Abstract
CK2 is a highly conserved protein kinase with growth-promoting and oncogenic properties. It is known to activate RNA polymerase III (PolIII) transcription in Saccharomyces cerevisiae and is shown here to also exert a potent effect on PolIII in mammalian cells. Peptide and chemical inhibitors of CK2 block PolIII transcription in human cell extracts. Furthermore, PolIII transcription in mammalian fibroblasts is decreased significantly when CK2 activity is compromised by chemical inhibitors, antisense oligonucleotides, or kinase-inactive mutants. Coimmunoprecipitation and cofractionation show that endogenous human CK2 associates stably and specifically with the TATA-binding protein-containing factor TFIIIB, which brings PolIII to the initiation site of all class III genes. Serum stimulates TFIIIB phosphorylation in vivo, an effect that is diminished by inhibitors of CK2. Binding to TFIIIC2 recruits TFIIIB to most PolIII promoters; this interaction is compromised specifically by CK2 inhibitors. The data suggest that CK2 stimulates PolIII transcription by binding and phosphorylating TFIIIB and facilitating its recruitment by TFIIIC2. CK2 also activates PolI transcription in mammals and may therefore provide a mechanism to coregulate the output of PolI and PolIII. CK2 provides a rare example of an endogenous activity that operates on the PolIII system in both mammals and yeasts. Such evolutionary conservation suggests that this control may be of fundamental importance.
Protein kinase CK2 (formerly known as casein kinase II) is ubiquitous and highly conserved in eukaryotes (reviewed in references 1 and 29). It phosphorylates proteins on serine and threonine in both the nucleus and the cytoplasm. CK2 exists as a tetramer, composed of two isozymic catalytic subunits, α and α′, and two copies of a regulatory β subunit or one copy each of β and the closely related β′. The CK2α and CK2α′ subunits are nearly 90% identical and can compensate for each other, but there is also some functional specialization (57, 69). The β subunits allow optimal kinase activity and can regulate substrate specificity; they form a stable dimer linking the two catalytic subunits, which do not contact each other (35).
Although its signaling function has long remained obscure, CK2 has been shown recently to form part of the Wnt pathway in both Drosophila and mammals (54, 65). Many studies have found that increases in the level and/or activity of CK2 are associated with cell growth and proliferation (for example, references 3, 4, 7, 25, 30, 34, and 39). Thus, CK2 expression can be increased by mitogens (7, 39), and CK2 is most abundant in cells with high mitotic activity, such as transformed cells and normal colorectal mucosa (34). Indeed, microinjection of CK2 can induce immediate-early gene expression in the absence of growth factors (11). Conversely, inactivation of CK2 by specific antibodies or antisense oligonucleotides can arrest the proliferation of primary human fibroblasts (41, 42). Similarly, cell cycle progression is blocked in Saccharomyces cerevisiae when temperature-sensitive CK2 mutants are cultured at the nonpermissive temperature (17). Inactivation of CK2 is also lethal in Schizosaccharomyces pombe and Caenorhabitis elegans (10, 53).
A growth-promoting role for CK2 is consistent with reports that link it with tumorigenesis. One of the first came from analysis of theileriosis, a bovine leukemia-like condition caused by the protozoan Theileria parva. This parasite infects and transforms lymphocytes, causing them to proliferate out of control, eventually killing the infected animal due to overwhelming lymphocytosis (37). The infected cells show a marked and specific elevation of CK2 activity, but tyrosine kinases and other signaling pathways are not deregulated (38). CK2 is abnormally active in a variety of human cancers, including leukemias and solid tumors (9, 34, 36). Both of the CK2 catalytic subunits were found to cooperate with Ras in transforming primary rat fibroblasts (39), although the converse result was reported for CK2α in NIH 3T3 cells (19). Direct evidence that CK2 has oncogenic activity comes from transgenic mice overexpressing the α subunit, which develop lymphomas from 6 months of age with an incidence of 15 to 20% per year (49). The latency of onset and the monoclonality of the lymphomas indicate that other mutations are required for transformation in a multistep process. However, combining the CK2α transgene with a Myc transgene results in polyclonal neonatal leukemia, suggesting that deregulation of these two genes can be sufficient to transform lymphoid cells (67). Similarly, leukemogenesis is accelerated dramatically by transgenic coexpression of CK2α and tal-1 (24). A CK2α transgene also accelerates lymphoma development in p53-deficient transgenic mice (26, 67).
A variety of substrates have been identified for CK2, including several oncogene products (31). In S. cerevisiae, CK2 has been shown to stimulate the synthesis of tRNA and 5S rRNA by RNA polymerase III (PolIII) (20). This activity may contribute to its growth-promoting activity, since a high rate of tRNA and rRNA production is associated with rapid growth. When a yeast strain carrying a temperature-sensitive version of CK2α′ is shifted to the nonpermissive temperature, both growth and PolIII transcription are inhibited specifically (20). The transcriptional defect can be reproduced in vitro by using extracts prepared from the mutant strain (14, 15, 20). The addition of the PolIII-specific factor TFIIIB is sufficient to restore transcription in CK2α′ mutant cell extracts (14). This effect is abolished when TFIIIB is pretreated with phosphatase (14). These observations led to the proposal that CK2 stimulates PolIII transcription in yeast cells by phosphorylating and activating TFIIIB (14). In support of this proposal, a population of CK2 molecules cofractionates and coimmunoprecipitates with yeast TFIIIB (15).
TFIIIB is a complex composed of the TATA-binding protein (TBP) and the two associated subunits, B" and BRF (reviewed in references 12, 40, and 59). It plays an essential role in PolIII transcription, by recruiting the polymerase and positioning it over the initiation site (23). The β subunit of Saccharomyces CK2 binds to TBP (15). Furthermore, CK2 phosphorylates yeast TBP efficiently and enhances its ability to stimulate transcription in a CK2α′ mutant cell extract (14, 15). Ghavidel and Schultz concluded that CK2 regulates PolIII activity by phosphorylating the TBP subunit of TFIIIB (14). Recruitment of TFIIIB to a tRNA gene promoter is deficient in an extract from CK2α′ mutant cells (15). Promoter association is also severely impaired following phosphatase treatment of TFIIIB (15), suggesting that phosphorylation by CK2 in yeast cells stimulates TFIIIB assembly into a transcription complex.
We demonstrate that CK2 is also required for active mammalian PolIII transcription. Inhibiting human CK2 specifically compromises the binding of TFIIIB to the assembly factor TFIIIC2, an interaction which is necessary to bring TFIIIB onto most PolIII templates. This scenario can explain the observation with yeast cells that CK2 is required for promoter recruitment of TFIIIB. We also demonstrate that human CK2 interacts stably with TFIIIB. We provide the first evidence that BRF is phosphorylated in cells and show that CK2 inhibitors can decrease this phosphorylation. The data suggest that CK2 plays a major role in stimulating the synthesis of PolIII products in mammals by binding and phosphorylating TFIIIB, thereby promoting transcription complex assembly. These data provide a rare example of a transcriptional control mechanism that operates on the PolIII system in both yeast and mammals. The fact that it has been conserved through evolution argues strongly for its functional importance. PolIII is responsible for about 10% of all nuclear transcription, including the synthesis of tRNA and 5S rRNA; through its potent effect on PolIII activity, CK2 is likely to have a very major impact on the biosynthetic capacity of cells. This scenario may help explain the oncogenic properties of CK2 in mammalian systems.
MATERIALS AND METHODS
Cell culture, transfection, and labeling.
A31 and Rat1A are mouse and rat fibroblast cell lines, respectively, which were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal calf serum (FCS) and antibiotics. IMR-90 cells are human diploid fibroblasts which were grown in McCoy's medium supplemented with 20% FCS and antibiotics. RS2.31, RS3.22, GV7.21, and GV13.35 are derivatives of the human osteosarcoma line U2OS which stably express combinations of CK2 subunits; their production and culturing have been described elsewhere (57).
CHO cells were cultured in Ham's F-12 medium supplemented with 10% heat-inactivated FCS and antibiotics. Hemagglutinin (HA)-tagged BRF was introduced by transient transfection of pcDNA3HA.BRF with Lipofectamine 48 h prior to labeling. Labeling was carried out with 0.25 mCi of [32P]orthophosphate/ml in phosphate-free medium for 3 h. After incubation, cells were washed twice in ice-cold phosphate-buffered saline and then solubilized in 0.5 ml of IP buffer (50 mM HEPES [pH 7.5], 5 mM EDTA, 10 mM sodium phosphate, 10 mM NaF, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 0.5 mM phenylmethylsulfonyl fluoride, 0.5 μg of leupeptin/ml, 0.7 μg of pepstatin/ml, 0.5 μg of aprotinin/ml, 40 μg of bestatin/ml, 1 mM sodium vanadate, 50 mM β-glycerophosphate). After 60 min on a rotating wheel, insoluble material was removed by centrifugation at 14,000 × g for 15 min prior to immunoprecipitation.
