Abstract
Bim (Bcl-2-interacting mediator of cell death) is a member of the BH3 domain-only subgroup of Bcl-2 family members, for which three splice variants have been described. Bim is expressed in many healthy cell types, where it is maintained in an inactive conformation through binding to the microtubule-associated dynein motor complex. Upon certain apoptotic stimuli, Bim is released from microtubules and mediates caspase-dependent apoptosis through a mechanism that is still unclear. Here, we have identified and characterized novel splice variants of human Bim mRNA. In particular, we show that a newly discovered, small protein isoform, BimAD, is also able to induce apoptosis strongly in several human cell lines. BimAD and the previously characterized isoform BimS are shown to be capable of heterodimerizing in vivo with both death antagonists (Bcl-2 and Bcl-XL) and death agonists (Bax). Mutants of BimAD that bind to Bax but not to Bcl-2 still promote apoptosis, indicating that Bim can regulate apoptosis through direct activation of the Bax-mediated cell death pathway without interaction with antiapoptotic Bcl-2 family members. Furthermore, we have shown that the interaction of the BimS and BimAD isoforms with Bax leads to a conformational change in this protein analogous to that triggered by the BH3-only protein Bid.
Members of the Bcl-2 family of proteins have come to be regarded as central players in the “decision” step triggering apoptosis. Some proteins within this family, such as Bcl-2, Bcl-XL, and Mcl-1, inhibit apoptosis, while others, including Bax, Bak, Bad, and Bim, promote cell death. Members of this family contain at least one of four conserved motifs known as the Bcl-2 homology (BH) domains, termed BH1, BH2, BH3, and BH4 (reviewed in references 1 and 2). These domains are important in determining the pro- and antiapoptotic properties of the various family members and in mediating interactions between them. Structural studies and mutational analyses have highlighted the key role of the BH3 domain in the death agonist activity of proapoptotic members. Furthermore, the increasing number of cell death agonists with no similarity to Bcl-2 beyond their BH3 homology has confirmed the key role of this domain in triggering apoptosis. The proapoptotic proteins fall into two groups, those having only the BH3 domain in common, such as Bad, Bid, and Bim, and those having the BH1, BH2, and BH3 domains in common, such as Bax and Bak. Several of these proteins appear to exist in an inactive conformation in viable cells, and depending on the specific death signals and the particular cell type, one or more death agonists can undergo a posttranslational modification resulting in an active conformation. This then determines the intracellular localization and defines the pattern of interactions with other family members, allowing the activated protein to exert its proapoptotic activity (reviewed in reference 17).
Although it has been suggested that BH3-only proteins promote apoptosis by binding and inhibiting the activity of antiapoptotic members of the family, recent data also point to their potential role as direct enhancers of the proapoptotic function of Bax and Bak. In support of this view, Bid has been shown to interact with either survival factors or proapoptotic Bax and Bak, in which it produces a conformational change that promotes their death activity (9, 37).
Among the different Bcl-2 family members, we were interested in characterizing in more detail the function of the BH3-only death agonist, Bim. Bim was originally identified as a Bcl-2-interacting protein by screening a bacteriophage λ cDNA expression library constructed from a mouse thymic lymphoma (21). Three isoforms were characterized, BimEL, BimL, and BimS. These differed from each other in cytotoxicity, with BimS being the most potent. This is partly explained by the sequestration of BimEL and BimL in the cytoskeleton-associated motor complex bound to dynein light chain LC8, from which they are released upon various apoptotic stimuli (26). The expression of forkhead transcription factors may also contribute to Bim expression and activity (10). BimS protein has been only recently detected in 293 human embryonic kidney cells (25), while BimEL and BimL have been found in a variety of tissues and cell types (22). Inhibition of Bim expression has been shown to have specific effects on hematopoietic homeostasis, and Bim proteins appear to play an important role in the prevention of autoimmunity (6).
In this report, we describe the cloning and functional characterization of novel Bim isoforms which are the products of alternative splicing. Apoptosis assays reveal clear differences in the activities of the new isoforms, and only one, named BimAD, has a proapoptotic activity comparable to that of BimS. Analysis of the human Bim gene indicates that BimAD results from the splicing out of exons 3 and 4 during mRNA processing. The BimAD protein retains the BH3 domain but lacks the predicted carboxyl-terminal hydrophobic region found in the previously characterized Bim isoforms due to an early stop codon in a newly discovered exon. In transient transfection assays, BimAD induces apoptosis that is suppressible by coexpression of Bcl-2 or the pan-caspase inhibitor zVAD-fmk. Binding studies show that BimS and BimAD are able to bind not only to the antiapoptotic proteins Bcl-2 and Bcl-XL but also to proapoptotic Bax. Engineering mutations in Bim that distinguish between Bcl-2 and Bax binding indicate that Bim can induce apoptosis through direct interaction with Bax as well as through interaction with Bcl-2 and Bcl-XL. Furthermore, we show that the expression of BimS and BimAD results in a conformational change in Bax that has been proposed as a critical step in Bax activation (9, 20). The data presented here provide new insights for understanding the cell death pathway triggered by Bim.
MATERIALS AND METHODS
Cloning of Bim cDNAs.
An oligonucleotide primer (5′-TGATATCAATGCATTCTCCACACCAGGCGGAC-3′) complementary to the human BimEL sequence was designed and used for reverse transcription-PCR, employing reagents from a 5′/3′RACE cDNA amplification kit (Boehringer Mannheim) and RNA from 293 human embryonal kidney cells or OVCAR3 ovarian cancer cells. The cDNA products were amplified by PCR with the reverse oligonucleotide primer (see above) and a forward oligonucleotide (5′-AGAATTCATGGCAAAGCAACCTTCTGATGTAAG-3′) complementary to the human BimEL sequence, and the amplification products were subcloned into the pcDNA3.1/V5-His-TOPO vector by using the TOPO TA cloning kit (Invitrogen). The sequences of derived clones were verified by automated sequencing with a SequiTherm EXCEL II DNA sequencing kit-LC kit (Epicentre Technologies). The sequences were determined with a Li Cor DNA sequencer 4200 series.
Site-directed mutagenesis of Bim.
Site-directed mutagenesis of Bim was performed with a QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's recommended procedure. Bim-BH3 mutants were generated as follows: the G156E substitution mutation was generated with the mutagenic forward primer 5′-GAGTTGCGGCGTATCGAAGACGAGTTTAACGC-3′ and a complementary reverse primer; the G156A point mutation was generated with the forward primer 5′-GAGTTGCGGCGTATCGCAGACGAGTTTAACGC-3′ and its complementary reverse primer; N160A was generated with the forward primer 5′-CGGAGACGAGTTTGCCGCTTACTATGC-3′ and its complementary reverse primer. The pcDNA3.1/BimSTaptag and pcDNA3.1/BimADTaptag plasmids (see below) were used as DNA templates. Mutations were confirmed by DNA sequencing (as described above).
Cell culture.
Human embryonal kidney 293 cells and HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 50 μg of streptomycin/ml, and 50 IU of penicillin/ml. Mouse NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium supplemented with 5% donor calf serum, 50 μg of streptomycin/ml, and 50 IU of penicillin/ml. The cultures were incubated at 37°C in a humidified atmosphere of 10% CO2.
The Burkitt lymphoma-derived Daudi cell line, the human HL60 leukemia cell line, the Jurkat T-cell line, and HeLa-S3 cervical carcinoma cells were cultured in RPMI medium supplemented with 7.5% fetal calf serum, 50 μg of streptomycin/ml, and 50 IU of penicillin/ml. The cultures were incubated at 37°C in a humidified atmosphere of 5% CO2.
Apoptosis assays.
NIH 3T3 cells in a logarithmic phase of growth were seeded at a density of 2.6 × 105 per 35-mm-diameter dish the day before transfection. The cells were transiently cotransfected with 1.3 μg of each of the expression plasmids or a vector (negative control) and 0.7 μg of pcDNA3.1/V5-His-TOPO/lacZ (Invitrogen) containing the gene for β-galactosidase (β-Gal) by using Superfect (Qiagen). At 14 h after transfection, the cells were washed once with phosphate-buffered saline (PBS) and then fixed in 3.7% formaldehyde for 10 min on ice. The cells were washed twice again with PBS, stained in X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) buffer (0.1% X-gal [Bioline], 4 mM potassium ferricyanide, 4 mM potassium ferrocyanide, 2 mM MgCl in PBS) to indicate the β-Gal expressing cells, and incubated at 37°C overnight. The percentage of apoptotic cells was quantified by microscopically counting the number of apoptotic (round) blue cells versus the total number of blue cells counted (500 to 800 cells) from five randomly chosen fields.
