Abstract
Transcriptional silencing at the budding yeast silent mating type (HM) loci and telomeric DNA regions requires Sir2, a conserved NAD-dependent histone deacetylase, Sir3, Sir4, histones H3 and H4, and several DNA-binding proteins. Silencing at the yeast ribosomal DNA (rDNA) repeats requires a complex containing Sir2, Net1, and Cdc14. Here we show that the native Sir2/Sir4 complex is composed solely of Sir2 and Sir4 and that native Sir3 is not associated with other proteins. We further show that the initial binding of the Sir2/Sir4 complex to DNA sites that nucleate silencing, accompanied by partial Sir2-dependent histone deacetylation, occurs independently of Sir3 and is likely to be the first step in assembly of silent chromatin at the HM loci and telomeres. The enzymatic activity of Sir2 is not required for this initial binding, but is required for the association of silencing proteins with regions distal from nucleation sites. At the rDNA repeats, we show that histone H3 and H4 tails are required for silencing and rDNA-associated H4 is hypoacetylated in a Sir2-dependent manner. However, the binding of Sir2 to rDNA is independent of its histone deacetylase activity. Together, these results support a stepwise model for the assembly of silent chromatin domains in Saccharomyces cerevisiae.
The packaging of DNA into chromatin appears to play a central role in the regulation of nearly every aspect of gene transcription. In particular, an increasing number of complexes associated with the activation or repression of transcription have been shown to contain chromatin remodeling activity or enzymes that covalently modify histones or both (25, 30). The silent mating type loci (HML and HMR) and telomeric DNA regions in the budding yeast Saccharomyces cerevisiae are packaged into a specialized chromatin structure that silences transcription and, like heterochromatin in metazoans, is epigenetically inherited (29; reviewed in reference 19). Moreover, a distinct example of silencing occurs at the highly repetitive yeast ribosomal DNA genes (rDNA) where RNA polymerase II reporter genes inserted within the repeats are inactivated (8, 13, 53). Silent chromatin is also associated with DNA regions that are involved in chromosome maintenance and transmission, such as telomeres in budding yeast and centromeres in many other eukaryotes. In these regions, silencing appears to play a structural role that is independent of transcriptional repression (1).
Silencing at each of the above loci requires Sir2, a highly conserved protein which possesses an intrinsic NAD-dependent protein and histone deacetylation activity (2, 8, 13, 24, 31, 36, 48, 53, 54). Sir2 is an unusual deacetylase in that it couples deacetylation to the hydrolysis of a high-energy bond in NAD and transfers the acetyl group from its protein substrate to ADP-ribose to generate a novel compound, 2′,3′-O-acetyl-ADP-ribose (35, 49, 59, 61). Mutations that abolish the in vitro activity of Sir2 result in a complete loss of silencing in vivo, suggesting that direct deacetylation and/or another aspect of this activity is required for silencing (24, 60, 61).
Genetic and biochemical evidence suggests that Sir2 is a component of two distinct complexes that carry out its telomeric/mating type and rDNA-silencing activities, respectively (16, 40-42, 57). Sir2 binds to Sir4 affinity columns and coimmunoprecipitates with Sir4 from yeast extracts (40, 41, 55). Although Sir2 or Sir4 immunoprecipitates contain only trace amounts of Sir3 (41), Sir3 and Sir4 interact in two-hybrid assays (42), and truncations of the Sir4 protein can associate efficiently with Sir3 (41), suggesting that the three proteins interact physically at some stage during the assembly of telomeric/HM silent chromatin (22, 23, 41, 42). At the rDNA repeats, Sir2 assembles with a different set of proteins into a second complex called RENT (regulator of nucleolar silencing and telophase exit) (52, 57). In addition to Sir2, RENT contains at least two other proteins, Net1 and Cdc14 (52, 57), and Net1 is required for the localization of Sir2 to rDNA and for rDNA silencing (57). Despite their central role in gene silencing, neither Sir2-containing complex has been purified to homogeneity. In particular, the molecular composition of the complex containing the Sir2 and Sir4 proteins or the putative Sir3 complex has not been defined.
The amino termini of histones H3 and H4 play an essential role in silencing at the HM loci and telomeres (2, 29). At these loci, the H3 and H4 N termini are fully hypoacetylated, and a number of studies suggest that this hypoacetylated state is critical for silencing and provides a binding site for the Sir3 and Sir4 proteins (6, 7, 22, 26, 58). The discovery of histone deacetylation activity in Sir2 provides a direct link between Sir2-containing silencing complexes and the hypoacetylated state of histone tails in silent chromatin domains (24, 36, 54). However, the idea that Sir2 deacetylates histones in vivo has not been tested. Furthermore, while a strong genetic link exists between silencing and histone hypoacetylation at HM loci and telomeres, the role of histone tails in rDNA silencing is unknown. Thus, it is unclear whether Sir2 activity at rDNA is required to deacetylate histones or other proteins.
Little is known about how the Sir proteins assemble onto chromatin and what role, if any, each Sir protein might have in the initial nucleation, stable association, and spreading of silent domains. Immunoprecipitation of chromatin from in vivo cross-linked cells has shown that Sir2 and its associated proteins are structural components of silent chromatin domains (17, 23, 57). Moreover, the association of each Sir2, Sir3, and Sir4 with extended silent chromatin regions at the HM loci and telomeres is disrupted in cells that carry a single deletion of either gene, suggesting that the three proteins are recruited to chromatin in a cooperative fashion or as components of a single complex (55). However, these studies do not distinguish between interactions that may be involved in nucleation of silent chromatin domains from those that may be required for the spreading of silencing proteins away from nucleation sites.
To gain better insight into the nature of the yeast silencing complexes, we purified each of the Sir proteins to near homogeneity using a tandem-affinity purification (TAP) approach. Despite their large apparent sizes, the composition of the purified complexes is surprisingly simple. A large complex of approximately 700 kDa, purified using affinity tags on Sir4, is composed of Sir2 and Sir4. A smaller complex of approximately 450 kDa, purified using tagged Sir3, is composed primarily of Sir3. Having defined the composition of these complexes, we determined the requirement for individual silencing proteins and the NAD-dependent deacetylase activity of Sir2 for assembly of each of the above proteins on chromatin. Our results show that the enzymatic activity of Sir2 is not absolutely required for the association of the Sir proteins with DNA sites that initiate silencing at the HM loci and telomeres or for the binding of Sir2 itself to rDNA. However, at the HM loci and telomeres, Sir2 and it enzymatic activity are required for the efficient association of the Sir proteins with DNA regions that are distal from nucleation sites.
Furthermore, by testing the requirement for each Sir protein in the assembly of silencing complexes on chromatin, we found that the Sir2 and Sir4 proteins could partially associate with silencers and DNA regions immediately adjacent to telomeric repeats, independently of Sir3. These results define independent steps in assembly of the Sir proteins at sites that nucleate the initiation of silent chromatin and suggest that Sir4 is the most upstream protein in the assembly pathway. Finally, we show that histone H4 is hypoacetylated in silent chromatin domains in a Sir2 activity-dependent manner and that, similar to what has been described previously for the HM loci and telomeres, the N termini of histones H3 and H4 play a critical role in rDNA silencing.
MATERIALS AND METHODS
Plasmids and yeast strains.
pDM271 contains an EagI-HindIII fragment corresponding to the region coding for green fluorescent protein (GFP) in a pRS304 backbone. To create SIR2-GFP alleles, a portion of the SIR2 open reading frame was PCR amplified from plasmids that contained either wild-type SIR2 or sir2-H364Y. The PCR products were digested with SacI (cuts uniquely in SIR2) and EagI (introduced by PCR) and fused in frame to the N terminus of GFP in plasmid pDM271 to create plasmids pJT18 (SIR2-GFP) and pJT19 (sir2-H364Y-GFP). pDM267 was constructed by subcloning a PCR-generated Asp718-EagI fragment containing the C-terminal 150 amino acids of Sir3 and an EagI-HindIII fragment containing GFP(S65T, V167A) into the Asp718- and HindIII-digested Yplac111, a yeast LEU2-marked integrating vector. A SacI-HindIII fragment containing the SIR3-GFP fragment from pDM267 was subcloned into pMR52, a derivative of pRS306 containing the ACT1 terminator, to generate pDM272 (SIR3-GFP-URA3). pDM274 was constructed by subcloning a PCR-generated GFP fragment into the EcoRI site of pDM158 to replace the six-histidine-three hemagglutinin (6HIS-3HA) tag with GFP. pDM158 contains the entire SIR4 open reading frame fused to 6HIS-3HA. pSIR2-LEU2, pSIR2-H364Y-LEU2, and pSIR2-G262A-LEU2 have been described previously (61). pDM554 (psir4Δ::LEU2) was a gift from A. Kahana and Dan Gottschling.
pDM311 (HIS3-mURA3::rDNA) was constructed by ligation of a 1-kb EcoRI-XhoI fragment containing a portion of the rDNA nontranscribed spacer region and an EcoRI-EagI fragment containing the mURA gene (53) into pRS303 digested with XhoI and EagI.