Northern blotting.
Total cellular RNA was extracted with TRI reagent (Sigma) according to the manufacturer's instructions. Agarose gel electrophoresis, Northern transfer, and hybridization were carried out as previously described (6). The B2 gene and acidic ribosomal phosphoprotein P0 (ARPP P0) gene probes have been described elsewhere (47).
Cell extraction and fractionation.
HeLa cell nuclear extracts were purchased from the Computer Cell Culture Center (Mons, Belgium). Whole-cell extracts were prepared from U2OS derivatives by a previously described method (61). Phosphocellulose columns were run as previously described; PC-B is a 0.1 to 0.35 M KCl step fraction containing TFIIIB and PolIII, and PC-C is a 0.35 to 0.6 M KCl step fraction containing TFIIIC and PolIII (48). Bacterially expressed recombinant CK2 was obtained from New England Biolabs.
Transcription assays.
Transcription reactions were carried out as previously described (62, 63), except that pBR322 was not included and the incubations were done for 60 min at 30°C. The pVA1 and pLeu template plasmids contain the adenovirus VA1 gene and a human tRNALeu gene, respectively (61).
Antisense treatment and RT-PCR.
Phosphorothioate oligonucleotides (sense, 5′-GACGTGAAGATGAGCAGCTC-3′; antisense, 5′-GAGCTGCTCATCTTCACGTC-3′) were incubated with cells for 20 h at 100 μg/ml prior to harvesting of RNA. Reverse transcription (RT)-PCR was carried out as previously described (66).
Antibodies and Western blotting.
Peptide antisera 128 and 330 against BRF and antiserum 4286 against TFIIICβ were described and characterized previously (2, 6, 28, 55). Antiserum against the BN51 subunit of PdIII was generously provided by Michael Ittmann (22). CK2β antibody C40420 was purchased from Affiniti Research Products. Retinoblastoma protein (RB) antibody C-15, CK2α antibody H-286, HA tag antibody F-7, Myc antibody 9E10, and TAFI48 antibody M-19 were obtained from Santa Cruz Biotechnology. Western immunoblotting was performed as previously described (61).
Immunoprecipitation.
Whole-cell extract (150 μg) was incubated at 4°C on an orbital shaker with 20 μl of protein A-Sepharose beads carrying equivalent amounts of prebound immunoglobulin G. Samples were then pelleted, supernatants were removed, and the beads were washed five times with 150 μl of LDB buffer. The bound material was analyzed by Western blotting. Reticulocyte lysate (Promega) was used to synthesize BRF in the presence of [35S]Met and [35S]Cys, according to the manufacturer's specifications, and then was mixed with nuclear extract prior to immunoprecipitation (see Fig. 8A). In this instance, the precipitated material was analyzed by autoradiography rather than Western blotting.
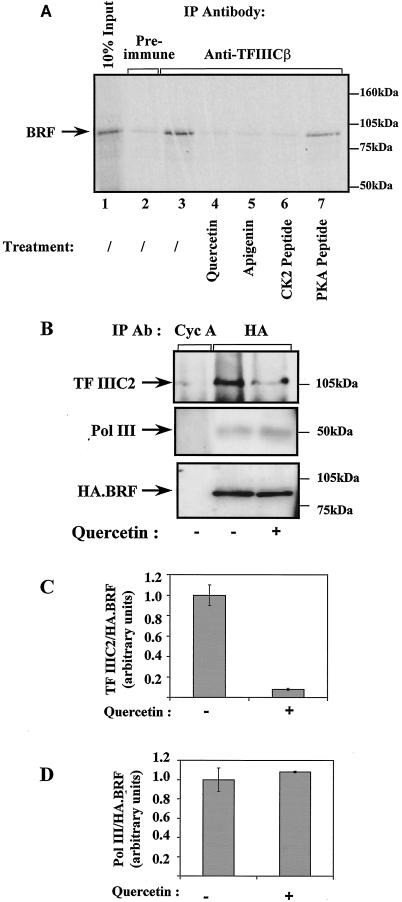
FIG. 8.
CK2 inhibitors specifically compromise the binding of TFIIIB to TFIIIC2. (A) A reticulocyte lysate (10 μl) containing in vitro-translated BRF was mixed with a HeLa cell extract (150 μg) in the presence of buffer (lanes 2 and 3), 100 μM quercetin (lane 4), 80 μM apigenin (lane 5), 40 μg of CK2 phosphoacceptor peptide (lane 6), or 40 μg of PKA phosphoacceptor peptide (control) (lane 7). The mixture was then immunoprecipitated (IP) with anti-TFIIICβ antiserum 4286 (lanes 3 to 7) or the corresponding preimmune serum (lane 2). Proteins retained after extensive washing were resolved by SDS-polyacrylamide gel electrophoresis and visualized by autoradiography. Lane 1 shows 10% of the input reticulocyte lysate. (B) Rat1A cells stably transfected with pcDNA3HA.BRF were lysed and immu-noprecipitated with anti-cyclin A (Cyc A) antibody (Ab) BF683 (lane 1) or anti-HA antibody F-7 (lanes 2 and 3). Immunoprecipitated material was resolved by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting with antiserum 4286 against TFIIIC2 (top), anti-BN51 antiserum against PolIII (middle), and antibody F-7 against the HA tag on transfected BRF (bottom). (C) Coprecipitated TFIIIC2 was quantitated by densitometry and normalized against the amount of immunoprecipitated HA-BRF. Nonspecific binding to control antibody was subtracted. The value obtained for vehicle-treated cells was arbitrarily assigned as 1.0. Values are the means of three experiments; error bars indicate standard deviations. (D) As for panel C, except that coprecipitated PolIII was quantitated instead of TFIIIC2.
When immunoprecipitation was carried out after transfection, Rat1A cells were washed with phosphate-buffered saline and then scraped into microextraction buffer (20 mM HEPES [pH 7.8], 450 mM NaCl, 50 mM NaF, 25% glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 μg of leupeptin/ml, 0.7 μg of pepstatin/ml, 1.0 μg of trypsin inhibitor/ml, 0.5 μg of aprotinin/ml, 40 μg of bestatin/ml) containing 1 mM sodium vanadate, 50 mM β-glycerophosphate, and 0.5% Triton X-100. After incubation on ice for 15 min, the lysates were cleared by centrifugation at 4°C for 10 min in a microcentrifuge and then used for immunoprecipitation.
RESULTS
CK2 inhibitors abolish PolIII transcription in human cell extracts.
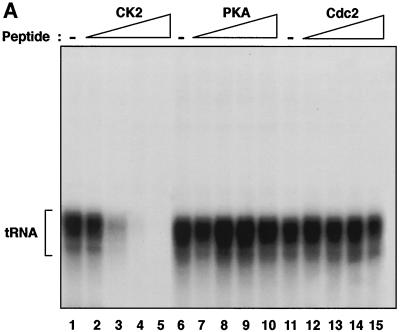
To begin to investigate whether CK2 regulates mammalian PolIII transcription, we tested whether blocking CK2 activity could influence the level of tRNA synthesis by using fractionated HeLa cell extracts. For this, we used a substrate peptide carrying a consensus CK2 phosphorylation site which has been shown to act as a competitive inhibitor (58). We confirmed that recombinant human CK2 phosphorylates this peptide efficiently, without recognizing two control peptides containing phosphoacceptor sites of alternative serine/threonine kinases (data not shown). As further evidence of specificity, we confirmed that the CK2 substrate peptide is not recognized by recombinant glycogen synthase kinase 3 (data not shown). PolIII transcription was found to decrease by up to 90-fold after preincubation of HeLa cell fractions with the CK2 phosphoacceptor peptide (Fig. 1A). This effect was highly specific, since expression was virtually unaffected by equal amounts of peptides bearing consensus phosphoacceptor sites for Cdc2 or protein kinase A (PKA). Efficient and specific PolIII repression was also observed when the CK2 peptide was used with a VA1 gene template and unfractionated HeLa cell extracts (Fig. 1B). As well as the lack of effect with PKA or Cdc2 peptides, specificity was also indicated by the fact that repression with the CK2 peptide could be reversed by using recombinant CK2 (Fig. 1B, lanes 3 to 5). Although blocking endogenous CK2 activity clearly inhibited PolIII transcription, little or no stimulation was observed when recombinant CK2 was added to extracts in the absence of the peptide (Fig. 1B, lanes 1 to 3). This result can be explained if the endogenous CK2 activity is already saturating. The data suggest that HeLa cell extracts contain an excess of CK2 that contributes significantly to the level of PolIII transcription in vitro.
