Abstract
Nicotiana plumbaginifolia UBP1 is an hnRNP-like protein associated with the poly(A)+ RNA in the cell nucleus. Consistent with a role in pre-mRNA processing, overexpression of UBP1 in N. plumabaginifolia protoplasts enhances the splicing of suboptimal introns and increases the steady-state levels of reporter mRNAs, even intronless ones. The latter effect of UBP1 is promoter specific and appears to be due to UBP1 binding to the 3′ untranslated region (3′-UTR) and protecting the mRNA from exonucleolytic degradation (M. H. L. Lambermon, G. G. Simpson, D. A. Kirk, M. Hemmings-Mieszczak, U. Klahre, and W. Filipowicz, EMBO J. 19:1638-1649, 2000). To gain more insight into UBP1 function in pre-mRNA maturation, we characterized proteins interacting with N. plumbaginifolia UBP1 and one of its Arabidopsis thaliana counterparts, AtUBP1b, by using yeast two-hybrid screens and in vitro pull-down assays. Two proteins, UBP1-associated proteins 1a and 2a (UBA1a and UBA2a, respectively), were identified in A. thaliana. They are members of two novel families of plant-specific proteins containing RNA recognition motif-type RNA-binding domains. UBA1a and UBA2a are nuclear proteins, and their recombinant forms bind RNA with a specificity for oligouridylates in vitro. As with UBP1, transient overexpression of UBA1a in protoplasts increases the steady-state levels of reporter mRNAs in a promoter-dependent manner. Similarly, overexpression of UBA2a increases the levels of reporter mRNAs, but this effect is promoter independent. Unlike UBP1, neither UBA1a nor UBA2a stimulates pre-mRNA splicing. These and other data suggest that UBP1, UBA1a, and UBA2a may act as components of a complex recognizing U-rich sequences in plant 3′-UTRs and contributing to the stabilization of mRNAs in the nucleus.
Newly synthesized pre-mRNAs associate with a set of proteins known as heterogeneous nuclear RNA-binding proteins (hnRNPs). In human cells, more than 20 abundant hnRNPs, designated A1 through U, have been identified (reviewed in references 15, 33, 57, and 61). Similar proteins are also present in other vertebrates. In Drosophila melanogaster, 12 major proteins, designated hrp36 through hrp75, have been identified (26, 27, 28, 40, 42). They share considerable sequence similarity with human hnRNPs (26, 27, 28, 40, 42). Some hnRNP-like proteins have been identified in Saccharomyces cerevisiae (1, 10, 32, 54, 64).
Originally, hnRNPs were believed to be responsible only for packaging and proper folding of processing substrates (reviewed in reference 57). However, more recently, many of the hnRNPs have been identified as factors involved in different stages of mRNA maturation (reviewed in references 33, 56, and 61). They have been shown to play roles in pre-mRNA splicing; 3′-end formation; mRNA transport, translation, and stability; and transcription regulation (reviewed in reference 33). Notably, the same hnRNP can influence mRNA maturation at different levels. For example, hnRNP A1 is involved in alternative splicing (8, 12, 44) and mRNA export (30, 45, 60; reviewed in reference 47). It is generally accepted that hnRNPs bind to the nascent RNA in a transcript-specific way, resulting in a unique combination of hnRNPs present on each mRNA precursor (2). Both protein-RNA and protein-protein interactions appear to contribute to the structures of ribonucleoprotein complexes acting as processing substrates (2, 41).
Previous work suggested that nuclear extracts originating from plant cells contain relatively few proteins which interact with pre-mRNA transcripts in vitro (20). However, recent experiments and database searches indicate that a complex set of hnRNP-like proteins is expressed in plant cells (34, 36; reviewed in reference 37). Among hnRNP-like proteins identified in Arabidopsis thaliana are homologues of the metazoan hnRNPs A/B, I (PTB), H, and F (reviewed in reference 37).
In higher plants, introns and 3′ untranslated regions (3′-UTRs) of nuclear pre-mRNAs are generally enriched in A+U or U residues, and considerable evidence exists that this property is essential for accurate and efficient pre-mRNA processing (reviewed in references 6, 25, 37, 38, 52, 53, and 55). When characterizing proteins which might recognize U-rich sequences in plant pre-mRNAs, Lambermon et al. previously identified a nuclear RNA-binding protein from Nicotiana plumbaginifolia, called UBP1 (hereafter referred to as NpUBP1), and showed that this protein meets many criteria of hnRNPs (34). NpUBP1 is the major product of UV cross-linking reactions carried out with introns and 3′-UTRs in vitro, and it is also found in association with poly(A)+ RNA in plant cell nuclei in vivo. Overexpression of NpUBP1 in N. plumbaginifolia protoplasts greatly increases the splicing efficiency of suboptimal introns and augments the steady-state levels of reporter mRNAs that contain suboptimal introns or are intronless. The enhanced accumulation is not caused by the stimulation of transcription but appears to be due to NpUBP1 interacting with the mRNA 3′-UTR, thereby protecting the mRNA from exonucleolytic degradation (34). Interestingly the effect on mRNA accumulation, but not on mRNA splicing, was found to be promoter specific. NpUBP1 induces the accumulation of mRNAs when transcription is driven by the cauliflower mosaic virus (CaMV) 35S promoter but not the cellular β-glucanase (GLB) promoter, indicating that NpUBP1 may interact with the transcription machinery at the step of initiation. Taken together, these experiments indicated that NpUBP1 is an hnRNP-like protein that functions at different steps of mRNA maturation in the nucleus (34).
At the primary sequence level, NpUBP1 and related proteins from other plants are most similar to the mammalian protein TIA-1, which was recently shown to facilitate recognition by U1 small nuclear ribonucleoprotein particles (snRNPs) of suboptimal 5′ splice sites by associating with intronic U-rich sequences positioned downstream (13, 17). The function of TIA-1 is similar to that of the yeast U1 snRNP protein Nam8p, which is required for the recruitment of U1 snRNP to 5′ splice sites which are either suboptimal or present in noncapped pre-mRNAs (24, 51). There is an obvious parallel among UBP1, TIA-1, and Nam8p in that they are all structurally related proteins containing three RNA recognition motifs (RRMs), they bind RNA with a specificity for oligouridylates, and they stimulate the processing of introns containing suboptimal splicing signals. In addition, TIA proteins, like UBP1, regulate gene expression at posttranscriptional levels other than splicing (references 31 and 49 and references therein).
To gain more insight into UBP1 function in pre-mRNA maturation in plants, we set out to characterize proteins interacting with NpUBP1 and one of its A. thaliana counterparts, AtUBP1b. We identified two novel RNA-binding proteins, which we named UBP1-associated proteins 1a and 2a (UBA1a and UBA2a, respectively). They are members of two protein families which appear to be present only in higher plants. Available evidence suggests that UBP1, UBA1a, and UBA2a are components of a complex regulating the turnover of mRNAs in the nucleus.
MATERIALS AND METHODS
Plasmid constructions.
pDEDH/Nco, pUBP1, pUBP1-HA, pGLB/Syn3, pGUS, and pGLB/GUS were described by Lambermon et al. (34). pSynGC/ClaU was described by Gniadkowski et al. (20). pSyn3 corresponds to CaMV cDNA and was described by Connelly and Filipowicz (9). pSyn7 was described by Goodall and Filipowicz (21). pDEmac (pZmAct) was described by Goodall and Filipowicz (22). pBD-UBP1 was constructed by cloning the UBP1 coding sequence into the SmaI and PstI sites of pGBT9 (Clontech).
(i) Cloning of UBA1a, UBA2a, and AtUBP1b into pDEDH-HA.
To clone full-length UBA1a into a plant expression vector, the coding region was PCR amplified by using the plasmid from the two-hybrid positive clone as a template and the following primers: 5′ primer, GCGGATCCAAACCATGGCGAAAACCCTAGACAAATCTAAGAAGCGCAAACTCGTG, and 3′ primer, GCGGATCCTGAATAGTTTGAGTAGCCATG (introduced BamHI sites are in boldface). The 1,082-bp BamHI fragment was ligated into the BamHI site of pDEDH-HA (19), resulting in pUBA1a-HA. The coding region of UBA2a was PCR amplified from the plasmid isolated in the yeast two-hybrid screen by using the following primers: 5′ primer, TGCAGGTCGACAATAAACCATGACAAAGAAGAGAAAGCTCGAAGG, and 3′ primer, ACGAGTCGGATCCGTGACCCATGTAAGGAGGTA. The 5′ primer introduced a SalI restriction site (in bold type), followed by the plant consensus translation initiation sequence (in italic type), whereas the 3′ primer introduced a BamHI restriction site (in bold type) in place of the stop codon. The PCR fragment was cut with SalI and BamHI and ligated into vector pDEDH-HA opened with the same restriction enzymes, resulting in pUBA2a-HA. The coding region of AtUBP1b was PCR amplified from the expressed sequence tag (EST) T88403 by using the following primers: 5′ primer, TGCGGGTCGACAATAAACCATGCAGAGGTTGAAGCAGCA, and 3′ primer, TATTGCAGATCTCTGGTAGTACATGAGCTGC; these primers introduced SalI and BglII restriction sites (in bold type) in front of the start codon and in place of the stop codon, respectively. The PCR product was cut with SalI and BglII and ligated into SalI- and BamHI-opened pDEDH-HA, resulting in pAtUBP1b-HA.
