Abstract
Notch genes encode a family of transmembrane proteins that are involved in many cellular processes, such as differentiation, proliferation, and apoptosis. It is well established that all four Notch genes can act as oncogenes; however, the mechanism by which Notch proteins transform cells remains unknown. Previously, we reported that both nuclear localization and transcriptional activation are required for neoplastic transformation of RKE cells. Furthermore, we identified cyclin D1 as a direct transcriptional target of constitutively active Notch molecules. In an effort to understand the mechanism by which Notch functions in the nucleus, we sought to determine if Notch formed stable complexes using size exclusion chromatography. Herein, we report that the Notch intracellular domain (Nic) forms distinct high-molecular-weight complexes in the nuclei of transformed RKE cells. The largest complex is approximately 1.5 MDa and contains both endogenous CSL (for CBF1, Suppressor of Hairless, and Lag-1) and Mastermind-Like-1 (Maml). Nic molecules that do not have the high-affinity binding site for CSL (RAM) retain the ability to associate with CSL in a stable complex through interactions involving Maml. However, Maml does not directly bind to CSL. Furthermore, Maml can rescue ΔRAM transcriptional activity on a CSL-dependent promoter. These results indicate that deletion of the RAM domain does not equate to CSL-independent signaling. Moreover, in SUP-T1 cells, Nic exists exclusively in the largest Nic-containing complex. SUP-T1 cells are derived from a T-cell leukemia that harbors the t(7;9)(q34;q34.3) translocation and constitutively express Nic. Taken together, our data indicate that complex formation is likely required for neoplastic transformation by Notchic.
The Notch pathway is an evolutionarily conserved signaling mechanism that has been utilized throughout metazoan development to influence cell fate decisions (for a review, see reference 1). Notch signaling has been shown to both promote and inhibit key cellular functions such as proliferation, apoptosis, and differentiation, depending upon the cellular context of Notch activation (8, 33-35, 37, 38, 43, 45, 49). Remarkably, it is thought that Notch is able to exert these pleiotropic effects by the utilization of common signaling effectors. Direct binding of a DSL (Delta, Serrate, Lag-2) ligand from a signaling cell to a Notch receptor on the plasma membrane of the receiving cell initiates a series of proteolytic processing events at the membrane interface, which ultimately results in the release of the intracellular domain (Nic) from its plasma membrane tether (1, 10, 25, 30, 36, 53, 56, 57). By an unknown mechanism, Nic translocates into the nucleus, where it directly functions in the regulation of gene expression. In the nucleus, Nic interacts with the transcriptional repressor CSL (CBF1, Suppressor of Hairless, Lag-1) and converts it from a repressor to an activator (20, 24, 58). Nic is thought to accomplish this by displacing CSL corepressors such as SMRT and HDAC1 and by providing an intrinsic activation domain (21, 23, 31, 32, 39). A considerable debate persists within the Notch field concerning CSL-dependent versus CSL-independent mechanisms of Notch signaling (1, 28, 54). CSL interacts with Nic through a high-affinity binding site on Nic termed the RAM domain. Nic proteins that have a deletion of the RAM domain (ΔRAM) retain activity in biological assays even though the CSL binding site has been deleted. These results have been interpreted as evidence for a CSL-independent signaling mechanism mediated by ΔRAM proteins (4, 13, 22, 31, 37). For example, we have shown that the RAM domain is dispensable for neoplastic transformation of RKE cells, even though an interaction between ΔRAM proteins and CSL was not detected in reporter and GST pull-down assays (26).
A substantial body of evidence implicates the mammalian Notch gene family in oncogenesis (2, 3, 5-7, 14, 15, 41, 47, 48, 60). The human Notch1 gene was cloned from SUP-T1 cells, which were derived from a T-cell acute lymphoblastic leukemia (T-ALL) that harbors the t(7,9)(q34;q34.3) chromosomal translocation. This translocation joins the T-cell receptor β locus with the Notch1 gene and results in the constitutive expression of Nic-like molecules (14). Although this is the only documented genetic alteration of Notch1 in a human cancer, aberrant expression of Notch1, Notch2, and Notch3 has been detected in several other human neoplasms (9, 44, 60). Furthermore, activated forms of all four Notch proteins have been shown to contribute to oncogenesis in both animal and in vitro model systems (5-7, 11, 13, 16, 17, 41, 47, 48, 55). We have shown a direct role for Notch in the transformation of cells by demonstrating that constitutive expression of Nic cooperates with E1A to transform primary baby rat kidney cells (7). In addition, expression of N1ic or N2ic can transform an E1A-immortalized baby rat kidney cell line, RKE (7, 26, 50). The molecular mechanism by which Nic transforms cells is unknown, but we have recently shown that both nuclear localization and transcriptional activation are required for neoplastic transformation of RKE cells (26). In agreement with this model, cyclin D1 has been shown to be an important target gene upregulated by Nic in transformed RKE cells. Cyclin D1 transcription is induced by Nic proteins, but it is not induced by a nontransforming, transcriptionally inactive Nic molecule (49).
We report the identification of protein complexes containing Nic in the nuclei of Nic-transformed RKE cells and in the human T-ALL cell line SUP-T1. Nic integrates into two stable high-molecular-weight complexes, the largest of which contains both endogenous CSL and Mastermind-Like-1 (Maml). Deletion of the RAM domain does not significantly affect association with CSL and Maml in RKE cells, demonstrating that the RAM domain is not required to interact with CSL in vivo. We present data that Maml functions, in part, to promote association between Nic and CSL and that, as a result, Maml can rescue the transcriptional activity of Nic ΔRAM on a CSL-responsive promoter. However, Maml is not the exclusive mediator of Nic transcriptional activity, since excess Maml is unable to compensate for a mutation in Nic that abolishes transactivation but does not affect association with Maml. We propose that complex formation is important for both transcriptional activity and transformation of RKE cells by Nic. In addition, constitutive accumulation of this complex in the nuclei of SUP-T1 cells might account for the neoplastic conversion of these cells by Nic.
MATERIALS AND METHODS
Cell culture and generation of stable RKE cell lines.
RKE, HeLa, and 293T cells were propagated in Dulbecco's modified Eagle medium (Life Technologies) under standard conditions. SUP-T1 cells were propagated in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (Life Technologies).
Cell lines described in this study were generated by retroviral infection of RKE cells. Retroviral particles were packaged in 293T cells by cotransfection of Nic derivative retroviral expression vector DNA (3 μg) with simian virus 40 ψ− ecotropic helper virus DNA (3 μg) by N-N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid-calcium phosphate precipitation. At 48 h posttransfection, retrovirus-containing supernatants were harvested and used to infect 1.0 × 105 RKE cells at a 1:10 dilution. Following infection, RKE cells were trypsinized and split into selection media (2 μg of puromycin per ml). Drug-resistant colonies were pooled into polyclonal cell lines, and protein expression was verified by Western blot analysis. Cell lines were propagated in Dulbecco's modified Eagle medium supplemented with the appropriate selectable marker (2 μg of puromycin per ml).
Subcellular fractionation.
Nuclear protein extracts were prepared from cells essentially as described by Dignam et al. (12). Cells were rinsed twice with ice-cold 1× PBS, scraped into centrifuge tubes, and collected by centrifugation. The cell pellet was washed once with hypotonic lysis buffer (40 mM Tris·HCl [pH 7.4], 10 mM NaCl, 1 mM EDTA, 0.5 mM dithiothreitol [DTT]) supplemented with the protease inhibitors aprotinin (2 μg/ml), leupeptin (5 μg/ml), and Pefabloc (2 mM) and then resuspended in five packed cell volumes of hypotonic lysis buffer and allowed to swell on ice for 10 min. Cells were burst by Dounce homogenization with 20 strokes of a type B pestle. Crude nuclei were collected by centrifugation for 10 min at 1,600 × g in a Heraeus bench top 4°C centrifuge. The resulting supernatant was transferred to a fresh tube and centrifuged at 100,000 × g for 1 h in a Sorvall microultracentrifuge to separate cytosolic (S100) and membrane fractions. The crude nuclear pellet was washed three times in hypotonic lysis buffer. Nuclei were resuspended in an equal volume of nuclear extraction buffer (420 mM NaCl, 40 mM Tris·HCl [pH 7.4], 1 mM EDTA, 0.5 mM DTT) supplemented with protease inhibitors and was allowed to extract on ice for 30 min. Debris was pelleted at 25,000 × g for 30 min at 4°C; the resulting supernatant contained the nuclear protein extract that was used for subsequent analysis.
Size exclusion chromatography and TCA precipitation.
Typically, 400 μg of nuclear extract from each indicated stable RKE cell line was fractionated by size exclusion chromatography on a Superose 6 HR 10/30 resin by fast protein liquid chromatography (AKTA; Amersham-Pharmacia Biotech). The predicted size exclusion maximum for this resin is 40 MDa, with a void volume of 7.35 ml, which corresponds to fraction 21. The column was equilibrated in 2 column volumes of either high-salt column buffer (600 mM NaCl, 40 mM Tris·HCl [pH 7.4], 1 mM EDTA, 0.5 mM DTT, 5% glycerol, 0.001% NP-40) or physiological-salt column buffer (150 mM NaCl, 40 mM Tris·HCl [pH 7.4], 1 mM EDTA, 0.5 mM DTT, 5% glycerol, 0.001% NP-40) prior to sample loading. Nuclear lysates were run in high-salt column buffer unless otherwise indicated. High-molecular-weight protein column standards were used to define the column resolution (Pharmacia). Each standard was prepared at a concentration of 1 mg/ml in physiological-salt column buffer, and standards were sequentially loaded onto the column. Protein peaks were detected by both UV monitoring and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. Thyroglobulin (Mr, 669,000) peaked in fraction 34, ferritin (Mr, 440,000) peaked in fraction 40, catalase (Mr, 232,000) peaked in fraction 43, and bovine serum albumin (Mr, 67,000) peaked in fraction 45. Prior to loading, each nuclear lysate was adjusted to the appropriate column conditions and centrifuged at 100,000 × g for 30 min. A 200-μl portion of each sample was loaded onto the column and collected into 350-μl fractions; fractions were then subjected to trichloroacetic acid (TCA) precipitation for Western blot analysis, or individual fractions were pooled for immunoprecipitations. For TCA precipitation, the sample volume was adjusted to 500 μl with the corresponding column buffer followed by the addition of 50 μl of 0.15% sodium deoxycholate; tubes were vortexed and incubated at room temperature for 10 min. Protein was precipitated by the addition of 25 μl of 100% TCA (Sigma, St. Louis, Mo.) followed by a 20-min incubation at −20°C. Precipitated proteins were collected by centrifugation at 10,000 × g for 10 min at room temperature. Protein pellets were washed with acetone and air dried. The protein pellet was solubilized in 1× sample buffer (62.5 mM Tris [pH 6.8], 0.72 M β-mercaptoethanol or 0.1 M DTT, 10% glycerol, 2% SDS, and 0.05% bromophenol blue) and resolved by SDS-8% PAGE. Proteins were subsequently transferred to polyvinylidene difluoride membranes (Millipore) and processed for Western blot analysis by using standard protocols. αNic-927 is a purified rabbit polyclonal antibody directed at residues 1759 through 2095 of human Nic and was used at 0.1 μg/ml for Western blot analysis. Anti-CSL polyclonal antiserum was provided by Emery Bresnick (University of Wisconsin—Madison), and anti-Maml hybridoma tissue culture supernatant was provided by James D. Griffin (Dana Farber Cancer Institute, Harvard Medical School) (59). Spryos Artavanis-Tsakonas (Harvard University) provided the bTAN15A hybridoma (60). Antihemagglutinin (anti-HA) polyclonal antibody (Y-11) was purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.), and anti-Flag M2 monoclonal antibody was purchased from Sigma.
