Abstract
Nuclear factor of activated T cells (NFAT) is implicated in multiple biological processes, including cytokine gene expression, cardiac hypertrophy, and adipocyte differentiation. A conserved NFAT homology domain is identified in all NFAT members. Dephosphorylation of the NFAT homology region is critical for NFAT nuclear translocation and transcriptional activation. Here we demonstrate that NFATc4 is phosphorylated by p38 mitogen-activated protein (MAP) kinase but not by JNK. The p38 MAP kinase phosphorylates multiple residues, including Ser168 and Ser170, in the NFAT homology domain of NFATc4. Replacement of Ser168,170 with Ala promotes nuclear localization of NFATc4 and increases NFAT-mediated transcription activity. Stable expression of Ala168,170 NFATc4, but not of wild-type NFATc4, in NIH 3T3 cells promotes adipocyte formation under differentiation conditions. Molecular analysis indicates that peroxisome proliferator-activated receptor γ2 (PPARγ2) is a target of NFAT. Two distinct NFAT binding elements are located in the PPARγ2 gene promoter. Stable expression of Ala168,170 NFATc4, but not of wild-type NFATc4, increases the expression of PPARγ, which contributes in part to increased adipocyte formation. Thus, NFAT regulates PPARγ gene expression and has a direct role in adipocyte differentiation.
Nuclear factor of activated T cells (NFAT) is a group of transcription factors that was first identified to play an important role in cytokine gene expression (22). Subsequent studies demonstrated that NFATs are present in numerous tissues (33, 34, 40). The wide tissue distribution of the NFAT isoforms suggests that NFAT may participate in multiple physiological processes. Recently, NFAT activity has been implicated in adipocyte differentiation, cardiac hypertrophy, and learning and memory (30, 32, 41). Thus, elucidation of mechanisms that regulate NFAT is critical for understanding these biological processes.
Four distinct genes encoding closely related NFAT proteins (NFATc1/NFATc/NFAT2, NFATc2/NFATp/NFAT1, NFATc3/NFAT4/NFATx, and NFATc4/NFAT3) have been identified (reviewed in references 18 and 47). Alternative mRNA splicing of these four genes further generates at least 10 different NFAT polypeptides. The function of these alternatively spliced NFAT isoforms remains elusive. However, all NFAT members contain a highly conserved NH2-terminal regulatory NFAT homology domain and a COOH-terminal Rel homology region for DNA binding. Thus, understanding the function of these conserved domains will provide new insights on NFAT regulation.
The NH2-terminal NFAT homology domain encodes several distinct sequences, including the PXIXIT motif, the Ser-rich region (SRR), and the Ser-Pro (SP)-rich boxes for NFAT regulation (18, 47). These sequences are found in all NFAT members. The PXIXIT motif is recognized by the calcineurin phosphatase (3, 15), which dephosphorylates NFAT upon activation. Sequestration of the calcineurin phosphatase by overexpression of the PXIXIT motif blocks NFAT activation. The SRR and the SP boxes are major targets for NFAT phosphorylation (4-6, 12-14, 44, 46, 58). Dephosphorylation of Ser residues in the SRR and the SP boxes promotes nuclear localization of NFAT. Thus, dephosphorylation of the NFAT homology domain, which is mediated by the calcineurin phosphatase, plays an important role in NFAT activation.
Once NFAT is dephosphorylated and translocated into the nucleus, activated NFAT interacts with other transcription factors to induce gene expression. The interaction of NFAT with Fos-Jun (AP-1 complex), GATA, and MEF2 suggests that NFAT often functions at composite DNA elements (7, 41, 43, 54, 56). Formation of a ternary complex induces expression of NFAT targets, such as interleukin-2 (IL-2), IL-4, IL-5, and tumor necrosis factor alpha. However, physiological function of NFAT in nonimmune tissues remains to be established.
Multiple protein kinases, including the mitogen-activated protein (MAP) kinase group (ERK, JNK, and p38 kinase), glycogen synthase kinase 3β (GSK3β), protein kinase A (PKA), and casein kinase 1α (CK1α), have been shown to phosphorylate NFAT (4, 6, 12-14, 29, 46, 58). NFAT is phosphorylated on multiple Ser residues located in the conserved SRR and the SP boxes. Phosphorylation of these Ser residues opposes nuclear localization of NFAT either by promoting nuclear export or by impeding nuclear import. For example, phosphorylation at Ser269 of NFATc1 (6) and Ser289 of NFATc4 (12) could be mediated by PKA. Sequence comparisons indicate that Ser269 of NFATc1 corresponds to Ser289 of NFATc4. Phosphorylation at Ser269 of NFATc1 promotes its subsequent phosphorylation mediated by GSK3β, which is critical to enhance NFATc1 nuclear export. Phosphorylation at Ser289 of NFATc4, in conjunction with phosphorylation at Ser272, recruits 14-3-3, a signaling modulator, to mask the function of an adjacent nuclear localization sequence; thus, phosphorylated NFATc4 is located in the cytosol. Additional phosphorylation at other Ser residues present in the NFAT homology domain may promote intramolecular interactions to mask the nuclear localization sequence (6, 14, 58) or to obstruct calcineurin binding (12) and hence maintain NFAT in a phosphorylated and inactive state.
The MAP kinase group of signaling proteins also phosphorylates members of the NFAT family. NFATc3 was identified as a substrate for JNK in a yeast two-hybrid assay (14). Phosphorylation of Ser163 and Ser165 of NFATc3 by JNK opposes calcineurin-mediated nuclear localization. Ser172 of NFATc1, which is located in a position analogous to that of Ser165 of NFATc3, is also phosphorylated by JNK (13, 46). Replacement of Ser172 with Ala, to prevent JNK phosphorylation, promotes nuclear localization of NFATc1. Importantly, gene targeting studies that disrupt the JNK1 locus also promote NFATc1 nuclear localization and enhance the expression of IL-4, a target of NFAT in T cells (21). Thus, JNK negatively regulates NFATc1 and NFATc3 but not other NFAT isoforms.
The purpose of this study was to examine the phosphorylation and function of NFATc4. Since NFATc4 is primarily expressed in nonimmune tissues and has been implicated in multiple biological processes, understanding the regulation of NFATc4 phosphorylation is an important goal. We report that NFATc4 is differentially phosphorylated by MAP kinases. Ser168 and Ser170 of NFATc4, which are analogously located at Ser172 of NFATc1 and Ser163,165 of NFATc3, are targets of the p38 MAP kinase but not of the JNK MAP kinase. Replacement of Ser168 and Ser170 with Ala promotes NFATc4 nuclear localization and enhances NFAT-mediated transcription activity. Stable expression of Ala168,170 NFATc4 (but not wild-type NFATc4) in NIH 3T3 cells promotes adipocyte formation under differentiation conditions. Molecular analysis indicates that NFATc4 binds to two distinct DNA elements on the peroxisome proliferator-activated receptor γ2 (PPARγ2) promoter. Increased PPARγ expression caused by NFATc4 accounts, in part, for the increased adipocyte differentiation.
MATERIALS AND METHODS
Cell culture.
BHK fibroblasts were cultured in minimal essential medium. NIH 3T3 fibroblasts and COS and 293T cells were cultured in Dulbecco modified Eagle medium. All media were supplemented with 10% fetal calf serum, 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml) (Invitrogen). Cells were transfected by using Lipofectamine (Invitrogen). The expression vectors for wild-type and Ala168,170 NFATc4 were linearized and transfected into NIH 3T3 cells. The empty expression vector was also transfected as a control. Three days after transfection, cells were selected in the presence of G418 (0.5 mg/ml). G418-resistant clones were pooled, from two independent transfections, after 3 weeks of selection. G418-resistant cells were also propagated to confluence, and insulin (5 μg/ml), dexamethasone (1 μM), and the phosphodiesterase inhibitor isobutylmethylxanthine (IBMX) (0.5 mM) were administered to initiate adipocyte differentiation. Adeno-associated virus (AAV) (Stratagene) was produced by following the manufacturer's protocol. Subconfluent (70 to 80%) NIH 3T3 cells were infected with AAV expressing enhanced green fluorescent protein (EGFP), NFATc4, or the NFATc4 Ala168,170 mutant. Infected cells were cultured to confluence and subjected to adipocyte differentiation as described previously.
Reagents.
The NFAT-luciferase reporter plasmid and the expression vectors for JNK1, p38 isoforms, MLK3, MKK6-Glu, calcineurin, and NFATc4 have been described previously (12-15, 25, 26, 34, 53). Ala168,170 NFATc4 was created by PCR and sequenced with an Applied Biosystems machine. The PPARγ2 (−1 to −1900) gene promoter was amplified from human genomic DNA and subcloned into the basic pGL3-luciferase reporter plasmid (Promega) by using MluI and XhoI sites. Deletions and mutations on the PPARγ2 promoter were generated by PCR. Bacterially expressed NFAT proteins were purified by glutathione affinity chromatography as described previously (13). Monoclonal antibody M2 was obtained from Sigma. Lipid droplets were stained with Oil red O (Sigma).
Kinase assays.
Hemagglutinin epitope-tagged p38α MAP kinase and JNK1 were coexpressed in COS cells with and without MKK6-Glu and MLK3, respectively. Cell extracts were prepared with Triton-lysis buffer (20 mM Tris [pH 7.4], 137 mM NaCl, 2 mM EDTA, 1% Triton X-100, 25 mM β-glycerophosphate, 1 mM sodium vanadate, 2 mM sodium pyrophosphate, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml) 48 h after transfection. Immune complex kinase assays were performed with recombinant NFATc3 and NFATc4 proteins (1 μg) as the substrates.
Phosphopeptide analysis.
COS cells were transfected with a Flag epitope-tagged NFAT expression vector (6 μg) and incubated for 24 h. The transfected cells were then incubated with [32P]phosphate (1 mCi/ml) for 5 h. The NFAT proteins were isolated by immunoprecipitation with anti-Flag monoclonal antibody M2. Immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electrotransferred to a polyvinylidene difluoride membrane (Millipore), and visualized by autoradiography. The band containing 32P-labeled NFAT was excised from the membrane and digested with trypsin, and the peptides obtained were examined by phosphopeptide mapping.
