Abstract
Mona/Gads is a Grb2-related, Src homology 3 (SH3) and SH2 domain-containing adapter protein whose expression is restricted to cells of hematopoietic lineage (i.e., monocytes and T lymphocytes). During monocyte/macrophage differentiation, Mona is induced and interacts with the macrophage colony-stimulating factor receptor, M-CSFR (also called Fms), suggesting that Mona could be involved in developmental signaling downstream of the M-CSFR by recruiting additional signaling proteins to the activated receptor. Our present results identify Mona as a specific partner protein for the DOS/Gab family member Gab3 in monocytic/macrophage development. Mona does not interact with Gab2; however, Gab3 also forms a complex with the Mona-related adapter Grb2. Glutathione S-transferase pull-down experiments demonstrate that the Mona and Gab3 interaction utilizes the carboxy-terminal SH3 domain of Mona and the atypical proline-rich domain of Gab3. Mona is known to interact with the phosphorylated Y697 site of the M-CSFR. The M-CSFR mutation Y697F exhibited qualitative and quantitative abnormalities in receptor and Gab3 tyrosine phosphorylation, and Mona induction was greatly reduced. The Y807F M-CSFR mutation is defective in differentiation signaling, but not growth signaling, and also fails to induce Mona protein expression. During M-CSF-stimulated macrophage differentiation of mouse bone marrow cells, Mona and Gab3 expression is coinduced, these proteins interact, and Mona engages in multimolecular complexes. These data suggest that association of Mona and Gab3 plays a specific role in mediating the M-CSFR differentiation signal.
Macrophage colony-stimulating factor (M-CSF) controls the survival, proliferation, and differentiation of cells belonging to the monocyte/macrophage lineage (48, 52). Injection of M-CSF into mice or nonhuman primates causes increased numbers of macrophages along with their progenitor cells (2, 39, 58), and osteopetrotic (op/op) mice, which are naturally deficient in M-CSF, show severely impaired macrophage production that can be cured by M-CSF (24). Therefore, M-CSF is thought to control cellular responses throughout the monocyte/macrophage pathway in vivo. Consistent with this notion, M-CSF receptor (M-CSFR, also called Fms), a member of the tyrosine kinase receptor family, is prominently expressed on monocytic cells and their progenitors (12, 36). Numerous in vitro studies have supported the key role of M-CSF in macrophage production (6, 7, 38, 41-43, 51). M-CSF was initially defined as the growth factor stimulating production of macrophage colonies from bone marrow cells (51). Myeloid bipotential progenitors, or granulocyte-macrophage colony-forming cells, express cell surface M-CSFR and stimulation with M-CSF results in their proliferation and differentiation to macrophages only, whereas granulocyte-macrophage colony-stimulating factor stimulation results in production of both granulocytes and macrophages (6, 41).
Studies on ectopically expressed M-CSFR in several cell systems have revealed structure-function relationships initiating the early stages of M-CSF signaling. Following M-CSF dimerization of receptors, the adjacent cytoplasmic tyrosine kinase domains trans-phosphorylate specific tyrosine residues, which serve as anchoring sites for Src homology 2 (SH2)-containing signaling molecules (18, 49). Recruitment of each SH2-containing protein initiates a downstream intracellular signaling sequence thought to control the specific biological effects of M-CSF on monocytic cell growth and development. These known pathways include those activating Ras/mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase/SHIP/Akt, and phospholipase C/protein kinase C (reviewed in reference 10).
Recently, additional signaling molecules have been identified that transduce proliferation and differentiation signals upon M-CSF stimulation. These proteins are members of the DOS/Gab family which rely on interactions with additional signaling proteins and thus are termed scaffolding proteins. Genetic studies in Drosophila indicate that these proteins regulate differentiation (19, 46) and studies of the Gab1 family member in mammalian liver cell development support this notion (21). The primary effector protein necessary for developmental signaling through DOS is the tyrosine phosphatase SHP-2 (16, 19). Both Gab2 and the newly identified Gab3 protein participate in macrophage development downstream of the M-CSFR, and binding of SHP-2 to Gab2 and Gab3 is an essential function for macrophage development (31, 56). The DOS/Gab family of proteins also requires localization to the cytoplasmic face of the plasma membrane and association with a receptor, which may provide the tyrosine kinase activity for phosphorylation of regulatory and SH2-binding motifs. Each DOS/Gab protein contains an N-terminal pleckstrin homology domain providing membrane localization through phosphatidylinositol polyphosphate lipids (34, 47). Adapter proteins, such as Grb2, link the DOS/Gab protein to specific receptors (4, 15, 17, 20, 32, 40). In M-CSF signaling, the SH3 domains of Grb2 bind polyproline motifs in Gab2 or Gab3, and the Grb2 SH2 domain interacts with phosphorylated Y697 within the kinase insert region of the activated M-CSFR (31, 56). Thus, signals from the activated M-CSFR may direct the close association with Gab2 and its phosphorylation, association with SHP-2, and dephosphorylation of yet unidentified target proteins, which subsequently effect differentiation signaling. Gab3, a newer member of the Gab family, also becomes strongly phosphorylated in response to M-CSF and is induced during macrophage differentiation of FDC-P1 cells. However, its exact role in differentiation signaling is not well understood (56).
Adapter proteins participate in assembling the Gab family signaling complexes and are therefore essential for maintaining the specificity of many steps in signaling pathways. Grb2 is a broadly expressed adapter protein containing one SH2 domain flanked by two SH3 domains. The monocytic adapter protein (Mona, also called Gads, GrpL, Grap2, Grf40, and GRID) is hematopoietic cell specific, with a domain structure resembling Grb2 but exhibiting a unique proline-rich region between the SH2 domain and the carboxy-terminal SH3 domain (1, 9, 13, 25, 28, 44). Mona expression was detected in circulating monocytes and also during differentiation of NFS-60 cells towards monocytes/macrophages (9). Moreover, the carboxy-terminal SH3 domain of Mona can bind to an atypical polyproline motif found in all members of the DOS/Gab family, including Gab3 (26, 32, 56). These observations led us to explore potential relationships between Mona and Gab3 during macrophage differentiation. In this report, we present data demonstrating the M-CSF-dependent expression of Mona and Gab3 in macrophage differentiation signaling and provide evidence for the Mona-Gab3 complex functioning in late M-CSFR signaling.
MATERIALS AND METHODS
Cell culture, stimulation, and retroviral infections.
Two sources (clone 19 and UW cells) of FDC-P1 cells (FD cells) were used in these studies. Following retroviral expression of M-CSFR cDNA, both cell lines, referred to as FD/Fms cells, are able to differentiate into macrophages in response to M-CSF (7, 56). FDC-P1 UW cells were used in all experiments except for those shown in Fig. 1B, 2A, 6, and 7, for which cells were obtained from clone 19. FD/Fms cells expressing Mona (FD/Fms/Mona) or V5-tagged Gab3 (FD/Fms/Gab3V5), FD/Fms Y697F cells, and FD/Fms Y807F cells have been previously described (7, 9, 27, 56). To obtain FD/Fms/Gab3V5 cells expressing Mona (FD/Fms/Gab3V5/Mona), FD/Fms/Gab3V5 cells were cultivated on a layer of ψ2/L(Mona)SN producer cells (9) for 48 h and then selected for 1 week by 1 mg of G418 (Life Technologies) per ml. The FD/Fms Y697F cells expressing Gab3V5 and Mona (FD/Fms Y697F/Gab3V5/Mona) were obtained following retroviral infection of each cDNA.
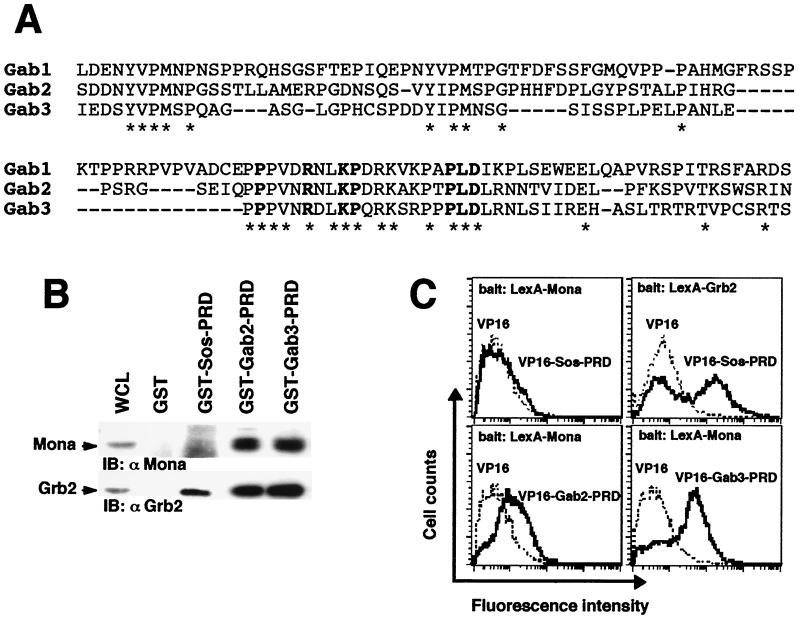
FIG. 1.
Gab2 and Gab3 are potential Mona partners. (A) Alignment of the carboxy-terminal PRDs of Gab family members. Bold characters show amino acid residues featuring the atypical SH3-binding domain as defined by Lock et al . (32). (B) The PRD is sufficient to mediate binding of Grb2 and Mona to either Gab2 or Gab3. Lysates from FD/Fms/Mona cells were incubated with purified, immobilized GST fused to protein segments encompassing Gab2- or Gab3-PRD. Bound proteins were run on an SDS-12% PAGE gel and transferred onto nitrocellulose membranes that were immunoblotted (IB) with anti-Mona (1:1,000) or anti-Grb2 (1:5,000) antibody (left panel). The GST-Sos-PRD fusion protein was used as a specificity control, known to bind Grb2 only (right panel). (C) Interaction between Mona and Gab proteins assessed in the yeast two-hybrid system. Bait and target proteins were expressed in yeast as described in Materials and Methods. Positive interactions of bait and target proteins were detected by fluorescence due to transcriptional activation of the GFP reporter gene. Data are representative of three independent experiments (except for Grb2, for which n = 2). For LexA-Mona experiments, average fluorescence intensities were 7.6 ± 1.0 (empty VP16), 8.0 ± 0.6 (VP16-Sos-PRD), 14.7 ± 1.3 (VP16-Gab2-PRD), and 44.2 ± 3.0 (VP16-Gab3-PRD). For LexA-Grb2 experiments, average fluorescence intensities were 9.0 (empty VP16) and 125.2 (VP16-Sos-PRD). Values are given as means ± standard errors of the means.