Immunoprecipitations.
RKE cell lines were harvested in standard lysis buffer (150 mM NaCl, 50 mM HEPES [pH 7.4], 1 mM EDTA, 0.5 mM DTT, 10% glycerol, 1% NP-40) supplemented with aprotinin (2 μg/ml), leupeptin (5 μg/ml), and Pefabloc (2 mM). Cells were lysed on ice for 20 min and then centrifuged at 10,000 × g for 30 min. Protein lysates were further clarified by centrifugation at 100,000 × g for 30 min at 4°C. The sample was then split into two equal aliquots; one received 10 μl of anti-Myc antibody (9E10) coupled to protein A-Sepharose (∼4 μg) to immunoprecipitate Nic protein complexes, and the other was mock precipitated by addition of a nonspecific mouse immunoglobulin G (IgG) control antibody and protein A-Sepharose (αGSK-3; Transduction Laboratories). Immune complexes were collected by centrifugation, washed three times in standard lysis buffer, and separated by SDS-PAGE.
For immunoprecipitation of protein complexes from Superose 6 gel filtration column fractions, three fractions encompassing the relevant peak of immunoreactivity were pooled into a microcentrifuge tube (final volume, approximately 1 ml) followed by the addition of 4 μg of the appropriate antibody. Immune complexes were allowed to form for 2 h at 4°C with constant rocking and then collected by the addition of protein A-Sepharose. Immunoprecipitates were washed three times in column buffer and separated by SDS-PAGE, and proteins were detected by Western blot analysis.
Immunoprecipitation from 293T cells was performed as follows. Cells were transfected with 3 μg of Nic expression plasmid, 3 μg of Flag-Maml vector, and 0.5 μg of HA-CSL expression vector. At 48 h posttransfection, cells were harvested in standard lysis buffer supplemented with 50 mM NaF, 1 mM sodium orthovanadate, aprotinin (2 μg/ml), leupeptin (5 μg/ml), and Pefabloc (2 mM). Cells were lysed on ice for 20 min, followed by centrifugation at 10,000 × g for 30 min. Lysates were immunoprecipitated with 1 μg of anti-HA antibody for 1 h at 4°C with rocking. Complexes were collected by addition of protein A-Sepharose beads, washed three times in lysis buffer, and separated by SDS-PAGE.
Nic-containing complexes were reconstituted in 293T cells by transfection with 2 μg of Nic expression plasmid, 3 μg of Flag-Maml vector, and 0.5 μg of HA-CSL plasmid. Following transfection, nuclei were isolated by hypotonic lysis and lysed in physiological-salt column buffer containing 1% NP-40 for 30 min on ice. Nuclear debris was removed by centrifugation at 25,000 × g for 30 min at 4°C. The lysate was centrifuged at 100,000 × g for 30 min at 4°C and subsequently loaded onto a Superose 6 column equilibrated in physiological-salt column buffer containing 1% NP-40. Immunoprecipitation from various pooled column fractions was performed as described above.
Luciferase reporter assay.
For luciferase assays, 1.0 × 105 HeLa cells were seeded in six-well plates and were cotransfected with 0.4 μg of reporter vector, 0.2 μg of CMV-β-gal (internal transfection control plasmid; Clontech), 0.1 μg of the indicated pBabePuro 283 Nic expression plasmid and either 0.2 or 0.4 μg of the pcDNA 3.1 Maml expression plasmid. Luciferase reporter constructs used contained either a genomic fragment from the mouse HES-1 promoter (HES-1-luc) or eight copies of the CSL DNA binding consensus sequence (8x-CSL-luc) cloned into the pGL3 (HES-1-luc) or pGL2 (8x-CSL-luc) reporter vector (Promega). Cells were transfected with 10 μl of Lipofectamine in a total volume of 2 ml of OptiMEM. At 48 h posttransfection, lysates were prepared and luciferase activities were determined in a Xylux Femtomaster FB 12 luminometer according to the manufacturer's suggested protocol (Promega). Luciferase values were corrected for transfection efficiency by normalizing to β-galactosidase activity.
Plasmid constructions.
Nic deletion constructs used in this study have been described (26). ΔRAMΔ2105 was generated by PCR with Δ2105 as the DNA template. The PCR product was digested with BamHI and XhoI and ligated into pBabePuro 283 as described previously (26). Details concerning plasmid constructions will be provided upon request. Human full-length Flag-Maml was kindly provided by James D. Griffin (59). Maml-305 was generated by PCR, and the resulting product was cloned into pBabePuro 283 as described above.
The HES-1-luc luciferase reporter vector was constructed by amplification of a genomic fragment of the murine HES-1 promoter containing bases −190 through +160 relative to the start site by PCR. The resulting PCR product was cut with the appropriate restriction enzymes and cloned into pGL3 basic (Promega).
RESULTS
Nic and CSL exist in a high-molecular-weight protein complex.
A schematic of the Nic constructs used in this study is presented in Fig. 1A. Nic consists of residues 1759 through 2556 of human Notch1. ΔRAM has an N-terminal deletion that removes the high-affinity binding site for CSL yet retains the ability to transform RKE cells (26). NicΔ2444 has a deletion of 102 C-terminal residues that encode the entire PEST domain. ΔRAMΔ2444 has a composite deletion of both the RAM domain and the PEST domain. Δ2105 has a deletion of 10 residues (amino acids 2105 through 2114) of Nic and is incapable of transforming RKE cells, even though it retains the ability to bind CSL through the RAM domain (26). ΔRAMΔ2105 has deletions of both the RAM domain and residues 2105 to 2114 of Nic and is also nontransforming (data not shown). The minimal transformation domain (TFD) of Nic has both the RAM domain and 354 C-terminal residues deleted. A seventh ankyrin repeat is disrupted by the Δ2105 mutation (see Discussion for details). All constructs contain a C-terminal Myc epitope tag for detection with the anti-Myc monoclonal antibody 9E10.
FIG. 1.
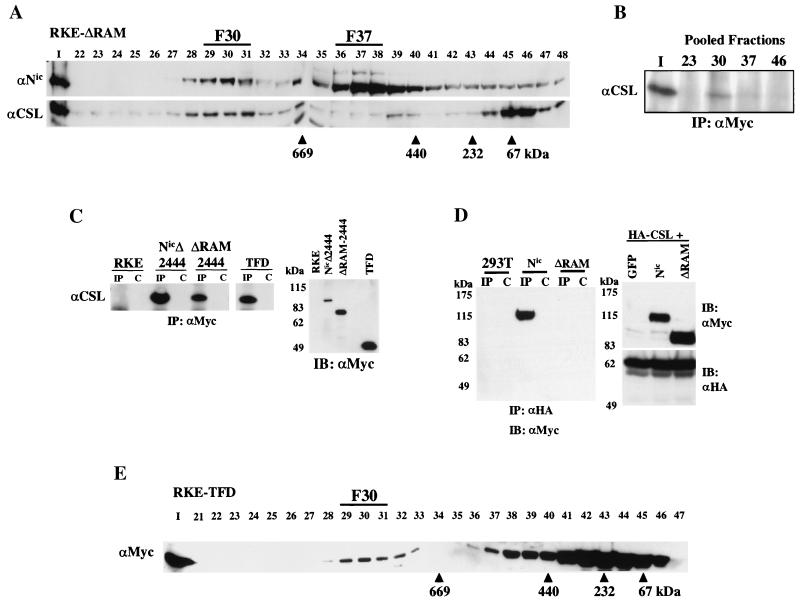
Nic exists in high-molecular-weight nuclear protein complexes with CSL. (A) Schematic representation of Nic derivatives used in this study. Numbers indicate the N- and C-terminal residues for each construct. The seventh ankyrin repeat is highlighted in silver. All Nic constructs are engineered with a C-terminal Myc epitope tag (M). (B) Nuclear lysate from a Nic-transformed RKE cell line was fractionated by size exclusion chromatography. Nic was visualized by Western blotting with αNic-927 (top). Numbers above the lanes indicate collected fractions (I, 10% of total protein input). Arrowheads indicate the native molecular masses of known protein standards: thyroglobulin, 669 kDa; ferritin, 440 kDa; catalase, 232 kDa; and bovine serum albumin, 67 kDa. CSL was detected with anti-CSL antisera. (C) Nic and CSL are physically associated in the high-molecular-weight complex. Three fractions encompassing the indicated gel filtration peaks were pooled and immunoprecipitated with an αNic-927. Immunoprecipitates were detected with the indicated antibody.
To begin to elucidate how Nic might function in the nucleus, we sought to determine if Nic formed stable complexes in transformed RKE cells. Nuclear extracts were prepared from a Nic-transformed RKE cell line, and proteins were fractionated by size exclusion chromatography on a Superose 6 column as described in Materials and Methods. When the fractions were analyzed by Western blotting using a Notch1 polyclonal antibody (αNic-927), we found that Nic immunoreactivity elutes in two high-molecular-weight peaks with apparent molecular masses of approximately 1.5 MDa and 500 to 600 kDa, as calculated from the elution peaks of known protein size standards (Fig. 1B). The largest complex is referred to as the fraction 30 complex (F30), and the smaller complex is referred to as the fraction 37 complex (F37); each number represents the peak of Nic immunoreactivity in each complex. Since CSL is a principal nuclear effector for Notch signaling, we investigated whether CSL coeluted with Nic in F30 and/or F37 by probing the same immunoblots with antisera specific for CSL (Fig. 1B). A portion of the endogenous CSL coelutes with Nic in F30, while the majority of CSL immunoreactivity elutes in fraction 46, with some CSL immunoreactivity detectable between each peak. The fraction 46 peak is consistent with the monomeric size of CSL (∼59 kDa) indicating the majority of CSL in RKE cells might not be stably associated with other proteins.