(ii) Cloning of UBA1a and UBA2a into pDEDH-GFP.
The green fluorescent protein (GFP) coding region was cut out from pUBP1-GFP (34) with BamHI and PstI and cloned into pDEDH (pDEDH/Nco or pGGS2). The UBA1a open reading frame (ORF) was PCR amplified by using the following primers: 5′ primer, CCCGGGGTCGACAATAAACCATGGCGAAAACCCTAGACAAATCA, and 3′ primer, GCGGATCCTGAATAGTTTGAGTAGCCATG. The PCR product was cut with SalI and BamHI (in bold) and ligated into pDEDH-GFP opened with the same enzymes, resulting in pUBA1a-GFP. The ORF of UBA2a was cut out from pUBA2a-HA with SalI and BamHI and ligated into pDEDH-GFP opened with the same enzymes, resulting in pUBA1a-GFP.
(iii) Cloning of UBA1a and UBA2a into pGEX-3X.
To create pGST-UBA1a, the 1,082-bp PCR product described above was cloned into the BamHI site of pGEX-3X (Pharmacia). The UBA2a ORF was PCR amplified from pAD-UBA2a by using the following primers: 5′ primer, TGCAGGGATCCCCACAAAGAAGAGAAAGCTCGAAGG, and 3′ primer, ACGAGTCGGATCCGTGACCCATGTAAGGAGGTA. Both primers introduced BamHI restriction sites (in bold) in place of the start and stop codons, respectively. The PCR product was digested with BamHI and ligated into BamHI-opened and dephosphorylated pGEX-3X, resulting in pGST-UBA2a.
(iv) Cloning of UBA1a, UBA2a, and AtUBP1 into yeast two-hybrid vectors.
pBD-UBA1a was created by cloning of the 1,082-bp PCR product described above into the BamHI site of pGBT9. The insert for cloning UBA2a into pGEX-3X was ligated into BamHI-opened and dephosphorylated pGBT9, resulting in pBD-UBA2a. Coding regions of AtUBP1a and AtUBP1b were PCR amplified from EST clones T21032 and T88403, respectively. AtUBP1c was PCR amplified from pUBP1c-HA (to be described elsewhere). Gene-specific primers introduced EcoRI sites in place of start and stop codons. PCR products were cut with EcoRI and ligated into pGAD424 and pGBT9, resulting in pAD-AtUBP1a, pAD-AtUBP1b, pAD-AtUBP1c, pBD-AtUBP1a, pBD-AtUBP1b, and pBD-AtUBP1c.
Site-directed mutagenesis.
Site-directed mutagenesis was performed on plasmids pGST-UBA1a and pGST-UBA2a as described by Nasr and Filipowicz (48). The oligonucleotides used for UBA1a were as follows: UBA1a-WT, CCCTTTAGCCTTACCAGTAGCCA; UBA1a-M1, GCTGGAGCTGTTATGTTCAAGACTAGGAAAG; and UBA1a-M2, GCTGGAGCTGTTATGGCCAAGACTAGGAAAG. The oligonucleotides used for RRM1 of UBA2a were as follows: UBA2a-WT1, ACCTTTAGATTTCCCTGAGATCTTATC; UBA2a-M1, GCTGGCGCTATTCTGTACAAGTCTCGTTCAGGTGCTCGC; and UBA2a-M2, GCTGGCGCTATTCTGGCCAAGTCTCGTTCAGGTGCTCGC. Those used for RRM2 of UBA2a were as follows: UBA2a-WT2, ACCTTTAGGTCTCCCAGTATACTTATC; UBA2a-M3, GCCTGTGCCTTTGTTTATAAGTCGTCTGAAAGCGCGA; and UBA2a-M4, GCCTGTGCCTTTGTTGCTAAGTCGTCTGAAAGCGCGA. In the above sequences, the mutagenized codons are underlined. Mutagensis of UBA1a and UBA2a was done in two and four steps, respectively. The resulting clones, pGST-UBA1aM and pGST-UBA2aM, respectively, were checked by sequencing of the entire UBA1a and UBA2a coding regions.
Yeast two-hybrid screening.
The A. thaliana cDNA library, yeast strains, and yeast plasmids pGBT9 and pGAD424 were from Clontech. Manipulation of the yeast cells and library screening were carried out according to the conditions suggested by the manufacturer. In brief, the S. cerevisiae reporter strain HF7c was transformed first with pBD-UBP1 and subsequently with the A. thaliana cDNA library. Approximately 3 × 106 transformants were plated onto 150-mm plates containing synthetic medium lacking histidine, leucine, and tryptophan. A total of 104 His+ transformants were assayed for β-galactosidase activity by filter assays as described by the manufacturer. The library plasmid from the His+ LacZ+ clones was recovered into Escherichia coli strain DH5α. True positive clones were confirmed by their ability to transactivate HIS3 and LacZ reporters when cotransforming HF7c with pBD-UBP1.
The cDNA insert encoding UBA1a was missing four N-terminal amino acids, as revealed by 5′ rapid amplification of the cDNA ends. PCR amplification of the 5′ end of the cDNA encoding UBA2a from the A. thaliana lambda cDNA library revealed that the original cDNA insert obtained in the two-hybrid screening was missing the N-terminal methionine.
Two-hybrid screening with NpUBP1 lacking RRM1, with AtUBP1b, and with UBA1a as bait was performed as described above.
Plant protoplasts, cellular fractionation, preparation of total cellular extracts, and pull-down assays.
N. plumbaginifolia mesophyll protoplasts were prepared from leaves grown in tissue cultures as described by Goodall et al. (23). Cellular fractionation was performed as described by Lambermon et al. (34). Total extracts from transfected protoplasts were prepared as follows. Protoplasts were washed in W5 medium (23), pelleted by centrifugation at 100 × g for 5 min, and resuspended (2 × 106/ml) in binding buffer (50 mM HEPES [pH 7.5], 200 mM NaCl, 2.5 mM MgCl2, 1 mM dithiothreitol [DTT], 0.5% Nonidet P-40) supplemented with protease inhibitor complete cocktail (Boehringer Mannheim). Protoplasts were disrupted by passing the suspension through a 25-gauge needle, and the lysates were centrifuged for 45 min at 4°C and 17,000 × g. The supernatant was added to glutathione-Sepharose 4B beads (Pharmacia), already loaded with glutathione S-transferase (GST), GST-UBA1a, GST-UBA2a, GST-UBA1aM, or GST-UBA2aM. The mixture was incubated for 5 h at 4°C. The beads were washed eight times with 1.5 ml of binding buffer. One additional wash was done with phosphate-buffered saline. Beads were then resuspended in 2× sodium dodecyl sulfate (SDS) loading buffer and boiled for 5 min. Retained proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 12% gels and analyzed by Western blotting with monoclonal antibody (MAb) DG6 against UBP1 or anti-hemagglutinin (HA) MAb 3F10 against UBA1a-HA and UBA2a-HA.
Preparation of recombinant proteins and in vitro UV cross-linking.
GST-UBA1a, GST-UBA1aM, GST-UBA2a, and GST-UBA2aM proteins used in pull-down experiments were overexpressed in E. coli strain BL21(DE3). GST-UBA1a and GST-UBA2a used in cross-linking experiments (see Fig. 6) were overexpressed in E. coli strain BL21 CodonPlus (Stratagene). Recombinant proteins were purified on glutathione-Sepharose 4B beads according to the manufacturer's instructions. In vitro cross-linking and homoribopolymer competition assays were performed as described by Domon et al. (14).
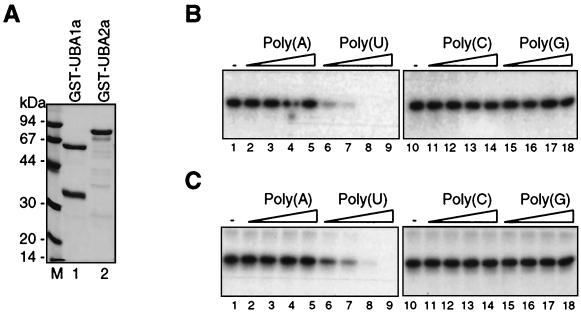
FIG. 6.
Determination of RNA-binding specificities of UBA1a and UBA2a. (A) Coomassie blue-stained gel of overexpressed GST-UBA1a and GST-UBA2a proteins purified on glutathione-Sepharose 4B. Each lane was loaded with 1.5 μg of protein. Positions of molecular mass markers (Amersham Pharmacia Biotech) are indicated on the left (lane M). (B and C) Competition of homoribopolymers with UV cross-linking of the 32P-labeled CaMV 3′-UTR RNA to recombinant GST-UBA1a (B) and GST-UBA2a (C). Polymers poly(C), poly(G), poly(A), and poly(U), indicated at the top of each panel, were added at 2-fold (lanes 2, 6, 11, and 15), 5-fold (lanes 3, 7, 12, and 16), 25-fold (4, 8, 13, and 17), and 100-fold (lanes 5, 9, 14, and 18) excesses (calculated in moles of nucleotides) over labeled RNA. Lanes 1 and 10, cross-linking with no competitor added. Only relevant gel fragments are shown. The approximately 35-kDa abundant polypeptide present in the GST-UBA1a preparation, most likely corresponding to the GST portion of the fusion protein, does not cross-link to RNA.
