Abstract
To identify novel genes involved in DNA double-strand break (DSB) repair, we previously isolated Schizosaccharomyces pombe mutants which are hypersensitive to methyl methanesulfonate (MMS) and synthetic lethals with rad2. This study characterizes one of these mutants, rad60-1. The gene that complements the MMS sensitivity of this mutant was cloned and designated rad60. rad60 encodes a protein with 406 amino acids which has the conserved ubiquitin-2 motif found in ubiquitin family proteins. rad60-1 is hypersensitive to UV and γ rays, epistatic to rhp51, and defective in the repair of DSBs caused by γ-irradiation. The rad60-1 mutant is also temperature sensitive for growth. At the restrictive temperature (37°C), rad60-1 cells grow for several divisions and then arrest with 2C DNA content; the arrested cells accumulate DSBs and have a diffuse and often aberrantly shaped nuclear chromosomal domain. The rad60-1 mutant is a synthetic lethal with rad18-X, and expression of wild-type rad60 from a multicopy plasmid partially suppresses the MMS sensitivity of rad18-X cells. rad18 encodes a conserved protein of the structural maintenance of chromosomes (SMC) family (A. R. Lehmann, M. Walicka, D. J. Griffiths, J. M. Murray, F. Z. Watts, S. McCready, and A. M. Carr, Mol. Cell. Biol. 15:7067-7080, 1995). These results suggest that S. pombe Rad60 is required to repair DSBs, which accumulate during replication, by recombination between sister chromatids. Rad60 may perform this function in concert with the SMC protein Rad18.
DNA double-strand breaks (DSBs) cause cellular lethality if not repaired and can lead to chromosomal aberrations such as deletions or translocations if repaired improperly. DSBs are repaired mainly through two mechanisms in eukaryotes: homologous recombination (HR) and nonhomologous end joining (NHEJ) (20). Studies of X-ray-sensitive mutants of the budding yeast Saccharomyces cerevisiae identified a group of genes involved in HR that contribute to the process of DSB repair (9). These genes include RAD50, MRE11, XRS2, RAD51, RAD52, RAD54, RAD55, and RAD57. The first three of these genes are involved in processing DSB ends, and the other genes facilitate DNA strand exchange. The Rad51 protein is structurally and functionally homologous to the Escherichia coli RecA protein (44). In S. cerevisiae, HR is the major pathway for DSB repair and NHEJ plays a role in tolerance to ionizing radiation only when there is a deficiency in HR (45). In contrast, both HR and NHEJ play important roles in the repair of ionizing-radiation-induced DNA damage in vertebrates (20).
In the fission yeast Schizosaccharomyces pombe, the rad32, rhp51, rad22, rhp54, rhp55, and rhp57 genes are homologous to the S. cerevisiae genes MRE11, RAD51, RAD52, RAD54, RAD55, and RAD57, respectively, and strains with a mutation in any of these genes are hypersensitive to ionizing radiation (23, 33, 34, 40, 51, 53). In addition to these genes, the rad18 and rad21 genes have been implicated in DSB repair (25). rad21 is essential for growth (3) and homologous to the S. cerevisiae MCD1/SCC1/RHC21 gene. rad18 is essential for growth and encodes a protein that belongs to the structural maintenance of chromosomes (SMC) superfamily (26). The SMC family proteins are structurally related to each other and include N- and C-terminal globular domains, which possess Walker A and B motifs for ATP binding, respectively, and two central coiled-coil segments, which are separated by a flexible hinge (for reviews, see references 12 and 48). The SMC family proteins of eukaryotes form heterodimers with several non-SMC proteins. The SMC protein complexes regulate higher-order chromosome structures involving chromosome cohesion, condensation, and dosage compensation. The Rad18 protein of S. pombe forms a complex with the SMC family protein Spr18 and five other unidentified proteins (8).
The rad18-X mutant is sensitive to UV irradiation and removes UV-induced DNA damage less efficiently than does the wild type (26). rad18 is not epistatic to the conserved nucleotide excision repair pathway but is involved in a secondary nucleotide excision repair pathway that requires the rad2 and rhp51 genes. rad2 encodes a structure-specific endonuclease homologous to mammalian Fen-1, which is required for Okazaki fragment maturation (37), and is involved in a second excision repair pathway, in which repair is initiated by UV-damage endonuclease in S. pombe (57). Nevertheless, rad18-X is not epistatic to UV-damage endonuclease, and hence, rad18 is implicated in both a DNA damage tolerance pathway and the second excision repair pathway (26, 36). A rad18 mutant, the rad18-74 mutant, was isolated in a genetic screen for mutants defective in DNA damage checkpoint control; this mutant is impaired in the maintenance of checkpoint arrest (55). Therefore, rad18 is also implicated in a DNA damage checkpoint.
This study is part of our ongoing effort to characterize novel genes involved in recombinational repair in S. pombe. Our strategy is based on the fact that strains with mutations in recombination genes are often synthetic lethals with rad2 (37, 51). In an earlier study, seven methyl methanesulfonate (MMS)-sensitive mutants were isolated which were synthetical lethals with rad2. The rhp57 gene, a homolog of S. cerevisiae RAD57, complements the MMS sensitivity of one of the previously identified mutants (53). In this study, we isolated a second gene, designated rad60, that complements one of the previously identified MMS-sensitive mutants. rad60 interacts genetically with rad18. The rad60-1 mutant is defective in DSB repair, and the rad60 gene is essential for growth. Hence, rad60 appears to play a role in a cellular process that is required for both DSB repair and normal DNA replication. rad60 may act in concert with rad18.
MATERIALS AND METHODS
S. pombe media and methods.
S. pombe cells were grown in yeast extract-supplemented (YES) medium or Edinburgh minimal medium (EMM), and standard genetic and molecular procedures were employed as described previously (32). To measure the sensitivity of cells to γ rays, exponential-phase culture was harvested and irradiated with γ rays from a 60Co source at a dose of 21 Gy/min. After irradiation, appropriately diluted samples were spread on YES plates and incubated at 30°C for 5 days, and the colonies were counted. To measure the sensitivity of cells to UV, exponential-phase cultures diluted to appropriate concentrations were spread on YES plates. The plates were irradiated with the indicated doses of UV light and incubated at 30°C for 5 days, and the colonies were counted.
S. pombe strains and plasmids.
The S. pombe strains used in this study are listed in Table 1. Strains 972, NCYC1979, and NCYC1982 were obtained from the National Collection of Yeast Cultures (NCYC; Norwich, United Kingdom). Strain HM248 (39) was kindly provided by O. Niwa. All other strains were constructed for this study. pUR19 (1) and pJK148 (21) were described previously. pU19 was constructed by deleting the ClaI fragment containing ars1 from pUR19.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| 972 | h− | NCYC |
| NCYC1979 | h−rad18-X | NCYC |
| NCYC1982 | h−rad21-45 | NCYC |
| HM248 | h+his2 ade6-M210 Ch16 | Niwa et al. (39) |
| MP2 | h+ | This work |
| MP10 | h−leu1-32 ura4-D18 | This work |
| MP11 | h+leu1-32 ura4-D18 | This work |
| MPR101 | h−leu1-32 ura4-D18 rad60-1 | This work |
| MPR104 | h+rad60-1 | This work |
| MPR105 | h+leu1-32 ura4-D18 rad18-X | This work |
| MPR111 | h+leu1-32 ura4-D18 rad60-1 | This work |
| MPR113 | h−leu1-32 ura4-D18 rad60-1 rad2::ura4+ | This work |
| MPR114 | h+leu1-32 ura4-D18 rad60-1 | This work |
| MPR115 | h+leu1-32 ura4-D18 rad2::ura4+ | This work |
| MPR116 | h−leu1-32 ura4-D18 | This work |
| MPR117 | h−smt-0 leu1-32 ura4-D18 | This work |
| MPR118 | h−smt-0 leu1-32 ura4-D18 rad60-1 | This work |
| MPR119 | h−smt-0 leu1-32 ura4-D18 rad51::ura4+ | This work |
| MPR120 | h−smt-0 leu1-32 ura4-D18 rad60-1 rhp51::ura4+ | This work |
| MPR121 | h+mat1PΔ17::LEU2 leu1-32 ura4-D18 rad60-1 rhp51::ura4+ | This work |
| MPR122 | h+mat1PΔ17::LEU2 leu1-32 ura4-D18 rad60-1 | This work |
| MPR123 | h+mat1PΔ17::LEU2 leu1-32 ura4-D18 | This work |
| MPR124 | h+mat1PΔ17::LEU2 leu1-32 ura4-D18 rhp51::ura4+ | This work |
| MPR125 | h+leu1-32 ura4-D18 ade6-M210 Ch16 | This work |
| MPR126 | h+leu1-32 ura4-D18 ade6-M210 rad60-1 Ch16 | This work |
| MPD1 | h+/h−leu1-32/leu1-32 ura4-D18/ura4-D18 his7-366/+ ade6-M210/ade6-M216 | This work |
| MPDR101 | h+/h−leu1-32/leu1-32 ura4-D18/ura4-D18 his7-366/+ ade6-M210/ade6-M216 rad60::ura4/+ | This work |
Chromosome segregation assay.
