Abstract
Aberrant expression of PU.1 inhibits erythroid cell differentiation and contributes to the formation of murine erythroleukemias (MEL). The molecular mechanism by which this occurs is poorly understood. Here we show that PU.1 specifically and efficiently inhibits CBP-mediated acetylation of several nuclear proteins, including the hematopoietic transcription factors GATA-1, NF-E2, and erythroid Krüppel-like factor. In addition, PU.1 blocks acetylation of histones and interferes with acetylation-dependent transcriptional events. CBP acetyltransferase activity increases during MEL cell differentiation as PU.1 levels decline and is inhibited by sustained PU.1 expression. Finally, PU.1 inhibits the differentiation-associated increase in histone acetylation at an erythroid-specific gene locus in vivo. Together, these findings suggest that aberrant expression of PU.1 and possibly other members of the Ets family of oncoproteins subverts normal cellular differentiation in part by inhibiting the acetylation of critical nuclear factors involved in balancing cellular proliferation and maturation.
Cellular transformation can result from deregulated expression of nuclear transcription factors. The Ets family oncoprotein PU.1 (Spi-1) is normally expressed in myeloid and lymphoid cells. Aberrant PU.1 expression in erythroid precursor cells frequently results from integration of the spleen focus-forming virus, a component of the Friend virus complex, near the PU.1 gene and causes the formation of murine erythroleukemias (MEL) (50; for reviews see references 16 and 49). Studies showing that infection of bone marrow cultures with a PU.1-containing retrovirus efficiently immortalizes erythroblasts (69) and that transgenic mice overexpressing PU.1 develop erythroleukemias (51) demonstrate that PU.1 is a bone fide oncoprotein.
In MEL cells PU.1 levels decline upon differentiation induction (20, 62, 63, 68, 81). Sustained expression of PU.1 prevents MEL cell differentiation (62, 81), suggesting that transformation by PU.1 is achieved by maintaining erythroid precursor cells in an undifferentiated, proliferative state.
Several recent reports showed that PU.1 binds to the erythroid transcription factor GATA-1 and inhibits its activity (40, 54, 63, 83). GATA-1 is essential for differentiation and survival of erythroid precursor cells (19, 79) and participates in the regulation of all erythroid-expressed genes tested to date (for a review see reference 78). Thus, GATA-1 represents a potential target for oncoproteins that interfere with erythroid cell differentiation. Of note, the differentiation block resulting from forced PU.1 expression occurs at the same stage at which GATA-1-deficient erythroid cells are arrested (50, 77), suggesting that GATA-1 is a biologically relevant target of PU.1-mediated inhibition. In agreement with this interpretation, overexpression of GATA-1 can rescue the PU.1-induced differentiation block (63).
In the normal hematopoietic system, PU.1 is essential for the formation of the myeloid and lymphoid cell lineages (41, 71; for reviews see references 16 and 49). PU.1 levels increase during granulocytic/monocytic differentiation of immature progenitor cells but remain low or decline further during erythroid differentiation (13, 20, 76). The balance between PU.1 and GATA-1 appears to be important in determining myeloid versus erythroid cell fate. Forced expression of PU.1 in multipotent progenitor cells leads to myeloid differentiation at the expense of erythroid cell formation and GATA-1 expression (53). Conversely, expression of GATA-1 in these cells triggers erythroid differentiation with a concomitant reduction in PU.1 expression and a block in myeloid differentiation (31, 42). Of note, inhibition of myeloid gene expression by GATA-1 does not require a decrease in PU.1 expression, suggesting that GATA-1 can directly inhibit PU.1 activity (54; see below).
The coactivator CBP is an acetyltransferase (AT) that interacts with numerous nuclear proteins (for reviews see references 8, 12, and 21). While acetylation of histones is generally associated with transcriptional activation, acetylation of transcriptional regulators can result in stimulation or inhibition of transcription. CBP and its close relative p300 are targets of several viral oncoproteins, including adenovirus E1A, simian virus 40 T, human papillomavirus E6, Epstein-Barr virus Zta, and the Kaposi's sarcoma-associated herpesvirus protein viral interferon regulatory factor (for a review see reference 21). The ability of E1A to block the differentiation of a variety of cell lines and to inhibit the activity of numerous transcription factors correlates with its ability to bind to CBP and p300. Thus, E1A has been frequently used to examine the requirement of CBP and p300 for cellular functions. For example, E1A blocks terminal differentiation of MEL cells, implicating CBP and p300 as critical cofactors for erythroid transcriptional regulators (9). Indeed, three erythroid-expressed transcription factors, GATA-1, erythroid Krüppel-like factor (EKLF), and NF-E2, which are important for erythroid differentiation and globin gene expression, interact with CBP, and their activities are inhibited by E1A (9, 14, 18, 28, 85).
Our previous work showed that CBP binds to GATA-1 and stimulates its transcriptional activity (9). CBP acetylates GATA-1 at two highly conserved lysine-rich motifs near each of the two zinc fingers (10, 27). Mutations in the acetylation sites impair the ability of GATA-1 to trigger differentiation of erythroid cells (27), suggesting that acetylation is important for GATA-1 function in vivo.
There are notable similarities between the effects of E1A and PU.1 in erythroid cells. Both proteins can block GATA-1 activity and cellular differentiation. Moreover, both molecules bind to the CH3 domain of CBP (21, 82; our own observations), and their respective CBP-binding domains are required for blocking erythroid differentiation and for inhibition of GATA-1 activity (9, 40, 63, 82). Since it has been proposed previously that the transforming potential of E1A is linked to its ability to inhibit the CBP AT activity (11, 25, 57) and since E1A can block GATA-1 acetylation in vivo (27), we examined whether PU.1 might also inhibit acetylation of GATA-1 and other nuclear factors.
Here we demonstrate that PU.1 inhibits CBP-mediated acetylation of GATA-1 in vitro and in vivo. PU.1 also inhibits acetylation of other nuclear proteins, including histones. PU.1 inhibits CBP-mediated acetylation of endogenous cellular proteins and inhibits AT-dependent transcription in vivo. In addition, PU.1 inhibits stimulation of CBP AT activity associated with MEL cell differentiation. Finally, chromatin immunoprecipitation (ChIP) experiments show that sustained expression of PU.1 in MEL cells prevents the increase in histone acetylation at the globin gene locus that normally occurs during terminal differentiation. Together, these results suggest that the mechanism by which overexpressed PU.1 contributes to differentiation arrest and malignant transformation involves changes in protein acetylation in a manner similar to that for certain viral oncoproteins.
MATERIALS AND METHODS
Plasmids.
GST-GATA-1 constructs, mammalian GATA-1 expression vectors, GST-CBP-AT (amino acids 1196 to 1718), and GST-PCAF-AT (amino acids 352 to 832) have been described previously (27). GST-CH3 (amino acids 1680 to 1915) and GST-AT-CH3 (amino acids 1196 to 1896) were generated by PCR and subcloned into pGEX-4T. GST-EKLF (residues 272 to 376) and pCMV5-CBP are gifts from M. Crossley and M. Rosenfeld, respectively. GST-PU.1 and CMV-PU.1 constructs have been described previously (58, 60, 61). C-terminally His-tagged CBP-CH3 (amino acids 1680 to 1910) was generated by PCR and subcloned between the BamHI and HindIII sites of pET-21a(+) (Novagen). All constructs were sequenced. The Bax promoter construct is a gift from John Reed.
GST-Ets-1 and GST-Fli-1 were gifts from B. Graves and S. Baker, respectively. GST-p53 was described by Liu et al. (36), and GST-MafG was generated by subcloning PCR-amplified MafG between the BamHI and EcoRI sites of pGEX2TK (Amersham Pharmacia). GAL4-CBP-AT and GAL4-CBP-ΔAT expression vectors and the adenovirus major late promoter reporter construct containing GAL4-binding sites were gifts from T. Kouzarides (39). In transient transfections of MEL cells the GAL4-dependent reporter pGL2-luciferase (Promega) containing five GAL4-binding sites was used. Tethered NF-E2 fused to glutathione S-transferase (GST) was a gift from V. Blank (7). For EKLF transactivation assays we used GAL4-EKLF (gift from J. Bieker) (5), since high levels of cellular proteins that bind to EKLF elements mask activation by EKLF.
AT assays.
Purification of baculovirus-expressed Flag-tagged p300 was performed as reported previously (11). In vitro AT assays were performed in the presence of [14C]acetyl coenzyme A as described in reference 3. All nonhistone substrates were GST fusion proteins expressed in Escherichia coli DH5α. Control reaction mixtures included GST alone. If added proteins varied in amount or concentration, buffer in which proteins are dissolved was added to ensure that all reactions were carried out under the exact same conditions. GST-PU.1 or mutant derivatives were preincubated in the reaction mix for 15 min at 4°C prior to addition of substrates. Acetylation of transcription factors was quantitated by phosphorimager analysis. Histone acetylation was quantitated by spotting the acetylation reaction mixture on a filter. Following repeated washes, filters were analyzed by scintillation counting. A control reaction mixture lacking histones was analyzed in parallel, and the obtained counts (which largely reflect autoacetylation of CBP AT) were subtracted from the total counts obtained for reaction mixtures where histones were present. Free histones were purchased from Sigma.
Transfections, immunoprecipitations, and Western blotting.
Dishes (10-cm diameter) of NIH 3T3 cells were transfected with Lipofectamine (Gibco/BRL) with 1 μg of GATA-1 expression vector, 2 μg of CBP-expressing vector, and 5 μg of PU.1 expression vector. Acetylation of GATA-1 in NIH 3T3 cells was determined as described previously (27), except for the acetyllysine antibodies, which were obtained from Upstate Biotechnology Inc., Lake Placid, N.Y. Prior to immunoprecipitation with acetyllysine antibodies, cell lysates were analyzed by anti-GATA-1 Western blotting to ensure that equal amounts of GATA-1 protein were present in each sample. In addition, pilot experiments were performed to determine possible effects of PU.1 on CBP expression.
To monitor acetylation of cellular proteins in vivo, 293 cells were transiently transfected with vectors expressing CBP-ΔE1A-BD and/or PU.1. Whole-cell lysates were generated by boiling washed cell pellets in sample buffer and analyzed by Western blotting with broad-specificity antiacetyllysine antibodies from Upstate Biotechnology.
