Abstract
The pathway leading to BRCA1-dependent tumor suppression is not yet clear but appears to involve activities in DNA repair as well as gene transcription. Moreover, it has been shown that BRCA1 can regulate p53-dependent transcription. Because BRCA1 overexpression stabilizes wild-type p53 but does not lead to apoptosis of most cell lines, we investigated the selectivity of BRCA1 for p53-dependent target gene activation. We find that BRCA1-stabilized p53 regulates transcription of DNA repair and growth arrest genes while p53 stabilized by DNA-damaging agents induces a wide array of genes, including those involved in apoptosis. This differential expression profile was reflected in the treatment outcome—apoptosis following DNA damage and growth arrest after expression of BRCA1. Depletion of BRCA1 in wild-type-p53-expressing cells abolished the induction of such repair genes as p53R2, while the expression of PIG3, an apoptosis-inducing gene, was still induced. BRCA1 also conferred diminished cell death in a p53-dependent manner in response to adriamycin compared to that conferred by controls. These results suggest that BRCA1 selectively coactivates the p53 transcription factor towards genes that direct DNA repair and cell cycle arrest but not towards those that direct apoptosis.
The BRCA1 gene is commonly mutated in hereditary breast and ovarian cancer but not in sporadic breast cancer (7). The P53 tumor suppressor gene is mutated in sporadic breast cancers as well as in Li-Fraumeni syndrome, in which patients have an increased risk of premenopausal breast cancer (19, 27). P53 mutations also occur commonly in tumors derived from patients with BRCA1 mutations (9). The occurrence of P53 mutations in BRCA1-mutated tumors suggests that the two genes might function in separate pathways to suppress tumorigenesis (35). Alternatively, when BRCA1 mutation occurs first, as it does in familial inherited breast cancer cases, it is not sufficient to promote tumor progression if a functional p53 acts downstream of BRCA1 in the same pathway. This is a possibility that has been suggested by results obtained with mice that derive breast neoplasia much more effectively when both BRCA1 and P53 are deleted (8, 10). In addition, novel p53 mutations are found in BRCA1-linked breast cancers, indicating a difference between the inactivation of p53 in these cases and inactivation in cases of other cancers (36, 40). The suppression of genomic instability by BRCA1 is considered a possible function as a cancer-inhibitory gene (33), in part because BRCA1-null cells exhibit genomic instability (43, 48), a feature also observed in P53-deficient cells (49). In early embryogenesis, chromosomal damage of BRCA1-null cells activates a p53-dependent checkpoint, leading to growth suppression and embryonic lethality, a phenotype partially rescued by deletion of the P53 tumor suppressor gene (13).
Other evidence has linked the pathways through which p53 and BRCA1 function. Both p53 and BRCA1 become phosphorylated by checkpoint kinases when cells are exposed to DNA-damaging agents, including UV, infrared, mitomycin C, and adriamycin (53). The expression levels of both p53 and BRCA1 proteins increase in cells exposed to DNA-damaging agents (22, 25). Both p53 and BRCA1 appear to transcriptionally regulate the expression of the growth arrest and DNA damage-inducible gene GADD45 (14, 18, 23), and both BRCA1-deficient and GADD45-deficient cells possess a G2/M cell cycle checkpoint defect as well as genomic instability (16, 48). Both p53 and BRCA1 have also been reported to regulate the expression of the cell cycle inhibitor p21WAF1, leading to cell cycle arrest (12, 38). These observations have suggested a possible mechanism for BRCA1 to maintain genomic integrity involving the arrest of cells through a checkpoint in order to allow time for DNA repair. Some support for this transcriptional pathway has come from studies that revealed interactions between BRCA1 and proteins involved in the regulation of gene expression (10) and from evidence that BRCA1 is a nuclear protein that contains a transactivation domain (51).
BRCA1 and p53 are also known to physically interact, with BRCA1 acting as a coactivator for p53 (30, 50). Interaction domains of p53 on BRCA1 were identified both at the N terminus, overlapping in the Rad50-binding domain, and in the second BRCT domain at the C terminus. BRCA1 interacts with the C terminus of p53. While BRCA1 is a potent enhancer of p53-mediated transcription, BRCA1-mediated stabilization of wild-type p53 protein occurs through transcriptional activation of the p14ARF gene and effects on the phosphorylation of mouse p53 on serine 18 (the equivalent of serine 15 in humans) through the presence of the exon 11 region (37, 47). Indeed, cells with deletions of exon 11 of BRCA1 are defective in the rapid stabilization of p53 following DNA damage (47). As some studies have found that BRCA1 is transiently induced following DNA damage by agents such as UV (22), it is likely that BRCA1 participates in p53 stabilization in vivo. Like p14ARF, p53 has also been shown to repress the transcription of BRCA1 (2, 22, 32), acting in a negative feedback loop of its own stabilization.
The connections between BRCA1 and p53 have raised questions of relevance to their mechanisms of action. Because p53 may be required for the tumor suppressor function of BRCA1, we hypothesized that BRCA1 may signal DNA repair in part through a transcriptional mechanism involving p53. As BRCA1-mediated stabilization of p53 does not lead to apoptosis in most cell lines (23) and in some cases is able to inhibit apoptosis (17), it is possible that BRCA1 may direct a specific p53 response in terms of transcriptional targets, leading to cell cycle arrest and DNA repair but not apoptosis. If true, this would be a novel finding relevant to our understanding of how p53 functions in terms of causing cell cycle arrest or apoptosis, a major unanswered question. We investigated whether, at identical levels of p53 expression, a given cell would undergo either cell cycle arrest or apoptosis in response to p53 stabilization following different signals. Using arrays containing genes that are known transcriptional targets of p53, we analyzed the expression of such genes in the presence of amplified expression of BRCA1 and observed alterations in growth arrest and DNA repair genes but not apoptosis genes. We also found that BRCA1- and DNA damage-stabilized p53 led to different outcomes and transcriptional responses. The results reveal that similar levels of p53 stabilization in response to different signals can lead to the activation of different transcriptional targets that could explain the observed phenotypes of cell cycle arrest versus apoptosis. The results have implications for the mechanisms of action of BRCA1 and p53 and further unravel functional relationships between these tumor suppressors.
MATERIALS AND METHODS
Array screening.
Total RNA was isolated from Ad-GFP- or Ad-BRCA1-infected HCT116 p53+/+ (multiplicity of infection [MOI], 40), HCT116 p53−/− (MOI, 40), H460-Neo (MOI, 20), H460-E6 (MOI, 20), H460 (MOI, 20), and SW480 (MOI, 50) cells with an RNEasy kit from Qiagen (Chatsworth, Calif.). RNA was amplified, labeled, and hybridized to the Affymetrix (Santa Clara, Calif.) human cancer 110 GeneChip as previously described (21, 44). For each RNA sample, the average intensity of all genes on the chip was adjusted to 1,500 to allow for comparison and subsequent analysis (21, 44). Analysis was performed with Affymetrix Microarray Suite 4.0 according to the manufacturer's protocol. The Clontech (Mountain View, Calif.) Atlas human cancer array was hybridized and analyzed as described previously (23). Custom-made membrane arrays were prepared and hybridized as described previously (39).
Cell culture.
The culture conditions of SW480, PA1-Neo, PA1-E6, H460, H460-Neo, and H460-E6 cells have been previously described (22, 23). Adriamycin was added to the culture media at a concentration of 200 ng/ml at the indicated time points.
Adenovirus infection and propagation.
Ad-LacZ, Ad-GFP, Ad-p53, and Ad-BRCA1 have been described previously (22, 23, 37). Ad-AntisenseBRCA1 was developed by subcloning a KpnI/XhoI fragment of BRCA1 (amino acids 1 to 500) [BRCA1(1-500)] from pCR3-BRCA1(1-500) into pAdTrack-CMV (15). pAdTrack-BRCA1(1-500) was cotransformed into BJ5183 bacteria with pAdEasy to obtain a homologously recombined pAd-BRCA1(1-500) vector. This vector was transfected into 293 cells for the production of viral plaques. Adenoviruses expressing truncated BRCA1 were purified from 293 cells over a CsCl gradient and stored in a glycerol buffer (12). Viruses were propagated, their titers were determined, and they were amplified as described previously (12).