Chromosome segregation was measured by monitoring the transmission of linear minichromosome Ch16 (39) carrying the ade6-M216 allele, which complements the ade6-M210 allele of chromosome III. Cells that have lost Ch16 appear pink in color on medium containing a low concentration of adenine. To determine the rate of minichromosome loss, rad60-1 (MPR126) or rad60+ (MPR125) cells carrying Ch16 were cultured on YES plates at 30°C for 3 days. A single colony was used to inoculate 5 ml of YES medium and cultured at 30°C until cell density reached 3 × 107 to 5 × 107/ml. The cultures were diluted and spread on YES plates containing 10 μg of adenine sulfate/ml. The plates were incubated at 30°C for 5 days, and total and pink colonies were counted. The rate of minichromosome loss (p) was determined by the equation 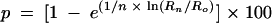 , where R0 and Rn are the percentages of Ade+ cells at generations 0 and n after transfer to nonselective medium, respectively (28). In this case, R0 was 100, since each of the cultures was derived from a single cell.
, where R0 and Rn are the percentages of Ade+ cells at generations 0 and n after transfer to nonselective medium, respectively (28). In this case, R0 was 100, since each of the cultures was derived from a single cell.
Cloning of the rad60 gene.
rad60-1 cells were transformed with the S. pombe genomic library constructed from vector pUR19 (1) and spread on EMM plates containing leucine (200 μg/ml) and MMS (0.004%). Transformants were examined for plasmid-dependent MMS resistance. Plasmids which complemented the MMS sensitivity of rad60-1 cells were isolated, transformed into E. coli DH5α cells, and recovered from the transformant.
Cloning of the rad60 cDNA.
The rad60 cDNA was constructed with RNA from the wild-type strain 972 by using the RNeasy mini kit (Qiagen) and amplified by using the RNA LA PCR kit, version 1.1 (Takara Shuzo). Primers had the sequences 5′-TCACATATGGACAACCTAGATGAAG-3′ and 5′-TCAGGATCCTTAATCCAAAACAACACTAACTTG-3′, corresponding to the 5′ and 3′ ends of the rad60 coding region, respectively. The PCR fragment was cloned into pUC19 after cleavage with NdeI and BamHI, whose recognition sites were engineered near the 5′ ends of the PCR primers.
Disruption of the rad60 gene.
pUC118 carrying the 2.6-kbp BamHI-NruI region of the rad60 gene was used to generate a construct for gene disruption. The 0.8-kbp XbaI-SphI fragment of the coding region was replaced with the 1.8-kbp HindIII fragment containing the ura4 gene from pREP2 (29). The 3.6-kbp fragment for gene disruption was released from the plasmid by digestion with SacI and SalI and used to transform the MPD1 cells and create the strain MPDR101. Gene disruption was confirmed by Southern blot analysis, and MPDR101 cells heterozygous for the disruption were sporulated and subjected to tetrad analysis.
Recovery of the rad60-1 mutant allele.
The rad60-1 mutant gene was recovered by the eviction method (56). pU19 carrying the 3.6-kbp BamHI fragment including the wild-type rad60 gene was linearized at the unique MunI site in the rad60 flanking region and used to transform the rad60-1 mutant. Genomic DNA of the resulting transformant was extracted, digested with SalI, ligated, and used to transform E. coli strain DH5α. The 3.6-kbp BamHI fragment of the recovered plasmid was subcloned into pU19, and the nucleotide sequence was analyzed.
Pulsed-field gel electrophoresis.
DNA plugs were prepared as described previously (46), with the exception that cells were lysed by incubation with Zymolyase 100T (Seikagaku Corporation) (0.5 mg/ml) for 30 min. Pulsed-field gel electrophoresis was carried out with 0.6% chromosomal-grade agarose (Bio-Rad) in 0.5× TAE buffer (20 mM Tris-acetate, 0.5 mM EDTA) by using a CHEF Mapper apparatus (Bio-Rad). The settings were as follows: 2 V/cm; pulse time, 30-min; angle, 106°; 72 h.
Localization of Rad60 protein in the cell.
A plasmid carrying a gene encoding an enhanced green fluorescence protein (EGFP)-Rad60 fusion protein (EGFP-rad60 fusion gene) was constructed as follows. An NdeI site was engineered at the first ATG codon of the rad60 gene with a QuickChange site-directed mutagenesis kit (Stratagene). The NdeI cassette of the EGFP-encoding gene from pGEM-T-EGFP (5) was inserted into the NdeI site to create an EGFP-rad60 fusion. The NcoI-SalI region of pREP42EGFPN (5) was replaced with the NcoI-SalI fragment of the EGFP-rad60 fusion gene. The resulting plasmid, pErad60, was transformed into wild-type S. pombe MP11. The transformants were recovered on an EMM plate containing leucine (200 μg/ml) and thiamine (15 μM). Expression of the EGFP-Rad60 fusion protein was induced in EMM medium containing leucine (200 μg/ml) for 20 h. Cells were fixed with 2.5% glutaraldehyde, treated with 0.1% sodium borohydride three times for 5 min each time, stained with 1 μg of 4′,6′-diamidino-2-phenylindole (DAPI)/ml, and observed under an epifluorescence microscope (38).
RESULTS
rad60-1 cells are defective in DNA repair and temperature sensitive for growth.