Immunofluorescence analysis.
Expression plasmids for Flag-tagged NFATc4 (0.3 μg) or activated calcineurin (0.2 μg) were transfected into BHK cells. Expression plasmids for constitutively active MKK6 (MKK6-Glu) (0.2 μg) or dominant-negative p38α (0.2 μg) were also cotransfected as indicated. NFATc4 was detected by immunofluorescence analysis with anti-Flag monoclonal antibody M2 (1:500; Sigma) or NFATc4 rabbit polyclonal antibody (1:100; Affinity Bioreagents). The secondary antibody was Texas red-conjugated anti-mouse or anti-rabbit immunoglobulin antibody (1:100; Jackson ImmunoResearch), and nuclei were visualized with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma).
Luciferase assays.
An NFAT expression vector (0.1 μg) was cotransfected with an NFAT-luciferase reporter plasmid (0.3 μg) and the control plasmid pRSV β-galactosidase (0.2 μg) into BHK cells. Luciferase and β-galactosidase activities were measured 48 h after transfection. Cells were stimulated with ionomycin (2 μM) plus phorbol myristate acetate (PMA) (100 nM) as indicated. The data were presented as relative luciferase activity, which was calculated as the ratio of the luciferase activity to the activity of β-galactosidase (mean ± standard error [n = 4]).
Protein immunoblotting analysis.
Expression plasmids for Flag-tagged NFATc4 (2 μg) were cotransfected with dominant-negative p38α (3 μg) into COS cells. Transfected cells (48 h posttransfection) were treated with ionomycin for 1 h before UV irradiation (2 min). Cell extracts were isolated in Triton-lysis buffer after 30 min of recovery, separated by SDS-7% PAGE, and electrotransferred to a polyvinylidene difluoride membrane (Millipore). Immunoblot analysis was performed with the M2 monoclonal antibody (Sigma) and visualized by enhanced chemiluminescence.
Semiquantitative reverse transcription-PCR (RT-PCR).
Total RNA was isolated from stably transfected cells after 6 days of differentiation with Trizol reagents (Invitrogen). Isolated RNA (1 μg) was reverse transcribed with mouse mammary tumor virus reverse transcriptase (Promega). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (sense, 5′-CTG ACG TGC CGC CTG GAG AAA-3′; antisense, 5′-TTG GGG GCC GAG TTG GGA TAG-3′) was amplified (16, 18, 20, and 24 cycles) by PCR as a control. A normalized amount of cDNA was used to determine the expression of PPARγ (sense, 5′-CAC AGG CCG AGA AGG AGA AGC-3′; antisense, 5′-AGG GAG GCC AGC ATC GTG TAG-3′), caveolin-1 (sense, 5′-AGG CCA TGG CAG ACG AGG TG-3′; antisense, 5′-GCT GAT GCG GAT GTT GCT GAA TA-3′), and fatty acid synthase (FAS) (sense, 5′-CCA GCC CCG ACC CAC AAC AA-3′; antisense, 5′-TAG CCC TCC CGT ACA CTC ACT CGT-3′). PCR products were separated by agarose gel electrophoresis and transferred to a Zeta-Probe membrane (Bio-Rad) for Southern blotting. PCR products were detected by autoradiography and measured by PhosphorImager (Molecular Dynamics) analysis.
Gel mobility shift assays.
Nuclear extracts were prepared from transfected COS cells or differentiated 3T3-L1 adipocytes as described previously (11). Double-stranded oligonucleotides encoding NFAT binding sites from the IL-2 gene promoter (5′-AGA AAG GAG GAA AAA CTG TTT CAT ACA GAA GG-3′) and the PPARγ2 gene promoter (proximal NFAT site, 5′-GAG ACA GTG TGG CAA TAT TTT CCC TGT AAT-3′; distal NFAT site, 5′-AGC AAG AGA TTT AAG TTT TCC ATT TAA GAA-3′) were labeled with [α-32P]dCTP. The binding reactions were carried out at room temperature in gel-shift buffer [1 mM CaCl2, 1 mM MgCl2, 10 mM HEPES (pH 7.9), 50 mM NaCl, 15 mM β-mercaptoethanol, 10% glycerol, 0.1 mg of bovine serum albumin/ml, and 1 mg of poly(dI-dC)/ml] for 30 min. Protein-DNA complexes were separated in a 5% nondenaturing polyacrylamide gel in Tris-glycine-EDTA buffer (25 mM Tris, 200 mM glycine, and 1 mM EDTA) and visualized by autoradiography. For competition analysis, unlabeled oligonucleotides (1, 5, and 10 pmol) were incubated with the labeled probe before the addition of nuclear extract. For supershift analysis, the antibody was preincubated with nuclear extract for 30 min at room temperature before the addition of the labeled probe.
RESULTS
Differential phosphorylation of NFAT by MAP kinases.
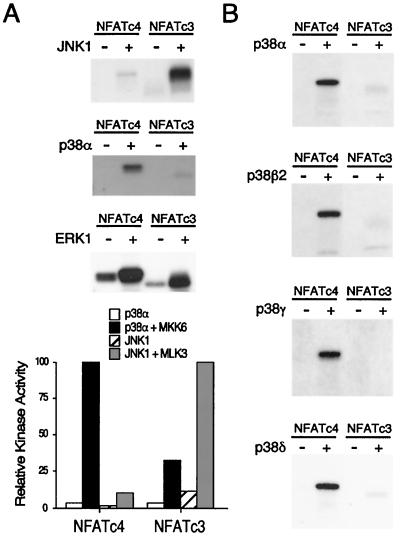
Multiple protein kinases have been shown to phosphorylate NFAT (4, 6, 12-14, 29, 46, 58). Among the protein kinases identified, the JNK MAP kinases interact with and phosphorylate NFATc1 and NFATc3 (13, 14, 46). The p38 MAP kinase phosphorylates NFATc2 (29). Phosphorylation of NFATc4, however, remains elusive. We tested whether MAP kinases phosphorylate NFATc4. Immune complex kinase assays were performed with activated epitope-tagged MAP kinases. Activation of JNK1 with upstream kinase MLK3 increased NFATc3 phosphorylation (Fig. 1A). However, only minimal phosphorylation was detected on NFATc4. Activation of p38α by coexpression with constitutively active MKK6 (MKK6-Glu) increased NFATc4 phosphorylation but not NFATc3 phosphorylation (Fig. 1A). Activation of ERK1 by constitutively active MEK phosphorylated NFATc3 and NFATc4. These data indicate that NFATc3 and NFATc4 are preferentially phosphorylated by JNK and p38α MAP kinase, respectively.
FIG. 1.
Differential phosphorylation of NFAT by MAP kinases. (A) Differential phosphorylation of NFATc3 and NFATc4 by JNK1 and p38α MAP kinases. Epitope-tagged JNK1 and p38α MAP kinases were activated by coexpression with (+) and without (−) MLK3 and MKK6-Glu mutant, respectively. Phosphorylation of NFATc3 and NFATc4 by ERK MAP kinase was also shown. Immune complex kinase assays were performed with recombinant NFATc3 and NFATc4 as the substrates. Phosphorylated NFATc3 and NFATc4 were detected by autoradiography and quantitated by PhosphorImager analysis. (B) All four p38 MAP kinase isoforms phosphorylate NFATc4 in vitro. Four different p38 MAP kinase isoforms (p38α, p38β2, p38γ, and p38δ) were expressed in COS cells in the presence (+) and absence (−) of the MKK6-Glu mutant. Immune complex kinase assays were performed with recombinant NFATc3 and NFATc4 as the substrates. Phosphorylated NFATc4 was detected by autoradiography.
The p38 group of MAP kinase includes four different members (p38α, p38β2, p38γ, and p38δ). The activation of these four p38 MAP kinase isoforms is differentially regulated (25, 26). Furthermore, the pyridinyl imidazole drug SB203580 inhibits the p38α and p38β2 isoforms but not the p38γ and p38δ isoforms (19, 26, 28, 37). To test whether these four p38 MAP kinase isoforms phosphorylate NFATc4, we performed immune complex kinase assays. Activation of the four different p38 MAP kinase isoforms by MKK6-Glu increased NFATc4 phosphorylation (Fig. 1B). However, NFATc3 was only weakly phosphorylated by all four activated p38 MAP kinase isoforms. These data further support the conclusion that JNK and p38 MAP kinases differentially phosphorylate NFATc3 and NFATc4, respectively.
p38α MAP kinase phosphorylates Ser168 and Ser170 of NFATc4.
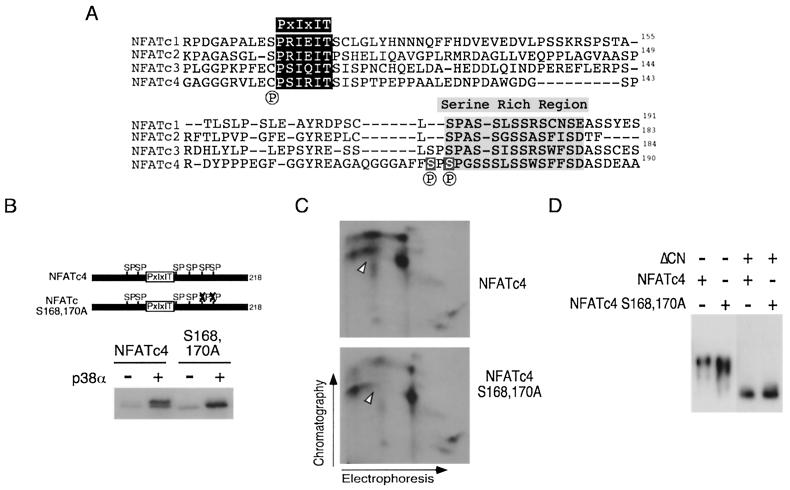
Next, we mapped the p38α MAP kinase phosphorylation sites on NFATc4. Previous studies demonstrated that the conserved NH2-terminal NFAT homology domain, especially the SRR and the SP boxes, was the primary targets of phosphorylation (4, 6, 12-14, 29, 44, 46, 58). Ser168 and Ser170 are the most prominent sites in the SRR of NFATc4 because similar Ser residues are found in other NFAT members (Fig. 2A). We tested whether p38α MAP kinase phosphorylates Ser168 and Ser170 of NFATc4. Phosphorylation by the p38α MAP kinase caused decreased electrophoretic mobility of NFATc4 during SDS-PAGE (Fig. 2B). Mutational replacement of Ser168,170 with Ala eliminated the decreased electrophoretic mobility, although the Ala168,170 NFATc4 remained phosphorylated by the p38α MAP kinase upon activation. These data indicate that p38α MAP kinase phosphorylates multiple sites, including residues Ser168 and Ser170, on the NFATc4 proteins.
