Abstract
Sphingomyelin is a major lipid in the bilayer of subcellular membranes of eukaryotic cells. Different sphingomyelinases catalyze the initial step in the catabolism of sphingomyelin, the hydrolysis to phosphocholine and ceramide. Sphingomyelinases have been postulated to generate ceramide as a lipophilic second messenger in intracellular signaling pathways involved in cell proliferation, differentiation, or apoptosis. To elucidate the function of the first cloned Mg2+-dependent, neutral sphingomyelinase (nSMase 1) in sphingomyelin catabolism and its potential role in signaling processes in a genetic and molecular approach, we have generated an nSMase 1-null mutant mouse line by gene targeting. The nSMase 1-deficient mice show an unconspicuous phenotype and no accumulation or changed metabolism of sphingomyelin or other lipids, despite grossly reduced nSMase activity in all organs except brain. We also addressed the recent proposal that nSMase 1 possesses lysophospholipase C activity. The unaltered metabolism of lysophosphatidylcholine or lyso-platelet-activating factor excludes the proposed role of nSMase 1 as a lysophospholipase C.
Acid sphingomyelinases (aSMases) and neutral sphingomyelinases (nSMase) (sphingomyelin [SPM]-phosphodiesterase, EC 3.1.4.12) cleave the phosphodiester bond of SPM to ceramide and phosphocholine. SPM is distributed in a gradient fashion in membranes of the endoplasmic reticulum, Golgi apparatus, and lysosomes to the plasma membrane. Sphingomyelinases differ in their tissue and subcellular distribution, enzymatic properties, and regulation.
The lysosomal aSMase (unigene nomenclature, SMPD1; SPM-phosphodiesterase 1) was the first sphingomyelinase to be cloned and characterized biochemically (16, 17, 20, 21, 31). In gene-targeting experiments by homologous recombination, asmase-null allelic mice were generated, which, like the human neurovisceral form (type A) of Niemann-Pick disease accumulate SPM in the reticuloendothelial system, predominantly in the liver, spleen, lung, bone marrow, and brain, leading to death in early childhood (9, 19). SPM storage also leads to unbalanced cholesterol-sphingolipid ratios in aSMase-deficient mice in the plasma membrane, where raft formation and raft-associated processes are severely perturbed (18).
nSMase activity was described first in brain (5, 22) and later found in several mammalian tissues and cell lines (12). The most active membrane-bound Mg2+-dependent sphingomyelinase activity occurs in the central nervous system and to a lesser extent in other tissues. nSMase activity is believed to be responsible for stress-induced ceramide generation and for relaying antiapoptotic signals from cell surface receptors (12, 42). The recent debate on the interpretation of these experiments (7, 40, 41) underscores the need for a genetic and molecular approach to unravel the role of the neutral mammalian nSMases in their genuine environment.
We recently identified and characterized the first two mammalian nSMases (nSMase 1 and nSMase 2, in unigene nomenclature SMPD2 and SMPD3), integral membrane proteins with remote similarity to the secreted bacterial sphingomyelinases. nSMase 1 is ubiquitously expressed with the highest mRNA and protein amounts in kidney (34). nSMase 2, which has a different domain structure, is expressed mainly in brain (8).
We found that the nSMase 1 protein is located in the endoplasmic reticulum (35), which was confirmed by other laboratories (15, 24). This subcellular localization makes a signaling function of this isoform unlikely, which was underscored by studies unable to confirm the influence of overexpressed nSMase 1 in tumor necrosis factor alpha-induced signaling pathways (34) and on Fas-induced apoptosis (33). Recently, analyses of the lipid pattern of nSMase 1-overexpressing cells led to the conclusion that the in vivo function of the enzyme might be that of a lysophospholipase C with lysophosphatidylcholine (lyso-PC) and lyso-platelet-activating factor (lyso-PAF) as substrates (28).
The yeast homolog of nSMase 1 called ISC1 (inositol phosphosphingolipid phospholipase C) uses inositol phosphosphingolipids, while the yeast SPM analogs are substrates (29). This enzyme does not hydrolyze the lysophospholipids, but instead the yeast null mutant develops a storage of inositol phosphosphingolipids. This result points to an essential role of the mammalian nSMase 1 homolog in SPM metabolism.
In order to elucidate the function of nSMase 1 in vivo, we generated an nSMase 1-deficient mouse line by gene targeting in embryonic stem (ES) cells. Surprisingly, nsmase 1−/− mice are phenotypically normal and show, unlike the yeast mutant, neither lipid accumulation nor changes in SPM levels detectable with the presently available analytical tools, despite a gross reduction of nSMase activity in all organs except in the central nervous system.
MATERIALS AND METHODS
Cloning of the murine nsmase 1 gene and construction of the targeting vector.
Genomic nsmase 1 sequences were cloned from a λ-phage mouse genomic library (129/SvEvTacfBR; Stratagene) screened with a 770-bp HindIII cDNA fragment from the murine nSMase 1 cDNA (34).
Two overlapping 8-kb clones were shown to contain the complete gene. They were mapped and sequenced to yield the information about the structure of the genomic locus.