Western blotting.
Western blotting was carried out essentially as described previously (23) with mouse anti-human p53 monoclonal antibody (Pab1801; Oncogene Research, San Diego, Calif.), mouse anti-human p21WAF1 monoclonal antibody (EA10; Oncogene Research), and mouse anti-human actin (C-2; Santa Cruz Biotechnology, Santa Cruz, Calif.).
Northern blotting.
Total RNA isolation and Northern blotting were carried out as described previously (23). A HindIII/NotI fragment of 5.6 kb was excised from pCR3.1-BRCA1 and used as a probe for BRCA1 Northern blots. A HindIII/XhoI fragment of 2.0 kb was excised from pCEP4-p53 as a probe for p53 Northern blots. All other probes were amplified from reverse-transcribed cDNA, sequence verified, random-primer labeled, and hybridized to Northern blots.
Flow cytometry.
Cells were plated in six-well culture dishes and treated as described previously. Cells were harvested by trypsinization and fixed in cold 70% ethanol. Genomic DNA was stained with 5 μg of propidium iodide/ml and analyzed on a Coulter Epics elite counter. DNA content analysis was performed with MacCycle software (Phoenix Flow Systems, San Diego, Calif.).
RESULTS
Equal stabilization of wild-type p53 with adriamycin or BRCA1.
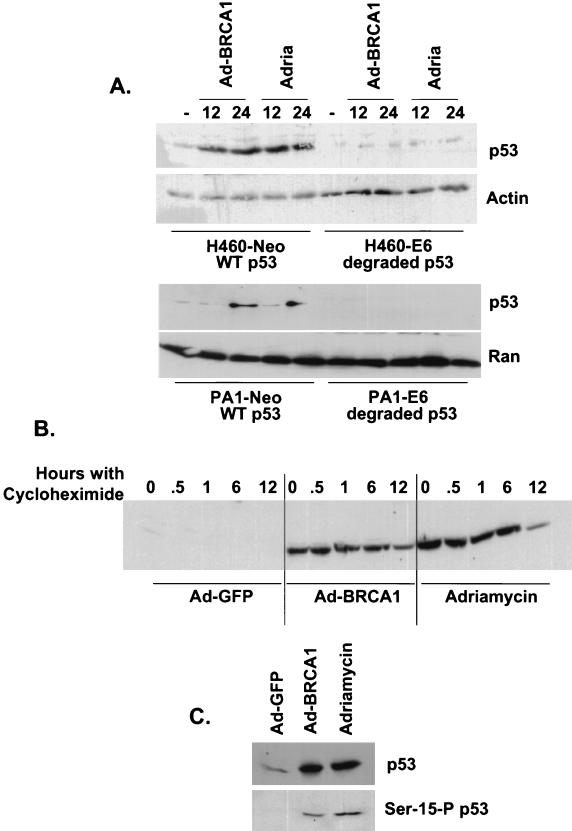
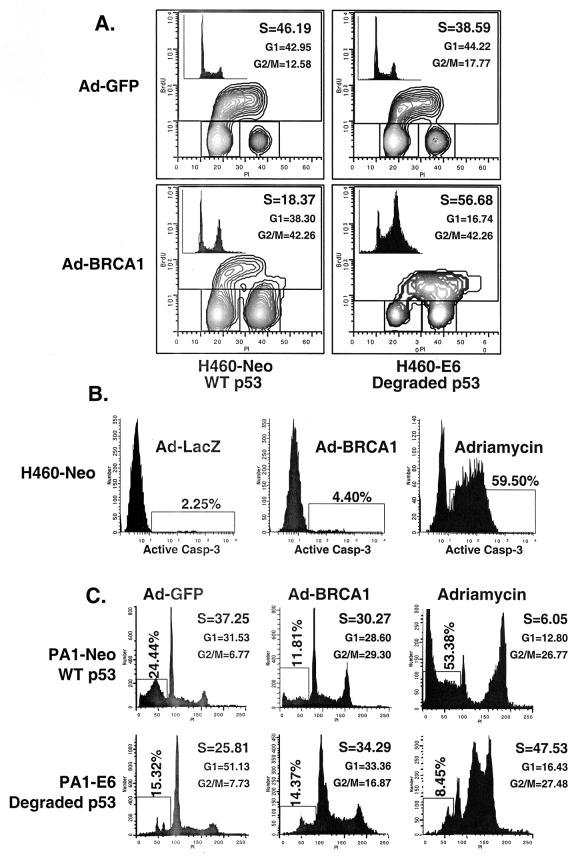
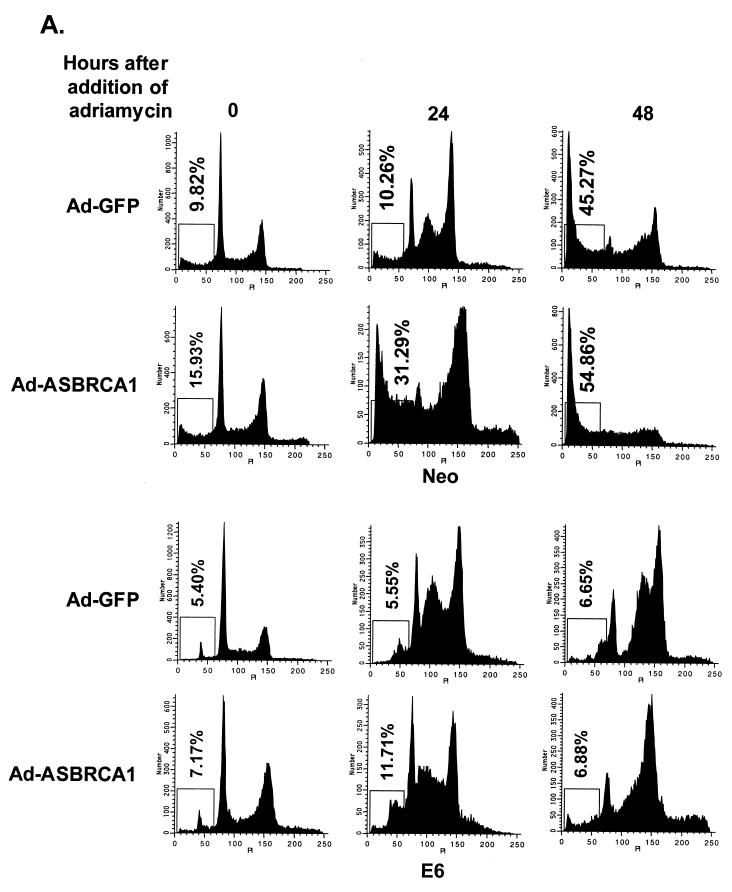
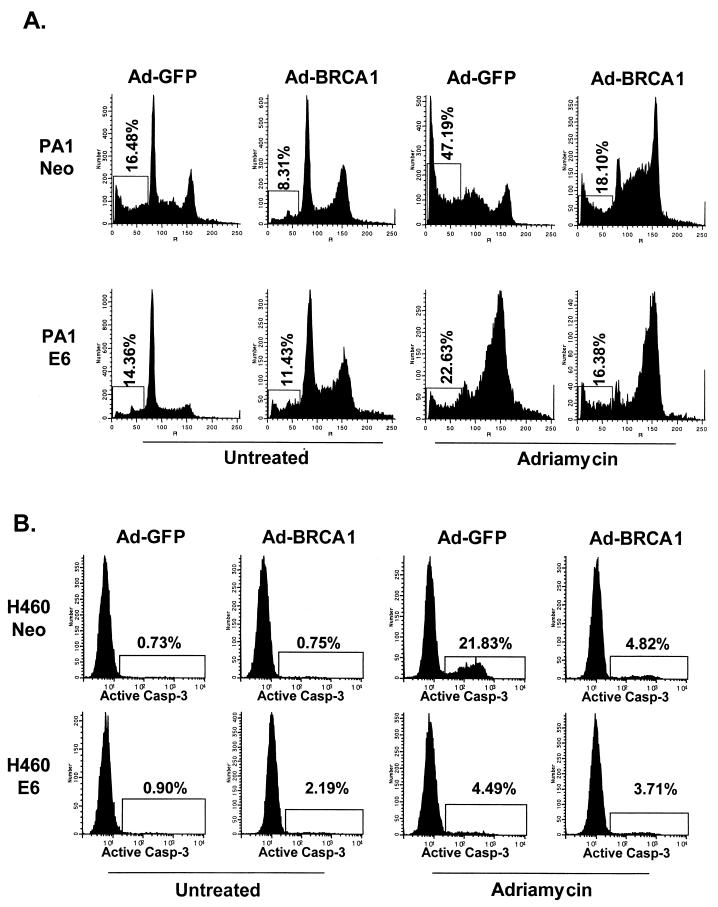
We infected the wild-type-p53-expressing cells H460 and PA1—which had been stably transfected with a control vector (Neo) or with one that expressed the human papilloma virus E6 protein, which degrades endogenous p53 protein (46)—with Ad-BRCA1 or treated them with adriamycin. Following 24 h of treatment, we prepared nuclear extracts and immunoblotted for p53 protein. While neither BRCA1 nor adriamycin was able to stabilize p53 in the presence of E6, adriamycin and Ad-BRCA1 stabilized p53 to roughly the same extent in the H460- and PA1-Neo cells (Fig. 1A). This is essential, as it can be argued that simply the expression level of p53 could account for differences seen between the two methods of p53 stabilization. The stability of both populations of p53 appeared roughly equal, as cycloheximide treatment of both stabilizing agents resulted in the disappearance of p53 by about 12 h (Fig. 1B). In addition, serine 15 phosphorylation of p53, a key modification in the stabilization of p53 following DNA damage, was virtually identical between BRCA1- and adriamycin-stabilized p53 (Fig. 1C). This may indicate another pathway to p53 stabilization by BRCA1 in addition to that of p14ARF, which has previously been reported (37). Concurrently with these expression analyses, we performed fluorescence-activated cell sorting (FACS) to determine the level of apoptosis and S-phase content of these cells. Bromodeoxyuridine uptake, an indicator of cells actively replicating genomic DNA, was decreased by more than half in H460-Neo cells infected with Ad-BRCA1 compared to the level in controls, while H460-E6 cells infected with Ad-BRCA1 not only had decreased S-phase content but also continued into a second replication phase before dividing (Fig. 2A). This indicated a p53 dependence on BRCA1-induced cell cycle arrest in this cell line. The entry into another DNA replication phase may reflect the involvement of BRCA1 in this process, and as p53 is lacking in the E6 line, the cells are incapable of imposing a G2/M phase arrest in response. In addition, we measured active