Abstract
We utilized a site-specific cross-linking technique to investigate the mechanism of nucleosome remodeling by hSWI/SNF. We found that a single cross-link between H2B and DNA virtually eliminates the accumulation of stably remodeled species as measured by restriction enzyme accessibility assays. However, cross-linking the histone octamer to nucleosomal DNA does not inhibit remodeling as monitored by DNase I digestion assays. Importantly, we found that the restriction enzyme-accessible species can be efficiently cross-linked after remodeling and that the accessible state does not require continued ATP hydrolysis. These results imply that the generation of stable remodeled states requires at least transient disruption of histone-DNA interactions throughout the nucleosome, while hSWI/SNF-catalyzed disruption of just local histone-DNA interactions yields less-stable remodeled states that still display an altered DNase I cleavage pattern. The implications of these results for models of the mechanism of SWI/SNF-catalyzed nucleosome remodeling are discussed.
The eukaryotic genome is packaged by histone and nonhistone proteins to form chromatin (34). The assembly of nucleosomes, the basic building block of chromatin, as well as compaction of nucleosomal arrays into higher-order chromatin structures create a highly restrictive environment for critical nuclear processes that require access to DNA (16, 39). To ensure the efficient progression of these processes, cells have developed multiple strategies to facilitate access to DNA in chromatin by trans-acting factors (15, 42). One general strategy involves targeted posttranslational modifications of the histone proteins, such as acetylation, phosphorylation, or methylation, which can either directly or indirectly lead to a more permissive chromatin environment (10, 32, 41). A second strategy involves complexes such as the hSWI/SNF complex, which directly couple the energy of ATP hydrolysis to alterations of chromatin structure that facilitate the activity of trans-acting factors (15, 35, 42).
The hSWI/SNF complex is a multisubunit protein complex that is known to play a key role in regulation of chromatin accessibility (25, 27, 36). SWI/SNF was originally identified in yeast as required for activation of a variety of genes (26, 38). Interestingly, although none of the subunits of the SWI/SNF complex are essential for growth in yeast, a related RSC chromatin-remodeling complex contains essential subunits (4). The SWI/SNF and other ATP-dependent remodeling factors have been identified in a wide variety of organisms, including humans (15, 17, 33). The hSWI/SNF complexes have been shown to be essential for regulation of several developmentally specific gene expression programs (27).
Purified SWI/SNF complexes from yeast and human cells have been shown to perturb nucleosome structure and facilitate binding of trans-acting factors in an ATP-dependent fashion in many in vitro assays. These perturbations increase susceptibility of nucleosomal DNA to DNase I and restriction endonucleases (6, 7, 12, 13) and result in changes in cross-linking between histones and nucleosomal DNA (20, 31) and alterations in the number of DNA supercoils constrained by nucleosomes (2, 14, 17). Moreover, SWI/SNF can catalyze ATP-dependent nucleosome translocation (sliding) in which DNA sequences previously occluded by core histones are exposed in internucleosomal regions (14, 30, 37). Biochemical evidence and recent atomic force microscopy studies indicate that SWI/SNF remodeling involves both sliding and disruption of histone-DNA interactions (14, 30).
Despite extensive study, the mechanism by which SWI/SNF complexes remodel nucleosomes is not well understood. The increase in accessibility of nucleosomal DNA to trans-acting factors occurs without large changes in either the configuration or gross changes in core histone stoichiometry, as shown by remodeling of nucleosomes containing disulfide-linked (H3-H4)2 tetramer or fluorescently tagged H3 (3) and isolation of remodeled structures (22, 29). Moreover, remodeling appears to proceed unimpeded with nucleosomes in which the core histones have been cross-linked together, indicating that most native histone protein-protein interactions are maintained during remodeling (2). In addition, inclusion of a single cross-link between histone H2A and nucleosomal DNA does not impede remodeling, as determined by disruption of the canonical DNase I digestion pattern of the nucleosome (20). Interestingly, the SWI/SNF complex has been shown to induce topological stress within a linear DNA fragment in an ATP-dependent fashion (11). Moreover, nucleosomes placed within a topologically restrained environment were found to be refractory to remodeling, and this inhibition could be relieved by topoisomerase (8). However, remodeling-generated DNA torsional stress may not be directly involved in nucleosome sliding (18).
Analysis of the kinetics of ATP hydrolysis and nucleosome disruption by hSWI/SNF indicates that perhaps ≥50 ATPs are required to expose relatively short stretches of DNA within nucleosomes (24). In order to further define the pathway or pathways by which nucleosomes are remodeled and to determine if global disruption of histone-DNA interactions is an obligatory step for remodeling, we tethered the histone octamer to nucleosomal DNA via a single cross-link at multiple positions throughout the nucleosome. We found that cross-linking did not significantly restrict remodeling as judged by a standard DNase I digestion assay. However, the cross-link drastically changed the ability of SWI/SNF to stimulate cleavage of nucleosomal DNA by restriction enzymes. These data suggest that the generation of an extensively remodeled state in which the DNA is accessible to restriction enzymes requires at least transient disruption of most histone-DNA interactions throughout the nucleosome.
MATERIALS AND METHODS
DNA fragments.
Either the 154-bp EcoRI-RsaI fragment or the 215-bp EcoRI-DdeI fragment containing a Xenopus borealis somatic 5S RNA gene derived from the plasmid pXP-10 was used for nucleosome reconstitutions (40). These fragments contained a SacI restriction enzyme site at position −5 (5). Fragments were radioactively labeled at the 5′ end at the EcoRI restriction site by standard techniques.
Preparation of unmodified core histones and H2B26C-APB.
Xenopus H2A, H2B, and a mutant H2B protein containing a glycine-to-cysteine substitution at position 26 (H2BG26C) were expressed in bacteria and purified as preformed dimers as described previously (19). H2BG26C was modified with 4-azidophenacylbromide (APB) (Sigma), and the extent of modification was determined by reaction of a portion of the sample with excess 14C-labeled N-ethylmaleimide (NEM) (Amersham) as described previously (19).
Reconstitution and glycerol gradient purification of mononucleosomes.
Nucleosomes were reconstituted with either the H2A/H2BG26C-APB dimer or wild-type H2A/H2B and H3/H4 tetramers prepared from chicken erythrocyte nuclei. Reconstitution with the 5S DNA fragments described above yields a relatively homogeneous population of nucleosomes with the dyad axis of symmetry (−3) positioned near the transcription start site of the 5S gene (+1; Fig. 1) (5, 40). Reconstitutions were loaded onto 10-ml 5 to 30% glycerol gradients (in 10 mM Tris-Cl [pH 8.0]), and nucleosomes were sedimented at 198,000 × g for 18 h at 4°C. Fractions containing the purified nucleosomes were identified by running a small portion of the samples on a 0.7% agarose nucleoprotein gels with 1/2× Tris-borate-EDTA (TBE). Fractions containing mononucleosomes were dialyzed for 3 h against a buffer containing 10 mM Tris-Cl (pH 8.0).