Yeast strains used in this study are listed in Table 1. Transformations of all yeast strains were performed by the lithium chloride method (21). To create SIR2-GFP strains, plasmids pJT18 and pJT19 were integrated at the SIR2 locus in strain BJ5459 after digestion with BglII. This yielded strains DMY1247 (SIR2-GFP) and DMY1249 (sir2-H364Y-GFP). To create the SIR3-GFP strains used in the experiments shown in Fig. 5, plasmid pDM272 was integrated at the SIR3 locus in strain JRY3433 after digestion with XhoI. This yielded strain DMY1943. This strain was then transformed with plasmid pSIR2-LEU2 or pSIR2-H364Y-LEU2, generating strains DMY2165 and DMY2166, respectively.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| SF1 | W303-1a, MATaade2-1 can1-100 his3-11 leu2-3,112 trp1 ura3-1 | R. Rothstein |
| SF3 | SF1 with sir2Δ::HIS3 | J. Rine |
| SF4 | SF1 with sir3Δ::TRP1 | J. Rine |
| SF5 | SF1 with sir4Δ::HIS3 | J. Rine |
| SF10 | BJ5459, MATaura3-52 trp1 lys2-801 leu2 Δ1 pep4Δ::HIS3 prb1Δ1.6R can1 | E. Jones |
| DMY1864 | SF3 with pSIR2-LEU2 | This work |
| DMY1865 | SF3 with pRS315 | This work |
| DMY1866 | SF3 with pSIR2-H364Y-LEU2 | This work |
| DMY1867 | SF3 with pSIR2-G262A-LEU2 | This work |
| DMY1247 | SF10 with SIR2-GFP::URA3 | This work |
| DMY1249 | SF10 with sir2-H364Y-GFP::URA3 | This work |
| DMY1942 | SF3 with SIR3-GFP::URA3 | This work |
| DMY1944 | SF3 with GFP-SIR4::URA3 | This work |
| DMY2165 | DMY1942 with pSIR2-LEU2 | This work |
| DMY2166 | DMY1942 with pSIR2-H364Y-LEU2 | This work |
| DMY2167 | DMY1944 with pSIR2-LEU2 | This work |
| DMY2168 | DMY1944 with pSIR2-H364Y-LEU2 | This work |
| DMY1928 | MATα ade2 his3 leu2 trp1 ura3 lys2 dam::LYS2 hht2,hhf2::HIS3 hht1,hhf1::LEU2 pRM200(CEN/ARS HHT2 HHF2-TRP1) | M. Grunstein |
| DMY2129 | DMY1928 with sir2Δ::kanr | This work |
| DMY1493 | MATα ade2 his3 leu2 trp1 ura3 lys2 dam::LYS2 hht2,hhf2::HIS3 hht1,hhf1::LEU2 pRM200 (CEN/ARS hht2Δ4-30 HHF2-TRP1) | M. Grunstein |
| DMY1503 | MATα ade2 his3 leu2 trp1 ura3 lys2 dam::LYS2 hht2,hhf2::HIS3 hht1,hhf1::LEU2 pYXL409 (CEN/ARS HHT2 hhf2K16Q-TRP1) | M. Grunstein |
| DMY2137 | DMY1928 with mURA3::rDNA | This work |
| DMY2139 | DMY1928 with mURA3::HIS3 | This work |
| DMY2141 | DMY2129 with mURA3::rDNA | This work |
| DMY2143 | DMY2129 with mURA3::HIS3 | This work |
| DMY2145 | DMY1493 with mURA3::rDNA | This work |
| DMY2147 | DMY1493 with mURA3::HIS3 | This work |
| DMY2149 | DMY1503 with mURA3::rDNA | This work |
| DMY2151 | DMY1503 with mURA3::HIS3 | This work |
| DMY1704 | SF10 with SIR4::TAP::TRP1Kl | This work |
| DMY1737 | SF10 with SIR3::TAP::TRP1Kl | This work |
FIG. 5.
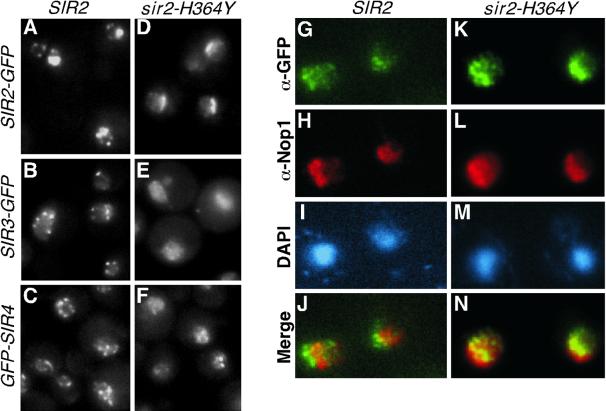
Enzymatic activity of Sir2 is required for localization of Sir2, Sir3, and Sir4 proteins to telomeric foci but not for localization of Sir2 to the nucleolus. The localization of Sir2-GFP, Sir3-GFP, and Sir4-GFP in cells containing either wild-type SIR2 (A to C, DMY1247, DMY2165, and DMY2167, respectively) or the enzymatically inactive sir2-H364Y (D to F, DMY1249, DMY2166, and DMY2168, respectively) is shown. Panels A and D show the localization of Sir2-GFP and sir2-H364Y-GFP, respectively. Colocalization of Sir2-GFP (G to J, DMY1247) and sir2-H363Y-GFP (K to N, DMY2164) with the nucleolar marker Nop1 confirms that Sir2-H364Y-GFP is localized to the nucleolus. See Table 1 for strains.
To create the GFP-SIR4 strains, plasmid pDM274 was integrated at the SIR4 locus in strain JRY3433 after digestion with SphI. This yielded strain DMY1944, which was then transformed with plasmid pSIR2-LEU2 or pSIR2-H364Y-LEU2 to generate strains DMY2167 and DMY2168, respectively. rDNA silencing strains (see Fig. 4C) were made by transforming plasmid pDM311 cut with either SmaI or NheI for integration into RDN1 and HIS3, respectively. Construction of sir2-H364Y and sir2-G262A has been described previously (61). DMY1706 and DMY1737 were constructed by integration of the TAP tag immediately before the stop codon of the SIR3 and SIR4 genes, respectively, as described previously (47). Correct integration was confirmed by PCR.
FIG. 4.
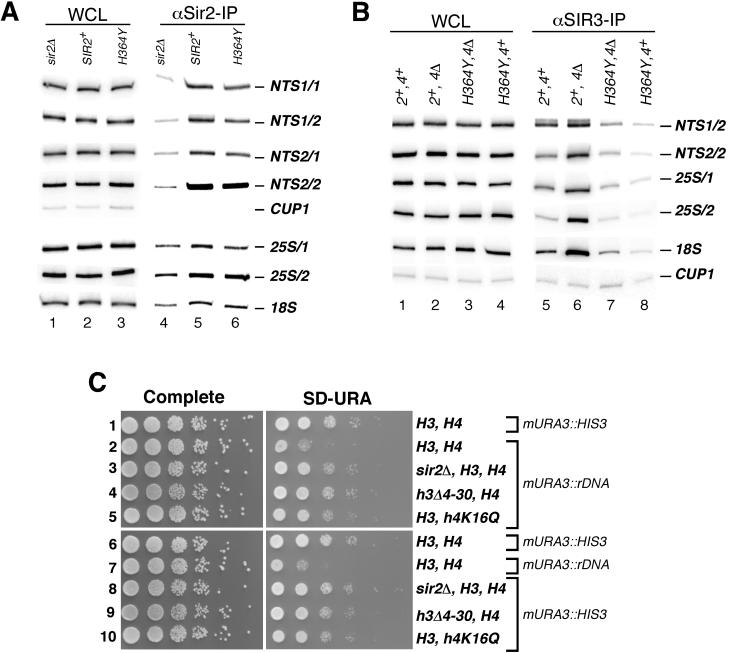
Requirement for the enzymatic activity of Sir2 in localization of Sir2 and Sir3 to rDNA and the role of histones H3 and H4 in rDNA silencing. (A) Chromatin immunoprecipitation was carried out from sir2Δ, SIR2+, and sir2-H364Y strains using an anti-Sir2 antibody. See Fig. 3 legend for strain names. (B) Chromatin immunoprecipitation experiments showing that the association of Sir3 with rDNA requires the enzymatic activity of Sir2. Association of Sir3 with rDNA fragments in sir2Δ, SIR2+, and sir2-H364Y strains carrying either wild-type SIR4 or sir4Δ is shown. (C) Loss of rDNA silencing in histone H3 and H4 mutants. Tenfold serial dilutions of wild-type or histone mutant strains were plated on complete medium or medium lacking uracil (SD−Ura), and plates were photographed after 2 to 3 days of growth at 30°C.
Purification of Sir proteins.
Sir3-TAP was purified from 10 to 20 g of yeast cells lysed by grinding with dry ice in a coffee grinder for 5 min, followed by grinding under liquid nitrogen with a mortar and pestle for 20 min. The yeast powder was resuspended in 2 volumes of lysis buffer (10 mM Tris [pH 7.5], 500 mM NaCl, 50 mM NaF, 50 mM sodium β-glycerophosphate [pH 7.4], 1 mM EDTA, 1 mM EGTA, 1 mM NaVO4, 5% glycerol, 0.5% NP-40, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], and one tablet of protease inhibitor cocktail [Roche] per 50 ml of lysis buffer), stirred for 30 min on ice, and then centrifuged at 30,000 × g for 15 min, and 100,000 × g for 1 h. Sir3-TAP was purified from the resultant 40 ml of 12-mg/ml clarified extract as previously described except that tobacco etch virus cleavage was performed at 4°C overnight (47).