caspase-3 content, a marker of apoptotic cells, in these infected cells as well as in those treated with adriamycin for 24 h and found that BRCA1 did not induce an appreciable amount of cell death (Fig. 2B). We also utilized the wild-type-p53-expressing PA1 (ovarian cancer)-Neo-PA1-E6 cell line system (46) in addition to H460 both to provide a physiologically relevant cell line to BRCA1 in these analyses and to allow detection of apoptosis by pre-G1 cell content by adriamycin treatment, a phenotype not seen in H460 cells. While adriamycin treatment of PA1-Neo cells led to more than 50% of the cells undergoing apoptosis, Ad-BRCA1 did not have a significant effect on the induction of cells possessing lower than 2N DNA content, indicating the presence of apoptotic cells (Fig. 2C). FACS profiles of Ad-BRCA1-infected cells revealed a slight decrease in cells in S phase as well as an increase in cells in the G2 and M phases of the cell cycle, suggesting a growth arrest, consistent with previously obtained results with similar cell lines (23). Interestingly, the absence of p53 in the PA1-E6 cell line resulted in a lack of apoptosis with adriamycin and an increase in cells in S phase with Ad-BRCA1, indicating a dependence of p53 on both of these effects. Therefore, adriamycin treatment and Ad-BRCA1 infection, both p53-stabilizing procedures, appeared to have differential effects on the resulting cellular phenotype.
FIG. 1.
Equal stabilization of p53 by BRCA1 and adriamycin. (A) H460-Neo (wild-type-p53-expressing) and H460-E6 (degraded-p53-expressing) cells were either infected with Ad-GFP (−) or Ad-BRCA1 at an MOI of 20 or treated with 0.2 μg of adriamycin (Adria)/ml for 12 and 24 h, after which nuclear protein was harvested. The protein was immunoblotted with antibodies to p53, p21WAF1, and actin or Ran. (B) H460 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20 or treated with 200 ng of adriamycin/ml for 18 h, treated with 10 μg of cycloheximide/ml, and then harvested for total protein at the indicated time points. Total protein was immunoblotted for p53 expression. (C) H460 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20 or treated with 200 ng of adriamycin/ml for 18 h and then harvested for total protein. Total protein was immunoblotted for p53 and serine 15-phosphorylated (Ser-15-P) p53. WT, wild-type.
FIG. 2.
Different consequences of p53 stabilization by BRCA1 and adriamycin. (A) H460-Neo and -E6 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20 or treated with 200 ng of adriamycin/ml for 24 h. Cells were then harvested and analyzed for bromodeoxyuridine (BrdU) uptake for detection of cells in S phase. The insets in the bromodeoxyuridine FACS analyses are two-dimensional views of the propidium iodide (PI) stain from the same cells. (B) H460-Neo cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20 or treated with 200 ng of adriamycin/ml for 24 h. Cells were then harvested and analyzed for the presence of active caspase 3 enzyme for the detection of cells in apoptosis. (C) PA1-Neo (wild-type-p53-expressing) and PA1-E6 (degraded-p53-expressing) cells were either infected with Ad-GFP (−) or Ad-BRCA1 at an MOI of 20 or treated with 200 ng of adriamycin/ml for 18 h, after which the cells were harvested, fixed, and stained for cellular DNA content with propidium iodide. Samples were analyzed for propidium iodide fluorescence by flow cytometry. WT, wild-type.
Differential expression of p53-regulated genes.