The apoptosis assays were repeated with incubation of the cells with the broad-spectrum caspase inhibitor zVAD-fmk (Calbiochem; 50 μM).
Antibodies.
The antibodies used in the study of Bim protein isoforms and members of the Bcl-2 family were as follows: a rabbit polyclonal antibody against amino acids 22 to 40 of human Bim protein (anti-Bim; N terminal; Calbiochem), a rat monoclonal antibody against human Bim [Bim (Ab-1); Calbiochem], a rabbit polyclonal antibody against amino acids 11 to 30 of human Bax (Bax N20: sc-493; Santa Cruz Biotechnology), a mouse monoclonal antibody against human Bax (YTH-6A7; Trevigen), a mouse monoclonal antibody against human Bcl-2 [Bcl-2 (100): sc-509; Santa Cruz Biotechnology], a goat polyclonal antibody against actin [actin (C-11): sc-1615; Santa Cruz Biotechnology], a rabbit polyclonal antibody against amino acids 18 to 233 of human Bcl-XS/L (B22630; Transduction Laboratories), and a goat polyclonal antibody against glutathione S-transferase (GST) (Amersham Pharmacia Biotech).
TAP tag purification and immunoblot analysis.
Each Bim variant was subcloned as a fusion product with the C-terminal tandem affinity purification (TAP) tag sequence (24, 28) in pcDNA3.1, and derived clones were verified by automated sequencing (as described above). 293 cells in a logarithmic phase of growth were seeded on 100-mm-diameter tissue culture dishes at a density of 2 × 105 cells/ml in fresh medium the day before transfection. The cells were transiently cotransfected with 10 μg of each of the expression plasmids by using a CalPhos mammalian transfection kit (Clontech) according to the manufacturer's recommended procedure. Adherent cells were lysed 48 h after transfection in 1 ml of 1% Triton buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100) supplemented with the protease inhibitor cocktail Complete (Boehringer Mannheim). After centrifugation at 13,000 × g for 15 min at 4°C, the supernatant was recovered and added to 100 μl of immunoglobulin G Sepharose bead suspension (Amersham) previously washed once in 1 ml of IPP150 (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.1% NP-40). The samples were rotated for 2 h at 4°C to allow the binding of the fusion protein. The beads were washed three times with 1 ml of IPP150 and once with 1 ml of Tev cleavage buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, 1 mM dithiothreitol) and then resuspended in 200 μl of Tev cleavage buffer containing 20 U of Tev protease (Gibco Life Technologies). The samples were rotated at room temperature for 2 h to cleave the tagged protein. One hundred microliters of calmodulin binding buffer (Cbb) (10 mM β-mercaptoethanol, 10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40) was added to the suspension and centrifuged at 13,000 × g for 1 min. The supernatant was collected and added to a calmodulin bead suspension (Amersham) previously washed in 1 ml of Cbb and resuspended in 900 μl of Cbb. Ten microliters of 100 mM CaCl2 solution was added, and the samples were rotated for 1 h at 4°C to allow binding to the beads. After being washed three times in Cbb, 200 μl of calmodulin elution buffer (10 mM β-mercaptoethanol, 10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM EGTA, 0.1% NP-40) was added and the samples were incubated at room temperature for 5 min followed by centrifugation (13,000 × g, 1 min). The supernatant was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the resolved proteins were transferred onto Immobilon-P membranes (Millipore). After the blocking of nonspecific binding for 1 h at room temperature with 1.5% bovine serum albumin in TPBS (0.1% Tween 20 in PBS), the membrane was incubated for 1 h at room temperature with the appropriate primary antibody (1:2,000 dilution). To ensure equal loading and transfer, the membranes were also probed for actin. The immunoreactive proteins were visualized with the appropriate (antirabbit or antimouse) horseradish peroxidase-linked secondary antibody, and the antigen-antibody complex was visualized by using an ECL system (Amersham).
Parallel experiments were performed by lysing 293 cells in 1% CHAPS buffer (150 mM NaCl, 10 mM HEPES [pH 7.4], 1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}) supplemented with the protease inhibitor cocktail Complete. NP-40 was replaced with the same concentration of CHAPS in all of the buffers used in the purification. Western blot analysis was performed as described above.
Purification of recombinant GST-BimS, GST-BimAD, and His-tagged BaxΔTM.
BimS and BimAD were cloned in pGEX-5X-1 (Amersham), and GST fusion proteins were expressed in competent BL21/pLysS Escherichia coli. An overnight Luria broth culture was diluted 1/10 and grown to an optical density at 600 nm of 0.6. Expression of GST-Bim protein was induced with 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h, and cells were then harvested by centrifugation at 4,000 × g at 4°C. The pellet from a 1-liter culture was resuspended in 50 ml of lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100) supplemented with Complete protease inhibitor cocktail. The lysate was centrifuged at 13,000 × g for 30 min, and the supernatant was recovered. A 2-ml aliquot of Glutathione-Uniflow Resin (Clontech) was washed in washing buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.1% NP-40) and then incubated with the supernatant for 1 h at 4°C. The beads were washed three times with washing buffer, and the Bim-GST product was eluted with 1.5 ml of reduced glutathione (10 mM). The eluted proteins were dialyzed against PBS.
His-tagged Bax-α lacking 21 amino acids at the COOH terminus (His-BaxΔTM) cloned in the pTrcHis vector (Invitrogen) was kindly donated by Ingram Iaccarino. This construct was expressed in E. coli BL21(DE3)/pLysS cells. An overnight Luria broth culture was diluted 1/10 and grown to an optical density at 600 nm of 0.6. Expression of His-tagged Bax protein was induced with 2 mM IPTG for 2 h, and cells were then harvested by centrifugation at 4°C. The pellet from a 100-ml culture was resuspended in 5 ml of lysis buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 10% Glycerol, 1% Triton X-100) supplemented with Complete protease inhibitor cocktail. The lysate was sonicated for 10 s and centrifuged at 13,000 × g for 30 min. The supernatant was incubated with 0.5 ml of washed His beads (Qiagen) for 1 h at 4°C. The beads were washed three times with washing buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 20 mM imidazole, 0.1% Triton), and the His-tagged Bax product was eluted with 0.5 ml of elution buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 250 mM imidazole, 0.1% Triton). The eluted protein was dialyzed against PBS.
In vitro binding assay.
Fifty microliters of GST-Sepharose beads (Amersham) was washed in PBS and resuspended in 0.5 ml of PBS. Recombinant Bim-GST proteins or control GST (final concentration of 100 nM each) were added to different bead preparations, and the samples were rotated for 1 h at 4°C. The beads were recovered by centrifugation at 4,000 × g for 1 min and washed three times in 1 ml of IPP150 washing buffer and one time in 1% Triton buffer. Recombinant His-Bax (final concentration of 10 nM) was added to the beads resuspended in 1 ml of 1% Triton buffer, and the samples were rotated for 1 h at 4°C. The beads were washed five times in IPP150, each time followed by rotation for 5 min at 4°C. Material bound to the beads was eluted by the addition of 100 μl of reduced glutathione (10 mM), and the eluted proteins were analyzed by SDS-PAGE and Western blotting for Bax immunoreactivity as described above. The intensity of each band was quantitated with a Typhoon 8600 optical scanner and ImageQuant 5.1 image analysis software (Molecular Dynamics). Data were normalized to the appropriate GST intensity.
Immunoprecipitation.