To demonstrate that comigration of Nic and CSL in F30 is due to a physical association between the two proteins, immunoprecipitation assays were performed. Column fractions 22 to 24, 29 to 31, 36 to 38, and 45 to 47 were separately pooled and immunoprecipitated with αNic-927 (Fig. 1C). CSL was coimmunoprecipitated with Nic from pooled fractions 29 to 31 (Fig. 1C, lane 30), demonstrating that Nic and CSL are physically associated in F30. A small percentage of CSL could be coimmunoprecipitated with Nic from F37 (lane 37). However, no detectable CSL was immunoprecipitated from the fraction 46 pool, although this fraction contains most of the CSL protein, indicating that Nic specifically associates with CSL in F30 and F37.
The RAM domain is dispensable for complex formation in RKE cells.
Previously we demonstrated that the RAM domain of Nic is dispensable for transformation of RKE cells (26). This domain of Nic is required for high-affinity association with CSL both in vivo and in vitro (4, 26, 28, 53, 58). Consistent with this, ΔRAM neither bound CSL in vitro nor activated a CSL-responsive reporter in HeLa cells (26). Given that Nic and CSL associate in a high-molecular-weight complex in RKE cells, we wanted to determine the significance of the RAM domain in forming F30.
Nuclear lysate from ΔRAM-transformed RKE cells was fractionated through the Superose 6 gel filtration column. ΔRAM displayed a column profile similar to that of Nic, with major peaks of immunoreactivity eluting in both F30 and F37 (Fig. 2A). Endogenous CSL also coeluted with ΔRAM in F30 (Fig. 2A). A physical association between ΔRAM and CSL was demonstrated by coimmunoprecipitation. The indicated column fractions were immunoprecipitated with 9E10 and CSL was detected by immunoblotting with anti-CSL polyclonal antisera (Fig. 2B). CSL was coimmunoprecipitated with ΔRAM in both the input nuclear lysate and the fraction 30 pool, demonstrating that ΔRAM and endogenous CSL are physically associated in F30 independent of the RAM domain.
FIG. 2.
The RAM domain of Nic is dispensable for complex formation with CSL in RKE cells. (A) Nuclear lysate from ΔRAM-transformed RKE cells was fractionated by size exclusion chromatography. ΔRAM protein was visualized by Western blot analysis with bTAN15A (αNic). CSL protein was detected with anti-CSL antisera. (B) CSL stably integrates into the high-molecular-weight complex in the absence of the RAM domain. Fifty percent of the column load (I) and the three fractions encompassing the indicated gel filtration peaks were individually pooled and immunoprecipitated with anti-Myc antibody. CSL was detected with anti-CSL antisera. (C) CSL coimmunoprecipitates with the TFD of Nic. Whole-cell protein lysates from the indicated RKE cell lines were immunoprecipitated with anti-Myc antibody. CSL is found in the lanes precipitated with anti-Myc antibody (IP) but not in IgG control lanes (C). Expression of Nic derivative proteins was detected by Western blot analysis with anti-Myc antibody. Molecular mass markers are shown on the left. (D) The RAM domain is required for Nic and CSL interaction in 293T cells. 293T cells were cotransfected with the indicated expression plasmids. Whole-cell lysates were made and immunoprecipitated with the anti-HA antibody directed against CSL (lanes IP) or preimmune serum (lanes C); Nic proteins were detected by Western blot analysis with anti-Myc antibody. Five percent of each 293T lysate was separated by SDS-PAGE and processed for immunoblot analysis; proteins were detected with the indicated antibody (right). Molecular mass markers are shown on the left. (E) The TFD forms F30 in RKE cells. Cytoplasmic lysate from RKE cells expressing the TFD was fractionated by size exclusion chromatography, and the TFD was visualized by Western blot analysis with anti-Myc antibody.
To determine if C-terminal sequences present in ΔRAM were required for the association with endogenous CSL in RKE cells, we performed coimmunoprecipitations with Nic deletion derivatives that had both the RAM domain and C-terminal residues deleted. ΔRAMΔ2444 has deletions of both the RAM domain and 102 C-terminal residues that include the PEST domain, whereas NicΔ2444 contains an intact RAM domain and is used as a positive control for CSL binding (26). The TFD has deletions of 354 C-terminal residues of Nic as well as the RAM domain. Whole-cell lysates from the RKE cell lines were immunoprecipitated with 9E10, and CSL was detected by immunoblotting with anti-CSL polyclonal antisera. C-terminal truncations did not affect the ability of ΔRAM mutants to bind to endogenous CSL within RKE cells; each protein tested could coimmunoprecipitate CSL irrespective of the RAM domain (Fig. 2C, lanes IP). CSL was not immunoprecipitated from cellular lysates by a nonspecific mouse antibody, indicating the association was specific for the presence of Nic (Fig. 2C, lanes C). Expression of each Nic deletion derivative was verified by Western blot analysis with 9E10 (Fig. 2C, right). In contrast to the results in RKE cells, the RAM domain is essential for CSL interaction with Nic in 293T cells. When CSL is coexpressed with either Nic or ΔRAM in 293T cells, only Nic can be coimmunoprecipitated with CSL (Fig. 2D, lanes IP) (4, 26, 53). Even when highly expressed in 293T cells, ΔRAM is unable to associate with CSL. This indicates that perhaps 293T cells are missing a protein(s) or a modification(s) of Nic that is important to promote formation of F30 in the absence of the RAM domain.
Since the TFD retains binding to CSL in the absence of the RAM domain, we wanted to determine if the TFD was able to integrate into F30 and/or F37. Cytoplasmic lysate from a TFD-transformed RKE cell line was fractionated by size exclusion chromatography. Western blot analysis revealed that the 40-kDa TFD was able to integrate into F30, indicating that the TFD retains the protein contacts required for F30 formation (Fig. 2E). However, when we salt extracted the nuclei at 420 mM NaCl, the TFD F30 was unstable and could be recovered only as a peak at fraction 33, indicating that C-terminal sequences might be required for complex stability within the nucleus (data not shown). F37 is shifted to the second peak of TFD immunoreactivity that elutes over a broad range peaking at fraction 43, indicating that F37 depends somewhat on the molecular weight of Nic or on sequences C terminal to residue 2202 which might be required for some protein interactions.
Maml associates with Nic and CSL in F30.
Recently, the human homologue of Drosophila Mastermind, Maml, was cloned and shown to form a trimeric complex with Nic and CSL in vitro and to potentiate Nic transcriptional activation on a CSL-responsive reporter (29, 42, 59). Given this, we hypothesized that Maml could be a potential binding partner for Nic and CSL in F30.
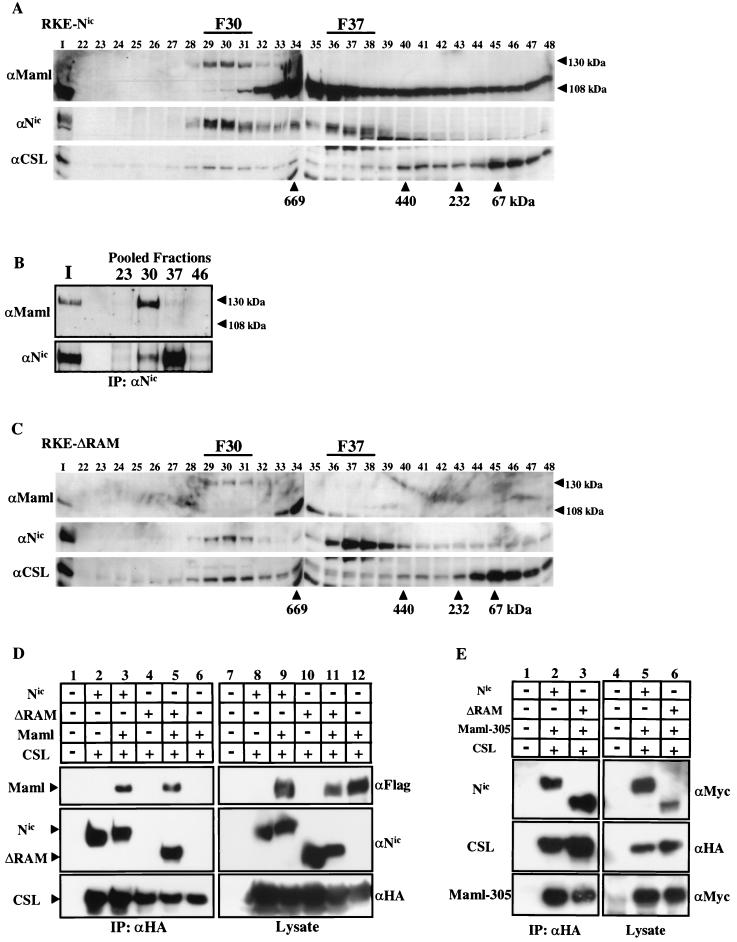
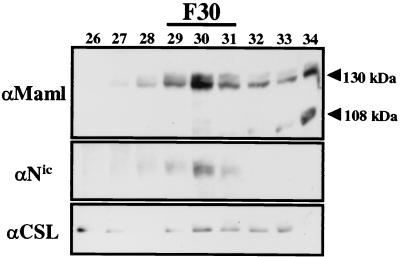
Nuclear extract from a Nic transformed RKE cell line was fractionated on a Superose 6 column, and Nic and CSL were detected by Western blot analysis. As previously demonstrated, both Nic and CSL coelute in F30 (Fig. 3A). To determine if endogenous Maml associates with Nic and CSL, the membrane was then probed with anti-Maml antibody (Fig. 3A). The antibody detected two bands that correspond to endogenous Maml. The most prominent band corresponds to the calculated molecular mass for Maml (108 kDa) and eluted in a broad profile that peaked in fractions 34 and 35 and trailed down the column profile to fraction 48. The second detectable band migrated in SDS-PAGE gels with an apparent molecular mass of approximately 130 kDa and precisely coeluted with both Nic and CSL in fraction 30. The 130-kDa form of endogenous Maml in RKE cell nuclear extracts is equivalent to the size reported for transfected Maml expressed in both 293T and COS-7 cells (29, 59). The molecular basis for the alternate species of Maml protein detected in RKE cells is currently not known. However, when a Flag-tagged Maml construct is expressed in 293T cells, both forms are detected in whole-cell lysates with the Flag antibody. We note that when cellular lysates are made in the absence of phosphatase inhibitors, primarily the 108-kDa species is observed, indicating that phosphorylation might be partially responsible for the migration of the 130 kDa species (S. Jeffries and A. J. Capobianco, unpublished observations).