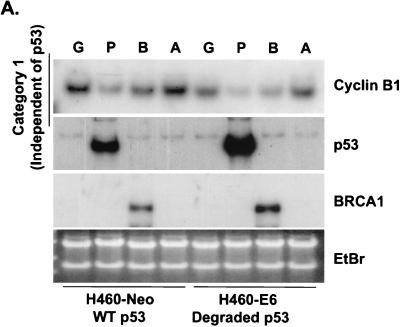
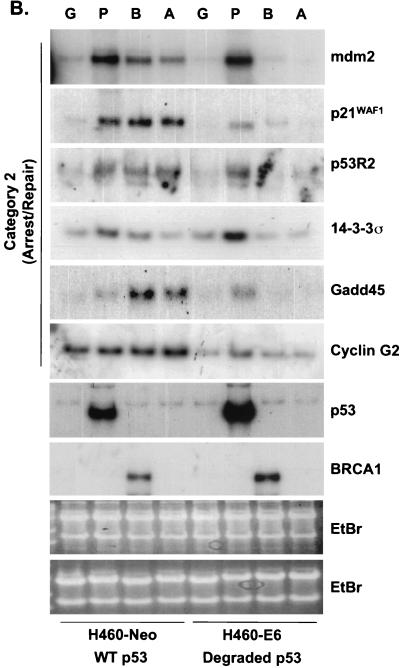
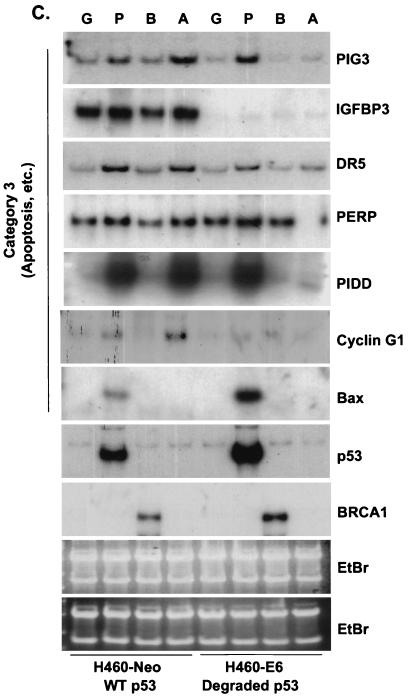
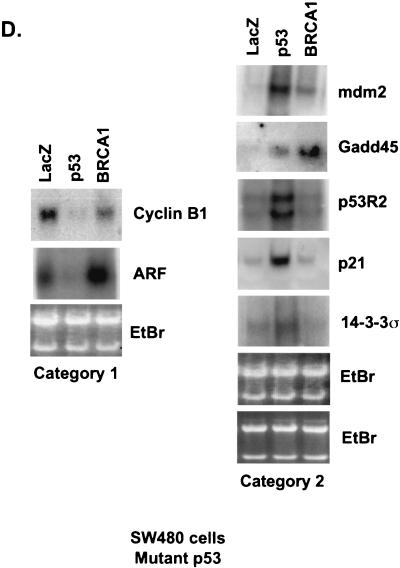
Because BRCA1 stabilizes endogenous p53, it was predicted that p53 targets would be transcriptionally upregulated. However, while the levels of stabilized p53 were equal between Ad-BRCA1-infected and adriamycin-treated cells, the observed cellular phenotypes were different. Adriamycin treatment of H460-Neo and PA1-Neo cells caused a potent apoptotic response, whereas Ad-BRCA1 infection in the same cell lines caused growth arrest (Fig. 2A and B) (23, 46). Given these differences, we employed array screening to search for p53-regulated genes that were altered in expression upon BRCA1 infection. Affymetrix GeneChips as well as custom-made membrane arrays were screened with a number of different systems comparing a p53 wild-type background and mutant or null background, including HCT116 p53+/+ versus HCT116 p53−/−, H460-Neo (wild-type p53) versus H460-E6 (degraded p53), and H460 (wild-type p53) versus SW480 (mutant p53). While there existed differences in the total expression profile from cell line to cell line, trends in the type of p53 genes that were found, regulated by BRCA1, were seen in each cell line. The results are presented in confirmatory Northern blots in Fig. 3. H460-Neo and -E6 cells were each infected with green fluorescent protein (GFP)-, p53-, or BRCA1-expressing adenoviruses at an MOI of 20 or treated with 0.2 μg of adriamycin/ml. Gene expression changes were clustered in one of three categories: (i) expression changes induced by BRCA1 regardless of p53 status (i.e., changes detected in both wild-type-p53-expressing and p53-null cells), (ii) expression changes induced by BRCA1 dependent on the presence of wild-type p53 (i.e., changes observed in wild-type-p53-expressing cells but not in p53-null cells), and (iii) p53-regulated genes that were not affected by BRCA1-mediated stabilization of p53. Results similar to those obtained with the H460-Neo-H460-E6 cell line system were also found with the PA1-Neo-PA1-E6 cell line system (data not shown). Category 1 (Fig. 3A) included genes that have been previously described as BRCA1 transcriptional targets, including cyclin B1 (23). Category 2 included genes that are known to be involved in the growth arrest and DNA damage response (Fig. 3B). p21WAF1 is a well-characterized BRCA1 target gene (38). However, to our knowledge, the induction of endogenous p21WAF1 mRNA by BRCA1 overexpression has not been investigated to date; study on this effect has strictly involved the activity of the p21WAF1 promoter luciferase constructs and protein expression. Here we find that significant induction of p21WAF1 mRNA by BRCA1 depends on the presence of wild-type p53 in the cell. In addition, the p53R2 gene, encoding a ribonucleotide reductase, was also induced by BRCA1 in a p53-dependent manner (26, 41). Another growth arrest and DNA repair gene, GADD45, was also induced in a p53-dependent manner in this system; however, we and others have shown induction of GADD45 by BRCA1 in mutant-p53 backgrounds. This contradiction may indicate that BRCA1 controls gene expression in a different manner in mutant-p53-expressing cell lines. Cyclin G2, a gene proposed to be involved in DNA repair, was weakly induced by BRCA1 but was nevertheless not changed in expression in a cell line lacking p53 (4). Not shown here is another gene regulated by BRCA1 in a p53-dependent manner, the nucleotide excision repair gene DDB2, which is described elsewhere (40a). The control of category 1 and lack of control of category 2 genes by BRCA1 in a p53-null background were found to be similarly regulated in a mutant-p53 background in the cell line SW480 (Fig. 3D). Category 3 contained the largest number of genes, those that were not affected by BRCA1-induced stabilization of p53 (Fig. 3C). The most significant component of this category was a group of genes known to be involved in the p53-mediated apoptotic response. This included KILLER/DR5, PIDD, PERP, and the PIG genes (3, 20, 31, 45). We have also previously found that Bax is either not affected or in some cases repressed by BRCA1 (23); interestingly, though, adriamycin did not induce Bax in these cell lines, perhaps reflecting a different pathway to cell death by adriamycin in these cells. Cyclin G1, a gene that has recently been described to facilitate cell death (29), was induced in a p53-dependent manner by adriamycin but not by BRCA1. While Ad-p53 infection adequately induced the genes in this category, BRCA1 expression failed to have any appreciable effect on the expression of these genes. Importantly, adriamycin, which induces similar levels of p53 protein in the H460-Neo line, was capable of activating expression of these apoptotic p53 transcriptional targets. This difference of p53 target gene activation seems then to underlie the differential phenotypes induced by Ad-BRCA1 infection versus adriamycin treatment in a wild-type-p53 background.
FIG. 3.
BRCA1 controls a subset of p53-regulated genes. H460-Neo and -E6 cells were infected with GFP (G), p53 (P), or BRCA1 (B) adenoviruses at an MOI of 20 or treated with 0.2 μg of adriamycin (A)/ml for 24 h and harvested for total RNA. RNA was Northern blotted for the indicated genes. The expression pattern of the effect of BRCA1 on p53 transcriptional targets fell into three categories: genes that were affected by BRCA1 expression regardless of the status of p53 (A), genes that were affected by BRCA1 only in cells that express wild-type p53 (B), and genes that were not affected by BRCA1 expression (C). (D) SW480 cells were infected with Ad-LacZ, Ad-p53, or Ad-BRCA1 at an MOI of 20 for 24 h and then harvested for total RNA. Total RNA was Northern blotted for expression of the indicated genes. EtBr, ethidium bromide.
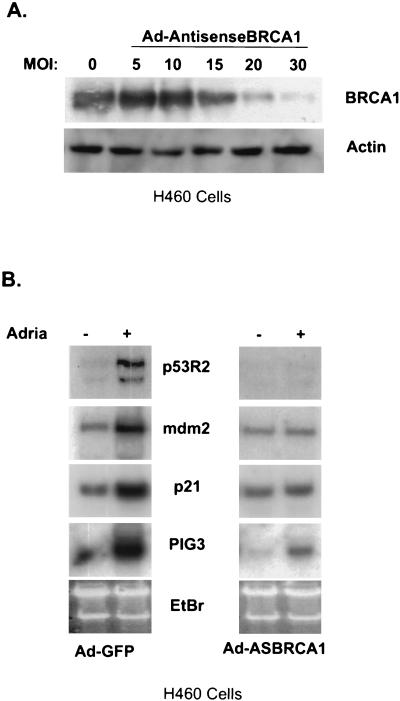
Elimination of BRCA1 abolishes the DNA repair pathway and hastens cell death.