FIG. 1.
DNA fragments used for nucleosome reconstitution. The 215-bp fragment is an extension of the 154-bp fragment. The positions of relevant restriction enzyme sites and the region of DNA assembled into the nucleosome (oval) are shown.
hSWI/SNF reactions.
The human SWI/SNF complex was prepared as described previously (29). In a 200-μl reaction mixture, approximately 5 ng of nucleosomes was incubated with the indicated amounts of hSWI/SNF (see figure legends) in 12 mM HEPES (pH 7.9), 60 mM KCl, 7 mM MgCl2, 0.6 mM dithiothreitol (DTT), 60 μM EDTA, and 100 ng of bovine serum albumin per μl in the presence or absence of 4 mM ATP (20). Reaction mixtures were incubated at 30°C for 15 min. For determination of positions of H2BG26C-APB cross-linking, the reaction mixtures were irradiated with UV light of 365 nm (VWR LM20E transilluminator) for 25 s after incubation with hSWI/SNF. The reactions were then directly loaded onto a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (6% polyacrylamide). The gel was exposed to Kodak XO-Mat AR film overnight, and the cross-linked complexes were gel isolated. DNA from the cross-linked samples was purified and then treated with 10% piperidine at 95°C for 10 min, and sites of cross-linking were visualized on autoradiographs of sequencing gels. To determine the effect of cross-linking on hSWI/SNF remodeling, H2BG26C-APB-containing samples were irradiated with UV light as previously described before incubation with hSWI/SNF. The samples were then digested with 4 U of DNase I (Worthington) for 3 min at room temperature. The samples were loaded onto an SDS gel, and both the uncross-linked and cross-linked species were isolated. The samples were ethanol precipitated and loaded onto sequencing gels (20). Some nucleosomes were irradiated with UV light prior to incubation with hSWI/SNF and then subjected to restriction digestion (SacI or EcoRV from New England Biolabs) at 10 U/μl for the times indicated in the figure legends. Aliquots were taken at various times, digestion was terminated with EDTA-SDS, and samples were loaded for SDS-PAGE as described above. The restriction enzyme digestion of uncross-linked and cross-linked nucleosomal DNA was monitored independently by phosphorimager.
RESULTS
A nucleosome containing site-specifically modified histone H2B is efficiently remodeled by hSWI/SNF.
Before employing H2B-DNA cross-linking to examine the mechanism of hSWI/SNF-catalyzed nucleosome remodeling, we first examined if the modification itself affected nucleosome formation or SWI/SNF activity. Nucleosomes were reconstituted with an H2B site-specifically modified with APB (H2BG26C-APB; see Materials and Methods), wild-type histones H2A, H3, and H4 and a radioactively end-labeled 154-bp DNA fragment containing the X. borealis 5S nucleosome positioning sequence (Fig. 1). The modified residue is located near where the N-terminal tail of H2B exits the nucleosome through stacked DNA superhelical gyres (23). The ability of hSWI/SNF to remodel nucleosomes containing the modified H2B was tested by DNase I digestion (12). DNase I cleavage of the reconstituted nucleosomes reveals a typical 10-bp cleavage ladder in the absence of ATP for nucleosomes containing either wild-type or modified H2B (Fig. 2, lanes 5 and 13, respectively). Incubation of nucleosomes with hSWI/SNF in the presence of ATP results in the disruption of the 10-bp DNase I cleavage ladder, indicating that nucleosomes containing wild-type and APB-modified H2B are remodeled by hSWI/SNF with similar efficiencies in an ATP-dependent manner (Fig. 2, lanes 6 to 10 and 14 to 18, respectively). Similar results were obtained with nucleosomes assembled on a related 215-bp DNA fragment (Fig. 1) (results not shown).
FIG. 2.
Nucleosomes containing APB-modified H2B (H2BG26C-APB) are remodeled efficiently by hSWI/SNF. Nucleosomes were reconstituted with either wild-type (WT) H2B (lanes 4 to 10) or H2B26C-APB (lanes 12 to 18) and then subjected to hSWI/SNF remodeling. Lane 1 shows the G-specific reaction of the radiolabeled 154-bp 5S DNA fragment used for reconstitution. Lane 2 shows the DNase I digestion pattern of the naked 5S DNA fragment (uncross-linked DNA [FD]). Lanes 3 and 11 show 5S DNA prior to DNase I digestion. Lanes 4 and 12 and 5 and 13, respectively, show the DNase I cleavage pattern of nucleosomes incubated in the absence of hSWI/SNF or in the presence of 1.3 μg of SWI/SNF, but without ATP. Lanes 6 to 10 and 14 to 18, respectively, show the DNase I footprint of nucleosomes incubated with 270, 405, 540, and 810 ng and 1.3 μg of hSWI/SNF in the presence of ATP.
Effect of SWI/SNF remodeling on H2B-DNA interactions.
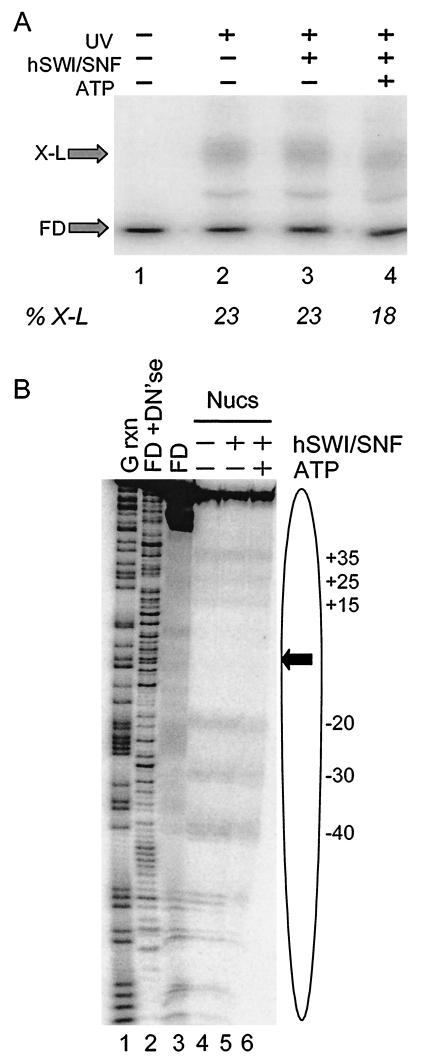
To determine if hSWI/SNF remodeling causes a net loss of histone H2B-DNA interactions, nucleosomes containing APB-modified H2B and radioactively labeled DNA were incubated with hSWI/SNF in the absence or presence of ATP and then irradiated with UV light to cross-link H2B to DNA. Irradiated nucleosomes were then mixed with SDS to dissociate uncross-linked proteins, and the extent of cross-link formation was evaluated by electrophoresis of the sample by preparative SDS-PAGE (6% polyacrylamide) (Fig. 3A). DNA cross-linked to histones migrates more slowly than uncross-linked DNA in these gels (19). SWI/SNF remodeling caused a small but detectable reduction in efficiency of cross-linking within the population of remodeled nucleosome of about 20% (Fig. 3A, lane 4). Dissociation of SWI/SNF from nucleosomes after remodeling by the addition of excess competitor DNA did not affect the amount of cross-link formation (results not shown).