FIG. 3.
Endogenous Maml is associated with Nic and CSL in F30. (A) Nuclear lysate from Nic-transformed RKE cells was fractionated on a Superose 6 size exclusion column. Maml protein was visualized by Western blot analysis with anti-Maml antibody. Arrowheads indicate the different forms of Maml species detected in RKE cells. Nic protein was detected with αNic-927, and CSL protein was detected with anti-CSL antiserum. (B) Maml physically associates with Nic. Ten percent of the column load (I) and the three fractions encompassing the indicated gel filtration peaks were individually pooled and immunoprecipitated with a αNic-927. Western blots were probed with the indicated antibodies. (C) Maml coelutes with ΔRAM in F30. Nuclear lysate from RKE cells stably expressing ΔRAM was fractionated by gel filtration and Western blotted with the indicated antibodies as previously described. (D) Maml tethers ΔRAM to CSL in 293T cells. 293T cells were cotransfected with the indicated plasmids and proteins were immunoprecipitated with the anti-HA antibody directed against CSL (lanes 1 to 6). Proteins were detected with the indicated antibodies. Expression of each protein was verified by Western blot analysis with the indicated antibody (lanes 7 to 12). (E) Maml-305 (amino acids 1 to 305) is sufficient to tether CSL to ΔRAM in 293T cells. 293T cells were cotransfected with the indicated plasmids. Nuclear extract for each transfection was immunoprecipitated with the anti-HA antibody directed against CSL (lanes 1 to 3). Expression of each protein was verified by Western blot analysis with the indicated antibody (lanes 4 to 6).
To determine if Maml is physically associated with Nic in F30, column fractions encompassing each peak of Nic immunoreactivity were individually pooled and immunoprecipitated by αNic-927 (Fig. 3B). Probing the immunoblot with anti-Maml antibody indicated that the 130-kDa form of Maml could be coimmunoprecipitated with Nic from F30 and in the input nuclear lysate (Fig. 3B, lane I). A small amount of Maml immunoreactivity could also be detected by coimmunoprecipitation from F37. However, the 108-kDa Maml species was not detectably coimmunoprecipitated with Nic in either the F30 or F37. Moreover, none of the Maml 108-kDa form was coimmunoprecipitated with Nic in the input nuclear lysate, even though this is the prominent Maml species observed in the lysate prior to loading the column (Fig. 3A, lane I). This data indicates that either there is a preference for Notch to bind the 130-kDa species of Maml or there is potentially a modification of the 108-kDa species upon formation of F30.
Considering this result, we reasoned that it is possible that Maml could function as a tether between ΔRAM and CSL. When ΔRAM gel filtration profile blots were probed with anti-Maml antibody, we found that the 130-kDa Maml also comigrates with ΔRAM and CSL in F30 (Fig. 3C). This result indicates that Maml is associated with both ΔRAM and CSL in RKE cells and might be responsible for tethering ΔRAM to CSL. To directly test the hypothesis that Maml was the tether between ΔRAM and CSL, 293T cells were cotransfected with CSL and either Nic or ΔRAM in the presence or absence of Maml. Whole-cell lysates were made, and protein complexes were immunoprecipitated with anti-HA antibody, specific for the epitope tag on transfected CSL (Fig. 3D, lanes 1 to 6). Protein expression for each transfection was verified by Western blot analysis of protein lysates with the appropriate antibody (Fig. 3D, lanes 7 to 16). This analysis demonstrated that ΔRAM could only be coimmunoprecipitated by CSL in the presence of Maml (Fig. 3D, lane 5), whereas Nic is bound to CSL through the RAM domain and can be immunoprecipitated in either the presence or absence of Maml (lanes 2 and 3). Furthermore, CSL does not interact directly with Maml in the absence of Nic, as determined by failure of Maml to coimmunoprecipitate with CSL (Fig. 3D, lane 6). Since Maml does not directly bind to CSL, it cannot be the direct tether between Nic and CSL. However, it is clear that Maml promotes the association between Nic and CSL, possibly through the recruitment of an additional factor or by the stabilization of CSL binding to a RAM-independent site on Nic.
Wu et al. reported that a N-terminal fragment of Maml (amino acids 1 to 302), which contains the Nic binding domain, was unable to form a complex with ΔRAM and CSL in vitro (59). This indicates that C-terminal sequences of Maml (i.e., residues 303 to 1016) might be required to stabilize binding of ΔRAM to CSL. To determine if these C-terminal 713 residues were also required for complex formation in vivo, 293T cells were transfected with expression vectors for CSL, either Nic or ΔRAM and Maml-305. Maml-305 encodes residues 1 through 305 of human Maml and is appended with a C-terminal Myc epitope tag. Nuclear proteins were extracted from each set of transfections and processed for immunoprecipitation with anti-HA antibody, specific for the epitope tag on transfected CSL (Fig. 3E, lanes 1 to 3). Western blot analysis demonstrated that coexpression of Maml-305 was sufficient to promote association of ΔRAM to CSL (Fig. 3E, lane 3). This result indicates that either another factor binds to this region of Maml and facilitates its tethering function or a modification in this region that does not occur in bacteria is responsible for the ability of the Maml N-terminal region to bridge ΔRAM to CSL.
Nic integrates into F30 in a human T-ALL cell line.
Given that Nic, Maml, and CSL associate to form F30 in RKE cells, we reasoned that if this complex was required for Nic neoplastic signaling, then a human T-ALL cell line that expresses Nic should also contain F30. SUP-T1 cells are derived from a human T-ALL line that constitutively expresses Nic-like molecules (14, 46). To determine if Nic formed stable complexes in SUP-T1 cells, nuclear proteins were extracted and fractionated by size exclusion chromatography. Western blot analysis with αNic-927 revealed that Nic elutes in a stable complex at fraction 30 and was not detected in any other fractions (Fig. 4). Probing the membrane with antibodies for endogenous Maml and CSL demonstrated that these proteins coelute with Nic at fraction 30 in SUP-T1 nuclear lysate (Fig. 4). The 130-kDa species of Maml coeluted with Nic at fraction 30, while the 108-kDa species peaked at fractions 34 and 35, consistent with data obtained from RKE cells (Fig. 3A). These results indicate that formation of F30 by Nic, Maml, and CSL is common to both transformed RKE and SUP-T1 cells and that this might represent the active Nic signaling complex which promotes the neoplastic transformation of these cells.
FIG. 4.
F30 is formed in a human T-cell leukemia cell line that constitutively expresses Nic. Nuclear lysate from SUP-T1 cells was fractionated by size exclusion chromatography in buffer containing 500 mM NaCl, and proteins were detected with the indicated antibodies.
F30 can be reconstituted in a heterologous cell system.
Both Nic and ΔRAM associate with CSL and Maml to form F30 in RKE cells; however, ΔRAM does not bind CSL in 293T cells unless Maml is coexpressed. Considering this result, we wanted to determine if Maml was the missing component for F30 assembly in 293T cells. This possibility was tested by transient transfection of 293T cells with Nic alone or in combination with CSL and Maml.
Nuclear lysate from Nic-transfected 293T cells was fractionated by gel filtration, and Nic was detected by Western blot analysis with 9E10. When expressed alone in 293T cells, Nic elutes only in F37 and does not form an F30 (Fig. 5A), indicating that 293T cells are missing a component(s) required for F30 assembly. In addition, this result demonstrates that F30 in RKE cells is likely not due to aggregation of Nic proteins, since only F37 forms in 293T cells and Nic is not detected in F30 even when expressed at levels much greater than those in RKE cells. When Nic and CSL are coexpressed in 293T cells the column profile for Nic is not altered, demonstrating that CSL is not sufficient to promote F30 formation (data not shown).
FIG. 5.
F30 can be reconstituted in a heterologous cell system. (A) 293T cells were individually transfected with expression vectors for Flag-Maml, Nic-Myc, or HA-CSL, and nuclear lysate was fractionated on a Superose 6 size exclusion column. Transfected proteins are indicated on the left, and the antibodies used for immunodetection are listed on the right. (B) Nic and Maml form F30 with endogenous CSL in 293T cells. Cells were cotransfected with expression vectors encoding Flag-Maml and Nic-Myc, and nuclear lysate was processed as before. Each transfected protein was detected by Western blotting as previously described. Endogenous CSL was detected with anti-CSL polyclonal antisera. (C) Formation of F30 by cotransfection of Nic, Maml, and CSL in 293T cells. Cells were cotransfected with expression vectors encoding Flag-Maml, Nic-Myc, and HA-CSL, and nuclear lysate was fractioned by size exclusion chromatography. Each transfected protein was detected by Western blotting as previously described. (D) Nic, Maml, and CSL are physically associated in F30 when transfected into 293T cells. Ten percent of the column load (Input) and three fractions covering the indicated complex peaks (F30 and F37) were individually pooled and immunoprecipitated with αNic-927 (lanes IP) or IgG control (lanes C). Proteins were detected by Western blotting with the indicated antibody. (E) ΔRAM incorporates into F30 in 293T cells when coexpressed with Maml and CSL. 293T cells were cotransfected with expression vectors encoding Flag-Maml, ΔRAM-Myc, and HA-CSL, and proteins were processed and detected as previously described.
The column profiles for individually expressed CSL and Maml were determined to control for migration differences seen in the presence and absence of Nic coexpression. When expressed alone in 293T cells, CSL elutes primarily as a monomer but can be detected in all column fractions, which is likely a result of overexpression (Fig. 5A). Unlike in RKE cells, transfected Maml is detected only as a 130-kDa species in 293T nuclear extract and, when expressed alone, peaks in fractions 34 and 35, with some immunoreactivity trailing down the column profile from fractions 36 through 48 (Fig. 5A).