Transfection of protoplasts and RNA analysis.
Protoplasts of N. plumbaginifolia were transfected by the polyethylene glycol method (23). Unless indicated otherwise, 5 μg of the reporter plasmid and various amounts (see the legends to Fig. 7 and 8) of plasmids pUBP1-HA, pUBA1a-HA, and pUBA2a-HA were used. The total amount of DNA was kept constant by the addition of the empty expression vector pDEDH/Nco. RNA was extracted from the protoplasts by using an RNeasy plant mini kit (Qiagen) and treated with DNase I as described by Goodall et al. (23). Probes for RNase A or T1 mapping were prepared by in vitro transcription with appropriately linearized plasmids containing cDNA or gene inserts as templates (34). All probes were labeled with [α-32P]UTP (800 Ci/mmol; NEN). RNase A or T1 protection assays and quantification of the data were performed as described earlier (20, 22). Endogenous U2 snRNA was mapped to provide a reference facilitating quantification of the data. RNA yields are given as ratios of transcript levels seen in the presence and absence of pUBP1, pUBA1a-HA, and pUBA2a-HA.
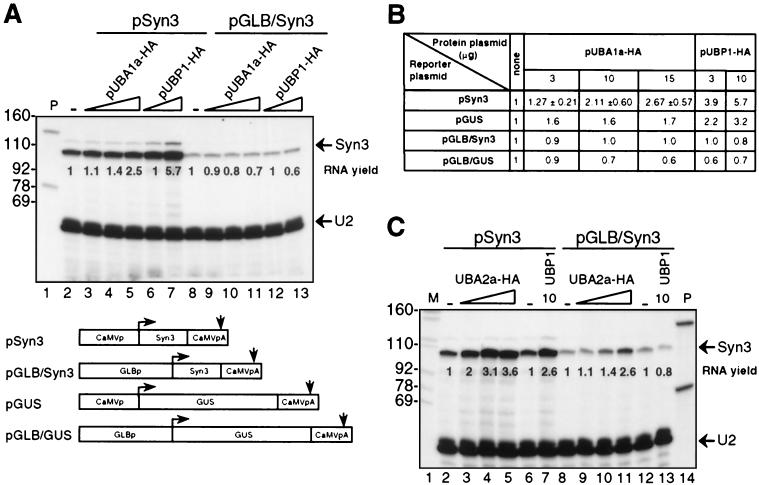
FIG. 7.
Effects of overexpression of UBA1a and UBA2a on the accumulation of reporter RNAs transiently expressed in N. plumbaginifolia protoplasts. (A) Effect of UBA1a overexpression on the accumulation of the intronless Syn3 reporter RNA. Protoplasts were cotransfected with 5 μg of the reporter plasmid and with empty vector alone (lanes 2 and 8), with increasing amounts of pUBA1a-HA (3, 10, and 15 μg; lanes 3 to 5 and 9 to 11), or with increasing amounts of pUBP1-HA (3 and 10 μg; lanes 6, 7, 12, and 13). Lanes 2 to 7, Syn3 RNA transcribed from the CaMV promoter; lanes 8 to 13, Syn3 RNA transcribed from the GLB promoter. The total amount of plasmid was kept constant (20 μg) by the addition of appropriate amounts of empty expression vector pDEDH/Nco. Mappings with the U2 probe, which detects a 50-nucleotide-long 5′-terminal fragment of endogenous U2 RNA, were performed in the same reactions; U2-protected bands are indicated. Lane 1, aliquot of undigested probes. Size markers (in nucleotides) are indicated on the left. Relative RNA yields are indicated in the lanes. Diagrams of relevant portions of plasmids expressing Syn3 and GUS reporter RNAs from CaMV and GLB promoters are shown below the gel. Horizontal arrows indicate transcription start sites, and vertical arrows indicate polyadenylation sites. (B) Summary of the effect of UBA1a on the abundance of different reporter RNAs initiated at the CaMV or GLB promoter. Values for expression from pSyn3 are means and standard deviations of three (plasmid concentrations, 3 and 15 μg) or four (plasmid concentration, 10 μg) independent transfection experiments. Remaining values are means of two independent transfection experiments. Quantification is based on PhosphorImager data corrected for endogenous U2 snRNA recovery. (C) Effect of overexpression of UBA2a on the accumulation of the intronless Syn3 reporter RNA expressed in N. plumbaginifolia protoplasts. Protoplasts were cotransfected with 5 μg of the reporter plasmid and with empty vector alone (lanes 2, 6, 8, and 12), with increasing amounts of pUBA2a-HA (1, 3, and 10 μg; lanes 3 to 5 and 9 to 11), or with 10 μg of pUBP1-HA (lanes 7 and 13). Lanes 2 to 7, Syn3 RNA transcribed from the CaMV promoter; lanes 8 to 13, Syn3 RNA transcribed from the GLB promoter. The total amount of plasmid was kept constant (15 μg) by the addition of appropriate amounts of empty expression vector pDEDH/Nco. Mappings with the U2 probe, which detects a 50-nucleotide-long 5′-terminal fragment of the endogenous U2 RNA, were performed in the same reactions. Relative RNA yields are indicated in the lanes. Size markers (in nucleotides) are indicated on the left (lane 1). Lane 14, aliquot of undigested probes. In an additional experiment with 3, 10, and 15 μg of plasmid expressing UBA2a-HA, the respective yields for Syn3 RNA transcribed from the CaMV promoter were 3.4, 5.6, and 7.4, and those for Syn3 RNA transcribed from the GLB promoter were 1.3, 1.1, and 2.8.
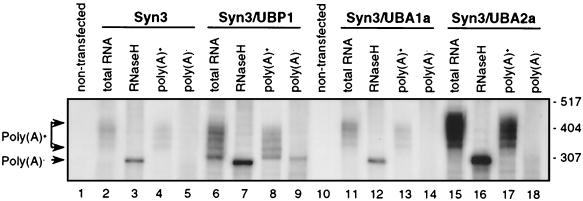
FIG. 8.
Effects of UBA1a, UBA2a, and UBP1 on the accumulation of Syn3 reporter RNA, as analyzed on a high-resolution polyacrylamide gel. RNA from protoplasts cotransfected with 5 μg of pSyn3 and either 10 μg of empty vector pDEDH-HA (lanes 2, 3, 4, and 5), pUBP1 (lanes 6, 7, 8, and 9), pUBA1a-HA (lanes 11, 12, 13, and 14), or pUBA2a-HA (lanes 15, 16, 17, and 18) was resolved on a 6% polyacrylamide-8 M urea gel and analyzed by Northern blotting. Antisense Syn3 RNA was used as a probe. Lanes 1 and 10, RNA from nontransfected protoplasts; lanes 2, 6, 11, and 15, total RNA; lanes 3, 7, 12, and 16, RNA treated with RNase H and oligo(dT); lanes 4, 8, 13, and 17, poly(A)+ RNA; lanes 5, 9, 14, and 18, poly(A)− RNA. Positions of size markers (in nucleotides) are indicated on the right. Only the relevant portion of the gel is shown.
Northern analysis (see Fig. 8) and treatment of RNA with RNase H and its fractionation on oligo(dT) Dynabeads were performed as described earlier (34).
Western analysis.
Proteins separated by SDS-12% PAGE were electroblotted onto a polyvinylidene difluoride membrane (Millipore). Antibodies were diluted as follows: anti-UBP1 MAb DG6 (34), 1:200; mouse anti-HA MAb 12CA5 (Boehringer Mannheim), 1:100; rat anti-HA MAb 3F10 (Boehringer Mannheim), 1:5,000; goat anti-mouse horseradish peroxidase-conjugated antibody (Jackson ImmunoResearch Laboratories), 1:5,000; and goat anti-rat horseradish peroxidase-conjugated antibody (Boehringer Mannheim), 1:5,000.
Nucleotide sequence accession numbers.
The sequences of the AtUBA1a and AtUBA2a cDNAs have been deposited in the EMBL database under accession numbers AJ439403 and AJ439404, respectively.
RESULTS
Two-hybrid screens with UBP1.
A fusion of NpUBP1 to the GAL4 DNA-binding domain was used to screen an A. thaliana cDNA library in the yeast strain HF7c. About 104 His+ transformants (from a total of 3 × 106 primary transformants) were tested for β-galactosidase activity. One true positive was isolated. Reporter gene activation by the positive clone was strictly dependent on the expression of the UBP1-GAL4 DNA-binding domain fusion; no interaction with the GAL4 DNA-binding domain alone was observed. Sequencing of the cDNA insert revealed that it encodes a novel protein, UBA1a (Fig. 1) (see also Materials and Methods). Subsequent screening of the A. thaliana cDNA library with the NpUBP1 deletion mutant lacking RRM1 or one of the three A. thaliana homologues of NpUBP1 (AtUBP1b) (34) (see also below) as bait resulted in the isolation of a clone encoding the same protein. In the latter screen, about 300 His+ transformants (from a total of 106 primary transformants) were tested for β-galactosidase activity. In addition to the UBA1a clone, a second positive clone was isolated. Sequencing of the cDNA insert revealed that it encodes another novel protein, UBA2a (Fig. 2) (see also below).
