Abstract
Zellweger syndrome is a lethal neurological disorder characterized by severe defects in peroxisomal protein import. The resulting defects in peroxisome metabolism and the accumulation of peroxisomal substrates are thought to cause the other Zellweger syndrome phenotypes, including neuronal migration defects, hypotonia, a developmental delay, and neonatal lethality. These phenotypes are also manifested in mouse models of Zellweger syndrome generated by disruption of the PEX5 or PEX2 gene. Here we show that mice lacking peroxisomal membrane protein PEX11β display several pathologic features shared by these mouse models of Zellweger syndrome, including neuronal migration defects, enhanced neuronal apoptosis, a developmental delay, hypotonia, and neonatal lethality. However, PEX11β deficiency differs significantly from Zellweger syndrome and Zellweger syndrome mice in that it is not characterized by a detectable defect in peroxisomal protein import and displays only mild defects in peroxisomal fatty acid β-oxidation and peroxisomal ether lipid biosynthesis. These results demonstrate that the neurological pathologic features of Zellweger syndrome can occur without peroxisomal enzyme mislocalization and challenge current models of Zellweger syndrome pathogenesis.
Peroxisomes are single-membrane bound metabolic organelles that are present in all eukaryotic cells. In their lumen reside the enzyme systems responsible for a wide variety of metabolic pathways, including the β-oxidation of very long-chain fatty acids (VLCFAs), α- and β-oxidation of branched-chain fatty acids, biosynthesis of ether linked lipids and cholesterol, synthesis of bile acids, metabolism of polyunsaturated fatty acids, and H2O2 metabolism (25, 36). The importance of peroxisomes for human health is best demonstrated by the existence of Zellweger syndrome, a lethal neurological disorder characterized by defects in peroxisomal matrix enzyme import (30). This defect negatively impacts virtually all peroxisomal metabolic functions, which leads, in turn, to the accumulation of peroxisomal α- and β-oxidation substrates (e.g., phytanic acid and VLCFAs, respectively) and reduced levels of ether-linked lipids (e.g., plasmalogens) (11). Zellweger syndrome is also associated with severe defects in mitochondrial structure and function (5, 10), as well as a pleiotropic set of clinical phenotypes, including a developmental delay, hypotonia, neuronal migration defects, enhanced neuronal apoptosis, and an array of hepatic and renal abnormalities (11).
There is uncertainty regarding the etiologic agent(s) and mechanisms responsible for the neuronal migration defect and other phenotypes of Zellweger syndrome. However, the accumulation of toxic peroxisomal α- and β-oxidation substrates or depletion of peroxisomal products, such as ether-linked lipids, have been proposed to cause its pathologic features (28). In contrast to the uncertainty regarding Zellweger syndrome pathogenesis, the molecular genetics of Zellweger syndrome and its milder variants (neonatal adrenoleukodystrophy and infantile Refsum disease) are well understood (11, 30). These peroxisome biogenesis disorders are inherited in an autosomal recessive fashion and are caused by mutations in any of at least 12 distinct PEX genes (11). Approximately 20 PEX genes are required for peroxisome biogenesis, and with the exception of PEX11, all are required for peroxisomal matrix enzyme import (30). PEX11 proteins are components of the peroxisomal membrane in a wide array of species, including yeast, protozoan parasites, and mammals (1, 2, 6, 19, 21, 27, 32, 34). They appear to play an important role in peroxisome division, although the nature of their role is currently the subject of debate. In one model, PEX11 proteins are thought to play a direct role in peroxisome division (6, 12, 21, 27, 34). Recently, another model was proposed in which PEX11 proteins play a direct role in medium-chain fatty acid oxidation and only affect peroxisome division indirectly through this metabolic role (35).
Mammals express at least two PEX11 genes, the inducible PEX11α gene and the constitutively expressed PEX11β gene (1, 2, 27, 34). Here we report an analysis of mice that lack the PEX11β gene. Quite unexpectedly, we found that PEX11β−/− mice exhibit numerous Zellweger syndrome pathologic features, including a developmental delay, hypotonia, neuronal migration defects, and enhanced neuronal apoptosis, even though they have no apparent defect in peroxisomal protein import and have only mild defects in peroxisomal metabolic function.
MATERIALS AND METHODS
Cloning and disruption of the murine PEX11β gene.
A bacterial artificial chromosome (BAC) clone (no. 17747; Incyte Genomics Inc., St. Louis, Mo.) with the complete PEX11β gene was obtained by screening a BAC library of 129/svJ mouse genomic DNA with the full-length murine PEX11β cDNA (34). EcoRI-XbaI, XbaI, or HindIII-fragments spanning a 13-kb region containing the PEX11β gene were subcloned into pLITMUS vectors (New England BioLabs, Beverly, Mass.) and sequenced. The targeting vector, designed to disrupt the first three exons of the PEX11β gene (Fig. 1A), was generated by insertion of ApaLI-SmaI (3.1-kb PEX11β 5′ untranslated region) and HindIII-NotI (3.4 kb of intron 3) fragments into pGT-N29 as the 5′- and 3′-flanking regions of the pgk-Neo cassette. A 200-μg sample of targeting vector-DNA was linearized with SwaI and electroporated into 6.5 × 106 R1 embryonic stem (ES) cells as previously described (40). After selection with G418 at 200 μg/ml, recombined ES cell clones were identified by mini-Southern blot hybridization analysis of EcoRI- or EcoRV-SseI-digested ES cell genomic DNA with two flanking probes. Four PEX11β+/− ES cell clones were injected into blastocysts of C57BL/6 host mice. Chimeric males from three different ES cell lines were intercrossed with C57BL/6 mice (Jackson Laboratory, Bar Harbor, Maine), and agouti offspring were tested for the presence of the gene disruption by Southern blotting (33). Heterozygous F1 mice were backcrossed with C57BL/6 mice for five generations. Genotypes of mice from generation F2 and beyond were determined by PCR with primer 8 (5′-GTCTAGGACAGGCTTCTGCTGTTC-3′), primer 9 (5′-GTTTCCCCATCTTTCCCTTGAG-3′), and primer Neo (5′-ATATTGCTGAAGAGCTTGGCGGC-3′). Amplification reactions were done with 0.1 μg of DNA for the wild-type allele (primers 8 and 9 → 590 bp) or the targeted allele (primers 8 and Neo → 980 bp). For RNA blots, total RNA was isolated from mouse livers with the Purescript RNA isolation kit (Gentra Systems, Minneapolis, Minn.). RNA (10 μg/lane) was separated by electrophoresis on 1.5% formaldehyde-agarose gels, transferred to GeneScreen Plus membranes (NEN Life Science Products, Boston, Mass.), and hybridized in accordance with standard procedures (33).