Fifty microliters of protein A Sepharose beads (Amersham) was washed, resuspended in 1 ml of Triton buffer, and then incubated with rotation for 1 h at 4°C with 3 μl of polyclonal anti-Bim antibody (500 μg/ml). Similarly, 50 μl of protein G Sepharose beads (Amersham) was incubated with 8 μl of monoclonal anti-Bim antibody (100 μg/ml). The beads were recovered by centrifugation and washed twice with 1 ml of Triton buffer. 293 cells seeded at 2 × 105 cells/ml on two 100-mm-diameter dishes were lysed in 1 ml of 1% Triton buffer with freshly added protease inhibitors. The lysates were centrifuged at 13,000 × g for 5 min to remove nuclei and cellular debris, and the supernatant was added to the bead suspension and then mixed by rotation for 2 h at 4°C. Immunoprecipitates were collected by a brief spin and washed three times with 1 ml of lysis buffer before fractionation by SDS-PAGE, which was followed by Western blot analysis with the polyclonal anti-Bim antibody. For immunoprecipitation of Bax, a 50-μl aliquot of protein G Sepharose beads (Amersham) was incubated with 7.5 μl of polyclonal anti-Bax antibody (200 μg/ml) followed by incubation with 293 cell lysates as described above. For immunoprecipitation of Bim from HL60 and Daudi cells, approximately 5 ml of cell pellet was lysed in 10 ml of 1% Triton buffer with freshly added protease inhibitors. The lysates were incubated with 5 μl of polyclonal anti-Bim antibody (500 μg/ml) or 50 μl of monoclonal anti-Bim antibody (100 μg/ml) and mixed by rotation for 2 h at 4°C. Protein G Sepharose beads (50-μl aliquot; Amersham) were added to pull down immune complexes and then mixed by rotation for 1 h at 4°C. Immunoprecipitates were collected and washed as described above before fractionation by SDS-PAGE, which was followed by Western blot analysis with the polyclonal anti-Bax antibody.
Immunostaining.
The day before transfection, HeLa cells were plated at a density of 1.2 × 105 cells/ml on 24-mm glass coverslips in 35-mm-diameter culture dishes. The cells were transiently cotransfected with 0.3 μg of each of the expression plasmids or a vector (negative control) and 0.1 μg of pEGFP-Actin (Clontech) by using Effectene (Qiagen). At 12 h after transfection, the cells were washed twice with PBS and fixed for 15 min in 1% paraformaldehyde at room temperature. After being washed with PBS, the cells were incubated in PBS containing the permeabilizing agent digitonin (500 μg/ml; Fluka) and the mouse anti-Bax 6A7 monoclonal antibody (1:100 dilution) overnight at 4°C. The coverslips were washed three times with PBS and incubated with cy3-conjugated antimouse antibody (Amersham) diluted 1:200 in 1% bovine serum albumin-PBS for 1 h in the dark. After being washed three times with PBS, the coverslips were mounted with ProLong Antifade mounting medium (Molecular Probes) on microscope slides and analyzed with a fluorescence microscope (Zeiss UK). The percentage of Bax-positive cells was quantified by microscopically counting the number of cells showing a red punctate distribution versus the total number of green (green fluorescent protein [GFP]-expressing) cells. Approximately 500 cells were counted from 20 randomly chosen fields for each experiment.
As a positive control for the induction of Bax immunoreactivity, the cells were treated with staurosporine (Sigma) at 1.5 μM for 5 h.
RESULTS
Identification of Bim splice variants and isolation of BimAD.
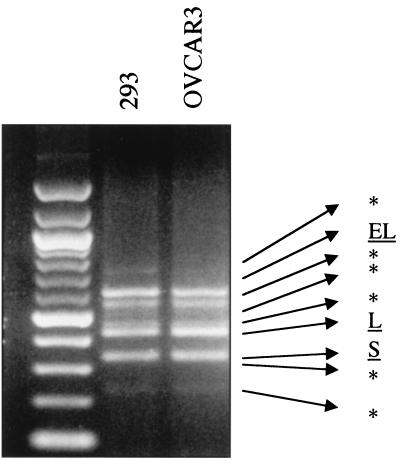
Bim-specific cDNA was amplified from both 293 human embryonic kidney cells and OVCAR3 ovarian cancer cells following reverse transcription-PCR with specific Bim primers (see Materials and Methods). Several transcripts were amplified together with the previously described BimEL, BimL, and BimS (Fig. 1). Negative control reactions with only a single primer, with RNA without reverse transcription, or in the presence of RNase did not generate any product (data not shown).
FIG. 1.
Isolation of human Bim isoforms. Agarose gel electrophoresis of Bim cDNAs after reverse transcription-PCR on 293 or OVCAR3 cell lines is shown, and the three known Bim isoforms together with six other related cDNAs (∗) are identified. Leftmost lane, 100-bp DNA marker (New England Biolabs).
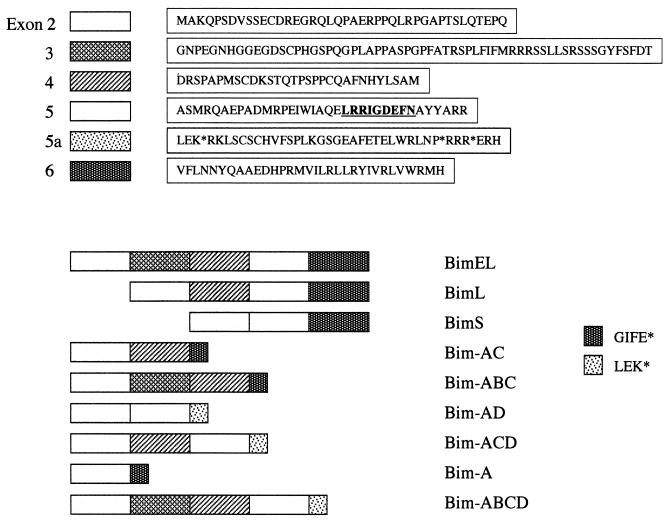
Sequence analysis of the six unknown variants revealed the presence of a novel sequence, 130 bp long, in three of them, BimAD, BimACD, and BimABCD (Fig. 2). The position of the insertion corresponded exactly with the junction of the previously described exons 5 and 6 of the Bim gene (7) and was therefore designated 5a. Although the variants contained exon 6 in the cDNA, a stop codon at the beginning of the novel insert resulted in truncated proteins, all lacking the carboxyl-terminal hydrophobic domain identified in exon 6; only a short C-terminal amino acid stretch (LEK) was present as a result of translation of the novel insert. Similarly, the other three variants, named BimAC, BimABC, and BimA, also lacked the exon 6-encoded C terminus due to a stop codon resulting from translational frameshift.
FIG. 2.
Bim proteins and predicted amino acid sequences of human Bim variants (the BH3 domain is underlined). The names of the new variants were assigned by reference to the domain junctions. The novel exon (5a), identified in the human Bim gene, is present in BimAD, BimACD, and BimABCD (short product LEK). Termination codon positions are indicated (∗) according to the reading frame predicted by the junction between exons 5 and 5a. The short product (GIFE) in BimAC, BimABC, and BimA comes from a translational frameshift due to the junction between exons 4 and 6 or 2 and 6.
Comparison of the cDNA sequences of each variant with genomic data indicates that they are produced by alternative splicing events. The coding sequence of BimAD, the principal subject of this study, is derived from the second and fifth exons. BimAD therefore contains the BH3 domain found in BimEL, BimL, and BimS.
Induction of apoptosis by Bim proteins.
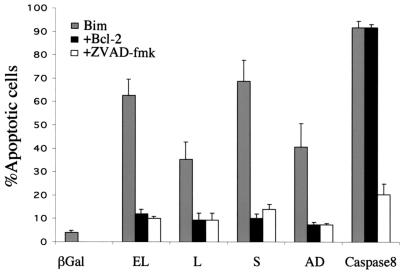
The function of the Bim variants was explored by transient transfection in NIH 3T3 mouse fibroblasts, where apoptotic cells are readily detectable by a change in morphology. The cells were cotransfected with expression plasmids encoding each Bim isoform along with pCMV-β-Gal as a marker for transfected cells. At 14 h posttransfection, the cells were fixed, stained for β-Gal expression, and examined for the morphological features of cell death. As shown in Fig. 3A, BimEL, BimL, and BimS all induced apoptosis, as evidenced by the appearance of shrunken and rounded blue cells. The new variants BimAC, BimABC, and BimACD lacked apoptotic activity; BimABCD had only a weak effect, whereas BimAD had a higher proapoptotic activity (Fig. 3A and data not shown). The percentage of apoptotic β-Gal-positive (blue) cells cotransfected with BimAD was intermediate between those for BimS (the most potent) and BimL. No morphological changes were observed in parallel cultures transfected with the control pcDNA3 vector (Fig. 3A). Immunoblot analysis of lysates from transfected cells that were cultured in the presence of the caspase inhibitor zVAD-fmk to inhibit apoptosis induced by the transgenes demonstrated that the differential effects of the isoforms did not arise from different levels of protein expression (Fig. 3C).