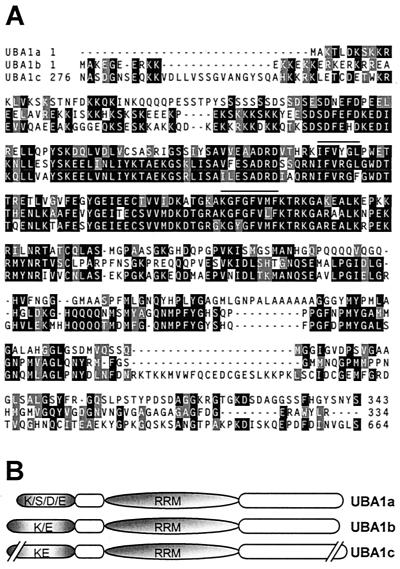
FIG. 1.
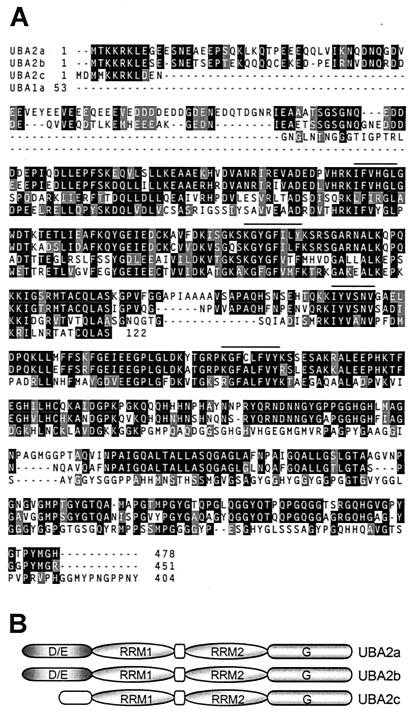
Sequence alignment (A) and schematic structure (B) of the UBA1 proteins. Alignment was generated by using the CLUSTAL W program (59) with the default parameters and was improved manually. Conserved amino acids were shaded by using the BOXSHADE program available at http://www.ch.embnet.org/software/BOX_form.html. Amino acids identical or similar in two of the three analyzed sequences have black and gray backgrounds, respectively. Dashes indicate gaps. The RNP2 sequence (six amino acids) and the RNP1 sequence (eight amino acids) of the RRM are overlined. Genomic sequences encoding UBA1a (GenBank accession number AC007232; protein identification: AAD25815), UBA1b (GenBank accession number AC007232; protein identification: AAD25814), and UBA1c (GenBank accession number AC003058; protein identification: AAC16468) have recently been deposited in GenBank. The predicted size of the UBA1c protein is 849 amino acids. In the alignment, we included only the middle part of the predicted UBA1c protein, which is homologous to UBA1a and UBA1b. K/S/D/E, domain rich in lysine, serine, and aspartic and glutamic acid residues; K/E, domain rich in lysine and glutamic acid residues. Open ovals represent protein regions without any obvious characteristic features.
FIG. 2.
Sequence alignment (A) and schematic structure (B) of the AtUBA2 proteins. The alignment also includes 120 amino acids of UBA1a (RRM plus 39 amino acids upstream of the RRM) which share strong sequence similarity with the UBA2 RRM1, including the corresponding upstream region. Genomic sequences encoding UBA2a (GenBank accession number T44669; protein identification: CAC00749), UBA2b (GenBank accession number AC004261; protein identification: AAD12005), and UBA2c (GenBank accession number AP000370; protein identification: BAA97064) have recently been deposited in GenBank. Which of the 5′-proximal AUG codons in UBA2c is used for translation initiation is not clear. However, the first methionine codon has the best initiation consensus sequence. The nucleotide sequence of the relevant region is CTGTAACCATGGATATGATG. D/E, domain rich in aspartate and glutamate residues; G, domain rich in glycine residues. For other details, see the legend to Fig. 1.
UBA1a and UBA2a are members of novel families of RNA-binding proteins.
Conceptual translation of the UBA1a cDNA revealed that it encodes a 343-amino-acid protein containing one RRM domain (Fig. 1). Furthermore, UBA1a contains a basic region at the N terminus, followed by a stretch of seven serines and an acidic domain (Fig. 1). UBA1a has a calculated molecular mass of 36 kDa, but Western analysis of lysates prepared from protoplasts transfected with a plasmid encoding HA-tagged UBA1a indicated that it migrates with an apparent mass of ∼45 kDa (see below). Northern analysis indicated that the 1.8-kb UBA1a mRNA is expressed at similar levels in A. thaliana leaves, stems, roots, and flowers (data not shown). Database searches identified two additional A. thaliana genes encoding proteins related to UBA1a, referred to as UBA1b and UBA1c, with calculated molecular masses of 43 and 95 kDa, respectively. Notably, the serine stretch is not present in these two members of the UBA1 family. Instead, they have longer basic regions at the N terminus (Fig. 1). We must mention here, however, that we could not find any EST sequence which would have confirmed the existence of the predicted UBA1c protein (849 amino acids). It is possible that the gene is either incorrectly predicted or represents a pseudogene.
The second identified UBP1-interacting protein, UBA2a, contains two putative RRMs and an acidic domain rich in glutamate and aspartate residues, positioned upstream of RRM1 (Fig. 2). UBA2a has a calculated molecular mass of 51 kDa, but HA-tagged UBA2a migrates with an apparent mass of ∼66 kDa (see below). Database searches revealed that UBA2a is also a member of a family of proteins in Arabidopsis comprising two additional members, UBA2b and UBA2c, with calculated molecular masses of 49 and 42 kDa, respectively (Fig. 2A). The sequence conservation between UBA2a and UBA2b is particularly high (72% identity and 78% similarity).
Comparison of the proteins belonging to the UBA1 and UBA2 families indicated that they all share similarity over a region of approximately 130 amino acids. An example of this sequence conservation is shown in the alignment of UBA2 proteins in Fig. 2A, including also the conserved region of UBA1a. The identity and similarity between the UBA1 and UBA2 proteins in this region are approximately 40 and 53%, respectively. The region of similarity comprises the RRM of UBA1 and RRM1 of UBA2 as well as the 40-amino-acid N-terminal region of these RRMs (Fig. 2A). Other regions of the proteins show no significant interfamily similarity.
UBA1a and UBA2a interact with UBP1 in vitro.
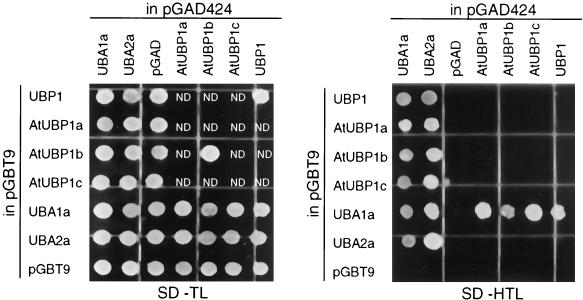
An additional yeast two-hybrid screen with UBA1a as bait resulted in the isolation of clones encoding UBA2a and UBA1a, suggesting that UBA1a interacts not only with NpUBP1 but also with UBA2a and with itself. In a two-hybrid reconstitution experiment, we found that both UBA1a and UBA2a interact with NpUBP1 and with all three Arabidopsis UBP1 isoforms (Fig. 3) (34). Furthermore, this experiment confirmed the interaction of UBA1a with UBA2a and with itself and revealed the interaction of UBA2a with itself (Fig. 3). We note, however, that interactions between UBA2a and UBP1 (from both N. plumbaginifolia and A. thaliana) were observed only with UBA2a cloned in pGAD424 and UBP1 cloned in pGBT9 and not with the reciprocal combination.
FIG. 3.
Reconstitution of the yeast two-hybrid interactions. Combinations of the bait and prey plasmids (indicated on both panels) were transformed into the yeast reporter strain, and the cells were grown on a synthetic medium lacking tryptophan and leucine (SD-TL) or histidine, tryptophan, and leucine (SD-HTL). Positive interactions obtained on SD-HTL medium were verified by β-galactosidase assays (data not shown). Interactions between UBA2a and UBP1 (from both N. plumbaginifolia and A. thaliana) were observed only when UBA2a and UBP1 were expressed from pGAD424 and pGBT9, respectively, and not in the reciprocal combination. ND, not determined.