FIG.3.
hSWI/SNF activity results in a marginal loss of interactions between H2B and nucleosomal DNA. (A) Effect of hSWI/SNF remodeling on total cross-link formation. Nucleosomes were reconstituted with H2B26C-APB and the 154-bp 5S DNA fragment and then incubated with hSWI/SNF in the presence or absence of ATP. Samples were then irradiated with UV light as indicated, and the amounts of H2B-DNA cross-linked species (X-L) and uncross-linked DNA (FD) were analyzed by SDS-PAGE and autoradiography. Lanes 1 and 2 show cross-linking in nucleosomes incubated in the absence of SWI/SNF, without or with subsequent UV irradiation, respectively. Lanes 3 and 4 show cross-linking in nucleosomes incubated with hSWI/SNF in the absence or presence of ATP, respectively, followed by UV irradiation. The error on determination of percent cross-linking (%X-L) is ±5%. (B) Sites of interaction between H2B and nucleosomal DNA before and after hSWI/SNF remodeling. Nucleosomes were prepared as in panel A, and the sites of H2B26C-APB cross-linking to nucleosome DNA were determined by mapping piperidine-induced DNA strand breaks on sequencing gels (see Materials and Methods). Lanes 1 and 2 show the G-specific and DNase I cleavage reactions of the naked 154-bp 5S DNA fragment (FD+DN′se); lane 3 shows piperidine-treated, uncross-linked 5S DNA (FD) as a background control. Lanes 4 to 6 show cross-links generated in nucleosomes incubated in the absence of hSWI/SNF, in the presence of SWI/SNF, but without ATP, and in the presence of hSWI/SNF and ATP. Numbers indicate the locations of major sites of cross-linking within the 5S sequence (Fig. 1). The locations of the nucleosome (oval) and nucleosome dyad (arrow) are indicated
To determine if remodeling also caused a redistribution of H2B-DNA interactions within the nucleosome, the cross-linked DNA was isolated from the preparative SDS-PAGE gel (6% polyacrylamide), and the positions of the cross-links were determined by piperidine cleavage and analysis by sequencing gels (Fig. 3B). In the absence of ATP, cross-links to DNA were observed at positions −40, −30, −20, +15, +25, and +35 in the 5S sequence (Fig. 3B, lane 4). These correspond to positions ±37, ±27, and ±17 bp from the predicted location of the nucleosomal dyad at position −3 (40) (Fig 3B). Given the relatively homogeneous translational positioning of nucleosomes assembled with this 154-bp DNA fragment (5), the wide distribution of cross-links is likely to be indicative of dynamic movements of this section of H2B with respect to DNA within the complex (C. Zheng and J. J. Hayes, unpublished results). When the nucleosomes were incubated with hSWI/SNF in the presence of ATP, there were no apparent changes in the pattern of cross-linking (Fig. 3B, lane 6). However, there was some loss of intensity of specific cross-linking bands upon remodeling compared to the overall extent of cross-linking within the sample, suggesting that some H2B-DNA interactions had been redistributed to other sites within the DNA (compare Fig. 3B, lanes 5 and 6).
Site-specific cross-linking of H2B to DNA does not inhibit hSWI/SNF remodeling as detected by DNase I assays.
We next investigated the effect of cross-linking H2B to nucleosomal DNA on remodeling by hSWI/SNF. The covalent cross-link was expected to effectively immobilize the H2B on the DNA and, given the robust nature of histone-histone interactions, cross-linked nucleosomes were expected to be refractory to remodeling if nucleosome sliding or global disruption of histone-DNA interactions is an obligatory component of the remodeling mechanism. To test this possibility, reconstituted nucleosomes containing the APB-modified H2B (H2BG26C-APB) were purified by centrifugation through glycerol gradients and then UV irradiated to cross-link a portion (approximately 5%) of the H2B to the nucleosomal DNA. The cross-linked sample was then incubated with hSWI/SNF in the presence or absence of ATP, exposed to DNase I, and loaded for preparative SDS-PAGE to separate cross-linked from uncross-linked nucleosomal DNA. DNA from each band was isolated, and the DNase I cleavage pattern for cross-linked nucleosomes was compared to that of the uncross-linked nucleosomes from the same reaction. As expected, cross-linking of the H2B itself does not result in significant alteration of the 10-bp DNase I ladder typically obtained for a nucleosome reconstituted on the 215-bp 5S DNA fragment (Fig. 4, compare lanes 3 and 9) or the 154-bp template (results not shown). Importantly, when nucleosomes were incubated with hSWI/SNF in the presence of ATP for 30 min, we found that the cross-linked nucleosomes were remodeled as efficiently as the uncross-linked species (Fig. 4, compare lanes 8 and 14). Since partial inhibition of the rate of SWI/SNF remodeling might not be revealed in such single-time-point assays, we carried out DNase I digestions at various times after the addition of ATP to the hSWI/SNF-nucleosome sample. The time course shows that disruption of the DNase I nucleosome ladder occurs rapidly and is detectable by the earliest time point in the experiment (Fig. 4, compare lanes 5 to 8 to lanes 11 to 14). Moreover, remodeling apparently occurs to the same overall extent in the cross-linked and uncross-linked samples.
FIG. 4.
Prior cross-linking of H2B to nucleosomal DNA does not inhibit hSWI/SNF remodeling as detected by DNase I digestion assay. Nucleosomes reconstituted with H2BG26C-APB and the radiolabled 215-bp 5S DNA fragment were irradiated with UV and then incubated with hSWI/SNF in the presence or absence of ATP, and remodeling of cross-linked and uncross-linked nucleosomes was analyzed by DNase I footprinting. Lane 1, G-specific reaction of naked 5S DNA. Lane 2, DNase I digestion of naked 5S DNA. Lanes 3 and 9 show DNase I digestion of uncross-linked and cross-linked 5S nucleosomal DNA, respectively. Lanes 4 and 10 show DNase I digestion of uncross-linked nucleosomes incubated with 245 ng of hSWI/SNF in the absence of ATP, respectively. Lanes 5 to 8 and 11 to 14 show DNase I digestion of cross-linked and uncross-linked 5S nucleosomal DNA as indicated, incubated with 245 ng of hSWI/SNF and ATP. DNase I was added either at the same time as hSWI/SNF (lanes 5 and 11) or 2 (lanes 6 and 12), 7 (lanes 7 and 13), or 27 (lanes 8 and 14) min after the addition of hSWI/SNF, and the digestions were allowed to proceed for 3 min.