FIG. 1.
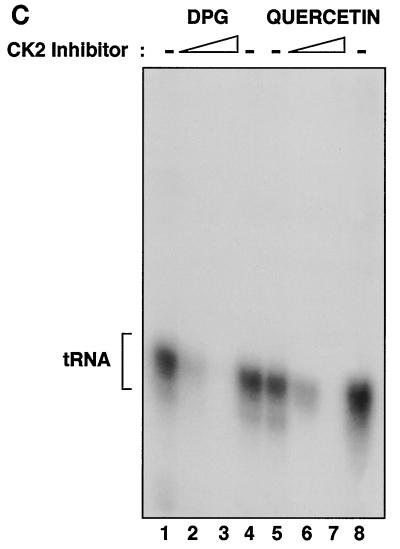
Human PolIII transcription is repressed specifically by peptide and chemical inhibitors of CK2. (A) Transcription using HeLa cell PC-B (2 μg) and PC-C (0.7 μg) fractions and a pLeu template (250 ng) after preincubation for 15 min at 30°C with buffer (lanes 1, 6, and 11) or 10, 20, 30, or 40 μg of CK2 phosphoacceptor peptide (RRREEETEEE; lanes 2 to 5, respectively), PKA phosphoacceptor peptide (LRRASLG; lanes 7 to 10, respectively), or Cdc2 phosphoacceptor peptide (RRRPMSPKKKA; lanes 12 to 15, respectively). (B) Transcription using a HeLa cell nuclear extract (15 μg) and a pVA1 template (250 ng) after preincubation for 15 min at 30°C with buffer (lanes 1, 3, and 6) or 30 μg of CK2 phosphoacceptor peptide (lanes 4 and 5) or PKA phosphoacceptor peptide (lanes 7 and 8). Reaction mixtures also contained 1 μl of recombinant CK2 (lanes 2, 5, and 8) or the corresponding buffer. (C) Transcription using a HeLa cell nuclear extract (15 μg) and a pLeu template (250 ng) after preincubation for 15 min at 30°C with buffer (lanes 1, 4, 5, and 8), with 6 and 12 mM 2,3-diphosphoglycerate (lanes 2 and 3, respectively), or with 50 and 100 μM quercetin (lanes 6 and 7, respectively).
As an alternative to using peptides, we also examined the effect of several well-characterized chemical reagents that are known to inhibit CK2. Transcription of a human tRNALeu gene was severely repressed by 2,3-diphosphoglycerate (DPG) and quercetin (Fig. 1C). This response was dose dependent and was observed at concentrations that have been shown to inhibit CK2 phosphorylation of established natural substrates, including yeast TFIIIB (20, 56). PolIII transcription of tRNAPro, B2, and 5S rRNA genes was also repressed by DPG and quercetin (data not shown). A third compound that inhibits CK2 activity is the ATP analogue 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (52). Like DPG and quercetin, DRB was found to reduce the rate of PolIII transcription in a HeLa cell extract (data not shown). Again, the response was dose dependent and occurred within a micromolar concentration range that is known to be effective against CK2 (52). Indeed, we confirmed that human CK2 activity was inhibited by these chemicals at the concentrations used in the transcription assays (data not shown). The selective CK2 inhibitor apigenin (54) also suppressed transcription by PolIII (data not shown). We conclude that the expression of class III genes with human factors in vitro can be influenced strongly by four unrelated compounds that function as inhibitors of CK2 kinase. Indeed, very little expression is observed under conditions in which the kinase is inhibited.
CK2 inhibitors reduce the expression of class III genes in mammalian fibroblasts.
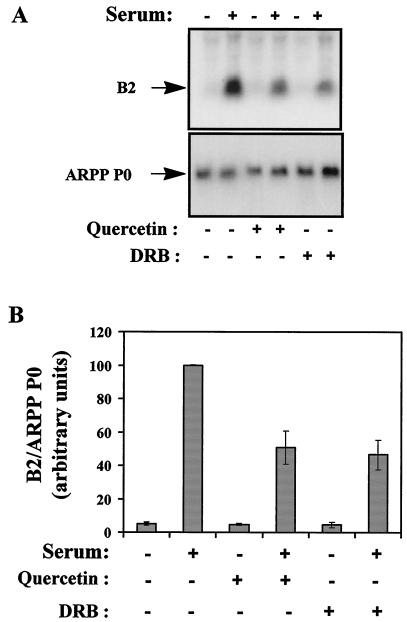
Low doses of quercetin and DRB were also applied to living cells to test for an effect on PolIII activity in vivo. The addition of 10 μM quercetin to proliferating murine fibroblasts caused a marked decrease in the expression of PolIII transcripts from the B2 middle repetitive gene family (Fig. 2A, top). This effect was specific, since there was little or no change in the expression of a control PolII transcript encoding ARPP P0 (Fig. 2A, bottom). Very similar results were obtained with 10 μM DRB, suggesting that these inhibitors influence PolIII activity through the same molecular mechanism. After normalization to the ARPP P0 control, B2 RNA levels in proliferating cells were found to drop by 2.3-fold in response to quercetin and by 2.2-fold in response to DRB (Fig. 2B). In contrast, neither compound made more than a 12% difference in the low basal level of B2 expression detected in resting cells. Measurements of thymidine incorporation into newly synthesized DNA showed that the low doses of inhibitors used in these experiments were insufficient to cause substantial cell cycle arrest when added to proliferating fibroblasts (data not shown). Additional experiments extended these observations to include HeLa cells, tRNA genes, and the CK2 inhibitor apigenin (data not shown).
FIG. 2.
Chemical inhibitors of CK2 reduce the expression of PolIII transcripts in proliferating murine fibroblasts. (A) Northern blot analysis of total RNA (10 μg) from A31 cells cultured in 0.5% serum (lanes 1, 3, and 5) or 20% serum (lanes 2, 4, and 6) and treated with 10 μM quercetin (lanes 3 and 4) or 10 μM DRB (lanes 5 and 6). The upper panel shows the blot probed with a B2 gene; the lower panel shows the same blot that has been stripped and reprobed with an ARPP P0 gene. (B) The B2 signals were quantified and normalized against the ARPP P0 signals. Means and standard deviations from three independent experiments are represented graphically.
Overexpression of kinase-dead CK2 mutants reduces PolIII transcription.
Vilk and coworkers constructed a series of stable human osteosarcoma cell lines which overexpress, in a tetracycline-regulatable manner, CK2β along with either CK2α or CK2α′; in addition, they made corresponding lines in which the catalytic subunits carry an inactivating substitution (57). We prepared extracts from these cells grown in the presence or absence of tetracycline. Western blotting confirmed that in each instance, tetracycline induced the regulatory β subunit and the wild-type or mutant α or α′ subunit (data not shown). Transcription assays carried out with these extracts showed that overexpressing wild-type CK2 made little difference to VA1 gene transcription by PolIII (Fig. 3, lanes 1 to 4). This result may reflect the fact that these tumor cells have high endogenous CK2 levels even without induction of the transfected constructs (57). However, induction of the kinase-dead mutants resulted in a significant reduction in VA1 gene transcriptional activity (Fig. 3, lanes 5 to 8). This result probably reflects interference of the mutants with the activity of the endogenous kinase. These data provide further evidence that mammalian PolIII transcription is sensitive to CK2 kinase activity. Overproduction of the CK2α′ mutant causes a significant decrease in the rate of cell proliferation (57), an effect which may contribute to the reduction in PolIII activity. However, cells overexpressing the CK2α mutant proliferate normally (57), a fact which rules out the possibility of an indirect growth effect influencing PolIII transcription in this situation. The observed reduction in class III gene transcription might be expected to slow cell growth and proliferation; however, parental osteosarcoma cells have abnormally elevated levels of PolIII activity, which may allow them to tolerate the reduction due to the CK2α mutant without it becoming limiting for growth.