Since transfection of Nic alone or with CSL does not result in F30 formation, the effect of cotransfection with Maml on complex assembly was tested (Fig. 5B). When Nic and Maml were coexpressed in 293T cells, a significant portion of Nic immunoreactivity was shifted from fraction 37 to a peak at fraction 30 (Fig. 5B). Maml immunoreactivity was also shifted, indicating that transfection of Nic and Maml resulted in formation of F30 (Fig. 5B). To determine if F30 in 293T cells contained endogenous CSL, the gel filtration profile for this double transfection was probed with the anti-CSL antibody. Endogenous CSL also coelutes in both F30 and F37 and is not detected in any other fractions tested (Fig. 5B). This result indicates that Maml is a necessary component to promote F30 assembly with Nic and endogenous CSL in 293T cells.
Since endogenous CSL could also be a limiting factor for F30 formation when Nic and Maml are ectopically coexpressed, we performed a triple transfection of Nic, Maml, and CSL in 293T cells to see if the majority of Nic immunoreactivity could be shifted to F30 in the presence of excess CSL and Maml (Fig. 5C). When all three proteins are simultaneously expressed, nearly all of the Nic is shifted from F37 to F30 (Fig. 5C). The elution profiles for Maml and CSL are also altered, and they coelute with Nic at F30 peak. In these transfection assays the inability to shift all of the Nic to F30 is likely due to the relative abundance of each protein composing F30. A physical interaction between the three proteins in F30 was verified by coimmunoprecipitation from pooled column fractions (Fig. 5D). The column fractions were immunoprecipitated with either αNic-927 (Fig. 5D, lanes IP) or an irrelevant rabbit IgG (lanes C), and proteins were detected by immunoblotting with the appropriate antibody. Both CSL and Maml could be coimmunoprecipitated from the fraction 30 pool with Nic (Fig. 5D, lane F30) and a fraction of CSL was also coimmunoprecipitated with Nic in F37 (Fig. 5D, lane F37). Nic and Maml could also be coimmunoprecipitated from fraction 30 by the anti-HA antibody, directed at the epitope tag of transfected CSL, indicating that F30 is composed of a Nic-CSL-Maml complex rather than a mixture of separate Nic-CSL and Nic-Maml complexes (data not shown). These analyses demonstrate that expression of all three complex members in 293T cells can recapitulate the binding characteristics of F30 and F37 observed in RKE cells.
Since Maml appears to function as a tether between ΔRAM and CSL in 293T cells (Fig. 3D), we sought to determine if ΔRAM would form F30 when coexpressed with Maml and CSL. 293T cells were cotransfected with expression vectors encoding ΔRAM, CSL, and Maml, and the nuclear lysate was fractionated by gel filtration. Proteins were detected by Western blot analysis with the appropriate antibody (Fig. 5E). ΔRAM elutes with both Maml and CSL in F30, further demonstrating that Maml functions to promote association between ΔRAM and CSL to induce F30 formation in 293T cells.
F30 formation requires sequences within the TFD.
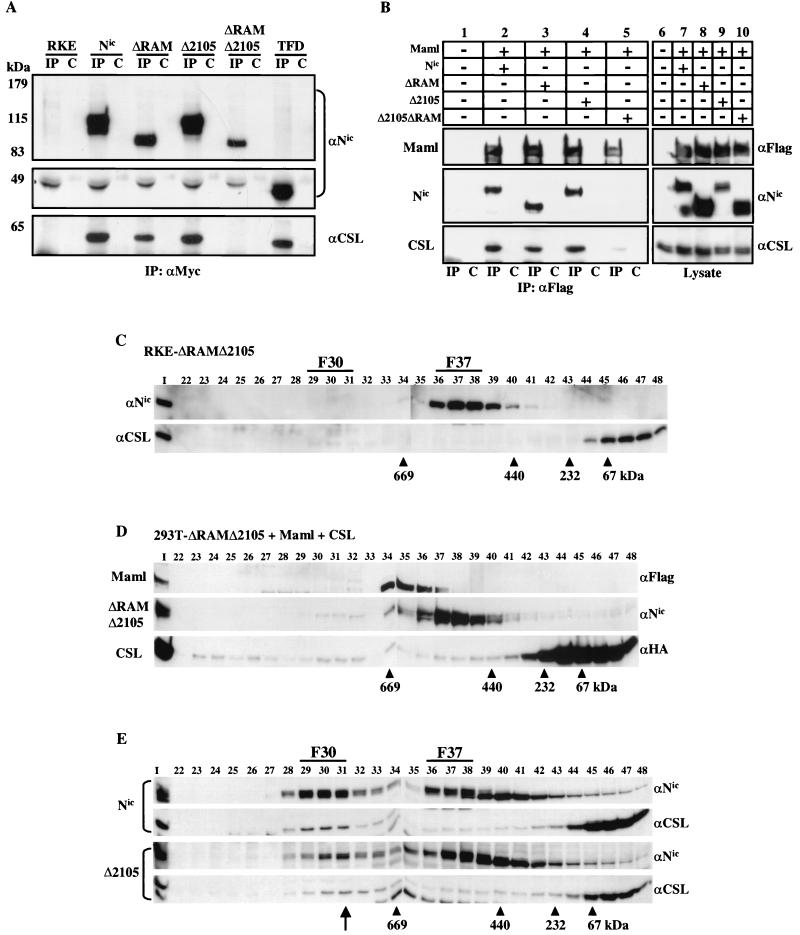
If CSL is a required component of F30, then loss of association with CSL could indicate that F30 has been disrupted. We have shown previously that deletion of residues 2105 to 2114 (Δ2105) in the context of full-length Nic results in a molecule that is unable to transform RKE cells and cannot activate transcription from a CSL-responsive promoter even though it can bind to CSL (26). We hypothesized that if residues 2105 to 2114 function to stabilize association with CSL in the absence of the RAM domain, then engineering the Δ2105 mutation into the ΔRAM mutation might be sufficient to disrupt interaction with CSL. The Δ2105 construct retains binding to Maml (Fig. 6B), indicating that if complex formation is disrupted in the double mutant it is likely due to another protein(s) binding to this region. To determine if these sequences of Nic are required for complex formation, Nic mutants were assayed for interaction with endogenous CSL in RKE cells. Whole-cell lysates were made from RKE cell lines expressing the various Nic derivatives, and proteins were immunoprecipitated with 9E10 (Fig. 6B, lanes IP) or with an irrelevant mouse IgG (lanes C). Immune complexes were resolved by SDS-PAGE, and proteins were detected by Western blot analysis with the appropriate antibody (Fig. 6A). CSL was coimmunoprecipitated with Nic, ΔRAM, Δ2105, and the TFD irrespective of the RAM domain as previously demonstrated. In contrast, ΔRAMΔ2105 did not detectably associate with CSL in RKE cells, indicating that residues 2105 to 2114 are required for either complex stability or formation. CSL was not detected in immunoprecipitates from parental RKE cells or in the irrelevant IgG control for each cell line tested (Fig. 6A).
FIG. 6.
Deletion of the RAM domain and residues (amino acids 2105 to 2114) within the TFD results in loss of F30 formation. (A) Whole-cell lysates were made from the indicated RKE cell lines and were immunoprecipitated with either the anti-Myc antibody (lanes IP) or a nonspecific mouse IgG antibody control (lanes C). Proteins were detected with the indicated antibody. CSL is immunoprecipitated when the RAM domain or the Δ2105 region is present but not when both regions are deleted. Since the TFD is approximately 45 kDa, the membrane was split to be probed with both anti-Notch and anti-CSL antibodies. (B) ΔRAMΔ2105 is unable to appreciably associate with Maml and endogenous CSL in 293T cells. 293T cells were cotransfected with the indicated plasmids, and whole-cell lysates were immunoprecipitated with anti-Flag directed against Maml (lanes IP) or with a control mouse IgG (lanes C). Immunoprecipitated proteins were detected with the indicated antibodies (lanes 1 to 5).Expression of each protein was verified by Western blot analysis with the indicated antibodies (lanes 6 to 10). (C) ΔRAMΔ2105 derivative does not integrate into F30. Nuclear lysate from RKE cells expressing ΔRAMΔ2105 was fractionated by size exclusion chromatography. ΔRAMΔ2105 protein was visualized by Western blot analysis with bTAN15A (αNic). CSL protein was detected with anti-CSL polyclonal antisera. (D) ΔRAMΔ2105 is impaired for F30 reconstitution in 293T cells. 293T cells were cotransfected with the indicated expression vectors and processed as previously described. (E) A 10-residue deletion within the TFD that abolishes transformation also disrupts protein complex formation. Gel filtration profiles for Nic and Δ2105 are compared. Proteins were detected by Western blotting with the indicated antibodies (right). Note that the position of F30 in the Δ2105 cell line is shifted down one column fraction compared to the Nic profile (arrow).
Complex reconstitution in 293T cells was used to verify the loss of association between ΔRAMΔ2105 and CSL observed in RKE cells and to determine if ΔRAMΔ2105 retained binding to Maml. 293T cells were cotransfected with expression vectors for Flag-Maml and the relevant Nic construct. Whole-cell lysates were harvested and immunoprecipitated with anti-Flag antibody, specific for the epitope on the Maml construct or a mouse IgG control antibody (Fig. 6B, lanes 1 to 5). Western blot analysis indicated that Nic, ΔRAM, and Δ2105 could all associate with Maml (Fig. 6B). However, ΔRAMΔ2105 was not appreciably coimmunoprecipitated with Maml even when highly expressed in 293T cells (Fig. 6B, lane 5). When the blot was probed for endogenous CSL with anti-CSL polyclonal antibody, we found that ΔRAMΔ2105 also did not appreciably bind to CSL (Fig. 6B, lane 5). In contrast, Nic, ΔRAM, and Δ2105 were all able to associate with endogenous CSL (Fig. 6B, lanes 2 to 4), although ΔRAM bound less CSL than either Nic or Δ2105. Protein expression in each lysate was determined with the appropriate antibody (Fig. 6B, lanes 6 to 10). The results obtained with 293T cells verify the data from RKE cells (Fig. 6A) and indicate that residues from 2105 to 2114 act in concert with the RAM domain for stable association of Nic with both Maml and CSL.
To examine if ΔRAMΔ2105 retained the ability to form stable complexes within RKE cells, nuclear extract from an RKE cell line expressing this construct was fractionated by gel filtration chromatography and proteins were detected by Western blot analysis with the indicated antibody (Fig. 6C). ΔRAMΔ2105 does not form detectable levels of F30 in RKE cells but does form a stable F37 (Fig. 6C); however, no detectable CSL coelutes with ΔRAMΔ2105 (Fig. 6C). Furthermore, when we attempted to reconstitute Nic complexes in 293T cells by triple cotransfection of ΔRAMΔ2105, Maml, and CSL, we found that even when overexpressed, ΔRAMΔ2105 did not appreciably form F30 (Fig. 6D). These results indicate that both the RAM domain and residues 2105 to 2114 of Nic are required for F30 formation and stable association with CSL.