After purification, 5% of each fraction was separated on a polyacrylamide gel and silver stained. Purified Sir3-calmodulin-binding peptide (CBP) (20% of fraction 3; Fig. 1A) and 3 mg of the starting clarified extract were diluted into 0.5 ml of SC buffer (20 mM Tris [pH 8.0], 150 mM NaCl, 1 mM EDTA, 5% glycerol, 0.1% NP-40, 1 mM DTT, 0.1 mM PMSF, and 2 μg of purified glutathione S-transferase [GST] per ml for the column run of purified Sir3-CBP) and injected onto a Superose 6 (Pharmacia) sizing column that had been equilibrated in SC buffer. Then 1-ml fractions were collected and either diluted into 2× sample buffer (extract) or precipitated with trichloroacetic acid (purified), and 1% of the extract fractions and 25% of the purified fractions were blotted with an anti-Sir3 antibody (1:2,500). Similar results were obtained when Sir3-TAP was purified using lower ionic strength buffers (≈150 mM NaCl), but the background of nonspecific proteins in these experiments was higher.
FIG. 1.
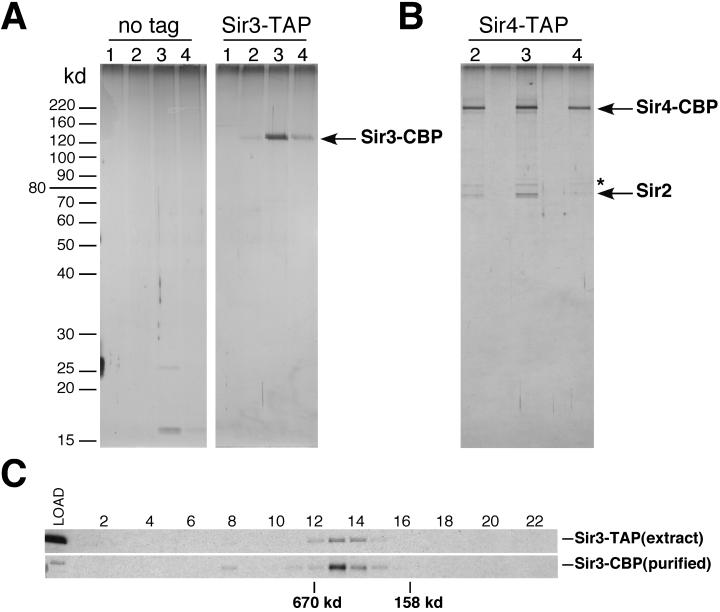
Purification of Sir3 and Sir2/Sir4 from yeast extracts. Silver stained SDS-polyacrylamide gels showing the purification of TAP-tagged Sir3 (A) and Sir4 (B) from strains DMY1737 and DMY1704, respectively. The left panel in A shows a control purification from the parental strain lacking the TAP tag (SF10). Proteins were identified by mass spectrometry. The band marked with an asterisk (∗) in B was identified as Ssb1, a yeast Hsp70 protein, which is a common contaminant of TAP purifications. Lanes 1 to 4 show successive elution fractions from calmodulin-Sepharose, the second column used in the purifications. (C) Western blots showing the migration of Sir3 in a Superose 6 gel filtration column in crude yeast extracts (Sir3-TAP, top panel) and after purification on IgG-Sepharose and calmodulin-Sepharose (Sir3-CBP, top panel, same material as in A, lane 3). Molecular size markers used for calibration of the sizing column were thyroglobulin (670 kDa) and aldolase (158 kDa).
Sir4-TAP was purified under similar conditions from 50 to 100 g of yeast cells resuspended in 1 to 2 volumes of lysis buffer (50 mM HEPES [pH 7.6], 100 mM NaCl, 50 mM NaF, 80 mM β-glycerophosphate, 1 mM EDTA, 1 mM EGTA, 5% glycerol, 0.1% NP-40, 10 mM β-mercaptoethanol, 1 mM PMSF, and one tablet of protease inhibitor cocktail per 50 ml of lysis buffer) and lysed either by grinding under liquid nitrogen or by agitation with glass beads using a BioSpec bead beater.
Protein identification: in-gel trypsin digestion, nanoscale microcapillary liquid chromatography-mass spectrometry, and computer database spectral matching.
An adaptation of previous methods was used for protein identification (45). Briefly, silver-stained gels were dried in a cellophane sandwich without heat prior to analysis. Bands selected for identification were excised from the dried gel with a scalpel and rehydrated with purified water. The cellophane was removed, followed by in-gel digestion with trypsin as previously described (50). Digested samples were pressure loaded onto a fused silica microcapillary C18 column (Magic; Michrom BioResources, Auburn, Calif.) prepared in-house (75 μm inner diameter, 9.5 cm long). An Agilent 1100 high-pressure liquid chromatography (HPLC) system (Agilent Technologies, Palo Alto, Calif.) was used to deliver a gradient across a flow splitter to the column over 40 min. The column eluant was directed into an LCQ-DecaXP electrospray ion-trap mass spectrometer (ThermoFinnigan, San Jose, Calif.), and eluting peptides were dynamically selected for fragmentation by the operating software. The peptide fragmentation spectra were data searched against the nonredundant protein database using the Sequest computer algorithm (10). We typically obtained 25 to 50% sequence coverage for each protein.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was performed as described previously with the following modifications (33, 55, 58): 100 ml of yeast cells were grown to an optical density at 660 nm (OD660) of 1.5 and cross-linked with 1% formaldehyde for 15 to 20 min at room temperature. Cross-linking was stopped, cells were washed and resuspended in lysis buffer (50 mM HEPES-KOH [pH 7.5], 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 0.1% sodium deoxycholate, 1 mM PMSF, protease inhibitor cocktail) at 5 × 109 cells per 500 μl. Lysis was performed using zirconia-silica beads (BioSpec) in a mini-bead beater (BioSpec) with 4 pulses of 90 s followed by 90 s on ice. The lysate was sonicated three times at 4°C for 20 s using a Branson digital sonifier 450 equipped with a microtip to generate a mean DNA size of 0.1 to 1 kb. After centrifugation, the whole-cell lysate (WCL) was frozen in liquid nitrogen for storage.
Immunoprecipitations were carried out with either 1 μl of rabbit anti-Sir2 antibody, 0.5 μl of rabbit anti-Sir3 antibody (60), or 1 μl of anti-Sir4 antibody per 150 μl of WCL or 1.5 μl of anti-acetyl-H4 per 50 μl of WCL (Upstate Biotechnology) overnight at 4°C. Immune complexes were collected and washed, cross-links were reversed, and DNA was precipitated as previously described. PCRs were carried out in a volume of 12.5 μl with 1/50 to 1/200 of the immunoprecipitated material and 1/12,500 to 1/25,000 of input material. The reaction was carried out with 0.5 μM primers, 0.1 mM deoxynucleoside triphosphates, and 0.1 mCi of [α-32P]dCTP (specific activity, 3,000 Ci/mmol) per ml. The number of cycles was determined in initial experiments using twofold dilutions to ensure linearity of the signal. Cycling parameters were 1 cycle of 95°C for 2 min, 55°C for 30 s, and 72°C for 1 min, followed by cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and a final step at 72°C for 4 min. Typically, 20 cycles were used for rDNA and 26 cycles for telomeres and HM loci. Samples were run on 6 or 8% polyacrylamide gels at 100 V for 1 h. The dried gel was exposed to a Phosphorimager screen and screened on a Bio-Rad Phosphorimager. Quantification was performed using QuantityOne software (Bio-Rad). The sequences of the PCR primers used in the chromatin immunoprecipitation experiments are shown in Table 2.
TABLE 2.
List of PCR primers used in chromatin immunoprecipitation experiments in this study
| Locus | Primer pair |
|---|---|
| TEL0.35 | TAACAAGCGGCTGGACTACTTT and CCTAATAATCACCGTTAAACTCAGC |
| TEL0.6 | CAGGCAGTCCTTTCTATTTC and GCTTGTTAACTCTCCGACAG |
| TEL1.4 | AATGTCTTATCAAGACCGAC and TACAGTCCAGAAATCGCTCC |
| TEL2.8 | CTGATCTGATGTTCTCACGC and TCTGTATGAGTCATCGAAGC |
| TEL4.7 | GGTTCGTGACTACAAAGG and CTACCAACAGATGAGGTC |
| NTS1/1 | CTCCTCCGATATTCCTAC and TGCAAGATGAATAGCCAG |
| NTS1/2 | GCT TCC TAT GCT AAA TCC and AGAAGCAACTAAACGAGG |
| NTS2/1 | GGTAACCCAGTTCCTCAC and CTTTCCTGTTATGGCATGG) |
| NTS2/2 | GCATGAAGTACCTCCCAACT and CGCTTCCGCTTCCGCAGTAA |
| 25S/1 | GACGTCATAGAGGGTGAGAA and TTGACTTACGTCGCAGTCCR |
| 25S/2 | ATTTCACTGGCGCCGAAGCT and TACGGACAAGGGGAATCTGA |
| 18S | TAGAGTGTTCAAAGCAGGCG and CCCAGAACCCAAAGACTTTG |
| HML-E | GGATGGATCTAGGGTTTTATGCC and TTTGGCCCCCGAAATCG |
| HMLα | AGACGGCCAGAAACCTC and TCGCCTACCTTCTTGAAC |
| HMR-E | TGCAAAAACCCATCAACCTGG and ACCAGGAGTACCTGCGCTTA |
| HMRa/MATa | CCATCCGCCGATTTATTTT and CAGTTTCCCCGAAAGAACAA |
| CUP1 | TTTTCCGCTGAACCGTTCCA and CATTGGCACTCATGACCTTC or GAGAAGCAAATAACTCCTTGTC |
| ACT1 | CCAATTGCTCGAGAGATTTC and CATGATACCTTGGTGTCTTG |
| GAL1 | TGCTAGATCGCCTGGTAGAG and GCAAACCTTTCCGGTGCAAG |
Microscopy.