The nsmase 1 gene spans approximately 3.3 kb and contains 10 exons. Because of the lack of unique restriction sites, we decided to delete a large portion of the gene from the end of exon II to the beginning of exon VIII.
For the construction of the targeting vector, the EcoRI site of pPNT (36) was deleted by EcoRI digestion and blunt-end religation. Then a 2.2-kb KpnI-SmaI fragment isolated originally from the λ clone was inserted blunt end into the BamHI site of pPNT. A 4.5-kb 5′ fragment was amplified by PCR with the primers 5′-Not-s (5′-GCGGCCGCACAGCCTGAGATGCCCTGTCCT-3′) introducing a NotI restriction site and ExonIIXho-as (5′-CTCGAGTCCGCCGGCAGTCCTGGAAGGAAT-3′) introducing an XhoI restriction site. After the 4.5-kb fragment was subcloned into pCRII (Invitrogen), the fragment was introduced into the NotI and XhoI sites of pPNT (Fig. 1).
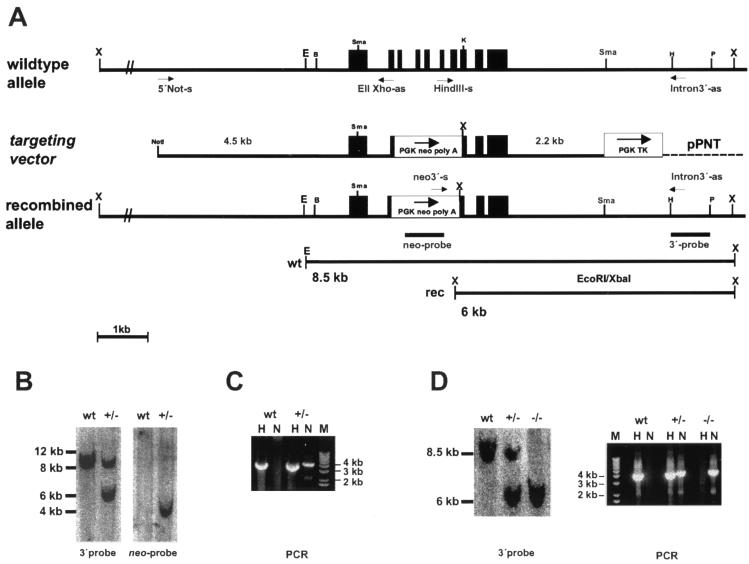
FIG. 1.
Gene targeting of nsmase 1. (A) Strategy for the targeted disruption of the gene locus. The wild-type allele, the targeting vector, the recombined gene locus, and a schematic view of the occurring restriction fragment length polymorphism and the probes and primers used are shown. (B) Southern blot analysis of EcoRI-XbaI-digested ES cell DNA. The blot was probed with the 3′ probe and the neo probe, showing the fragments indicated in panel A. wt, wild type. (C) PCR analysis of ES cell DNA with primers specific for the wild-type allele (H) and the targeted allele (N). (D) Analysis of mouse tail DNA showing the expected restriction fragments and PCR fragments for each genotype. X, XbaI; E, EcoRI, B, BamHI; Sma, SmaI; K, KpnI; H, HindIII; and P, PstI.
ES cell culture and production of chimeric mice.
ES cell lines R1 (14), Cj7 (32), and MpiII (39) were cultured, electroporated, and selected as described before (19). Geneticin (G418)-resistant clones were plated on gelatin-coated 24 wells and were analyzed by Southern blotting and PCR. Homologously recombined clones were expanded and used for production of chimeric males by blastocyst injection. Backcrossing of the chimeric males with C57BL/6 females resulted in germ line transmission and establishment of the nsmase 1-deficient mouse line.
Genotyping by Southern blotting and PCR.
Genotyping was performed with total DNA from ES cells and tail biopsy samples. For Southern blotting, the XbaI-EcoRI-digested DNA was hybridized with a 0.9-kb PstI-HindIII fragment indicated in Fig. 1. The wild-type allele yielded a 8.5-kb fragment, while the targeted allele produced a 6-kb fragment due to the restriction site introduced with the PGK neo-pA cassette. Radioactive signals were detected by phosphorimaging.
Homologous recombination was proven additionally by PCR using either the primer combination HindIII-sense (5′-GCCCAAGCTTGGGAACTGGCCCAG-3′, located in exon VI) and 3′Intron-as (5′-GGAGGGAGAGGGAGACTGGACAATAAGTAG-3′, located externally) for the wild-type allele, indicated as H, or neo3′-sense (5′-AGTGTAGCGCCTTCTATCGTTTTCTTG-ACG- 3′, located at the 3′end of the PGK neo box) and 3′Intron-as (indicated as N), respectively, for the targeted allele, yielding ∼4-kb fragments each (Fig. 1).
RNA analysis.
