Abstract
The mitochondrial genome of Trypanosoma brucei does not encode tRNAs. Consequently, all mitochondrial tRNAs are imported from the cytosol and originate from nucleus-encoded genes. Analysis of all currently available T. brucei sequences revealed that its genome carries 50 tRNA genes representing 40 different isoacceptors. The identified set is expected to be nearly complete since all but four codons are accounted for. The number of tRNA genes in T. brucei is very low for a eukaryote and lower than those of many prokaryotes. Using quantitative Northern analysis we have determined the absolute abundance in the cell and the mitochondrion of a group of 15 tRNAs specific for 12 amino acids. Except for the initiator type tRNAMet, which is cytosol specific, the cytosolic and the mitochondrial sets of tRNAs were qualitatively identical. However, the extent of mitochondrial localization was variable for the different tRNAs, ranging from 1 to 7.5% per cell. Finally, by using transgenic cell lines in combination with quantitative Northern analysis it was shown that import of tRNALeu(CAA) is independent of its 5′-genomic context, suggesting that the in vivo import substrate corresponds to the mature, fully processed tRNA.
Sequence analysis of a large number of mitochondrial genomes has shown that in many organisms a variable number of apparently essential tRNA genes are lacking. It has been postulated that these missing genes are compensated for by import of the corresponding cytosolic tRNAs (41). Mitochondrial tRNA import is predicted to occur in most protozoa and plants, some fungi, and a few invertebrates and was experimentally confirmed to occur in Saccharomyces cerevisiae (47) and several protozoal (14, 35, 44) and plant (10, 13) species. In most cases, only a subset of tRNAs are imported and some mitochondrial tRNA genes are retained (39). The yeast S. cerevisiae imports only a single tRNA species. Interestingly, import of this tRNA appears to be redundant, as the yeast mitochondrial genome encodes all tRNAs necessary for organellar translation (47). The most extreme situation is found in two unrelated groups of protozoa, the trypanosomatids and the apicomplexans, which do not have any mitochondrial tRNA genes and therefore have to import the whole set from the cytosol.
The mechanism of tRNA import has been best characterized for yeast. By using in vitro and in vivo import systems as well as mutants deficient in the mitochondrial protein import pathway it was shown that the single imported tRNALys complexes with a cytosolic factor (the precursor of the mitochondrial lysyl-tRNA synthetase) and is subsequently coimported across the protein import channel (46). Recently, three groups have reported the reconstitution of tRNA import by using isolated mitochondria from trypanosomatids (Trypanosoma brucei and two Leishmania species) (28, 34, 48). There is a consensus that in these systems tRNA import requires external and probably internal ATP, as well as protein components at the surface of the mitochondrion. Furthermore, and in contrast to the situation in yeast, no cytosolic factors were required, suggesting that the import mechanisms are different in yeast and in trypanosomatids.
Whereas in Leishmania it has convincingly been shown by in vivo and in vitro experiments that tRNAs are imported as mature, fully processed molecules (21, 26, 34), it is unclear whether this also applies for T. brucei. Mature tRNAs have been proposed as import substrates based on the fact that in vivo import of heterologous tRNAs occurs independently of their genomic context (18). On the other hand, it has also been suggested that tRNAs require long 5′ extensions for import (16). Indeed, a tRNASer(CGA)/tRNALeu(CAA) dicistronic transcript separated by a 59-nucleotide spacer has been detected in T. brucei (25). Furthermore, it was shown that only the dicistronic transcript, but not its mature derivative, was imported into mitochondria in an in vitro system (48). Interestingly, however, even though an RNase P-like activity has been identified in T. brucei mitochondria (36), the imported precursors were not processed.
All mitochondrial tRNAs in trypanosomatids are of cytosolic origin, which makes these organisms excellent systems for studying tRNA import (40). In vivo studies have shown that the vast majority of tRNAs are found in both the cytosol and mitochondria (14, 44). Although individual tRNAs and their intracellular localization in T. brucei have been studied, a global study is still lacking. Based on information derived from the T. brucei genome sequencing project, we have identified the nearly complete set of predicted trypanosomal tRNA genes. Furthermore, we have determined the expression, mitochondrial localization, and stability of a subset of 15 tRNAs by quantitative Northern analysis. Finally, using transgenic cell lines in combination with quantitative Northern blots, we have shown that tRNALeu(CAA) does not require a specific 5′-flanking sequence to be imported in vivo, in contrast to what was predicted from in vitro import experiments (48).
MATERIALS AND METHODS
Computational methods.
All publicly available DNA sequence data for T. brucei were used. The majority of these data were produced as part of the T. brucei genome project by the sequencing centers (The Institute of Genomic Research and the Sanger Centre). In a first approach the entire T. brucei database (59 Mb total) was analyzed with the BLAST software using default parameter settings to search for tRNA genes by homology to the corresponding tRNAs in other organisms. Potential coding regions were further analyzed with tRNAScan-SE (27) to confirm the presence of tRNA genes on the individual fragments. tRNAScan-SE detects tRNA genes based on the predicted secondary structures of their corresponding gene products. In a second approach, tRNAScan-SE was used directly on all available T. brucei sequences. The same set of genes was found with the two approaches. All fragments containing predicted tRNA genes were then assembled by AutoAssembler 2.1 (Applied Biosystems) to obtain a nonredundant set of tRNA genes (see Table 2). An online reference to all the T. brucei tRNAs discussed here may be obtained at http://zoosun00.unifr.ch/Trypanos/MITBIO.html.
TABLE 2.
Potential codon recognition pattern of tRNA isoacceptors predicted from the genomic sequence of T. brucei
| Amino acid | No. of genesa (predicted anticodon) | Anticodonb | Codonc | Amino acid | No. of genesa (predicted anticodon) | Anticodonb | Colonc | |
|---|---|---|---|---|---|---|---|---|
| Ala | 1 (AGC) | IGC | GCU | |||||
| IGC | GCC | |||||||
| 1 (UGC) | UGC igc | GCA | ||||||
| 1 (CGC) | CGC ugc | GCG | ||||||
| Arg | 3 (ACG) | ICG | CGU | |||||
| ICG | CGC | |||||||
| 1 (UCG) | UCG icg | CGA | ||||||
| 1 (CCU) | CCU ucg | CGG | ||||||
| 1 (UCU) | UCU | AGA | ||||||
| ucu | AGG | |||||||
| Asn | guu | AAU | ||||||
| 2 (GUU) | GUU | AAC | ||||||
| Asp | guc | GAU | ||||||
| 1 (GUC) | GUC | GAC | ||||||
| Cys | gca | UGU | ||||||
| 1 (GCA) | GCA | UGC | ||||||
| Gln | 1 (UUG) | UUG | CAA | |||||
| uug | CAG | |||||||
| Glu | 1 (UUC) | UUC | GAA | |||||
| 2 (CUC) | CUC uuc | GAG | ||||||
| Gly | gcc | GGU | ||||||
| 2 (GCC) | GCC | GGC | ||||||
| 1 (UCC) | UCC | GGA | ||||||
| 1 (CCC) | CCC | GGG | ||||||
| His | gug | CAU | ||||||
| 1 (GUG) | GUG | CAC | ||||||
| Ile | ?d | AUU | ||||||
| ? | AUC | |||||||
| 1 (UAU) | UAU | AUA | ||||||
| Leu | 1 (AAG) | IAG | CUU | |||||
| IAG | CUC | |||||||
| 1 (UAG) | UAG | CUA | ||||||
| 1 (CAG) | CAG uag | CUG | ||||||
| ? | UUA | |||||||
| 1 (CAA) | CAA | UUG | ||||||
| Lys | 2 (UUU) | UUU | AAA | |||||
| 3 (CUU) | CUU uuu | AAG | ||||||
| Met (i and e)e | 1 + 2 (CAU) | CAU | AUG | |||||
| Phe | gaa | UUU | ||||||
| 1 (GAA) | GAA | UUC | ||||||
| Pro | 1 (AGG) | IGG | CCU | |||||
| IGG | CCC | |||||||
| 1 (UGG) | UGG igg | CCA | ||||||
| ugg | CCG | |||||||
| Ser | 1 (AGA) | IGA | UCU | |||||
| IGA | UCC | |||||||
| 1 (UGA) | UGA iga | UCA | ||||||
| 1 (CGA) | CGA uga | UCG | ||||||
| gcu | AGU | |||||||
| 1 (GCU) | GCU | AGC | ||||||
| Thr | 1 (AGU) | IGU | ACU | |||||
| IGU | ACC | |||||||
| 1 (UGU) | UGU igu | ACA | ||||||
| 1 (CGU) | CGU ugu | ACG | ||||||
| Trp | ? | UGG | ||||||
| Tyr | gua | UAU | ||||||
| 1 (GUA) | GUA | UAC | ||||||
| Val | 2 (AAC) | IAC | GUU | |||||
| IAC | GUC | |||||||
| 1 (UAC) | UAC iac | GUA | ||||||
| 1 (CAC) | CAC uac | GUG |
Nonredundant gene number of indicated isoacceptors per haploid genome of T. brucei.