MEL cells were transiently transfected with DMRIE-C reagent (Invitrogen) according to the manufacturer's instructions. At 24 h after transfection, cells were split into regular medium and into medium containing 2% dimethyl sulfoxide (DMSO). Luciferase assays were performed following incubation for 72 h. Fold activation reflects increase in activity with GAL4 alone as a reference.
ChIP assays.
ChIP assays were performed exactly as described previously (17) except for the following modifications. DNA-protein-antibody complexes were recovered by eluting them twice with 100 μl of 0.1 M NaHCO3. Pooled eluates were diluted with Tris-EDTA buffer to 400 μl of total volume before phenol-chloroform extraction. Five micrograms of glycogen and 5 μg of tRNA, but no additional salt, were added for DNA precipitation. PCRs were performed in the presence of [32P]dCTP. Products were separated on a 5% Tris-buffered EDTA polyacrylamide gel. Band intensities were quantitated by phosphorimager analysis.
RESULTS
PU.1 inhibits GATA-1 acetylation in vitro.
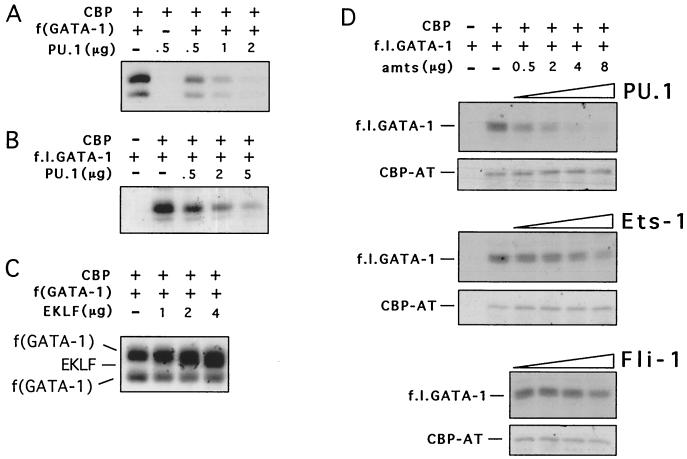
To test whether PU.1 inhibits CBP-mediated acetylation of GATA-1, we performed in vitro acetylation assays with purified GST fusion proteins containing the AT domain of CBP (CBP-AT), the zinc finger region of GATA-1 [f(GATA-1)], and increasing amounts of full-length PU.1. f(GATA-1) is efficiently acetylated by CBP-AT, consistent with our previous results (Fig. 1A) (27). When increasing amounts of PU.1 protein were added to the reaction mixture, f(GATA-1) acetylation was inhibited in a dose-dependent manner (Fig. 1A). Almost-complete inhibition of GATA-1 acetylation was observed when PU.1 was in fivefold molar excess over GATA-1. PU.1 also inhibited acetylation of full-length GATA-1 with comparable efficiency (Fig. 1B). The half-maximal inhibitory concentration (50% inhibitory concentration [IC50]) of PU.1 for full-length GATA-1 was approximately 185 nM (average of three measurements), indicating that PU.1 is at least as potent as previously described AT inhibitors (11, 72). PU.1 also inhibited acetylation of GATA-1 by full-length baculovirus-expressed p300 with comparable efficiency (data not shown).
FIG. 1.
Inhibition of GATA-1 acetylation by PU.1 in vitro. (A) Representative experiment in which 0.5 μg of GST-f(GATA-1) was acetylated in the presence of indicated amounts of GST-PU.1 protein. (B) PU.1 inhibits acetylation of full-length GST-GATA-1 (f.l.GATA-1). In the experiment shown, 1.5 μg of f.l.GATA-1 was used. (C) GST-EKLF did not substantially alter acetylation of GATA-1. EKLF is acetylated in these reactions as described previously (85). (D) Acetylation of f.l.GATA-1 in the presence of indicated amounts of PU.1, Ets-1, and Fli-1. A 1.5-μg quantity of f.l.GATA-1 was used. Note that even in the presence of approximately a 6.5 M excess of Fli-1 (8 μg) little or no inhibition of GATA-1 acetylation was observed. Autoacetylation of CBP-AT was not affected by PU.1, Ets-1, and Fli-1.
To determine whether inhibition of GATA-1 acetylation is specific to PU.1, we examined the erythroid transcription factor EKLF. Similar to PU.1, EKLF can bind both GATA-1 (43) and CBP (85). The GST-EKLF fragment that we used (amino acids 272 to 376) contains the zinc fingers, which bind to GATA-1, and all lysine residues acetylated by CBP (86). Neither GST alone (data not shown) nor GST-EKLF inhibited GATA-1 acetylation, indicating that PU.1 action is specific (Fig. 1C). Autoacetylation of CBP was not affected by PU.1 (Fig. 1D), suggesting that PU.1 action is specific toward certain substrates and that PU.1 does not inhibit the AT activity of CBP per se but might instead regulate accessibility of CBP substrates. PU.1 itself was not acetylated to a significant degree in these experiments (data not shown). Furthermore, PU.1 showed no deacetylase activity (data not shown), indicating that PU.1 inhibits acetylation of GATA-1 by CBP rather than deacetylating it.
We next examined whether other Ets family proteins inhibit GATA-1 acetylation. Both Ets-1 and Fli-1 have been implicated previously in erythroleukemic transformation (4, 44, 55). Ets-1 inhibited GATA-1 acetylation, although with lower efficiency than that of PU.1. In contrast, Fli-1 showed little or no inhibitory activity even at up to 6.5-fold molar excess over GATA-1 (Fig. 1D). Together, these results indicate that the inhibitory activity of PU.1 is specific and that a subset of Ets family proteins might share this activity.
PU.1 inhibits GATA-1 acetylation in vivo.
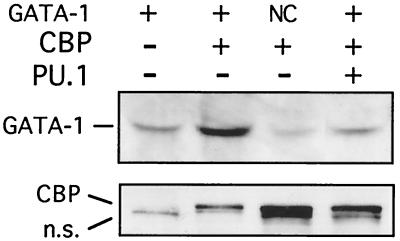
To examine whether PU.1 can inhibit CBP-mediated GATA-1 acetylation in intact cells, 3T3 cells were transiently transfected with GATA-1 alone or together with CBP and PU.1. To ensure that equal amounts of GATA-1 protein were present in all samples, cell extracts were analyzed by Western blotting with anti-GATA-1 antibodies. Samples adjusted to contain equal amounts of GATA-1 protein were immunoprecipitated with antibodies directed against acetyllysine, followed by Western blotting with anti-GATA-1 antibodies. We found that cotransfection of CBP strongly stimulated acetylation of wild-type GATA-1 (Fig. 2, compare first and second lanes) but not an acetylation-defective form of GATA-1 (Fig. 2, third lane [NC]), indicating that acetylation of GATA-1 by CBP occurs at the relevant sites in vivo. Coexpression of PU.1 reduced GATA-1 acetylation to basal levels (Fig. 2, fourth lane). Control Western blots showed that PU.1 did not inhibit the expression of CBP (Fig. 2, bottom). From these results we conclude that PU.1 inhibits CBP-mediated GATA-1 acetylation in intact cells. Since PU.1 inhibits GATA-1 transcriptional activity (40, 63, 83), these findings establish a correlation between the transcriptional activity of GATA-1 and its acetylation status in the presence or absence of PU.1.
FIG. 2.
PU.1 inhibits GATA-1 acetylation in vivo. Wild-type GATA-1 or a mutant construct bearing alanine substitutions in both major acetylation sites (NC) was transfected alone or together with CBP and/or PU.1. (Top) Cell lysates were immunoprecipitated with antiacetyllysine antibodies, followed by Western blotting with a monoclonal GATA-1 antibody. Prior to immunoprecipitation GATA-1 protein levels were determined and cell lysates were adjusted to ensure that equal amounts of GATA-1 protein were present in each reaction mixture. (Bottom) Nonprecipitated extracts were analyzed for CBP expression with anti-Flag antibodies. n.s., nonspecific band that is present in all lanes.
PU.1 inhibits acetylation of a broad spectrum of substrates.
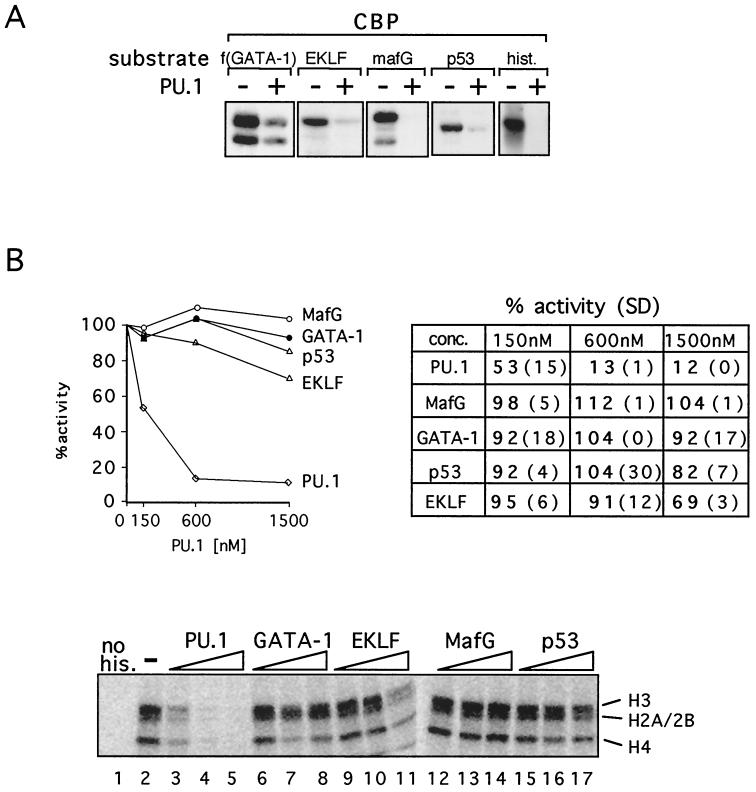
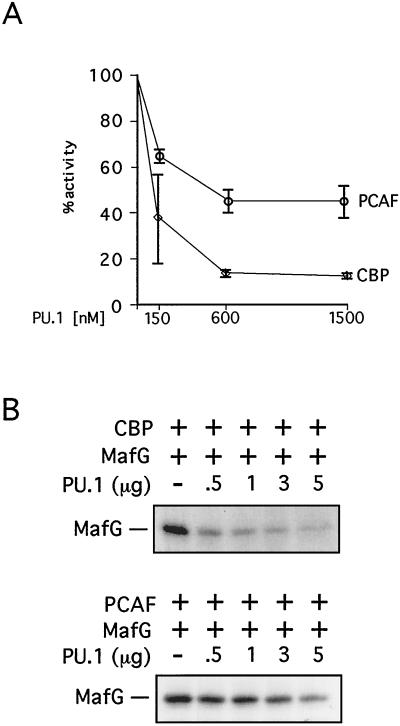
Overexpression of PU.1 inhibits erythroid differentiation and contributes to malignant transformation. While GATA-1 represents an attractive target for a PU.1-mediated differentiation block, it is likely not the only factor inhibited by PU.1 expression. Therefore, we examined whether PU.1 inhibits acetylation of other nuclear proteins known to be substrates for CBP, including the MafG subunit of the hematopoietic transcription factor NF-E2 (28), EKLF (85), p53 (22, 36, 65), and histones (3, 56). Both MafG and EKLF play important roles in hematopoietic gene expression. Using the in vitro AT assay described above, we found that PU.1 efficiently blocks CBP-mediated acetylation of all proteins examined, including histones (Fig. 3A). The IC50s of PU.1 for EKLF and MafG acetylation were approximately 600 and 125 nM, respectively. Thus, PU.1 appears to be a more general inhibitor of CBP-mediated protein acetylation and might block erythroid differentiation by inhibiting acetylation of multiple nuclear factors essential for cell differentiation.