FIG. 3.
Overexpression of kinase-inactive CK2 mutants reduces PolIII transcriptional activity. Transcription was analyzed with the pVA1 template (250 ng) and extracts (20 μg) of RS2.31 (lanes 1 and 2), RS3.22 (lanes 3 and 4), GV7.21 (lanes 5 and 6), and GV13.35 (lanes 7 and 8) cells grown in the presence (even-numbered lanes) or absence (odd-numbered lanes) of tetracycline (Tet). The CK2 catalytic subunit that is overexpressed in each sample is indicated; in each sample, CK2β is overexpressed in parallel. mut, mutant.
Depletion of CK2β from human cells reduces primary tRNA transcript levels.
The data above provide several independent lines of evidence that CK2 has a substantial influence on the mammalian PolIII machinery. Since each of these approaches involved manipulating kinase activity, it implicated the catalytic subunits of CK2 but did not address whether the regulatory β subunit also influences PolIII. To this end, we adopted a published antisense approach to target CK2β specifically in vivo (42, 49). An antisense oligonucleotide directed against mRNA encoding CK2β depleted this subunit from IMR-90 human diploid fibroblasts, whereas the complementary sense oligonucleotide had little effect (Fig. 4, right). Although the antisense depletion was incomplete, it exceeded the modest decrease in CK2β observed after serum starvation. RNA was harvested from these cells and used to produce cDNAs for RT-PCR. To assess PolIII activity, we used primers which hybridize to the introns within unprocessed tRNA precursors; since these short-lived primary transcripts are spliced very rapidly, their levels in a cell provide a reliable indication of ongoing transcription (66). The levels of the RT-PCR products were not diminished significantly when cells were treated with the sense oligonucleotide. In contrast, a substantial decrease was observed for both tRNAArg and tRNATyr following treatment with the antisense oligonucleotide (Fig. 4, left). These effects were specific, since there was no change in the level of glyceraldehyde phosphate dehydrogenase mRNA. The data suggest that the production of PolIII transcripts in vivo is sensitive to the level of CK2β.
FIG. 4.
Depletion of CK2β with an antisense oligonucleotide results in decreased tRNA synthesis in human fibroblasts. (A) PCR amplification was carried out with tRNAArg (top), tRNATyr (middle), or glyceraldehyde phosphate dehydrogenase (GAPDH) (bottom) primers and cDNAs prepared from RNAs extracted from IMR-90 cells treated with sense (lane 1), antisense (lane 2), or no (lanes 3 and 4) oligonucleotides and grown in the presence (lanes 1 to 3) or absence (lane 4) of serum. (B) Extracts (30 μg) prepared from IMR-90 cells treated with sense (lane 1), antisense (lane 2), or no (lanes 3 and 4) oligonucleotides and grown in the presence (lanes 1 to 3) or absence (lane 4) of serum were resolved by SDS-polyacrylamide gel electrophoresis and then blotted with CK2β antibody C40420.
Preassembled PolIII transcription complexes are resistant to CK2 inhibitors.
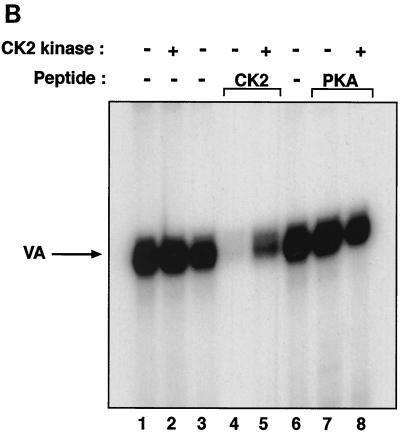
We used order-of-addition experiments to test whether CK2 acts before or after transcription complex assembly. When a HeLa cell extract was preincubated with peptide inhibitors, as in Fig. 1, the CK2 competitor caused a specific repression of VA1 expression that was not seen with the PKA control (Fig. 5, lanes 1 to 3). However, in parallel assays in which the extract was preincubated with template DNA to allow initiation complex assembly prior to peptide addition, transcription was no longer inhibited by the CK2 competitor (Fig. 5, lanes 4 to 6). Similarly, quercetin blocked VA1 expression very efficiently when added before the extract was mixed with the template but had only a slight effect when the complex was allowed to preassemble (Fig. 5, lanes 7 to 10). Indeed, quercetin was ∼11-fold less effective as an inhibitor when added to the preformed transcription complex. Identical results were obtained with DRB as an unrelated CK2 inhibitor or with a tRNA gene as an alternative template (data not shown). These observations suggest that CK2 acts specifically during assembly of the class III machinery; its inhibition appears not to affect the subsequent initiation, elongation, or termination steps of transcription. Furthermore, since our assay measures multiple rounds of transcription from stable complexes, the data suggest that polymerase recruitment is unaffected by blocking CK2.
FIG. 5.
Preassembled PolIII transcription complexes are resistant to CK2 inhibitors. Transcription was analyzed with a HeLa cell nuclear extract (15 μg) and the pVA1 template (250 ng) in the presence of buffer (lanes 1, 4, 7, and 9), 20 μg of CK2 phosphoacceptor peptide (lanes 2 and 5), 20 μg of PKA phosphoacceptor peptide (lanes 3 and 6), or 100 μM quercetin (lanes 8 and 10). Lanes 1 to 3, 7, and 8 show reactions in which the extract was preincubated for 15 min at 30°C with peptide, buffer, or quercetin prior to the addition of pVA1 and nucleotides. Lanes 4 to 6, 9, and 10 show reactions in which the extract was preincubated for 15 min at 30°C with pVA1 before peptide, buffer, or quercetin was added with the nucleotides.
Mammalian CK2 interacts stably with endogenous TFIIIB.
Initial experiments to test whether human CK2 associates with the PolIII machinery revealed the presence of CK2 kinase activity in TFIIIB fractions that had been affinity purified on antibody columns; in contrast, fractions containing immunoaffinity-purified TFIIIC2 showed only a low level of CK2 activity that was close to the background level found in mock-purified controls (data not shown). Since this result suggested that CK2 may interact stably with TFIIIB, we tested for cofractionation during gradient chromatography. When fractions containing TFIIIB were subjected to gradient chromatography on hydroxyapatite, heparin-Sepharose, or Mono Q columns, TFIIIB activity was found to coincide with a clear peak of CK2 activity (data not shown). These effects were specific, since TFIIIB did not copurify on these columns with PKA, glycogen synthase kinase 3, or DNA-dependent protein kinase (data not shown). Although it could be fortuitous, the consistent cofractionation of CK2 with TFIIIB is suggestive of a stable interaction between these proteins.
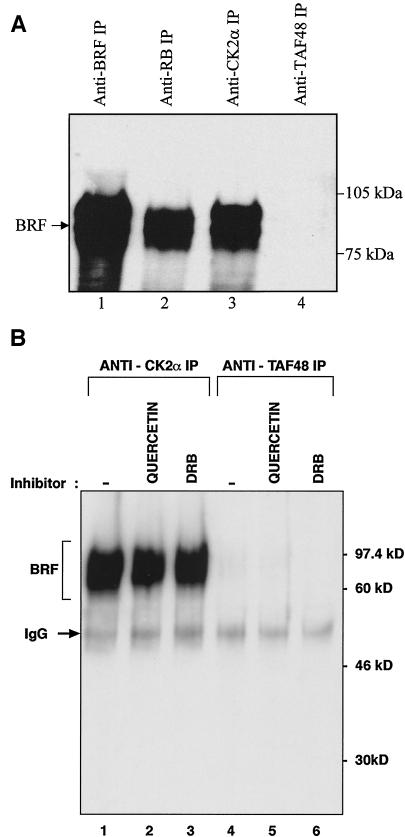
As an independent test of this association, immunoprecipitation experiments were carried out with a HeLa cell extract and an antibody against CK2α. As positive controls, we used an antibody against the BRF subunit of TFIIIB and an antibody against the RB protein, which is known to bind to TFIIIB (27). The negative control was an antibody against the TAFI48 subunit of the PolI-specific factor SL1/TIF-IB. Western blotting revealed that BRF was coimmunoprecipitated with CK2α and RB but not with the TAFI48 subunit antibody (Fig. 6A). This result was confirmed by using a second antibody raised against a different region of BRF (data not shown). The interaction between CK2α and TFIIIB is not dependent on the kinase activity of CK2, since it was undiminished by the presence of either quercetin or DRB (Fig. 6B). A stable association between TFIIIB and CK2 was also observed in the converse experiment, where an antibody against BRF was consistently found to coimmunoprecipitate CK2 activity from cell extracts (data not shown). These observations again indicate a stable and specific association between human TFIIIB and CK2.