Anticodon predicted to decode the codons listed in the last column. Wobble position A was presumed to be posttranscriptionally converted to inosine (I). In cases where more than one anticodon is capable of decoding a single codon, the second one is indicated in lowercase.
Codon read by the anticodon in the previous column.
Apparently missing isoacceptors are indicated by a question mark.
Both initiator (i) and elongator (e) tRNAMet were found.
Cells.
Procyclic T. brucei, stock 427, was grown at 27°C in SDM-79 medium supplemented with 5% fetal bovine serum. Cells were harvested at late log phase, corresponding to 2.5 × 107 to 3.5 × 107 cells/ml, and washed once in cold 20 mM sodium phosphate buffer (pH 7.9) containing 150 mM NaCl and 20 mM glucose. The resulting cellular pellets were used either to isolate total RNA or to prepare mitochondria.
Isolation of mitochondria.
If large amounts of mitochondria were needed (e.g., for quantitative Northern analyses of wild-type cells), they were isolated by the hypotonic lysis procedure as described elsewhere (5, 17). The final mitochondrial pellet (yield, ca. 50 mg of mitochondrial proteins from 5 liters of cell culture) was directly resuspended either in guanidinium isothiocyanate to isolate the RNA or in the buffer used for the tRNA degradation experiments.
In the case where only a limited quantity of cells was available (e.g., for quantitative Northern analyses of transgenic T. brucei), mitochondrial fractions were prepared by digitonin extractions. Washed cells (108 cells each) were resuspended in 0.5 ml of SoTE (0.6 M sorbitol, 20 mM Tris-HCl [pH 7.5], and 2 mM EDTA). Five percent of the sample (25 μl) was removed to isolate the total RNA. After the addition of 0.475 ml of SoTE containing 0.05% (wt/vol) of digitonin, the samples were mixed by pipetting and incubated on ice for 5 min. The suspension was centrifuged (8,000 × g for 5 min at 4°C), and the supernatants were discarded. Next, the resulting pellets were resuspended in 500 μl of SoTE containing 1 μg of RNase A and incubated on ice for 15 min. After a final centrifugation, the supernatants were discarded and RNA was isolated from the pellets. At a concentration of 0.05%, digitonin selectively disrupts the cell membrane but does not affect mitochondria and therefore results in the digestion of contaminating cytosolic RNA; the obtained pellet therefore corresponds to a crude mitochondrial fraction and is essentially free of cytosolic RNAs. The hypotonic and the digitonin preparations yielded fractions of comparable quality, since very similar import efficiencies for initiator tRNAMet (tRNAMet-i), elongator tRNAMet (tRNAMet-e), and tRNALeu(CAA) were obtained when mitochondrial fractions from either source were used (see Table 3).
TABLE 3.
Total cellular and mitochondrial abundance of 15 tRNA isoacceptors of T. brucei
| tRNA (anticodon) | No. of molecules per cell | No. of molecules per mitochondrion (% of total) |
|---|---|---|
| Asn (GUU) | 13,500 | 370 (2.7) |
| Asp (GUC) | 40,600 | 2,430 (6.0) |
| Cys (GCA) | 36,800 | 1,770 (4.8) |
| Gln (UUG) | 61,500 | 1,210 (2.0) |
| Glu (CUC) | 52,300 | 1,420 (2.7) |
| Ile (UAU) | 47,000 | 3,520 (7.5) |
| Leu (AAG) | 130,000 | 3,580 (2.8) |
| Leu (CAA) | 220,100 | 7,700 (3.5) |
| Leu (CAG) | 121,300 | 1,210 (1.0) |
| Leu (UAG) | 60,700 | 1,820 (3.0) |
| Lys (CUU, UUU) | 113,700 | 4,090 (3.6) |
| Met-i (CAU) | 1,900 | 4 (0.2) |
| Met-e (CAU) | 15,600 | 880 (5.6) |
| Phe (GAA) | 36,400 | 1,350 (3.7) |
| Tyr (GUA) | 6,100 | 410 (6.8) |
| Met-i | ND | ND (0.3)a |
| Met-e | ND | ND (5.0)a |
| Leu (CAA) | ND | ND (3.2)a |
| 0-Leu∗ (CAA)b | 3,600 | 100 (2.9)a |
| 10-Leu∗ (CAA)b | 15,200 | 460 (3.0)a |
| 59-Leu∗ (CAA)b | 39,900 | 1,160 (2.9)a |
| 216-Leu∗ (CAA)b | 49,200 | 1,480 (3.0)a |
Mitochondria were isolated by digitonin extraction (see Materials and Methods).
Tagged tRNALeu(CAA) expressed in transgenic T. brucei (see Fig. 6). The abundance of the tRNA isoacceptor was determined by quantitative Northern analysis using the corresponding in vitro-transcribed tRNAs as marker. Unless otherwise indicated, mitochondria were isolated by the hypotonic procedure outlined in Materials and Methods.
RNA isolation.
RNA from total cells or isolated mitochondrial fractions was purified by the acidic guanidinium isothiocyanate method as described previously (9). The mitochondrial RNA fraction was routinely treated with RQ-DNase (Promega) (ca. 2 U/100 μg of nucleic acids) for 20 min at 37°C in 35 mM Tris-HCl (pH 7.6)-5 mM MgCl2-2.5 mM dithiothreitol (DTT) containing 8 U of RNasin (Promega). Different dilutions of isolated RNA fractions were quantified by measurements of optical density at 260 nm.
In vitro transcription.