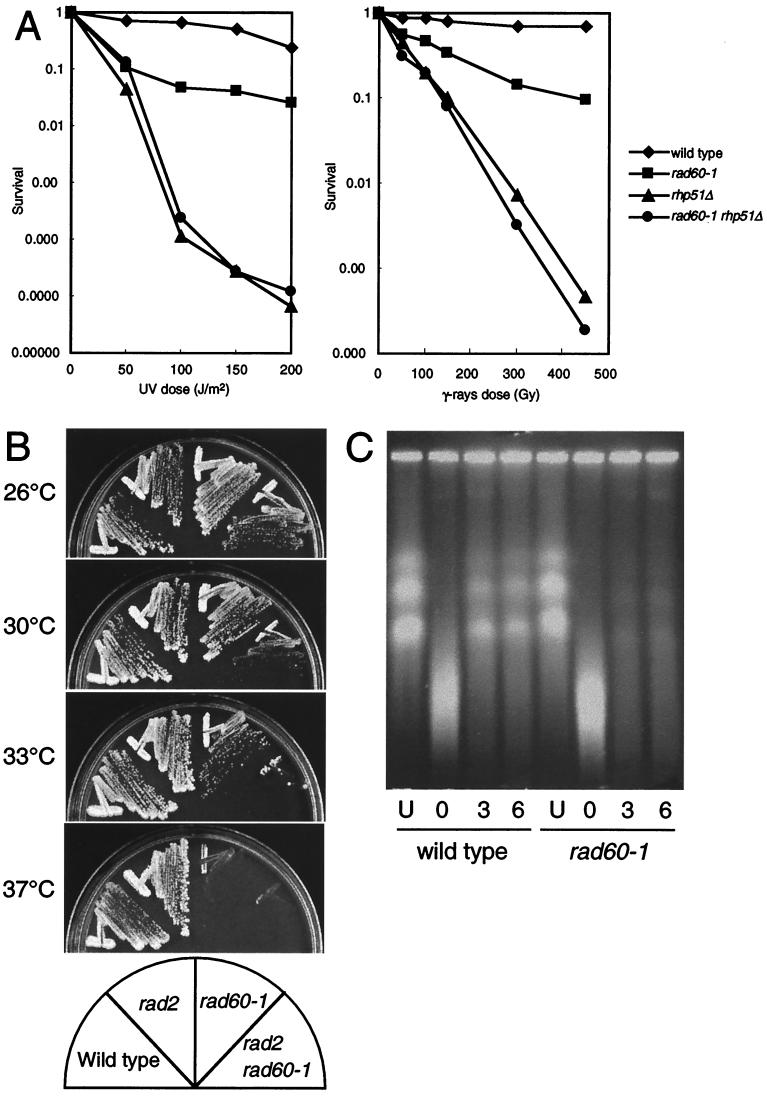
This study characterizes a previously identified MMS- and γ-ray-sensitive mutant of S. pombe that is a synthetic lethal with rad2 (53). This mutant, designated rad60-1, was repeatedly backcrossed with wild-type strains, and an MMS-sensitive segregant clone was selected. As shown in Fig. 1A, the rad60-1 mutant is more sensitive to DNA-damaging agents, UV and γ rays, than is the wild type. The rad60-1 mutant is temperature sensitive for growth on YES plates at 37°C but grows normally at 26°C.
FIG.1.
The rad60-1 mutant is defective in DSB repair. (A) Epistasis between rad60-1 and rhp51Δ. Midexponential phase cells were irradiated with UV or γ rays at the indicated doses, and relative plating efficiencies were determined. (B) Synthetic lethality of rad60-1 and rad2Δ. Cells of wild-type (MPR116), rad2 (MPR115), rad60-1 (MPR114), and rad60-1 rad2 (MPR113) strains were streaked on YES plates. The plates were incubated at the indicated temperature for 6 days and photographed. (C) Repair of DSBs in wild-type and rad60-1 cells. Genomic DNA of wild-type (MP2) and rad60-1 (MPR104) strains was subjected to pulsed-field gel electrophoresis. Samples were taken from unirradiated cells (U) or cells incubated in YES medium at 30°C for the indicated number of hours after irradiation with 500 Gy of γ rays.
A rad60-1 rad2 double mutant was obtained by crossing rad60-1 with rad2 and culturing the spores on YES plates at 26°C. The growth of the double mutant was examined at different temperatures (Fig. 1B). The rad60-1 rad2 double mutant grows at 26°C, but growth is severely impaired at 30°C and completely inhibited at 33 and 37°C. On the other hand, rad60-1 grows at 33°C, and the rad2 mutant grows at all temperatures tested. Thus, rad60-1 is a synthetic lethal with rad2 at 33°C and higher.
rad60-1 is epistatic to rhp51.
Homologous recombination is a major pathway for the repair of radiation-induced DSBs in S. pombe. rhp51, the S. pombe ortholog of S. cerevisiae RAD51, is a key player in this process (19, 33). Therefore, the relationship between rad60 and rhp51 was examined. As shown in Fig. 1A, the rhp51 mutant is more sensitive to UV and γ radiation than is the rad60-1 mutant, but the rad60-1 rhp51 double mutant is as sensitive as the single rhp51 mutant. These results indicate that rad60-1 is epistatic to rhp51 with respect to DNA repair.
The rad60-1 mutant is defective in repairing DSBs.
rad60-1 is hypersensitive to MMS and γ rays, suggesting that the rad60 gene is involved in repairing DSBs. This possibility was tested by directly monitoring the efficiency of DSB repair in this mutant. DSBs were induced by γ-irradiation at 500 Gy, and DNA isolated at various times after irradiation was analyzed by pulsed-field gel electrophoresis. In wild-type cells, fragmented chromosomes were repaired and undamaged chromosomes were detected within 3 h after irradiation; in contrast, fragmented chromosomes were repaired very inefficiently in rad60-1 cells (Fig. 1C). This result indicates that rad60-1 cells are defective in repairing DSBs.
The rad60-1 mutant is defective in maintaining chromosome structure.
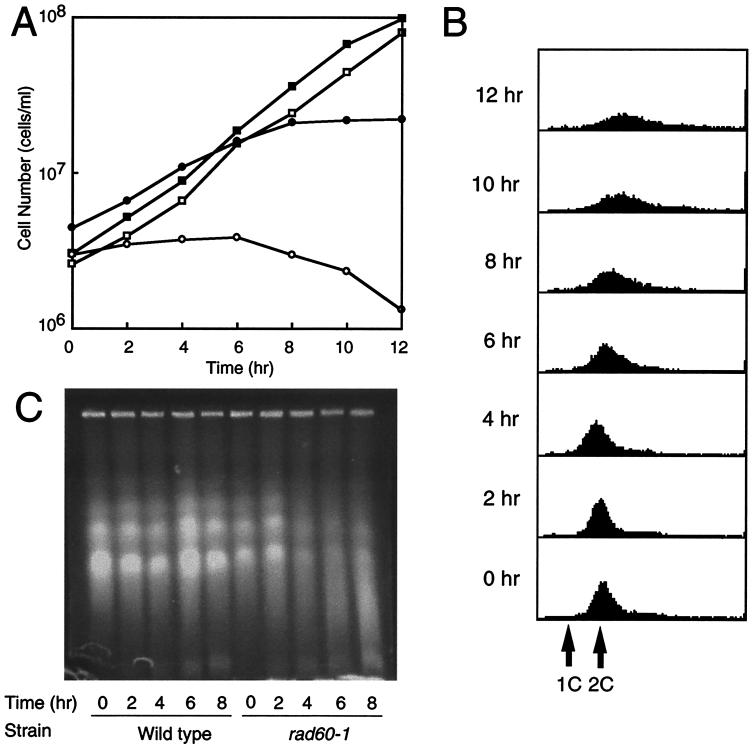
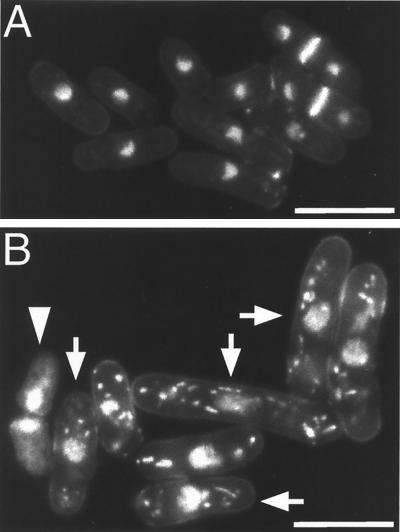
Experiments were carried out to determine the cause of the growth defect in the rad60-1 mutant. rad60-1 cells were grown logarithmically at 26°C and shifted to the restrictive temperature of 37°C, and cell growth and viability (measured in CFU) were monitored (Fig. 2A). The rad60-1 mutant stopped growing 8 h after the temperature shift, at which time the cell number had increased 4.7-fold. The number of CFU did not increase appreciably after the temperature shift and started to decrease after approximately 6 h at 37°C. Consequently, after 8 h at the restrictive temperature, more than 80% of the cells lost their ability to form colonies. rad60-1 cells growing at 26°C or for 8 h at 37°C were fixed, stained with DAPI to visualize chromosomal DNA, and observed under an epifluorescence microscope. Hemisphere morphology of the nuclear chromosomal domain (i.e., interphase morphology) was observed in most of the rad60-1 cells at 26°C (Fig. 3A). The nuclear chromosomal domain of most of the rad60-1 cells grown at 37°C was more diffuse than that of cells grown at 26°C (compare Fig. 3A and B), and nearly half of the cells had aberrant morphology (Fig. 3B). In these cells, the chromosomal domain was extended and often dappled. This result suggests that rad60 is required to maintain proper chromosome structure. In rad60-1 cells grown for 8 h at 37°C, about 5% had a “cut” phenotype (Fig. 3B), in which a septum forms without complete chromosomal segregation (13), and about 12% had a biased nuclear position which would occur when cut cells detached. Therefore, rad60-1 appears to be deficient in preventing septum formation when DNA replication and/or chromosomal segregation is incomplete.