FIG. 2.
p38α MAP kinase phosphorylates Ser168 and Ser170 of NFATc4. (A) Sequence comparison of the NH2-terminal phosphorylation sites in NFAT members. Previously identified phosphorylation sites on NFATc1 (Ser117 and Ser172), NFATc2 (Ser168), and NFATc3 (Ser163 and Ser165) are indicated (by a circled P below the sequence). The conserved PXIXIT motif (dark shaded box) and the SRR (light shaded boxes) are shown. Ser168 and Ser170 of NFATc4 are also highlighted. (B) Ser168 and Ser170 of NFATc4 are phosphorylated in vitro. Immune complex kinase assays were performed by using p38α MAP kinase activated with (+) and without (−) the MKK6-Glu mutant. Phosphorylation of recombinant NFATc4 proteins was examined. Phosphorylated NFATc4 was separated by SDS-PAGE and detected by autoradiography. (C) Ser168 and Ser170 of NFATc4 are phosphorylated in vivo. Epitope-tagged wild-type and Ala168,170 NFATc4 were expressed in COS cells. The cells were labeled with [32P]phosphate, and the labeled NFATc4 proteins were isolated by immunoprecipitations. The phosphorylation of NFATc4 was examined by tryptic phosphopeptide mapping. Replacement of Ser168 and Ser170 by Ala caused the loss of a phosphopeptide (arrowheads). (D) Mutational replacement of Ser168 and Ser170 with Ala increases the electrophoretic mobility of NFATc4 during SDS-PAGE. Epitope-tagged wild-type and mutated Ala168,170 NFATc4 were expressed in COS cells without (−) and with (+) activated calcineurin (ΔCN). NFATc4 proteins were detected in immunoblot analysis by using monoclonal antibody M2.
To test whether Ser168 and Ser170 of NFATc4 were phosphorylated in vivo, we performed tryptic phosphopeptide analysis of [32P]phosphate-labeled wild-type and Ala168,170 NFATc4 proteins (Fig. 2C). Tryptic digestion of the in vivo-labeled NFATc4 proteins generated several phosphopeptides. Replacement of Ser168 and Ser170 with Ala caused the loss of a phosphopeptide (Fig. 2C). These data indicate that Ser168 and Ser170 of NFATc4 are phosphorylated in vivo.
Dephosphorylation causes an increase in the electrophoretic mobility of NFAT during SDS-PAGE (12, 38). We therefore examined the electrophoretic mobility of wild-type and Ala168,170 NFATc4. Immunoblot analysis demonstrated that Ala168,170 NFATc4 exhibited an increase in electrophoretic mobility compared to the wild-type NFATc4 (Fig. 2D). Activated calcineurin dephosphorylated both wild-type and Ala168,170 NFATc4 and greatly increased the electrophoretic mobility of NFATc4. Importantly, dephosphorylated wild-type and Ala168,170 NFATc4 exhibited similar electrophoretic mobilities. These data further indicate that Ser168 and Ser170 of NFATc4 are phosphorylated in vivo.
Mutational replacement of Ser168,170 with Ala promotes NFATc4 nuclear localization and increases NFATc4-mediated transcription activity.
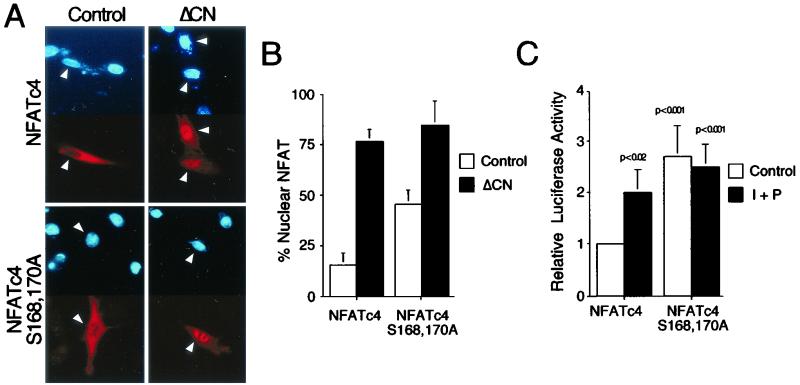
Phosphorylation of NFAT plays an important role in the regulation of NFAT subcellular distribution. Replacement of Ser172 of NFATc1, Ser168 of NFATc2, or Ser163,165 of NFATc3 with Ala promotes nuclear localization (13, 14, 44, 46). Next, we examined subcellular distribution of wild-type and Ala168,170 NFATc4 by using immunofluorescence analysis (Fig. 3A and B). Epitope-tagged wild-type and Ala168,170 NFATc4 were expressed in cells in the presence or absence of activated calcineurin. In resting cells, wild-type NFATc4 was located predominantly in the cytosol. Replacement of Ser168,170 with Ala promoted nuclear localization of NFATc4. Coexpression of activated calcineurin caused the majority of NFATc4 to be located in the nucleus. These data indicate that phosphorylation at Ser168 and Ser170 regulates the subcellular distribution of NFATc4.
FIG. 3.
Mutational replacement of Ser168,170 with Ala promotes NFATc4 nuclear localization and increases NFAT-mediated transcription activity. (A and B) Epitope-tagged wild-type and Ala168,170 NFATc4 proteins were expressed in BHK cells and detected by immunofluorescence microscopy with monoclonal antibody M2. The subcellular distribution of NFATc4 was examined in the presence and absence of activated calcineurin (ΔCN). The images of representative cells are illustrated (A). The nuclei (blue) of transfected cells expressing NFATc4 (red) are indicated with arrowheads. The percentage of cells with NFATc4 in the nucleus (n = 300) is also presented (B). (C) BHK cells were transfected with an NFAT-luciferase reporter plasmid together with an expression vector for wild-type or Ala168,170 NFATc4. The cells were stimulated without (control) and with PMA (P) (100 nM) plus ionomycin (I) (2 μM) for 16 h before harvest. P values are in comparison with unstimulated NFATc4-mediated transcription activity.
To examine whether nuclear NFATc4 mediates transcription activity, we performed transfection assays with an NFAT reporter plasmid. The expression of Ala168,170 NFATc4 increased NFAT-mediated transcription compared to wild-type NFATc4 (Fig. 3C). Stimulation of transfected cells with PMA and calcium ionophore ionomycin further increased transcription mediated by NFATc4. Together, these data indicate that replacement of Ser168,170 with Ala of NFATc4 promotes nuclear localization and increases NFAT-mediated transcription activity.
p38α MAP kinase regulates NFATc4 phosphorylation and nuclear localization.
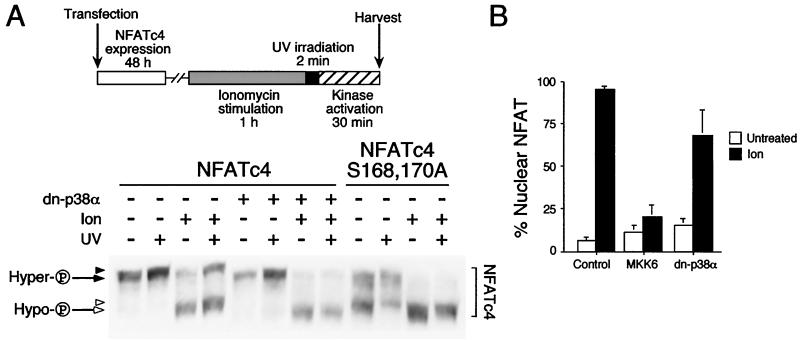
In vitro phosphorylation by p38 MAP kinases decreased NFATc4 electrophoretic mobility during SDS-PAGE (Fig. 2B). Conversely, dephosphorylation mediated by the calcineurin phosphatase or replacement of Ser168,170 by Ala, to prevent phosphorylation, increased NFATc4 electrophoretic mobility (Fig. 2D). These data indicate that the electrophoretic mobility of NFATc4 mirrors NFAT phosphorylation. To examine in vivo phosphorylation of NFATc4 by p38 MAP kinases, we expressed NFATc4 in the presence or absence of dominant-negative p38α (Fig. 4A). Transfected cells were irradiated with UV to activate endogenous p38 MAP kinases and to stimulate NFATc4 phosphorylation. Phosphorylated NFATc4 was separated by SDS-PAGE to determine electrophoretic mobility. Coexpression with dominant-negative p38α did not affect the electrophoretic mobility of NFATc4. UV irradiation slightly decreased the electrophoretic mobility of NFATc4 under unstimulated conditions (Fig. 4A). These data indicate that NFATc4 is phosphorylated under unstimulated conditions.
FIG. 4.
p38α MAP kinase regulates NFATc4 phosphorylation and nuclear localization. (A) Phosphorylation of NFATc4 by p38 MAP kinases in vivo. NFATc4 and Ala168,170 NFATc4 were coexpressed without (−) or with (+) dominant-negative p38α (dn-p38α) in COS cells. Transfected cells were treated without (−) or with (+) ionomycin (Ion) to generate hyperphosphorylated (hyper-P) (filled arrow) and hypophosphorylated (hypo-P) (open arrow) NFATc4. After 1 h of ionomycin treatment, cells were irradiated (+) or not (−) with UV for 2 min. After a 30-min recovery, cell extracts were harvested and examined by immunoblotting analysis with M2 monoclonal antibody. A decrease in the electrophoretic mobility of NFATc4 was indicated (filled and open arrowheads). (B) Expression plasmids for NFATc4 were cotransfected with or without MKK6-Glu or dominant-negative p38α (dn-p38α) into BHK cells. The subcellular distribution of NFATc4 was examined in the presence and absence of ionomycin by immunofluorescence microscopy. The percentage of cells with NFATc4 in nucleus (n = 300) is presented.