Site-specific cross-linking of H2B to DNA inhibits hSWI/SNF remodeling as detected by restriction enzyme assay.
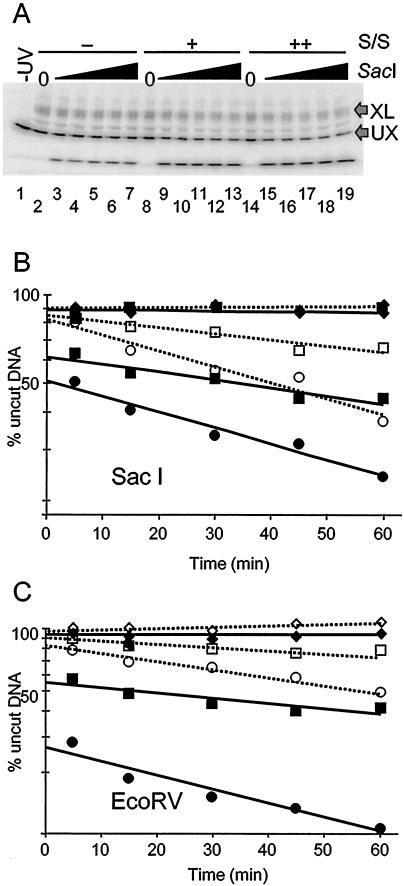
In addition to DNase I, digestion with restriction enzymes has provided a useful and often more quantitative assay for DNA accessibility during hSWI/SNF remodeling (14, 21, 24). The DNA fragments we used for reconstitution contain a SacI restriction enzyme site at position −5, close to the nucleosome dyad (−3), and an EcoRV site at position +35 (Fig. 1). Nucleosomes were reconstituted with the 215-bp fragment and APB-modified H2B and then irradiated with UV light to cause cross-linking in a fraction of the sample. The nucleosomes were incubated with hSWI/SNF in the presence or absence of ATP for 15 min and then subjected to cleavage by SacI or EcoRV restriction enzymes, and the extent of digestion in uncross-linked and cross-linked nucleosomal DNA was determined by SDS-PAGE and autoradiography (Fig. 5A).
FIG. 5.
A single cross-link severely inhibits SWI/SNF-catalyzed nucleosome remodeling as detected by restriction enzyme digestion analysis. Nucleosomes reconstituted with H2B26C-APB and the 215-bp 5S DNA fragment were irradiated with UV light and then digested with either SacI or EcoRV restriction enzymes as described in Materials and Methods. (A) Time course of SacI digestion for cross-linked and uncross-linked nucleosomal DNA. Digested nucleosome samples were loaded onto an SDS-PAGE gel to separate cross-linked (XL) and uncross-linked (UX) DNAs, and the extent of enzyme digestion was determined by autoradiography. Lane 1 shows 5S nucleosomes without UV irradiation or SacI digestion. Lanes 2 to 7 show products of SacI digestion of UV-irradiated nucleosomes incubated without hSWI/SNF. Lanes 8 to 13 and 14 to 19, respectively, show SacI digests ofUV-irradiated 5S nucleosomes incubated with either 270 or 570 ng of hSWI/SNF. (B) Plot of SacI digestion of 5S nucleosomal DNA. The amount of undigested nucleosomal DNA remaining at each time point was calculated from the gel shown in panel A, and the natural log of the percent remaining uncut was plotted for cross-linked and uncross-linked fractions versus time of digestion. The data were fit to linear trend lines calculated in Excel. Solid symbols with solid trend lines are SacI digestion profiles of uncross-linked nucleosomes, and open symbols with dotted trend lines are SacI digestion profiles of cross-linked nucleosomes. Diamonds represent SacI digestions in the absence of hSWI/SNF; squares and circles, respectively, represent SacI digestions in the presence of 270 or 570 ng of hSWI/SNF. (C) Plot of EcoRV digestion of 5S nucleosomal DNA. A plot of the loss of nucleosomal DNA substrate (cross-linked and uncross-linked) by EcoRV cleavage over time is shown as in panel B. Symbols and trend lines are as in panel B.
As expected, in the absence of hSWI/SNF, the 215-bp nucleosomal DNA fragment is very resistant to SacI cleavage (Fig. 5B), in accordance with previous measurements of the probability of DNA site exposure at the nucleosome dyad (5, 28). Typically, the activities of restriction enzymes, which require full exposure of a DNA site for cleavage, are reduced approximately ∼10,000-fold for sites located near the nucleosome dyad when compared to naked DNA (28). We also find that in the presence of hSWI/SNF only (+SWI/SNF, −ATP), nucleosomes are equally resistant to cleavage by SacI (results not shown). Importantly, in the absence of hSWI/SNF, DNA within the cross-linked nucleosomes is cleaved at approximately the same low rate as that within the uncross-linked nucleosome fraction (Fig. 5B). This implies that conformational transitions leading to spontaneous site exposure are not abolished when the nucleosome contains a cross-link.
We also note that approximately 5 to 10% of the DNA in both cross-linked and uncross-linked unremodeled samples is rapidly cleaved within the first 30 s of digestion (Fig. 5B) (results not shown). This is most clearly demonstrated by the extrapolation of a straight line fit to the plot of the ln (fraction of substrate remaining undigested) versus time for both digests, which intercepts the y axis at about 90% (Fig. 5B and C). This suggests that these samples contain about 10% nucleosomes in which the restriction enzyme site is not occluded by histones and/or by minor contamination with histone-free DNA (5, 28). The presence of this rapidly digesting component does not affect subsequent analyses.
As expected, incubation of nucleosomes with hSWI/SNF and ATP for 15 min resulted in major changes in the SacI digestion profile of uncross-linked nucleosomes (Fig. 5B). First, remodeling by hSWI/SNF greatly increased the initial amount of SacI cleavage such that about 50% of the nucleosomes were digested within the initial phase of the reaction (Fig. 5B and Table 1). The rate of SacI digestion in this initial, rapid phase is comparable to the rate at which the enzyme digests naked DNA (results not shown). In addition, the rate of the second phase of cleavage after the initial burst is increased by approximately 10-fold compared to the unremodeled nucleosome digest (Fig. 5B and Table 1). The increase in the rate of SacI digestion, both within this phase and to the extent of the initial burst phase, is dependent upon the amount of hSWI/SNF present in the reaction (Fig. 5B).
