Abstract
The mouse genome contains two Sim genes, Sim1 and Sim2. They are presumed to be important for central nervous system (CNS) development because they are homologous to the Drosophila single-minded (sim) gene, mutations in which cause a complete loss of CNS midline cells. In the mammalian CNS, Sim2 and Sim1 are coexpressed in the paraventricular nucleus (PVN). While Sim1 is essential for the development of the PVN (J. L. Michaud, T. Rosenquist, N. R. May, and C.-M. Fan, Genes Dev. 12:3264-3275, 1998), we report here that Sim2 mutant has a normal PVN. Analyses of the Sim1 and Sim2 compound mutants did not reveal obvious genetic interaction between them in PVN histogenesis. However, Sim2 mutant mice die within 3 days of birth due to lung atelectasis and breathing failure. We attribute the diminished efficacy of lung inflation to the compromised structural components surrounding the pleural cavity, which include rib protrusions, abnormal intercostal muscle attachments, diaphragm hypoplasia, and pleural mesothelium tearing. Although each of these structures is minimally affected, we propose that their combined effects lead to the mechanical failure of lung inflation and death. Sim2 mutants also develop congenital scoliosis, reflected by the unequal sizes of the left and right vertebrae and ribs. The temporal and spatial expression patterns of Sim2 in these skeletal elements suggest that Sim2 regulates their growth and/or integrity.
The transcription factors SIM1 and SIM2 are presumed to play important roles during mammalian development based on their region-specific expression patterns throughout development (reviewed in reference 24). In fact, Sim1 has been shown to be essential in hypothalamus development by targeted gene inactivation in the mouse (25). In the absence of Sim1 function, the paraventrivular nucleus (PVN) and supraoptic nucleus (SON) of the hypothalamus fail to develop. PVN and SON play important roles in modulating hormone secretion of the pituitary to maintain homeostasis. SIM2, on the other hand, was first identified by exon trapping of a region of the human chromosome 21 (2, 9) that is known to be associated with many of the pathological features of Down syndrome (DS) (10). Its homolog was subsequently found to be located at the syntenic region on chromosome 16 in the mouse (15, 29, 47). The expression pattern of mouse Sim2 in the brain, craniofacial structures, and muscles suggests that it may contribute to the mental retardation, facial dysmorphology, and hypotonia of DS patients (9, 15, 47). Transgenic mice overexpressing Sim2 display learning impairment (4, 13) and reduced fear response (4)—but not other phenotypes related to DS. Further insight into the normal function of Sim2 requires gene inactivation studies.
Both SIM1 and SIM2 are members of a family of transcription factors characterized by the presence of basic helix-loop-helix (bHLH) and PAS (for Per, ARNT, and Sim) domains (reviewed in references 8, 18, and 36). They are highly homologous to fly (Drosophila melanogaster) SIM domain (6). In the fly, the sim gene directs the development of the cells located at the midline of the developing central nervous system (CNS) (5, 6). Many downstream transcription targets of Sim have been identified (7). Although unique in their own category due to the PAS domain, the bHLH-PAS proteins form selective heterodimers (43), as do their related bHLH proteins (45). For example, Sim and the bHLH-PAS protein Tango heterodimerize to bind to their responsive element, called the CME (for CNS-midline element), to activate transcription (40). In this complex, Tango is a general dimerization partner and Sim is a tissue-specific partner. Similarly in the mouse, SIM1 and SIM2 can heterodimerize with the Tango-related proteins ARNT, ARNT2, and BMAL1 (12, 20, 26, 31, 35, 43). Importantly, Sim1 and Arnt2 mutant mice have identical PVN and SON defects in the hypothalamus (19, 22, 26), demonstrating their partnership. While most bHLH-PAS proteins are transcriptional activators, as assessed by their ability to activate reporter genes via their cognitive binding sites in cultured cells, SIM2, in conjunction with ARNT or ARNT2, acts as a repressor in this assay, indicating its unique property (12, 30, 31, 35). The repressor activity has been localized to the C-terminal half of SIM2 (12, 30, 31), which has no clear similarity to other proteins, not even to the C termini of Sim and SIM1 (3, 12, 15, 31).
In view of the unique repressor activity of SIM2 and the intriguing phenotype found in Sim2 overexpression studies, a mutant animal with loss of Sim2 is needed to assess its function. Here, we describe phenotypic analyses of mutant animals carrying a targeted null Sim2 allele. Despite its overlapping expression with Sim1 in the PVN, Sim2 function appears dispensable for PVN development. Instead, Sim2 mutants die perinatally due to a failure to maintain efficient breathing, as indicated by lung atelectasis. Our data show that the dyspnea and poor breathing efficacy are not due to an intrinsic lung defect. Rather, we suggest that multiple defects in the vertebrae, ribs, pleural wall, and diaphragm—structures that mechanically inflate the lung—contribute to the gradual deterioration in breathing efficacy in the mutant.
MATERIALS AND METHODS
Generation of Sim2 mutant mice.
Sim2 mutant mice were generated by homologous recombination in ES cells. The targeting construct is diagrammed in Fig. 1A and summarized below. An 8-kb BamHI fragment containing the first exon of Sim2 was used. Four hundred twenty-two base pairs of the 5′ untranslated region (UTR) and the first 144 bp of Sim2 coding region were replaced with a PGK-neo cassette, resulting in a 1.2-kb 5′ arm and a 6.2-kb 3′ arm. The PGK-tk cassette was placed at the 5′ end. The construct was linearized by NotI and transfected into ES cells. The frequency of recombination was 1/70 to 1/72 from three independent tries, but no germ line transmission was obtained in these early efforts. Later, M. Shamblott, A. Lawer, and J. Gearhart at Johns Hopkins Medical School obtained and used this construct to generate one germ line-transmitting recombined ES clone. An initial report was given as a poster presentation at an American Society of Human Genetics meeting (M. J. Shamblott, E. M. Bugg, A. M. Lawler, C. M. Fan, M. E. Blue, and J. D. Gearhart, Meeting Am. Soc. Hum. Genet., abstr. A393, 1997). One heterozygous female was provided by Shamblott et al. for our analysis in conjunction with the Sim1 mutant. This female was crossed to a BL6 male. Backcrosses to BL6 mice were performed for five successive generations. The colony was maintained by sib-matings of the backcrossed animals. The genotype was determined by Southern blotting or PCR. Two sets of primers were used in PCR. One set amplifies a 189-bp fragment of the neo coding region (5′-CTCGGCAGGAGCAAGGTGAGATG and 5′-GTCAAGACCGACCTGTCCGGTGC), and the other set amplifies a 135-bp fragment containing the wild-type Sim2 bHLH domain that is deleted in the mutant allele (5′-AGCCGAGGCGCGATGAAGGAGAA and 5′-TGGACGCTTTGTCCAGCTGCGAG).
FIG. 1.