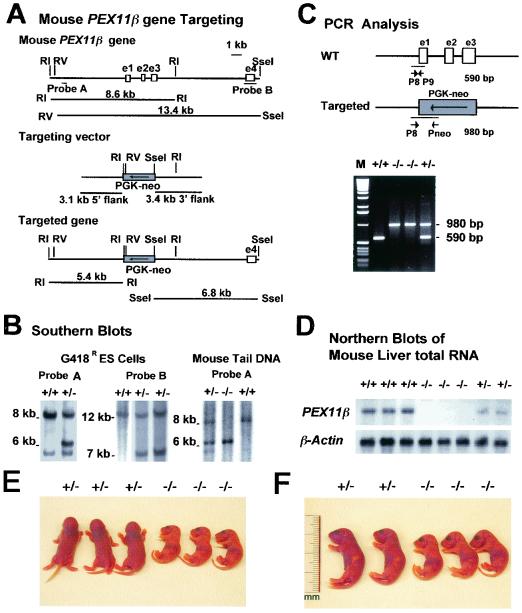
FIG. 1.
Targeting of PEX11β. (A) Schematic representation of the PEX11β wild-type locus (top), the targeting vector (middle), and the targeted allele (bottom). Two flanking Southern blot probes, A and B, are indicated. (B) Southern blot analyses of the G418r ES clone DNA with probes A and B and the mouse tail DNA with probe A. Probe A detects an 8.6-kb EcoRI fragment in the wild-type allele and a 5.4-kb fragment in the targeted allele. Probe B detects a 13.4-kb EcoRV/SseI fragment in the wild-type allele and a 6.8-kb fragment in the targeted allele. (C) PCR analysis of mouse tail DNA. Positions of the primers are indicated. The wild-type (WT) allele product is 590 bp, and the targeted-allele product is 980 bp. M, molecular size markers. (D) Northern blot analysis of total liver RNA from wild-type (+/+), homozygous (−/−), and heterozygous (+/−) animals. The Northern blot shown was probed with a radioactively labeled murine PEX11β cDNA probe (top), stripped, and probed with labeled β-actin cDNA (bottom). (E and F) PEX11β−/− mice display intrauterine growth retardation and are hypotonic. One litter of newborn mice with heterozygous (+/−) and homozygous (−/−) offspring is depicted either in the conscious state (E) or under ether anesthesia (F).
MEF preparation and immunofluorescence assay.
Mouse embryo fibroblasts (MEF) were isolated from E14.5 embryos as previously described (13). For indirect immunofluorescence assay, cells were fixed for 20 min in 3% formaldehyde in Dulbecco’s phosphate-buffered saline (Life Technologies), pH 7.1; permeabilized for 5 min in 1% Triton X-100-Dulbecco’s phosphate-buffered saline; and further processed as previously described (31). Affinity-purified rabbit anti-human PEX14 antibody has been described previously (31), sheep anti-human catalase antibodies were obtained from The Binding Site (Birmingham, United Kingdom), mouse monoclonal anti-myc antibodies were obtained from the supernatant of hybridoma 1-9E10 (7), and labeled secondary antibodies were obtained from standard commercial sources.
Histology and electron microscopy.
Under ether anesthesia, mouse embryos (embryonic day 17.5 [E17.5] and E18.5) and newborn mice (postnatal day 0.5 [P0.5]) were perfusion fixed via the heart with 4% paraformaldehyde in phosphate-buffered saline, pH 7.4. Whole-mouse sections were obtained following overnight immersion fixation (OIF) at 4°C and paraffin embedding. Five-micrometer sagittal sections of whole mice were stained with hematoxylin and periodic acid-Schiff (PAS) for pathological analysis (3). For brain analysis, OIF was performed with opened skulls and the brains were removed and embedded in paraffin. Serial coronal sections (5 μm) were cut and stained with cresyl fast violet for pathological analysis (3). More than 200 carefully matched sections were examined for neuronal migration defects and apoptosis. For electron microscopy, cardiac perfusion fixation of newborn mice was performed with 4% paraformaldehyde-0.05% glutaraldehyde-2% sucrose-0.05% CaCl2-0.1 M piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer, pH 7.4, followed by OIF in the same fixative without glutaraldehyde. Livers were removed, cut into 100-μm sections, and postfixed for 15 min with 1.0% glutaraldehyde in 0.1 M PIPES buffer. Catalase cytochemistry (30 min of preincubation and 2 h of incubation at 45°C) was performed with alkaline DAB medium as described by Fahimi (8). DAB-stained sections were postfixed with either aqueous or reduced osmium, embedded in Epon 812, and examined by electron microscopy.