The Δ2105 mutant retains the ability to associate with CSL through the RAM domain and can bind Maml, but it is incapable of transforming RKE cells or transactivating a CSL-responsive promoter (26). If Δ2105 no longer functions as a transcriptional activator and fails to transform RKE cells due to loss of a physical association with other proteins, then it should display an altered gel filtration profile compared to Nic. Nuclear lysate from an RKE cell line expressing Δ2105 was generated and fractionated by gel filtration chromatography as previously described. When the elution profile was analyzed by immunoblotting for Δ2105 and for CSL, the peak of immunoreactivity for each protein was shifted down one column fraction in F30 (from fraction 30 to fraction 31) compared to the profile for Nic (Fig. 6E). The elution of F37 and the fraction 46 peak of monomeric CSL was not changed in the Δ2105 nuclear lysate compared to Nic, indicating that the altered elution profile was specific for the fraction 30 complex. A shift from fraction 30 to fraction 31 on this Superose 6 column equates to a reduction in size by approximately 200 to 400 kDa as calculated from the elution peaks of known protein standards. This indicates that residues 2105 to 2114 of Nic might serve as a binding site for a protein(s) that is required for F30 formation, transcriptional activation, and transformation of RKE cells.
Exogenous Maml can rescue ΔRAM but not Δ2105 transactivation in HeLa cells.
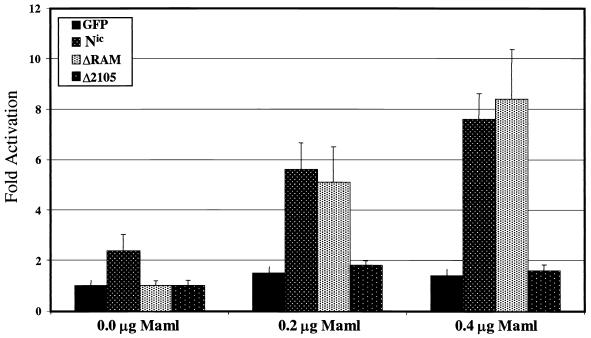
The RAM domain is required for Nic transactivation of CSL-dependent reporters in HeLa cells (22, 24, 26, 37, 40, 54). Based on our results that Maml is responsible for tethering ΔRAM to CSL in 293T and RKE cells, it is a possibility that ΔRAM is inactive in this assay due to insufficient expression of endogenous Maml in HeLa cells. We proposed that by increasing the level of Maml in HeLa cells through ectopic expression, the transcriptional activity of ΔRAM on a CSL responsive reporter could be rescued. In addition, if Maml is exclusively responsible for Nic-mediated transactivation, then excess Maml should elevate the transcriptional activity of Δ2105, since this protein retains the ability to physically interact with Maml and CSL. To test these hypotheses, HeLa cells were transfected with a HES-1 luciferase reporter gene, a constant amount of Nic, ΔRAM, or Δ2105 (0.1 μg), and either 0.2 or 0.4 μg of Maml. Luciferase values were determined and are presented as activation over a green fluorescent protein (GFP) control transfection (Fig. 7). When transfected without Maml, Nic is able to activate transcription approximately threefold over background levels, whereas ΔRAM and Δ2105 exhibited basal activity. When Maml is included in the transfection, transcriptional activity for both Nic and ΔRAM is stimulated approximately fivefold (0.2 μg of Maml) and eightfold (0.4 μg of Maml) over background. At both levels of Maml tested, the transcriptional activity of ΔRAM can be restored to a level equivalent to that of Nic. In contrast, exogenously added Maml has no effect on the transcriptional activity of the Δ2105 construct (Fig. 6). Both the GFP-transfected control and Δ2105 displayed basal levels of activity when cotransfected with Maml. This indicates that Maml is likely not the sole effector of Nic transcriptional activity and implies the existence of an additional coactivator(s) that requires the Δ2105 sequences. Given the 200- to 400-kDa size difference between F30 formed by Nic, ΔRAM, and the TFD and that formed by Δ2105, we propose the existence of an additional protein(s) that is responsible for the transcriptional activity of Nic.
FIG. 7.
Exogenous Maml rescues transactivation by ΔRAM in HeLa cells but cannot rescue Δ2105 activity. HeLa cells were transfected with the indicated Nic derivatives (0.1 μg), increasing amounts of Maml (0.2 and 0.4 μg), Hes-1 luciferase plasmid (0.4 μg), and CMV-βgal (0.2 μg). Luciferase values were determined 48 h posttransfection and are expressed as average increase in activation over cells transfected with GFP, as determined in three independent experiments performed in duplicate. Error bars represent the standard deviations from each set. Maml augments transcriptional activity of both Nic and ΔRAM but has no effect on Δ2105 activity.
Dominant negative Maml inhibits Nic transcriptional activity and disrupts F30 formation.
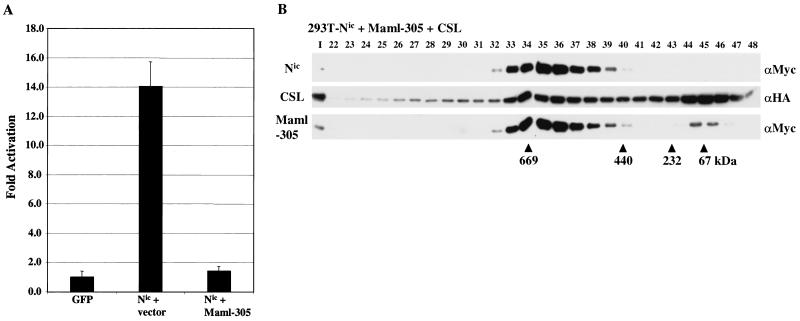
To further understand the role of Maml in Nic-mediated transcriptional transactivation and F30 assembly, we sought to characterize the effects of Maml-305 on Nic activity. Analogous Maml mutations have been shown to behave in a dominant inhibitory fashion to Nic signaling in both Drosophila and mammalian systems (19, 29, 59). We hypothesized that Maml-305 functions as a dominant negative factor by binding to Nic and CSL without recruiting an essential Maml coactivator. To examine if Maml-305 can inhibit Nic signaling, HeLa cells were transfected with the 8x-CSL-luc reporter, Nic (0.1 μg), and either empty vector or Maml-305 (0.4 μg) (Fig. 8A). When transfected with empty vector, Nic stimulates luciferase activity 14-fold over background levels. Transfection of Maml-305 with Nic results in a complete loss of Nic transcriptional activity. These results indicate that Maml-305 does inhibit Nic-mediated transcriptional activity in this assay, which might be due to disruption of Nic transcriptional complexes.
FIG. 8.
Dominant negative mutations in Maml disrupt both Nic-mediated transactivation and F30 formation. (A) HeLa cells were transfected with the 8x-CSL-luc reporter (0.4 μg), CMV-βgal (0.2 μg), Nic expression plasmid (0.1 μg), and either empty vector or a Maml-305 expression vector (0.4 μg). Luciferase values are expressed as average increase in activation over background. (B) Maml-305 disrupts F30 formation in transfected 293T cells. Cells were cotransfected with expression vectors encoding the indicated proteins (left). Following transfection, nuclear lysate was fractionated by size exclusion chromatography and proteins were detected by Western blot analysis with the indicated antibodies (right).
Complex reconstitution in 293T cells was performed to examine the effect of Maml-305 expression on F30 assembly. Cells were cotransfected with expression vectors encoding Nic, Maml-305, and CSL and nuclear proteins were fractionated by size exclusion chromatography. Western blot analysis demonstrated that coexpression of Maml-305 with Nic and CSL dramatically altered the column elution profile of Nic (Fig. 8B; compare Fig. 5C). All three transfected proteins form a peak of coelution in fractions 35 and 36 which is equivalent to a reduction in complex size of more than 700 kDa from F30 (Fig. 5B and 5C). We propose that the size reduction results from the loss of protein interactions with the C-terminal portion of Maml. However, it is possible that the altered migration is due solely to the deletion of 712 residues from Maml, although we think this unlikely. The inability to recruit this unknown factor(s) to Nic probably accounts for the dominant negative effect of Maml-305.
DISCUSSION
Previously we demonstrated that Nic must accumulate in the nucleus in order to induce neoplastic transformation of RKE cells. In an effort to understand how Nic might function in the nucleus, we sought to determine if Nic formed stable protein complexes. Using size exclusion chromatography, we found that Nic forms two stable high-molecular-weight protein complexes in the nuclei of transformed RKE cells. In the larger of these complexes, Nic is physically associated with both endogenous Maml and CSL. In addition, we find that Nic also forms the largest complex in the human leukemia SUP-T1 cell line, indicating that complex formation is common to Nic-mediated neoplastic transformation. Our data indicate that deletion of the RAM domain is not equivalent to CSL-independent signaling, since ΔRAM proteins retain the ability to associate with CSL in a stable complex as well as transform RKE cells. We propose that Maml functions in part to bridge Nic to CSL and, as a consequence of this property, is able to rescue ΔRAM transcriptional activity. In addition, we demonstrate that F30 is likely to be required for neoplastic transformation of RKE cells, since the TFD can integrate into F30 while nontransforming Nic mutants are deficient for F30 formation.
Nic forms stable high-molecular-weight complexes in RKE and SUP-T1 cells.
Many proteins have been demonstrated to associate with Nic; however, it is unknown if these proteins assemble to form higher-order protein complexes with Nic in vivo. We describe the existence of two high-molecular-weight complexes that can be extracted from the nuclei of Nic-transformed RKE cells and from cells derived from a human leukemia (SUP-T1 cells). The largest of these complexes is approximately 1.5 MDa and contains both endogenous Maml and CSL in addition to ectopically expressed Nic. The smaller (500- to 600-kDa) complex is composed of Nic and contains a minor amount of Maml and CSL immunoreactivity. Currently we do not understand the relationship between these complexes. Since F37 is composed predominately of Nic and contains potentially substoichiometric amounts of both Maml and CSL, it is possible that F37 represents either a precursor to or a breakdown of F30. In addition, both F30 and F37 are detected in cytoplasmic extract (S100; data not shown) from transformed RKE cells. Since Nic is predominantly localized to the nucleus by indirect immunofluorescence, it is possible that Nic isolated from the S100 fraction might be due to partial disruption of the nuclei during lysis. Further analysis will be required to elucidate the relationship between the two complexes and their subcellular localization.
