Abstract
Induction of low-density lipoprotein (LDL) receptor transcription in response to depletion of cellular sterols in animal cells is well established. The intracellular signal or signals involved in regulating this process, however, remain unknown. Using a specific inhibitor of protein kinase C (PKC), calphostin C, we show the requirement of this kinase in the induction process in human hepatoma HepG2 cells. Overexpression of PKCɛ, but not PKCα, -γ, -δ, or -ζ was found to dramatically induce (approximately 18-fold) LDL receptor promoter activity. Interestingly, PKCɛ-mediated induction was found to be sterol resistant. To further establish that PKCɛ is involved in the sterol regulation of LDL receptor gene transcription, endogenous PKCɛ was specifically inhibited by transfection with antisense PKCɛ phosphorothionate oligonucleotides. Antisense treatment decreased endogenous PKCɛ protein levels and completely blocked induction of LDL receptor transcription following sterol depletion. PKCɛ-induced LDL receptor transcription is independent of the extracellular signal-regulated kinase 1 and 2 (p42/44MAPK) cascade, because the MEK-1/2 inhibitor, PD98059 did not inhibit, even though it blocked p42/44MAPK activation. Finally, photoaffinity labeling studies showed an isoform-specific interaction between PKCɛ and sterols, suggesting that sterols may directly modulate its function by hampering binding of activators. This was confirmed by PKC activity assays. Altogether, these results define a novel signaling pathway leading to induction of LDL receptor transcription following sterol depletion, and a model is proposed to account for a new function for PKCɛ as part of a sterol-sensitive signal transduction pathway in hepatic cells.
Protein kinase C (PKC) represents a family of serine/threonine protein kinases that plays a key role in signal transduction and regulation of gene expression (11, 39, 40, 43). Molecular cloning and biochemical studies have revealed at least 12 PKC subspecies, which can be classified into three groups—classic (cPKC), novel (nPKC), and atypical (aPKC)—on the basis of their structures. The PKC family comprises a regulatory domain in the amino terminus and a catalytic domain in the carboxyl terminus. The cPKCs, including PKCα, -βI, -βII, and -γ, have two common regions, C1 and C2, in the regulatory domain. The C1 region has two cysteine-rich loops that are the binding site for diacylglycerol (DAG) and phorbol ester (tetradecanoyl phorbol acetate [TPA]). The C2 region binds to calcium. The nPKCs, including PKCδ, -ɛ, -η, and -θ, lack the C2 region. The aPKCs, including PKCζ and -ι/λ, lack the C2 region and have only one cysteine-rich loop in the C1 region. Key lipid cofactors of PKCs are DAG, phosphatidylserine (PS), and calcium. All of these are required to activate cPKCs, whereas calcium is not required to activate nPKCs. aPKCs are insensitive to both DAG and calcium. Other lipid species, such as cholesterol sulfate, fatty acids, lysophospholipids, and phosphatidylinositols, also modulate the activity of certain PKC isoforms (3, 12, 24, 41). Several PKC family members displaying responsiveness to calcium, DAG, and fatty acids are expressed in hepatic cells (16, 26), but their precise functional roles in cholesterol uptake and biosynthesis are not yet clearly defined.
Animal cells regulate their cholesterol content by fine-tuning of the supply of exogenous and endogenous cholesterol (5). Both cholesterol and fatty acids have been demonstrated to be important regulators of plasma low-density lipoprotein (LDL) cholesterol levels due to alterations in LDL receptor activity (6, 7, 20). The mechanism by which sterols modulate LDL receptor gene transcription has been examined in great detail. Involved in this pathway are a family of proteins, designated sterol regulatory element binding proteins (SREBPs), that play an integral role in the feedback pathway by which cholesterol suppresses transcription of LDL receptor gene. The SREBPs are transcription factors that are bound to the endoplasmic reticulum and nuclear envelope by virtue of two membrane-spanning regions. When these membranes are depleted of sterols, a two-step proteolytic process releases the amino terminal of SREBPs, which then travel to the nucleus and activate LDL receptor transcription via interaction with multiple nuclear factors and coactivators (7, 42). When cells are overloaded with sterols, cleavage of SREBPs is inhibited, resulting in suppression of the LDL receptor gene. A similar mechanism has been shown for the regulation of other sterol-responsive genes and genes involved in fatty acid biosynthesis. Three isoforms of SREBPs have been identified. Interestingly, expression and proteolytic processing of two of these proteins, designated SREBP-1a and -1c, are more sensitive to feedback control by fatty acids, whereas expression and processing of SREBP-2 are under feedback control by cholesterol. Because cholesterol does not directly bind to purified SREBPs or to any component involved in its processing, considerable uncertainty remains concerning the precise mechanisms by which cellular cholesterol or fatty acids communicate with the SREBP processing machinery. Activation of transcription factors is usually regulated by signaling pathways, and recent studies from our laboratory as well as by others have focused on the roles of mitogen-activated protein kinase (MAPK) signaling cascades in the regulation of LDL receptor transcription in human hepatoma HepG2 cells by cytokines, growth factors, TPA, and insulin (14, 25, 31, 34, 55). Recently, insulin-induced LDL receptor induction has been linked to extracellular signal-regulated kinase 1 and 2 (p42/44MAPK)-mediated phosphorylation of SREBP-1c (49).
There is virtually no information available on the underlying signaling kinase(s) initiated by the alterations in cellular cholesterol leading to the regulation of the LDL receptor gene. Previous studies have shown that LDL induces a rapid but transient cytosol-to-membrane translocation of TPA-sensitive PKC isoforms in human vascular smooth muscle cells or skin fibroblasts (37, 52). Furthermore, suppression of TPA-sensitive PKC activity in fibroblasts from Nieman-Pick C patients has been linked to accumulation of naturally occurring PKC modulators, such as sterols and sphingosine (35, 48). In view of these observations, we investigated the role of PKC in the induction of LDL receptor transcription following depletion of sterols in HepG2 cells. We show that the PKC signaling mechanism controls LDL receptor transcription and specifically define PKCɛ as the single isoform responsible for LDL receptor induction in response to depletion of sterols. We propose that PKCɛ is a candidate physiologic modulator of LDL receptor gene expression in response to the depletion of sterols and may play an important role in mediating specific effects of other lipids on expression of this receptor gene.
MATERIALS AND METHODS
Materials.
Recombinant human PKC isoforms and catalytic subunit were obtained from Calbiochem. Cholesterol, all-trans-retinoic acid (atRA), PS, histone type III, DAG, and TPA were purchased from Sigma Corp. 25-Hydroxycholesterol was obtained from Steraloids, Inc. TRIzol and tissue culture supplies were purchased from Life Technologies. Fetal bovine lipoprotein-deficient serum (LPDS) was obtained from PerImmune, Inc. Zeta-Probe blotting membrane and the protein assay reagent were purchased from Bio-Rad Laboratories. PKCɛ peptide substrate and the chemiluminescent reporter gene assay system for the detection of luciferase were purchased from Tropix, Inc. PD98059, indomethacin, nordihydroguaiaretic acid, and phospholipase C (PLC) inhibitors were purchased from Calbiochem-Novabiochem Corp. Antibodies to phosphorylation-independent and phospho-specific activated forms of p42/44MAPK (Thr202/Tyr204), as well as antibodies to different PKC isoforms, were purchased from Cell Signaling Technology, Inc. Antisense or scrambled phosphorothionate oligonucleotides were synthesized by Gibco-BRL (Invitrogen, Calif.). [11,12-3H]all-trans-retinoic acid ([3H]atRA) was purchased from Perkin-Elmer Life Science. 3′-[α-32P]dCTP (3,000 Ci/mmol) was obtained from DuPont, and 5′-[γ-32P]dATP was obtained from Amersham Pharmacia Biotech. The enhanced chemiluminescence (ECL) detection kit was obtained from Amersham Pharmacia Biotech (Piscataway, N.J.).
Plasmid.
Wild-type and constitutively active PKC isoform constructs were gifts from Peter Parker (Imperial Cancer Research Fund, London, United Kingdom) (51). The cDNAs of the constitutive active (CA) PKC mutants carry deletions or point mutations in the N-terminal pseudosubstrate region. In PKCα, amino acids 22 to 28 are deleted; in PKCδ, Ala-147 is exchanged for Glu; PKCɛ lacks amino acids 156 to 162; and PKCζ carries a deletion from amino acids 116 to 122. A dominant-negative (DN) mutant of PKCα containing a mutation in the phosphorylation sites (Thr-494, Thr-495, and Thr-497) in the activation loop of the kinase domain was also obtained from Peter Parker (17). The LDL receptor promoter-luciferase construct A has been described previously (33). The construction of plasmids containing single point mutations in Sp1 (C to A), SRE-1 (A to C), and FP1 (GG to TT) has been described previously (13). Plasmid DNA was prepared by purification through columns, as instructed by the manufacturer (Qiagen, Inc., Valencia, Calif.).