TABLE 1.
Effect of cross-linking on SWI/SNF complex-stimulated cleavage of nucleosomal DNA by restriction enzymesa
| SWI/SNF stimulation |
SacI digestion
|
EcoRV digestion
|
||||||
|---|---|---|---|---|---|---|---|---|
| % Uncleaved (1st phase)
|
Relative rate (2nd phase)
|
% Uncleaved (1st phase)
|
Relative rate (2nd phase)
|
|||||
| Cross-linking
|
Cross-linking
|
Cross-linking
|
Cross-linking
|
|||||
| − | + | − | + | − | + | − | + | |
| − | 83 ± 2 | 86 ± 2 | 1 | 0.9 ± 0.8 | 94 ± 2 | 99 ± 1 | 1 | 0.4 ± 1.1 |
| + | 47 ± 6 | 78 ± 4 | 10 ± 3 | 9 ± 3 | 26 ± 4 | 82 ± 7 | 31 ± 4 | 22 ± 5 |
Rate values shown correspond to relative first-order rate constants derived from slopes from fits to plots of the restriction enzyme cleavage data as described in Materials and Methods. The percentage of nucleosomal DNA cleaved in the first phase was determined by extrapolation of the lines fitted to the second phase of the reaction to zero time (i.e., total amount digested in the second phase).
The large SWI/SNF-dependent increase in the initial phase of SacI cleavage is consistent with the accumulation of stably remodeled nucleosomes in which the SacI site is accessible to the enzyme (24). Likewise, the slower second phase of the digest is likely due to continued exposure of sites within nucleosomes that exist in unremodeled or restriction enzyme-inaccessible states. To test this, we digested uncross-linked nucleosomes with SacI at various times after the addition of SWI/SNF and ATP (Fig. 6). Addition of restriction enzyme simultaneously with SWI/SNF yields a digestion profile that can be fit well by a single first-order exponential decay for a majority of the nucleosome substrates (Fig. 6), similar to that observed with purified BRG1 (24). Addition of SacI at later times results in profiles that are described well by a combination of a decreasing amount of this component and an increasing amount of a rapidly digesting component, suggesting the accumulation of a stably remodeled, restriction enzyme-accessible species over time. In addition, we find that reducing the amount of SacI enzyme fivefold in the reaction does not affect the apparent rate of digestion in the second phase of the reaction, indicating that the rate is limited by generation of remodeled species rather than the restriction enzyme cleavage step (24).
FIG. 6.
Accumulation of stably remodeled nucleosomes. Nucleosomes reconstituted with the 215-bp 5S DNA fragment were treated with hSWI/SNF and ATP for various times and then subjected to EcoRV digestion, and the extent of cleavage was determined as in Fig. 5. The percentage of DNA uncut versus the time of digestion is plotted for reactions without hSWI/SNF (diamonds) and with 254 ng of hSWI/SNF for 0.5, 2, 5, and 10 min (squares, triangles, solid circles, and open circles, respectively). The solid line represents the single exponential fit to 0.5-min data.
In order to determine the effect of cross-linking on the restriction enzyme assay (REA), nucleosomes were cross-linked before the remodeling reaction, and then the extent of SacI digestion was determined. In striking contrast to the DNase I results, we found that the presence of a cross-link drastically altered the SacI cleavage profile of nucleosomes subsequently remodeled by SWI/SNF (Fig. 5B). The presence of the cross-link causes an almost complete loss of the rapid initial phase of cleavage, suggesting that the cross-link inhibits the accumulation of stably remodeled species (described above). However, the rate of restriction enzyme digestion in the second slower phase of digestion appears similar to that observed in the uncross-linked, remodeled nucleosomes, approximately 10 times faster than that observed in the unremodeled samples at the maximum amount of hSWI/SNF used in the assay (Fig. 5 and Table 1). Thus, some transient disruption of histone-DNA interactions is detected in cross-linked nucleosomes when the restriction enzyme is continually present in the remodeling reaction.
In order to substantiate the results obtained with SacI, we performed the REA experiment with the enzyme EcoRV, which cleaves the 5S DNA approximately 40 bp away from the dyad axis of symmetry of the nucleosome (Fig. 1B). As in the SacI enzyme assay, the nucleosomal DNA was effectively resistant to EcoRV cleavage in the absence of hSWI/SNF, regardless of the presence or absence of cross-linking. Moreover, as observed with SacI, upon hSWI/SNF remodeling, the EcoRV site within the nucleosome became much more accessible (Fig. 5C). Quantification of these data revealed that remodeling activity caused a substantial fraction of uncross-linked nucleosomes (∼70%) to be digested during the rapid initial phase of the cleavage reaction (Table 1). In addition, the rate of digestion of the nucleosomes remaining after the rapid phase was increased ∼30-fold as a result of SWI/SNF remodeling. As before, cross-linking H2B to DNA within the nucleosome severely diminished the amount of nucleosomes digested during the initial phase, from 76% to 18%, while the rate of digestion of cross-linked nucleosomes remaining after this initial phase was still substantially increased as a result of remodeling (Fig. 5C and Table 1).
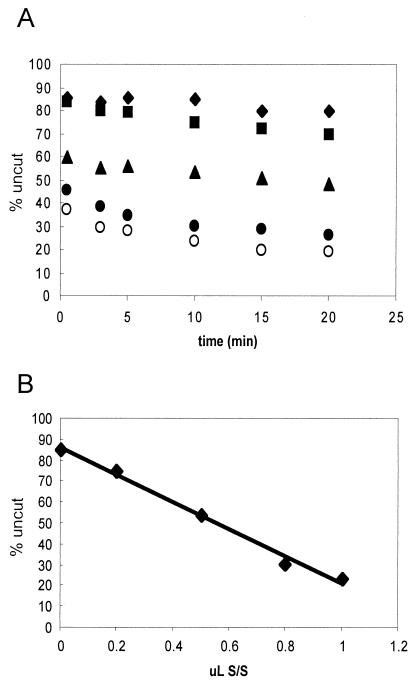
Previous reports indicate that remodeled nucleosomes detected by a supercoiling assay are stabilized by continued interaction with the hSWI/SNF complex (9). We noted that accumulation of the remodeled state rapidly digested by restriction enzymes appears to reach a limit after ∼5 min of exposure to hSWI/SNF (Fig. 6) (described above). To test if this limit is related to the amount of hSWI/SNF present in the reaction, we titrated increasing amounts of hSWI/SNF, allowed remodeling for 15 min, and measured the kinetics of SacI digestion. An examination of the kinetics of these digestions (Fig. 7A) and of a plot of the total fraction of the sample left undigested after 10 min (Fig. 7B) indicates that the extent of nucleosomes rapidly digested by SacI is linearly related to the amount of SWI/SNF present in the reaction mixture. We also asked if the stability of the restriction enzyme-accessible species depends on continued ATP hydrolysis. Remodeled nucleosomes were treated with apyrase and then digested with restriction enzymes. The results show that continued ATP hydrolysis is not required for maintaining this remodeled state within the 15-min time frame tested (results not shown; see reference 9).