In vitro transcripts corresponding to the 15 selected tRNAs (see Fig. 2) and the tagged tRNALeu(CAA) (see Fig. 6) were prepared in order to obtain the standards for the quantitative Northern analysis. PCR products, linearized plasmids containing the desired tRNA genes, or synthetically synthesized tRNA genes, all carrying a 5′-flanking T7 promoter, were used as templates for the transcription reactions. The tRNA transcripts were either identical in size to the corresponding mature tRNAs or maximally 150 nucleotides in length. In vitro transcriptions were performed with 20 μl of 40 mM Tris-HCl (pH 7.9)-6 mM MgCl2-2 mM spermidine-20 mM DTT-50 μg of bovine serum albumin per ml-15 U of RNasin-0.02% NP-40-2 mM concentrations of each nucleotide-ca. 0.2 μg of template DNA-150 U of T7-RNA polymerase (New England Biolabs) and incubated for 1 h at 37°C. For quantification of the transcripts, 0.013 μM [α-32P]GTP (400 Ci/mmol) was added. The reaction was stopped by a phenol extraction, and the transcripts were analyzed on an 8 M urea-10% polyacrylamide gel. The bands corresponding to the in vitro transcripts were visualized with ethidium bromide, cut out, and eluted with 0.5 M ammonium acetate-0.4 mM EDTA-0.1% sodium dodecyl sulfate (SDS). The eluate was precipitated and quantified by scintillation counting. The amount of [α-32P]GTP which was incorporated into the transcripts was determined by comparing the counts in the transcripts with the ones of 0.1 μl of [α-32P]GTP stock solution, which corresponds to 2.7 pmol. The amount of the labeled GTP needs to be multiplied by the ratio between cold (2 mM) and labeled (0.013 μM) GTP, which is ca. 154,000, in order to obtain the amount of all incorporated GTP. Finally, the resulting number has to be divided by the number of Gs which occur in the transcript in order to obtain the quantity of the in vitro-synthesized tRNAs.
FIG. 2.
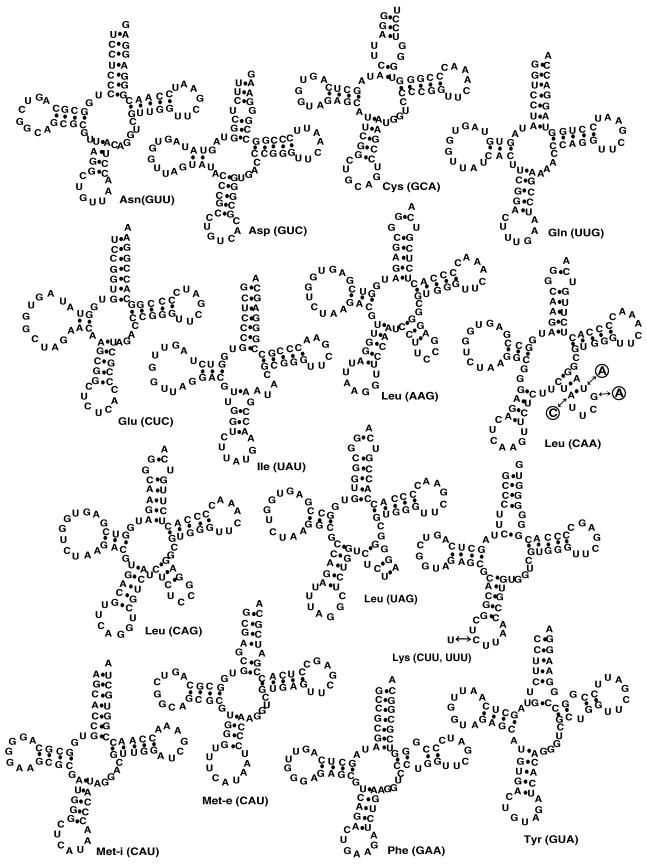
Secondary structures of T. brucei tRNAs. Primary sequences and potential secondary structures as predicted by tRNAScan-SE (27) of the 15 tRNA isoacceptors of T. brucei which were used to measure tRNA abundance and mitochondrial localization are shown. Nucleotide substitutions which were introduced into the tRNALeu(CAA) gene of the transgenic cell lines (Fig. 6) are indicated in circles.
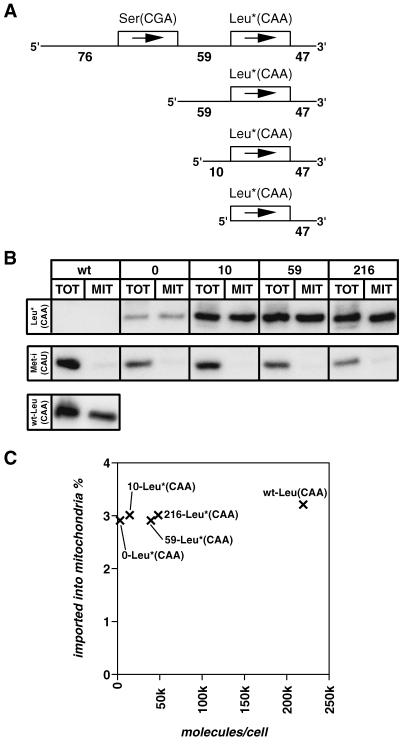
FIG. 6.
In vivo import of tagged tRNALeu(CAA) is independent of its 5′-genomic context. (A) Schematic representation of the inserts of the pHD-437 plasmid-based (3) constructs containing the tagged tRNALeu(CAA) gene (indicated by an asterisk) and the indicated length (0, 10, 59, or 214 nucleotides) of endogenous 5′-flanking region. The three mutations in the variable loop which were introduced as a tag are shown in Fig. 2. (B) Northern blot analysis of wild-type (wt) cells and of cell lines expressing the tagged tRNALeu(CAA) in the context of 0, 10, 59, or 214 nucleotides of their natural 5′-flanking sequence. RNAs isolated from total cells (TOT) (0.5 × 107 cells) and from digitonin-extracted mitochondrial fraction (MIT) (108 cell equivalents) were analyzed. Leu*(CAA), Northern blot probed with an oligonucleotide specifically recognizing the tagged tRNALeu(CAA); Met-i, same blot reprobed with an oligonucleotide specific for the cytosol-specific tRNAMet-i; Leu(CAA), the blot for wild-type cells was reprobed with an oligonucleotide which specifically recognizes the wild-type tRNALeu(CAA). The oligonucleotides used as probes are listed in Table 1. (C) The numbers of tagged tRNALeu(CAA) molecules per cell in the four transgenic cell lines and of the unchanged tRNALeu(CAA) in wild-type cells, as determined by quantitative Northern analysis, were plotted against the percentage of each which is imported into mitochondria.
Quantitative Northern analysis.
Known amounts of RNA (of total cells, mitochondria, or in vitro-transcribed tRNAs) were separated on an 8 M urea-10% polyacrylamide gel and blotted onto GeneScreen Plus membranes (NEN, Life Science Products). Blotting was performed with 40 mM Tris-acetic acid (pH 8)-1 mM EDTA for an initial 30 min at 200 mA followed by 2.5 h at 1.5 to 2 A. After blotting, the damp filters were UV irradiated at 312 nm using the 250-mJ setting of the UV chamber (GS Genelinker from Bio-Rad) and air dried at room temperature. Hybridization was done overnight at 55°C in 6× SSPE (60 mM NaH2PO4, 0.9 M NaCl, 6 mM EDTA) containing 5× Denhardt's solution and 1% SDS using the labeled oligonucleotides listed in Table 1. Finally, the blots were washed twice for 3 min in 2× SSPE-0.5% SDS at room temperature followed by two 3-min washes in the same buffer at 55°C before they were exposed and analyzed on a phosphorimager.
TABLE 1.
