Abstract
RAD26 in the yeast Saccharomyces cerevisiae is the counterpart of the human Cockayne syndrome group B (CSB) gene. Both RAD26 and CSB act in the preferential repair of UV lesions on the transcribed strand, and in this process, they function together with the components of nucleotide excision repair (NER). Here, we examine the role of RAD26 in the repair of DNA lesions induced upon treatment with the alkylating agent methyl methanesulfonate (MMS). MMS-induced DNA lesions include base damages such as 3-methyl adenine and 7-methyl guanine, and these lesions are removed in yeast by the alternate competing pathways of base excision repair (BER), which is initiated by the action of MAG1-encoded N-methyl purine DNA glycosylase, and NER. Interestingly, a synergistic increase in MMS sensitivity was observed in the rad26Δ strain upon inactivation of NER or BER, indicating that RAD26 promotes the survival of MMS-treated cells by a mechanism that acts independently of either of these repair pathways. The galactose-inducible transcription of the GAL2, GAL7, and GAL10 genes is reduced in MMS-treated rad26Δ cells and also in mag1Δ rad14Δ cells, whereas a very severe reduction in transcription occurs in MMS-treated mag1Δ rad14Δ rad26Δ cells. From these observations, we infer that RAD26 plays a role in promoting transcription by RNA polymerase II through damaged bases. The implications of these observations are discussed in this paper.
Cockayne syndrome (CS) in humans is characterized by severe growth retardation, with the outward appearance of cachectic dwarfism, and individuals with CS suffer from impaired neurological development and mental retardation. Mutations in the CSA and CSB genes account for ∼90% of CS cases, and the mean age of death in these patients is ∼12 years (13). Mutations in the CSA and CSB genes abolish preferential repair of UV-induced DNA lesions (27), a phenomenon known as transcription-coupled repair (TCR) (10). Because of the defect in the TCR of UV lesions, CS patients display mild sun sensitivity; however, they do not suffer from the high incidence of skin cancers characteristic of xeroderma pigmentosum patients.
The Saccharomyces cerevisiae RAD26 gene is the counterpart to the human CSB gene, and its inactivation creates a defect in the TCR of UV lesions in yeast (26). The CSB and Rad26 proteins are members of the SWI2/SNF2 family of ATPases, and both these proteins have DNA-dependent ATPase activity (4, 21). The mechanism of the TCR of UV lesions is best understood in Escherichia coli, where the Mfd protein, which is a DNA-dependent ATPase, displaces RNA polymerase (Pol) stalled at the lesion site in a reaction dependent upon ATP hydrolysis. Subsequently, via its interaction with UvrA, Mfd recruits the UvrA-UvrB-UvrC enzyme complex to the lesion site (22). The CSB and Rad26 proteins resemble Mfd in their DNA-dependent ATPase activity, but how these eukaryotic proteins bring about TCR is not known.
For the repair of UV lesions in both yeast and humans, TCR represents one subpathway of the nucleotide excision repair (NER) of UV lesions; the other subpathway of global genomic repair promotes the repair of UV lesions from nontranscribed regions of the genome. Since both these subpathways require NER proteins, such as XPA, ERCC1, and XPF in humans and their respective counterparts Rad14, Rad10, and Rad1 in yeast, TCR of UV lesions is abolished in both yeast and humans in the absence of any of these or other essential NER proteins.
Recently, evidence of the preferential removal of the oxidative DNA lesions thymine glycol (TG) and 8-oxoguanine (8-oxoG) from the transcribed strands in human cells has been presented (3, 9). TCR of these lesions, however, is not affected in cells defective in NER, but TCR is defective in cells derived from CS patients with mutations in the CSB, XPB, XPD, or XPG gene. From these observations, CSB, XPG, and TFIIH (since the XPB and XPD DNA helicases are components of TFIIH) have been inferred to act in the displacement of RNA Pol II arrested at the sites of these damaged bases and subsequently to aid in the recruitment to the lesion sites of the components of base excision repair (BER), such as DNA glycosylases and apurinic endonucleases. Thus, in the TCR of UV lesions, CS proteins function with the NER proteins, but in the TCR of 8-oxoG- and TG-damaged bases, CS proteins have been proposed to act specifically in the recruitment of BER proteins.
Recent studies have indicated that RAD26 plays a role in Pol II-dependent transcription elongation in yeast cells in the absence of any exogenous DNA damage (8) and that purified CSB protein increases the rate of elongation by Pol II on oligo(dC)-tailed DNA templates in the absence of any additional transcription factors (20). The involvement of Rad26/CSB proteins in transcription elongation through naturally occurring arrest sites raised the possibility that these proteins might also enable Pol II to transcribe through damaged bases. In that case, by freeing the lesion from stalled Pol II, the Rad26 and CSB proteins would promote the accessibility of DNA lesions to repair enzymes. In such a scenario, the Rad26 and CSB proteins would act independently of any of the repair processes.