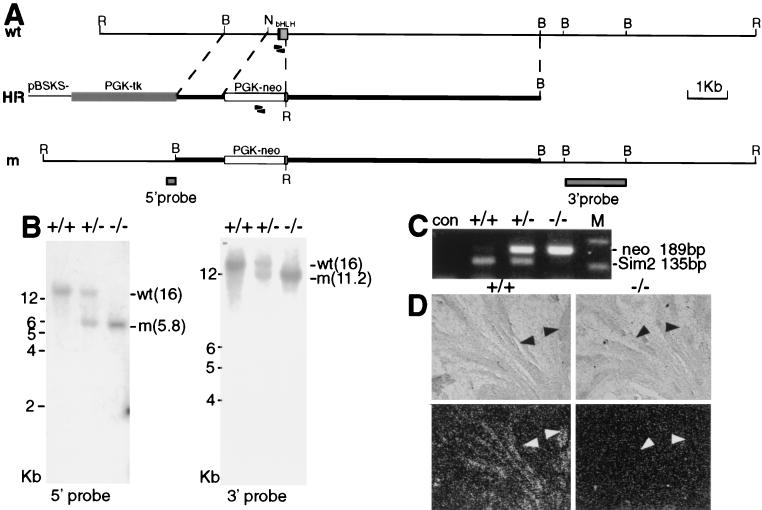
Generation of the Sim2 mutant. (A) The top line shows the genomic organization of the wild-type (wt) Sim2 locus, the middle line shows the homologous recombination vector (HR), and the bottom line shows the recombined mutant allele (m). PGK-neo and PGK-tk are cassettes for positive and negative selections. BamHI, B; EcoRI, R; NotI, N. The size bar = 1 kb. Arrowheads indicate the positions of the primers used for PCR, and shaded boxes indicate the 5′ and 3′ probes. (B) Southern blot analyses with the 5′ (left) and 3′ (right) probes in panel A. EcoRI was used to digest the genomic DNA of the wild type (+/+), heterozygous (+/−), and mutant (−/−) mice. The size markers (kilobases) and the wild-type and mutant bands (in kilobases) are as labeled. (C) PCR of the genomic DNA to show that Sim2 bHLH region is deleted in the mutant. The neo sequence is detected as a 189-bp fragment, while the Sim2 bHLH domain is detected as a 135-bp fragment. Con, without DNA input; M, 123-bp marker. (D) Section in situ hybridization of wild-type and mutant kidneys with a 35S-UTP-labeled Sim2 3′UTR (not deleted in the mutant) probe. The top panels show the phase images, and the bottom panels show the corresponding dark-field images. Sim2 is normally expressed in the collecting ducts, but not in the mutant (black and white arrowheads).
Generation of double mutants.
The Sim1 mutant was described in reference 25. Sim1 and Sim2 heterozygotes were mated to generate double heterozygotes. Double heterozygotes, identified by Southern blotting, were mated to obtain double-null mutants of Sim1 and Sim2. Descendants of these matings were also genotyped by Southern blotting.
Histology and electron microscopy.
All protocols described below for tissue harvesting and physiological tests of experimental animals were reviewed and approved by the Institutional Animal Care and Use Committee. Embryos or newborns were fixed in Bouin's or Carnoy's solution, dehydrated through an ethanol series, treated with toluene, embedded in paraffin, and sectioned at a thickness of 8 μm. Mallory's reagent, PAS reagent, and hematoxylin, and eosin (Surgipath) were used as histological stains (48). Skeleton preparation with alcian blue and alizarin red was performed according to standard procedures. For electron microscopy, lungs were fixed in 2% formaldehyde-1% glutaraldehyde overnight, processed for plastic sections, and subjected to microscopy (48).
Immunocytochemistry.
Lungs and brains were fixed in 4% paraformaldehyde for 2 h at 4°C, processed through 30% sucrose, and embedded in OTC compound (VWR) for frozen sections at a thickness of 12 μm. 2H3 and MF-20 antibodies (Abs) were obtained from the Hybridoma Bank and used at dilutions of 1:3 and 1:5, respectively. Bromodeoxyuridine (BrdU), CGRP (Sigma), and VP and SS (Peninsula Laboratories) Abs were used according to the manufacturer's instructions. Secondary Abs were horseradish peroxidase (HRP) conjugated, and the Vecta-stain kit was used for color reaction.
In situ hybridization.
35S-UTP-labeled antisense RNA probes to Sim1 and Sim2 were generated, and the in situ hybridization was performed according to the protocol previously described (15). The Trh and Crh probes were described previously (25, 37).
RT-PCR assay.
One microgram of total RNA was used as starting material for reverse transcription-PCR (RT-PCR). To detect Sim2 mRNA (5′-AGCCGAGGCGCGATGAAGGAGAA and 5′-ATGGGATCCCAGCTCTTTTG; product, 260 bp) in the lung, 35 cycles of PCR were used. To detect surfactants A, B, C, and D (SP-A, -B, -C, and -D, respectively) and CC-10, 30 cycles of PCR were used. The primer sequences are described in reference 46 and are available upon request. β-Actin (25 cycles of PCR) was assayed as a control (1).
Glucocorticoid injection.
Pregnant females were injected twice at post-plug days 15.5 and 16.5. Two nanograms of glucocorticoid per g of body weight was injected each time (39). The newborn pups were monitored for their breathing and for lethality.
RESULTS
Sim2 mutants die perinatally.
The configuration of the targeting vector and the predicted recombination event are diagrammed in Fig. 1A. Southern blot analyses with the appropriate 5′ and 3′ probes to the Sim2 locus on the wild-type (+/+), heterozygous (+/−), and mutant (−/−) genomic DNA samples (Fig. 1B) showed that the targeted allele is a result of homologous recombination. Since the initiation methionine and the first 48 amino acids of the 56-amino-acid bHLH domain were deleted, this allele is most likely a null. PCR analysis confirmed that the deleted bHLH portion is absent in the mutant genome (Fig. 1C). In situ hybridization with a probe to the 3′UTR (not deleted) detected Sim2 mRNA in the collecting ducts of the Sim2+/+ kidney, but not in those of the Sim2−/− kidney (Fig. 1D). (Note that Sim2−/− kidney appears normal and contains collecting ducts.) This further establishes that the mutant allele is a null, because the mRNA is not detectable.
Sim2 mutants were born alive at a Mendelian ratio (58:133:50 for +/+:+/−:−/−) and could not be distinguished from their siblings initially. However, most mutants died within 3 days of birth (Table 1). Prior to death, mutants displayed severe respiratory distress (manifested by dyspnea and aerophagia) and cyanosis, suggesting that poor oxygenation due to breathing difficulty is the cause of death. The longest-surviving mutant was 4 months old and died of unknown causes.
TABLE 1.
Sim2 mutants die perinatally
| Agea | No. of live pups | Genotype | % |
|---|---|---|---|
| E18.5 | 4 | +/+ | 17 |
| 14 | +/− | 58 | |
| 6 | −/− | 25 | |
| P0 | 58 | +/+ | 24 |
| 133 | +/− | 55 | |
| 50 | −/− | 21 | |
| P1 | 13 | +/+ | 29 |
| 28 | +/− | 62 | |
| 4 | −/− | 9 | |
| P2 | 18 | +/+ | 35 |
| 33 | +/− | 63 | |
| 1 | −/− | 2 |
P0 is defined as the day of birth, and P1 and P2 represent 1 and 2 days after birth, respectively.
Sim2 is expressed in the CNS and the mesodermal tissues.
