Abstract
In a gene trap screen we recovered a mouse mutant line in which an insertion generated a null allele of the Brd4 gene. Brd4 belongs to the Fsh/Brd family, a group of structurally related proteins characterized by the association of two bromodomains and one extraterminal domain. Members of this family include Brd2/Ring3/Fsrg1 in mammals, fs(1)h in Drosophila, and Bdf1 in Saccharomyces cerevisiae. Brd4 heterozygotes display pre- and postnatal growth defects associated with a reduced proliferation rate. These mice also exhibit a variety of anatomical abnormalities: head malformations, absence of subcutaneous fat, cataracts, and abnormal liver cells. In primary cell cultures, heterozygous cells also display reduced proliferation rates and moderate sensitivity to methyl methanesulfonate. Embryos nullizygous for Brd4 die shortly after implantation and are compromised in their ability to maintain an inner cell mass in vitro, suggesting a role in fundamental cellular processes. Finally, sequence comparisons suggest that Brd4 is likely to correspond to the Brd-like element of the mediator of transcriptional regulation isolated by Y. W. Jiang, P. Veschambre, H. Erdjument-Bromage, P. Tempst, J. W. Conaway, R. C. Conaway, and R. D. Kornberg (Proc. Natl. Acad. Sci. USA 95:8538-8543, 1998) and the Brd4 mutant phenotype is discussed in light of this result. Together, our results provide the first genetic evidence for an in vivo role in mammals for a member of the Fsh/Brd family.
The bromodomain is a conserved 110-amino acid motif which specifically interacts with acetyl-lysines, at least in the context of short histone H3 and H4 peptides (reference 9; see also reference 39). Although bromodomains have now been found in more than 40 different proteins (13, 17, 39), the function of this motif is poorly understood. The association of two N-terminal bromodomains with a C-terminal extraterminal (ET) domain defines the Fsh/Brd subgroup (23), which includes members in many species: Saccharomyces cerevisiae (Bdf1 and Bdf2) (5, 23, 24), Caenorhabditis elegans (cefsh) (36), Drosophila [fs(1)h, previously called fsh (Flybase nomenclature)] (14), and hagfish (hffsh; GenBank accession no. AF191032). In vertebrates, four members of the Fsh/Brd subgroup have now been identified: Brd2/RING3/fsrg1 (1, 32, 36), Brd3/ORFX/fsrg2 (37), Brd4/HUNK1/MCAP (8), and Brd5/BRDT (19).
The yeast Bdf1 protein interacts with the general transcription factor TFIID (24) as well as with histones H3 and H4 (28), and the Bdf1 mutant phenotype shares features with those of mutants affected in general transcription (23). Bdf1 mutants display a reduced rate of vegetative growth, failure to undergo one or both meiotic divisions, and sensitivity to the DNA-damaging agent methyl methanesulfonate (MMS). Some alleles of Drosophila fs(1)h cause partial or complete loss of segments and homeotic transformations in progeny of mutant females (10, 14), but little is known about the molecular properties of its product. In vertebrates, the human Brd2 protein has been identified as a mitogen-activated nuclear protein associated with a kinase activity (6, 30, 32) and a regulator of E2F-dependent cell cycle genes (7).
In a gene trap screen designed to identify genes involved in mouse development, we recovered an integration producing a null allele of Brd4/MCAP, another member of the Brd/Fsh family (8). We showed that the Brd4 nullizygous condition results in early embryonic lethality. We also demonstrated that only one functional allele of Brd4 is not able to sustain normal development. Heterozygosity for Brd4 leads to pre- and postnatal growth defects that are associated with reduced proliferation in vitro and in vivo. Together, our results provide the first genetic evidence of an in vivo role for a Fsh/Brd family member in mammals.
MATERIALS AND METHODS
Gene trap and rapid amplification of cDNA ends (RACE).
Production of gene trap embryonic stem (ES) cells and the gene trap vector (a kind gift of W. Skarnes) was performed as described previously (4). The vector contained a splice acceptor sequence upstream of the CD4 transmembrane domain sequence, a lacZ-neomycin-phosphotransferase fusion (β-geo), and a polyadenylation signal such that integrations into introns of transcriptionally active genes produce a β-galactosidase fusion protein that contains an endogenous sequence amino terminal to the site of insertion. The inclusion of a transmembrane domain-encoding region in the vector was designed to enrich for insertions downstream of secretory signal sequences of type I transmembrane domains, because β-galactosidase activity is only preserved when the enzyme is retained in the cytoplasm (34). Therefore, an unexpected aspect of our results while using this vector was the isolation of a nuclear protein (34). In a systematic screen using a similar vector, J. Brennan and W. Skarnes isolated genes coding for nonsecreted proteins in 5 out of 12 cases, 3 of these involving insertion of the vector into the part of the gene corresponding to the 5′ untranslated region (UTR), as is the case for Brd4. In these cases, a functional β-geo protein was presumably produced via internal initiation of translation (reference 33 and J. Brennan, personal communication).
RACE was performed as described previously (4). The ST132 RACE fragment was used to screen an embryonic day 13 (E13) testis cDNA library (27) by standard procedures. The cDNA sequence matched precisely the MCAP sequence described in the study cited in reference 8. The 3′ UTR of the 3.5-kb Brd4 clone was obtained by 3′ RACE using a Marathon-Ready cDNA kit (Clontech) and was cloned into a Topo II vector (Invitrogen). Reverse transcription-PCR (RT-PCR) was performed using an Advantage One-Step RT-PCR Kit (Clontech) (primer 1f, AAA TCA GCT CAC CAG GCTG T; primer 1r, TCT TGG GCT TGT TAG GGT TG; primer 2r, CAC CAC CAG GTT CAC TTC CT). Sequence comparison of the different members of the Brd family was performed with ClustalX software (17, 35). Sequence alignment of the Ring3-like peptides of the mediator with the different members of the Brd family was performed with Lasergene software (DNASTAR).
Mouse strains.
CGR8 ES cells (129/Ola) were used to generate the gene trap cell lines (4, 34). Chimeras and their heterozygous progeny were backcrossed for five to eight generations onto a C57BL6/J background. Heterozygous individuals were obtained by mating a Brd4gt/Brd4+ male with a C57BL6/J female in order to avoid possible maternal effects of the Brd4gt allele. The day of detection of the vaginal plug was defined as E0.
Radiation hybrid mapping.