Biochemical assays.
Livers and brains (telencephalon) of newborn mice (P0.5) were weighed and homogenized in a tissue grinder with chloroform-methanol (2:1). Total lipids were extracted with 2:1:0.8 chloroform-methanol-water, converted to their methyl esters, dissolved in hexane at a concentration of ∼1 μg/μl, and then separated by gas chromatography (DB-1 and SP-2560 columns) as previously described (24). The levels of VLCFAs and plasmalogens were calculated as percentages of the total fatty acids. Plasmalogen synthesis activity was determined in cultured MEF with the double-substrate, double-isotope method (with [14C]hexadecanol and [3H]hexadecyl-glycerol as substrates) (29). Fatty acid β-oxidation assays were carried out with [1-14C]palmitic acid (C16:0) and [1-14C]lignoceric acid (C24:0) as substrates in intact MEF (41). Branched-chain fatty acid (phytanic and pristanic acids) oxidation assays (41, 42) were done with cultured MEF with [2,3-3H]phytanic acid or [1-14C]pristanic acid as the substrate.
RESULTS
PEX11β−/− mice display neonatal lethality, intrauterine growth retardation, and hypotonia.
Exons 1 to 3 of the mouse PEX11β gene were disrupted in R1 ES cells by gene conversion with a PEX11β/pgk-Neor cassette (Fig. 1A). Correctly targeted PEX11β+/− cell lines were identified and injected into C57BL/6 blastocysts, and chimeric animals were generated. Chimeras were mated with C57BL/6 mice, and PEX11β+/− heterozygote animals were obtained from several chimeras and two independently generated PEX11β+/− ES cell lines (Fig. 1B and C). These mice were backcrossed to C57BL/6 mice five times to place the PEX11β disruption allele over a relatively homogeneous background. PEX11β+/+, PEX11β+/−, and PEX11β−/− fetuses were obtained in the expected Mendelian ratios in intercrosses between PEX11β+/− animals. PEX11β−/− animals lack detectable PEX11β mRNA (Fig. 1D). Animals lacking PEX11β died shortly after birth on day 1, were undersized (∼80% of wild-type size at P0.5) (Fig. 1E and F), were underweight (∼60% of wild-type body weight at P0.5), were hypotonic (Fig. 1E), and suckled only poorly, as was evident from the reduced amounts of milk in their gastrointestinal tracts (Fig. 1F). No heterozygote phenotype was observed.
PEX11β−/− mice exhibit neuronal migration defects and a developmental delay.
Neonatal hypotonia and neonatal lethality caused by defects in a peroxisomal membrane protein are unique characteristics of Zellweger syndrome. In addition, prior mouse models of Zellweger syndrome, which reproduce virtually all of the hallmarks of the human disease, also display the intrauterine growth defects we observed in PEX11β−/− mice (3, 9). Therefore, we investigated the possibility that PEX11β-deficient mice may display other pathologic characteristics of Zellweger syndrome. Zellweger syndrome is characterized by defective neuronal migration, and Zellweger syndrome mice display both a neuronal migration defect and enhanced apoptosis within the neocortex (11).
The neuronal migration defect of PEX5−/− mice is focal in nature and can be observed only in coronal sections through the neocortex (3) (E. Baumgart, unpublished data). In PEX11β−/− mice, we also detected focal areas of decreased neuronal migration in coronal sections of the neocortex. This neuronal migration defect is evident from the increased numbers of neurons in the intermediate zone and layer V, as well as structural alterations and slightly reduced thickness of the cortical plate (Fig. 2A and B). This was observed in all three PEX11β−/− mice that were examined and was not observed any of the three PEX11β+/+ littermate control mice. Thus, the neuronal migration defect appeared to be a fully penetrant phenotype of PEX11β−/− mice. In addition, because PEX11β−/− mice display growth retardation and a developmental delay, we examined the neuronal migration defect in four PEX11β−/− embryos and four size-matched PEX11β+/+ embryos. The neuronal migration defect was apparent in all four PEX11β−/− embryos and was not detected in any of the size-matched control embryos (data not shown).
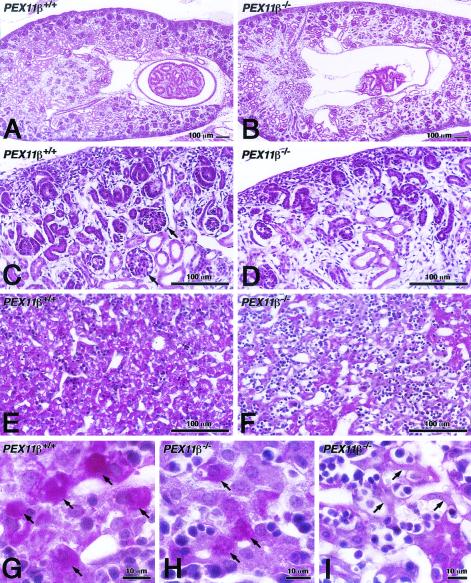
FIG. 2.
Neuronal defects in PEX11β−/− mice. Carefully matched coronal (A and B; stained with cresyl fast violet) or sagittal (C to F; stained with PAS and hematoxylin) sections of control (A and C) and PEX11β−/− (B and D to F) mice in medial regions of the neocortex. In panel B, the neuronal migration defect is revealed by higher cell density in the intermediate zone (IZ) and layer V, as well as the altered structure and slightly lesser thickness of the cortical plate (CP). GZ, germinative zone; SP, subplate. Neuronal apoptoses of different stages are indicated by arrows (D to F). Insets 1 and 2 in panel E depict higher magnifications of early stages of chromatin condensation with typical nuclear cap structures of apoptotic cells, indicated by white arrowheads. Black arrowheads indicate macrophages (MP) (slightly more PAS positive) with phagocytosed material. Empty spaces are clearly visible where apoptotic neurons were located (D to F). In contrast, empty spaces in panel C represent endothelium-lined capillaries.