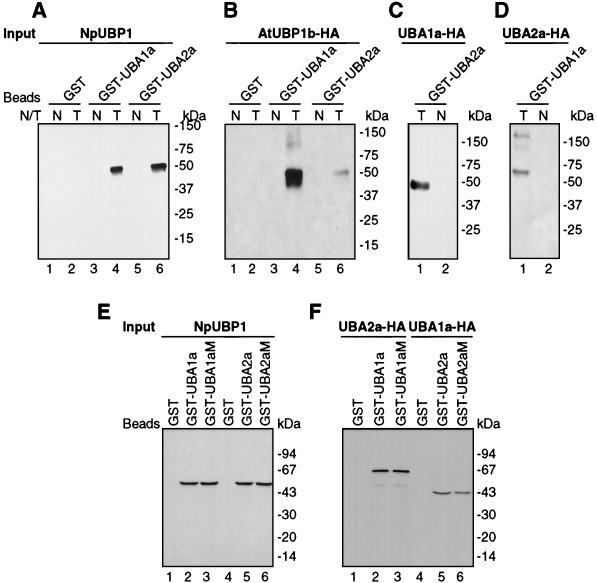
Interactions between the proteins were also studied by in vitro pull-down assays. To test the binding of UBA1a and UBA2a to UBP1, UBA1a and UBA2a were overexpressed in E. coli as GST fusion proteins and immobilized on glutathione-Sepharose beads. Beads coated with GST alone served as a control. Pull-down experiments were performed with protein extracts prepared from N. plumbaginifolia protoplasts transfected with plasmids expressing either NpUBP1 (Fig. 4A) or AtUBP1b (Fig. 4B). Protein extracts were treated with RNase A to eliminate indirect interactions mediated by RNA. Western analysis of proteins retained on the beads, performed with anti-UBP1 MAb DG6 (Fig. 4A) or anti-HA MAb 3F10 (Fig. 4B), showed that both NpUBP1 and AtUBP1b interact with GST-UBA1a and GST-UBA2a (Fig. 4A and B, lanes 4 and 6) but not with GST alone (Fig. 4A and B, lane 2).
FIG. 4.
Protein interactions studied by in vitro pull-down assays. GST-UBA1a and GST-UBA2a wild-type or mutant fusion proteins and GST alone were overexpressed in E. coli and immobilized on glutathione-Sepharose 4B beads. The beads were incubated with RNase-treated whole-cell protein extracts prepared from nontransfected N. plumbaginifolia protoplasts (lanes N) or protoplasts transfected (lanes T) with pUBP1 (encoding NpUBP1) (A), pAtUBP1b-HA (B), pUBA1a-HA (C), or pUBA2a-HA (D). (A and B) Pull-down assays with GST, GST-UBA1a, and GST-UBA2a. (C and D) Extracts were incubated with glutathione beads with immobilized GST-UBA2a or GST-UBA1a. (E and F) UBP1, UBA1a, and UBA2a interactions are not RNA mediated. Protein extracts from protoplasts overexpressing the proteins indicated were incubated with glutathione beads coated with GST, GST-UBA1a, GST-UBA1aM, GST-UBA2a, or GST-UBA2aM. Proteins retained on the beads were analyzed by SDS-PAGE and subsequent Western blotting with UBP1 MAb DG6 (A and E) and anti-HA MAb 3F10 (B, C, D, and F).
A similar approach was used to confirm the interaction between UBA1a and UBA2a. Glutathione-Sepharose beads were coated with GST-UBA2a or GST-UBA1a and subsequently incubated with protein extracts prepared from protoplasts overexpressing either UBA1a-HA (Fig. 4C) or UBA2a-HA (Fig. 4D). Proteins retained on the beads were then analyzed by Western blotting with anti-HA antibody. The results shown in Fig. 4C and D demonstrated that GST-UBA2a and GST-UBA1a interact in vitro with UBA1a-HA and UBA2a-HA, respectively.
In order to definitively rule out the possibility that the interactions among UBP1, UBA1a, and UBA2a are mediated by RNA, we mutated the aromatic residues to alanines in the RNP1 motif of the RRM of UBA1a (mutation KGFGFVMF→KGAGAVMA) and both RRMs of UBA2a (mutations KGYGFILY→KGAGAILA and KGFCLFVY→KGACAFVA). It was previously shown that aromatic amino acids in RNP1 are essential for RNA binding (43). Mutant proteins GST-UBA2aM and GST-UBA2aM were overexpressed in E. coli and purified. Their activity for binding RNA, as measured by UV cross-linking to the CaMV 3′-UTR, was decreased by approximately 90% (data not shown). Both mutant proteins were then used for in vitro pull-down assays. As shown in Fig. 4E and F, the mutant proteins associated with UBP1 and with each other as efficiently as wild-type proteins, indicating that the interactions are not mediated by RNA.
In summary, the results of the pull-down experiments confirmed the interactions observed in the yeast two-hybrid system, thus demonstrating that UBP1, UBA1a, and UBA2a associate with each other and that these associations are independent of the RNA-binding potential of UBA1a and UBA2a. Since the pull-down experiments were performed with protoplast extracts, we cannot rule out the possibility that some of the interactions observed were mediated by other proteins. However, this possibility is very unlikely, since our two-hybrid screens have never resulted in the identification of plant proteins other than the three proteins studied in this work. It is also important to note that no obvious homologues of UBP1, UBA1a, and UBA2a are encoded in the yeast genome. It is therefore highly improbable that a yeast protein could bind to all three plant proteins, mediating the two-hybrid interactions identified in this study.
UBA1a and UBA2a localize to the nucleus.
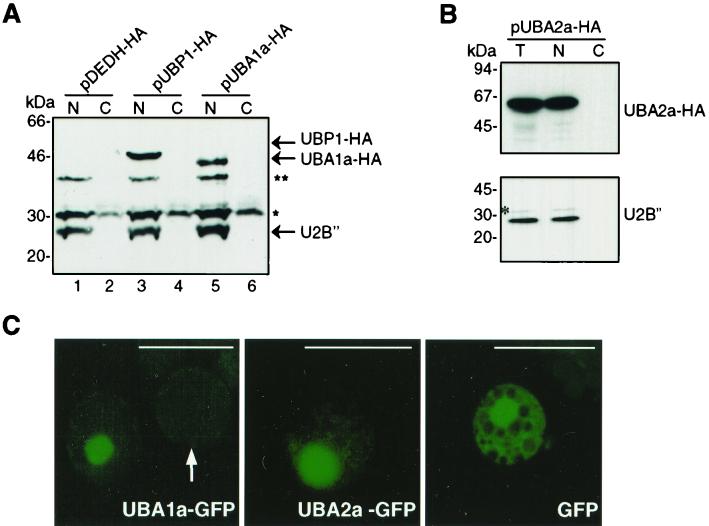
To study the intracellular distributions of UBA1a and UBA2a, the proteins were transiently expressed in N. plumbaginifolia protoplasts as fusions with an HA tag. Lysates of the protoplasts were separated into nuclear and cytoplasmic fractions, which were analyzed by Western blotting with anti-HA epitope MAbs (Fig. 5). To assess the quality of the fractionation procedure, Western analysis of the nuclear spliceosomal U2 snRNP U2B" was performed. These experiments showed that, similar to the distribution of NpUBP1 (Fig. 5A, lanes 3 and 4) (see also reference 34), both UBA1a and UBA2a are found in the nuclear fraction, without any detectable signal in the cytoplasmic fraction (Fig. 5A, lanes 5 and 6, and 5B).
FIG. 5.
Subcellular localization of UBA1a and UBA2a by cellular fractionation (A and B) or imaging of protoplasts expressing GFP fusions (C). (A) N. plumbaginifolia protoplasts were transfected with the empty vector pDEDH-HA or plasmids pUBP1-HA and pUBA1a-HA. (B) Protoplasts were transfected with pUBA2a-HA. Cell extracts were fractionated as described before (34). Lanes N, C, and T, nuclear, cytoplasmic, and total cellular protein fractions, respectively. Proteins were resolved by SDS-PAGE and analyzed by Western blotting with mouse anti-HA MAb 12CA5 (A), anti-U2B" MAb 4G3 (A and B, lower panel), or rat anti-HA antibody 3F10 (B, upper panel). The protein marked with an asterisk cross-reacts nonspecifically with the anti-U2B" MAb 4G3. The 40-kDa nuclear protein, marked with a double asterisk, cross-reacts nonspecifically with the anti-HA MAb 12CA5. (C) GFP imaging of protoplasts transfected with pUBA1a-GFP, pUBA2a-GFP, and pDEDH-GFP (expressing GFP alone). Protoplasts were analyzed by using a Zeiss Axioplan epifluorescence microscope. The arrow indicates an untransfected protoplast. Bars, 50 μm.
We also studied the cellular distribution of UBA1a and UBA2a expressed in protoplasts as GFP fusion proteins. Fluorescence microscopy analysis showed that the UBA1a (Fig. 5A) and UBA2a (Fig. 5B) fusion proteins are mainly localized in the nucleus, in contrast to GFP alone, which is also present in the cytoplasm (Fig. 5C). Taken together, these results indicate that UBA1a and UBA2a are predominantly nuclear proteins.
UBA1a and UBA2a bind RNA in a uridylate-specific manner.
We investigated the RNA-binding properties of UBA1a and UBA2a. The proteins were overexpressed in E. coli as GST fusion proteins and purified by use of gluthatione-Sepharose columns (Fig. 6A). Radiolabeled RNAs corresponding to the CaMV 3′-UTR or a synthetic AU-rich intron, Syn7 (21), were used in UV cross-linking experiments. Both proteins could be efficiently cross-linked to the CaMV 3′-UTR (Fig. 6B and C) and Syn7 intron (data not shown) sequences. The nucleotide specificity of UBA1a and UBA2a binding to the CaMV 3′-UTR was assessed by competition with different homoribopolymers (Fig. 6B and C). These experiments revealed that UBA1a and UBA2a are, like NpUBP1, oligouridylate-binding proteins (Fig. 6B and C, lanes 6 to 9). No competition with CaMV 3′-UTR RNA binding was observed in the presence of ribopolymers other than poly(U), even at a 400-fold molar excess (Fig. 6B and C and data not shown).