The mouse-hamster radiation hybrid (RH) panel was used according to the supplier's instructions (Research Genetics). Notch31f (AGG GAC CTG TGT GGA TGG CGT GAG C) was used as an upstream primer and Notch31r (CCA TTC TGA CAG GGT GCC TGG CTG C) was used as a downstream primer to specifically amplify a 228-bp fragment from the mouse Notch3 locus. 18f (CAA TCT GAC TGA GCT GTT CAT ATA GGT C) was used as an upstream primer and 18r (GTT GTA GAC ATT TGG GAG GTT TCT AGT C) was used as a downstream primer to specifically amplify a 414-bp fragment from the mouse Brd4 locus. Maps and extensive information on mouse RH can be found in The Jackson Laboratory RH database site (http://lena.jax.org/resources/documents/cmdata/).
Histology, antibody staining, and in situ hybridization.
The Brd4 common probe contains the first 1,820 nucleotides of the published cDNA (NCBI accession number NM 0205508). X-Gal staining was performed as described previously (16). For in situ hybridization on blastocyst explants, blastocysts were grown on gelatinized Lab-Tek chamber slide (catalog no. 177445; Nunc) in ES cell culture medium without leukemia-inhibiting factor (LIF) (15) for 2 to 5 days. The resulting culture was rinsed twice in phosphate-buffered saline, fixed for 4 h in fresh 4% paraformaldehyde in phosphate-buffered saline, washed twice in phosphate-buffered saline, and hybridized as previously described (25). The anti-phospho-histone H3 antibody (catalog no. 06570; Upstate Biotechnology) was used according to the specifications of the supplier (5 μg/ml on paraffin sections with antigen retrieval). For eosin-hematoxylin staining, organs were fixed in Bouin's fixative. After fixation, samples were processed, embedded in paraffin, sectioned, and stained according to standard procedures. For skeleton preparations, skulls from 2-month-old males were skinned, dehydrated in 95% ethanol for 24 h, fat dissolved in acetone for 24 h, cleared in 2% KOH for about 3 h, and stained for 3 days in 0.015% alcian blue-0.005% alizarin red-5% acetic acid in 60% ethanol. The skulls were then cleared in 2% KOH.
Mouse embryonic fibroblast culture and proliferation.
Mouse embryonic fibroblasts were isolated and cultured as described in the study cited in reference 15 except that minced embryos were only trypsinized once at 37°C for 10 min and spun down for 5 min and the pellet was plated. For the proliferation test, 1.00 × 105 cells were harvested at day 0 and cells were counted at different time points by using a hemocytometer. The MMS was supplied by Aldrich (catalog no. 12,992-5). UV irradiation was done in a Stratalinker apparatus using 254-nm bulbs.
RESULTS
Isolation and characterization of the Brd4 gene.
Isolation of the Brd4 gene was performed as follows. ST132 was an ES cell clone selected from a gene trap screen originally designed to identify integrations into genes expressed during early murine organogenesis (4, 34). After demonstration of the expression of the lacZ reporter in midgestation chimeras, a mouse strain carrying the ST132 integration was established (see Materials and Methods).
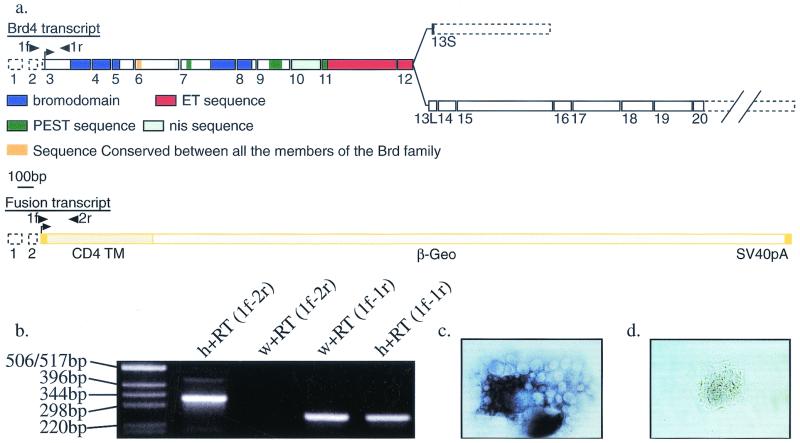
5′ RACE was used to clone a 209-bp fragment of the trapped gene from the ST132 cell line. This fragment showed no open reading frame or obvious homology to any sequence from the databases. A probe corresponding to the ST132 5′ RACE product was used to isolate a 1,890-bp clone from a mouse embryonic testis cDNA library (27). The sequencing of the complete cDNA was achieved by a combination of expressed sequence tag alignments, RT-PCR, and 3′ RACE (Fig. 1a). The junction between the 5′ RACE fragment and the cDNA sequence, as well as the junction between the 5′ RACE fragment and the gene trap sequence, was confirmed by RT-PCR (Fig. 1b). While this work was in progress, the same cDNA was reported under the name MCAP (mitotic chromosome-associated protein (8). However, we have designated it Brd4 for reasons of consistency with the nomenclature used for the members of this family, as recommended by the Mouse Gene Informatics Nomenclature Committee.
FIG. 1.
The gene trap strategy and Brd4 gene. (a) Structure of the Brd4 and Brd4gt transcripts. A long Brd4 mRNA (6 kb, encoding a 1,400-amino acid protein) is transcribed from 20 exons (1 to 20), and a short Brd4 mRNA (3.5 kb, encoding a 722-amino acid protein) is formed by the use of an alternative 13th exon (13S). The fusion transcript containing the first two noncoding exons from Brd4 fused to the gene trap sequences is transcribed from the Brd4gt allele. 1f, 1r, and 2r are the specific primers used for RT-PCR in Fig. 1b. Dotted boxes, UTR; plain boxes, coding exons; nis, nuclear localization signal; TM, transmembrane domain. (b) RT-PCR with primers 1f (5′ RACE fragment specific), 1r (Brd4-specific coding sequence), and 2r (gene trap-specific primer). Primers 1f and 2r (1f-2r) amplified the expected 338-bp product in total RNA from Brd4gt/Brd4+ (h) but not Brd4+/Brd4+(w) individuals. Primers 1f and 1r (1f-1r) amplified the expected 256-bp fragment in total RNA from both wild types and heterozygotes. (c and d) Brd4+/Brd4+ (c) and Brd4gt/Brd4gt (d) blastocyst explants cultured for 2 days and hybridized with a probe covering exons 1 to 9 and part of exon 10. No transcript could be detected in the homozygote.