It has recently been shown that the expression of an antisense-BRCA1 adenovirus is able to abolish endogenous BRCA1 expression (40a) (Fig. 4A). This elimination of BRCA1 is able to block the induction of another category 2 gene, DDB2 (40a). We sought to find other genes that require the presence of BRCA1 for transcriptional regulation following DNA damage by adriamycin. We infected H460 cells with Ad-GFP or an antisense-BRCA1-expressing adenovirus and subsequently treated them with 0.2 μg of adriamycin/ml or left the cells untreated. While the induction of p53R2 was eliminated compared to that of controls when the antisense construct of BRCA1 was infected into cells, the apoptosis-inducing gene PIG3, while somewhat muted, was still induced upon adriamycin treatment (Fig. 4B). This suggests that BRCA1 may have more of a role in the coactivation of p53 target genes that induce DNA repair than in the coactivation of those that allow apoptosis to occur.
FIG. 4.
Antisense BRCA1 blocks BRCA1 protein expression and alters the induction of p53 targets. (A) H460 cells were infected with an adenovirus that expresses the first 1,500 nucleotides of BRCA1 in the antisense orientation at increasing MOI, harvested for total protein, and immunoblotted for BRCA1. (B) H460 cells were infected with Ad-AntisenseBRCA1 at an MOI of 20 or control infected and exposed to 0.2 μg of adriamycin (Adria)/ml. Total RNA was harvested 12 h later and Northern blotted for the indicated genes. EtBr, ethidium bromide.
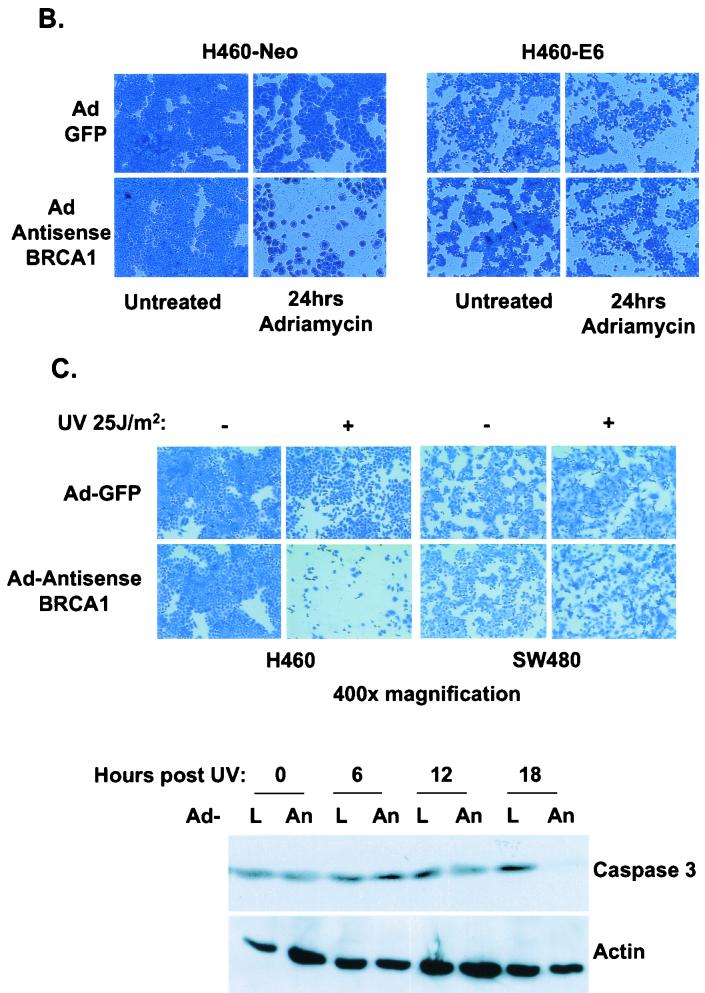
If BRCA1 indeed causes p53 to shunt its transcriptional functions toward a DNA repair and growth arrest pathway and/or away from its apoptosis targets, then the elimination of BRCA1 from a cell that is exposed to a p53-stabilizing agent should undergo apoptosis at a higher rate than it would if BRCA1 were present as a coactivator of p53. To test this hypothesis, the PA1-Neo-PA1-E6 cell system was infected with Ad-GFP or Ad-AntisenseBRCA1 and exposed to 200 ng of adriamycin/ml. Cells were collected at 18 and 36 h after treatment and analyzed by flow cytometry for DNA content. While control-infected cells underwent apoptosis by 36 h, a greater proportion of cells infected with Ad-AntisenseBRCA1 had undergone cell death by the earlier time point of 18 h (Fig. 5A ). Similar effects were seen with H460-Neo and -E6 cells following adriamycin treatment and staining at 18 h posttreatment to detect attached cells (Fig. 5B). We also looked at another type of DNA damage, UV irradiation, to see if this had a similar effect. We chose this type of stress because it has recently been reported that BRCA1 regulates the expression of DNA repair genes that are involved in nucleotide excision repair of UV-induced DNA adducts (40a). We see that in a wild-type-p53-expressing cell line, Ad-AntisenseBRCA1 allows more apoptosis to occur and at a higher rate than do controls in response to UV treatment (Fig. 5C), indicating sensitivity to apoptosis stimuli.
FIG. 5.
Antisense BRCA1 sensitizes cells to apoptosis stimuli. (A) PA1-Neo and -E6 cells were infected with Ad-GFP or Ad-AntisenseBRCA1 at an MOI of 20, treated with 0.2 μg of adriamycin/ml or left untreated, and then measured for the presence of cells containing less than 2N genomic DNA content by FACS analysis. (B) H460-Neo and -E6 cells were infected with Ad-GFP or Ad-AntisenseBRCA1 at an MOI of 20, treated with 0.2 μg of adriamycin/ml or left untreated, and then stained with Coomassie blue to detect attached cells after 18 h of treatment. (C) H460 and SW480 cells were infected with Ad-GFP or Ad-AntisenseBRCA1 at an MOI of 20 or 50, respectively, and exposed to 25 J of UV/m2. Total protein was harvested at 0, 6, 12, and 18 h following irradiation and immunoblotted for caspase-3 expression, and separate wells were stained with Coomassie blue 3 days later. L, Ad-LacZ; An, Ad-Antisense BRCA1.
These data suggest that the depletion of BRCA1 from cells made them more susceptible to apoptosis. Taken together, these results point to a pathway in which BRCA1 participates in p53 transcription by allowing predominantly survival genes to be expressed first while apoptosis-inducing genes are expressed later. Inhibition of this pathway leads to immediate apoptosis induction.
BRCA1 enhances survival of adriamycin-treated cells in a p53-dependent manner.
