Abstract
The cloning and functional characterization of a novel interferon regulatory factor (IRF), IRF-10, are described. IRF-10 is most closely related to IRF-4 but differs in both its constitutive and inducible expression. The expression of IRF-10 is inducible by interferons (IFNs) and by concanavalin A. In contrast to that of other IRFs, the inducible expression of IRF-10 is characterized by delayed kinetics and requires protein synthesis, suggesting a unique role in the later stages of an antiviral defense. Accordingly, IRF-10 is involved in the upregulation of two primary IFN-γ target genes (major histocompatibility complex [MHC] class I and guanylate-binding protein) and interferes with the induction of the type I IFN target gene for 2′,5′-oligo(A) synthetase. IRF-10 binds the interferon-stimulated response element site of the MHC class I promoter. In contrast to that of IRF-1, which has some of the same functional characteristics, the expression of IRF-10 is not cytotoxic for fibroblasts or B cells. The expression of IRF-10 is induced by the oncogene v-rel, the proto-oncogene c-rel, and IRF-4 in a tissue-specific manner. Moreover, v-Rel and IRF-4 synergistically cooperate in the induction of IRF-10 in fibroblasts. The level of IRF-10 induction in lymphoid cell lines by Rel proteins correlates with Rel transformation potential. These results suggest that IRF-10 plays a role in the late stages of an immune defense by regulating the expression some of the IFN-γ target genes in the absence of a cytotoxic effect. Furthermore, IRF-10 expression is regulated, at least in part, by members of the Rel/NF-κB and IRF families.
The interferon (IFN) regulatory factor (IRF) family is a group of transcription factors that have extensive homology in the DNA-binding domain (DBD) (36, 62). In mammals, nine family members have been identified: IRF-1 to -7, IRF-8 (IFN consensus sequence-binding protein), and IRF-9 (IFN-stimulated gene factor 3γ/p48). In avian species, homologues of IRF-1, IRF-2, IRF-4, and IRF-8 have been described (39, 48, 63). An additional chicken IRF, cIRF-3, has been cloned that has the highest level of homology to IRF-7 but is not an IRF-7 orthologue (29). IRF family members have also been described in fish and amphibians (37, 106). Moreover, viral IRF-encoding genes are present in the genomes of several rhadinoviruses, including human herpesvirus 8, the virus associated with Kaposi's sarcoma (10, 61, 70).
The major function of the IRF family of transcription factors is the regulation of immune responses, especially those directed against viral infection. The immune response consists of innate defenses and an adaptive defense (22). The innate response is the first line of defense against a virus infection and involves the production of IFNs and other cytokines, the activation of complement, and natural killer cells. These events, in turn, stimulate the adaptive immune response, which enables recognition of antigens with a high degree of structural specificity. IRF family members play a crucial role in both the innate and adaptive responses. In response to viral infection, IRF-3, -5, and -7 rapidly translocate to the nucleus and activate the transcription of type I IFNs and other cytokines (58, 108). Type I IFNs are produced by a variety of cells and bind cell surface receptors activating the induction of genes with direct antiviral activity. The induction of genes such as those for Mx, 2′,5′-oligo(A) synthetase (OAS), and guanylate-binding protein (GBP) may contribute to the establishment of the antiviral state in infected and neighboring cells (47). In addition, several IRFs are expressed directly in response to viral infection or type I IFN, resulting in amplification of the overall antiviral response and activation of the adaptive defense. The adaptive response is induced by antigens presented by major histocompatibility complex (MHC) class I or II molecules and leads to the clonal expansion of specific lymphoid populations. The expression of both MHC class antigens is also regulated, in part, by IRF transcription factors (13, 38, 104). Antigen-stimulated T cells and natural killer cells produce IFN-γ, which functions to coordinate the innate and adaptive immune responses (7). IFN-γ is an important inducer of IRF genes, MHC class genes, the gene for inducible nitric oxide synthetase, and other genes. Several IRFs are activated directly by antigen stimulation or by proliferation stimuli, which mimics activation by an antigen, such as concanavalin A (ConA) (62, 76). The targets of IRF transcription factors during the antiviral response are genes directly involved not only in virus elimination but also in differentiation, proliferation, and apoptosis of cells of the immune system. Disruption of this regulation leads to tumorigenesis, as documented by the association of cancer with failure to express IRF-1 or IRF-8, with mutations of IRF-2, and with the up-regulation of IRF-4 (43, 87, 102, 103).
The DBD of all IRFs, like that of c-myb, contains repeated tryptophan residues (62, 76). The DBD recognizes a GAAA sequence that is part of the motifs termed the IFN-stimulated response element (ISRE) and the IRF element (20). All IRFs, except IRF-1 and IRF-2, have an IRF association domain (IAD) that is responsible for interaction with other family members or transcription factors such as PU.1, E47, and STAT (78, 92, 93, 98). Another association domain (IAD2) that is present in IRF-1 and IRF-2 is important for their interaction with IRF-8 (68, 86). A nuclear localization signal has been identified in IRF-1, and similar sequences are present in other family members (39, 86). A bipartite nuclear retention signal located within the N terminus of the DBD has been identified in IRF-4, IRF-8, and IRF-9 (57). IRFs possess a transactivation domain in the middle of the protein (62).
The transcription factors of the Rel/NF-κB family share a conserved N-terminal Rel homology region that contains the DNA-binding and dimerization domains conferring DNA binding to κB sites (25, 26, 31). Most of the genes regulated by Rel/NF-κB are involved in immune, inflammatory, or stress responses (11, 31, 49). The avian and mammalian Rel/NF-κB family members are c-Rel, RelA, NF-κB1 (p50), NF-κB2 (p52), and RelB. Altered regulation of Rel/NF-κB activities is associated with oncogenesis (23, 28, 81). v-Rel, the naturally occurring, acutely oncogenic member of the Rel/NF-κB family, was formed as a result of a nonhomologous recombination event between the c-rel proto-oncogene and an avian retrovirus (27). v-Rel rapidly induces an invariably fatal lymphoma in avian species (8). v-rel transforms cells by inappropriately activating or repressing genes coding for cytokines, transcription factors, chaperons, receptors, inhibitors of apoptosis, and adhesion molecules, which are normally regulated by c-Rel and other Rel/NF-κB family members (40).
In this report, we describe the characterization of a novel IRF (IRF-10). IRF-10 is expressed principally in cells of hematopoietic origin, but its level of expression in bursal cells is very low. The inducible expression of IRF-10 by type I and II IFNs in fibroblasts and by ConA in T cells has delayed kinetics and is dependent on protein synthesis. The cells exogenously expressing IRF-10 have a prolonged induction of GBP expression after treatment with IFNs. IRF-10 induces the expression of MHC class I in fibroblasts. IRF-10 also binds to the ISRE site of the MHC class I promoter. The expression of IRF-10 is induced in bursal cell lines by v-Rel and c-Rel and in fibroblasts by IRF-4. v-Rel and IRF-4 synergistically cooperate in the induction of IRF-10 expression.
MATERIALS AND METHODS
Cloning of chicken IRF-10 cDNA.
A computer homology search for genes similar to IRF-4 identified a chicken expressed sequence tag (EST) clone, pat.pk0019.d2 (GenBank accession number A1980252), that encodes the C-terminal region of a novel IRF protein that we designated IRF-10. Clone pat.pk0019.d2 was isolated from a ConA-activated chicken splenic T-cell cDNA library as a part of the University of Delaware's chicken EST project (97). The 5′ sequence encoding the N terminus of IRF-10 was cloned by 5′ rapid amplification of cDNA ends (RACE). The first-strand cDNA was obtained by using Powerscript reverse transcriptase (Clontech Laboratories, Inc., Palo Alto, Calif.) from 5 μl of total RNA isolated from splenic lymphocytes activated for 48 h by ConA (50 μg/ml). SMART-RACE (Clontech) was performed with 2.5 μl of the first-strand substrate with a specific primer (5′-CCTCGTGGTATTTGCCCTTGTAGACGG-3′), and the PCR products were cloned into pGEM-T Easy (Promega, Madison, Wis.). The longest 5′ RACE clone, p8N2, contained a translation start site and the 5′ untranslated region of the IRF-10 cDNA. To verify that the partial cDNA sequences from p8N2 and pat.pk0019.d2 were from the same gene, the complete open reading frame (ORF) of IRF-10 was amplified with an Advantage-GC cDNA PCR kit (Clontech) and primers that flanked the ORF on both sides (5′-AGCCGGGATGGCGGAGCCGGGGTCTCCCAT-3′, 5′-CCAGGCTTCGCGTCCCCTCAGCTGCC-3′) and cloned into pGEM-T Easy. The plasmids designated pGE.IRF10-1 to -4 containing cloned PCR fragments were sequenced. Because the pat.pk0019.d2 cDNA clone has an incomplete 3′ untranslated region, the sequence of the 3′ terminus of the IRF-10 mRNA was determined from another University of Delaware EST clone (pnl-b.pk0007.e2), which contains a sequence overlapping the sequence of pat.pk0019.d2 in 482 nucleotides (nt).
Plasmids.
pTZ-IRF10 was constructed by cloning an EcoRI-AccI fragment of pGE.IRF10-4 containing the 5′ end of the IRF-10 ORF and the AccI-NotI fragment of pat.pk0019.d2 containing the 3′ region of the IRF-10 ORF into pTZ-A10 via a NotI-MluI oligonucleotide adaptor (5′-GGCCGCGGTCGACGA-3′, 5′-CGCGTCGTCGACCGC-3′). pTZ-A10 was derived by cloning an XhoI-EcoRI-AccI-MluI oligonucleotide adaptor (5′-AATTGCTCGAGGCGAATTCGAGTCTACGAGCGGCCGCACGCGT-3′, 5′-AGCTACGCGTGCGGCCGCTCGTAGACTCGAATTCGCCTCGAGC-3′) between the EcoRI and HindIII sites of pTZ-18R. pTZ-IRF4 was described previously (39). pTZ-IRF8 was created by cloning a 1,370-bp EcoRI-BamHI fragment of a chicken IRF-8 (IFN consensus sequence-binding protein) cDNA clone into pTZ-18R (48).
The REV-T-based retroviral vectors pREV-TW, pREV-C, pREV-CΔXba, pREV-IRF-4, and pREV-IRF-4ΔE6, which expressed v-rel, c-rel, c-relΔXba, IRF-4, and IRF-4ΔE6, were described previously (39, 42, 75). pREV-IRF-10, expressing IRF-10, was derived by cloning the XhoI-MluI fragment from pTZ-IRF10 into the XhoI and BssHII sites of retroviral vector pREV-0. pCSV11S3 contains an infectious genome of chicken syncytial virus (CSV) (21). pDS3+pREP is a replication-competent retroviral vector derived from a genomic clone of the Schmidt-Ruppin strain of Rous sarcoma virus (subgroup A) by deletion of v-src (14, 77). pDSv-Rel, derived from pDS3, was used for the expression of v-Rel in fibroblasts (54). pcDNAchIFN1 and pcDNAchIFN-γ were used for the production of IFN1 and IFN-γ in COS-1 cells as previously described (89, 94).
The pATH-I10#8 bacterial expression plasmid encodes a TrpE/IRF-10 fusion protein. This plasmid was created by cloning the PvuII-HindIII fragment of the IRF-10 PCR fragment between the SmaI and HindIII sites of pATH1 (52). This fragment was PCR amplified from cDNA prepared from ConA-stimulated spleen cells with IRF-10-specific primer IRF10AB-1 (5′-AGTCCCGGGCAGCTGGACATCTCCGAGCCTTAC-3′), containing an SmaI site, and primer IRF10AB-2 (5′-CTGAAGCTTCTAGGGGTACTCCTCACCGAAGCACAGGT-3′), containing a STOP codon and a HindIII site. The sequence fidelity of the IRF-10 fragment cloned into pATH-1 was verified by sequencing.
Chickens, cell lines, viruses, and tissue culture.
Embryonated eggs obtained from pathogen-free White Leghorn chickens were obtained from Hy-Vac, Adel, Iowa (SPF-SC strain), and Charles River SPAFAS, North Franklin, Conn. (SPAFAS strain). Chicken embryonic fibroblasts (CEF) were prepared from 10- or 11-day-old embryos of the SPF-SC strain. Cells were cultured with Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% newborn calf serum (Atlanta Biologicals, Norcross, Ga.), 5% chicken serum (Gibco BRL Life Technologies, Grand Island, N.Y.), 100 U of penicillin, and 50 μg of streptomycin per ml. Secondary cultures of CEF were used for transfection of retroviral plasmid DNA by a calcium phosphate method as described previously (39). Retroviruses were harvested between 5 and 7 days after transfection, and infectious titers of Rel-expressing viruses were determined by an immunochemical titration assay with a monoclonal antibody (HY87) (42, 95). Vesicular stomatitis virus (VSV) (Indiana strain) was a gift from E. Ulug.
The simian COS-1 cell line was used for production of recombinant IFN1 and IFN-γ (67, 89). The supernatant fluids from the COS-1 cell line contained 104 IU of antiviral activity/ml, as determined by a VSV cytopathic effect inhibition assay (89). All of the other cell lines used were of chicken origin. DT40 is a B-cell line established from an avian leukosis virus (ALV)-infected chicken bearing a bursa-derived lymphoid tumor (5). DT40 cells are transformed bursal stem cells that are continuously undergoing immunoglobulin gene diversification (51). The DT95 cell line, which is also derived from a chicken with an ALV-induced lymphoid leukosis, exhibits a more mature phenotype and secretes immunoglobulin M (IgM) (5). MSB-1 and RP-1 are T-cell lines established from a Marek's disease virus-induced lymphoma (1, 73). AEV-1 is an avian erythroblastosis virus-transformed erythroid cell line (80). BM-2 is a macrophage-like cell line derived by transformation of yolk sac cells by avian myeloblastosis virus (72). 123/12, 160/2, and 123/6T are B-cell, T-cell, and macrophage-like v-rel-transformed cell lines (42). The cell lines TWB2, TWB4, and TWB5 were obtained by transformation of bursal lymphocytes in vitro by REV-TW (CSV).