FIG. 3.
(A) PU.1 inhibits acetylation of various proteins by CBP. In vitro acetylation reactions with indicated substrates were carried out in the presence or absence of PU.1. (B) Inhibition of histone acetylation is specific for PU.1. (Top) Histone acetylation in the presence of indicated proteins was quantitated by scintillation counting. Acetylation in the absence of PU.1 was defined as 100%. Data shown are averages of two to four independent experiments. The table shows scintillation counts with standard deviations (SD) in parentheses. (Bottom) Representative autoradiogram showing histone acetylation in the presence of added GST fusion proteins. Concentrations are 150, 600, and 1,500 nM. “no his.” indicates that no histones were added.
Potent inhibition by PU.1 of CBP-mediated histone acetylation was a surprising result. To determine whether this activity is specific for PU.1 or whether other proteins that bind to CBP have a similar activity, we examined PU.1 in parallel with GATA-1, MafG, EKLF, and p53. PU.1 binds to the CH3 domain of CBP (82) (Fig. 5D). Similarly, GATA-1 (9), MafG (28), and p53 (2, 23, 70) bind at or near the CH3 domain. While the EKLF-binding site has not been mapped in detail, our preliminary data indicate that the CH3 domain of CBP can bind EKLF efficiently in vitro (data not shown). The results in Fig. 3B show that PU.1 strongly inhibits histone acetylation with an IC50 of approximately 110 nM, while little or no inhibition was observed with the other proteins examined. PU.1 also inhibited histone acetylation by full-length baculovirus-expressed p300, although with lower efficiency than that of CBP (data not shown). Together, these results indicate that PU.1 is a potent and specific inhibitor of CBP-mediated histone acetylation and that PU.1 has an activity not shared by several other transcription factors that bind to the CH3 domain of CBP.
FIG. 5.
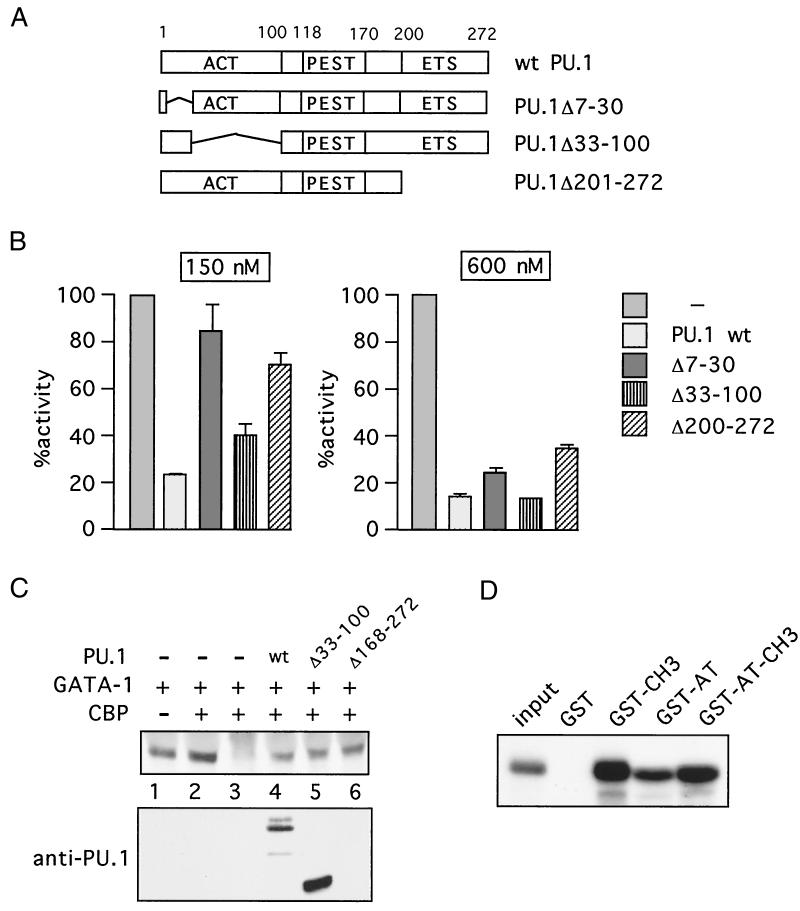
The activation domain and the ETS domain of PU.1 contribute to CBP inhibition. (A) PU.1 constructs. Amino acids 1 to 100 comprise the activation domain, and amino acids 200 to 272 contain the ETS domain. (B) In vitro acetylation of histones in the presence of indicated proteins was quantitated by scintillation counting. Acetylation in the absence of added proteins was defined as 100%. Data shown are averages of two independent experiments. (C) (Top) In vivo acetylation of GATA-1 in the presence of PU.1 constructs. Lysates were precipitated with antiacetyllysine antibodies and blotted with anti-GATA-1 antibodies, except in lane 3, where control serum was used. (Bottom) Anti-PU.1 Western blot. Construct PU.1Δ168-272 lacks the epitope recognized by the anti-PU.1 antibody. (D) PU.1 binds to both the CH3 domain and the AT domain. Indicated GST fusion proteins or GST alone was used in the binding reactions. Input, 10% of in vitro-translated [35S]methionine-labeled PU.1. wt, wild type.
Since E1A can inhibit the AT activities of both CBP and PCAF (11, 25, 57), we examined whether the same might also be true for PU.1. In vitro AT assays with various concentrations of PU.1 protein revealed that PU.1 inhibits both CBP- and PCAF-mediated histone acetylation (Fig. 4A). However, maximal inhibition of CBP and PCAF approximated 90 and 55%, respectively, indicating that CBP is the preferred target for PU.1 inhibition (Fig. 4A). Similarly, Fig. 4B shows that PU.1 also inhibits PCAF-mediated acetylation of MafG (IC50 of ∼640 nM) but is more effective in inhibiting CBP-mediated MafG acetylation (IC50 of ∼125 nM). Together with the above findings, these results show that PU.1 is a potent and specific inhibitor of protein acetylation.
FIG. 4.
(A) PU.1 inhibits histone acetylation by CBP and PCAF with different efficiencies. Error bars denote standard deviations. (B) PU.1 inhibits CBP-mediated acetylation of MafG (top) more efficiently than it does PCAF-mediated acetylation (bottom).
The activation domain and the ETS domain of PU.1 contribute to CBP inhibition.
To determine which domains of PU.1 are required for CBP inhibition, a series of PU.1 deletion constructs were examined in AT assays with histones as substrates. The constructs that were tested (Fig. 5A) include PU.1Δ7-30, which lacks amino acids 7 to 30, which are part of the N-terminal activation domain (29). Construct PU.1Δ30-100 lacks most of the transcriptional activation domain. PU.1Δ201-272 lacks the DNA-binding (ETS) domain. Although only the ETS domain of PU.1 is required for GATA-1 binding, both the activation and ETS domains are required for inhibiting GATA-1 activity (40, 54, 63, 83). All constructs were tested in AT assays using CBP-AT as enzyme and histones as substrate. The results show that deletion of part of the activation domain of PU.1 (PU.1Δ7-30) reduced but did not abolish the inhibitory activity of PU.1 (Fig. 5B). PU.1Δ33-100 also inhibited histone acetylation less efficiently than did wild-type PU.1, consistent with its reduced affinity for CBP (82; our own observations). The difference between wild-type PU.1 and PU.1Δ33-100 was noticeable at protein concentrations near the IC50 (150 nM). Deletion of the ETS domain (PU.1Δ201-272) also led to a reduction but not complete loss of activity. Preliminary data suggest that a C-terminal fragment of PU.1 (amino acids 242 to 272) is dispensable for activity (data not shown). Essentially the same results were obtained when GATA-1 was used as substrate instead of histones or when p300 was used as enzyme (data not shown).
To determine whether the activation and ETS domains of PU.1 are required for inhibition of protein acetylation in vivo, we assayed GATA-1 acetylation in transfected 3T3 cells as described above. Wild-type PU.1 inhibited GATA-1 acetylation (Fig. 5C, lane 4), consistent with the results in Fig. 2. Deletion of portions of the ETS domain (amino acids 168 to 272) reduced but did not eliminate PU.1 inhibitory activity (Fig. 5C, lane 6). Similarly, deletion of part of the activation domain (amino acids 33 to 100) did not abolish PU.1 activity. However, PU.1Δ33-100 likely has less specific activity than does wild-type PU.1, since control Western blots consistently revealed higher expression levels (Fig. 5C).
