promoters in the absence of hormone, with ER binding generally increasing after estradiol addition. Early evidence for in vivo binding of receptor in the absence of hormone came from promoter interference studies with mammalian cells (38). More recently in vivo footprinting of the endogenous pS2 promoter in breast cancer cells revealed that the consensus half site of the imperfect pS2 ERE was equally well protected with or without hormone (24). In addition, chromatin immunoprecipitation data conclusively demonstrated significant levels of estrogen receptor bound at the endogenous pS2 and cathepsin D promoters in mammalian cells in the absence of hormone (45, 56).
The discovery of unliganded ER at certain ERE-containing promoters raises the possibility that the chromatin unfolding activity we have described in this paper might play a physiological role. One possibility is that the unfolded structure represents a poised, transcriptionally competent chromatin state still dependent on hormone to recruit coactivators for gene activation. This transcriptionally inactive but competent state could prepare genes for the rapid response to hormone that has been observed in studies of the kinetics of endogenous estradiol-responsive genes. This essentially represents a two-stage model of transcriptional activation, in which changes in higher-order and large-scale chromatin structure precede recruitment of transcriptional coactivators and the general transcriptional machinery.
A second possibility is that the unfolded structure could be involved in transcriptional repression by unliganded ER. Recent experiments have demonstrated that ER binds to endogenous ERE-containing promoters in the absence of ligand but its transcriptional activity is repressed (56). A genetic screen for proteins required for long-range enhancer-promoter interactions has identified Chip (34) and the adherin-related Nipped-B (40), proteins which may be involved in cross-linking chromatin and in chromosome adhesion, respectively. This work has led to speculation that higher-order or large-scale chromatin condensation may be required for enhancer-stimulated transcriptional activation (12), thereby suggesting that dispersion of chromatin may be a mechanism of transcriptional repression.
Finally, the large-scale chromatin unfolding produced by unliganded ER might produce a remodeled, permissive state, allowing activators and/or repressors full access to the gene locus. Thus, like chromatin remodeling at the lower levels of folding, large-scale chromatin unfolding may not be inherently activating or repressing but rather may present a window of opportunity for trans factors to determine a promoter's response.
Addition of ligand partially disrupts ER's chromatin unfolding activity.
When estradiol was added to A03_1 cells with unfolded GFP-lac rep-ER-produced structures, chromatin unfolding was rapidly, though only partially, reversed. Structures after long-term estradiol treatment were smaller than those produced in the absence of hormone but larger than those produced by GFP-lac rep alone. This partial reversal of decondensation activity was dependent on hormone binding and recruitment of functional coactivators and/or corepressors by ER's LXXLL binding pocket. Our present view is that the recondensation we observed is related to a downregulation of the estrogen receptor. It is known that several ER-responsive genes are rapidly induced after estradiol addition but show a decrease in activity over time, despite the continued presence of estradiol (6, 7, 10, 13). One potential mechanism for this downregulation involves the inactivation of some coactivators via acetylation by other coactivators (7).
Relationship between local chromatin structure and large-scale chromatin structure.
The remodeling of nucleosomes and histone acetylation are frequently proposed to alter higher levels of chromatin structure (23, 52). Some correlations have been observed; for example, GFP-lac rep-VP16 produces activated transcription, acetylated histones, and unfolded large-scale chromatin structure (51). However, a causal relationship has not been established. In fact, large-scale decondensation of chromatin by the BRCT domains of BRCA1 occurs in the absence of detectable histone hyperacetylation (55). In addition, recent unpublished work in our laboratory has provided an example in which large-scale chromatin decondensation by VP16 AAD is inhibited even in the presence of histone hyperacetylation (S. Pop and A. S. Belmont, personal communication). In this work, we found that the decondensation induced by ER does not correlate with histone hyperacetylation. The most dramatic unfolding was produced under conditions where histone hyperacetylation was undetectable (unliganded GFP-lac rep-ER and CFP-ER recruited by YFP-lac rep-SRC570-780). It is therefore unlikely that the proteins involved in large-scale chromatin unfolding are among the histone acetyltransferases more strongly recruited to ER after estradiol addition, although we cannot rule out the involvement of proteins modifying histones in other ways, e.g., methylation or phosphorylation.
Observations of the recruitment of various proteins cannot establish their involvement in large-scale chromatin unfolding. Nevertheless, we observed that the chromatin remodeling complex component brahma is recruited significantly to GFP-lac rep-ER in the absence of hormone, and this recruitment increases as chromatin recondenses in response to estradiol. These observations do not provide definite evidence that chromatin remodeling complexes are involved in large-scale chromatin unfolding, but neither do they rule out their involvement.
Experimental considerations and future directions.