FIG. 3.
Effect of Bim variants after transient transfection into NIH 3T3 cells. (A) NIH 3T3 cells stained for β-Gal expression 14 h after transfection with 1.3 μg of the various Bim constructs together with 0.7 μg of the β-Gal expression vector. (B) Percentages of round blue cells relative to the total population of blue cells. Bars indicate the standard deviation of the results of three independent experiments. (C) Expression of the Bim variants in NIH 3T3 cells.
Overexpression of Bcl-2 effectively blocked the apoptotic effect of BimAD as well as those of BimEL, BimL, and BimS but not that of caspase 8, which is known to mediate apoptosis independent of the Bcl-2 checkpoint. zVAD-fmk, a wide-spectrum caspase inhibitor, was able to inhibit the proapoptotic activity of the Bim isoforms as well as that of caspase 8, as shown in Fig. 4. These data suggest that the novel isoform BimAD also mediates apoptosis through a caspase-mediated pathway, as has been shown for the previously characterized Bim variants (21), and that this can be blocked by the overexpression of antiapoptotic Bcl-2 family proteins.
FIG. 4.
Bim-induced cell death is blocked by Bcl-2 and zVAD-fmk. Bcl-2 cotransfection blocks Bim-induced apoptosis but not caspase 8-mediated killing. The caspase inhibitor zVAD-fmk inhibits apoptosis induced by Bim as well as by caspase 8, further validating the data obtained. The data shown are the percentages of round blue cells relative to the total population of blue cells. The apoptotic activities of the different variants in the presence or absence of Bcl-2 and zVAD-fmk are compared. Bars indicate the standard deviation of the results of three independent experiments.
The effect of BimAD in promoting apoptosis is not restricted to NIH 3T3 cells, since overexpression of BimAD was able to induce apoptosis in both the HeLa and 293 cell lines (data not shown).
BimS and BimAD heterodimerize with both death antagonists (Bcl-2 and Bcl-XL) and death agonists (Bax).
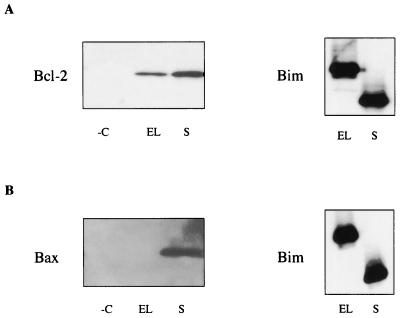
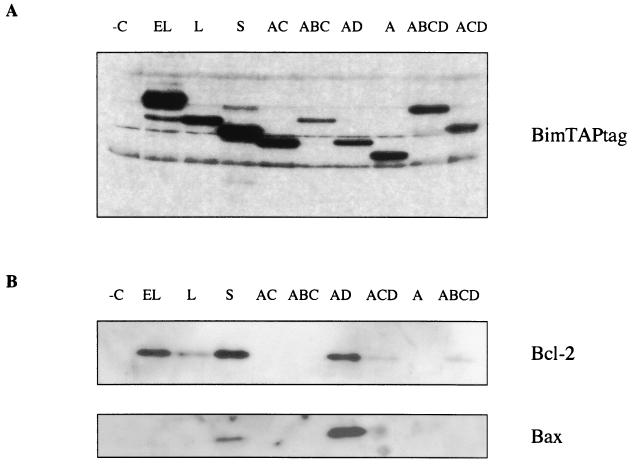
It has been suggested that BH3-only members of the Bcl-2 family exert their proapoptotic activity by heterodimerizing with antiapoptotic Bcl-2 family members, antagonizing their protective function. We therefore explored whether Bim isoforms are capable of associating with other Bcl-2 family proteins by using a TAP method (24, 28). This is a two-step strategy to purify efficiently and rapidly complexes between TAP-tagged proteins and associated components present in the cell at their endogenous levels. Initially, we studied the isoforms BimEL and BimS by subcloning them fused to the C-terminal TAP tag sequence in pcDNA3.1, and the derived clones were transiently transfected into 293 cells. The fusion proteins and associated endogenous proteins were recovered from extracts by two affinity steps and subjected to SDS-PAGE-immunoblot assays with antibodies specific for several Bcl-2 family members. As shown in Fig. 5, both isoforms were able to associate with Bcl-2. BimS was also shown to interact with the proapoptotic protein Bax. In view of these data, the study was extended to include the other Bim isoforms. All of the variants containing the BH3 domain were able to associate with Bcl-2, thus correlating with the data demonstrating that Bcl-2 can inhibit Bim-induced apoptosis (Fig. 6B). Importantly, the novel isoform BimAD was also capable of binding to Bax. Furthermore, no binding was observed with the empty TAP tag parental vector, confirming the specificity of these interactions (Fig. 6B).
FIG. 5.
Association of BimEL and BimS with various Bcl-2 family members. Immunoblot analysis with anti-Bcl-2 antibody (A) or anti-Bax antibody (B) of 293 cells transiently transfected with BimEL-TAPtag, BimS-TAPtag, or TAPtag empty vector (−C) followed by TAP tag purification (left panels) is shown. Equal loading of the two Bim isoforms was confirmed with the polyclonal anti-Bim antibody (right panels).
FIG. 6.
Proteins associating with Bim variants. 293 cells were transiently transfected with each of the Bim-TAPtag-expressing plasmids followed by TAP tag purification and immunoblot analysis. (A) Expression of each variant detected by Western blot analysis with an anti-Bim polyclonal antibody. (B) BimEL, BimL, BimS, BimAD, BimACD, and BimABCD, all containing the BH3 domain, heterodimerize with Bcl-2. Both BimS and BimAD associate with Bax. Negative control (−C) cells were transfected with TAP tag parental vector and purified by the same TAP tag procedure.
It has been shown that in the presence of nonionic detergents, such as NP-40 or Triton-X, Bax can undergo a conformational change, leading to the exposure of the amino-terminal epitope. Hence, we performed the protein complex purification in transient transfection analysis by permeabilizing 293 cells in a buffer containing 1% CHAPS, a zwitterionic detergent that has been shown to maintain Bax in its native conformation (13, 14). Under these conditions, no specific signal was detected, although BimS and BimAD could still bind to Bcl-2 (data not shown).
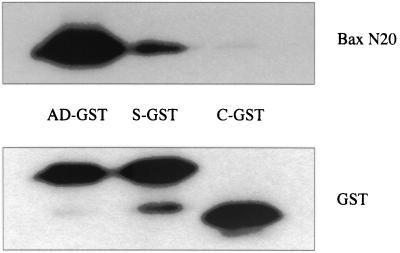
To explore whether the interaction between BimS or BimAD with activated Bax occurs directly, we generated recombinant BimS-GST and BimAD-GST proteins as well as a His-tagged BaxΔTM protein to test for binding in an in vitro assay. Recombinant GST was used as a negative control. Consistent with the coimmunoprecipitation data, both BimAD-GST and BimS-GST were shown to bind to His-BaxΔTM in the activated conformation (Fig. 7). The intensity of the immunoreactive bands was quantitated and normalized to the appropriate GST intensity (Fig. 7). The amount of His-BaxΔTM pulled down by BimAD-GST was 12.8-fold greater than that pulled down by BimS-GST.
FIG. 7.
Interactions between BimAD-GST, BimS-GST, or control GST (C-GST) with His-tagged BaxΔTM in vitro. Immunoblotting was performed with the polyclonal anti-Bax antibody or anti-GST antibody.
Expression of endogenous BimAD and association of endogenous Bim proteins with Bax.