All FDC-P1-derived cells were maintained in Iscove's modified Dulbecco's medium (IMDM; Life Technologies) supplemented with 10% fetal bovine serum (FBS; Sigma) and 5% X63-IL-3 cell conditioned medium as a source of growth factor (23). For M-CSF stimulation, cells were washed twice in phosphate-buffered saline (PBS) to remove interleukin-3 (IL-3), starved in IMDM containing 1% FBS for 3 h at 37°C, and then stimulated with 2,500 U of M-CSF per ml at 37°C. The source of M-CSF was a conditioned medium from Sf9 insect cells expressing M-CSF (55). To induce differentiation, cells were washed twice in PBS and seeded at a density of 5 × 104 cells/ml in IMDM containing 10% FBS and 2,500 U of M-CSF per ml; the medium was changed daily and cell density was adjusted to 105 cells/ml. For obtaining normal macrophage precursors, murine bone marrow cells were cultivated in IMDM supplemented with 15% FBS (Dutscher) and 5 ng of Flt3 ligand (FL; R&D Systems) per ml for 6 days. Terminal differentiation was obtained by cultivating the cells (day 6 FL cells) for 3 days in IMDM-15% FBS supplemented with 1,000 U of M-CSF per ml.
Antibodies.
Polyclonal rabbit antisera to Mona (#L1), Gab3, Gab2, and Shc have been previously described (9, 27, 31, 56). Monoclonal antibody to phosphotyrosine (clone 4G10) was kindly provided by P. Dubreuil (INSERM U119, Marseille, France). Monoclonal anti-Grb2 antibody was from Transduction Laboratories; monoclonal antibody against the V5 epitope was purchased from Invitrogen. Anti-M-CSFR was from Santa Cruz Biotechnologies.
GST constructs, production of GST fusion proteins and GST pull-down assays.
The glutathione S-transferase (GST)-Mona domains (N-SH3, amino acids 1 to 62; C-SH3, amino acids 256 to 322; ProR, amino acids 153 to 258) were constructed by PCR from mouse Mona cDNA (9) and the amplified fragments were digested with BamHI and EcoRI and ligated in the same sites of the pGEX-3X vector (Pharmacia). PCR-based mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene) was used to generate inactivating P47L and P313L mutations in the amino- and carboxy-terminal SH3 domains of Mona; the mutants were sequenced and cloned into the pGEX-3X vector. GST-Mona and GST-Mona SH2 fusion proteins were produced as previously described (9). For construction of GST-Sos PRD, the proline-rich domain (PRD) (amino acids 1123 to 1337) was inserted into the BamHI and EcoRI sites of the pGEX-3X vector. GST-Grb2 was a kind gift from P. Dubreuil. GST-Gab3-PRD (amino acids 383 to 479) and GST-Gab2-PRD (amino acids 392 to 496) were previously described (31, 56). Recombinant GST fusion proteins were affinity purified from cell lysates of Escherichia coli BL21 (Pharmacia) by adsorption to glutathione-Sepharose 4B beads (Pharmacia) as previously described (9). For GST pull-down assays, cell lysates (1.2 mg) were incubated with 20 μg of GST fusion protein coupled on glutathione beads for 4 h at 4°C. Precipitated proteins were washed, eluted, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting analysis as described below.
Yeast two-hybrid analysis.
For construction of LexA-Mona and LexA-Grb2 baits, the complete cDNA encoding each of the molecules (Mona, amino acids 1 to 322; Grb2, amino acids 1 to 217) was inserted into the pBTM116 vector, resulting in production of LexA-Mona and LexA-Grb2 fusion proteins. The VP16-Sos-PRD (amino acids 1123 to 1337), VP16-Gab2-PRD (amino acids 392 to 496), and VP16-Gab3-PRD (amino acids 383 to 479) proteins contain the sequence of the PRD of each molecule cloned in the BamHI and EcoRI sites of the pVP16 vector. The Saccharomyces cerevisiae YRN974 strain containing an integrated LexA-operator-green fluorescent protein (GFP) cassette (33) was transformed with two plasmids: pBTM116, encoding LexA-Mona or LexA-Grb2, and pVP16, either empty (as a negative control) or containing the PRD from Gab2, Gab3, or Sos. Three individual clones were isolated on selective agar plates for each transfection and expanded in similar selective liquid medium. Exponentially growing cultures were then analyzed for fluorescence intensity using a Becton Dickinson FACScalibur flow cytometer. Plasmids, selective media, and transformation protocol have been described previously (54).
Immunoprecipitation and immunoblotting.
Cells were collected by centrifugation and lysed in ice-cold lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 5 mM KCl, 1 mM EDTA, 2 mM Na3VO4, 10% glycerol, 1% Triton X-100) containing protease inhibitor cocktail (Roche). After 10 min on ice, lysates were spun at 25,000 × g for 10 min at 4°C, and pellets containing nuclei and other insoluble material were discarded. For immunoprecipitation, antibody (1 to 5 μg) and 10 μl of protein G-Sepharose (Pharmacia) were added to cell lysates for a 2-h incubation at 4°C. Immunoprecipitates were washed four times with lysis buffer and eluted by boiling in Laemmli sample buffer. Samples were electrophoresed on SDS-7.5 or 12% PAGE gels and transferred to a nitrocellulose membrane (Protan 85; Schleicher and Schuell) on a semidry blotting apparatus (Amersham-Pharmacia). The membranes were blocked in blocking buffer (10 mM Tris-HCl, 150 mM NaCl [pH 9.5], 0.25% Tween 20, 3% nonfat milk, except for antiphosphotyrosine antibody, for which 3% nonfat milk was replaced with 3% bovine serum albumin) for a minimum of 1 h at room temperature, incubated with primary antibodies for 1 h, and then incubated for 45 min with horseradish peroxidase-conjugated anti-mouse (Bio-Rad) or anti-rabbit antibody (Sigma) diluted in blocking buffer. Blots were developed by enhanced chemiluminescence reagent (ECL+; Amersham-Pharmacia).
Flow cytometry analysis.
For cell morphology analysis, cells were washed with ice-cold PBS-5% FBS and resuspended with PBS-5% FBS containing 1 μg of propidium iodide (Sigma) per ml for staining and exclusion of dead cells. Samples were directly analyzed for light scatter intensity to monitor cellular size (forward scatter) and cellular granularity (side scatter) using a FACScan apparatus. For analysis of Mac-1 and F4/80 antigen cell surface expression, all incubations were performed in PBS-5% FBS containing 40 μg of normal goat immunoglobulin G (IgG) (Sigma) per ml. Cells were incubated for 45 min on ice with anti-Mac1 or anti-F4/80 antibody (Caltag Laboratories), washed, and stained for 30 min on ice with fluorescein isothiocyanate-conjugated F(ab′)2 fragment goat anti-IgG antibody (Caltag Laboratories). Cells were washed again, resuspended in PBS-5% FBS containing 1 μg of propidium iodide per ml, and then analyzed for fluorescence intensity. Controls were cells first incubated in PBS-5% FBS containing 40 μg of normal goat IgG per ml alone and then in the same secondary antibody solution as that used with test antibody.
Gel filtration chromatography.
Gel filtration was performed using a fast protein liquid chromatography system (Pharmacia). A Superose 12 HR10/60 gel filtration column was calibrated with a molecular weight marker kit for gel filtration (Bio-Rad). Cell lysates (100 μl) containing 2 mg of cytosolic protein were applied at a flow rate of 0.3 ml/min onto the column, which had been previously equilibrated in PBS. Proteins were collected in 300-μl fractions and 30 μl of each fraction was loaded in an SDS-12% PAGE gel for immunoblotting analysis.
RESULTS
Mona interacts with both Gab2 and Gab3 in vitro but only with Gab3 in myeloid progenitor cells.
Mona is structurally related to Grb2 and contains one SH2 domain and two SH3 domains that share approximately 50% homology with the corresponding domains of Grb2 (9). The carboxy-terminal SH3 domains of both adapters have been shown to recognize an unusual proline-containing motif whose consensus sequence is P-X3-R-X2-K-P-X7-P-L-D (26, 32). This motif is present and highly conserved in the primary sequences of all Gab family members (Fig. 1A). However, only in vitro association of Mona with Gab1 has been reported so far (32, 44). Therefore our initial approach sought to verify the Mona interaction with the other two Gab family members, Gab2 and Gab3, which are strongly and preferentially expressed in hematopoietic cells (16, 56). As a cellular model, we used various derivatives of the FD/Fms cells, a myeloid progenitor cell line (FDC-P1) expressing exogenous M-CSFR (7, 56). These cells express endogenous Grb2 and Gab2, and for these experiments, both Mona and V5-tagged Gab3 were ectopically expressed from retroviral constructs (9, 27, 31, 56).