Oligonucleotides used for Northern analyses
| Target RNAa | Complementary position in RNA sequence | Sequence (5′-3′) |
|---|---|---|
| Asn (GUU) | 73-56 | CTCCTCCCGTTGGATTCG |
| Asp (GUC) | 72-55 | CTTCCCGGCCGGGAATTG |
| Cys (GCA) | 72-55 | AGGACCCACCCGGGTTTG |
| Gln (UUG) | 72-58 | GTGGTGGTCCTACCAGGAT |
| Glu (CUC) | 72-55 | TTCCGGTGCCGGGGATCG |
| Ile (UAU) | 72-54 | TGCTCCCGGCGGGTTCGAA |
| Leu (AAG) | 59-42 | ACGCCCTCGGAAGGATTG |
| Leu (CAA) | 62-45 | CCCACGCCTACGAATAGA |
| Leu (CAG) | 56-39 | CCTCCGGAGAGATGACGA |
| Leu (UAG) | 57-40 | ACGCCCCTAGAGACCAGA |
| Lys (CUU, UUU) | 73-50 | GTGGCACCCCCCGTGGGGCT CGAACCCA |
| Met-i (CAU) | 64-47 | GTTGGTTTCGATCCAACG |
| Met-e (CAU) | 65-48 | GTGAGGCTCGAACTCACG |
| Phe (GAA) | 73-62 | AGGTGTGCCGCGACCCG |
| Tyr (GUA) | 70-53 | TTCCGGCCGGAATCGAAC |
| 12S rRNAb | 869-848 | AGGAGAGTAGGACTTGCCCT |
| gCYb (560A)c | 47-28 | TTATCCTCCCCATTACTCAG |
| Leu∗ (CAA)d | 61-43 | CCACGCCTTTGAAGAGAAG |
Oligonucleotides (10 to 30 pmol) were 5′ end labeled in 20 μl of 70 mM Tris-HCl (pH 7.6)-10 mM MgCl2-10 mM spermidine-5 mM DTT with a 2.4 μM concentration of end-labeling grade [γ-32P]ATP (7,000 Ci/mmol) from ICN Pharmaceuticals and 12 U of T4 polynucleotide kinase (New England Biolabs). After 1 h of incubation at 37°C, the reaction was stopped by a phenol extraction step and unincorporated nucleotides were removed by using a Sephadex G-25 spin column.
Calculation of tRNA abundance.
Our own experiments and published data (33) have shown that approximately 13 μg of RNA corresponds to 107 procyclic T. brucei cells. The absolute amounts of the different tRNA isoacceptors per microgram of total RNA were determined by quantitative Northern analysis as described above. Sequence information from the genome project was used to design oligonucleotide probes specific for the corresponding tRNA(s) or, in the case of the tRNAsLeu, for the different isoacceptors. The molecular weights of the different tRNA species are known, and their number per cell can therefore be calculated. To determine the percentage of a given tRNA that is localized in mitochondria, it was necessary to determine the fraction of total RNA which is mitochondrial. Hybridizations of Northern blots containing different dilutions of total cellular and isolated mitochondrial RNA with two mitochondrion-specific probes, directed against 12S rRNA and a guide RNA, respectively (gCYb560A) (33) (Table 1), have shown that 1.8 to 2.2% of total cellular RNA is mitochondrial. With this value it is possible to extrapolate the signal on Northern blots, which were obtained with the same oligonucleotide probes used to measure total tRNA abundance, to a cellular basis. This means that comparing the same amounts of RNA in total and mitochondrial fractions actually corresponds to a comparison of one cell equivalent of total RNA to 50 cell equivalents of mitochondrial RNA.
Intramitochondrial tRNA degradation.
In order to compare the intramitochondrial stabilities of different tRNA isoacceptors, isolated mitochondria were incubated for 0, 0.5, 1, 2, and 4 h at 25°C in 20 mM Tris-HCl (pH 7.2)-15 mM KH2PO4-0.6 M sorbitol-20 mM MgSO4-2.5 mg of fatty acid-free bovine serum albumin per ml-4 mM ATP-1 mM concentrations (each) of CTP, UTP, and GTP. At each time point a 200-μg sample was removed, mitochondria were reisolated, and RNA was extracted from the pellets. Isolated RNA was fractionated on 8 M urea-10% acrylamide gels and analyzed by quantitative Northern blotting using the oligonucleotides listed in Table 1.
Transfection of T. brucei.
For expression of the tagged tRNAsLeu(CAA) (see Fig. 6), derivatives of the pHD-437 plasmid (3) which allow stable integration into trypanosomal ribosomal DNA loci were used. The plasmid was digested with KpnI and BamHI, and the region containing the procycline promoter and the luciferase gene was replaced by inserts containing the tagged tRNALeu(CAA) genes. The tag was introduced by PCR-mediated mutagenesis. Four constructs containing the tagged tRNALeu(CAA) together with either 0, 10, 59, or 216 nucleotides of genomic 5′-flanking region were prepared. All inserts carried 47 nucleotides of the natural 3′-flanking region. The identity of each insert was verified by sequence analysis. Finally, the constructs were linearized with NotI and electroporated into T. brucei, and transformants were selected with phleomycine as previously described (3).
RESULTS
tRNA genes.
A survey of the T. brucei sequence project has resulted in the identification of 50 trypanosomal tRNA genes representing 40 different isoacceptor species (one tRNA with undetermined specificity was also detected [2] but will not further be discussed here) (Table 2). Several of these genes have been characterized before by different research groups (6, 7, 15, 25, 29-31, 45). It is clear that the trypanosomal genome is not yet completely represented in the sequence database and therefore some tRNA genes may have escaped detection. However, the fraction of undetected tRNA genes is expected to be small for the following reasons. If an A at the first position of the anticodon is modified to inosine, which is known to decode U, C, and A (23), the 40 isoacceptors are sufficient to decode 94% of all 61 sense codons (Table 2). The four codons which cannot be read are UGG (Trp), AUU and AUC (Ile), and UAA (Leu). Based on this observation, the 50 tRNA genes are expected to represent about 94% of the total genomic tRNA gene complement. An independent estimate of the size of the trypanosomal tRNA gene complement was obtained based on the information that each tRNA gene was detected on average 2.46 times. Fifteen genes were found once, and the remaining 35 were found two to six times each. From a statistical analysis of the obtained histogram, we estimate that an additional 3 to 13 genes (8 to 33%, P = 0.05) might be present in the still-missing part of the genome. This analysis assumes that the entire genome was sequenced at random and that therefore the frequencies with which each gene was found should follow a Poisson distribution. Sequencing, however, has also included a chromosome-by-chromosome strategy. Thus, the sequence coverage is higher for some chromosomes than expected for an ideally random approach. After taking these considerations into account, we conclude that more than 80% of all trypanosomal tRNA genes were detected in the present work. The estimated total number of trypanosomal tRNA genes is therefore ca. 62 (representing at least 43 different isoacceptors). Whereas the number of isoacceptors is in the range expected for a genome with a GC content of 45 to 50% (20), the number of tRNA genes is far smaller than for any other eukaryotic genome characterized so far with the exception of the microsporidian parasite Encephalitozoon cuniculi, which has 44 tRNA genes (22). Preliminary analysis of the Leishmania genome (data not shown) indicates that in this organism the number of tRNA genes is also very low, suggesting that this might be a general feature of trypanosomatids or maybe even parasites in general.
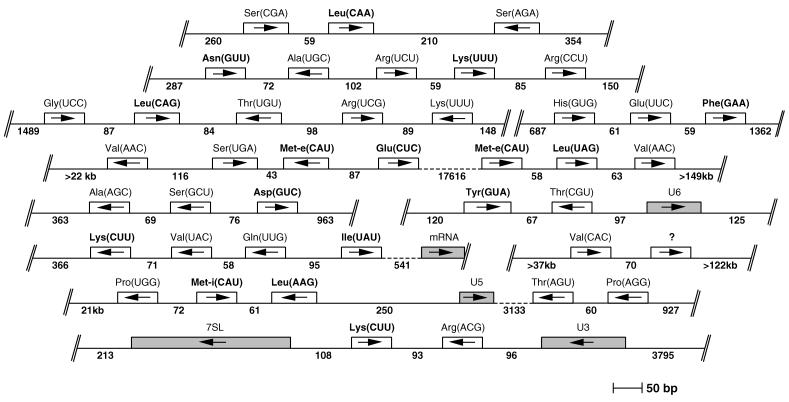
Eighty percent of the detected tRNA genes are found in clusters of two to five genes separated by very short intergenic regions of 79 nt on average (Fig. 1). Within these clusters, the tRNA genes appear to be randomly arranged: head-to-head and tail-to-tail arrangements occur as frequently as tandem repeats. The remaining 20% are found dispersed throughout the whole genome. Looking at the predicted tRNA-coding regions, it appears that trypanosomal tRNAs fold into secondary structures that are essentially similar to bona fide eukaryotic tRNAs. The tRNATyr is the only one that contains an intron (43). Two distinct tRNAsMet were found, one of which has an AU base pair at the end of the acceptor stem and therefore corresponds to the eukaryotic tRNAMet-i (32); the other shows all features of a tRNAMet-e. Their predicted secondary structures are shown in Fig. 2.