Total RNA of kidney, liver, and brain was prepared with Trizol (Gibco BRL) and poly(A)+ mRNA from total RNA using Oligotex mRNA Midi Kits (Qiagen). Northern blots were probed with a 770-bp HindIII mouse nSMase 1 cDNA fragment, a 350-bp mouse β-actin fragment generated by reverse transcriptase PCR with the primers β-actin-s (5′-TGGAATCCTGTGGCATCCATGAA-3′) and β-actin-as (5′-TAAAACGCAGCTCAGTAACAGTC-3′) and a 700-bp PstI fragment isolated from the neo cassette of pPNT. Radioactive signals were detected by phosphorimaging.
Protein analysis.
For Western blotting and enzymatic assays, membrane proteins were extracted from different organs. Organs were homogenized with a Potter-Elvejhem in homogenization buffer (50 mM Tris HCl, pH 7.4, 10 mM EDTA, 10 mM EGTA, 5 mM dithiothreitol, 0.32 M sucrose, and 2× Complete without EDTA [Roche]). Nuclei were sedimented for 10 min at 900 × g (4°C). The supernatant was centrifuged for 1 h (100,000 × g, 4°C). Membrane proteins were extracted from the sediment for 1 h (4°C) with extraction buffer (homogenization buffer + 0.5% Triton X-100). Insoluble material was sedimented (100,000 × g, 20 min, 4°C), and the protein amount was determined by Bradford assay (2).
For Western blot analysis, 100- to 200-μg membrane proteins were separated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis and were electroblotted onto a nitrocellulose membrane (Schleicher & Schuell). After blocking overnight with phosphate-buffered saline (PBS)-3% fatty acid-free milk powder-0.2% Tween 20 at 4°C, blots were incubated with anti-mouse nSMase 1 antibody (35) (2 μg/ml in PBS-3% bovine serum albumin). Blots were washed three times with PBS-0.1% NP-40 and PBS alone each. After incubation with the second antibody (anti-rabbit immunoglobulin G, peroxidase conjugated) (Sigma) and subsequent washings, the chemiluminescent signals were detected after incubation with LumiLight Western blotting substrate (Roche) on X-ray films (Kodak).
Specific nSMase activities from membrane protein fractions were measured as described before (35).
Lipid analysis.
Lipids were extracted from organs and cells (1). Briefly, tissues were homogenized in 1 ml of PBS, extracted with 3 ml of chloroform/methanol (1:2), and 0.8 ml of 1 M NaCl for 10 min at room temperature by magnetic stirring. After the addition of 1 ml of chloroform and 1 ml of 1 M NaCl, phases were separated by centrifugation. The organic phase was evaporated in a stream of nitrogen.
Insaponifiable sphingolipids, etherlipids, and plasmalogens were obtained by alkaline hydrolysis of total lipids in 1 ml of 0.5 N methanolic KOH at 37°C for 2 h. Water (1 ml) was added, and the mixture was extracted three times with chloroform. Phases were separated by centrifugation. The combined chloroform extracts were evaporated to dryness and were dissolved in chloroform-methanol (2:1). Total and alkali-stable lipids were separated on silica gel high-performance thin-layer chromatography (HPTLC) plates (Merck) using the solvent system chloroform-methanol-water (65:25:4, vol/vol/vol), chloroform/methanol (15:1, vol/vol), or a combination of both. Phospholipid bands were visualized with Zinzadze reagent (3) and glycosphingolipids according to Ledeen and Yu (11). Charring with 50% H2SO4 at 120°C for 15 min visualized all lipid bands. Individual lipid classes were identified by comigration with standards.
Cells were labeled with [1-14C]acetic acid (52 mCi/mmol), [1-14C-palmitoyl]lyso-PC (55 mCi/mmol), and [1-o-octadecyl-3H]lyso-PAF (167 mCi/mmol) (Amersham Pharmacia Biotech) for the times indicated in the text by adding the substances to the culture medium; lipids were extracted and separated as stated above. Radioactive signals were detected by phosphorimaging and quantified with the corresponding software (Image Quant; Molecular Dynamics).
Expression profiling.
Total pooled RNA from kidney was isolated from four age-matched wild-type and nsmase 1−/− mice with Trizol reagent and was further purified by using RNA Easy kits (Qiagen). Ten micrograms of total RNA was reverse transcribed, amplified, and labeled as described by the manufacturer (Affymetrix). Biotin-labeled cRNA was hybridized to oligonucleotide arrays containing probes for ∼12,000 murine genes (Mouse MG-U74Av; Affymetrix). Arrays were washed, stained with antibiotin streptavidin-phycoerythrin-labeled antibody, and scanned by using the GeneChip system (Hewlett-Packard) and GENECHIP 4.1. software (Affymetrix) to determine the expression of each gene. Intensity values were scaled by calculating the overall signal for each array type.
RESULTS
Targeting of the murine nsmase 1 gene.
To elucidate the biological function of nSMase 1, we inactivated the nsmase 1 gene in the mouse.