To further examine NFATc4 phosphorylation, calcium ionophore ionomycin was administered to dephosphorylate NFATc4 prior to UV irradiation (Fig. 4A). Treatment with ionomycin greatly increased the electrophoretic mobility of NFATc4 (Fig. 4A). The slower migrating hyperphosphorylated (i.e., higher apparent molecular weight) NFATc4 was dephosphorylated and became a faster migrating hypophosphorylated form (i.e., lower apparent molecular weight). UV irradiation promoted a conversion of the faster migrating hypophosphorylated NFATc4 to a slower migrating hyperphosphorylated form, even in the presence of ionomycin stimulation. Importantly, UV irradiation further decreased the electrophoretic mobility of both hypophosphorylated and hyperphosphorylated NFATc4 (Fig. 4A). The decrease in electrophoretic mobility of the hypophosphorylated NFATc4 was blocked by coexpression with the dominant-negative p38α. In addition, the faster migrating hypophosphorylated NFATc4 did not revert to the hyperphosphorylated form in the presence of the dominant-negative p38α. Together, these data indicate that p38 MAP kinases phosphorylate NFATc4 and decrease NFATc4 electrophoretic mobility in SDS-PAGE.
Next, we examined the electrophoretic mobility of Ala168,170 NFATc4 upon UV irradiation (Fig. 4A). The NFATc4 Ala168,170 mutant has increased electrophoretic mobility compared to wild-type NFATc4. In the presence or absence of ionomycin, UV irradiation slightly decreased Ala168,170 NFATc4 electrophoretic mobility (Fig. 4A). Unlike the wild-type NFATc4, UV irradiation did not revert the faster migrating hypophosphorylated Ala168,170 NFATc4 to the slower migrating hyperphosphorylated form. The predominance of the faster migrating hypophosphorylated Ala168,170 NFATc4 upon costimulation with ionomycin and UV irradiation recapitulated the inhibition by coexpression of the dominant-negative p38α. These data further indicate the importance of phosphorylation at Ser168,170 in NFATc4.
Phosphorylation at Ser168,170 regulated the subcellular distribution of NFATc4 (Fig. 3). We further performed immunofluorescence to examine whether activation of p38 MAP kinases affects NFATc4 nuclear localization (Fig. 4B). NFATc4 was expressed in cells in the presence or absence of MKK6-Glu or dominant-negative p38α. Coexpression with MKK6-Glu or dominant-negative p38α did not affect NFATc4 subcellular distribution. Stimulation with ionomycin increased NFATc4 nuclear localization. Coexpression with MKK6-Glu, however, opposed nuclear accumulation of NFATc4 under ionomycin stimulation. Coexpression of the dominant-negative p38α did not affect the subcellular distribution of NFATc4 in the presence or absence of ionomycin. Together, these data indicate that p38 MAP kinases phosphorylate NFATc4 and oppose NFATc4 nuclear localization.
NIH 3T3 cells stably expressing Ala168,170 NFATc4 exhibit adipocyte formation.
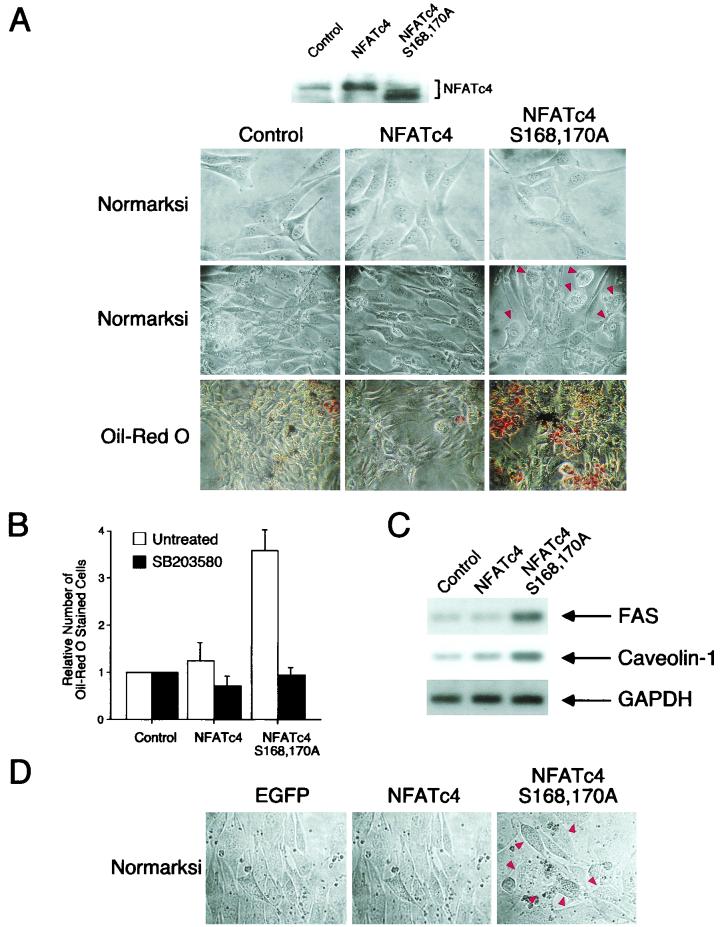
Previous studies indicate that certain fibroblastic cell lines can be differentiated into adipocytes under appropriate hormonal stimuli and/or treatment of pharmacological agents (reviewed in references 17, 39, and 49). For example, treatment of confluent 3T3-L1 fibroblasts with insulin, dexamethasone, and the phosphodiesterase inhibitor IBMX induces adipocyte differentiation (31). Alternatively, overexpression of transcription factors, such as members of the CCAAT enhancer binding protein (C/EBP) or PPARγ, promotes adipocyte differentiation from NIH 3T3 cells (8, 9), which in general do not exhibit adipocyte phenotypes under treatment with insulin, dexamethasone, and IBMX. Since the calcineurin-NFAT signaling pathway has been implicated in adipogenesis in studies using the drug cyclosporine (CsA) (32), we examined whether NFAT has a direct role in adipogenesis by using the stably transfected NIH 3T3 fibroblasts. Confluent NIH 3T3 cells were treated with insulin, dexamethasone, and IBMX to induce adipocyte formation (Fig. 5A). Control cells cultured in the differentiation media did not induce adipocyte formation. These observations are in agreement with previous reports that NIH 3T3 cells in general did not differentiate into adipocytes. Constitutive expression of wild-type NFATc4 also did not induce adipocyte formation. However, treatment with insulin, dexamethasone, and IBMX induced accumulation of shiny vacuoles in NIH 3T3 cells that constitutively express Ala168,170 NFATc4 (Fig. 5A). In addition, the elongated shapes of Ala168,170 NFATc4 NIH 3T3 fibroblasts were altered and became round upon differentiation. These morphological changes resemble those that occur during adipocyte differentiation. Together, these data indicate that in the presence of insulin, dexamethasone, and IBMX, confluent NIH 3T3 cells expressing Ala168,170 NFATc4 exhibit characteristics of adipocytes.
FIG.5.
NIH 3T3 cells stably expressing Ala168,170 NFATc4 exhibit adipocyte formation. (A and B) Expression vectors for wild-type and Ala168,170 NFATc4 were transfected into NIH 3T3 cells. An empty vector was also transfected as a control. G418-resistant cells were pooled, from two rounds of transfections, after 3 weeks of antibiotic selection. The images of representative undifferentiated cells are illustrated (top row, panel A). Confluent NIH 3T3 cells were cultured in media containing insulin, dexamethasone, and IBMX to promote adipocyte differentiation. The morphologies of differentiated cells, cultured for 6 days in differentiation media, were examined under light microscopy (middle row, panel A). Cells with shiny vacuoles are indicated (arrowheads). The accumulation of oil droplets was detected by Oil red O stain (bottom row, panel A). The number of Oil-red-O-stained cells was observed under a light microscope (30 fields) and presented (B). The effect of p38 MAP kinase inhibitor SB203580 on adipocyte differentiation was also examined (B). The expression of NFATc4 proteins was detected by M2 monoclonal antibody (A). (C) Total RNA was harvested from differentiated NIH 3T3 cells, and semiquantitative RT-PCR was performed to determine the expression levels of FAS and caveolin-1. The expression of GAPDH was used as a control. (D) NIH 3T3 cells were infected with AAV expressing EGFP, wild-type NFATc4, and Ala168,170 NFATc4. Infected cells were cultured to confluence and subjected to differentiation. The morphologies of differentiated cells, after culture for 6 days in differentiation media, were examined under light microscopy.
Shiny vacuoles observed in Ala168,170 NFATc4 NIH 3T3 cells are indicative of accumulated lipid droplets. To determine whether cells expressing Ala168,170 NFATc4 accumulate lipid, we stained differentiated cells with Oil red O. Positive cells were observed in the differentiated Ala168,170 NFATc4 NIH 3T3 cells (Fig. 5A). However, very few Oil-red-O-positive cells were observed in the control or the wild-type NFATc4 differentiated cells. The number of Oil-red-O-positive cells was increased in Ala168,170 NFATc4 NIH 3T3 cells compared to control or wild-type NFATc4 cells (Fig. 5B). Together, these data indicate that expression of Ala168,170 NFATc4 in NIH 3T3 cells promotes adipocyte formation.