The experimental approach in this paper allows the targeting of large amounts of an activator to a visible chromosome region so that effects on large-scale chromatin structure can be observed. While it would be preferable to observe the effects of the activation of a single estrogen-responsive gene on nearby chromatin, suitable electron microscopy methods do not yet exist. Chromatin changes at arrays of a viral promoter have recently been observed (36), and we have been developing methods towards observing natural promoters, including ER-responsive promoters. The method we have used in this work is a useful first step and provides a few advantages over studying a natural promoter. The lac repressor targeting allows direct comparison of the effects of any protein using the same target chromatin. This system has revealed the effects of various other nuclear receptors, including thyroid hormone receptor, progesterone receptor, and Pit-1 (M. G. Mancini, K. X. Patel, Z. D. Sharp, and M. A. Mancini, unpublished observations). Most importantly, this approach provides a rapid method for mapping the specific protein domains responsible for this decondensation activity.
A key question remaining, however, is whether the observed results using this experimental methodology are physiologically relevant given the high numbers of lac operator repeats involved. It is unlikely that each lac operator binds a lac repressor molecule, as there is reason to believe that lac repressor binding may be significantly limited by steric constraints and phasing of lac operators relative to the nucleosome linker DNA. Our present model is that the structural changes we observe using this targeting methodology represent an amplification of similar structural perturbations produced over much smaller neighborhoods surrounding endogenous promoters.
Recently changes in large-scale chromatin structure were observed using a cell line containing an array of roughly 200 copies of a vector, each of which contains one copy of the mouse mammary tumor virus promoter upstream of the ras and bovine papillomavirus genes. The six glucocorticoid receptor (GR) binding sites in each copy of that promoter (16) were sufficient to produce large-scale chromatin decondensation within 3 h of hormone treatment (36). Within the following 5 h, arrays recondensed despite the continued presence of hormone (36). It is interesting to compare these results with GR with our study of ER, since the two steroid receptors are in many ways similar. Significantly, one difference between the receptors is that in the absence of ligand, GR is restricted to the cytoplasm whereas ER is located primarily in the nucleus. Therefore, GR would not be expected to affect large-scale chromatin structure in the absence of hormone, while ER's localization makes it possible that it could, particularly in light of recent evidence that ER under some conditions may bind to EREs in the absence of hormone (24, 45, 56). Interestingly, both studies showed down-regulation of the unfolding activity over time, although the timing differed: ER's activity was dramatically reduced within 30 min and gradually reduced over the following 48 h, while GR's unfolding activity was gradually reduced over the course of about 5 h (from 3 to 8 h after hormone treatment). The differences in timing could reflect functional differences between the two receptors; for example, GR may be regulated differently in order to allow a period of unfolding before downregulation begins since it does not enter the nucleus until hormone is added. We cannot rule out, however, that the timing observed for ER may by altered due to some aspect of our artificial system.
Most previous studies of the estrogen receptor have used transcription as a readout of ER function. By applying an assay which directly focuses on ER's effect on large-scale chromatin structure, we have demonstrated that ER is capable of unfolding chromatin. This assay should allow identification of the ER subdomain(s) and the interacting proteins required for this activity.
Acknowledgments
This work was supported by grants from the National Institutes of Health to A. S. Belmont (R01-GM58460 and R01-GM42516), M. A. Mancini (R01-DK55622), and B. S. Katzenellenbogen (CA 18119 and CA 60514); a National American Heart Association Scientist Development Award to M. A. Mancini; an NIH postdoctoral fellowship (1F32DK09787) to D. Stenoien; an NIH Bioacoustics and Bioengineering in Radiation Oncology Traineeship (NIH T32 CA09067) and Department of Defense Predoctoral Traineeship (DAMD17-99-1-9219) to R. R. Rajendran; and an NIH Cell and Molecular Biology Traineeship (T32-GM07283) and a Howard Hughes Medical Institute Predoctoral fellowship to A. C. Nye.
We thank Sevi Pop for immunostaining protocols and Rong Li for helpful comments.