Here, we use the yeast system to examine the role of RAD26 in promoting transcription through DNA lesions induced by the alkylating agent methyl methanesulfonate (MMS). MMS alkylates bases in DNA, particularly adenine at the N3 position (3meA) and guanine at the N7 position (7meG). We show that transcription through such damaged bases is severely inhibited in rad26Δ cells lacking both the NER and BER pathways required for the removal of these lesions. Consequently, sensitivity to MMS is greatly enhanced in rad26Δ cells lacking NER, BER, or both of these repair mechanisms. These results implicate the Rad26 protein in the promotion of transcription through damaged bases, and they indicate that in this role Rad26 functions independently of these two DNA repair processes.
MATERIALS AND METHODS
Yeast strains.
In this study, the wild-type strain EMY74.7 (MATa his3-Δ1 leu2-3,112 trp1Δ ura3-52) and its isogenic derivative strains YRP296, YR14.42, YR14.80, YR26.1, and YR14.82, which carry the mag1Δ, rad14Δ, mag1Δ rad14Δ, rad26Δ, and mag1Δ rad14Δ rad26Δ mutations, respectively, were used. The other strains used in this study, which were also derived from EMY74.7, were YR1.63, YR4.1, YR1.139, YR4.20, YR14.24, and YR26.25, and they carried the rad1Δ, rad4Δ, rad1Δ rad26Δ, rad4Δ rad26Δ, rad14Δ rad26Δ, and mag1Δ rad26Δ mutations, respectively.
MMS treatment.
For the determination of survival rates after MMS treatment, cells were grown overnight in YEPD (yeast extract-peptone-dextrose) medium. The cells were washed with distilled water and resuspended in 0.05 mM KPO4 at a density of 3 × 108 cells per ml. Aliquots (0.5 ml) of cells were treated with MMS at 30°C for 20 min with vigorous shaking. MMS was neutralized with 1 ml of 10% Na thiosulfate, and appropriate dilutions were plated on YEPD for viability determinations.
Transcription analyses.
For the examination of GAL2, GAL7, and GAL10 transcription, cells were grown at 30°C to log phase in YPL (1% yeast extract, 2% peptone, 3.7% lactate) medium. The cells were diluted to an optical density at 600 nm of 0.5 in YPL medium containing 2% galactose and 0.25% MMS. Samples were taken at selected time points after the cells were transferred to the medium containing galactose and MMS. The cells were pelleted and frozen quickly in crushed dry ice. Frozen cells were maintained at −80°C until RNA isolation.
Total RNA was isolated by the hot-phenol method (1) and fractionated by electrophoresis on 1.4% agarose-6% formaldehyde gels, followed by transfer to Hybond nylon membranes (Amersham). Each DNA probe was 32P labeled by the Multiprime DNA-labeling system (Amersham). Hybridization was performed at 42°C in a solution containing 40% formamide, 5% dextran sulfate, 1% sodium dodecyl sulfate (SDS), 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's solution, and 1 M KPO4 containing 100 μg of denatured herring sperm DNA per ml. The blots were washed twice at room temperature with 2× SSC-0.1% SDS for 5 and 10 min, once with 0.5× SSC-0.1% SDS for 30 min at 50°C, and once with 0.1× SSC-0.1% SDS for 15 min at 50°C. Quantitation of mRNA levels was performed in a PhosphorImager with ImageQuant software.
RESULTS
RAD26 functions independently of NER and BER in restoring survival to MMS-treated yeast cells.
For the preferential repair of UV lesions on the transcribed DNA strand, Rad26 functions together with components of the NER machinery. Consequently, introduction of the rad26Δ mutation into a strain with deletions of any of the genes indispensable for NER, such as RAD14, confers no additional increase in the UV sensitivity of the NER-defective mutant (28).