The potential interaction of Mona with Gab2 or Gab3 was first examined by an in vitro pull-down assay. GST proteins fused to Gab2-PRD or to Gab3-PRD were used to precipitate PRD-binding proteins from lysates of Mona-expressing FD/Fms cells (FD/Fms/Mona cells) (9). Both PRDs contain the atypical P-X3-R-X2-K-P-X7-P-L-D motif, but not the classic binding motif for Grb2, PXXP (31, 56). Figure 1B shows that GST-Gab2-PRD and GST-Gab3-PRD are equally capable of interacting with Mona or Grb2. The specificity of these interactions was validated by the demonstration that GST-Sos-PRD could precipitate Grb2, but not Mona, from the same cell lysates, as reported previously (28). These results indicate that Mona is a potential partner for the Gab2 and Gab3 proteins. We next asked whether these interactions are direct or indirect. For that purpose, interactions were analyzed by flow cytometry in a modified yeast two-hybrid assay using the YRN974 yeast strain in which the reporter gene, driven by a positive LexA/VP16 interaction, encodes GFP (33). An interaction between target and bait proteins fused to LexA and VP16, respectively, results in GFP expression. As shown in Fig. 1C, yeast clones expressing either LexA-Mona or LexA-Grb2, together with VP16 alone, show only background autofluorescence (dotted lines). However, coexpression of LexA-Mona with either VP16-Gab2-PRD or VP16-Gab3-PRD increased the fluorescence intensity (thick lines). Likewise, a positive interaction was observed by coexpression of LexA-Grb2 with its known binding site on the Sos protein (VP16-Sos-PRD), whereas coexpression of LexA-Mona with the same VP16-Sos-PRD served as a negative control (Fig. 1C). It is noteworthy that the Mona-Gab2 interaction yielded lower fluorescence levels than the Mona-Gab3 interaction, suggesting a weaker interaction in this assay.
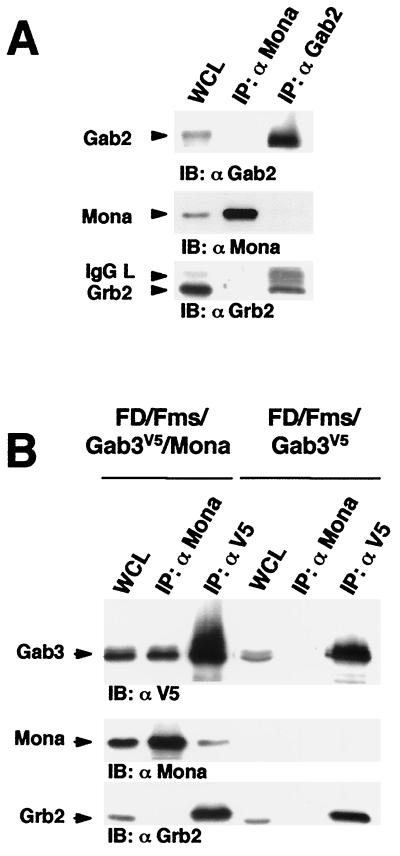
We then performed coimmunoprecipitation assays to further examine the interaction between Mona and Gab family proteins in vivo. Within lysates of FD/Fms/Mona cells, Mona antiserum could not pull down Gab2 along with Mona, whereas Gab2 antiserum coimmunoprecipitated Grb2 but not Mona (Fig. 2A). This demonstrates that Mona is not able to associate with Gab2 in myeloid cells even though it binds Gab2-PRD in vitro or in yeast. To examine the Mona-Gab3 interaction, FD/Fms/Gab3V5/Mona cells were generated by overexpressing Mona in FD/Fms cells, which already expressed the V5-tagged Gab3 protein (FD/Fms/Gab3V5 cells). In lysates from the FD/Fms/Gab3V5/Mona cells, Mona antiserum could coimmunoprecipitate the Gab3 protein but not Grb2. The reciprocal experiment using anti-V5 antibody recognized the Gab3 protein directly and coimmunoprecipitated both Mona and Grb2 (Fig. 2B). In FD/Fms/Gab3V5 cells, the anti-V5 antibody coimmunoprecipitated Grb2, but not Mona, and the interaction of Mona and Gab3 was not observed in these cells (e.g., without exogenous Mona expression). These results indicate that Mona interacts specifically with Gab3 (and not Gab2), but that Gab3 also interacts with Grb2 (56).
FIG. 2.
Gab3, but not Gab2, interacts with Mona in vivo. (A) Cell lysates from FD/Fms/Mona cells were immunoprecipitated (IP) using anti-Mona or anti-Gab2 antibody. Whole cell lysates (WCL; 50 μg) and immunoprecipitates were separated by SDS-12% PAGE and immunoblotted (IB) with anti-Gab2 (1:2,000), anti-Mona (1:1,000), or anti-Grb2 (1:5,000) antibody. (B) Cell lysates from FD/Fms/Gab3V5/Mona and FD/Fms/Gab3V5 cells were immunoprecipitated using antibodies against Mona or the V5 epitope detecting Gab3. WCL (50 μg) and immunoprecipitates were separated by SDS-12% PAGE and then immunoblotted with anti-V5 (1:5,000), anti-Mona, or anti-Grb2 antibody.
Mona associates with Gab3 through its C-terminal SH3 domain.
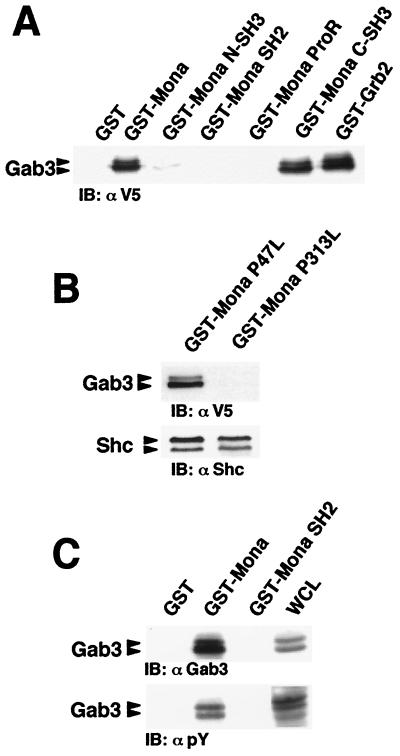
The results in Fig. 1B demonstrated the interaction between Mona and the Gab3-PRD, suggesting a role for at least one of the two Mona SH3 domains in this interaction. Therefore, GST fusion proteins containing either the full-length Mona protein, each of its potential SH3 domains, the SH2 domain, or the unique proline-rich (ProR) linker domain were used to pull down Gab3 from lysates of IL-3-maintained FD/Fms/Gab3 cells. As shown in Fig. 3A, the carboxy-terminal SH3 domain, as well as full-length Mona, pulled down Gab3, whereas neither the amino-terminal SH3 domain, the SH2 domain, nor the ProR domain bound Gab3. Moreover, the P313L mutation in the carboxy-terminal SH3 domain totally prevented association of Mona with Gab3, whereas the equivalent P47L mutation in the amino-terminal SH3 domain had no effect on the Mona-Gab3 interaction (Fig. 3B). Both mutations preserved the ability of Mona to bind Shc, which occurs through the Mona SH2 domain (28), indicating that these mutations did not grossly affect the Mona structure (Fig. 3B). We conclude that the carboxy-terminal SH3 domain of Mona mediates the interaction with Gab3.
FIG. 3.
The carboxy-terminal Mona SH3 domain mediates binding to Gab3. (A) Lysates from FD/Fms/Gab3V5 cells were incubated with purified, immobilized GST fused to the full-length Mona or Grb2 protein or to the individual Mona domains, including amino-terminal SH3 domain (N-SH3), SH2 domain, ProR, and carboxy-terminal SH3 domain (C-SH3). Bound proteins were run on an SDS-12% PAGE gel and immunoblotted (IB) with anti-V5 (1:5,000) antibody. (B) Mona C-SH3 domain mutant fails to bind Gab3. Purified, immobilized GST fusion proteins containing full-length Mona protein harboring the P47L mutation in the amino-terminal SH3 domain or the P313L mutation in the carboxy-terminal SH3 domain were mixed with FD/Fms/Gab3V5 cell lysates. Bound proteins were analyzed on an SDS-12% PAGE gel by immunoblotting with anti-V5 or anti-Shc (1:2,000) antibody. (C) Mona SH2 domain does not bind to tyrosine phosphorylated Gab3. Cell lysates from M-CSF-stimulated FD/Fms/Gab3V5 cells (1 min 30 s, 37°C) were incubated with purified, immobilized GST fused with full-length Mona or Mona SH2 domain. WCL (50 μg) and protein complexes were separated by SDS-7.5% PAGE and immunoblotted with anti-Gab3 (1:100) or antiphosphotyrosine (αpY; 1:10,000) antibody.
The Gab3 primary amino acid sequence contains one potential binding site for the Grb2 SH2 domain that might also be recognized by the Mona SH2 domain (56). We therefore wondered whether Mona could interact with phosphorylated Gab3 through its SH2 domain. To address this question, GST-Mona SH2 fusion protein was incubated with cell lysates from M-CSF-stimulated FD/Fms/Gab3V5 cells. Figure 3C shows that no Gab3 protein could be precipitated using GST-Mona SH2 protein, whereas in the same experiment, the GST-Mona full-length protein readily detected Gab3 (also shown in Fig. 3A). Collectively, these data point to the exclusive involvement of the carboxy-terminal SH3 domain in the binding of Mona to Gab3.
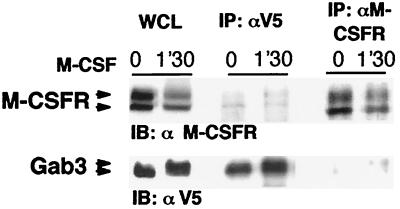
Our previous results indicated that the Mona SH2 domain associates with the activated M-CSFR through the tyrosine-phosphorylated motif encompassing Y697 within the receptor kinase insert region (9). Therefore, we looked for a potential physical interaction between M-CSFR and Gab3. FD/Fms/Gab3V5/Mona cells were stimulated for 1 min 30 s with M-CSF and cell lysates were used for anti-V5 or anti-M-CSFR immunoprecipitation, followed by Western blotting using the alternate antibody. Regardless of which precipitating antibody was used, the association between Gab3 and M-CSFR was only weakly detectable, even after M-CSF stimulation (Fig. 4). The weak interaction is often characteristic of indirect interactions; however, low avidity or high turnover could also explain these results. Whether this interaction is mediated by Mona, Grb2, or both proteins has yet to be determined. The low efficiency of Gab3 and M-CSFR coimmunoprecipitation hampered reliable comparison between FD/Fms/Gab3 cells expressing Mona or not (i.e., Mona and Grb2 or Grb2 alone, respectively) or expressing M-CSFR that is mutated or not on tyrosine 697, the common binding site of Mona and Grb2 (11, 53).