Preparation of white blood cells, lymphocytes, and bone marrow cells.
Peripheral white blood cells and lymphocyte-enriched fractions from the bursa, thymus, and spleen used to prepare RNA for Northern analyses were isolated with Histopaque (Sigma Chemical Co., St. Louis, Mo.). Bone marrow, including the stroma, was isolated from the femur and tibiotarsus bones and used for RNA preparation.
LipofectAMIN-mediated delivery of pI · pC.
Cells were seeded 48 h before transfection in 150-mm-diameter plates in DMEM without antibiotics. The cells were washed with Opti-MEM I medium (Gibco BRL Life Technologies) and overlaid with 10 μg of pI · pC (Sigma Chemical Co.) per ml and 10 μl of LipofectAMIN 2000 (Gibco BRL Life Technologies) per ml in 8 ml of Opti-MEM I per plate. After 3 h, 20 ml of DMEM with serum lacking antibiotics was added to the cultures.
In vitro transcription-translation.
Proteins were synthesized by using pTZ-18R-based plasmids pTZ-IRF10, pTZ-IRF4, and pTZ-IRF8, which are described above. Transcription templates were prepared by linearization of the plasmids with MluI, BssHII, or BamHI, respectively. The proteins were produced in a TNT T7 Quick Coupled Transcription/Translation system (Promega), which employs a rabbit reticulocyte lysate. A 1-μg sample of linearized transcription templates was used per 50-μl reaction mixture. Proteins were cotranslationally labeled with [35S]methionine (Perkin-Elmer, Inc., Boston, Mass.), separated on sodium dodecyl sulfate (SDS)-8% acrylamide gels, fixed for 1 h in 45% methanol-10% acetic acid, dried, and subjected to autoradiography.
Electrophoretic mobility shift assay.
Nuclear extracts were prepared from 2 × 107 DT40 cells infected with REV-IRF-10 (CSV) or with CSV by a procedure described previously (88). The concentration of total protein was determined with Bio-Rad Protein Assay Kit II (Bio-Rad Laboratories, Inc., Richmond, Calif.). Nuclear extracts were adjusted to the same protein concentration with extraction buffer C (88). A double-stranded DNA oligonucleotide probe was prepared by annealing of a primer (5′-AGCTCAAGC-3′) to an oligonucleotide template, followed by primer extension with the Klenow fragment of DNA polymerase I (Promega) in the presence of [α-32P]dCTP. The template used (5′-TCCGCCTTTCGCTTTCGCTTCACAACGCTTGAGCT-3′) contained an ISRE-binding site (underlined) of the chicken MHC class I α-chain gene promoter (55).
For DNA-binding assays, aliquots of nuclear extract containing 8 μg of total protein were combined with 50 fmol of the probe in a total volume of 20 μl. The binding buffer used contained 10 mM Tris-HCl [pH 7.8], 30 mM KCl, 0.1 mM EDTA, 5% glycerol, 0.5 mg of acetylated bovine serum albumin per ml, 0.05% Nonidet P-40, 0.3 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 0.05 μg of poly(dC-dI)-poly(dC-dI) (Sigma Chemical Company) per μl. The nuclear extracts were first preincubated with poly(dC-dI)-poly(dC-dI) in ice-cold buffer for 10 min, and after the addition of a radioactively labeled oligonucleotide, the reaction mixture was incubated for 30 min on ice. For supershift experiments, 2 μl of rabbit antiserum was added and the incubation was continued for an additional 15 min at 25°C. The complexes were separated from the free DNA probe by electrophoresis through a 4% polyacrylamide gel in 0.5× TBE (0.09 M Tris-borate, 2 mM EDTA [pH 8.0]). The gels were run at room temperature at 125 V, fixed for 5 min in 45% methanol-10% acetic acid, dried, and then analyzed by autoradiography.
The specificity of DNA binding was verified by the addition of competitor oligonucleotides added to the binding reaction mixture. The double-stranded competitor oligonucleotide containing the MHC class I ISRE site was obtained by annealing of the oligonucleotide template with this site (described above) with complementary oligonucleotide 5′-AGCTCAAGCGTTGTGAAGCGAAA GCGAAAGGCGGA-3′. The double-stranded oligonucleotide containing a mutated ISRE site was obtained by annealing of oligonucleotide 5′-TCCGCCTTTCAGCTTTCAGCTTCACAACGCTTGAGCT-3′ with complementary oligonucleotide 5′-AGCTCAAGCGTTGTGAAGCTGAAAGCTGAAAGGCGGA-3′.
Antisera.
The AI10-8 polyclonal antiserum was raised in a rabbit immunized with a TrpE/IRF-10 fusion protein purified from C600 bacteria transformed by the pATH-I10#8 expression plasmid. The TrpE/IRF-10 fusion protein contains amino acids 101 to 356 of chicken IRF-10. The goat anti-rabbit IgG biotinylated antibody was purchased from Kirkegaard & Perry Laboratories (Gaithersburg, Md.).
Western analysis.
Western analysis was performed as described previously (42). Briefly, harvested cells were washed, resuspended, and boiled in SDS sample buffer and proteins were separated on an SDS-polyacrylamide gel with a Mini-PROTEAN II apparatus (Bio-Rad Laboratories). Proteins were transferred to a PolyScreen polyvinylidene difluoride membrane (Perkin-Elmer Life Science) and sequentially reacted with rabbit polyclonal antiserum AI10-8, goat anti-rabbit IgG biotinylated antibody, and streptavidin-linked alkaline phosphatase (Roche Molecular Biochemicals, Indianapolis, Ind.). Proteins were visualized by an enzymatic reaction with 5-bromo-4-chloro-3-indolyl phosphate and 4-nitroblue tetrazolium chloride as substrates (Roche Molecular Biochemicals). Calibrated prestained molecular weight markers from Bio-Rad Laboratories were used. Nuclear and cytoplasmic fractions of CEF were prepared in hypotonic buffer (50 mM Tris-HCl [pH 8], 1.1 mM MgCl2, 0.5% Triton X-100) as previously described (71).
Northern analysis and probes.
Total RNA was isolated by RNAwiz (Ambion, Austin, Tex.). RNA was separated by electrophoresis in a 1% agarose gel in 20 mM MOPS [3-(N-morpholino)propanesulfonic acid buffer] and transferred to a Hybond N+ membrane (Amersham Pharmacia Biotechnology, Piscataway, N.J.). Parallel samples with ethidium bromide were analyzed under identical conditions, and the gel was photographed. Filters were hybridized with DNA fragments of the chicken genes labeled with [α-32P]dCTP by nick translation as described previously (42). The following list of probes includes the name of the gene product, followed, in parentheses, by the GenBank accession number, the positions of the first and last nucleotides of the fragment based on the GenBank sequence, and the reference describing the isolation of the gene used: cIRF-3 (U20338; 59 to 767; 29), c-Rel (X52193; 16 to 1865; 12), GBP (X92112.2; 1499 to 1876; 91), IRF-1 (L39766.1; 1 to 1935; 48), IRF-2 (X95478.1; 1 to 837; 63), IRF-4 (AF320331; 473 to 1357; 39), IRF-8 (L39767.1; 1 to 1640; 48), IRF-10 (AF380350; 186 to 1334; this report), Mx (Z23168; 1 to 2545; 6), and MHC class II β chain (M26306.1; 673 to 936; 105). A probe for OAS was derived from University of Delaware EST clone ptr1c.pk002.b9 (AW240080.1). The fragment used corresponds to nt 831 to 1272 of the GenBank OAS sequence (accession number AB002585). The probe for MHC class I α chain was prepared by PCR amplification of cDNA from a v-Rel-transformed cell line. The sequence of the region amplified corresponded to nt 360 to 896 of the chicken MHC class I α chain (B2 allele) mRNA (GenBank accession number AF013492.1). There was only a single nucleotide difference in the sequence of the amplified fragment from the GenBank sequence.
Sequence analysis and phylogenetic tree construction.
The nucleotide sequence of the gene for chicken IRF-10 was compared with the GenBank, EMBL, DDBJ, and PDB databases by using the BLAST 2.0 search engine, which is available at http://www.ncbi.nlm.nih.gov/BLAST (2, 3). The same search engine was used to compare the sequence encoded by the ORF for chicken IRF-4 with the protein sequence databases. All IRF sequences encoding the full-length proteins were retrieved on the basis of their similarity to chicken IRF-10. Additionally, a sequence derived from an EST clone (gb|AU090774.1) that encodes Japanese flounder IRF-8 and is likely incomplete at the C terminus was also included in a further analysis. A protein sequence alignment was then constructed by the ClustalX program (46). The alignment was visualized with the MacBoxshade 2.15 analysis tool (http://www.imtech.res.in/pub/oth/macboxshade/mac/macboxshade215.hqx). A phylogenetic tree was constructed by the neighbor-joining method as implemented in ClustalX (83). The tree was plotted by the tree-drawing program Unrooted (http://pbil.univ-lyon1.fr/software/unrooted.html). One thousand bootstrap replicates were generated by ClustalX for the bootstrap tests.
Nucleotide sequence of IRF-10.
The sequence of chicken IRF-10 is a composite of the sequences of three cDNA clones: p8N2, pat.pk0019.d2, and pnl-b.pk0007.e2. Nucleotide 118, which was an A in the 5′ RACE clone (p8N2), was changed to a G because other, independently cloned sequences (five clones) contained a G at this position and we concluded that the A at this position was a PCR artifact. The pat.pk0019.d2 and pnl-b.pk007.e2 (GenBank accession numbers AI980252 and AW198398) are University of Delaware EST clones. These two clones overlap at 482 nt and differ by a single nucleotide (the C at position 1117 of composite sequence is a T in pnl-b.pk0007.e2). This difference does not change the amino acid sequence.
Nucleotide sequence accession no.
The sequence of chicken IRF-10 was submitted to the GenBank database under accession number AF380350.
RESULTS
Cloning and sequence characterization of a novel IRF (IRF-10).
The expression of IRF-4 and several other members of the IRF family has been shown to be upregulated in v-Rel-expressing cells, and an increased level of IRF-4 plays a role in the v-Rel-mediated transformation process (39). A computer homology search of EST databases identified a chicken EST clone (pat.pk0019.d2) isolated from activated T cells that encodes an N-terminally truncated protein similar to IRF-4 (97). The sequence of this clone was, however, significantly different from those of IRF-4 and other known IRF family members, indicating that this gene encodes a novel IRF. The new gene product was designated IRF-10 in accordance with the nomenclature employed for the mammalian IRF-encoding genes (84). Since Northern blot analysis demonstrated that IRF-10 expression is induced in lymphoid cells by v-Rel, the function of this gene product may be relevant to the v-Rel transformation process; therefore, the gene was further characterized.
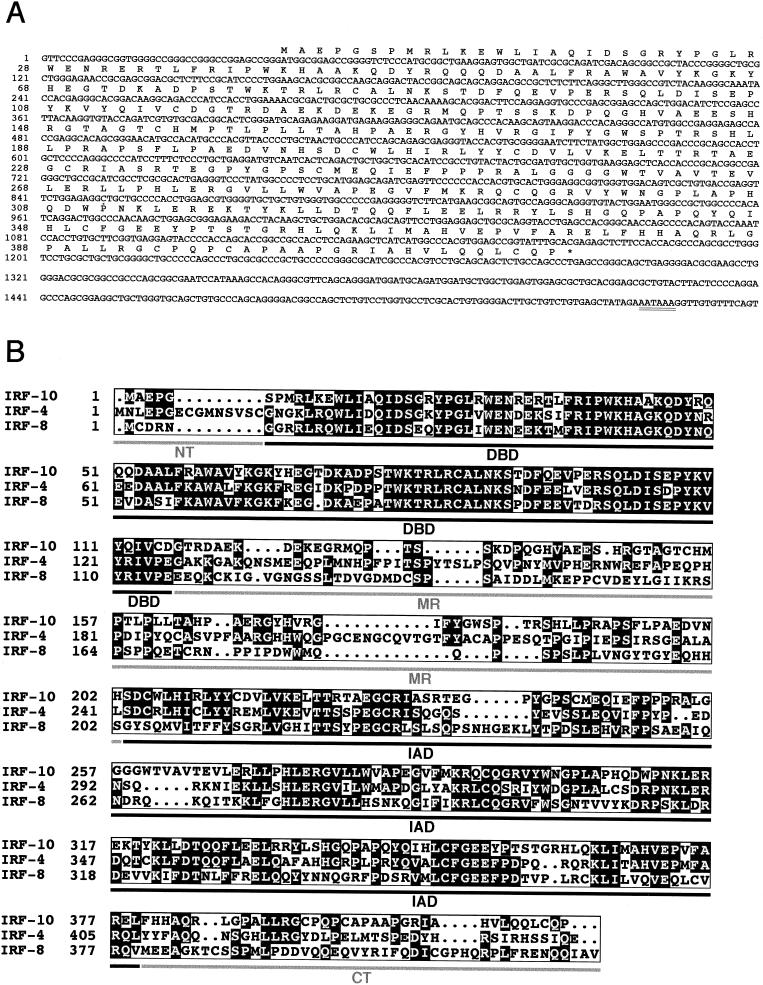
The region encoding the missing N terminus of IRF-10 was obtained by 5′ RACE, and the continuity of its sequence with the sequence of the EST clone was verified by PCR amplification of the entire ORF. The sequence of the 3′-terminal untranslated region was obtained from another EST clone (pnl-b.pk0007.e2) from liver cDNA (J. Burnside, unpublished data). The composite sequence of the avian IRF-10 cDNA is 1,560 nt long (Fig. 1A). The sequence contains a polyadenylation signal (AATAAA) at nt 1541 to 1546 located upstream of the poly(A) tract that follows immediately after nt 1560. The longest ORF encodes a protein of 416 amino acids. The start codon of this ORF is located at nt 41 to 43 and lies in favorable sequence context for the initiation of translation with a purine in the −3 position and a G in the +4 position (53).