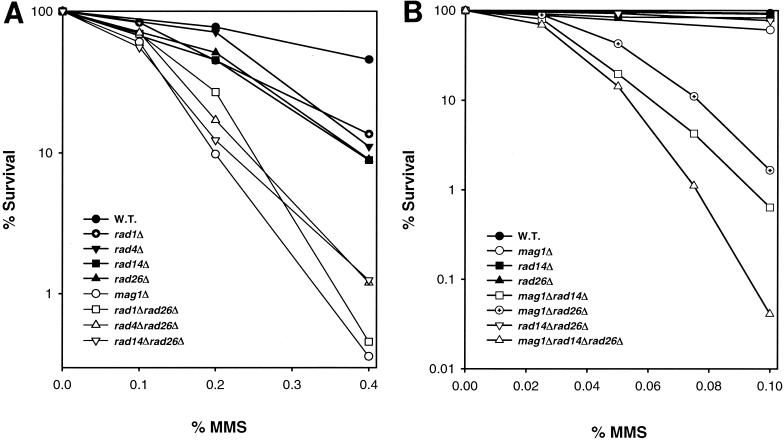
To determine if Rad26 functions similarly with the NER machinery for the repair of MMS-induced DNA lesions on the transcribed strand, we examined the MMS sensitivity of the NER-defective rad1Δ, rad4Δ, and rad14Δ mutants singly and in combination with the rad26Δ mutation. The Rad1 and Rad10 proteins form a DNA endonuclease which incises the damaged strand on the 5′ side of the lesion, while the Rad4-Rad23 complex and the Rad14 protein function at the lesion recognition step (14). As shown in Fig. 1A, sensitivity to MMS was enhanced to about the same extent in the rad1Δ, rad4Δ, rad14Δ, and rad26Δ strains. Interestingly, a synergistic increase in MMS sensitivity was observed when the rad26Δ mutation was combined with the rad1Δ, rad4Δ, or rad14Δ mutation. These observations suggest a role for RAD26 in promoting the survival of MMS-treated cells by a mechanism that functions independently of NER.
FIG. 1.
Effect of the rad26Δ mutation on the survival of MMS-treated yeast cells lacking NER or BER. RAD26 functions independently of NER (A) and MAG1-initiated BER (B) in promoting the survival of MMS-treated cells. The cells were treated with the indicated concentrations of MMS for 20 min at 30°C, and then MMS was inactivated with sodium thiosulfate and appropriate dilutions of cells were plated on YEPD for viability determinations. A concentration of 0.1% MMS corresponds to a 11.8 mM concentration of the chemical. The survival curves represent results from an average of three or more experiments for each strain. W.T., wild type.
The alkylated bases, such as 3meA and 7meG, formed in DNA by the action of MMS are removed by an N-methyl purine DNA glycosylase encoded in yeast by the MAG1 gene. This gene was previously referred to as MAG (1a); however, in keeping with the nomenclature for yeast genes, we refer to it as MAG1. Deletion of MAG1 confers a much higher degree of MMS sensitivity than does deletion of any of the NER genes (Fig. 1A), indicating a more prominent role for MAG1 than for NER in the removal of MMS-induced DNA damage. A synergistic enhancement in MMS sensitivity occurs in the absence of both MAG1 and RAD14 (Fig. 1B), indicating that NER competes with Mag1-dependent BER for the removal of MMS-induced DNA lesions.
Next, we examined the effect of the rad26Δ mutation on the MMS sensitivity of the mag1Δ and mag1Δ rad14Δ strains. Compared to the mag1Δ and rad26Δ single mutant strains, the mag1Δ rad26Δ double mutant strain exhibits a synergistic enhancement in MMS sensitivity, and the mag1Δ rad14Δ rad26Δ triple mutant strain displays a higher level of MMS sensitivity than the mag1Δ rad14Δ, mag1Δ rad26Δ, or rad14Δ rad26Δ double mutant strain (Fig. 1B). The synergistic enhancement in MMS sensitivity in the rad26Δ strain upon the inactivation of NER or BER implies that RAD26 promotes the survival of MMS-treated cells by a mechanism that acts independently of both of these repair processes.
Inhibition of transcription in MMS-treated rad26Δ cells.
Genetic studies of yeast have indicated a role for RAD26 in the promotion of transcription elongation by Pol II through intrinsic arrest sites in undamaged cells (8). Because of the independence of the function of RAD26 from NER and BER in restoring survival to MMS-treated cells, we considered the possibility that Rad26 can also promote transcription through MMS-induced DNA lesions. In that case, rather than promoting coupling to either of these repair processes following the removal of stalled Pol II from the lesion, Rad26 would aid in the repair of the transcribed strand by preventing the inhibitory effect of stalled Pol II on the ability of repair enzymes to gain access to the lesion.