The induction of specific genes is required during adipocyte differentiation (17, 39, 49). Previous studies demonstrated that expression levels of caveolin-1 and FAS were increased upon adipocyte differentiation (42, 45, 48, 50). Expression of caveolin-1 and FAS facilitates lipid accumulation in differentiating adipocytes. We performed semiquantitative RT-PCR to examine the expression levels of caveolin-1 and FAS in differentiated Ala168,170 NFATc4 cells (Fig. 5C). The expression of caveolin-1 and FAS was similar in control cells and in cells constitutively expressing wild-type NFATc4. However, caveolin-1 and FAS expression was increased in cells expressing Ala168,170 NFATc4. As a control, GAPDH expression was similar in all three stably transfected cells. These data further confirm adipogenic characteristics in cells expressing Ala168,170 NFATc4.
We also infected parental NIH 3T3 cells with AAV expressing EGFP, wild-type NFATc4, or the NFATc4 Ala168,170 mutant (Fig. 5D). Infected cells were cultured to confluence and subjected to adipocyte differentiation. Accumulations of shiny vacuoles were found in infected cells expressing Ala168,170 NFATc4. Viral infection to express EGFP or wild-type NFATc4 did not promote adipocyte formation. These data further support that the expression of Ala168,170 NFATc4 promotes adipocyte formation.
Previous studies demonstrated that the p38 MAP kinase inhibitor SB203580 blocks adipocyte differentiation (23, 24). Since p38 MAP kinases phosphorylated NFATc4, we tested whether SB203580 affects adipocyte differentiation in cells expressing Ala168,170 NFATc4. Administration of SB203580 in differentiation media inhibited adipocyte formation (Fig. 5B). These data indicate that the pyridinyl imidazole drug SB203580 strongly inhibits adipocyte differentiation. In addition, these data indicate that NFATc4 is not the only step dependent on p38 MAP kinases during adipocyte differentiation.
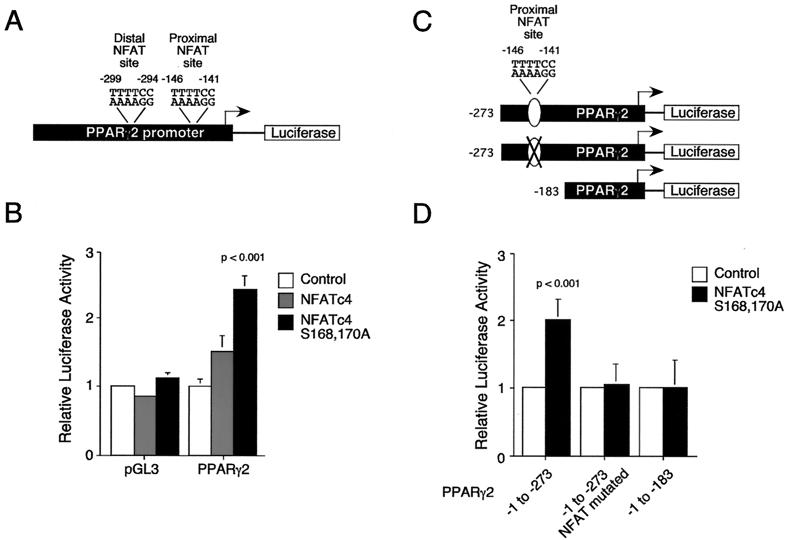
Expression of Ala168,170 NFATc4 increases PPARγ2 promoter activity.
Induction of adipocyte formation in NIH 3T3 cells by Ala168,170 NFATc4, but not by wild-type NFATc4, suggests that increased NFAT nuclear localization and subsequent transcriptional activity are critical for adipogenesis. Previous studies indicated that the transcription factor PPARγ is induced as an early event in adipogenesis (55). Importantly, overexpression of PPARγ in NIH 3T3 cells promotes adipocyte differentiation (51). These data indicate that the level of PPARγ gene expression is important to determine adipocyte formation. Since the pharmacological inhibitor CsA effectively blocks adipocyte differentiation only at the early stage of differentiation (32), we tested whether NFATc4 regulates PPARγ promoter activity. A fragment of the PPARγ2 gene promoter was cloned into a promoterless luciferase reporter plasmid (Fig. 6A) and transfected into NIH 3T3 cells. Coexpression of Ala168,170 NFATc4 increased PPARγ2 promoter reporter activity (Fig. 6B). However, coexpression of wild-type NFATc4 did not increase the PPARγ2 promoter reporter activity. As a control, coexpression of wild-type or Ala168,170 NFATc4 did not increase the promoterless reporter activity. These data indicate that constitutive nuclear NFAT increases PPARγ2 gene promoter activity.
FIG. 6.
Expression of Ala168,170 NFATc4 increases PPARγ2 promoter activity. (A) Schematic representation of the PPARγ2 gene promoter. Two putative NFAT binding sites (proximal and distal) are also indicated. (B) Promoterless (pGL3) and PPARγ2 (PPARγ2) promoter luciferase plasmids were cotransfected with expression vector for wild-type or Ala168,170 NFATc4 into NIH 3T3 cells. Cells were harvested 36 h after transfection. The transfection efficiency was monitored by measurement of β-galactosidase activity. P was <0.001 for comparison with the basal activity of the PPARγ2 gene promoter. (C) Schematic representation of the deletion and mutational constructs of the PPARγ2 gene promoter. The mutation of the proximal NFAT binding element is indicated (X). (D) Deletion and mutational analysis of the PPARγ2 promoter. PPARγ2 (−1 to −273), PPARγ2 (−1 to −273, with the proximal NFAT site mutated), and PPARγ2 (−1 to −183) promoter luciferase plasmids were cotransfected with Ala168,170 NFATc4. The cells were harvested 36 h after transfection. The transfection efficiency was monitored by measurement of β-galactosidase activity. P was <0.001 for comparison with the Ala168,170 NFATc4-mediated transcription activity from the PPARγ2 (−1 to −273, with the proximal NFAT site mutated) or the PPARγ2 (−1 to −183) promoters.
Sequence comparisons indicate that two putative NFAT binding sites (proximal and distal) are located in the human PPARγ2 gene promoter (Fig. 6A). Similar sequences encompassing these two putative NFAT binding elements are also found in the mouse PPARγ2 gene promoter (27). We generated PPARγ2 promoter deletion mutants that encompassed (or not) the putative proximal NFAT site (PPARγ2 −1 to −273 and −1 to −183) (Fig. 6C). We also performed site-directed mutagenesis to abolish the proximal NFAT binding site on PPARγ2 (−1 to −273) (Fig. 6C). Coexpression with Ala168,170 NFATc4 increased the PPARγ2 (−1 to −273) gene promoter activity (Fig. 6D). However, deletion or mutational removal of the proximal NFAT site abolished the induction of the PPARγ2 gene promoter activity by Ala168,170 NFATc4. These data further indicate that NFAT regulates the PPARγ2 gene promoter.
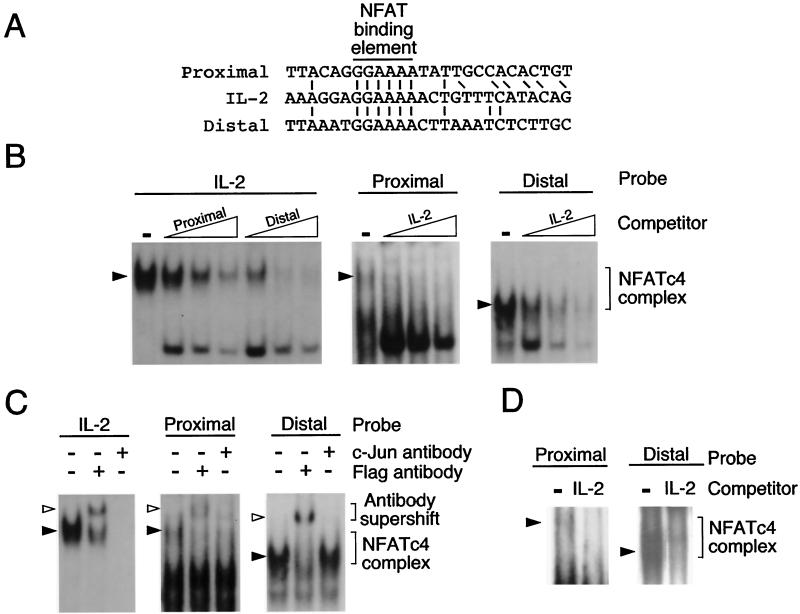
NFATc4 binds to the PPARγ2 promoter on two distinct sites.
Induction of the PPARγ2 gene transcription activity by Ala168,170 NFATc4 indicates that the two putative NFAT binding elements are functional. Sequence analysis indicated that the two putative NFAT binding sites have sequences matching exactly to the core binding element found in the canonical NFAT site of the IL-2 gene promoter (Fig. 7A). To test whether these two putative NFAT binding sites interact with NFAT, we performed gel mobility shift assays with Flag epitope-tagged NFATc4. Specific NFAT-DNA complexes were detected by using the canonical NFAT element from the IL-2 gene as a probe (Fig. 7B). Competition analysis indicated that the proximal and the distal NFAT sites of the PPARγ2 gene diminished formation of the IL-2 NFAT-DNA complex in a dose-dependent manner. Conversely, the NFAT-DNA complexes resulting from the proximal and the distal NFAT sites of the PPARγ2 gene were competed by the IL-2 NFAT element. These data indicate that the two NFAT binding elements present on the PPARγ2 gene promoter form NFAT-DNA complexes.
FIG. 7.