The Brd4 gene is transcribed from 20 exons.
The Brd4 gene encodes a putative 1,400-amino acid protein characterized by the presence of two bromodomains in the N-terminal region of the protein and an ET domain in the more C-terminal region. The presence of several nuclear localization sequences is in agreement with the reported nuclear localization of the protein (8). The presence of several PEST sequences suggests that Brd4 may be unstable (31).
Sequence comparison of the murine Brd4 cDNA (NCBI accession number NM 0205508) with murine genomic sequences (GenBank accession no. AC074208, AC073765, and AC073779) revealed that Brd4 is transcribed from 20 exons spanning at least 110 kb (GenBank accession no. AF461395) (Fig. 1a). As noted in reference 8, the Brd4 gene shares the highest sequence identity with the uncharacterized human HUNK1 cDNA (NCBI accession number NM 014299), which encodes a protein of 722 amino acids, which is much smaller than Brd4. Using 3′ RACE performed on the E11 cDNA library and RT-PCR procedures performed on adult tissues, we were able to reveal the existence of an alternative 13th exon (13S [short]) that encodes three C-terminal amino acids as well as an alternative 3′ UTR. Use of the exon 13L (long) is predicted to lead to the synthesis of a 6-kb transcript coding for a 1,400-amino acid protein. Use of the exon 13S is predicted to lead to the synthesis of a 3.5-kb transcript coding for a 722-amino acid protein with 96% identity with the HUNK1 protein (Fig. 1a) (GenBank accession no. AF461396). Sequence comparison of the murine and human Brd4 cDNAs to human genomic sequences (NT 011290) revealed that the human gene contains the same number of exons as the mouse gene and covers 43.6 kb. In both species, exon 13S is located between exons 12 and 13L and is highly conserved (85% identity over 414 bp). However, although we were able to reveal the existence of this short transcript by RT-PCR and 3′ RACE, we were unable to detect it by Northern blotting or in situ hybridization, a result in agreement with the single-size protein detected by Western blotting (8). Therefore, the biological significance of this short transcript remains elusive. In conclusion, both the high sequence identity and the similarities in genomic organization strongly suggest that Brd4 is the mouse orthologue of HUNK1.
Brd4 belongs to a group of structurally related proteins.
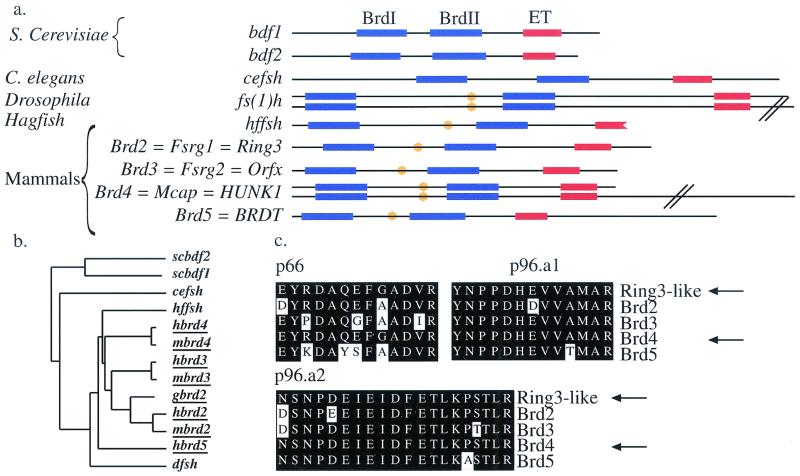
Several proteins that are characterized by the association of two N-terminal bromodomains with a more C-terminal ET domain have now been identified. We compared the motif organizational structures (Fig. 2a) and sequences (Fig. 2b) of the different members of this family. Besides their bromodomains and ET domains, all the vertebrate members of the Fsh/Brd family as well as the Drosophila fs(1)h protein contain identical short stretches of 11 amino acids (GVKRKADTTTP) (Fig. 2a). This motif was not encountered in any other protein present in the databases, and its function is presently unknown.
FIG. 2.
Structure of the Brd4 gene and conservation of the Brd family. (a) Domain organizations of the different members of the Brd/fs(1)h family. A convenient nomenclature for the vertebrate members of this family based on the root fsrg (female sterile homeotic related) was proposed by Rhee et al. (32). However, here we use the nomenclature retained by the Mouse Gene Informatics Nomenclature Committee, who proposed the root Brd to designate the members of this bromodomain-containing family. Bromodomains are shown in blue, ET domains are shown in red, and the conserved 10-amino acid motif is shown in orange. Note that the hffsh sequence is only partial and terminates in the ET domain. (b) Similarity tree of the members of the Brd family (vertebrate members of the family are underlined). Note that Brd2/RING3 and Brd3/ORFX group together, in agreement with the important conservation of synteny observed between human chromosomes 6 and 9 (see reference 21 for review). Brd5/BRDT, the only member of the Brd family that exhibits a nonubiquitous pattern of expression (19), is found outside the mammalian group, suggesting a possible divergent function. Bdf1 and Bdf2 group together, suggesting a duplication after the separation between yeast and metazoans. Alignments of bromodomains also confirmed this result (data not shown). (c) Sequence comparison of the Ring3-like peptides (p66, p96a1, and p96a2) (isolated in the study cited in reference 18) with those of the four mammalian members of the Brd family. Arrows point at the sequences of the peptides and Brd4.
The Brd2/RING3 protein was originally described as a mitogen-activated nuclear kinase (6), although the existence of kinase activity of Brd2 remains controversial (7, 30, 32). We compared the sequences of the kinase motifs described for Brd2 with the Brd4 sequence. Neither the Brd2 putative ATP binding motif (LGPSGFGPS) nor the putative catalytic lysine (ATK) was conserved in Brd4. Therefore, if Brd2 has a kinase activity, it is unlikely to be conserved in Brd4. However, it remains possible that Brd4 is able to recruit another protein with kinase activity, since such a function has been proposed for Brd2 (30).