PU.1 inhibited protein acetylation by the AT domain of CBP. Therefore, we examined whether PU.1 could directly interact with this domain by in vitro protein-binding assays. GST fused to the CH3 domain (GST-CH3), the AT domain (GST-AT), or both (GST-AT-CH3) was analyzed for binding to in vitro-translated [35S]methionine-labeled PU.1. The results show that PU.1 bound to GST-CH3 as well as to GST-AT-CH3 (Fig. 5D), consistent with previous results (82). Surprisingly, PU.1 also bound to GST-AT (Fig. 5D), suggesting that PU.1 might inhibit protein acetylation by interfering with the enzyme-substrate interaction.
Together, these results suggest that at least two domains of PU.1, the activation domain and the ETS domain, are required for maximal CBP inhibition and that PU.1 directly binds to the AT domain of CBP. It is noteworthy that both the activation and DNA-binding domains of PU.1 are required for inhibition of MEL cell differentiation (63), thus establishing a correlation between inhibition of protein acetylation and inhibition of cellular differentiation. The observation that the ETS domain alone has inhibitory activity might explain the inhibitory effects of Ets-1, which shares sequence similarity with PU.1 throughout the ETS domain.
PU.1 inhibits CBP-mediated acetylation of endogenous cellular proteins.
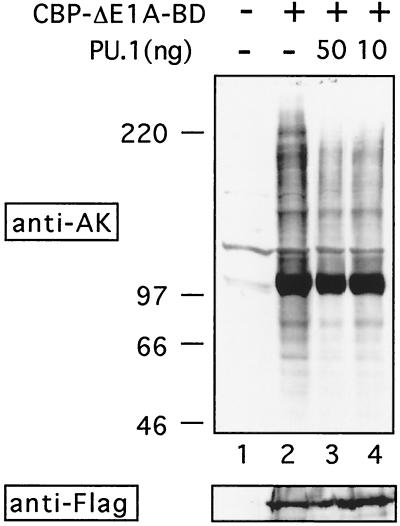
To determine whether PU.1 inhibits acetylation of endogenous cellular proteins by CBP, we transfected 293 cells with a mutant form of CBP lacking the E1A-binding domain (amino acids 1805 to 1890, CBP-ΔE1ABD). Since 293 cells express E1A, we expected that CBP-ΔE1ABD might be partially resistant to the inhibitory activity of E1A and allow for global increases in protein acetylation. When whole-cell lysates were analyzed by Western blotting with antibodies against acetyllysine, a dramatic increase in general protein acetylation was observed over that for untransfected cells (Fig. 6, lanes 1 and 2). Expression of wild-type CBP had no effect (data not shown). Coexpression of PU.1 led to reduced protein acetylation of numerous proteins in a dose-dependent fashion (Fig. 6, lanes 3 and 4). Of note, acetylation of several proteins appeared unaffected by the presence of PU.1, suggesting that inhibition by PU.1 is selective. Western analysis showed that PU.1 did not interfere with CBP expression (Fig. 6, lower panel). These results indicate that PU.1 can inhibit acetylation by CBP of numerous cellular proteins. In addition, they suggest that PU.1 can inhibit AT activity of CBP lacking the E1A-binding domain, as was observed in the in vitro acetylation experiments. Consistent with this finding, in vitro protein-binding experiments revealed that PU.1 binds to CBP and to CBP-ΔE1ABD with comparable efficiency (data not shown).
FIG. 6.
PU.1 inhibits acetylation of endogenous cellular proteins. (Top) Whole-cell extracts of 293 cells transiently transfected with indicated plasmids were analyzed by Western blotting with antiacetyllysine antibodies. Note that acetylation of many but not all proteins was reduced by PU.1. (Bottom) Equal amounts of Flag-tagged CBP-ΔE1ABD protein were present in each lane.
PU.1 inhibits the activity of CBP/p300-dependent transcription factors.
If PU.1 inhibits acetylation of various nuclear factors, it might be expected to inhibit their activity in transactivation assays. In the case of GATA-1, the acetylation status correlates with its transcriptional activity (27). To determine whether this correlation applies to other proteins, we examined the effects of PU.1 on the activities of NF-E2, EKLF, and the tumor suppressor protein p53.
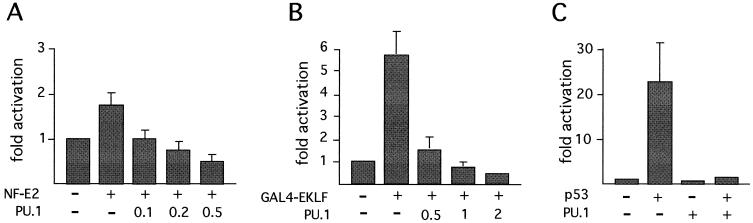
NF-E2 is a heterodimer consisting of the hematopoietic subunit p45 and a widely expressed member of the small Maf protein family (6, 52). NF-E2 elements are important for the expression of the globins and other erythroid-expressed genes. We previously showed that MafG is acetylated by CBP and PCAF, whereas p45 is acetylated by PCAF only (28; unpublished observations). To assay NF-E2 activity without interference by endogenous proteins that heterodimerize with either partner, we used a version of NF-E2 in which p45 and MafG are physically tethered by a linker peptide. This construct is fully active and can restore NF-E2 activity in NF-E2-deficient erythroid cells (7). NF-E2 activated transcription of a luciferase reporter gene driven by the NF-E2-responsive promoter of the porphobilinogen deaminase gene (45) (Fig. 7A). Activation by NF-E2 is moderate, since the NF-E2-binding site can be occupied by AP-1-like transcription factors, leading to high basal-level activation. However, coexpression of PU.1 reduced NF-E2 activity in a dose-dependent manner (Fig. 7A). Control Western blots showed that PU.1 did not reduce expression of NF-E2 from transfected plasmids (data not shown).
FIG. 7.
PU.1 inhibits CBP-dependent transcription factors. (A) Tethered NF-E2 was transfected into 3T3 cells with a luciferase reporter gene driven by the porphobilinogen deaminase promoter. (B) EKLF was analyzed as a GAL4 fusion on a reporter containing a GAL4-binding site upstream of the simian virus 40 promoter. (C) Saos-2 cells that lack p53 were transfected with a construct containing the Bax promoter driving the human growth hormone gene together with vectors expressing p53 and PU.1. Numbers indicate fold activation. Error bars denote standard deviations.
EKLF is an erythroid-restricted transcription factor essential for globin gene expression. Recent work showed that CBP binds and acetylates EKLF (85, 86). GAL4-EKLF activates transcription of a GAL4-binding site-containing reporter gene, while PU.1 expression inhibits GAL4-EKLF activity in a dose-dependent manner (Fig. 7B). Together, these results suggest that PU.1 may block erythroid differentiation by targeting not only GATA-1 but also multiple CBP-dependent erythroid transcription factors.
p53 is acetylated by both PCAF and CBP, and acetylation strongly stimulates DNA binding to naked DNA in vitro (22, 36, 65). To test the effects of PU.1 on p53 activity, p53-deficient Saos-2 cells were transiently transfected with the p53-responsive promoter of the Bax gene fused to the human growth hormone gene as reporter. Cotransfection with p53 strongly activated the reporter construct in a manner dependent on an intact p53-binding site (Fig. 7C), consistent with previous experiments (48). Upon coexpression of PU.1, reporter activity was dramatically reduced, whereas the basal activity of the Bax promoter was not inhibited (Fig. 7C). PU.1 also inhibited activity of a reporter construct bearing an isolated p53-binding site, although inhibition was less pronounced than that with the Bax promoter (data not shown). These results show that PU.1 inhibits the activity of several CBP-regulated transcription factors.
PU.1 inhibits AT-dependent transcriptional activation by CBP.
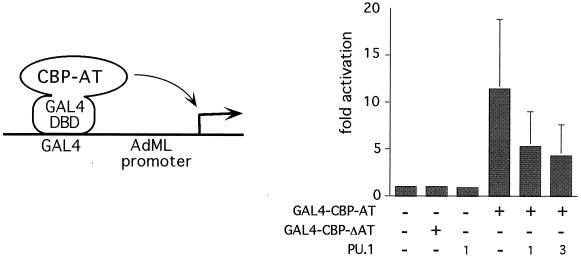
Our results are consistent with a model in which forced expression of PU.1 inhibits CBP/p300- and PCAF-dependent transcriptional events. However, it remains possible that PU.1 inhibits CBP/p300 and PCAF function by a mechanism independent of AT activity. To measure the effects of PU.1 on acetylation-dependent transcriptional activation, we used a construct in which the AT domain of CBP is fused to the DNA-binding domain of the yeast transcription factor GAL4 (GAL4-CBP-AT [Fig. 8 ]). GAL4-CBP-AT activates a reporter gene containing GAL4-binding sites in transfected U2OS cells (39). Activation is dependent on AT activity, since a series of mutations across the AT domain revealed a tight correlation between AT activity and transcriptional output (39). By this assay, PU.1 was examined for its ability to block CBP-AT function. GAL4-CBP-AT strongly activated the reporter gene, while an enzymatically defective mutant (GAL4-CBP-ΔAT) was inactive, consistent with previous results (Fig. 8) (39). Cotransfection of increasing amounts of PU.1 resulted in a dose-dependent inhibition of transcriptional activity (Fig. 8). Basal-level promoter activity was not affected in the presence of high amounts of coexpressed PU.1. Together, these results further support a model in which PU.1 inhibits AT activity of CBP in intact cells.
FIG. 8.
PU.1 inhibits AT-dependent transcriptional activation. (Left) The schematic illustrates the assay. The AT domain of CBP fused to the DNA-binding domain (DBD) of GAL4 (GAL4-AT) activates transcription of a reporter gene containing a GAL4-binding site and the adenovirus major late (AdML) promoter driving the chloramphenicol AT reporter gene. (Right) U2OS cells were transfected with the reporter construct and the indicated expression plasmids. CBP-AT contains the CBP AT domain, and CBP-ΔAT contains an 18-residue deletion (amino acids 1458 to 1475) and lacks AT activity. Error bars denote standard deviations.