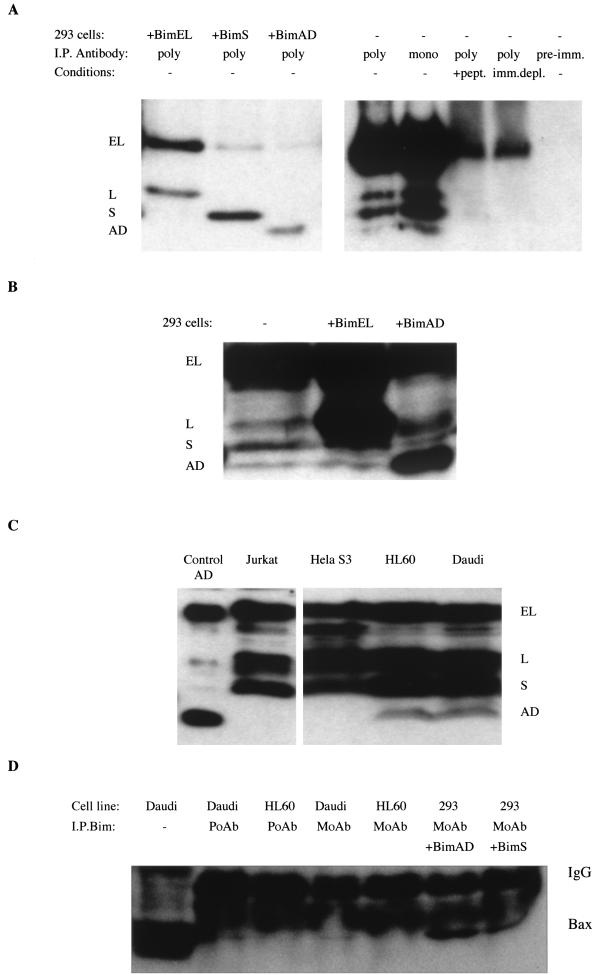
To demonstrate that endogenous BimAD mRNA is translated into protein in 293 cells, immunoprecipitation assays were performed with anti-Bim antibodies. Immunoprecipitates from either untransfected cells or cells transfected with BimEL, BimS, or BimAD were obtained by using either a polyclonal or a monoclonal anti-Bim antibody, and the products were subjected to SDS-PAGE-immunoblot assay with the polyclonal anti-Bim antibody. A protein migrating at the same level as BimAD was immunoprecipitated from the untransfected 293 cells (Fig. 8A). Control experiments with preimmune serum and a specific peptide to block the polyclonal anti-Bim antibody showed that this represented specific Bim immunoreactivity. Furthermore, Bim was not detected in 293 cell lysates previously immunodepleted with the polyclonal antibody (Fig. 8A). In addition, immunoblot analysis of total lysates from parental 293 cells or 293 cells transiently transfected with pcDNA-BimEL or pcDNA-BimAD revealed the presence of a band comigrating with BimAD (Fig. 8B).
FIG. 8.
Expression of BimAD. (A) Immunoprecipitation (I.P.) of lysates from 293 cells after transient transfection of BimEL, BimS, or BimAD or of lysates from untransfected cells. Both polyclonal (poly) and monoclonal (mono) anti-Bim antibodies were employed in the study. Western blot analysis was performed with the polyclonal anti-Bim antibody. Control experiments involved immunoprecipitation with preimmune serum (pre-imm.), blocking of the anti-Bim polyclonal antibody with a specific peptide (+pept.), or immunodepletion of 293 cell lysates with the polyclonal antibody (imm.depl.). (B) Western blot of whole-cell lysate from 293 cells or 293 cells transiently transfected with BimEL or BimAD as controls. The expression of Bim was detected by immunoblotting with an anti-Bim polyclonal antibody. (C) Expression of Bim isoforms in Jurkat, HelaS3, HL60, and Daudi cells. (D) Immunoprecipitation of endogenous Bim from Daudi and HL60 cells and of exogenous Bim in transfected 293 cells with either a polyclonal antiserum (PoAb) or a monoclonal antibody (MoAb). The presence of Bax in the immunoprecipitated complexes was detected with the polyclonal Bax N20 antiserum. IgG, immunoglobulin G.
Since the loss of Bim in Bim−/− mice was shown to affect markedly the hematopoietic compartment (6), we also analyzed the expression of the Bim variants in a panel of hematopoietic cell lines. While the longer isoforms EL, L, and S were expressed in all of the lines tested, the predicted Bim(AD) protein was found in HL60 and Daudi (Fig. 8C). As these two cell lines express BimAD (and BimS) in readily detectable amounts, lysates from either HL60 or Daudi were subjected to immunoprecipitation studies to assess whether Bim could coimmunoprecipitate with Bax when they were expressed at their endogenous levels. Bim could be immunoprecipitated with Bax in Daudi but not in HL60 cells under the experimental conditions used (Fig. 8D). Control immunoprecipitation experiments with 293 cells transfected with exogenous BimAD or BimS showed coimmunoprecipitation of Bax under the same conditions (Fig. 8D).
Interaction with Bax requires the BH3 domain of both BimS and BimAD.
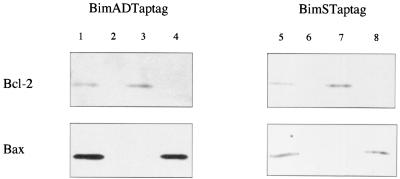
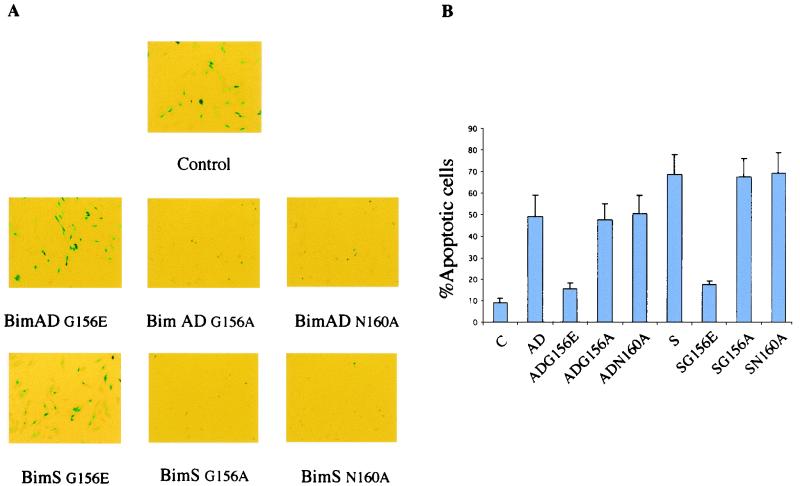
Since Bid, another proapoptotic BH3-only member of the Bcl-2 family, has been shown to interact with Bax through its BH3 domain in a manner distinct from that of its interaction with Bcl-2 (36), we made mutants of Bim that might be able to bind differentially to Bax and Bcl-2. BH3 mutants of BimS-TAPtag and BimAD-TAPtag were tested for their binding to Bcl-2 and Bax in vivo by using the TAP tag method. We derived three mutants with different specificities: Bim G156E does not bind to either Bax or Bcl-2; Bim G156A binds to Bcl-2 but not to Bax; and Bim N160A binds to Bax but not to Bcl-2 (Fig. 9). These mutants or control wild-type Bim plasmids were transiently transfected into NIH 3T3 cells together with pCMV-β-Gal as a reporter gene, and the β-Gal assay was performed as previously described. Neither of the Bim G156E mutants (of BimS and BimAD) was able to promote apoptosis in this system, while both the Bim G156A and Bim N160A mutants still retained death agonist activity (Fig. 10). In view of these data, we postulate a model in which Bim can induce apoptosis by two distinct mechanisms. It can bind directly to Bax, leading to its activation and targeting to mitochondria, and it can also bind to Bcl-2, neutralizing its ability to sequester Bax in an inactive state.
FIG. 9.
The BH-3 domain of Bim is required for its association with both Bcl-2 and Bax. Western blot analysis was performed with anti-Bcl-2 antibody (upper panels) or anti-Bax antibody (lower panels). Lanes 1 and 5, control from BimAD-TAPtag and BimS-TAPtag, respectively, after TAP tag purification; lanes 2 and 6, G156E mutants are impaired in their binding to both Bcl-2 and Bax; lanes 3 and 7, G156A mutants retain binding to Bcl-2 but not to Bax; lanes 4 and 8, N160A mutants retain binding to Bax but not to Bcl-2.
FIG. 10.
Effect of Bim mutants after transient transfection into NIH 3T3 cells. (A) A β-Gal assay after transient transfection of NIH 3T3 cells shows differential effects of single or double mutations. Control cells were transfected with empty vector along with β-Gal expression vector. (B) Percentages of round blue cells relative to the total population of blue cells. Bars indicate the standard deviation of the results of three independent experiments. C, control.
Bim induces Bax conformational change.