The nsmase 1 gene was isolated from a mouse genomic phage library on two independent overlapping clones and sequenced. The gene spans only 3.3 kb of genomic sequence and contains 10 exons. Exons II to VIII were replaced by the neo cassette, and a targeting vector was constructed consisting of (i) a 4.5-kb 5′ fragment with the promoter, the entire first exon, and a portion of exon II; (ii) the neo cassette under the control of the pgk promoter as positive selection marker; (iii) a 2.2-kb 3′ fragment containing the remaining exons VIII to X; and (iv) the HSV-tk cassette under the control of the pgk promoter as a negative selection marker (Fig. 1A). The targeting construct was linearized with NotI for transfection into different ES cell lines. Three homologously recombined clones were isolated by combined G418-ganciclovir double selection and were characterized (i) by the truncated genomic sequence recognized by the 3′ probe, (ii) by the neo sequence with the neo probe (Fig. 1B), and (iii) by PCR with primers specific for the recombined allele (Fig. 1C).
Generation of nSMase 1-deficient mice.
The three ES cell clones, one derived from R1 and two derived from Cj7, were used for blastocyst injection, and highly chimeric males were produced. However, only one Cj7 clone showed germ line transmission but with low frequency (∼4%). Genotyping of offspring of heterozygous crosses by Southern blotting and PCR (Fig. 1D) showed a normal Mendelian distribution of genotypes and sexes. Homozygous animals of both sexes were fertile as proven by crossing with wild-type animals. This excludes an important role of nSMase 1 in mouse embryogenesis or development.
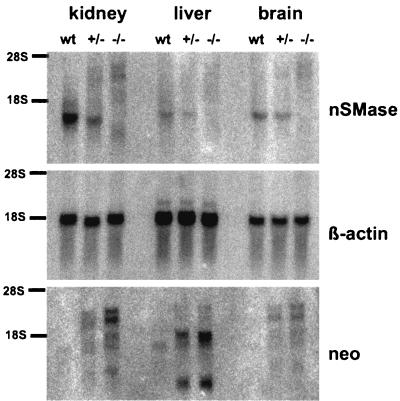
Northern blot analyses of kidney, liver, and brain mRNA proved the absence of the nSMase-specific mRNA with the correct size in homozygous animals and an approximately 50% reduction in heterozygous animals. Several weak signals became visible most prominently in homozygous animals by hybridization with the neo probe (Fig. 2). Reverse transcriptase PCR and sequencing identified them as splice products of the first exon with fragments of the selection marker and exons IX and X (data not shown). However, these chimeric mRNAs can only be translated to polypeptides with 29 amino acid residues or less.
FIG. 2.
Northern blot analysis of kidney, liver, and brain poly(A) RNA. The blot was hybridized with an nSMase 1-specific cDNA probe and a β-actin probe for normalization. The nSMase 1-specific signal disappears in homozygous animals, but several other signals are visible, most prominently in kidney. These signals hybridized also with the neo probe, indicating splice products of the gene with the neo box. wt, wild type.
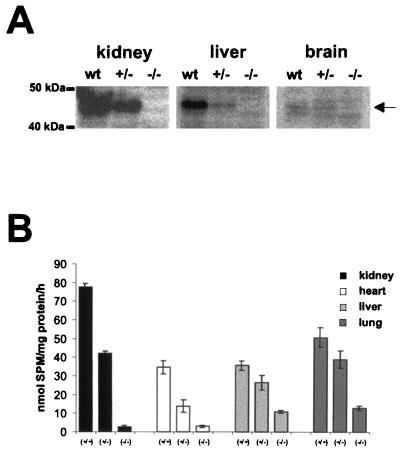
We next explored the presence of nSMase 1 protein in the membrane fractions of organs of homozygous animals by Western blotting using the nSMase 1-specific antibody (35). No specific signal was detectable in homozygous animals. The blot also reveals the different amounts of nSMase 1 protein in the organs of wild-type animals (Fig. 3). Note that 1.5 times more liver and brain proteins were used in contrast to kidney. This indicates the low degree of expression of nSMase 1, especially in brain.
FIG. 3.
(A) Western blot analysis of kidney, liver, and brain membrane proteins showing the absence of nSMase 1 protein in homozygous animals. wt, wild type. (B) nSMase activities of different organs. Different amounts of membrane protein extracts were used for the in vitro nSMase assay, and specific enzymatic activities were determined. The residual activity in kidney and heart from nsmase 1−/− animals corresponds to the background of the assay, whereas liver and lung showed significant residual activities.
Absence of nSMase 1 protein leads to reduced nSMase activity in organs but leaves lipid composition unaltered.
We explored the specific nSMase enzymatic activities in the membrane fraction of different organs of wild-type and nsmase 1-null allelic mice. The analysis revealed organs with (i) no residual activity like kidney and heart, (ii) a significant residual activity, like liver, lung, thymus, and testes (Fig. 4) and (iii) those like brain and intestine, with nSMase activity that was still very high (not shown). These results clearly indicate the existence of one or more additional nSMases, at least in brain.