One member of the Fsh/Brd family has been isolated in the mouse counterpart of the yeast mediator of transcriptional regulation, the role of which is to integrate the positive and negative regulatory information originating from enhancer and silencer elements. It then transmits this information to the RNA polymerase II, modulating its activity in promoter-dependent transcription (18). At the time, Jiang et al. could not identify the member of the family that they had purified. When we compared the protein sequences of the three Brd-like peptides that they isolated with those of the four members of the Fsh/Brd family, we found that they were 100% identical to parts of the bromodomain II (peptides p66 and p96a1) and ET domains (peptide p96a2) of Brd4 (46 amino acids out of 46) (Fig. 2c). The other members of the mammalian Brd family did not share 100% identity with the peptides (41-of-46 amino acid identity for Brd2; 40 of 46 for Brd3; 40 of 46 for Brd5). These peptides are not predicted to be encoded by any other putative open reading frame presently identified in the human or mouse genome. Therefore, we conclude that Brd4 is very likely to be the protein identified by Jiang et al. as a component of the mediator.
Brd4 is located on mouse chromosome 17 and reveals a conservation of synteny with human chromosome 19p13.1.
BRD4/HUNK1 is located on human chromosome 19p13.1, 35.6 kb from the Notch3 gene (NT 011290). The mouse-hamster RH panel was used to map both mouse Brd4 and Notch3 on the mouse genome. We localized Brd4 and Notch3 on mouse chromosome 17 between markers D17Mit181 and D17Mit191, a result confirmed by the presence of both genes on the draft genome sequence (GenBank accession no. AC074208). This localization reveals a short conservation of synteny between Brd4/Notch3 on chromosome 17 on the mouse genome and HUNK1/NOTCH3 on chromosome 19p13.1 on the human genome. This conservation of synteny gives further evidence for the orthology between Brd4 and HUNK1. Interestingly, the three other members of the Fsh/Brd family also map close to members of the Notch family, at least in humans. Brd2 maps next to Notch4 on human chromosome 6, Brd3 next to Notch1 on human chromosome 9, and Brd5 next to Notch2 on human chromosome 1 (reviewed in reference 20).
Brd4gt is a null allele of Brd4.
The gene trap vector integrated within the second intron of Brd4. A fusion mRNA containing the noncoding exons 1 and 2 from Brd4 spliced to the gene trap sequences is predicted to be transcribed from the trapped locus (Fig. 1a and b). Therefore, the transcript encoded by the gene trap allele (designated Brd4gt) should not contain any Brd4 coding sequences. In order to confirm that we had indeed generated a null allele of Brd4, we cultured blastocysts from Brd4gt/Brd4+ × Brd4gt/Brd4+ matings. Using in situ hybridization with a probe spanning Brd4 exons 1 to 9 and part of exon 10, we showed that blastocyst explants homozygous for Brd4gt are clearly recognizable by the absence of Brd4 transcript, even after extended periods of staining (compare Fig. 1c and d). In order to ensure a homogeneous genetic background and to eliminate any independent mutation that may have occurred in the ES cell clone, we backcrossed the heterozygotes onto a C57BL6/J background for five to eight generations. In parallel, fluorescent in situ hybridization analysis of the G-banded metaphase spread of ST132 ES cells revealed that only a single gene trap vector insertion was evident and that it coincided grossly with the position of Brd4 (data not shown). We conclude from these results that the phenotype in Brd4gt mice (see below) is attributable to a null allele of Brd4.
Expression pattern of Brd4.
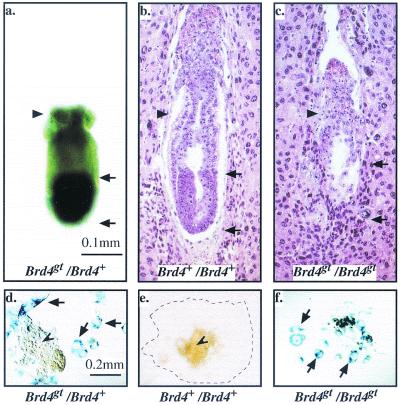
In Brd4gt/Brd4+ embryos, we detected a faint expression of the lacZ reporter gene from the blastocyst stage onwards. We also showed that the gene is strongly expressed in blastocyst explants (Fig. 1c and Fig. 6d and f). In E6 embryos, the expression of the reporter gene was ubiquitous but the intensity of the signal appeared to be stronger in the embryonic than in the extraembryonic part of the conceptus (see Fig. 6a). The highest level of expression was observed in E10 and E11 embryos (data not shown). At later stages of development and after birth, Brd4 remains expressed in all tissues tested, although the levels of expression differ (reference 8 and D. Houzelstein, unpublished data).
FIG. 6.
The homozygote phenotype. (a to c) E6 embryos. Black arrows demarcate the embryonic part of the conceptus, whereas the black arrowhead points at the extraembryonic part. Whole-mount β-galactosidase staining of a Brd4gt/Brd4+ embryo (a) and hematoxylin-eosin staining of sections of Brd4+/Brd4+ (b) and Brd4gt/Brd4gt (c) embryos are shown. Note that the embryonic part which normally expresses the gene strongly is missing in the homozygote. (d to f) Blastocyst explants stained with β-galactosidase after 5 days in culture. The arrows point at the giant trophoblastic cells, and the arrowhead points at the inner cell mass. In Fig. 6e, the limit of trophoblast giant cell spread is indicated by a dotted line. In Fig. 6f, the inner cell mass of the homozygote has degenerated and is missing.
Postnatal death of Brd4gt/Brd4+ individuals.