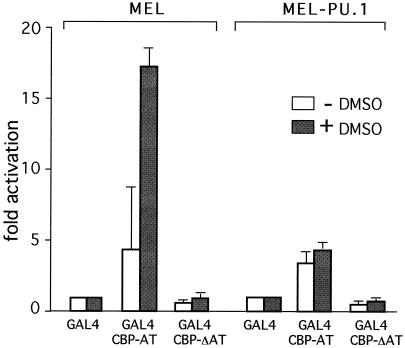
CBP-AT activity increases during MEL cell differentiation and is inhibited by PU.1.
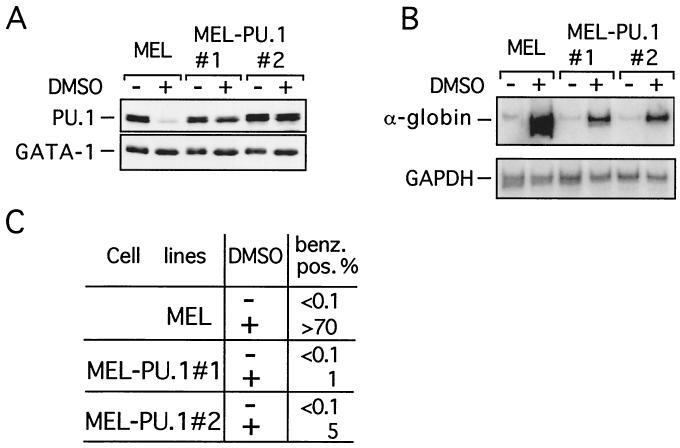
If PU.1 inhibits CBP-AT activity, the decline in PU.1 levels that accompanies MEL cell differentiation might relieve CBP-AT inhibition. To test this directly, MEL cells were transfected with a GAL4-dependent reporter gene, GAL4-CBP-AT, and GAL4-CBP-ΔAT. Cells were induced to differentiate with DMSO, and reporter gene activity was determined. GAL4 alone was used as a reference. In undifferentiated MEL cells GAL4-CBP-AT activated transcription between four- and fivefold (Fig. 9), while GAL4-CBP-ΔAT had no activity, indicating that transcription reflects AT activity. Upon differentiation induction GAL4-CBP-AT activity increased up to 17-fold, while GAL4-CBP-ΔAT activity remained essentially unchanged. These results show that GAL4-CBP-AT activity increases upon differentiation. If extinction of PU.1 expression accounts for this increase, sustained expression of PU.1 would be expected to block the increase in GAL4-CBP-AT activity. To test this possibility, we generated stable PU.1-expressing MEL cell lines (MEL-PU.1). In order to minimize the risk of obtaining artifacts due to unphysiologically high PU.1 levels, we selected MEL-PU.1 clones that expressed the lowest possible levels of PU.1 without losing the differentiation blocking activity. Two lines (MEL-PU.1#1 and MEL-PU.1#2) were selected that expressed PU.1 at levels very similar to those found in uninduced control MEL cells (Fig. 10A). Upon differentiation induction, these cells maintained PU.1 levels that were clearly above those found in control cells but were equal to or lower than those in uninduced cells (Fig. 10A). We next determined whether MEL-PU.1#1 and MEL-PU.1#2 were blocked in differentiation by two criteria. First, we determined α-globin mRNA levels by reverse transcription-PCR. Second, cells were stained with the dye benzidine, which stains hemoglobin, and the percentage of benzidine-positive cells was determined. As shown in Fig. 10B and C, while DMSO treatment of control MEL cells led to efficient differentiation as reflected in increased α-globin gene expression and increased numbers of benzidine-positive cells, both MEL-PU.1#1 and MEL-PU.1#2 displayed an almost-complete differentiation block. MEL-PU.1 cells were transfected with GAL4-CBP-AT, GAL4-CBP-ΔAT, or GAL4 and analyzed as described above. In contrast to control MEL cells, DMSO treatment of MEL-PU.1 cells failed to stimulate GAL4-CBP-AT activity (Fig. 9). Together, these results show that PU.1 inhibits the differentiation-associated increase in CBP-AT activity and reveal an inverse correlation between CBP-AT activity and PU.1 levels in MEL cells.
FIG. 9.
AT activity of CBP increases during differentiation and is inhibited by PU.1. MEL or MEL-PU.1 cells were transfected with a GAL4-dependent reporter plasmid and plasmids expressing GAL4, GAL4-CBP-AT, or GAL4-CBPΔAT. Error bars denote standard deviations. Note the absence of induction of GAL4-CBPΔAT activity in DMSO-treated MEL-PU.1 cells.
FIG. 10.
Sustained PU.1 expression inhibits MEL cell differentiation. (A) Generation of PU.1-expressing MEL cells. (Top) Anti-PU.1 Western blot. (Bottom) The above Western blot was stripped and reprobed with anti-GATA-1 antibodies. DMSO indicates DMSO-treated cells. (B) (Top) Reverse transcription-PCR was performed using primers for α-globin mRNA in the presence of [32P]dCTP. (Bottom) Expression of the housekeeping gene GAPDH was used as a control. (C) Differentiation of MEL cell lines was monitored by determining the percentage of benzidine-positive cells.
PU.1 inhibits differentiation-induced histone acetylation at the β-globin gene locus.
During MEL cell differentiation histone H3 acetylation increases at the transcribed globin genes and at the locus control region (LCR) (17, 66). Hyperacetylation of histone H3 at the active globin genes and the LCR is not simply the consequence of activated transcription, since it can also occur at transcriptionally silent globin genes (17, 26, 67). Furthermore, in Saccharomyces cerevisiae histone acetylation precedes transcription initiation and does not require active transcription (30, 33). Thus, it is believed that histone acetylation is an early event during gene activation and sets the stage for subsequent transcriptional events.
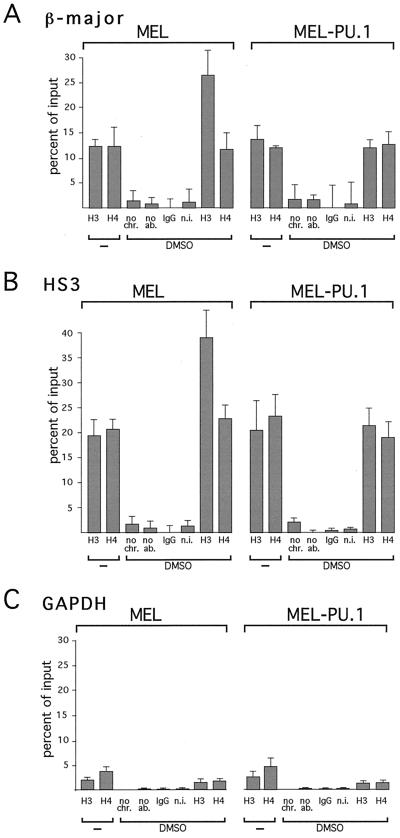
If PU.1 inhibits histone acetylation by CBP, sustained expression of PU.1 might be predicted to interfere with differentiation-associated histone acetylation at the globin gene locus. To test this hypothesis, ChIP assays were performed to measure histone acetylation at the β-globin locus in MEL and MEL-PU.1 cells. Upon differentiation induction, control MEL cells showed a twofold increase in acetylation of histone H3 at the promoter of the β-major globin gene (Fig. 11A), consistent with published reports (17, 66). In contrast, in MEL-PU.1 cells, stimulation of histone H3 acetylation was completely inhibited (Fig. 11A). The second site that we examined for histone acetylation was DNase I-hypersensitive site 3 (HS3) of the LCR, since it contains GATA-1-binding sites that are essential for HS formation (59). While histone H3 acetylation increased approximately twofold in control MEL cells, no change in histone H3 acetylation was observed in both MEL-PU.1 clones (Fig. 11B). Thus, PU.1 potently inhibits differentiation-induced histone acetylation at the β-globin gene promoter and the LCR. These results further indicate that relatively small increases in PU.1 protein levels have dramatic effects on histone acetylation in vivo.
FIG. 11.
PU.1 inhibits differentiation-induced histone acetylation at the β-globin gene locus. (A and B) ChIP assays were performed using antibodies against diacetylated histone H3 and tetra-acetylated histone H4. Controls include no added chromatin (no chr.), no added antibodies (no ab.), nonimmune immunoglobulin G (IgG), and nonimmune serum (n.i.). DMSO treatment increased histone H3 acetylation at the β-major globin gene promoter (A) and at HS3 (B) in MEL cells but not in MEL-PU.1 cells. The results shown are averages of three independent experiments using two separate clones of MEL-PU.1. (C) As a control, histone acetylation was monitored at the GAPDH gene.
We next examined the levels of histone acetylation at the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which is neither induced during differentiation (Fig. 10B) nor regulated by GATA-1 (75). Acetylation of histones H3 and H4 at the GAPDH gene was lower than that at the β-globin locus in control MEL cells, perhaps reflecting lower expression levels than those for the β-globin gene (Fig. 11C). No increase in histone H3 or H4 acetylation was observed upon differentiation induction (Fig. 11C). Importantly, PU.1 expression had no effect on histone acetylation at these genes in untreated and DMSO-treated cells (Fig. 11C). Thus, it appears that PU.1 specifically inhibits differentiation-induced increases in histone acetylation.
Normal DNA binding of GATA-1 in PU.1-expressing MEL cells.
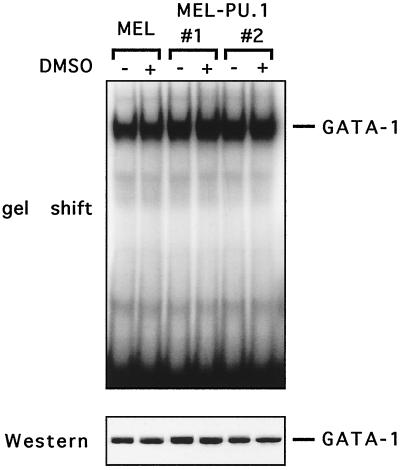
PU.1's ability to inhibit histone acetylation at the β-globin gene locus might be explained if PU.1 inhibited the activity of erythroid transcription factors. In this regard, it is noteworthy that PU.1 has been reported to inhibit DNA binding of GATA-1 (40, 80, 84). If PU.1 inhibits DNA binding of a critical transcription factor that interacts with CBP, this, rather than inhibition of CBP-AT activity, might account for the inhibition of histone acetylation. To examine whether PU.1-expressing cell lines display altered GATA-1 DNA-binding activity, gel shift experiments were performed. The results show that GATA-1 DNA binding is unchanged in MEL cells before and after differentiation, despite a significant decline in PU.1 protein levels (Fig. 12). Furthermore, DNA binding in both MEL-PU.1 clones was equal to that in control MEL cells, regardless of the presence of DMSO. This suggests that the PU.1 levels present in MEL-PU.1#1 and MEL-PU.1#2 were insufficient to alter GATA-1 DNA binding and that the inhibitory effects of PU.1 are instead the result of inhibiting CBP activity.