REFERENCES
- 1.Agard, D. A., Y. Hiraoka, P. Shaw, and J. W. Sedat. 1989. Fluorescence microscopy in three dimensions. Methods Cell. Biol. 30:353-377. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 3.Belmont, A. S., and K. Bruce. 1994. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell Biol. 127:287-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belmont, A. S. 2001. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 11:250-257. [DOI] [PubMed] [Google Scholar]
- 5.Brown, M., and P. A. Sharp. 1990. Human estrogen receptor forms multiple protein-DNA complexes. J. Biol. Chem. 265:11238-11243. [PubMed] [Google Scholar]
- 6.Cavailles, V., P. Augereau, M. Garcia, and H. Rochefort. 1988. Estrogens and growth factors induce the mRNA of the 52K-pro-cathepsin-D secreted by breast cancer cells. Nucleic Acids Res. 16:1903-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J., and K. S. Matthews. 1992. Deletion of lactose repressor carboxyl-terminal domain affects tetramer formation. J. Biol. Chem. 267:13843-13850. [PubMed] [Google Scholar]
- 9.Chiba, H., M. Muramatsu, A. Nomoto, and H. Kato. 1994. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 22:1815-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cicatiello, L., C. Ambrosino, B. Coletta, M. Scalona, V. Sica, F. Bresciani, and A. Weisz. 1992. Transcriptional activation of jun and actin genes by estrogen during mitogenic stimulation of rat uterine cells. J. Steroid Biochem. Mol. Biol. 41:523-528. [DOI] [PubMed] [Google Scholar]
- 11.Collingwood, T. N., F. D. Urnov, and A. P. Wolffe. 1999. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J. Mol. Endocrinol. 23:255-275. [DOI] [PubMed] [Google Scholar]
- 12.Dorsett, D. 1999. Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 9:505-514. [DOI] [PubMed] [Google Scholar]
- 13.Dubik, D., and R. P. Shiu. 1988. Transcriptional regulation of c-myc oncogene expression by estrogen in hormone-responsive human breast cancer cells. J. Biol. Chem. 263:12705-12708. [PubMed] [Google Scholar]
- 14.Ekena, K., K. Weis, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 1996. Identification of amino acids in the hormone binding domain of the human estrogen receptor important in estrogen binding. J. Biol. Chem. 271:20053-20059. [DOI] [PubMed] [Google Scholar]
- 15.Ekena, K., K. E. Weis, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 1997. Different residues of the human estrogen receptor are involved in the recognition of structurally diverse estrogens and antiestrogens. J. Biol. Chem. 272:5069-5075. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher, T. M., B. W. Ryu, C. T. Baumann, B. S. Warren, G. Fragoso, S. John, and G. L. Hager. 2000. Structure and dynamic properties of a glucocorticoid receptor-induced chromatin transition. Mol. Cell. Biol. 20:6466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fryer, C. J., and T. K. Archer. 1998. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88-91. [DOI] [PubMed] [Google Scholar]
- 18.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 19.Hiraoka, Y., J. R. Swedlow, M. R. Paddy, D. A. Agard, and J. W. Sedat. 1991. Three-dimensional multiple-wavelength fluorescence microscopy for the structural analysis of biological phenomena. Semin. Cell Biol. 2:153-165. [PubMed] [Google Scholar]
- 20.Htun, H., L. T. Holth, D. Walker, J. R. Davie, and G. L. Hager. 1999. Direct visualization of the human estrogen receptor alpha reveals a role for ligand in the nuclear distribution of the receptor. Mol. Biol. Cell 10:471-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichinose, H., J. M. Garnier, P. Chambon, and R. Losson. 1997. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene 188:95-100. [DOI] [PubMed] [Google Scholar]
- 22.Ince, B. A., Y. Zhuang, C. K. Wrenn, D. J. Shapiro, and B. S. Katzenellenbogen. 1993. Powerful dominant negative mutants of the human estrogen receptor. J. Biol. Chem. 268:14026-14032. [PubMed] [Google Scholar]
- 23.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J., L. N. Petz, Y. S. Ziegler, J. R. Wood, S. J. Potthoff, and A. M. Nardulli. 2000. Regulation of the estrogen-responsive pS2 gene in MCF-7 human breast cancer cells. J. Steroid Biochem. Mol. Biol. 74:157-168. [DOI] [PubMed] [Google Scholar]
- 25.Klinge, C. M. 2000. Estrogen receptor interaction with co-activators and co-repressors. Steroids 65:227-251. [DOI] [PubMed] [Google Scholar]
- 26.Klinge, C. M. 2001. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 29:2905-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraus, W. L., and J. T. Kadonaga. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12:331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, G., G. Sudlow, and A. S. Belmont. 1998. Interphase cell cycle dynamics of a late replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and intranuclear positioning. J. Cell Biol. 140:975-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llopis, J., S. Westin, M. Ricote, Z. Wang, C. Y. Cho, R. Kurokawa, T. M. Mullen, D. W. Rose, M. G. Rosenfeld, R. Y. Tsien, C. K. Glass, and J. Wang. 2000. Ligand-dependent interactions of coactivators steroid receptor coactivator-1 and peroxisome proliferator-activated receptor binding protein with nuclear hormone receptors can be imaged in live cells and are required for transcription. Proc. Natl. Acad. Sci. USA 97:4363-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margeat, E., N. Poujol, A. Boulahtouf, Y. Chen, J. D. Muller, E. Gratton, V. Cavailles, and C. A. Royer. 2001. The human estrogen receptor alpha dimer binds a single SRC-1 coactivator molecule with an affinity dictated by agonist structure. J. Mol. Biol. 306:433-442. [DOI] [PubMed] [Google Scholar]