For the TCR of UV-induced DNA lesions, RAD26 functions with the NER machinery, where it may act in a manner analogous to that of the E. coli Mfd protein. In this role, following the displacement of stalled Pol II from the lesion site, Rad26 may promote the recruitment of the NER enzyme complex to the lesion site. For UV lesions, then, we expect transcription to remain inhibited in the absence of any of the essential NER protein factors, since in spite of the displacement of stalled Pol II from the lesion site, in the absence of any lesion removal, Pol II would continue to stall at the lesion site because of the persistence of such lesions. In agreement with this hypothesis, recovery of mRNA synthesis in the inducible GAL10 and RNR3 genes is abolished in UV-irradiated cells from NER-defective mutants whereas recovery of RNR3 mRNA synthesis is abolished, and that of GAL10 mRNA is very considerably slowed, in the UV-irradiated rad26Δ strain (15). In contrast, if Rad26 enabled Pol II to transcribe through MMS-induced DNA lesions, then we would expect a considerable level of transcription to persist even in the absence of both NER and BER. Transcription should decline precipitously, however, in the rad26Δ strain lacking both NER and BER.
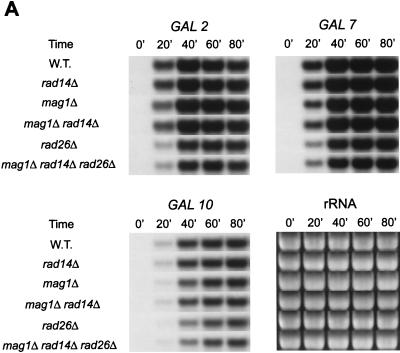
To investigate the role of RAD26 in promoting transcription through MMS-induced DNA lesions, we examined the synthesis of GAL2, GAL7, and GAL10 mRNAs in the wild-type and rad14Δ, mag1Δ, mag1Δ rad14Δ, rad26Δ, and mag1Δ rad14Δ rad26Δ strains in MMS-treated and untreated cells. In the absence of MMS treatment, transcription of these GAL genes, which is induced upon the addition of galactose, was not reduced in the rad14Δ, mag1Δ, and mag1Δ rad14Δ mutant strains compared to that in the wild-type strain (Fig. 2). The levels of these transcripts, however, were lower in the rad26Δ strain than in the wild-type or rad14Δ, mag1Δ, and mag1Δ rad14Δ mutant strains, and the pattern of transcript accumulation in the mag1Δ rad14Δ rad26Δ strain was virtually identical to that in the rad26Δ strain (Fig. 2). The lack of any inhibition of transcription in the mag1Δ rad14Δ cells and the absence of any further inhibition of transcription in mag1Δ rad14Δ rad26Δ cells beyond the level of that in the rad26Δ cells suggest that there is no significant accumulation of lesions such as 3meA and 7meG in cells not treated with MMS.
FIG. 2.
Transcription of GAL2, GAL7, and GAL10 genes in wild-type (W.T.) and mutant strains in the absence of MMS treatment. Total RNAs from cells grown in YPL medium containing galactose were subjected to Northern analyses. (A) Transcript levels of GAL2 (top left panel), GAL7 (top right panel), and GAL10 (bottom left panel) genes. The ethidium bromide-stained gel (bottom right panel) indicates the levels of RNAs loaded. mRNA levels were examined at the indicated time periods after the cells were transferred to galactose-containing medium. (B) Quantitation of GAL2, GAL7, and GAL10 mRNA levels. mRNA units at each time point are relative to the highest mRNA level in the rad14Δ strain. Symbols: •, wild-type strain; ▪, rad14Δ strain; ▴, mag1Δ strain; ○, rad26Δ strain; □, mag1Δ rad14Δ strain; ▵, mag1Δ rad14Δ rad26Δ strain.
In MMS-treated cells, however, the levels of these GAL gene transcripts were consistently lower in the mag1Δ rad14Δ strain than in the wild-type strain or the rad14Δ or mag1Δ single mutant strain (Fig. 3). This suggests that under the experimental conditions used the number of DNA lesions is such that Rad26 becomes limiting and is unable to promote transcription through all of the lesion sites in the mag1Δ rad14Δ strain. Importantly, the levels of all three transcripts were much lower in MMS-treated rad26Δ cells than in similarly treated mag1Δ rad14Δ cells, and a very severe reduction in transcription occurred in the mag1Δ rad14Δ rad26Δ strain (Fig. 3).
FIG. 3.
Transcription of GAL2, GAL7, and GAL10 genes in wild-type (W.T.) and mutant strains in the presence of MMS treatment. Total RNAs from cells grown in YPL medium containing galactose and MMS were subjected to Northern analyses. (A) Transcript levels of GAL2 (top left panel), GAL7 (top right panel), and GAL10 (bottom left panel) genes. The ethidium bromide-stained gel (bottom right panel) indicates the levels of RNAs loaded. mRNA levels were examined at the indicated time periods after the cells were transferred to medium containing galactose and MMS. (B) Quantitation of GAL2, GAL7, and GAL10 mRNA levels. mRNA units at each time point are relative to the highest mRNA level in the rad14Δ strain. Symbols: •, wild-type strain; ▪, rad14Δ strain; ▴, mag1Δ strain; ○, rad26Δ strain; □, mag1Δ rad14Δ strain; ▵, mag1Δ rad14Δ rad26Δ strain.