Cell culture.
The human hepatoma cell line HepG2 was maintained as monolayer cultures in Eagle's minimum essential medium (BioWhittaker, Inc., Walkersville, Md.) supplemented with 10% fetal calf serum (Life Technologies, Rockville, Md.). Cells were grown at 37°C in a humidified 5% carbon dioxide-95% air atmosphere. For suppression and induction of LDL receptor expression, the cells were switched to medium containing 10% LPDS alone (induced) or together with sterols (10 μg of 25-hydroxycholesterol per ml plus 10 μg of cholesterol per ml; suppressed) and incubated for an additional 12 to 16 h.
Transient transfection.
HepG2 cells were transfected by the Lipofectamine (Life Technologies) method. For transfection experiments, cells (105) were seeded at a density of 2.5 × 105 cells in a six-well plate 1 day in advance. Transfections were performed in duplicate with 0.6 μg of DNA for the LDL receptor promoter-luciferase construct containing the bp −173/+35 sequences of the human LDL receptor promoter (plasmid A) (13, 33) and with the indicated amounts of the relevant expression vector or the corresponding empty vector (14, 55). After 5 h, the cells were washed with basic medium and refed with Eagle's minimum essential medium containing 10% fetal calf serum. Approximately 12 to 16 h later, transfected cells were switched to medium supplemented with either 10% LPDS or 10% LPDS plus 25-hydroxycholesterol/cholesterol and were incubated for an additional 16 to 20 h. Finally, dishes were washed with phosphate-buffered saline and lysed with luciferase lysis buffer (100 mM potassium phosphate [pH 7.8], 0.2% Triton X-100 Σ, 0.5 mM dithiothreitol). Luciferase activity was measured according to the Tropix protocol. Data are representative of at least four independent experiments performed in duplicate and are expressed as X-fold increases in luciferase activity, which was calculated relative to the basal level of LDL receptor promoter activity (set to 1 U) and corrected for empty vector effects for each expression vector.
Treatment of cells with phosphorothionate-modified oligonucleotides.
HepG2 cells were seeded at 2 × 105 cells per well in six-well plates and transfected with 10 μg of oligonucleotides with the use of 40 μl of Lipofectamine according to the manufacturer's protocol. Cells were left to incubate for 24 h and then switched to medium containing 10% LPDS and the indicated concentrations of oligonucleotides. After 12 h, transfected cells were harvested to determine luciferase expression levels. Antisense oligonucleotides to PKC isoforms were complementary to the translation initiation region of mRNA specific for humans. The antisense sequences used for PKCɛ were 5′-GAACACTACCATGGTCGG-3′. Sense oligonucleotide for PKCɛ (5′-CCGACCATGGTAGTGTTC-3′) was used as a control. Sequences were complementary to the translation initiation region, nucleotides −6 to 12 for PKCɛ (29). Oligonucleotides were dissolved in sterile deionized water to a final concentration of 1 mM, aliquoted, and stored at −20°C until ready for use.
Western blot analysis.
Cell extracts (10 to 20 μg) were resolved on a 10% acrylamide separating gel by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to a nitrocellulose membrane. Membrane blocking, washing, antibody incubation, and detection by ECL were performed as previously described (55). Quantitative analysis of protein levels was performed by densitometric scanning of the autoradiograms from three or more independent experiments. To quantify the signals, membranes were scanned by a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.), and image and quantification analyses were carried out with ImageQuant 5.0 software (Molecular Dynamics). All values are reported as normalized to that of the control, which was set to 1.
Northern blot analysis.
Total RNA was isolated with TRIzol, and Northern blotting was done essentially as described earlier (55). Briefly, 10 μg of total cellular RNA was fractionated on a 1% formaldehyde agarose gel and transferred to Zeta-Probe membrane by capillary blotting. RNA blots were hybridized with LDL receptor-specific single-stranded M13 probe labeled with [α-32P]dCTP. In most cases, the same blot was rehybridized with 32P-labeled single-stranded M13 probe specific for β-actin. Hybridized filters were washed and exposed to Kodak X-ray film. The relative intensities of specific bands were determined densitometrically within the linear range of the film on a model 300A laser densitometer (Molecular Dynamics) with ImageQuant software. LDL receptor mRNA was normalized to the β-actin mRNA level, and data for each point were plotted as the percentage of LDL receptor mRNA relative to controls.
Photoaffinity labeling with [3H]atRA.
Direct photoaffinity labeling with [3H]atRA was done with modifications of the method described by Radominska-Pandya et al. (46). PKCα and PKCɛ (7 pmol each in 20 mM HEPES at pH 7) were incubated for 10 min at room temperature with or without increasing concentrations (0, 5, 50, and 100 μM) of 25-hydroxycholesterol. [3H]atRA (30 Ci/mmol) was added to a final concentration of 1.5 μM (0.03 nmol [1 μCi] in a final volume of 20 μl). 25-Hydroxycholesterol, atRA, and [3H]atRA were added in ethanol with a final concentration of 2% ethanol in all samples. Samples were incubated for 10 min on ice prior to photolabeling with a handheld long-wave (366 nm) UV lamp (UVP-21; UV Products, San Gabriel, Calif.) for 15 min. Proteins were denatured by addition of NuPAGE denaturing buffer (Invitrogen), followed by sonication, boiling for 1 min, and, finally, centrifugation at 13,000 rpm in an Eppendorf microcentrifuge (Brinkman Instruments, Westbury, N.Y.). Proteins were separated by SDS-PAGE on a NuPAGE mini-gel (1.5 mm).
Following electrophoresis, gels were stained with Coomassie blue, destained, washed thoroughly with water, treated with Autofluor autoradiography enhancer, (National Diagnostics, Manville, N.J.), dried, and subjected to autoradiography at −80°C for 4 to 7 days. Results were quantitated by densitometric analysis of the autoradiograms by using an AlphaInnotech IS-100 Digital Imaging System (AlphaInnotech, San Leandro, Calif.).
PKC enzymatic assay.
PKCα activity was assayed with Triton X-100 mixed micelles containing PS and DAG, with histone 1 as the substrate for PKCα phosphorylation, as described by Radominska-Pandya et al. (46). For PKCɛ activity, PKCɛ peptide substrate was substituted for histone 1. Unless stated otherwise, the reaction mixture contained 20 mM Tris (pH 7.5), 20 mM MgCl2, 1.0 mM CaCl2, 0.25 mM EGTA, 0.25 mM EDTA, 0.28 mg of PS per ml, 10 μM phorbol myristate acetate (PMA), 1 mM β-mercaptoethanol, 0.3% Triton X-100, 50 μM histone 1 or PKCɛ peptide substrate, 20 μM [γ-32P]ATP, and 150 mU of PKC in a final volume of 25 μl. Mixed micelles were prepared by drying PS (2.5 mg) and PMA (final concentration, 100 μM), dissolved in chloroform in a glass tube, under a stream of nitrogen. One milliliter of freshly prepared 3% Triton X-100 (prepared fresh) was added, and the mixture was sonicated.
Seven picomoles each of PKCα or PKCɛ in 20 mM HEPES (pH 7) was incubated for 10 min at room temperature with or without increasing concentrations of 25-hydroxycholesterol (0, 25, 50, 75, 100, and 200 μM for PKCɛ and 0, 25, 75, 150, and 250 μM for PKCα). [γ-32P]ATP (20 μM) was added, and the samples were incubated for 10 min at 30°C. The reactions were terminated by spotting an aliquot of reaction mixture on phosphocellulose disks (P-81, 2.3 cm in diameter; Whatman, Clifton, N.J.). After washing the disks with 1% phosphoric acid, γ-32P incorporation into the substrate was determined by liquid scintillation spectrometry. Non-PKC-specific activity (background) was determined in samples incubated in the absence of the enzyme.