FIG. 4.
Examination of M-CSFR and Gab3 association. FD/Fms/Gab3V5 cells were stimulated, where indicated, for 1 min 30 s with 2,500 U of M-CSF per ml. Proteins in cell lysates were immunoprecipitated (IP) using anti-M-CSFR or anti-V5 antibody as specified. WCL (50 μg) and immunoprecipitates were separated by SDS-7.5% PAGE and proteins were visualized by immunoblotting (IB) using anti-M-CSFR (1:500) or anti-V5 (1:5,000) antibody.
Biphasic Gab3 phosphorylation in response to M-CSF.
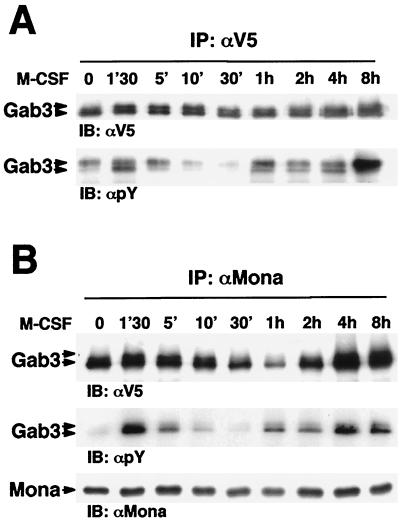
Stimulation of myeloid cells by M-CSF results in the activation of multiple intracellular signaling pathways, such as the Raf/MAP kinase pathway and those controlled by phosphoinositides (10). We recently reported that M-CSF evoked two waves of MAP kinase phosphorylation in FD/Fms cells (11). Interestingly, the second wave occurs after several hours of M-CSF stimulation, correlating with the onset of macrophage differentiation (11). Since M-CSF triggers phosphorylation of Gab3 in FD/Fms cells and Gab3 expression facilitates their differentiation in response to M-CSF (56), we looked for longer-term effects of Gab3 signaling. Thus, Gab3 phosphorylation was studied over extended times after M-CSF stimulation in FD/Fms/Gab3V5/Mona cells. Gab3 immunoprecipitation followed by phosphotyrosine detection clearly showed two waves of Gab3 phosphorylation after shifting the cells to M-CSF (Fig. 5A).
FIG. 5.
M-CSF regulates Gab3 phosphorylation and association with Mona. (A) FD/Fms/Gab3V5/Mona cells were washed in IMDM, starved, and then stimulated (except the zero time point) with 2,500 U of M-CSF per ml for the indicated times at 37°C. Cell lysates from each time were subjected to immunoprecipitation (IP) using anti-V5 antibody. Immunoprecipitates were separated by SDS-12% PAGE and proteins were visualized by immunoblotting (IB) using antiphosphotyrosine (αpY; 1:10,000) or anti-V5 (1:5,000) antibody. (B) As for panel A, but proteins in cell lysates were immunoprecipitated with anti-Mona antibody. Immunoprecipitates were separated by SDS-12% PAGE and visualized by immunoblotting using anti-V5, antiphosphotyrosine, or anti-Mona (1:1,000) antibody.
We have previously shown that Mona and Gab3 association is mediated through interaction of the Mona carboxy-terminal SH3 domain and the Gab3 atypical proline-rich motif (Fig. 1B and 3). Such interactions are thought to be constitutive, as they do not require protein modifications such as phosphorylation. However, intracellular signaling usually results in protein translocation or conformational changes that may modify constitutive protein-protein interactions. Since Gab3 phosphorylation varies with M-CSF stimulation time (Fig. 5A), we followed Mona and Gab3 association over time in FD/Fms/Gab3V5/Mona cells stimulated with M-CSF. Figure 5B shows that Mona and Gab3 interaction was slightly increased after 1 min 30 s of M-CSF stimulation and was then decreased by 1 h of stimulation. Large amounts of Gab3 protein were again detected in Mona immunoprecipitates after 4 or 8 h of M-CSF stimulation. These results indicate that Mona and Gab3 interaction is modulated in a time-course-dependent manner after M-CSF stimulation.
The M-CSFR mutated at the Mona binding site exhibits qualitative and quantitative alterations in signaling.
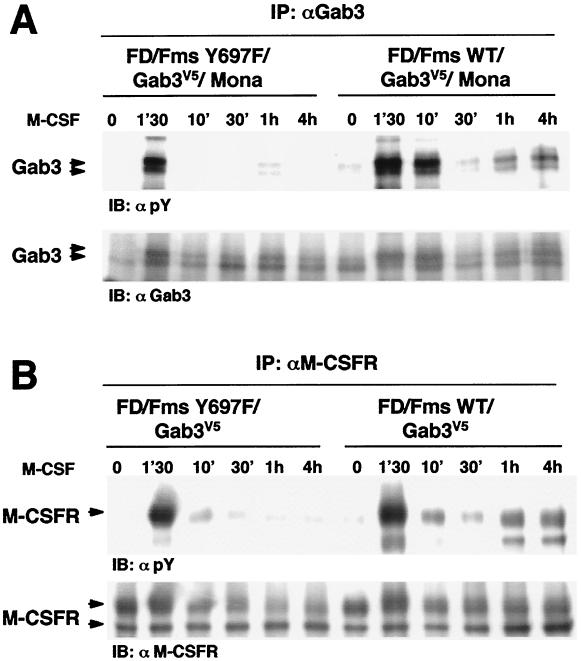
The early and late phases of M-CSF-induced tyrosine phosphorylation of Gab3 were examined utilizing cells expressing the M-CSFR mutated at the Y697 binding site. The SH2 domain of either Mona or Grb2 can bind this site and either adapter could serve as a linker to Gab3 (reference 56 and this study). We therefore examined the potential role of this site in signaling by studying Gab3 phosphorylation in FD/Fms Y697F/Gab3V5/Mona cells, which contain the M-CSFR Y697F mutation. Rapid and intense Gab3 phosphorylation similar to that observed in FD/Fms WT/Gab3V5/Mona cells was observed in FD/Fms Y697F/Gab3V5/Mona cells (Fig. 6A). Furthermore, whereas late Gab3 phosphorylation was clearly detected in FD/Fms WT/Gab3V5/Mona cells, it was strikingly reduced in FD/Fms Y697F/Gab3V5/Mona cells (Fig. 6A). Similar results were obtained when cells did not express exogenous Mona (data not shown). Thus, mutation of M-CSFR tyrosine 697 resulted in reduced or absent Gab3 phosphorylation in the second phase of the response to M-CSF.
FIG. 6.
The Y697F mutation of M-CSFR results in altered M-CSFR and Gab3 phosphorylation. (A) Effects of Y697F mutation on Gab3 phosphorylation. FD/Fms WT/Gab3V5/Mona and FD/Fms Y697F/Gab3V5/Mona cells were stimulated with 2,500 U of M-CSF per ml at 37°C for the indicated times. Proteins in cell lysates were immunoprecipitated (IP) using anti-Gab3 antibody. Immunoprecipitates were separated by SDS-7.5% PAGE and visualized by immunoblotting (IB) using antiphosphotyrosine (αpY; 1:10,000) or anti-Gab3 (1:100) antibody. (B) Effects of Y697F mutation on M-CSFR phosphorylation. FD/Fms WT/Gab3V5 and FD/Fms Y697F/Gab3V5 cells were stimulated with M-CSF as described above, and proteins in cell lysates were immunoprecipitated using anti-M-CSFR antibody. Immunoprecipitates were separated by SDS-7.5% PAGE and visualized by immunoblotting using antiphosphotyrosine (1:10,000) or anti-M-CSFR (1:500) antibody.
Impairment of late-phase Gab3 phosphorylation in FD/Fms Y697F/Gab3V5 cells might be the consequence of deficient M-CSFR kinase and/or autophosphorylation activity. This prompted us to compare M-CSFR phosphorylation in FD/Fms WT/Gab3V5 cells versus that in FD/Fms Y697F/Gab3V5 cells. The results, shown in Fig. 6B, demonstrate that similar peaks of M-CSFR phosphorylation were observed in both cell lines at 1 min 30 s after M-CSF stimulation, indicating that the Y697F mutation does not significantly alter receptor kinase activity. However, the second wave of M-CSFR phosphorylation appeared to be severely affected by the Y697F mutation. We have consistently observed tyrosine phosphorylation, 1 to 4 h after M-CSF stimulation, of the mature form of the M-CSFR but also of a smaller receptor-related product comigrating with the immature form of the M-CSFR (Fig. 6B, FD/Fms WT/Gab3V5 cells, at 1 or 4 h). We are presently exploring the nature of this smaller protein. Overall, these results further support a link between M-CSFR signaling and Gab3 phosphorylation at an early and a late stage of exposure to M-CSF.
Temporal expression of Mona and Gab3 correlates with the onset of macrophage differentiation.