NFATc4 binds to the PPARγ2 promoter on two distinct sites. (A) Sequence comparison of the two putative NFAT elements (proximal and distal) found on the PPARγ2 promoter with the canonical NFAT binding motif located on the IL-2 gene. The NFAT binding element (5′-GGAAAA-3′) is aligned. Adjacent conserved nucleotides are also indicated. (B) NFATc4 binds to the proximal and the distal NFAT elements of the PPARγ2 gene promoter. Flag epitope-tagged NFATc4 proteins were expressed in COS cells and nuclear extracts were prepared for gel mobility shift assays. Oligonucleotides encoding the canonical NFAT binding site from the IL-2 gene promoter (IL-2) were labeled with [α-32P]dCTP and used as a probe (left). NFATc4-DNA complexes (filled arrowheads) were competed with unlabeled (1, 5, and 10 pmol) proximal and distal NFAT elements of the PPARγ2 gene promoter. The formation of NFATc4-DNA complexes with labeled proximal and distal NFAT elements of the PPARγ2 gene promoter is also shown (middle and right). The specificity of the NFATc4-DNA complex was examined by competition by using unlabeled (1, 5, and 10 pmol) NFAT element from the IL-2 gene. (C) Distinct protein complex binds to the proximal and the distal NFAT sites of the PPARγ2 gene promoter. NFATc4-DNA complexes (filled arrowheads) from the NFAT binding sites of the IL-2 and the PPARγ2 genes were supershifted with c-Jun or Flag antibody. Antibody supershifted complexes are indicated (open arrowheads). (D) Nuclear extracts prepared from differentiated 3T3-L1 adipocytes form NFAT-DNA complexes on the proximal and the distal NFAT binding elements of the PPARγ2 gene. The specificity of the NFATc4-DNA complex (filled arrowheads) was examined by competition with unlabeled (10 pmol) NFAT element from the IL-2 gene.
Gel mobility shift analysis indicated that there were two NFAT binding elements present in the PPARγ2 gene promoter. However, the electrophoretic mobility of the NFAT-DNA complexes was markedly different with the proximal and the distal NFAT sites as probes (Fig. 7B). In addition, sequences surrounding the core NFAT binding element were different (Fig. 7A). These observations suggest that the proximal and the distal NFAT sites form different NFAT-DNA complexes. Sequence comparisons indicate that the proximal, but not the distal, NFAT sites have additional sequences that resemble adjacent nucleotides present on the NFAT site of the IL-2 gene (Fig. 7A). Previous studies indicated that members of the transcription factor Fos and Jun families were part of the complexes found in the NFAT element of the IL-2 gene (36). Thus, we tested whether transcription factor c-Jun is bound to the proximal NFAT site of the PPARγ2 gene. Antibody supershift analysis indicated that administration of c-Jun antibody abolished the formation of NFAT-DNA complexes when the IL-2 gene NFAT site was used as a probe (Fig. 7C). A partial elimination of the NFAT-DNA complex by the c-Jun antibody was exhibited when the proximal PPARγ2 NFAT site was used as a probe. However, the presence of c-Jun antibody did not affect NFAT-DNA complex formation when the distal NFAT site was used as a probe. As a control, epitope-tagged NFATc4 present in the NFAT-DNA complexes was supershifted by Flag antibody (Fig. 7C). Together, these data indicate that the proximal and the distal NFAT binding elements form different nuclear complexes to regulate the PPARγ2 gene promoter activity.
We also performed gel mobility shift assays with nuclear extracts prepared from differentiated adipocytes. Both proximal and distal NFAT binding elements form specific DNA-protein complexes (Fig. 7D). Competition analysis with the canonical NFAT element from the IL-2 gene further indicated specificity of NFAT binding. These data confirm that the proximal and the distal NFAT binding elements in the PPARγ2 gene promoter are functional.
Expression of Ala168,170 NFATc4 in NIH 3T3 cells increases PPARγ2 gene expression.
Previous studies indicated that overexpression of PPARγ induced adipocyte formation in NIH 3T3 cells (51). These data indicate that the expression of PPARγ positively regulates adipocyte differentiation. The presence of two NFAT binding sites and the induction of the PPARγ2 gene promoter activity suggest that PPARγ gene expression is altered in cells that constitutively express Ala168,170 NFATc4. Increased PPARγ gene expression may, in part, account for the induction of adipocyte formation in NIH 3T3 cells that stably express Ala168,170 NFATc4 (Fig. 5). Expression of PPARγ was similar in control cells and in cells that constitutively express wild-type NFATc4 (Fig. 8). However, expression of Ala168,170 NFATc4 increased PPARγ gene expression. As a control, GAPDH expression was similar in all three stably transfected cells. Together, these data indicate that increased NFAT activity by mutation at the conserved Ser residues induces PPARγ gene expression and, in part, accounts for the increase in adipocytes formation in NIH 3T3 cells.
FIG. 8.
Expression of Ala168,170 NFATc4 in NIH 3T3 cells increases PPARγ gene expression. Stably transfected confluent NIH 3T3 cells were cultured in differentiation media to promote adipocyte formation. Total RNA was harvested, and expression of the PPARγ and GAPDH genes was examined by semiquantitative RT-PCR (A) and measured by PhosphorImager analysis (B).
DISCUSSION
Conserved NFAT phosphorylation sites.
NFAT is a highly phosphorylated protein in resting cells. Primary phosphorylation sites are located at the NH2-terminal NFAT homology domain (4-6, 12-14, 46, 58). In this study, we demonstrate that phosphorylation on the NFAT homology domain plays an important role in NFATc4 subcellular distribution. Importantly, phosphorylation of the conserved Ser residues is shown to be critical. The location of Ser168,170 in NFATc4 is similar to those of Ser172 in NFATc1, Ser168 in NFATc2, and Ser163,165 in NFATc3. This report, in conjunction with previous studies (13, 14, 44, 46), demonstrates that these conserved Ser residues are phosphorylated in vivo. The presence of conserved Ser phosphorylation suggests a common regulatory mechanism to activate or inhibit NFAT function. NFAT has been shown to interact with other phosphorylating kinases such as GSK3β and CK1α (6, 58). Under certain circumstances, both GSK3β and CK1α have been termed hierarchical protein kinases as phosphorylation of their target sites requires prior phosphorylation by other protein kinases at a residue in the vicinity. Conserved phosphorylation at Ser168,170 of NFATc4, Ser163,165 of NFATc3, Ser168 of NFATc2, and Ser172 of NFATc1 may facilitate subsequent phosphorylation at the Ser-rich domain mediated by GSK3β and CK1α. Hence, Ser168,170 of NFATc4, Ser163,165 of NFATc3, Ser168 of NFATc2, and Ser172 of NFATc1 may be switches that initiate the phosphorylation and subsequent inactivation of NFAT function.
Conversely, these conserved Ser residues may be the primary targets of the calcineurin phosphatase. This report, together with previous studies (13, 14, 44, 46), indicates that mutational replacement of these conserved Ser residues with Ala promotes nuclear localization of NFAT. Further mutational replacement of other Ser residues in the SRR with Ala enhances NFAT nuclear localization (5, 44). These observations suggest that phosphorylation at Ser168,170 of NFATc4, Ser163,165 of NFATc3, Ser168 of NFATc2, and Ser172 of NFATc1 is the primary target for calcineurin-mediated dephosphorylation and subsequent activation.
Besides the presence of conserved Ser residues in the SRR, there are other Ser residues in the NFAT homology domain that are phosphorylated by other kinases. For example, PKA phosphorylates Ser269 of NFATc1 and Ser289 of NFATc4 (6, 12). Sequence comparisons indicate that Ser269 of NFATc1 and Ser289 of NFATc4 are located in analogous positions. Similar Ser residues could be found in NFATc2 (Ser255) and NFATc3 (Ser277). Phosphorylation at Ser255 of NFATc2 and Ser277 of NFATc3 has not been demonstrated. However, it is likely that these Ser residues are also targets of phosphorylation. Whether PKA or other protein kinases mediate the phosphorylation of Ser255 of NFATc2 and Ser277 of NFATc3 remains to be investigated. These observations further indicate that there are functionally conserved Ser residues present in the NH2-terminal homology domain to regulate NFAT activity.
Differential phosphorylation of NFAT by MAP kinases.
In this report, we show that NFATs are differentially phosphorylated by MAP kinases. NFATc1 and NFATc3 both contain sequences for JNK binding and are phosphorylated by JNK (13, 14). However, similar interaction of NFATc2 and NFATc4 with JNK was not detected (data not shown). Since interaction has been implicated as a prerequisite for subsequent JNK phosphorylation, the lack of binding suggests that NFATc2 and NFATc4 are not JNK substrates. Indeed, we show that NFATc4 is poorly phosphorylated by JNK in vitro. Interestingly, the p38 MAP kinase phosphorylates NFATc4, but not NFATc3, in vitro. Differential phosphorylation of NFAT by the JNK and the p38 MAP kinases suggests that these kinases are critical for NFAT regulation. Importantly, conserved Ser residues of NFAT (Ser168,170 of NFATc4, Ser163,165 of NFATc3, and Ser172 of NFATc1) are targets of JNK and p38 MAP kinase phosphorylation.
Homozygous disruption of the JNK1 gene increases IL-4 expression and promotes TH2 cell differentiation (13, 21). Induction of IL-4 expression is, in part, due to increased NFATc1 nuclear localization. Thus, JNK1 negatively regulates NFATc1 activity. The present study indicates that NFATc4 is a substrate for p38 MAP kinases. Targeted disruption of the p38 MAP kinase gene may cause similar increases in the nuclear localization of NFATc4. Nuclear NFATc4 may then induce NFAT target genes and promote cell growth and differentiation.
Three distinct MAP kinase pathways (ERK, JNK, and p38) have been identified (reviewed in references 16 and 20). The JNK and the p38 MAP kinase pathways are frequently associated with stress activation (e.g., UV irradiation and cytokine stimulation). On the other hand, the ERK MAP kinase pathway is regulated by growth factors and mitogens. Thus, the JNK and the p38 MAP kinase pathways are distinct from the ERK MAP kinase pathway. In this report, we show that the JNK and the p38 MAP kinases differentially phosphorylate NFATc3 and NFATc4, respectively. However, the ERK MAP kinase phosphorylates both NFATc3 and NFATc4. These results suggest that although the JNK and the p38 MAP kinase pathways differentially regulate NFAT members, a common signal may be derived from the ERK MAP kinase pathway to regulate general NFAT activity. Differential phosphorylation by the JNK and the p38 MAP kinases may achieve specific negative regulation of particular NFAT members in different cell types. On the contrary, the ERK phosphorylation may provide a general signal, positively and/or negatively, to regulate NFAT activity. A goal for further research will be to identify the ERK phosphorylation sites on NFAT to further delineate molecular mechanisms of NFAT regulation.