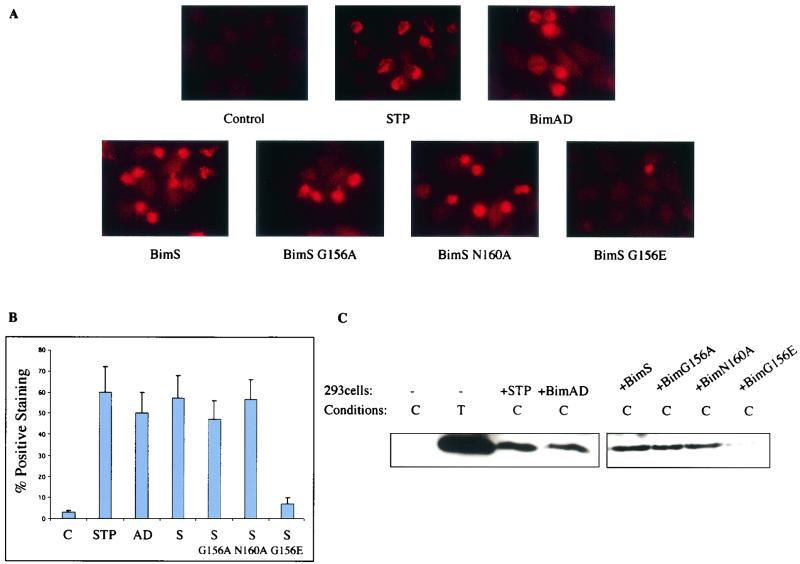
Since one of the mechanisms proposed for Bax activation is the conversion to an active conformation (9), we explored whether the binding of Bim to Bax could correlate with Bax conformational change. For this purpose, HeLa cells were transiently transfected with BimS or BimAD together with a GFP-expressing vector as a marker for transfected cells. At 12 h after transfection, Bax immunostaining was assessed by microscopy by employing a monoclonal antibody that specifically recognizes the epitope exposed by the conformational change in the Bax protein (6A7). The assay was performed at 12 h, since this time corresponds with the onset of Bim expression (data not shown) and Bax conformational change has been shown to be an early event in the apoptotic cascade (11, 19). As shown in Fig. 11A, BimS or BimAD expression leads to the appearance of activated Bax immunoreactivity, showing a punctate distribution within the cytoplasm. This result was similar to that observed in HeLa cells treated with staurosporine, which were previously reported to induce Bax activation in this system (Fig. 11A) (9). Control cells transfected with the empty vector together with GFP-expressing plasmid did not display positive immunoreactivity for Bax (Fig. 11A). Similarly, BimS mutants were assessed for Bax immunoreactivity. Mutations in BimS that block its binding either to Bax (BimS-G156A) or to Bcl-2 (BimS-N160A) did not affect its ability to induce Bax conformational change. However, the mutant BimS-G156E, which inhibits binding to both Bax and Bcl-2, was unable to activate Bax (Fig. 11A).
FIG. 11.
BimS and BimAD promote Bax conformational change. (A) HeLa cells growing on glass coverslips were transiently transfected with BimAD, BimS, or BimS mutants. Twelve hours after transfection, the cells were immunostained with a monoclonal antibody that specifically recognizes the conformationally active Bax (6A7). Bax-associated fluorescence was visualized with a cy3-conjugated secondary antibody (red). As a positive control, the cells were treated with staurosporine (1 μM) for 6 h. As a negative control, the cells were transiently transfected with the parental pcDNA3 plasmid. (B) Percentages of positive (red) cells relative to the total population of GFP-positive (green) cells. In the case of staurosporine, positive cells were counted relative to the total population. Bars indicate the standard deviation of the results of three independent experiments. (C) 293 cells were transiently transfected with BimAD, BimS, BimS-G156A, BimS-N160A, BimS-G156E, or control empty vector. Cells were lysed in 1% CHAPS (C) and subjected to immunoprecipitation with the conformation-specific Bax antibody (N20). Conformationally active Bax was detected by immunoblotting with polyclonal anti-Bax antibody. As a positive control, 293 cells were lysed in 1% Triton (T) and treated as described above. STP, staurosporine.
The effect of Bim on Bax conformation was further confirmed by immunoprecipitation assays with another Bax antibody (N20) that recognizes only the activated conformation (Fig. 11C). 293 cells were transiently transfected with BimS, BimAD, or the BimS mutants and lysed in buffer containing 1% CHAPS. Consistent with the microscopic data, Bax was detected in the immunoprecipitates from BimS- or BimAD-transfected cells but not in control untransfected cells (Fig. 11C). Both the BimS-G156A and BimS-N160A mutants were also able to induce Bax conformational change, while the mutant BimS-G156E was impaired in this activity (Fig. 11C). Bax could also be immunoprecipitated in cells lysed in the presence of Triton X-100, a nonionic detergent previously shown to expose the N-terminal epitope of Bax (14).
DISCUSSION
We have identified six new splice variants of Bim in human cells and investigated in detail the function of one variant, BimAD. BimAD encodes a protein of 80 amino acids and results from an alternative splicing event in which exons 3 and 4 of the Bim gene are lost while a unique 130-bp sequence is inserted after exon 5. This generates a change in the reading frame with a premature termination codon and elimination of the C-terminal hydrophobic tail. Therefore, except for the C-terminal deletion, the amino acid sequence predicted for the variant matches that of the previously described BimS. While this manuscript was in revision, a number of nonapoptotic splice variants of Bim were described (35), but our study is the first to identify BimAD, the novel proapoptotic form.
To assess the nature of the unknown sequence, a Blast search was performed against the latest draft of the human genome. The sequence was located in chromosome 2 within an intron of the human Bim gene, as deduced from sequence accession numbers AC013332 and AC046192. The 130-bp sequence was found in the intronic region flanked by intron-exon junction sequences corresponding to the GT-AG rule (27). In addition, we identified a good candidate for the branch point (CCTGAC) lying 24 nucleotides upstream of the 3′ splice junction, which has been found to be required for lariat formation during splicing (27). Finally, a pyrimidine stretch is located between the branch site and the AG at the 3′ splice site, although this deviates from the respective consensus sequence (Pyr)12CAG, with several purines interrupting the polypyrimidine tract. Weak 3′ splice sites with higher purine content have been shown to be common in alternatively spliced exons (30).
The formation of BimAD is analogous to that of N-Bak, which has been shown to result from the insertion of a 20-bp sequence that causes a translational frameshift. This in turn leads to a truncated protein due to a premature stop codon (31).
It has become clear that alternative splicing plays an important role in the expression of many genes in the programmed cell death pathway (15). Within the Bcl-2 family, splicing variants have also been reported for Bcl-2, Bcl-X, Bcl-W, Bax, Mcl-1, Bok, Bak, and Bcl-G. As we have shown for Bim, in some cases variants of the same gene may exhibit different biological functions. The long form of Bcl-X (L) is a strongly antiapoptotic protein, while Bcl-X (S) and Bcl-X-β are proapoptotic (5, 29). Further, Mcl-1s is a splicing variant of the antiapoptotic Bcl-2 family member Mcl-1 but exhibits proapoptotic activity (3, 4). It has recently been shown that the BH-3 domain-only splice variant of proapoptotic Bak, N-Bak, is antiapoptotic in neurons (31). Similarly, the five Bim isoforms we isolated that do not show death-promoting potential could have intrinsic antiapoptotic activity or, alternatively, could operate as transdominant inhibitors of the proapoptotic forms.
As with BimS, BimAD contains the BH3 region but lacks the dynein-binding domain responsible for the reduced proapoptotic activity of the longer forms. However, BimAD is less efficient in inducing apoptosis than BimS when overexpressed in cells. This might be explained by the fact that BimAD lacks the C-terminal hydrophobic tail, which has been shown to have proapoptotic potential for other Bcl-2 family members (2, 16). For the same reason, overexpression of BimEL in NIH 3T3 results in a more potent killing effect than that of BimAD. Surprisingly, BimL was less efficient in inducing apoptosis in our system, raising the possibility that some region in exon 3 might also be important for the death agonist activity of these molecules. To summarize the data on the apoptotic potency of the different splice products, it appears that the BH3 domain (exon 5) is required for killing and the dynein binding domain (exon 4) reduces the strength of apoptosis induction, presumably by causing sequestration of Bim to the cytoskeleton, while the hydrophobic C-terminal domain (exon 6) increases the apoptotic strength.