FIG. 1.
Characterization of the IRF-10 sequence. (A) Nucleotide and predicted amino acid sequences of chicken IRF-10 cDNA. The polyadenylation signal is doubly underlined. The last T nucleotide at position 1560 is followed in the pnl-b.pk0007.e2 clone by a tract of multiple A nucleotides. The 1,560-nt IRF-10 sequence plus the usual 200-nt poly(A) tract corresponds approximately to the 1.8- to 1.9-kb size of the IRF-10 mRNA estimated by Northern blot analysis (data not shown). (B) Sequence alignment of the predicted amino acid sequences of chicken IRF-10, IRF-4, and IRF-8. Light letters on a dark background identify identical amino acids. The lines under the sequences show the locations of the five functional regions of IRF family members. NT, N terminus; MR, middle region that includes the transactivation domain; CT, C terminus.
The nucleotide and predicted amino acid sequences of chicken IRF-10 were compared with all of the sequences in the combined nucleotide and protein sequence databases. One additional chicken sequence—EST clone pgf1n.pk008.d16 from fat tissue cDNA (L. Cogburn et al., unpublished data)—was found that encodes a protein identical to IRF-10. All of the additional sequences found represented other IRF family members. Compared to these IRF family members, the IRF-10 protein sequence showed the greatest homology to chicken, human, and mouse IRF-4 and IRF-8, being 43 to 44% identical to IRF-4 and 36 to 37% identical to IRF-8 proteins (Fig. 1B; data not shown). These homology scores are far lower than the scores between the chicken and mammalian orthologues of IRF-4 or IRF-8 (84 or 73% identity, respectively). IRF-10 had 30% or less sequence identity with the other IRF proteins. We also compared the homology between the individual functional domains of IRF-10 with those of its closest relatives, IRF-4 and IRF-8 (Table 1). The level of sequence identity in the less conserved domains (the IAD and especially the middle region and C terminus) demonstrated a closer relationship of IRF-10 to IRF-4 than to IRF-8. The sequences of the individual domains showed much less similarity between IRF-10 and IRF-4 or -8 than between avian and mammalian orthologues of IRF-4 or IRF-8, suggesting that IRF-10 has a long separate evolutionary history.
TABLE 1.
Percentages of identical amino acids within predicted functional domains of IRF-10, IRF-4, and IRF-8
| IRF proteins compared | % Sequence identity in individual functional domainsa
|
||||
|---|---|---|---|---|---|
| NT | DBD | MR | IAD | CT | |
| G. gallus IRF-10 × G. gallus IRF-4 | 20 | 69 | 19 | 53 | 21 |
| G. gallus IRF-10 × G. gallus IRF-8 | 20 | 71 | 10 | 34 | 9 |
| G. gallus IRF-4 × G. gallus IRF-8 | 0 | 78 | 12 | 41 | 9 |
| G. gallus IRF-4 × H. sapiens IRF-4 | 53 | 99 | 68 | 88 | 87 |
| G. gallus IRF-8 × H. sapiens IRF-8 | 100 | 97 | 58 | 72 | 51 |
Boundaries of individual domains are shown in Fig. 1B.
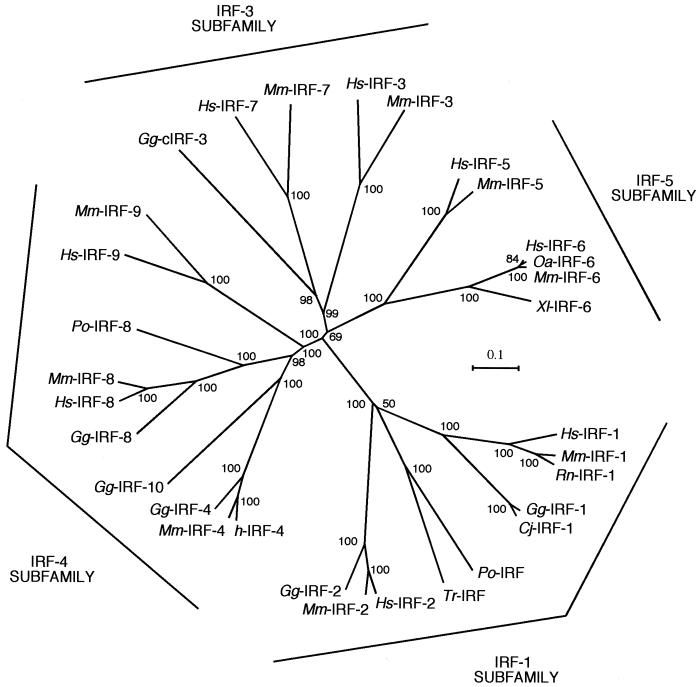
To better understand the position of IRF-10 in the evolutionary history of the IRF family, a phylogenetic tree was constructed from the sequences of the IRF proteins from mammals, birds, amphibians, and fish (Fig. 2; Table 2). The tree indicates that the IRF proteins can be classified into four subfamilies, IRF-1, IRF-3, IRF-4, and IRF-5. IRF-10 belongs to the IRF-4 subfamily together with IRF-4, IRF-8, and IRF-9. The avian IRFs (IRF-1, -2, -4, and -8) are more closely related to their mammalian orthologues than IRF-10 is to its closest relative, IRF-4. The tree also suggests the existence of a fish homologue of IRF-8 (Paralichthys olivaceus IRF-8) that is distinct from IRF-10. Additional fish members of the IRF-4 subfamily not included in Fig. 2 were tentatively identified by analysis of EST databases, i.e., an IRF-4 encoded by a clone from Japanese flounder (P. olivaceus) leukocytes (dbj|AU091159.1) and an IRF-9 encoded by a clone isolated from a zebra fish (Danio rerio) kidney (gb|BG737308.1). Early diversification also took place in the other three IRF groups, as the GenBank database contains fish EST sequences similar to those of avian and mammalian IRF-2 (gb|AF146733.1), cIRF-3 (gb|BE605317.1), IRF-5 (gb|AW174576.1 and gb|AW203040.1), and IRF-6 (gb|AW422721.1).
FIG. 2.
Phylogenetic relationship of the IRF protein family. A neighbor-joining tree of the entire family was constructed based on alignment of protein sequences by the computer program ClustalX. Chicken (Gg), clawed toad (Xl), human (Hs), Japanese flounder (Po), mouse (Mm), quail (Cj), rat (Rn), sheep (Oa), and torafugu (Tr) IRF sequences were analyzed. The full-length sequences of the individual IRF family members and a sequence of P. olivaceus IRF-8 with an incomplete C terminus derived from the EST database entry were included (see Table 2). The four IRF subfamilies are indicated in the margins. The bootstrap confidence values shown at the nodes of the tree are based on 1,000 bootstrap replications.
TABLE 2.
Protein sequences used for phylogenetic tree constructiona
| Class, species | Protein | Database | Accession no. | Reference(s) |
|---|---|---|---|---|
| Mammalia | ||||
| Homo sapiens (human) | IRF-1 | SwissProt | P10914 | 69 |
| IRF-2 | SwissProt | P14316 | 45 | |
| IRF-3 | SwissProt | Q14653 | 4 | |
| IRF-4 | SwissProt | Q15306 | 30 | |
| IRF-5 | SwissProt | Q13568 | Unpublished | |
| IRF-6 | SwissProt | O14896 | Unpublished | |
| IRF-7 | SwissProt | Q92985 | 110 | |
| IRF-8 | SwissProt | Q02556 | 101 | |
| IRF-9 | SwissProt | Q00978 | 99 | |
| Mus musculus (mouse) | IRF-1 | SwissProt | P15314 | 69 |
| IRF-2 | SwissProt | P23906 | 34 | |
| IRF-3 | SwissProt | P70671 | Unpublished | |
| IRF-4 | SwissProt | Q64287 | 18, 65 | |
| IRF-5 | SwissProt | P56477 | Unpublished | |
| IRF-6 | GenBank | AAB36714.1 | Unpublished | |
| IRF-7 | SwissProt | P70434 | Unpublished | |
| IRF-8 | SwissProt | P23611 | 17 | |
| IRF-9 | SwissProt | Q61179 | 96 | |
| Ovis aries (sheep) | IRF-6 | GenBank | AAF34782.1 | Unpublished |
| Rattus norvegicus (rat) | IRF-1 | SwissProt | P23570 | 109 |
| Aves | ||||
| Coturnix japonica (quail) | IRF-1 | EMBL | CAB91630.1 | 112 |
| Gallus gallus (chicken) | IRF-1 | SwissProt | Q90876 | 48 |
| IRF-2 | SwissProt | Q98925 | 63 | |
| IRF-4 | GenBank | AAK08198.1 | 39 | |
| IRF-8 | SwissProt | Q90871 | 48 | |
| IRF-10 | GenBank | AAK55444.1 | This work | |
| cIRF-3 | SwissProt | Q90643 | 29 | |
| Amphibia, Xenopus laevis (toad) | IRF-6 | DDBJ | BAA24349.1 | 37 |
| Teleostei | ||||
| Paralichthys olivaceus (flounder) | IRF | DDBJ | BAA83468.1 | 106 |
| IRF-8 | GenBank | AU090774.1 | Unpublished | |
| Takifugu rubipes (tarafugu) | IRF | GenBank | AAK28340.1 | 82 |
All sequences are those of full-length proteins, with the exception of the sequence of P. olivaceus IRF-8, which is likely incomplete at the C terminus. This sequence, derived from an EST clone (gblAU090774.1), encodes 315 amino acids (with the assumption of one sequence error breaking the reading frame between codons 8 and 9) and represents about 75% of the presumed length of the protein.
Characterization of the IRF-10 protein.
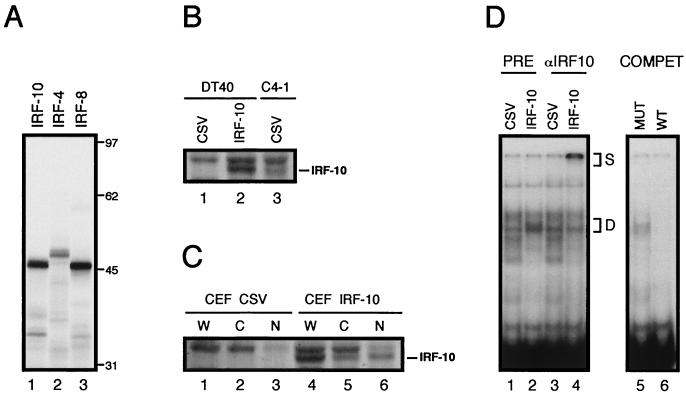
The protein product of the cloned IRF-10 cDNA was translated in vitro and compared with proteins, IRF-4 and IRF-8, encoded by the two most closely related genes (Fig. 3A). The IRF-10 cDNA encoded a protein with a molecular mass of 46 kDa, which closely agrees with the calculated theoretical molecular mass. IRF-10 was also analyzed in vivo by Western blotting. The coding region of IRF-10 was inserted into a retroviral vector and expressed in the B-cell line DT40 and fibroblast cultures (Fig. 3B and C). Exogenous IRF-10 was abundantly expressed in both cell types, while no endogenous protein was detected in control cells infected with CSV alone. Retrovirus-expressed IRF-10 had a size similar to that of the endogenous protein detected in the C4-1 B-cell line transformed by the S2A3v-rel oncogene (Fig. 3B). In contrast to IRF-4, which was previously shown to have two closely migrating species, IRF-10 was expressed as a single species (39). IRF-10 also differed from IRF-4 in its subcellular localization (Fig. 3C). While most of the IRF-4 protein exogenously expressed in CEF was found in the cytoplasm, approximately two-thirds of the IRF-10 protein was located in the nucleus (39). Like that of IRF-4, the expression of IRF-10 was maintained for several weeks in fibroblasts and in the DT40 cell line. This is in contrast to fibroblast cultures or DT40 cells exogenously expressing either IRF-1 or IRF-8, where the cells expressing these IRF family members were rapidly eliminated due to their antiproliferative or apoptotic effect on the cells (data not shown).
FIG. 3.