We compared the levels of GAL2, GAL7, and GAL10 transcripts (Tables 1, 2, and 3, respectively) in MMS-treated and untreated mutant cells to those in similarly treated wild-type cells. While there is little or no evidence of a reduction in the levels of these transcripts in MMS-treated rad14Δ and mag1Δ cells, the levels of transcripts of all three GAL genes were consistently lower in the MMS-treated mag1Δ rad14Δ strain than in the wild-type or the rad14Δ and mag1Δ mutant strains that were treated similarly. The levels of GAL gene transcripts were reduced even more in rad26Δ cells treated with MMS than in similarly treated mag1Δ rad14Δ cells, and a much more severe inhibition of transcription occurred in the mag1Δ rad14Δ rad26Δ strain than in the mag1Δ rad14Δ or rad26Δ strain.
TABLE 1.
Levels of GAL2 mRNA in rad14Δ, mag1Δ, mag1Δ rad14Δ, rad26Δ, and mag1Δ rad14Δ rad26Δ mutant strains in the absence and presence of MMS
| Time (min)b |
GAL2 mRNA level in strain with indicated deletiona
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
rad14Δ
|
mag1Δ
|
mag1Δ rad14Δ
|
rad26Δ
|
mag1Δ rad14Δ rad26Δ
|
||||||
| − | + | − | + | − | + | − | + | − | + | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 117 | 123 | 60 | 92 | 97 | 62 | 23 | 21 | 37 | 2.6 |
| 40 | 120 | 128 | 110 | 82 | 110 | 65 | 54 | 35 | 58 | 7.7 |
| 60 | 129 | 114 | 154 | 82 | 131 | 49 | 83 | 42 | 85 | 8.8 |
| 80 | 140 | 103 | 163 | 78 | 151 | 47 | 95 | 44 | 86 | 12.5 |
mRNA levels in the mutant strains are given relative to the mRNA levels in the wild-type strain times 100. −, MMS absent; +, MMS present.
Time point of sample removal.
DISCUSSION
RAD26 promotes survival of MMS-damaged cells by a process that functions independently of NER or BER.
RAD26 functions in the preferential repair of UV lesions on the transcribed strand in conjunction with NER, and consequently, introduction of the rad26Δ mutation into any of the NER-defective mutants, for example, the rad14Δ strain, causes no further increase in UV sensitivity. Here, we show that a synergistic increase in MMS sensitivity occurs in the rad14Δ rad26Δ double mutant strain compared to that in the rad14Δ or rad26Δ single mutant strain. A synergistic enhancement in MMS sensitivity also occurs upon the incorporation of the rad26Δ mutation into the mag1Δ strain. A synergistic rise in MMS sensitivity is also seen in the mag1Δ rad14Δ strain compared to that that in the mag1Δ and rad14Δ strains, and the mag1Δ rad14Δ rad26Δ strain displays a higher level of MMS sensitivity than any of the double mutants carrying any two of these three deletion mutations. From these observations, we infer that the NER and BER systems compete for the repair of MMS-induced DNA lesions and that Rad26 promotes the survival of MMS-damaged cells by a mechanism that acts independently of either of these repair processes.
RAD26 promotes transcription through damaged bases.
The levels of GAL gene transcripts are reduced in MMS-treated rad26Δ cells, and a reduction in transcript levels also occurs in MMS-treated mag1Δ rad14Δ double mutant strains. Further, and importantly, a more precipitous decline in transcription occurs in the MMS-treated mag1Δ rad14Δ rad26Δ strain than in the similarly treated mag1Δ rad14Δ or rad26Δ strain. The accentuation of transcriptional defects that occurs in the mag1Δ rad14Δ strain upon the inactivation of RAD26 implies that Rad26 enables Pol II to resume transcription through MMS-induced DNA lesions in the absence of both NER and BER, the two repair systems available for the removal of these lesions. If Rad26 had promoted transcription by displacing stalled Pol II from the lesion site followed by the recruitment of the NER or BER proteins, then there should have been no further decline in the level of transcription in the mag1Δ rad14Δ rad26Δ strain beyond that seen in the mag1Δ rad14Δ strain. This is because, in the absence of both the repair systems, Pol II would stall at the site of the DNA lesions, and even if Pol II were to be removed from the lesion site by the action of Rad26, due to the persistent stalling of Pol II, this process would continue to yield incompletely formed transcripts which would then be subject to degradation. On the other hand, if Rad26 were to promote transcription through MMS-induced DNA lesions, then even in the absence of any removal of these lesions, full-length transcripts would be formed.