FIG. 1.
Genomic organization of T. brucei tRNA genes. The 12 clusters which were identified in the genome of T. brucei and which contain in total 40 tRNA genes are shown and drawn to scale (some of these clusters have been analyzed before [6, 7, 15, 25, 29-31, 45]). The directions of transcription are indicated by arrows, and the predicted anticodons are shown in parentheses. The question mark indicates a tRNA of unknown identity. The numbers indicate the lengths of the known 5′- and 3′-flanking and intergenic sequences. Double slashes denote the ends of each contig. Broken lines represent large intergenic regions. Genes for structural RNAs (U2, U5, U6, 7SL) or mRNAs which are found adjacent to tRNA genes are also shown. The tRNA genes which are found dispersed elsewhere in the genome are not shown. tRNA genes whose gene products have been analyzed in this study are shown in bold.
Expression of tRNAs.
In order to obtain an overview of the steady-state expression level of tRNAs, we have selected 15 different tRNA species specific for 12 amino acids (predicted secondary structures are shown in Fig. 2) and determined their absolute abundance by quantitative Northern analysis. Different amounts of in vitro transcripts were used as standards and analyzed together with known quantities of total cellular RNA by specific oligonucleotide hybridization. The hybridization signals were quantified on a phosphorimager and allowed us to calculate the number of molecules per cell (see Materials and Methods). A potential drawback of the quantitative Northern analysis is the possible presence of nucleotide modifications in regions of the tRNA which are complementary to the oligonucleotide probes. Modifications will be absent from the in vitro-synthesized tRNAs. It is therefore possible that in such cases the hybridization to the cellular tRNA is less efficient than to the in vitro-produced marker tRNAs, which would result in an underestimation of the tRNA abundance.
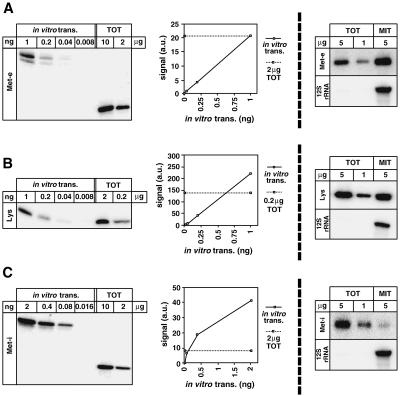
Three representative Northern blots with the corresponding quantifications are shown on the left panels of Fig. 3. A summary of the results on tRNA abundance per cell is presented in Table 3 (first column). It shows that on average 64,000 molecules of each of the selected tRNA species, ranging from 1,850 molecules for the tRNAMet-i to 220,000 for the tRNAsLeu(CAA), are found in a cell.
FIG. 3.
Quantitative Northern analysis. Specific oligonucleotide hybridization was used to detect tRNAMet-e(CAU) (A), tRNALys(CUU, UUU) (B), and tRNAMet-i(CAU) (C). (Left panels) The abundance of tRNAs in total cellular RNA (TOT) was determined by comparison with known quantities of the corresponding in vitro-transcribed tRNA (in vitro trans.). The graphs show the quantification of the blots shown on the right using a phosphorimager. Signal intensities are indicated in arbitrary units (a.u.). (Right panels) Mitochondrial localization was determined by hybridization of the corresponding specific oligonucleotides to known quantities of total and mitochondrial (MIT) RNAs. Lower panels show hybridizations using a probe specific for mitochondrion-encoded rRNA (12S rRNA).
Mitochondrial import.
The same 15 tRNAs whose intracellular quantities have been determined were analyzed for mitochondrial import. Northern blots containing known amounts of total cellular and isolated mitochondrial RNAs were hybridized with mitochondrion-specific probes (directed against a guide RNA and 12S rRNA). The data (not shown) indicated that ca. 2% of total RNA is mitochondrial. The cytosolic contamination of the mitochondrial RNA was shown to be ca. 0.33 to 0.40% as determined by hybridization to 7SL RNA (not shown). This RNA is a component of the signal recognition particle and expected to be exclusively cytosolic, although previous studies have established that this marker overestimates the extent of cytosolic contamination (18, 37). Determining the intracellular distribution of tRNAs is expected to be less error prone than measuring total tRNA abundance, since modified nucleotides in the region recognized by the probe will interfere with the interpretation of the results only if they are compartment specific. Only very few mitochondrion-specific nucleotide modifications are known for T. brucei, and they appear to occur at the same relative positions in most tRNA molecules (42).
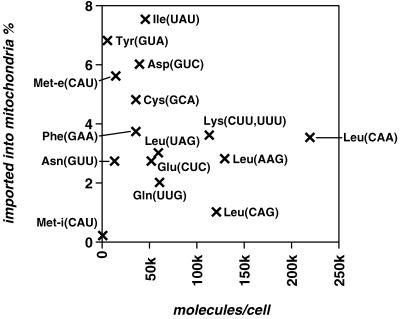
The right panels of Fig. 3 show Northern blots representing the intracellular distribution of three tRNAs, exhibiting high (tRNAMet-e) and intermediate (tRNALys) levels of mitochondrial localization as well as the only tRNA with an exclusive cytosolic localization (tRNAMet-i). The number of imported molecules per cell for each of the selected tRNAs is summarized in Table 3 (second column) and ranges from 370 to 7,700. Abundance and intracellular distribution of all 15 tRNAs are summarized on the graph in Fig. 4, in which the percentage of each tRNA recovered in mitochondria is plotted against the total number of the molecules per cell. The following conclusions can be drawn from this graph.
FIG. 4.
tRNA abundance in total cell and mitochondria. This is a graphical representation of the results shown in Table 3. The numbers of tRNA molecules per cell were plotted against the percentage of those found in mitochondria. Identities of the tRNAs and their anticodons are indicated. No correlation between expression level and extent of mitochondrial localization was observed (R = −0.29, P = 0.310).
(i) Only 0.2% of the tRNAMet-i is found in mitochondria, a proportion which is less than for cytosolic markers and therefore is most likely the result of cytosolic contamination. The tRNAMet-i therefore represents the first cytosol-specific tRNA to be characterized for T. brucei. The existence of cytosolic tRNAsMet in both T. brucei and Leishmania tarentolae has been shown before, but their identities have not been determined (14, 44). The cytosolic localization of the tRNAMet-i is in agreement with its eukaryotic initiator function, since this tRNA is not expected to be functional in the bacterial-type translation system of the mitochondria (32). In contrast, the tRNAMet-e, which is homologous to the tRNAMet-i (Fig. 2), is efficiently imported into mitochondria.
(ii) If we assume that the selected 15 tRNAs are representative of the whole population, the complements of mitochondrial and cytosolic tRNAs might be identical. With the exception of the tRNAMet-i, all tRNAs are to some extent imported into mitochondria. However, no tRNA showing an exclusive mitochondrial localization has been detected. This is reminiscent of all other organisms in which tRNA import has been studied and suggests that nucleus-encoded tRNAs which are specifically localized to mitochondria do not exist (41).