GFP fluorescence was monitored in live cells grown to log phase in synthetic medium containing 22 mM sodium citrate (56). Cells were visualized on a Nikon Eclipse E800 microscope, and images were captured using a monochrome charge-coupled device camera (Princeton Instruments). Images were processed using the Metamorph software. Immunofluorescence experiments were carried out as previously described using a 1:2,000 dilution of rabbit anti-GFP and 1:1,000 dilution of mouse anti-Nop1 (3, 57). Secondary antibodies, fluorescein isothiocyanate-anti-rabbit immunoglobulin (Ig) conjugate and rhodamine-anti-mouse immunoglobulin conjugate, were used at a 1:100 dilution.
Silencing assays.
rDNA silencing assays were performed as previously described (53). Overnight cultures were diluted 1:25 and grown to log phase at 30°C. Three microliters of 10-fold serial dilutions of each culture in water were spotted on yeast extract-peptone-dextrose (YEPD) and SD−Ura plates. Plates were incubated at 30°C for 2 to 3 days and photographed using a Nikon digital camera.
RESULTS
Purification of Sir3 and Sir4 proteins.
In order to define the composition of silencing complexes and identify other proteins that may be associated with the Sir proteins, we used the TAP approach to isolate each of the Sir proteins from yeast extracts. In this approach, two protein A repeats and a CBP, interrupted by the tobacco etch virus protease recognition site, are fused to the protein of interest and used for sequential affinity purification on resins containing immobilized immunoglobulin G and calmodulin, respectively (47).
We constructed yeast strains in which the endogenous copies of SIR2, SIR3, or SIR4 were replaced with modified versions encoding a TAP tag fused to the C terminus of each protein. The tagged proteins were fully functional as assayed for telomeric, mating type, or rDNA silencing. Whole-cell yeast extracts were prepared from strains carrying the tagged proteins and an untagged control strain under mild lysis conditions. Purification from the Sir3-TAP strain yielded a single doublet of approximately 130 kDa, which was identified by mass spectrometry analysis as Sir3 (Fig. 1A). Under our purification conditions, nearly no contaminating proteins were present in the final purified fractions after elution from the calmodulin column, as shown in the no-tag control purification (Fig. 1A). Occasional contaminants, such as the 26- and 17-kDa bands in the control purification, were identified as tobacco etch virus protease and ribosomal protein L12 (Rpl12A) (Fig. 1A, left panel, lane 3). The Sir3-TAP preparation contained substoichiometric amounts of at least one other protein, the possible physiological significance of which is being investigated (A. D. Rudner, unpublished results). However, no other proteins copurified with Sir3 at near stoichiometric ratios, suggesting that under our extraction conditions (0.15 to 0.5 M NaCl and 0.5% NP-40), Sir3 is not present in a complex with other proteins.
Purification from the Sir4-TAP strain yielded two major proteins of approximately 190 and 64 kDa and a minor 70-kDa species (Fig. 1B). Mass spectrometry sequencing identified the 190-kDa species as Sir4 and the doublet at 64 kDa as Sir2. The minor species at 70 kDa was identified as a yeast hsp70 family protein (Ssb1), which we believe is a contaminant (Fig. 1B, denoted by asterisk). Purification of Sir2-TAP resulted in isolation of both Sir4 and components of the RENT complex (Net1 and Cdc14; data not shown). Purified Sir3-TAP and Sir4-TAP were analyzed by gel filtration chromatography and found to migrate with relatively large apparent molecular sizes, ≈450 kDa and ≈700 kDa, respectively, similar to their sizes in yeast extracts (Fig. 1C and data not shown). Together with previous observations, these experiments show that the core telomeric/HM silencing complex in budding yeast is composed of a Sir2/Sir4 heteromer that associates with a Sir3 multimer during assembly on chromatin. These data also suggest that the binding of the Sir2/Sir4 complex to chromatin may be separable from Sir3 binding during silent chromatin assembly.
FIG. 3.
Enzymatic activity of Sir2 is required for association of Sir2, Sir3, and Sir4 proteins with telomeric DNA regions and the HML mating type locus. (A and B) Chromatin immunoprecipitation was carried out from a sir2 deletion strain (sir2Δ), a SIR2 wild-type strain (SIR2+), and strains containing sir2 alleles that encode enzymatically inactive Sir2 proteins (sir2-H364Y and sir2-G262A) with anti-Sir2, anti-Sir3, and anti-Sir4 antibodies. Panels show phosphorimager data of PCR amplifications corresponding to input (WCL) and immunoprecipitated chromatin for the indicated regions of the VI-R telomere and the HML locus. The following strains were used: DMY1865 (sir2Δ), DMY1866 (SIR2+), DMY1865 (sir2-H364Y), and DMY1867 (sir2-G262A). See Fig. 2A for locations of primers.
Requirement for SIR2, SIR3, and SIR4 in assembly of yeast silent chromatin.
To further study the mechanisms of silent chromatin assembly, we performed chromatin immunoprecipitation experiments using affinity-purified antibodies that recognize the Sir2, Sir3, and Sir4 proteins. Our experiments were aimed at studying the interaction of each protein with a number of silent chromatin regions, including silencers (Fig. 2A). Silencers are discrete DNA sites that bind to a combination of DNA-binding proteins that directly recruit the Sir proteins to DNA (12, 51, 63). By investigating the association of silencing proteins with silencers, our experiments distinguish between requirements for initial association with chromatin from requirements for the more extensive association of silencing factors with chromatin that involves their spreading along the chromatin fiber.
FIG. 2.
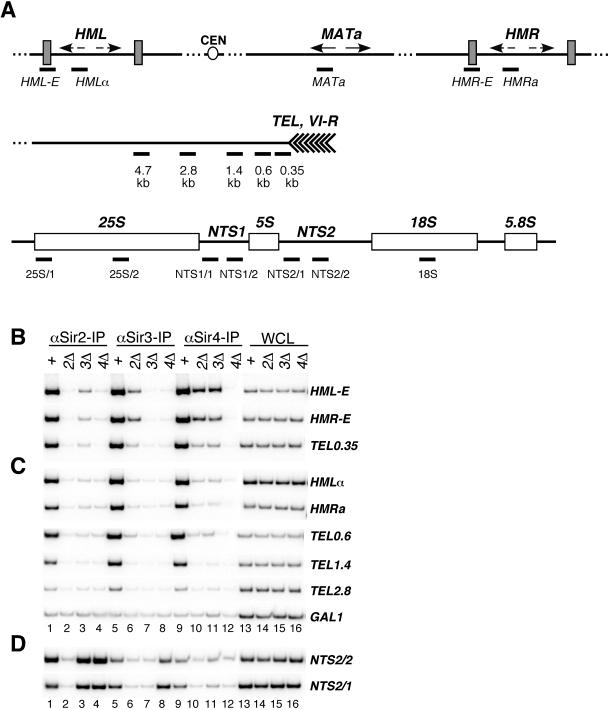
Requirement for SIR2, SIR3, and SIR4 for assembly of silent chromatin. (A) Schematic diagram showing the location of PCR primers corresponding to the HML, HMR, and MATa loci on chromosome III, the subtelomeric region on the right arm of chromosome VI (TEL and VI-R), and the rDNA repeats on chromosome XII used in chromatin immunoprecipitation experiments. The locations of PCR primers are indicated under each region as thick bars. The telomeric primers amplify DNA fragments from 0.35, 0.6, 1.4, 2.8, and 4.7 kb from the telomeric repeats. The HML and HMR primers flank the E silencers (HML-E and HMR-E) or are located within HMLα or HMRa, respectively. The rDNA primers amplify fragments within the nontranscribed spacer regions (NTS1 and NTS2) and the 35S rRNA coding region. Strains used were the wild type (W303-1a) and sir2Δ (SF3), sir3Δ (SF4), and sir4Δ (SF5) mutants. (B) Phosphorimager data of chromatin immunoprecipitation experiments showing the association of the Sir2, Sir3, and Sir4 proteins with silencers (HML-E and HMR-E) and DNA immediately adjacent to telomere VI-R in wild-type (lanes +), sir2Δ (2Δ), sir3Δ (3Δ), and sir4Δ (4Δ) strains. PCR amplifications of anti-Sir2, anti-Sir3, and anti-Sir4 chromatin immunoprecipitations and input DNA from WCL are shown. (C) Association of Sir2, Sir3, and Sir4 in the same genetic backgrounds as in B with silent chromatin regions that are more distal from initiation sites and with a control nonsilenced locus (GAL1). (D) Association of Sir2, Sir3, and Sir4 with rDNA.
The loci investigated in our experiments included DNA regions surrounding the E silencer elements of HML and HMR, regions within each HML and HMR, regions from 0.35 to 2.8 kb from the right telomere of chromosome VI, and DNA regions throughout the rDNA (Fig. 2A). The nonsilenced MAT locus, ACT1, GAL1, and CUP1 were used as controls. The association of each protein with the above regions was assessed by quantification of phosphorimager data of DNA products obtained by PCR amplification using region-specific pairs of primers in the presence of a 32P-labeled nucleotide (23, 33, 58).
Previous chromatin immunoprecipitation experiments have shown that the Sir2, Sir3, and Sir4 proteins are associated with DNA fragments throughout silent chromatin domains (23, 55). We found that Sir2, Sir3, and Sir4 were associated with HMLα and HMRa and with DNA fragments at distances of 0.6, 1.4, and 2.8 kb from the telomere, in agreement with previous results (23, 55) (Fig. 2C, lanes 1, 5, and 9). Neither protein showed a significant association above background with control DNA fragments such as GAL1 (Fig. 2C and data not shown). Also, consistent with previous results (55), the association of each Sir2, Sir3, and Sir4 with HMLα, HMRa, and telomeric DNA fragments from 0.6 to 2.8 kb on the right arm of chromosome VI depended on the presence of a wild-type copy of SIR2, SIR3, and SIR4, as deletion of each gene resulted in a nearly complete loss of binding of the other two proteins to DNA (Fig. 2C, lanes 2 to 4, 5 to 7, and 9 to 12).