Our data indicate that there are other proteins in F30. We do not know the identities of additional members, but we have attempted to place other known Nic binding proteins within these complexes. Recently p300 was shown to bind Nic in 293 cell transfection assays, and the binding site on Nic maps to the Δ2105 region (40). We probed column elution profiles and column fraction immunoprecipitation blots from both RKE and 293T cells with anti-p300 antibody but found no evidence that p300 is a member of either F30 or F37, since it did not coimmunoprecipitate with Nic. While we cannot rule out the association of p300 with either of the Nic complexes, our data do not support a role for p300 as a stable component of Nic complex formation in RKE cells. At this time, we do not understand the basis for the discrepancy between our data and those of Oswald et al. (40).
Nic has been shown to interact with transcription factors other than CSL, including Nur-77, NF-κB, and most recently LEF-1 (18, 27, 51). It remains to be determined if F30 functions only with CSL or if the complex can be targeted to DNA by other transcription factors. One possibility is that ubiquitous expression of CSL permits Nic to activate a core set of target genes in all tissues and that Nic utilizes tissue-specific transcription factors to modulate expression of other genes in a tissue-specific manner. For example, expression of both Nur-77 and LEF-1 is restricted to T cells, indicating that in this cellular context, Nic might complex with these transcription factors to modulate target genes specific for thymocyte development and function, in addition to genes targeted by CSL.
The RAM domain is not required for stable association with CSL.
A critical target in Nic signal transduction is the transcriptional repressor CSL. Deletion of the RAM domain from Nic results in a loss of physical and functional association with CSL and has been used to propose that Nic can function in a CSL-independent pathway (4, 13, 22, 31, 37, 54). In our previous work, we found that ΔRAM proteins were able to transform RKE cells; however, they did not bind to CSL in vitro and did not activate a CSL-responsive promoter (26). This finding was consistent with published reports and led us to propose that transformation of RKE cells might be a CSL-independent event. Similar findings were reported by Dumont et al., who also demonstrated that transformation of RKE cells did not require the RAM domain (13). We now conclusively demonstrate that Nic constructs with the RAM domain deleted are able to form a higher-order complex with endogenous CSL in RKE cells. RAM-independent binding to CSL is not unique to RKE cells, since we can recapitulate ΔRAM association with CSL in 293T cells by coexpression of Maml. However, it is likely that Maml requires other associated factors to perform this tethering function. Wu et al. demonstrated using in vitro binding assays that a bacterially expressed ΔRAM and an N-terminal fragment of Maml (amino acids 1 to 302) were not able to bind to CSL that was expressed in 293 cells. This indicates that either the C-terminal 713 residues of Maml are required to tether ΔRAM to CSL or another cellular factor(s) or a modification(s) is required for this function (59). We demonstrate that in 293T cells, Maml-305 is sufficient to promote association between ΔRAM and CSL, indicating that if additional cellular factors are required for this function, they must interact with the N-terminal region of Maml and that the C-terminal region might provide an additional activity other than bridging CSL and Nic. We and others have shown that Maml proteins analogous to Maml-305 behave as antagonists of Nic activity (29, 59). Our results demonstrate that expression of Maml-305 inhibits the transcriptional activity of Nic and disrupts formation of F30 with Nic in 293T cells. It is a possibility that the dramatic reduction in F30 size caused by coexpression of Maml-305 with Nic and CSL in 293T cells is due solely to the loss of Maml C-terminal sequences. This remains to be determined, but we propose that the loss is more likely due to the deletion of a binding site(s) from Maml for a protein(s) that is required for F30 formation. The inability of Maml-305 to recruit this protein(s) results in disruption of F30 and likely accounts for the dominant negative effect of Maml-305 on Nic transcriptional activity.
In further support of our hypothesis that Maml tethers ΔRAM to CSL, we show that ectopic expression of Maml in HeLa cells is able to restore the transcriptional activity of ΔRAM on a CSL-responsive promoter. These functional data are in agreement with our biochemical data and indicate that a lack or low level of endogenous Maml expression within HeLa cells accounts for the inability of ΔRAM to activate the HES-1 promoter construct. Given these results, it is possible that ΔRAM can associate with endogenous CSL in other cell systems that have been used as evidence for a CSL-independent pathway, such as C2C12 cells; whether this is the case deserves reexamination. It is likely that cell lines expressing Maml can support Notch signaling through ΔRAM proteins, whereas cell lines that do not express (or express low levels of) Maml cannot. For instance, Dievart et al. found that transformation of mouse mammary epithelial HC11 cells by Nic required the RAM domain, perhaps indicating that HC11 cells do not express sufficient levels of endogenous Maml (11).
A seventh ankyrin repeat is required for F30 formation and transcriptional activity.
We have shown that the Δ2105 mutant has a disruption of F30 formation and is unable to activate transcription from a CSL-responsive reporter. Sequence analysis reveals that the region of Nic surrounding and including the Δ2105 mutation shares a high degree of conservation with the fourth ankyrin repeat of p16INK4a (52). It is probable that this region in Nic adopts a similar structure and forms a seventh Nic ankyrin repeat. This region in Notch is also highly conserved throughout evolution, with very few amino acid substitutions being introduced from Drosophila Notch to human Notch1. Furthermore, this region is well conserved among all four mammalian Notch proteins. The high degree of conservation indicates that this region plays an important role in the function of Nic. Zweifel and Barrick have recently demonstrated by solution dynamics that, consistent with the homology to p16INK4a, the ankyrin repeat domain in Drosophila Notch extends through these sequences (61, 62).
F30 is likely required for Nic neoplastic signaling.
We have presented data showing that the Δ2105 mutant is disrupted in F30 formation, potentially missing components that account for approximately 200 to 400 kDa of total complex size. This loss is sufficient to render the Δ2105 mutant incapable of transforming RKE cells and activating transcription. When the RAM domain and the Δ2105 region are both deleted, F30 formation is completely disrupted and only F37 can form. This indicates that formation of F37 is not sufficient for Nic biological activity and that F30 formation might be required to induce transformation of RKE cells. The protein(s) binding to the Δ2105 region likely functions to stabilize F30 in concert with Maml and CSL as well as provide transcriptional activation for Nic. In addition, we have demonstrated that the TFD of Nic, consisting primarily of the ankyrin repeat region, is capable of forming F30. Our data indicate that this minimal domain maintains all the protein contacts required to form F30. We do note, however, that F30 formed by the TFD is sensitive to conditions for nuclear extraction, indicating that additional C-terminal residues of Nic might function to stabilize the complex. Although this is the minimum domain of Nic required to transform RKE cells in culture, Aster et al. demonstrated that leukemia failed to develop in mice reconstituted with bone marrow cells expressing a Nic derivative analogous to the TFD (5), indicating that the C-terminal region of Nic is required for the full neoplastic potential of Nic in vivo, perhaps due to stability of the F30 signaling complex.
Acknowledgments
We thank members of the Capobianco lab for intellectual support and technical assistance during this work. We also thank Melanie Stegman and other members of the Robbins lab for assistance with the fast protein liquid chromatography. We are grateful to Emery Bresnick (University of Wisconsin—Madison) for providing anti-CSL antibody and to James D. Griffin (Dana Farber Cancer Institute, Harvard Medical School) for supplying anti-Maml antibody and the cDNA for human Maml. We also thank Spryos Artavanis-Tsakonas (Harvard University) for providing the bTAN15A hybridoma.
This work was supported by NIH grant RO1CA83736-02 (to A.J.C.). A.J.C. is a scholar of the Leukemia and Lymphoma Society (award 1298-02). This work was also supported in part by predoctoral awards to S.J. from the DOD Breast Cancer Research Program, DAMD17-01-1-0202, and the Albert J. Ryan Foundation.