(iii) The extent of mitochondrial localization is distinct for different isoacceptors and ranges from 1 to 7.5%. There is no apparent correlation between the overall concentration of a given tRNA and the extent of its mitochondrial localization (R = −0.29, P = 0.310).
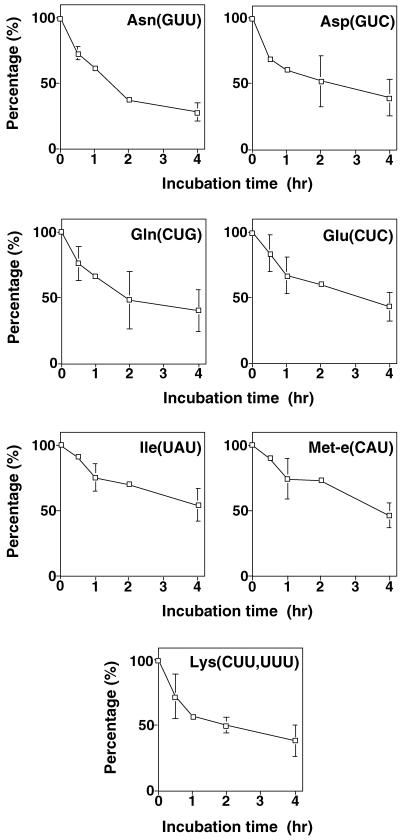
Northern analysis measures only steady-state levels of tRNAs in the different compartments under investigation. As there is no tRNA synthesis in the mitochondria of T. brucei, tRNA abundance is expected to be determined solely by the import efficiency and the rate of tRNA degradation inside mitochondria. In order to exclude that differential degradation of distinct tRNA species significantly interferes with our attempts to determine the import levels from steady-state quantities of tRNAs, we measured the degradation of selected tRNAs by incubation of isolated mitochondria at 27°C. Time course experiments show an approximately 50% loss of full-length tRNAs in a 4-h incubation period (Fig. 5). The observed loss can be attributed to two processes: (i) RNase protection assays (18) using mitochondrially encoded 12S rRNA as a marker (not shown) indicate that 33% (± 4.6% [standard deviation]) of mitochondria lyse during the incubation, resulting in a complete degradation of all released RNA; (ii) the remaining ca. 17% reflect intramitochondrial tRNA degradation. Most importantly in the context of this work, however, is the fact that no significant differences in the kinetics of the degradation of the different tRNA species were observed. It is therefore concluded that the steady-state levels of imported tRNAs indeed correlate with their import efficiencies.
FIG. 5.
Intramitochondrial degradation of tRNAs. Isolated mitochondria were incubated at 27°C, and the degradation of the indicated tRNAs was determined by Northern analysis using the same oligonucleotides that were used to determine tRNA abundance. Mean values of two independent experiments are shown. For time points where three or more independent values were available, the standard deviation is indicated.
In vivo import substrate.
All results discussed so far were obtained from wild-type cells. However, quantitative Northern analysis can also be used in transgenic cells, in which tRNA genes and their genomic contexts can be manipulated. We have used this possibility to address the question of whether mature or 5′-extended tRNAs are the in vivo import substrates in T. brucei. The best-studied import substrate is the tRNALeu(CAA) encoded on the tRNASer(CGA)/tRNALeu(CAA) cluster (Fig. 1). It has previously been shown that this region is transcribed as a dicistronic precursor (25). Furthermore, in vitro experiments have established that only the tRNALeu(CAA) precursor but not its mature derivative was imported into mitochondria (48). In order to confirm these results, we performed the corresponding in vivo experiments using the same tRNA substrate. A tag corresponding to three nucleotide substitutions in the variable loop was introduced into the tRNALeu(CAA) gene (Fig. 2). The tagged gene containing either 216, 59, 10, or 0 nucleotides of its original 5′-flanking sequence was cloned into an expression plasmid and stably integrated into a ribosomal DNA locus of T. brucei (3) (Fig. 6A). Finally, by using the same methods as for wild-type cells (Fig. 4), expression of the tagged tRNALeu(CAA) was quantified in all four cell lines. In order to analyze import, mitochondria of the transgenic cell lines were isolated by digitonin extractions followed by RNase digestions (see Materials and Methods). This procedure has the advantage that it requires only small quantities of cells. It yields crude mitochondrial preparations which are essentially free of cytosolic RNAs. Mitochondrial fractions obtained by digitonin extractions can directly be compared to the ones isolated by the hypotonic isolation procedure as evidenced by the fact that very similar import efficiencies for tRNAMet-i, tRNAMet-e, and tRNALeu(CAA) were obtained irrespectively of which of the two procedures was used (Table 3). Figure 6B shows that in all four transgenic cell lines the tagged tRNALeu(CAA) is imported into mitochondria with an efficiency of 2.9 to 3.0%, which is essentially identical to the 3.2 to 3.5% observed for the wild-type tRNALeu(CAA). The cytosol-specific tRNAMet-i, as expected, is found only in the cytosolic fraction. Thus, these results show that unlike what can be predicted from in vitro experiments, import of the tagged tRNALeu(CAA) is independent of its endogenous 5′-flanking region. Even complete removal of all of the natural 5′-flanking sequence results in an import efficiency identical to that of the wild-type molecule. In all four constructs the tagged tRNA is expressed at a lower level than that of the wild-type tRNA. tRNALeu(CAA) (Fig. 6C and Table 3). One reason for this is probably the presence of the tag in the variable loop. Furthermore, expression is strongly influenced by the 5′ context of the tagged tRNA gene in that longer 5′-flanking regions result in a higher expression of the corresponding gene. Even though expression of the tagged tRNALeu(CAA) in the different cell lines varies more than 10-fold, the same extent of import is observed. These results therefore allow us to extend the conclusion that there is no apparent correlation between the overall concentration of a given tRNA and the extent of its mitochondrial localization (Fig. 4) to a single tRNA species.
DISCUSSION
Most tRNA genes in T. brucei are clustered and have very short intergenic sequences (Fig. 1). Tight clustering of tRNA genes is common in prokaryotes, in which the clusters correspond to transcription units (19). The genomic organization of eukaryotic tRNA genes depends on the organism: in S. cerevisiae and Caenorhabditis elegans they are found dispersed over the whole genome, while clustering of tRNA genes is observed in Drosophila and rat (24). As in T. brucei, clustered tRNA genes of other eukaryotes can be oriented in both directions; however, their intergenic regions are generally longer. It has been shown that the two tandemly linked genes for tRNASer(CGA) and tRNALeu(CAA) can be cotranscribed (25). However, many clustered trypanosomal tRNA genes are transcribed individually, since they do not show uniform orientations within the clusters.
The cellular amounts of distinct tRNA isoacceptors are generally very different. In E. coli, for example, the difference between the most and the least abundant tRNAs is 46-fold (11). It has been shown for many organisms that the quantities of the tRNAs are typically adapted to the needs of the cell (11, 12, 20). This is reflected by the positive correlation between the abundance of individual tRNAs and the frequency of the corresponding codons that are used in abundant proteins (e.g., ribosomal proteins). This characteristic appears to hold true for T. brucei in that the relative abundance of the tested tRNAs varies by a factor of 116 and correlates with the codon usage. The correlation was best (R = 0.68, P = 0.008) when a codon usage table representing proteins known to be expressed at high levels in procyclic T. brucei (tubulin, procyclins, and ribosomal proteins) was used (not shown).