However, for DNA fragments that encompassed silencers, we observed significant binding in strains that carried single deletions of each of the SIR genes (Fig. 2B). Specifically, we detected weak binding of Sir2 to both the HML-E and HMR-E silencers in sir3Δ cells but not in sir4Δ cells, and weak binding of Sir3 to the silencers in sir2Δ cells but not in sir4Δ cells (Fig. 2B, lanes 2 to 4 and 6 to 8). Strikingly, Sir4 was significantly associated with silencers in the absence of either Sir2 or Sir3. Similar but weaker association of each protein was observed with a DNA fragment 0.35 kb from the end of chromosome VI (Fig. 2B, bottom panel), the closest chromosome VI-R DNA fragment not including the repetitive Rap1 binding sites (Fig. 2A). Thus, Sir4 can partially associate with silencers in the absence of Sir2 and Sir3. Moreover, partial association of each Sir2 or Sir3 with silencers can occur independently of each other but requires Sir4.
At the rDNA repeats, as reported previously, association of Sir2 with DNA occurred independently of either Sir3 or Sir4 (17) (Fig. 2D, lanes 1 to 4). In sir4Δ cells, the Sir3 protein has been shown to localize to the nucleolus in a Sir2-dependent manner (17). Localization of Sir3 to rDNA can also be detected by chromatin immunoprecipitation and requires Sir2 (Fig. 2D, lanes 5 to 8). However, we also observed low levels of association of Sir3 with rDNA in cells carrying a wild-type copy of SIR4 (Fig. 2D, lane 5); similarly, low levels of Sir4 were associated with rDNA in a Sir2-dependent fashion (Fig. 2D, compare lanes 9 and 10).
Requirement for enzymatic activity of Sir2 in association of Sir proteins with chromatin.
We wished to know whether the requirement for Sir2 in Sir3 and Sir4 binding reflected a requirement for an enzymatic function of Sir2 or a structural one. To distinguish among these possibilities, we analyzed the association of each Sir protein with the DNA fragments shown in Fig. 2A in cells containing either wild-type SIR2 or the sir2-H364Y allele, which encodes an enzymatically inactive Sir2 protein that is still able to associate with Sir4 and Net1 (60). Consistent with our previous observations (60), in cells containing the sir2-H364Y allele, the association of Sir2 with a DNA fragment at 0.6 kb from the telomere was greatly reduced (Fig. 3A, lanes 10 to 12). Moreover, in the H364Y mutant strain, the association of Sir3 and Sir4 with this telomeric DNA fragment and the association of all three proteins with DNA fragments at 1.4 and 2.8 kb from the telomere and with the HML locus was greatly diminished to levels that were the same or slightly higher than that observed in sir2Δ cells (Fig. 3A and B, lanes 10 to 21).
Similar results were obtained with a different catalytic site mutation in Sir2 that converts the conserved glycine 262 to alanine (sir2-G262A) (Fig. 3A and B, lanes 1 to 9, and data not shown). Like sir2-H364Y, this mutation abolishes the enzymatic activity of Sir2 in vitro and all silencing in vivo (61; J. C. Tanny, unpublished observations). In both sir2-H364Y and sir2-G262A cells, higher-than-background levels of Sir2 were associated with DNA fragments encompassing the HML-E silencer or at 0.6 kb from the telomere (Fig. 3A and B, compare lanes 3 and 1 and lanes 12 and 10), suggesting that activity is not absolutely required for initial binding of Sir2 to the silencer. Quantification of the chromatin immunoprecipitation data from at least two independent experiments showed that, on average, the Sir2, Sir3, and Sir4 proteins bound three to five times more DNA at 0.6 to 1.4 kb from the telomere in SIR2 strains compared to sir2Δ and sir2-H364Y strains. Thus, the enzymatic activity of Sir2 is required for the efficient binding of the Sir2, Sir3, and Sir4 proteins to the HM and telomeric silent chromatin regions, especially when these regions are distal from nucleation sites.
In contrast to what we observed for the telomeres and the HM loci (Fig. 3), the Sir2-H364Y protein associated with rDNA with an efficiency close to that of wild-type Sir2 (Fig. 4A, compare lanes 5 and 6). Quantification of the phosphorimager data in Fig. 4A showed that the efficiency of cross-linking for Sir2-H364Y to various rDNA fragments was between 69 and 88% of that of wild-type Sir2. Based on these and previous observations demonstrating that the Net1 and Cdc14 proteins localize to rDNA independently of Sir2 (57; D. Moazed, unpublished observations), we conclude that the RENT complex localizes to rDNA independently of interactions involving Sir2 or its enzymatic activity.
Since enzymatically inactive Sir2 associated with rDNA with wild-type efficiency, we wished to test whether association of Sir2 with rDNA was sufficient for recruitment of Sir3 to rDNA or, alternatively, whether Sir2 activity played a role in Sir3 recruitment. As expected from previous immunofluorescence localization studies (17), deletion of SIR4 resulted in an increase in association of Sir3 with rDNA fragments (Fig. 4B, lanes 5 and 6; also see Fig. 2D, lanes 5 and 8). This association was abolished in both sir2Δ and sir2-H364Y cells (Fig. 4B, compare lane 6 with lanes 7 and 8; Fig. 2D, lane 6). Thus, the enzymatic activity of Sir2, not just its binding to rDNA, is required for the association of Sir3 with rDNA.
The requirement for the enzymatic activity of Sir2 in the localization of Sir3, a histone H3 and H4 binding protein, to rDNA raised the possibility that, similar to their role in telomeric/HM silencing, histone H3 and H4 N termini may be required for rDNA silencing. In order to test this possibility, we assessed the efficiency of rDNA silencing in cells in which the endogenous copies of genes encoding the H3 and H4 histones have been deleted and replaced with H3 and H4 mutants containing either an H3 N-terminal deletion removing amino acids 4 to 30 (h3Δ4-30) or a lysine 16 to glutamine mutation in H4 (h4K16Q). rDNA silencing was assessed using cells in which a modified URA3 reporter gene (mURA3) is inserted within the rDNA locus (53).
In wild-type cells, this reporter is silenced, resulting in loss of growth on medium lacking uracil (Fig. 4C, row 2) (53). In contrast, when the same reporter gene was inserted at a nonsilenced locus (within the HIS3 gene), growth on medium lacking uracil was observed (Fig. 4C, rows 1 and 6). Strains containing a deletion of the H3 tail (h3Δ4-30) or the lysine 16 to glutamine (h4K16Q) mutation in histone H4 each displayed a dramatic loss of rDNA silencing, as evidenced by increased growth on medium lacking uracil (Fig. 4C, compare rows 4 and 5 with row 2). Loss of rDNA silencing in these strains was similar to the loss observed in sir2Δ cells (Fig. 4C, compare rows 4 and 5 with row 3) (53). As additional controls, all strains grew well on complete medium (Fig. 4C, left panels), and the histone mutations had no effect on growth rate on medium lacking uracil when the reporter gene was inserted at the nonsilenced HIS3 locus (Fig. 4C, rows 8 to 10). These results indicate that histone H4-lysine 16 and the N terminus of histone H3 are absolutely required for rDNA silencing.
Enzymatic activity of Sir2 and subnuclear localization of the Sir proteins.
The Sir2, Sir3, and Sir4 proteins are localized to regions near the nuclear periphery that reflect their association with clusters of several telomeres (17, 44). The characteristic telomeric foci corresponding to Sir2-GFP were lost in strains containing the enzymatically inactive Sir2-H364Y-GFP protein (compare Fig. 5A and D). Similarly, the telomeric foci of Sir3-GFP and GFP-Sir4 observed in SIR2 cells (Fig. 5B and C) became dispersed throughout the nucleus in cells containing the sir2-H364Y allele (Fig. 5E and F). However, although Sir4-GFP appeared more dispersed in sir2-H364Y cells, some Sir4-GFP foci remained visible (Fig. 5F), possibly reflecting the partial association of Sir4 with silencers and DNA fragments immediately adjacent to the chromosome end that occurs independently of Sir2 (Fig. 2 and 3). In contrast to loss from telomeric foci, Sir2-H364Y-GFP remained localized to the nucleolus in a fashion similar to wild-type Sir2 (Fig. 5G to N). Together with the chromatin immunoprecipitation data in Fig. 3 and 4, these results show that the enzymatic activity of Sir2 is dispensable for its localization to rDNA and the nucleolus but is required for efficient localization of the Sir2, Sir3, and Sir4 proteins to telomeric foci.
Deacetylation of histone H4 in silent chromatin correlates with the binding of an enzymatically active Sir2 to DNA.