Enhanced neuronal apoptosis was also observed in PEX11β−/− mice (Fig. 2C to F). Zellweger syndrome mice also exhibit a delay in renal development, and this too was observed in PEX11β−/− mice (Fig. 3A to D). Furthermore, we detected a focal mosaic pattern of a developmental delay and decreased glycogen in PEX11β−/− livers (Fig. 3E to I), although this could be due to the inability of PEX11β−/− mice to suckle properly. One other important feature of Zellweger syndrome is facial dysmorphism (25), but this was not observed in Zellweger syndrome mice. However, these mice did exhibit a developmental delay in calvarium ossification (9). PEX11β−/− animals also lack facial dysmorphism and exhibit a delay in the ossification of calvaria (data not shown).
FIG. 3.
Developmental delay in kidneys and livers of PEX11β−/− mice. Histological analysis of PAS- and hematoxylin-stained kidney (A to D) and liver (E to I) sections (5 μm) of P0.5 control (A, C, E, and G) and PEX11β−/− (B, D, F, H, and I) mice, depicting the strong delay in the development of both organs in PEX11β−/− animals. Well-developed glomeruli are present in control animals (C, arrows) but absent from PEX11β−/− animals (D). Tubules are also reduced in panel D. Livers of PEX11β−/− mice show regions of underdeveloped hepatocytes (F) with reduced glycogen (reduced PAS staining). Even areas with well-developed PEX11β−/− hepatocytes contained less glycogen (H) than did those of control animals (G). Arrows in panel H depict underdeveloped hepatocytes in PEX11β−/− mouse liver, which were not observed in controls.
Peroxisomal protein import and mitochondrial structure are not affected in PEX11β−/− mice.
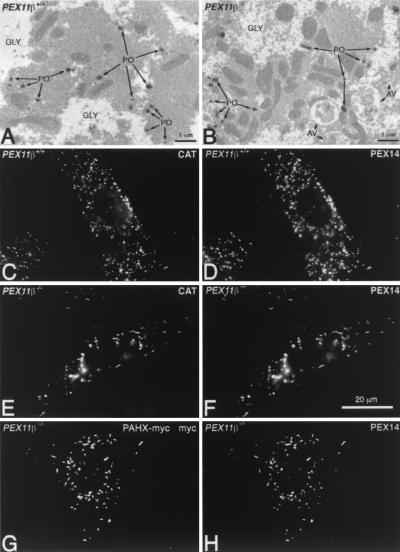
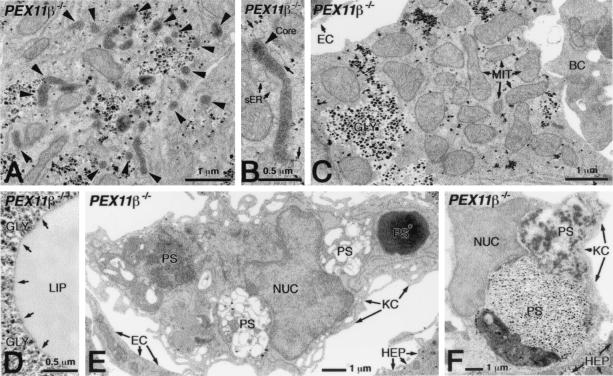
Zellweger syndrome and its corresponding mouse models display a number of cellular abnormalities. Chief among these is the inability to import peroxisomal matrix enzymes (11). However, enzyme import appears to be unaffected in PEX11β−/− cells. This is shown here by the cytochemical detection of the peroxisomal marker enzyme catalase (imported through the PTS1 pathway) within hepatocyte peroxisomes of PEX11β−/− mice (Fig. 4A and B), as well as the colocalization of the peroxisomal enzyme catalase and the peroxisomal membrane protein PEX14 in PEX11β−/− MEF (Fig. 4C to F). An enzyme imported through the PTS2 pathway, phytanoyl coenzyme A hydroxylase, was also targeted into peroxisomes when expressed as a myc-tagged form in PEX11β−/− MEF (Fig. 4G and H), demonstrating that all peroxisomal protein import pathways are functional in cells lacking PEX11β. Nevertheless, the loss of PEX11β did appear to reduce peroxisome abundance and increase peroxisome clustering (Fig. 5A) and elongation (Fig. 4F and 5B). Defects in mitochondrial structure have also been reported in Zellweger syndrome patients and Zellweger syndrome mice (3, 10), but these too were absent from PEX11β−/− hepatocytes (Fig. 5C), although we did detect mitochondrial proliferation in some cells (Fig. 4B and 5C). Zellweger syndrome is also characterized by hepatic accumulation of VLCFAs (36, 38), a substrate of the peroxisomal β-oxidation pathway, which is thought to explain the presence of needle-like lipid crystals on the surface of hepatocyte lipid droplets and within phagosomes of Kupffer cells (the resident macrophages of the liver) in mouse models of Zellweger syndrome (3). We found no evidence of such needle-like lipid crystals in the livers of PEX11β−/− mice (Fig. 5D to F).
FIG. 4.