FIG. 7.
Extent of accumulation of remodeled species is related to SWI/SNF concentration. Nucleosomes reconstituted with the 215-bp 5S DNA fragment were incubated with various amounts of hSWI/SNF for 15 min, and then the extent of remodeling was examined by digestion with EcoRV. (A) Plot of digests. Digests of reaction mixtures incubated with 0, 0.2, 0.5, 0.8, and 1.0 μl of 245 ng of hSWI/SNF per μl, plotted as diamonds, squares, triangles, solid circles, and open circles, respectively, are shown. (B) Plot of extent of EcoRV digestion at the 10-min time point versus the amount of hSWI/SNF present in the incubation.
Cross-linked, remodeled nucleosomes retain partial restriction enzyme accessibility.
We observed a drastic effect of cross-linking on the generation of stably remodeled nucleosomes, as measured by REAs. However, we also found that hSWI/SNF remodeling has only a marginal effect (∼20% reduction) on the overall yield of cross-links within the entire nucleosome sample. Thus, we more closely investigated the extent to which H2B-DNA interactions are disrupted in remodeled nucleosomes by measuring the extent of restriction enzyme accessibility in nucleosomes cross-linked after remodeling. For example, if restriction enzyme-accessible stably remodeled nucleosomes can be cross-linked as efficiently as unremodeled nucleosomes, then the digestion profile of nucleosomes cross-linked after remodeling will be identical to the profile of uncross-linked remodeled nucleosomes. On the other hand, if restriction enzyme-accessible remodeled nucleosomes are not able to be cross-linked, then the fraction of cross-linked nucleosomes should be relatively resistant to digestion, and the digestion profile will be identical to that of the unremodeled control.
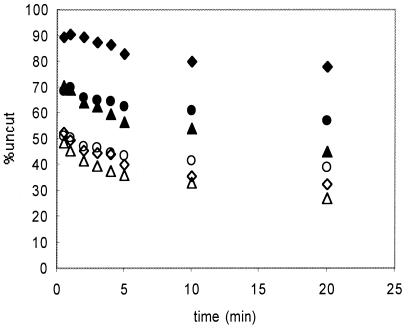
To examine this issue, we first incubated nucleosomes with hSWI/SNF and ATP for 15 min and then briefly irradiated the sample to instigate cross-linking. The sample was then treated with EcoRV, and the extent of digestion in cross-linked and uncross-linked nucleosomes was plotted as described above (Fig. 8). As expected, the digestion profile of remodeled, uncross-linked nucleosomes from the irradiated sample was identical to that of the unirradiated, remodeled control (Fig. 8). Treatment of the sample with apyrase prior to EcoRV digestion did not substantially alter the profile (Fig. 8). Interestingly, we found that nucleosomes cross-linked after remodeling had an intermediate degree of EcoRV accessibility (Fig. 8) compared to nucleosomes remaining uncross-linked or nucleosomes from the unirradiated control. Moreover, as observed with uncross-linked nucleosomes, this accessibility does not require the continued presence of ATP (Fig. 8). This result implies that cross-links can coexist with restriction enzyme-accessible regions within remodeled nucleosomes—i.e., that substantial H2B-DNA contacts are present within stably remodeled nucleosomes. We note that this observation is in striking contrast to the drastic inhibition of remodeling observed in nucleosomes that already contain a cross-link (Fig. 5) and suggests that the presence of the cross-link is not compatible with some intermediate transition state on the pathway to formation of stably remodeled, restriction enzyme-accessible nucleosomes.
FIG. 8.
Cross-linking occurs within remodeled nucleosomes. Nucleosomes containing cross-linkable H2B and the 215-bp 5S DNA fragment were treated with hSWI/SNF and ATP and then irradiated, and the extent of EcoRV accessibility in cross-linked and uncross-linked fractions was determined. In some cases, remodeling reaction mixtures were treated with apyrase before the irradiation step. The digestion profiles for unirradiated samples incubated with SWI/SNF without ATP and with ATP (solid and open diamonds, respectively) are shown for reference. The profiles for cross-linked and uncross-linked fractions from the SWI/SNF remodeled, irradiated sample (solid and open triangles, respectively) and cross-linked and uncross-linked fractions from remodeled samples treated with apyrase before irradiation (solid and open circles, respectively) are shown.
DISCUSSION
Digestion of hSWI/SNF remodeled 5S nucleosomes with either of two different restriction enzymes results in the rapid digestion of a significant fraction of the sample (Fig. 5 and 6). Based on previous findings (9, 24), it is likely that these rapidly digested species correspond to stably remodeled nucleosomes in which the DNA is accessible to restriction enzymes, perhaps stabilized by continued interaction with the hSWI/SNF complex (Fig. 7). Our data indicate that formation of a single cross-link between H2B and nucleosomal DNA at sites located throughout the central 80 bp of the nucleosome nearly completely eliminates the hSWI/SNF-dependent accumulation of stably remodeled nucleosomes (Fig. 5). In contrast, cross-linking does not appear to inhibit hSWI/SNF activity when remodeling is monitored by a standard DNase I assay (Fig. 4). DNase I cuts DNA nearly randomly with regard to sequence and requires only an exposed minor groove of DNA on the nucleosome surface for endonucleolytic cleavage (34). On the other hand, restriction enzymes such as SacI and EcoRV require complete liberation of their respective cognate sequences from the histone surface before DNA site-specific recognition, binding, and cleavage can occur (28). Thus, the different effects of cross-linking on the DNase I and REAs are likely due to the different requirements of these nucleases for cleavage of nucleosomal DNA.
It is interesting to consider the drastic effect of cross-linking on REA in light of the fact that in many cases cross-linking occurs some distance from the actual site being probed. Cross-links occur within a 74-bp region encompassing the nucleosomal dyad, while the EcoRV site is located about 35 bp to one side of the dyad (Fig. 1 and 3B). Thus, half of the cross-links causing inhibition of hSWI/SNF-dependent EcoRV cleavage occur on the opposite side of the nucleosome, 50 to 70 bp distant from the EcoRV recognition site, yet cleavage is inhibited in nearly all cross-linked species. This suggests that generation of a restriction enzyme-accessible state within nucleosomal DNA requires at least transient disruption of distant histone-DNA interactions throughout the nucleosome.