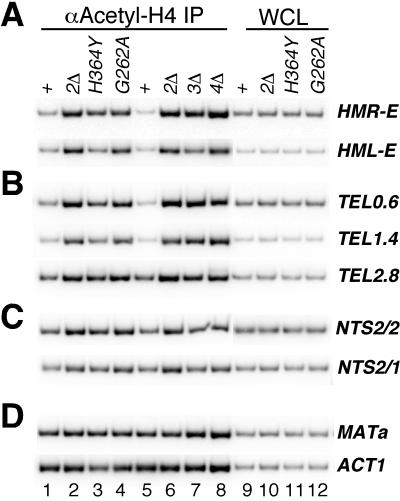
The ability of Sir2 to deacetylate histones in vitro together with the association of Sir2 with silent chromatin domains and with histone-binding proteins Sir3 and Sir4 strongly suggest that histones are in vivo targets of deacetylation by Sir2. We used antibodies that recognize the acetylated tail of histone H4 in order to test whether the enzymatic activity of Sir2 is required for H4 hypoacetylation in silent chromatin. We observed a marked increase in levels of HMR-E, HML-E, and subtelomeric DNA (0.6, 1.4, and 1.4 kb from telomeres) associated with the anti-acetyl-H4 immunoprecipitates from sir2Δ, sir2-H364Y, and sir2-G262A strains compared to immunoprecipitates from the isogenic SIR2 wild-type strain, indicating that histone H4 associated with these regions was hypoacetylated in a Sir2- and Sir2 activity-dependent fashion (Fig. 6A and B, lanes 1 to 8).
FIG. 6.
Hypoacetylation of histone H4 associated with HMR-E, HML-E, telomeric DNA regions, and rDNA requires Sir2. Chromatin immunoprecipitations were carried out using an anti-acetylated H4 antibody from SIR2+ (W303-1a), sir2Δ (SF3 or DMY1865), sir2-H364Y (DMY1866), sir2G262A (DMY1867), sir3Δ (SF4), and sir4Δ (SF5) strains. PCR amplifications of immunoprecipitated DNA (lanes 1 to 8) and WCL (lanes 9 to 12) for the HM silencers (A), telomeric DNA regions (B), rDNA (C), and nonsilenced MATa and ACT1 loci (D) are shown.
Acetylation of H4 in the sir2-H364Y strain was consistently lower than in the sir2Δ and sir2-G262A strains (Fig. 6A and B, lanes 2 to 4). This difference is likely to indicate that the Sir2-H364Y protein retains some activity in vivo and is consistent with the trace amounts of activity that are observed with this mutant protein in in vitro deacetylation assays (61). Interestingly, we observed weak but reproducible hypoacetylation of H4 associated with DNA that encompassed the HMR-E or HML-E silencer in sir3Δ strains (Fig. 6A, lane 7). Low levels of Sir2 and Sir4 were bound to silencers in a Sir3-independent manner (Fig. 3B), and this may be sufficient for partial deacetylation of silencer-proximal nucleosomes. Hypoacetylation of histone H4 therefore correlates with the binding of an enzymatically active Sir2 to DNA.
Since enzymatically inactive Sir2 localizes to rDNA with nearly wild-type efficiency (Fig. 4A), the possible role of Sir2 in hypoacetylation of histones associated with rDNA chromatin can be evaluated independently of the localization of Sir2 itself to rDNA. About twofold higher levels of rDNA nontranscribed spacer region fragments were precipitated with an anti-acetyl-H4 antibody using chromatin isolated from sir2Δ, sir2-H364Y, or sir2-G262A cells compared to a SIR2 wild-type strain, indicating that histone H4 was hypoacetylated in rDNA in a SIR2- and Sir2 activity-dependent manner (Fig. 6C and data not shown). As controls, the anti-acetyl-H4 immunoprecipitates from sir2Δ cells did not contain higher levels of the transcriptionally active ACT1 and MAT loci (Fig. 6D). Thus, the enzymatic activity of Sir2, not just its binding to DNA, is required for hypoacetylation of H4 in rDNA chromatin.
DISCUSSION
The results presented here provide new insight into the nature of yeast silencing complexes and their mechanism of assembly on chromatin. Specifically, our data (i) demonstrate that the apparently large Sir3 and Sir4 complexes are composed of Sir3 and of Sir2 and Sir4, respectively, (ii) suggest that silencing proteins assemble at HM loci and telomeres in a stepwise fashion, with binding of the Sir2/Sir4 complex to sites that initiate silencing as the first step in this process, (iii) show that the enzymatic activity of Sir2 is required for association of each Sir2, Sir3, and Sir4 with HM and telomeric DNA regions that are distal from nucleation sites but not for the association of Sir2 with rDNA, (iv) show that the N termini of histones H3 and H4 are required for rDNA silencing, and (v) show that hypoacetylation of histone H4 within silent chromatin domains correlates with the binding of an enzymatically active Sir2 to DNA. The implications of these results for the mechanism of gene silencing in yeast and its relationship to silencing in other systems will be discussed below.
Purification of Sir3 and the Sir2/4 complex.
The first clues for the existence of a Sir complex came from genetic interactions among the SIR2, SIR3, and SIR4 genes (48). Although subsequent biochemical and two-hybrid experiments uncovered a number of protein-protein interactions involving the Sir2, Sir3, and Sir4 proteins, a Sir complex had never been purified to homogeneity and its composition has remained in question. For example, based on fractionation of yeast extracts and partial purification of Sir2-containing complexes, a recent study proposed that Sir2 exists in a complex with Sir4 that may contain several other proteins (16). Affinity purification of the Sir proteins shows that the composition of these complexes is surprisingly simple (Fig. 1). The large apparent size of the Sir proteins in gel filtration experiments may result from either a large molecular radius or the presence of multiple copies of each protein in these complexes (Fig. 1C) (also see reference 15).
Consistent with previous studies, our purification of Sir3 and Sir4 shows that the Sir2/Sir4 complex (heretofore referred to as the Sir2/4 complex) does not contain any Sir3 and that purified Sir3 is devoid of Sir2 and Sir4. Therefore, although the Sir3 and Sir4 proteins can interact physically, they do not form a stable complex in solution and may only interact during assembly on DNA (16, 41). The demonstration that Sir2/4 can bind to silencers independently of Sir3 is also consistent with these biochemical purifications (Fig. 2, and see below). In addition to Sir2 and Sir3, the Sir4 protein has been shown to interact with a number of other proteins in two-hybrid or affinity column experiments, which are involved in either recruiting the Sir2/4 complex to DNA or regulating the efficiency of silencing, including Rap1, Ubp3, Dot4, and Sif2 (9, 28, 40, 42). None of these proteins is stably associated with Sir4 even under the relatively mild and rapid purification conditions employed in our experiments.
Distinct steps in assembly of yeast silent chromatin.
Our studies suggest the existence of distinct steps in the assembly of silent chromatin domains in yeast. These steps are revealed by examining the interaction of the Sir2, Sir3, and Sir4 proteins with silencers in cells that carried deletions of individual SIR genes. Silencers are ideally suited for this purpose because they are distinct, nonrepetitive sites for nucleation of silent chromatin. Similar experiments examining the role of individual Sir proteins in nucleation of silent chromatin at telomeres are problematic because telomeric repeats, which are required for nucleation of silent chromatin, are composed of short repetitive sequences not suitable for detection by PCR-based assays. Previous studies examining the role of individual Sir proteins in assembly of silencing factors on chromatin did not include sites that are involved in nucleation of silent chromatin (23, 55). As a result, in these studies, deletion of any one of the SIR genes completely abolished binding of all three, Sir2, Sir3, and Sir4, to DNA sites distal from silencers and chromosome ends.
We found that the association of Sir2, Sir3, and Sir4 proteins with silencers can occur in steps that do not require all three proteins (Fig. 2). First, Sir4 can bind to silencers independently of both Sir2 and Sir3, while the binding of Sir2 and Sir3 to silencers is absolutely Sir4 dependent. This result places Sir4 as the most upstream factor in the assembly process. The role of Sir4 in HM/telomeric silencing is reminiscent of the role of Net1 in rDNA silencing. Like Net1, Sir4 can associate with chromatin independently of Sir2, but Sir2 requires Sir4 for its association with silencers. Second, Sir2 and Sir3 can each weakly bind to silencers independently of each other. However, this binding absolutely requires Sir4.
Since Sir2 is part of a stable complex with Sir4, the simplest explanation for these observations is that the Sir2/Sir4 complex is recruited to DNA via interactions that involve Sir4 (Fig. 7A, step 1). The Sir4 protein has previously been shown to interact with silencer and telomere binding proteins such as Rap1 (42). Although the Sir3 protein can interact with Rap1 in two-hybrid and pulldown assays, it does not detectably associate with silencers or telomeres in the absence of Sir4 (Fig. 2A). We therefore propose that Sir4 plays a primary role in recruitment of Sir3 to DNA. Initial Sir3 recruitment is Sir2 independent, as revealed by the weak association of Sir3 with silencers in sir2Δ cells, indicating that this Sir3-silencer association is unlikely to require Sir2-dependent histone deacetylation.
FIG. 7.
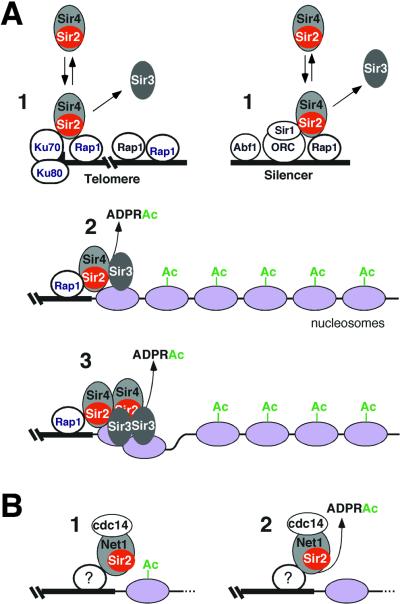
Model for assembly of silent chromatin domains in yeast. (A) Assembly of the Sir complex at silencers and telomeres is proposed to occur in a stepwise fashion involving at least three steps: transient binding, stable association, and spreading of the Sir proteins. In step 1, the Sir2/4 heterodimer transiently binds to the silencer or the telomere via interactions with Rap1/Sir1 and Rap1/yKu70, respectively. This binding does not require the enzymatic activity of Sir2 and is independent of Sir3. In fact, Sir4 can bind to silencers in sir2Δ and sir3Δ cells (see Fig. 2). The association of Sir3 with the silencer can also occur independently of Sir2 but requires Sir4. The Sir4 interactions with the silencer- and telomere-bound factors are based on two-hybrid assays (42, 63, 64). Stable association of the complex with chromatin (step 2) and spreading (binding to regions distal from nucleation sites, step 3) requires all three Sir proteins and the NAD-dependent deacetylation activity of Sir2 (23; this study). (B) Stable association of Sir2 with rDNA requires Net1 but does not require the NAD-dependent deacetylase activity of Sir2 and probably occurs in the absence of deacetylation (step 1) (57; this study). An unknown protein(s) is likely responsible for targeting Net1/Sir2/Cdc14 to rDNA and plays a role analogous to that of silencer and telomere binding proteins. Sir2 activity is required for rDNA silencing and hypoacetylation of histone H4 associated with rDNA (step 2). ADPRAc, O-acetyl-ADP-ribose.