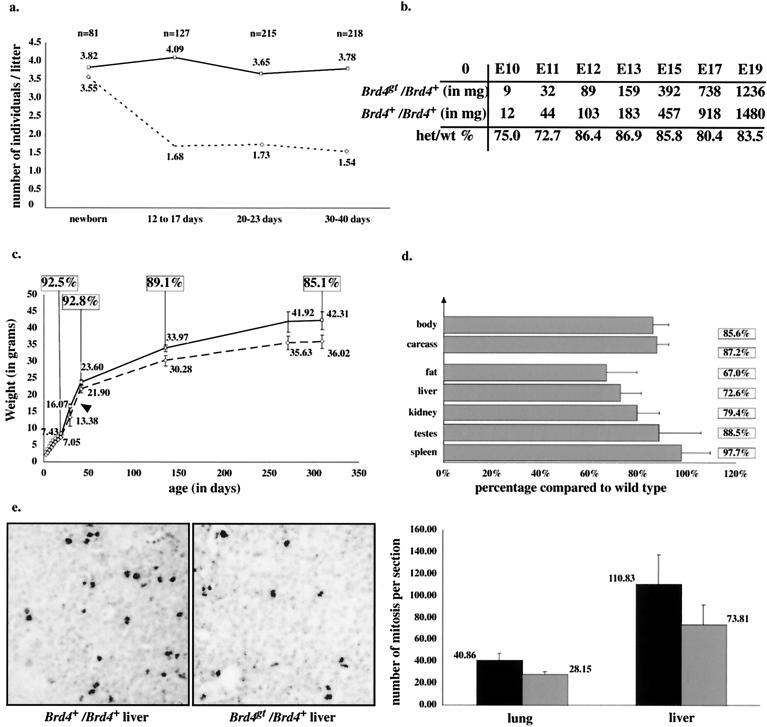
Shortly after birth, the expected 1:1 ratio of heterozygous to wild-type individuals was observed from Brd4gt/Brd4+ × Brd4+/Brd4+ matings (Fig. 3a). However, 12 to 17 days after birth (P12 to P17), about 50% of the heterozygotes were missing, whereas after P17 the proportions of heterozygous individuals remained relatively constant, indicating a critical period for survival during the first 2 weeks of postnatal life. No significant numbers of deaths occurred in wild-type C57BL6/J littermates over this period.
FIG. 3.
In vivo phenotype of the heterozygotes. (a) Postnatal lethality: comparison between the average numbers of Brd4+/Brd4+ (plain line) and Brd4gt/Brd4+ (dotted line) individuals per litter from matings of Brd4gt/Brd4+ males with Brd4+/Brd4+ females, from birth to adulthood. The number indicated above each point represents the average number of individuals per litter. Note that the expected ratio of heterozygous to wild-type individuals was observed at birth. n, number of individuals counted. (b) Growth defect before birth: average weights of Brd4+/Brd4+ and Brd4gt/Brd4+ individuals from E10 to E19. At least three littermate individuals from each genotype were counted at every point. Heterozygote versus wild-type weight percentages are indicated. Note that the difference in weight between heterozygous and wild-type individuals is most pronounced between E10 and E11. (c) Growth defect after birth. Six Brd4gt/Brd4+ and five Brd4+/Brd4+ Brd4gt/Brd4+ males were weighed over a period of 320 days. Wild-type individuals (plain line) were significantly heavier than their heterozygous (dotted line) littermates. Weights of both males and females from several litters gave results quantitatively and qualitatively similar. The average weight is indicated over each point, and heterozygote versus wild-type weight percentages are indicated in the squares. (d) Growth defect of individual organs. Individual organs from Brd4+/Brd4+ and Brd4gt/Brd4+ 10-month-old females were weighed separately. Every organ was affected by the mutation. (e) Reduced numbers of mitotic cells in the Brd4gt/Brd4+ individuals. An anti-phospho-histone H3 antibody was used to reveal mitotic cells from E15 liver and lung sections. An enlargement of part of such a section is shown. The average numbers of mitotic cells per section are indicated. There were significantly fewer mitotic cells in sections from heterozygous embryos than in those from wild-type embryos, although the cell density was unaffected.
Prenatal and postnatal growth defects of the Brd4gt/Brd4+ individuals.
While the cause of heterozygous death remained unclear, these individuals appeared consistently smaller than wild-type littermates. Therefore, heterozygous and wild-type individuals were systematically weighed before and after birth. At E10, the heterozygotes weighed, on average, 25% less than their wild-type littermates. This growth defect was maintained throughout the prenatal period until birth (Fig. 3b). After birth, the defect was less pronounced, since heterozygotes were only 10% lighter than the wild types (Fig. 3c). The weights of several organs from heterozygous adults were reduced in similar proportion compared to those of wild types (Fig. 3d). Since the growth defect was maintained throughout life, it is unlikely to result from an abnormal growth hormone pathway, which acts only after birth. A defect in nutrient supply from the mother also seems unlikely, since the growth defect was maintained after birth and weaning (Fig. 3a). To address the cause of the growth defect, we performed immunolabeling experiments on lung and liver sections by using an antibody directed against phospho-histone H3, a specific marker of mitotic cells (14) (Fig. 3e). We found a significantly reduced number of mitotic cells in heterozygotes compared to that in wild-type tissue (Fig. 3c). Taken together, these results indicate that the general growth restriction observed in heterozygous tissues is likely to be due to a reduction in the proliferation rate.
Anatomical defects of the Brd4gt/Brd4+ individuals.
A few weeks after birth, the heterozygotes were easily recognizable because of the abnormal shape of their heads. Skeleton preparations from 8-week-old heterozygous and wild-type animals revealed that this recognizable facies was due to a shorter incisive bone as well as a shorter and bent nasal bone (Fig. 4a and b). The mandible was also reduced in length. No discernible differences between the axial and appendicular skeletons of the wild types and heterozygotes could be found (data not shown).
FIG. 4.
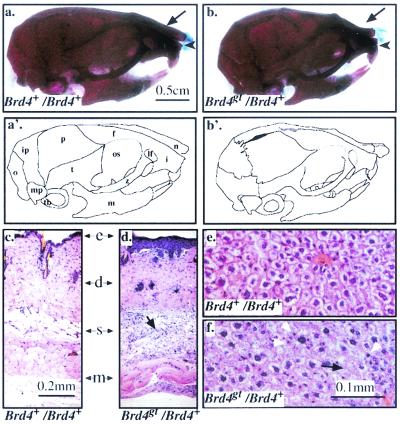
Anatomical and histological defects in the heterozygotes. (a, a′, b, and b′) Skeletal preparations of two-month-old Brd4+/Brd4+ (a) and Brd4gt/Brd4+ (b) male skulls, respectively; bone is stained in red and cartilage in blue. Note the shorter and bent nasal bone (arrow), the shorter incisor bone (arrowhead), and the shorter mandible of the heterozygote. f, frontal bone; i, incisive bone; ip, interparietal bone; lf, lacrimal foramen; m, mandible; mp, mastoid process; n, nasal bone; o, occipital bone; os, orbital surface of the frontal bone; p, parietal bone; t, temporal bone; tb, tympanic bulla; z, zygomatic bone. (c and d) Sections through the skin of two Brd4+/Brd4+ (c) and Brd4gt/Brd4+ (d) 1-month-old individuals. In the heterozygote, the epidermis (e) was thickened and the amount of subcutaneous adipose tissue (s) was reduced and contained degenerating adipose cells surrounded by phagocytic cells (black arrow). d, dermis; m, muscle. (e and f) Sections through the liver of two Brd4+/Brd4+ (e) and Brd4gt/Brd4+ (f) 2-month-old individuals. The black arrow in Fig. 4f points to degenerating hepatocytes, and the white arrowheads point at surrounding phagocytes in the heterozygote.