Phosphorylation cycle of NFAT.
NFAT is phosphorylated under basal, unstimulated conditions. Dephosphorylation is mediated by the calcineurin phosphatase, which promotes nuclear translocation and subsequent transcriptional activation of NFAT. Rephosphorylation of the dephosphorylated NFAT terminates NFAT activation. Thus, NFAT phosphorylation is a reversible event. Whether the kinases that phosphorylate NFAT under the basal state conditions are similar to the kinases that mediate rephosphorylation remains to be determined. In this report, we show that p38 MAP kinases phosphorylate NFATc4. Under the basal state conditions, the activated p38 MAP kinases only slightly affect NFATc4 phosphorylation. However, activation of the p38 MAP kinases dramatically affects the dephosphorylated NFATc4. These data suggest that the p38 MAP kinases are more likely to mediate rephosphorylation of NFATc4.
Upon UV irradiation, the dramatic decrease in electrophoretic mobility of the dephosphorylated NFATc4 suggests that multiple rephosphorylations have occurred. The p38 MAP kinases phosphorylate Ser168,170 and potentially a few other Ser-Pro motifs of NFATc4. Phosphorylation by the p38 MAP kinases at these Ser-Pro motifs would not make up for the dramatic decrease in NFATc4 electrophoretic mobility. Thus, additional rephosphorylation events mediated by other protein kinases are likely to account for the dramatic change in electrophoretic mobility. These additional kinases may include GSK3β, CK1α, or unidentified protein kinases.
The dramatic decrease in the electrophoretic mobility of NFATc4 upon UV irradiation is blocked by coexpression of the dominant-negative p38α or mutational replacement of Ser168,170 with Ala. These observations suggest that the p38 MAP kinases are required to initiate the rephosphorylation of NFATc4. In addition, the initiation of rephosphorylation requires intact phosphorylation sites at Ser168,170. These data support the hypothesis that phosphorylation at the conserved Ser residues (Ser168,170 of NFATc4 and similar Ser residues in other NFAT members) is a switch that initiates NFAT inactivation. Together, we propose the following model for NFATc4 rephosphorylation. The p38 MAP kinases phosphorylate Ser168,170 of the dephosphorylated NFATc4. This phosphorylation event is likely to take place in the nucleus, where dephosphorylated NFATc4 is located. Ser168,170-phosphorylated NFATc4 may remain active and mediate transcriptional activation until additional kinases (e.g., GSK3β, CK1α, or unidentified kinases) bind to and complete the rephosphorylation of NFATc4. Rephosphorylated NFATc4 is exported out of the nucleus, and NFATc4-mediated transcription is terminated.
Role of NFAT in adipocyte differentiation.
Previous studies indicated that treatment with CsA effectively blocked adipocyte differentiation (32). Presumably, CsA inhibits calcineurin phosphatase activity and, as a consequence, NFAT activation is attenuated. However, CsA has been shown to increase transforming growth factor β production (35), which negatively regulates adipocyte differentiation (52). Thus, whether the CsA-NFAT pathway has a direct effect on adipogenesis is unclear. On the contrary, another report indicated that CsA did not affect adipogenesis (57). In this report, we show that expression of Ala168,170 NFATc4 promotes adipocyte formation under differentiation conditions. These observations indicate that increased NFAT-dependent transcriptional activity, as a consequence of increased nuclear localization, has a direct role in adipocyte formation.
Effective inhibition of adipogenesis by CsA requires early intervention during differentiation (32). These observations suggest that NFAT is critical in the initiation stage of adipocyte differentiation. NFAT may act as an activator and regulate certain gene expression that is required for adipocyte differentiation. In this report, we show that NFAT increases PPARγ gene expression. Increased expression of PPARγ may then commit cells to initiate adipogenesis. Interestingly, PPARγ expression is also upregulated by the transcription factor C/EBP (55). Whether both NFAT and C/EBP, or other factors, are required for optimal expression of PPARγ remains to be investigated. Nonetheless, the regulation of the PPARγ gene expression by multiple transcription factors suggests that the expression of PPARγ is critical for the initiation of adipocyte differentiation. Thus, multiple transcription complexes relay different signals, in parallel or in synergy, to the PPARγ gene promoter to set off adipogenesis.
The presence of two distinct NFAT complexes in the PPARγ gene promoter suggests that NFAT conveys different signals to promote adipogenesis. Previous studies indicated that NFAT interacted with multiple nuclear factors to activate gene transcription. For example, NFAT interacts with Fos-Jun complexes to mediate IL-2 gene expression (36). In addition, NFAT interacts with transcription factor MEF2 to determine muscle differentiation (7, 10). Besides acting as a transcriptional activator, NFAT has been shown to bind to DNase I hypersensitive sites on the IL-4-IL-13 gene locus (1, 2). DNase I hypersensitive sites are indicative of chromatin remodeling. The presence of binding elements in the DNase I hypersensitive sites suggests that NFAT participates in the opening of the chromatin structure on specific gene loci upon activation. Transcriptional activation and chromatin remodeling are likely events during initiation of adipogenesis. Thus, increased NFAT nuclear localization may promote the opening in the condensed chromatin and/or activation of the PPARγ gene promoter through the two distinct NFAT complexes.
In summary, differential phosphorylation of the conserved Ser residues on NFAT by MAP kinases provides new insights to understand NFAT regulations. Future investigations with phosphorylation defective NFAT proteins will provide additional avenues to investigate the biological roles of NFAT.
Acknowledgments
The first three authors contributed equally to this work.
We thank T. Hoey, B. M. Spiegelman, and T. Soderling for providing reagents; T. Barrett for technical assistance; and K. Gemme for administrative assistance.
This work was supported, in part, by grants from the National Cancer Institute (to R.J.D.) and the American Health Assistance Foundation (to C.-W.C.). R.J.D. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Agarwal, S., O. Avni, and A. Rao. 2000. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity 12:643-652. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, S., and A. Rao. 1998. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9:765-775. [DOI] [PubMed] [Google Scholar]
- 3.Aramburu, J., F. Garcia-Cozar, A. Raghavan, H. Okamura, A. Rao, and P. G. Hogan. 1998. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol. Cell 1:627-637. [DOI] [PubMed] [Google Scholar]
- 4.Avots, A., M. Buttmann, S. Chuvpilo, C. Escher, U. Smola, A. J. Bannister, U. R. Rapp, T. Kouzarides, and E. Serfling. 1999. CBP/p300 integrates Raf/Rac-signaling pathways in the transcriptional induction of NF-ATc during T cell activation. Immunity 10:515-524. [DOI] [PubMed] [Google Scholar]
- 5.Beals, C. R., N. A. Clipstone, S. N. Ho, and G. R. Crabtree. 1997. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 11:824-834. [DOI] [PubMed] [Google Scholar]
- 6.Beals, C. R., C. M. Sheridan, C. W. Turck, P. Gardner, and G. R. Crabtree. 1997. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science 275:1930-1934. [DOI] [PubMed] [Google Scholar]
- 7.Blaeser, F., N. Ho, R. Prywes, and T. A. Chatila. 2000. Ca(2+)-dependent gene expression mediated by MEF2 transcription factors. J. Biol. Chem. 275:197-209. [DOI] [PubMed] [Google Scholar]
- 8.Brun, R. P., P. Tontonoz, B. M. Forman, R. Ellis, J. Chen, R. M. Evans, and B. M. Spiegelman. 1996. Differential activation of adipogenesis by multiple PPAR isoforms. Genes Dev. 10:974-984. [DOI] [PubMed] [Google Scholar]
- 9.Cao, Z., R. M. Umek, and S. L. McKnight. 1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 5:1538-1552. [DOI] [PubMed] [Google Scholar]
- 10.Chin, E. R., E. N. Olson, J. A. Richardson, Q. Yang, C. Humphries, J. M. Shelton, H. Wu, W. Zhu, R. Bassel-Duby, and R. S. Williams. 1998. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 12:2499-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow, C. W., M. P. Clark, J. E. Rinaldo, and R. Chalkley. 1995. Multiple initiators and C/EBP binding sites are involved in transcription from the TATA-less rat XDH/XO basal promoter. Nucleic Acids Res. 23:3132-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow, C.-W., and R. J. Davis. 2000. Integration of calcium and cyclic AMP signaling pathways by 14-3-3. Mol. Cell. Biol. 20:702-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow, C.-W., C. Dong, R. A. Flavell, and R. J. Davis. 2000. c-Jun NH2-terminal kinase inhibits targeting of the protein phosphatase calcineurin to NFATc1. Mol. Cell. Biol. 20:5227-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow, C. W., M. Rincon, J. Cavanagh, M. Dickens, and R. J. Davis. 1997. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science 278:1638-1641. [DOI] [PubMed] [Google Scholar]
- 15.Chow, C.-W., M. Rincón, and R. J. Davis. 1999. Requirement for transcription factor NFAT in interleukin-2 expression. Mol. Cell. Biol. 19:2300-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cobb, M. H. 1999. MAP kinase pathways. Prog. Biophys. Mol. Biol. 71:479-500. [DOI] [PubMed] [Google Scholar]