Enzymatic activity of Sir2 and assembly of silencing complexes on chromatin.
At telomeres and the HM loci, one primary outcome of the enzymatic activity of Sir2 is to promote the stable association of the Sir2/4 complex and Sir3 with chromatin (Fig. 3). Our findings are consistent with a model in which the enzymatic activity of Sir2 is required for steps beyond the initial interaction of silencing proteins with silencers or sites that are immediately adjacent to telomeric repeats. In this model, initial binding to nucleation sites would occur independently of Sir2 (Fig. 7A, step 1), but spreading of the Sir proteins along the chromatin fiber would require Sir2 and its enzymatic activity (Fig. 7A, steps 2 and 3).
Sir3 and particularly Sir4 can associate with silencers in sir2Δ cells (Fig. 2), but the magnitude of Sir3 and Sir4 binding to silencers in sir2Δ cells is reduced compared to that in SIR2+ cells. This may indicate that Sir2 activity also contributes to the stable binding of silencing complexes to silencers. However, increased binding of Sir4 to silencers in SIR2+ cells is likely to result from the limited resolution of the chromatin immunoprecipitation assay. Since chromatin immunoprecipitation is performed from samples containing DNA fragments with an average size of ≈500 bp (100 to 1,000 bp), some increase in the immunoprecipitation of silencer DNA is expected to result from the spreading of the Sir proteins to DNA regions that flank the silencers. For example, binding of Sir4 to DNA regions adjacent to the silencer in SIR2+ cells would result in immunoprecipitation of these DNA regions as well as silencer DNA.
A direct role for histone tails in promoting the association of the Sir2, Sir3, and Sir4 proteins with chromatin is supported by previous studies showing that histone tail mutations disrupt the binding of the Sir proteins to telomeric DNA regions (23). In the model presented in Fig. 7A, a primary role of histone deacetylation (and other enzymatic activities that are coupled to deacetylation) is to promote the stable binding and spreading of the Sir proteins to the HM loci and telomeres. In this model, although deacetylation may directly promote chromatin condensation and gene repression, it also functions to control the stepwise association of histone-binding proteins with chromatin. In fact, this latter function may be the critical outcome of histone deacetylation and other covalent modifications that are coupled to histone-binding proteins.
The assembly model proposed here is similar to the recently described assembly process for the HP1/Swi6-based silencing mechanisms in metazoans and fission yeast, where methylation of histone H3 lysine 9 by Clr4 in Schizosaccharomyces pombe and SUV39H1 in mammalian cells creates binding sites for the Swi6 and HP1 proteins, respectively (4, 34, 43, 46; reviewed in reference 25). Therefore, a conserved aspect of gene-silencing mechanisms may involve the direct physical association of histone-modifying enzymes with proteins that recognize and bind to such modified histones (39).
In contrast to the situation at the HM loci and telomeres, the enzymatic activity of Sir2 appears to play only a minor role in the stable association of Sir2 with rDNA. For DNA fragments throughout most of the rDNA repeat, we observed a similar level of cross-linking for wild-type Sir2 and Sir2-H364Y, an enzymatically inactive version of Sir2. We have previously shown that the Net1 subunit of the RENT complex associates with rDNA independently of Sir2 (57). Together with our present observations, these results suggest a distinct mechanism for assembly of silencing complexes at rDNA, which is presented schematically in Fig. 7B. In rDNA, the RENT complex assembles on chromatin independently of Sir2 activity (Fig. 7B, step 1). Deacetylation of histones and/or another outcome of the activity of Sir2 (Fig. 7B, step 2) then results in chromatin structural changes that either are directly responsible for silencing or promote a later step required for this process. Sir2 activity may also regulate the spreading of the RENT complex in rDNA, since in sir2-H364Y cells the cross-linking of Sir2 to a DNA fragment within the 25S rRNA coding regions is diminished (Fig. 3A). However, the magnitude of any such spreading is relatively small, as the amount of Sir2 bound to rDNA regions outside of the nontranscribed spacer region is only about twofold above background (Fig. 3A).
The validity of the above conclusions requires that the point mutations employed in our experiments abolish the enzymatic activity of Sir2 without affecting its stability or interaction with other proteins. Three lines of evidence support a specific role for histidine 364, one of the point mutations used in our studies, in substrate binding. First, this mutation has no effect on the levels of Sir2 in yeast extracts or its interaction with Sir4 and Net1, two proteins with which Sir2 is known to interact (60). Second, histidine 364 is conserved in all Sir2-like proteins (5, 14). Such conservation suggests that this residue is involved in important catalytic or structural functions rather than in species- or locus-specific targeting interactions. Third, and perhaps most important, in the recently described crystal structures of two Sir2-like proteins (the archaeal Sir2Af and the human SIRT2), the corresponding histidine residue is one of the conserved amino acids that forms the binding site for NAD in these enzymes (11, 38). In the archaeal Sir2Af, histidine 116, which is analogous to H364 in yeast Sir2, makes a hydrogen bond with the 2′-hydroxyl of nicotinamide ribose in NAD (38). We therefore believe that the Sir2-H364Y mutant protein used in our studies is unlikely to perturb protein-protein interactions that involve Sir2, but instead has a specific defect in substrate binding or catalysis.
Finally, in support of the above arguments, we obtained similar results using a different point mutation that converts the conserved glycine 262 within the NAD binding site of Sir2 to alanine (Fig. 3D) (G. J. Hoppe, J. C. Tanny, and D. Moazed, unpublished observations). As is the case with the sir2-H364Y mutation, the sir2-G262A mutation abolishes both the in vitro NAD-dependent deacetylase activity of Sir2 and in vivo silencing (61; J. C. Tanny, unpublished observations).
Sir2-dependent hypoacetylation of histone H4.
A large body of evidence suggests that histones are among the in vivo targets of deacetylation by Sir2 (6, 20, 24, 55). However, acetylation of a number of nonhistone proteins has recently been shown to play a role in the regulation of transcription (37, 65; reviewed in reference 32). For the Rpd3 class of histone deacetylases, the strongest evidence that histones are their in vivo targets comes from studies in which Rpd3 is artificially tethered to DNA via fusion to a heterologous DNA-binding protein and shown to deacetylate nucleosomes that are proximal to the tethering site (27). An enzymatically inactive version of Rpd3 is used as a control for indirect effects that may be caused by steric hindrance (27). In our experiments, we observed an analogous correlation between the binding of an enzymatically active Sir2 to its natural sites of action and histone H4 deacetylation in the same regions. For example, Sir2 can bind weakly to silencers in the absence of Sir3, and this weak binding correlates with weak histone H4 hypoacetylation, which is particularly evident for the HML-E silencer (Fig. 6A, lane 7). This result separates the effect of Sir2 binding from the requirement for its enzymatic activity in H4 deacetylation. Maximal deacetylation, however, correlates with maximal binding, which is only observed in cells containing wild-type copies of Sir2, Sir3, and Sir4 (Fig. 6A, lanes 1 and 5).
A similar situation occurs in the rDNA, where an enzymatically inactive version of Sir2 binds to rDNA with nearly wild-type efficiency but fails to deacetylate H4. Here again, deacetylation correlates with Sir2 activity and not Sir2 binding, supporting the hypothesis that H4 is a direct target of deacetylation by Sir2 in vivo. The magnitude of the Sir2-dependent hypoacetylation of H4 in rDNA is relatively small (1.6- to 2.4-fold). We believe that this is due to the fact that in exponentially growing cells, about half of the rDNA repeats are actively transcribed, accounting for nearly 60% of total cellular transcription activity. Histones associated with these active repeats are expected to be highly acetylated in a manner that is unlikely to be affected by Sir2. Therefore, any effect of Sir2 on histone acetylation levels in rDNA is assessed against this background, which would limit the maximum magnitude of the effect to about twofold.
Role of histones H3 and H4 in rDNA silencing.
The requirement for the enzymatic activity of Sir2 in rDNA silencing (24), the sensitivity of rDNA silencing to a reduction in the dosage of histones H2A and H2B (8), and increased accessibility of rDNA to micrococcal nuclease and dam methyltransferase in sir2Δ cells (13) all suggest that rDNA silencing involves changes in chromatin structure. The role of the histone H3 and H4 amino termini in rDNA silencing has not been evaluated despite their requirement for silencing at telomeres and the HM mating type loci. The results presented here show that the amino termini of histones H3 and H4 are required for rDNA silencing. Their role in rDNA silencing, however, particularly the requirement for the N terminus of histone H3, appears to be distinct from that described for telomeres and the HM loci.