Various morphological abnormalities were seen in the skin, liver, testis (epididymal duct and vas deferens), and brain of heterozygotes. In the skin, the epidermis appeared abnormally thick, although the layers were clearly recognizable. The subcutaneous adipose tissue was considerably reduced and contained degenerating cells surrounded by infiltrating macrophages. The dermis also contained an increased number of neutrophils and mature fibrocytes (Fig. 4c and d). In the heterozygous liver, many hepatocytes contained one large aberrant nucleus or even two nuclei. Degenerated and necrotic hepatocytes surrounded by phagocytes (Küpffer cells and neutrophils) were also detected (Fig. 4e and f). In the testis, several abnormal seminiferous tubules lacking spermatogenic cells that might indicate impaired spermatogenesis were also noted. In the epididymis and vas deferens, many cells of the epithelium lining the lumen of ducts had abnormal polyploid nuclei and were altered in size and shape. Some of the epithelial cells also showed features of degeneration. In the brain, a few necrotic neurons were observed but classical histological procedures could not reveal any defect in myelination (data not shown).
In a sample population of 34 heterozygotes between 6 weeks and 6 months of age, four individuals had one or both eyes reduced in size and seven had a cataract. No eye defects were evident in a control group of 48 wild-type animals (data not shown).
Heterozygous mouse embryonic fibroblasts (MEFs) have a reduced proliferation rate and increased sensitivity to MMS.
To investigate further the cellular impact of the Brd4 heterozygous condition, we isolated fibroblasts from E13 embryos. The doubling time of heterozygous MEFs was significantly longer (4 days) than that of wild-type control MEFs (2 days) (Fig. 5a). We conclude that the growth restriction observed in heterozygous embryos was maintained in cell culture. Therefore, MEFs constitute an in vitro system suitable for the further analysis of the Brd4 growth phenotype. We showed that medium conditioned with wild-type MEFs could not rescue the phenotype of heterozygous MEFs, suggesting that the growth restriction phenotype is cell autonomous (data not shown).
FIG. 5.
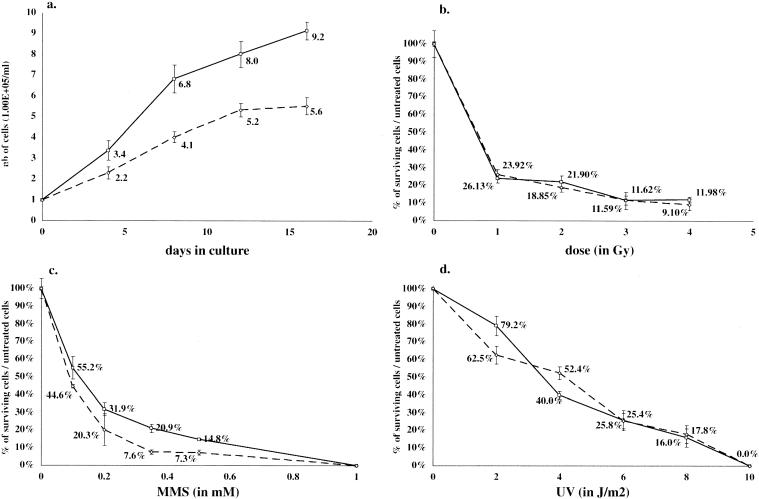
Effect of DNA damaging agents on survival of MEFs. (a) 1.00 × 105 Brd4+/Brd4+ (plain line) or Brd4gt/Brd4+ (dotted line) MEFs were harvested on individual plates at day 0 and counted at 4-day intervals. (b to d) MEFs were plated as for Fig. 5a. After they adhered to the plate, they were submitted to increasing doses of IR (b), MMS (c), or UV (d), cultured for 8 days, and counted.
Some aspects of the heterozygous growth restriction phenotype are reminiscent of those observed in various mutants affected in different DNA damage repair pathways (see reference 11 for review). Thus, we tested the survival of heterozygous cells after exposure to ionizing radiation (IR). IR is known to induce lesions that are the substrate for the nonhomologous end joining repair pathway and the transcription-coupled repair pathway (21). For the doses tested, no difference could be seen between heterozygous and wild-type MEFs (Fig. 5b). When heterozygous thymocytes were tested, the relative insensitivity to IR was also observed and the absence of nonhomologous end joining deficiency in these cells was corroborated by the normal generation of T-cell receptor variable region diversity (data not shown).
We then measured the survival of heterozygous cells following exposure to MMS. MMS induces lesions, such as N-methylpurines (29), which are substrates for the base excision repair pathway (see reference 22 for a recent review). We detected up to a twofold increase in sensitivity to MMS of heterozygous cells compared to that detected in wild-type cells (Fig. 5c).
Finally, the survival of heterozygous cells following exposure to UV radiation was assayed. Lesions induced by UV radiation, such as pyrimidine dimers, are substrates for the nucleotide excision repair pathway. No clear difference could be detected between heterozygous and wild-type cells in these conditions (Fig. 5d).
Characterization of the homozygous phenotype in vivo and in vitro.
To investigate the phenotype of individuals nullizygous for Brd4, heterozygous individuals were mated. No Brd4gt/Brd4gt homozygous progeny could be identified by in situ hybridization performed on midgestation litters (0 of 22). Out of 43 decidua recovered between E7 and E9, wild-type (10 of 43) and heterozygous embryos (24 of 43) were observed in the expected proportions. The remaining 25% of decidua (9 of 43), likely to correspond to the missing homozygotes, were empty, suggesting postimplantation lethality. When heterozygous females were mated with wild-type males, no empty site was recovered (0 of 23), indicating that the lethality observed in heterozygous matings was not due to a maternal defect. A number of E6 decidua were processed for histological examination. In 7 out of 37 cases, the embryonic region was absent (Fig. 6c; compare with a wild type in Fig. 6b). The embryonic region was the part of the conceptus where expression of the reporter gene was predominantly detected (Fig. 6a), whereas the extraembryonic region, which normally expresses Brd4 at lower levels, was less affected in the homozygous mutants (Fig. 6a to c).