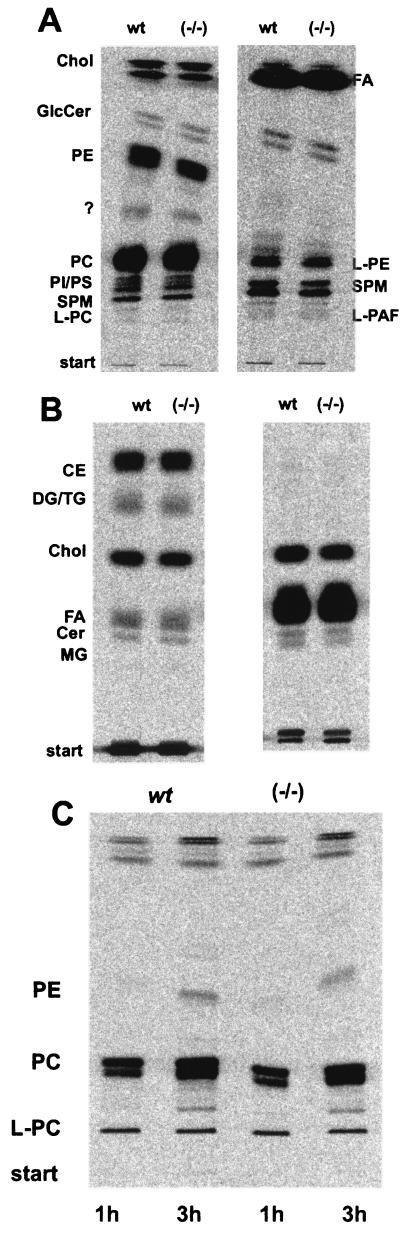
FIG. 4.
Lipid analysis of [1-14C]acetate-labeled EMFIs. Cells were labeled for 24 h, and total lipids were extracted. Equal amounts of radioactive lipids were separated on TLC plates in different solvent systems. Labeled lipids were detected and quantified by phosphorimaging. (A) Total lipids (left lanes) and alkali-stable lipids (right lanes) separated in chloroform-methanol-water (65:25:4, vol/vol/vol). No differences were found between wild-type and nsmase 1−/− cells. (B) Total lipids (left) and alkali-stable lipids (right) separated in chloroform-methanol (15:1, vol/vol); the lipid patterns of wild-type and nsmase 1−/− cells were identical. (C) Lipid analysis of lyso-PC-labeled EMFIs.Cells were labeled for the indicated times with [1-14C-palmitoyl]lyso-PC, and lipids were separated in chloroform-methanol-water (65:25:4, vol/vol/vol). Neither storage nor an altered metabolism of the lipids was detected in nsmase 1−/− cells. Times are given in hours at the bottoms of the lanes. CE, cholesterol-ester; Chol, cholesterol; Cer, ceramide; GlcCer, glycosylceramide; PE, phosphatidylethanolamine; PC, phosphatidylcholine, PI, phosphatidylinositol; PS, phosphatidylserine; MG, monoglycerol, DG, diglycerol; TG, triglycerol; FA, fatty acid; L-PC, lyso-PC; L-PAF, lyso-PAF; L-PE, lysophosphatidylethanolamine; ?, not identified; PAF, platelet-activating factor; PC, phosphatidylcholine; HEK, human embryonic kidney; wt, wild type; and ER, endoplasmic reticulum.
The composition of total lipids of kidney, liver, and brain and of alkali-stable lipids (data not shown) analyzed by HPTLC was equivalent in wild-type and homozygous animals. In contrast to the aSMase (asmase−/−) deficiency, no lipid storage disease occurred in nSMase 1-deficient (nsmase 1−/−) mice. Plasma cholesterol and plasma triglyceride levels were unchanged (data not shown). We therefore exclude a generalized lipid disorder.
We next investigated the lipid metabolism in embryonic fibroblasts (EMFIs) derived from nSMase 1-deficient mice. Homozygous cells showed a decreased nSMase activity (7.6 ± 0.1 nmol of SPM/mg of protein/h) in contrast to wild-type cells (44 ± 5 nmol of SPM/mg of protein/h).
Cells were metabolically labeled with [1-14C]acetate or [14C]palmitate, and lipids were isolated and analyzed by HPTLC. Separation of the radioactive lipids in several solvent systems showed no significant changes in any of the saponifiable and alkali-stable lipid classes.
The lipid catabolism in nSMase 1-deficient EMFIs was investigated by pulse-chase experiments. Cells were labeled for 16 h with [1-14C]acetate followed by a chase of up to 12 h. No changes in total or saponified lipids were detectable (data not shown).
nSMase 1-overexpressing HEK293 cells revealed no changes in SPM or ceramide mass but revealed minute lyso-PC and lyso-PAF concentrations with simultaneous increases of monoacyl-glycerol and monoalkyl-glycerol, respectively. These lysolipids could also be hydrolyzed by His-tagged purified mouse nSMase 1 enzyme in the in vitro assay but at a rate reduced more than 100-fold from that of SPM (data not shown; see Discussion). A role for our cloned nSMase 1 as a lysophospholipase C has been proposed recently (13, 24, 28).
We addressed this possibility by labeling nSMase 1-deficient EMFIs with [1-palmitoyl-14C]lyso-PC or [1-o-octadecyl-3H]lyso-PAF. Lipids were separated by HPTLC and were quantified as described before. We could not detect any changes in the metabolism of these lysophospholipids in the two genotypes. The in vitro and in vivo results disprove the proposed function of nSMase 1 as a lysophospholipase C.
nSMase 1-deficient mice exhibit no obvious phenotype.