Our results can be interpreted in the context of a recently developed kinetic and thermodynamic framework for nucleosome remodeling by hSWI/SNF and its catalytic subunit, BRG1 (24). This framework suggests that remodeling involves formation of a partially disrupted intermediate state or states that collapse to a number of remodeled products, each having a different stretch of DNA completely exposed (24). Given the less stringent requirements of DNase I for cleavage, it is possible that this enzyme detects remodeling intermediates, the formation of which is not inhibited by cross-linking. Likewise, in the context of this model, our results indicate that either the final remodeled state or an intermediate on the pathway to its formation is incompatible with an H2B-DNA cross-link at a range of sites within the nucleosome. This final remodeled state may involve looping out of short stretches of nucleosomal DNA or some other disruption of histone-DNA interactions (24). It is possible that a single histone-DNA cross-link would block the propagation of topological stress through the nucleosome (8, 11), thereby inhibiting remodeling. We also show that H2B-DNA interactions are at least partially compatible with the final, stably remodeled state, suggesting that the presence of the cross-link inhibits the formation of some intermediate species in the pathway to the final remodeled state (Fig. 8).
The results may also be at least partly interpreted within the framework of a nucleosome sliding model of remodeling. ATP-dependent chromatin remodeling complexes, including the SWI/SNF complex, can catalyze the translocation of histone octamers along DNA in cis (37). Recently, many ATP-dependent chromatin remodeling complexes, including the SWI/SNF complex, were shown to impart topological stress onto DNA, and nucleosomes present within a topologically constrained environment were shown to be relatively resistant to remodeling (8, 11). Thus, torsional stress imparted to the DNA by remodeling complexes may cause DNA twist diffusion or looping, resulting in nucleosome sliding (8, 11, 37; however, see reference 18). In the context of a sliding model, we assume that cross-linking does not inhibit hSWI/SNF-induced local changes in histone-DNA interactions, which can be detected by DNase I. However, given the robust nature of histone-histone interactions, cross-linking would be expected to severely inhibit translocation of the histone octamer along the DNA. Indeed cross-linking does inhibit uncatalyzed nucleosome sliding in vitro (S.A. and J.H., unpublished observations). Thus, it is possible that small alterations in histone-DNA interactions normally leading to nucleosome sliding occur within cross-linked nucleosomes, while actual sliding and the associated restriction enzyme site exposure are prevented by the cross-linking.
As mentioned above, virtually all remodeling activities, including the SWI/SNF complex, can catalyze changes in nucleosome translational position (sliding). However, it is unclear whether remodeling-catalyzed nucleosome sliding is an obligatory component of the remodeling mechanism. Thus, despite the ability of SWI/SNF to catalyze nucleosome sliding, the exact molecular mechanism of remodeling remains undefined. Indeed there are many mechanistic possibilities that can lead to nucleosome sliding (concerted movements of entry and exit points, twist diffusion, loop recapture, and collapse from a more general altered conformation). While it is possible that every productive remodeling event catalyzed by SWI/SNF involves translational repositioning, currently available data are also consistent with a model in which nucleosome sliding is instead one of several possible outcomes of a conformational change introduced into the nucleosome via the action of SWI/SNF. Indeed, recent results suggest that hSWI/SNF causes both sliding and disruption of nucleosome structure on nucleosome arrays (30) and that hSWI/SNF remodeling of a mononucleosome results in exposure of cognate sites for restriction enzymes at rates that are not easily explained by a sliding mechanism (24). Thus, an intermediate model in which conformational changes induced by remodeling may yield either translationally repositioned nucleosomes or stable altered structures is possible as well. Our data are consistent with either a strictly disruption model or a strictly sliding model. However, the observation that restriction enzyme activity is not altered at all during the nonstable part of the reaction (slopes of lines in Fig. 5) is easier to reconcile with the idea that sliding is not an obligatory component of remodeling (20), since cross-link-impeding translational movement would be expected to slow the overall reaction.
Despite the large inhibition of hSWI/SNF-dependent accumulation of restriction enzyme-accessible complexes by cross-linking, we found remodeling activity still significantly increased the kinetics of restriction enzyme cleavage during the second phase of the digestion (Table 1). The slope of fits to this phase was increased 10- to 30-fold over the slow rate at which the restriction enzymes cleaved the unremodeled nucleosomes, regardless of cross-linking. This may reflect the production of either moderately accessible complexes that are prohibited from conversion to the stable, accessible state(s) or fully remodeled complexes that quickly relax to the intermediately remodeled state because of the presence of the cross-link.
We previously demonstrated that cross-linking of either of two different locations in the H2A tail domain to nucleosomal DNA did not hinder hSWI/SNF nucleosome remodeling as monitored by DNase I digestion (20). Interestingly, small ∼5- to 10-bp stretches of nucleosome-like DNase I cleavages surrounding the sites of H2A cross-linking seemed to be retained in the remodeled, cross-linked complexes (20). In the present study, we did not detect a similar effect of cross-linking in the DNase I digestion patterns, probably because the H2B-DNA interactions probed here are more flexible, with cross-linking to nucleosomal DNA occurring over a range of three sites separated by ∼10 bp. H2B is intimately dimerized with H2A in the nucleosome, and the 26th residue in H2B occupies roughly analogous positions to the 12th residue in H2A with respect to location within the proteins and proximity to DNA. Our results indicate that there is some loss of interaction between the 26th amino acid residue of H2B and DNA upon hSWI/SNF remodeling, but no new sites of cross-linking were detected. This result parallels results obtained with cross-linking of the 12th amino acid residue position in H2A (20). In addition, SWI/SNF remodeling resulted in a significant alteration in the location of cross-links formed by the second amino acid position near the tip of the H2A tail domain (20). These results support the interpretation that remodeling causes disruption of interactions between H2A/H2B histone fold domains and DNA, while the tail domains remain in contact with DNA (1, 20). Cross-linking at other sites in the histone tail domains will further clarify these issues.
Acknowledgments
This work was supported by NIH grant RO1GM52426 and American Cancer Society grant RPG-00-080-01-GMC (J.J.H.), as well as grants from NIH and Hoechst AG to R.E.K and an NRSA award to S.S.