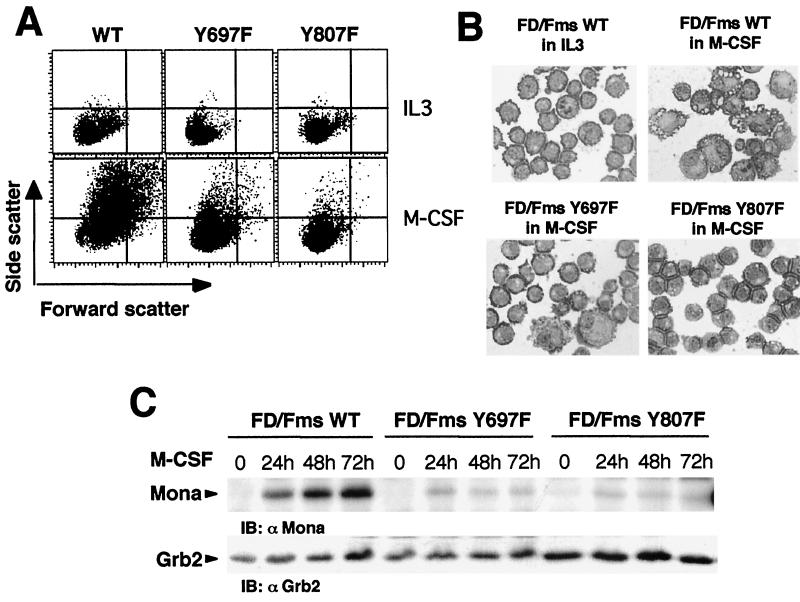
Gab3 expression is induced during M-CSF stimulation of FD/Fms cells. This expression correlates with macrophage differentiation because a single mutation (Y807F) in the M-CSFR reduces differentiation, but not growth-promoting functions, and also greatly reduces the M-CSF induction of Gab3 expression (56). M-CSF-induced expression of Mona or Gab3 has been observed during macrophage differentiation of two different myeloid cell lines (NFS-60 and FDC-P1) (9, 56). Collectively, these results raise the possibility that Mona and Gab3 expression are coincident and possibly linked with macrophage differentiation. This possibility was addressed experimentally using the FD/Fms cell model. FD/Fms cells cultivated for 3 days in the presence of M-CSF showed increased cell size (forward scatter) and granularity (side scatter) compared to the nondifferentiating IL-3-stimulated cells (Fig. 7A). This was accompanied by dramatic morphological changes that are typical of macrophage differentiation (Fig. 7B, compare FD/Fms WT in IL-3 and FD/Fms WT in M-CSF). Mona protein expression was induced as early as 8 h after shifting FD/Fms cells to M-CSF (not shown) and progressively increased from day 1 to day 3 of M-CSF stimulation (Fig. 7C, FD/Fms WT). Next, we looked at Mona induction in FDC-P1 cells expressing M-CSFR mutated at tyrosine 697 (FD/Fms Y697F cells) or tyrosine 807 (FD/Fms Y807F cells). It has been shown previously that FD/Fms Y807F cells do not differentiate in response to M-CSF, whereas a less severe defect was suggested for FD/Fms Y697F cells (7). Therefore, we reasoned that such cells might indicate whether Mona induction in FD/Fms cells is associated with macrophage differentiation rather than growth pathways of M-CSF signaling. After 3 days in M-CSF, FD/Fms Y697F cells showed increased forward and side scatter parameters, but at a much lesser degree than FD/Fms WT cells (Fig. 7A, compare left and middle panels). Morphology analysis confirmed a low propensity of FD/Fms Y697F cells to undergo macrophage differentiation in response to M-CSF (Fig. 7B, lower left panel). In agreement with previous reports (7, 8), FD/Fms Y807F cells lose the ability to differentiate in response to M-CSF, as shown by flow cytometric (Fig. 7A, Y807F) and morphology (Fig. 7B, lower right panel) analyses. Interestingly, although FD/Fms Y807F cells appeared more refractory to M-CSF-induced differentiation than FD/Fms Y697F cells, Mona protein expression was drastically decreased in both cell lines, and only marginal expression could be detected after M-CSF stimulation (Fig. 7C). Based on these data and previous reports (7-9, 56), we conclude that Mona and Gab3 expression are directly linked to the macrophage differentiation program in FD/Fms cells.
FIG. 7.
Comparison of Mona expression in FD/Fms WT, FD/Fms Y697F, and FD/Fms Y807F cells following M-CSF stimulation. (A) Fluorescence-activated cell sorter analysis of cell morphology. Cells were either maintained in IL-3 or incubated with 2,500 U of M-CSF per ml for 3 days and then analyzed by flow cytometry for light scatter intensity to monitor cellular size (forward scatter) and cellular granularity (side scatter). (B) As for panel A, but cells were stained with May-Grünwald Giemsa reagent. (C) Induction of Mona expression in FD/Fms cells is dependent on macrophage differentiation. Cells were cultivated as indicated above for indicated times. Cell lysates from each time point were analyzed for Mona and Grb2 expression on an SDS-12% PAGE gel by immunoblotting (IB) using anti-Mona (1:1,000) or anti-Grb2 (1:5,000) antibody.
Mona and Gab3 are induced and associate during macrophage differentiation of bone marrow-derived myeloid precursor cells.
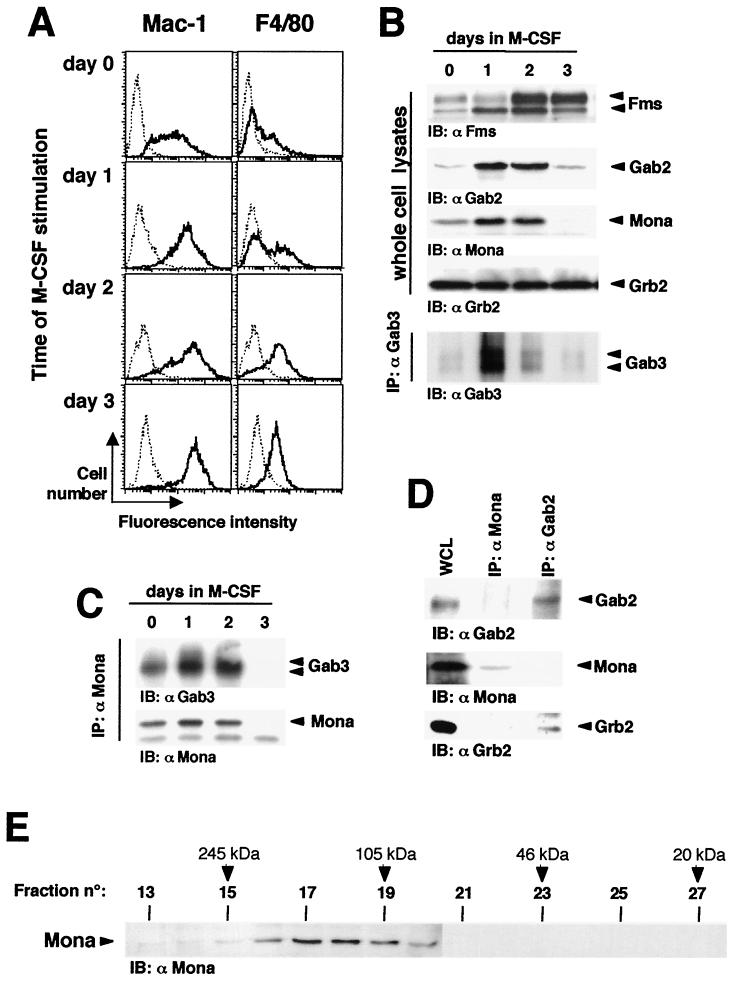
We next examined Gab3-Mona in a more physiological model of macrophage differentiation, taking advantage of a bone marrow culture system that permits massive in vitro expansion of monocytic cells from mouse bone marrow (unpublished data). Cultivation of mouse bone marrow cells in the presence of 5 ng of FL per ml for 6 days results in a cell population (day 6 FL cells) composed mainly of morphologically immature precursors which express the M-CSFR. Shifting these cells to M-CSF-containing medium allows their differentiation towards macrophages, as shown by morphology, adherence, phagocytosis, and increased expression of Mac-1 and the specific macrophage marker, F4/80 (3). In the experiment presented in Fig. 8A, flow cytometry was performed to examine Mac-1 and F4/80 expression on day 6 FL cells (i.e., day 0 cells in Fig. 8) or after a 1-, 2-, or 3-day cultivation in the presence of M-CSF. The day 0 cells showed low to moderate Mac-1 expression, whereas most were negative for F4/80 expression. Significant changes in Mac-1 and F4/80 expression were observed 2 days after M-CSF stimulation and reached a maximum at day 3, when most cells were adherent and acquired typical macrophage morphology (data not shown).
FIG. 8.
Mona/Gab3 complexes are induced during differentiation of normal macrophage precursors. (A) Differentiation of bone marrow-derived macrophage precursors. Day 6 FL cells (day 0) were obtained following cultivation of normal mouse bone marrow cells for 6 days in the presence of FL and then shifted to M-CSF-containing medium (1,000 U/ml) for various times as specified in the figure. Differentiation was detected by flow cytometry based on expression of the macrophage markers Mac-1 and F4/80. Dotted lines represent control cells labeled with secondary antibody alone. Bold lines show cells labeled for the differentiation markers specified. (B) Upregulated expression of Fms, Mona, Gab2, and Gab3. Bone marrow cells were cultivated in the presence of FL for 6 days (day 6 FL, day 0 in M-CSF) and then in the presence of 1,000 U of M-CSF per ml for 1, 2, or 3 days. Fms, Gab2, Mona, and Grb2 expression was detected in WCL following SDS-12% PAGE and immunoblotting (IB) using anti-Fms (1:500), anti-Gab2 (1:2,000), anti-Mona (1:1,000), or anti-Grb2 (1:5,000) antibody. The same lysates (1 mg) were used for Gab3 immunoprecipitation, run on an SDS-7.5% PAGE gel, and immunoblotted with anti-Gab3 antibody. (C) Mona associates with Gab3 during macrophage differentiation. Cell lysates were prepared from cells cultivated as described above, immunoprecipitated using anti-Mona antibody, separated on an SDS-12% PAGE gel, and immunoblotted with anti-Gab3 or anti-Mona antibody. (D) Mona does not associate with Gab2 during macrophage differentiation. Cell lysates (day 6 FL, day 2 in M-CSF) were obtained as described above, and proteins were immunoprecipitated using anti-Mona or anti-Gab2 antibodies, run on an SDS-12% PAGE gel, and immunoblotted with anti-Gab2, anti-Mona, or anti-Grb2 antibody. (E) Mona is found in multimolecular complexes in monocytes/macrophages. Day 6 FL cells were cultivated for 1 day in the presence of 1,000 U of M-CSF per ml. Protein complexes in WCL were separated on a Superose 12 HR10/60 gel filtration column. Protein complexes from fractions 13 to 31 were then separated by SDS-12% PAGE and analyzed for Mona expression by immunoblotting with anti-Mona antibody. Estimated molecular masses were calculated based on the standard curve produced from the molecular size markers as indicated in Materials and Methods.