PEX11β deficiency does not affect peroxisomal protein import. Ultrastructural analysis of hepatocytes from P0.5 control (A) and PEX11β−/− mice (B) shows normal import of the peroxisomal enzyme catalase, as determined by DAB staining (post-fixation with aqueous osmium). GLY, glycogen. (C to F) Indirect immunofluorescence analysis of control (C and D) and PEX11β−/− (E and F) MEF with antibodies to catalase (C and E) and PEX14 (D and F), a peroxisomal membrane protein. (G to H) Phytanoyl coenzyme A hydroxylase (PAHX)-myc targets to peroxisomes in PEX11β−/− fibroblasts. Cells were transfected with pcDNA3-PAHXmyc (22), grown for 2 days, and then processed for indirect immunofluorescence assay with a mouse monoclonal antibody to the myc epitope and rabbit anti-PEX14 antibodies.
FIG. 5.
No mitochondrial defects or lipid crystals in PEX11β−/− mouse liver. Ultrastructural analysis of DAB-stained preparations (postfixation and with aqueous osmium) revealed elevated peroxisome clustering (A) and elongation (B) in some hepatocytes of PEX11β−/− mice. Arrowheads indicate the positions of peroxisomes, and arrows in panel B show the position of smooth endoplasmic reticulum (sER). (C) Hepatocytes of PEX11β−/− mice had no mitochondrial structural abnormalities, although in some cells they did appear to be more abundant than in those of controls. MIT, mitochondria; GLY, glycogen; EC, endothelial cell; BC, blood cell. (D) Hepatocytes of PEX11β−/− mice lacked needle-like crystals on the surface of their lipid droplets (D; arrows show where large, needle-like crystals would be seen in Zellweger syndrome patients and mice). (E and F) VLCFA crystals were also absent from phagosomes (PS) of Kupffer cells (KC). NUC, nucleus of a Kupffer cell; EC, endothelial cell; HEP, hepatocyte; PS*, red blood cell in a phagosome. Note the high level of phagocytic degradation of different blood cells in phagosomes (E) and the occasional phagocytosis of hepatocytes (F) in Kupffer cells of PEX11β−/− mice.
VLCFA accumulation is not responsible for neuronal abnormalities in PEX11β−/− mice.
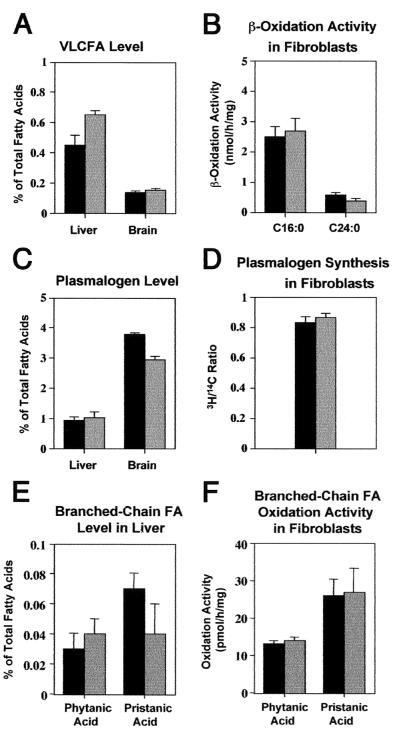
The diagnosis of Zellweger syndrome is typically confirmed by biochemical tests (24, 38). In Zellweger syndrome patients and prior mouse models of Zellweger syndrome, VLCFA levels are 10 times higher than normal and plasmalogen levels are 10 to 100 times lower than normal (3, 9, 24). By contrast, PEX11β−/− mice have normal levels of brain VLCFAs, no alteration of hepatic plasmalogens, and only slight alterations of hepatic VLCFAs (1.4×) and brain plasmalogens (0.8×) (Fig. 6A and C). We also measured rates of VLCFA β-oxidation and plasmalogen synthesis in cultured fibroblasts. Relative to controls, we detected no change in plasmalogen synthesis rates and only a slight decrease in VLCFA β-oxidation, to 0.6× that of normal controls (Fig. 6B and D). Some Zellweger syndrome patients accumulate the branched-chain fatty acids phytanic acid and pristanic acid, which are substrates of the peroxisomal fatty acid α- and β-oxidation pathways, respectively, and the oxidation of these substrates is typically impaired in Zellweger syndrome fibroblasts (11, 25, 37). PEX11β−/− animals, on the other hand, do not accumulate these fatty acids in the liver (Fig. 6E) or other tissues (data not shown), and PEX11β−/− fibroblasts have normal oxidation activities toward both phytanic and pristanic acids (Fig. 6F).
FIG. 6.
PEX11β−/− mice have a mild, generalized defect in peroxisome metabolism. In all of the graphs, black bars represent average values obtained with PEX11β+/+, gray bars represents average values obtained with PEX11β−/−, and brackets represent 1 standard deviation. Abundance of VLCFAs (A) and activities of mitochondrial (C16:0) and peroxisomal (C24:0) fatty acid β-oxidation (B) in cultured MEF. (C) Plasmalogens in control and PEX11β−/− mice, expressed as percentages of the total fatty acids. (D) Plasmalogen synthesis activities in cultured MEF, expressed as the ratio of peroxisomal incorporation of [14C]hexadecanol to endoplasmic reticulum incorporation of [3H]hexadecyl-glycerol. (E) Levels of the branched-chain fatty acids phytanic acid and pristanic acid in the liver. (F) Rates of phytanic acid and pristanic acid oxidation in cultured fibroblasts from mutant and control animals.