RESULTS
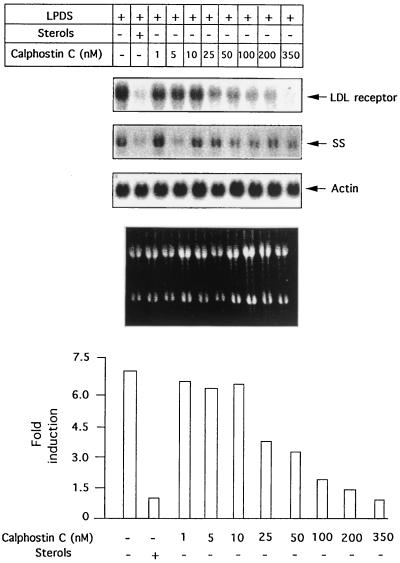
LDL receptor induction in response to sterol depletion blocked by calphostin C.
To assess the role of PKC in the induction of LDL receptor transcription in response to depletion of sterols, we examined the effects of inhibition of this kinase by using a specific inhibitor, calphostin C (15). Cells were pretreated for 30 min with increasing concentrations of calphostin C and were then switched to medium containing 10% LPDS in the presence or absence of sterols. After 16 h, total RNA was isolated, and the effects on LDL receptor induction were examined by Northern blotting (Fig. 1). Interestingly, calphostin C suppressed LDL receptor expression in a dose-dependent manner without significantly affecting expression of a housekeeping gene coding for the protein actin (Fig. 1). A significant decrease in LDL receptor gene expression was apparent at 50 nM, and maximal suppression was observed at 350 nM. In fact, the expression levels of LDL receptor seen on treatment with 350 nM calphostin C were similar to or lower than suppressed levels in the presence of sterols. We also measured expression of another sterol-sensitive gene of the cholesterol biosynthetic pathway, coding for squalene synthase (SS). As shown in Fig. 1, unlike the LDL receptor, induction of SS expression in response to sterol deprivation is slightly reduced (approximately twofold) on calphostin C treatment, showing that LDL receptor expression is much more sensitive to PKC inhibition than SS. This observation suggests basic differences in the regulatory mechanisms controlling their expression either due to the involvement of nuclear factors that bind adjacent to SREBP or due to the participation of different SREBP isoforms.
FIG. 1.
Effect of calphostin C on the endogenous LDL receptor gene induction in response to sterol depletion. HepG2 cells were either untreated or pretreated with indicated concentrations of calphostin C and then incubated for 16 h in medium containing 10% LPDS or 10% LPDS supplemented with 10 μg of cholesterol per ml and 10 μg of 25-hydroxycholesterol per ml. Total cellular RNA was subjected to Northern blotting to determine the amounts of LDL receptor, SS, and actin mRNAs. Autoradiographs were quantitated densitometrically, as described previously (55). LDL receptor mRNA levels were normalized by comparison with levels of actin mRNA. In the bottom panel, results are expressed as X-fold induction by 10% LPDS in the presence of calphostin C compared with untreated cells incubated in 10% LPDS plus sterols (given an arbitrary value of 1). Values shown are the averages of two different experiments. The experiment was repeated two times with similar results.
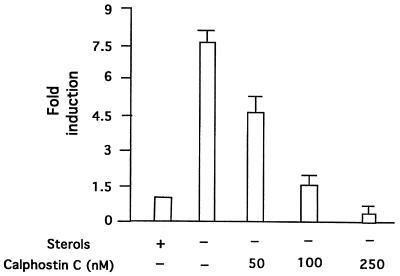
To support the possibility that calphostin C inhibits LDL receptor transcription, we examined the effects of this inhibitor on human LDL receptor promoter activity in HepG2 cells. LDL receptor transcription was monitored by transfecting HepG2 cells with a previously cloned fragment of the human LDL receptor promoter fused to a luciferase reporter gene (34) in the absence and presence of calphostin C. As shown in Fig. 2, calphostin C completely blocked induction of LDL receptor transcription following depletion of sterols, suggesting that PKC activity is required for induction to occur. As a complementary approach, cells were transfected with a kinase-inactive mutant of PKCα, which is a DN inhibitor of all the endogenous PKC isoforms (18). The LDL receptor promoter luciferase construct was cotransfected with DN PKCα, and the cells were grown in the absence or presence of sterols. As shown in Fig. 3, expression of this mutant also inhibited the induction of the LDL receptor promoter in response to sterol depletion.
FIG. 2.
Effect of calphostin C on induction of human LDL receptor promoter transcription in response to sterol depletion in HepG2 cells. HepG2 cells were transfected with a human LDL receptor-luciferase reporter construct A (0.6 μg/plate), as described earlier (33). After 24 h, the indicated concentration of calphostin C was added, and the cells were incubated for an additional 12 h in 10% LPDS medium before harvesting. Luciferase activity was determined and normalized to the protein content of each extract. The X-fold suppression in luciferase activity by treatment with calphostin C was calculated relative to the expression level of the untreated cells. Results are expressed as X-fold induction compared with untreated cells incubated in 10% LPDS plus sterols (given an arbitrary value of 1). The values obtained are the mean ± standard error of three separate experiments.
FIG. 3.
Effects of a DN mutant of PKCα on the induction of human LDL receptor promoter transcription in response to sterol depletion. HepG2 cells were cotransfected with a human LDL receptor-luciferase reporter plasmid (0.6 μg/plate) with or without the indicated concentrations of PKCα mutant plasmid or empty vector. After 24 h, cells were incubated for an additional 12 h in medium containing 10% LPDS. Cells were harvested, and luciferase activity was determined and normalized to the protein content of each extract. The X-fold induction in luciferase activity was calculated relative to the maximal expression level of the control vector-transfected cells. Luciferase activity expressed by cells transfected with empty vector in the presence of sterols was given an arbitrary value of 1. Values shown are the mean ± standard error of three separate experiments.
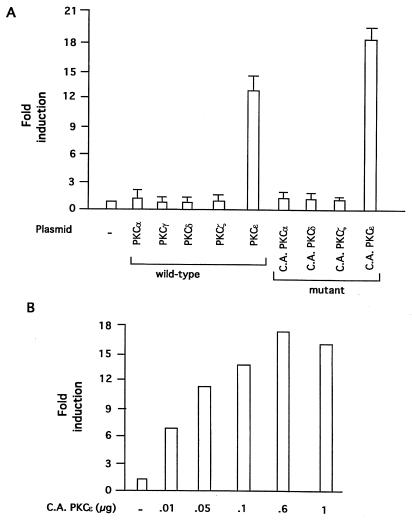
Effects of overexpression of constitutively active PKC isoforms on LDL receptor induction and its regulation by sterols.
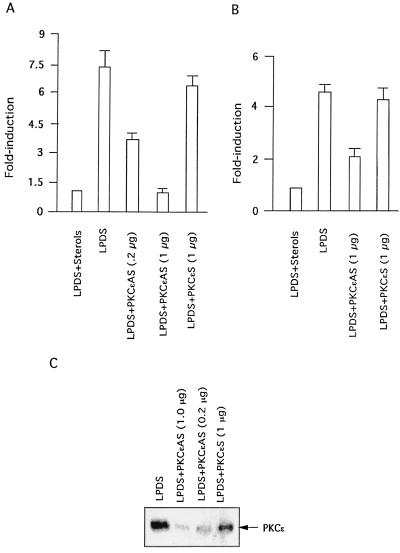
To further resolve which of the PKC isoforms may be involved in mediating the induction of the LDL receptor transcription, we determined the effects of transient overexpression of PKC isoforms on induction of LDL receptor promoter in response to depletion of sterols. HepG2 cells were cotransfected with a human LDL receptor promoter-luciferase construct and an expression vector encoding a specific PKC isoform. Transfected cells were then shifted to either 10% LPDS or 10% LPDS plus sterols for 12 h. As shown in Fig. 4A, overexpression of either wild-type or a CA mutant of PKCɛ dramatically increased LDL receptor promoter activity by approximately 18-fold, whereas overexpression of either wild-type or CA mutants of PKCα, PKCγ, PKCδ, or PKCζ did not appreciably alter the level of luciferase expression relative to the level determined with the control empty vector. The increase in LDL receptor promoter activity in response to overexpression of a CA mutant of PKCɛ was found to be dose dependent (Fig. 4B).
FIG. 4.
Effects of overexpression of different PKC isoforms on the induction of human LDL receptor promoter transcription. (A) HepG2 cells were grown as described and cotransfected with LDL receptor-luciferase reporter (14, 33) and a vector control plasmid or each of the PKC isoform wild-type or CA cDNA expression vectors. After 24 h, cells were incubated in medium containing 10% LPDS, as described in the legend to Fig. 2. After 12 h, cells were harvested, and luciferase activity was determined and normalized to the protein content of each extract. Luciferase activity expressed by cells transfected with empty vector was given an arbitrary value of 1. The amounts of DNA used were as follows: LDL receptor-luciferase reporter, 0.6 μg per well; and expression vector, 0.3 μg per well. The results are presented as means ± standard error and represent at least four individual experiments. (B) HepG2 cells were cotransfected with LDL receptor-luciferase reporter plasmid (0.6 μg per well) and the indicated amounts of expression vector encoding CA PKCɛ. Transfected cells were incubated in 10% LPDS, as mentioned above, and luciferase activity and protein contents were measured after 12 h. Each column represents the mean ± standard error of three independent experiments performed in duplicate.