FIG. 12.
PU.1 does not inhibit DNA binding of GATA-1. (Top) Gel shift experiment using an oligonucleotide containing a GATA-1-binding site. (Bottom) Control Western blot with anti-GATA antibodies.
DISCUSSION
Our results demonstrate that PU.1 is a potent inhibitor of CBP-AT activity toward histone and nonhistone proteins. Inhibition of protein acetylation occurs in vitro and in intact cells. Furthermore, PU.1 inhibits acetylation-dependent transcriptional events. During differentiation of MEL cells there is an inverse correlation between PU.1 levels and CBP-AT activity. These findings provide a potential mechanism for PU.1-mediated inhibition of cellular differentiation and have broad implications regarding the role of Ets proteins during malignant transformation.
Our data indicate that inhibition of CBP-mediated histone acetylation by PU.1 is specific, as several proteins that can bind to the CH3 domain of CBP, including EKLF, GATA-1, p53, and MafG, did not inhibit CBP-AT activity at concentrations where PU.1-mediated inhibition was virtually complete. Thus, simple binding to CBP is not sufficient for inhibition. Specificity of PU.1 is further indicated by our observation that PU.1 but not EKLF inhibits GATA-1 acetylation, although both proteins can bind to CBP and GATA-1. However, since the sites in CBP to which PU.1 and EKLF bind have not yet been accurately mapped, it is possible that inhibition of CBP requires specific contacts within or outside the CH3 domain. Both E1A and PU.1 bind to the CH3 domain of CBP but require additional contacts for full CBP inhibition. In the case of PU.1, these contacts likely reside within the AT domain of CBP, since PU.1 inhibits a CBP construct that contains the AT domain but lacks an intact CH3 domain (amino acids 1196 to 1718). Similar observations were made with E1A and the basic helix-loop-helix protein Twist (11, 25).
Structure-function analysis of PU.1 in vitro and in vivo suggests that both the activation domain and the ETS domain are required for full inhibitory activity toward CBP and p300. These domains are also required for inhibition of MEL cell differentiation (63). Therefore, these results are consistent with a model in which inhibition of CBP/p300-mediated protein acetylation contributes to a differentiation arrest. Furthermore, these results might explain the moderate inhibitory effects of Ets-1, since the similarity of PU.1 and Ets-1 is largely limited to their ETS domains. Similar to E1A, PU.1 also inhibited AT activity of PCAF. However, inhibition of PCAF required higher PU.1 concentrations and was less complete than the inhibition of CBP, suggesting a degree of specificity.
Haploinsufficiency of CBP predisposes humans and mice to various malignancies including those derived from the hematopoietic system (32, 46), suggesting that CBP is a tumor suppressor protein and that cellular CBP levels are limiting. Thus, maintenance of full CBP dosage is critical for normal cellular functions. As a tumor suppressor protein, CBP is a potential target for both viral and cellular oncoproteins. Our results showing that PU.1 inhibits CBP function are consistent with this hypothesis and reveal striking similarities between PU.1 and E1A. Of note, both E1A and PU.1 bind RB, further extending their functional similarities (24). However, it remains to be seen what role, if any, the PU.1 interaction with RB plays during cellular transformation.
Recent work showed that E1A can inhibit (11, 25, 57) or stimulate (1) CBP-AT activity. The discrepancy between these reports might be the result of differences in E1A dosage where inhibition of CBP-AT activity requires high concentrations of E1A (35). During DMSO-induced differentiation of MEL cells we observed an inverse relationship between PU.1 levels and CBP-AT activity. Furthermore, sustained expression of PU.1 blocked the DMSO-induced increase in CBP-AT activity. These results further strengthen our model in which PU.1 targets CBP to inhibit erythroid differentiation, similar to what we observed with E1A (9). It remains possible that the effects of PU.1 on CBP-AT activity in MEL cells are an indirect consequence of the differentiation block. However, PU.1 specifically inhibited CBP-mediated acetylation of GATA-1 in transfected cells (Fig. 2), indicating that PU.1 is a direct inhibitor of CBP in intact cells. Furthermore, PU.1 inhibits GAL4-CBP-AT activity both in U2OS cells and in MEL cells. Since GAL4-CBP-AT consists largely of the AT domain of CBP, this further suggests a direct effect of PU.1 on CBP function. Together with the data showing that PU.1 inhibits CBP-AT activity in vitro, these results strongly support a model in which PU.1 is a direct inhibitor of CBP-AT activity in vivo. In contrast to PU.1, several CBP-bound transcription factors such as GATA-1, p53, EKLF, and MafG did not inhibit CBP-AT activity. Therefore, we speculate that factors that inhibit CBP-AT activity have oncogenic potential while nononcogenic CBP-binding proteins would fall in the group of proteins that fail to inhibit CBP-AT activity.
Previous work has indicated that acetylation of GATA-1 is important for GATA-1 function in differentiating erythroid cells (27). Thus, inhibition of GATA-1 acetylation by PU.1 provides an attractive mechanism by which GATA-1 function might be controlled. In addition, it has been reported previously that high levels of PU.1 can inhibit DNA binding of GATA-1 (40, 80, 84). However, our gel shift experiments using extracts from MEL and MEL-PU.1 cells showed no detectable differences in GATA-1 DNA-binding activity before and after differentiation despite significant changes in PU.1 protein levels. This is consistent with our model in which inhibition of GATA-1 activity results from inhibition of its coactivator (i.e., CBP) rather than from blocking its DNA contacts. It is possible that inhibition of GATA-1 DNA binding requires very high concentrations of PU.1. In this regard, our experiments show that a relatively small increase in PU.1 protein in DMSO-treated MEL cells is sufficient to inhibit CBP-AT activity and histone acetylation at the β-globin gene locus. Together, these findings demonstrate that histone acetylation is exquisitely sensitive to PU.1 levels and suggest that inhibition of protein acetylation is an important mechanism by which PU.1 inhibits erythroid cell differentiation.
It is unlikely that lack of a DMSO-induced increase in histone acetylation in PU.1-expressing cells is simply the consequence of inhibited transcription. Both in yeast and in mammalian cells histone acetylation precedes transcription and occurs even in the absence of transcription (17, 26, 30, 33, 67). Thus, our results are consistent with a model in which inhibition of histone acetylation by PU.1 is the primary event, followed by inhibition of gene expression and cellular differentiation.
Since CBP and PCAF interact with numerous transcriptional regulators, it appeared unlikely that GATA-1 is the only target during the PU.1-induced erythroid differentiation block. Indeed, PU.1 inhibited acetylation and transcriptional activity of the hematopoietic transcription factors NF-E2 and EKLF. In erythroid cells the combination of such inhibitory events would be a powerful means to inhibit globin gene expression and cell differentiation. However, it is important to note that loss of GATA-1 function not only results in a differentiation block but also leads to apoptotic cell death (79). Thus, if GATA-1 were the sole target of PU.1-mediated inhibition, bone marrow-derived erythroid precursor cells expressing PU.1 might be expected to undergo apoptosis rather than proliferate. Therefore, the ability of PU.1 to immortalize primary erythroid cells likely requires that PU.1 interfere with the function of additional factors. In this context it is noteworthy that PU.1 inhibits acetylation and activity of the tumor suppressor protein p53. Inhibition of p53 by PU.1 might play a role during the transformation of primary erythroid cells.
Inhibition of CBP-mediated protein acetylation by PU.1 could occur by several mechanisms. For example, PU.1 might directly inhibit the enzymatic activity of CBP. However, our finding that PU.1 did not inhibit autoacetylation of CBP strongly argues against this possibility. A more likely mechanism is that PU.1 interferes with substrate recognition by CBP, possibly by masking critical residues in the AT domain. This possibility is supported by our finding that PU.1 forms direct contacts with the AT of CBP. The failure of PU.1 to inhibit autoacetylation of CBP might therefore be related to the concentrations of PU.1 used in the AT assays. Thus, the PU.1 levels required for inhibition of autoacetylation would be predicted to be substantially higher than those required for acetylation of other substrates. Another prediction from this mechanism would be that the potency of acetylation inhibition by PU.1 might be determined by the relative affinities of PU.1 and acetylation substrates for the AT domain of CBP.
Preliminary protein-binding studies showed that PU.1 competes with GATA-1 and NF-E2 for binding to the CH3 domain of CBP (unpublished observation). Similarly, the mutually repressive effects between c-Myb and GATA-1 were attributed to the failure of these proteins to simultaneously bind to CBP (74). However, if PU.1 were to inhibit GATA-1, EKLF, MafG, and p53 acetylation simply by competing for a single binding site within CBP, all these factors would consequently be expected to inhibit histone acetylation. However, our results show that this is not the case. Therefore, it is likely that the additional PU.1-binding sites in the AT domain of CBP contribute to the inhibitory effects of PU.1. Transcription factors that bind CBP but fail to inhibit CBP-AT activity might interact with the AT domain with much lower affinity or not at all.
It is also possible that PU.1 interacts with substrate proteins to prevent their interaction with CBP. For example, the direct interaction between PU.1 and GATA-1 (40, 54, 63, 83) might prevent GATA-1 from interacting with CBP. Thus, it is possible that the high efficiency of PU.1 in inhibiting GATA-1 acetylation is the result of PU.1's ability to bind to both CBP and GATA-1. Ultimately, the relative affinities between CBP and CBP-binding proteins are likely to be important determinants of their sensitivity to PU.1.