All our observations—the reduction in the levels of transcription in the rad26Δ and mag1Δ rad14Δ strains and the even greater reduction in the level of transcription in the mag1Δ rad14Δ rad26Δ strain—are in accord with the inference that Rad26 plays a role in the promotion of transcription through MMS-induced DNA lesions. Transcription is affected to a lesser degree in the rad26Δ or mag1Δ rad14Δ strain than in the mag1Δ rad14Δ rad26Δ strain, because lesions can still be removed by the NER and BER systems in the rad26Δ strain and Rad26 can still promote the passage of Pol II through lesions in the mag1Δ rad14Δ strain, although these lesions cannot be removed. In the mag1Δ rad14Δ rad26Δ triple mutant strain, however, DNA lesions persist and transcription will continue to stall at these lesion sites.
Lesion bypass by Pol II.
Transcription by RNA polymerases is not as accurate as DNA synthesis by replicative polymerases. In contrast to the high fidelity of nucleotide incorporation by the prokaryotic and eukaryotic replicative DNA polymerases, RNA polymerases incorporate an incorrect nucleotide with frequencies of 10−3 to 10−5 (24). Also, in contrast to the replicative DNA polymerase, E. coli RNA polymerase can carry out efficient bypass of lesions such as O6-methyl guanine (m6G), 8-oxoG, and abasic sites located on the transcribed strand of a duplex DNA, and it incorporates a uracil opposite m6G, either an adenine or cytosine opposite 8-oxoG, and an adenine opposite abasic sites (29, 30). T7 RNA polymerase also transcribes through an 8-oxoG lesion present on the template strand by incorporating an adenine or cytosine opposite the lesion (2). In contrast to the efficient bypass of various damaged bases, E. coli RNA polymerase stalls at cyclobutane pyrimidine dimers (CPD) and has to be displaced by the Mfd protein, which subsequently recruits the NER proteins to the dimer site (22). A CPD, then, is more inhibitory to E. coli RNA polymerase than are the various base damages.
Like the prokaryotic RNA polymerases, eukaryotic Pol II is also unable to transcribe through CPDs, and it stalls at such lesion sites (19). Similar to Mfd in E. coli, the Rad26 and CSB proteins may be crucial for the removal of stalled Pol II from dimer sites. However, the Rad26 and CSB factors may enable Pol II to transcribe through various base damages, including oxidative DNA lesions such as 8-oxoG and TG, and such transcriptional lesion bypass may additionally require TFIIH and XPG.
Other considerations.
NER is the primary mechanism available in eukaryotes for the repair of UV-induced DNA lesions, such as CPDs and (6-4) photoproducts, and for the removal of other distorting DNA lesions, such as intrastrand and interstrand cross-links and bulky adducts formed upon treatment with various chemical agents (14, 17). NER, however, also functions in the removal of a variety of damaged bases, where it competes with a diverse array of BER pathways. In vitro studies with human cell extracts have indicated that NER can remove a variety of damaged bases, including N6-methyladenine, m6G, 8-oxoG, and TG, and abasic sites (5, 16). Genetic studies of yeast have provided in vivo evidence of the involvement of NER in the removal of abasic sites (23) and 8-oxoG lesions (18), and the genetic evidence presented here supports the hypothesis that NER plays a role in the removal of damaged bases such as 3meA and 7meG.
Consistent with the involvement of NER in the removal of a variety of damaged bases and abasic sites, xeroderma pigmentosum patients exhibit an increase in the frequency of internal cancers, particularly those in the brain and other parts of the central nervous system (6, 7). Our observation that Rad26 functions independently of NER or BER in promoting transcription through damaged bases suggests that the developmental problems of CS patients would worsen in the absence of NER. Although there are no known cases of such double mutations in humans, mice lacking both the XPA and CSB genes display severe growth retardation, ataxia, and motor dysfunction during early postnatal development, and they die before weaning (11). In contrast, XPA-deficient mice do not show any obvious developmental or neurological abnormalities (12), and CSB-deficient mice exhibit only mild growth and neurological abnormalities (25). Our findings for yeast provide an explanation for the more severe developmental and neurological defects seen in XPA−/− CSB−/− mice than in mice with single mutations, as they predict that in the absence of CSB, transcription would stall at the sites of naturally occurring base damages and at abasic sites that would accumulate in DNA in the absence of XPA. The stalling of transcription at such lesion sites would then be the cause of developmental defects in XPA−/− CSB−/− mice.
TABLE 2.