FIG. 6.
Endogenous TFIIIB is coimmunoprecipitated from HeLa cells with endogenous CK2. (A) HeLa cell extract (150 μg) was immunoprecipitated (IP) with anti-BRF antibody 128 (lane 1), anti-RB antibody C-15 (lane 2), anti-CK2α antibody H-286 (lane 3), or anti-TAFI48 antibody M-19 (lane 4). Precipitates were resolved by SDS-polyacrylamide gel electrophoresis and then analyzed by Western blotting with anti-BRF antibody 330. (B) HeLa cell extract (150 μg) was immunoprecipitated in the presence of buffer (lanes 1 and 4), 100 μM quercetin (lanes 2 and 5), or 400 μM DRB (lanes 3 and 6) with anti-CK2α antibody H-286 (lanes 1 to 3) or anti-TAFI48 antibody M-19 (lanes 4 to 6). Precipitates were resolved by SDS-polyacrylamide gel electrophoresis and then blotted with anti-BRF antibody 128. IgG, immunoglobulin G.
CK2 phosphorylates the BRF subunit of TFIIIB.
The data indicate that CK2 binds stably to TFIIIB and that its kinase activity has a strong stimulatory effect on PolIII transcription both in vitro and in vivo. The most likely explanation is that CK2 phosphorylates one or more components of the PolIII machinery. Indeed, consensus CK2 phosphoacceptor sites are present in TFIIIC2 and all three subunits of TFIIIB. Furthermore, preliminary analyses showed that TBP, BRF, and B" can all be phosphorylated by purified recombinant CK2 in vitro (data not shown). Although TBP has already been shown to be a substrate for phosphorylation (14, 18, 61), there have been no reports that the TAF components of TFIIIB are also phosphorylated. We therefore investigated whether BRF is subject to phosphorylation in vivo by transfecting into CHO cells a vector encoding HA-tagged BRF. The transfected cells were cultured in the presence of high or low serum concentrations. They were labeled with [32P]orthophosphate, harvested, and subjected to immunoprecipitation with an anti-HA antibody. The precipitated material was resolved on a denaturing gel and subjected to both autoradiography and Western blotting. This experiment revealed that BRF was phosphorylated in vivo and that the level of phosphorylation increased ∼4-fold when the cells were stimulated to proliferate with serum (Fig. 7, lanes 1 and 2). However, when low doses of quercetin were used to reduce the activity of CK2 in these fibroblasts, BRF labeling was decreased by ∼56% in the proliferating cells (Fig. 7, lanes 4). In contrast, quercetin made only an ∼15% difference in the level of BRF phosphorylation in serum-starved cells (Fig. 7, lanes 3). Western blotting confirmed that equal amounts of BRF were immunoprecipitated in each instance. Similar results were obtained when DRB was used instead of quercetin to inhibit the activity of endogenous CK2 (data not shown). These findings suggest that CK2 contributes to the phosphorylation of BRF in vivo and that its contribution increases when cells proliferate.
FIG. 7.
BRF is phosphorylated in vivo. (A) CHO cells in 10% FCS were transiently transfected with pcDNA3HA.BRF (10 μg) and labeled 48 h later with [32P]orthophosphate for 3 h in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 20 μM quercetin. Cells in reactions 1 and 3 were transferred to FCS-free medium 24 h before labeling. Transfected BRF was immunoprecipitated with anti-HA antibody F-7, resolved on an SDS-7.8% polyacrylamide gel, transferred to nitrocellulose, and then visualized by autoradiography (top) and Western blotting with F-7 (bottom). (B) Phosphorylated BRF was quantified and normalized against total immunoprecipitated BRF. Means and standard deviations from three independent experiments are represented graphically.
CK2 promotes interactions between TFIIIB and TFIIIC2.
TFIIIB makes important functional contacts with both PolIII and TFIIIC2 (12, 40, 59). Coimmunoprecipitation experiments were carried out to test whether CK2 activity influences these interactions. The BRF subunit of TFIIIB was 35S labeled by translation in vitro and then mixed with a HeLa cell extract. In immunoprecipitation reactions carried out with antiserum against TFIIIC2, radiolabeled BRF was coprecipitated by virtue of the interaction between these proteins; this effect was specific, since BRF was not immunoprecipitated when the corresponding preimmune serum was used (Fig. 8A, lanes 2 and 3). The association of BRF with TFIIIC2 in this assay was not DNA mediated, as it was unaffected by the inclusion of ethidium bromide (data not shown). When quercetin was added to inhibit CK2 activity, BRF was no longer coprecipitated with TFIIIC2 (Fig. 8A, lane 4). In contrast, quercetin did not prevent the immunoprecipitation of BRF when anti-BRF antiserum was used (data not shown). The same effect was observed with the unrelated CK2 inhibitors apigenin and DRB (Fig. 8A, lane 5, and data not shown). Furthermore, a CK2 inhibitory peptide also blocked the coprecipitation of BRF with antiserum against TFIIIC2, whereas an equal amount of control PKA peptide did not (Fig. 8A, lanes 6 and 7). Similarly, quercetin and DRB could block the coimmunoprecipitation of TFIIIC2 with antibodies against TFIIIB (data not shown). These results suggest that phosphorylation by CK2 may be required for BRF to interact efficiently with TFIIIC2.
To find out whether similar effects occur in vivo, Rat1A cells were stably transfected with a vector encoding HA-tagged BRF. When an anti-HA antibody was used to immunoprecipitate lysates of these cells, both TFIIIC2 and PolIII were coprecipitated with HA-tagged BRF, as revealed by Western blotting (Fig. 8B, lane 2). This result was due to specific interactions, since little or none of these proteins was detected when an irrelevant antibody against cyclin A was used in a negative control immunoprecipitation (Fig. 8B, lane 1). Furthermore, the anti-HA antibody failed to coprecipitate TFIIIC2 or PolIII when cells were transfected with an empty vector encoding the HA tag without BRF attached (data not shown). When HA-tagged BRF-transfected cells were treated with quercetin in order to reduce CK2 activity, the amount of coprecipitated TFIIIC2 was decreased by fivefold (Fig. 8B and C). This effect was specific, since the interaction between HA-tagged BRF and PolIII was undiminished (Fig. 8B and D). The same behavior was seen when DRB was used instead of quercetin to inhibit CK2 (data not shown). The data suggest that CK2 activity specifically promotes the interaction between TFIIIB and TFIIIC2 in living mammalian cells.
DISCUSSION
Several independent lines of evidence indicate that CK2 activity is required for efficient transcription of many PolIII templates in mammals. Unrelated chemical inhibitors specifically diminish the expression of class III genes in proliferating fibroblasts and virtually abolish their transcription in vitro when used at concentrations that block CK2. Transcription by PolIII in human cell extracts can also be ablated specifically by a peptide inhibitor of CK2, an effect reversed by recombinant CK2. Overexpression in human osteosarcoma cells of kinase-dead mutants of CK2α or CK2α′ reduces PolIII transcriptional activity. Furthermore, antisense-mediated depletion of CK2β from human diploid fibroblasts decreases the level of primary tRNA transcripts. Additional evidence is provided by cofractionation and coimmunoprecipitation experiments, which reveal a stable and specific interaction between HeLa cell CK2 and TFIIIB. On the basis of these combined data, we suggest that transcription by mammalian PolIII is highly dependent on CK2. Clearly, this notion has not been established for all types of class III genes. However, it is likely to be a very general effect, since it has been found in every example tested (VA1, 5S rRNA, B2, and several tRNA genes) and involves the interaction between BRF and TFIIIC2, which is considered necessary for most PolIII transcription. Possible exceptions might be provided by the U6 and 7SK genes, which do not use either BRF or TFIIIC2 (12, 40, 46).