Our results indicate that the effects of overexpressed PU.1 are much broader than originally thought. By inhibiting the multifunctional coactivator CBP, transformation by PU.1 and possibly other Ets oncoproteins might involve alteration of protein acetylation patterns in the cell. The importance of global changes in protein acetylation during erythroid maturation is underscored by the observation that treatment of MEL cells with the deacetylase inhibitors trichostatin A, suberoylanilide hydroxaminic acid, and m-carboxycinamic acid bishydroxamide is a powerful inducer of differentiation (64). Since various transformed lines of diverse cellular origins differentiate in response to deacetylase inhibitors, it is conceivable that other viral or cellular transforming proteins might similarly trigger broad changes in protein acetylation patterns. An example to support this hypothesis is provided by the viral interferon regulatory factor of Kaposi's sarcoma-associated herpesvirus, which has transforming activity and has been shown to inhibit p300-mediated histone acetylation (34). In addition, the cellular protein Twist, which can inhibit p300-AT and PCAF-AT activities (25), has been shown to have oncogenic potential (38). A recently identified cellular protein called E1A-like inhibitor of differentiation (EID-1) can inhibit CBP/p300-AT activity and block myogenic differentiation (37, 47). Also, components of the INHAT complex, which can inhibit p300-mediated histone acetylation, are potentially oncogenic (72). Together, these studies indicate that the net result of inhibition of CBP-AT activity is a cellular differentiation block. A recent report showed that neurodegeneration triggered by expression of the polyglutamine-containing protein Httex1p involves inhibition of CBP/p300- and PCAF-AT activities (73). The progression of neurodegeneration is slowed by treatment with deacetylase inhibitors, further emphasizing the importance of a controlled balance of protein acetylation and deacetylation in cellular functions in diverse tissues (73).
In the normal hematopoietic system PU.1 is essential for myeloid and lymphoid cell development (41, 71). PU.1 activates numerous myeloid and lymphoid tissue-expressed genes (16, 49), and CBP stimulates PU.1 activity in transient transfections (82). How can our results showing that PU.1 is a potent inhibitor of CBP-dependent transcriptional events be best reconciled with the above studies? There are at least two possible explanations. First, PU.1 function depends on promoter context. In the regulatory regions of myeloid and lymphoid tissue-expressed genes PU.1 binds DNA directly and cooperates with other transcription factors to activate gene expression. In contrast, PU.1 can inhibit the activity of transcription factors such as GATA-1 in the absence of PU.1-binding sites (40, 63, 83). Thus, the presence or absence of PU.1-binding sites, as well as promoter architecture, might determine whether PU.1 activates or inhibits transcription of a given gene. Second, the function of PU.1 likely depends on its expression level. Previous studies showed that low concentrations of PU.1 in hematopoietic precursor cells are permissive for a developmental path toward the erythroid cell fate, while higher levels favor differentiation toward the myeloid and lymphoid lineage and inhibit erythroid differentiation (see the introduction). Furthermore, PU.1 levels appear to be critical in determining myeloid versus lymphoid cell fate (15). At high concentrations PU.1 might be able to occupy and activate myeloid and lymphoid promoters while leaving enough PU.1 protein available to inhibit the activities of erythroid transcription factors. Our observation that the PU.1 concentrations required for inhibition of protein acetylation vary among CBP substrates further emphasizes the importance of PU.1 dosage. This leads to the speculation that, at high levels of PU.1, such as those observed during transformation, PU.1 might inhibit acetylation of a wider spectrum of CBP/p300 substrates. Finally, the high sensitivity of cells to CBP/p300 dosage supports the importance of a regulated balance between CBP and PU.1 during cellular differentiation.
Acknowledgments
We thank Suzanne Baker, Volker Blank, Barbara Graves, Tony Kouzarides, and John Reed for plasmids and Hsiao-Ling Hung for providing recombinant GST-CBP protein. We are grateful to Margaret Chou, Merlin Crossley, Tom Kadesch, and Mitch Weiss for critically reading the manuscript and to Ed Scott, Celeste Simon, Dan Tenen, and Pu Zhang for helpful discussions.
This work was supported by NIH grant 1RO1 DK54937-01 (G.A.B.).
REFERENCES
- 1.Ait-Si-Ali, S., S. Ramirez, F. X. Barre, F. Dkhissi, L. Magnaghi-Jaulin, J. A. Girault, P. Robin, M. Knibiehler, L. L. Pritchard, B. Ducommun, D. Trouche, and A. Harel-Bellan. 1998. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature (London) 396:184-186. [DOI] [PubMed] [Google Scholar]
- 2.Avantaggiati, M. L., V. Ogryzko, K. Gardner, A. Giordano, A. S. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89:1175-1184. [DOI] [PubMed] [Google Scholar]
- 3.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature (London) 384:641-643. [DOI] [PubMed] [Google Scholar]
- 4.Ben-David, Y., E. B. Giddens, K. Letwin, and A. Bernstein. 1991. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 5:908-918. [DOI] [PubMed] [Google Scholar]
- 5.Bieker, J. J., and C. M. Southwood. 1995. The erythroid Krüppel-like factor transactivation domain is a critical component for the cell-specific inducibility of a β-globin promoter. Mol. Cell. Biol. 15:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blank, V., and N. C. Andrews. 1997. The maf transcription factors: regulators of differentiation. Trends Biochem. Sci. 22:437-441. [DOI] [PubMed] [Google Scholar]
- 7.Blank, V., M. J. Kim, and N. C. Andrews. 1997. Human MafG is a functional partner for p45 NF-E2 in activating globin gene expression. Blood 89:3925-3935. [PubMed] [Google Scholar]
- 8.Blobel, G. A. 2000. CBP/p300: molecular integrators of hematopoietic transcription. Blood 95:745-755. [PubMed] [Google Scholar]
- 9.Blobel, G. A., T. Nakajima, R. Eckner, M. Montminy, and S. H. Orkin. 1998. CREB-binding protein (CBP) cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl. Acad. Sci. USA 95:2061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyes, J., P. Byfield, Y. Nakatani, and V. Ogryzko. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594-598. [DOI] [PubMed] [Google Scholar]
- 11.Chakravarti, D., V. Ogryzko, H. Y. Kao, A. Nash, H. Chen, Y. Nakatani, and R. M. Evans. 1999. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96:393-403. [DOI] [PubMed] [Google Scholar]
- 12.Chan, H. M., and N. B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363-2373. [DOI] [PubMed] [Google Scholar]
- 13.Chen, H. M., P. Zhang, M. T. Voso, S. Hohaus, D. A. Gonzalez, C. K. Glass, D. E. Zhang, and D. G. Tenen. 1995. Neutrophils and monocytes express high levels of PU.1 (Spi-1) but not Spi-B. Blood 85:2918-2928. [PubMed] [Google Scholar]
- 14.Cheng, X., M. J. Reginato, N. C. Andrews, and M. A. Lazar. 1997. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol. Cell. Biol. 17:1407-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeKoter, R. P., and H. Singh. 2000. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 288:1439-1441. [DOI] [PubMed] [Google Scholar]
- 16.Fisher, R. C., and E. W. Scott. 1998. Role of PU.1 in hematopoiesis. Stem Cells 16:25-37. [DOI] [PubMed] [Google Scholar]
- 17.Forsberg, E. C., K. M. Downs, H. M. Christensen, H. Im, P. A. Nuzzi, and E. H. Bresnick. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 97:14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsberg, E. C., K. Johnson, T. N. Zaboikina, E. A. Mosser, and E. H. Bresnick. 1999. Requirement of an E1A-sensitive coactivator for long-range transactivation by the β-globin locus control region. J. Biol. Chem. 274:26850-26859. [DOI] [PubMed] [Google Scholar]
- 19.Fujiwara, Y., C. P. Browne, K. Cunniff, S. C. Goff, and S. H. Orkin. 1996. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 93:12355-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galson, D. L., J. O. Hensold, T. R. Bishop, M. Schalling, A. D. D'Andrea, C. Jones, P. E. Auron, and D. E. Housman. 1993. Mouse beta-globin DNA-binding protein B1 is identical to a proto-oncogene, the transcription factor Spi-1/PU.1, and is restricted in expression to hematopoietic cells and the testis. Mol. Cell. Biol. 13:2929-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 22.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 23.Gu, W., X.-L. Shi, and R. G. Roeder. 1997. Synergistic activation of transcription by CBP and p53. Nature (London) 387:819-823. [DOI] [PubMed] [Google Scholar]
- 24.Hagemeier, C., A. J. Bannister, A. Cook, and T. Kouzarides. 1993. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc. Natl. Acad. Sci. USA 90:1580-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamamori, Y., V. Sartorelli, V. Ogryzko, P. L. Puri, H. Y. Wu, J. Y. Wang, Y. Nakatani, and L. Kedes. 1999. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell 96:405-413. [DOI] [PubMed] [Google Scholar]
- 26.Hebbes, T. R., A. W. Thorne, A. L. Clayton, and C. Crane-Robinson. 1992. Histone acetylation and globin gene switching. Nucleic Acids Res. 20:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung, H.-L., J. Lau, M. J. Weiss, and G. A. Blobel. 1999. CREB-binding protein (CBP) acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol. 19:3496-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung, H. L., A. Y. Kim, W. Hong, C. Rakowski, and G. A. Blobel. 2001. Stimulation of NF-E2 DNA binding by CREB-binding protein (CBP)-mediated acetylation. J. Biol. Chem. 276:10715-10721. [DOI] [PubMed] [Google Scholar]
- 29.Klemsz, M. J., and R. A. Maki. 1996. Activation of transcription by PU.1 requires both acidic and glutamine domains. Mol. Cell. Biol. 16:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krebs, J. E., M. H. Kuo, C. D. Allis, and C. L. Peterson. 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 13:1412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulessa, H., J. Frampton, and T. Graf. 1995. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9:1250-1262. [DOI] [PubMed] [Google Scholar]