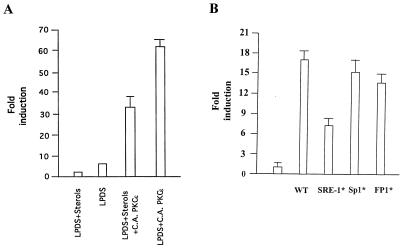
We next examined the effect of overexpression of the PKCɛ isoform on suppression of LDL receptor transcription by sterols. HepG2 cells were cotransfected with the LDL receptor promoter construct and a CA form of PKCɛ. After transient transfection, cells were grown in the absence and presence of sterols, and luciferase activity was determined as described in Materials and Methods. The luciferase activity is expressed relative to the activity in the presence of sterols. Most importantly, in response to overexpression of CA PKCɛ, sterol suppression of LDL receptor promoter activity was significantly reduced (Fig. 5A), suggesting that sterols may reduce LDL receptor promoter activity by modulating PKCɛ function. Similar results were obtained with the wild-type PKCɛ vector (results not shown).
FIG. 5.
Possible involvement of SRE-1 in PKCɛ-induced LDL receptor transcription. (A) Overexpression of a CA PKCɛ imparts resistance to suppression of LDL receptor transcription by sterols in HepG2 cells. HepG2 cells were cotransfected with LDL receptor-luciferase reporter plasmid (0.6 μg per plate) and CA PKCɛ vector (0.3 μg per plate). After 24 h, transfected cells were further incubated with either 10% LPDS or 10% LPDS supplemented with 10 μg of cholesterol per ml plus 10 μg of 25-hydroxycholesterol per ml. After 12 h, cells were harvested, and luciferase activity was determined and normalized to the protein content of each extract. Corrected luciferase activities were calculated as described for Fig. 2. Luciferase activity expressed by untransfected cells incubated in the presence of 10% LPDS supplemented with sterols was given an arbitrary value of 1. (B) HepG2 cells were cotransfected with the indicated reporter construct (0.5 μg per well) and CA PKCɛ (0.3 mg per well). After transfection, cells were cultured in medium containing 10% LPDS for 20 h prior to lysis. The relative fold induction represents the increase in luciferase activity of constructs stimulated by PKCɛ relative to the construct transfected with the control vector.
To evaluate the contribution of already known regulatory elements in PKCɛ-induced LDL receptor transcription, transient transfections were performed with wild-type and different point-mutated LDL receptor promoter variants described previously for their responsiveness to PKCɛ (13). The human LDL receptor promoter constructs containing wild-type and mutant promoter sequences fused upstream of the luciferase reporter gene along with CA PKCɛ were used for transfection of HepG2 cells for functional analysis. As shown in Fig. 5B, coexpression of exogenous PKCɛ resulted in a dramatic increase in luciferase activity with the wild-type construct. Interestingly, none of the point mutations in FP1 or the Sp1 site significantly affected transactivation, whereas mutation in SRE-1 reduced transactivation, suggesting its role in the induction process.
Effect of PKCɛ-specific antisense oligonucleotides on LDL receptor induction.
To further support the finding that PKCɛ is indeed the isoform involved in mediating LDL receptor induction by sterol depletion, studies were carried out with PKCɛ-selective antisense oligonucleotides to specifically inhibit the endogenous isoform in the cell. Cells were cultured in the presence of antisense oligonucleotides to PKCɛ for 24 h. Optimal concentrations of oligonucleotides and Lipofectamine were determined from dose-response curves. Transfected cells were grown for an additional 12 h in 10% LPDS medium containing 10 μM oligonucleotides, and luciferase activity was measured to monitor LDL receptor promoter activity. As shown in Fig. 6, sense-strand control oligonucleotides to PKCɛ did not affect the induction process. On the other hand, antisense oligonucleotides to PKCɛ abolished induction of the LDL receptor promoter. We also confirmed the action of antisense PKCɛ oligonucleotides by demonstrating a decrease in PKCɛ protein on Western blot analysis of cell lysates obtained from the same experiment. PKCɛ protein was reduced by 77% (to 23% of the control level) in cells treated with antisense PKCɛ oligonucleotides (Fig. 6B). We confirmed by immunoblotting with isoform-specific antibodies (data not shown) that antisense PKCɛ oligonucleotides did not decrease expression of other PKC isoforms. Furthermore, to examine the effects of inhibition of PKCɛ on the induction of endogenous LDL receptor expression, HepG2 cells were transfected with antisense oligonucleotides to PKCɛ, and LDL receptor expression was determined in the absence and presence of sterols. As shown in Fig. 6A, reduced stimulation of LDL receptor expression in response to depletion of sterols was observed in cells transfected with PKCɛ antisense oligonucleotides, compared to that in cells treated with sense oligonucleotides.
FIG. 6.
Effect of PKCɛ-specific antisense oligonucleotide treatment of HepG2 cells on induction of LDL receptor transcription in response to sterol depletion. (A) Treatment of HepG2 cells with PKCɛ-specific antisense oligonucleotides inhibited induction of the LDL receptor promoter plasmid following depletion of sterols. HepG2 cells were cotransfected with LDL receptor-luciferase reporter plasmid (0.6 μg) and either PKCɛ-specific sense (S) or PKCɛ-specific antisense (AS) oligonucleotides at the indicated concentrations. After 24 h, cells were grown for an additional 12 h in the presence of the introduced oligonucleotides in 10% LPDS. Data are presented to show loss of induction in the presence of oligonucleotides in response to sterol depletion. Results are given as the average ± standard error of three separate experiments performed in duplicate. (B) Effect of antisense PKCɛ oligonucleotides on the induction of endogenous LDL receptor expression in response to depletion of sterols. Cells pretreated with the indicated concentration of antisense PKCɛ oligonucleotides for 24 h were incubated in the absence or presence of sterols for 12 h. Total cellular RNA was prepared and subjected to Northern blot analysis with radiolabeled cDNA probe for LDL receptor. (C) Selective decrease in PKCɛ protein levels with treatment of cells with PKCɛ-specific antisense oligonucleotide. Cells treated with the indicated concentrations of PKCɛ-specific antisense or sense control oligonucleotides for 24 h were incubated in the absence of sterols for an additional 12 h and in the continuous presence of the oligonucleotides. Cells were harvested by scraping into lysis buffer. Aliquots of the cell lysates containing 50 μg of total protein were analyzed for changes in the levels of specific PKC isoforms by Western blotting.
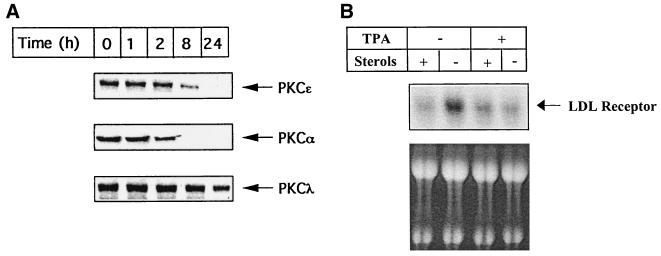
Additional evidence for the involvement of PKCɛ was obtained by using HepG2 cells in which PKC isoforms (α and ɛ) have been depleted by chronic treatment with a high concentration (1 μM) of TPA for 24 h (Fig. 7). There was no reduction observed under these conditions in the TPA-insensitive, atypical PKCλ isoform. Considering that transfection of PKCα cDNA had no dramatic effect on LDL receptor transcription, a decrease in PKCɛ protein (Fig. 7A) probably correlates with reduced induction of the endogenous LDL receptor expression (Fig. 7B). Thus, depletion of PKCɛ caused a significant reduction in the stimulation of LDL receptor expression by low cellular levels of sterols. In short, the results presented above strongly support a role for PKCɛ in regulating LDL receptor expression in HepG2 cells.
FIG. 7.