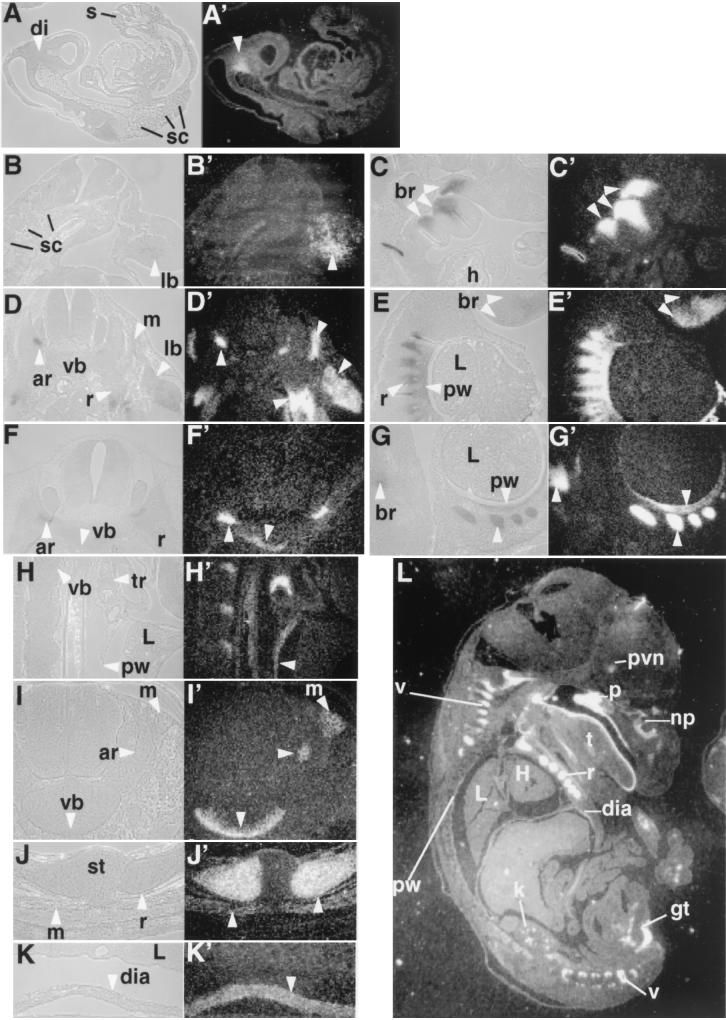
Perinatal lethality suggests that a defect or defects arose embryonically. To facilitate the localization of the tissue defect or defects that may cause breathing failure in the mutant, we reexamined the Sim2 expression pattern during development. From embryonic development day 8.0 (E8.0) to E10.0, Sim2 expression is predominantly detected in the ventroposterior diencephalon (15, 47) (Fig. 2A and A′, at E9.5) where it overlaps with the Sonic Hedgehog (Shh) expression domain (14). Although ectopic expression of Sim2 activates Shh, Sim2−/− and Sim1−/−/Sim2−/− embryos have normal Shh expression (14), indicating that Sim1 and Sim2 are not essential for Shh expression. Sim2 expression in the diencephalon persists until birth (Fig. 2L) (see below).
FIG. 2.
Expression pattern of Sim2. In situ hybridization with 35S-labeled Sim2 probe was used to assess the Sim2 expression pattern. Embryos were Carnoy's solution fixed, paraffin embedded, and sectioned at 8 μm. The histology of the sections is on the left next to the corresponding dark-field image revealing Sim2 expression (the silver granules). (A and A′) Midsagittal section of an E9.5 embryo. di, diencephalon. (B and B′) Transverse sections of an E10.5 embryo. (C and C′) Sagittal sections of an E10.5 embryo. sc, sclerotome; br, branchial arches; h, heart; lb, limb. (D and D′) Transverse sections of an E11.5 embryo. (E and E′) Sagittal sections of an E11.5 embryo. ar, vertebral arch; vb, vertebral body; r, ribs; L, lung; pw, pleural wall. (F and F′) Transverse sections of an E12.5 embryo. (G and G′) Sagittal sections of an E12.5 embryo. (L) Sagittal section of an E16.5 embryo. Sim2 shows expression in the PVN, kidney (k), muscles in the genital tubercles (gt), palate (p), nasal pit (np), tongue (t), ribs (r), vertebra (v), diaphragm (dia), and trachea (tr). st, sternum; H, heart. Higher magnifications of sagittal sections of E16.5 pleural cavity (H and H′) and diaphragm (K and K′) and transverse sections of vertebra and erector muscles (I and I′) and ribs (J and J′) are also shown. Arrowheads indicate Sim2-positive structures.
Sim2 is also expressed in mesoderm-derived tissues after midgestation (15). At E10.5, Sim2 mRNA is first detected in the limb mesenchyme and branchial arches, but not in the somites (Fig. 2B to C′). The limb and branchial arch expression persists until birth (Fig. 2E, E′, G, G′, and L) (data not shown) and is detectable in epithelium, mesenchyme, muscle, and cartilage cell types. At E11.5, its expression is found in the mesenchyme and mesothelium lining the pleural cavity (Fig. 2E and E′), distal rib cells, and hypaxial myocytes (Fig. 2D and D′). At E12.5 (Fig. 2F to G′), E14.5 (data not shown), and E16.5 (Fig. 2H to L), these sites of expression become organized into the mature pleural wall, rib cartilage, diaphragm, a subset of erector spinae muscles (i.e., the longissimus thoracis and lumborum) of the trunk (but not the intercostal or intervertebral muscles), and the cartilage and muscles in the appendages. Expression in the ventral centrum (i.e., the vertebral body) and the ventral tip of the neural arch is seen at E12.5 and onward. In the skeleton, Sim2 expression is detected in the cartilage, but not in the ossified bone. Finally, Sim2 transcript is not detected in the hindbrain, carotid body, or lung proper in the above stages and newborns (data not shown).
Sim2 mutants have normal craniofacial structures.
Sim2 is highly expressed in the nasal pit, palate, mandible, tongue, and trachea (Fig. 2) (15). We therefore examined whether defects in these regions may obstruct the airway and cause the breathing difficulty of the mutant. Skeletal preparation (with alcian blue and alizarin red dyes) of the Sim2−/− newborn head indicated there were no obvious defects in the mandible (Fig. 3A and B), trachea, and epiglottis (Fig. 3C to D′). When the lower jaw was removed, the mutant palate was found to be joined at the midline like the wild-type palate (Fig. 3E and F). Finally, the isolated mutant nasal cartilage showed no obvious difference from the wild type (Fig. 3G and H). These whole-mount studies (n = 7) were confirmed by histological sections of paraffin-embedded newborn heads (Fig. 3I and J; n = 10). Thus, the cause of respiratory distress in Sim2 mutants is not due to abnormal upper airway structure.
FIG.3.
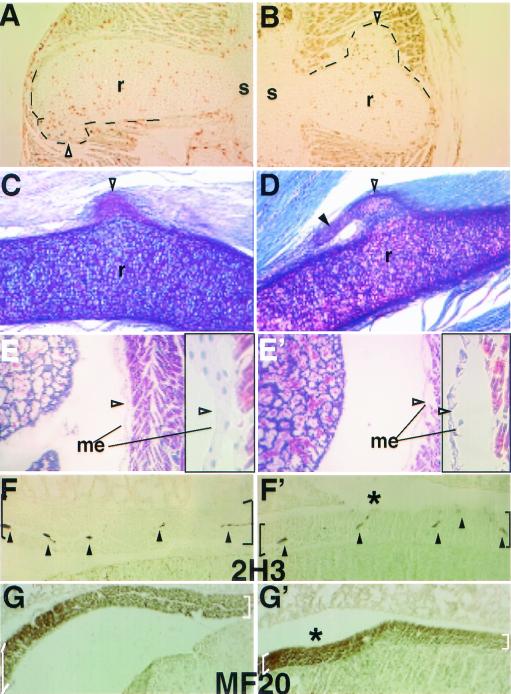
Axial skeleton phenotype of the Sim2 mutant: Shown are skeleton preparations of the newborn wild-type (+/+) (A, C, C′, E, G, I, and K) and mutant Sim2 (−/−) (B, D, D′, F, H, J, and L) siblings with alcian blue and alizarin red. Panels A and B show sagittal views of the heads. Panels C and D show frontal views of the trachea. Panels C′ and D′ show top views of the trachea and epiglottis (open arrowheads). Panels E and F show ventral views of the palate. Asterisks label the palatal bones, and arrowheads indicate the fusion midlines. Panels G and H show dorsal views of the isolated nasal cartilage (nc). Panels I and J show coronal sections of the heads stained with hematoxylin. ob, olfactory bulb; p, palate; t, tongue; np, nasal pit. Panels K and L show frontal partial views of the rib cages. White arrowheads indicate the outgrowths on the ribs in panel L. Panel M shows skeleton preparation illustrating the scoliosis of the Sim2 mutant on the right (−/−); the wild-type littermate is on the left (+/+). Panel N shows an example of an asymmetric T2 vertebra/rib element from a mutant. The bent line through the center of the vertebral body and the spinal process indicate that the left arch is larger, a cause of scoliosis.