Lysates were prepared from day 0 cells (i.e., day 6 FL cells) and their M-CSF-induced differentiating progeny in order to follow Mona and Gab3 expression and interaction during macrophage differentiation. The results are shown in Fig. 8B. The M-CSFR, Fms, was expressed in day 0 cells and expression increased throughout the 3 days. Mona was expressed in lysates from day 0 cells and its expression increased during the first 2 days of M-CSF stimulation and then rapidly disappeared after completion of macrophage differentiation at day 3. Gab3 expression followed similar kinetics, with maximal expression after cultivating day 0 cells for 1 day in the presence of M-CSF. The similar kinetics of Mona and Gab3 expression reflected an association, because Mona antiserum could readily coimmunoprecipitate Mona and Gab3 from day 6 FL cells grown for 1 or 2 days in the presence or absence of M-CSF (Fig. 8C). As might be expected from their respective time-course expression patterns, the Gab3-Mona complexes were more abundant following shifting of cells to M-CSF. However, such complexes were no longer detectable after 3 days of growth in M-CSF, which may be attributable to the sharp decrease in Gab3 and Mona expression at day 3 (Fig. 8B). Interestingly, Gab2 expression also increased during macrophage differentiation of day 6 FL cells grown in M-CSF, but Gab2 protein associated with Grb2 and not with Mona in the differentiating cells (Fig. 8D). These results are consistent with those obtained with FD/Fms cells (Fig. 2B).
M-CSF stimulates the formation of chromatographically distinct multimeric signaling complexes (22, 59). Thus, we performed gel filtration chromatography in order to assess the distribution of Mona in differentiating myeloid cells. Figure 8E shows an immunoblot analysis of fractions 13 to 31 obtained from lysates of day 6 FL cells stimulated for 24 h with M-CSF, i.e., committing to macrophage differentiation. Since Mona is a 38- to 40-kDa protein, it was expected that Mona monomers would be found in fractions 23 to 25. In fact, most Mona proteins were detected in higher-molecular-size fractions, especially in fractions 15 to 20 (100 to 245 kDa). However, we failed to detect Gab3 in these fractions, probably due to low Gab3 expression. Indeed, only immunoprecipitation enabled us to detect Gab3 protein in differentiating day 6 FL cells (Fig. 8A). These data indicate that most Mona protein is found in large molecular complexes, strengthening the idea that Mona complexes have a major role in monocyte/macrophage differentiation.
DISCUSSION
Structurally, Mona is a Grb2-related hematopoietic-specific adapter molecule originally cloned through its ability to interact with activated M-CSFR (9). Mona is expressed predominantly in T cells and myeloid lineage cells and shares 49% sequence identity with Grb2. The SH2 and SH3 domains of Mona are highly homologous to those of Grb2, and the major difference encompasses a discontinuous insert of about 100 amino acids between the central SH2 domain and the C-terminal SH3 domain in Mona. Mona was independently cloned as Gads, a partner for phosphorylated Shc protein, GRID, GRAP-2, a Gab1-interacting protein, GrpL, and Grf40 (1, 13, 25, 28, 44). Gads was shown to play a key role in T-cell activation and development by recruiting SLP76 to LAT (5, 28, 29). Furthermore, mice deficient in Gads exhibit a severe alteration of T-cell development due to a block in proliferation of CD4− CD8− thymocytes (60). Mona overexpression in normal myeloid progenitors prevents macrophage differentiation in vitro, and in FD/Fms cells, it results in augmented MAP kinase activation in response to M-CSF (9, 11). Therefore, Mona may have important roles in both T-cell and myeloid cell signaling.
Gab3 belongs to the DOS/Gab family of scaffolding proteins, whose members contain an N-terminal pleckstrin homology domain, multiple tyrosine-containing motifs for SH2-domain binding, and at least two conserved proline-rich regions whose amino acid sequences suggest potential Grb2 or Mona SH3 domain binding sites (16, 20, 56). Gab3 is expressed primarily in hematopoietic cells, with lower expression levels found in the heart, kidneys, brain, and lungs. The Gab3 protein is tyrosine phosphorylated after M-CSF stimulation of appropriate target cells and, like other members of the DOS/Gab family of proteins, may function in differentiation of the myeloid lineage cells (19, 31, 56).
Previously, we had collectively identified Mona and Gab3 proteins and shown that the expression of each is induced in M-CSF-stimulated FD/Fms cells (9, 56). We have now demonstrated that the induction of Mona and Gab3 is linked directly to the signaling pathway for macrophage differentiation. M-CSF stimulation of FD/Fms cells and day 6 FL cells results in macrophage development over a 3-day time period. Within the first 24 h of this stimulation, induced expression of Mona and Gab3 is detectable, and protein levels increase over the following days. Two different mutations in the M-CSFR (Y807F and Y697F) block, to varying degrees, receptor differentiation signaling ability, and each has a profound effect on expression of Mona and Gab3 in FD/Fms cells (references 7 and 56 and this study). The Y807F receptor mutation presumably prevents a conformational alteration in the receptor kinase domain needed for differentiation. Biologically, this mutation dramatically ablates differentiation of FD/Fms cells, but growth signaling remains as good as, or better than, that of the wild-type receptor (7). Similarly, the Y697F mutation destroys the potential Mona/Grb2 SH2 domain binding site (9, 53) and, although less severe than the Y807F mutation, also decreases morphological differentiation. M-CSF stimulation of the FD cells expressing either mutant M-CSFR fail to induce Mona expression (Fig. 7C), and similar results were seen for Gab3 expression in FD cells expressing the Y807F M-CSFR (56). Therefore, differentiation signaling rather than growth signaling is required for the concurrent induction of both Mona and Gab3.
In addition to the M-CSF-dependent coexpression of Mona and Gab3, we also have demonstrated that these two induced proteins exhibit a specific and possibly functional interaction. In vitro, Mona and Grb2 show an unbiased interaction toward the PRD of either Gab2 or Gab3 (Fig. 1), consistent with conservation of the P-X3-R-X2-K-P-X7-P-L-D sequence in both PRD. Similar assays in the yeast two-hybrid system showed a preference of Mona for the PRD of Gab3 over the PRD of Gab2 (Fig. 1). Strikingly, Mona/Gab2 complexes could not be detected in an immunoprecipitation assay (Fig. 2), suggesting that structurally the Gab2 protein does not permit the in vivo association of Mona even though Gab2 contains the same conserved atypical polyproline-binding site found in Gab3. Preferential signaling complexes involving Mona/Gads and Grb2 have also been shown to occur in the T-cell system. Grb2 and Mona/Gads are able to interact with SLP76 in vitro, but only Mona/Gads-SLP76 complexes could be detected in activated T lymphocytes (25, 29); moreover, even in the absence of Mona/Gads in Gads−/− mice, no Grb2-SLP76 could be detected (56). The atypical proline-rich motif in Gab3 PRD may represent a common docking site for the carboxy-terminal domains of Mona and Grb2. Therefore, within M-CSFR signaling, it is also possible that Gab3/Mona and Gab3/Grb2 interactions are mutually exclusive. However, Grb2, but not Mona, may also interact with the typical proline-rich motif common to all Gab family members found in the putative Met-binding domain (32, 56). Thus, whether this motif is actually used by Grb2 for binding to Gab3 should be determined before considering a possible competition between Mona and Grb2 for Gab3 association.
An additional unique observation on the role of Mona/Gab3 proteins in M-CSF signaling is that tyrosine phosphorylation and signaling were observed during two temporally distinct phases or waves. These were observed initially in the FD/Fms cells expressing exogenous Mona and Gab3 (Fig. 5). The first wave occurred early, reaching a peak at 1 min 30 s after stimulation, and was essentially completed by 10 min after M-CSF addition. The mature form of the M-CSFR was rapidly and transiently tyrosine phosphorylated in this phase (Fig. 6), as was Gab3, which also associated with Mona. Surprisingly, a late and persistent wave of signaling was detected from 4 to 24 h after M-CSF stimulation. The second wave of M-CSFR tyrosine phosphorylation also evoked tyrosine phosphorylation of the receptor. Likewise, a second wave of Gab3 tyrosine phosphorylation, concurrent with Mona/Gab3 association, was observed. During the same time period, the increased expression of both Gab3 and Mona proteins occurred, and therefore, this phase may be due more to the induced Mona/Gab3 protein expression than the early wave. In any case, without exogenous expression of Mona and Gab3, the first phase does not involve Mona or Gab3, but rather Gab2 and Grb2 interaction, primarily (reference 56 and personal observations).
The second wave of Gab3 phosphorylation, and presumably its association with Mona, was coincident with the onset of macrophage differentiation signaling and sensitive to mutations in the M-CSFR which block or impair differentiation. Previous results have shown that the differentiation-defective M-CSFR mutation, Y807F, fails to induce Gab3 protein expression even after 3 days of M-CSF stimulation in FDC-P1 cells (56). The results presented in Fig. 7 demonstrate that the induced expression of Mona is, likewise, defective in the same cells over the 3-day time period. The Y697F mutation of the M-CSFR also is defective for Mona induction and the second wave of signaling. The Y697 site on the M-CSFR is the most plausible binding site for the SH2 domain of Grb2 or Mona; however, a second similar motif at Y920 might also serve the same function (33). Therefore, the conclusion that defective binding of either Grb2 or Mona to M-CSFR is the cause of the phenotype must await further studies on how and when these proteins interact in signaling.