Analysis of IRF-10 protein. (A) Protein product of IRF-10 ORF translated in vitro with rabbit reticulocyte lysate. The mobility of the protein is compared with those of chicken IRF-4 and IRF-8. Molecular mass markers in kilodaltons are shown on the right. (B) Comparison of retrovirus-expressed IRF-10 in the DT40 B-cell line with endogenous IRF-10 present in C4-1 B cells transformed by S2A3v-rel. Western blot analysis of whole-cell lysates. (C) Expression of IRF-10 in CEF cultures. Western blot analysis of whole-cell lysates (W) and lysates of cytoplasmic (C) and nuclear (N) fractions. For experiments shown in both panels B and C, cells were infected either with a retrovirus expressing IRF-10 or with the CSV helper only. The lysates from 4 × 105 cells were loaded in each lane of the gel. Detection was performed with IRF-10 antiserum AI10-8. The positions of IRF-10 bands are shown on the right. Just above the IRF-10 band is a prominent background band that was present in all of the cells analyzed. (D) DNA-binding complexes in the nuclei of DT40 cells infected with a retrovirus expressing IRF-10 in comparison with complexes from control CSV-infected cells. Oligonucleotide probe containing the ISRE site from the promoter of the chicken gene encoding the α chain of the MHC class I protein was used for the analysis. Binding reaction mixtures shown in lanes 1 to 4 contained preimmune rabbit antiserum (PRE) or IRF-10-specific antiserum AI10-8 (αIRF10). Supershifted high-molecular-weight complexes (S) and complexes partially depleted by IRF-10-specific antiserum (D) are indicated. To verify that IRF-10 binding is specific, competitor oligonucleotides (COMPET) with a wild-type (WT) or mutated (MUT) ISRE site were added in 50-fold excess to the reaction mixtures containing nuclear extract from DT40 cells expressing IRF-10 (lanes 5 and 6).
IRF-10 was further tested in vivo for the ability to bind to the ISRE present in the α chain of the MHC class I promoter. Electrophoretic mobility shift assays revealed differences in DNA-binding complexes in the nuclei of DT40 cells expressing IRF-10 in comparison with complexes detected in the nuclei of control cells (Fig. 3D, lanes 1 and 2). The presence of IRF-10 in the DNA-binding complexes from IRF-10-expressing cells was verified with IRF-10 specific antiserum (Fig. 3D, lane 4).
Tissue distribution of IRF-10.
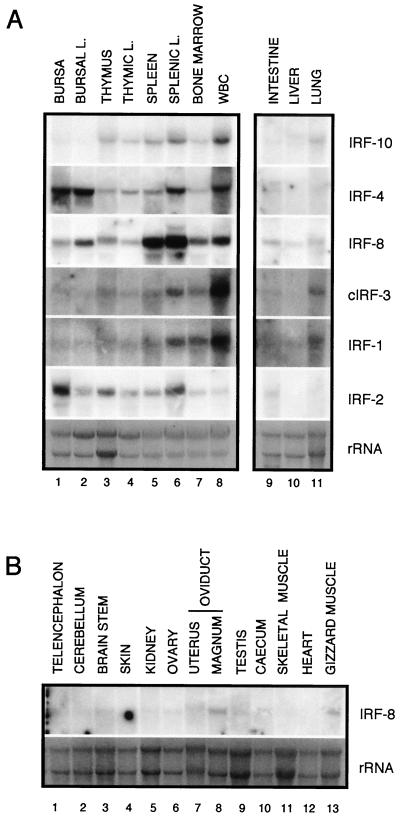
The expression of IRF-10 in various organs and cells was determined by Northern blot analysis (Fig. 4A). The highest level of expression was detected in cells of hematopoietic origin. A very low level of IRF-10 expression was detected, however, in bursal cells. IRF-10 was also expressed in lung tissue, but it was not detected in other organs (data not shown). In order to compare the expression pattern of IRF-10 with those of other family members, parallel blots were hybridized with IRF-4, IRF-8, cIRF-3, IRF-1, and IRF-2 probes. All of the members of the IRF family examined differed in tissue distribution. With the exception of IRF-8, their expression was limited to hematopoietic cells and lung, liver, and intestine tissues. In striking contrast to IRF-10, IRF-4 was expressed at the highest level in bursae and bursal lymphocytes. The highest level of IRF-8 expression was detected in the spleen and splenic lymphocytes, followed by other hematopoietic cells and organs. Surprisingly, IRF-8 expression was also detected in most other organs, including oviduct, testis, gizzard muscle, intestine, lung, liver, and brain tissues (Fig. 4B). The IRF-1 and cIRF-3 expression pattern was most similar to that of IRF-10; however, their overall expression level in hematopoietic organs was much lower than that of IRF-10 (IRF-1 and cIRF-3 blots were exposed 3.7 times longer). IRF-2 tissue distribution was unique among the IRFs analyzed, with the highest expression in bursae and splenic lymphocytes. In conclusion, IRF-10, like IRF-4 and IRF-8, is highly expressed in organs of hematopoietic origin but, in contrast to them, its expression level in bursal cells is low.
FIG. 4.
Expression pattern of chicken IRF-10, IRF-4, IRF-8, cIRF-3, IRF-1, and IRF-2 in various tissues from a 1-month-old chicken. (A) Total RNA (10 μg) isolated from bursa, bursal lymphocytes (BURSAL L.), thymus, thymic lymphocytes (THYMIC L.), spleen, splenic lymphocytes (SPLENIC L.), bone marrow, white blood cells (WBC), intestine, liver, and lung was subjected to Northern analysis. (B) Total RNA (10 μg) isolated from the various organs was hybridized with a probe against IRF-8. The blots hybridized with IRF-10, IRF-4, IRF-8, and IRF-2 were exposed for 15 h, and the blots hybridized with IRF-1 and cIRF-3 were exposed for 56 h. The specific activity of all of the probes was approximately 2 × 105 cpm/ng. The probes are described in Materials and Methods. The intensity of the rRNA stained with ethidium bromide is shown at the bottom (rRNA).
IRF-10 expression is inducible by IFNs and ConA.
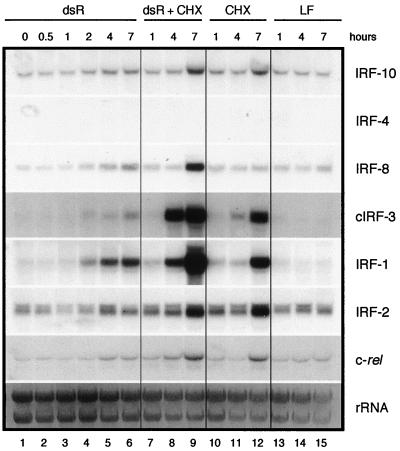
The expression of IRFs may be induced by some or all of the following stimuli: virus infection, double-stranded RNA (dsRNA), IFNs, or proliferative signals (62). Therefore, the kinetics of induction of IRF-10 expression by dsRNA, type I and II IFNs, and ConA was analyzed and compared with that of IRF-4, IRF-8, cIRF-3, IRF-1, and IRF-2 (Fig. 5; see also Fig. 6 and 7). CEF cultures were treated with dsRNA (10 μg of pI · pC per ml) with LipofectAMIN 2000, and samples for Northern blot analysis were taken at 0.5, 1, 2, 4, and 7 h after treatment (Fig. 5). Parallel cultures were treated simultaneously with dsRNA and cycloheximide, cycloheximide alone, or LipofectAMIN 2000. IRF-10 transcription was not elevated 4 and 7 h after dsRNA treatment above the background of LipofectAMIN 2000 alone (Fig. 5, lanes 5, 6, and 13 to 15). IRF-4 mRNA was not detected in fibroblasts in this experiment. The expression of IRF-8 and cIRF-3 was slightly induced 2 and 7 h after treatment and was not inhibited by cycloheximide. The expression of IRF-1 was strongly induced beginning at 2 h after treatment and was not inhibited by cycloheximide. The expression of IRF-2 did not significantly change during the experiment. The expression of c-rel increased 4 to 7 h after treatment of cells with dsRNA, and this increase was independent of cycloheximide. In conclusion, in contrast to IRF-8, IRF-1, and cIRF-3, IRF-10 did not respond to dsRNA.
FIG. 5.
Induction of IRF-10, IRF-4, IRF-8, cIRF-3, IRF-1, IRF-2, and c-rel expression by dsRNA (dsR). Fibroblasts cultivated for 2 weeks after explantation from chicken embryos were treated with pI · pC (10 μg/ml) (lanes 1 to 6) by using LipofectAMIN 2000 (LF) as described in Materials and Methods. Parallel cultures were simultaneously treated with cycloheximide (CHX; 10 μg/ml) (lanes 7 to 9). The control cells were treated with cycloheximide (lanes 10 to 12) from Sigma or LipofectAMIN 2000 (lanes 13 to 15) alone. RNA was isolated from these cells at the time points indicated. Total RNA (10 μg per lane) was subjected to Northern analysis and hybridized with probes described in Materials and Methods. The intensity of the rRNA staining with ethidium bromide is shown at the bottom (rRNA).
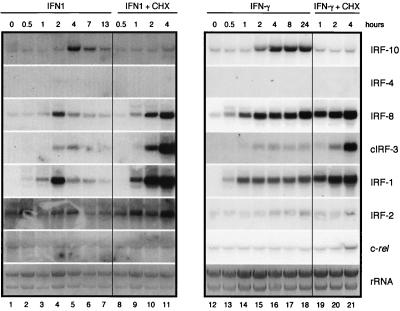
FIG. 6.
Induction of IRF-10, IRF-4, IRF-8, cIRF-3, IRF-1, IRF-2, and c-rel expression by IFN1 and IFN-γ. Fibroblasts cultivated for 2 weeks after explantation from chicken embryos were treated with 200-fold-diluted supernatant fluids from COS-1 cells expressing IFN1 or IFN-γ (IFN1, lanes 1 to 7; IFN-γ, lanes 13 to 19). Parallel cultures were simultaneously treated with cycloheximide at 10 μg/ml (IFN1 + CHX, lanes 8 to 12; IFN-γ + CHX, lanes 20 to 22). RNA was isolated at the time points indicated. Total RNA (10 μg per lane) was subjected to Northern analysis and hybridized with the probes described in Materials and Methods. The intensity of the rRNA stained with ethidium bromide is shown at the bottom (rRNA).
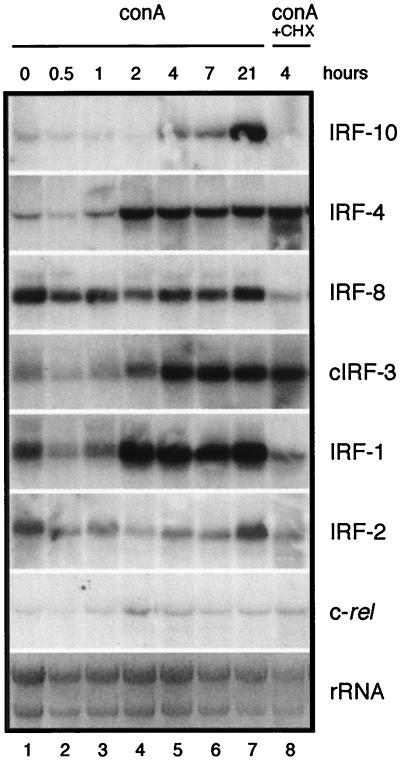
FIG. 7.
Induction of IRF-10, IRF-4, IRF-8, cIRF-3, IRF-1, IRF-2, and c-rel expression by ConA. The spleens from 2-month-old chickens were used as a source of lymphocytes. The splenic cells were purified with Histopaque. Lymphocytes (80 × 106/ml) were treated with ConA (Sigma Chemical Co.) at 25 μg/ml (lanes 1 to 7). A parallel sample was simultaneously treated with cycloheximide at 10 μg/ml (ConA + CHX, lane 8). RNA was isolated at the time points indicated. Total RNA (10 μg per lane) was subjected to Northern analysis and hybridized with the probes described in Materials and Methods. The intensity of rRNA staining with ethidium bromide is shown at the bottom (rRNA).
CEF cultures were treated with IFN1 or IFN-γ, and samples were taken for Northern blot analysis at the various time points indicated (Fig. 6). Parallel cultures were treated simultaneously with cycloheximide and either IFN1 or IFN-γ. The increases in IRF expression induced by IFN1 were transient and peaked at 2 to 4 h after treatment (Fig. 6, lanes 4 and 5). The expression of IRF-10 was induced relatively late, at 4 h, and was dependent on protein synthesis (Fig. 6, lanes 5 and 11). IRF-4 expression was not detected after IFN1 treatment. IRF-8 was rapidly induced by IFN1 treatment after 1 h, with a peak after 2 h (Fig. 6, lanes 3 and 4). Induction of cIRF-3 expression was more rapid than that of IRF-10, but its peak at 4 h coincided with the peak of IRF-10 expression (Fig. 6, lane 5). The induction of IRF-1 expression was similar to that of IRF-8 but could be detected as early as 30 min following exposure to IFN1 (Fig. 6, lane 2). The level of IRF-2 induction was very low (Fig. 6, lanes 3 and 4). CEF cultures treated with recombinant IFN-γ also increased the expression of IRF-10. The level of IRF-10 mRNA increased 2 h after treatment and reached a plateau at 4 h (Fig. 6, lanes 15 and 16). IRF-10 expression was not induced with IFN-γ in the presence of cycloheximide (Fig. 6, lanes 15, 16, 20, and 21). As expected, IRF-4 expression was not induced by IFN-γ. However, the expression of IRF-8 and IRF-1 was strongly and rapidly induced by IFN-γ starting as early as 30 min after treatment (Fig. 6, lane 13). Expression of cIRF-3 began 1 h after treatment, and its level remained relatively low (Fig. 6, lane 14). The induction of IRF-2 expression was very low in level and delayed (Fig. 6, lane 16). The expression of c-rel did not change after treatment with either of the two IFNs. With the exception of IRF-10, the induction of all of the other IRFs with IFNs was independent of protein synthesis.