Overexpression of UBA1a and UBA2a increases the steady-state levels of reporter RNAs in transfected protoplasts.
It was previously shown that the overexpression of NpUBP1 in protoplasts increases the steady-state levels of reporter RNAs in a promoter-specific manner. In addition, it enhances the splicing of the inefficiently processed introns in a promoter-independent mode (34) (see the introductory paragraphs). Having identified UBA1a and UBA2a as proteins interacting with UBP1, we sought to determine whether they have comparable effects on pre-mRNA maturation.
We first examined the effect of the overexpression of UBA1a and UBA2a on the steady-state levels of the intronless Syn3 and β-glucuronidase (GUS) reporter RNAs expressed either from the CaMV promoter (plasmids pSyn3 and pGUS) or from the cellular GLB promoter (plasmids pGLB/Syn3 and pGLB/GUS). The respective pairs of genes containing the different promoters were constructed such that they yielded transcripts with identical sequences (34). Steady-state levels of the reporter RNAs were assessed by RNase A or T1 protection assays. Endogenous U2 snRNA was mapped to provide a reference facilitating quantification of the data. An example of RNase A or T1 analysis of Syn3 RNA expressed from plasmids containing different promoters is shown in Fig. 7A, and quantification of the data originating from different transfection experiments is shown in Fig. 7B.
By analogy with NpUBP1, the steady-state levels of Syn3 and GUS RNAs expressed from the CaMV promoter were increased upon cotransfection with pUBA1a-HA (Fig. 7A, lanes 2 to 5, and 7B), but not when expression was driven by the GLB promoter (Fig. 7A, lanes 8 to 11, and 7B). The steady-state level of Syn3 RNA expressed from the CaMV promoter increased 2.7-fold upon cotransfection with 15 μg of pUBA1a-HA and 5.7-fold upon cotransfection with 10 μg of pUBP1-HA (Fig. 7A and B). The respective values for GUS RNA expressed from the CaMV promoter were 1.7- and 3.2-fold (Fig. 7B). Since Western analyses indicated that NpUBP1 and UBA1a are expressed at comparable levels (Fig. 5 and data not shown), the effect of UBA1a on RNA levels is apparently less pronounced than the effect of UBP1 (Fig. 7B). Neither protein increased the steady-state levels of Syn3 and GUS RNAs when transcription was directed by the GLB promoter. In fact, with the GUS RNA reporter, a slight decrease in expression was observed (Fig. 7B). From these data, we conclude that, by analogy with UBP1, the overexpression of UBA1a can increase the steady-state levels of reporter transcripts in a manner dependent on the promoter driving gene expression, although the effect of UBA1a is less pronounced than that of UBP1.
The effect of UBA2a-HA overexpression on the steady-state level of Syn3 reporter RNA was investigated by a similar approach. As shown in Fig. 7C, the overexpression of UBA2a, in contrast to that of UBP1 and UBA1a, increased the steady-state level of Syn3 RNA in a promoter-independent manner. Cotransfection with 10 μg of pUBA2a-HA resulted in a 3.6- or 2.6-fold increase in the Syn3 RNA level when transcription was driven by the CaMV or GLB promoter, respectively (Fig. 7C, lanes 5 and 11). Of the three proteins studied, UBA2a appears to have the most pronounced effect on the steady-state levels of reporter RNAs. In some transfection experiments, its overexpression increased the level of Syn3 RNA expressed from the CaMV promoter up to 12-fold (Fig. 8 and data not shown).
Previous Northern analyses of Syn3 and β-globin reporter RNAs revealed that NpUBP1 overexpression not only increases the yield of poly(A)+ RNA but also leads to the appearance of an additional, shorter RNA, corresponding to the deadenylated “body” of the mRNA. These results suggested that NpUBP1 may bind to the 3′-UTR and protect the mRNA against exonucleolytic degradation (34). We used similar high-resolution Northern analyses to evaluate the effects of UBA1a and UBA2a on the accumulation of Syn3 RNA. The effect of UBP1 was also analyzed as a control. RNA isolated from protoplasts transfected with different plasmid combinations was separated on a 6% polyacrylamide gel, electroblotted to a nitrocellulose membrane, and hybridized to a Syn3-specific probe. Consistent with the results of RNase mapping experiments (34; this work), the overexpression of UBP1 and UBA2a increased the yield of Syn3 transcripts very strongly, while the effect of UBA1a was much less pronounced (Fig. 8). As reported before, the overexpression of UBP1, in addition to increasing the yield of poly(A)+ RNA, led to the appearance of an additional, shorter RNA with a size similar to that of the deadenylated body of the mRNA (Fig. 8, lanes 6 to 9). Notably, this short RNA was not present in cells overexpressing either UBA1a or UBA2a. Instead, the expression of these proteins resulted in a general increased accumulation of RNAs containing poly(A) tails of different lengths (Fig. 8, lanes 11 to 18). We conclude that the overexpression of all three proteins, UBP1, UBA1a, and UBA2a, has a stimulatory effect on the level of poly(A)+ RNA in transfected protoplasts but that only the expression of UBP1 leads to the accumulation of the poly(A)-free RNA form.
Overexpression of UBA1a and UBA2a has no effect on the splicing of the reporter RNA.
We also examined how the overexpression of UBA1a-HA and UBA2a-HA affects the processing of an intron-containing reporter RNA coexpressed in protoplasts of N. plumbaginifolia. Plasmids pSynGC/ClaU and pDEmac, which express synthetic pre-mRNA containing a GC-rich inefficiently spliced intron (20, 34) and intron 1 of the maize actin gene (ZmAct) (22), respectively, were cotransfected with increasing amounts of plasmids encoding UBA1a-HA and UBA2a-HA. The RNA processing efficiency was assessed by either reverse transcription-PCR or RNase A or T1 mapping. The overexpression of UBA1a-HA or UBA2a-HA did not stimulate the splicing of the SynGC/ClaU or ZmAct introns. In a parallel control experiment, the addition of the plasmid encoding NpUBP1-HA increased the splicing efficiencies of the SynGC/ClaU and ZmAct introns from 15 to 65% and from 40 to 75%, respectively (34) (data not shown). We conclude that, in contrast to that of NpUBP1, the overexpression of UBA1a or UBA2a does not stimulate splicing.
DISCUSSION
We previously characterized a nuclear protein from N. plumbaginifolia, NpUBP1, associated with the nuclear poly(A)+ RNA in vivo, as demonstrated by UV cross-linking experiments performed with intact cells. NpUBP1 has binding specificity for oligouridylates and interacts with U-rich intron and 3′-UTR sequences in vitro. Transfection experiments indicated that the overexpression of NpUBP1 has two independent posttranscriptional effects on reporter mRNAs. It enhances the splicing of suboptimal introns and increases steady-state levels of transcripts, even intronless ones (34). To gain more insight into how UBP1 functions in pre-mRNA maturation, we have now characterized two A. thaliana proteins interacting with NpUBP1 and its A. thaliana counterparts. These proteins, UBA1a and UBA2a, are members of two novel families of RRM domain-containing proteins which appear to be expressed only in higher plants. Like NpUBP1, UBA1a and UBA2a are RNA-binding proteins localized in the nucleus. Both proteins interact with 3′UTR sequences and have strong binding preference for oligouridylates in vitro.
Analysis of the effects of UBA1a and UBA2a overexpression on reporter RNA maturation revealed some similarities with but also substantial differences from UBP1. Like that of UBP1, the expression of either of the two new proteins in transfected protoplasts led to increased accumulation of Syn3 and GUS reporter mRNAs, with the effect of UBA2a being significantly more pronounced (Fig. 7 and 8). Previous efforts to elucidate the mechanism of the stimulatory effect of UBP1 on mRNA abundance suggested that UBP1 may bind to the 3′-UTR of reporter mRNAs and prevent exonucleolytic degradation from continuing beyond the poly(A) tail. This conclusion was based on the observations that UBP1 efficiently cross-links to the 3′-UTR region in vitro and that its overexpression in protoplasts results in the accumulation of transcripts corresponding to the poly(A)-free form of mRNAs (34). Northern analysis performed in this work indicated that, in marked contrast to UBP1 overexpression, neither UBA1a nor UBA2a overexpression leads to the appearance of the poly(A)-free mRNA (Fig. 8). In light of the results demonstrating various interactions among the three proteins, it is plausible that UBP1, UBA1a, and UBA2a participate in different complexes, with specificity defined by their precise compositions. Overexpression of one of the components could promote the formation of unique complexes with distinct properties. It is possible, for example, that UBA1a and UBA2a interact with the poly(A)-binding protein and, when overexpressed, predominantly stabilize its interaction with poly(A) tails, leading to higher stability of poly(A)+ RNA. In contrast, UBP1 may not contribute to such interactions and may affect mainly the level of the poly(A)-free mRNA. It will be interesting to investigate whether UBA1a and UBA2a interact with the poly(A)-binding protein.