The use of a primary culture system for bone marrow cell cultivation allowed us to examine the relevance of the Mona and Gab3 expression in a more physiological background of macrophage development. For this purpose, we cultivated murine bone marrow cells in the presence of FL to produce large numbers of macrophage precursors. After shifting these cells to growth on M-CSF, we observed mature macrophages after 3 days in culture, and Mona and Gab3 were coexpressed and interacted during days 1 and 2 of culture but were absent at day 3. These results are similar to those found in cell lines in culture but suggest that the Mona and Gab3 complex is active only during early macrophage development. In addition, most Mona protein was present in multimolecular complexes during the early developmental period (Fig. 8). Mature macrophages lacked both proteins. These results are consistent with a role for Mona in macrophage differentiation of bone marrow cells; however, the present experiments did not differentiate between early and late waves of signaling, nor did they confirm our previous result of a constant level of Gab2 but inducible Gab3 expression (56). Certainly, differences in biological behavior exist between cell lines and primary cultures, and each may be useful in different ways.
Gel filtration experiments performed on lysates of M-CSF-stimulated bone marrow-derived macrophage precursors led to the conclusion that Mona proteins massively participate in multimolecular complexes (Fig. 8E). Therefore, Mona and Gab3 expression at a specific step of macrophage differentiation may modify M-CSF signaling by creating specific multiprotein signaling complexes. It is now necessary to identify the molecules recruited by Mona to Gab3 and associated proteins. At least six different proteins have been shown to interact with the Mona/Gads carboxyl-terminal SH3 domain, but none have been shown to interact with its amino-terminal SH3 domain (1, 13, 25, 29, 30, 44, 57). Current experiments are aimed at identifying new Mona partners in order to determine how Mona/Gab3 complexes contribute to the M-CSFR differentiation signal.
The biphasic nature of M-CSFR signaling in macrophage differentiation, as shown in this paper, suggests that the late wave of tyrosine phosphorylation may have some unique function in development. Biological events such as proliferation and commitment (14, 37) are thought of in the context of temporal differentiation, as are such biochemical phenomena as the late persistent phase of MAPK activation in relationship to megakaryocyte, neuronal, or yeast cell differentiation (35, 45, 50). Our future experiments, therefore, will be directed toward understanding the M-CSFR signaling mechanisms in relation to these other aspects of differentiation.
Acknowledgments
We thank Marie-France Grasset and Maud Renon for technical assistance.
During the course of this work, C.B. was supported by fellowships from the Ligue Nationale contre le Cancer. L.R.R. was a fellow of the John Simon Guggenheim Memorial Foundation on sabbatical in the laboratory of G.M. during a portion of these studies. This work was supported by grants from the Ligue Nationale contre le Cancer and from the Centre National de la Recherche Scientifique.
REFERENCES
- 1.Asada, H., N. Ishii, Y. Sasaki, K. Endo, H. Kasai, N. Tanaka, T. Takeshita, S. Tsuchiya, T. Konno, and K. Sugamura. 1999. Grf40, a novel Grb2-family member, is involved in T-cell signaling through interaction with SLP76 and LAT. J. Exp. Med. 189:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asakura, E., T. Yamauchi, A. Umemura, T. Hanamura, and T. Tanabe. 1997. Intravenously administered macrophage colony-stimulating factor (M-CSF) specifically acts on the spleen, resulting in the increasing and activating spleen macrophages for cytokine production in mice. Immunopharmacology 37:7-14. [DOI] [PubMed] [Google Scholar]
- 3.Austyn, J. M., and S. Gordon. 1981. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur. J. Immunol. 11:805-815. [DOI] [PubMed] [Google Scholar]
- 4.Bardelli, A., P. Longati, D. Gramaglia, M. C. Stella, and P. M. Comoglio. 1997. Gab1 coupling to the HGF/Met receptor multifunctional docking site requires binding of Grb2 and correlates with the transforming potential. Oncogene 15:3103-3111. [DOI] [PubMed] [Google Scholar]
- 5.Berry, D. M., S. J. Benn, A. M. Cheng, and C. J. McGlade. 2001. Caspase-dependent cleavage of the hematopoietic specific adapter Gads alters signaling from the T cell receptor. Oncogene 20:1203-1211. [DOI] [PubMed] [Google Scholar]
- 6.Bicknell, D. C., D. E. Williams, and H. E. Broxmeyer. 1988. Correlation between CSF-1 responsiveness and expression of (CSF-1 receptor) c-fms in purified murine granulocyte-macrophage progenitor cells (CFU-GM). Exp. Hematol. 16:240-243. [PubMed] [Google Scholar]
- 7.Bourette, R. P., G. M. Myles, K. Carlberg, A. R. Chen, and L. R. Rohrschneider. 1995. Uncoupling of the proliferation and differentiation signals mediated by the murine macrophage colony-stimulating factor receptor expressed in myeloid FDC-P1 cells. Cell. Growth Differ. 6:631-645. [PubMed] [Google Scholar]
- 8.Bourette, R. P., G. M. Myles, J. L. Choi, and L. R. Rohrschneider. 1997. Sequential activation of phosphatidylinositol 3-kinase and phospholipase C-gamma 2 by the M-CSF receptor is necessary for differentiation signaling. EMBO J. 16:5880-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourette, R. P., S. Arnaud, G. M. Myles, J. P. Blanchet, L. R. Rohrschneider, and G. Mouchiroud. 1998. Mona, a novel hematopoietic-specific adapter interacting with the macrophage colony-stimulating factor receptor, is implicated in monocyte/macrophage development. EMBO J. 17:7273-7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourette, R. P., and L. R. Rohrschneider. 2000. Early events in M-CSF receptor signaling. Growth Factors 17:155-166. [DOI] [PubMed] [Google Scholar]
- 11.Bourgin, C., R. P. Bourette, G. Mouchiroud, and S. Arnaud. 2000. Expression of Mona (monocytic adapter) in myeloid progenitor cells results in increased and prolonged MAP kinase activation upon macrophage colony-stimulating factor stimulation. FEBS Lett. 480:113-117. [DOI] [PubMed] [Google Scholar]
- 12.Byrne, P. V., L. J. Guilbert, and E. R. Stanley. 1981. Distribution of cells bearing receptors for a colony-stimulating factor (CSF-1) in murine tissues. J. Cell Biol. 91:848-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis, J., C. Ashman, M. Burden, K. Kilpatrick, M. Morse, and P. Hamblin. 2000. GRID: a novel Grb-2-related adapter protein that interacts with the activated T cell costimulatory receptor CD28. J. Immunol. 164:317-321. [DOI] [PubMed] [Google Scholar]
- 14.Enver, T., C. M. Heyworth, and T. M. Dexter. 1998. Do stem cells play dice? Blood 92:348-351. [PubMed] [Google Scholar]
- 15.Fixman, E. D., M. Holgado-Madruga, L. Nguyen, D. M. Kamikura, T. M. Fournier, A. J. Wong, and M. Park. 1997. Efficient cellular transformation by the Met oncoprotein requires a functional Grb2 binding site and correlates with phosphorylation of the Grb2-associated proteins, Cbl and Gab1. J. Biol. Chem. 272:20167-20172. [DOI] [PubMed] [Google Scholar]
- 16.Gu, H., J. C. Pratt, S. J. Burakoff, and B. G. Neel. 1998. Cloning of p97/Gab2, the major SHP2-binding protein in hematopoietic cells, reveals a novel pathway for cytokine-induced gene activation. Mol. Cell 2:729-740. [DOI] [PubMed] [Google Scholar]
- 17.Gu, H., H. Maeda, J. J. Moon, J. D. Lord, M. Vookim, B. H. Nelson, and B. G. Neel. 2000. New role for Shc in activation of the phosphatidylinositol 3-kinase/Akt pathway. Mol. Cell. Biol. 20:7109-7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton, J. A. 1997. CSF-1 signal transduction. J. Leukoc. Biol. 62:145-155. [DOI] [PubMed] [Google Scholar]
- 19.Herbst, R., X. Zhang, J. Qin, and M. A. Simon. 1999. Recruitment of the protein tyrosine phosphatase CSW by DOS is an essential step during signaling by the Sevenless receptor tyrosine kinase. EMBO J. 18:6950-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holgado-Madruga, M., D. R. Emlet, D. K. Moscatello, A. K. Godwin, and A. J. Wong. 1996. A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature 379:560-564. [DOI] [PubMed] [Google Scholar]
- 21.Itoh, I., Y. Yoshida, K. Nishida, M. Narimatsu, M. Hibi, and T. Hirano. 2000. Role of Gab1 in heart, placenta, and skin development and growth factor-and-cytokine-induced extracellular signal-regulated kinase mitogen-activated protein kinase activation. Mol. Cell. Biol. 20:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanagasundaram, V., A. Jaworowski, R. Byrne, and J. A. Hamilton. 1999. Separation and characterization of the activated pool of colony-stimulating factor 1 receptor forming distinct multimeric complexes with signaling molecules in macrophages. Mol. Cell. Biol. 19:4079-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karasuyama, H., and F. Melchers. 1988. Establishment of mouse cell lines which constitutively secrete large quantities of interleukin 2, 3, 4, or 5 using modified cDNA expression vectors. Eur. J. Immunol. 18:97-104. [DOI] [PubMed] [Google Scholar]