FIG. 2.
Growth arrest of rad60-1 cells at 37°C. Wild-type (MP2) or rad60-1 (MPR104) cells growing exponentially at 26°C in YES medium were shifted to 37°C. (A) Cell number (filled symbols) and CFU (open symbols) were measured for wild-type (squares) or rad60-1 (circles) cells at the indicated time points. (B) Cells of the rad60-1 strain were fixed at the indicated time points, stained with propidium iodide, and processed by FACS to determine DNA content. (C) Genomic DNA of wild-type and rad60-1 strains was subjected to pulsed-field gel electrophoresis. Samples were taken at the indicated time points.
FIG. 3.
Nuclear morphology of the rad60-1 cells at 37°C. rad60-1 cells growing at 26°C (A) or after 8 h at 37°C (B) were fixed with 2.5% glutarardehyde and stained with 1 μg of DAPI/ml and 20 μg of calcofluor white/ml. Fluorescence images obtained with an epifluorescence microscope are shown. In panel B, arrows indicate cells with disorganized nuclear morphology and an arrowhead indicates a cut cell. Scale bars, 10μ m.
The DNA content of rad60-1 cells was measured in cells incubated for different lengths of time at 37°C by using a fluorescence-activated cell sorter (FACS) (Fig. 2B). The DNA content of most cells was approximately 2C until at least 6 h at the restrictive temperature. At later time points, DNA content increased slightly and the peak of the FACS analysis broadened. This change was probably due to an increase in mitochondrial DNA or in cell size. Most rad60-1 cells divided several times before arresting growth at 37°C (Fig. 2A), so the 2C DNA content at arrest indicates that DNA synthesis proceeds relatively normally in these cells.
The rad60-1 mutant accumulates DSBs at the restrictive temperature.
Chromosomal DNA in wild-type and rad60-1 cells was examined by pulsed-field gel electrophoresis after incubation at 37°C (Fig. 2C). In wild-type cells, three chromosomes were detected at 37°C. In contrast, the three chromosomes in rad60-1 cells decreased in size and became a smeared band of DNA after 6 or 8 h at 37°C. This result indicates that DSBs accumulate in rad60-1 cells at 37°C and that chromosomes become fragmented.
Elevated rate of minichromosome loss in the rad60-1 mutant.
The results presented above suggest that the rad60-1 mutant is defective in maintaining proper chromosome structure at the restrictive temperature. The role played by rad60 in chromosome stability was examined further by measuring the rate of loss of the nonessential minichromosome Ch16 in rad60-1 cells. Ch16 was lost at a much higher rate in rad60-1 cells (0.17% per generation) than in wild-type cells (<0.005% per generation) at 30°C. Incubation of rad60-1 cells at 37°C for 6 h caused a fourfold increase in the fraction of cells without Ch16 (data not shown). This result indicates that chromosome loss occurs more frequently in rad60-1 cells than in wild-type cells and that chromosome loss in rad60-1 cells is elevated at the restrictive temperature.
Cloning and sequencing of the rad60 gene.
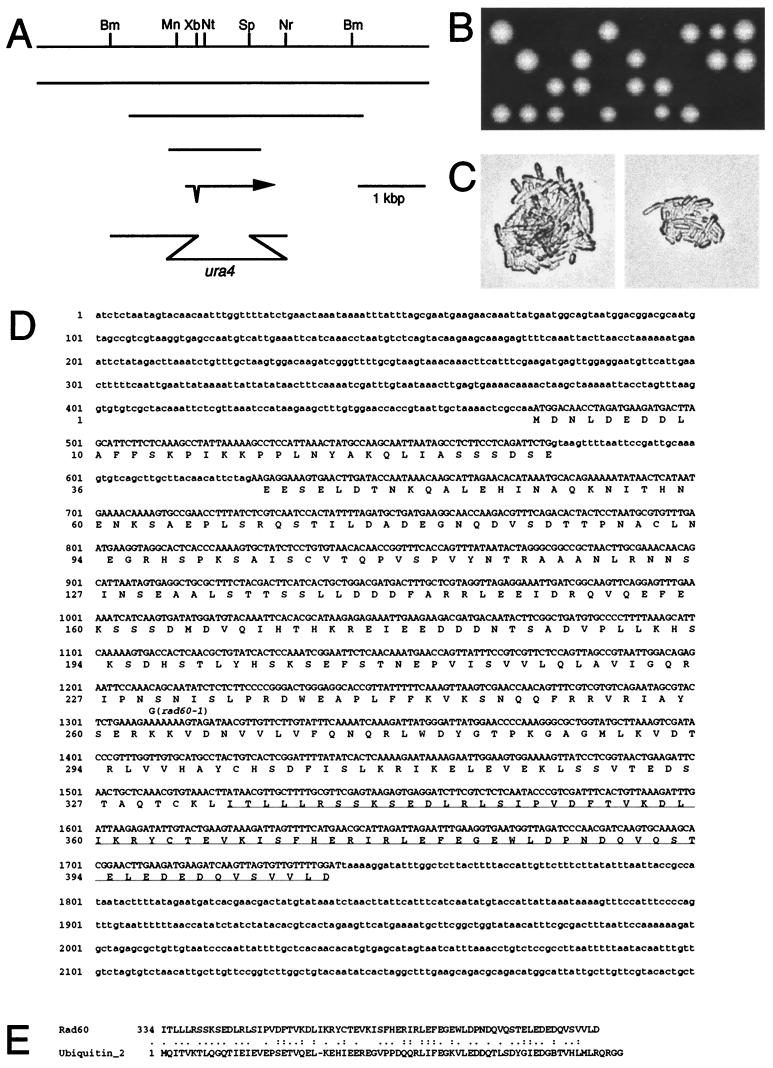
The rad60 gene was cloned by complementing the MMS sensitivity of the rad60-1 mutant. rad60-1 cells were transformed with an S. pombe genomic library, and plasmids were recovered from MMS-resistant transformants and characterized in E. coli. Three complementing plasmids carried an overlapping genomic region from S. pombe (Fig. 4A). The nucleotide sequence of a 2.2-kbp region encompassing an MunI site and an NruI site was determined (Fig. 4D). Putative exons and introns were identified and confirmed by amplifying and analyzing rad60 cDNA by PCR (Fig. 4D). The cloned region includes a sequence coding for a protein of 406 amino acids with a calculated molecular weight of 46,077. This nucleotide sequence is identical to a portion of cosmid c1920 (accession number AL122033 in EMBL, GenBank, and DDBJ), and the predicted amino acid sequence matches that of SPBC1921.02 (EMBL accession number T397868). No homologous proteins were found by the SSEARCH program of DDBJ (http://www.ddbj.nig.ac.jp/). However, a search for amino acid sequence motifs in the PROSITE profile library (http://motif.genome.ad.jp/) (14) revealed a significant match between amino acids 336 and 406 of Rad60 and the ubiquitin-2 motif (PROSITE ID PS50053) (Fig. 4E).