The sizes of PolIII transcripts do not change appreciably when CK2 is inhibited; this fact suggests that inactivation of the kinase does not interfere with selection of the sites of transcription initiation or termination. Indeed, primer extension revealed no change in start site selection following the addition of CK2 inhibitors (data not shown). We also found no evidence for effects on the elongation phase of transcription, as might be revealed by the appearance of short transcripts that result from premature pausing. These observations are consistent with the results of order-of-addition experiments, which showed that CK2 acts prior to initiation. We demonstrated that CK2 inhibitors compromise the interaction between TFIIIB and TFIIIC2 without affecting the association of TFIIIB with PolIII. We also found that blocking CK2 activity had no effect on DNA binding by TFIIIC2 (data not shown). Our data therefore implicate a single step in the transcription cycle that responds to CK2, namely, the recruitment of TFIIIB by DNA-bound TFIIIC2. Coimmunoprecipitation assays show that CK2 inhibitors severely compromise the binding of endogenous TFIIIC2 to BRF in vitro and in fibroblasts. This effect is very selective, since the binding of PolIII to BRF is not compromised by the same treatment. Studies with Saccharomyces have shown that the binding of BRF to TFIIIC is a rate-limiting step in PolIII transcription. Moir and colleagues and Rameau and colleagues demonstrated that transcription can be stimulated, both in vitro and in vivo, by mutations in TFIIIC131, the TFIIIC subunit that binds to BRF (33, 44). These mutations facilitate the recruitment of the latter by a complex mechanism which, in the case of the PCF1-1 allele, appears to involve a conformational change in TFIIIC131 that increases its affinity for BRF (32, 33; R. Moir and I. Willis, personal communication). The mutations cluster in a discrete region of TFIIIC131 that contains a tetratricopeptide repeat which is conserved in the human homologue of this subunit (21). Since BRF is also conserved through evolution (21), a similar rearrangement may occur in mammals. If so, it would be likely that such a rate-limiting step is targeted for regulation. Indeed, it was shown previously that the binding of TFIIIB to TFIIIC2 is subject to repression by the RB protein in mammalian cells and that this effect helps mediate cell cycle control (47, 55). The current study suggests that the same step in preinitiation complex assembly is also regulated in a positive fashion through phosphorylation by CK2. This effect appears to have a very substantial influence on the activity of the mammalian PolIII machinery.
Several investigators have provided unequivocal evidence that CK2 stimulates PolIII transcription in Saccharomyces (14, 15, 20). The fact that this role has been maintained through evolution is consistent with the very high level of phylogenetic conservation that is displayed by CK2. For example, mouse CK2α′ is 99 and 98% identical to the human and chicken proteins, respectively (39, 68). TFIIIB is the target for CK2 within the yeast PolIII machinery (14), reflecting a stable interaction between these proteins (15). Furthermore, CK2 function is necessary for yeast TFIIIB to be recruited effectively to a tRNA gene (15). These findings are all consistent with our observations of mammals; we found that CK2 associates with TFIIIB and stimulates its binding to TFIIIC2, the interaction responsible for recruiting TFIIIB to tRNA genes. However, within the Saccharomyces TFIIIB complex, only TBP is phosphorylated efficiently by purified CK2 in vitro, and Ghavidel and Schultz (14) concluded that the phosphorylation of TBP is likely to be responsible for the activation of TFIIIB by CK2. This conclusion is strongly supported by recent experiments with TBP mutants in vivo (15). It remains to be determined how this phosphorylation can influence TFIIIB recruitment or why the effect is specific to PolIII. In contrast, preliminary data suggest that all three subunits of human TFIIIB can be phosphorylated directly by CK2 in vitro. This notion is consistent with the presence of consensus CK2 phosphoacceptor sequences in human BRF and B", as well as TBP. Furthermore, we have provided the first evidence that BRF is subject to phosphorylation in cells. Endogenous CK2 is likely to be partly responsible, since in vivo labeling of BRF decreases by more than twofold when proliferating fibroblasts are treated with low doses of CK2 inhibitors. Since BRF binds directly to TFIIIC, it is easy to imagine how its phosphorylation might regulate the recruitment of TFIIIB. However, the phosphorylation of TBP or, indeed, B" may also influence complex assembly, directly or indirectly.
A previous study found that CK2 inhibits the ability of human La to stimulate RNA synthesis from isolated PolIII transcription complexes (8). However, our work indicates that CK2 promotes rather than inhibits human PolIII transcription, both in vitro and in vivo. The reason for this discrepancy is unclear. The stimulatory effect that we observed with CK2 is supported by the observations made in yeast studies (14, 15, 20). It is also more consistent with the role of CK2 in promoting proliferation (17, 41, 42), since repression of PolIII transcription would not be expected in situations of active growth.
PolIII transcription in mammals is subject to a wide variety of regulatory influences (reviewed in references 5 and 59). For example, it can respond strongly to cell differentiation (63) and displays marked cell cycle fluctuations, being repressed during mitosis and much of G1 and peaking during S and G2 (60). However, there has been little evidence to date for instances of PolIII control that cross the evolutionary divide. This fact seems surprising, since the polymerase and much of the basal machinery have been fairly well conserved (12, 21). A striking illustration is provided by growth arrest, which involves the repression of PolIII transcription both in mammals and in yeast. When logarithmically growing S. cerevisiae reaches stationary phase, there is a marked decrease in the level of the BRF subunit of TFIIIB (50). Even though BRF is conserved through evolution, its abundance is unaffected when mammalian fibroblasts arrest due to serum deprivation (47). Instead, genetic and biochemical evidence shows that the RB protein is primarily responsible for suppressing PolIII transcription in growth-arrested fibroblasts (47). Thus, very different molecular mechanisms are used by mammals and yeast to achieve the same end. In contrast, CK2 displays a potent capacity to activate PolIII transcription in both yeast and humans. The conservation of this control suggests that it may be of fundamental importance. Indeed, this notion may help explain why CK2 has been so well conserved through evolution. In light of this important precedent, we anticipate that some of the other regulatory mechanisms that operate on PolIII in S. cerevisiae may also prove to be relevant to humans. For example, the TOR kinases stimulate PolIII transcription in yeast (70), and we have evidence that this signaling pathway can also influence PolIII activity in mammals (data not shown). In S. cerevisiae, the PolIII enzyme is bound and repressed by Maf1p, a protein which displays strong phylogenetic conservation and so may well prove to function in a similar capacity in metazoans (43). Unveiling the controls that have been maintained through evolution is likely to provide important insights into the fundamentals of PolIII regulation.
Ghavidel and Schultz (15) recently showed that PolIII transcription is repressed when yeast is exposed to genotoxic stresses (UV or methane methylsulfonate). This result reflects a loss of TFIIIB activity due to the dissociation of CK2α′ (15). Substitution of a CK2 consensus site on the surface of TBP (S128) compromises the PolIII stress response, suggesting that TBP phosphorylation provides a major part of this control (15). Although a serine is found at the corresponding position of human TBP (S222), its surrounding residues do not match the CK2 consensus (13). It is possible that this site is nevertheless phosphorylated by CK2, but it is also plausible that alternative residues and/or polypeptides are functionally significant targets for human CK2. When bound to TFIIIB, CK2 may be ideally located to phosphorylate multiple proteins within the transcription complex. Indeed, CK2 phosphoacceptor sites are common, and all five subunits of TFIIIC2 are phosphorylated in human cells, although the kinases responsible have yet to be identified (51). The stimulatory effect of CK2 on the interaction between TFIIIB and TFIIIC2 may involve the phosphorylation of either or both of these factors. Further work is required to investigate these possibilities and to test whether CK2 allows mammalian PolIII to respond to DNA damage.
CK2 has also been found to stimulate rRNA synthesis by PolI both in mammals and in yeast (3, 15, 58). This finding may be of significance for its ability to promote proliferation, since a high cellular content of rRNA and ribosomes is a prerequisite for rapid growth (16, 45). As 5S rRNA is required in equimolar amounts with the large rRNA molecules, PolIII is usually coregulated with PolI (40). The fact that CK2 stimulates transcription by both PolI and PolIII suggests that this kinase may be involved in coordinating the activities of these two polymerase systems in mammalian cells, perhaps helping to balance the production of large rRNAs with that of 5S rRNA. Since PolI and PolIII together are responsible for ∼80% of nuclear transcription, such a role might make CK2 an important player in coordinating biosynthetic activity in mammals, perhaps helping to ensure that the supply of tRNA and rRNA is appropriate for the physiological status of the cells.