- 17.Cowherd, R. M., R. E. Lyle, and R. E. McGehee, Jr. 1999. Molecular regulation of adipocyte differentiation. Semin. Cell Dev. Biol. 10:3-10. [DOI] [PubMed] [Google Scholar]
- 18.Crabtree, G. R. 1999. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell 96:611-614. [DOI] [PubMed] [Google Scholar]
- 19.Cuenda, A., P. Cohen, V. Buee-Scherrer, and M. Goedert. 1997. Activation of stress-activated protein kinase-3 (SAPK3) by cytokines and cellular stresses is mediated via SAPKK3 (MKK6); comparison of the specificities of SAPK3 and SAPK2 (RK/p38). EMBO J. 16:295-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis, R. J. 1995. Transcriptional regulation by MAP kinases. Mol. Reprod. Dev. 42:459-467. [DOI] [PubMed] [Google Scholar]
- 21.Dong, C., D. D. Yang, M. Wysk, A. J. Whitmarsh, R. J. Davis, and R. A. Flavell. 1998. Defective T cell differentiation in the absence of Jnk1. Science 282:2092-2095. [DOI] [PubMed] [Google Scholar]
- 22.Durand, D. B., J. P. Shaw, M. R. Bush, R. E. Replogle, R. Belagaje, and G. R. Crabtree. 1988. Characterization of antigen receptor response elements within the interleukin-2 enhancer. Mol. Cell. Biol. 8:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelman, J. A., A. H. Berg, R. Y. Lewis, A. Lin, M. P. Lisanti, and P. E. Scherer. 1999. Constitutively active mitogen-activated protein kinase kinase 6 (MKK6) or salicylate induces spontaneous 3T3-L1 adipogenesis. J. Biol. Chem. 274:35630-35638. [DOI] [PubMed] [Google Scholar]
- 24.Engelman, J. A., M. P. Lisanti, and P. E. Scherer. 1998. Specific inhibitors of p38 mitogen-activated protein kinase block 3T3-L1 adipogenesis. J. Biol. Chem. 273:32111-32120. [DOI] [PubMed] [Google Scholar]
- 25.Enslen, H., D. M. Brancho, and R. J. Davis. 2000. Molecular determinants that mediate selective activation of p38 MAP kinase isoforms. EMBO J. 19:1301-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enslen, H., J. Raingeaud, and R. J. Davis. 1998. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J. Biol. Chem. 273:1741-1748. [DOI] [PubMed] [Google Scholar]
- 27.Fajas, L., D. Auboeuf, E. Raspe, K. Schoonjans, A. M. Lefebvre, R. Saladin, J. Najib, M. Laville, J. C. Fruchart, S. Deeb, A. Vidal-Puig, J. Flier, M. R. Briggs, B. Staels, H. Vidal, and J. Auwerx. 1997. The organization, promoter analysis, and expression of the human PPARgamma gene. J. Biol. Chem. 272:18779-18789. [DOI] [PubMed] [Google Scholar]
- 28.Goedert, M., A. Cuenda, M. Craxton, R. Jakes, and P. Cohen. 1997. Activation of the novel stress-activated protein kinase SAPK4 by cytokines and cellular stresses is mediated by SKK3 (MKK6); comparison of its substrate specificity with that of other SAP kinases. EMBO J. 16:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez del Arco, P., S. Martinez-Martinez, J. L. Maldonado, I. Ortega-Perez, and J. M. Redondo. 2000. A role for the p38 MAP kinase pathway in the nuclear shuttling of NFATp. J. Biol. Chem. 275:13872-13878. [DOI] [PubMed] [Google Scholar]
- 30.Graef, I. A., P. G. Mermelstein, K. Stankunas, J. R. Neilson, K. Deisseroth, R. W. Tsien, and G. R. Crabtree. 1999. L-type calcium channels and GSK-3 regulate the activity of NF-ATc4 in hippocampal neurons. Nature 401:703-708. [DOI] [PubMed] [Google Scholar]
- 31.Green, H., and O. Kehinde. 1975. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 5:19-27. [DOI] [PubMed] [Google Scholar]
- 32.Ho, I.-C., J. H.-J. Kim, J. W. Rooney, B. M. Spiegelman, and L. H. Glimcher. 1998. A potential role for the nuclear factor of activated T cells family of transcriptional regulatory proteins in adipogenesis. Proc. Natl. Acad. Sci. USA 95:15537-15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho, S. N., D. J. Thomas, L. A. Timmerman, X. Li, U. Francke, and G. R. Crabtree. 1995. NFATc3, a lymphoid-specific NFATc family member that is calcium-regulated and exhibits distinct DNA binding specificity. J. Biol. Chem. 270:19898-19907. [DOI] [PubMed] [Google Scholar]
- 34.Hoey, T., Y. L. Sun, K. Williamson, and X. Xu. 1995. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity 2:461-472. [DOI] [PubMed] [Google Scholar]
- 35.Hojo, M., T. Morimoto, M. Maluccio, T. Asano, K. Morimoto, M. Lagman, T. Shimbo, and M. Suthanthiran. 1999. Cyclosporine induces cancer progression by a cell-autonomous mechanism. Nature 397:530-534. [DOI] [PubMed] [Google Scholar]
- 36.Jain, J., P. G. McCaffrey, V. E. Valge-Archer, and A. Rao. 1992. Nuclear factor of activated T cells contains Fos and Jun. Nature 356:801-804. [DOI] [PubMed] [Google Scholar]
- 37.Lee, J. C., J. T. Laydon, P. C. McDonnell, T. F. Gallagher, S. Kumar, D. Green, D. McNulty, M. J. Blumenthal, J. R. Heys, S. W. Landvatter, et al. 1994. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature 372:739-746. [DOI] [PubMed] [Google Scholar]
- 38.Loh, C., K. T. Shaw, J. Carew, J. P. Viola, C. Luo, B. A. Perrino, and A. Rao. 1996. Calcineurin binds the transcription factor NFAT1 and reversibly regulates its activity. J. Biol. Chem. 271:10884-10891. [DOI] [PubMed] [Google Scholar]
- 39.Mandrup, S., and M. D. Lane. 1997. Regulating adipogenesis. J. Biol. Chem. 272:5367-5370. [DOI] [PubMed] [Google Scholar]
- 40.Masuda, E. S., Y. Naito, H. Tokumitsu, D. Campbell, F. Saito, C. Hannum, K. Arai, and N. Arai. 1995. NFATx, a novel member of the nuclear factor of activated T cells family that is expressed predominantly in the thymus. Mol. Cell. Biol. 15:2697-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molkentin, J. D., J. R. Lu, C. L. Antos, B. Markham, J. Richardson, J. Robbins, S. R. Grant, and E. N. Olson. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93:215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moustaid, N., and H. S. Sul. 1991. Regulation of expression of the fatty acid synthase gene in 3T3-L1 cells by differentiation and triiodothyronine. J. Biol. Chem. 266:18550-18554. [PubMed] [Google Scholar]
- 43.Musaro, A., K. J. McCullagh, F. J. Naya, E. N. Olson, and N. Rosenthal. 1999. IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature 400:581-585. [DOI] [PubMed] [Google Scholar]
- 44.Okamura, H., J. Aramburu, C. Garcia-Rodriguez, J. P. Viola, A. Raghavan, M. Tahiliani, X. Zhang, J. Qin, P. G. Hogan, and A. Rao. 2000. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol. Cell 6:539-550. [DOI] [PubMed] [Google Scholar]
- 45.Paulauskis, J. D., and H. S. Sul. 1988. Cloning and expression of mouse fatty acid synthase and other specific mRNAs. Developmental and hormonal regulation in 3T3-L1 cells. J. Biol. Chem. 263:7049-7054. [PubMed] [Google Scholar]
- 46.Porter, C. M., M. A. Havens, and N. A. Clipstone. 2000. Identification of amino acid residues and protein kinases involved in the regulation of NFATc subcellular localization. J. Biol. Chem. 275:3543-3551. [DOI] [PubMed] [Google Scholar]
- 47.Rao, A., C. Luo, and P. G. Hogan. 1997. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15:707-747. [DOI] [PubMed] [Google Scholar]
- 48.Razani, B., T. P. Combs, X. B. Wang, P. G. Frank, D. S. Park, R. G. Russell, M. Li, B. Tang, L. A. Jelicks, P. E. Scherer, and M. P. Lisanti. 2002. Caveolin-1 deficient mice are lean, resistant to diet-induced obesity, and show hyper-triglyceridemia with adipocyte abnormalities. J. Biol. Chem. 277:8635-8647. [DOI] [PubMed] [Google Scholar]
- 49.Rosen, E. D., C. J. Walkey, P. Puigserver, and B. M. Spiegelman. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14:1293-1307. [PubMed] [Google Scholar]
- 50.Scherer, P. E., M. P. Lisanti, G. Baldini, M. Sargiacomo, C. C. Mastick, and H. F. Lodish. 1994. Induction of caveolin during adipogenesis and association of GLUT4 with caveolin-rich vesicles. J. Cell Biol. 127:1233-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tontonoz, P., E. Hu, and B. M. Spiegelman. 1994. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 79:1147-1156. [DOI] [PubMed] [Google Scholar]
- 52.Torti, F. M., S. V. Torti, J. W. Larrick, and G. M. Ringold. 1989. Modulation of adipocyte differentiation by tumor necrosis factor and transforming growth factor beta. J. Cell Biol. 108:1105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitmarsh, A. J., and R. J. Davis. 1998. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem. Sci. 23:481-485. [DOI] [PubMed] [Google Scholar]
- 54.Wu, H., F. J. Naya, T. A. McKinsey, B. Mercer, J. M. Shelton, E. R. Chin, A. R. Simard, R. N. Michel, R. Bassel-Duby, E. N. Olson, and R. S. Williams. 2000. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 19:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, Z., Y. Xie, N. L. Bucher, and S. R. Farmer. 1995. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 9:2350-2363. [DOI] [PubMed] [Google Scholar]
- 56.Yang, X. Y., L. H. Wang, T. Chen, D. R. Hodge, J. H. Resau, L. DaSilva, and W. L. Farrar. 2000. Activation of human T lymphocytes is inhibited by peroxisome proliferator-activated receptor gamma (PPARgamma) agonists. PPARgamma co-association with transcription factor NFAT. J. Biol. Chem. 275:4541-4544. [DOI] [PubMed] [Google Scholar]
- 57.Yeh, W. C., B. E. Bierer, and S. L. McKnight. 1995. Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc. Natl. Acad. Sci. USA 92:11086-11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, J., F. Shibasaki, R. Price, J. C. Guillemot, T. Yano, V. Dotsch, G. Wagner, P. Ferrara, and F. McKeon. 1998. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell 93:851-861. [DOI] [PubMed] [Google Scholar]