FIG.4.
Physical map of the rad60 genomic region and disruption and nucleotide sequence of the rad60 gene. (A) Restriction map of the rad60 region. Abbreviations for restriction enzymes are as follows: Bm, BamHI; Mn, MunI; Xb, XbaI; Nt, NotI; Sp, SphI; Nr, NruI. Lines represent three overlapping genomic fragments which complement rad60-1. The arrow represents the rad60 coding region. The single intron is represented by a protrusion downward within the arrow. The 2.6-kbp BamHI-NruI region of the rad60 gene was used to disrupt the chromosomal rad60 gene. To prepare this construct, the 0.8-kbp XbaI-SphI segment of the coding region was replaced with a 1.8-kbp HindIII fragment containing the ura4 gene. (B) The diploid strain MPDR101 (heterozygous for rad60 disruption) was sporulated and subjected to tetrad analysis. The segregants from the dissected spores were grown on a YES plate at 30°C for 7 days and photographed. (C) Procedures were carried out as described for panel B, with the exception that the segregants from the dissected spores were grown for 2 days. Images of microcolonies of two putative rad60::ura4+ segregants are shown. (D) Nucleotide and predicted amino acid sequences of the rad60 gene are shown. The exon sequence is shown in uppercase, and the intron sequence is shown in lowercase. The amino acid sequence with homology to the ubiquitin-2 motif is underlined. (E) The sequence of the C-terminal 73 amino acids of the Rad60 protein was aligned with the ubiquitin-2 motif. Identical amino acids are shown with a double dot, and similar amino acids are shown with a single dot.
The rad60-1 allele and its flanking genomic region were recovered from the rad60-1 mutant by the eviction method, and the nucleotide sequence was determined. rad60-1 has a single A-to-G nucleotide change altering the AAA codon for lysine 263 to a GAA codon for glutamic acid. This nucleotide substitution was reconstructed in a chimeric rad60 gene in which the 0.6-kbp NotI-SphI region of the wild-type gene was replaced with the corresponding region of the rad60-1 gene. The chimeric construct was cloned into the S. pombe integrating vector pJK148 and linearized at the unique NdeI site in the leu1 gene. The linear plasmid was transformed into the diploid strain MPDR101, which is heterozygous for rad60::ura4+, such that the plasmid was integrated into the leu1 gene. The diploid transformant was sporulated and subjected to tetrad analysis. Leu+ Ura+ segregants were obtained which carry the rad60::ura4+ allele and the chimeric rad60-1 construct described above. These cells do not grow at 37°C or in the presence of 0.005% MMS at 26°C. This result demonstrates that the lysine-to-glutamic acid substitution at codon 263 of rad60 is sufficient to cause the abnormal phenotype of rad60-1 cells.
rad60 is an essential gene.
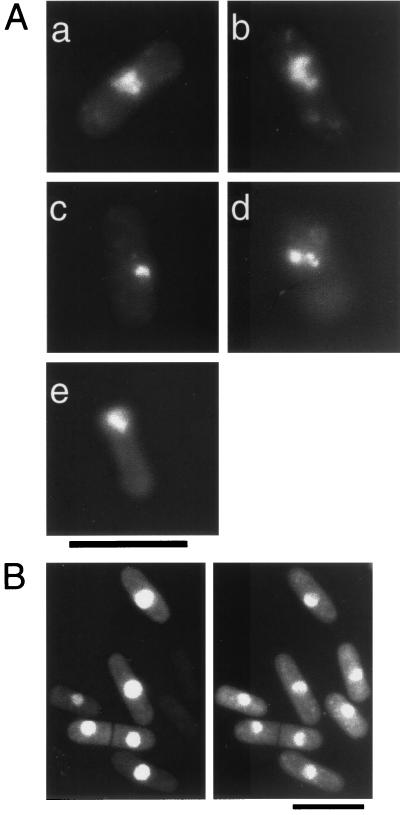
The 0.8-kbp XbaI-SphI region containing about two-thirds of the rad60 coding region was replaced with the DNA fragment containing the ura4 gene (Fig. 4A), and the resulting DNA fragment was used to disrupt one of the chromosomal rad60 genes in a diploid strain. The heterozygous diploid cells obtained were sporulated and subjected to tetrad analysis. Twelve asci were dissected and allowed to germinate on YES medium at 30°C, and only two viable segregants arose from each of the 12 asci (Fig. 4B). All 24 viable segregants required uracil for growth, indicating that these segregants did not carry the rad60::ura4+ allele. These results indicate that rad60::ura4+ cells are lethal and that the rad60 gene is essential for growth. Of the 24 nonviable putative rad60::ura4+ segregants, 21 formed microcolonies of more than 10 cells and then stopped growing (Fig. 4C); the remaining 3 segregants stopped growing at the two-cell stage. One possible explanation for the limited and variable amount of growth of these cells is that residual Rad60 in the mutant spores supports several cycles of cell division. In most of the rad60::ura4+ cells, the morphology of the nuclear chromosomal domain was aberrant. The following morphologies were observed: amorphously extended (37%; Fig. 5Aa and b), shrunk (17%; Fig. 5Ac), fragmented (9%; Fig. 5Ad), and biased in position (27%; Fig. 5Ae). The latter morphology could result from the detachment of cut cells. This nuclear morphology appears to be similar to but more extreme than the morphology of rad60-1 cells at 37°C.
FIG. 5.
Nuclear morphology of the cells from the rad60Δ microcolonies and cellular localization of the EGFP-Rad60 fusion protein. (A) The diploid strain MPDR101 was sporulated and subjected to tetrad analysis, and segregants from the dissected spores were grown on a YES plate at 30°C for 2 days. Cells of putative rad60Δ microcolonies were collected, fixed with 70% ethanol, and stained with DAPI (1 μg/ml). Panels a and b show fluorescence images of representative cells with extended, aberrantly shaped nuclear chromosomal domains. Other aberrant morphologies shown are as follows: shrunk (panel c), fragmented (panel d), and biased in position (panel e). (B) Cells of the wild type (MP11) carrying pErad60 were fixed, stained with DAPI (1 μg/ml), and observed under an epifluorescence microscope. Fluorescence images of EGFP (left) and DAPI (right) are shown. Scale bars, 10 μm.
Rad60 protein is localized to the nucleus.
Cellular localization of the Rad60 protein was studied by examining S. pombe expressing a fusion protein of Rad60 and EGFP. When the EGFP-rad60 fusion gene was expressed from the rad60 promoter, no fluorescent signal was detected (data not shown). However, fluorescence was detected when the EGFP-rad60 fusion gene was expressed from the modified nmt1 promoter on pREP42 (2, 29). In the absence of thiamine, the EGFP signal colocalized with DAPI-stained chromosomal DNA (Fig. 5B). The EGFP-rad60 fusion gene produces a functional protein that complements the MMS sensitivity and temperature sensitivity of rad60-1 cells (data not shown). Thus, these results indicate that Rad60 protein is localized to the nucleus and suggest that Rad60 carries out its biological function in the nucleus.
Evidence that the rad60 gene is functionally related to the rad18 gene.