Sim2 mutants have abnormal ribs, vertebrae, and diaphragm.
We next examined the structures (which express Sim2) surrounding the pleural cavity that mechanically drive breathing motion. The skeletal preparation showed small protrusions on the ribs of newborn Sim2 mutants (Fig. 3L). They occurred close to the sternum and randomly at all levels. This phenotype was also found in the heterozygotes, although it was less frequent and with a smaller number of protrusions (Table 2). In addition, we found that incompletely penetrant scoliosis, defined by spine curvature greater than 10°, occurred in both the heterozygotes and mutants (Fig. 3M). The scoliosis was found mainly at the thoracic level and was never more than 30°. The curvature was either deviated to the right or the left without predominance (Table 2). Each vertebra or rib element was isolated and examined. In the more severe cases, we found macroscopic differences in the sizes of the left and right ribs and neural arches at the thoracic level (Fig. 3N). The visibly longer ribs, when found, were located on the concave side of the curvature. We did not observe misshapen vertebral body, hemivertebra, or missing vertebra.
TABLE 2.
Axial skeletal defects in newborn Sim2 mutants
| Defect | Genotype | No. affected/examined | Effecta |
|---|---|---|---|
| Scoliosis | +/+ | 1/14 | 1R |
| +/− | 10/21 | 4L/6R | |
| −/− | 11/24 | 5L/6R | |
| Rib protrusions | +/+ | 0/14 | |
| +/− | 5/21 | 2-5 | |
| −/− | 24/24 | 14-23 |
For scoliosis, the number of affected animals whose scoliosis displays left (L) or right (R) curvature is shown. For the rib protrusion defect, the range of numbers of protrusions found in these affected animals is shown. The numbers are based on visible protrusions per animal by alcian blue staining.
Even though Sim2 is expressed in the ribs as early as E11.5, we did not find visible protrusions on the mutant ribs until E16.5 (two out of six examined). To determine whether these rib protrusions are due to abnormal proliferation, we performed 12-h BrdU incorporation between E18.5 and E19.0. In and near the base of all protrusions, we found more (although highly variable) BrdU-positive cells (Fig. 4A and B) than in other regions of the ribs, consistent with increased proliferation. PAS staining of the sectioned ribs indicated that the protrusions can form attachments to the intercostal muscles (Fig. 4C and D), implying a potential to disrupt their integrity and function.
FIG. 4.
Multiple phenotypes of the Sim2 mutant may contribute to lung atelectasis. Panels A and B show two examples of 12-h BrdU-labeled E18.5 ribs. Proliferating cells (HRP-positive brown nuclei) were found in the rib protrusions. r, rib; s, sternum. Black lines outline the protrusions. Panels C and D show two examples of the PAS-stained rib protrusions (open arrowheads) and their connections to the intercostal muscle. A solid arrowhead marks the extension off of some protrusions. Panels E (+/+) and E′ (−/−) show Mallory's staining of the pleural mesothelium basement membrane in the mutant indicating it was disrupted after the mutant had shown severe dyspnea. Before severe dyspnea, however, the mutant mesothelium appeared normal (data not shown). Higher magnification of the mesothelium (arrowhead) is to the right of each figure in black frames. Panels F (+/+) and F′ (−/−) show 2H3 (antineurofilament) staining of the innervated nerves (arrowheads) in the diaphragm (black brackets). Panels G (+/+) and G′ (−/−) show MF20 (antimyosin) staining of muscle fibers in the diaphragm (white brackets). Table 2 provides a summary of the frequency and phenotype of rib protrusions and scoliosis.
The mutant diaphragm was thinner than those of the wild type and heterozygote (Fig. 4F to G′) (Sim2−/−, 395 ± 60.0 μm; Sim2+/−, 522 ± 51.4 μm; Sim2+/+, 539± 80.2 μm; n = 6 each; P < 0.01 by t test), suggesting a compromised breathing action. By using 2H3 (antineurofilament) and MF20 (anti-muscle myosin) Abs, we found that the mutant and wild-type diaphragms contained similar nerve densities (Fig. 4F and F′) and organized striated muscle (Fig. 4G and G′). The density of the mutant intercostal muscle innervation was also normal by 2H3 staining (data not shown). Finally, although there was no observable difference in pleural mesothelium basement membrane between the mutants and controls between E12.5 and birth, the basement membrane appeared disrupted in the mutants after they already displayed severe dyspnea (Fig. 4E′; n = 3). This defect is consistent with a mechanical disruption—possibly a secondary consequence of irregular pleural pressure.
Sim2 mutant displays lung atelectasis.
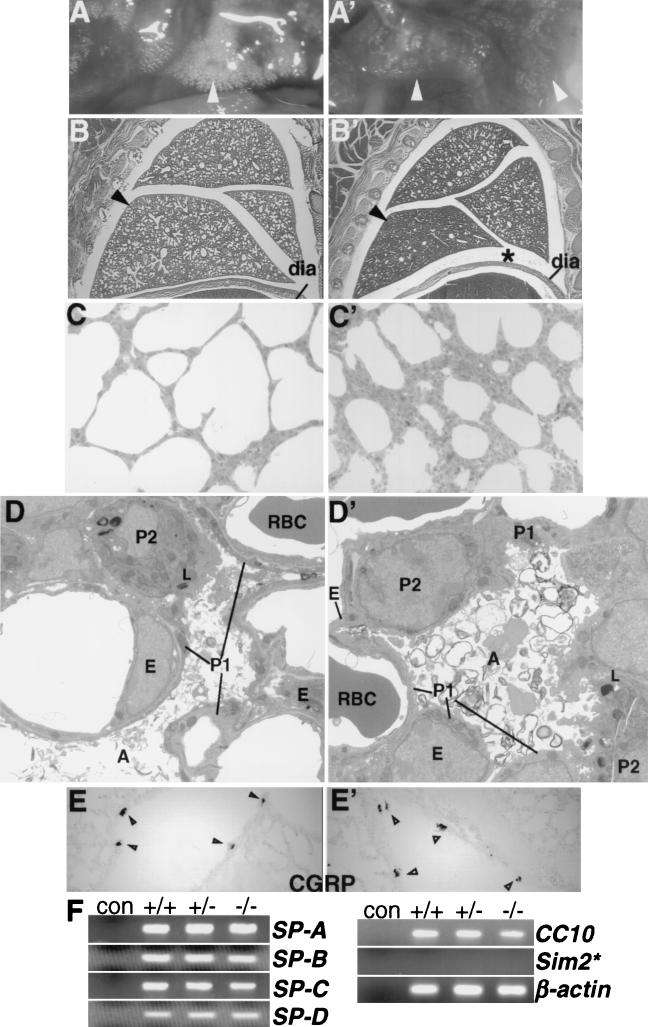
In order to characterize the consequences of the observed respiratory failure of Sim2 mutant mice, we autopsied their lungs. The mutant lungs showed decreased inflation, and the stomach and intestine were sometimes distended with air. Even prior to any signs of respiratory distress, the mutant lungs were already only partially filled with air (Fig. 5A ′). Histological analysis confirmed that the pulmonary alveoli were not fully inflated (atelectasis) (Fig. 5B′ and C′). From E12.5 to E18.5, however, the mutant lungs were indistinguishable from the control lungs in size, morphology, and histology (data not shown). The fact that alveolar atelectasis was observed while Sim2 mutants appeared vigorous and healthy indicates that their lungs can sustain survival for a period of time.