Most transformed cell lines and tumors have abnormally high levels of PolIII activity (5). Various mechanisms may contribute to this deregulation, but probably the most frequent is the loss of repression by the tumor suppressor RB, which binds TFIIIB and blocks its interactions with TFIIIC2 and PolIII (55). This function is compromised by mutations which arise naturally in cancers (5, 55, 64), by the binding of viral oncoproteins (28, 64), and by RB hyperphosphorylation due to overexpression of cyclin D1 or loss of p16 (47). The discovery that mammalian CK2 activates PolIII transcription suggests another potential mechanism that contributes to the overexpression of class III genes in certain types of malignancy. It is notable that the interaction between TFIIIB and TFIIIC2 is subject to opposing controls by the tumor suppressor RB and the putative oncoprotein CK2. The balance between these antagonistic forces will have a major impact on PolIII transcriptional activity and perhaps also on the growth potential of cells.
Acknowledgments
We thank David Litchfield for CK2-overexpressing cell lines, Debbie Johnson for BRF-overexpressing Rat1A cells, Michael Ittmann for antiserum against BN51, and Nouria Hernandez for constructs encoding human BRF and B".
This work was funded by project grant 17/C11067 to R.J.W. from the Biotechnology and Biological Sciences Research Council. P.H.S. is a Wellcome Trust Research Fellow, and R.J.W. is a Jenner Research Fellow of the Lister Institute of Preventive Medicine.
REFERENCES
- 1.Allende, J. E., and C. C. Allende. 1995. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 9:313-323. [DOI] [PubMed] [Google Scholar]
- 2.Alzuherri, H. M., and R. J. White. 1998. Regulation of a TATA-binding protein-associated factor during cellular differentiation. J. Biol. Chem. 273:17166-17171. [DOI] [PubMed] [Google Scholar]
- 3.Belenguer, P., V. Baldin, C. Mathieu, H. Prats, M. Bensaid, G. Bouche, and F. Amalric. 1989. Protein kinase NII and the regulation of rDNA transcription in mammalian cells. Nucleic Acids Res. 17:6625-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosc, D. G., B. Luscher, and D. W. Litchfield. 1999. Expression and regulation of protein kinase CK2 during the cell cycle. Mol. Cell. Biochem. 191:213-222. [PubMed] [Google Scholar]
- 5.Brown, T. R. P., P. H. Scott, T. Stein, A. G. Winter, and R. J. White. 2000. RNA polymerase III transcription: its control by tumor suppressors and its deregulation by transforming agents. Gene Expr. 9:15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns, C. A., and R. J. White. 1998. p53 is a general repressor of RNA polymerase III transcription. EMBO J. 17:3112-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll, D., and D. R. Marshak. 1989. Serum-stimulated cell growth causes oscillations in casein kinase II activity. J. Biol. Chem. 264:7345-7348. [PubMed] [Google Scholar]
- 8.Fan, H., A. L. Sakulich, J. L. Goodier, X. Zhang, J. Qin, and R. J. Maraia. 1997. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell 88:707-715. [DOI] [PubMed] [Google Scholar]
- 9.Faust, R. A., M. Gapany, P. Tristani, A. Davis, G. L. Adams, and K. Ahmed. 1996. Elevated protein kinase CK2 activity in chromatin of head and neck tumours: association with malignant transformation. Cancer Lett. 101:31-35. [DOI] [PubMed] [Google Scholar]
- 10.Fraser, A. G., R. S. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann, and J. Ahringer. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408:325-330. [DOI] [PubMed] [Google Scholar]
- 11.Gauthier-Rouviere, C., M. Basset, J.-M. Blanchard, J.-C. Cavadore, A. Fernandez, and N. J. C. Lamb. 1991. Casein kinase II induces c-fos expression via the serum response element pathway and p67SRF phosphorylation in living fibroblasts. EMBO J. 10:2921-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geiduschek, E. P., and G. A. Kassavetis. 2001. The RNA polymerase III transcription apparatus. J. Mol. Biol. 310:1-26. [DOI] [PubMed] [Google Scholar]
- 13.Ghavidel, A., D. J. Hockman, and M. C. Schultz. 1999. A review of progress towards elucidating the role of protein kinase CK2 in polymerase III transcription: regulation of the TATA binding protein. Mol. Cell. Biochem. 191:143-148. [PubMed] [Google Scholar]
- 14.Ghavidel, A., and M. C. Schultz. 1997. Casein kinase II regulation of yeast TFIIIB is mediated by the TATA-binding protein. Genes Dev. 11:2780-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghavidel, A., and M. C. Schultz. 2001. TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell 106:575-584. [DOI] [PubMed] [Google Scholar]
- 16.Grummt, I. 1999. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acids Res. Mol. Biol. 62:109-154. [DOI] [PubMed] [Google Scholar]
- 17.Hanna, D. E., A. Rethinaswamy, and C. V. C. Glover. 1995. Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae. J. Biol. Chem. 270:25905-25914. [DOI] [PubMed] [Google Scholar]
- 18.Heix, J., A. Vente, R. Voit, A. Budde, T. M. Michaelidis, and I. Grummt. 1998. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 17:7373-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heriche, J.-K., F. Lebrin, T. Rabilloud, D. Leroy, E. M. Chambaz, and Y. Goldberg. 1997. Regulation of protein phosphatase 2A by direct interaction with casein kinase 2α. Science 276:952-955. [DOI] [PubMed] [Google Scholar]
- 20.Hockman, D. J., and M. C. Schultz. 1996. Casein kinase II is required for efficient transcription by RNA polymerase III. Mol. Cell. Biol. 16:892-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, Y., and R. J. Maraia. 2001. Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res. 29:2675-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ittmann, M., J. Ali, A. Greco, and C. Basilico. 1993. The gene complementing a temperature-sensitive cell cycle mutant of BHK cells is the human homologue of the yeast RPC53 gene, which encodes a subunit of RNA polymerase C (III). Cell Growth Differ. 4:503-511. [PubMed] [Google Scholar]
- 23.Kassavetis, G. A., B. R. Braun, L. H. Nguyen, and E. P. Geiduschek. 1990. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 60:235-245. [DOI] [PubMed] [Google Scholar]
- 24.Kelliher, M. A., D. C. Seldin, and P. Leder. 1996. Tal-1 induces T cell acute lymphoblastic leukemia accelerated by casein kinase IIα. EMBO J. 15:5160-5166. [PMC free article] [PubMed] [Google Scholar]
- 25.Klarlund, J. K., and M. P. Czech. 1988. Insulin-like growth factor I and insulin rapidly increase casein kinase II activity in BALB/c 3T3 fibroblasts. J. Biol. Chem. 263:15872-15875. [PubMed] [Google Scholar]
- 26.Landesman-Bollag, E., P. L. Channavajhala, R. D. Cardiff, and D. C. Seldin. 1998. p53 deficiency and misexpression of protein kinase CK2α collaborate in the development of thymic lymphomas in mice. Oncogene 16:2965-2974. [DOI] [PubMed] [Google Scholar]
- 27.Larminie, C. G. C., C. A. Cairns, R. Mital, K. Martin, T. Kouzarides, S. P. Jackson, and R. J. White. 1997. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 16:2061-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larminie, C. G. C., J. E. Sutcliffe, K. Tosh, A. G. Winter, Z. A. Felton-Edkins, and R. J. White. 1999. Activation of RNA polymerase III transcription in cells transformed by simian virus 40. Mol. Cell. Biol. 19:4927-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Litchfield, D. W., and B. Luscher. 1993. Casein kinase II in signal transduction and cell cycle regulation. Mol. Cell. Biochem. 127/128:187-199. [DOI] [PubMed] [Google Scholar]
- 30.Marais, R. M., J. J. Hsuan, C. McGuigan, J. Wynne, and R. Treisman. 1992. Casein kinase II phosphorylation increases the rate of serum response factor-binding site exchange. EMBO J. 11:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meisner, H., and M. P. Czech. 1991. Phosphorylation of transcriptional factors and cell-cycle-dependent proteins by casein kinase II. Curr. Opin. Cell Biol. 3:474-483. [DOI] [PubMed] [Google Scholar]
- 32.Moir, R. D., K. V. Puglia, and I. M. Willis. 2000. Interactions between the tetratricopeptide repeat-containing transcription factor TFIIIC131 and its ligand, TFIIIB70. J. Biol. Chem. 275:26591-26598. [DOI] [PubMed] [Google Scholar]