Effects of high-dose TPA treatment on the time-dependent degradation of selective PKC isoforms and on the level of induction of endogenous LDL receptor expression in response to depletion of sterols. HepG2 cells were grown in either 10% LPDS or 10% LPDS plus sterols in the continued presence of 1 μM TPA. Expression of PKC isoforms was detected at the indicated time periods by Western blot analysis of HepG2 cell lysates. Total RNA was isolated after 24 h of treatment and subjected to Northern blotting to determine amounts of LDL receptor and actin mRNAs. Autoradiographs obtained after longer exposures were quantitated densitometrically. LDL receptor mRNA levels were normalized to actin mRNA levels. Results are expressed as X-fold induction by depletion of sterols as compared with cells grown in the presence of sterols (10% LPDS plus sterols).
Independence of PKCɛ-mediated induction from p42/44MAPK, PLC, and eicosanoid pathways.
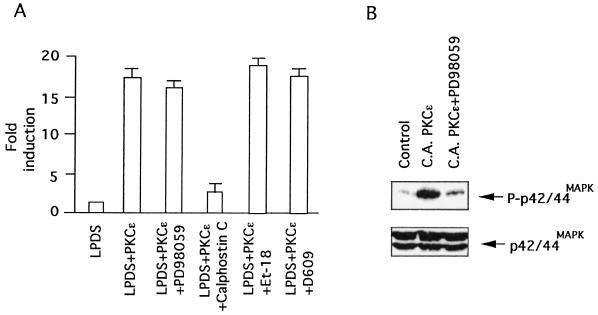
In addition to Ras, PKCɛ is a known activator of Raf-1 kinase (9) and thus has the potential to induce LDL receptor transcription via p42/44MAPK activation. In view of our earlier observation that activation of the p42/44MAPK cascade alone is sufficient to induce LDL receptor transcription (14, 55), we evaluated the role of this pathway by examining the effects of the MEK-1/2 inhibitor, PD98059, on the induction process. As shown in Fig. 8, PKCɛ can induce LDL receptor promoter activity in the presence of this inhibitor, even though p42/44MAPK phosphorylation is blocked, suggesting that PKCɛ mediates induction of LDL receptor expression independently of p42/44MAPK-dependent mechanisms.
FIG. 8.
PKCɛ-mediated induction of LDL receptor transcription does not require p42/44MAPK, PLC, and eicosanoid signaling cascades. (A) HepG2 cells were transiently cotransfected with LDL receptor-luciferase reporter plasmid (0.6 μg) and CA PKCɛ (0.3 μg) as described in the Materials and Methods. Following transfection, transiently transfected cells were grown in the absence or presence of either PD98059 (50 μg/ml), calphostin C (250 nM), or Et-18 (15 μM) in 10% LPDS medium. After 12 h, cells were harvested, and luciferase activity was determined and normalized to the protein content of each extract. Luciferase activity expressed by cells transfected with empty vector was given an arbitrary value of 1. The results are presented as means ± standard error and represent at least four individual experiments. (B) Western blot analysis of transfected HepG2 cells grown under the conditions described above with either phospho-specific or phosphorylation-independent p42/44MAPK antibodies.
In view of earlier studies showing that PKC regulates PLC activity (45) and cholesterol can directly modulate PLC activity (50), we examined whether the PLC cascade is directly involved in regulating sterol-sensitive LDL receptor expression. Transfected cells pretreated with a PLC inhibitor were subjected to growth in the absence or presence of sterols, and induction of LDL receptor expression in response to depletion of sterols was measured. As shown in Fig. 8, pretreatment with either the phosphatidylcholine-specific PLC inhibitor D609 (tricyclodecan-9-yl-xanthogenate) or the phosphatidylinositol-specific PLC inhibitor Et-18 (1-O-octadecyl-2-O-methyl-rac-glycero-3-phosphorylcholine) did not affect induction of LDL receptor expression in response to depletion of sterols, suggesting that the PLC cascade is also not involved in the signaling pathway controlling PKCɛ-mediated LDL receptor induction.
We also examined the involvement of cyclooxygenase metabolites in sterol regulation of LDL receptor expression because 25-hydroxycholesterol has been shown to increase expression of prostaglandin G/H synthase 2 (57). Inhibition of this enzyme by indomethacin or of lipoxygenase by nordihydroguaiaretic acid had no effect on induction of LDL receptor expression by depletion of sterols (results not shown), thus ruling out their involvement in the induction process.
Sterols interact with PKCɛ in an isoform-specific manner.
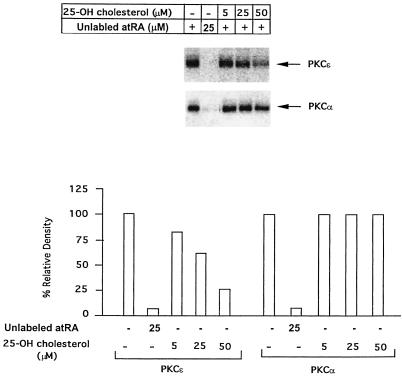
In order to get some insight into the possible mechanism of interaction between sterols and PKCɛ, we employed a photoaffinity labeling assay with [11,12-3H]atRA as a photoprobe as described previously (46). The purpose of this experiment was to check whether there is a direct interaction between the PKC and sterols. We have demonstrated previously that atRA binds directly to a PS-binding site localized in the C2 domains of several PKCs and modulates their catalytic activity. We have also shown that for PKCs that lack the C2 domain, atRA may bind to the C1 domain either in or in close proximity to the fatty acid binding site. Based on this information, we examined the ability of 25-hydroxycholesterol, the most active cholesterol derivative in suppressing LDL receptor transcription, to compete with [3H]atRA for binding to PKCɛ (C1 domain) and PKCα (C2 domain). Direct comparison of the photoaffinity labeling of PKCɛ and PKCα by atRA and protection by various concentrations of 25-hydroxycholesterol is presented in Fig. 9. Both the PKCs were photolabeled by atRA to an equal efficiency, and the labeling was not only light sensitive (Fig. 9, lane 2), but also showed protection by unlabeled atRA (data not shown), demonstrating that atRA binding to both isoforms is specific. Interestingly, as shown in Fig. 9, in contrast to PKCα, there was a strong competition between 25-hydroxycholesterol and atRA for binding to PKCɛ. These results indicate that sterols do not interact with PKCα and supported the possibility that sterols bind specifically to PKCɛ in the C1 domain, most likely at the PS/fatty acid-binding site.
FIG. 9.
Photoaffinity labeling of PKCα and PKCɛ by [3H]atRA and specific protection of PKCɛ labeling by 25-hydroxycholesterol. Purified PKCα and PKCɛ were photolabeled with [3H]atRA in the absence and presence of increasing concentrations (0, 5, 25, and 50 μM) of 25-hydroxycholesterol (lanes 3 to 6); lane 2, control with no UV exposure. The photoaffinity labeling was performed as described in the Materials and Methods. The relative photoincorporation of [3H]atRA into the PKC isoforms was determined by densitometry and is shown in the bottom panel. Incorporation in the absence of unlabeled atRA or 25-hydroxycholesterol was assigned a value of 100%. The graph shows the result of quantitation of the protection experiments by densitometry.
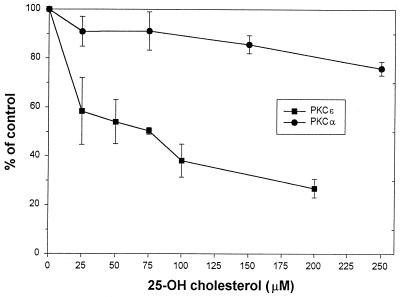
To further examine whether sterols directly affect PKC catalytic activity, the activities of purified recombinant PKCɛ and PKCα were determined in the presence or absence of various concentrations of 25-hydroxycholesterol. Preincubation of either PKCα or PKCɛ with increasing concentrations of 25-hydroxycholesterol followed by the addition of two PKC activators, PS and TPA, had no significant effects on PKCα activity, whereas PKCɛ activity was significantly reduced by 25-hydroxycholesterol in a concentration-dependent manner. These results directly support specific modulation of PKCɛ function by sterols (Fig. 10). The lack of effect of 25-hydroxycholesterol on PKCα activity further validates the specificity of our transfection studies.
FIG. 10.
Effects of 25-hydroxycholesterol on the activities of PKCα and PKCɛ isoforms. To assess the overall effect of 25-hydroxycholesterol on PKC activity, prior to the measurement of enzymatic activity, PKCα or PKCɛ was incubated at 30°C in the presence of increasing concentrations of 25-hydroxycholesterol followed by 10 min of incubation with mixed micelles containing Triton X-100, PS, and TPA. The results are expressed as the percentage of PKC activity in the absence of 25-hydoxycholesterol and the presence of vehicle (ethanol, 2% final concentration). The results shown are the average of three independent experiments. Similar results were obtained in all of the experiments.