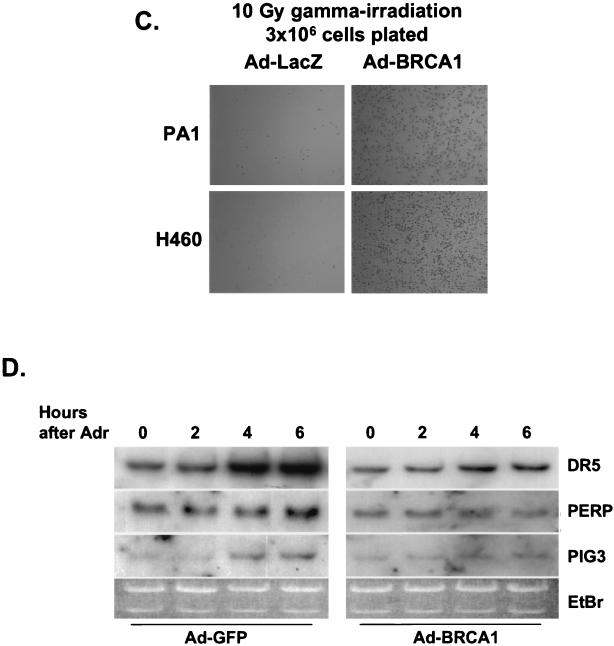
It is possible that BRCA1 may limit the transcriptional activity of p53 to genes that allow growth arrest and repair and/or may direct p53 activity away from apoptosis regulators. Given these possibilities, we postulated that BRCA1 could control the p53 response in terms of cellular phenotype in response to a DNA-damaging agent such as adriamycin. We infected PA1-Neo and -E6 cells with either GFP- or BRCA1-expressing adenoviruses 12 h prior to exposing them to 200 ng of adriamycin/ml. At 18 and 36 h following treatment with adriamycin, cells were collected and analyzed by flow cytometry for apoptotic cells with less than 2N DNA content. PA1-Neo cells expressing BRCA1 possessed less than half as many apoptotic cells as those cells infected with a control adenovirus at both time points (23) (Fig. 6A ). We also performed a similar analysis of the H460-Neo-H460-E6 lines, where we found a reduction of apoptotic cells by over 75% in cells that had been preinfected with Ad-BRCA1 compared to the level observed for control infections with adriamycin after 18 h (Fig. 6B). These data are consistent with previously published results that showed fewer H460 cells that had been preinfected with Ad-BRCA1 and treated with adriamycin staining for trypan blue than those that had been treated with adriamycin alone, as well as reports that have shown that the reintroduction of BRCA1 into the mutant BRCA1-expressing cell line HCC1937 allows for increased cell survival following gamma irradiation. We also saw a similar effect in the wild-type-p53-expressing cell lines H460 and PA1 infected with Ad-BRCA1; infection prior to gamma irradiation allowed increased survival compared to that of controls 1 week after treatment (Fig. 6C). To determine whether BRCA1 not only induced a growth arrest and/or DNA repair response from p53 but also repressed an apoptotic response, we performed Northern blotting on H460 cells preinfected with control or BRCA1-expressing adenoviruses and then treated with adriamycin for increasing amounts of time for apoptosis regulators controlled by p53. The induction of genes such as DR5 and PERP was delayed or muted in cells infected with Ad-BRCA1 compared to the level in controls, indicating that BRCA1 acts to repress apoptosis inducers as well as to activate growth arrest and repair targets (Fig. 6D). It is apparent, then, that the continuous presence of BRCA1 enhances the growth arrest and DNA repair transcriptional pathway of p53 and at least delays apoptosis until a later time.
FIG. 6.
BRCA1 infection enhances cell survival in a p53-dependent manner. (A) PA1-Neo and -E6 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20, treated with 200 ng of adriamycin/ml or left untreated, and then measured for the presence of cells containing less than 2N genomic DNA content by FACS analysis at 24 h after treatment with adriamycin. (B) H460-Neo and -E6 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20, treated with 200 ng of adriamycin/ml or left untreated, and then measured for the presence of active caspase-3 to detect cells undergoing apoptosis. (C) PA1 and H460 cells were infected with Ad-LacZ or Ad-BRCA1 and then plated to T25 flasks at a concentration of 3 × 106 cells/flask. Cells were exposed to 10-γ gamma irradiation and then stained with Coomassie blue 1 week later. (D) H460 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20 for 12 h, treated with 200 ng of adriamycin/ml or left untreated, and then harvested for total RNA at 2, 4, and 6 h after adriamycin treatment. RNA was blotted for the expression of the indicated genes. EtBr, ethidium bromide.
DISCUSSION
The method by which p53 determines whether a cell should undergo apoptosis or reversible growth arrest has been a topic of intense study. Here we present evidence that BRCA1 may contribute to this decision. We have found that BRCA1 is able to stabilize p53 in wild-type-p53-expressing cells but affects the expression only of a subset of known p53-regulated genes. We hypothesize that after DNA damage, BRCA1 associates with and stabilizes p53 alongside other stabilizing events, such as phosphorylation, and directs p53 transcription towards growth arrest and DNA repair genes. In the event that the damage is minimal, repair commences and the cells continue to proliferate. However, if the damage to the genome is significant and the repair process continues for longer periods of time, p53 then overcomes the BRCA1-mediated effect through repression of the BRCA1 gene, a known but until now puzzling effect of p53 (2, 22), and proceeds to activate the apoptosis pathway. Previous results with the p53AIP1 protein have shown that the growth arrest-DNA repair p53 pathway is activated first after UVC DNA damage followed by the induction of apoptosis-inducing genes (28). This transition is accompanied by the phosphorylation of p53 at serine 46. It will be of interest in the future to determine whether this phosphorylation event has any effect on the binding of p53 to BRCA1.
Studies involving mouse embryo fibroblasts derived from mice that have deletions of exon 11 of BRCA1, a mutation that leaves only the N- and C-terminal ends of the protein and results in a mutant lacking the p53-binding domain, have given insight into the interactions between BRCA1 and p53 (47). The BRCA1Δ11 protein, which lacks the p53-binding domain, causes a defect in the normal stabilization of p53 following DNA damage by gamma irradiation. In addition to this, the phosphorylation of mouse p53 on serine 18 is diminished in cells that carry the BRCA1Δ11 deletion. These results show that stabilization of p53 through a BRCA1-mediated pathway is indeed an event that occurs in vivo. p53 protein does, however, eventually increase through alternative mechanisms.
In these fibroblasts and BRCA1-null embryonic stem cells, a robust induction of p21WAF1, a gene that is described here to be induced by BRCA1 in a p53-dependent manner, is seen. While BRCA1 is able to cause such an upregulation, in the system described here, we found that p21WAF1 was still induced 1.5- to 2-fold in the presence of an antisense-BRCA1 adenovirus (Fig. 4) (40a). This would indicate alternative pathways to the induction of p21WAF1, a hypothesis that has much support in the current literature (11). Regulation of p21WAF1 independent of p53 and control of its transcription have been described in numerous cell lines and with several transcription factors. Therefore, while we point here to an involvement of BRCA1 in p53-dependent p21WAF1 induction, by no means is this the sole mechanism of p21WAF1 gene regulation.
The phenotypes of mice with deletions of exon 11 of BRCA1 have lent credence to the involvement of BRCA1 in suppressing p53 transcriptional activity towards apoptosis genes. Mak et al. targeted a BRCA1Δ11 knockout to T cells and found severely defective development of this cell type (24). Although these cells and BRCA1−/− embryos have been shown to possess an activated p53 pathway with abundant p21WAF1 protein, the T cells in this particular study were not affected by coknockout of BRCA1 exon 11 and p21WAF1. The T cells affected were found to undergo widespread cell death. In accordance with these results, Xu et al. found that BRCAΔ11 embryos did not survive to term unless p53 was also deleted, in which case they were viable at birth (47). Interestingly, codeletion with p21WAF1 had no effect on the survival of the embryos. Therefore, it is possible that the BRCAΔ11 embryos and T cells in the targeted knockout possessed diminished survival capabilities due to the enhanced apoptotic activities of p53 and not due to prolonged cell cycle arrest.