- 24.Kodama, H., M. Nose, S. Niida, and A. Yamasaki. 1991. Essential role of macrophage colony-stimulating factor in the osteoclast differentiation supported by stromal cells. J. Exp. Med. 173:1291-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law, C. L., M. K. Ewings, P. M. Chaudhary, S. A. Solow, T. J. Yun, A. J. Marshall, L. Hood, and E. A. Clark. 1999. GrpL, a Grb2-related adapter protein, interacts with SLP76 to regulate nuclear factor of activated T cell activation. J. Exp. Med. 189:1243-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewitzky, M., C. Kardinal, N. H. Gehring, E. K. Schmidt, B. Konkol, M. Eulitz, W. Birchmeier, U. Schaeper, and S. M. Feller. 2001. The C-terminal SH3 domain of the adapter Grb2 binds with high affinity to sequences in Gab1 and SLP76 which lack the SH3-typical P-x-x-P core motif. Oncogene 20:1052-1062. [DOI] [PubMed] [Google Scholar]
- 27.Lioubin, M. N., G. M. Myles, K. Carlberg, D. Bowtell, and L. R. Rohrschneider. 1994. Shc, Grb2, Sos1, and a 150-kilodalton tyrosine-phosphorylated protein form complexes with Fms in hematopoietic cells. Mol. Cell. Biol. 14:5682-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, S. K., and C. J. McGlade. 1998. Gads is a novel SH2 and SH3 domain-containing adapter protein that binds to tyrosine-phosphorylated Shc. Oncogene 17:3073-3082. [DOI] [PubMed] [Google Scholar]
- 29.Liu, S. K., N. Fang, G. A. Koretzky, and C. J. McGlade. 1999. The hematopoietic-specific adapter protein Gads functions in T-cell signaling via interaction with the SLP76 and LAT adapters. Curr. Biol. 9:67-75. [DOI] [PubMed] [Google Scholar]
- 30.Liu, S. K., C. A. Smith, R. Arnold, F. Kiefer, and C. J. McGlade. 2000. The adaptor protein Gads (Grb2-related adaptor downstream of Shc) is implicated in coupling hematopoietic progenitor kinase-1 to the activated TCR. J. Immunol. 165:1417-1426. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Y., B. Jenkins, J. L. Shin, and L. R. Rohrschneider. 2001. Scaffolding protein Gab2 mediates differentiation signaling downstream of Fms receptor tyrosine kinase. Mol. Cell. Biol. 21:3047-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lock, L. S., I. Royal, M. A. Naujokas, and M. Park. 2000. Identification of an atypical Grb2 carboxyl-terminal SH3 domain binding site in Gab docking proteins reveals Grb2-dependent and -independent recruitment of Gab1 to receptor tyrosine kinases. J. Biol. Chem. 275:31536-31545. [DOI] [PubMed] [Google Scholar]
- 33.Mancini, A., R. Niedenthal, H. Joos, A. Koch, S. Trouliaris, H. Niemann, and T. Tamura. 1997. Identification of a second Grb2 binding site in the v-Fms tyrosine kinase. Oncogene 15:1565-1572. [DOI] [PubMed] [Google Scholar]
- 34.Maroun, C. R., D. K. Moscatello, M. A. Naujokas, M. Holgado-Madruga, A. J. Wong, and M. Park. 1999. A conserved inositol phospholipid binding site within the pleckstrin homology domain of the Gab1 docking protein is required for epithelial morphogenesis. J. Biol. Chem. 274:31719-31726. [DOI] [PubMed] [Google Scholar]
- 35.Matsumura, I., K. Nakajima, H. Wakao, S. Hattori, K. Hashimoto, H. Sugahara, T. Kato, H. Miyazaki, T. Hirano, and Y. Kanakura. 1998. Involvment of prolonged ras activation in thrombopoietin-induced megakaryocytic differentiation in a human factor-dependent hematopoietic cell line. Mol. Cell. Biol. 18:4282-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKinstry, W. J., C. L. Li, J. E. Rasko, N. A. Nicola, G. R. Johnson, and D. Metcalf. 1997. Cytokine receptor expression on hematopoietic stem and progenitor cells. Blood 89:65-71. [PubMed] [Google Scholar]
- 37.Metcalf, D. 1998. Lineage commitment and maturation in hematopoietic cells: the case for extrinsic regulation. Blood 92:345-347. [PubMed] [Google Scholar]
- 38.Metcalf, D., N. A. Nicola, N. M. Gough, M. Elliott, G. McArthur, and M. Li. 1992. Synergistic suppression: anomalous inhibition of the proliferation of factor-dependent hemopoietic cells by combination of two colony-stimulating factors. Proc. Natl. Acad. Sci. USA 89:2819-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munn, D. H., M. B. Garnick, and N. K. Cheung. 1990. Effects of parental recombinant human macrophage colony-stimulating factor on monocyte number, phenotype, and antitumor cytotoxicity in nonhuman primates. Blood 75:2042-2048. [PubMed] [Google Scholar]
- 40.Nguyen, L., M. Holgado-Madruga, C. Maroun, E. D. Fixman, D. Kamikura, T. Fournier, A. Charest, M. L. Tremblay, A. J. Wong, and M. Park. 1997. Association of the multisubstrate docking protein Gab1 with the hepatocyte growth factor receptor requires a functional Grb2 binding site involving tyrosine 1356. J. Biol. Chem. 272:20811-20819. [DOI] [PubMed] [Google Scholar]
- 41.Nicholls, S. E., S. Winter, R. Mottram, J. A. Miyan, and A. D. Whetton. 1999. Flt3 ligand can promote survival and macrophage development without proliferation in myeloid progenitor cells. Exp. Hematol. 27:663-672. [DOI] [PubMed] [Google Scholar]
- 42.Pawlak, G., C. Valadoux-Delplanque, V. Revol, R. P. Bourette, J. P. Blanchet, and G. Mouchiroud. 1999. Colony-stimulating factor-1 impairs both proliferation and differentiation signals of erythropoietin during the commitment of bipotential NFS-60 cell line to the monocytic lineage. Exp. Hematol. 27:797-805. [DOI] [PubMed] [Google Scholar]
- 43.Pawlak, G., M. F. Grasset, S. Arnaud, J. P. Blanchet, and G. Mouchiroud. 2000. Receptor for macrophage colony-stimulating factor transduces a signal decreasing erythroid potential in the multipotential hematopoietic EML cell line. Exp. Hematol. 28:1164-1173. [DOI] [PubMed] [Google Scholar]
- 44.Qiu, M., S. Hua, M. Agrawal, G. Li, J. Cai, E. Chan, H. Zhou, Y. Luo, and M. Liu. 1998. Molecular cloning and expression of human GRAP-2, a novel leukocyte-specific SH2- and SH3-containing adapter-like protein that binds to Gab-1. Biochem. Biophys. Res. Commun. 253:443-447. [DOI] [PubMed] [Google Scholar]
- 45.Qui, M. S., and S. H. Green. 1992. PC12 cell neuronal differentiation is associated with prolonged p21ras activity and consequent prolonged ERK activity. Neuron 9:705-717. [DOI] [PubMed] [Google Scholar]
- 46.Raabe, T., J. Riesgo-Escovar, X. Liu, B. S. Bausenwein, P. Deak, P. Maröy, and E. Hafen. 1996. DOS, a novel pleckstrin homology domain-containing protein required for signal transduction between sevenless and Ras1 in Drosophila. Cell 85:911-920. [DOI] [PubMed] [Google Scholar]
- 47.Rodrigues, D. A., M. Falasca, Z. Zhang, S. H. Ong, and J. Schlessinger. 2000. A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Mol. Cell. Biol. 20:1448-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohrschneider, L. R. 1994. The macrophage colony-stimulating factor (M-CSF) receptor, p. 168-170. In N. A. Nicola (ed.), Guidebook to cytokines and their receptors. Oxford University Press, Oxford, United Kingdom.
- 49.Rohrschneider, L. R., R. P. Bourette, M. N. Lioubin, P. A. Algate, G. M. Myles, and K. Carlberg. 1997. Growth and differentiation signals regulated by the M-CSF receptor. Mol. Reprod. Dev. 46:96-103. [DOI] [PubMed] [Google Scholar]
- 50.Sabbagh, W., Jr., L. J. Flatauer, A. J. Bardwell, and L. Bardwell. 2001. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol. Cell 8:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanley, E. R., D. M. Chen, and H. S. Lin. 1978. Induction of macrophage production and proliferation by a purified colony-stimulating factor. Nature 274:168-170. [DOI] [PubMed] [Google Scholar]
- 52.Stanley, E. R., K. L. Berg, D. B. Einstein, P. S. Lee, and Y. G. Yeung. 1994. The biology and action of colony-stimulating factor 1. Stem Cells 12:15-24. [PubMed] [Google Scholar]
- 53.van der Geer, P., and T. Hunter. 1993. Mutation of Tyr697, a GRB2-binding site, and Tyr721, a PI 3-kinase binding site, abrogates signal transduction by the murine CSF-1 receptor expressed in Rat-2 fibroblasts. EMBO J. 12:5161-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vojtek, A. B., and S. M. Hollenberg. 1995. Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 255:331-342. [DOI] [PubMed] [Google Scholar]
- 55.Wang, Z. E., G. M. Myles, C. S. Brandt, M. N. Lioubin, and L. Rohrschneider. 1993. Identification of the ligand-binding regions in the macrophage colony-stimulating factor receptor extracellular domain. Mol. Cell. Biol. 13:5348-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolf, I., B. J. Jenkins, Y. Liu, M. Seiffert, J. M. Custodio, P. Young, and L. R. Rohrschneider. 2002. Gab3, a new DOS/Gab family member, facilitates macrophage differentiation. Mol. Cell. Biol. 22:231-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia, C., Z. Bao, F. Tabassam, W. Ma, M. Qiu, S. Hua, and M. Liu. 2000. GCIP, a novel human Grap2 and cyclin D interacting protein, regulates E2F-mediated transcriptional activity. J. Biol. Chem. 275:20942-20948. [DOI] [PubMed] [Google Scholar]
- 58.Yamauchi, T., K. Yada, A. Umemura, E. Asakura, T. Hanamura, and T. Tanabe. 1996. Effect of recombinant human macrophage-colony stimulating factor on marrow, splenic, and peripheral hematopoietic progenitor cells in mice. J. Leukoc. Biol. 59:296-301. [DOI] [PubMed] [Google Scholar]
- 59.Yeung, Y.-G., Y. Wang, D. B. Einstein, P. S. W. Lee, and E. R. Stanley. 1998. Colony-stimulating factor-1 stimulates the formation of multimeric cytosolic complexes of signaling proteins and cytoskeletal components in macrophages. J. Biol. Chem. 273:17128-17137. [DOI] [PubMed] [Google Scholar]
- 60.Yoder, J., C. Pham, Y. M. Iizuka, O. Kanagawa, S. K. Liu, C. J. McGlade, and A. M. Cheng. 2001. Requirement for the SLP76 adapter GADS in T cell development. Science 291:1987-1991. [DOI] [PubMed] [Google Scholar]