REFERENCES
- 1.Angelov, D., M. Chara, M. Seve, J. Côté, S. Khochbin, and S. Dimitrov. 2000. Differential remodeling of the HIV-1 nucleosome upon transcription activators and SWI/SNF complex binding. J. Mol. Biol. 302:315-326. [DOI] [PubMed] [Google Scholar]
- 2.Bazett-Jones, D. P., J. Côté, C. C. Landel, C. L. Peterson, and J. L. Workman. 1999. The SWI/SNF complex creates loop domains in DNA and polynucleosome arrays and can disrupt DNA-histone contacts within these domains. Mol. Cell. Biol. 19:1470-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer, L. A., X. Shao, R. H. Ebright, and C. L. Peterson. 2000. Roles of the histone H2A-H2B dimers and the (H3-H4)2 tetramer in nucleosome remodeling by the SWI-SNF complex. J. Biol. Chem. 275:11545-11552. [DOI] [PubMed] [Google Scholar]
- 4.Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. [DOI] [PubMed] [Google Scholar]
- 5.Chafin, D. R., J. M. Vitolo, L. A. Henricksen, R. A. Bambara, and J. J. Hayes. 2000. Human DNA ligase I efficiently seals nicks in nucleosomes. EMBO J. 19:5492-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Côté, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53-60. [DOI] [PubMed] [Google Scholar]
- 7.Côté, J., C. L. Peterson, and J. L. Workman. 1998. Perturbation of nucleosome core structure by the SWI/SNF complex persists following its detachment, enhancing subsequent transcription factor binding. Proc. Natl. Acad. Sci. USA 95:4947-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gavin, I., P. J. Horn, and C. L. Peterson. 2001. SWI/SNF chromatin remodeling requires changes in DNA topology. Mol. Cell 7:97-104. [DOI] [PubMed] [Google Scholar]
- 9.Guyon, J. R., G. J. Narlikar, E. K. Sullivan, and R. E. Kingston. 2001. Stability of a human SWI-SNF remodeled nucleosomal array. Mol. Cell. Biol. 21:1132-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen, J. C., C. Tse, and A. P. Wolffe. 1998. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry 37:17037-17040. [DOI] [PubMed] [Google Scholar]
- 11.Havas, K., A. Flaus, M. Phelan, R. E. Kingston, P. A. Wade, D. M. Lilley, and T. Owen-Hughes. 2000. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 103:1133-1142. [DOI] [PubMed] [Google Scholar]
- 12.Imbalzano, A. N., H. Kwon, M. R. Green, and R. E. Kingston. 1994. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370:481-485. [DOI] [PubMed] [Google Scholar]
- 13.Imbalzano, A. N., G. R. Schnitzler, and R. E. Kingston. 1996. Nucleosome disruption by human SWI/SNF is maintained in the absence of continued ATP hydrolysis. J. Biol. Chem. 271:20726-20733. [DOI] [PubMed] [Google Scholar]
- 14.Jaskelioff, M., I. M. Gavin, C. L. Peterson, and C. Logie. 2000. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell. Biol. 20:3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 16.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:587-598. [DOI] [PubMed] [Google Scholar]
- 17.Kwon, H., A. N. Imbalzano, P. A. Khavari, R. E. Kingston, and M. R. Green. 1994. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature 370:477-481. [DOI] [PubMed] [Google Scholar]
- 18.Längst, G., and P. B. Becker. 2001. ISWI induces nucleosome sliding on nicked DNA. Mol. Cell 8:1085-1092. [DOI] [PubMed] [Google Scholar]
- 19.Lee, K. M., and J. J. Hayes. 1997. The N-terminal tail of histone H2A binds to two distinct sites within the nucleosome core. Proc. Natl. Acad. Sci. USA 94:8959-8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, K. M., S. Sif, R. E. Kingston, and J. J. Hayes. 1999. hSWI/SNF disrupts interactions between the H2A N-terminal tail and nucleosomal DNA. Biochemistry 38:8423-8429. [DOI] [PubMed] [Google Scholar]
- 21.Logie, C., and C. L. Peterson. 1997. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 16:6772-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorch, Y., M. Zhang, and R. D. Kornberg. 2001. RSC unravels the nucleosome. Mol. Cell 7:89-95. [DOI] [PubMed] [Google Scholar]
- 23.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 24.Narlikar, G. J., J. R. Guyon, and R. E. Kingston. 2001. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell 8:1219-1230. [DOI] [PubMed] [Google Scholar]
- 25.Peterson, C. L. 1996. Multiple SWItches to turn on chromatin? Curr. Opin. Genet. Dev. 6:171-175. [DOI] [PubMed] [Google Scholar]
- 26.Peterson, C. L., and I. Herskowitz. 1992. Characterization of the yeast SWI1, SWI2 and SWI3 genes, which encode a global activator of transcription. Cell 68:573-583. [DOI] [PubMed] [Google Scholar]
- 27.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 28.Polach, K. J., and J. Widom. 1995. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 254:130-149. [DOI] [PubMed] [Google Scholar]
- 29.Schnitzler, G., S. Sif, and R. E. Kingston. 1998. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell 94:17-27. [DOI] [PubMed] [Google Scholar]
- 30.Schnitzler, G. R., C. L. Cheung, J. H. Hafner, A. J. Saurin, R. E. Kingston, and C. M. Lieber. 2001. Direct imaging of human SWI/SNF-remodeled mono- and polynucleosomes by atomic force microscopy employing carbon nanotube tips. Mol. Cell. Biol. 21:8504-8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sengupta, S. M., M. VanKanegan, J. Persinger, C. Logie, B. R. Cairns, C. L. Peterson, and B. Bartholomew. 2001. The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J. Biol. Chem. 276:12636-12644. [DOI] [PubMed] [Google Scholar]
- 32.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 33.Tamkun, J. W., R. Deuring, M. P. Scott, M. Kissinger, A. M. Pattatucci, T. C. Kaufman, and J. A. Kennison. 1992. Brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SWI2/SNF2. Cell 68:561-572. [DOI] [PubMed] [Google Scholar]
- 34.van Holde, K. E. 1989. Chromatin. Springer-Verlag, New York, N.Y.
- 35.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wade, P. A., and A. P. Wolffe. 1999. Transcriptional regulation: SWItching circuitry. Curr. Biol. 9:R221-R224. [DOI] [PubMed] [Google Scholar]
- 37.Whitehouse, I., A. Flaus, B. R. Cairns, M. F. White, J. L. Workman, and T. Owen-Hughes. 1999. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400:784-787. [DOI] [PubMed] [Google Scholar]
- 38.Winston, F., and M. Carlson. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8:387-391. [DOI] [PubMed] [Google Scholar]
- 39.Wolffe, A. P. 1998. Chromatin: structure and function. Academic Press, London, England.
- 40.Wolffe, A. P., and J. J. Hayes. 1993. Transcription factor interactions with model nucleosomal templates. Methods Mol. Genet. 2:314-330. [Google Scholar]
- 41.Wolffe, A. P., and J. J. Hayes. 1999. Chromatin disruption and modification. Nucleic Acids Res. 27:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]