Induction of IRF-10 and other IRF family members by ConA was analyzed in splenic cells by Northern blot analysis at different times after treatment (Fig. 7). Parallel cultures were treated with ConA and cycloheximide, and RNA was isolated 4 h after treatment. Exposure of splenic lymphocytes to ConA led to induction of IRF-10, IRF-4, IRF-1, and cIRF-3, which continued during the 21-h time frame evaluated. IRF-10 expression was induced at 4 h after exposure of splenic cells to ConA and was not observed in the presence of cycloheximide (Fig. 7, lanes 5 and 8). In contrast, induction of IRF-4 expression started earlier (2 h after treatment), immediately reached a plateau, and was cycloheximide independent (Fig. 7, lanes 4 and 8). Surprisingly, IRF-8 expression was unresponsive to ConA treatment. cIRF-3 expression was induced by ConA at 2 h (Fig. 7, lane 4). IRF-1 expression was strongly induced by ConA 2 h after treatment, while IRF-2 expression was not changed. Expression of c-rel transiently increased between 2 and 4 h after ConA treatment (Fig. 7, lanes 4 and 5). With the exception of that of IRF-10 and IRF-1, induction was independent of protein synthesis. Another distinct feature of IRF-10 ConA-mediated induction of expression was the continued increase at the later time points.
These experiments demonstrated that IRF-10, in contrast to IRF-4, is induced by both types of IFNs in primary fibroblasts. IRF-10, like IRF-4, is also induced by ConA in splenic cultures. The kinetics of induction of IRF-10 by IFNs and ConA is delayed relative to that of other IRFs and depends on protein synthesis.
IRF-10 modulates the induction of the IFN-induced genes for GBP and OAS.
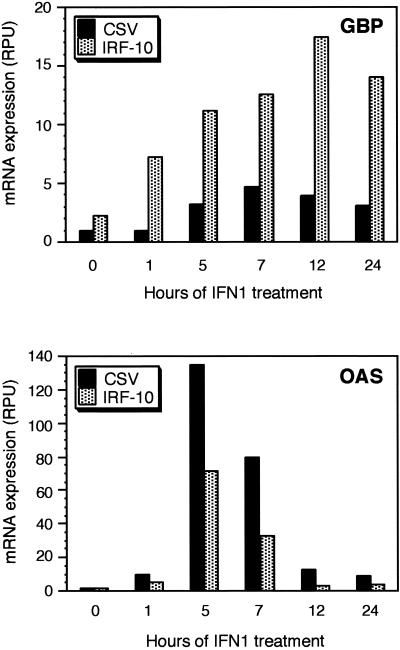
IRF family members play an important role in antiviral responses; therefore, the ability of IRF-10 to induce some IFN-responsive genes was evaluated. Fibroblast cultures expressing high levels of IRF-10 from retroviral vectors were established (Fig. 3C). The induction of several IFN-responsive genes, of which GBP, OAS, and Mx are the best characterized, is regulated, in part, by IRF family members (9, 64, 90). To characterize the IFN induction pattern of these genes in IRF-10-expressing cells, control and IRF-10-expressing cultures were treated with IFN1 and the kinetics of induction of these IFN target genes was evaluated by Northern blot analyses (Fig. 8). Overexpression of IRF-10 resulted in a slight increase in GBP expression even without IFN treatment, and IFN1-mediated induction of GBP in these cells was stronger than in control cells and was prolonged over a 24-h period. Surprisingly, IFN1-mediated induction of OAS was weaker and briefer in IRF-10-expressing cells than in control cells. Mx induction by IFN1 was unaffected in cells overexpressing IRF-10 in comparison to normal CEF (data not shown).
FIG. 8.
IRF-10 modulates the expression of GBP- and OAS-encoding genes involved in an antiviral response. CEF expressing IRF-10 or control cells expressing the helper virus (CSV) were treated with 1,000-fold-diluted supernatant fluids from COS-1 cells expressing IFN1. RNA was isolated at the time points indicated. Total RNA (10 μg per lane) was subjected to Northern analysis with the indicated probes (described in Materials and Methods). The radioactive signal on Northern blots was quantified by a Phosphorimager 225SI (Molecular Dynamics, Sunnyvale, Calif.), and the intensities of the hybridizing bands are shown in relative phosphorimager units (RPU).
IRF-10 contributes to the regulation of expression of MHC class I.
IRF-10 is unique among IRFs in its delayed induction and dependency on protein synthesis, suggesting that it may play a role in the late stages of an antiviral defense. The purpose of the delayed defense is to present viral antigens to stimulate a specific immune response, which can ultimately recognize and eliminate infected cells. Viral antigens are presented together with MHC class I and II molecules on antigen-presenting cells. The regulation of MHC class I is partially under the control of IFNs, and some IRF family members have been shown to mediate this antiviral action at the promoter level (13, 38, 104). Therefore, we investigated whether IRF-10 also participates in the regulation of MHC class I expression.
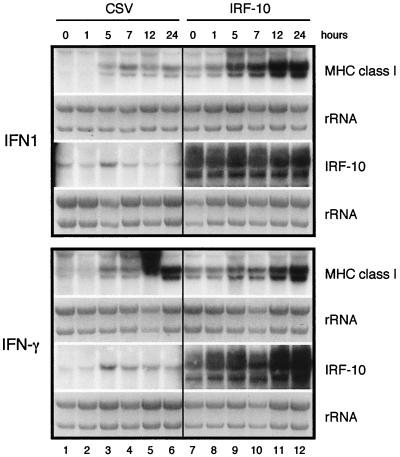
The expression of MHC class I and IRF-10 in fibroblasts after treatment with IFNs was analyzed (Fig. 9). IFN1 and IFN-γ induced the expression of MHC class I transcription beginning at 5 h after treatment, and the steady-state level of RNA increased at the later time points (Fig. 9, lanes 3 to 6). At that time, endogenous IRF-10 already reached its highest levels (Fig. 9, lane 3). To determine if IRF-10 may mediate the induction of MHC class I by IFNs, the expression of MHC class I in IRF-10-overexpressing cells before and after treatment with IFN1 and IFN-γ was analyzed (Fig. 9, lanes 7 to 12). When IRF-10 was expressed in fibroblasts exogenously, the levels of MHC class I were significantly elevated (Fig. 9, lane 7). Moreover, in these cells, the expression of MHC class I was further enhanced by treatment with IFN1, suggesting that IRF-10 contributes to MHC class I regulation in fibroblasts (Fig. 9, lanes 9 to 12). MHC class II expression was not detected in control or IRF-10-expressing fibroblasts before or after treatment with IFNs (data not shown). Taken together, these results suggest that IRF-10 contributes to the regulation of MHC class I in the response to IFNs.
FIG. 9.
IRF-10 modulates the expression of MHC class I. CEF cultures expressing IRF-10 or control cells expressing helper virus (CSV) were treated with 1,000-fold-diluted supernatant fluids from COS-1 cells expressing IFN1 or IFN-γ. RNA was isolated at the time points indicated. Total RNA (10 μg) was subjected to Northern analysis with MHC class I and IRF-10 probes. The sample from CSV-infected, IFN-γ-treated CEF cultures (bottom, lane 5) exhibits a defect when hybridized with an MHC class I probe. Northern blots hybridized with the IRF-10 probe show endogenous IRF-10 mRNA (1.8 to 1.9 kb) in CSV-infected CEF cultures (lanes 1 to 6), while two retroviral REV-IRF-10 RNAs (approximately 6 and 3 kb) are shown in blots from REV-IRF-10-infected CEF cultures (lanes 7 to 12). The hybridized blots showing endogenous IRF-10 mRNA were exposed to film six times longer than the blots with REV-IRF-10 RNA. The intensity of rRNA staining with ethidium bromide is shown below for each Northern blot (rRNA).
Expression of IRF-10 in transformed cell lines.
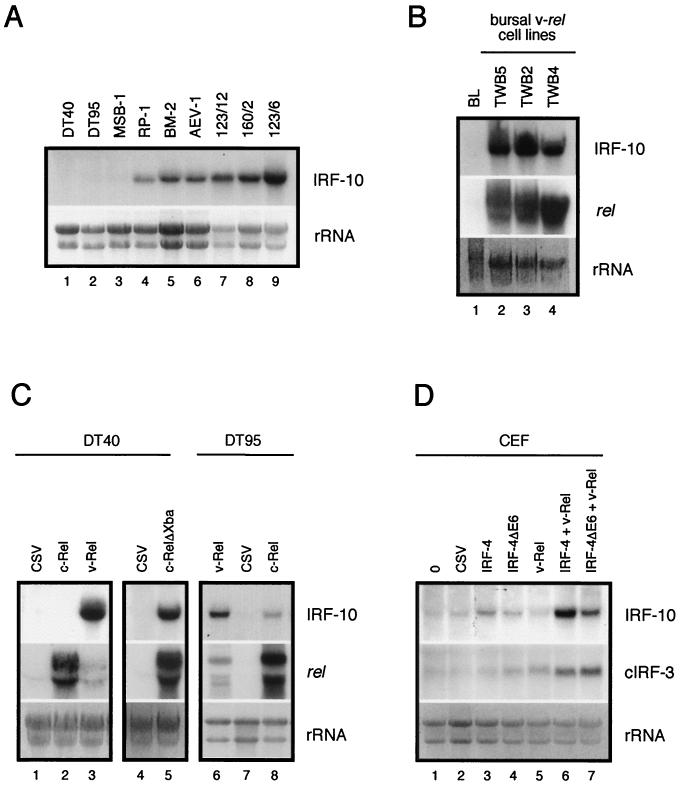
Several transformed cell lines were analyzed for expression of IRF-10 mRNA (Fig. 10A). Bursal B-cell lines DT40 and DT95 failed to express IRF-10. The expression pattern in T cells transformed by Marek's disease virus, RP-1 and MSB-1, was variable. IRF-10 was strongly expressed in RP-1, but its expression in MSB-1 cells was very weak. IRF-10 expression correlated with the expression of MHC class II in these cell lines; MSB-1 cells express very low levels of MHC class II, while RP-1 express high levels (79; data not shown), suggesting that IRF-10 may contribute to regulation of the expression of the gene for MHC class II. Strong expression of IRF-10 was also detected in the BM-2 macrophage cell line and an AEV-1 erythroblastoid cell line.
FIG. 10.
Constitutive and induced expression of IRF-10 mRNA in transformed cell lines and CEF. Total RNA (10 μg per lane) was subjected to Northern analysis and hybridized with IRF-10, cIRF-3, and c-rel probes. The intensity of rRNA staining with ethidium bromide is shown at the bottom (rRNA). (A) Constitutive IRF-10 expression in B-cell lines DT40 and DT95, T-cell lines MSB-1 and RP-1, myeloblastoid cell line BM-2, erythroblastoid cell line AEV-1, and v-rel-transformed cell lines with the B-cell (123/12), T-cell (160/2), and non-B-, non-T-cell phenotypes (123/6T) was analyzed. (B) Constitutive expression of IRF-10 in bursal lymphocytes (BL) and v-rel-transformed cell lines of bursal origin (TWB5, TWB2, and TWB4). The expression of v-rel from a retroviral vector is shown in the middle. Endogenous c-rel mRNA is not visible at this exposure. (C) Induction of IRF-10 expression in DT40 and DT95 B-cell lines by v-Rel, c-Rel, and c-RelΔXba and comparison with a CSV control. The expression of rel genes from retroviral vectors is shown in the middle. Endogenous c-rel mRNA is at the detection threshold at this exposure and is visible in lanes 1, 4, and 6. (D) Cooperation of IRF-4 with v-Rel in the induction of IRF-10 and cIRF-3 in CEF. Secondary cultures of CEF were infected with REV-IRF-4 (IRF-4), REV-IRF-4ΔE6 (IRF-4ΔE6), or DSv-Rel (v-Rel) or coinfected with DSv-Rel and REV-IRF-4 or REV-IRF-4ΔE6 at a multiplicity of infection of 3. Control cells were left uninfected or infected with CSV. The expression of IRF-4 and v-Rel proteins in these fibroblasts was confirmed by Western blot analysis at 3 weeks after infection as described previously (39), and the expression of IRF-10 and cIRF-3 mRNA was then analyzed by Northern blot analysis as described in the legend to Fig. 4.
The highest level of IRF-10 expression was detected in v-rel-transformed cell lines of splenic origin, irrespective of their phenotype; IgM-expressing line 123/12, TCR2-expressing 160/2, and 123/6T, which expresses macrophage markers CMTD I and II, all had high levels of IRF-10 mRNA (42). Because IRF-10 levels were low in bursal lymphocytes and were not detected in bursal ALV-derived cell lines DT40 and DT95, it was of interest to determine whether IRF-10 is expressed in v-rel-transformed cell lines of bursal origin. The expression of IRF-10 in three v-rel-transformed cell lines derived from the bursa was examined (Fig. 10B). In contrast to bursal lymphocytes, all of these cell lines expressed high levels of IRF-10 mRNA. In summary, IRF-10 is expressed in a variety of hematopoietic transformed cell lines but the highest expression level was detected in v-rel-transformed cell lines.
v-Rel and c-Rel induce IRF-10 expression in transformed lymphoid cell lines.