FIG. 5.
Lung phenotype of the Sim2 mutant. Panels A (+/+) and A′ (−/−) show whole-mount live lungs in the chest cavities of newborn wild-type and Sim2 mutant mice. Panels B (+/+) and B′ (−/−) show sagittal sections of newborn lungs stained with hematoxylin and eosin. dia, diaphragm. Asterisks indicate the space between the lung and the diaphragm in the mutant. Arrowheads indicate the part of the lung that is the most different in histology. Panels C (+/+) and C′ (−/−) are 1-μm plastic sections of the lungs at magnifications of ×100. Atelectasis in the mutant is evident by the smaller alveolar opening and thicker alveolar wall. Panels D (+/+) and D′ (−/−) show electron microscopy of the alveolus (A), endothelium (E), capillary, erythrocyte (RBC), type I (P1) and II (P2) pneumocytes, lamina body (L), and the surfactant (×1,650). Atmagnifications of ×2,100 and ×4,400, intact basement membranes, cell junctions, smooth muscle actins, and lamina body structures were seen in the mutant. In other fields, macrophages and fibroblast supporting cells also were normal in their position and integrity (data not shown). Panels E (+/+) and E′ (−/−) show CGRP staining of the neuroendocrine cells in the airway epithelium (arrowheads). (F) RT-PCR assay for expression of SP-A, -B, -C, and -D and CC-10, (30 cycles), SIM2 (35 cycles), and β-actin (25 cycles) in the lung (see Materials and Methods). The primers for Sim2 could detect the correct size of product in the kidney RNA samples with 30 cycles of PCR (not shown). β-Actin was assayed for normalization. con, no RNA control.
The lack of Sim2 expression in the lung proper suggests that Sim2 does not contribute to lung development, and the mutant lung develops normally. To demonstrate this, we performed extensive histological and electron microscopic analyses of the mutant lung: The architecture (alveoli, capillaries, basement membranes, and surfactant) and major cell types (erythrocyte, macrophage, endothelium, type I and II pneumocytes, smooth muscle, and supporting fibroblast) were normal in the mutant lung (Fig. 5B to D′) (data not shown) (48). Sim2 mutant lung also contained the same number of CGRP-positive pulmonary neuroendocrine cells as the wild-type lung (Fig. 5E and E′). Finally, Sim2 mutant lung expressed normal levels of surfactants and CC10 transcripts (Fig. 5F) (46), indicating that type II pneumocytes and Clara and endothelial secretory cells are functionally normal.
Alternatively, Sim2 may be required for the maturation of the lung. To explore this possibility, at E15.5 and E16.5, we conducted maternal glucocorticoid administration, a treatment used to accelerate lung maturation and prevent lung atelectasis of premature babies (33, 39). However, the treated mutants still displayed breathing difficulty and died in the same time frame as the nontreated mice (Table 3). This finding is consistent with the normal levels of surfactant mRNAs found in the mutant lung. Thus, instead of an intrinsic lung defect, we suggest that pulmonary atelectasis results from reduced mechanical force to inflate the lung, likely caused by the rib and diaphragm defects.
TABLE 3.
Sim2 mutants are not rescued by glucocorticoid injection
| Agea | No. of live pups | Genotype | % |
|---|---|---|---|
| P0 | 4 | +/+ | 27 |
| 8 | +/− | 53 | |
| 3 | −/− | 20 | |
| P1 | 4 | +/+ | 29 |
| 8 | +/− | 57 | |
| 2 | −/− | 14 | |
| P2 | 9 | +/+ | 39 |
| 13 | +/− | 61 | |
| 0 | −/− | 0 |
P0, P1, and P2 are defined as in Table 1.
Sim2 mutant displays no additive phenotype in the Sim1+/− background for PVN development and obesity.
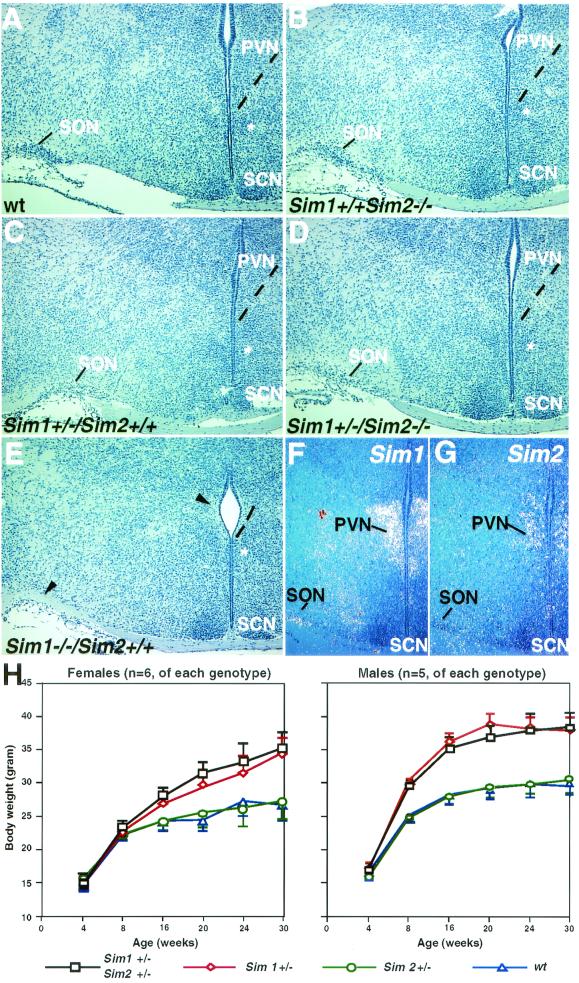
Sim1 is essential for the development of the PVN and SON (24). Sim1−/− mice die perinatally, presumably due to the lack of PVN and SON. Sim2 is also expressed in the PVN, though at a lower level and in a smaller domain than Sim1 (Fig. 2L and Fig. 6F and G) (44). We examined whether Sim2−/− mice also displayed PVN deficiency. In contrast to Sim1−/− mice, Sim2−/− mice had histologically normal PVN and SON (Fig. 6B), indicating that Sim2 does not play an essential role independent of Sim1 here. Furthermore, Sim2 mutant PVN contained neurons, major PVN cell types expressing thyrotropin-releasing hormone, corticotropin-releasing hormone, somatostatin, and vasopressin (data not shown).
FIG. 6.