DISCUSSION
Peroxisomal enzyme import defects are the root cause of all known instances of Zellweger syndrome and prior mouse models of Zellweger syndrome (11). PEX11β deficiency has most of the pathological hallmarks of Zellweger syndrome, including a neuronal migration defect, enhanced neuronal apoptosis, a developmental delay, neonatal hypotonia, and neonatal lethality, without the peroxisomal enzyme import defects that are the cellular hallmarks of this disease. PEX11β deficiency also differs from Zellweger syndrome with regard to peroxisomal metabolic defects. Zellweger syndrome patients and prior mouse models of Zellweger syndrome show 1,000% increases in VLCFAs, a marker substrate of the peroxisomal β-oxidation pathway, and 90 to 99% decreases in plasmalogens, a marker product of the peroxisomal ether lipid synthesis pathway (3, 9, 24). In contrast, PEX11β−/− mice have no alteration in brain VLCFAs or liver plasmalogens, only a slight increase (∼40%) in liver VLCFAs, a slight decrease (∼20%) in brain plasmalogens, and no accumulation of peroxisomal fatty acid α-oxidation substrates. In cultured cells, PEX11β deficiency has no effect on plasmalogen synthesis and no effect on phytanic acid and pristanic acid oxidation and causes only a slight decrease (∼40%) in VLCFA β-oxidation. Thus, PEX11β deficiency represents a novel peroxisomal disorder that mimics major neurological and developmental pathologic features of Zellweger syndrome but lacks many of its cellular and biochemical features.
It is important to note that PEX11β-deficient mice lack several other features of Zellweger syndrome, such as facial dysmorphism, enlarged cranial fontanelles, and renal cysts (23). However, these pathologic features were also absent from prior mouse models of Zellweger syndrome (3, 9). One reasonable explanation for these differences is that both Zellweger syndrome mice and PEX11β-deficient mice die very shortly after birth, precluding the development of what may be later-stage pathologic features. The severe kidney and bone development delay we observed in PEX11β-deficient mice may be an early manifestation of these deficiencies.
Although there is no firm hypothesis about exactly which substrate, product, or combination of substrates and products is the etiologic agent of Zellweger syndrome, patients who are defective in the peroxisomal β-oxidation enzyme D-bifunctional protein (D-BP) (14, 38) have clinical phenotypes in common with Zellweger syndrome patients. In contrast, patients with defects in ether lipid synthesis or fatty acid α-oxidation are associated with distinct phenotypes (38, 39). These and other observations have led to the hypothesis that Zellweger syndrome pathologic features might be induced primarily by their severe fatty acid β-oxidation defect and perhaps by VLCFA toxicity (28). With regard to VLCFA toxicity, PEX11β−/− mice have only a very mild accumulation of VLCFAs (∼40%), which is far less severe than the VLCFA accumulation of Zellweger syndrome mice (∼1,000%). VLCFA accumulation in PEX11β−/− mice is also less severe than the 200 to 300% increase in VLCFAs observed in ALD-deficient mice, which have no detectable pathologic features (20, 26). Thus, it appears that the developmental delay, neuronal migration defects, enhanced neuronal apoptosis, neonatal hypotonia, and neonatal lethality of PEX11β deficiency cannot be explained by VLCFA toxicity. With regard to the hypothesis that Zellweger syndrome pathologic features are caused primarily by defects in peroxisomal β-oxidation, the phenotypes of PEX11β−/− mice are distinct from those of mice lacking the peroxisomal β-oxidation enzyme D-BP (4). In particular, PEX11β-deficient mice do not display the nearly total defect in VLCFA oxidation and branched-chain (pristanic) fatty acid oxidation of D-BP-deficient mice.
The exceedingly mild metabolic defects in PEX11β−/− mice raise the interesting possibility that the pathologic features of these mice, and perhaps Zellweger syndrome mice and Zellweger syndrome patients, are not caused by nonspecific metabolite toxicity. Rather, it may be that the metabolic abnormalities of these diseases impair animal development through a subtle, yet inappropriate, activation or inactivation of one or more signaling pathways in the body. Peroxisomal metabolic pathways play critical roles in the synthesis of many signaling lipids, including ligands of nuclear hormone receptors RXR, PPARα, and PPARγ (15-17). If true, this pathogenic hypothesis raises the possibility of therapeutic interventions that could normalize the putative signaling pathways and the development of these animals. Future studies of animals should allow us to test this hypothesis directly, as well as its relevance to Zellweger syndrome pathogenesis.
Our results also have implications for the disparate topics of Zellweger syndrome diagnosis and PEX11 function. Current diagnostic procedures for Zellweger syndrome and other peroxisomal disorders (24, 38) would incorrectly exclude a generalized peroxisomal dysfunction in a patient with PEX11β deficiency, even though these patients display defects in two unrelated peroxisomal metabolic pathways. Therefore, we propose that current diagnostic protocols be modified to include the possibility of PEX11β deficiency in the human population. With regard to the molecular role of PEX11β and other PEX11 proteins in peroxisome biogenesis, there are two competing hypotheses. One predicts that PEX11 proteins play an important role in peroxisome division (6, 12, 21), and the other holds that PEX11 proteins play a direct role in medium-chain fatty acid oxidation and affect peroxisome abundance only indirectly (35). This issue was addressed in detail in a previous report, which concluded that PEX11 proteins act directly in peroxisome division (18). This conclusion is supported by the observation here that PEX11β−/− mice are defective in two unrelated peroxisomal metabolic pathways, a strong indicator that PEX11β plays a more direct role in peroxisome biogenesis than peroxisomal metabolism.
Although the loss of PEX11β is lethal, PEX11β−/− mice still contain an intact PEX11α gene and do not lack PEX11 activity altogether. In fact, the loss of PEX11β induces a slight increase (∼1.5×) in hepatic expression of PEX11α (data not shown). A more complete understanding of PEX11 function in mammals requires the generation and analysis of mice lacking all PEX11 genes.
Acknowledgments
We thank the Johns Hopkins University Transgenic Core Facility for blastocyst injections and generation of chimeric animals, Ann and Hugo Moser for the use of their laboratory for biochemical experiments, and Stephanie Mihalik and Paul Watkins for help with the peroxisomal α- and β-oxidation activity measurements.