Experiments carried out with yeast and mammalian cells demonstrated that substantial fractions of pre-mRNA transcripts undergo rapid turnover in the nucleus (references 5 and 62 and references therein) and that the 3′→5′ exonuclease complex (exosome) is involved in this process (5). Do the UBP1, UBA2a, and UBA2a proteins regulate the turnover of mRNAs or their precursors in the nuclei of plant cells? Localization studies have shown that UBP1, UBA2a, and UBA2a are nuclear proteins (34; this work). In addition, it was previously demonstrated that UBP1 is associated with the nuclear poly(A)+ RNA in plant cells in vivo (34) and that the poly(A)− RNA accumulating upon the overexpression of UBP1 is localized in the nucleus (63). Taking these and other established properties of UBP1, UBA1a, and UBA2a into account, it is tempting to propose that all three proteins, either individually or as a complex, interact with different regions of the generally U-rich 3′-UTRs in plant cell nuclei. Such interactions may prevent 3′→5′ exonuclease-mediated degradation from continuing beyond the poly(A) tail and may make the RNAs available for readenylation. It is also possible that the proteins facilitate some steps in nuclear pre-mRNA processing or mRNA export to the cytoplasm. However, it is important to note that we have found no evidence of UBP1 having a stimulatory effect on the 3′-end cleavage or polyadenylation reaction in transfected plant protoplasts (34). It has also been demonstrated that UBP1 does not shuttle between the nucleus and the cytoplasm in a transcription-dependent manner (63). However, the possibility that the protein shuttles between these compartments in a process independent of transcription has not been eliminated by our experiments.
It was previously shown that the stimulatory effect of NpUBP1 on mRNA abundance in transfected protoplasts is promoter specific. NpUBP1 increased the level of mRNA when transcription was driven by the CaMV 35S promoter but not the cellular GLB promoter, indicating that NpUBP1 may interact with the transcription machinery at the step of initiation. We have now found that the effect of UBA1a, but not that of UBA2a, also depends on the nature of the promoter driving the transcription of the reporter gene. Hence, the association of both UBP1 and UBA1a with transcripts apparently requires their prior interaction with the transcribing RNA polymerase II (PolII) complex. It is now generally accepted that mRNA transcription and processing are coupled in vivo. Many protein factors participating in various RNA processing reactions in mammalian and yeast cells associate with the CTD domain of the large subunit of PolII at transcription initiation (16; reviewed in references 3, 46, and 50). Notably, the recruitment of some mammalian SR proteins to pre-mRNA was shown to be modulated by the promoter structure. As reported previously, we did not find evidence of an interaction between UBP1 and the CTD, either in vitro or in the yeast two-hybrid system (34). Likewise, the yeast two-hybrid experiments did not reveal interactions of UBA1a and UBA2a with the A. thaliana CTD (unpublished results). Despite these negative results, UBP1 and UBA1a may still associate with the CTD, but the interaction may require the simultaneous presence of UBP1 and UBA1a and possibly also of UBA2a or other proteins. Alternatively, the association of the investigated proteins with the PolII complex may depend upon prior interactions with specific transcription factors, as has been described for the ASF/SF2 splicing factor in mammalian cells (11, 18).
In contrast to that of UBP1, the overexpression of UBA1a and UBA2a has no effect on the splicing efficiency of reporter RNAs containing suboptimal introns, such as intron SynGC/ClaU (20) or intron 1 of the maize actin gene (22). Hence, the interactions among UBP1, UBA1a, and UBA2a described in this work are apparently related to the functions of these proteins in mRNA accumulation or stabilization but probably not in the splicing-promoting activity of UBP1. Establishing the mechanism by which UBP1 stimulates intron processing will require additional experimentation. However, the structural similarity of UBP1 to the TIA-1 and Nam8p proteins (34, 36) (see also above) makes it plausible that, by binding to U-rich sequences in introns, UBP1 helps to recruit U1 snRNP or other splicing factors to pre-mRNA.
The previous finding that the overexpression of UBP1 stimulates both pre-mRNA splicing and mRNA accumulation raised the possibility that the effect on splicing is indirect, resulting from increased transcript stability in the nucleus. However, the demonstration that the effect of UBP1 on RNA accumulation is promoter specific and that the effect on splicing is not indicated that the two effects are independent (34). This conclusion is further supported by data on UBA1a and UBA2a. The expression of either protein in protoplasts stimulated the accumulation of reporter mRNAs but not their splicing. Hence, an effect on RNA accumulation need not necessarily result in an apparent increase in pre-mRNA splicing. It should be noted that the effects of UBP1, UBA1a, and UBA2a on RNA accumulation are clearly not splicing dependent, since they were observed with several different intronless reporter RNAs.
The presence of introns can dramatically increase the levels of expression of mRNAs both in mammals and in plants (4, 7, 35; reviewed in reference 55). To compensate for the lack of introns, many natural intronless mRNAs in mammalian cells contain sequence elements that enable efficient intron-independent processing and nuclear export (29, 35, 39). In A. thaliana and probably in other plants, approximately 20% of genes lack introns (58). Previous observations that UBP1 affects the accumulation of only intronless or otherwise suboptimal reporter pre-mRNAs and that this effect occurs in a promoter-dependent manner indicated that UBP1 plays a role in the expression of certain classes of pre-mRNAs (34). The identification of UBP1-interacting proteins, UBA1a and UBA2a, which stimulate the accumulation of reporter mRNAs in a manner similar (although not identical) to that of UBP1 suggests that complexes of these proteins regulate pre-mRNA metabolism in plants. Like UBP1 (34; unpublished results), UBA1a and UBA2a are both members of multiprotein families. Different isoforms of the UBP1, UBA1, and UBA2 proteins may form complexes with related but not identical properties important for the expression of specific classes of mRNAs. In the future, it will be important to inactivate individual variants of UBP1 and its associated proteins and to determine the effect on the expression of intronless genes in A. thaliana.
Acknowledgments
We thank Jirina Petruska for technical assistance. We are grateful to Andrea Barta and Dorothee Staiger for providing chemicals for experiments performed during revision of the manuscript. The Arabidopsis Biological Resource Center, The Ohio State University, is acknowledged for providing A. thaliana EST clones.
Z.J.L. was the recipient of a long-term fellowship from the European Molecular Biology Organization (EMBO). The Friedrich Miescher Institute for Biomedical Research is part of the Novartis Research Foundation.
REFERENCES
- 1.Anderson, J. T., S. M. Wilson, K. V. Datar, and M. S. Swanson. 1993. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol. 73:2730-2741. [DOI] [PMC free article] [PubMed]
- 2.Bennett, M., S. Michaud, J. Kingston, and R. Reed. 1992. Protein components specifically associated with pre-spliceosome and spliceosome complexes. Genes Dev. 6:1986-2000. [DOI] [PubMed] [Google Scholar]
- 3.Bentley, D. 1999. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 11:347-351. [DOI] [PubMed] [Google Scholar]
- 4.Bourdon, V., A. Harvey, and D. M. Lonsdale. 2001. Introns and their positions affect the translational activity of mRNA in plant cells. EMBO Rep. 2:394-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bousquet-Antonelli, C., C. Presutti, and D. Tollervey. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102:765-775. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. W. S., and C. G. Simpson. 1998. Splice site selection in plant pre-mRNA splicing. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:77-95. [DOI] [PubMed] [Google Scholar]
- 7.Callis, J., M. From, and V. Walbot. 1987. Introns increase gene expression in cultured maize cells. Genes Dev. 1:1183-1200. [DOI] [PubMed] [Google Scholar]
- 8.Chabot, B., M. Blanchette, I. Lapierre, and H. La Branche. 1997. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol. Cell. Biol. 17:1776-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connelly, S., and W. Filipowicz. 1993. Activity of chimeric U small nuclear RNA (snRNA)/mRNA genes in transfected protoplasts of Nicotiana plumbaginifolia: U snRNA 3′-end formation and transcription initiation can occur independently in plants. Mol. Cell. Biol. 13:6403-6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad, N. K., S. M. Wilson, E. J. Steinmetz, M. Patturajan, D. A. Brow, M. S. Swanson, and J. L. Corden. 2000. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154:557-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cramer, P., J. F. Caseres, D. Cazalla, S. Kadener, A. F. Muro, F. E. Baralle, and A. R. Kornblihtt. 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9D8 effects on an exonic splicing enhancer. Mol. Cell 4:251-258. [DOI] [PubMed] [Google Scholar]