To test whether homozygous embryos survive to the blastocyst stage, embryos were isolated at E3, at which point they showed no overt developmental delay and were morphologically indistinguishable from wild-type blastocysts (data not shown). The growth potential of wild-type and mutant blastocysts was monitored in vitro over a period of 2 to 5 days. During the first 2 days, both wild-type and homozygous blastocysts hatched out of the zona pellucida and attached to the slide. During this period, explants grown from homozygous embryos, as assayed by the absence of Brd4 transcript, appeared morphologically normal, albeit smaller than wild types (compare Fig. 1c and d). After 5 days in culture, two cell populations were clearly recognizable morphologically in explants from wild-type blastocysts: flat, spread-out, giant trophoblast cells (Fig. 6d) and the smaller inner-cell-mass cells (Fig. 6d and 6e). In explants grown from homozygous embryos, trophoblast giant cells were clearly recognizable and did not appear necrotic, whereas cells from the inner cell mass completely degenerated (Fig. 6f). Therefore, at least one copy of Brd4 is required for the maintenance of the inner cell mass.
DISCUSSION
We have examined the consequences of a gene trap insertion resulting in a null mutation in the Brd4 gene. We showed that Brd4 is expressed ubiquitously from the blastocyst stage and that nullizygous mutant embryos implant in the uterine wall but arrest in their development shortly thereafter, indicating that Brd4 exerts cellular functions essential for early postimplantation development. We have also found that individuals with only one functional allele of the Brd4 gene are growth restricted both before and after birth. Growth restriction is also observed in primary cell cultures and is therefore not dependent on diffusible growth factors, either in the medium or from wild-type cells. Thus, the defect appears to be cell autonomous, consistent with a predicted nuclear location for Brd4. We showed that, in vivo, deletion of only one Brd4 allele results in a reduction of the number of mitotic cells in at least two embryonic tissues. This result is consistent with the in vitro observation that microinjection of anti-Brd4 antibodies into HeLa cell nuclei completely inhibits entry into mitosis (8). The reduced mitotic activity is likely to underlie the growth restriction during the whole life of Brd4gt heterozygotes. This growth restriction is most pronounced at E10 and E11 (i.e., the earliest stage tested), when cell division is relatively rapid and the reduced proliferative ability of the Brd4gt cells is likely to be most detrimental.
The complex set of anatomical defects exhibited by the Brd4gt/Brd4+ mutants is reminiscent of some of the defects observed in two related human syndromes, Cockayne syndrome (CS) and trichothiodystrophy (TTD). They include partial lethality, impaired growth, peculiar facies, lack of subcutaneous fat, and cataracts (see references 2 and 3 for recent reviews). Patients suffering from these syndromes are affected in DNA repair pathways resulting, in particular, in severe sensitivity to UV radiation. However, we demonstrate that Brd4-deficient cells do not display sensitivity to ionizing and UV radiation, excluding a major role for Brd4 in these pathways. Heterozygous cells appeared significantly more sensitive to MMS than wild-type cells, suggesting a defect in DNA damage repair. However, this could be a primary or secondary consequence of the mutation. Interestingly, a similar phenotype (slow growth, sensitivity to MMS, and relative insensitivity to IR) was also described for the yeast Bdf1 mutant.
However, since TTD and CS are now known to be the consequence of defects in both DNA repair and transcription (see references 2 and 3 for recent reviews), impaired transcription in the Brd4gt mutants remains a possibility to explain the phenotypic similarities between the Brd4gt mutants and TTD-CS patients. Consistent with this possibility is our demonstration that Brd4 is very likely to be a component of the mediator of transcriptional regulation isolated in the study cited in reference 18. However, the transcriptional coactivator function of the murine mediator has yet to be demonstrated (18) and many other mammalian mediator-related complexes have been characterized extensively and do not appear to contain any Brd family member as a constitutive unit (see references 12 and 26 for reviews). Therefore, the question of whether Brd4 or other members of the Fsh/Brd family act as a constitutive unit of the mediator awaits detailed biochemical analyses. Nevertheless, it is noteworthy that the defects in Brd4 nullizygous embryos are very similar to those found when Srb7, which encodes the only other protein isolated as part of the mediator whose function has been inactivated in mice, is deleted. Srb7 nullizygous embryos also die shortly after implantation, and the inner cell mass of blastocysts grown in culture degenerates (38).
Summary.
Our results suggest that a fundamental cellular process is impaired in Brd4gt cells and that the complex pathological features observed in Brd4gt/Brd4+ individuals might be explained by a global deficiency in a basic mechanism, resulting in reduced proliferative ability. Indeed, the most remarkable defect of Brd4gt/Brd4+ mice is their pre- and postnatal growth defect. Given that this phenotype is associated with haploinsufficiency in the mouse, heterozygous mutations in the Brd human homologues could also be associated with pre- and postnatal growth defects. Therefore, Brds may be candidate genes for some human disorders characterized by a short stature. Thus, the Brd4gt mouse mutant will be of great value, not only in elucidating Brd protein function during pre- and postnatal life but also as a model for human pathology.
Acknowledgments
We are especially grateful to Françoise Poirier for her invaluable help during the preparation of the manuscript. We also thank Margaret Buckingham, Alain Sarasin, Thierry Magnaldo, and Jean-Pierre Rousset for comments on the manuscript. We thank Dimitris Kioussis and Owen Williams for thymocyte cultures and Judy Fletcher for FISH.
This work was supported by the MRC and by a Marie Curie training grant from the European Commission to D.H.
D.H. and S.L.B. contributed equally to this work.
Footnotes
This article is dedicated to the memory of Rosa Beddington.