This work was supported by grants from the National Institutes of Health to S.J.G. (DK59479) and D.V. (HD10981). D.V. is an investigator of the Howard Hughes Medical Institute. E.B. was supported by a Max-Kade Scholarship (Max-Kade Foundation, New York, N.Y., and Deutsche Forschungsgemeinschaft).
REFERENCES
- 1.Abe, I., and Y. Fujiki. 1998. cDNA cloning and characterization of a constitutively expressed isoform of the human peroxin Pex11p. Biochem. Biophys. Res. Commun. 252:529-533. [DOI] [PubMed] [Google Scholar]
- 2.Abe, I., K. Okumoto, S. Tamura, and Y. Fujiki. 1998. Clofibrate-inducible, 28-kDa peroxisomal integral membrane protein is encoded by PEX11. FEBS Lett. 431:468-472. [DOI] [PubMed] [Google Scholar]
- 3.Baes, M., P. Gressens, E. Baumgart, P. Carmeliet, M. Casteels, M. Fransen, P. Evrard, D. Fahimi, P. E. Declercq, D. Collen, P. P. van Veldhoven, and G. P. Mannaerts. 1997. A mouse model for Zellweger syndrome. Nat. Genet. 17:49-57. [DOI] [PubMed] [Google Scholar]
- 4.Baes, M., S. Huyghe, P. Carmeliet, P. E. Declercq, D. Collen, G. P. Mannaerts, and P. P. Van Veldhoven. 2000. Inactivation of the peroxisomal multifunctional protein-2 in mice impedes the degradation of not only 2-methyl-branched fatty acids and bile acid intermediates but also of very long chain fatty acids. J. Biol. Chem. 275:16329-16336. [DOI] [PubMed] [Google Scholar]
- 5.Baumgart, E., I. Vanhorebeek, M. Grabenbauer, M. Borgers, P. E. Declercq, H. D. Fahimi, and M. Baes. 2001. Mitochondrial alterations caused by defective peroxisomal biogenesis in a mouse model for Zellweger syndrome (PEX5 knockout mouse). Am. J. Pathol. 159:1477-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erdmann, R., and G. Blobel. 1995. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 128:509-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evan, G. E., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fahimi, H. D. 1969. Cytochemical localization of peroxidatic activity of catalase in rat hepatic microbodies (peroxisomes). J. Cell Biol. 43:275-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faust, P. L., and M. E. Hatten. 1997. Targeted deletion of the PEX2 peroxisome assembly gene in mice provides a model for Zellweger syndrome, a human neuronal migration disorder. J. Cell Biol. 139:1293-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldfischer, S., C. L. Moore, A. B. Johnson, A. J. Spiro, M. P. Valsmis, H. K. Wisniewski, R. H. Ritch, W. T. Norton, I. Rapin, and L. M. Gartner. 1973. Peroxisomal and mitochondrial defects in the cerebro-hepato-renal syndrome. Science 182:62-64. [DOI] [PubMed] [Google Scholar]
- 11.Gould, S. G., D. Valle, and G. V. Raymond. 2001. The peroxisome biogenesis disorders, p. 3181-3217. In C. R. Scriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic and molecular bases of inherited disease, 8th ed., vol. 2. McGraw-Hill, New York, N.Y. [Google Scholar]
- 12.Gould, S. J., and D. Valle. 2000. The genetics and cell biology of the peroxisome biogenesis disorders. Trends Genet. 16:340-344. [DOI] [PubMed] [Google Scholar]
- 13.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Kaufmann, W. E., C. Theda, S. Naidu, P. A. Watkins, A. B. Moser, and H. W. Moser. 1996. Neuronal migration abnormality in peroxisomal bifunctional enzyme defect. Ann. Neurol. 39:268-271. [DOI] [PubMed] [Google Scholar]
- 15.Kersten, S., B. Desvergne, and W. Wahli. 2000. Roles of PPARs in health and disease. Nature 405:421-424. [DOI] [PubMed] [Google Scholar]
- 16.Kitareewan, S., L. T. Burka, K. B. Tomer, C. E. Parker, L. J. Deterding, R. D. Stevens, B. M. Forman, D. E. Mais, R. A. Heyman, T. McMorris, and C. Weinberger. 1996. Phytol metabolites are circulating dietary factors that activate the nuclear receptor RXR. Mol. Biol. Cell 7:1153-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemotte, P. K., S. Keidel, and C. M. Apfel. 1996. Phytanic acid is a retinoid X receptor ligand. Eur. J. Biochem. 236:328-333. [DOI] [PubMed] [Google Scholar]
- 18.Li, X., and S. G. Gould. 2002. PEX11 promotes peroxisome division independently of peroxisome metabolism. J. Cell Biol. 156:643-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenz, P., A. G. Maier, E. Baumgart, R. Erdmann, and C. Clayton. 1998. Elongation and clustering of glycosomes in Trypanosoma brucei overexpressing the glycosomal Pex11p. EMBO J. 17:3542-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, J. F., A. M. Lawler, P. A. Watkins, J. M. Powers, A. B. Moser, H. W. Moser, and K. D. Smith. 1997. A mouse model for X-linked adrenoleukodystrophy. Proc. Natl. Acad. Sci. USA 94:9366-9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marshall, P., Y. Krimkevich, R. Lark, J. Dyer, M. Veenhuis, and J. Goodman. 1995. Pmp27 promotes peroxisomal proliferation. J. Cell Biol. 129:345-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mihalik, S., J. Morrell, D. Kim, K. Sacksteder, P. Watkins, and S. Gould. 1997. Identification of PAHX as a Refsum disease gene. Nat. Genet. 17:185-189. [DOI] [PubMed] [Google Scholar]