- 12.Del Gatto-Konczak, F., M. Olive, M. C. Gesnel, and R. Breathnach. 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 19:251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Gatto-Konczak, F., C. F. Bourgeois, C. Le Guiner, L. Kister, M. C. Gesnel, J. Stevenin, and R. Breathnach. 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domon, C., Z. J. Lorković, J. Valcarcel, and W. Filipowicz. 1998. Multiple forms of the U2 small nuclear ribonucleoprotein auxiliary factor U2AF subunits expressed in higher plants. J. Biol. Chem. 273:34603-34610. [DOI] [PubMed] [Google Scholar]
- 15.Dreyfuss, G., M. J. Matunis, S. Pinol-Roma, and C. G. Burd. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62:289-321. [DOI] [PubMed] [Google Scholar]
- 16.Fong, N., and D. L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 15:1783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forch, P., O. Puig, N. Kedersha, C. Martinez, S. Granneman, B. Seraphin, P. Anderson, and J. Valcarcel. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 6:1089-1098. [DOI] [PubMed] [Google Scholar]
- 18.Ge, H., Y. Si, and A. P. Wolffe. 1998. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell 2:751-759. [DOI] [PubMed] [Google Scholar]
- 19.Genschik, P., J. Hall, and W. Filipowicz. 1997. Cloning and characterization of the Arabidopsis cyclic phosphodiesterase which hydrolyzes ADP-ribose 1",2"-cyclic phosphate and nucleoside 2′,3′-cyclic phosphates. J. Biol. Chem. 272:13211-13219. [DOI] [PubMed] [Google Scholar]
- 20.Gniadkowski, M., M. Hemmings-Mieszczak, U. Klahre, H.-X. Liu, and W. Filipowicz. 1996. Characterization of intronic uridine-rich sequence elements acting as possible targets for nuclear proteins during pre-mRNA splicing in Nicotiana plumbaginifolia. Nucleic Acids Res. 24:619-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodall, G. J., and W. Filipowicz. 1989. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58:473-483. [DOI] [PubMed] [Google Scholar]
- 22.Goodall, G. J., and W. Filipowicz. 1991. Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO J. 10:2635-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodall, G. J., K. Wiebauer, and W. Filipowicz. 1990. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 181:148-161. [DOI] [PubMed] [Google Scholar]
- 24.Gottschalk, A., J. Tang, O. Puig, J. Salgado, G. Neubauer, H. V. Colot, M. Mann, B. Seraphin, M. Rosbash, R. Luhrmann, and P. A. Fabrizio. 1998. Comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4:374-393. [PMC free article] [PubMed] [Google Scholar]
- 25.Graber, J. H., C. R. Cantor, S. C. Mohr, and T. F. Smith. 1999. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. USA 96:14055-14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haynes, S. R., D. Jonson, G. Raychaudhuri, and A. L. Beyer. 1991. The Drosophila Hrb87F gene encodes a new member of the A and B hnRNP protein group. Nucleic Acids Res. 19:25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haynes, S. R., G. Raychaudhuri, and A. L. Beyer. 1990. The Drosophila Hrb98DE locus encodes four protein isoforms homologous to the A1 protein of mammalian heterogeneous nuclear ribonucleoprotein proteins. Mol. Cell. Biol. 10:316-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haynes, S. R., G. Raychaudhuri, D. Jonson, S. Amero, and A. L. Beyer. 1990. The Drosophila Hrb loci: a family of hnRNA binding proteins. Mol. Biol. Rep. 14:93-94. [DOI] [PubMed] [Google Scholar]
- 29.Huang, Y., K. M. Wimler, and G. G. Carmichael. 1999. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 18:1642-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izzauralde, E., A. Jarmolowski, C. Beisel, I. W. Mattaj, and G. Dreyfuss. 1997. A role of the M9 transport signal of hnRNP A1 in mRNA export. J. Cell Biol. 137:27-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kedersha, N., M. R. Cho, W. Li, P. W. Yacono, S. Chen, N. Gilks, D. E. Golan, and P. Anderson. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kessler, M. M., M. F. Henry, E. Shen, J. Zhao, S. Gross, P. A. Silver, and C. L. Moore. 1997. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 11:2545-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krecic, A., and M. S. Swanson. 1999. HnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11:363-371. [DOI] [PubMed] [Google Scholar]
- 34.Lambermon, M. H. L., G. G. Simpson, D. A. Kirk, M. Hemmings-Mieszczak, U. Klahre, and W. Filipowicz. 2000. UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 19:1638-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, X., and J. E. Mertz. 1995. HnRNP L binds a cis-acting RNA sequence element that enables intron-dependent gene expresison. Genes Dev. 9:1766-1780. [DOI] [PubMed] [Google Scholar]
- 36.Lorković, Z. J., D. A. Wieczorek Kirk, U. Klahre, M. Hemmings-Mieszczak, and W. Filipowicz. 2000. RBP45 and RBP47, two oligouridylate-specific hnRNP-like proteins interacting with poly(A)+ RNA in nuclei of plant cells. RNA 6:1610-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorković, Z. J., D. A. Wieczorek Kirk, M. H. L. Lambermon, and W. Filipowicz. 2000. Pre-mRNA splicing in higher plants. Trends Plant Sci. 5:160-167. [DOI] [PubMed] [Google Scholar]
- 38.Luehrsen, K. R., S. Taha, and V. Walbot. 1994. Nuclear pre-mRNA processing in higher plants. Prog. Nucleic Acid Res. Mol. Biol. 47:149-193. [DOI] [PubMed] [Google Scholar]
- 39.Matsumoto, K., K. M. Wassarman, and A. P. Wolffe. 1998. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 17:2107-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matunis, E. L., M. J. Matunis, and G. Dreyfuss. 1992. Characterization of the major hnRNP proteins from Drosophila melanogaster. J. Cell Biol. 116:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matunis, E. L., M. J. Matunis, and G. Dreyfuss. 1993. Association of individual hnRNP proteins and snRNPs with nascent transcripts. J. Cell Biol. 121:219-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matunis, M. J., E. L. Matunis, and G. Dreyfuss. 1992. Isolation of hnRNP complexes from Drosophila melanogaster. J. Cell Biol. 116:245-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayeda, A., S. H. Munroe, J. F. Caseres, and A. R. Krainer. 1994. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 13:5483-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayeda, A., and A. R. Krainer. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68:365-375. [DOI] [PubMed] [Google Scholar]
- 45.Michael, W. M., M. Choi, and G. Dreyfuss. 1995. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83:415-422. [DOI] [PubMed] [Google Scholar]
- 46.Minvielle-Sebastia, L., and W. Keller. 1999. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 11:352-357. [DOI] [PubMed] [Google Scholar]
- 47.Nakielny, S., and G. Dreyfuss. 1997. Nuclear export of proteins and RNAs. Curr. Opin. Cell Biol. 9:420-429. [DOI] [PubMed] [Google Scholar]
- 48.Nasr, F., and W. Filipowicz. 2000. Characterization of the Saccharomyces cerevisiae cyclic nucleotide phosphodiesterase involved in the metabolism of ADP-ribose 1",2"-cyclic phosphate. Nucleic Acids Res. 28:1676-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piecyk, M., S. Wax, A. R. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19:4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Proudfoot, N. 2000. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 25:290-293. [DOI] [PubMed] [Google Scholar]
- 51.Puig, O., A. Gottschalk, P. Fabrizio, and B. Seraphin. 1999. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 13:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothnie, H. 1996. Plant mRNA 3′-end formation. Plant Mol. Biol. 32:43-61. [DOI] [PubMed] [Google Scholar]
- 53.Schuler, M. A. 1998. Plant pre-mRNA splicing, p. 1-19. In J. Bailey-Serres and D. Gallie (ed.), A look beyond transcription: mechanisms determining mRNA stability and translation in plants, vol. 1. American Society of Plant Physiologists, Rockville, Md. [Google Scholar]
- 54.Shen, E. C., T. Stage-Zimmermann, P. Chui, and P. A. Silver. 2000. The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J. Biol. Chem. 275:23718-23724. [DOI] [PubMed] [Google Scholar]
- 55.Simpson, G. G., and W. Filipowicz. 1996. Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol. 32:1-41. [DOI] [PubMed] [Google Scholar]
- 56.Siomi, H., and G. Dreyfuss. 1997. RNA-binding proteins as regulators of gene expression. Curr. Opin. Genet. Dev. 7:345-353. [DOI] [PubMed] [Google Scholar]
- 57.Swanson, M. S. 1995. Functions of nuclear pre-mRNA/mRNA binding proteins, p. 17-33. In A. I. Lamond (ed.), Pre-mRNA processing. R. G. Landes Company, Austin, Tex.
- 58.The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796-815. [DOI] [PubMed] [Google Scholar]
- 59.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Visa, N., A. T. Alzahnova-Ericsson, X. Sun, E. Kiseleva, B. Bjorkroth, T. Wurtz, and B. Daneholt. 1996. A pre-mRNA binding protein accompanies the RNA from the gene through the nuclear pores into polysomes. Cell 84:253-264. [DOI] [PubMed] [Google Scholar]
- 61.Weighardt, F., G. Biamonti, and S. Riva. 1996. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays 18:747-756. [DOI] [PubMed] [Google Scholar]
- 62.Weil, D., S. Boutain, A. Audibert, and F. Dautry. 2000. Mature mRNAs accumulated in the nucleus are neither the molecules in transit to the cytoplasm nor constitute a stockpile for gene expression. RNA 6:962-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wieczorek Kirk, D. A. 2000. Novel hnRNP-like proteins that function at multiple steps of pre-mRNA maturation in plants. Ph.D. thesis. University of Basel, Basel, Switzerland.
- 64.Wilson, S. M., K. V. Datar, M. R. Paddy, J. R. Swedlow, and M. S. Swanson. 1994. Characterisation of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J. Cell Biol. 127:1173-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]