In some prokaryotes, and all eukaryotes investigated so far, an important regulatory component of the intracellular tRNA concentration is gene dosage (12, 20). The situation is very different for T. brucei; even though its genome is 1.8- and 2.8-fold larger than the ones from S. cerevisiae and Schizosaccharomyces pombe, respectively, it encodes 5- and 3-fold-fewer tRNA genes (http://rna.wustl.edu/GtRDB/) (Table 2). Even some prokaryotes such as Bacillus subtilis or Escherichia coli have ca. 50% more tRNA genes than what is predicted for T. brucei (20). Only 8 of 40 isoacceptor genes are found in more than one copy in the haploid genome, and there is no apparent correlation between gene copy number and tRNA abundance. So, clearly, regulation of tRNA abundance in T. brucei cannot be achieved by gene dosage.
To our knowledge, there are no recent studies which have measured concentrations of individual tRNAs in eukaryotes. There is, however, a recent publication in which the abundance of the complete set of E. coli tRNAs has been determined (11). Between 104 and 4,752 tRNA molecules were found in an E. coli cell, the average intracellular concentration of an E. coli tRNA being 5.8 μM (11). If the volume of a procyclic T. brucei is ca. 49 μm3 (4), the average intracellular concentration of the tested tRNAs can be calculated to be 2 μM (range, 0.063 to 7.4 μM). These values are in a range similar to that of E. coli.
Except for tRNAMet-i, discussed below, the cytosolic and the mitochondrial sets of trypanosomal tRNAs are qualitatively identical. Organisms which do not import tRNAs and therefore rely on mitochondrion-encoded tRNAs only (e.g., all vertebrates) use a reduced set of isoacceptors (e.g., 22 for vertebrates) compared to the cytosol to perform mitochondrial translation (38). Mitochondria of T. brucei, however, are expected to have ca. 40 different isoacceptors, which can all be used for organellar translation. The sets of tRNAs found in the two compartments might be qualitatively identical, but each tRNA is not imported to the same extent. Interestingly, the steady-state expression level of the tRNAs does not correlate with the extent of their import (Fig. 4). Thus, there are quantitative differences between the cytosolic and the mitochondrial tRNA complements. The cytosolic set appears to be adapted to cytosolic codon usage (see above). However, the differential import efficiencies of the tested tRNAs do not result in an organellar set which in composition is adapted to the mitochondrial codon usage. So, whereas the T. brucei mitochondrial translation system might have the advantage of a large set of isoacceptors, their abundance inside mitochondria appears not to be optimally adapted.
Mitochondrial tRNA import in T. brucei, unlike in other organisms such as S. cerevisiae or plants, is not very specific. From the data presented here, all except one of the tested tRNAs are partially imported into mitochondria. tRNAMet-i is the only cytosol-specific tRNA detected in this study. A homologue of this tRNA was found in L. tarentolae and shown to be cytosol specific as well (data not shown). The cytosol-specific tRNAMet-i is highly homologous to the elongator-type tRNAMet-e, which is efficiently imported into mitochondria. Both tRNAs are charged by the same aminoacyl-tRNA synthetase. It is therefore unlikely that the methionyl-tRNA synthetase is involved in import. How the cytosol-specific localization of the tRNAMet-i is achieved is not known at present.
A previous in vivo study has shown that the import of two heterologous tRNAs (yeast tRNAHis and human tRNALys) was independent of genomic 5′-flanking sequences of trypanosomal origin. A similar situation was found for a tagged version of the trypanosomal tRNATyr which was still imported even though all but 25 nucleotides of its 5′-genomic context had been replaced by a heterologous sequence (18). Removing the entire 5′-flanking sequence, however, was not possible since it abolished expression of tRNATyr. These experiments suggest that the in vivo import substrates in T. brucei, at least in the artificial situation of heterologous tRNAs, are mature tRNAs. However, recent in vitro results showed that import of the trypanosomal tRNALeu(CAA) into isolated mitochondria required long (more than 59-nucleotide) homologous 5′-flanking sequences (48). In order to resolve this discrepancy, we have performed in vivo import experiments using the same tRNA substrate—tRNALeu(CAA)—that was used for the in vitro assays. Interestingly, these experiments show that wild-type import levels of the tagged version of the tRNALeu(CAA) can be obtained independently of a specific 5′ sequence (Fig. 6). These results are therefore in full agreement with the previously published in vivo study and suggest that in T. brucei, as in Leishmania (1, 21), tRNAs are imported as fully processed molecules. It therefore appears that the in vitro requirements for tRNA import are more stringent than in the living cells. A possible explanation for this might be the lack of cytosolic factors in the reconstituted system. Furthermore, whereas in vitro import of precursor tRNAs required a membrane potential and ATP, no processing of the imported tRNA precursors was observed (48). This is surprising since a mitochondrial RNase P activity has recently been characterized for T. brucei (36) and may suggest that RNAs other than tRNA precursors are the in vivo substrates for this enzyme. Indeed, it is known that the RNase P of yeast and that of humans, besides mediating tRNA processing, are also involved in rRNA maturation (8).
In summary, the presented work provides a comprehensive overview of trypanosomal tRNA genes. Furthermore, a quantitative analysis of tRNA expression and mitochondrial localization is presented. Finally, this analysis has been extended to transgenic cell lines in order to show that in vivo mitochondrial import of tRNALeu(CAA) is independent of its 5′-genomic context.
Acknowledgments
Timothy H. P. Tan and Roland Pach contributed equally to this work.
We thank C. Clayton for the gift of pHD-437, E. K. Horn for excellent technical assistance, D. Meyer for extensive help with the statistical analyses, and T. Seebeck for critical review of the manuscript.
This study was supported by grants 31-056825.99 and 4037-55154 from the Swiss National Foundation and by a fellowship of the Prof. Dr. Max Cloëtta Foundation.
REFERENCES
- 1.Aphasizhev, R., U. Karmarkar, and L. Simpson. 1998. Are tRNAs imported into mitochondria of kinetoplastid protozoa as 5′-extended precursors? Mol. Biochem. Parasitol. 93:73-80. [DOI] [PubMed] [Google Scholar]
- 2.Béja, O., E. Ullu, and S. Michaeli. 1993. Identification of a tRNA-like molecule that copurifies with the 7SL RNA of Trypanosoma brucei. Mol. Biochem. Parasitol. 57:223-230. [DOI] [PubMed] [Google Scholar]
- 3.Biebinger, S., L. E. Wirtz, P. Lorenz, and C. Clayton. 1996. Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 85:99-112. [DOI] [PubMed] [Google Scholar]
- 4.Böhringer, S., and H. Hecker. 1975. Quantitative ultrastructural investigations of the life cycle of Trypanosoma brucei: a morphometric analysis. J. Protozool. 22:463-467. [DOI] [PubMed] [Google Scholar]
- 5.Braly, P., L. Simpson, and F. Kretzer. 1974. Isolation of kinetoplast-mitochondrial complexes from Leishmania tarentolae. J. Protozool. 21:782-790. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, D. A. 1989. Tandemly linked tRNAGln, tRNAVal and tRNALys genes in Trypanosoma brucei. Nucleic Acids Res. 17:9479.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, D. A., Y. Suyama, and L. Simpson. 1989. Genomic organisation of nuclear tRNA-Gly and tRNA-Leu genes in Trypanosoma brucei. Mol. Biochem. Parasitol. 37:257-262. [DOI] [PubMed] [Google Scholar]
- 8.Chamberlain, J. R., Y. Lee, W. S. Lane, and D. R. Engelke. 1998. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 12:1678-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczyinski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 10.Dietrich, A., J. H. Weil, and L. Maréchal-Drouard. 1992. Nuclear-encoded transfer RNAs in plant mitochondria. Annu. Rev. Cell Biol. 8:115-131. [DOI] [PubMed] [Google Scholar]
- 11.Dong, H., L. Nilsson, and C. G. Kurland. 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260:649-663. [DOI] [PubMed] [Google Scholar]
- 12.Duret, L. 2000. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 16:287-289. [DOI] [PubMed] [Google Scholar]
- 13.Glover, K. E., D. F. Spencer, and M. W. Gray. 2001. Identification and structural characterization of nucleus-encoded transfer RNAs imported into wheat mitochondria. J. Biol. Chem. 276:639-648. [DOI] [PubMed] [Google Scholar]
- 14.Hancock, K., and S. L. Hajduk. 1990. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J. Biol. Chem. 265:19208-19215. [PubMed] [Google Scholar]
- 15.Hancock, K., and S. L. Hajduk. 1992. Sequence of Trypanosoma brucei tRNA genes encoding cytosolic tRNAs. Nucleic Acids Res. 20:4567.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock, K., A. J. LeBlanc, D. Donze, and S. L. Hajduk. 1992. Identification of nuclear encoded precursor tRNAs within the mitochondrion of Trypanosoma brucei. J. Biol. Chem. 267:23963-23971. [PubMed] [Google Scholar]
- 17.Harris, M. E., D. R. Moore, and S. L. Hajduk. 1990. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 265:11368-11376. [PubMed] [Google Scholar]
- 18.Hauser, R., and A. Schneider. 1995. tRNAs are imported into mitochondria of Trypanosoma brucei independent of their genomic context and of their genetic origin. EMBO J. 14:4212-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inokuchi, H., and F. Yamao. 1995. Structure and expression of prokaryotic tRNA genes, p. 17-30. In D. Söll and U. L. RajBhandary (ed.), tRNA: structure, biosynthesis, and function. ASM Press, Washington, D.C.