- 33.Moir, R. D., I. Sethy-Coraci, K. Puglia, M. D. Librizzi, and I. M. Willis. 1997. A tetratricopeptide repeat mutation in yeast transcription factor IIIC131 (TFIIIC131) facilitates recruitment of TFIIB-related factor TFIIIB70. Mol. Cell. Biol. 17:7119-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munstermann, U., G. Fritz, G. Seitz, Y. P. Lu, H. R. Schneider, and O. G. Issinger. 1990. Casein kinase II is elevated in solid human tumours and rapidly proliferating nonneoplastic tissue. Eur. J. Biochem. 189:251-257. [DOI] [PubMed] [Google Scholar]
- 35.Niefind, K., B. Guerra, I. Ermakowa, and O.-G. Issinger. 2001. Crystal structure of human protein kinase CK2: insights into basic properties of the CK2 holoenzyme. EMBO J. 20:5320-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Notterman, D. A., U. Alon, A. J. Sierk, and A. J. Levine. 2001. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 61:3124-3130. [PubMed] [Google Scholar]
- 37.Ole-MoiYoi, O. K. 1989. Theileria parva: an intracellular protozoan parasite that induces reversible lymphocyte transformation. Exp. Parasitol. 69:204-210. [DOI] [PubMed] [Google Scholar]
- 38.Ole-MoiYoi, O. K., W. C. Brown, K. P. Iams, A. Nayar, T. Tsukamoto, and M. D. Macklin. 1993. Evidence for the induction of casein kinase II in bovine lymphocytes transformed by the intracellular protozoan parasite Theileria parva. EMBO J. 12:1621-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orlandini, M., F. Semplici, R. Ferruzzi, F. Meggio, L. A. Pinna, and S. Oliviero. 1998. Protein kinase CK2α′ is induced by serum as a delayed early gene and cooperates with Ha-ras in fibroblast transformation. J. Biol. Chem. 273:21291-21297. [DOI] [PubMed] [Google Scholar]
- 40.Paule, M. R., and R. J. White. 2000. Transcription by RNA polymerases I and III. Nucleic Acids Res. 28:1283-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pepperkok, R., P. Lorenz, W. Ansorge, and W. Pyerin. 1994. Casein kinase II is required for transition of G0/G1, early G1, and G1/S phases of the cell cycle. J. Biol. Chem. 269:6986-6991. [PubMed] [Google Scholar]
- 42.Pepperkok, R., P. Lorenz, R. Jakobi, W. Ansorge, and W. Pyerin. 1991. Cell growth stimulation by EGF: inhibition through antisense-oligodeoxynucleotides demonstrates important role of casein kinase II. Exp. Cell Res. 197:245-253. [DOI] [PubMed] [Google Scholar]
- 43.Pluta, K., O. Lefebvre, N. C. Martin, W. J. Smagowicz, D. R. Stanford, S. R. Ellis, A. K. Hopper, A. Sentenac, and M. Boguta. 2001. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:5031-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rameau, G., K. Puglia, A. Crowe, I. Sethy, and I. Willis. 1994. A mutation in the second largest subunit of TFIIIC increases a rate-limiting step in transcription by RNA polymerase III. Mol. Cell. Biol. 14:822-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeder, R. H. 1999. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog. Nucleic Acids Res. Mol. Biol. 62:293-327. [DOI] [PubMed] [Google Scholar]
- 46.Schramm, L., P. S. Pendergrast, Y. Sun, and N. Hernandez. 2000. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev. 14:2650-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott, P. H., C. A. Cairns, J. E. Sutcliffe, H. M. Alzuherri, A. Mclees, A. G. Winter, and R. J. White. 2001. Regulation of RNA polymerase III transcription during cell cycle entry. J. Biol. Chem. 276:1005-1014. [DOI] [PubMed] [Google Scholar]
- 48.Segall, J., T. Matsui, and R. G. Roeder. 1980. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J. Biol. Chem. 255:11986-11991. [PubMed] [Google Scholar]
- 49.Seldin, D. C., and P. Leder. 1995. Casein kinase IIα transgene-induced murine lymphoma: relation to theileriosis in cattle. Science 267:894-897. [DOI] [PubMed] [Google Scholar]
- 50.Sethy, I., R. D. Moir, M. Librizzi, and I. M. Willis. 1995. In vitro evidence for growth regulation of tRNA gene transcription in yeast. J. Biol. Chem. 270:28463-28470. [DOI] [PubMed] [Google Scholar]
- 51.Shen, Y., M. Igo, P. Yalamanchili, A. J. Berk, and A. Dasgupta. 1996. DNA binding domain and subunit interactions of transcription factor IIIC revealed by dissection with poliovirus 3C protease. Mol. Cell. Biol. 16:4163-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shugar, D. 1994. Development of inhibitors of protein kinases CK1 and CK2 and some related aspects, including donor and acceptor specificities and viral protein kinases. Cell. Mol. Biol. Res. 40:411-420. [PubMed] [Google Scholar]
- 53.Snell, V., and P. Nurse. 1994. Genetic analysis of cell morphogenesis in fission yeast—a role for casein kinase II in the establishment of polarized growth. EMBO J. 13:2066-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song, D. H., D. J. Sussman, and D. C. Seldin. 2000. Endogenous protein kinase CK2 participates in Wnt signaling in mammary epithelial cells. J. Biol. Chem. 275:23790-23797. [DOI] [PubMed] [Google Scholar]
- 55.Sutcliffe, J. E., T. R. P. Brown, S. J. Allison, P. H. Scott, and R. J. White. 2000. Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Mol. Cell. Biol. 20:9192-9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuazon, P. T., and J. A. Traugh. 1991. Casein kinase I and II--multipotential serine protein kinases: structure, function, and regulation. Adv. Second Messenger Phosphoprotein Res. 23:123-164. [PubMed] [Google Scholar]
- 57.Vilk, G., R. B. Saulnier, R. St. Pierre, and D. W. Litchfield. 1999. Inducible expression of protein kinase CK2 in mammalian cells. J. Biol. Chem. 274:14406-14414. [DOI] [PubMed] [Google Scholar]
- 58.Voit, R., A. Schnapp, A. Kuhn, H. Rosenbauer, P. Hirschmann, H. G. Stunnenberg, and I. Grummt. 1992. The nucleolar transcription factor mUBF is phosphorylated by casein kinase II in the C-terminal hyperacidic tail which is essential for transactivation. EMBO J. 11:2211-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White, R. J. 1998. RNA polymerase III transcription. Springer-Verlag KG, Berlin, Germany.
- 60.White, R. J., T. M. Gottlieb, C. S. Downes, and S. P. Jackson. 1995. Cell cycle regulation of RNA polymerase III transcription. Mol. Cell. Biol. 15:6653-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White, R. J., T. M. Gottlieb, C. S. Downes, and S. P. Jackson. 1995. Mitotic regulation of a TATA-binding-protein-containing complex. Mol. Cell. Biol. 15:1983-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White, R. J., and S. P. Jackson. 1992. Mechanism of TATA-binding protein recruitment to a TATA-less class III promoter. Cell 71:1041-1053. [DOI] [PubMed] [Google Scholar]
- 63.White, R. J., D. Stott, and P. W. J. Rigby. 1989. Regulation of RNA polymerase III transcription in response to F9 embryonal carcinoma stem cell differentiation. Cell 59:1081-1092. [DOI] [PubMed] [Google Scholar]
- 64.White, R. J., D. Trouche, K. Martin, S. P. Jackson, and T. Kouzarides. 1996. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature 382:88-90. [DOI] [PubMed] [Google Scholar]
- 65.Willert, K., M. Brink, A. Wodarz, H. Varmus, and R. Nusse. 1997. Casein kinase 2 associates with and phosphorylates Dishevelled. EMBO J. 16:3089-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winter, A. G., G. Sourvinos, S. J. Allison, K. Tosh, P. H. Scott, D. A. Spandidos, and R. J. White. 2000. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumours. Proc. Natl. Acad. Sci. USA 97:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu, X., E. Landesman-Bollag, P. L. Channavajhala, and D. C. Seldin. 1999. Murine protein kinase CK2: gene and oncogene. Mol. Cell. Biochem. 191:65-74. [PubMed] [Google Scholar]
- 68.Xu, X., E. S. Rich, Jr., and D. C. Seldin. 1998. Murine protein kinase CK2α′: cDNA and genomic cloning and chromosomal mapping. Genomics 48:79-86. [DOI] [PubMed] [Google Scholar]
- 69.Xu, X., P. A. Toselli, L. D. Russell, and D. C. Seldin. 1999. Globozoospermia in mice lacking the casein kinase II α′ catalytic subunit. Nat. Genet. 23:118-121. [DOI] [PubMed] [Google Scholar]
- 70.Zaragoza, D., A. Ghavidel, J. Heitman, and M. C. Schultz. 1998. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18:4463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]