- 23.Moser, A., M. Rasmussen, S. Naidu, P. Watkins, M. McGuinness, A. Hajra, G. Chen, G. Raymond, A. Liu, D. Gordon, K. Garnaas, D. Walton, O. Okjeldal, M. Guggenheim, L. Jackson, E. Elias, and H. Moser. 1995. Phenotype of patients with peroxisomal disorders subdivided into sixteen complementation groups. J. Pediatr. 127:13-22. [DOI] [PubMed] [Google Scholar]
- 24.Moser, A. B., N. Kreiter, L. Bezman, S. Lu, G. V. Raymond, S. Naidu, and H. W. Moser. 1999. Plasma very long chain fatty acids in 3,000 peroxisome disease patients and 29,000 controls. Ann. Neurol. 45:100-110. [DOI] [PubMed] [Google Scholar]
- 25.Moser, H. W. 1999. Genotype-phenotype correlations in disorders of peroxisome biogenesis. Mol. Genet. Metab. 68:316-327. [DOI] [PubMed] [Google Scholar]
- 26.Moser, H. W., K. D. Smith, P. A. Watkins, J. Powers, and A. B. Moser. 2001. X-linked adrenoleukodystrophy, p. 3257-3301. In C. R. Scriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic and molecular bases of inherited disease, 8th ed., vol. 2. McGraw-Hill, New York, N.Y. [Google Scholar]
- 27.Passreiter, M., M. Anton, D. Lay, R. Frank, C. Harter, F. T. Wieland, K. Gorgas, and W. W. Just. 1998. Peroxisome biogenesis: involvement of ARF and coatomer. J. Cell Biol. 141:373-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powers, J. M., and H. W. Moser. 1998. Peroxisomal disorders: genotype, phenotype, major neuropathologic lesions, and pathogenesis. Brain Pathol. 8:101-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roscher, A., B. Molzer, H. Bernheimer, S. Stockler, I. Mutz, and F. Paltauf. 1985. The cerebrohepatorenal (Zellweger) syndrome: an improved method for the biochemical diagnosis and its potential value for prenatal detection. Pediatr Res. 19:930-933. [DOI] [PubMed] [Google Scholar]
- 30.Sacksteder, K. A., and S. J. Gould. 2000. The genetics of peroxisome biogenesis. Annu. Rev. Genet. 34:623-652. [DOI] [PubMed] [Google Scholar]
- 31.Sacksteder, K. A., J. M. Jones, S. T. South, X. Li, Y. Liu, and S. J. Gould. 2000. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J. Cell Biol. 148:931-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakai, Y., P. A. Marshall, A. Saiganji, K. Takabe, H. Saiki, N. Kato, and J. M. Goodman. 1995. The Candida boidinii peroxisomal membrane protein Pmp30 has a role in peroxisomal proliferation and is functionally homologous to Pmp27 from Saccharomyces cerevisiae. J. Bacteriol. 177:6773-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Schrader, M., B. E. Reuber, J. C. Morrell, G. Jimenez-Sanchez, C. Obie, T. Stroh, D. Valle, T. A. Schroer, and S. J. Gould. 1998. Expression of PEX11β mediates peroxisome proliferation in the absence of extracellular stimuli. J. Biol. Chem. 273:29607-29614. [DOI] [PubMed] [Google Scholar]
- 35.van Roermund, C. W., H. F. Tabak, M. van Den Berg, R. J. Wanders, and E. H. Hettema. 2000. Pex11p plays a primary role in medium-chain fatty acid oxidation, a process that affects peroxisome number and size in Saccharomyces cerevisiae. J. Cell Biol. 150:489-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wanders, R. J., and J. M. Tager. 1998. Lipid metabolism in peroxisomes in relation to human disease. Mol. Aspects Med. 19:69-154. [DOI] [PubMed] [Google Scholar]
- 37.Wanders, R. J., P. Vreken, S. Ferdinandusse, G. A. Jansen, H. R. Waterham, C. W. van Roermund, and E. G. Van Grunsven. 2001. Peroxisomal fatty acid alpha- and beta-oxidation in humans: enzymology, peroxisomal metabolite transporters and peroxisomal diseases. Biochem. Soc. Trans. 29:250-267. [DOI] [PubMed] [Google Scholar]
- 38.Wanders, R. J. A., P. G. Barth, and H. S. A. Heymans. 2001. Single peroxisomal enzyme deficiencies, p. 3219-3256. In C. R. Scriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic and molecular bases of inherited disease, 8th ed., vol. 2. McGraw-Hill, New York, N.Y. [Google Scholar]
- 39.Wanders, R. J. A., C. Jakobs, and O. H. Skejldal. 2001. Refsum disease, p. 3303-3322. In C. R. Scriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic and molecular bases of inherited disease, 8th ed., vol. 2. McGraw-Hill, New York, N.Y. [Google Scholar]
- 40.Wang, T., A. M. Lawler, G. Steel, and D. Valle. 1995. Paradoxical hypoornithenemia and neonatal lethality in mice with targeted disruption of the OAT gene. Nat. Genet. 11:185-190. [DOI] [PubMed] [Google Scholar]
- 41.Watkins, P., E. J. Ferrell, J. Pedersen, and G. Hoefler. 1991. Peroxisomal fatty acid β-oxidation in HepG2 cells. Arch. Biochem. Biophys. 289:329-366. [DOI] [PubMed] [Google Scholar]
- 42.Zenger-Hain, J., D. A. Craft, and W. B. Rizzo. 1992. Diagnosis of inborn errors of phytanic acid oxidation with tritiated phytanic acid. Prog. Clin. Biol. Res. 375:399-407. [PubMed] [Google Scholar]