- 20.Kanaya, S., Y. Yamada, Y. Kudo, and T. Ikemura. 1999. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238:143-155. [DOI] [PubMed] [Google Scholar]
- 21.Kapushoc, S. T., J. D. Alfonzo, M. A. T. Rubio, and L. Simpson. 2000. End processing precedes mitochondrial importation and editing of tRNAs in Leishmania tarentolae. J. Biol. Chem. 275:37907-37914. [DOI] [PubMed] [Google Scholar]
- 22.Katinka, M. D., S. Duprat, E. Cornillot, G. Metenier, F. Thomarat, G. Prensier, V. Barbe, E. Peyretaillade, P. Brottier, P. Wincker, F. Delbac, H. E. Alaoui, P. Peyret, W. Saurin, M. Gouy, J. Weissenbach, and C. P. Vivares. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414:450-453. [DOI] [PubMed] [Google Scholar]
- 23.Knight, R. D., S. J. Freeland, and L. F. Landweber. 2001. Rewriting the keyboard: evolvability of the genetic code. Nat. Rev. Genet. 2:49-58. [DOI] [PubMed] [Google Scholar]
- 24.Kubli, E. 1981. The structure and function of tRNA genes of higher eukaryotes. Experientia 37:1-9. [DOI] [PubMed] [Google Scholar]
- 25.LeBlanc, A. J., A. E. Yermovsky-Kammerer, and S. L. Hajduk. 1999. A nuclear encoded and mitochondrial imported dicistronic tRNA precursor in Trypanosoma brucei. J. Biol. Chem. 274:21071-21077. [DOI] [PubMed] [Google Scholar]
- 26.Lima, B. D., and L. Simpson. 1996. Sequence-dependent in vivo import of tRNAs into the mitochondrion of Leishmania tarentolae. RNA 2:429-440. [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahapatra, S., and S. Adhya. 1996. Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane-bound receptors. J. Biol. Chem. 271:20432-20437. [DOI] [PubMed] [Google Scholar]
- 29.Marchetti, M. A., C. Tschudi, E. Silva, and E. Ullu. 1998. Physical and transcriptional analysis of the Trypanosoma brucei genome reveals a typical eukaryotic arrangement with close interspersion of RNA polymerase II- and III-transcribed genes. Nucleic Acids Res. 26:3591-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mottram, J. C., S. D. Bell, R. G. Nelson, and J. D. Barry. 1991. tRNAs of Trypanosoma brucei: unusual gene organization and mitochondrial importation. J. Biol. Chem. 266:18313-18317. [PubMed] [Google Scholar]
- 31.Mottram, J. C., Y. Shafi, and J. D. Barry. 1991. Sequence of a tRNA gene cluster in Trypanosoma brucei. Nucleic Acids Res. 19:3995.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.RajBhandary, U. L. 1994. Initiator transfer RNAs. J. Bacteriol. 176:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riley, G. R., P. J. Myler, and K. Stuart. 1995. Quantitation of RNA editing substrates, products and potential intermediates: implications for developmental regulation. Nucleic Acids Res. 23:708-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubio, M. A., X. Liu, H. Yuzawa, J. D. Alfonzo, and L. Simpson. 2000. Selective importation of RNA into isolated mitochondria from Leishmania tarentolae RNA. RNA 6:988-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rusconi, C. P., and T. R. Cech. 1996. The anticodon is the signal sequence for mitochondrial import of glutamine tRNA in Tetrahymena. Genes Dev. 10:2870-2880. [DOI] [PubMed] [Google Scholar]
- 36.Salavati, R., A. K. Panigrahi, and K. D. Stuart. 2001. Mitochondrial ribonuclease P activity of Trypanosoma brucei. Mol. Biochem. Parasitol. 115:109-117. [DOI] [PubMed] [Google Scholar]
- 37.Sbicego, S., C. Nabholz, R. Hauser, B. Blum, and A. Schneider. 1998. In vivo import of unspliced tyrosyl-tRNA containing synthetic introns of variable length into mitochondria of Leishmania tarentolae. Nucleic Acids Res. 26:5251-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheffler, I. E. 1999. Mitochondria, p. 106-107. Wiley-Liss, Inc., New York, N.Y.
- 39.Schneider, A. 2001. Does the evolutionary history of aminoacyl-tRNA synthetases explain the loss of mitochondrial tRNA genes? Trends Genet. 17:557-559. [DOI] [PubMed] [Google Scholar]
- 40.Schneider, A. 2001. Unique aspects of mitochondrial biogenesis in trypanosomatids. Int. J. Parasitol. 31:1403-1415. [DOI] [PubMed] [Google Scholar]
- 41.Schneider, A., and L. Marechal-Drouard. 2000. Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell Biol. 10:509-513. [DOI] [PubMed] [Google Scholar]
- 42.Schneider, A., K. P. McNally, and N. Agabian. 1994. Nuclear-encoded mitochondrial tRNAs of Trypanosoma brucei have a modified cytidine in the anticodon loop. Nucleic Acids Res. 22:3699-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider, A., K. P. McNally, and N. Agabian. 1993. Splicing and 3′-processing of tyrosine tRNA in Trypanosoma brucei. J. Biol. Chem. 268:21868-21874. [PubMed] [Google Scholar]
- 44.Simpson, A. M., Y. Suyama, H. Dewes, D. A. Campbell, and L. Simpson. 1989. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 17:5427-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suyama, Y., S. Wong, and D. A. Campbell. 1998. Regulated tRNA import in Leishmania mitochondria. Biochim. Biophys. Acta 1396:138-142. [DOI] [PubMed] [Google Scholar]
- 46.Tarassov, I., N. Entelis, and R. P. Martin. 1995. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 14:3461-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarassov, I., and R. Martin. 1996. Mechanisms of tRNA import into yeast mitochondria: an overview. Biochimie 78:502-510. [DOI] [PubMed] [Google Scholar]
- 48.Yermovsky-Kammerer, A. E., and S. Hajduk. 1999. In vitro import of a nuclearly encoded tRNA into the mitochondrion of Trypanosoma brucei. Mol. Cell. Biol. 19:6253-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]