The high level of IRF-10 expression in v-rel-transformed cell lines, especially in bursal cell lines, suggested that IRF-10 may be regulated, in part, by Rel/NF-κB transcription factors. Therefore, the expression of IRF-10 in the DT40 and DT95 cell lines and primary CEF expressing v-Rel or c-Rel was compared to the expression of IRF-10 in parental cells (Fig. 10C and D). The expression of IRF-10 was strongly elevated in the DT40 cell line expressing v-Rel in comparison to that in the parental cell line, but it was not detected in DT40 cells expressing c-Rel (Fig. 10C). However, endogenous c-Rel, unless activated by an external signal, resides predominantly in the cytoplasm (41). Deletion of the cytoplasmic anchoring sequence located in the C terminus in mutant c-Rel proteins allows nuclear access in the absence of an appropriate external signal. These mutant proteins would be expected to be better transcriptional activators than wild type c-Rel (75). Therefore, c-RelΔXba, a transforming mutant form of c-Rel lacking the C terminus, was evaluated for induction of IRF-10 in DT40 cells. While c-RelΔXba induced high levels of IRF-10, this level did not reach the level induced by v-Rel even though c-RelΔXba was more abundant in cells than was v-Rel. Similarly, in the DT95 cell line expressing v-Rel, the level of IRF-10 was higher than in c-Rel-expressing DT95 cells although the level of v-Rel was lower than that of c-Rel in these cells (Fig. 10C). Surprisingly, the expression of IRF-10 was not elevated in fibroblast cultures overexpressing v-Rel or c-Rel (Fig. 10D, data not shown). These results indicate that IRF-10 expression is inducible by v-Rel and a to lesser degree by c-Rel in lymphoid cells.
IRF-4 induction of IRF-10 is enhanced by v-Rel.
The failure of v-Rel to induce IRF-10 in fibroblasts suggests that some cell-specific transcription factor(s) necessary for IRF-10 induction may be absent in these cells. Fibroblasts and the B-cell lines DT40 and DT95 differ in the level of expression of IRF-4. The endogenous level of IRF-4 mRNA in fibroblasts is very low; by contrast, the levels of IRF-4 in DT40 and DT95 are high (39). To test the hypothesis that the difference in the levels of IRF-4 is responsible for the tissue-specific induction of IRF-10 expression by v-Rel, fibroblasts were infected with a retrovirus expressing v-Rel, IRF-4, or IRF-4 splice variant IRF-4ΔE6 or coinfected with retroviruses expressing v-Rel and IRF-4 or v-Rel and IRF-4ΔE6 (Fig. 10D). In contrast to v-Rel, IRF-4 alone induced the expression of IRF-10 in fibroblasts. However, when v-Rel was coexpressed with IRF-4 or IRF-4ΔE6 in fibroblasts, expression of IRF-10 mRNA was highly elevated, suggesting that IRF-4 and v-Rel cooperate in the induction of IRF-10. Similar cooperation between v-Rel and IRF-4 was observed in induction of the expression of cIRF-3 but not with other IRF family members (Fig. 10D; data not shown).
DISCUSSION
Evolution of the IRF family.
This report describes the characterization of a novel member of the IRF family, IRF-10. IRF-encoding genes are known only in vertebrates. All IRF family members are characterized by a DBD with similarly spaced tryptophan residues, as in the DBD of the myb transcription factors (99). The myb family functions in proliferation, and myb homologues have been identified in both vertebrates and invertebrates (59). The relatively recent evolution of the IRF family, therefore, likely occurred from a prototypical protein with a myb-like DNA-binding motif. All IRFs, except the members of the IRF-1 subfamily, also contain an IAD protein-protein interaction domain, which has a high level of similarity to the transactivation domain of Smad morphogens (19). The acquisition of this protein-protein interaction module of the Smad family of transcription factors resulted in further branching of the IRF network.
The phylogenetic analysis presented in this report suggests that the family of IRF transcription factors appeared early in vertebrate evolution and was followed by rapid diversification since all four IRF subfamilies are present in Teleostei fish. The majority of the IRF family members identified in nonmammalian vertebrates are orthologues of the mammalian IRFs. However, the fish IRF-1 subfamily members (P. olivaceus IRF and Takifugu rubipes IRF) and avian cIRF-3 differ in sequence to such an extent that it is difficult to consider them orthologues of any specific mammalian IRF (29, 82, 106). A detailed analysis of the sequences of P. olivaceus IRF and T. rubipes IRF suggests greater similarity to mammalian IRF-1 than to IRF-2. Moreover, a fish IRF with strong homology to IRF-2 has been recently identified (GenBank accession number AF146733.1). Therefore, it is unlikely that P. olivaceus IRF and T. rubipes IRF is a common precursor of IRF-1 and IRF-2, suggesting that the difference between P. olivaceus IRF or T. rubipes IRF and mammalian IRF-1 is the result of extensive diversification. The situation is different with cIRF-3. The gene with strong homology to avian cIRF-3 has been identified in fish (GenBank accession number BE605317.1), but a cIRF-3 homologue is not present in the human genome. By contrast, the orthologues of mammalian IRFs most closely related to cIRF-3 (IRF-3 and IRF-7) have not been identified in nonmammalian vertebrates (data not shown). Therefore, it is plausible that IRF-3 and IRF-7 evolved from a cIRF-3-like precursor by duplication and subsequent diversification. The appearance of most of the IRF family members in lower vertebrates correlates with the appearance of adaptive immunity in these species. However, the evolution by duplication and diversification further modified the IRF family members in individual vertebrate branches.
IRF-10 evolved as one of the last IRF family members from a precursor it has in common with IRF-4. Comparison of the lengths of the branches of the IRF phylogenetic tree suggests that unless the IRF-10 diversification rate was substantially higher than for other IRF family members, IRF-10 evolved in the ancestors of Sauropsida and mammals. Its origin may even predate the divergence of Teleostei fish and tetrapods. Although we were unable to identify an IRF-10 protein in fish, we have identified a tentative P. olivaceus IRF-4 orthologue (dbj|AU091159.1), suggesting that IRF-10 may exist in recently evolved fish. More confusing is the fate of mammalian IRF-10. Last year, sequences from two almost complete human genome projects became available (56, 100). All nine human IRF family members can be identified in both databases. Only one additional IRF-like sequence was found close to the centromeric region of chromosome 20 (hypothetical protein products of this gene have GenBank accession numbers CAC28981 and CAC28982). This sequence has the greatest homology to IRF-10, but the locus diverged to such an extent that it cannot produce a full-length protein. Accordingly, we were not able to detect mRNA (in human spleen or in HeLa cells in response to human IFN-γ) with probes or primers representing the region, which contains the DBD in other IRF family members (data not shown). Recently, a cDNA clone from a carcinoma cell line was identified that contains mRNA formed by splicing of the last three exons of the putative human IRF-10-encoding locus (GenBank accession number AK000652). However, the predicted ORF is incomplete and contains at least one interruption; therefore, it is unlikely that a functional IRF protein could be translated from that mRNA. Our current hypothesis is that IRF-10 is not expressed in the more recently evolved mammals. We hypothesize that elimination of IRF-10 expression in mammals is connected with the branching of cIRF-3 into two closely related but functionally different IRF-3 and IRF-7 family members. IRF-3 is considered to be the first IRF to function in response to viral infection whereby the protein located in the cytoplasm translocates to the nucleus and activates the expression of IFN-β (62). Subsequently, the transcriptional activation of IRF-7 by IFN-β serves to amplify the expression of IFN-β and activates the IFN-α genes in lymphoid cells (107). This two-step induction model may be similar to the avian system, with IRF-10 employed in the delayed phase.
IRF-10 has a unique function in the immune response.
IRF-10 differs in constitutive and inducible expression from its closest relatives, IRF-4 and IRF-8, suggesting a different biological function. In contrast to that of IRF-4, expression of IRF-10 is induced by IFN type I. IFN type I-inducible IRFs, like IRF-1, play a role in the protection of cells against most viruses by regulating expression of IFN target genes (84). The IFN target gene that encodes GBP is regulated by mammalian IRF-1, but overexpression of avian IRF-1 has no effect on GBP levels (9, 111). Therefore, it is possible that IRF-10, which enhanced the induction of GBP by IFN1, may compensate for avian IRF-1. IRF-1 is also involved in the regulation of MHC class I expression in both avian and mammalian cells (38, 111). Similarly, IRF-10 is able to induce the expression and enhance the induction of MHC class I in fibroblasts exposed to type I IFN. The genes for GBP and MHC class I are induced by both types of IFNs, which is in contrast to the genes for OAS and Mx, which are only induced by type I IFN (15). It is interesting that OAS expression is reduced after treatment of IRF-10-espressing cells with IFN1. The ability of IRF-10 to regulate the expression of GBP and MHC class I and interfere with OAS expression suggests that IRF-10 may selectively regulate type II over type I IFN target genes.
The role of IRF-10 in the induction of GBP and MHC class I resembles the function of mammalian IRF-1. There are, however, two major differences between IRF-10 and IRF-1. In contrast to IRF-10, IRF-1 is the first IRF induced in response to the activators whereas IRF-10 has a very delayed kinetics of induction. The other difference is in their effects on cell proliferation. Both fibroblasts and lymphoid B-cell lines support the propagation of retrovirus overexpressing IRF-10 but not IRF-1. IRF-1 is known to induce a cytotoxic effect in several cell types and upregulates the expression of proapoptotic genes (84). In conclusion, IRF-10 has a unique role in regulating the switch from an innate to an adaptive immune response against virus without an antiproliferative effect.
The Rel/NF-κB and IRF families regulate IRF-10 expression.
v-Rel, c-Rel, and IRF-4 regulate IRF-10 expression in a cell-specific manner. The expression of other avian IRFs, including IRF-4, IRF-8, cIRF-3, and IRF-1, is also induced by v-Rel and c-Rel (39, 40). Several lines of evidence indicate that IRF-encoding genes are direct transcriptional targets of Rel/NF-κB and IRF proteins. Rel/NF-κB factors regulate IRF-4 and IRF-1 at the promoter level (32, 44). The cIRF-3 promoter contains κB sites responsible for dsRNA inducibility (66). Furthermore, both the avian and mammalian IRF-8 promoters contain a κB site and the κB site in the mammalian promoter is bound by Rel/NF-κB factors (16, 50). Similarly, the regulation of several IRFs by IFNs is mediated directly through IRF-binding sites in their promoters (16, 35, 60, 66). Therefore, it is likely that IRF-10 regulation by v-Rel, c-Rel, and IRF-4 also takes place at the promoter level.
IRF-10 expression was strongly induced by v-Rel in lymphoid cells. However, wild type c-Rel induced IRF-10 expression considerably less efficiently than did v-Rel. c-Rel is a critical Rel/NF-κB transcriptional regulator in B cells (33). Therefore, the low efficiency with which c-Rel induces the expression of IRF-10 may be one reason why IRF-10 is present at such low levels in bursal cells. v-Rel is a highly mutated form of c-Rel (27). v-Rel has a C-terminal deletion that removes a cytoplasmic retention domain, allowing nuclear translocation in the absence of an appropriate exogenous signal (12). By contrast, c-Rel is retained in the cytoplasm until cells receive appropriate signals. A c-Rel C-terminal deletion mutant protein that is expressed predominantly in the nucleus induces IRF-10 expression to significantly higher levels than does wild-type c-Rel. Therefore, c-Rel, if activated and translocated to the nucleus, is also capable of efficiently inducing IRF-10 expression. However, v-Rel apparently is still a more efficient inducer of IRF-10 expression than C-terminally truncated c-Rel. This is likely due the difference in the DNA-binding specificities of c-Rel and v-Rel, which, in turn, also contribute to their different transcriptional activities (74). Induction of IRF-10 expression by c-Rel, its C-terminally deleted mutant, and v-Rel correlates with their transformation potential, suggesting a possible role of IRF-10 in v-Rel transformation (39).
The IRF and Rel/NF-κB families participate in regulation of the immune response, and certain members of these two families cooperate in the regulation of the transcription of some genes (24, 85). v-Rel failed to induce the expression of IRF-10 in fibroblasts, but when IRF-4 or IRF-4ΔE6 was expressed at high levels together with v-Rel, expression of IRF-10 was induced in a synergistic manner. Synergism between the IRF-4 lymphocyte-specific factors and Rel/NF-κB family members may explain why IRF-10 is predominantly expressed constitutively in hematopoietic cells. The same synergism was also observed in the regulation of cIRF-3 by these factors.
In summary, a novel IRF family member has been identified and characterized. Although it is most closely related to IRF-4 and IRF-8, IRF-10 has a distinct constitutive and inducible expression pattern. Unlike that of other IRF family members, the induction of IRF-10 is delayed and requires protein synthesis. IRF-10 functions in the later stages of an antiviral defense by regulating IFN-γ target genes.
Acknowledgments
We are grateful to many colleagues for providing chicken cDNA clones. We thank to Robin Morgan (University of Delaware, Newark) for cDNA clone ptr1c.pk002.b9, which was isolated as a part of the University of Delaware chicken EST project. We thank Igor Dawid (National Institutes of Health) for IRF-1 and IRF-8, Roger Deeley and Caroline Grant (Queen's University, Kingston, Ontario, Canada) for cIRF-3, Thomas Gilmore (Boston University, Boston, Mass.) for c-rel, Christoph Jungwirth (Universität Würzburg, Würzburg, Germany) for IRF-2, Christine Sick and Peter Staeheli (Universität Freiburg, Freiburg, Germany) for IFN1 and IFN-γ, Kirsten Weining (Universität Freiburg) for Mx and GBP, and Y. Xu (Iowa State University, Ames) for MHC class II β-chain cDNA clones. We thank Emin Ulug (University of Texas, Austin) for the kind gift of VSV.
This study was supported by Public Health Service grant CA33192 from the National Cancer Institute.