DISCUSSION
The present study defines for the first time a component of the signaling pathway that is required for sterol-regulated LDL receptor transcription and implicates PKCɛ as a critical kinase in the induction process. The major finding of this study is that this pathway is specific for PKCɛ, because PKCα, -γ, -ζ, and -δ are not required for the induction of LDL receptor transcription in response to depletion of sterols. These results, combined with earlier observations from other investigators that sterol depletion stimulates changes in cellular signaling cascades (17, 23), provide direct evidence that low cholesterol levels can activate signal transduction pathways that affect gene expression in a manner similar to that of conventional agonist-receptor-initiated signaling events.
PKC isoforms are a family of serine/threonine kinases with different cofactors and substrate specificities. The tissue distribution of PKC isoforms varies considerably, with PKCα, -δ, and -ζ being widespread, while others are localized in a tissue- or cell-specific manner. Earlier studies have shown that HepG2 cells express at least six PKC isoforms representative of three major isoform types (16, 26). The classic PKC isoforms PKCα and -β; the novel isoforms PKCɛ, -μ, and -δ; and the atypical isoform PKCλ were identified by immunoblotting. Among six PKC isoforms reported, PKCɛ seems to play the most crucial role in the induction of LDL receptor transcription following depletion of sterols. We employed multiple approaches to establish the role of PKCɛ in the mechanism of sterol-mediated regulation of LDL receptor promoter activity. First, a highly selective PKC inhibitor, calphostin C, prevented induction of LDL receptor expression in response to depletion of sterols. It has previously been shown that calphostin C inhibits TPA binding to the DAG/TPA-binding region in the regulatory domain (15). Thus, calphostin C preferentially inhibits novel PKCs, including PKCɛ, rather than the DAG-independent PKCs, such as PKCλ. Second, we also found that overexpression of either the wild-type or CA mutant of PKCɛ alone dramatically induced LDL receptor transcription following depletion of sterols. Third, PKCɛ induces LDL receptor transcription in a sterol-resistant manner. Inactivation of SRE-1 reduced PKCɛ-mediated LDL receptor induction. Fourth, specific downregulation of endogenous PKCɛ by an antisense approach inhibited the induction process. Using this approach, various laboratories have established a specific role for PKC isoforms in various cellular processes (28, 29, 44, 56). Fifth, depletion of PKCɛ upon chronic treatment with TPA correlated with reduced expression of the endogenous LDL receptor. Finally, a specific direct physical interaction between 25-hydroxycholesterol and PKCɛ, but not PKCα, was observed. Our photoaffinity labeling experiments demonstrated that 25-hydroxycholesterol binds to the PS/fatty acid-binding site. As is evident from Fig. 10, a significant effect of 25-hydroxycholesterol on PKCɛ catalytic activity was observed under in vitro conditions. This indicates that direct binding of cholesterol to the PS/fatty acid-binding site modulates PKCɛ catalytic activity. Taken together, these results indicate that PKCɛ directly participates in the signaling cascade, leading to induction of LDL receptor transcription in response to cholesterol depletion in hepatic cells. In view of earlier demonstrations that PKC function is modulated in Niemann-Pick C disease due to accumulation of naturally occurring transcriptional modulators, such as cholesterol and sphingosine (48), our study raises an interesting possibility of the involvement of PKCɛ in the pathophysiology of this disease.
How is the induction of LDL receptor transcription mediated through PKCɛ in response to sterol depletion? Cholesterol can modulate the function of PKCɛ by two distinct mechanisms, either by changing membrane fluidity or by a highly specific interaction that is not based on the overall physical state of the plasma membrane. Evidence has accrued that cholesterol is not evenly distributed within membranes, but is instead localized into cholesterol-rich and cholesterol-poor domains. Biological membranes have been viewed as a “mosaic of lipid domains” rather than as a homogenous fluid mosaic. These specialized microdomains in mammalian cells are commonly known as lipid rafts (2, 4, 21, 54). These are enriched in glycosphingolipids and cholesterol, which form lateral lipid assemblies in an unsaturated glycerophospholipid environment. Lipid rafts harbor a number of components of the various intracellular signal transduction pathways, including G protein-coupled receptors, heterotrimeric and small G proteins, nonreceptor tyrosine kinases, components of the Ras/MAPK pathways, PKC isoforms, and glycosylphosphatidylinositol-anchored plasma membrane proteins, which play pivotal roles in coupling cell surface receptors to intracellular signaling cascades. It is possible that depletion of sterols changes the fluidity of membranes, inducing the activation of multiple membrane receptors. This then leads to the activation of PLC, resulting in the cleavage of phosphatidylinositol bisphosphate and generation of inositol 1,4,5-triphosphate and DAG and thus modulating PKCɛ activity. This notion is supported by two studies, one of which shows that sterol depletion activates phosphorylation of PLCγ1 (23) and the other of which shows that cholesterol modulates the activity of several membrane receptors (8, 10, 19, 27). Alternatively, other modulators of PKCɛ, such as phosphatidylinositol 3,4-bisphosphate and phosphatidylinositol 3,4,5-triphosphate, may be increased in response to sterol depletion. Reports show that both of these phosphoinositides generated by phosphatidylinositol 3-kinase activity are potent and selective activators of the novel PKCs and have little or no effect on classic or atypical PKC isoforms (30). To date, no studies have been published regarding changes in phosphatidylinositol 3-kinase activity in response to sterol depletion. Moriya et al. (38) reported that both the phosphatidylinositol 3-kinase and the PLC pathways can activate PKCɛ in a cell- and stimulus-specific manner. While the above studies are suggestive, direct physical evidence for the above signaling cascade in response to cholesterol is lacking. Furthermore, our observation that inhibition of PLC does not affect LDL receptor induction or its sterol regulation also argues against the above mechanism. A clue to a second mechanism by which 25-hydroxycholesterol modulates PKCɛ function has been provided by our demonstration that there is a direct physical interaction between PKCɛ and 25-hydroxycholesterol. We have shown that the PS/fatty acid-binding site in PKCɛ may function as a site for direct interaction with 25-hydroxycholesterol, and regulation of its activity strongly supports such modulation. Compatible with this conclusion is the finding that PKCɛ is abundant in membranes, possibly in lipid rafts, which allows its close proximity to the cellular structural and kinetic cholesterol pool (26, 36). It remains to be seen whether oxysterol receptors (47) may directly or indirectly modulate sterol-sensitive gene expression through interaction with PKCɛ.
The mechanisms by which PKCɛ induces LDL receptor transcription in a sterol-resistant manner remain unclear. Our observation that the overexpression of PKCɛ alone is sufficient to induce LDL receptor transcription in a sterol-resistant manner may be explained if the substrate involved in sterol regulation is phosphorylated by endogenous PKCɛ in response to sterol depletion, so that its overexpression alters the phosphorylation state of the substrate. There are at least two potential mechanisms by which PKCɛ may function. First, it may directly mediate the signal pathway to the LDL receptor promoter (i.e., may serve a causal role). Modulation of transactivation potential of Sp1, SREBPs, and CREB-binding protein through phosphorylation by multiple kinases, including PKC and p42/44MAPK (1, 32, 58), strongly supports such a possibility. Our results demonstrating that the PKC inhibition decreased induction of both LDL receptor and SS expression, and the inactivation of SRE-1 reduced LDL receptor induction, implicates SREBP-dependent mechanisms in the induction process. Alternatively, PKCɛ may be necessary for other reactions and processes required for efficient transmission of the signal to the effector molecules, i.e., serve a permissive role. These two mechanisms may also operate in a synergistic fashion to sustain transactivation of LDL receptor transcription in a sterol-resistant manner. The locations to which the PKCs translocate upon depletion of sterols are probably critical to their ultimate function, but these sites may vary from cell to cell. Shirai et al. (53) used an overexpressed, green fluorescent protein-linked PKC construct to show that high concentrations of lipid, mainly fatty acids, transiently induced translocation of PKCɛ from the cytoplasm to the Golgi network in Chinese hamster ovary cells. It is important to note here that the Golgi network plays a crucial role in sterol-sensitive processing of SREBPs. Translocation of PKCɛ in response to sterol depletion will link this isoform with the maturation of SREBPs. On the other hand, studies with cardiac myocytes showed that fatty acids stimulate specific translocation of PKCɛ from the cytoplasm to a filament/nuclear fraction (22). Future experiments have to address these possibilities.