The nSMase 1-deficient mice behave normally and have a regular life span. Histological examinations of organs of 3-month-old mice showed no remarkable differences between them and wild-type animals. We excluded possible compensatory mechanisms by gene expression profiling. Using Affymetrix oligonucleotide arrays, we analyzed the expression pattern of kidney, the organ with the highest nSMase 1 message and protein in wild-type and mutant mice.
Among the more than 12,000 genes on the array, the only significantly differently expressed mRNA was the nSMase 1 mRNA, which was called present with a fold change of 3.8. This agrees well with the result of the RNA analyses, where we found different splice products of the nSMase 1 mRNA with the selection marker. The data of the array make a compensatory gene regulation masking an obvious phenotype unlikely.
DISCUSSION
In this report we describe the organization of the 3.3-kb genomic sequence of the nSMase 1 enclosing 10 exons. An nSMase 1-deficient mouse mutant with a deletion of exons II to VIII was constructed and characterized. The small nsmase 1 gene locus and the large deletion strongly reduced the recombination frequency (1:800).
The data reported here indicate that nSMase 1 deficiency does not affect the metabolism of SPM or of any lysophospholipid and causes no storage of SPM or of any other lipid. Furthermore, the nSMase 1 deficiency leaves the development and longevity of the nSMase 1-deficient mouse mutant unimpaired.
The determination of nSMase enzymatic activities in the different organs of the nsmase 1−/− mouse clearly indicated that nSMase 1 is not responsible for the high nSMase activity in brain, in support of our previous immunoprecipitation experiments (35) and other chromatographic studies on mouse brain sphingomyelinases (4). nSMase 2 is the likeliest candidate responsible for the high nSMase activity in the central nervous system and the residual activity in the other organs.
The presence of additional nSMases could explain the unchanged lipid composition in organs of the nSMase 1-deficient mouse mutant. Another possibility would be limitation by the available detection methods of the minute amount of SPM.
However, the sensitive labeling experiments with EMFIs were also unable to detect any changes in lipid metabolism. Whether lipid analyses of aging animals and of subcellular fractions may reveal differences remains to be seen.
The presence of monoglycerols and of decreased levels of lyso-PC and/or lyso-PAF (data not shown and reference 28) in highly nsmase 1-overexpressing HEK cells led to the suggestion that the in vivo function of our cloned nSMase is that of a lysophospholipase C. The results of our experiments clearly demonstrate an identical metabolism of lyso-PC and lyso-PAF in the wild type and in nSMase 1-deficient EMFIs and make the proposed function of nSMase 1 as a lysophospholipase C unlikely.
nSMase 1 is an integral membrane protein of the ER membrane using SPM, a component of the bilayer as substrate. In the in vitro assay, hydrolysis of the lyso-PAF and lyso-PC, which possess detergent properties and are water-soluble subtrates, is minute. The cross-reactivity could be explained by their related structure around the phosphodiester bond, the free 3-hydroxy-group of the sphingosine moiety of SPM and the free 2-hydroxy group of the lysocompound.
At this stage, the biochemical and cell biological data derived from the comparison of the wild type and the nsmase 1−/− mutant are insufficient to define the function of nSMase 1 in cellular metabolism. Therefore, we may only speculate on possible functions of nSMases, e.g., the regulation of the SPM/cholesterol ratio in membranes and their metabolism; nSMase 1 may contribute to the modification of local microdomains in the membrane organization (6, 27, 37, 38). Modification of the lipid bilayer by e.g., lysophosphatidic acid acyl transferase endophilin (30), phospholipase D (10), phospholipase C, and sphingomyelinase (26, 44), is a prerequisite for effective protein functions involved in vesicle formation, transport, and fusion (43). nSMase 1, localized in the membranes of the endoplasmic reticulum, might be an additional modifying enzyme involved in these processes, including the disassembly of the Golgi membrane stacks during mitosis (25).
The analysis of the nSMase 2 knockout (nsmase 2−/−) mouse might provide insights into the function of the nSMase enzyme family on the molecular level. Finally, the generation of an nSMase 1/nSMase 2 double-knockout mouse will answer the question whether the two nSMases cloned so far are the only ones in mammals or if additional isoforms contribute to the (sub)cellular SPM metabolism.
Acknowledgments
This work was supported by the Federal Ministry of Education, Science, Research and Technology (BMBF) (projects 0311-686 and 01 KS 9502) within the Interdisciplinary Center for Molecular Medicine, Cologne (ZMMK) (project 24) and by the Deutsche Forschungsgemeinschaft (project Sto32/36-1).