REFERENCES
- 1.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 2.Aster, J., W. Pear, R. Hasserjian, H. Erba, F. Davi, B. Luo, M. Scott, D. Baltimore, and J. Sklar. 1994. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harbor Symp. Quant. Biol. 59:125-136. [DOI] [PubMed] [Google Scholar]
- 3.Aster, J. C., and W. S. Pear. 2001. Notch signaling in leukemia. Curr. Opin. Hematol. 8:237-244. [DOI] [PubMed] [Google Scholar]
- 4.Aster, J. C., E. S. Robertson, R. P. Hasserjian, J. R. Turner, E. Kieff, and J. Sklar. 1997. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J. Biol. Chem. 272:11336-11343. [DOI] [PubMed] [Google Scholar]
- 5.Aster, J. C., L. Xu, F. G. Karnell, V. Patriub, J. C. Pui, and W. S. Pear. 2000. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by Notch1. Mol. Cell. Biol. 20:7505-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellavia, D., A. F. Campese, E. Alesse, A. Vacca, M. P. Felli, A. Balestri, A. Stoppacciaro, C. Tiveron, L. Tatangelo, M. Giovarelli, C. Gaetano, L. Ruco, E. S. Hoffman, A. C. Hayday, U. Lendahl, L. Frati, A. Gulino, and I. Screpanti. 2000. Constitutive activation of NF-κB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 19:3337-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capobianco, A. J., P. Zagouras, C. M. Blaumueller, S. Artavanis-Tsakonas, and J. M. Bishop. 1997. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol. Cell. Biol. 17:6265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlesso, N., J. C. Aster, J. Sklar, and D. T. Scadden. 1999. Notch1-induced delay of human hematopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. Blood 93:838-848. [PubMed] [Google Scholar]
- 9.Daniel, B., A. Rangarajan, G. Mukherjee, E. Vallikad, and S. Krishna. 1997. The link between integration and expression of human papillomavirus type 16 genomes and cellular changes in the evolution of cervical intraepithelial neoplastic lesions. J. Gen. Virol. 78:1095-1101. [DOI] [PubMed] [Google Scholar]
- 10.De Strooper, B., W. Annaert, P. Cupers, P. Saftig, K. Craessaerts, J. S. Mumm, E. H. Schroeter, V. Schrijvers, M. S. Wolfe, W. J. Ray, A. Goate, and R. Kopan. 1999. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518-522. [DOI] [PubMed] [Google Scholar]
- 11.Dievart, A., N. Beaulieu, and P. Jolicoeur. 1999. Involvement of notch1 in the development of mouse mammary tumors. Oncogene 18:5973-5981. [DOI] [PubMed] [Google Scholar]
- 12.Dignam, J. D., P. L. Martin, B. S. Shastry, and R. G. Roeder. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582-598. [DOI] [PubMed] [Google Scholar]
- 13.Dumont, E., K. P. Fuchs, G. Bommer, B. Christoph, E. Kremmer, and B. Kempkes. 2000. Neoplastic transformation by Notch is independent of transcriptional activation by RBP-J signalling. Oncogene 19:556-561. [DOI] [PubMed] [Google Scholar]
- 14.Ellisen, L. W., J. Bird, D. C. West, A. L. Soreng, T. C. Reynolds, S. D. Smith, and J. Sklar. 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66:649-661. [DOI] [PubMed] [Google Scholar]
- 15.Gallahan, D., and R. Callahan. 1997. The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4). Oncogene 14:1883-1890. [DOI] [PubMed] [Google Scholar]
- 16.Gallahan, D., C. Jhappan, G. Robinson, L. Hennighausen, R. Sharp, E. Kordon, R. Callahan, G. Merlino, and G. H. Smith. 1996. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 56:1775-1785. [PubMed] [Google Scholar]
- 17.Girard, L., Z. Hanna, N. Beaulieu, C. D. Hoemann, C. Simard, C. A. Kozak, and P. Jolicoeur. 1996. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 10:1930-1944. [DOI] [PubMed] [Google Scholar]
- 18.Guan, E., J. Wang, J. Laborda, M. Norcross, P. A. Baeuerle, and T. Hoffman. 1996. T cell leukemia-associated human Notch/translocation-associated Notch homologue has IκB-like activity and physically interacts with nuclear factor-κB proteins in T cells. J. Exp. Med. 183:2025-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helms, W., H. Lee, M. Ammerman, A. L. Parks, M. A. Muskavitch, and B. Yedvobnick. 1999. Engineered truncations in the Drosophila mastermind protein disrupt Notch pathway function. Dev. Biol. 215:358-374. [DOI] [PubMed] [Google Scholar]
- 20.Honjo, T. 1996. The shortest path from the surface to the nucleus: RBP-Jκ/Su(H) transcription factor. Genes Cells 1:1-9. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh, J. J., T. Henkel, P. Salmon, E. Robey, M. G. Peterson, and S. D. Hayward. 1996. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16:952-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh, J. J., D. E. Nofziger, G. Weinmaster, and S. D. Hayward. 1997. Epstein-Barr virus immortalization: Notch2 interacts with CBF1 and blocks differentiation. J. Virol. 71:1938-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh, J. J., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarriault, S., C. Brou, F. Logeat, E. H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of activated mammalian Notch. Nature 377:355-358. [DOI] [PubMed] [Google Scholar]
- 25.Jarriault, S., O. Le Bail, E. Hirsinger, O. Pourquie, F. Logeat, C. F. Strong, C. Brou, N. G. Seidah, and A. Israël. 1998. Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol. Cell. Biol. 18:7423-7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeffries, S., and A. J. Capobianco. 2000. Neoplastic transformation by notch requires nuclear localization. Mol. Cell. Biol. 20:3928-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jehn, B. M., W. Bielke, W. S. Pear, and B. A. Osborne. 1999. Protective effects of notch-1 on TCR-induced apoptosis. J. Immunol. 162:635-638. [PubMed] [Google Scholar]
- 28.Kato, H., Y. Taniguchi, H. Kurooka, S. Minoguchi, T. Sakai, S. Nomura-Okazaki, K. Tamura, and T. Honjo. 1997. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development 124:4133-4141. [DOI] [PubMed] [Google Scholar]
- 29.Kitagawa, M., T. Oyama, T. Kawashima, B. Yedvobnick, A. Kumar, K. Matsuno, and K. Harigaya. 2001. A human protein with sequence similarity to Drosophila Mastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters. Mol. Cell. Biol. 21:4337-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopan, R., E. H. Schroeter, H. Weintraub, and J. S. Nye. 1996. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. USA 93:1683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurooka, H., K. Kuroda, and T. Honjo. 1998. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 26:5448-5455. (Erratum, 27:following 1407, 1999.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lecourtois, M., and F. Schweisguth. 1995. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 9:2598-2608. [DOI] [PubMed] [Google Scholar]
- 33.Miele, L., and B. Osborne. 1999. Arbiter of differentiation and death: notch signaling meets apoptosis. J. Cell. Physiol. 181:393-409. [DOI] [PubMed] [Google Scholar]
- 34.Milner, L. A., and A. Bigas. 1999. Notch as a mediator of cell fate determination in hematopoiesis: evidence and speculation. Blood 93:2431-2448. [PubMed] [Google Scholar]
- 35.Morimura, T., R. Goitsuka, Y. Zhang, I. Saito, M. Reth, and D. Kitamura. 2000. Cell cycle arrest and apoptosis induced by Notch1 in B cells. J. Biol. Chem. 275:36523-36531. [DOI] [PubMed] [Google Scholar]
- 36.Mumm, J. S., E. H. Schroeter, M. T. Saxena, A. Griesemer, X. Tian, D. J. Pan, W. J. Ray, and R. Kopan. 2000. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5:197-206. [DOI] [PubMed] [Google Scholar]
- 37.Nofziger, D., A. Miyamoto, K. M. Lyons, and G. Weinmaster. 1999. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development 126:1689-1702. [DOI] [PubMed] [Google Scholar]
- 38.Nye, J. S., R. Kopan, and R. Axel. 1994. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development 120:2421-2430. [DOI] [PubMed] [Google Scholar]
- 39.Olave, I., D. Reinberg, and L. D. Vales. 1998. The mammalian transcriptional repressor RBP (CBF1) targets TFIID and TFIIA to prevent activated transcription. Genes Dev. 12:1621-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oswald, F., B. Tauber, T. Dobner, S. Bourteele, U. Kostezka, G. Adler, S. Liptay, and R. M. Schmid. 2001. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 21:7761-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pear, W. S., J. C. Aster, M. L. Scott, R. P. Hasserjian, B. Soffer, J. Sklar, and D. Baltimore. 1996. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 183:2283-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petcherski, A. G., and J. Kimble. 2000. Mastermind is a putative activator for Notch. Curr. Biol. 10:R471-R473. [DOI] [PubMed] [Google Scholar]
- 43.Pui, J. C., D. Allman, L. Xu, S. DeRocco, F. G. Karnell, S. Bakkour, J. Y. Lee, T. Kadesch, R. R. Hardy, J. C. Aster, and W. S. Pear. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11:299-308. [DOI] [PubMed] [Google Scholar]
- 44.Rae, F. K., S. A. Stephenson, D. L. Nicol, and J. A. Clements. 2000. Novel association of a diverse range of genes with renal cell carcinoma as identified by differential display. Int. J. Cancer 88:726-732. [DOI] [PubMed] [Google Scholar]
- 45.Rangarajan, A., C. Talora, R. Okuyama, M. Nicolas, C. Mammucari, H. Oh, J. C. Aster, S. Krishna, D. Metzger, P. Chambon, L. Miele, M. Aguet, F. Radtke, and G. P. Dotto. 2001. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynolds, T. C., S. D. Smith, and J. Sklar. 1987. Analysis of DNA surrounding the breakpoints of chromosomal translocations involving the beta T cell receptor gene in human lymphoblastic neoplasms. Cell 50:107-117. [DOI] [PubMed] [Google Scholar]
- 47.Robbins, J., B. J. Blondel, D. Gallahan, and R. Callahan. 1992. Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. J. Virol. 66:2594-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohn, J. L., A. S. Lauring, M. L. Linenberger, and J. Overbaugh. 1996. Transduction of Notch2 in feline leukemia virus-induced thymic lymphoma. J. Virol. 70:8071-8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronchini, C., and A. J. Capobianco. 2001. Induction of cyclin D1 transcription and CDK2 activity by Notchic: implication for cell cycle disruption in transformation by Notchic. Mol. Cell. Biol. 21:5925-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ronchini, C., and A. J. Capobianco. 2000. Notchic-ER chimeras display hormone-dependent transformation, nuclear accumulation, phosphorylation and CBF1 activation. Oncogene 19:3914-3924. [DOI] [PubMed] [Google Scholar]
- 51.Ross, D. A., and T. Kadesch. 2001. The Notch intracellular domain can function as a coactivator for LEF-1. Mol. Cell. Biol. 21:7537-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russo, A. A., L. Tong, J. O. Lee, P. D. Jeffrey, and N. P. Pavletich. 1998. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 395:237-243. [DOI] [PubMed] [Google Scholar]
- 53.Schroeter, E. H., J. A. Kisslinger, and R. Kopan. 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393:382-386. [DOI] [PubMed] [Google Scholar]
- 54.Shawber, C., D. Nofziger, J. J. Hsieh, C. Lindsell, O. Bogler, D. Hayward, and G. Weinmaster. 1996. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development 122:3765-3773. [DOI] [PubMed] [Google Scholar]
- 55.Smith, G. H., D. Gallahan, F. Diella, C. Jhappan, G. Merlino, and R. Callahan. 1995. Constitutive expression of a truncated INT3 gene in mouse mammary epithelium impairs differentiation and functional development. Cell Growth Differ. 6:563-577. [PubMed] [Google Scholar]
- 56.Struhl, G., and I. Greenwald. 1999. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398:522-525. [DOI] [PubMed] [Google Scholar]
- 57.Struhl, G., and I. Greenwald. 2001. Presenilin-mediated transmembrane cleavage is required for Notch signal transduction in Drosophila. Proc. Natl. Acad. Sci. USA 98:229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tamura, K., Y. Taniguchi, S. Minoguchi, T. Sakai, T. Tun, T. Furukawa, and T. Honjo. 1995. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-Jκ/Su(H). Curr. Biol. 5:1416-1423. [DOI] [PubMed] [Google Scholar]
- 59.Wu, L., J. C. Aster, S. C. Blacklow, R. Lake, S. Artavanis-Tsakonas, and J. D. Griffin. 2000. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26:484-489. [DOI] [PubMed] [Google Scholar]
- 60.Zagouras, P., S. Stifani, C. M. Blaumueller, M. L. Carcangiu, and S. Artavanis-Tsakonas. 1995. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc. Natl. Acad. Sci. USA 92:6414-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zweifel, M. E., and D. Barrick. 2001. Studies of the ankyrin repeats of the Drosophila melanogaster Notch receptor. 1. Solution conformational and hydrodynamic properties. Biochemistry 40:14344-14356. [DOI] [PubMed] [Google Scholar]
- 62.Zweifel, M. E., and D. Barrick. 2001. Studies of the ankyrin repeats of the Drosophila melanogaster Notch receptor. 2. Solution stability and cooperativity of unfolding. Biochemistry 40:14357-14367. [DOI] [PubMed] [Google Scholar]