Levels of GAL7 mRNA in rad14Δ, mag1Δ, mag1Δ rad14Δ, rad26Δ, and mag1Δ rad14Δ rad26Δ mutant strains in the absence and presence of MMS
| Time (min)b |
GAL7 mRNA level in strain with indicated deletiona
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
rad14Δ
|
mag1Δ
|
mag1Δ rad14Δ
|
rad26Δ
|
mag1Δ rad14Δ rad26Δ
|
||||||
| − | + | − | + | − | + | − | + | − | + | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 86 | 97 | 65 | 100 | 61 | 69 | 30 | 24 | 52 | 14 |
| 40 | 101 | 100 | 100 | 107 | 97 | 77 | 56 | 45 | 55 | 29 |
| 60 | 93 | 101 | 120 | 122 | 101 | 77 | 74 | 55 | 73 | 33 |
| 80 | 110 | 95 | 119 | 118 | 96 | 73 | 73 | 59 | 72 | 37 |
mRNA levels in the mutant strains are given relative to the mRNA levels in the wild-type strain times 100. −, MMS absent; +, MMS present.
Time point of sample removal.
TABLE 3.
Levels of GAL10 mRNA in rad14Δ, mag1Δ, mag1Δ rad14Δ, rad26Δ, and mag1Δ rad14Δ rad26Δ mutant strains in the absence and presence of MMS
| Time (min)b |
GAL10 mRNA level in strain with indicated deletiona
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
rad14Δ
|
mag1Δ
|
mag1Δ rad14Δ
|
rad26Δ
|
mag1Δ rad14Δ rad26Δ
|
||||||
| − | + | − | + | − | + | − | + | − | + | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 67 | 91 | 33 | 94 | 50 | 59 | 17 | 29 | 33 | 22 |
| 40 | 98 | 91 | 79 | 86 | 88 | 58 | 38 | 31 | 43 | 18 |
| 60 | 100 | 88 | 104 | 89 | 100 | 52 | 57 | 36 | 58 | 16 |
| 80 | 119 | 87 | 116 | 87 | 100 | 51 | 70 | 39 | 70 | 18 |
mRNA levels in the mutant strains are given relative to the mRNA levels in the wild-type strain times 100. −, MMS absent; +, MMS present.
Time point of sample removal.
Acknowledgments
This work was supported by National Institutes of Health grants CA35035 and CA41261.
REFERENCES
- 1.Ausubel, F., R. Brent, R. E. Kingston, and D. Moore (ed.). 1993. Current protocols in molecular biology, vol. 2. Preparation of yeast RNA. Wiley Interscience, New York, N.Y. [CD-ROM.]
- 1a.Chen, J., B. Derfler, and L. Samson. 1990. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 9:4569-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, Y.-H., and D. F. Bogenhagen. 1993. Effects of DNA lesions on transcription elongation by T7 RNA polymerase. J. Biol. Chem. 268:5849-5855. [PubMed] [Google Scholar]
- 3.Cooper, P. K., T. Nouspikel, S. G. Clarkson, and S. A. Leadon. 1997. Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science 275:990-993. [DOI] [PubMed] [Google Scholar]
- 4.Guzder, S. N., Y. Habraken, P. Sung, L. Prakash, and S. Prakash. 1996. RAD26, the yeast homolog of human Cockayne's syndrome group B gene, encodes a DNA dependent ATPase. J. Biol. Chem. 271:18314-18317. [DOI] [PubMed] [Google Scholar]
- 5.Huang, J.-C., D. S. Hsu, A. Kazantsev, and A. Sancar. 1994. Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc. Natl. Acad. Sci. USA 91:12213-12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kraemer, K. H., L. L. Lee, and J. Scotto. 1984. DNA repair protects against cutaneous and internal neoplasia: evidence from xeroderma pigmentosum. Carcinogenesis 5:511-514. [DOI] [PubMed] [Google Scholar]
- 7.Kraemer, K. H., M.-M. Lee, A. D. Andrews, and W. C. Lambert. 1994. The role of sunlight and DNA repair in melanoma and non-melanoma skin cancer. Arch. Dermatol. 130:1018-1021. [PubMed] [Google Scholar]
- 8.Lee, S.-K., S.-L. Yu, L. Prakash, and S. Prakash. 2001. Requirement for yeast RAD26, a homolog of the human CSB gene, in elongation by RNA polymerase II. Mol. Cell. Biol. 21:8651-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Page, F., E. E. Kwoh, A. Avrutskaya, A. Gentil, S. A. Leadon, A. Sarasin, and P. K. Cooper. 2000. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell 101:159-171. [DOI] [PubMed] [Google Scholar]
- 10.Mellon, I., G. Spivak, and P. C. Hanawalt. 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51:241-249. [DOI] [PubMed] [Google Scholar]