Two previously identified S. pombe rad genes, rad18 and rad21, and rad60 are required for DSB repair and are essential for growth. The relationship between rad60, rad18, and rad21 was examined by constructing double mutants between rad60 and these two genes. rad60-1 was crossed with rad18-X or rad21-45, and the spores were subjected to tetrad analysis at 26°C. In the cross between rad60-1 and rad18-X, the numbers of viable colonies were 4, 3, 2, and 1 for 1, 8, 3, and 0 tetrads, respectively. Each clone with a four-viable-tetrad showed parental ditype segregation, and all four segregants were hypersensitive to MMS. Each of the three-viable-colony tetrads had one wild-type segregant. One of the two-viable-colony tetrads had two wild-type segregants and is likely to be a nonparental ditype with two nonviable rad60-1 rad18-X segregants. These results strongly support the conclusion that the rad60-1 rad18-X double mutant is nonviable. In contrast, the rad60-1 rad21-45 double mutant is viable.
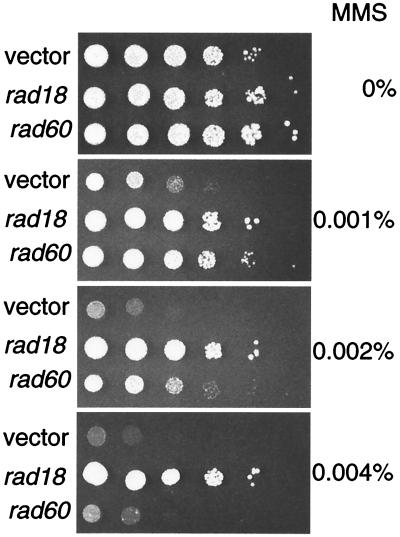
When the rad60 gene was expressed in a rad18-X background from a multicopy plasmid, the MMS hypersensitivity of rad18-X was partially suppressed (Fig. 6). This result suggests that the rad60 and rad18 genes are functionally related. Expression of rad60 from a multicopy plasmid did not suppress the leaky growth defect of the rad18-X mutant at 37°C (data not shown).
FIG. 6.
Suppression of rad18-X by a multicopy plasmid expressing rad60. rad18-X cells (MPR105) carrying the pUR19 vector or pUR19 with the rad18 or the rad60 gene were grown to saturation on EMM medium with leucine (200 μg/ml). Serial 10-fold dilutions were spotted on YES plates with the indicated concentrations of MMS. The plates were incubated at 30°C for 3 days and photographed.
DISCUSSION
S. pombe radiation-sensitive mutants have mutations in genes involved in nucleotide excision repair, checkpoint control, and recombinational repair (for a review, see reference 25). Earlier studies identified five DNA repair mutants that are encoded by genes that are essential for growth in S. pombe: these essential genes are rad4/cut5, rad11, rad15, rad18, and rad21. rad4/cut5 is homologous to S. cerevisiae DPB11 and is required for DNA replication and checkpoint control (7, 43). rad11 encodes the largest subunit of replication protein A, which is essential for DNA replication (41). rad15 is an ortholog of S. cerevisiae RAD3, encoding a component of general transcription factor TFIIH which is required for nucleotide excision repair (35, 42). rad21 is an ortholog of S. cerevisiae MCD1/SCC1/RHC21 which encodes a subunit of the cohesin complex (11, 30, 50). rad18 encodes an SMC family protein (26) which forms a complex with another SMC family protein, Spr18, and five other unidentified proteins (8). This study describes rad60, another essential S. pombe gene that plays a role in DNA repair.
The rad60-1 mutation changes lysine 263 of the Rad60 protein to a glutamic acid residue. The rad60-1 mutant is sensitive to MMS and γ rays (Fig. 1A) and synergistically impaired in growth when combined with the rad2 mutation (Fig. 1B). These phenotypes suggest that rad60-1 is defective in DSB repair, because γ rays and MMS cause DSBs (4, 16). The rad2 gene is the S. pombe ortholog of the mammalian FEN-1 gene (37). The FEN-1 protein is thought to play a role in Okazaki fragment maturation by removing RNA from the RNA-DNA hybrid (27). In E. coli cells, DNA polymerase I encoded by polA performs an analogous role, and a polA recA double mutant is lethal (10, 31).
It has been postulated that a DSB forms in a polA mutant cell when a replication fork reaches a nick or gap in the template DNA, because this mutant has a defect in maturation of Okazaki fragments; it is possible that DNA recombination is required to repair the DSB and to restart replication in the polA mutant (24). Consistent with this idea, recB, recG, and ruv mutations are also synthetic lethals with polA (15, 17, 18, 31).
An analogous situation might occur in eukaryotic cells. In S. cerevisiae, mutations that disrupt the FEN-1 ortholog RAD27 are synthetically lethal with mutations in genes of the RAD52 epistasis group (6, 49, 52). In S. pombe, the rad2 rhp51, rad2 rhp54, rad2 rad32, and rad2 rhp57 double mutants are lethal (37, 51, 53). Therefore, the synergistic growth defect of rad60-1 and rad2 supports the hypothesis that rad60 is required for the repair of DSBs caused by the collapse of the replication fork in S. pombe.
DSBs in γ-irradiated rad60-1 cells are not repaired after further incubation (Fig. 1C), and this observation directly shows that rad60-1 is defective in DSB repair. The phenotype of rad60-1 also involves an elevated rate of minichromosome loss at 30°C (see “Elevated rate of minichromosome loss in the rad60-1 mutant” above); this defect may result from the failure to repair spontaneous DSBs in the minichromosome. An increased rate of minichromosome loss has been observed for other S. pombe DNA repair-deficient mutants (34, 37, 51, 55).
The fact that rad60 is essential for growth suggests that rad60 may play a role in some cellular process other than the repair of DNA lesions caused by exogenous DNA-damaging treatments. rad60-1 cells stop growing after 8 h at the restrictive temperature of 37°C (Fig. 2A). After 8 h at 37°C, chromosomes become highly fragmented (Fig. 2C) and cells arrest with 2C DNA content (Fig. 2B). These results suggest that in the absence of functional Rad60 protein, spontaneous DSBs occur during replication, remain unrepaired, and cause DNA fragmentation. Nevertheless, DNA replication proceeds relatively normally, with doubling of DNA content to 2C. The nuclear chromosomal domain appears diffuse and has aberrant morphology in the rad60-1 cells grown at 37°C (Fig. 3B), which may be the consequence of accumulating DSBs. DSBs may interfere with the compaction of interphase chromosomes and cause a diffuse and apparently aberrantly shaped nuclear chromosomal domain to exist. Extensive minichromosome loss (see “Elevated rate of minichromosome loss in the rad60-1 mutant” above) could also be the consequence of DSBs. Many rad60-1 cells may undergo checkpoint arrest, because approximately 70% of the cells did not form a septum after 8 h at 37°C, and a small portion of the cells (17%) seem to escape checkpoint arrest, because they develop the morphology of an aberrant mitosis (i.e., cut or biased nuclear position). Spores carrying a deletion of the rad60 gene germinate and in most cases cease to grow after forming a microcolony of 10 cells or more. DAPI staining of the rad60::ura4+ cells revealed the presence of cells with aberrant nuclear morphology, some of which were cut cells or cells with biased nuclear position (Fig. 5A). These arrest phenotypes largely coincide with those of the rad60-1 cells at 37°C. These observations suggest that rad60 is required for replication to proceed normally and to prevent the accumulation of spontaneous DSBs.
rad60-1 is a synthetic lethal with rad18-X, and overexpression of rad60 suppresses the MMS sensitivity of rad18-X (Fig. 6); these results suggest that there may be a functional link between rad60 and rad18. Both rad60 and rad18 are essential for growth, and some rad mutants (e.g., rad60-1, rad18-X, and rad18-74 mutants) are defective in the repair of DSBs (26, 55). Cells that overexpress dominant-negative rad18 alleles or have a disruption in spr18, an SMC partner of Rad18 (8), and rad60-1 cells at 37°C often contain aberrant chromosomes or have a cut phenotype. In addition, some rad18-disrupted cells show aberrant mitosis (55). rad60-1, rad18-X (37), and rad18-74 (55) mutants have a higher rate of minichromosome loss than wild-type S. pombe. Both rad60-1 and rad18-X (26) are epistatic to rhp51Δ. Therefore, rad60 and rad18 appear to act in a cellular process that is required for cell proliferation and DNA repair.