Sim2 has no obvious genetic interaction with Sim1 in PVN development and energy homeostasis. Brains of wild-type (Sim1+/+/Sim2+/+) (A), Sim2 mutant (Sim1+/+/Sim2−/−) (B), Sim1 heterozygous (Sim1+/−/Sim2+/+) (C), Sim1 heterozygote/Sim2 mutant (Sim1+/−/Sim2−/−) (D), and Sim1 mutant (Sim1−/−) (E) mice were embedded in paraffin, sectioned, and stained with hematoxylin. SCN, suprachiasmatic nucleus. An asterisk labels the medial preoptic nucleus as a reference. Dashed lines outline the ventral boundary of the PVN. Arrowheads in panel E indicate the lack of PVN and SON in Sim1−/− mice. (F and G) 35S radioactive in situ hybridization of the PVN/SON region with Sim1 and Sim2 probes. (H) Six females (left) and five males (right) of each of the following genotypes were used in this study: wild type (wt, triangles), Sim1+/− (diamonds), Sim2+/− (circles), and Sim1+/−/Sim2+/− (squares). Animals were weighed every 4 weeks after birth, continuing on until 30 weeks. The average weight (in grams) was plotted, and the error bars represent standard errors. By Student's t test, there is no significant difference between wild-type and Sim2 heterozygous mice (P > 0.75) or between Sim1+/− and Sim2+/−/Sim1+/− mice (P > 0.8).
To test whether there is a genetic interaction between Sim1 and Sim2 in directing PVN development, we analyzed the Sim1 and Sim2 compound mutants. Since Sim1−/− mouse has no PVN (Fig. 6E), we examined whether in the Sim1+/− background, loss of Sim2 causes a discernible PVN deficit. However, Sim1+/−/Sim2+/− and Sim1+/−/Sim2−/− mice have PVN and SON structures comparable to those of Sim1+/− mice (Fig. 6C and D), suggesting a lack of genetic interaction between them.
Sim1+/− animals are obese (27). This provides us with an additional assay for possible genetic interaction between Sim1 and Sim2. We measured and compared the body weights among the wild-type, Sim1+/−, Sim2+/−, and Sim1+/−/Sim2+/− animals. Both females (n = 6 of each genotype) and males (n = 5 of each genotype) were used. There was no significant difference in weight gains between Sim2+/− and wild-type animals, nor was there a difference between Sim1+/−/Sim2+/− and Sim1+/− animals (Fig. 6H). At the end of the 7-month period, the average weight of Sim2+/− animals was similar to that of the wild-type animals in both sexes (Sim2+/− versus wild type, respectively: females, 27.2 ± 2.7 versus 26.7 ± 2.3 g; males, 30.1 ± 2.1 versus 29.9 ± 1.4 g). The average weight of the Sim1+/−/Sim2+/− animals was similar to that of the Sim1+/− animals (Sim2+/−/Sim1+/− versus Sim1+/−, respectively: females, 35.5 ± 2.4 versus 34.4 ± 2.5 g; males, 38.4 ± 2.1 versus 38.1 ± 1.8 g). Thus, loss of one allele of Sim2 does not cause obesity on its own, nor does it enhance the Sim1+/− phenotype.
DISCUSSION
Sim2 and DS.
Based on overexpression animal studies, Sim2 has been implicated in neurological defects of DS (4, 13). Whether Sim2 normally mediates cognitive function remains to be determined. Craniofacial abnormality is another hallmark of DS (10). Although Sim2 is highly expressed in the developing branchial arches, neither overexpression (4, 13) nor mutant studies reveal its role in craniofacial development. If SIM2 indeed contributes to craniofacial development, it may do so in conjunction with other genes on chromosome 21.
Sim2 and the regulation of rib growth.
The timing of Sim2 expression in the ribs and vertebral arches corresponds to the migration and proliferation phase of these tissues, not their specification (Fig. 2). Consistently, only size difference was observed in the mutant. Despite the visible size variation, we were unable to document a difference in BrdU incorporation between the left and right ribs and vertebral arches, possibly because of low proliferation (sparse BrdU-positive nuclei even after 12 h of incorporation), low penetrance, random side preference, and the animal's bulk. However, the mutant rib protrusions do contain more proliferating cells by BrdU labeling. In conjunction with the increased length of the ribs, this observation suggests that Sim2 inhibits rib growth during development.
Rib protrusions and exostoses.
The rib protrusions observed in Sim2 mutant mice are reminiscent of exostoses (also named osteochondromas), benign protrusions on cartilage and bones with the potential for malignant transformation (34). When found in the thorax, exostoses are typically located at the sternal ends of ribs as seen in the Sim2 mutant. However, some features distinguish the Sim2 mutant protrusions from the typical exostoses. First, exostoses are rarely observed at birth. Second, when multiple, exostoses are not restricted to the ribs. Finally, in humans, SIM2 does not map to the autosomal dominant regions associated with multiple hereditary exostoses (MHE). It has also been proposed that exostoses arise through clonal expansion of a single mutant chondrocyte (34). The pathogenic origin of the Sim2 mutant rib protrusions is not yet known, but it does involve proliferation as shown by BrdU labeling. Of note, a newborn has been described with several exostoses only found in the ribs (11). It is thus possible that Sim2 represents a distinct genetic component for rib-specific exostoses.
Sim2 and the common form of scoliosis.
Congenital scoliosis is typically characterized by abnormally shaped vertebra or vertebrae resulting from a segmentation defect (17). This was not found in the Sim2 mutant, thus distinguishing it from a patterning defect. Patients with idiopathic scoliosis, which presents later in life, tend to show asymmetric ribs, with the longer ribs found on the concave side (32, 41). Three-dimensional modeling of these patients suggests that growth asymmetry of the ribs can initiate scoliosis (38, 41, 42). It remains unclear whether the rib asymmetry observed in these cases is the primary cause or a secondary event. The fact that Sim2 mutant's scoliosis is found at the thoracic level and that the longer ribs are at the concave side of the curvature strongly supports that its scoliosis results from the asymmetry of the ribs. Whether the few erector muscles expressing Sim2 (which are present and appear normal in the mutant) directly contribute to scoliosis has been difficult to assess. Nonetheless, our data suggest that asymmetric growth of the ribs can arise from a single genetic event and cause congenital scoliosis. It is tempting to speculate that SIM2 haploinsufficency in humans contributes to the pathogenesis of congenital or idiopathic scoliosis.
Complexity of the Sim2 mutant pulmonary atelectasis.
Sim2 mutant mice develop severe respiratory distress accompanied by classical alveolar atelectasis of the lung. Several strains of mutant mice also display alveolar atelectasis of perinatal onset. Some mutants are characterized by type II pneumocyte immaturity, a decrease of surfactant level, and pulmonary hypoplasia: e.g., transforming growth factor β3 (TGF-β3) (21), epidermal growth factor receptor (EGFR) (28), and NDST-1 (16) mutants. This is typical of the respiratory distress syndrome of premature babies (33). In other models, such as Sim2 and Gsh4 (23) mutants, alveolar collapse is not associated with decreased surfactants. The physiological basis for this form of atelectasis appears heterogeneous. A decrease of respiratory movements (at birth) secondary to neural involvement has been evoked to account for the atelectasis of mice homozygous for a null allele of Gsh4 (23), which is expressed in the brainstem regions that control breathing. This does not apply to the Sim2 mutant. First, the breathing pattern of Sim2 mutants can be normal for up to 3 days. Second, Sim2 mutant mice display gasping or aerophagia behavior, indicating that they have an intact neural feedback system responding to O2 deprivation. Third, Sim2 expression is not found in the brainstem or the phrenic nerve. Thus, the Sim2 mutant lung phenotype does not appear to be a simple consequence of neurological impairment.
Of note, there appears to be reduced pleural movement and increased abdominal movement in breathing behavior of Sim2 mutants. Therefore, we propose that the thinner diaphragm, abnormal thoracic cage, and compromised intercostal muscle movement together contribute to Sim2 mutant's pulmonary atelectasis. A thin diaphragm of the mutant implies compromised mechanical force to inflate the lung. Abnormal muscle-rib protrusion junctions are likely to contribute to reduced rib cage movement. Scoliosis may also limit pleural expansion (17). We imagine that as the Sim2 mutant brainstem senses low O2 concentration due to poorly inflated alveoli, it triggers compensatory exertion in breathing behavior (dyspnea and aerophagia). The timing of such events varies, dependent on the severity of the rib and diaphragm defects. The continuous exertion eventually leads to pleural mesothelium tearing. Once the pleural wall is torn and the negative pressure is lost, the breathing failure follows, as was observed in the Sim2 mutants.