- 11.Murai, M., Y. Enokido, N. Inamura, M. Yoshino, Y. Kakatsu, G. T. J. van der Horst, J. H. J. Hoeijmakers, K. Tanaka, and H. Hatanaka. 2001. Early postnatal ataxia and abnormal cerebellar development in mice lacking xeroderma pigmentosum group A and Cockayne syndrome group B DNA repair genes. Proc. Natl. Acad. Sci. USA 98:13379-13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakane, H., S. Takeuchi, S. Yuba, M. Saijo, Y. Nakatsu, H. Murai, Y. Nakatsuru, T. Ishikawa, S. Hirota, Y. Kitamura, Y. Kato, Y. Tsunoda, H. Miyachi, T. Horio, T. Tokunaga, T. Matsunaga, O. Nikaido, Y. Nishimune, Y. Okada, and K. Tanaka. 1995. High incidence of ultraviolet-B- or chemical-carcinogen-induced skin tumours in mice lacking the xeroderma pigmentosum group A gene. Nature 377:165-168. [DOI] [PubMed] [Google Scholar]
- 13.Nance, M. A., and S. A. Berry. 1992. Cockayne syndrome: review of 140 cases. Am. J. Med. Genet. 42:68-84. [DOI] [PubMed] [Google Scholar]
- 14.Prakash, S., and L. Prakash. 2000. Nucleotide excision repair in yeast. Mutat. Res. 451:13-24. [DOI] [PubMed] [Google Scholar]
- 15.Reagan, M. S., and E. C. Friedberg. 1997. Recovery of RNA polymerase II synthesis following DNA damage in mutants of Saccharomyces cerevisiae defective in nucleotide excision repair. Nucleic Acids Res. 25:4257-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reardon, J. T., T. Bessho, H. C. Kung, P. H. Bolton, and A. Sancar. 1997. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. USA 94:9463-9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43-81. [DOI] [PubMed] [Google Scholar]
- 18.Scott, A. D., M. Neishabury, D. H. Jones, S. H. Reed, S. Boiteux, and R. Waters. 1999. Spontaneous mutation, oxidative DNA damage, and the roles of base and nucleotide excision repair in the yeast Saccharomyces. Yeast 15:205-218. [DOI] [PubMed] [Google Scholar]
- 19.Selby, C. P., R. Drapkin, D. Reinberg, and A. Sancar. 1997. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 25:787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selby, C. P., and A. Sancar. 1997. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. USA 94:11205-11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selby, C. P., and A. Sancar. 1997. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem. 272:1885-1890. [DOI] [PubMed] [Google Scholar]
- 22.Selby, C. P., and A. Sancar. 1993. Molecular mechanism of transcription-repair coupling. Science 260:53-58. [DOI] [PubMed] [Google Scholar]
- 23.Torres-Ramos, C. A., R. E. Johnson, L. Prakash, and S. Prakash. 2000. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol. Cell. Biol. 20:3522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uptain, S. M., C. M. Kane, and M. J. Chamberlain. 1997. Basic mechanisms of transcript elongation and its regulation. Annu. Rev.Biochem. 66:117-172. [DOI] [PubMed] [Google Scholar]
- 25.van der Horst, G. T. J., H. van Steeg, R. J. W. Berg, A. J. van Gool, J. de Wit, G. Weeda, H. Morreau, R. B. Beems, C. F. van Kreijl, F. R. de Gruijl, D. Bootsma, and J. H. J. Hoeijmakers. 1997. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell 89:425-435. [DOI] [PubMed] [Google Scholar]
- 26.van Gool, A. J., R. Verhage, S. M. A. Swagemakers, P. van de Putte, J. Brouwer, C. Troelstra, D. Bootsma, and J. H. J. Hoeijmakers. 1994. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 13:5361-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Hoffen, A., A. T. Natarajan, L. V. Mayne, A. A. van Zeeland, L. H. F. Mullenders, and J. Venema. 1993. Deficient repair of the transcribed strand of active genes in Cockayne's syndrome cells. Nucleic Acids Res. 21:5890-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verhage, R. A., A. J. van Gool, N. de Groot, J. H. J. Hoeijmakers, P. van de Putte, and J. Brouwer. 1996. Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol. Cell. Biol. 16:496-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viswanathan, A., and P. W. Doetsch. 1998. Effects of nonbulky DNA base damages on Escherichia coli RNA polymerase-mediated elongation and promoter clearance. J. Biol. Chem. 273:21276-21281. [DOI] [PubMed] [Google Scholar]
- 30.Zhou, W., and P. W. Doetsch. 1993. Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases. Proc. Natl. Acad. Sci. USA 90:6601-6605. [DOI] [PMC free article] [PubMed] [Google Scholar]