A protein complex has been purified which includes Rad18, Spr18, and five other unidentified proteins (8). Rad60 may be one of the components of this Rad18 complex. We examined the direct interaction between Rad60 and Rad18 proteins by immunoprecipitation experiments. Epitope-tagged Rad60 and Rad18 proteins were overexpressed in wild-type cells, but no coprecipitaion of Rad60 and Rad18 by either antibody was observed even though each protein was efficiently precipitated (data not shown). Moreover, no interaction was detected between Rad60 and Rad18 by two-hybrid analysis. Therefore, Rad60 and Rad18 are not likely to be directly associated. It is still possible that Rad60 is included in the Rad18-Spr18 complex described by Fousteri and Lehmann (8).
The brc1 gene has been isolated as a multicopy suppressor of rad18-74 (55). Cells with a deletion of brc1 have a partial defect in chromosome segregation, and double brc1Δ and rad18-X or rad18-74 mutants are lethal, suggesting a functional relationship between rad18 and brc1 (55). It has been proposed that brc1 plays a role downstream of rad18 and is needed for a subfunction of rad18. However, the functional relationship between brc1 and rad60 remains to be elucidated.
As discussed above, rad60 and rad18 appear to participate in the same cellular function, but the exact nature of that function is not clear. rad18 encodes an SMC family protein, and proteins in this family form protein complexes that may regulate higher-order chromosome structure. Examples of these protein complexes are the cohesin, condensin, and dosage compensation complexes (12, 48). Fousteri and Lehmann propose that the Rad18/Spr18 complex may be analogous to these complexes. They suggest that the Rad18/Spr18 complex may facilitate recombinational repair by holding and/or bringing together damaged DNA and allowing the Rhp51 protein to initiate strand exchange (8). This model explains the defect of rad18 mutants in DNA repair.
In addition to their role in DNA repair, both rad60 and rad18 are essential for growth. The rad60-1 mutant causes DSBs at the restrictive temperature in cells not exposed to exogenous DNA-damaging agents. Therefore, rad60 appears to play a role in repairing spontaneous DSBs that occur during DNA replication. By analogy to SMC complexes, Rad60 and Rad18 may be required to hold newly replicated DNA duplexes together at a collapsed replication fork to facilitate efficient recombinational repair. In E. coli, when a DSB occurs at a stalled replication fork, homologous recombination is required to repair the DSB and assemble an alternative primosome (24). In vertebrate cells, RAD51 is essential for growth (54), and chicken DT40 cells with a conditional RAD51 transgene accumulate chromosomal breaks when RAD51 expression is suppressed (47). This result suggests that DSBs occur during normal cell cycle progression and that the recombination machinery is needed to repair them to avoid their lethality. The S. pombe RAD51 homolog rhp51 is not essential for growth; however, S. pombe rhp51Δ cells grow slowly even in the absence of exogenous DNA-damaging agents (34). Therefore, a similar situation is likely to exist for S. pombe. In particular, a stalled replication fork could lead to DSBs in dividing S. pombe cells, and rhp51Δ cells may be partially impaired in repairing DSBs and reassembling the replication fork. In contrast to rhp51Δ cells, which are viable, rad60Δ and rad18Δ cells are lethal. It is possible that an inefficient rhp51-independent recombination pathway repairs the breaks at the stalled replication fork in the absence of rhp51. In contrast, rad60Δ and rad18Δ cells might be defective in rhp51-dependent and rhp51-independent recombination pathways, and these cells might not survive the spontaneous DSBs that occur during normal DNA replication.
The above model does not explain the fact that a small fraction of cut cells are observed in the rad60-1 mutant at 37°C. Verkade et al. (55) isolated a rad18 mutant, rad18-74, which is defective in the maintenance of checkpoint arrest after DNA damage. Therefore, in addition to their roles in repairing DNA damage, rad18 and rad60 may play roles in maintaining checkpoint arrest until damage is repaired, and the cut cells in rad60-1 might result from failure to maintain the checkpoint arrest. Several proteins play roles in DNA replication and replication checkpoint response, including replication factor C, Dpb11/Cut5, S. pombe DNA polymerase α, and S. cerevisiae Orc1 and DNA polymerase ɛ (22). Therefore, Rad60 and Rad18 could play roles in DNA repair, DNA replication, and checkpoint arrest in response to DNA damage.
Functional and physical interactions between Rad60 and Rad18 should be further explored to better understand their roles in DNA recombination and replication. These proteins may contribute to unique functional features of eukaryotic chromosomes, including maintenance of their complex higher-order structure and organization. It is possible that novel results could emerge from these studies, because these enzymes may help maintain chromosome stability and integrity by mechanisms that are unique to eukaryotic cells.
Acknowledgments
We thank Osami Niwa and Antony Carr for providing materials and Toshiji Ikeda for γ-irradiation.
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan and by a grant from the Human Frontier Science Program Organizaion (HFSPO) to H.S. Y.T. was supported by a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
REFERENCES
- 1.Barbet, N., W. J. Muriel, and A. M. Carr. 1992. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene 114:59-66. [DOI] [PubMed] [Google Scholar]
- 2.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. [DOI] [PubMed] [Google Scholar]
- 3.Birkenbihl, R. P., and S. Subramani. 1992. Cloning and characterization of rad21, an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 20:6605-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chlebowicz, E., and W. J. Jachymczyk. 1979. Repair of MMS-induced DNA double-strand breaks in haploid cells of Saccharomyces cerevisiae, which requires the presence of a duplicate genome. Mol. Gen. Genet. 167:279-286. [DOI] [PubMed] [Google Scholar]
- 5.Craven, R. A., D. J. Griffiths, K. S. Sheldrick, R. E. Randall, I. M. Hagan, and A. M. Carr. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 221:59-68. [DOI] [PubMed] [Google Scholar]
- 6.Debrauwere, H., S. Loeillet, W. Lin, J. Lopes, and A. Nicolas. 2001. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. USA 98:8263-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenech, M., A. M. Carr, J. Murray, F. Z. Watts, and A. R. Lehmann. 1991. Cloning and characterization of the rad4 gene of Schizosaccharomyces pombe; a gene showing short regions of sequence similarity to the human XRCC1 gene. Nucleic Acids Res. 19:6737-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fousteri, M. I., and A. R. Lehmann. 2000. A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J. 19:1691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Game, J. C. 2000. The Saccharomyces repair genes at the end of the century. Mutat. Res. 451:277-293. [DOI] [PubMed] [Google Scholar]
- 10.Gross, J. D., J. Grunstein, and E. M. Witkin. 1971. Inviability of recA− derivatives of the DNA polymerase mutant of De Lucia and Cairns. J. Mol. Biol. 58:631-634. [DOI] [PubMed] [Google Scholar]
- 11.Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano, T. 1999. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 13:11-19. [DOI] [PubMed] [Google Scholar]
- 13.Hirano, T., S. Funahashi, T. Uemura, and M. Yanagida. 1986. Isolation and characterization of Schizosaccharomyces pombe cut mutants that block nuclear division but not cytokinesis. EMBO J. 5:2973-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The PROSITE database, its status in 1999. Nucleic Acids Res. 27:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong, X., G. W. Cadwell, and T. Kogoma. 1995. Escherichia coli RecG and RecA proteins in R-loop formation. EMBO J. 14:2385-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iliakis, G. 1991. The role of DNA double strand breaks in ionizing radiation-induced killing of eukaryotic cells. Bioessays 13:641-648. [DOI] [PubMed] [Google Scholar]