- 32.Kung, A. L., V. I. Rebel, R. T. Bronson, L. E. Ch'ng, C. A. Sieff, D. M. Livingston, and T. P. Yao. 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 14:272-277. [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 34.Li, M., B. Damania, X. Alvarez, V. Ogryzko, K. Ozato, and J. U. Jung. 2000. Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol. Cell. Biol. 20:8254-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, Q., A. Imhof, T. N. Collingwood, F. D. Urnov, and A. P. Wolffe. 1999. p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J. 18:5634-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacLellan, W. R., G. Xiao, M. Abdellatif, and M. D. Schneider. 2000. A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol. Cell. Biol. 20:8903-8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maestro, R., A. P. Dei Tos, Y. Hamamori, S. Krasnokutsky, V. Sartorelli, L. Kedes, C. Doglioni, D. H. Beach, and G. J. Hannon. 1999. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 13:2207-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Balbas, M., A. Bannister, K. Martin, P. Haus-Seuffert, M. Meisternst, and T. Kouzarides. 1998. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 17:2886-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumura, I., A. Kawasaki, H. Tanaka, J. Sonoyama, S. Ezoe, N. Minegishi, K. Nakajima, M. Yamamoto, and Y. Kanakura. 2000. Biologic significance of GATA-1 activities in Ras-mediated megakaryocytic differentiation of hematopoietic cell lines. Blood 96:2440-2450. [PubMed] [Google Scholar]
- 41.McKercher, S. R., B. E. Torbett, K. L. Anderson, G. W. Henkel, D. J. Vestal, H. Baribault, M. Klemsz, A. J. Feeney, G. E. Wu, C. J. Paige, and R. A. Maki. 1996. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15:5647-5658. [PMC free article] [PubMed] [Google Scholar]
- 42.McNagny, K., M. H. Sieweke, G. Doderlein, T. Graf, and C. Nerlov. 1998. Regulation of eosinophil-specific gene expression by a C/EBP-Ets complex and GATA-1. EMBO J. 17:3669-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merika, M., and S. H. Orkin. 1995. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with Krüppel family proteins Sp1 and EKLF. Mol. Cell. Biol. 15:2437-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metz, T., and T. Graf. 1991. Fusion of the nuclear oncoproteins v-Myb and v-Ets is required for the leukemogenicity of E26 virus. Cell 66:95-105. [DOI] [PubMed] [Google Scholar]
- 45.Mignotte, V., L. Wall, E. deBoer, F. Grosveld, and P.-H. Romeo. 1989. Two tissue-specific factors bind the erythroid promoter of the human porphobilinogen deaminase gene. Nucleic Acids Res. 17:37-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, R. W., and J. H. Rubinstein. 1995. Tumors in Rubinstein-Taybi syndrome. Am. J. Med. Gen. 56:112-115. [DOI] [PubMed] [Google Scholar]
- 47.Miyake, S., W. R. Sellers, M. Safran, X. Li, W. Zhao, S. R. Grossman, J. Gan, J. A. DeCaprio, P. D. Adams, and W. G. Kaelin, Jr. 2000. Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol. Cell. Biol. 20:8889-8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyashita, T., and J. C. Reed. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80:293-299. [DOI] [PubMed] [Google Scholar]
- 49.Moreau-Gachelin, F. 1994. Spi-1/PU.1: an oncogene of the Ets family. Biochim. Biophys. Acta 1198:149-163. [DOI] [PubMed] [Google Scholar]
- 50.Moreau-Gachelin, F., A. Tavitian, and P. Tambourin. 1988. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature 331:277-280. [DOI] [PubMed] [Google Scholar]
- 51.Moreau-Gachelin, F., F. Wendling, T. Molina, N. Denis, M. Titeux, G. Grimber, P. Briand, W. Vainchenker, and A. Tavitian. 1996. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol. Cell. Biol. 16:2453-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motohashi, H., J. A. Shavit, K. Igarashi, M. Yamamoto, and J. D. Engel. 1997. The world according to Maf. Nucleic Acids Res. 25:2953-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nerlov, C., and T. Graf. 1998. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 12:2403-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nerlov, C., E. Querfurth, H. Kulessa, and T. Graf. 2000. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 95:2543-2551. [PubMed] [Google Scholar]
- 55.Nunn, M. F., and T. Hunter. 1989. The ets sequence is required for induction of erythroblastosis in chickens by avian retrovirus E26. J. Virol. 63:398-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogryzko, V. V., L. R. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 57.Perissi, V., J. S. Dasen, R. Kurokawa, Z. Wang, E. Korzus, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1999. Factor-specific modulation of CREB-binding protein acetyltransferase activity. Proc. Natl. Acad. Sci. USA 96:3652-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perkel, J. M., and M. L. Atchison. 1998. A two-step mechanism for recruitment of Pip by PU.1. J. Immunol. 160:241-252. [PubMed] [Google Scholar]
- 59.Pomerantz, O., A. J. Goodwin, T. Joyce, and C. H. Lowrey. 1998. Conserved elements containing NF-E2 and tandem GATA binding sites are required for erythroid-specific chromatin structure reorganization within the human β-globin locus control region. Nucleic Acids Res. 26:5684-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pongubala, J. M., and M. L. Atchison. 1997. PU.1 can participate in an active enhancer complex without its transcriptional activation domain. Proc. Natl. Acad. Sci. USA 94:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pongubala, J. M., S. Nagulapalli, M. J. Klemsz, S. R. McKercher, R. A. Maki, and M. L. Atchison. 1992. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3′ enhancer activity. Mol. Cell. Biol. 12:368-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rao, G., N. Rekhtman, G. Cheng, T. Krasikov, and A. I. Skoultchi. 1997. Deregulated expression of the PU.1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene 14:123-131. [DOI] [PubMed] [Google Scholar]
- 63.Rekhtman, N., F. Radparvar, T. Evans, and A. I. Skoultchi. 1999. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 13:1398-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richon, V. M., S. Emiliani, E. Verdin, Y. Webb, R. Breslow, R. A. Rifkind, and P. A. Marks. 1998. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc. Natl. Acad. Sci. USA 95:3003-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sawado, T., K. Igarashi, and M. Groudine. 2001. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc. Natl. Acad. Sci. USA 98:10226-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schubeler, D., C. Francastel, D. M. Cimbora, A. Reik, D. I. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 14:940-950. [PMC free article] [PubMed] [Google Scholar]
- 68.Schuetze, S., R. Paul, B. C. Gliniak, and D. Kabat. 1992. Role of the PU.1 transcription factor in controlling differentiation of Friend erythroleukemia cells. Mol. Cell. Biol. 12:2967-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schuetze, S., P. E. Stenberg, and D. Kabat. 1993. The Ets-related transcription factor PU.1 immortalizes erythroblasts. Mol. Cell. Biol. 13:5670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scolnick, D. M., N. H. Chehab, E. S. Stavridi, M. C. Lien, L. Caruso, E. Moran, S. L. Berger, and T. D. Halazonetis. 1997. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 57:3693-3696. [PubMed] [Google Scholar]
- 71.Scott, E. W., M. C. Simon, J. Anastasi, and H. Singh. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265:1573-1577. [DOI] [PubMed] [Google Scholar]
- 72.Seo, S. B., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104:119-130. [DOI] [PubMed] [Google Scholar]
- 73.Steffan, J. S., L. Bodai, J. Pallos, M. Poelman, A. McCampbell, B. L. Apostol, A. Kazantsev, E. Schmidt, Y. Z. Zhu, M. Greenwald, R. Kurokawa, D. E. Housman, G. R. Jackson, J. L. Marsh, and L. M. Thompson. 2001. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413:739-743. [DOI] [PubMed] [Google Scholar]
- 74.Takahashi, T., N. Suwabe, P. Dai, M. Yamamoto, S. Ishii, and T. Nakano. 2000. Inhibitory interaction of c-Myb and GATA-1 via transcriptional co-activator CBP. Oncogene 19:134-140. [DOI] [PubMed] [Google Scholar]
- 75.Tsang, A. P., J. E. Visvader, C. A. Turner, Y. Fujiwara, C. Yu, M. J. Weiss, M. Crossley, and S. H. Orkin. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109-119. [DOI] [PubMed] [Google Scholar]
- 76.Voso, M. T., T. C. Burn, G. Wulf, B. Lim, G. Leone, and D. G. Tenen. 1994. Inhibition of hematopoiesis by competitive binding of transcription factor PU.1. Proc. Natl. Acad. Sci. USA 91:7932-7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weiss, M. J., G. Keller, and S. H. Orkin. 1994. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1-embryonic stem cells. Genes Dev. 8:1184-1197. [DOI] [PubMed] [Google Scholar]
- 78.Weiss, M. J., and S. H. Orkin. 1995. GATA transcription factors: key regulators of hematopoiesis. Exp. Hematol. 23:99-107. [PubMed] [Google Scholar]
- 79.Weiss, M. J., and S. H. Orkin. 1995. Transcription factor GATA-1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc. Natl. Acad. Sci. USA 92:9623-9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamada, T., F. Kihara-Negishi, H. Yamamoto, M. Yamamoto, Y. Hashimoto, and T. Oikawa. 1998. Reduction of DNA binding activity of the GATA-1 transcription factor in the apoptotic process induced by overexpression of PU.1 in murine erythroleukemia cells. Exp. Cell Res. 245:186-194. [DOI] [PubMed] [Google Scholar]
- 81.Yamada, T., N. Kondoh, M. Matsumoto, M. Yoshida, A. Maekawa, and T. Oikawa. 1997. Overexpression of PU.1 induces growth and differentiation inhibition and apoptotic cell death in murine erythroleukemia cells. Blood 89:1383-1393. [PubMed] [Google Scholar]
- 82.Yamamoto, H., F. Kihara-Negishi, T. Yamada, Y. Hashimoto, and T. Oikawa. 1999. Physical and functional interactions between the transcription factor PU.1 and the coactivator CBP. Oncogene 18:1495-1501. [DOI] [PubMed] [Google Scholar]
- 83.Zhang, P., G. Behre, J. Pan, A. Iwama, N. Wara-Aswapati, H. S. Radomska, P. E. Auron, D. G. Tenen, and Z. Sun. 1999. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl. Acad. Sci. USA 96:8705-8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang, P., X. Zhang, A. Iwama, C. Yu, K. A. Smith, B. U. Mueller, S. Narravula, B. E. Torbett, S. H. Orkin, and D. G. Tenen. 2000. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood 96:2641-2648. [PubMed] [Google Scholar]
- 85.Zhang, W., and J. J. Bieker. 1998. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl. Acad. Sci. USA 95:9855-9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang, W., S. Kadam, B. M. Emerson, and J. J. Bieker. 2001. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol. 21:2413-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]