REFERENCES
- 1.Akiyama, Y., and S. Kato. 1974. Two cell lines from lymphomas of Marek's disease. Biken J. 17:105-116. [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Au, W. C., P. A. Moore, W. Lowther, Y. T. Juang, and P. M. Pitha. 1995. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc. Natl. Acad. Sci. USA 92:11657-11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baba, T. W., B. P. Giroir, and E. H. Humphries. 1985. Cell lines derived from avian lymphomas exhibit two distinct phenotypes. Virology 144:139-151. [DOI] [PubMed] [Google Scholar]
- 6.Bernasconi, D., U. Schultz, and P. Staeheli. 1995. The interferon-induced Mx protein of chickens lacks antiviral activity. J. Interferon Cytokine Res. 15:47-53. [DOI] [PubMed] [Google Scholar]
- 7.Boehm, U., T. Klamp, M. Groot, and J. C. Howard. 1997. Cellular responses to interferon-γ. Annu. Rev. Immunol. 15:749-795. [DOI] [PubMed] [Google Scholar]
- 8.Bose, H. R., Jr. 1992. The Rel family: models for transcriptional regulation and oncogenic transformation. Biochim. Biophys. Acta 1114:1-17. [DOI] [PubMed] [Google Scholar]
- 9.Briken, V., H. Ruffner, U. Schultz, A. Schwarz, L. F. Reis, I. Strehlow, T. Decker, and P. Staeheli. 1995. Interferon regulatory factor 1 is required for mouse Gbp gene activation by gamma interferon. Mol. Cell. Biol. 15:975-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burýšek, L., W.-S. Yeow, B. Lubyová, M. Kellum, S. L. Schafer, Y. Q. Huang, and P. M. Pitha. 1999. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J. Virol. 73:7334-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushdid, P. B., D. M. Brantley, F. E. Yull, G. L. Blaeuer, L. H. Hoffman, L. Niswander, and L. D. Kerr. 1998. Inhibition of NF-κB activity results in disruption of the apical ectodermal ridge and aberrant limb morphogenesis. Nature 392:615-618. [DOI] [PubMed] [Google Scholar]
- 12.Capobianco, A. J., D. L. Simmons, and T. D. Gilmore. 1990. Cloning and expression of a chicken c-rel cDNA: unlike p59v-rel, p68c-rel is a cytoplasmic protein in chicken embryonic fibroblasts. Oncogene 5:257-265. [PubMed] [Google Scholar]
- 13.Chang, C. H., J. Hammer, J. E. Loh, W. L. Fodor, and R. A. Flavell. 1992. The activation of major histocompatibility complex class I genes by interferon regulatory factor-1 (IRF-1). Immunogenetics 35:378-384. [DOI] [PubMed] [Google Scholar]
- 14.DeLorbe, W. J., P. A. Luciw, H. M. Goodman, H. E. Varmus, and J. M. Bishop. 1980. Molecular cloning and characterization of avian sarcoma virus circular DNA molecules. J. Virol. 36:50-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Der, S. D., A. Zhou, B. R. G. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon α, β, or γ with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dosch, E., B. Zöller, I. Redmann-Müller, I. Nanda, M. Schmid, A. Viciano-Gofferge, and C. Jungwirth. 1998. The genomic structure of the chicken ICSBP gene and its transcriptional regulation by chicken interferon. Gene 210:265-275. [DOI] [PubMed] [Google Scholar]
- 17.Driggers, P. H., D. L. Ennist, S. L. Gleason, W. H. Mak, M. S. Marks, B. Z. Levi, J. R. Flanagan, E. Appella, and K. Ozato. 1990. An interferon γ-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc. Natl. Acad. Sci. USA 87:3743-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenbeis, C. F., H. Singh, and U. Storb. 1995. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 9:1377-1387. [DOI] [PubMed] [Google Scholar]
- 19.Eroshkin, A., and A. Mushegian. 1999. Conserved transactivation domain shared by interferon regulatory factors and Smad morphogens. J. Mol. Med. 77:403-405. [DOI] [PubMed] [Google Scholar]
- 20.Escalante, C. R., J. Yie, D. Thanos, and A. K. Aggarwal. 1998. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature 391:103-106. [DOI] [PubMed] [Google Scholar]
- 21.Filardo, E. J., M. F. Lee, and E. H. Humphries. 1994. Structural genes, not the LTRs, are the primary determinants of reticuloendotheliosis virus A-induced runting and bursal atrophy. Virology 202:116-128. [DOI] [PubMed] [Google Scholar]
- 22.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and, A. M. Skalka. 2000. Principles of virology: molecular biology, pathogenesis, and control. ASM Press, Washington, D.C.
- 23.Foo, S. Y., and G. P. Nolan. 1999. NF-κB to the rescue: RELs, apoptosis and cellular transformation. Trends Genet. 15:229-235. [DOI] [PubMed] [Google Scholar]
- 24.Genin, P., M. Algarte, P. Roof, R. Lin, and, J. Hiscott. 2000. Regulation of RANTES chemokine gene expression requires cooperativity between NF-κB and IFN-regulatory factor transcription factors. J. Immunol. 164:5352-5361. [DOI] [PubMed] [Google Scholar]
- 25.Gerondakis, S., R. Grumont, I. Rourke, and M. Grossmann. 1998. The regulation and roles of Rel/NF-κB transcription factors during lymphocyte activation. Curr. Opin. Immunol. 10:353-359. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225-260. [DOI] [PubMed] [Google Scholar]
- 27.Gilmore, T. D. 1999. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-Rel. Oncogene 18:6925-6937. [DOI] [PubMed] [Google Scholar]
- 28.Gilmore, T. D., M. Koedood, K. A. Piffat, and D. W. White. 1996. Rel/NF-κB/IκB proteins and cancer. Oncogene 13:1367-1378. [PubMed] [Google Scholar]
- 29.Grant, C. E., M. Z. Vasa, and R. G. Deeley. 1995. cIRF-3, a new member of the interferon regulatory factor (IRF) family that is rapidly and transiently induced by dsRNA. Nucleic Acids Res. 23:2137-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grossman, A., H.-W. Mittrücker, J. Nicholl, A. Suzuki, S. Chung, L. Antonio, S. Suggs, G. R. Sutherland, D. P. Siderovski, and T. W. Mak. 1996. Cloning of human lymphocyte-specific interferon regulatory factor (hLSIRF/hIRF4) and mapping of the gene to 6p23-p25. Genomics 37:229-233. [DOI] [PubMed] [Google Scholar]
- 31.Grossmann, M., Y. Nakamura, R. Grumont, and S. Gerondakis. 1999. New insights into the roles of ReL/NF-κB transcription factors in immune function, hemopoiesis and human disease. Int. J. Biochem. Cell Biol. 31:1209-1219. [DOI] [PubMed] [Google Scholar]
- 32.Grumont, R. J., and S. Gerondakis. 2000. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: modulation of interferon-regulated gene expression by Rel/nuclear factor κB. J. Exp. Med. 191:1281-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gugasyan, R., R. Grumont, M. Grossmann, Y. Nakamura, T. Pohl, D. Nesic, and S. Gerondakis. 2000. Rel/NF-κB transcription factors: key mediators of B-cell activation. Immunol. Rev. 176:134-140. [DOI] [PubMed] [Google Scholar]
- 34.Harada, H., T. Fujita, M. Miyamoto, Y. Kimura, M. Maruyama, A. Furia, T. Miyata, and T. Taniguchi. 1989. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 58:729-739. [DOI] [PubMed] [Google Scholar]
- 35.Harada, H., E.-I. Takahashi, S. Itoh, K. Harada, T.-A. Hori, and T. Taniguchi. 1994. Structure and regulation of the human interferon regulatory factor 1 (IRF-1) and IRF-2 genes: implications for a gene network in the interferon system. Mol. Cell. Biol. 14:1500-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harada, H., T. Taniguchi, and N. Tanaka. 1998. The role of interferon regulatory factors in the interferon system and cell growth control. Biochimie 80:641-650. [DOI] [PubMed] [Google Scholar]
- 37.Hatada, S., M. Kinoshita, S. Takahashi, R. Nishihara, H. Sakumoto, A. Fukui, M. Noda, and M. Asashima. 1997. An interferon regulatory factor-related gene (xIRF-6) is expressed in the posterior mesoderm during the early development of Xenopus laevis. Gene 203:183-188. [DOI] [PubMed] [Google Scholar]
- 38.Hobart, M., V. Ramassar, N. Goes, J. Urmson, and P. F. Halloran. 1997. IFN regulatory factor-1 plays a central role in the regulation of the expression of class I and II MHC genes in vivo. J. Immunol. 158:4260-4269. [PubMed] [Google Scholar]
- 39.Hrdličková, R., J. Nehyba, and H. R. Bose, Jr. 2001. Interferon regulatory factor 4 contributes to transformation of v-Rel-expressing fibroblasts. Mol. Cell. Biol. 21:6369-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hrdličková, R., J. Nehyba, and H. R. Bose. 1999. Reticuloendotheliosis viruses (Retroviridae), p. 1496-1503. In A. Granoff and R. G. Webster (ed.), Encyclopedia of virology, vol. 3. Academic Press, London, United Kingdom. [Google Scholar]
- 41.Hrdličková, R., J. Nehyba, and E. H. Humphries. 1994. In vivo evolution of c-rel oncogenic potential. J. Virol. 68:2371-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hrdličková, R., J. Nehyba, and E. H. Humphries. 1994. v-rel induces expression of three avian immunoregulatory surface receptors more efficiently than c-rel. J. Virol. 68:308-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iida, S., P. H. Rao, M. Butler, P. Corradini, M. Boccadoro, B. Klein, R. S. K. Chaganti, and R. Dalla-Favera. 1997. Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat. Genet. 17:226-230. [DOI] [PubMed] [Google Scholar]
- 44.Imanishi, D., K. Yamamoto, H. Tsushima, Y. Miyazaki, K. Kuriyama, M. Tomonaga, and, T. Matsuyama. 2000. Identification of a novel cytokine response element in the human IFN regulatory factor-1 gene promoter. J. Immunol. 165:3907-3916. [DOI] [PubMed] [Google Scholar]
- 45.Itoh, S., H. Harada, T. Fujita, T. Mimura, and T. Taniguchi. 1989. Sequence of a cDNA coding for human IRF-2. Nucleic Acids Res. 17:8372.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 47.Joklik, W. K. 1991. Interferons, p. 343-370. In B. N. Fields and D. M. Knipe (ed.), Fundamental virology, second ed. Raven Press, New York, N.Y.
- 48.Jungwirth, C., M. Rebbert, K. Ozato, H. J. Degen, U. Schultz, and I. B. Dawid. 1995. Chicken interferon consensus sequence-binding protein (ICSBP) and interferon regulatory factor (IRF) 1 genes reveal evolutionary conservation in the IRF family. Proc. Natl. Acad. Sci. USA 92:3105-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanegae, Y., A. T. Tavares, J. C. I. Belmonte, and I. M. Verma. 1998. Role of Rel/NF-κB transcription factors during the outgrowth of the vertebrate limb. Nature 392:611-614. [DOI] [PubMed] [Google Scholar]
- 50.Kanno, Y., C. A. Kozak, C. Schindler, P. H. Driggers, D. L. Ennist, S. L. Gleason, J. E. Darnell, Jr., and K. Ozato. 1993. The genomic structure of the murine ICSBP gene reveals the presence of the gamma interferon-responsive element, to which an ISGFα subunit (or similar) molecule binds. Mol. Cell. Biol. 13:3951-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim, S., E. H. Humphries, L. Tjoelker, L. Carlson, and C. B. Thompson. 1990. Ongoing diversification of the rearranged immunoglobulin light-chain gene in a bursal lymphoma cell line. Mol. Cell. Biol. 10:3224-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koerner, T. J., J. E. Hill, A. M. Myers, and A. Tzagoloff. 1991. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 194:477-490. [DOI] [PubMed] [Google Scholar]
- 53.Kozak, M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kralova, J., A. S. Liss, W. Bargmann, and H. R. Bose, Jr. 1998. AP-1 factors play an important role in transformation induced by the v-rel oncogene. Mol. Cell. Biol. 18:2997-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kroemer, G., R. Zoorob, and C. Auffray. 1990. Structure and expression of a chicken MHC class I gene. Immunogenetics 31:405-409. [DOI] [PubMed] [Google Scholar]
- 56.Lander, E. S., L. M. Linton, B. Birren, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 57.Lau, J. F., J. P. Parisien, and C. M. Horvath. 2000. Interferon regulatory factor subcellular localization is determined by a bipartite nuclear localization signal in the DNA-binding domain and interaction with cytoplasmic retention factors. Proc. Natl. Acad. Sci. USA 97:7278-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin, R., C. Heylbroeck, P. Genin, P. M. Pitha, and J. Hiscott. 1999. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol. Cell. Biol. 19:959-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lipsick, J. S., J. Manak, N. Mitiku, C.-K. Chen, P. Fogarty, and E. Guthrie. 2001. Functional evolution of the Myb oncogene family. Blood Cells Mol. Dis. 27:456-458. [DOI] [PubMed] [Google Scholar]
- 60.Lu, R., W.-C. Au, W.-S. Yeow, N. Hageman, and P. M. Pitha. 2000. Regulation of the promoter activity of interferon regulatory factor-7 gene. J. Biol. Chem. 275:31805-31812. [DOI] [PubMed] [Google Scholar]
- 61.Lubyova, B., and P. M. Pitha. 2000. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J. Virol. 74:8194-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mamane, Y., C. Heylbroeck, P. Genin, M. Algarte, M. J. Servant, C. LePage, C. DeLuca, H. Kwon, R. Lin, and J. Hiscott. 1999. Interferon regulatory factors: the next generation. Gene 237:1-14. [DOI] [PubMed] [Google Scholar]
- 63.Marienfeld, R., I. Nanda, B. Zöller, M. Schmid, M. Rebbert, and C. Jungwirth. 1997. Cloning of chicken interferon regulatory factor-2 (IRF-2) cDNA: expression and mapping of the IRF-2 gene. J. Interferon Cytokine Res. 17:219-227. [DOI] [PubMed] [Google Scholar]
- 64.Massa, P. T., L. W. Whitney, C. Wu, S. L. Ropka, and K. W. Jarosinski. 1999. A mechanism for selective induction of 2′-5′ oligoadenylate synthetase, anti-viral state, but not MHC class I genes by interferon-beta in neurons. J. Neurovirol. 2:161-171. [DOI] [PubMed] [Google Scholar]
- 65.Matsuyama, T., A. Grossman, H. W. Mittrucker, D. P. Siderovski, F. Kiefer, T. Kawakami, C. D. Richardson, T. Taniguchi, S. K. Yoshinaga, and T. W. Mak. 1995. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE). Nucleic Acids Res. 23:2127-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.May, D. L., C. E. Grant, and, R. G. Deeley. 2000. Cloning and promoter analysis of the chicken interferon regulatory factor-3 gene. DNA Cell Biol. 19:555-566. [DOI] [PubMed] [Google Scholar]
- 67.Mellon, P., V. Parker, Y. Gluzman, and T. Maniatis. 1981. Identification of DNA sequences required for transcription of the human α1-globin gene in a new SV40 host-vector system. Cell 27:279-288. [DOI] [PubMed] [Google Scholar]
- 68.Meraro, D., S. Hashmueli, B. Koren, A. Azriel, A. Oumard, S. Kirchhoff, H. Hauser, S. Nagulapalli, M. L. Atchison, and B. Z. Levi. 1999. Protein-protein and DNA-protein interactions affect the activity of lymphoid-specific IFN regulatory factors. J. Immunol. 163:6468-6478. [PubMed] [Google Scholar]
- 69.Miyamoto, M., T. Fujita, Y. Kimura, M. Maruyama, H. Harada, Y. Sudo, T. Miyata, and T. Taniguchi. 1988. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-β gene regulatory elements. Cell 54:903-913. [DOI] [PubMed] [Google Scholar]
- 70.Moore, P. S., C. Boshoff, R. A. Weis, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 71.Morrison, L. E., G. Boehmelt, and P. J. Enrietto. 1992. Mutations in the rel-homology domain alter the biochemical properties of v-rel and render it transformation defective in chicken embryo fibroblasts. Oncogene 7:1137-1147. [PubMed] [Google Scholar]
- 72.Moscovici, C., N. Zeller, and M. G. Moscovici. 1982. Continuous lines of AMV-transformed nonproducer cells: growth and oncogenic potential in the chick embryo, p. 435-449. In R. F. Revoltella et al. (ed.), Expression of differentiated functions in cancer cells. Raven Press, New York, N.Y.