The suggestion that different PKC isoforms play distinct functional roles in the cell by phosphorylating either isoform-specific or subcellular compartment-specific substrates is widely accepted. Few studies, however, have been reported establishing that a specific PKC isoform may selectively regulate a given biological function. The evidence reported here indicates for the first time that PKCɛ is involved in mediating primary regulation of LDL receptor transcription by cellular cholesterol and hence in controlling plasma cholesterol levels. On the basis of our results, we propose a hypothetical model that accounts for direct interaction between 25-hydroxycholesterol and PKCɛ in order to explain the role of this isoform in sterol-mediated regulation of LDL receptor transcription. The central hypothesis of our model is that PKCɛ may act to coordinate the interaction of sterols with a variety of downstream signal-transducing molecules and sense steady-state membrane cholesterol levels through direct binding to the PS/fatty acid-binding site. High endogenous cholesterol levels result in an increased binding of sterols to the C2 domain of PKCɛ, thus, hampering binding of PKC activators and inhibition of its activity, whereas depletion of endogenous sterol levels results in increased binding of activators, resulting in modulation of its function with a significant effect on catalytic activity. Considering that formation of an active transcriptional complex at the LDL receptor promoter is a highly regulated process, the modified PKCɛ may target the transcriptional machinery in an SREBP-dependent manner. The understanding of these mechanisms will be the focus of future investigation.
Our findings are potentially relevant for elucidation of novel approaches to the therapy of hypercholesterolemia. Small molecules that specifically modulate PKCɛ function may be useful for hypercholesterolemia therapy. Furthermore, reduced responsiveness and sensitivity of PKCɛ or components immediately downstream may explain differential development of atherosclerosis within a human population after exposure to high-cholesterol and/or high-fat diets.
Acknowledgments
This work was supported by Established Investigator grant 9940163N of the American Heart Association (K.D.M.) and National Institutes of Health grants R01 HL67760 (K.D.M.) and DK56226 (A.R.-P).
We thank Jing Xu for her expert technical assistance and Joanna Little for careful proofreading of the manuscript.
REFERENCES
- 1.Ait-Si-Ali, S., D. Carlisi, S. Ramirez, L. C. Upegui-Gonzalez, A. Duquet, P. Robin, B. Rudkin, A. Harel-Bellan, and D. Trouche. 1999. Phosphorylation by p44 MAP kinase/ERK1 stimulates CBP histone acetyl transferase activity in vitro. Biochem. Biophys. Res. Commun. 262:157-162. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. G. 1998. The caveolae membrane system. Annu. Rev. Biochem. 67:199-225. [DOI] [PubMed] [Google Scholar]
- 3.Bottega, R., and R. M. Epand. 1992. Inhibition of protein kinase C by cationic amphiphiles. Biochemistry 31:9025-9030. [DOI] [PubMed] [Google Scholar]
- 4.Brown, D. A., and E. London. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14:111-136. [DOI] [PubMed] [Google Scholar]
- 5.Brown, M. S., and J. L. Goldstein. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science 232:34-47. [DOI] [PubMed] [Google Scholar]
- 6.Brown, M. S., and J. L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331-340. [DOI] [PubMed] [Google Scholar]
- 7.Brown, M. S., and J. L. Goldstein. 1999. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA 96:11041-11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burger, K., G. Gimpl, and F. Fahrenholz. 2000. Regulation of receptor function by cholesterol. Cell Mol. Life Sci. 57:1577-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, H., U. Smola, V. Wixler, I. Eisenmann-Tappe, M. T. Diaz-Meco, J. Moscat, U. Rapp, and G. M. Cooper. 1997. Role of diacylglycerol-regulated protein kinase C isotypes in growth factor activation of the Raf-1 protein kinase. Mol. Cell Biol. 17:732-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, P. C., A. Cherukuri, M. Dykstra, S. Malapati, T. Sproul, M. R. Chen, and S. K. Pierce. 2001. Floating the raft hypothesis: the roles of lipid rafts in B cell antigen receptor function. Semin. Immunol. 13:107-114. [DOI] [PubMed] [Google Scholar]
- 11.Dekker, L. V., and P. J. Parker. 1994. Protein kinase C—a question of specificity. Trends Biochem. Sci. 19:73-77. [DOI] [PubMed] [Google Scholar]
- 12.Denning, M. F., M. G. Kazanietz, P. M. Blumberg, and S. H. Yuspa. 1995. Cholesterol sulfate activates multiple protein kinase C isoenzymes and induces granular cell differentiation in cultured murine keratinocytes. Cell Growth Differ. 6:1619-1626. [PubMed] [Google Scholar]
- 13.Dhawan, P., R. Chang, and K. D. Mehta. 1997. Identification of essential nucleotides of the FP1 element responsible for enhancement of low density lipoprotein receptor gene expression. Nucleic Acids Res. 25:4132-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhawan, P., A. Bell, A. Kumar, C. Golden, and K. D. Mehta. 1999. Critical role of p42/44(MAPK) activation in anisomycin and hepatocyte growth factor-induced LDL receptor expression: activation of Raf-1/Mek-1/p42/44(MAPK) cascade alone is sufficient to induce LDL receptor expression. J. Lipid Res. 40:1911-1919. [PubMed] [Google Scholar]
- 15.Dubyak, G. R., and S. B. Kertesy. 1997. Inhibition of GTP gamma S-dependent phospholipase D and Rho membrane association by calphostin is independent of protein kinase C catalytic activity. Arch. Biochem. Biophys. 341:129-139. [DOI] [PubMed] [Google Scholar]
- 16.Ducher, L., F. Croquet, S. Gil, J. Davy, J. Feger, and A. Brehier. 1995. Differential expression of five protein kinase C isoenzymes in FAO and HepG2 hepatoma cell lines compared with normal rat hepatocytes. Biochem. Biophys. Res. Commun. 217:546-553. [DOI] [PubMed] [Google Scholar]
- 17.Furuchi, T., and R. G. Anderson. 1998. Cholesterol depletion of caveolae causes hyperactivation of extracellular signal-related kinase (ERK). J. Biol. Chem. 273:21099-21104. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Paramio, P., Y. Cabrerizo, F. Bornancin, and P. J. Parker. 1998. The broad specificity of dominant inhibitory protein kinase C mutants infers a common step in phosphorylation. Biochem. J. 333:631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gimpl, G., K. Burger, and F. Fahrenholz. 1997. Cholesterol as modulator of receptor function. Biochemistry 36:10959-10974. [DOI] [PubMed] [Google Scholar]
- 20.Hannah, V. C., J. Ou, A. Luong, J. L. Goldstein, and M. S. Brown. 2001. Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 276:4365-4372. [DOI] [PubMed] [Google Scholar]
- 21.Hannun, Y. A., and R. M. Bell. 1987. Lysosphingolipids inhibit protein kinase C: implications for the sphingolipidoses. Science 235:670-674. [DOI] [PubMed] [Google Scholar]
- 22.Huang, X. P., Y. Pi, A. J. Lokuta, M. L. Greaser, and J. W. Walker. 1997. Arachidonic acid stimulates protein kinase C-epsilon redistribution in heart cells. J. Cell Sci. 110:1625-1634. [DOI] [PubMed] [Google Scholar]
- 23.Kabouridis, P. S., J. Janzen, A. L. Magee, and S. C. Ley. 2000. Cholesterol depletion disrupts lipid rafts and modulates the activity of multiple signaling pathways in T lymphocytes. Eur. J. Immunol. 30:954-963. [DOI] [PubMed] [Google Scholar]
- 24.Khan, W. A., G. Blobe, A. Halpern, W. Taylor, W. C. Wetsel, D. Burns, C. Loomis, and Y. A. Hannun. 1993. Selective regulation of protein kinase C isoenzymes by oleic acid in human platelets. J. Biol. Chem. 268:5063-5068. [PubMed] [Google Scholar]