The effect of BRCA1 selectivity on p53 transcriptional activity would suggest that in the constant presence of BRCA1, cells would not undergo or would be delayed in undergoing p53-mediated apoptosis. Indeed, we found that more than half of adriamycin-treated wild-type-p53-expressing cells underwent apoptosis after treatment; however, if they were first infected with a constitutively expressed BRCA1 construct, they had a greater survival rate. In a p53-null cell line (PA1-E6), preinfection with Ad-BRCA1 not only increased S-phase content, it also led to only slightly decreased numbers of apoptotic cells compared to those in control infections following adriamycin treatment. Recent work performed by Scully et al. and Abbott et al. has described similar effects of BRCA1 on cell survival in the face of genomic DNA damage (1, 34, 42). The studies utilized a cell line that was mutant in the one remaining allele of Brca1 and was especially sensitive to DNA damage. The colony survival of cells following gamma irradiation was increased 2 to 3% once wild-type BRCA1 was introduced. One major difference between the cell lines used in these reports and in the work presented here is that HCC1937 cells harbor a mutation in p53 as well, while H460 and PA1 cells possess wild-type p53. In our analysis of apoptosis following adriamycin treatment, we saw more than 20% of cells rescued or delayed from apoptosis by BRCA1 preinfection, a much higher percentage than that seen with HCC1937. We propose that while the introduction of BRCA1 into HCC1937 allows the reestablishment of the direct DNA repair functions of BRCA1, a second pathway to enhanced DNA repair and survival through p53 is not present, due to a mutation in p53. When purified from cells, BRCA1 has recently been found to be present in different complexes (6), indicating that the protein very likely participates in several independent functions. The introduction of BRCA1 into a wild-type-p53-expressing cell line allows more BRCA1 not only to be present in the DNA repair complexes of which it is known to be a part, such as Rad51-BRCA2 and Mre11-Rad50-Nbs1 (5, 52), but also to hyperactivate the p53 DNA repair and growth arrest targets that it coactivates and to direct p53 away from a cell death pathway. Such an activation of two separate survival pathways could account for the differences in survival rates between mutant- and wild-type-p53-expressing cell lines.
In conclusion, we have presented evidence that BRCA1, while assisting in stabilizing wild-type p53 protein, directs p53 transcription towards growth arrest and DNA repair genes and away from an apoptotic pathway. Forced expression of BRCA1 allows wild-type p53 cells that would normally undergo apoptosis after adriamycin treatment to survive longer. These data shed light both on the importance of BRCA1 in the p53 transcriptional response and on the factors that contribute to the life-or-death transcriptional program that p53 initiates once stabilized.
Acknowledgments
We thank David Dicker for excellent flow cytometry expertise, Bert Vogelstein for providing pAd-TRACK and pAd-Easy constructs, and Magda Sgagias and Ken Cowan for providing Ad-BRCA1.
REFERENCES
- 1.Abbott, D. W., M. E. Thompson, C. Robinson-Benion, G. Tomlinson, R. A. Jensen, and J. T. Holt. 1999. BRCA1 expression restores radiation resistance in BRCA1-defective cancer cells through enhancement of transcription-coupled DNA repair. J. Biol. Chem. 274:18808-18812. [DOI] [PubMed] [Google Scholar]
- 2.Arizti, P., L. Fang, I. Park, Y. Yin, E. Solomon, T. Ouchi, S. A. Aaronson, and S. W. Lee. 2000. Tumor suppressor p53 is required to modulate BRCA1 expression. Mol. Cell. Biol. 20:7450-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attardi, L. D., E. E. Reczek, C. Cosmas, E. G. Demicco, M. E. McCurrach, S. W. Lowe, and T. Jacks. 2000. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 14:704-718. [PMC free article] [PubMed] [Google Scholar]
- 4.Bates, S., S. Rowan, and K. H. Vousden. 1996. Characterisation of human cyclin G1 and G2: DNA damage inducible genes. Oncogene 13:1103-1109. [PubMed] [Google Scholar]
- 5.Chen, J., D. P. Silver, D. Walpita, S. B. Cantor, A. F. Gazdar, G. Tomlinson, F. J. Couch, B. L. Weber, T. Ashley, D. M. Livingston, and R. Scully. 1998. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell 2:317-328. [DOI] [PubMed] [Google Scholar]
- 6.Chiba, N., and J. D. Parvin. 2001. Redistribution of BRCA1 among four different protein complexes following replication blockage. J. Biol. Chem. 276:38549-38554. [DOI] [PubMed] [Google Scholar]
- 7.Couch, F. J., M. L. DeShano, M. A. Blackwood, K. Calzone, J. Stopfer, L. Campeau, A. Ganguly, T. Rebbeck, and B. L. Weber. 1997. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N. Engl. J. Med. 336:1409-1415. [DOI] [PubMed] [Google Scholar]
- 8.Cressman, V. L., D. C. Backlund, E. M. Hicks, L. C. Gowen, V. Godfrey, and B. H. Koller. 1999. Mammary tumor formation in p53- and BRCA1-deficient mice. Cell Growth Differ. 10:1-10. [PubMed] [Google Scholar]
- 9.Crook, T., L. A. Brooks, S. Crossland, P. Osin, K. T. Barker, J. Waller, E. Philp, P. D. Smith, I. Yulug, J. Peto, G. Parker, M. J. Allday, M. R. Crompton, and B. A. Gusterson. 1998. p53 mutation with frequent novel condons but not a mutator phenotype in BRCA1- and BRCA2-associated breast tumours. Oncogene 17:1681-1689. [DOI] [PubMed] [Google Scholar]
- 10.Deng, C. X., and F. Scott. 2000. Role of the tumor suppressor gene Brca1 in genetic stability and mammary gland tumor formation. Oncogene 19:1059-1064. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry, W. S. 1998. p21/p53, cellular growth control and genomic integrity. Curr. Top. Microbiol. Immunol. 227:121-137. [DOI] [PubMed] [Google Scholar]
- 12.El-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 13.Hakem, R., J. L. de la Pompa, C. Sirard, R. Mo, M. Woo, A. Hakem, A. Wakeham, J. Potter, A. Reitmair, F. Billia, E. Firpo, C. C. Hui, J. Roberts, J. Rossant, and T. W. Mak. 1996. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85:1009-1023. [DOI] [PubMed] [Google Scholar]
- 14.Harkin, D. P., J. M. Bean, D. Miklos, Y. H. Song, V. B. Truong, C. Englert, F. C. Christians, L. W. Ellisen, S. Maheswaran, J. D. Oliner, and D. A. Haber. 1999. Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell 97:575-586. [DOI] [PubMed] [Google Scholar]
- 15.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollander, M. C., M. S. Sheikh, D. V. Bulavin, K. Lundgren, L. Augeri-Henmueller, R. Shehee, T. A. Molinaro, K. E. Kim, E. Tolosa, J. D. Ashwell, M. P. Rosenberg, Q. Zhan, P. M. Fernandez-Salguero, W. F. Morgan, C. X. Deng, and A. J. Fornace, Jr. 1999. Genomic instability in Gadd45a-deficient mice. Nat. Genet. 23:176-184. [DOI] [PubMed] [Google Scholar]
- 17.Irminger-Finger, I., W.-C. Leung, J. Li, M. Dubois-Dauphin, J. Harb, A. Feki, C. E. Jefford, J. V. Soriano, M. Jaconi, R. Montesano, and K.-H. Krause. 2001. Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis. Mol. Cell 8:1255-1266. [DOI] [PubMed] [Google Scholar]
- 18.Kastan, M. B., Q. Zhan, W. S. El-Deiry, F. Carrier, T. Jacks, W. V. Walsh, B. S. Plunkett, B. Vogelstein, and A. J. Fornace, Jr. 1992. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71:587-597. [DOI] [PubMed] [Google Scholar]