Although our data support pulmonary atelectasis being secondary to structural defects, it is possible that Sim2 is expressed transiently, at very low levels, or in very few cells of the lung or that surrounding tissues (e.g., the pleural mesothelium, which expresses Sim2) influence lung function. The inserted PGK-neo cassette may also disrupt the regulation of a nearby gene or genes that are important for lung function.
Sim2 mutant displays distinct phenotype from that of Sim1.
Sim1 is essential for PVN development (25). Our analysis of the brains of Sim2−/−, Sim1+/−/Sim2+/−, and Sim1+/−/Sim2−/− mice did not reveal any obvious requirement of Sim2 nor a genetic interaction between Sim1 and Sim2 in PVN development. Sim2, unlike Sim1, also may not contribute to weight regulation. Since we cannot assess the weight gains in Sim2 mutants due to their lethality, the heterozygous test is not definitive to exclude its involvement. Only when Sim2 can be conditionally inactivated to bypass the lethality, will we be able to assess its contribution in body weight regulation decisively.
Acknowledgments
We thank M. Shamblott, A. Lawer, and J. Gearheart for providing the Sim2 heterozygous animal for our analysis with the Sim1 mutant. We also thank members of the Fan laboratory for reading the manuscript. J. Michaud is a clinician-scientist of the Canadian Institutes of Health Research.
This work is supported by NIH grant RO1 HD35596 to C.-M. Fan.
E. Goshu and H. Jin contributed equally to this study.
REFERENCES
- 1.Byragyn, A., S. Arkins, and K. W. Kelley. 1994. Riboprobe expression cassettes for measuring IGF-I, b-actin, and glyceraldehyde 3-phospate dehydrogenase transcripts. J. Immunol. Methods 168:235-244. [DOI] [PubMed] [Google Scholar]
- 2.Chen, H., R. Chrast, C. Rossier, A. Gos, S. E. Antonarakis, J. Kudoh, A. Yamaki, N. Shindoh, H. Maeda, and S. Minoshima. 1995. Single-minded and Down syndrome? Nat. Genet. 10:9-10. [DOI] [PubMed] [Google Scholar]
- 3.Chrast, R., H. S. Scott, H. Chen, J. Kudoh, C. Rossier, S. Minoshima, Y. Wang, N. Shimizu, and S. E. Antonarakis. 1997. Cloning of two human homologs of the Drosophila single-minded gene SIM1 on chromosome 6q and SIM2 on 21q within the Down syndrome chromosomal region. Genome Res. 7:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chrast, R., H. S. Scott, R. Madani, L. Huber, D. P. Wolfer, M. Prinz, A. Aguzzi, H. P. Lipp, and S. E. Antonarakis. 2000. Mice trisomic for a bacterial artificial chromosome with the single-minded 2 gene (Sim2) show phenotypes similar to some of those present in the partial trisomy 16 mouse models of Down syndrome. Hum. Mol. Genet. 9:1853-1864. [DOI] [PubMed] [Google Scholar]
- 5.Crews, S., R. Franks, S. Hu, B. Matthews, and J. Nambu. 1992. Drosophila single-minded gene and the molecular genetics of CNS midline development. J. Exp. Zool. 261:234-244. [DOI] [PubMed] [Google Scholar]
- 6.Crews, S. T., J. B. Thomas, and C. S. Goodman. 1988. The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell 52:143-151. [DOI] [PubMed] [Google Scholar]
- 7.Crews, S. T. 1998. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 12:607-620. [DOI] [PubMed] [Google Scholar]
- 8.Crews, S. T., and C.-M. Fan. 1999. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr. Opin. Genet. Dev. 9:580-587. [DOI] [PubMed] [Google Scholar]
- 9.Dahmane, N., G. Charron, C. Lopes, M. L. Yaspo, C. Maunoury, L. Decorte, P. M. Sinet, B. Bloch, and J. M. Delabar. 1995. Down syndrome-critical region contains a gene homologous to Drosophila sim expressed during rat and human central nervous system development. Proc. Natl. Acad. Sci. USA 92:9191-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delabar, J. M., D. Theophile, Z. Rahmani, Z. Chettouh, J. L. Blouin, M. Prieur, B. Noel, and P. M. Sinet. 1993. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur. J. Hum. Genet. 1:114-124. [DOI] [PubMed] [Google Scholar]
- 11.De Turckheim, M. C., J. M. Clavert, and M. Paira. 1991. Costal exostoses complicated in the neonatal period by brachial plexus paralysis. A distinct entity of exotoses? Ann. Pediatr. 38:23-25. [PubMed] [Google Scholar]
- 12.Ema, M., M. Morita, S. Ikawa, M. Tanaka, Y. Matsuda, O. Gotoh, Y. Saijoh, H. Fujii, H. Hamada, Y. Kikuchi, and Y. Fujii-Kuriyama. 1996. Two new members of the murine Sim gene family are transcriptional repressors and show different expression patterns during mouse embryogenesis. Mol. Cell. Biol. 16:5865-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ema, M., S. Ikegami, T. Hosoya, J. Mimura, H. Ohtani, K. Nakao, K. Inokuchi, M. Katsuki, and Y. Fujii-Kuriyama. 1999. Mild impairment of learning and memory in mice overexpressing the mSim2 gene located on chromosome 16: an animal model of Down's syndrome. Hum. Mol. Genet. 8:1409-1415. [DOI] [PubMed] [Google Scholar]
- 14.Epstein, D. J., L. Matinu, J. L. Michaud, K. M. Losos, C.-M. Fan, and A. L. Joyner. 2000. Members of the bHLH-PAS family regulate Shh transcription in forebrain regions of the mouse CNS. Development 127:1701-1709. [DOI] [PubMed] [Google Scholar]
- 15.Fan, C.-M., E. Kuwana, A. Bulfone, C. F. Fletcher, N. G. Copeland, N. A. Jenkins, S. Crews, S. Martinez, L. Puelles, J. L. Rubenstein, and M. Tessier-Lavigne. 1996. Expression patterns of two murine homologs of Drosophila single-minded suggest possible roles in embryonic patterning and in the pathogenesis of Down syndrome. Mol. Cell Neurosci. 7:1-16. [DOI] [PubMed] [Google Scholar]
- 16.Fan, G., L. Xiao, L. Cheng, X. Wang, B. Sun, and G. Hu. 2000. Targeted disruption of NDST-1 gene leads to pulmonary hypoplasia and neonatal respiratory distress in mice. FEBS Lett. 467:7-11. [DOI] [PubMed] [Google Scholar]
- 17.Giampietro, P. F., C. L. Raggio, and R. D. Blank. 1999. Synteny-defined candidate genes for congenital and idiopathic scoliosis. Am. J. Med. Genet. 83:164-177. [PubMed] [Google Scholar]
- 18.Hankinson, O. 1995. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 35:307-340. [DOI] [PubMed] [Google Scholar]
- 19.Hosoya, T., Y. Oda, S. Takahashi, M. Morita, S. Kawauchi, M. Ema, M. Yamamoto, and Y. Fujii-Kuriyama. 2001. Defective development of secretory neurones in the hypothalamus of Arnt2-knockout mice. Genes Cells 6:361-374. [DOI] [PubMed] [Google Scholar]