- 17.Ishioka, K., A. Fukuoh, H. Iwasaki, A. Nakata, and H. Shinagawa. 1998. Abortive recombination in Escherichia coli ruv mutants blocks chromosome partitioning. Genes Cells 3:209-220. [DOI] [PubMed] [Google Scholar]
- 18.Ishioka, K., H. Iwasaki, and H. Shinagawa. 1997. Roles of the recG gene product of Escherichia coli in recombination repair: effects of the delta recG mutation on cell division and chromosome partition. Genes Genet. Syst. 72:91-99. [DOI] [PubMed] [Google Scholar]
- 19.Jang, Y. K., Y. H. Jin, E. M. Kim, F. Fabre, S. H. Hong, and S. D. Park. 1994. Cloning and sequence analysis of rhp51+, a Schizosaccharomyces pombe homolog of the Saccharomyces cerevisiae RAD51 gene. Gene 142:207-211. [DOI] [PubMed] [Google Scholar]
- 20.Kanaar, R., J. H. Hoeijmakers, and D. C. van Gent. 1998. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 8:483-489. [DOI] [PubMed] [Google Scholar]
- 21.Keeney, J. B., and J. D. Boeke. 1994. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136:849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 23.Khasanov, F. K., G. V. Savchenko, E. V. Bashkirova, V. G. Korolev, W. D. Heyer, and V. I. Bashkirov. 1999. A new recombinational DNA repair gene from Schizosaccharomyces pombe with homology to Escherichia coli RecA. Genetics 152:1557-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kogoma, T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61:212-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehmann, A. R. 1996. Molecular biology of DNA repair in the fission yeast Schizosaccharomyces pombe. Mutat. Res. 363:147-161. [DOI] [PubMed] [Google Scholar]
- 26.Lehmann, A. R., M. Walicka, D. J. Griffiths, J. M. Murray, F. Z. Watts, S. McCready, and A. M. Carr. 1995. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 15:7067-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lieber, M. R. 1997. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays 19:233-240. [DOI] [PubMed] [Google Scholar]
- 28.Matsumoto, T., S. Murakami, O. Niwa, and M. Yanagida. 1990. Construction and characterization of centric circular and acentric linear chromosomes in fission yeast. Curr. Genet. 18:323-330. [Google Scholar]
- 29.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 30.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 31.Monk, M., and J. Kinross. 1972. Conditional lethality of recA and recB derivatives of a strain of Escherichia coli K-12 with a temperature-sensitive deoxyribonucleic acid polymerase I. J. Bacteriol. 109:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 33.Muris, D. F., K. Vreeken, A. M. Carr, B. C. Broughton, A. R. Lehmann, P. H. Lohman, and A. Pastink. 1993. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 21:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muris, D. F., K. Vreeken, A. M. Carr, J. M. Murray, C. Smit, P. H. Lohman, and A. Pastink. 1996. Isolation of the Schizosaccharomyces pombe RAD54 homologue, rhp54+, a gene involved in the repair of radiation damage and replication fidelity. J. Cell Sci. 109:73-81. [DOI] [PubMed] [Google Scholar]
- 35.Murray, J. M., C. L. Doe, P. Schenk, A. M. Carr, A. R. Lehmann, and F. Z. Watts. 1992. Cloning and characterisation of the S. pombe rad15 gene, a homologue to the S. cerevisiae RAD3 and human ERCC2 genes. Nucleic Acids Res. 20:2673-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray, J. M., H. D. Lindsay, C. A. Munday, and A. M. Carr. 1997. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 17:6868-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray, J. M., M. Tavassoli, R. al-Harithy, K. S. Sheldrick, A. R. Lehmann, A. M. Carr, and F. Z. Watts. 1994. Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage. Mol. Cell. Biol. 14:4878-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nabeshima, K., S. Saitoh, and M. Yanagida. 1997. Use of green fluorescent protein for intracellular protein localization in living fission yeast cells. Methods Enzymol. 283:459-471. [DOI] [PubMed] [Google Scholar]
- 39.Niwa, O., T. Matsumoto, and M. Yanagida. 1986. Construction of a mini-chromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol. Gen. Genet. 203:397-405. [Google Scholar]
- 40.Ostermann, K., A. Lorentz, and H. Schmidt. 1993. The fission yeast rad22 gene, having a function in mating-type switching and repair of DNA damages, encodes a protein homolog to Rad52 of Saccharomyces cerevisiae. Nucleic Acids Res. 21:5940-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker, A. E., R. K. Clyne, A. M. Carr, and T. J. Kelly. 1997. The Schizosaccharomyces pombe rad11+ gene encodes the large subunit of replication protein A. Mol. Cell. Biol. 17:2381-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds, P. R., S. Biggar, L. Prakash, and S. Prakash. 1992. The Schizosaccharomyces pombe rhp3+ gene required for DNA repair and cell viability is functionally interchangeable with the RAD3 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 20:2327-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saka, Y., and M. Yanagida. 1993. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+. Cell 74:383-393. [DOI] [PubMed] [Google Scholar]
- 44.Shinohara, A., and T. Ogawa. 1995. Homologous recombination and the roles of double-strand breaks. Trends Biochem. Sci. 20:387-391. [DOI] [PubMed] [Google Scholar]
- 45.Siede, W., A. A. Friedl, I. Dianova, F. Eckardt-Schupp, and E. C. Friedberg. 1996. The Saccharomyces cerevisiae Ku autoantigen homologue affects radiosensitivity only in the absence of homologous recombination. Genetics 142:91-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, C. L., T. Matsumoto, O. Niwa, S. Klco, J. B. Fan, M. Yanagida, and C. R. Cantor. 1987. An electrophoretic karyotype for Schizosaccharomyces pombe by pulsed field gel electrophoresis. Nucleic Acids Res. 15:4481-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonoda, E., M. S. Sasaki, J. M. Buerstedde, O. Bezzubova, A. Shinohara, H. Ogawa, M. Takata, Y. Yamaguchi-Iwai, and S. Takeda. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17:598-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strunnikov, A. V., and R. Jessberger. 1999. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur. J. Biochem. 263:6-13. [DOI] [PubMed] [Google Scholar]
- 49.Symington, L. S. 1998. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26:5589-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatebayashi, K., J. Kato, and H. Ikeda. 1998. Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics 148:49-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tavassoli, M., M. Shayeghi, A. Nasim, and F. Z. Watts. 1995. Cloning and characterisation of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 23:383-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tishkoff, D. X., N. Filosi, G. M. Gaida, and R. D. Kolodner. 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88:253-263. [DOI] [PubMed] [Google Scholar]
- 53.Tsutsui, Y., T. Morishita, H. Iwasaki, H. Toh, and H. Shinagawa. 2000. A recombination repair gene of Schizosaccharomyces pombe, rhp57, is afunctional homolog of the Saccharomyces cerevisiae RAD57 gene and is phylogenetically related to the human XRCC3 gene. Genetics 154:1451-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsuzuki, T., Y. Fujii, K. Sakumi, Y. Tominaga, K. Nakao, M. Sekiguchi, A. Matsushiro, Y. Yoshimura, and T. Morita. 1996. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 93:6236-6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verkade, H. M., S. J. Bugg, H. D. Lindsay, A. M. Carr, and M. J. O'Connell. 1999. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell 10:2905-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winston, F., F. Chumley, and G. R. Fink. 1983. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 101:211-228. [DOI] [PubMed] [Google Scholar]
- 57.Yonemasu, R., S. J. McCready, J. M. Murray, F. Osman, M. Takao, K. Yamamoto, A. R. Lehmann, and A. Yasui. 1997. Characterization of the alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe. Nucleic Acids Res. 25:1553-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]