Most BH3-only proteins are thought to induce apoptosis by interacting with antiapoptotic Bcl-2 family members, neutralizing their ability to antagonize the function of the proapoptotic Bax and Bak. Our binding experiments with the TAP tag method showed that BimS and BimAD are capable of heterodimerizing in vivo with Bcl-2 and Bcl-XL as well as with Bax in the presence of nonionic detergents. Furthermore, immunoprecipitation studies showed that Bim can associate with Bax even when expressed at the endogenous level. These interactions require an intact BH3 domain, as point mutations within this domain disrupt the binding activity of BimS and BimAD to the other proteins. The observation that Bim cannot bind Bax stably in the presence of 1% CHAPS, while it can still bind Bcl-2 and induce the exposure of the N terminus of Bax, prompted us to review the available literature in more detail. To date, only three proapoptotic proteins have been shown to associate directly with Bax: the BH3-only proteins Bid and MAP-1 (34, 36) and the SH-3 domain-containing protein Bif-1 (8). Notably, Bif-1 was reported to bind to Bax only in its native conformation, and the addition of nonionic detergents was able to disrupt this interaction. On the other hand, the study of the interaction of MAP-1 with Bax was performed only in the presence of 0.2% NP-40, rendering it difficult to discern the relevance of this interaction under native conditions. Surprisingly, previous reports of Bid interaction with Bax or Bak also used nonionic detergents in the lysis of cells expressing ectopic Bid and in the subsequent purification steps (9, 23, 36, 37). Only in two cases was this interaction studied in the presence of CHAPS, and there was no indication that either full-length or cleaved Bid could bind to Bax under these conditions (19, 32). As in the case of Bim, Bid has also been extensively reported to induce a change in Bax conformation that is detectable through immunofluorescence and immunoprecipitation studies (9, 23). Hence, the scenario for Bid appears to be very similar to that which we have found for Bim.
Recent reports on Bax and Bak activation highlight a possible model to explain our results (12, 32). These data imply that the change at the N terminus of Bax and Bak is not the only critical event in the activation of these molecules but that a second and distinct conformational change at the C terminus is necessary for complete Bax activation and dissociation of the inhibitory Bcl-2-Bcl-XL interaction. In this scenario, Triton X-100 could only trigger the exposure of an epitope at the amino terminus, leaving the carboxy terminus unaltered (32) and Bcl-XL still able to bind (12). Only in the presence of a damage signal could both of the epitopes be exposed (12, 32), and in this situation, Bcl-XL binding is lost (12). Importantly, Bcl-XL overexpression did not alter the kinetics of the Bak N-terminal conformational change after etoposide treatment, although it was found to delay the second change (12). Similarly, expression of the viral Bcl-2 homolog E1B19K protein did not prevent the first N-terminal change in Bax after treatment with tumor necrosis factor plus cycloheximide, although it prevented the conformational change at the C terminus and the formation of Bax cross-linked products (32).
These data agree with those of previous studies on the solution structure of monomeric Bax (33), which suggest that the N terminus is highly mobile and remains flexible and solvent-exposed in the presence of detergents and that this event does not necessarily induce exposure of the BH3 domain, which remains occluded by the C terminus. Indeed, the exposure of the N terminus of Bax is rapid, reversible, and not sufficient alone to commit the cell to apoptosis (18).
In this scenario, Bim (and possibly Bid) could represent the necessary signal for Bax activation, and such a role has already been proposed in Taxol-induced damage in SH-SY5Y cells (18). In our model (Fig. 12), the Bax N terminus exposure induced by Bim (or Bid) is a reversible event kept under control by the inhibitory interaction of Bcl-2 (indeed, Bim associated with Bcl-2 even in the presence of CHAPS). A stable interaction between Bim and Bax could become detectable only if the N terminus remained exposed for a sufficient period of time, as we have seen in the presence of Triton as well as in the in vitro binding experiment where His-BaxΔTM was constitutively active. Indeed, the amount of Bax that could be pulled down by the Bax N20 conformation-specific antibody was severalfold higher in the presence of Triton than in the presence of CHAPS, even in control cells treated with staurosporine (Fig. 11C). The formation of the active Bax (step II), with the BH3 domain exposed and Bim bound, could be a rapid event leading to Bax oligomerization and translocation to mitochondria. Indeed, the interactions between the C-terminal helix and the BH3 binding pocket are mainly hydrophobic, and rotation at this level would lead to a very energetically unfavorable conformation (33). The formation of oligomers could be the energy-driven process, and Bim is unlikely to bind to Bax when oligomers are found. In another report, Bid did not cofractionate with the 500-kDa Bax complex in cells treated with tumor necrosis factor plus cycloheximide (32), and a hit-and-run model for truncated Bid-dependent Bax activation was proposed in that study. Further studies are needed to elucidate such a model for Bim.
FIG. 12.
Model for the apoptotic pathway triggered by Bim. See the text for details.
From the behavior of the BimS and BimAD BH3 domain point mutants, it appears that the ability of these Bim isoforms to cause apoptosis can occur via two pathways (Fig. 12). Bim and the mutant G156A can bind to Bcl-2 and shift the equilibrium towards the first step (indirect pathway). In addition, Bim and the mutant N160A may bind directly to Bax and induce an activated conformation. There are thus direct and indirect pathways to Bax activation. While most BH3-only proteins have been suspected to use the indirect pathway, Bid activates Bax directly and Bim appears to act in both ways. Bim is only the second BH3-only protein to be shown to be able to activate Bax directly. In the case of the indirect mechanism through binding to Bcl-2 or other antiapoptotic family members, the activation of Bax still results with high efficiency. While the exact mechanism for this is unknown, it is likely that the effective removal of Bcl-2 and Bcl-XL leads to the accumulation of BH3 proteins (including Bim) in a free form that are able to target Bax directly. Alternatively, the formation of Bim-Bcl-2 heterodimers may lead to increased levels of free Bax that could be targeted for activation by proteins unrelated to the Bcl-2 family or that could become spontaneously activated.
The possibility has been raised that other molecules in addition to Bid could be responsible for Bax activation (38, 39). We show here that BimS and BimAD are likely candidates, but others may exist as well. At present it is not clear whether the more highly expressed isoforms of Bim, BimL and BimEL, also act directly on Bax. While it was not possible to see complexes of BimL and BimEL with Bax in the experiments performed here, it is possible that they may form under conditions where BimL and BimEL are released from microtubules following apoptotic stimuli. Similarly, other BH3-only proteins might be able to interact directly with Bax and Bak when appropriately stimulated.
Acknowledgments
This work was supported by Cancer Research UK and the Special Trustees of the Hammersmith and Acton Hospitals.
We thank Bim Laguda and Zoë Leech for expert technical assistance and Rita Lopes, Miguel Martins, Ingram Iaccarino, and Sophie Khanna for helpful discussions. We are grateful to Helen Hurst for critical reading of the manuscript.