- 20.Jain, S., E. Maltepe, M. M. Lu, C. Simon, and C. A. Bradfield. 1998. Expression of ARNT, ARNT2, HIF1 alpha, HIF2 alpha and Ah receptor mRNAs in the developing mouse. Mech. Dev. 73:117-123. [DOI] [PubMed] [Google Scholar]
- 21.Kaartinen, V., J. W. Voncken, C. Shuler, D. Warburton, D. Bu, N. Heisterkamp, and J. Groffen. 1995. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat. Genet. 11:415-421. [DOI] [PubMed] [Google Scholar]
- 22.Keith, B., D. M. Adelman, and M. C. Simon. 2001. Targeted mutation of the murine arylhydrocarbon receptor nuclear translocator 2 (Arnt2) gene reveals partial redundancy with Arnt. Proc. Natl. Acad. Sci. USA 98:6692-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, H., D. P. Witte, W. W. Branford, B. J. Aronow, M. Weinstein, S. Kaur, S. Wert, G. Singh, C. M. Schreiner, and J. A. Whitsett. 1994. Gsh-4 encodes a LIM-type homeodomain, is expressed in the developing central nervous system and is required for early post-natal survival. EMBO J. 13:2876-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michaud, J., and C.-M. Fan. 1997. Single-minded—two genes, three chromosomes. Genome Res. 7:569-571. [DOI] [PubMed] [Google Scholar]
- 25.Michaud, J. L., T. Rosenquist, N. R. May, and C.-M. Fan. 1998. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 12:3264-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaud, J. L., C. DeRossi, N. R. May, B. C. Holdener, and C.-M. Fan. 2000. ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mech. Dev. 90:253-261. [DOI] [PubMed] [Google Scholar]
- 27.Michaud, J. L., F. Boucher, A. Melnyk, F. Gauthier, E. Goshu, E. Levy, G. A. Mitchell, J. Himms-Hagen, and C.-M. Fan. 2001. Sim1 haploinsufficiency causes hyperphagia, obesity and reduction of the paraventricular nucleus of the hypothalamus. Hum. Mol. Genet. 10:1465-1473. [DOI] [PubMed] [Google Scholar]
- 28.Miettinen, P. J., D. Warburton, D. Bu, J. S. Zhao, J. E. Berger, P. Minoo, T. Koivisto, L. Allen, L. Dobbs, Z. Werb, and R. Derynck. 1997. Impaired lung branching morphogenesis in the absence of functional EGF receptor. Dev. Biol. 15:224-236. [DOI] [PubMed] [Google Scholar]
- 29.Moffett, P., M. Dayo, M. Reece, M. K. McCormick, and J. Pelletier. 1996. Characterization of msim, a murine homologue of the Drosophila sim transcription factor. Genomics 35:144-155. [DOI] [PubMed] [Google Scholar]
- 30.Moffett, P., M. Reece, and J. Pelletier. 1997. The murine Sim-2 gene product inhibits transcription by active repression and functional interference. Mol. Cell. Biol. 17:4933-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moffett, P., and J. Pelletier. 2000. Different transcriptional properties of mSim-1 and mSim-2. FEBS Lett. 466:80-86. [DOI] [PubMed] [Google Scholar]
- 32.Normelli, H., J. A. Sevastik, and J. Akrivos. 1985. The length and ash weight of the ribs of normal and scoliotic persons. Spine 10:590-592. [DOI] [PubMed] [Google Scholar]
- 33.Polin, R. A., and W. W. Fox. (ed.) 1998. Hormonal therapy for prevention of respiratory distress syndrome, chapter 119. Fetal and neonatal physiology. Penguin, Philadelphia, Pa.
- 34.Porter, D. E., and A. H. Simpson. 1999. The neoplastic pathogenesis of solitary and multiple osteochodromas. J. Pathol. 188:119-125. [DOI] [PubMed] [Google Scholar]
- 35.Probst, M. R., C.-M. Fan, M. Tessier-Lavigne, and O. Hankinson. 1997. Two murine homologs of the Drosophila single-minded protein that interact with the mouse aryl hydrocarbon receptor nuclear translocator protein. J. Biol. Chem. 272:4451-4457. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt, V. J., and C. A. Bradfield. 1996. Ah receptor signaling pathway. Annu. Rev. Cell Dev. Biol. 12:55-89. [DOI] [PubMed] [Google Scholar]
- 37.Seasholtz, A. F., S. A. Bourbonaiser, C. E. Harnden, and S. A. Camper. 1991. Nucleotide sequence and expression of mouse corticotropin releasing hormone gene. Mol. Cell. Neurosci. 2:266-273. [DOI] [PubMed] [Google Scholar]
- 38.Sevastik, B. 1996. Aspects on the influence of the thoracic wall on the pathogenesis of idiopathic scoliosis. Ph.D. dissertation. Kongl Carolinska Medico Chirurgiska Institutet, Stockholm, Sweden.
- 39.Shi, W., N. Heisterkamp, J. Groffen, J. Zhao, D. Warburton, and V. Kaartinen. 1999. TGF-beta3-null mutation does not abrogate fetal lung maturation in vivo by glucocorticoids. Am. J. Physiol. 277:1205-1213. [DOI] [PubMed] [Google Scholar]
- 40.Sonnenfeld, M., M. Ward, G. Nystrom, J. Mosher, S. Stahl, and S. Crews. 1997. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development 124:4571-4582. [DOI] [PubMed] [Google Scholar]
- 41.Stokes, I. A. F., J. Dansereau, and M. S. Moreland. 1989. Rib cage asymmetry in idiopathic scoliosis. J. Orthop. Res. 7:599-606. [DOI] [PubMed] [Google Scholar]
- 42.Stokes, I. A. F., and J. P. Laible. 1990. Three-dimensional osseo-ligamentous model of the thorax representing initiation of scoliosis by asymmetric growth. J. Biomech. 23:589-595. [DOI] [PubMed] [Google Scholar]
- 43.Swanson, H. I., W. K. Chan, and C. Bradfield. 1995. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J. Biol. Chem. 270:26292-26298. [DOI] [PubMed] [Google Scholar]
- 44.Wang, W., and T. Lufkin. 2000. The murine Otp homeobox gene plays an essential role in the specification of neuronal cell lineages in the developing hypothalamus. Dev. Biol. 227:432-449. [DOI] [PubMed] [Google Scholar]
- 45.Weintraub, H., R. Davis, S. Tapscott, M. Thayer, M. Krause, R. Benezra, T. K. Blackwell, D. Turner, R. Rupp, and S. Hollenberg. 1991. The myoD gene family: nodal point during specification of the muscle cell lineage. Science 251:761-766. [DOI] [PubMed] [Google Scholar]
- 46.Wuenschell, G. W., J. Zhao, J. D. Tefft, and D. Warburton. 1998. Nicotine stimulates branching and expression of SP-A and SP-C mRNAs in embryonic mouse lung culture. Am. J. Physiol. 274:L165-L170. [DOI] [PubMed] [Google Scholar]
- 47.Yamaki, A., S. Noda, J. Kudoh, N. Shindoh, H. Maeda, S. Minoshima, K. Kawasaki, Y. Shimizu, and N. Shimizu. 1996. The mammalian single-minded (SIM) gene: mouse cDNA structure and diencephalic expression indicate a candidate gene for Down syndrome. Genomics 35:136-143. [DOI] [PubMed] [Google Scholar]
- 48.Young, B., and J. W. Heath. 2000. Wheater's functional histology, 4th ed. Churchill Livingston, New York, N.Y.