- 19.Lehman, T. A., B. G. Haffty, C. J. Carbone, L. R. Bishop, A. A. Gumbs, S. Krishnan, P. G. Shields, R. Modali, and B. C. Turner. 2000. Elevated frequency and functional activity of a specific germ-line p53 intron mutation in familial breast cancer. Cancer Res. 60:1062-1069. [PubMed] [Google Scholar]
- 20.Lin, Y., W. Ma, and S. Benchimol. 2000. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat. Genet. 26:122-127. [DOI] [PubMed] [Google Scholar]
- 21.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 22.MacLachlan, T. K., B. C. Dash, D. T. Dicker, and W. S. El-Deiry. 2000. Repression of BRCA1 through a feedback loop involving p53. J. Biol. Chem. 275:31869-31875. [DOI] [PubMed] [Google Scholar]
- 23.MacLachlan, T. K., K. Somasundaram, M. Sgagias, Y. Shifman, R. J. Muschel, K. H. Cowan, and W. S. El-Deiry. 2000. BRCA1 effects on the cell cycle and the DNA damage response are linked to altered gene expression. J. Biol. Chem. 275:2777-2785. [DOI] [PubMed] [Google Scholar]
- 24.Mak, T. W., A. Hakem, J. P. McPherson, A. Shehabeldin, E. Zablocki, E. Migon, G. S. Duncan, D. Bouchard, A. Wakeham, A. Cheung, J. Karaskova, I. Sarosi, J. Squire, J. Marth, and R. Hakem. 2000. Brca1 required for T cell lineage development but not TCR loci rearrangement. Nat. Immunol. 1:77-82. [DOI] [PubMed] [Google Scholar]
- 25.Maltzman, W., and L. Czyzyk. 1984. UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol. Cell. Biol. 4:1689-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakano, K., E. Balint, M. Ashcroft, and K. H. Vousden. 2000. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene 19:4283-4289. [DOI] [PubMed] [Google Scholar]
- 27.Nichols, K. E., D. Malkin, J. E. Garber, J. F. Fraumeni, Jr., and F. P. Li. 2001. Germ-line p53 mutations predispose to a wide spectrum of early-onset cancers. Cancer Epidemiol. Biomarkers Prev. 10:83-87. [PubMed] [Google Scholar]
- 28.Oda, K., H. Arakawa, T. Tanaka, K. Matsuda, C. Tanikawa, T. Mori, H. Nishimori, K. Tamai, T. Tokino, Y. Nakamura, and Y. Taya. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102:849-862. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto, K., and C. Prives. 1999. A role of cyclin G in the process of apoptosis. Oncogene 18:4606-4615. [DOI] [PubMed] [Google Scholar]
- 30.Ouchi, T., A. N. Monteiro, A. August, S. A. Aaronson, and H. Hanafusa. 1998. BRCA1 regulates p53-dependent gene expression. Proc. Natl. Acad. Sci. USA 95:2302-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polyak, K., Y. Xia, J. L. Zweier, K. W. Kinzler, and B. Vogelstein. 1997. A model for p53-induced apoptosis. Nature 389:300-305. [DOI] [PubMed] [Google Scholar]
- 32.Robertson, K. D., and P. A. Jones. 1998. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol. Cell. Biol. 18:6457-6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scully, R. 2000. Role of BRCA gene dysfunction in breast and ovarian cancer predisposition. Breast Cancer Res. 2:324-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scully, R., S. Ganesan, K. Vlasakova, J. Chen, M. Socolovsky, and D. M. Livingston. 1999. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell 4:1093-1099. [DOI] [PubMed] [Google Scholar]
- 35.Scully, R., and D. M. Livingston. 2000. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408:429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, P. D., S. Crossland, G. Parker, P. Osin, L. Brooks, J. Waller, E. Philp, M. R. Crompton, B. A. Gusterson, M. J. Allday, and T. Crook. 1999. Novel p53 mutants selected in BRCA-associated tumours which dissociate transformation suppression from other wild-type p53 functions. Oncogene 18:2451-2459. [DOI] [PubMed] [Google Scholar]
- 37.Somasundaram, K., T. K. MacLachlan, T. F. Burns, M. Sgagias, K. H. Cowan, B. L. Weber, and W. S. El-Deiry. 1999. BRCA1 signals ARF-dependent stabilization and coactivation of p53. Oncogene 18:6605-6614. [DOI] [PubMed] [Google Scholar]
- 38.Somasundaram, K., H. Zhang, Y. X. Zeng, Y. Houvras, Y. Peng, G. S. Wu, J. D. Licht, B. L. Weber, and W. S. El-Deiry. 1997. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CiP1. Nature 389:187-190. [DOI] [PubMed] [Google Scholar]
- 39.Song, S., T. K. MacLachlan, R. D. Meng, and W. S. El-Deiry. 1999. Comparative gene expression profiling in response to p53 in a human lung cancer cell line. Biochem. Biophys. Res. Commun. 264:891-895. [DOI] [PubMed] [Google Scholar]
- 40.Sun, X. F., O. Johannsson, S. Hakansson, G. Sellberg, B. Nordenskjold, H. Olsson, and A. Borg. 1996. A novel p53 germline alteration identified in a late onset breast cancer kindred. Oncogene 13:407-411. [PubMed] [Google Scholar]
- 40a.Takimoto, R., T. K. MacLachlan, D. T. Dicker, Y. Niitsu, T. Mori, and W. S. El-Deiry. 2002. BRCA1 transcriptionally regulates damaged DNA binding protein (DDB2) in the DNA repair response following UV-irradiation. Cancer Biol. Ther. 1:177-186. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka, H., H. Arakawa, T. Yamaguchi, K. Shiraishi, S. Fukuda, K. Matsui, Y. Takei, and Y. Nakamura. 2000. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 404:42-49. [DOI] [PubMed] [Google Scholar]
- 42.Tomlinson, G. E., T. T. Chen, V. A. Stastny, A. K. Virmani, M. A. Spillman, V. Tonk, J. L. Blum, N. R. Schneider, I. I. Wistuba, J. W. Shay, J. D. Minna, and A. F. Gazdar. 1998. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 58:3237-3242. [PubMed] [Google Scholar]
- 43.Wang, H., Z. C. Zeng, T. A. Bui, S. J. DiBiase, W. Qin, F. Xia, S. N. Powell, and G. Iliakis. 2001. Nonhomologous end-joining of ionizing radiation-induced DNA double-stranded breaks in human tumor cells deficient in BRCA1 or BRCA2. Cancer Res. 61:270-277. [PubMed] [Google Scholar]
- 44.Wodicka, L., H. Dong, M. Mittmann, M. H. Ho, and D. J. Lockhart. 1997. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15:1359-1367. [DOI] [PubMed] [Google Scholar]
- 45.Wu, G. S., T. F. Burns, E. R. McDonald III, W. Jiang, R. Meng, I. D. Krantz, G. Kao, D. D. Gan, J. Y. Zhou, R. Muschel, S. R. Hamilton, N. B. Spinner, S. Markowitz, G. Wu, and W. S. El-Deiry. 1997. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat. Genet. 17:141-143. [DOI] [PubMed] [Google Scholar]
- 46.Wu, G. S., and W. S. El-Deiry. 1996. Apoptotic death of tumor cells correlates with chemosensitivity, independent of p53 or bcl-2. Clin. Cancer Res. 2:623-633. [PubMed] [Google Scholar]
- 47.Xu, X., W. Qiao, S. P. Linke, L. Cao, W. M. Li, P. A. Furth, C. C. Harris, and C. X. Deng. 2001. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat. Genet. 28:266-271. [DOI] [PubMed] [Google Scholar]
- 48.Xu, X., Z. Weaver, S. P. Linke, C. Li, J. Gotay, X. W. Wang, C. C. Harris, T. Ried, and C. X. Deng. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3:389-395. [DOI] [PubMed] [Google Scholar]
- 49.Yin, Y., M. A. Tainsky, F. Z. Bischoff, L. C. Strong, and G. M. Wahl. 1992. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell 70:937-948. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, H., K. Somasundaram, Y. Peng, H. Tian, D. Bi, B. L. Weber, and W. S. El-Deiry. 1998. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene 16:1713-1721. [DOI] [PubMed] [Google Scholar]
- 51.Zheng, L., S. Li, T. G. Boyer, and W. H. Lee. 2000. Lessons learned from BRCA1 and BRCA2. Oncogene 19:6159-6175. [DOI] [PubMed] [Google Scholar]
- 52.Zhong, Q., C. F. Chen, S. Li, Y. Chen, C. C. Wang, J. Xiao, P. L. Chen, Z. D. Sharp, and W. H. Lee. 1999. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science 285:747-750. [DOI] [PubMed] [Google Scholar]
- 53.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]