- 25.Kotzka, J., D. Muller-Wieland, G. Roth, L. Kremer, M. Munck, S. Schurmann, B. Knebel, and W. Krone. 2000. Sterol regulatory element binding proteins (SREBP)-1a and SREBP-2 are linked to the MAP-kinase cascade. J. Lipid Res. 41:99-108. [PubMed] [Google Scholar]
- 26.Kumar, A., T. C. Chambers, B. A. Cloud-Heflin, and K. D. Mehta. 1997. Phorbol ester-induced low density lipoprotein receptor gene expression in HepG2 cells involves protein kinase C-mediated p42/44 MAP kinase activation. J. Lipid Res. 38:2240-2248. [PubMed] [Google Scholar]
- 27.Lagane, B., G. Gaibelet, E. Meilhoc, J. M. Masson, L. Cezanne, and A. Lopez. 2000. Role of sterols in modulating the human mu-opioid receptor function in Saccharomyces cerevisiae. J. Biol. Chem. 275:33197-33200. [DOI] [PubMed] [Google Scholar]
- 28.Li, Y., K. L. Davis, and A. J. Sytkowski. 1996. Protein kinase C-epsilon is necessary for erythropoietin's up-regulation of c-myc and for factor-dependent DNA synthesis. Evidence for discrete signals for growth and differentiation. J. Biol. Chem. 271:27025-27030. [PubMed] [Google Scholar]
- 29.Liedtke, C. M., and T. S. Cole. 1998. Antisense oligonucleotide to PKC-epsilon alters cAMP-dependent stimulation of CFTR in Calu-3 cells. Am. J. Physiol. 275:C1357-C1364. [DOI] [PubMed] [Google Scholar]
- 30.Liscovitch, M., and L. C. Cantley. 1994. Lipid second messengers. Cell 77:329-334. [DOI] [PubMed] [Google Scholar]
- 31.Liu, J., T. E. Ahlborn, M. R. Briggs, and F. B. Kraemer. 2000. Identification of a novel sterol-independent regulatory element in the human low density lipoprotein receptor promoter. J. Biol. Chem. 275:5214-5221. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Y. Z., N. S. Thomas, and D. S. Latchman. 1999. CBP associates with the p42/p44 MAPK enzymes and is phosphorylated following NGF treatment. Neuroreport 10:1239-1243. [DOI] [PubMed] [Google Scholar]
- 33.Mehta, K. D., R. Chang, J. Underwood, J. Wise, and A. Kumar. 1996. Identification of a novel cis-acting element participating in maximal induction of the human low density lipoprotein receptor gene transcription in response to low cellular cholesterol levels. J. Biol. Chem. 271:33616-33622. [DOI] [PubMed] [Google Scholar]
- 34.Mehta, K. D., and L. Miller. 1999. Inhibition of stress-activated p38 mitogen-activated protein kinase induces low-density lipoprotein receptor expression. Trends Cardiovasc. Med. 9:201-205. [DOI] [PubMed] [Google Scholar]
- 35.Mendez, A. J., J. F. Oram, and E. L. Bierman. 1991. Protein kinase C as a mediator of high density lipoprotein receptor-dependent efflux of intracellular cholesterol. J. Biol. Chem. 266:10104-10111. [PubMed] [Google Scholar]
- 36.Mischak, H., J. A. Goodnight, W. Kolch, G. Martiny-Baron, C. Schaechtle, M. G. Kazanietz, P. M. Blumberg, J. H. Pierce, and J. F. Mushinski. 1993. Overexpression of protein kinase C-delta and -epsilon in NIH 3T3 cells induces opposite effects on growth, morphology, anchorage dependence, and tumorigenicity. J. Biol. Chem. 268:6090-6096. [PubMed] [Google Scholar]
- 37.Mollers, C., W. Drobnik, T. Resink, and G. Schmitz. 1995. High-density lipoprotein and low-density lipoprotein-mediated signal transduction in cultured human skin fibroblasts. Cell. Signal. 7:695-707. [DOI] [PubMed] [Google Scholar]
- 38.Moriya, S., A. Kazlauskas, K. Akimoto, S. Hirai, K. Mizuno, T. Takenawa, Y. Fukui, Y. Watanabe, S. Ozaki, and S. Ohno. 1996. Platelet-derived growth factor activates protein kinase C epsilon through redundant and independent signaling pathways involving phospholipase C gamma or phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA 93:151-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newton, A. C. 1997. Regulation of protein kinase C. Curr. Opin. Cell Biol. 9:161-167. [DOI] [PubMed] [Google Scholar]
- 40.Nishizuka, Y. 1988. The molecular heterogeneity of protein kinase C and its implications for cellular regulation. Nature 334:661-665. [DOI] [PubMed] [Google Scholar]
- 41.Nishizuka, Y. 1992. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science 258:607-614. [DOI] [PubMed] [Google Scholar]
- 42.Osborne, T. F. 2000. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J. Biol. Chem. 275:32379-32382. [DOI] [PubMed] [Google Scholar]
- 43.Parker, P. J., and L. V. Dekkers (ed.). 1997. Protein kinase C, p. 1-9. R. G. Landes, Austin, Tex.
- 44.Pike, L. J., and J. M. Miller. 1998. Cholesterol depletion delocalizes phosphatidylinositol bisphosphate and inhibits hormone-stimulated phosphatidylinositol turnover. J. Biol. Chem. 273:22298-22304. [DOI] [PubMed] [Google Scholar]
- 45.Radallah, D., M. Nogaro and B. Fournier. 1999. Protein kinase C stimulates PtdIns-4,5-P2-phospholipase C activity. Biochim. Biophys. Acta 1450:242-253. [DOI] [PubMed] [Google Scholar]
- 46.Radominska-Pandya, A., G. Chen, P. J. Czernik, J. M. Little, V. M. Samokyszyn, C. A. Carter, and G. Nowak. 2000. Direct interaction of all-trans-retinoic acid with protein kinase C (PKC). Implications for PKC signaling and cancer therapy. J. Biol. Chem. 275:22324-22330. [DOI] [PubMed] [Google Scholar]
- 47.Repa, J. J., G. Liang, J. Ou, Y. Bashmakov, J. M. Lobaccaro, I. Shimomura, B. Shan, M. S. Brown, J. L. Goldstein, and D. J. Mangelsdorf. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 14:2819-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez-Lafrasse, C., R. Rousson, S. Valla, P. Antignac, P. Louisot, and M. T. Vanier. 1997. Modulation of protein kinase C by endogenous sphingosine: inhibition of phorbol dibutyrate binding in Niemann-Pick C fibroblasts. Biochem. J. 325:787-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roth, G., J. Kotzka, L. Kremer, S. Lehr, C. Lohaus, H. E. Meyer, W. Krone, and D. Muller-Wieland. 2000. MAP kinases Erk1/2 phosphorylate sterol regulatory element-binding protein (SREBP)-1a at serine 117 in vitro. J. Biol. Chem. 275:33302-33307. [DOI] [PubMed] [Google Scholar]
- 50.Ruiz-Arguello, M. B., F. M. Goni, and A. Alonso. 1998. Phospholipase C hydrolysis of phospholipids in bilayers of mixed lipid compositions. Biochemistry 37:11621-11628. [DOI] [PubMed] [Google Scholar]
- 51.Schönwasser, D. C., R. M. Marais, C. J. Marshall, and P. J. Parker. 1998. Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel, and atypical protein kinase C isotypes. Mol. Cell. Biol. 18:790-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott-Burden, T., T. J. Resink, A. W. Hahn, U. Baur, R. J. Box, and F. R. Buhler. 1989. Induction of growth-related metabolism in human vascular smooth muscle cells by low density lipoprotein. J. Biol. Chem. 264:12582-12589. [PubMed] [Google Scholar]
- 53.Shirai, Y., K. Kashiwagi, K. Yagi, N. Sakai, and N. Saito. 1998. Distinct effects of fatty acids on translocation of gamma- and epsilon-subspecies of protein kinase C. J. Cell Biol. 143:511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 55.Singh, R. P., P. Dhawan, C. Golden, G. S. Kapoor, and K. D. Mehta. 1999. One-way cross-talk between p38(MAPK) and p42/44(MAPK). Inhibition of p38(MAPK) induces low density lipoprotein receptor expression through activation of the p42/44(MAPK) cascade. J. Biol. Chem. 274:19593-19600. [DOI] [PubMed] [Google Scholar]
- 56.Traub, O., B. P. Monia, N. M. Dean, and B. C. Berk. 1997. PKC-epsilon is required for mechano-sensitive activation of ERK1/2 in endothelial cells. J. Biol. Chem. 272:31251-31257. [DOI] [PubMed] [Google Scholar]
- 57.Wohlfeil, E. R., and W. B. Campbell. 1997. 25-Hydroxycholesterol enhances eicosanoid production in cultured bovine coronary artery endothelial cells by increasing prostaglandin G/H synthase-2. Biochim. Biophys. Acta 1345:109-120. [DOI] [PubMed] [Google Scholar]
- 58.Yuan, L. W., and J. E. Gambee. 2000. Phosphorylation of p300 at serine 89 by protein kinase C. J. Biol. Chem. 275:40946-40951. [DOI] [PubMed] [Google Scholar]