REFERENCES
- 1.Bligh, E., and W. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Dittmer, J., and R. Lester. 1964. A simple, specific spray for the detection of phospholipids on thin layer chromatography. J. Lipid Res. 5:126-127. [PubMed] [Google Scholar]
- 4.Fensome, A. C., F. Rodrigues-Lima, M. Josephs, H. F. Paterson, and M. Katan. 2000. A neutral magnesium-dependent sphingomyelinase isoform associated with intracellular membranes and reversibly inhibited by reactive oxygen species. J. Biol. Chem. 275:1128-1136. [DOI] [PubMed] [Google Scholar]
- 5.Gatt, S. 1976. Magnesium-dependent sphingomyelinase. Biochem. Biophys. Res. Commun. 68:235-241. [DOI] [PubMed] [Google Scholar]
- 6.Goni, F. M., and A. Alonso. 1999. Structure and functional properties of diacylglycerols in membranes. Prog. Lipid Res. 38:1-48. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann, K., and V. M. Dixit. 1998. Ceramide in apoptosis--does it really matter? Trends Biochem. Sci. 23:374-377. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann, K., S. Tomiuk, G. Wolff, and W. Stoffel. 2000. Cloning and characterization of the mammalian brain-specific, Mg2+-dependent neutral sphingomyelinase. Proc. Natl. Acad. Sci. USA 97:5895-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horinouchi, K., S. Erlich, D. P. Perl, K. Ferlinz, C. L. Bisgaier, K. Sandhoff, R. J. Desnick, C. L. Stewart, and E. H. Schuchman. 1995. Acid sphingomyelinase deficient mice: a model of types A and B Niemann-Pick disease. Nat. Genet. 10:288-293. [DOI] [PubMed] [Google Scholar]
- 10.Jones, D., C. Morgan, and S. Cockcroft. 1999. Phospholipase D and membrane traffic. Potential roles in regulated exocytosis, membrane delivery and vesicle budding. Biochim. Biophys. Acta 1439:229-244. [DOI] [PubMed] [Google Scholar]
- 11.Ledeen, R. W., and R. K. Yu. 1982. Gangliosides: structure, isolation, and analysis. Methods Enzymol. 83:139-191. [DOI] [PubMed] [Google Scholar]
- 12.Liu, B., L. M. Obeid, and Y. A. Hannun. 1997. Sphingomyelinases in cell regulation. Semin. Cell Dev. Biol. 8:311-322. [DOI] [PubMed] [Google Scholar]
- 13.Mizutani, Y., K. Tamiya-Koizumi, F. Irie, Y. Hirabayashi, M. Miwa, and S. Yoshida. 2000. Cloning and expression of rat neutral sphingomyelinase: enzymological characterization and identification of essential histidine residues. Biochim. Biophys. Acta 1485:236-246. [DOI] [PubMed] [Google Scholar]
- 14.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90:8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neuberger, Y., H. Shogomori, Z. Levy, M. Fainzilber, and A. H. Futerman. 2000. A lyso-platelet activating factor phospholipase C, originally suggested to be a neutral-sphingomyelinase, is located in the endoplasmic reticulum. FEBS Lett. 469:44-46. [DOI] [PubMed] [Google Scholar]
- 16.Newrzella, D., and W. Stoffel. 1996. Functional analysis of the glycosylation of murine acid sphingomyelinase. J. Biol. Chem. 271:32089-32095. [DOI] [PubMed] [Google Scholar]
- 17.Newrzella, D., and W. Stoffel. 1992. Molecular cloning of the acid sphingomyelinase of the mouse and the organization and complete nucleotide sequence of the gene. Biol. Chem. Hoppe-Seyler 373:1233-1238. [DOI] [PubMed] [Google Scholar]
- 18.Nix, M., and W. Stoffel. 2000. Perturbation of membrane microdomains reduces mitogenic signaling and increases susceptibility to apoptosis after T cell receptor stimulation. Cell Death Differ. 7:413-424. [DOI] [PubMed] [Google Scholar]
- 19.Otterbach, B., and W. Stoffel. 1995. Acid sphingomyelinase-deficient mice mimic the neurovisceral form of human lysosomal storage disease (Niemann-Pick disease). Cell 81:1053-1061. [DOI] [PubMed] [Google Scholar]
- 20.Quintern, L. E., E. H. Schuchman, O. Levran, M. Suchi, K. Ferlinz, H. Reinke, K. Sandhoff, and R. J. Desnick. 1989. Isolation of cDNA clones encoding human acid sphingomyelinase: occurrence of alternatively processed transcripts. EMBO J. 8:2469-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quintern, L. E., G. Weitz, H. Nehrkorn, J. M. Tager, A. W. Schram, and K. Sandhoff. 1987. Acid sphingomyelinase from human urine: purification and characterization. Biochim. Biophys. Acta 922:323-336. [DOI] [PubMed] [Google Scholar]
- 22.Rao, B. G., and M. W. Spence. 1976. Sphingomyelinase activity at pH 7.4 in human brain and a comparison to activity at pH 5.0. J. Lipid Res. 17:506-515. [PubMed] [Google Scholar]
- 23.Ridgway, N. D. 2000. Interactions between metabolism and intracellular distribution of cholesterol and sphingomyelin. Biochim. Biophys. Acta 1484:129-141. [DOI] [PubMed] [Google Scholar]