For example, the N terminus of histone H3 plays an essential and nonredundant role in rDNA silencing (Fig. 3C), but the same H3 N-terminal tail deletion (amino acids 4 to 30) results in only a modest decrease in telomeric silencing and causes a reduction in HM silencing only when combined with other mutations (62). Together, these results suggest that the H3 N terminus plays a more important role in rDNA silencing than in telomeric and HM silencing. Lysine 16 of histone H4, on the other hand, appears to be similarly important for both rDNA silencing (this study) and telomeric/HM silencing (26, 62). This is perhaps surprising, since this residue is thought to form part of the Sir3 binding site at the telomeres and HM loci (23), and although Sir3 can localize to rDNA, its deletion results in only a modest reduction in the efficiency of rDNA silencing (G. J. Hoppe and D. Moazed, unpublished observations) and does not affect rDNA recombination rates (18). These observations raise the possibility that Sir2-dependent deacetylation of histones H3 and H4 in rDNA creates a binding site for an as yet unidentified protein with a histone-binding activity that is similar to that of Sir3. We propose that the association of Sir3 with rDNA results from a competition between Sir3 and a histone-binding protein that may perform a redundant function with Sir3 at rDNA.
How could the enzymatic activity of Sir2 localize Sir3 to rDNA chromatin? Although the function of Sir3 at the nucleolus is not understood, knowledge of its mechanism of localization to the nucleolus and rDNA may provide general insight into its mode of assembly on silent chromatin domains. Here we have shown that Sir2 activity, and not just Sir2 localization to rDNA, is required for the association of Sir3 with rDNA chromatin (Fig. 4). An intriguing possibility is that the combination of specific lysine residues (on histones or other proteins) that are deacetylated by Sir2 creates a unique mark that can be specifically recognized by Sir3. Alternatively, another aspect of Sir2-dependent deacetylation activity, such as 2′,3′-O-acetyl-ADP-ribose synthesis, may trigger Sir3-chromatin association. At the rDNA repeats, one of these events may be sufficient for weak association of Sir3 with chromatin; at the HM loci and telomeres, the more stable and extensive binding of Sir3 to chromatin is likely to involve additional interactions, such as Sir3-Sir4 interactions (Fig. 7).
Acknowledgments
This work was supported by grants from the NIH (GM61641) and the Ellison Medical Foundation. A.D.R. is a fellow of the Jane Coffin Childs Memorial Fund for Medical Research.
We thank Michael Grunstein for plasmids and strains, Aaron Straight for help with microscopy, Tim Mitchison for the use of his microscopes, and Julie Huang and anonymous reviewers for helpful comments on the manuscript.
REFERENCES
- 1.Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio, O. M., B. L. Billington, and D. E. Gottschling. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66:1279-1287. [DOI] [PubMed] [Google Scholar]
- 3.Aris, J. P., and G. Blobel. 1988. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 107:17-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 5.Brachmann, C. B., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus, and J. D. Boeke. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888-2902. [DOI] [PubMed] [Google Scholar]
- 6.Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis, and J. R. Broach. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592-604. [DOI] [PubMed] [Google Scholar]
- 7.Braunstein, M., R. E. Sobel, C. D. Allis, B. M. Turner, and J. R. Broach. 1996. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 16:4349-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryk, M., M. Banerjee, M. Murphy, K. E. Knudsen, D. J. Garfinkel, and M. J. Curcio. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11:255-269. [DOI] [PubMed] [Google Scholar]
- 9.Cockell, M., H. Renauld, P. Watt, and S. M. Gasser. 1998. Sif2p interacts with Sir4p amino-terminal domain and antagonizes telomeric silencing in yeast. Curr. Biol. 8:787-790. [DOI] [PubMed] [Google Scholar]
- 10.Eng, J. K., A. L. McCormak, and J. R. Yates. 1994. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 11.Finnin, M. S., J. R. Donigian, and N. P. Pavletich. 2001. Structure of the histone deacetylase SIRT2. Nat. Struct. Biol. 8:621-625. [DOI] [PubMed] [Google Scholar]
- 12.Fox, C. A., A. E. Ehrenhofer-Murray, S. Loo, and J. Rine. 1997. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276:1547-1551. [DOI] [PubMed] [Google Scholar]
- 13.Fritze, C. E., K. Verschueren, R. Strich, and R. Easton Esposito. 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16:6495-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frye, R. A. 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273:793-798. [DOI] [PubMed] [Google Scholar]
- 15.Georgel, P. T., M. A. Palacios DeBeer, G. Pietz, C. A. Fox, and J. C. Hansen. 2001. Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc. Natl. Acad. Sci. USA 98:8584-8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghidelli, S., D. Donze, N. Dhillon, and R. T. Kamakaka. 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 20:4522-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotta, M., S. Strahl-Bolsinger, H. Renauld, T. Laroche, B. K. Kennedy, M. Grunstein, and S. M. Gasser. 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16:3243-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlieb, S., and R. E. Esposito. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56:771-776. [DOI] [PubMed] [Google Scholar]
- 19.Grunstein, M. 1997. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 9:383-387. [DOI] [PubMed] [Google Scholar]
- 20.Guarente, L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14:1021-1026. [PubMed] [Google Scholar]
- 21.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, Calif.
- 22.Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser, and M. Grunstein. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80:583-592. [DOI] [PubMed] [Google Scholar]
- 23.Hecht, A., S. Stahl-Bolsinger, and M. Grunstein. 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383:92-96. [DOI] [PubMed] [Google Scholar]
- 24.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 25.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, L. M., P. S. Kayne, E. S. Kahn, and M. Grunstein. 1990. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87:6286-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 28.Kahana, A., and D. E. Gottschling. 1999. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6608-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kayne, P. S., U. J. Kim, M. Han, J. R. Mullen, F. Yoshizaki, and M. Grunstein. 1988. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 55:27-39. [DOI] [PubMed] [Google Scholar]
- 30.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 31.Klar, A. J. S., S. Fogel, and K. MacLeod. 1979. MAR1—a regulator of HMa and HMα loci in Saccharomyces cerevisiae. Genetics 93:37-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 34.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 35.Landry, J., J. T. Slama, and R. Sternglanz. 2000. Role of NAD+ in the deacetylation activity of SIR2-like proteins. Biochem. Biophys. Res. Commun. 30:685-690. [DOI] [PubMed] [Google Scholar]
- 36.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Sternglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo, J., A. Y. Nikolaev, S. Imai, D. Chen, F. Su, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 38.Min, J., J. Landry, R. Sternglanz, and R. M. Xu. 2001. Crystal structure of a SIR2 homolog-NAD complex. Cell 105:269-279. [DOI] [PubMed] [Google Scholar]
- 39.Moazed, D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8:489-498. [DOI] [PubMed] [Google Scholar]
- 40.Moazed, D., and A. D. Johnson. 1996. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell 86:667-677. [DOI] [PubMed] [Google Scholar]
- 41.Moazed, D., A. Kistler, A. Axelrod, J. Rine, and A. D. Johnson. 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. USA 94:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moretti, P., K. Freeman, L. Coodly, and D. Shore. 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8:2257-2269. [DOI] [PubMed] [Google Scholar]
- 43.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 44.Palladino, F., T. Laroche, E. Gilson, A. Axelrod, L. Pillus, and S. M. Gasser. 1993. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75:543-555. [DOI] [PubMed] [Google Scholar]
- 45.Peng, J., and S. P. Gygi. 2001. Proteomics: the move to mixtures. J. Mass Spectrom. 36:1083-1091. [DOI] [PubMed] [Google Scholar]
- 46.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 47.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 48.Rine, J., and I. Herskowitz. 1987. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116:9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sauve, A. A., I. Celic, J. Avalos, H. Deng, J. D. Boeke, and V. L. Schramm. 2001. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry 40:15456-15463. [DOI] [PubMed] [Google Scholar]
- 50.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 51.Shore, D., and K. Nasmyth. 1987. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51:721-732. [DOI] [PubMed] [Google Scholar]
- 52.Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed, Z. W. S. Chen, J. Jang, A. Shevchenko, H. Charbonneau, and R. J. Deshaies. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97:233-244. [DOI] [PubMed] [Google Scholar]
- 53.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241-254. [DOI] [PubMed] [Google Scholar]
- 54.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 56.Straight, A. F., A. S. Belmont, C. C. Robinett, and A. W. Murray. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599-1608. [DOI] [PubMed] [Google Scholar]
- 57.Straight, A. F., W. Shou, G. J. Dowd, C. W. Turck, R. J. Deshaies, A. D. Johnson, and D. Moazed. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97:245-256. [DOI] [PubMed] [Google Scholar]
- 58.Suka, N., Y. Suka, A. A. Carmen, J. Wu, and M. Grunstein. 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8:473-479. [DOI] [PubMed] [Google Scholar]
- 59.Tanner, K. G., J. Landry, R. Sternglanz, and J. Denu. 2000. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 97:14178-14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanny, J. C., G. J. Dowd, J. Huang, H. Hilz, and D. Moazed. 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99:735-745. [DOI] [PubMed] [Google Scholar]
- 61.Tanny, J. C., and D. Moazed. 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson, J. S., X. Ling, and M. Grunstein. 1994. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369:245-247. [DOI] [PubMed] [Google Scholar]
- 63.Triolo, T., and R. Sternglanz. 1996. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381:251-253. [DOI] [PubMed] [Google Scholar]
- 64.Tsukamoto, Y., J. Kato, and H. Ikeda. 1997. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388:900-903. [DOI] [PubMed] [Google Scholar]
- 65.Vaziri, H., S. K. Dessain, E. Ng Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]