- 31.Mattick, S., K. Glenn, G. de Haan, and D. J. Shapiro. 1997. Analysis of ligand dependence and hormone response element synergy in transcription by estrogen receptor. J. Steroid Biochem. Mol. Biol. 60:285-294. [DOI] [PubMed] [Google Scholar]
- 32.McEwan, I. J. 2000. Gene regulation through chromatin remodelling by members of the nuclear receptor superfamily. Biochem. Soc. Trans. 28:369-373. [PubMed] [Google Scholar]
- 33.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 34.Morcillo, P., C. Rosen, M. K. Baylies, and D. Dorsett. 1997. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 11:2729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller, W. G., D. Walker, G. L. Hager, and J. G. McNally. 2001. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J. Cell Biol. 154:33-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olefsky, J. M. 2001. Nuclear receptor minireview series. J. Biol. Chem. 276:36863-36864. [DOI] [PubMed] [Google Scholar]
- 38.Reese, J. C., and B. S. Katzenellenbogen. 1992. Examination of the DNA-binding ability of estrogen receptor in whole cells: implications for hormone-independent transactivation and the actions of antiestrogens. Mol. Cell. Biol. 12:4531-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinett, C. C., A. Straight, G. Li, C. Willhelm, G. Sudlow, A. Murray, and A. S. Belmont. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rollins, R. A., P. Morcillo, and D. Dorsett. 1999. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152:577-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenfeld, M. G., and C. K. Glass. 2001. Coregulator codes of transcriptional regulation by nuclear receptors. J. Biol. Chem. 276:36865-36868. [DOI] [PubMed] [Google Scholar]
- 42.Sabbah, M., K. I. Kang, L. Tora, and G. Redeuilh. 1998. Oestrogen receptor facilitates the formation of preinitiation complex assembly: involvement of the general transcription factor TFIIB. Biochem. J. 336:639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitz, A., C. Coulondre, and J. H. Miller. 1978. Genetic studies of the lac repressor. V. Repressors which bind operator more tightly generated by suppression and reversion of nonsense mutations. J. Mol. Biol. 123:431-454. [DOI] [PubMed] [Google Scholar]
- 44.Schodin, D. J., Y. Zhuang, D. J. Shapiro, and B. S. Katzenellenbogen. 1995. Analysis of mechanisms that determine dominant negative estrogen receptor effectiveness. J. Biol. Chem. 270:31163-31171. [DOI] [PubMed] [Google Scholar]
- 45.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 46.Spencer, T. E., G. Jenster, M. M. Burcin, C. D. Allis, J. Zhou, C. A. Mizzen, N. J. McKenna, S. A. Onate, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194-198. [DOI] [PubMed] [Google Scholar]
- 47.Stenoien, D. L., M. G. Mancini, K. Patel, E. A. Allegretto, C. L. Smith, and M. A. Mancini. 2000. Subnuclear trafficking of estrogen receptor-alpha and steroid receptor coactivator-1. Mol. Endocrinol. 14:518-534. [DOI] [PubMed] [Google Scholar]
- 48.Stenoien, D. L., A. C. Nye, M. G. Mancini, K. Patel, M. Dutertre, B. W. O'Malley, C. L. Smith, A. S. Belmont, and M. A. Mancini. 2001. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor alpha-coactivator complexes in living cells. Mol. Cell. Biol. 21:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Straight, A. F., J. W. Sedat, and A. W. Murray. 1998. Time-lapse microscopy reveals unique roles for kinesin during anaphase in budding yeast. J. Cell Biol. 143:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tumbar, T., and A. S. Belmont. 2001. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat. Cell Biol. 3:134-139. [DOI] [PubMed] [Google Scholar]
- 51.Tumbar, T., G. Sudlow, and A. S. Belmont. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117-2130. [DOI] [PubMed] [Google Scholar]
- 54.Wrenn, C. K., and B. S. Katzenellenbogen. 1993. Structure-function analysis of the hormone binding domain of the human estrogen receptor by region-specific mutagenesis and phenotypic screening in yeast. J. Biol. Chem. 268:24089-24098. [PubMed] [Google Scholar]
- 55.Ye, Q., Y.-F. Hu, H. Zhong, A. C. Nye, A. S. Belmont, and R. Li. 2001. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J. Cell Biol. 155:911-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, L., L. A. Annab, C. A. Afshari, W. H. Lee, and T. G. Boyer. 2001. BRCA1 mediates ligand-independent transcriptional repression of the estrogen receptor. Proc. Natl. Acad. Sci. USA 98:9587-9592. [DOI] [PMC free article] [PubMed] [Google Scholar]