- 24.Rodrigues-Lima, F., A. C. Fensome, M. Josephs, J. Evans, R. J. Veldman, and M. Katan. 2000. Structural requirements for catalysis and membrane targeting of mammalian enzymes with neutral sphingomyelinase and lysophospholipid phospholipase C activities. Analysis by chemical modification and site-directed mutagenesis. J. Biol. Chem. 275:28316-28325. [DOI] [PubMed] [Google Scholar]
- 25.Rossanese, O. W., and B. S. Glick. 2001. Deconstructing golgi inheritance. Traffic 2:589-596. [DOI] [PubMed] [Google Scholar]
- 26.Ruiz-Arguello, M. B., F. M. Goni, and A. Alonso. 1998. Vesicle membrane fusion induced by the concerted activities of sphingomyelinase and phospholipase C. J. Biol. Chem. 273:22977-22982. [DOI] [PubMed] [Google Scholar]
- 27.Sankaram, M. B., and T. E. Thompson. 1990. Interaction of cholesterol with various glycerophospholipids and sphingomyelin. Biochemistry 29:10670-10675. [DOI] [PubMed] [Google Scholar]
- 28.Sawai, H., N. Domae, N. Nagan, and Y. A. Hannun. 1999. Function of the cloned putative neutral sphingomyelinase as lyso-platelet activating factor-phospholipase C. J. Biol. Chem. 274:38131-38139. [DOI] [PubMed] [Google Scholar]
- 29.Sawai, H., Y. Okamoto, C. Luberto, C. Mao, A. Bielawska, N. Domae, and Y. A. Hannun. 2000. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J. Biol. Chem. 275:39793-39798. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt, A., M. Wolde, C. Thiele, W. Fest, H. Kratzin, A. V. Podtelejnikov, W. Witke, W. B. Huttner, and H. D. Soling. 1999. Endophilin I mediates synaptic vesicle formation by transfer of arachidonate to lysophosphatidic acid. Nature 401:133-141. [DOI] [PubMed] [Google Scholar]
- 31.Schuchman, E. H., O. Levran, L. V. Pereira, and R. J. Desnick. 1992. Structural organization and complete nucleotide sequence of the gene encoding human acid sphingomyelinase (SMPD1). Genomics 12:197-205. [DOI] [PubMed] [Google Scholar]
- 32.Swiatek, P. J., and T. Gridley. 1993. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 7:2071-2084. [DOI] [PubMed] [Google Scholar]
- 33.Tepper, A. D., P. Ruurs, J. Borst, and W. J. van Blitterswijk. 2001. Effect of overexpression of a neutral sphingomyelinase on CD95-induced ceramide production and apoptosis. Biochem. Biophys. Res. Commun. 280:634-639. [DOI] [PubMed] [Google Scholar]
- 34.Tomiuk, S., K. Hofmann, M. Nix, M. Zumbansen, and W. Stoffel. 1998. Cloned mammalian neutral sphingomyelinase: functions in sphingolipid signaling? Proc. Natl. Acad. Sci. USA 95:3638-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomiuk, S., M. Zumbansen, and W. Stoffel. 2000. Characterization and subcellular localization of murine and human magnesium-dependent neutral sphingomyelinase. J. Biol. Chem. 275:5710-5717. [DOI] [PubMed] [Google Scholar]
- 36.Tybulewicz, V. L., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 37.van Meer, G., and J. C. Holthuis. 2000. Sphingolipid transport in eukaryotic cells. Biochim. Biophys. Acta 1486:145-170. [DOI] [PubMed] [Google Scholar]
- 38.Veiga, M. P., J. L. Arrondo, F. M. Goni, and A. Alonso. 1999. Ceramides in phospholipid membranes: effects on bilayer stability and transition to nonlamellar phases. Biophys. J. 76:342-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voss, A. K., T. Thomas, and P. Gruss. 1997. Germ line chimeras from female ES cells. Exp. Cell Res. 230:45-49. [DOI] [PubMed] [Google Scholar]
- 40.Watts, J. D., M. Gu, S. D. Patterson, R. Aebersold, and A. J. Polverino. 1999. On the complexities of ceramide changes in cells undergoing apoptosis: lack of evidence for a second messenger function in apoptotic induction. Cell Death Differ. 6:105-114. [DOI] [PubMed] [Google Scholar]
- 41.Watts, J. D., M. Gu, A. J. Polverino, S. D. Patterson, and R. Aebersold. 1997. Fas-induced apoptosis of T cells occurs independently of ceramide generation. Proc. Natl. Acad. Sci. USA 94:7292-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiegmann, K., S. Schutze, T. Machleidt, D. Witte, and M. Kroenke. 1994. Functional dichotomy of neutral and acidic sphingomyelinases in tumor necrosis factor signaling. Cell 78:1005-1015. [DOI] [PubMed] [Google Scholar]
- 43.Ybe, J. A., D. E. Wakeham, F. M. Brodsky, and P. K. Hwang. 2000. Molecular structures of proteins involved in vesicle fusion. Traffic 1:474-479. [DOI] [PubMed] [Google Scholar]
- 44.Zha, X., L. M. Pierini, P. L. Leopold, P. J. Skiba, I. Tabas, and F. R. Maxfield. 1998. Sphingomyelinase treatment induces ATP-independent endocytosis. J. Cell Biol. 140:39-47. [DOI] [PMC free article] [PubMed] [Google Scholar]