REFERENCES
- 1.Adams, J. M., and S. Cory. 2001. Life-or-death decisions by the Bcl-2 protein family. Trends Biochem. Sci. 26:61-66. [DOI] [PubMed] [Google Scholar]
- 2.Antonsson, B., and J. C. Martinou. 2000. The Bcl-2 protein family. Exp. Cell Res. 256:50-57. [DOI] [PubMed] [Google Scholar]
- 3.Bae, J., C. P. Leo, S. Y. Hsu, and A. J. Hsueh. 2000. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J. Biol. Chem. 275:25255-25261. [DOI] [PubMed] [Google Scholar]
- 4.Bingle, C. D., R. W. Craig, B. M. Swales, V. Singleton, P. Zhou, and M. K. Whyte. 2000. Exon skipping in Mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. J. Biol. Chem. 275:22136-22146. [DOI] [PubMed] [Google Scholar]
- 5.Boise, L. H., M. Gonzalez-Garcia, C. E. Postema, L. Ding, T. Lindsten, L. A. Turka, X. Mao, G. Nunez, and C. B. Thompson. 1993. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74:597-608. [DOI] [PubMed] [Google Scholar]
- 6.Bouillet, P., D. Metcalf, D. C. Huang, D. M. Tarlinton, T. W. Kay, F. Kontgen, J. M. Adams, and A. Strasser. 1999. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286:1735-1738. [DOI] [PubMed] [Google Scholar]
- 7.Bouillet, P., L. C. Zhang, D. C. Huang, G. C. Webb, C. D. Bottema, P. Shore, H. J. Eyre, G. R. Sutherland, and J. M. Adams. 2001. Gene structure alternative splicing, and chromosomal localization of pro-apoptotic Bcl-2 relative Bim. Mamm. Genome 12:163-168. [DOI] [PubMed] [Google Scholar]
- 8.Cuddeback, S. M., H. Yamaguchi, K. Komatsu, T. Miyashita, M. Yamada, C. Wu, S. Singh, and H. G. Wang. 2001. Molecular cloning and characterization of Bif-1. A novel Src homology 3 domain-containing protein that associates with Bax. J. Biol. Chem. 276:20559-20565. [DOI] [PubMed] [Google Scholar]
- 9.Desagher, S., A. Osen-Sand, A. Nichols, R. Eskes, S. Montessuit, S. Lauper, K. Maundrell, B. Antonsson, and J. C. Martinou. 1999. Bid-induced conformational change of Bax is responsible for mitochondrial cytochrome c release during apoptosis. J. Cell Biol. 144:891-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dijkers, P. F., R. H. Medemadagger, J. W. Lammers, L. Koenderman, and P. J. Coffer. 2000. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 10:1201-1204. [DOI] [PubMed] [Google Scholar]
- 11.Gilmore, A. P., A. D. Metcalfe, L. H. Romer, and C. H. Streuli. 2000. Integrin-mediated survival signals regulate the apoptotic function of Bax through its conformation and subcellular localization. J. Cell Biol. 149:431-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths, G. J., B. M. Corfe, P. Savory, S. Leech, M. Degli Esposti, J. S. Hickman, and C. Dive. 2001. Cellular damage signals promote sequential changes at the N-terminus and BH-1 domain of the pro-apoptotic protein Bak. Oncogene 20:7668-7675. [DOI] [PubMed] [Google Scholar]
- 13.Hsu, Y. T., and R. J. Youle. 1998. Bax in murine thymus is a soluble monomeric protein that displays differential detergent-induced conformations. J. Biol. Chem. 273:10777-10783. [DOI] [PubMed] [Google Scholar]
- 14.Hsu, Y. T., and R. J. Youle. 1997. Nonionic detergents induce dimerization among members of the Bcl-2 family. J. Biol. Chem. 272:13829-13834. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, Z. H., and J. Y. Wu. 1999. Alternative splicing and programmed cell death. Proc. Soc. Exp. Biol. Med. 220:64-72. [DOI] [PubMed] [Google Scholar]
- 16.Kataoka, T., N. Holler, O. Micheau, F. Martinon, A. Tinel, K. Hofmann, and J. Tschopp. 2001. Bcl-rambo, a novel Bcl-2 homologue that induces apoptosis via its unique C-terminal extension. J. Biol. Chem. 276:19548-19554. [DOI] [PubMed] [Google Scholar]
- 17.Kelekar, A., and C. B. Thompson. 1998. Bcl-2-family proteins: the role of the BH3 domain in apoptosis. Trends Cell Biol. 8:324-330. [DOI] [PubMed] [Google Scholar]
- 18.Makin, G. W., B. M. Corfe, G. J. Griffiths, A. Thistlethwaite, J. A. Hickman, and C. Dive. 2001. Damage-induced Bax N-terminal change, translocation to mitochondria and formation of Bax dimers/complexes occur regardless of cell fate. EMBO J. 20:6306-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy, K. M., U. N. Streips, and R. B. Lock. 2000. Bcl-2 inhibits a Fas-induced conformational change in the Bax N terminus and Bax mitochondrial translocation. J. Biol. Chem. 275:17225-17228. [DOI] [PubMed] [Google Scholar]
- 20.Nechushtan, A., C. L. Smith, I. Lamensdorf, S. H. Yoon, and R. J. Youle. 2001. Bax and Bak coalesce into novel mitochondria-associated clusters during apoptosis. J. Cell Biol. 153:1265-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Connor, L., A. Strasser, L. A. O'Reilly, G. Hausmann, J. M. Adams, S. Cory, and D. C. Huang. 1998. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 17:384-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Reilly, L. A., L. Cullen, J. Visvader, G. J. Lindeman, C. Print, M. L. Bath, D. C. Huang, and A. Strasser. 2000. The proapoptotic BH3-only protein bim is expressed in hematopoietic, epithelial, neuronal, and germ cells. Am. J. Pathol. 157:449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez, D., and E. White. 2000. TNF-alpha signals apoptosis through a bid-dependent conformational change in Bax that is inhibited by E1B 19K. Mol. Cell 6:53-63. [PubMed] [Google Scholar]
- 24.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (tap) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 25.Putcha, G. V., K. L. Moulder, J. P. Golden, P. Bouillet, J. A. Adams, A. Strasser, and E. M. Johnson. 2001. Induction of bim, a proapoptotic bh3-only bcl-2 family member, is critical for neuronal apoptosis. Neuron 29:615-628. [DOI] [PubMed] [Google Scholar]
- 26.Puthalakath, H., D. C. Huang, L. A. O'Reilly, S. M. King, and A. Strasser. 1999. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol. Cell 3:287-296. [DOI] [PubMed] [Google Scholar]
- 27.Reed, R. 1989. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 3:2113-2123. [DOI] [PubMed] [Google Scholar]
- 28.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 29.Shiraiwa, N., N. Inohara, S. Okada, M. Yuzaki, S. Shoji, and S. Ohta. 1996. An additional form of rat Bcl-x, Bcl-xbeta, generated by an unspliced RNA, promotes apoptosis in promyeloid cells. J. Biol. Chem. 271:13258-13265. [DOI] [PubMed] [Google Scholar]
- 30.Stamm, S., M. Q. Zhang, T. G. Marr, and D. M. Helfman. 1994. A sequence compilation and comparison of exons that are alternatively spliced in neurons. Nucleic Acids Res. 22:1515-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun, Y. F., L. Y. Yu, M. Saarma, T. Timmusk, and U. Arumae. 2001. Neuron-specific bcl-2 homology 3 domain-only splice variant of bak is anti-apoptotic in neurons, but pro-apoptotic in non-neuronal cells. J. Biol. Chem. 276:16240-16247. [DOI] [PubMed] [Google Scholar]
- 32.Sundararajan, R., and E. White. 2001. E1B 19K blocks Bax oligomerization and tumor necrosis factor alpha-mediated apoptosis. J. Virol. 75:7506-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki, M., R. J. Youle, and N. Tjandra. 2000. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell 103:645-654. [DOI] [PubMed] [Google Scholar]
- 34.Tan, K. O., K. M. Tan, S. L. Chan, K. S. Yee, M. Bevort, K. C. Ang, and V. C. Yu. 2001. MAP-1, a novel proapoptotic protein containing a BH3-like motif that associates with Bax through its Bcl-2 homology domains. J. Biol. Chem. 276:2802-2807. [DOI] [PubMed] [Google Scholar]
- 35.U, M., T. Miyashita, Y. Shikama, K. Tadokoro, and M. Yamada. 2001. Molecular cloning and characterization of six novel isoforms of human Bim, a member of the proapoptotic Bcl-2 family. FEBS Lett. 509:135-141. [DOI] [PubMed] [Google Scholar]
- 36.Wang, K., X. M. Yin, D. T. Chao, C. L. Milliman, and S. J. Korsmeyer. 1996. BID: a novel BH3 domain-only death agonist. Genes Dev. 10:2859-2869. [DOI] [PubMed] [Google Scholar]
- 37.Wei, M. C., T. Lindsten, V. K. Mootha, S. Weiler, A. Gross, M. Ashiya, C. B. Thompson, and S. J. Korsmeyer. 2000. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 14:2060-2071. [PMC free article] [PubMed] [Google Scholar]
- 38.Wei, M. C., W. X. Zong, E. H. Cheng, T. Lindsten, V. Panoutsakopoulou, A. J. Ross, K. A. Roth, G. R. MacGregor, C. B. Thompson, and S. J. Korsmeyer. 2001. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292:727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zong, W. X., T. Lindsten, A. J. Ross, G. R. MacGregor, and C. B. Thompson. 2001. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev. 15:1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]