REFERENCES
- 1.Beck, S., I. Hanson, A. Kelly, D. J. Pappin, and J. Trowsdale. 1992. A homologue of the Drosophila female sterile homeotic (fsh) gene in the class II region of the human MHC. DNA Sequence 2:203-210. [DOI] [PubMed] [Google Scholar]
- 2.Bergmann, E., and J. M. Egly. 2001. Trichothiodystrophy, a transcription syndrome. Trends Genet. 17:279-286. [DOI] [PubMed] [Google Scholar]
- 3.Berneburg, M., and A. R. Lehmann. 2001. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv. Genet. 43:71-102. [DOI] [PubMed] [Google Scholar]
- 4.Bullock, S. L., J. M. Fletcher, R. S. Beddington, and V. A. Wilson. 1998. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev. 12:1894-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chua, P., and G. S. Roeder. 1995. Bdf1, a yeast chromosomal protein required for sporulation. Mol. Cell. Biol. 15:3685-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denis, G. V., and M. R. Green. 1996. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 10:261-271. [DOI] [PubMed] [Google Scholar]
- 7.Denis, G. V., C. Vaziri, N. Guo, and D. V. Faller. 2000. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 11:417-424. [PMC free article] [PubMed] [Google Scholar]
- 8.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol. Cell. Biol. 20:6537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 10.Digan, M. E., S. R. Haynes, B. A. Mozer, I. B. Dawid, F. Forquignon, and M. Gans. 1986. Genetic and molecular analysis of fs(1)h, a maternal effect homeotic gene in Drosophila. Dev. Biol. 114:161-169. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg, E. C., and L. B. Meira. 2000. Database of mouse strains carrying targeted mutations in genes affecting cellular responses to DNA damage. Version 4. Mutat. Res. 459: 243-274. [DOI] [PubMed] [Google Scholar]
- 12.Hampsey, M., and D. Reinberg. 1999. RNA polymerase II as a control panel for multiple coactivator complexes. Curr. Opin. Genet. Dev. 9:132-139. [DOI] [PubMed] [Google Scholar]
- 13.Haynes, S. R., C. Dollard, F. Winston, S. Beck, J. Trowsdale, and I. B. Dawid. 1992. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 20:2603.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haynes, S. R., B. A. Mozer, D. N. Bhatia, and I. B. Dawid. 1989. The Drosophila fsh locus, a maternal effect homeotic gene, encodes apparent membrane proteins. Dev. Biol. 134:246-257. [DOI] [PubMed] [Google Scholar]
- 15.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Houzelstein, D., A. Cohen, M. E. Buckingham, and B. Robert. 1997. Insertional mutation of the mouse Msx1 homeobox gene by an nlacZ reporter gene. Mech. Dev. 65:123-133. [DOI] [PubMed] [Google Scholar]
- 17.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, Y. W., P. Veschambre, H. Erdjument-Bromage, P. Tempst, J. W. Conaway, R. C. Conaway, and R. D. Kornberg. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 95:8538-8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, M. H., M. Numata, and M. Shimane. 1997. Identification and characterization of BRDT: a testis-specific gene related to the bromodomain genes RING3 and Drosophila fsh. Genomics 45:529-534. [DOI] [PubMed] [Google Scholar]
- 20.Kasahara, M. 1999. Genome dynamics of the major histocompatibility complex: insights from genome paralogy. Immunogenetics 50:134-145. [DOI] [PubMed] [Google Scholar]
- 21.Le Page, F., E. E. Kwoh, A. Avrutskaya, A. Gentil, S. A. Leadon, A. Sarasin, and P. K. Cooper. 2000. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell 101:159-171. [DOI] [PubMed] [Google Scholar]
- 22.Lindahl, T., and R. D. Wood. 1999. Quality control by DNA repair. Science 286:1897-1905. [DOI] [PubMed] [Google Scholar]
- 23.Lygerou, Z., C. Conesa, P. Lesage, R. N. Swanson, A. Ruet, M. Carlson, A. Sentenac, and B. Seraphin. 1994. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res. 22:5332-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matangkasombut, O., R. M. Buratowski, N. W. Swilling, and S. Buratowski. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14:951-962. [PMC free article] [PubMed] [Google Scholar]
- 25.Myat, A., D. Henrique, D. Ish-Horowicz, and J. Lewis. 1996. A chick homologue of Serrate and its relationship with Notch and Delta homologues during central neurogenesis. Dev. Biol. 174:233-247. [DOI] [PubMed] [Google Scholar]
- 26.Myers, L. C., and R. D. Kornberg. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69:729-749. [DOI] [PubMed] [Google Scholar]
- 27.Nordqvist, K., and V. Tohonen. 1997. An mRNA differential display strategy for cloning genes expressed during mouse gonad development. Int. J. Dev. Biol. 41: 627-638. [PubMed] [Google Scholar]
- 28.Pamblanco, M., A. Poveda, R. Sendra, S. Rodriguez-Navarro, J. E. Perez-Ortin, and V. Tordera. 2001. Bromodomain factor 1 (Bdf1) protein interacts with histones. FEBS Lett. 496:31-35. [DOI] [PubMed] [Google Scholar]
- 29.Pieper, R. O. 1998. Cellular responses to methylation damage, p. 33-48. In J. A. Nickoloff and M. F. Hoekstra (ed.), DNA damage and repair, DNA repair in higher eukaryotes, vol. II. Humana Press Inc., Totowa, N.J. [Google Scholar]
- 30.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267-271. [PubMed] [Google Scholar]
- 32.Rhee, K., M. Brunori, V. Besset, R. Trousdale, and D. J. Wolgemuth. 1998. Expression and potential role of Fsrg1, a murine bromodomain-containing homologue of the Drosophila gene female sterile homeotic. J. Cell Sci. 111:3541-3550. [DOI] [PubMed] [Google Scholar]
- 33.Skarnes, W. C. 2000. Gene trapping methods for the identification and functional analysis of cell surface proteins in mice. Methods Enzymol. 328:592-615. [DOI] [PubMed] [Google Scholar]
- 34.Skarnes, W. C., J. E. Moss, S. M. Hurtley, and R. S. Beddington. 1995. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc. Natl. Acad. Sci. USA 92:6592-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorpe, K. L., S. Abdulla, J. Kaufman, J. Trowsdale, and S. Beck. 1996. Phylogeny and structure of the RING3 gene. Immunogenetics 44:391-396. [DOI] [PubMed] [Google Scholar]
- 37.Thorpe, K. L., P. Gorman, C. Thomas, D. Sheer, J. Trowsdale, and S. Beck. 1997. Chromosomal localization, gene structure and transcription pattern of the ORFX gene, a homologue of the MHC-linked RING3 gene. Gene 200:177-183. [DOI] [PubMed] [Google Scholar]
- 38.Tudor, M., P. J. Murray, C. Onufryk, R. Jaenisch, and R. A. Young. 1999. Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev. 13:2365-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winston, F., and C. D. Allis. 1999. The bromodomain: a chromatin-targeting module? Nat. Struct. Biol. 6:601-604. [DOI] [PubMed] [Google Scholar]