- 73.Nazerian, K., E. A. Stephens, J. M. Sharma, L. M. Lee, M. Gailitis, and R. I. Witter. 1977. A nonproducer T lymphoblastoid cell line from Marek's disease transplantable tumor (JMV). Avian Dis. 21:69-76. [PubMed] [Google Scholar]
- 74.Nehyba, J., R. Hrdličková, and H. R. Bose, Jr. 1997. Differences in κB DNA-binding properties of v-Rel and c-Rel are the result of oncogenic mutations in three distinct functional regions of the Rel protein. Oncogene 14:2881-2897. [DOI] [PubMed] [Google Scholar]
- 75.Nehyba, J., R. Hrdličková, and E. H. Humphries. 1994. Evolution of the oncogenic potential of v-rel: rel-induced expression of immunoregulatory receptors correlates with tumor development and in vitro transformation. J. Virol. 68:2039-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nguyen, H., J. Hiscott, and P. M. Pitha. 1997. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 8:293-312. [DOI] [PubMed] [Google Scholar]
- 77.Okuno, H., T. Suzuki, T. Yoshida, Y. Hashimoto, T. Curran, and H. Iba. 1991. Inhibition of jun transformation by a mutated fos gene: design of an anti-oncogene. Oncogene 6:1491-1497. [PubMed] [Google Scholar]
- 78.Ortiz, M. A., J. Light, R. A. Maki, and N. Assa-Munt. 1999. Mutation analysis of the Pip interaction domain reveals critical residues for protein-protein interactions. Proc. Natl. Acad. Sci. USA 96:2740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parcells, M. S., R. L. Dienglewicz, A. S. Anderson, and R. W. Morgan. 1999. Recombinant Marek's disease virus (MDV)-derived lymphoblastoid cell lines: regulation of a marker gene within the context of the MDV genome. J. Virol. 73:1362-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rao, A., K. Kline, and B. G. Sanders. 1990. Immune abnormalities in avian erythroblastosis virus-infected chickens. Cancer Res. 50:4764-4770. [PubMed] [Google Scholar]
- 81.Rayet, B., and C. Gelinas. 1999. Aberrant rel/nfkb genes and activity in human cancer. Oncogene 18:6938-6947. [DOI] [PubMed] [Google Scholar]
- 82.Richardson, M. P., B. H. Tay, B. Y. Goh, B. Venkatesh, and, S. Brenner. 2001. Molecular cloning and genomic structure of an interferon regulatory factor encoding gene in the pufferfish (Fugu rubripes). Mar. Biotechnol. 3:145-151. [DOI] [PubMed] [Google Scholar]
- 83.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 84.Sato, M., T. Taniguchi, and, N. Tanaka. 2001. The interferon system and interferon regulatory factor transcription factors—studies from gene knockout mice. Cytokine Growth Factor Rev. 12:133-142. [DOI] [PubMed] [Google Scholar]
- 85.Saura, M., C. Zaragoza, C. Bao, A. McMillan, and C. J. Lowenstein. 1999. Interaction of interferon regulatory factor-1 and nuclear factor κB during activation of inducible nitric oxide synthase transcription. J. Mol. Biol. 289:459-471. [DOI] [PubMed] [Google Scholar]
- 86.Schaper, F., S. Kirchhoff, G. Posern, M. Köster, A. Oumard, R. Sharf, B. Z. Levi, and H. Hauser. 1998. Functional domains of interferon regulatory factor I (IRF-1). Biochem. J. 335:147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmidt, M., S. Nagel, J. Proba, C. Thiede, M. Ritter, J. F. Waring, F. Rosenbauer, D. Huhn, B. Wittig, I. Horák, and A. Neubauer. 1998. Lack of Interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemia. Blood 91:22-29. [PubMed] [Google Scholar]
- 88.Schreiber, E., P. Matthias, M. M. Müller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’ prepared from a small number of cells. Nucleic Acids Res. 17:6419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schultz, U., C. Rinderle, M. J. Sekellick, P. I. Marcus, and P. Staeheli. 1995. Recombinant chicken interferon from Escherichia coli and transfected COS cells is biologically active. Eur. J. Biochem. 229:73-76. [DOI] [PubMed] [Google Scholar]
- 90.Schumacher, B., D. Bernasconi, U. Schultz, and P. Staeheli. 1994. The chicken Mx promoter contains an ISRE motif and confers interferon inducibility to a reporter gene in chick and monkey cells. Virology 203:144-148. [DOI] [PubMed] [Google Scholar]
- 91.Schwemmle, M., B. Kaspers, A. Irion, P. Staeheli, and U. Schultz. 1996. Chicken guanylate-binding protein. Conservation of GTPase activity and induction by cytokines. J. Biol. Chem. 271:10304-10308. [DOI] [PubMed] [Google Scholar]
- 92.Sharf, R., A. Azriel, F. Lejbkowicz, S. S. Winograd, R. Ehrlich, and B. Z. Levi. 1995. Functional domain analysis of interferon consensus sequence binding protein (ICSBP) and its association with interferon regulatory factors. J. Biol. Chem. 270:13063-13069. [DOI] [PubMed] [Google Scholar]
- 93.Sharf, R., D. Meraro, A. Azriel, A. M. Thornton, K. Ozato, E. F. Petricoin, A. C. Larner, F. Schaper, H. Hauser, and B. Z. Levi. 1997. Phosphorylation events modulate the ability of interferon consensus sequence binding protein to interact with interferon regulatory factors and to bind DNA. J. Biol. Chem. 272:9785-9792. [DOI] [PubMed] [Google Scholar]
- 94.Sick, C., U. Schultz, and P. Staeheli. 1996. A family of genes coding for two serologically distinct chicken interferons. J. Biol. Chem. 271:7635-7639. [DOI] [PubMed] [Google Scholar]
- 95.Stoker, A. W., and M. J. Bissell. 1987. Quantitative immunocytochemical assay for infectious avian retroviruses. J. Gen. Virol. 68:2481-2485. [DOI] [PubMed] [Google Scholar]
- 96.Suhara, W., M. Yoneyama, H. Yonekawa, and T. Fujita. 1996. Structure of mouse interferon stimulated gene factor 3 gamma (ISGF3 γ/p48) cDNA and chromosomal localization of the gene. J. Biochem. (Tokyo) 119:231-234. [DOI] [PubMed] [Google Scholar]
- 97.Tirunagaru, V. G., L. Sofer, J. Cui, and, J. Burnside. 2000. An expressed sequence tag database of T-cell-enriched activated chicken splenocytes: sequence analysis of 5251 clones. Genomics 66:144-151. [DOI] [PubMed] [Google Scholar]
- 98.Veals, S. A., T. Santa Maria, and D. E. Levy. 1993. Two domains of ISGF3γ that mediate protein-DNA and protein-protein interactions during transcription factor assembly contribute to DNA-binding specificity. Mol. Cell. Biol. 13:196-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Veals, S. A., C. Schindler, D. Leonard, X.-Y. Fu, R. Aebersold, J. E. Darnell, Jr., and D. E. Levy. 1992. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 12:3315-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Venter, J. C., M. D. Adams, E. W. Myers, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 101.Weisz, A., P. Marx, R. Sharf, E. Appella, P. H. Driggers, K. Ozato, and B. Z. Levi. 1992. Human interferon consensus sequence binding protein is a negative regulator of enhancer elements common to interferon-inducible genes. J. Biol. Chem. 267:25589-25596. [PubMed] [Google Scholar]
- 102.Willman, C. L., C. E. Sever, M. G. Pallavicini, H. Harada, N. Tanaka, M. L. Slovak, H. Yamamoto, K. Harada, T. C. Meeker, A. F. List, and T. Taniguchi. 1993. Deletion of IRF-1, mapping to chromosome 5q31.1, in human leukemia and preleukemic myelodysplasia. Science 259:968-971. [DOI] [PubMed] [Google Scholar]
- 103.Xi, H., and G. Blanck. 2000. Interferon regulatory factor-2 point mutations in human pancreatic tumors. Int. J. Cancer 87:803-808. [PubMed] [Google Scholar]
- 104.Xi, H., B. Goodwin, A. T. Shepherd, and, G. Blanck. 2001. Impaired class II transactivator expression in mice lacking interferon regulatory factor-2. Oncogene 20:4219-4227. [DOI] [PubMed] [Google Scholar]
- 105.Xu, Y., J. Pitcovski, L. Peterson, C. Auffray, Y. Bourlet, B. M. Gerndt, A. W. Nordskog, S. J. Lamont, and C. M. Warner. 1989. Isolation and characterization of three class II MHC genomic clones from the chicken. J. Immunol. 142:2122-2132. [PubMed] [Google Scholar]
- 106.Yabu, T., H. Hirose, I. Hirono, T. Katagiri, T. Aoki, and E. Yamamoto. 1998. Molecular cloning of a novel interferon regulatory factor in Japanese flounder, Paralichthys olivaceus. Mol. Mar. Biol. Biotechnol. 7:138-144. [PubMed] [Google Scholar]
- 107.Yeow, W.-S., W.-C. Au, Y.-T. Juang, C. D. Fields, C. L. Dent, D. R. Gewert, and P. M. Pitha. 2000. Reconstitution of virus-mediated expression of interferon α genes in human fibroblast cells by ectopic interferon regulatory factor-7. J. Biol. Chem. 275:6313-6320. [DOI] [PubMed] [Google Scholar]
- 108.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yu-Lee, L. Y., J. A. Hrachovy, A. M. Stevens, and L. A. Schwarz. 1990. Interferon-regulatory factor 1 is an immediate-early gene under transcriptional regulation by prolactin in Nb2 T cells. Mol. Cell. Biol. 10:3087-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang, L., and J. S. Pagano. 1997. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 17:5748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zoeller, B., M. Popp, A. Walter, I. Redmann-Müller, E. Lodemann, and C. Jungwirth. 1998. Overexpression of chicken interferon regulatory factor-1 (Ch-IRF-1) induces constitutive expression of MHC class I antigens but does not confer virus resistance to a permanent chicken fibroblast cell line. Gene 222:269-278. [DOI] [PubMed] [Google Scholar]
- 112.Zöller, B., I. Redman-Müller, I. Nanda, M. Guttenbach, E. Dosch, M. Schmid, R. Zoorob, and, C. Jungwirth. 2000. Sequence comparison of avian interferon regulatory factors and identification of the avian CEC-32 cell as a quail cell line. J. Interferon Cytokine Res. 20:711-717. [DOI] [PubMed] [Google Scholar]